Abstract
Objective
Patients with Sjӧgren's syndrome (SS) have an increased risk of developing malignant B cell lymphomas, particularly mucosa‐associated lymphoid tissue (MALT)–type lymphomas. We have previously shown that a predominant proportion of patients with SS‐associated salivary gland MALT lymphoma express somatically hypermutated IgM with strong amino acid sequence homology with stereotypic rheumatoid factors (RFs). The present study was undertaken in a larger cohort of patients with SS‐associated MALT lymphoma to more firmly assess the frequency of RF reactivity and the significance of somatic IGV‐region mutations for RF reactivity.
Methods
B cell antigen receptors (BCRs) of 16 patients with SS‐associated salivary gland MALT lymphoma were analyzed. Soluble recombinant IgM was produced of 12 MALT lymphoma samples, including 1 MALT lymphoma sample that expressed an IgM antibody fitting in a novel IGHV3‐30–encoded stereotypic IGHV subset. For 4 of the 12 IgM antibodies from MALT lymphoma samples, the somatically mutated IGHV and IGKV gene sequences were reverted to germline configurations. Their RF activity and binding affinity were determined by enzyme‐linked immunosorbent assay and surface plasmon resonance, respectively.
Results
Nine (75%) of the 12 IgM antibodies identified in patients with SS‐associated salivary gland MALT lymphoma displayed strong monoreactive RF activity. Reversion of the IGHV and IGKV mutations to germline configuration resulted in RF affinities for IgG that were significantly lower for 3 of the 4 somatically mutated IgM antibodies. In stereotypic IGHV3‐7/IGKV3‐15–encoded RFs, a recurrent replacement mutation in the IGKV3‐15–third complementarity‐determining region was found to play a pivotal role in the affinity for IgG‐Fc.
Conclusion
A majority of patients with SS‐associated salivary gland MALT lymphoma express somatically mutated BCRs that are selected for monoreactive, high‐affinity binding of IgG‐Fc. These data underscore the notion that soluble IgG, most likely in immune complexes in inflamed tissues, is the principal autoantigen in the pathogenesis of a variety of B cell lymphomas, particularly SS‐associated MALT lymphomas.
INTRODUCTION
Mucosa‐associated lymphoid tissue (MALT)–type lymphomas account for 5–10% of all B cell lymphomas. These lymphomas occur at various anatomic sites affected by chronic inflammation emerging from pathogens or autoimmunity 1, 2, 3, 4. Among all autoimmune diseases, Sjögren's syndrome (SS) is most strongly associated with lymphoma development, in particular salivary gland MALT lymphomas 1, 2, 3, 4, 5, 6, 7. Clinical predictors of lymphoma development are swelling of the salivary glands, splenomegaly, lymphadenopathy, and palpable purpura. The main biologic predictors are cryoglobulinemia, lymphopenia, low complement levels, and a monoclonal component in the serum or urine 3, 4. Recently, the presence of rheumatoid factors (RFs) in the serum was identified as an independent predictor of lymphoma development 8.
Previously, we have shown that ~40% of patients with salivary gland MALT lymphoma express near‐identical (also called stereotypic) B cell antigen receptors (BCRs), with a striking degree of VH–third complementarity‐determining region (CDR3) amino acid (aa) sequence homology with stereotypic RFs 9. Five groups of stereotypic RFs, each characterized by a canonical combination of IGHV and IGKV gene sequences and a distinct VH‐CDR3, have been identified. They are encoded by 2 distinct IGHV1‐69/JH4 rearrangements, designated as V1‐69‐RF and WOL‐RF (also known as RFs of the Wa idiotype), and by IGHV3‐7/JH3, IGHV4‐59/JH2, and IGHV4‐59/JH5 rearrangements, designated V3‐7‐RF, V4‐59‐RF, and V4‐59/JH5‐RF, respectively 9, 10. Stereotypic RF BCRs are also frequently expressed in gastric MALT lymphoma and hepatitis C virus (HCV)–related B cell lymphoma, and less frequently in ocular adnexal MALT lymphoma, splenic marginal zone B cell lymphoma, diffuse large B cell lymphoma, and B cell chronic lymphocytic leukemia (CLL) 9, 10, 11, 12, 13, 14, 15. Polyclonal stereotypic RFs are frequently present in HCV‐infected patients with type II mixed cryoglobulinemia and in donors immunized with mismatched red blood cells 16, 17.
In a previous study by our group 9 and by Martin et al 18, soluble IgM antibodies were recombinantly produced from SS‐associated salivary gland MALT lymphomas of a total of 8 patients, 6 of which showed strong monoreactive RF activity. Two of these 6 RFs were encoded by stereotypic RF IGHV rearrangements, i.e., V1‐69‐RF and V3‐7‐RF, respectively. Interestingly, all of the RFs expressed either an IGKV3‐15–encoded or IGKV3‐20–encoded light chain, both of which are also typically expressed by IGHV3‐7–encoded and IGHV1‐69–encoded stereotypic RFs.
In the present study, we assessed the configuration and specificity of the BCRs of 16 patients with SS‐associated salivary gland MALT lymphoma, and examined the contribution of the somatic mutations present in IGHV and IGKV on the affinity of RFs for IgG‐Fc.
PATIENTS AND METHODS
MALT lymphoma tissue samples
Frozen salivary gland tissue samples from patients with SS‐associated MALT lymphoma (patient samples M83, M86–M89, M91, and M93–M96) were obtained from the Departments of Pathology, Rheumatology, and Clinical Immunology at University Medical Center Groningen, The Netherlands. In addition, paraffin‐embedded salivary gland tissue from 1 patient (sample M83) was obtained from the Department of Pathology at Haaglanden Medical Center, The Hague, The Netherlands. The salivary gland samples from patients M5, M8, M11, M14, M21, and M22 have been described previously 9. Available serologic data from patients M86–M89, M91, and M93–M96 showed that all were positive for IgM‐RF, anti‐SSA, and, except for patients M87 and M91, anti‐SSB antibodies. Patients M91, M93, and M94 had low serum C4 levels (the patients’ immunohistochemical and serologic data are provided in Supplementary Table 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract). Presence of IgM‐RF and low levels of C4 are biologic predictors of lymphoma development 3, 4, 8.
The study was conducted in accordance with the ethics standards of our institutional medical ethics committee on human experimentation, as well as in accordance with the 1975 Declaration of Helsinki, as revised in 1983.
Immunoglobulin gene analysis
Of the salivary gland tissue from patients with SS‐associated MALT lymphoma, Ig clonality was assessed by polymerase chain reaction (PCR). In addition, immunohistochemical analyses were used to detect light‐chain restriction in those MALT lymphoma salivary gland samples showing plasmacytic differentiation, with monotypic IgMκ expression observed in 10 samples (M5, M8, M11, M21, M83, M86, M91, M93, M94, and M95), and IgAκ expression in 2 samples (M14 and M22). Salivary gland tissue from patient M96 expressed IgM, but immunohistochemical staining for the κ and λ light chains was indeterminate. In the samples from patients M87, M88, and M89, immunohistochemical staining for both the Ig heavy chain and Ig light chain was indeterminate (see Supplementary Table 1 [http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract]). However, in analyses by reverse transcription–PCR (RT‐PCR) aimed at IgM, IgG, and IgA transcripts, it was found that the salivary gland tissue from patients M87, M88, and M89 expressed monoclonal IgM (Table 1).
