Abstract
The Banff Digital Pathology Working Group (DPWG) was formed in the time leading up to and during the joint American Society for Histocompatibility and Immunogenetics/Banff Meeting, September 23‐27, 2019, held in Pittsburgh, Pennsylvania. At the meeting, the 14th Banff Conference, presentations directly and peripherally related to the topic of “digital pathology” were presented; and discussions before, during, and after the meeting have resulted in a list of issues to address for the DPWG. Included are practice standardization, integrative approaches for study classification, scoring of histologic parameters (eg, interstitial fibrosis and tubular atrophy and inflammation), algorithm classification, and precision diagnosis (eg, molecular pathways and therapeutics). Since the meeting, a survey with international participation of mostly pathologists (81%) was conducted, showing that whole slide imaging is available at the majority of centers (71%) but that artificial intelligence (AI)/machine learning was only used in ≈12% of centers, with a wide variety of programs/algorithms employed. Digitalization is not just an end in itself. It also is a necessary precondition for AI and other approaches. Discussions at the meeting and the survey highlight the unmet need for a Banff DPWG and point the way toward future contributions that can be made.
Keywords: basic (laboratory) research/science, biopsy, classification systems: Banff classification, clinical research/practice, informatics, organ transplantation in general, pathology/histopathology, rejection
Short abstract
The Banff Digital Pathology Working Group assesses the current state of digital pathology and presents next steps for digital transplantation pathology, including utilization of whole slide imaging and artificial intelligence and other algorithm types.
Abbreviations
- 3D
3‐dimensional
- AI
artificial intelligence
- ASHI
American Society for Histocompatibility and Immunogenetics
- DPWG
Digital Pathology Working Group
- WSI
whole slide image/imaging
1. INTRODUCTION
The American Society for Histocompatibility and Immunogenetics (ASHI)/Banff Joint Scientific Meeting was held September 23‐27, 2019, at the David L. Lawrence Convention Center, Pittsburgh, Pennsylvania, as a joint meeting between the Banff Foundation for Allograft Pathology and the American Society for Histocompatibility and Immunogenetics (ASHI). This was the 14th Banff Conference on Allograft Pathology. The meeting was preceded by a video trailer 1 by Kim Solez, which highlighted the digital pathology sessions and a video request for comments on a digital pathology white paper. 2
For purposes of the meeting and this paper, “digital pathology” refers to a broad collection of computerized techniques applied to pathology, particularly anatomic pathology, including whole slide imaging (WSI), algorithms for dedicated morphometric analysis, algorithms employing artificial intelligence (AI)/machine learning, natural language processing, and novel microscopic techniques (eg, multispectral, Fourier transform infrared and other infrared, and second harmonic generation imaging), which typically employ computerized interfaces. 3 , 4 , 5 , 6 , 7 This definition is in line with a white paper from the Digital Pathology Association, which defined “digital pathology” as “tools and systems to digitize pathology slides and associated meta‐data, their storage, review, analysis, and enabling infrastructure.” 8 Furthermore, “digital pathology” can be considered a topic in the larger field of “computational pathology.” 8 , 9
The meeting (Premeeting) began with the following presentations: “Regenerative Medicine: Introduction and Overview—Make No Small Plans” (with comments on digital pathology) by Kim Solez from the University of Alberta, “Deep Learning and Renal Transplant Pathology” by Jesper Kers from Amsterdam, “Single Cells and Human Cell Atlas Towards Banff Implementation” by Andrew Malone of Washington University in St. Louis, “Banff Interfacing AI Natural Language Processing and Virtual Biopsy Systems” by Alexandre Loupy from Paris, and a “Discussion and Proposal for a Digital Pathology White Paper” by Kim Solez from the University of Alberta and Yukako Yagi of Memorial Sloan Kettering Cancer Center in New York.
