Abstract
Noninvasive biomarker profiles of acute rejection (AR) could affect the management of liver transplant (LT) recipients. Peripheral blood was collected following LT for discovery (Northwestern University [NU]) and validation (National Institute of Allergy and Infectious Diseases Clinical Trials in Organ Transplantation [CTOT]‐14 study). Blood gene profiling was paired with biopsies showing AR or ADNR (acute dysfunction no rejection) as well as stable graft function samples (Transplant eXcellent—TX). CTOT‐14 subjects had serial collections prior to AR, ADNR, TX, and after AR treatment. NU cohort gene expression (46 AR, 45 TX) was analyzed using random forest models to generate a classifier training set (36 gene probe) distinguishing AR vs TX (area under the curve 0.92). The algorithm and threshold were locked and tested on the CTOT‐14 validation cohort (14 AR, 50 TX), yielding an accuracy of 0.77, sensitivity 0.57, specificity 0.82, positive predictive value (PPV) 0.47, and negative predictive value (NPV) 0.87 for AR vs TX. The probability score line slopes were positive preceding AR, and negative preceding TX and non‐AR (TX + ADNR) (P ≤ .001) and following AR treatment. In conclusion, we have developed a blood biomarker diagnostic for AR that can be detected prior to AR‐associated graft injury as well a normal graft function (non‐AR). Further studies are needed to evaluate its utility in precision‐guided immunosuppression optimization following LT.
Keywords: biomarker, clinical research/practice, clinical trial, genomics, immunobiology, immunosuppression/immune modulation, liver allograft function/dysfunction, liver transplantation/hepatology, rejection, translational research/science
Short abstract
This study reports on a novel peripheral blood gene signature intended to distinguish acute rejection from other causes and healthy graft function in liver transplant recipients and provide early detection to inform and enhance immunosuppression management.
Abbreviations
- ADNR
acute dysfunction no rejection
- ALT
alanine aminotransferase
- AP
alkaline phosphatase
- AR
acute rejection
- AUC
area under the curve
- CKD
chronic kidney disease
- CNI
calcineurin inhibitors
- CP
clinical phenotype
- CTOT
Clinical Trials in Organ Transplantation
- DCC
Data Coordinating Center
- HCV
hepatitis C virus
- IPA
ingenuity pathway analysis
- IS
immunosuppressive
- LT
liver transplant
- NIAID
National Institute of Allergy and Infectious Diseases
- NPV
negative predictive value
- NU
Northwestern University
- PPV
positive predictive value
- RNA
ribonucleic acid
- TB
total bilirubin
- TX
Transplant eXcellent
1. INTRODUCTION
The advent of calcineurin inhibitors (CNI) has resulted in more acceptable rates of acute rejection (AR) following liver transplantation (LT). 1 However, LT recipients are at high risk for immunosuppression (IS) complications and other issues related to adherence and cost. 2 , 3 Studies have addressed these issues by showing that IS minimization or even full IS withdrawal attempts may be feasible in select patients. 3 , 4 , 5 , 6 However, such strategies to withdraw CNI therapy early after LT, where the benefit on renal and metabolic parameters may be the greatest, have also been limited by the development of AR during these interventions. 7 , 8 , 9 In addition, approaches involving complete IS withdrawal require serial, invasive liver biopsies to confirm that the graft is free of rejection. 9 , 10 , 11
Despite a longstanding interest in immune monitoring strategies, there are currently no available assays other than IS drug levels, liver function tests (LFTs), or biopsies, which are either suboptimal or impractical for assessing individual IS requirements. 12 , 13 , 14 Several genomic biomarkers have been proposed to distinguish causes of graft dysfunction, 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 but these have not been subjected to robust validation and not analyzed before these events. Having a biomarker that can serially distinguish the onset of rejection versus continued stable graft function, before liver injury occurs, could more specifically enhance current “trial and error” IS practices and outcomes.
The Northwestern University (NU) Comprehensive Transplant Center biorepository study (NCT01644903) and the Clinical Trials in Organ Transplantation 14 (CTOT‐14; NCT01672164) study were designed to discover and validate molecular biomarkers for a number of clinical phenotypes following LT. Our focus was to evaluate the performance of a novel blood gene expression biomarker that can detect early, preclinical signs of rejection compared to stable graft function—ultimately to enhance clinical judgment during IS modifications following LT.
2. METHODS
2.1. Cohorts and subjects
From 2010 to 2015, we collected blood samples for biomarker studies in adult deceased or live donor LT recipients at the time of for‐cause graft biopsies. In addition, as our center does not perform routine surveillance biopsies akin to current practices, we randomly collected samples from LT recipients with longstanding normal LFTs for nonviral etiologies (Clinical Phenotypes discussed later). We characterized these samples as “Transplant eXcellent” (TX) and labeled them “virtual biopsies” as negative controls. Samples from NU biorepository subjects were used as the primary training set for biomarker discovery.
For the validation set, between October 2012 and December 2014, we enrolled 186 adult deceased or live donor LT recipients into the multicenter observational CTOT‐14 study. Subjects were followed for 12‐24 months. Similar to the NU discovery cohort, recipients underwent for‐cause biopsies for acute dysfunction per each participating site's standard of care. We collected blood samples for biomarker studies at the time of biopsy and also serially at week 2, month 1, 2, 3, 6, 9, 12, 15, 18, 21, 24 following LT. We limited our biomarker analyses to month 12 as only a small percentage of subjects were followed to month 24. Samples at the time of for‐cause or virtual TX biopsy (clinical phenotypes) were used to validate the classifiers from the NU discovery cohort. In addition, we analyzed the serial samples to detect the classifiers before liver test elevations (biopsy‐proven AR), following treatment of AR, and during the course of stable graft function over time.
Inclusion and exclusion criteria for both cohorts were identical and consisted of adult (≥18 years old) recipients of first LT from either a deceased or living donor. Prior or multiorgan transplants and HIV‐infected recipients were excluded. Although not excluded from enrollment, we excluded patients with hepatitis C virus (HCV) and active viremia from biomarker analysis to avoid HCV as a confounder and because of the diminishing clinical relevance in LT. 30 We did include seropositive nonviremic recipients. Clinical care followed standard practice at each site. All NU and CTOT‐14 biopsies were read locally for clinical care and later sent for independent, blinded central review (AJD). Recipients with Banff Rejection Activity Index ≥ 3 and no other etiology of graft dysfunction were classified as AR. 31 All had T cell–mediated rejection and none with antibody‐mediated rejection based on recent criteria. 32 Other causes, such as steatohepatitis, cholestasis, ischemia, or other etiology, were grouped together as acute dysfunction no rejection (ADNR). Clinical and histological data were also reviewed by a transplant hepatologist (JL) for phenotype designation.
For the NU cohort, demographic, clinical course data before and after liver or virtual TX biopsy were collected from the Northwestern Medicine Enterprise Data Warehouse, which is a comprehensive clinical repository. For CTOT‐14, similar electronic data were collected serially in MEDIDATA RAVE managed by the Data Coordinating Center (DCC ‐ Rho Federal Systems). Oversight included development of the study protocol, review of clinical site visits, classification of clinical phenotypes at the sample level, validation of analyses, and manuscript review. Both studies were both subject to institutional review board approval and informed consent was obtained from all patients.