Table 1.
IGHV and IGKV sequences of 10 salivary gland mucosa‐associated lymphoid tissue–type lymphomas of Sjögren’s syndrome patients
| Patient | Ig isotypea | IGHV; IGKV rearrangements | IGHV; IGKV somatic mutationsb | VH‐CDR3; VK‐CDR3 sequencec | VH‐CDR3; VK‐CDR3 length§ | IgM produced |
|---|---|---|---|---|---|---|
| M83 | IgM | V3‐74/D4‐23/JH4; VK1‐39/JK1 | 22; 9 | C AREYGNSRFFDY WGQG; C QQSYTSPRT FGQG | 12; 9 | No |
| M86 | IgM | V3‐30/D6‐13/JH4; VK3‐15/JK1 | 21; 11 | C AQGAFSNKWYSIGDY WGQG; C QQYNNWPPWT FGQG | 15; 10 | Yes |
| M87 | IgM | V1‐2/D5‐18/JH5 | 17 | C ARGQGTQVSHSWFDP WGQG | 15 | No |
| M88 | IgM | V1‐69/D5‐12/JH4 | 21 | C AREGKASVANGAFDY WGQG | 15 | No |
| M89 | IgM | V4‐59/D6‐6/JH3; VK3‐15/JK1 | 5; 5/7 | C ARDIANIATRRDDAFDI WGQG; C QHYNNWPPWT FGQG | 17; 10 | Yes |
| M91 | IgM | V1‐69/D5‐24/JH4; VK3‐20/JK2 | 7; 0 | C ARKGGDRGDAYDVFDY WGQG; C QQYGSSPYT FGQG | 16; 9 | Yes |
| M93‐I | IgM | V3‐7/D3‐22/JH3; VK3‐15/JK1 | 6/9; 5 | C ARGDYYDSGGS(S/N)YHDAFDV WGQG; C QHYNNWPPWT FGQG | 19; 10 | Yes |
| M93‐II | IgM | V4‐59/D3‐10/JH5; VK4‐1/JK2 | 12; 11 | C ACGGGGSGTYFRGWFDP WGQG; C QQYYSTLYT FGQG | 17; 9 | No |
| M94 | IgM | V3‐7/D3‐22/JH3; VK3‐15/JK1 | 10; 5 | C ARGDYYDSSGYYIDAFDI WGQG; C QHYNNWPPWT FGQG | 18; 10 | Yes |
| M95 | IgM | V3‐48/D3‐22/JH3; VK3‐15/JK4 | 18; 12 | C AREPYSDSSSFFPGSFDI WGQG; C QQYDNWPLT FGGG | 18; 9 | No |
| M96 | IgM | V3‐30.3/D2‐8/JH3; VK3D‐15/JK1 | 23/24; 4 | C ARAGQGSNGIW(G/D)GAFD(T/S) WGQG; C QHYNNWPPWT FGQG | 17; 10 | Yes |
Determined by reverse transcription–polymerase chain reaction.
Values are the number of mutations. Two values separated by a slash sign signify identification of molecular IGHV/IGKV clones harboring slightly different numbers of somatic mutations.
For the VH– and VK–third complementarity‐determining region (CDR3) sequences in which 2 amino acids separated by a slash sign are depicted, this indicates that 2 molecular IGHV clones with dissimilar amino acids at this position were identified.
Values are the number of amino acids.
In the paraffin‐embedded salivary gland tissue obtained from patient M83, DNA was isolated using a QIAamp DNA Mini kit (Qiagen). RNA from all other MALT lymphoma salivary gland samples was isolated using TRI Reagent (Sigma‐Aldrich, Merck), and complementary DNA was synthesized using Pd(N)6 random primers and Moloney murine virus RT (Invitrogen, Fisher Scientific), according to the manufacturer's protocol. The rearranged IGHV genes were amplified using IGHV primers specific for the framework region 1 (FR1) and FR2, in combination with fluorochrome‐labeled reverse primers specific for Ig JH and the constant regions of IgM (Cμ), IgG (Cγ), and IgA (Cα) 19. The IGHV PCR products were analyzed by gene scanning (VH‐CDR3 spectratyping). In salivary gland samples M86–M89, M91, and M93–M96, the FR1/FR2 PCR products with the Cμ primer yielded clonal IGHV PCR products. In the salivary gland tissue from patient M93, 2 clonal peaks were detected. The FR1/FR2 PCR with the Cγ primer yielded polyclonal IGHV PCR products in all MALT lymphoma samples. The FR1/FR2 PCR Cα primer yielded polyclonal or oligoclonal IGHV PCR products in most of the MALT lymphoma samples. An FR1/FR2 PCR with the Ig JH downstream primer was performed on the DNA sample from patient M83, which resulted in an IGHV PCR product with a monoclonal component. The clonal IGHV PCR products were sequenced using a Big Dye Terminator sequencing kit (ThermoFisher Scientific). The IGHV sequences were analyzed using V‐Quest on the IMGT web site (http://www.imgt.org/) 20. IGKV PCR was performed with IGKV family–specific leader primers, combined with either Ig JK or Ig JK –constant region primers. The IGKV sequences were analyzed using V‐Quest on the IMGT web site.
Production of recombinant MALT lymphoma–derived IgM antibodies
The IGHV sequences obtained using the FR1 × Cμ PCR products and appropriate IGHV family–specific leader primers, combined with an Ig JH primer, were used to produce full‐length IGHV gene products. The full‐length IGHV PCR products were obtained as described above. The complete IGHV and IGKV genes were cloned in pTOPO‐TA plasmids and subsequently in the pIGH(μ) and pIGL(κ) expression plasmids, which were then cotransfected in SP2/0 myeloma cells, as previously described 9. Supernatants of the transfected SP2/0 cells were screened for IgMκ production, using enzyme‐linked immunosorbent assays (ELISAs). The various IGHV and IGKV full‐length sequences in which somatic mutations were reverted to germline configuration (as seen in salivary gland samples M91, M93, M94, and M96) were ordered from GenScript.
Antigen‐specific ELISA and antigen affinity measurement
ELISA plates were coated overnight with human IgG (Sanquin), insulin from bovine pancreas, actin from bovine muscle, lipopolysaccharide (LPS) from Escherichia coli O55:B5 (Sigma‐Aldrich, Merck), and SSA/Ro52 or SSA/Ro60 (Diarect) in coating buffer (0.05M Na2CO3, 0.05M NaHCO3, pH 9.6). The ELISA plates were incubated with recombinant lymphoma–derived IgM in serial dilutions starting from 1 μg/ml, and then subsequently incubated with horseradish peroxidase–conjugated mouse anti‐human IgM (Southern Biotechnology), followed by development with tetramethylbenzidine as described previously 9, 21.
