Abstract
Super-resolution microscopy is transforming research in the life sciences by enabling the visualization of structures and interactions on the nanoscale. DNA-PAINT is a relatively easy-to-implement single-molecule-based technique, which uses the programmable and transient interaction of dye-labeled oligonucleotides with their complements for super-resolution imaging. However, similar to many imaging approaches, it is still hampered by the subpar performance of labeling probes in terms of their large size and limited labeling efficiency. To overcome this, we here translate the programmability and transient binding nature of DNA-PAINT to coiled coil interactions of short peptides and introduce Peptide-PAINT. We benchmark and optimize its binding kinetics in a single-molecule assay and demonstrate its super-resolution capability using self-assembled DNA origami structures. Peptide-PAINT outperforms classical DNA-PAINT in terms of imaging speed and efficiency. Finally, we prove the suitability of Peptide-PAINT for cellular super-resolution imaging by visualizing the microtubule and vimentin network in fixed cells.
Keywords: Super-resolution, coiled coil interactions, transient binding, DNA-PAINT, single-molecule imaging, Peptide-PAINT
Recently developed super-resolution fluorescence microscopy approaches allow researchers to characterize cellular structures on the nanoscale.1 A widely used super-resolution implementation is single molecule localization microscopy (SMLM),2 which uses fluorophore blinking to enable subdiffraction resolution imaging. Blinking can be achieved using photoswitchable proteins or organic dyes as in photoactivated localization microscopy (PALM)3 and stochastic optical reconstruction microscopy (STORM).4 In points accumulation in nanoscale topography (PAINT),5 transient binding interactions between freely diffusing dyes and their targets generate the required blinking pattern, making it easy to implement as photophysical switching of target-bound dye molecules from bright to dark states is not necessary. DNA-PAINT,6 an adaptation of this concept, uses short dye-labeled DNA oligonucleotides that can transiently, yet sequence-specifically, and repetitively interact with their target-bound complementary strands. Advantages of this approach include better than 5 nm spatial resolution6,7 and high multiplexing capability using DNA-barcoded probes on targets of interest.8
One of the main shortcomings of current super-resolution microscopy approaches is due to suboptimal labeling probes like antibodies, which are large in size and have limited labeling efficiency.9 While large probes introduce significant linkage artifacts,10,11 suboptimal binding efficiency may compromise sensitivity and quantitative detection.12 To overcome some of the limitations of antibodies, several different labeling probes have been developed for super-resolution microscopy, including nanobodies,9 genetically encoded self-labeling tags (e.g., SNAP and Halo),13,14 small protein scaffolds (e.g., affimers or iris probes),15,16 and aptamers.17,18 However, these methods fall short on delivering all prerequisites for an “ideal” probe, which are (1) site-specific target labeling, (2) smallest label size, (3) highest labeling efficiency, and (4) multiplexing capabilities. Recently, programmable designer peptides generating coiled coil interactions have been identified as potentially ideal probes for fluorescence microscopy with high specificity and small label size.19−23 Furthermore, the use of heterodimeric orthogonal coiled coils should allow multiplexing applications.24,25 The coiled coil motif involves α-helical peptides that wrap around each other and form a superhelix.26 An archetypical coiled coil-forming peptide comprises repeats of heptad sequences (hxxhcxc) arranging hydrophobic (h) and charged (x) amino acids residues in a way that enables mutual recognition of “complementary” peptide strands by means of hydrophobic and ionic interactions (x could be any amino acid but proline).27,28 We envisioned that the use of heterodimeric coiled coils enables a design in which one peptide serves as a landing hub for transient interactions with a fluorescence-labeled imager peptide. The mutual interactions between coiled coil peptides are tunable,29 potentially making them suitable for PAINT-based super-resolution microscopy.
Here, we rationally redesigned the well-studied, heterodimeric E/K coiled coil peptide pair30−32 for transient binding to be applicable for PAINT-type super-resolution microscopy and named it Peptide-PAINT. We tuned the coiled coil affinity by varying the length, achieving even faster association kinetics compared to classical DNA-PAINT. The resulting coiled coil pair of approximately 20 amino acids could considerably decrease target-linkage error when genetically fused to a protein of interest, furthermore leading to improved labeling efficiency. We quantitatively characterized the kinetics as well as labeling and imaging efficiency of Peptide-PAINT by single-molecule imaging. We then used DNA origami structures to demonstrate its suitability for super-resolution microscopy. Finally, we successfully demonstrated Peptide-PAINT in cells by super-resolving the microtubule and vimentin network.