2.2. Clinical phenotypes (CPs)
We used clinical, biochemical, and liver/virtual biopsy criteria to define 4 CPs: AR, ADNR, TX, and non‐AR (ADNR + TX). The CPs were confirmed by the NU team for discovery and by the CTOT‐14 DCC for validation. The biomarker team was blinded to the CPs for all analyses. Assignment of the diagnostic CPs for both cohorts utilized the following criteria:
AR: for‐cause biopsy for abnormal LFTs per site criteria; central biopsy read consistent with AR.
ADNR: for‐cause biopsy for abnormal LFTs per site criteria; central biopsy read consistent with non‐AR etiology.
TX: normal LFTs at the time of “virtual biopsy.” For both cohorts, we used the same definition: total bilirubin (TB) ≤1.5 mg/dL and direct bilirubin (DB) <0.5 mg/dL, alkaline phosphatase (AP) ≤200 U/L, and alanine transaminase (ALT) ≤60 U/L (males), ≤36 U/L (females). For NU subjects, we required normal LFTs 3 months before and after the “virtual biopsy” time point. As CTOT‐14 subjects had at least 12 months of follow‐up, we chose subjects with normal LFTs at the 6 month mid‐time point with the requirement that > 50% of their LFTs leading up to this time point were also normal.
Non‐AR: ADNR and TX combined (as defined above)
For the serial prediagnosis analysis before each CP (CTOT‐14 only), we used pre‐TX and pre‐ADNR as controls for pre‐AR to ensure specificity for detecting AR vs the other CPs. The following criteria were used:
Pre‐AR: >1 sample(s) collected prior to liver biopsy demonstrating AR as defined previously; at least 2 of the 3 liver tests (DB, AP, ALT) normal at each presample collection; samples collected when ALT > 100 U/L were excluded, even if AP and DB were normal.
Pre‐ADNR: same criteria as pre‐AR, except biopsy demonstrating ADNR as defined previously.
Pre‐TX: same criteria as pre‐AR, except virtual biopsy met TX criteria as defined previously.
Pre‐non‐AR: pre‐ADNR and pre‐TX samples combined.
For postdiagnosis, we focused only on gene expression changes following treatment of AR. We requested sample collections every 2 weeks for 8 weeks in CTOT‐14 subjects treated for AR per site protocol. As these collections were not strictly adhered to, all subjects with > 1 sample collected after treatment that coincided with resolution of AR – defined by normal LFTs – were analyzed.
2.3. Biomarker development
All blood samples were drawn directly into PAXgene tubes (BD BioSciences, San Jose, CA), processed, and subjected to a previous workflow 33 , 34 Specifically, globin‐reduced RNA (200 ng) was labeled using the Affymetrix 3’ IVT PLUS labeling kit Array hybridization. Subsequent washing, staining, and array scanning steps were done on the Affymetrix HG‐U133 Plus PM Array Plate arrays using the standard GeneTitan Gene Expression array workflow (Affymetrix, Santa Clara, CA). Raw expression data files (.CEL) generated by the GeneTitan were processed for QC metrics using the Affymetrix Expression Console software.
The genomic discovery and validation phases were performed in accordance with Institute of Medicine guidelines. 35 We focused our discovery model on distinguishing AR vs TX to best separate gene expression in rejection from healthy graft function and to be able to detect preclinical signs of each in serial monitoring. In addition, we included other causes of graft dysfunction (ADNR) to be more inclusive of the whole LT population (AR and non‐AR).
In the NU discovery cohort, the top 5000 variables probes were selected based on coefficient of variance. Features selection was performed using a random‐forest‐based algorithm with 10 000 trees. The variable selection algorithm uses out‐of‐bag error as minimization criterion and carry out variable elimination from random forest, by successively eliminating the least important variables. 36 The most informative genes were identified using random forests and Gini importance providing a relative ranking of the classifier features from which a final model was selected distinguishing AR vs TX. A performance threshold was selected favoring negative predictive value (NPV) over positive predictive value (PPV; AR), and the model and threshold were then locked for validation (CTOT‐14 cohort). The locked model and threshold were also used on pre‐AR, pre‐TX, and pre–non‐AR samples as well as post‐AR. As each subject had serial samples collected, a linear mixed effect model with random intercept was used to estimate the prebiopsy (or virtual TX biopsy) slope for each phenotype to account for within‐patient correlation. Data first stratified by phenotypes and coefficients were estimated and compared via linear mixed effect model. We used bootstrap method (n = 1000) to generate the 95% confidence interval for estimation of the earliest day prior to diagnosis of detecting differences between groups. Another linear mixed effect model was fitted to compare the pre‐ and post‐AR slopes. Analysis was performed using R version 3.5.1 (RStudio, Boston, MA).
Probes from the final locked models were then fed to Ingenuity Core Analysis (Qiagen, Inc, Hilden, Germany) that provides information about enriched pathways and allows comparison to literature data. 37 Enriched pathways were selected based on Fisher's exact test (P value < .05 statistically significant).
3. RESULTS
3.1. Northwestern University cohort
Recipients undergoing for‐cause liver biopsies (46 AR, 38 ADNR) and concomitant sample collections were enrolled. The 38 ADNR biopsies consisted of 11 biliary obstruction/cholestasis, 11 nonspecific “hepatitis,” 9 steatosis/steatohepatitis, and 7 other. In addition, 45 LT recipients with stable graft function had sample collections drawn in clinic and labeled as TX. The overall cohort was 50.8 ± 13.0 years old at transplant, 54% male, 73% Caucasian, 23% fatty liver/cryptogenic, 19% alcoholic, 30% immune mediated, 3% HCV nonviremic, and 24% other disease. AR subjects were younger, closer to transplant, and had LFT differences particularly compared to TX, as expected (Table 1).
TABLE 1.