Surface plasmon resonance assays were performed on an IBIS Mx96 instrument (IBIS Technologies) using a G‐type anti–human IgM chip. Data on the binding affinity of the IgM antibodies were processed as described previously 22.
RESULTS
IGHV and IGLV genes in salivary gland tissue from patients with SS‐associated MALT lymphoma
We studied a panel of 16 salivary gland samples from patients with SS‐associated MALT lymphoma; 6 of the IGHV and IGKV sequences had been determined previously 9. Of 10 remaining new MALT lymphoma salivary gland samples, IGHV and IGKV transcripts were amplified by RT‐PCR using IGHV– and IGKV–specific primers in combination with primers specific for the constant region of the μ heavy chain and κ light chain, respectively. All 10 MALT lymphoma samples expressed IgM antibodies, and their respective IGHV sequences were assessed (Table 1). In 1 of the samples (M93), we identified 2 distinct clonal IGHV sequences. In 8 of the 10 samples, we were able to identify the corresponding IGKV sequences as well. Remarkably, 6 MALT lymphoma salivary gland samples expressed an IGKV3‐15 rearrangement, and 1 expressed an IGKV3‐20 rearrangement. IGKV3‐15 and IGKV3‐20 are typically found in IGHV3‐7– and IGHV1‐69–encoded stereotypic RFs, respectively 9. In the salivary gland samples from patients M87 and M88, the IGKV PCR yielded polyclonal amplicons, most likely attributable to a relatively low tumor load, and thus we were prohibited from reliably identifying the expressed IGKV sequences. All of the IGHV and IGKV genes identified were somatically mutated, except for the IGKV gene in the salivary gland tissue from patient M91, which was unmutated (Table 1 and Supplementary Figure 1, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract).
Findings of VH‐CDR3 amino acid sequence homology studies
The VH‐CDR3 aa sequences in the salivary glands from patients with MALT lymphoma were compared with the VH‐CDR3 aa sequences from GenBank (release 228), obtained using the NCBI Protein‐BLAST algorithm. According to our arbitrary criteria, the VH‐CDR3s were considered homologous when they shared at least 60% aa sequence homology, with an allowed maximal gap of 3 aa. Except for samples M83, M91, M93‐II, and M96, the VH‐CDR3 aa sequences in all other samples showed VH‐CDR3 homology with MALT lymphomas, but also with other B cell lymphomas such as CLL (see Supplementary Table 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract). The VH‐CDR3s in salivary gland samples M88, M93‐I, M94, and M95 were found to be homologous with those of B cells derived from inflamed salivary glands and, except for sample M95, all were also homologous with stereotypic RFs, i.e., VH‐CDR3s in samples M93‐I and M94 showed homology with stereotypic V3‐7‐RFs, and VH‐CDR3 in sample M88 showed homology with stereotypic V1‐69‐RFs (Figure 1; see also Supplementary Tables 2 and 3, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract). Of note, sample M91 did not display a VH‐CDR3 sequence that was homologous with stereotypic V1‐69‐RFs, although this salivary gland tissue sample expressed the canonical V1‐69‐RF gene combination of IGHV1‐69/IGHJ4 and IGKV3‐20. Salivary gland sample M86 shared VH‐CDR3 homology with an IGHV3‐30–encoded HCV‐associated MALT lymphoma (Figure 1).
Figure 1.

VH–third complementarity‐determining region (CDR3) amino acid sequence homology of salivary gland mucosa‐associated lymphoid tissue (MALT)–type lymphomas of patients M86, M88, M93‐IB, M94, and M95 with 4 stereotypic rheumatoid factors (RFs) and with a MALT lymphoma (MALT‐L) from a hepatitis C virus (HCV)–infected patient. VH‐CDR3 homology is calculated as a percentage of identical (red) and similar (blue) amino acids.
Expression of monospecific RFs in most SS‐associated MALT lymphoma salivary gland samples
We produced, in total, 12 recombinant IgM antibodies from the MALT lymphoma salivary gland samples in our study, i.e., samples M86, M89, M91, M93‐I, M94, and M96, and from 6 MALT lymphoma salivary gland samples from our earlier study, i.e., samples M5, M8, M11, M14, M21, and M22 9. Of the salivary gland tissue from patient M93, we produced 2 recombinant IgM antibodies, M93‐IA and M93‐IB, which differed by 4 aa in IGHV. Of sample M96, we produced 2 recombinant IgM antibodies, M96‐A and M96‐B, which differed by 2 aa in the VH‐CDR3 (see Supplementary Figure 1 [http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract]).
The IgM antibodies produced from the MALT lymphoma samples were tested by ELISA for binding to insulin, actin, LPS, human IgG, SSA/Ro52, and SSA/Ro60. Except for samples M89, M8, and M14 in our earlier study, the IgM antibodies from all other MALT lymphoma salivary gland samples reacted with IgG and did not bind to any of the other antigens. The IgM antibodies from samples M91 and M96‐B showed the strongest IgG‐Fc binding (Figures 2A and B). It was noted that the M96‐A IgM antibody showed a much weaker binding to IgG‐Fc than the M96‐B IgM antibody.
Figure 2.
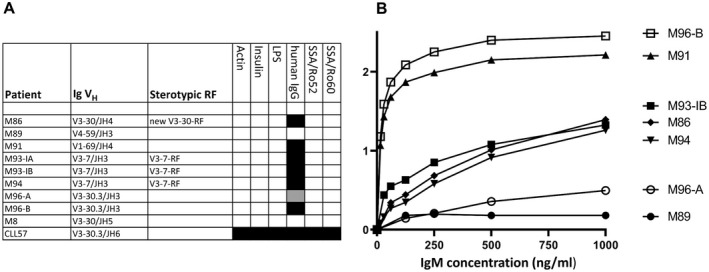
MALT lymphoma IgM antibodies characterized as high‐affinity monoreactive RFs. In enzyme‐linked immunosorbent assays (ELISAs), IgM derived from MALT lymphoma salivary gland samples M86, M91, M93‐I, M94, and M96 show specific binding to coated human IgG (A), with different binding affinities (B). Values on the y‐axis are the absorbance measured at 450 nm. Sample M8 is an IgM from a patient with Sjӧgren’s syndrome–associated MALT lymphoma who was analyzed previously and does not show RF activity. Sample CLL57 is an IGV‐gene–unmutated IgM derived from a patient with B cell chronic lymphocytic leukemia (CLL) and shows strong polyreactivity. Black boxes indicate positive ELISA results at all IgM concentrations tested; the gray‐shaded box indicates positive ELISA results at the 2 highest IgM concentrations tested. The IgMs were tested in at least 4 concentrations, i.e., 125, 250, 500, and 1,000 ng/ml. LPS = lipopolysaccharide (see Figure 1 for other definitions).