Results and Discussion
The PAINT process requires a dynamic equilibrium between bound and unbound states of the fluorescently labeled imager probe. In DNA-PAINT, this is achieved when the transiently formed duplexes have stabilities in the micromolar range (KD ≈ 1 μM). The interaction between the 21 amino acid (aa) long E3 and K3 peptides previously used for protein imaging20−23 leads to a coiled coil complex with nanomolar stability (KD ≈ 20 nM, Figure S1). Aiming for the identification of a suitable weaker affinity, we investigated the length dependence of coiled coil stability (Figure S1). This analysis suggested that complexes comprised of a 22 aa long E peptide and 19 or 18 aa long K peptides provide stability characteristics (KD ≈ 81 nM or KD ≈ 1.7 μM, respectively) suitable for PAINT.
Next, we developed a single-molecule imaging assay to characterize the kinetics of coiled coil interactions between the short peptides. In this assay, the 22 aa long K peptide (K22) served as a docking coil, which was linked to a DNA strand offering an additional docking site for DNA-PAINT. Hybridization with a second biotinylated-DNA strand allowed immobilization on a poly ethylene-glycol (PEG) passivated glass surface via biotin–neutravidin–biotin interactions (Figure 1a). We first located the positions of immobilized molecules via DNA-PAINT imaging of the P3 extension using complementary Cy3B-labeled imager strands (Figure 1b). After a washing step similar to Exchange-PAINT,8 we then performed Peptide-PAINT imaging via transient binding of Cy3B-labeled imager peptides (E19) to the immobilized complementary docking peptide (K22). A negatively charged E19-Cy3B imager peptide was used akin to the DNA-PAINT concept, where negatively charged imager strands bind to their targets. This assay enabled us to precisely characterize the kinetic properties and imaging efficiency of transient coiled coil interactions in direct comparison to DNA-PAINT. The multiplexed single-molecule data qualitatively showed a high degree of colocalization between the immobilized DNA extensions and that of the peptides (Figure 1b, see Figure S2 for whole field of view). To quantify Peptide-PAINT’s efficiency, we performed a colocalization analysis from the DNA- to the Peptide-PAINT signal, resulting in 86% colocalization (Figure 1c). As expected, in the absence of the K22 peptide on the surface-tethered DNA molecules, we observed no Peptide-PAINT signal, indicating that there is no unspecific interaction between the immobilized DNA and the peptide imager (Figure S3).
Figure 1.
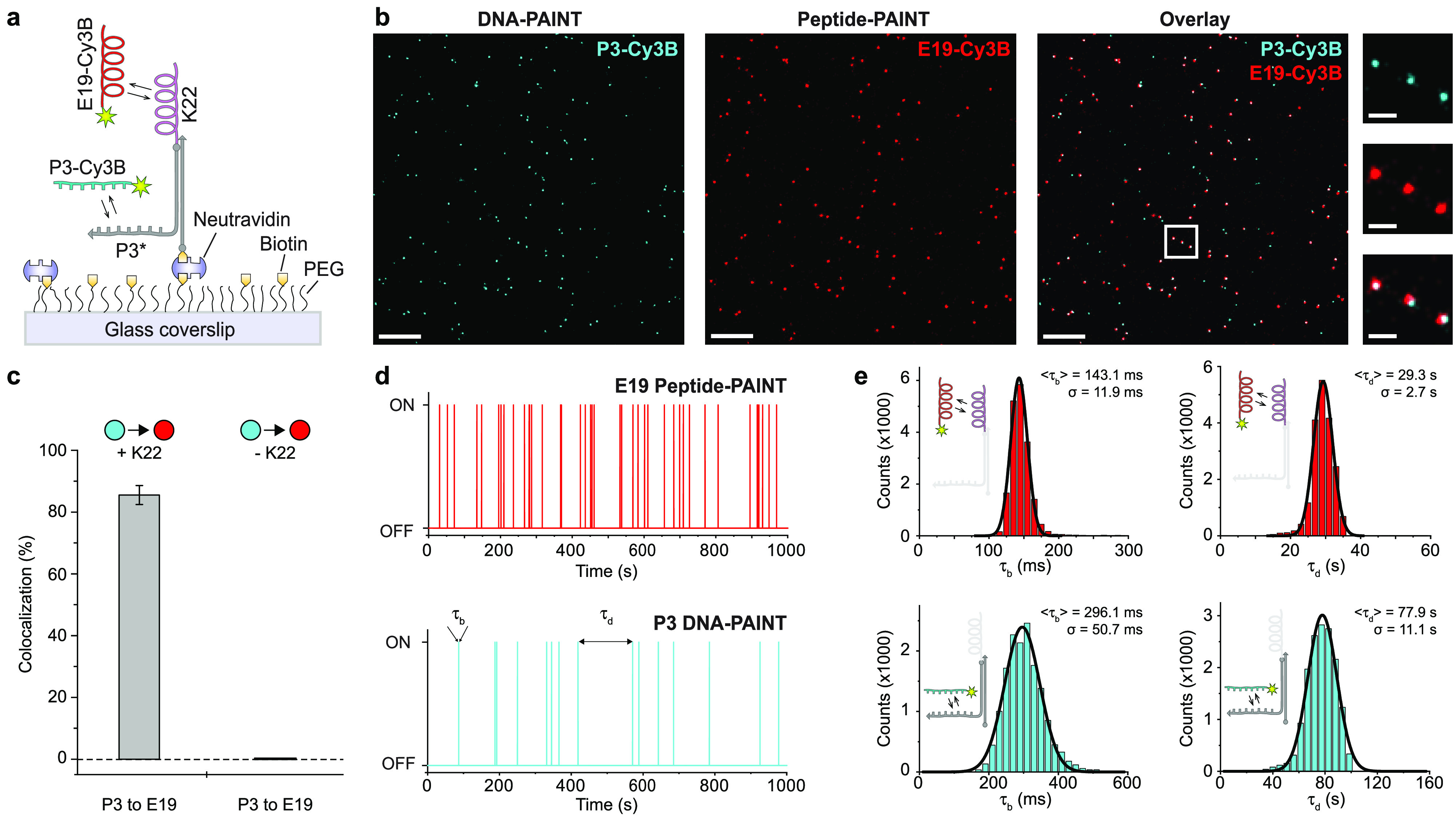