NU and CTOT‐14 phenotypes—patient characteristics
|
NU AR (n = 46) |
NU ADNR (n = 38) |
NU TX (n = 45) |
P value a |
CTOT‐14 AR (n = 14) |
CTOT‐14 ADNR (n = 28) |
CTOT‐14 TX (n = 50) |
P value a | |
|---|---|---|---|---|---|---|---|---|
| Age at transplant (y) | 46 ± 17.4 | 51.6 ± 13.0 | 54.9 ± 13.0 | .016 | 57 ± 12.9 | 52 ± 13.7 | 54 ± 11.4 | .489 |
| Caucasian race (%) | 30 (65%) | 28 (74%) | 36 (80%) | .276 | 12 (92%) | 17 (85%) | 38 (84%) | .769 |
| Male sex (%) | 18 (39%) | 20 (53%) | 14 (31%) | .143 | 8 (62%) | 12 (60%) | 27 (60%) | .995 |
| Primary liver diagnosis (%) | ||||||||
| Hepatitis C (nonviremic) | 1 (2%) | 1 (3%) | 2 (4%) | .248 | 4 (31%) | 2 (10%) | 0 | .065 |
| Alcohol | 6 (13%) | 7 (18%) | 12 (27%) | 2 (15%) | 4 (20%) | 12 (27%) | ||
| Nonalcoholic fatty liver or cryptogenic | 7 (15%) | 6 (16%) | 17 (38%) | 3 (23%) | 8 (40%) | 18 (40%) | ||
| Immune‐mediated (PSC, AIH, PBC) | 16 (35%) | 16 (42%) | 7 (16%) | 1 (8%) | 2 (10%) | 7 (16%) | ||
| Other | 16 (35%) | 8 (21%) | 7 (16%) | 3 (23%) | 4 (20%) | 8 (18%) | ||
| Months from LT b | 27.6 ± 34.8 | 60 ± 57.6 | 37 ± 37.2 | .003 | 8 ± 7.1 | 6 ± 5.4 | 7 ± 2.0 | .279 |
| Immunosuppression b | ||||||||
| CNI therapy | 42 (91%) | 31 (82%) | 41 (91%) | .357 | 12 (86%) | 22 (79%) | 50 (100%) | .004 |
| MMF or MPA | 28 (61%) | 17 (45%) | 26 (56%) | .322 | 9 (64%) | 10 (36%) | 36 (72%) | .007 |
| Other (SRL, EVL, AZA) | 6 (13%) | 2 (5%) | 2 (5%) | .292 | 3 (21%) | 3 (11%) | 2 (4%) | .114 |
| Prednisone | 22 (48%) | 17 (45%) | 20 (46%) | .95 | 7 (50%) | 18 (64%) | 37 (74%) | .222 |
| Laboratory values b | ||||||||
| ALT (U/L) | 322.2 ± 260.5 | 96.3 ± 74.9 | 21.4 ± 11.3 | <.001 | 219.9 ± 265.14 | 212.5 ± 251.80 | 21.6 ± 8.96 | <.001 |
| Alkaline phosphatase (U/L) | 332.3 ± 321.2 | 296.3 ± 241.9 | 79.3 ± 30.9 | <.001 | 269.3 ± 239.33 | 281.6 ± 233.40 | 96.6 ± 37.35 | <.001 |
| Total Bilirubin (mg/dL) | 4.9 ± 8.9 | 5.2 ± 18.5 | 0.6 ± 0.2 | <.001 | 4.1 ± 8.28 | 3.5 ± 4.49 | 0.6 ± 0.31 | .001 |
| Creatinine (mg/dL) | 1.2 ± 0.6 | 1.3 ± 0.4 | 1.3 ± 0.4 | .529 | 1.3 ± 0.44 | 1.5 ± 0.87 | 1.2 ± 0.31 | .056 |
| Rejection characteristics b | ||||||||
| Mild (RAI 3‐4) | 19 (41%) | — | — | — | 7 (50%) | — | — | — |
| Moderate ‐Severe (RAI 5‐9) | 27 (59%) | — | — | — | 7 (50%) | — | — | — |
Abbreviations: ADNR, acute dysfunction no rejection; AIH, autoimmune hepatitis; ALT, alanine aminotransferase; AR, acute rejection; AZA, azathioprine; CNI, calcineurin inhibitor; CTOT, Clinical Trials in Organ Transplantation; EVL, everolimus; LT, liver transplant; MMF, mycophenolate mofetil; MPA, mycophenolic acid; NU, Northwestern University; PBC, primary biliary cholangitis; PSC, primary sclerosing cholangitis; RAI, Rejection Activity Index; SRL, sirolimus; TX, Transplant eXcellent.
P value reported using the overall F‐test from analysis of variance for continuous variables and the Cochran–Mantel–Haenszel test of general association for categorical variables.
Reported characteristic corresponds to the time of diagnostic sample. For TX subjects in CTOT‐14, immunosuppression data reported using the month 3 time point.
3.2. CTOT‐14 cohort
A total of 186 LT recipients were enrolled. However, the final analysis included 75 patients who had 92 distinct phenotypes (14 AR biopsies, 28 ADNR biopsies, and 50 TX biopsy) with accompanying samples (Figure 1). The 28 ADNR biopsies consisted of 12 biliary obstruction/cholestasis, 6 ischemia/reperfusion injury, 5 nonspecific “hepatitis,” and 5 steatosis/steatohepatitis. The overall cohort was 54 ± 11.7 years old at transplant, 60% male, 87% Caucasian, 36% Fatty liver/cryptogenic, 24% alcoholic, 13% immune mediated, 8% HCV nonviremic, and 19% other disease. There were statistically significant differences in CNI and mycophenolic acid use and LFT differences between AR and TX, also as expected (Table 1).
FIGURE 1.
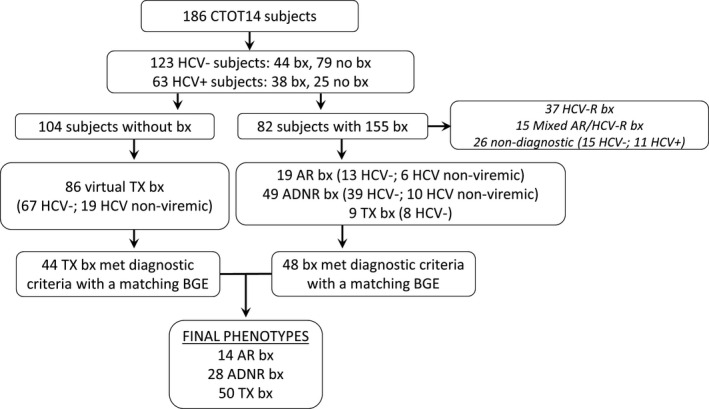
CTOT‐14 consort diagram—enrollment and clinical phenotypes with samples. ADNR, acute dysfunction no rejection; AR, acute rejection; BGE, blood gene expression; Bx, biopsy; CTOT, Clinical Trials in Organ Transplantation; HCV, hepatitis C virus; HCV‐R, hepatitis C virus recurrence; TX, Transplant eXcellent
3.3. Discovery and validation of AR vs TX genomic model (at diagnosis)
In the NU discovery set, blood samples at AR biopsy or the TX time point were analyzed by microarray. Selected probes generated the final 36‐gene probe model differentiating AR vs TX phenotypes. Given the NU cohort was not a prevalent population being enrolled only at biopsies later after LT, we used the late rejection prevalence averaged from the literature (15%) to report an adjusted PPV and NPV. 38 , 39 , 40 , 41 This resulted in a high area under the curve (0.92) for AR vs TX at diagnosis (Figure 2). At a threshold of 0.5, the accuracy was 0.84, sensitivity 0.85, specificity 0.82, prevalence‐adjusted PPV 0.46, and prevalence‐adjusted NPV 0.97. Using the probability threshold from the discovery cohort, the locked NU 36‐gene model (AR vs TX) was then applied to the CTOT‐14 cohort for validation. At the 0.5 threshold, the accuracy was 0.77, sensitivity 0.57, specificity 0.82, PPV 0.47, and NPV 0.87. We also analyzed the performance of the 36‐gene AR vs TX model and threshold in distinguishing AR from ADNR. In the NU and CTOT‐14 cohorts, 36 (55%) ADNR subjects classified as AR and 30 (45%) as TX.
FIGURE 2.