Overall, among our panel of 12 salivary gland MALT lymphoma IgM antibodies, 9 (75%) showed strong monoreactive RF activity. This panel included 5 stereotypic RFs, i.e., 1 V1‐69‐RF (sample M11), 3 V3‐7‐RFs (samples M5, M93‐I, and M94), and the newly recognized IGHV3‐30–encoded RF (sample M86).
Recurrent replacement mutations in the IGHV and IGKV genes of RFs
We aligned the aa sequences of the IGHV1‐69– and IGHV3‐7–encoded salivary gland MALT lymphoma RFs, including 3 IGHV1‐69–encoded RFs (samples M9, M11, and M22) and 2 IGHV3‐7–encoded RFs (samples M5 and M6) from our earlier study 9. Our results demonstrated that the IGHV1‐69–encoded RFs shared an A65P mutation (samples M88, M9, M11, and M22) or an I59L mutation (samples M91 and M11) in the VH‐CDR2, while an S36T mutation in the VH‐CDR1 was seen in 2 samples (M91 and M9). In the IGHV3‐7–encoded RFs, we found a Q58P mutation that was shared between salivary gland samples M93‐I and M6 only. Of note, samples M6 and M9 were from a patient with gastric MALT lymphoma and a patient with tonsillar MALT lymphoma 9.
Alignment of 8 IGKV3‐15 aa sequences, including IGKV3‐15 sequences of 2 stereotypic V3‐7‐RF–expressing MALT lymphoma salivary gland tissue samples from our previous study, demonstrated that all stereotypic V3‐7‐RFs (in samples M93, M94, M5, and M6) possessed a Q106H mutation in their VK‐CDR3 region. In addition, an IGHV4‐59–expressing MALT lymphoma (sample M89), which did not show RF activity, and an IGHV3‐30.3–expressing RF (sample M96) also shared this Q106H mutation (Figures 3A–C).
Figure 3.
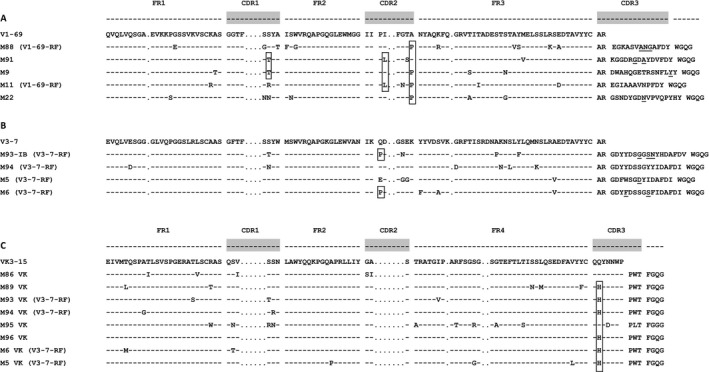
Alignment of amino acid sequences of IGHV1‐69–, IGHV3‐7–, and IGKV3‐15– encoded RFs of salivary gland MALT lymphomas. Boxes highlight shared somatic replacement mutations, i.e., S36T in the VH‐CDR1 of samples M91 and M9, I59L in the VH‐CDR2 of samples M91 and M11, and A65P in the VH‐CDR2 of samples M88, M9, M11, and M22 (A), Q58P in the VH‐CDR2 of samples M93IB and M6 (B), and Q106H in the VK‐CDR3 of samples M89, M93, M94, M96, M6, and M5 (C). Somatic replacement mutations in the VH‐CDR3 are underlined. FR1 = framework region 1 (see Figure 1 for other definitions).
Contribution of somatic mutations to the affinities of the MALT lymphoma RFs
We produced recombinant variants of the IgM antibodies of MALT lymphoma salivary gland samples M91, M93‐I, M94, and M96, in which somatic mutations in IGHV and/or IGKV were reverted to their respective germline configuration. Subsequently, we measured their affinity for IgG using surface plasmon resonance.
Reversion of the 4 replacement mutations in IGHV in MALT lymphoma salivary gland sample M91 had little effect on RF affinity (Figures 4A and B; compare the findings between bar A and bar B). However, when 2 replacement mutations in the VH‐CDR3 (Figure 3) were also reverted, the RF affinity was completely lost (Figures 4A and B; compare the findings of bars A and B with bar C).
Figure 4.
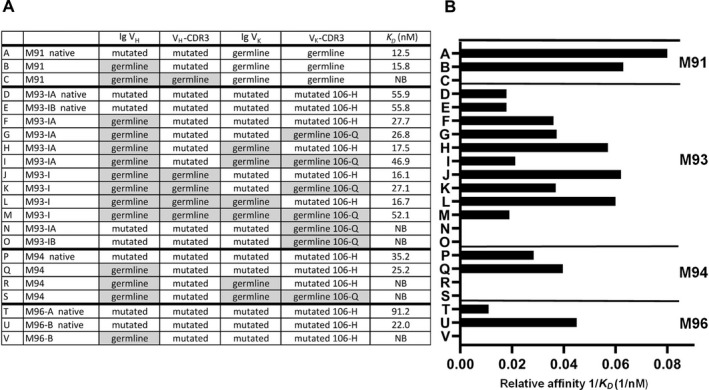
Binding affinities of MALT lymphoma RFs and germline‐reverted variants for human IgG, as measured by surface plasmon resonance assay. A, IGHV and IGKV configurations for the native lymphoma RFs and germline‐reverted variants of lymphoma RFs, are shown, along with their binding constants (K D) to indicate affinity for human IgG. B, Histograms display the 1/K D values of the native and germline‐reverted variants of lymphoma RFs. Each letter to the left of the bars for each sample represents an individual recombinantly produced IgM antibody. NB = no binding; 106‐H = histidine mutation on position 106 of VK‐CDR3; 106‐Q = germline glutamine on position 106 of VK‐CDR3 (see Figure 1 for other definitions).
The 2 native stereotypic V3‐7‐RF variants of sample M93‐I (variants M93‐IA and M93‐IB), which had 4 dissimilar aa in their IGHV sequences, had the same affinity for IgG‐Fc (Figures 4A and B; compare the findings in bar D with bar E). All IgM variants in sample M93‐I in which IGHV, with or without the VH‐CDR3 and with or without IGKV, were reverted to germline configuration showed slightly higher affinities for IgG‐Fc when compared to the natural IgM antibodies in sample M93‐I (M93‐IA and M93‐IB) (Figures 4A and B; compare the findings in bars F, G, H, I, J, K, and L with bars D and E). Even a completely reverted IgM of M93‐I, in which the H106Q mutation in VK‐CDR3 was also reverted, showed an affinity that was similar to that of the naturally mutated M93‐IA and M93‐IB antibodies (Figures 4A and B; compare bar M with bars D and E). The 106‐H mutation in the VK‐CDR3, however, proved to be as important for RF affinity, as was illustrated by the 3 variant pairs (H and I, J and K, and L and M in Figures 4A and B), which showed higher RF affinities in the mutated 106‐H than in the 106‐Q germline configuration. Most exemplary in this respect is the complete loss of RF affinity of M93‐IA and M93‐IB when the naturally mutated IgM antibodies were expressed with IGKV, in which only 106‐Q was in germline configuration (Figures 4A and B; compare bars N and O).