Characterization of transient coiled coil interactions using single-molecule imaging. (a) Schematic diagram depicting the single-molecule imaging assay for characterizing Peptide-PAINT. A peptide coil (K22) is conjugated to a DNA strand (DBCO-S1HP3H, see Table S4) for hybridization to a surface-bound complement. The strand on the coil is furthermore extended with a DNA-PAINT docking sequence for direct visualization. (b) Representative single-molecule localization data resulting from DNA-PAINT (left, cyan), Peptide-PAINT (middle, red), and their overlay (right) of a selected field of view. Zoom-ins highlight colocalization of DNA- and Peptide-PAINT. (c) Colocalization analysis (from DNA- to Peptide-PAINT signals) yields 86 ± 3% (mean and standard deviation) for the positive control and no colocalization for the negative control, which lacks the K22 peptide (n > 1500). (d) Exemplary binding traces showing transient interactions of E19 with surface immobilized K22 and P3 binding to its corresponding docking strand (with highlighted bright and dark times τb and τd). (e) τb and τd distributions for E19 (n = 18938) and P3 (n = 15544) and corresponding Gaussian fits (mean and standard deviation are stated). Scale bars: 300 nm (b, overview), 50 nm (b, zoom-ins).
We observed transient binding kinetics for Peptide-PAINT imaging, similar to DNA-PAINT, indicating that these coiled coil interactions are suitable for super-resolution imaging (Figure 1d, see Table S1 for a binding kinetics overview), yielding comparable localization precisions (3.6 nm for DNA-PAINT and 4 nm for Peptide-PAINT). Intriguingly, time traces of Peptide-PAINT qualitatively show more frequent binding than DNA-PAINT for the same imager concentration. Indeed, statistical analysis of bright and dark times for Peptide- and DNA-PAINT demonstrated that the E19 imager peptide manifests faster kinetics than its DNA-PAINT counterpart (Figure 1e), with more than two-times higher association rates (Peptide-PAINT average dark time = 30.0 ± 0.9 s, DNA-PAINT average dark time = 71.2 ± 5.5 s), ultimately leading to faster image acquisition.33 In addition, in an inverse configuration to what is presented in Figure 1a, we showed that E22 docking peptides (immobilized on a surface) can be imaged with K19 imager peptides with similar performance (Figure S4). Peptide-PAINT was also tested in this inverted configuration, in case a positively charged imager peptide (such as K19-Cy3B) would be preferred in future studies. In a negative control, where the surface-immobilized DNA molecules lacked the E22 peptide, no detectable binding of K19 imagers to the DNA was observed (Figure S5), once again indicating that the coiled coil interactions are specific and do not interact nonspecifically with DNA.