Jesper Kers presented the work by Radboud University Medical Center (UMC), Amsterdam UMC, and the Mayo Clinic on deep learning in the assessment of kidney specimens. With this work, multiclass semantic segmentation of periodic acid‐Schiff–stained kidney tissue sections was performed; and this could potentially be useful in quantitative studies of kidney specimens. For example, lesion quantifications by their convolutional neural network correlated well with the semiquantitative scores by renal pathologists without the issue of interobserver variability. 10
Andrew Malone presented his work on single‐cell transcriptomics and the kidney. 11 , 12 , 13 The importance of digital pathology to this work is illustrated by the fact that managing the data produced by these studies is a major bioinformatics challenge. A recent publication from Dr Malone's group highlights the utility of these techniques in characterizing a case of mixed cellular and antibody‐mediated rejection. In this case, diverse cell‐type populations were characterized, including a diversity of endothelial cells, monocytes, other immune cell subtypes, and a variety of native kidney cell types. 12 The quantitative data that emerge from these studies is quite compatible with that which emerges from digital pathology. Both contribute to computational biology approaches, and increasing clinical relevance will likely emerge.
Alexandre Loupy presented AI research being conducted by the Paris Transplant Group. These efforts add to their cutting edge work in the development of the iBox prediction system for renal allograft loss 14 and modeling allograft rejection injury, particularly antibody‐mediated rejection 15 and transplant glomerulopathy. 16 Their current work involves a number of AI efforts, including the use of natural language processing to mine discrete data in kidney transplant biopsy reports. He also presented methods for the integration of Banff diagnostic scores and diagnoses in systematic algorithms that can be executed by computer programs in a manner that extends upon prior efforts to assemble Banff scoring and diagnoses in centralized resources that can be easily accessed by the transplantation community. 17
Yukako Yagi presented work on digital 3‐dimensional (3D) approaches and multimodality imaging. Recent work has included extensive application of multiplex immunofluorescence and other methods to cancer research. 18 , 19 In the field of transplantation pathology, arteries and other anatomic features can be highlighted with 3D reconstruction of histological slides containing sequential serial sections. For example, “virtual coronary arteriography” of chronic allograft vasculopathy can be conducted, and it is likely that other disorders can also be structurally examined using these methods. 20 , 21 Dr Yagi’s group has also demonstrated the cost effectiveness of digital pathology. 22 , 23
Kim Solez presented concepts underlying the Banff Digital Pathology White Paper as well as larger issues. He reviewed the results of the video request for comment on the Digital Pathology White Paper. 2 Digital pathology can serve as a productivity tool in a field with projected workforce shortages. 24 When viewed as a new frontier for research and diagnostic use in the field of anatomic pathology, digital pathology could increase interest in the field of pathology as a specialty. Medical students are learning pathology from digital resources and could find it more appealing to contemplate joining a pathology department that is involved in the field of digital pathology. 25 Pathologists need to be charismatic role models emphasizing digital pathology. For trainees, going digital is appealing and consistent with the general goal of human flourishing. 26
It is not necessary that the digital pathology be complete end to end, applied uniformly to all specimens, and always used for primary diagnosis. Most pathology departments are positively inclined toward going digital. There is widespread support for at least digital archiving, but it seems too rigid to insist that digital pathology always be used for primary diagnosis, and be applied to every specimen without exceptions, particularly before validation of these tests. 8 , 27 , 28
A working lunch focusing on digital pathology including digital liver, kidney, and heart pathology algorithms was also conducted by Alton Brad Farris of Emory University in Atlanta and Andrew Lesniak of the University of Pittsburgh. After the presentations subsequently described in more detail, the working lunch resulted in goals for the DPWG that are covered more in the discussion of this paper.