The V3‐7‐RF–expressing IgM antibodies in salivary gland sample M94, in which the 6 replacement mutations in IGHV were reverted, displayed a slightly higher RF activity as compared to its native mutated variant. However, when the 2 replacement mutations in IGKV were also reverted, the RF activity was completely lost, which was already the case when the 106‐H mutation was preserved (Figures 4A and B; compare bar R and bar S).
Finally, of the natural variants in salivary gland sample M96, the M96‐B variant, which expressed dissimilar aa at 2 positions in the VH‐CDR3 as compared to variant M96‐A, showed a much higher affinity for IgG‐Fc. Reversion of the 12 replacement mutations in IGHV in the M96‐B variant resulted in the complete loss of RF activity (Figures 4A and B; compare bars T and U with bar V). Of note, the affinity measurements in MALT lymphoma salivary gland variants M96‐A and M96‐B correlated nicely with the results of the RF ELISA (as shown in Figure 2).
DISCUSSION
The results of this study demonstrate that in most, but not all, MALT lymphomas occurring in patients with SS, BCRs with strong RF reactivity are expressed. Taken together with the results of a previous study by Martin et al 18 on 2 SS‐associated MALT lymphomas, a total of 14 IgM antibodies of patients with SS‐associated salivary gland MALT lymphoma have been produced recombinantly, of which 11 showed RF specificity. Five of these were stereotypic RFs, i.e., 1 sample with V1‐69‐RF, 3 samples with V3‐7‐RFs, and 1 sample (M86) that shared VH‐CDR3 homology with an IGHV3‐30–encoded HCV‐associated MALT lymphoma, which could be considered as a newly identified stereotypic RF‐BCR group.
Remarkably, all MALT lymphoma IgM antibodies that showed RF activity expressed an Igκ light chain that was encoded either by IGKV3‐15 (samples M5, M86, M93‐I, M94, and M96) or by IGKV3‐20 (samples M11, M21, M22, and M91). Of the 3 MALT lymphoma IgM antibodies without RF activity (samples M8, M14, and M89), only those in the salivary gland tissue of patient M89 expressed an IGKV3‐15–encoded Igκ. In 5 MALT lymphoma samples, we did not produce IgM antibodies but we garnered information about their IGHV sequences with or without IGKV (samples M83, M87, M88, M93‐II, and M95); sample M95 also co‐expressed IGKV3‐15 and an IGHV3‐48–encoded Ig heavy chain with VH‐CDR3 homology with stereotypic V3‐7‐RFs. Sample M83 expressed an IGHV3‐74–encoded Ig heavy chain. IGHV3‐74 was also recently identified, by mass spectrometric sequencing of serum RFs, as one of the IGHV aa sequences present in 9 of 15 patients with SS 23. Finally, sample M88 expressed a stereotypic V1‐69‐RF Ig heavy chain. These data indicate that the MALT lymphoma salivary gland IgM antibodies in samples M83, M88, and M95 are likely also RFs.
Stereotypic RF reactivity is not unique to salivary gland MALT lymphomas. We have also demonstrated stereotypic RF reactivity in 2 samples from patients with gastric MALT lymphoma 9, 2 with CLL 12, 15, and 4 with HCV‐related B cell lymphoma 10. It is remarkable that whereas most SS‐associated salivary gland MALT lymphomas express RFs, the frequency of stereotypic RF B cells in inflamed salivary glands of SS patients was found to be low 24, 25. This suggests that RF B cells in SS, once recruited in an inflamed salivary gland and activated by IgG immune complexes, e.g., IgG‐SSA/SSB, have a growth advantage and are prone to transform 25.
Analysis of IGHV1‐69–encoded RFs demonstrated a limited number of recurrent replacement mutations, i.e., A65P and I59L in the VH‐CDR2, and S36T in the VH‐CDR1, which were present in 4 RFs, 2 RFs, and 2 RFs, respectively (of a total of 5). Among the 14 reported SS‐associated MALT lymphoma–derived IGHV1‐69 sequences, whose RF activity was not explored in vitro, the A65P mutation was not found, whereas I59L was found in 2 cases and S36T was found in 3 cases 26, 27. In the 11 IGHV1‐69–encoded RFs found in HCV‐infected patients with cryoglobulinemia, A65P was present in 3 cases, and both I59L and S36T were found in 2 cases 28.
In 2 of 4 stereotypic V3‐7‐RFs, we identified a recurrent Q58P mutation in the VH‐CDR2. In the literature, Q58P was reported to be present in 2 of 4 IGHV3‐7 sequences in the salivary glands of patients with SS‐associated MALT lymphoma 26, 27. Alignment of the corresponding IGKV3‐15 aa sequences of our stereotypic V3‐7‐RFs showed that all 4 carried a Q106H replacement mutation in their VK‐CDR3 region. Interestingly, the IGHV4‐59–expressing salivary gland MALT lymphoma from patient M89, which did not display RF activity, and the IGHV3‐30.3–expressing RF from patient M96 both harbored a Q106H mutation in the VK‐CDR3 as well. In the literature, it was reported that Q106H was found in an SS‐associated MALT lymphoma 18 and in a stereotypic V3‐7‐RF from a patient with CLL 12. For both of these lymphomas, in vitro RF activity was demonstrated 12, 18.
We observed that MALT lymphoma salivary gland sample M91 expressed the stereotypic combination of IGHV1‐69 and IGKV3‐20. However, this RF does not share VH‐CDR3 homology with stereotypic V1‐69‐RFs. Reversion of its 4 IGHV replacement mutations showed a small reduction in RF affinity, and when the 2 replacement mutations in the VH‐CDR3 were also reverted, IgG binding was completely lost.
These results were recently clarified in a description of the crystal structure of an IGHV1‐69/IGKV3‐20–encoded RF (YES8c) in complex with IgG1‐Fc 29. The YES8c RF also does not display VH‐CDR3 homology with V1‐69‐RFs. The VH‐CDR3 in sample M91 shared 47% aa sequence homology with the VH‐CDR3 of YES8c. In total, atoms of 12 IGHV aa of YES8c were shown to interact with the CH2 and CH3 regions of IgG1‐Fc (Figure 5A). Alignment of the sequences in samples M91 and YES8c demonstrates that 7 of these 12 aa are shared between M91 and YES8c. In sample M91, only 2 of the 4 replacement mutations in the IGHV region (S36T and I59L) were contact residues with IgG‐Fc, of which the I59L mutation, present both in M91 and in YES8c, led to a lower affinity in YES8c. Since serine and threonine are similar, the S36T mutation likely does not add much to the affinity in sample M91 29. It thus is envisioned that the reversion of these 4 replacement mutations in sample M91 will yield a small difference in RF affinity (Figure 4). Interestingly, the R111.1‐G mutation in the M91 VH‐CDR3 resulted in an identical aa sequence as that in YES8c. The 111.1‐G mutation was shown to interact with IgG1‐Fc (Figure 5B). The contribution of a patch of 4 or 5 contiguous aa in the VH‐CDR3 to the Fc‐IgG1 binding affinity was substantiated by the complete loss of RF activity upon germline reversion of 2 mutations within this patch. Eight IGKV aa of YES8c, of which 7 aa are identical to those in M91, were shown to interact with IgG1‐Fc (Figure 5A). Finally, the VH‐CDR3 of YES8c shared 73% homology with the VH‐CDR3 expressed by an SS‐associated salivary gland MALT lymphoma from GenBank (sample T580B) (Figure 5B).