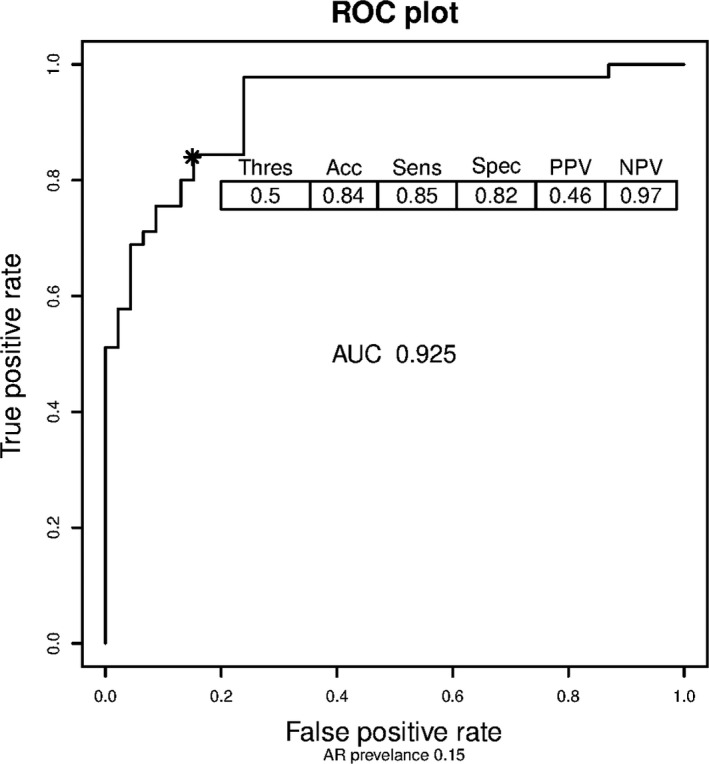
The receiver operating curves (ROC)—AR vs TX (NU discovery cohort). The area under the curve is displayed as well as the performance characteristics (15% AR prevalence adjustment) at the 0.5 threshold (asterisk). AR, acute rejection; NU, Northwestern University; TX, Transplant eXcellent
3.4. Serial prospective analysis of the AR vs TX genomic model (pre‐ and postdiagnosis)
To test the ability of the classifiers to detect AR vs non‐AR prior to these diagnoses, we used 33 CTOT‐14 serial samples collected before 12 AR, 24 prior to 11 ADNR, and 155 samples collected prior to 47 TX (virtual) biopsies. Of note, some in the diagnostic cohort (3 AR, 17 ADNR, and 3 TX) did not have prediagnosis samples that met the inclusion criteria and were excluded from the serial analysis. The timing of sample collections are displayed in Supplementary Figure 1. To compare changes in gene expression over time prediagnosis, we compared the slopes of each line of scores between the phenotypes. This showed a difference between the directions of the line slopes prior to AR (positive) vs TX (negative) (Figure 3A). When combining TX and ADNR to further distinguish AR trajectory from the overall cohort, AR was also different in direction than non‐AR (Figure 3B). The slopes for each analysis were also statistically different in the mixed model (AR vs TX, P < .001; AR vs non‐AR, P < .001). The earliest day prior to diagnosis for detecting significant gene expression score differences between AR vs TX was day −93 (95%CI: −130, −17.725), and day −86 (95%CI: −131.975, −12) for AR vs non‐AR. This divergence is also visually displayed in Figure 3A,B. Finally, we compared LFT trends over time to ensure there were no differences between AR and the other groups prior to the diagnoses, even though our criteria required normalcy over time. Starting at day −180 long before this gene expression divergence, there were no statistical differences between the AR vs TX and AR vs non‐AR groups in the slopes of bilirubin (P = .076, P = .94), ALT (P = .16, P = .205) or Alkphos (0.99, 0.949).
FIGURE 3.
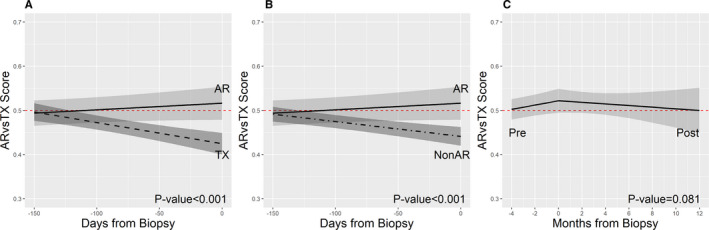
Serial changes in gene expression using line slopes prior to AR, TX, and non‐AR. A, Pre‐AR vs pre‐TX (P < .001). B, Pre‐AR vs pre‐non‐AR (P < .001). C, Pre‐AR vs post‐AR, P = .085). The P value result is the phenotype comparison of the entire line slope from the time of transplantation, whereas the figures visually display a more focused time period around AR, TX, and non‐AR diagnosis (Time 0). AR, acute rejection; TX, Transplant eXcellent [Color figure can be viewed at wileyonlinelibrary.com]
We performed a similar analysis for 53 samples following treatment of 12 AR cases. All responded to treatment defined by normalization of LFTs. This showed that the slope following treatment of AR turned negative, although not statistically different than pre‐AR (Figure 3C; P = .08), suggesting that the slope increase prior to AR was similar to the decrease with treatment. Interestingly, the gene expression profile reached the TX threshold for months following AR diagnosis and treatment. The fitted models displayed in Figure 3 are further specified in Table S1, including parameter estimates with confidence intervals, fixed and random effects, covariance structures, and covariates.
To evaluate for biological relevance, we tested the 36 probes using Ingenuity's pathway analysis. 37 Table 2A displays the identification (when reported) of the 36 genes and the direction and magnitude of gene expression. Most genes mapped to hepatic proliferation and inflammatory pathways—upregulated in AR and downregulated in TX. Tox analysis indicated that the highest percentage of genes have been previously reported to be involved in hepatic hyperplasia/hyper‐proliferation (Table 2B). The differential gene expression between AR and TX is also displayed in a heat map, hierarchical cluster plot, and 3‐dimensional principal component analysis plot (Figures S2A, B and S3, respectively).
TABLE 2.