Similar to the imager-docking strand interactions in DNA-PAINT, the binding (or bright) time of coiled coil interactions are also tunable based on the length of the peptide coils.34 Indeed, shortening the imager peptide by one amino acid (now called K18), resulted in a reduced bright time (59.8 ± 1.6 ms) compared to the 19 amino acid version (K19, 209.5 ± 2.8 ms), with similar dark times (Figure S6). In a negative control lacking E22 peptide, no detectable binding of K18 imagers to the DNA was observed (Figure S7). Due to the relatively short bright times of the K18 imager (potentially advantageous for live cell applications35), single binding events lead to a reduced number of collectable photons, resulting in suboptimal localization precisions (8.5 nm for Peptide-PAINT with K18) (Figure S6). Additionally, we note that coiled coil interactions can be tuned29,36 by modulating the sequence37 and overall charge of the peptide pairs24 or the ionic strength of the buffer.38 In fact, decreasing the salt concentration by half yields an increased dark time (average dark time for 5 nM E19-Cy3B imaging in a 0.5× PBS buffer is 57.3 ± 1.6 s), whereas increasing the salt concentration results in a reduced dark time, thus increasing acquisition speed (average dark time for 5 nM E19-Cy3B imaging in a 1× PBS buffer supplemented with 150 mM NaCl is 17.2 s ± 3.6 s).
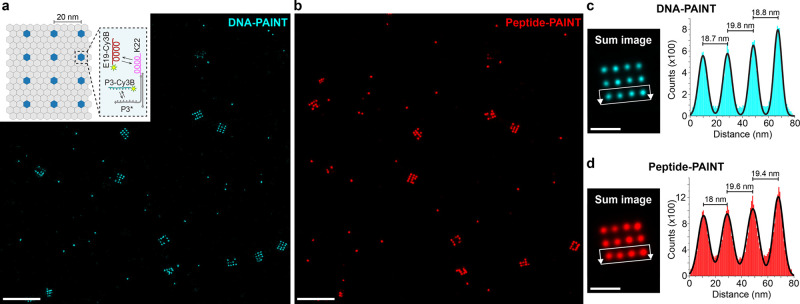
Next, we tested the achievable spatial resolution of Peptide-PAINT in a more controlled setting using programmable, self-assembled DNA nanostructures.6,39 We used 20 nm-grid origami, decorated with DNA-conjugated K22 docking peptides (Figure 2a), reminiscent of the single-molecule assay described above. Using the origami platform, we were able to image 20 nm-grid points with both DNA- and Peptide-PAINT with similar spatial resolution (Figure 2b and c). We used 100 origami structures for averaging (Figure 2d and Figure S8) and performed a cross-sectional histogram analysis on Peptide- and DNA-PAINT data, yielding the designed distances for the 20 nm-grid structures. Interestingly, when comparing the normalized number of localizations for both cases, Peptide-PAINT yielded approximately 20% more localizations, suggesting higher imaging efficiency compared to DNA-PAINT. We note that this difference in efficiency is unlikely to have originated from different accessibility of DNA versus Peptide location, as both were designed with sufficient spacing to the underlying DNA origami structure. In a negative control, where the DNA origami lacked the K22 peptide, no detectable binding of E19 imagers to the DNA nanostructures was observed (Figure S9). We observed similar performance with K19 imager peptides and E22-decorated DNA origami (Figures S10–S12).
Figure 2.
Peptide-PAINT super-resolution imaging of 20 nm DNA origami grids. (a) DNA-PAINT data (cyan) of 20 nm-grids (inset: schematic representation of 20 nm-grids with single-stranded extensions for DNA- and Peptide-PAINT, reminiscent of single-molecule assay in Figure 1). (b) Peptide-PAINT image (red) of the same region. (c) DNA-PAINT sum image obtained from 100 individual DNA origami (left) and cross-sectional histogram analysis of the localizations in the highlighted region. Gaussian fits yield the designed 20 nm-distances. (d) Corresponding Peptide-PAINT analysis. Scale bars: 300 nm (a, b), 50 nm (c, d). The experiment was repeated three times independently with similar results.
We also performed super-resolution imaging with the shorter K18 imager peptide on DNA origami with E22 docking peptides located at each corner of the origami (Figures S13 and S14). We chose these—easier to resolve—four-cornered structures to match the lower localization precision and thus reduced spatial resolution of the K18 imager peptide originating from its shorter binding time (Table S2).