Alton Brad Farris described prior Banff studies on renal fibrosis assessment, including computerized analysis for the quantification of renal fibrosis. 29 Farris and his colleagues have also quantitated renal fibrosis in other studies, 30 , 31 microvessel density, 31 and inflammation. 32 Similar methods can be applied to other organs to measure parameters such as steatosis in the liver. 33 Ultimately, whole slide imaging can be applied to provide reports that integrate these quantitative data into reports that improve patient care. 34
Andrew Lesniak described the studies that he, Anthony Demetris, and others at the University of Pittsburgh have performed in the past and have performed on transplant specimens. 35 , 36 , 37 , 38 , 39 In their laboratory, they utilize multiplex immunostaining to characterize various cells, particularly immune cells, 36 and use automated image analysis to perform “‐omics”‐type assessment of tissue. 38 , 39
2. MATERIALS AND METHODS
2.1. Survey
A survey was conducted after the 14th Banff Conference on Allograft Pathology in 2019 to determine current digital pathology practices within the international transplant pathology community. Questions were sent via SurveyMonkey 40 to both the NEPHROL and NEPHNPPT Discussion Groups moderated by Kim Solez and to The Transplantation Society mailing list via the Tribune Pulse October 31, 2019, Issue.
3. RESULTS
3.1. Survey of digital pathology routine practice
There were 42 participants in the Survey, and results are given in Figures 1, 2, 3, 4, 5. Survey respondents consisted of individuals from a variety of professions, with the majority (81%) coming from the field of pathology. A number of countries were also represented in the survey. Results of the study showed a diverse spectrum of current application of WSI and AI implementation. WSI is available at the majority of centers (71%). Customized/handcrafted and commercial programs are being implemented, and some groups report using multiple programs. Overall, AI/machine learning was only used in ≈12% of centers.
FIGURE 1.
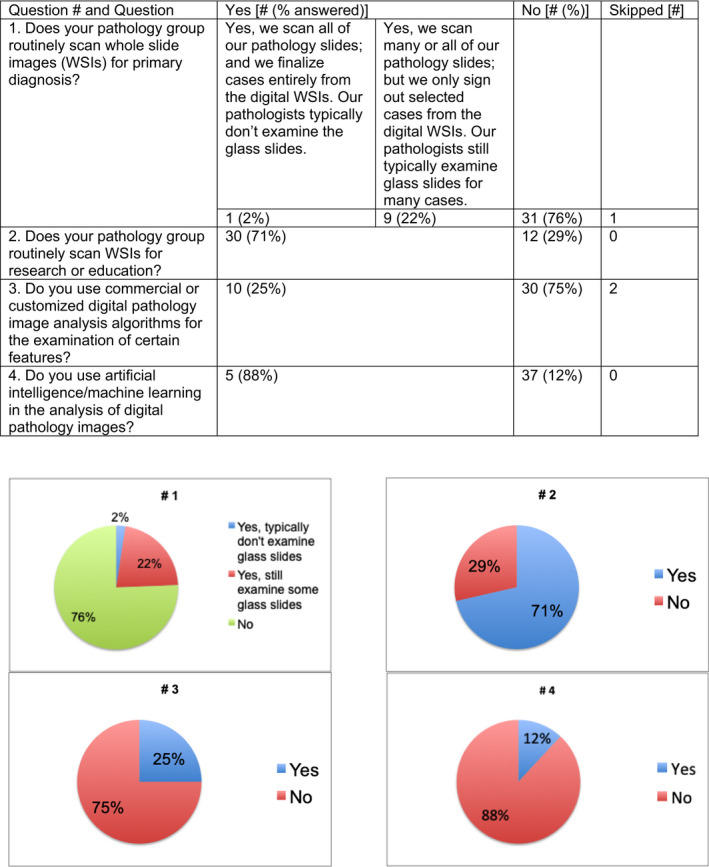
General questions regarding digital pathology were asked [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 2.