AR vs TX 36‐gene model
| (A) Differential gene expression | ||||
|---|---|---|---|---|
| Model probe | Symbol | Entrez gene name | Log fold change (AR/TX) | Log fold change (AR/ADNR) |
| 227671_PM_at | XIST | X inactive specific transcript | 2.556868022 | 1.325410826 |
| 205654_PM_at | C4BPA | complement component 4 binding protein alpha | 1.101224469 | 0.897920852 |
| 209773_PM_s_at | RRM2 | ribonucleotide reductase regulatory subunit M2 | 0.967267674 | 0.516531643 |
| 213060_PM_s_at | CHI3L2 | chitinase 3 like 2 | 0.653342435 | 0.364803907 |
| 218350_PM_s_at | GMNN | geminin DNA replication inhibitor | 0.600891795 | 0.192253388 |
| 217714_PM_x_at | STMN1 | stathmin 1 | 0.598102762 | 0.262734907 |
| 200878_PM_at | EPAS1 | endothelial PAS domain protein 1 | 0.588503631 | 0.48338859 |
| 1554696_PM_s_at | TYMS | thymidylate synthase | 0.558038596 | 0.372408001 |
| 202016_PM_at | MEST | mesoderm specific transcript | 0.543696437 | 0.259936021 |
| 227530_PM_at | AKAP12 | A‐kinase anchoring protein 12 | 0.511682439 | 0.405332578 |
| 210358_PM_x_at | GATA2 | GATA binding protein 2 | 0.511546109 | 0.477037794 |
| 219859_PM_at | CLEC4E | C‐type lectin domain family 4 member E | 0.398082121 | 0.207993257 |
| 218782_PM_s_at | ATAD2 | ATPase family AAA domain containing 2 | 0.389173008 | 0.191528324 |
| 206486_PM_at | LAG3 | lymphocyte activating 3 | 0.312979113 | 0.149092455 |
| 238281_PM_at | unidentified | n/a | −0.323326791 | −0.253762725 |
| 212478_PM_at | RMND5A | required for meiotic nuclear division 5 homolog A | −0.333262502 | −0.35407151 |
| 234431_PM_at | GSN | gelsolin | −0.36530257 | −0.112941939 |
| 240765_PM_at | unidentified | n/a | −0.447735132 | −0.29436907 |
| 236409_PM_at | unidentified | n/a | −0.44992291 | −0.249331692 |
| 233263_PM_at | unidentified | n/a | −0.509725068 | −0.312740615 |
| 237376_PM_at | unidentified | n/a | −0.525639819 | −0.250703138 |
| 231034_PM_s_at | NHSL1 | NHS like 1 | −0.534151178 | −0.227014882 |
| 1557685_PM_at | ASAP1‐IT2 | ASAP1 intronic transcript 2 | −0.563114239 | −0.004750863 |
| 232229_PM_at | SETX | senataxin | −0.593424521 | −0.172365985 |
| 241391_PM_at | unidentified | n/a | −0.611242782 | −0.242805807 |
| 236216_PM_at | unidentified | n/a | −0.635686036 | 0.013480067 |
| 244578_PM_at | LCP2 | lymphocyte cytosolic protein 2 | −0.651164906 | −0.257749646 |
| 242854_PM_x_at | DLEU2 | deleted in lymphocytic leukemia 2 | −0.654282894 | −0.274431034 |
| 242800_PM_at | NHS | NHS actin remodeling regulator | −0.668760971 | −0.055646547 |
| 243874_PM_at | unidentified | n/a | −0.720702961 | −0.167061836 |
| 233957_PM_at | unidentified | n/a | −0.725537325 | −0.240100487 |
| 238446_PM_at | NAIP | NLR family apoptosis inhibitory protein | −0.756993933 | −0.460497962 |
| 1560552_PM_a_at | unidentified | n/a | −0.819147799 | −0.391574931 |
| 243954_PM_at | LINC00877 | long intergenic nonprotein coding RNA 877 | −0.880361117 | −0.095247824 |
| 233700_PM_at | PPP1R12B | protein phosphatase 1 regulatory subunit 12B | −0.905378045 | −0.25976984 |
| 221874_PM_at | KIAA1324 | KIAA1324 | −1.001702606 | −0.692655875 |
| (B) Ingenuity Pathway (Tox) analysis | ||
|---|---|---|
| Pathway category | P value | Gene symbols |
| Liver Hyperplasia/Hyperproliferation | 8.75E‐05‐3.8E‐01 | SETX,NHSL1,RRM2,XIST,LCP2,TYMS,C4BPA,ATAD2,LAG3,KIAA1324 |
| Hepatocellular carcinoma | 8.6E‐04‐4.46E‐02 | RRM2,XIST,TYMS,ATAD2 |
| Cardiac inflammation | 1.09E‐03‐1.09E‐03 | TYMS |
| Glomerular injury | 4.34E‐03‐4.34E‐03 | STMN1 |
| Polycythemia | 4.34E‐03‐4.34E‐03 | EPAS1 |
| Renal fibrosis | 4.34E‐03‐4.34E‐03 | STMN1 |
| Kidney failure | 7.58E‐03‐7.58E‐03 | STMN1 |
| Renal proliferation | 7.58E‐03‐7.58E‐03 | STMN1 |
| Liver steatosis | 8.66E‐03‐8.66E‐03 | EPAS1 |
| Anemia | 1.08E‐02‐1.08E‐02 | EPAS1 |
| Renal inflammation | 1.19E‐02‐9.53E‐02 | TYMS,EPAS1 |
| Renal nephritis | 1.19E‐02‐9.53E‐02 | TYMS,EPAS1 |
| Hyperbilirubinemia | 1.83E‐02‐1.83E‐02 | EPAS1 |
| Cardiac enlargement | 3E‐02‐1.85E‐01 | AKAP12,GSN,EPAS1 |
| Pulmonary hypertension | 3.21E‐02‐3.21E‐02 | EPAS1 |
| Renal necrosis/cell death | 4.36E‐02‐2.84E‐01 | GSN,STMN1 |
| Cardiac arrhythmia | 8.44E‐02‐8.44E‐02 | LCP2 |
| Tachycardia | 8.44E‐02‐8.44E‐02 | LCP2 |
| Liver inflammation/hepatitis | 8.8E‐02‐8.8E‐02 | LCP2,AKAP12 |
| Renal damage | 9.14E‐02‐9.14E‐02 | LCP2 |
| Liver necrosis/cell death | 1.63E‐01‐1.63E‐01 | GSN |
| Cardiac necrosis/cell death | 2.25E‐01‐2.25E‐01 | GSN |
Abbreviations: ADNR, acute dysfunction no rejection; AR, acute rejection
4. DISCUSSION
The development of acute rejection after LT can significantly affect patient and graft survival, although there is still an impetus to lower IS therapy as much as possible to reduce its adverse effects. This “trial and error” IS minimization is typically paused only when complications such as rejection occur. However, there are no established noninvasive tests for serially detecting preclinical (eg, before LFT abnormalities) signs of rejection. In a recent similar publication, we described the validity of a biomarker in kidney transplant recipients in detecting silent rejection on biopsy in patients with stable graft function. 33 As surveillance biopsies are not routinely used in LT recipients, there is a great need for noninvasive serial monitoring of patients undergoing more aggressive early minimization, as these protocols are becoming more common. 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11
To address this, we have discovered and validated a blood biomarker profile diagnostic for AR that can be detected prior—when LFTs are normal—to AR diagnosis/graft injury. We recently reviewed the stringent criteria needed for biomarker development and adhered to these in designing the current study. 42 This profile became evident approximately 90 days before AR and gene expression scores continued to increase up to AR diagnosis, different than the decrease in patients who never developed AR. Resolution of the AR gene profile occurred long after biochemical normalization, similar to histological resolution seen in other liver diseases. 43 In addition, the AR genes mapped mainly to hepatic pathways, which are not specific to rejection itself but likely represent liver injury and regenerative responses. The strength in having a model that performs well with serial assessments lies in its potential to proactively monitor for early immune activation during interventions in clinical practice, such as IS minimization. This, however, needs to be proven in randomized interventional studies comparing biomarker‐guided vs standard IS management approaches.