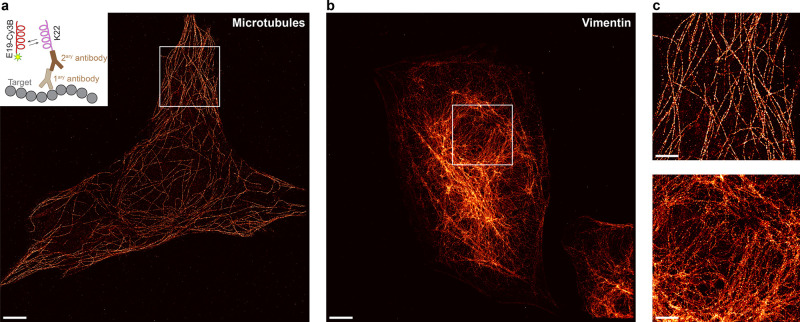
Finally, we sought to demonstrate the ability of Peptide-PAINT to image cellular protein structures. In a proof-of-principle experiment, we chose microtubules and vimentin as targets because of their unique and recognizable morphology. For this, we labeled U2OS cells with primary antibodies against α-tubulin or vimentin, followed by incubation with K22-peptide-conjugated secondary antibodies (inset in Figure 3a). Subsequently, Peptide-PAINT imaging was performed with E19 imagers. The reconstructed super-resolution images clearly demonstrate the faithful visualization of the microtubule (Figure 3a) and vimentin filament network (Figure 3b) with high specificity and spatial resolution, similar to previous DNA-PAINT imaging of those targets,10,33,40,41 which were not visible in the negative control (Figure S15).
Figure 3.
Peptide-PAINT super-resolution imaging of microtubules and vimentin in U2OS cells. (a) Exemplary super-resolved Peptide-PAINT image of microtubules labeled with primary and K22-conjugated secondary antibodies (inset: labeling scheme). (b) Representative super-resolved Peptide-PAINT data of the vimentin network. (c) Zoom-ins of highlighted areas from (a) and (b). Scale bars: 5 μm (a, b), 2 μm (c). Each experiment was repeated three times independently with similar results.
Conclusions
In conclusion, we developed Peptide-PAINT as a novel super-resolution imaging modality that uses transient interactions of short peptides. We have implemented a single-molecule assay to quantitatively characterize the binding kinetics of these programmable coiled coil interactions and observed that Peptide-PAINT exhibits faster association kinetics compared to classical DNA-PAINT, eventually allowing for faster imaging. We furthermore demonstrated the spatial resolution capabilities of Peptide-PAINT using DNA origami structures. Interestingly, we found that Peptide-PAINT yielded an approximately 20% improved imaging efficiency compared to DNA-PAINT. In a proof-of-principle experiment, we showed the applicability of Peptide-PAINT for cellular imaging by targeting the microtubule and vimentin network using primary and peptide-conjugated secondary antibodies.
We anticipate exciting future applications for Peptide-PAINT based on its improved imaging efficiency and speed compared to classical DNA-PAINT. The speed of Peptide-PAINT could be further increased by changing the salt concentration of the buffer33 or potentially by increasing the numbers of heptad repeats on the docking strand in combinations with a shorter peptide, similarly to recent speed improvements in DNA-PAINT.41 Alternatively, Peptide-PAINT kinetics could be fine-tuned for future applications by varying the sequence of the peptide pairs or the length of the imager peptide. Furthermore, Peptide-PAINT could be extended for multiplexing by adapting orthogonal coiled coil pairs, which have been reported in the literature,24 and testing them on our combined single-molecule and DNA origami characterization platform. We anticipate a multiplexing capability of at least six targets from orthogonal coiled coil pairs previously tested in cells.25 Due to the small size of the docking peptide, we expect that a genetically encoded peptide tag would be easier to clone and express while causing only minimal distortion to the physiological protein function, in contrast to a larger tag such as GFP (green fluorescent protein). Finally, with the combination of a short imager peptide, we also foresee that Peptide-PAINT will enable highest resolution and labeling efficiency by reducing the linkage error to a minimum and become the preferred labeling strategy for super-resolution microscopy.
Acknowledgments
We thank Alexander Auer and Philipp Nickels for initial experiments, Thomas Schlichthaerle for fruitful discussions and help with the data analysis, Luis Moroder for helpful discussion, and Sebastian Strauss for help with the cell imaging. We thank Stefan Pettera and Stephan Uebel from the MPIB peptide synthesis service for the production of the peptides and for their advice regarding the peptide modifications. A.S.E. acknowledges support from the International Max Planck Research School for Molecular and Cellular Life Sciences (IMPRS-LS). M.G. acknowledges funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement no. 796606.
