Abstract
An integrated system combining a magnetically‐driven micromotor and a synthetized protein‐based hyaluronic acid (HA) microflake is presented for the in situ selection and transport of multiple motile sperm cells (ca. 50). The system appeals for targeted sperm delivery in the reproductive system to assist fertilization or to deliver drugs. The binding mechanism between the HA microflake and sperm relies on the interactions between HA and the corresponding sperm HA receptors. Once sperm are captured within the HA microflake, the assembly is trapped and transported by a magnetically‐driven helical microcarrier. The trapping of the sperm‐microflake occurs by a local vortex induced by the microcarrier during rotation‐translation under a rotating magnetic field. After transport, the microflake is enzymatically hydrolyzed by local proteases, allowing sperm to escape and finally reach the target location. This cargo‐delivery system represents a new concept to transport not only multiple motile sperm but also other actively moving biological cargoes.
Keywords: enzymatic release, mature sperm selection, microflake synthesis, micromotor, sperm swarm transport
A cluster of motile and mature sperm previously immobilized on the BSA‐HA microflake is transported by a magnetically‐driven microhelix, which delivers the sperm‐loaded microflake close to the area of interest. Due to the presence of local proteases, the microflake is then hydrolyzed allowing the sperm release to finally reach the target location.
Introduction
Nano‐ and micromotors are being extensively developed toward numerous biomedical applications,1 ranging from targeted drug delivery2, 3 and single cell manipulation4 to biosensing.5, 6, 7 While much success has been achieved by employing chemical,8, 9, 10 physical and biohybrid micromotors,11, 12 the safety risks of toxic fuels or foreign micro‐organisms remain a barrier that restricts the micromotors’ application in vivo.2 Among all reported micromotors, magnetically‐driven ones are of great interest because of their fuel‐free actuation and controllable locomotion based on external magnetic fields,13 which can efficiently penetrate tissue without visible adverse effects.14 Particularly, helical micromotors powered by oscillating or rotating magnetic fields only need very low field strength15, 16 compared to those actuated by magnetic field gradients,17 making them safer for the human body and controllable in tissue, as it has been demonstrated in the vitreous,18 knee tissue19 and blood vessels.20 Bell et al. fabricated the first helical micromotor by rolled‐up nanotechnology that could transduce the rotational actuation induced by the rotating magnetic field into translational motion, relying on the anisotropic drag applied to the chiral structure of the micromotor.21 Since then, helical micromotors have been fabricated by different methods: strain engineering,21, 22 glancing angle deposition (GLAD),23 electrodeposition,24 self‐assembly,25 and 3D printing.15 They have been used for cargo‐delivery of particles,26 drugs27, 28 and single passive cells.29 They were utilized to manipulate micro‐objects by direct contact pushing26 or by locally induced hydrodynamic vortexes, a more gentle mechanism.30 However there is still a major challenge: the safe transport of multiple active cargoes, in particular of sensitive motile cells. An example of those cargoes is the sperm, which is hard to be controllably captured and released due to its forceful swimming. Additionally, any change in the medium composition or the transport itself could have important implications on the sperm function and motility.31 The transport of multiple sperm is crucial to ensure fertilization where tens to hundreds of sperm are necessary to reach the oocyte under normal conditions.32 Likewise, for any drug delivery application, the sperm amount directly determines the drug dosage and the anticancer efficiency of the treatment. Previous efforts have been mainly focused on the guidance or transport of single sperm29, 33 toward assisted fertilization34 and targeted drug delivery,35, 36 or on exploring intrinsic sperm taxis mechanism such as chemotaxis,37 