Although a number of key developments have been reported in kidney transplant and other organs, 28 , 33 , 34 , 44 , 45 , 46 , 47 , 48 , 49 , 50 , 51 , 52 , 53 , 54 there is a paucity of literature on similar developments in LT. One of the reasons may be that chronic rejection in the setting of medication adherence is rare following LT compared to other organ transplants, and therefore there is less incentive to detect subclinical rejection. On the other hand, as IS minimization strategies continue to be of great interest, particularly early CNI avoidance or withdrawal, there is a need to know whether or when an immune‐quiescent state turns active before biochemical graft dysfunction. Another reason is that prior work in LT biomarkers historically focused on distinguishing histological injury due to HCV versus AR. 25 , 26 , 27 , 29 Given the advent of oral antiviral therapy, most patients are cured of HCV, essentially erasing this issue in clinical practice.
Finally, the lack of progress in this area in LT may be related to difficulties inherent to the development of predictive biomarkers. Prior studies have demonstrated genetic polymorphisms, gene expression profiles, blood lymphocyte populations, and complement proteins associated with AR in the LT population. 15 , 16 , 17 , 18 , 20 , 21 , 22 , 23 , 24 , 28 , 55 , 56 , 57 , 58 The major limitation of these studies has been small sample sizes, lack of validation cohorts, and statistical rigor and biomarker analysis mostly performed only at diagnosis. Recent data have focused on rejection biomarkers in select patients enrolled in IS withdrawal studies. Results from a multicenter trial 9 involving early tapering to CNI monotherapy and randomization to withdrawal vs maintenance therapy between 1 and 2 years led to 2 discoveries: (1) 2 microRNAs increasing prior to AR during early IS minimization 28 ; and (2) detection of donor‐specific antibodies during the course of IS withdrawal predicted rejection. 59 Another smaller study demonstrated blood CXCL10 gene expression prior to rejection during IS withdrawal. 29 However, LFT increases were already occurring with gene expression changes, diminishing its clinical utility. Another study showed that graft mRNA expression was predictive of tolerance, although it was associated with iron homeostasis pathways and not clearly liver or immune related. 21 Taken together, blood and tissue biomarkers may have a role in predicting rejection in specific tolerance studies, but they are at this point applicable only to highly select patients and not the overall LT population. Blood‐based tests validated in routine serial monitoring, such as our model, may be more useful and generalizable.
As part of the CTOT‐14 study, we have also recently reported a novel clinical/protein model (PRESERVE) that can predict deterioration in renal function early after LT. 60 In future trials, we envision evaluating both tests to select patients for early CNI minimization/withdrawal (renal deterioration predicted by PRESERVE) and serially monitor with this biomarker to confirm that AR gene expression is not present before each IS reduction. This is why we selected the model and threshold optimizing for NPV because it is our intent to use the test in the future for reassurance of an immune‐quiescent state during IS minimization. This personalized approach may allow for more optimal decision‐making at each time point—for example, continue minimization if the blood test is below the threshold (eg, “TX”) vs stopping minimization if above the threshold (eg, “AR”). Other promising assays, such as microRNA and DSA, could also be evaluated to determine their utility. 29 , 59
This study has limitations that need to be addressed. The schedule of sample collections in our discovery and validation cohorts was different—single time points in the former and serial collections in the latter. However, we obtained samples at biopsies in both cohorts as part of routine clinical care. The serial CTOT‐14 analysis was also based on serial samples preceding only 12 AR and 58 non‐AR cases. Another limitation is that we did not have surveillance biopsies of normal histology that could be paired with blood samples to most accurately define TX. As a result, we used “virtual biopsy” samples in both cohorts. Recent studies have demonstrated subclinical graft injury, often despite relatively normal liver tests, in adult and pediatric LT recipients. 61 , 62 However, the reality is that few LT centers perform surveillance biopsies in patients with normal function as detecting subclinical graft injury has not been demonstrated to modify long‐term outcomes, which is different than other organ transplants. Thus we had to use a clinical definition that would parallel a healthy “normal” recipient in real clinical practice. The 36‐gene AR vs TX model also did not perform well in discriminating ADNR from AR at the time of biopsy. This is not surprising as ADNR is a mixture of graft injury causes which may represent an overlap of inflammatory (AR) and noninflammatory (TX) gene expressions. This issue has uncertain relevance as clinicians typically perform liver biopsies, not blood tests, to determine causes of graft dysfunction, further emphasizing that the utility of the biomarker lies in its serial monitoring prior to these events. Finally, non‐US patients and other variables like age, gender, race, ethnicity, time from transplant, and IS regimens were not included in our model given the difficulty in such bioinformatics adjustments, although having a biomarker independent of clinical variables is appealing in clinical practice.
In summary, the development of l biomarkers in LT could transform the field, particularly with the focus on avoiding adverse events from both under‐ and over‐immunosuppression. Our data represent an advance toward the development of clinically serviceable, blood‐based serial immune monitoring tests for use in liver transplantation, similar to other organs. 33 , 34 , 44 In combination with early kidney injury markers, 60 , 63 we are rapidly moving toward conducting biomarker‐based interventional studies to proactively detect and reduce deleterious complications.
Disclosures
The authors of this manuscript have conflicts of interest to disclose as described by the American Journal of Transplantation. Transplant Genomics Incorporated (Eurofins/Viracor): Josh Levitsky (Consultant, Stockholder), Sunil Kurian (Consultant, Stockholder), Michael Abecassis (Co‐Founder, Stockholder).
Supporting information
ACKNOWLEDGMENTS
The funding for this study was supported by the following sources: The National Institute of Allergy and Infectious Diseases Clinical Trials in Organ Transplantation U01AI084146 (Overall Principal Investigator: Abecassis; CTOT14 Protocol Chair: Levitsky), The Northwestern University Comprehensive Transplant Center, and Transplant Genomics, Inc.
Levitsky J, Asrani SK, Schiano T, et al; for the Clinical Trials in Organ Transplantation – 14 Consortium . Discovery and validation of a novel blood‐based molecular biomarker of rejection following liver transplantation. Am J Transplant. 2020;20:2173–2183. 10.1111/ajt.15953
DATA AVAILABILITY STATEMENT
The data and metadata used for development will be uploaded to an independently managed database dbGAP or GEO and will be made available to research community after publication.