Glossary
Abbreviations Used
- SMLM
single-molecule localization microscopy
- PALM
photoactivated localization microscopy
- STORM
stochastic optical reconstruction microscopy
- PAINT
points accumulation in nanoscale topography
- PEG
poly-ethelene glycol
- PBS
phosphate buffer saline
- GFP
green fluorescent protein
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.nanolett.0c02620.
Detailed experimental methods and materials; full field of view for the single-molecule experiment; negative controls for single-molecule, origami, and cell experiments; binding kinetics and imaging parameters for DNA-PAINT and Peptide-PAINT; Figures S1–S15 and Tables S1–S8 (PDF)
Author Contributions
# These authors contributed equally. A.S.E. conceived and performed experiments, analyzed data, and contributed to the writing of the manuscript. M.G. conceived experiments, analyzed data, wrote the manuscript, and supervised the study. G.G. developed the coiled coil pair and helped with general peptide design. O.S. conceived the study, supervised the study, and edited the manuscript. R.J. conceived and supervised the study, analyzed and interpreted data, and wrote the manuscript. All authors reviewed and approved the manuscript.
This work has been supported in part by the German Research Foundation through the Emmy Noether Program (DFG JU 2957/1-1), the SFB1032 (project A11), the European Research Council through an ERC Starting Grant (MolMap, grant agreement number 680241), the Allen Distinguished Investigator Program through The Paul G. Allen Frontiers Group, the Danish National Research Foundation (Centre for Cellular Signal Patterns, DNRF135), the Human Frontier Science Program through a Young Investigator Grant (HFSP RGY0065), and the Max Planck Foundation and the Max Planck Society.
The authors declare no competing financial interest.
Supplementary Material
References
- Sahl S. J.; Hell S. W.; Jakobs S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18 (11), 685–701. 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Sauer M.; Heilemann M. Single-Molecule Localization Microscopy in Eukaryotes. Chem. Rev. 2017, 117 (11), 7478–7509. 10.1021/acs.chemrev.6b00667. [DOI] [PubMed] [Google Scholar]
- Betzig E.; Patterson G. H.; Sougrat R.; Lindwasser O. W.; Olenych S.; Bonifacino J. S.; Davidson M. W.; Lippincott-Schwartz J.; Hess H. F. Imaging intracellular fluorescent proteins at nanometer resolution. Science 2006, 313 (5793), 1642–5. 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- Rust M. J.; Bates M.; Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 2006, 3 (10), 793–5. 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharonov A.; Hochstrasser R. M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (50), 18911–18916. 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzbauer J.; Strauss M. T.; Schlichthaerle T.; Schueder F.; Jungmann R. Super-resolution microscopy with DNA-PAINT. Nat. Protoc. 2017, 12 (6), 1198–1228. 10.1038/nprot.2017.024. [DOI] [PubMed] [Google Scholar]
- Dai M.; Jungmann R.; Yin P. Optical imaging of individual biomolecules in densely packed clusters. Nat. Nanotechnol. 2016, 11 (9), 798–807. 10.1038/nnano.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Woehrstein J. B.; Dai M.; Shih W. M.; Yin P. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 2014, 11 (3), 313–8. 10.1038/nmeth.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries J.; Kaplan C.; Platonova E.; Eghlidi H.; Ewers H. A simple, versatile method for GFP-based super-resolution microscopy via nanobodies. Nat. Methods 2012, 9 (6), 582–4. 10.1038/nmeth.1991. [DOI] [PubMed] [Google Scholar]
- Schlichthaerle T.; Ganji M.; Auer A.; Kimbu Wade O.; Jungmann R. Bacterially Derived Antibody Binders as Small Adapters for DNA-PAINT Microscopy. ChemBioChem 2019, 20 (8), 1032–1038. 10.1002/cbic.201800743. [DOI] [PubMed] [Google Scholar]