Figure 5.
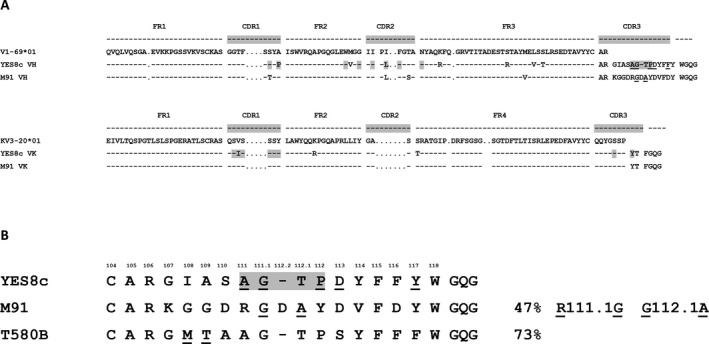
Alignment of the IGHV1‐69–encoded RFs of YES8c and M91. A, Somatic replacement mutations, as compared with the IGHV1‐69 and IGKV3‐20 germline sequences, are indicated. B, Alignment of the VH‐CDR3 amino acid sequences in YES8c, M91, and the homologous T580B, which originated from a patient with a Sjögren's syndrome–related MALT lymphoma, are indicated. Somatic replacement mutations are underlined. Gray‐shaded boxes highlight the amino acids of RFs in YES8c that are the contact residues with IgG‐Fc. FR1 = framework region 1 (see Figure 1 for other definitions).
The results of the IgG binding studies of the 12 constructed variants of the RFs in MALT lymphoma salivary gland sample M93‐I were, in part, more difficult to interpret. In sample M93‐I, a completely germline‐reverted IGHV/IGKV variant showed a similar RF affinity as that of the native IgM antibodies of M93‐IA and IB. All reverted variants of IGHV, with or without the VH‐CDR3 and with or without IGKV, displayed even slightly enhanced RF affinities. However, the 106‐H mutation of the VK‐CDR3 contributed to the RF affinity, which was illustrated by the variant pairs that differed only with respect to each other between 106‐H and 106‐Q. Most explanatory in this respect is the complete loss of RF affinity when the naturally mutated IgM antibodies of M93‐IA and IB were expressed with IGKV, in which only 106‐Q was in germline configuration (Figures 4A and B, bar N and bar O). Unfortunately, of the stereotypic V3‐7‐RFs, no crystal structure has as yet been reproduced. Our observation that reversion of particular replacement mutations in RFs can lead to higher IgG‐Fc affinities has been described before 28, 29.
A completely germline‐reverted IGHV/IGKV variant of V3‐7‐RF in sample M94 did not show any IgG binding. The VH‐CDR3 of M96‐A harbored 2 extra replacement mutations as compared to that of M96‐B, and yet it showed a much lower affinity for IgG, indicating that the germline aa at this position are important for RF affinity. Moreover, reversion of 12 IGHV replacement mutations both in M96‐A and in M96‐B resulted in a complete loss of IgG binding.
Taken together, these findings indicate that somatic IGV‐gene mutations contribute differently to the affinity of the RFs for IgG‐Fc. Germline reversion of replacement mutations in IGHV, including the VH‐CDR3 and, if applicable, also IGKV, resulted in lower RF affinities of the IgM antibodies of samples M91, M94, and M96. The affinities of the RFs varied, with binding constants (K D) ranging between 91 nM and 12 nM. These affinity values are lower than we have previously measured for tetanus toxoid–specific antibodies, which were found to have K D values between 1.9 nM and 0.1 nM 22. Overall, the experiments showing reversion of IGV‐gene mutations have demonstrated that the RFs are affinity‐selected. Even the germline‐converted RFs of M93‐IA and IB, which displayed an affinity similar to its naturally mutated variant, indicated that although the IGV‐gene mutations did not enhance the RF affinity, they still seemed to be selected to preserve the RF affinity and specificity.
Recently, it was shown that the RF epitopes of monomeric IgG in solution are shielded for soluble RFs and RF‐expressing B cells. The RF epitopes may become accessible, e.g., after physical adsorption on a hydrophobic surface in ELISA plates, by heat treatment or in vivo by binding of IgG to its antigen‐forming IgG immune complexes 30. The fact that RF B cells can be activated only by IgG in immune complexes was shown before 31, 32. Moreover, in a mouse model, RF B cells were shown to be activated by IgG–chromatin complexes through the synergistic engagement of the RF‐BCR and Toll‐like receptor 9 (TLR‐9) 33. As it is known that SSA/SSB are part of a ribonucleoprotein complex containing single‐stranded RNA stem‐loop structures, RF B cells in SS salivary glands may also be activated by IgG–SSA/SSB immune complexes via the RF‐BCR and TLR‐7. In support of this, it has recently been shown that IgG and IgA anti‐SSA/Ro52, Ro60, and SSB/La48 are produced by ~30% of salivary gland plasmablasts in patients with SS, and the corresponding autoantibodies are found in the serum 34. Similarly, using biotin‐labeled SSA/Ro60 in immunohistochemical analyses, we detected in situ anti‐SSA/Ro60–specific autoantibodies, as well as plasma cells, producing anti‐SSA/Ro60–specific autoantibodies in salivary gland samples from 3 patients with SS and in all 5 tested samples from patients with SS‐associated MALT lymphoma (samples M14 and M93–M96) (see Supplementary Figure 2, available on the Arthritis & Rheumatology web site at http://onlinelibrary.wiley.com/doi/10.1002/art.41263/abstract).
Somatic mutations in IGV genes are classically introduced during T helper cell–dependent germinal center (GC) reactions. However, it is unlikely that T helper cells specific for IgG peptides would escape negative selection during their development. The most obvious pathway for RF B cells to receive T cell help is that autoantigens that are contained in IgG–immune complexes are internalized and delivered to major histocompatibility complex class II–processing compartments, leading to the presentation of peptides of autoantigens, e.g., of SSA/Ro52/Ro60 and SSB/La48 35, 36. The fact that highly somatically mutated IgG autoantibodies against SSA/Ro52, Ro60, and SSB/La48 proteins are frequently found in patients with SS and in patients with systemic lupus erythematosus indicates that these proteins can initiate a GC reaction, in which the autoreactive B cells receive cognate help from autoreactive T cells that recognize peptides of SSA/Ro52, Ro60, and SS/La48 34, 37, 38. These same T helper cells may thus also permit indirectly, and perhaps less efficiently, a GC reaction of RF B cells. Normally, a proportion of B cells that have acquired high‐affinity BCRs undergo class switch recombination and become expressed as IgG. However, class‐switched RF B cells that express IgG will bind neighboring IgG molecules, which likely will result in overstimulation and autodeletion of these IgG‐expressing RF B cells. Similarly, IgM‐RF–expressing B cells in which the affinities are too high might become overstimulated by IgG bound to its antigen, and thereby become anergic or deleted. The possibility that chronic stimulation of RF B cells can lead to anergy has indeed been demonstrated in some patients with SS and patients with chronic HCV infection, whose peripheral blood harbored a population of expanded, unresponsive CD21−/low RF B cells 39, 40, 41.