REFERENCES
- 1. Wiesner RH, Batts KP, Krom RA. Evolving concepts in the diagnosis, pathogenesis, and treatment of chronic hepatic allograft rejection. Liver Transpl Surg. 1999;5(5):388‐400. [DOI] [PubMed] [Google Scholar]
- 2. Watt KD, Pedersen RA, Kremers WK, Heimbach JK, Charlton MR. Evolution of causes and risk factors for mortality post‐liver transplant: results of the NIDDK long‐term follow‐up study. Am J Transplant. 2010;10(6):1420‐1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Levitsky J, O'Leary JG, Asrani S, et al. Protecting the kidney in liver transplant recipients: practice‐based recommendations from the American society of transplantation liver and intestine community of practice. Am J Transplant. 2016;16(9):2532‐2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Levitsky J, Feng S. Tolerance in clinical liver transplantation. Hum Immunol. 2018;79(5):283‐287. [DOI] [PubMed] [Google Scholar]
- 5. Benítez C, Londoño M‐C, Miquel R, et al. Prospective multicenter clinical trial of immunosuppressive drug withdrawal in stable adult liver transplant recipients. Hepatology. 2013;58(5):1824‐1835. [DOI] [PubMed] [Google Scholar]
- 6. Feng S, Ekong UD, Lobritto SJ, et al. Complete immunosuppression withdrawal and subsequent allograft function among pediatric recipients of parental living donor liver transplants. JAMA. 2012;307(3):283‐293. [DOI] [PubMed] [Google Scholar]
- 7. De Simone P, Nevens F, De Carlis L, et al. Everolimus with reduced tacrolimus improves renal function in de novo liver transplant recipients: a randomized controlled trial. Am J Transplant. 2012;12(11):3008‐3020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Teperman L, Moonka D, Sebastian A, et al. Calcineurin inhibitor‐free mycophenolate mofetil/sirolimus maintenance in liver transplantation: the randomized spare‐the‐nephron trial. Liver Transpl. 2013;19(7):675‐689. [DOI] [PubMed] [Google Scholar]
- 9. Shaked A, DesMarais MR, Kopetskie H, et al. Outcomes of immunosuppression minimization and withdrawal early after liver transplantation. Am J Transplant. 2019;19(5):1397‐1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banff Working Group on Liver Allograft P . Importance of liver biopsy findings in immunosuppression management: biopsy monitoring and working criteria for patients with operational tolerance. Liver Transplantation. 2012;18(10):1154‐1170. [DOI] [PubMed] [Google Scholar]
- 11. Demetris AJ, Isse K. Tissue biopsy monitoring of operational tolerance in liver allograft recipients. Curr Opin Organ Transplant. 2013;18(3):345‐353. [DOI] [PubMed] [Google Scholar]
- 12. Kowalski RJ, Post DR, Mannon RB, et al. Assessing relative risks of infection and rejection: a meta‐analysis using an immune function assay. Transplantation. 2006;82(5):663‐668. [DOI] [PubMed] [Google Scholar]
- 13. Xue F, Zhang J, Han L, et al. Immune cell functional assay in monitoring of adult liver transplantation recipients with infection. Transplantation. 2010;89(5):620‐626. [DOI] [PubMed] [Google Scholar]
- 14. Cabrera R, Ararat M, Soldevila‐Pico C, et al. Using an immune functional assay to differentiate acute cellular rejection from recurrent hepatitis C in liver transplant patients. Liver Transpl. 2009;15(2):216‐222. [DOI] [PubMed] [Google Scholar]
- 15. Fan H, Li LX, Han DD, Kou JT, Li P, He Q. Increase of peripheral Th17 lymphocytes during acute cellular rejection in liver transplant recipients. Hepatobiliary Pancreat Dis Int. 2012;11(6):606‐611. [DOI] [PubMed] [Google Scholar]
- 16. Farid WRR, Pan Q, van der Meer AJP, et al. Hepatocyte‐derived microRNAs as serum biomarkers of hepatic injury and rejection after liver transplantation. Liver Transpl. 2012;18(3):290‐297. [DOI] [PubMed] [Google Scholar]
- 17. Gomez‐Mateo J, Marin L, Lopez‐Alvarez MR, et al. TGF‐beta1 gene polymorphism in liver graft recipients. Transpl Immunol. 2006;17(1):55‐57. [DOI] [PubMed] [Google Scholar]
- 18. Joshi D, Salehi S, Brereton H, et al. Distinct microRNA profiles are associated with the severity of hepatitis C virus recurrence and acute cellular rejection after liver transplantation. Liver Transpl. 2013;19(4):383‐394. [DOI] [PubMed] [Google Scholar]
- 19. Kamei H, Masuda S, Nakamura T, Oike F, Takada Y, Hamajima N. Association of transporter associated with antigen processing (TAP) gene polymorphisms in donors with acute cellular rejection in living donor liver transplantation. J Gastrointestin Liver Dis. 2013;22(2):167‐171. [PubMed] [Google Scholar]
- 20. Karimi MH, Daneshmandi S, Pourfathollah AA, et al. Association of IL‐6 promoter and IFN‐gamma gene polymorphisms with acute rejection of liver transplantation. Mol Biol Rep. 2011;38(7):4437‐4443. [DOI] [PubMed] [Google Scholar]
- 21. Bohne F, Martínez‐Llordella M, Lozano J‐J, et al. Intra‐graft expression of genes involved in iron homeostasis predicts the development of operational tolerance in human liver transplantation. J Clin Invest. 2012;122(1):368‐382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Massoud O, Heimbach J, Viker K, et al. Noninvasive diagnosis of acute cellular rejection in liver transplant recipients: a proteomic signature validated by enzyme‐linked immunosorbent assay. Liver Transpl. 2011;17(6):723‐732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Moya‐Quiles MR, Alvarez R, Miras M, et al. Impact of recipient HLA‐C in liver transplant: a protective effect of HLA‐Cw*07 on acute rejection. Hum Immunol. 2007;68(1):51‐58. [DOI] [PubMed] [Google Scholar]
- 24. Sindhi R, Higgs BW, Weeks DE, et al. Genetic variants in major histocompatibility complex‐linked genes associate with pediatric liver transplant rejection. Gastroenterology. 2008;135(3):830‐839.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Asaoka T, Kato T, Marubashi S, et al. Differential transcriptome patterns for acute cellular rejection in recipients with recurrent hepatitis C after liver transplantation. Liver Transpl. 2009;15(12):1738‐1749. [DOI] [PubMed] [Google Scholar]
- 26. Gehrau R, Maluf D, Archer K, et al. Molecular pathways differentiate hepatitis C virus (HCV) recurrence from acute cellular rejection in HCV liver recipients. Mol Med. 2011;17(7–8):824‐833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sreekumar R, Rasmussen DL, Wiesner RH, Charlton MR. Differential allograft gene expression in acute cellular rejection and recurrence of hepatitis C after liver transplantation. Liver Transpl. 2002;8(9):814‐821. [DOI] [PubMed] [Google Scholar]
- 28. Shaked A, Chang B‐L, Barnes MR, et al. An ectopically expressed serum miRNA signature is prognostic, diagnostic, and biologically related to liver allograft rejection. Hepatology. 2017;65(1):269‐280. [DOI] [PubMed] [Google Scholar]
- 29. Bonaccorsi‐Riani E, Pennycuick A, Londoño M‐C, et al. Molecular characterization of acute cellular rejection occurring during intentional immunosuppression withdrawal in liver transplantation. Am J Transplant. 2016;16(2):484‐496. [DOI] [PubMed] [Google Scholar]
- 30. Ishak K, Baptista A, Bianchi L, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696‐699. [DOI] [PubMed] [Google Scholar]
- 31. Banff schema for grading liver allograft rejection: an international consensus document. Hepatology. 1997;25(3):658‐663. [DOI] [PubMed] [Google Scholar]