- Thevathasan J. V.; Kahnwald M.; Cieslinski K.; Hoess P.; Peneti S. K.; Reitberger M.; Heid D.; Kasuba K. C.; Hoerner S. J.; Li Y.; Wu Y. L.; Mund M.; Matti U.; Pereira P. M.; Henriques R.; Nijmeijer B.; Kueblbeck M.; Sabinina V. J.; Ellenberg J.; Ries J. Nuclear pores as versatile reference standards for quantitative superresolution microscopy. Nat. Methods 2019, 16 (10), 1045–1053. 10.1038/s41592-019-0574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann R.; Avendano M. S.; Dai M.; Woehrstein J. B.; Agasti S. S.; Feiger Z.; Rodal A.; Yin P. Quantitative super-resolution imaging with qPAINT. Nat. Methods 2016, 13 (5), 439–42. 10.1038/nmeth.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A.; Gendreizig S.; Gronemeyer T.; Pick H.; Vogel H.; Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003, 21 (1), 86–9. 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Los G. V.; Encell L. P.; McDougall M. G.; Hartzell D. D.; Karassina N.; Zimprich C.; Wood M. G.; Learish R.; Ohana R. F.; Urh M.; Simpson D.; Mendez J.; Zimmerman K.; Otto P.; Vidugiris G.; Zhu J.; Darzins A.; Klaubert D. H.; Bulleit R. F.; Wood K. V. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol. 2008, 3 (6), 373–82. 10.1021/cb800025k. [DOI] [PubMed] [Google Scholar]
- Kiuchi T.; Higuchi M.; Takamura A.; Maruoka M.; Watanabe N. Multitarget super-resolution microscopy with high-density labeling by exchangeable probes. Nat. Methods 2015, 12 (8), 743–746. 10.1038/nmeth.3466. [DOI] [PubMed] [Google Scholar]
- Schlichthaerle T.; Eklund A. S.; Schueder F.; Strauss M. T.; Tiede C.; Curd A.; Ries J.; Peckham M.; Tomlinson D. C.; Jungmann R. Site-Specific Labeling of Affimers for DNA-PAINT Microscopy. Angew. Chem., Int. Ed. 2018, 57 (34), 11060–11063. 10.1002/anie.201804020. [DOI] [PubMed] [Google Scholar]
- Opazo F.; Levy M.; Byrom M.; Schafer C.; Geisler C.; Groemer T. W.; Ellington A. D.; Rizzoli S. O. Aptamers as potential tools for super-resolution microscopy. Nat. Methods 2012, 9 (10), 938–9. 10.1038/nmeth.2179. [DOI] [PubMed] [Google Scholar]
- Strauss S.; Nickels P. C.; Strauss M. T.; Jimenez Sabinina V.; Ellenberg J.; Carter J. D.; Gupta S.; Janjic N.; Jungmann R. Modified aptamers enable quantitative sub-10-nm cellular DNA-PAINT imaging. Nat. Methods 2018, 15 (9), 685–688. 10.1038/s41592-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano Y.; Yano A.; Oishi S.; Sugimoto Y.; Tsujimoto G.; Fujii N.; Matsuzaki K. Coiled-coil tag--probe system for quick labeling of membrane receptors in living cell. ACS Chem. Biol. 2008, 3 (6), 341–5. 10.1021/cb8000556. [DOI] [PubMed] [Google Scholar]
- Reinhardt U.; Lotze J.; Zernia S.; Morl K.; Beck-Sickinger A. G.; Seitz O. Peptide-templated acyl transfer: a chemical method for the labeling of membrane proteins on live cells. Angew. Chem., Int. Ed. 2014, 53 (38), 10237–41. 10.1002/anie.201403214. [DOI] [PubMed] [Google Scholar]
- Reinhardt U.; Lotze J.; Morl K.; Beck-Sickinger A. G.; Seitz O. Rapid Covalent Fluorescence Labeling of Membrane Proteins on Live Cells via Coiled-Coil Templated Acyl Transfer. Bioconjugate Chem. 2015, 26 (10), 2106–17. 10.1021/acs.bioconjchem.5b00387. [DOI] [PubMed] [Google Scholar]
- Lotze J.; Wolf P.; Reinhardt U.; Seitz O.; Morl K.; Beck-Sickinger A. G. Time-Resolved Tracking of Separately Internalized Neuropeptide Y2 Receptors by Two-Color Pulse-Chase. ACS Chem. Biol. 2018, 13 (3), 618–627. 10.1021/acschembio.7b00999. [DOI] [PubMed] [Google Scholar]
- Doh J. K.; White J. D.; Zane H. K.; Chang Y. H.; Lopez C. S.; Enns C. A.; Beatty K. E. VIPER is a genetically encoded peptide tag for fluorescence and electron microscopy. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (51), 12961–12966. 10.1073/pnas.1808626115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradisar H.; Jerala R. De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers. J. Pept. Sci. 2011, 17 (2), 100–6. 10.1002/psc.1331. [DOI] [PubMed] [Google Scholar]