where sperm are attracted by a chemical gradient present for example in the vicinity of the oocyte (see Table S1 in the Supporting Information). The latter mechanism, although promising for multiple sperm guidance, is not as strong in mammal sperm. Hence, a harmless micromotor that can transport collective motile sperm with no influence on their propulsion and viability is needed. While the collection of immotile sperm can be done by simple mechanical trapping,30 a more robust method with a sperm binding mechanism must be employed to select and transport the motile and mature ones. A possible strategy is making use of HA traps, which are widely used in clinical practice for sperm selection prior to intracytoplasmatic sperm injection (ICSI).38 HA is a non‐sulfated glycosaminoglycan secretion of the cervical mucus and the cumulus‐oophores complex, and sperm showing high HA affinity are known to present lower rates of aneuploidy and DNA fragmentation, which have a great influence on the pregnancy outcome.39 Selection based on HA is crucial as only sperm with intact DNA can successfully express HA receptors.39 Scheme 1 shows the concept of the transport of multiple motile sperm by a microflake (MF) made of bovine serum albumin (BSA) and hyaluronic acid (HA). First, motile sperm are co‐incubated with BSA‐HA microflakes. Once sperm are immobilized on and within the microflake, a magnetic helical microcarrier (MC) captures the assembly by means of a locally induced hydrodynamic vortex. After being precisely transported to the target location, sperm are released by hydrolyzing the microflake with local proteases present in the surrounding fluid. Physiologically, proteases are universally present in different organs of the human body.40 Specifically in the female reproductive system. Trypsin‐like proteases, also called “oviductal proteases” (e.g., “oviductin”), are secreted by the oviduct at concentrations of around 80 μg mL−1 for functions such as the external protein degradation,41 or the prevention of oviduct obstruction.42 Besides, sperm also release proteases such as acrosin and hyaluronidase which help to penetrate the oocyte cumulus and zona pellucida.43 Here we employ trypsin as a protease model at concentrations ranging from 50 to 200 μg mL−1 to demonstrate the sperm release in vitro. In the real scenario several proteases would serve as enzymatic triggers to catalyze the hydrolysis of the BSA‐HA microflake, thus releasing sperm at the desired location. By optimizing the size and cross‐linking degree of the microflake, the degradation time can be tuned according to the duration of the treatment and the time of delivery.
Scheme 1.
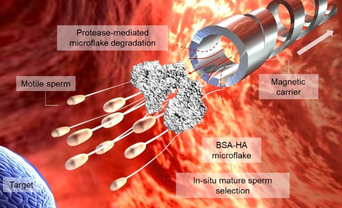
Concept of the immobilization of a cluster of motile sperm on the BSA‐HA microflake and their magnetic transport by a helical microcarrier.
Results and Discussion
The microcarrier has a helical body and a cylindrical cavity in its front. The helical body was designed as a screw thread with a length of 100 μm and a diameter of 50 μm. It was constructed layer‐by‐layer in a shape of spiral‐like stairs. Each structural line was exposed horizontally to enhance the mechanical stability of the structure as it can be seen in Video S1. The cylindrical front cavity was designed with the same diameter of the helical body to confine the cargo. Three prototypes were fabricated by two‐photon polymerization lithography, with different trapping front cavities: a cylinder with a solid bottom surface (MC1), a hollow cylinder (MC2), and a cylinder with a cross‐like bottom surface (MC3) (see Figure 1 a). The MCs were coated with a thin film of Fe and TiO2 for the magnetic actuation and biocompatibility, respectively (see details in the Supporting Information). When the helical body rotates synchronously with the external magnetic field, it converts the rotational actuation into translational motion. The MCs can be furthermore guided by changing the direction of the magnetic field. Hydrodynamic simulations of the three MCs designs are shown in Figure 1 b,c and Video S2. Due to the enclosed cavity of MC1, no fluid vortex was formed to efficiently capture a cargo, which