In conclusion, the majority of SS‐associated salivary gland MALT lymphomas express BCRs with monoreactive, high‐affinity RF activity. These RFs carry somatic mutations in their IGV genes and are affinity‐selected for IgG binding. In patients with SS, we hypothesize that IgG–autoantigen complexes in inflamed salivary glands provide a growth advantage to RF‐expressing B cells and are key in the pathogenesis of RF‐expressing MALT lymphoma.
AUTHOR CONTRIBUTIONS
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Bende had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design
Bende, Janssen, Haacke, Kroese, Guikema, van Noesel.
Acquisition of data
Bende, Janssen, Beentjes, Wormhoudt, Wagner, Haacke.
Analysis and interpretation of data
Bende, Janssen, Beentjes, Wormhoudt, Wagner, Haacke, Kroese, Guikema, van Noesel.
ADDITIONAL DISCLOSURES
Author Wagner is an employee of AIMM Therapeutics.
Supporting information
Supported by the Dutch Arthritis Foundation (grant 15‐2‐310) and the Dutch Cancer Society (grant UVA 2014‐6824).
1Richard J. Bende, PhD, Jerry Janssen, MSc, Anna Beentjes, BSc, Thera A. M. Wormhoudt, BSc, Jeroen E. J. Guikema, PhD, Carel J. M. van Noesel, MD, PhD: Amsterdam University Medical Center and University of Amsterdam, Amsterdam, The Netherlands; 2Koen Wagner, PhD: AIMM Therapeutics, Amsterdam, The Netherlands; 3Erlin A. Haacke, MD, MSc, Frans G. M. Kroese, PhD: University Medical Center Groningen and University of Groningen, Groningen, The Netherlands.
No potential conflicts of interest relevant to this article were reported.
References
- 1. Zucca E, Bertoni F. The spectrum of MALT lymphoma at different sites: biological and therapeutic relevance. Blood 2016;127:2082–92. [DOI] [PubMed] [Google Scholar]
- 2. Bende RJ, van Maldegem F, van Noesel CJ. Chronic inflammatory disease, lymphoid tissue neogenesis and extranodal marginal zone B‐cell lymphomas. Haematologica 2009;94:1109–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nocturne G, Pontarini E, Bombardieri M, Mariette X. Lymphomas complicating primary Sjögren's syndrome: from autoimmunity to lymphoma. Rheumatology (Oxford) 2019. E‐pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Goules AV, Tzioufas AG. Lymphomagenesis in Sjögren's syndrome: predictive biomarkers towards precision medicine. Autoimmun Rev 2019;18:137–43. [DOI] [PubMed] [Google Scholar]
- 5. Ekstrom‐Smedby K, Vajdic CM, Falster M, Engels EA, Martinez‐Maza O, Turner J, et al. Autoimmune disorders and risk of non‐Hodgkin lymphoma subtypes: a pooled analysis within the InterLymph Consortium. Blood 2008;111:4029–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kroese FG, Abdulahad WH, Haacke E, Bos NA, Vissink A, Bootsma H. B‐cell hyperactivity in primary Sjögren's syndrome [review]. Expert Rev Clin Immunol 2014;10:483–99. [DOI] [PubMed] [Google Scholar]
- 7. Pollard RP, Pijpe J, Bootsma H, Spijkervet FK, Kluin PM, Roodenburg JL, et al. Treatment of mucosa‐associated lymphoid tissue lymphoma in Sjögren's syndrome: a retrospective clinical study. J Rheumatol 2011;38:2198–208. [DOI] [PubMed] [Google Scholar]
- 8. Nocturne G, Virone A, Wan‐Fai Ng , Le Guern V, Hachulla E, Cornec D, et al. Rheumatoid factor and disease activity are independent predictors of lymphoma in primary Sjögren's syndrome. Arthritis Rheumatol 2016;68:977–85. [DOI] [PubMed] [Google Scholar]
- 9. Bende RJ, Aarts WM, de Jong D, Pals ST, van Noesel CJ. Among B‐cell non‐Hodgkin's lymphomas, MALT lymphomas express a unique antibody repertoire with frequent rheumatoid factor reactivity. J Exp Med 2005;201:1229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bende RJ, Janssen J, Wormhoudt TA, Wagner K, Guikema JE, van Noesel CJ. Identification of a novel stereotypic IGHV4‐59/IGHJ5‐encoded B‐cell receptor subset expressed by various B‐cell lymphomas with high affinity rheumatoid factor activity. Haematologica 2016;101:e200–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Van Maldegem F, Wormhoudt TA, Mulder MM, Oud ME, Schilder‐Tol E, Musler AR, et al. Chlamydia psittaci‐negative ocular adnexal marginal zone B‐cell lymphomas have biased VH4‐34 immunoglobulin gene expression and proliferate in a distinct inflammatory environment. Leukemia 2012;26:1647–53. [DOI] [PubMed] [Google Scholar]
- 12. Hoogeboom R, Wormhoudt TA, Schipperus MR, Langerak AW, Dunn‐Walters DK, Guikema JE, et al. A novel chronic lymphocytic leukemia subset expressing mutated IGHV3‐7‐encoded rheumatoid factor B‐cell receptors that are functionally proficient [letter]. Leukemia 2013;27:738–40. [DOI] [PubMed] [Google Scholar]
- 13. Mariette X. Lymphomas complicating Sjögren's syndrome and hepatitis C virus infection may share a common pathogenesis: chronic stimulation of rheumatoid factor B cells. Ann Rheum Dis 2001;60:1007–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. De Re V, De Vita S, Marzotto A, Rupolo M, Gloghini A, Pivetta B, et al. Sequence analysis of the immunoglobulin antigen receptor of hepatitis C virus‐associated non‐Hodgkin lymphomas suggests that the malignant cells are derived from the rheumatoid factor‐producing cells that occur mainly in type II cryoglobulinemia. Blood 2000;96:3578–84. [PubMed] [Google Scholar]
- 15. Kostareli E, Gounari M, Janus A, Murray F, Brochet X, Giudicelli V, et al. Antigen receptor stereotypy across B‐cell lymphoproliferations: the case of IGHV4‐59/IGKV3‐20 receptors with rheumatoid factor activity [letter]. Leukemia 2012;26:1127–31. [DOI] [PubMed] [Google Scholar]
- 16. Charles ED, Green RM, Marukian S, Talal AH, Lake‐Bakaar GV, Jacobson IM, et al. Clonal expansion of immunoglobulin M+CD27+ B cells in HCV‐associated mixed cryoglobulinemia. Blood 2008;111:1344–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Borretzen M, Chapman C, Natvig JB, Thompson KM. Differences in mutational patterns between rheumatoid factors in health and disease are related to variable heavy chain family and germ‐line gene usage. Eur J Immunol 1997;27:735–41. [DOI] [PubMed] [Google Scholar]