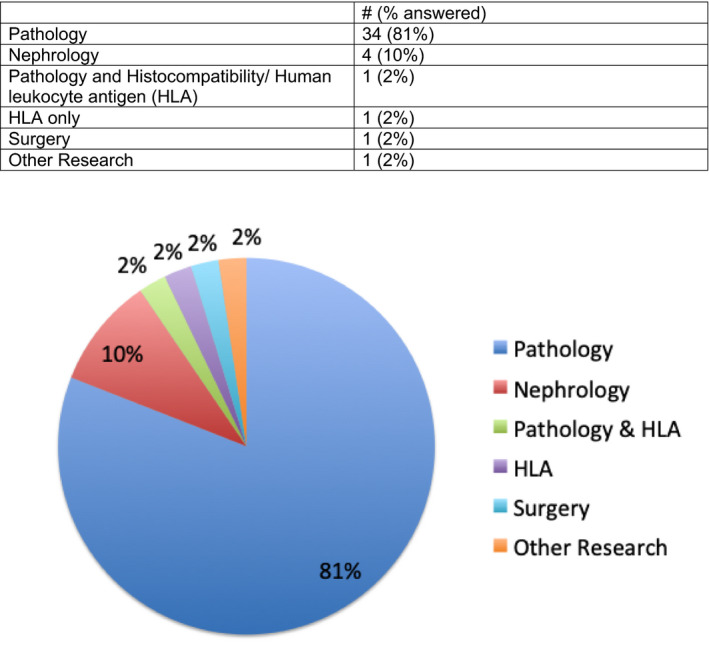
Question 5. What is your primary area of practice? [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
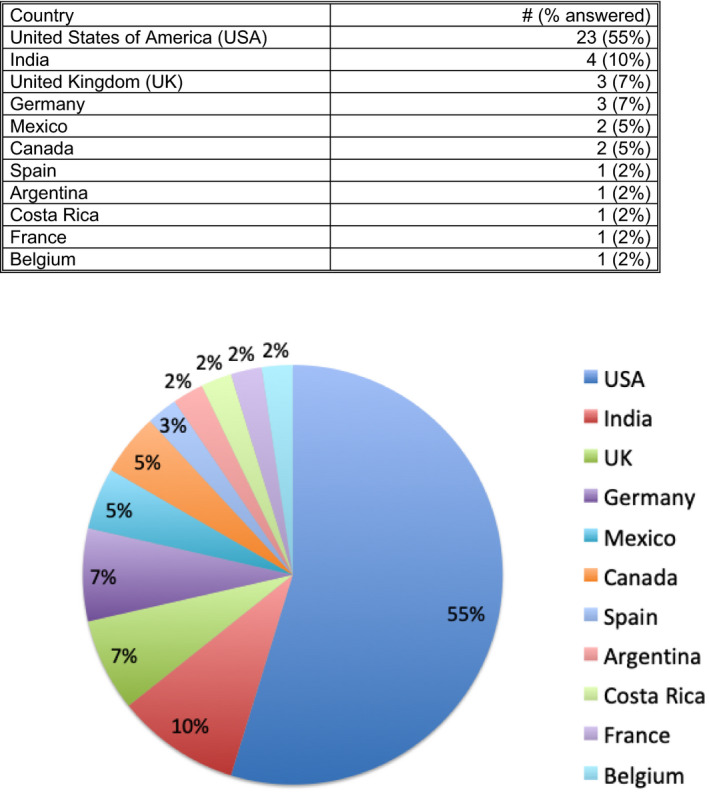
Question 6. What country do you primarily practice in? [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 4.
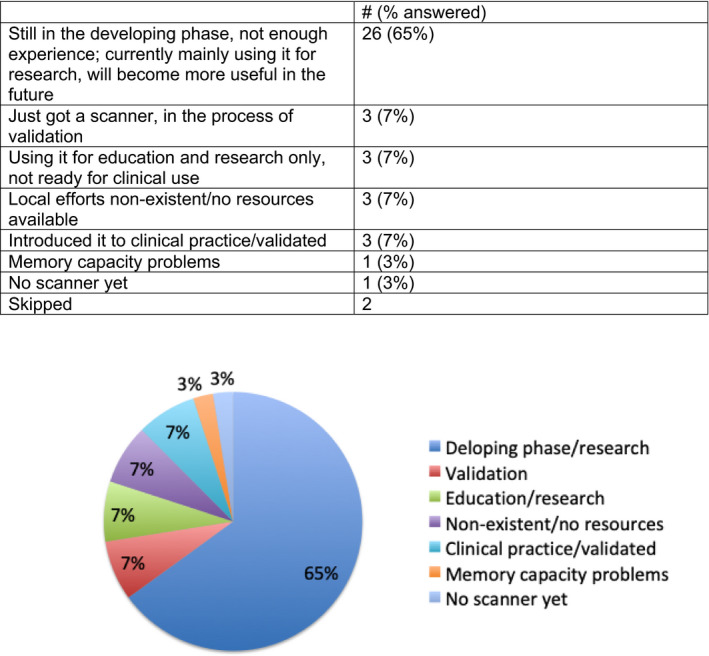
Question 7. Describe the current state of your digital pathology efforts and your view of efforts that will be useful to pathology in the future [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 5.