can also be seen by analyzing the pressure difference between P0 and P1 (Figure 1 b). This pressure difference generates a fluid stream from the tube opening to the outside part of the tube (P0<P1), pushing away the cargo (Figure S1a and Video S3). Thus, MC1 exhibited a divergent fluid (Figure 1 b, i–ii and c, i) leading to cargo loss during motion. In contrast, MC2 with a hollow tubular cavity created a micro‐vortex with P0>P1, which was sufficient to trap the cargo on its way (Figure 1 b, i–ii and c, ii). However, in our experiments we observed that small cargoes (i.e., small clusters of immotile sperm or small MF) could easily pass through the helix lumen and be pushed away by the rotating helix, in particular when the magnetic field frequency was higher than 40 Hz. This is attributed to the increased flow pressure difference during the rotation/translation of the MC2 helix (Figure 1 b i–ii). Detailed flow trajectories at different rotating frequencies are shown in Video S2. To avoid cargo loss, we added a mesh (cross structure) at the bottom surface of the cylindrical cavity in MC3 (see Figure 1 a). As observed in Figure 1 b i–ii and Figure 1 c iii, the increase pressure difference in the front cavity (P0>P1) was high enough to capture the cargo during transport (Figure 1 d and Video S3). This result was supported by the visualization of the flow field around the rotating MC3 (Figure S1b and Video S4). The flow field was obtained by tracking the tracer particles around MC3, using a Python implementation of the algorithms developed by J. C. Crocker and D. G. Grier,44 and the mean squared displacement (MSD) was calculated for all their trajectories, as shown for other passive and active particle trackings.45, 46 Therefore, we chose the MC3 design for the subsequent experiments. Figure 1 e illustrates the overlay motion sequence of MC3 under rotating magnetic field actuation (as also shown in Video S5). An open‐loop control over a square‐like trajectory was performed, reflecting a stable swimming behavior of the microcarrier and its fine synchronization with the applied magnetic field. As depicted in Figure 1 f, the swimming performance of the MC was strongly influenced by the strength and the rotating frequency of the applied field in line with previously reported data.47 We found that up to 4 Hz (when applying 4 mT), the MC responded to the magnetic field by a wobbling behavior. After the rotation reached the actuation frequency, the forward velocity of the MC almost linearly increased until the rotating frequency reached the so‐called step‐out frequency (at around 20 Hz).47 After that, the MC velocity declined rapidly as it was not able to follow the magnetic field. A higher field strength shifts the step‐out frequency and therefore increased the peak value of the forward velocity. The maximum swimming speed (ca. 125 μm s−1) was achieved when applying a magnetic field of 8 mT at 60 Hz. MC3 was used to transport a cluster of immotile sperm cells as the first demonstration. Figure 1 g and Video S6 show the trajectory of the transported immotile sperm by the microcarrier MC3. When the microcarrier approached the cluster of immotile sperm, sperm became trapped in the locally‐induced vortex in its front cavity. The cargo was carried so firmly that MC3 could perfectly follow the direction of the magnetic field, ensuring a stable steering during the whole MC journey. In order to evaluate the structural stability of the optimized carrier (MC3), we first applied a magnetic field of ca. 10 mT at 65 Hz on 31 of them, in sperm media for 20 min. The whole system was then incubated at 37 °C with 95 % humidity and 5 % CO2. After 3 days, no structural defects were observed in any microcarrier (Figure S2a). To confirm the MCs swimming capability, we applied the same magnetic field as before, observing the same behavior as in their initial state (Figure S2b). The good structural stability of our microcarriers is attributed to the outer TiO2 layer, which protects the underneath magnetic layer of the microhelix. TiO2 is a material which has been used for medical implants due to its biocompatibility and long‐term stability.48
Figure 1.