- 18. Martin T, Weber JC, Levallois H, Labouret N, Soley A, Koenig S, et al. Salivary gland lymphomas in patients with Sjögren's syndrome may frequently develop from rheumatoid factor B cells. Arthritis Rheum 2000;43:908–16. [DOI] [PubMed] [Google Scholar]
- 19. Van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T‐cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED‐2 Concerted Action BMH4‐CT98‐3936. Leukemia 2003;17:2257–317. [DOI] [PubMed] [Google Scholar]
- 20. Brochet X, Lefranc MP, Giudicelli V. IMGT/V‐QUEST: the highly customized and integrated system for IG and TR standardized V‐J and V‐D‐J sequence analysis. Nucleic Acids Res 2008;36:W503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bende RJ, Jochems GJ, Frame TH, Klein MR, van Eijk RV, van Lier RA, et al. Effects of IL‐4, IL‐5 and IL‐6 on growth and immunoglobulin production of Epstein‐Barr virus‐infected human B cells. Cell Immunol 1992;143:310–23. [DOI] [PubMed] [Google Scholar]
- 22. Slot LM, Wormhoudt TA, Kwakkenbos MJ, Wagner K, Ballering A, Jongejan A, et al. De novo gene mutations in normal human memory B cells. Leukemia 2019;33:1219–30. [DOI] [PubMed] [Google Scholar]
- 23. Wang JJ, Reed JH, Colella AD, Russell AJ, Murray‐Brown W, Chataway TK, et al. Molecular profiling and clonal tracking of secreted rheumatoid factors in primary Sjögren's syndrome. Arthritis Rheumatol 2018;70:1617–25. [DOI] [PubMed] [Google Scholar]
- 24. Visser A, Doorenspleet ME, de Vries N, Spijkervet FK, Vissink A, Bende RJ, et al. Acquisition of N‐glycosylation sites in immunoglobulin heavy chain genes during local expansion in parotid salivary glands of primary Sjögren patients. Front Immunol 2018;9:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bende RJ, Slot LM, Hoogeboom R, Wormhoudt TA, Adeoye AO, Guikema JE, et al. Stereotypic rheumatoid factors that are frequently expressed in mucosa‐associated lymphoid tissue–type lymphomas are rare in the labial salivary glands of patients with Sjögren's syndrome. Arthritis Rheumatol 2015;67:1074–83. [DOI] [PubMed] [Google Scholar]
- 26. Bahler DW, Swerdlow SH. Clonal salivary gland infiltrates associated with myoepithelial sialoadenitis (Sjögren's syndrome) begin as nonmalignant antigen‐selected expansions. Blood 1998;91:1864–72. [PubMed] [Google Scholar]
- 27. Miklos JA, Swerdlow SH, Bahler DW. Salivary gland mucosa‐associated lymphoid tissue lymphoma immunoglobulin V(H) genes show frequent use of V1‐69 with distinctive CDR3 features. Blood 2000;95:3878–84. [PubMed] [Google Scholar]
- 28. Charles ED, Orloff MI, Nishiuchi E, Marukian S, Rice CM, Dustin LB. Somatic hypermutations confer rheumatoid factor activity in hepatitis C virus‐associated mixed cryoglobulinemia. Arthritis Rheum 2013;65:2430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shiroishi M, Ito Y, Shimokawa K, Lee JM, Kusakabe T, Ueda T. Structure‐function analyses of a stereotypic rheumatoid factor unravel the structural basis for germline‐encoded antibody autoreactivity. J Biol Chem 2018;293:7008–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Maibom‐Thomsen SL, Trier NH, Holm BE, Hansen KB, Rasmussen MI, Chailyan A, et al. Immunoglobulin G structure and rheumatoid factor epitopes. PLoS One 2019;14:e0217624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Herlands RA, William J, Hershberg U, Shlomchik MJ. Anti‐chromatin antibodies drive in vivo antigen‐specific activation and somatic hypermutation of rheumatoid factor B cells at extrafollicular sites. Eur J Immunol 2007;37:3339–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Avalos AM, Busconi L, Marshak‐Rothstein A. Regulation of autoreactive B cell responses to endogenous TLR ligands. Autoimmunity 2010;43:76–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak‐Rothstein A. Chromatin‐IgG complexes activate B cells by dual engagement of IgM and Toll‐like receptors. Nature 2002;416:603–7. [DOI] [PubMed] [Google Scholar]
- 34. Takeshita M, Suzuki K, Kaneda Y, Yamane H, Ikeura K, Sato H, et al. Antigen‐driven selection of antibodies against SSA, SSB and the centromere ‘complex’, including a novel antigen, MIS12 complex, in human salivary glands. Ann Rheum Dis 2019;79:150–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shlomchik MJ. Sites and stages of autoreactive B cell activation and regulation. Immunity 2008;28:18–28. [DOI] [PubMed] [Google Scholar]
- 36. Roosnek E, Lanzavecchia A. Efficient and selective presentation of antigen‐antibody complexes by rheumatoid factor B cells. J Exp Med 1991;173:487–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mietzner B, Tsuiji M, Scheid J, Velinzon K, Tiller T, Abraham K, et al. Autoreactive IgG memory antibodies in patients with systemic lupus erythematosus arise from nonreactive and polyreactive precursors. Proc Natl Acad Sci U S A 2008;105:9727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Reed JH, Gorny MK, Li L, Cardozo T, Buyon JP, Clancy RM. Ro52 autoantibodies arise from self‐reactive progenitors in a mother of a child with neonatal lupus. J Autoimmun 2017;79:99–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Charles ED, Brunetti C, Marukian S, Ritola KD, Talal AH, Marks K, et al. Clonal B cells in patients with hepatitis C virus‐associated mixed cryoglobulinemia contain an expanded anergic CD21low B‐cell subset. Blood 2011;117:5425–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Glauzy S, Boccitto M, Bannock JM, Delmotte FR, Saadoun D, Cacoub P, et al. Accumulation of antigen‐driven lymphoproliferations in complement receptor 2/CD21−/low B cells from patients with Sjögren's syndrome. Arthritis Rheumatol 2018;70:298–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bende RJ, van Noesel CJ. Rheumatoid factor reactivity of expanded CD21−/low B cells in patients with Sjögren's syndrome: comment on the article by Glauzy et al [letter]. Arthritis Rheumatol 2019;71:169–70. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