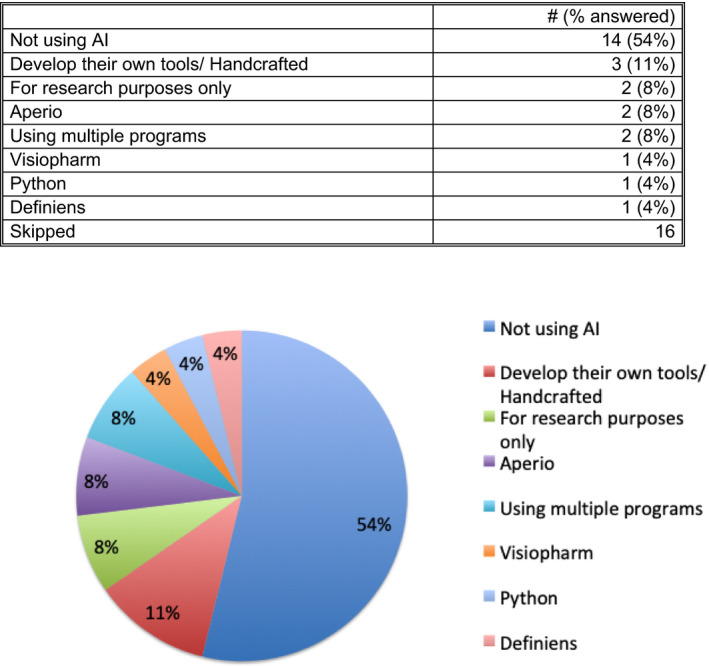
Question 8. If you are using artificial intelligence (AI) in the analysis of digital images, what programs and approaches are you using? [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
The Banff Digital Pathology Working Group (DPWG) consists of a community of individuals willing to collaborate with others and support the development and integration of digital pathology into clinical practice and research protocols. In the initial efforts, the current state has been surveyed. As also detailed in the upcoming Banff Meeting Report, the initial goals of the Banff DPWG are also detailed in Table 1. As discussed in the working lunch and other times at the Banff Conference, future plans include an image bank for testing algorithms so that research groups can test their AI and other algorithms in a manner similar to the CAMELYON challenges for breast cancer and lymph node metastasis. 41 , 42
TABLE 1.
The Banff Digital Pathology Working Group issues and future plans are depicted
| Topic | Items |
|---|---|
| Issues to address |
|
| Future plans |
|
Abbreviations: AI, artificial intelligence; IFTA, interstitial fibrosis and tubular atrophy.
Another future goal could be the release of “official” Banff algorithms that could be used by the Banff community. These include targeted, handcrafted, or “real intelligence” algorithms for parameters such as fibrosis, inflammation, steatosis, and so forth. AI also has the potential to increase diagnostic accuracy and inform enrollment and outcomes in pathology‐based clinical trials in transplantation. Eventually AI/machine learning algorithms that have been thoroughly benchmarked could be made available. Furthermore, data pipelines for the integration of “–omic” data could be made available so that centers could have mechanisms for mining data within their center as well as sharing data with other centers. Tools for integrating data such as natural language processing algorithms such as those discussed by Alexandre Loupy and being developed by the Paris Transplant group and others could eventually be distributed in the community. Of note, in addition to artificial intelligence, “AI” can refer to the term “augmented intelligence”; 43 , 44 and it is hoped that these technologies and others can help “augment” the skills of pathologists and other medical professions.