- 32. Demetris AJ, Bellamy C, Hübscher SG, et al. 2016 comprehensive update of the Banff Working Group on Liver Allograft Pathology: Introduction Of Antibody‐Mediated Rejection. Am J Transplant. 2016;16(10):2816‐2835. [DOI] [PubMed] [Google Scholar]
- 33. Friedewald JJ, Kurian SM, Heilman RL, et al. Development and clinical validity of a novel blood‐based molecular biomarker for subclinical acute rejection following kidney transplant. Am J Transplant. 2019;19(1):98‐109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kurian SM, Williams AN, Gelbart T, et al. Molecular classifiers for acute kidney transplant rejection in peripheral blood by whole genome gene expression profiling. Am J Transplant. 2014;14(5):1164‐1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Michell CM, Nass SJ, Ommen GS. Evolution of Translational Omics: Lessons Learned and the Path Forward. Washington, DC: Institute of Medicine, The National Academies Press;2012:0‐354. 10.17226/13297 [DOI] [PubMed] [Google Scholar]
- 36. Diaz‐Uriarte R, Alvarez de Andres S. Gene selection and classification of microarray data using random forest. BMC Bioinformatics. 2006;7:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chatzipetrou MA, Mathew JM, Kenyon NS, et al. Analysis of post‐transplant immune status in recipients of liver/bone marrow allografts. Hum Immunol. 1999;60(12):1281‐1288. [DOI] [PubMed] [Google Scholar]
- 38. Ramji A, Yoshida EM, Bain VG, et al. Late acute rejection after liver transplantation: the Western Canada experience. Liver Transpl. 2002;8(10):945‐951. [DOI] [PubMed] [Google Scholar]
- 39. Uemura T, Ikegami T, Sanchez EQ, et al. Late acute rejection after liver transplantation impacts patient survival. Clin Transplant. 2008;22(3):316‐323. [DOI] [PubMed] [Google Scholar]
- 40. Thurairajah PH, Carbone M, Bridgestock H, et al. Late acute liver allograft rejection; a study of its natural history and graft survival in the current era. Transplantation. 2013;95(7):955‐959. [DOI] [PubMed] [Google Scholar]
- 41. Levitsky J, Goldberg D, Smith AR, et al. Acute rejection increases risk of graft failure and death in recent liver transplant recipients. Clin Gastroenterol Hepatol. 2017;15(4):584‐593e582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kurian SM, Whisenant T, Mas V, et al. Biomarker guidelines for high‐dimensional genomic studies in transplantation: adding method to the madness. Transplantation. 2017;101(3):457‐463. [DOI] [PubMed] [Google Scholar]
- 43. Manns MP, Czaja AJ, Gorham JD, et al. Diagnosis and management of autoimmune hepatitis. Hepatology. 2010;51(6):2193‐2213. [DOI] [PubMed] [Google Scholar]
- 44. Modena BD, Kurian SM, Gaber LW, et al. Gene expression in biopsies of acute rejection and interstitial fibrosis/tubular atrophy reveals highly shared mechanisms that correlate with worse long‐term outcomes. Am J Transplant. 2016;16(7):1982‐1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mengel M, Sis B, Kim D, et al. The molecular phenotype of heart transplant biopsies: relationship to histopathological and clinical variables. Am J Transplant. 2010;10(9):2105‐2115. [DOI] [PubMed] [Google Scholar]
- 46. Sellarés J, Reeve J, Loupy A, et al. Molecular diagnosis of antibody‐mediated rejection in human kidney transplants. Am J Transplant. 2013;13(4):971‐983. [DOI] [PubMed] [Google Scholar]
- 47. Li L, Khatri P, Sigdel TK, et al. A peripheral blood diagnostic test for acute rejection in renal transplantation. Am J Transplant. 2012;12(10):2710‐2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Suthanthiran M, Schwartz JE, Ding R, et al. Urinary‐cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med. 2013;369(1):20‐31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muthukumar T, Dadhania D, Ding R, et al. Messenger RNA for FOXP3 in the urine of renal‐allograft recipients. N Engl J Med. 2005;353(22):2342‐2351. [DOI] [PubMed] [Google Scholar]
- 50. Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal‐allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344(13):947‐954. [DOI] [PubMed] [Google Scholar]
- 51. Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene‐expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362(20):1890‐1900. [DOI] [PubMed] [Google Scholar]
- 52. Rush DN, Gibson IW. Subclinical inflammation in renal transplantation. Transplantation. 2019;103(6):e139‐e145. [DOI] [PubMed] [Google Scholar]
- 53. Soma O, Hatakeyama S, Yoneyama T, et al. Serum N‐glycan profiling can predict biopsy‐proven graft rejection after living kidney transplantation. Clin Exp Nephrol. 2020;24(2):174‐184. [DOI] [PubMed] [Google Scholar]
- 54. Zhang W, Yi Z, Keung KL, et al. A peripheral blood gene expression signature to diagnose subclinical acute rejection. J Am Soc Nephrol. 2019;30(8):1481‐1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kamei H, Masuda S, Nakamura T, et al. Impact of glutathione S‐transferase T1 gene polymorphisms on acute cellular rejection in living donor liver transplantation. Transpl Immunol. 2013;28(1):14‐17. [DOI] [PubMed] [Google Scholar]
- 56. Evans PC, Smith S, Hirschfield G, et al. Recipient HLA‐DR3, tumour necrosis factor‐alpha promoter allele‐2 (tumour necrosis factor‐2) and cytomegalovirus infection are interrelated risk factors for chronic rejection of liver grafts. J Hepatol. 2001;34(5):711‐715. [DOI] [PubMed] [Google Scholar]
- 57. Hanvesakul R, Spencer N, Cook M, et al. Donor HLA‐C genotype has a profound impact on the clinical outcome following liver transplantation. Am J Transplant. 2008;8(9):1931‐1941. [DOI] [PubMed] [Google Scholar]
- 58. Toby TK, Abecassis M, Kim K, et al. Proteoforms in peripheral blood mononuclear cells as novel rejection biomarkers in liver transplant recipients. Am J Transplant. 2017;17(9):2458‐2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jucaud V, Shaked A, DesMarais M, et al. Prevalence and impact of de novo donor‐specific antibodies during a multicenter immunosuppression withdrawal trial in adult liver transplant recipients. Hepatology. 2019;69(3):1273‐1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Levitsky J, Asrani SK, Klintmalm G, et al. Discovery and validation of a biomarker model (PRESERVE) predictive of renal outcomes after liver transplantation. Hepatology. 2019; 10.1002/hep.30939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Londoño M‐C, Souza LN, Lozano J‐J, et al. Molecular profiling of subclinical inflammatory lesions in long‐term surviving adult liver transplant recipients. J Hepatol. 2018;69(3):626‐634. [DOI] [PubMed] [Google Scholar]
- 62. Feng S, Bucuvalas JC, Demetris AJ, et al. Evidence of chronic allograft injury in liver biopsies from long‐term pediatric recipients of liver transplants. Gastroenterology. 2018;155(6):1838‐1851e1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Levitsky J, Asrani SK, Abecassis M, Ruiz R, Jennings LW, Klintmalm G. External validation of a pretransplant biomarker model (REVERSE) predictive of renal recovery after liver transplantation. Hepatology. 2019;70(4):1349‐1359. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and metadata used for development will be uploaded to an independently managed database dbGAP or GEO and will be made available to research community after publication.