- Lebar T.; Lainscek D.; Merljak E.; Aupic J.; Jerala R. A tunable orthogonal coiled-coil interaction toolbox for engineering mammalian cells. Nat. Chem. Biol. 2020, 16 (5), 513–519. 10.1038/s41589-019-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. C. The packing of α-helices: simple coiled-coils. Acta Crystallogr. 1953, 6 (8–9), 689–697. 10.1107/S0365110X53001964. [DOI] [Google Scholar]
- McLachlan A. D.; Stewart M. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J. Mol. Biol. 1976, 103 (2), 271–98. 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]
- McLachlan A. D. The double helix coiled coil structure of murein lipoprotein from Escherichia coli. J. Mol. Biol. 1978, 121 (4), 493–506. 10.1016/0022-2836(78)90396-0. [DOI] [PubMed] [Google Scholar]
- Thomas F.; Boyle A. L.; Burton A. J.; Woolfson D. N. A set of de novo designed parallel heterodimeric coiled coils with quantified dissociation constants in the micromolar to sub-nanomolar regime. J. Am. Chem. Soc. 2013, 135 (13), 5161–6. 10.1021/ja312310g. [DOI] [PubMed] [Google Scholar]
- Chao H.; Houston M. E. Jr.; Grothe S.; Kay C. M.; O’Connor-McCourt M.; Irvin R. T.; Hodges R. S. Kinetic study on the formation of a de novo designed heterodimeric coiled-coil: use of surface plasmon resonance to monitor the association and dissociation of polypeptide chains. Biochemistry 1996, 35 (37), 12175–12185. 10.1021/bi9530604. [DOI] [PubMed] [Google Scholar]
- Litowski J. R.; Hodges R. S. Designing heterodimeric two-stranded alpha-helical coiled-coils. Effects of hydrophobicity and alpha-helical propensity on protein folding, stability, and specificity. J. Biol. Chem. 2002, 277 (40), 37272–9. 10.1074/jbc.M204257200. [DOI] [PubMed] [Google Scholar]
- Gröger K.; Gavins G.; Seitz O. Strand Displacement in Coiled-Coil Structures: Controlled Induction and Reversal of Proximity. Angew. Chem., Int. Ed. 2017, 56 (45), 14217–14221. 10.1002/anie.201705339. [DOI] [PubMed] [Google Scholar]
- Schueder F.; Stein J.; Stehr F.; Auer A.; Sperl B.; Strauss M. T.; Schwille P.; Jungmann R. An order of magnitude faster DNA-PAINT imaging by optimized sequence design and buffer conditions. Nat. Methods 2019, 16 (11), 1101–1104. 10.1038/s41592-019-0584-7. [DOI] [PubMed] [Google Scholar]
- De Crescenzo G.; Litowski J. R.; Hodges R. S.; O’Connor-McCourt M. D. Real-time monitoring of the interactions of two-stranded de novo designed coiled-coils: effect of chain length on the kinetic and thermodynamic constants of binding. Biochemistry 2003, 42 (6), 1754–63. 10.1021/bi0268450. [DOI] [PubMed] [Google Scholar]
- Oi C.; Gidden Z.; Holyoake L.; Kantelberg O.; Mochrie S.; Horrocks M. H.; Regan L. LIVE-PAINT: Super-Resolution Microscopy Inside Live Cells Using Reversible Peptide-Protein Interactions. bioRxiv 2020, 1. 10.1101/2020.02.03.932228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood C. W.; Woolfson D. N. CCBuilder 2.0: Powerful and accessible coiled-coil modeling. Protein Sci. 2018, 27 (1), 103–111. 10.1002/pro.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobnak I.; Gradisar H.; Ljubetic A.; Merljak E.; Jerala R. Modulation of Coiled-Coil Dimer Stability through Surface Residues while Preserving Pairing Specificity. J. Am. Chem. Soc. 2017, 139 (24), 8229–8236. 10.1021/jacs.7b01690. [DOI] [PubMed] [Google Scholar]
- Kohn W. D.; Kay C. M.; Hodges R. S. Salt effects on protein stability: two-stranded alpha-helical coiled-coils containing inter- or intrahelical ion pairs. J. Mol. Biol. 1997, 267 (4), 1039–52. 10.1006/jmbi.1997.0930. [DOI] [PubMed] [Google Scholar]
- Rothemund P. W. Folding DNA to create nanoscale shapes and patterns. Nature 2006, 440 (7082), 297–302. 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- Agasti S. S.; Wang Y.; Schueder F.; Sukumar A.; Jungmann R.; Yin P. DNA-barcoded labeling probes for highly multiplexed Exchange-PAINT imaging. Chem. Sci. 2017, 8 (4), 3080–3091. 10.1039/C6SC05420J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss S.; Jungmann R. Up to 100-fold speed-up and multiplexing in optimized DNA-PAINT. Nat. Methods 2020, 17, 789–791. 10.1038/s41592-020-0869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.