Economic concerns are likely relevant for many of the efforts of the Banff DPWG. Implementation of digital pathology can be rather costly, since it typically requires purchase of WSI scanners and computers and infrastructure for the acquisition, storage, and delivery of images. Such technology requires the financial support of departmental and institutional administrators, and business use case considerations must be addressed. Effects on workload must be considered, including potential redistribution of pathologist duties as well as the potential necessity for technicians to operate the WSI scanners and bioinformatics computer specialists to manage the data produced. In some cases, workload balancing and other factors could result in a return on investment. Cost considerations may also affect the selection of image analysis algorithms, since open source algorithms are essentially freely available as opposed to commercial algorithms that typically require financial investment for purchase and technical support. Because both radiology and pathology are subspecialties that deal with image‐intensive data, some authorities have advocated for joint radiology/pathology programs to manage the large images produced by WSI scanners. 27 , 28 , 45 , 46 For the Banff efforts proposed in this white paper, the working group will likely need to leverage the resources of departments and institutions that have excess capacity to support research collaboration. The Banff DPWG could eventually provide open source algorithms at essentially no cost, and this could help alleviate some of the economic burden for the implementation of digital pathology.
It is striking how much simpler the issues have become between the time of the September 10, 2019, video request for comments on the white paper 2 and the writing of this article, a reflection of the success of the feedback process. There is a further simplification one can envision reading Keith Kaplan's recent blog posting of January 14, 2020, 47 predicting the future of digital pathology where everything is digital so we stop using the word “digital” because it is redundant. We can all sense that that day will come; it is only a question of when. In that sense this white paper is only an interim step until we reach the point where we no longer have to say “digital” because everyone is thinking that way anyway.
Eventually, the integration of well‐characterized histopathologic data with molecular and other data will be facilitated by efforts such as those being conducted by the Banff DPWG; and these efforts will eventually contribute to improved research, education of trainees and practitioners, and, importantly, patient care.
DISCLOSURE
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
ACKNOWLEDGMENTS
We acknowledge the Banff Foundation for Allograft Pathology, the American Society for Histocompatibility and Immunogenetics (ASHI), their sponsors, and other funding organizations for making the recent Banff meeting possible.
Farris AB, Moghe I, Wu S, et al. Banff Digital Pathology Working Group: Going digital in transplant pathology. Am J Transplant. 2020;20:2392–2399. 10.1111/ajt.15850
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon request.
REFERENCES
- 1. Solez K. Pittsburgh Banff ASHI Meeting Trailer Greeting Final. https://www.youtube.com/watch?v=cqCeIRuDgEk. Accessed December 27, 2019.
- 2. Solez K. Request for comment on digital pathology white paper. https://www.youtube.com/watch?v=w21Atf0tGjg. Accessed December 27, 2019.
- 3. Fereidouni F, Griffin C, Todd A, Levenson R. Multispectral analysis tools can increase utility of RGB color images in histology. J Opt. 2018;20(4):pii:044007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Torres R, Velazquez H, Chang JJ, et al. Three‐dimensional morphology by multiphoton microscopy with clearing in a model of cisplatin‐induced CKD. J Am Soc Nephrol. 2016;27(4):1102‐1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Qian HS, Weldon SM, Matera D, et al. Quantification and comparison of anti‐fibrotic therapies by polarized SRM and SHG‐based morphometry in rat UUO model. PLoS ONE. 2016;11(6):e0156734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vuiblet V, Fere M, Gobinet C, Birembaut P, Piot O, Rieu P. Renal graft fibrosis and inflammation quantification by an automated fourier‐transform infrared imaging technique. J Am Soc Nephrol. 2016;27(8):2382‐2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Varma VK, Kajdacsy‐Balla A, Akkina SK, Setty S, Walsh MJ. A label‐free approach by infrared spectroscopic imaging for interrogating the biochemistry of diabetic nephropathy progression. Kidney Int. 2016;89(5):1153‐1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Abels E, Pantanowitz L, Aeffner F, et al. Computational pathology definitions, best practices, and recommendations for regulatory guidance: a white paper from the Digital Pathology Association. J Pathol. 2019;249(3):286‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Louis DN, Feldman M, Carter AB, et al. Computational pathology. Arch Pathol Lab Med. 2015;140(1):41‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hermsen M, de Bel T, den Boer M, et al. Deep learning‐based histopathologic assessment of kidney tissue. J Am Soc Nephrol. 2019;30(10):1968‐1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Malone AF, Wu H, Humphreys BD. Bringing renal biopsy interpretation into the molecular age with single‐cell RNA sequencing. Semin Nephrol. 2018;38(1):31‐39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wu H, Malone AF, Donnelly EL, et al. Single‐cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol. 2018;29(8):2069‐2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Malone AF, Humphreys BD. Single‐cell transcriptomics and solid organ transplantation. Transplantation. 2019;103(9):1776‐1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loupy A, Aubert O, Orandi BJ, et al. Prediction system for risk of allograft loss in patients receiving kidney transplants: international derivation and validation study. BMJ. 2019;366:l4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Loupy A, Lefaucheur C. Antibody‐mediated rejection of solid‐organ allografts. N Engl J Med. 2018;379(12):1150‐1160. [DOI] [PubMed] [Google Scholar]
- 16. Aubert O, Higgins S, Bouatou Y, et al. Archetype analysis identifies distinct profiles in renal transplant recipients with transplant glomerulopathy associated with allograft survival. J Am Soc Nephrol. 2019;30(4):625‐639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Roufosse C, Simmonds N, Clahsen‐van Groningen M, et al. A 2018 reference guide to the Banff classification of renal allograft pathology. Transplantation. 2018;102(11):1795‐1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yagi Y, Aly RG, Tabata K, et al. Three‐dimensional histologic, immunohistochemical and multiplex immunofluorescence analysis of dynamic vessel co‐option of spread through air spaces (STAS) in lung adenocarcinoma. J Thorac Oncol. 2020;15(4):589‐600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hossain MS, Hanna MG, Uraoka N, et al. Automatic quantification of HER2 gene amplification in invasive breast cancer from chromogenic in situ hybridization whole slide images. J Med Imaging (Bellingham). 2019;6(4):047501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fónyad L, Shinoda K, Farkash EA, et al. 3‐dimensional digital reconstruction of the murine coronary system for the evaluation of chronic allograft vasculopathy. Diagn Pathol. 2015;10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farahani N, Braun A, Jutt D, et al. Three‐dimensional imaging and scanning: current and future applications for pathology. J Pathol Inform. 2017;8:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanna MG, Reuter VE, Samboy J, et al. Implementation of digital pathology offers clinical and operational increase in efficiency and cost savings. Arch Pathol Lab Med. 2019;143(12):1545‐1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanna MG, Reuter VE, Hameed MR, et al. Whole slide imaging equivalency and efficiency study: experience at a large academic center. Mod Pathol. 2019;32(7):916‐928. [DOI] [PubMed] [Google Scholar]
- 24. Metter DM, Colgan TJ, Leung ST, Timmons CF, Park JY. Trends in the US and canadian pathologist workforces from 2007 to 2017. JAMA Netw Open. 2019;2(5):e194337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Glassy EF. The rise of the social pathologist: the importance of social media to pathology. Arch Pathol Lab Med. 2010;134(10):1421‐1423. [DOI] [PubMed] [Google Scholar]
- 26. VanderWeele TJ. On the promotion of human flourishing. Proc Natl Acad Sci USA. 2017;114(31):8148‐8156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zarella MD, Bowman D, Aeffner F, et al. A practical guide to whole slide imaging: a white paper from the Digital Pathology Association. Arch Pathol Lab Med. 2019;143(2):222‐234. [DOI] [PubMed] [Google Scholar]
- 28. Aeffner F, Zarella MarkD, Buchbinder N, et al. Introduction to digital image analysis in whole‐slide imaging: a white paper from the Digital Pathology Association. J Pathol Inform. 2019;10:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Farris AB, Chan S, Climenhaga J, et al. Banff fibrosis study: multicenter visual assessment and computerized analysis of interstitial fibrosis in kidney biopsies. Am J Transplant. 2014;14(4):897‐907. [DOI] [PubMed] [Google Scholar]
- 30. Farris AB, Adams CD, Brousaides N, et al. Morphometric and visual evaluation of fibrosis in renal biopsies. J Am Soc Nephrol. 2010;22(1):176‐186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farris AB, Ellis CL, Rogers TE, Lawson D, Cohen C, Rosen S. Renal medullary and cortical correlates in fibrosis, epithelial mass, microvascularity, and microanatomy using whole slide image analysis morphometry. PLoS ONE. 2016;11(8):e0161019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Moon A, Smith GH, Kong J, Rogers TE, Ellis CL, Farris ABB 3rd. Development of CD3 cell quantitation algorithms for renal allograft biopsy rejection assessment utilizing open source image analysis software. Virchows Arch. 2018;472(2):259‐269. [DOI] [PubMed] [Google Scholar]
- 33. Lee MJ, Bagci P, Kong J, et al. Liver steatosis assessment: correlations among pathology, radiology, clinical data and automated image analysis software. Pathol Res Pract. 2013;209(6):371‐379. [DOI] [PubMed] [Google Scholar]
- 34. Farris AB, Cohen C, Rogers TE, Smith GH. Whole slide imaging for analytical anatomic pathology and telepathology: practical applications today, promises, and perils. Arch Pathol Lab Med. 2017;141(4):542‐550. [DOI] [PubMed] [Google Scholar]
- 35. Wood‐Trageser MA, Lesniak AJ, Demetris AJ. Enhancing the value of histopathological assessment of allograft biopsy monitoring. Transplantation. 2019;103(7):1306‐1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Feng S, Bucuvalas JC, Demetris AJ, et al. Evidence of chronic allograft injury in liver biopsies from long‐term pediatric recipients of liver transplants. Gastroenterology. 2018;155(6):1838‐1851.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pantanowitz L, Wiley CA, Demetris A, et al. Experience with multimodality telepathology at the University of Pittsburgh Medical Center. J Pathol Inform. 2012;3:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Isse K, Lesniak A, Grama K, Roysam B, Minervini MI, Demetris AJ. Digital transplantation pathology: combining whole slide imaging, multiplex staining and automated image analysis. Am J Transplant. 2012;12(1):27‐37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Isse K, Grama K, Abbott IM, et al. Adding value to liver (and allograft) biopsy evaluation using a combination of multiplex quantum dot immunostaining, high‐resolution whole‐slide digital imaging, and automated image analysis. Clin Liver Dis. 2010;14(4):669‐685. [DOI] [PubMed] [Google Scholar]
- 40. SurveyMonkey . SurveyMonkey: the world’s most popular free online survey tool. http://www.surveymonkey.com. Accessed January 30, 2020.
- 41. Litjens G, Bandi P, Ehteshami Bejnordi B, et al. 1399 H&E‐stained sentinel lymph node sections of breast cancer patients: the CAMELYON dataset. Gigascience 2018;7(6). 10.1093/gigascience/giy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ehteshami Bejnordi B, Veta M, Johannes van Diest P, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA. 2017;318(22):2199‐2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. American Medical Association . Augmented Intelligence (AI) ‐ American Medical Association. https://www.ama‐assn.org/amaone/augmented‐intelligence‐ai. Accessed January 1, 2020.
- 44. Carter S, Nielsen M. Using artificial intelligence to augment human intelligence. https://distill.pub/2017/aia/. Accessed January 1, 2020.
- 45. Friedman BA. Orchestrating a unified approach to information management. Radiol Manage. 1997;19(6):30‐36. [PubMed] [Google Scholar]
- 46. Sorace J, Aberle DR, Elimam D, Lawvere S, Tawfik O, Wallace WD. Integrating pathology and radiology disciplines: an emerging opportunity? BMC Med. 2012;10:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kaplan K. What does 2020 and beyond mean for pathology? https://blog.corista.com/corista‐digital‐pathology‐blog/what‐does‐2020‐and‐beyond‐mean‐for‐pathology. Accessed January 31, 2020.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.