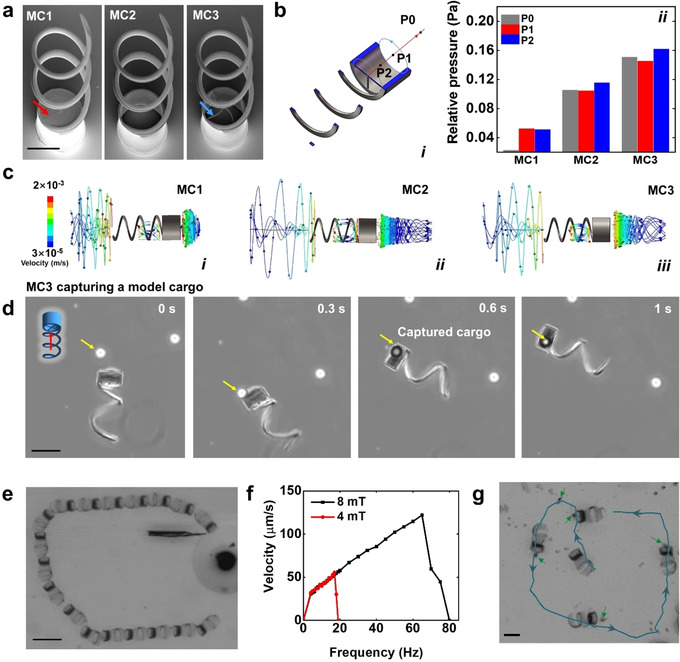
a) SEM images of three microcarrier designs (MC1, MC2, and MC3). Scale bar: 50 μm. Red arrow points at the bottom of MC1 and blue arrow points at the cross structure of MC3. b) Relative pressure of MCs: (i) Positions of three analyzed relative pressures. P0: 40 μm away from P1 along swimming direction; P1: center of the top opening of the cylinder; P2: 20 μm from P2 along the reverse direction of swimming. Blue arrow circles indicate the rotation of MCs. Red arrow line indicates the swimming direction. (ii) Relative pressure for the three different prototypes. c) Flow trajectories showing the fluid vortex of the three prototypes of MCs. Color bar indicates flow velocity (m s−1). Prototype with a trapping front cavity in form of: (i) cylinder with a solid bottom surface, (ii) hollow cylinder, and (iii) cylinder with a cross‐like bottom surface (parameters of the simulation can be found in the Supporting Information). d) MC3 capturing a model cargo (a 40 μm polystyrene particle). Scale bar 50 μm. e) Motion sequence of MC3 guided in a rectangular track. Scale bar: 200 μm. (f) Swimming velocity of MC3 at two magnetic field strengths (4 and 8 mT), in the frequency range from 0 to 80 Hz. g) Overlaid motion sequence and track of the transport of a cluster of immotile sperms by MC3. Blue line and arrows depict the track and directions of the MC movement. Green arrows point at the cluster of immotile sperms. Scale bar: 50 μm.
Furthermore, we assessed the swimming performance of MC3 in saliva, blood and mimicked oviduct fluid (0.4 % methylcellulose enriched sperm medium), which have typical viscosities of ca. 6,49 450 and 5.9 MPa s (Figure S3), respectively. Different from the motion in sperm media (ca. 0.9 MPa s),51 the viscoelasticity and cell composition of such biological fluids lead to a drifting behavior of the microcarrier during motion, which is only observed when the micromotor swims close to a surface. MC3 swam efficiently in saliva at 171±41 μm s−1 (Figure 2 a i and Video S7) while being magnetically guided to avoid big squamous cells.52 After 10 min, the microcarrier was found with mucus (mainly consisting of mucopolysaccharides and glycoproteins)53 as well as some small cells attached on its surface, disturbing its steering capability. In blood, the microcarriers showed a more intricate swimming behavior due to the complex blood composition and the presence of a large amount of red blood cells. 4× diluted whole blood was used in our experiment as a higher concentration of blood cells made it increasingly difficult to track the micromotors by optical microscopy. Figure 2 a ii and Video S7 show that during swimming, MC3 pulled the nearby blood cells into its front cavity by the abovementioned hydrodynamic vortex which then passed through the helix lumen, creating a cell‐free path behind the propeller. In contrast, MC1 with a closed bottom cylindrical cavity only pushed the cells forward and generated a spheroid‐shape accumulation of blood cells at the front. The accumulated cells increased the hydrodynamic resistance so much that MC1 was frequently stalled. As a result, MC3 moved faster (156±41 μm s−1) than MC1 (42±29 μm s−1). Figure 2 a iii shows the microcarrier swimming in mimicked oviduct fluid. Considering the low availability of oviduct fluid our lab recently developed a substitute which mimics the rheological properties of oviduct fluid (which has a viscosity between 2.3 and 5.9 MPa s at 1 Hz). In this work, we prepared mimicked oviduct fluid with 5.9 MPa s by supplementing sperm medium (SP‐TALP) with 0.4 % methylcellulose and microparticles of ca. 10 μm diameter, mimicking floating epithelial cells. As displayed in Video S7, the MC3 swam smoothly and controllably in such oviduct‐mimicking fluid at 197±22 μm s−1, with the nearby beads passing through the mesh bottom of the trapping cavity. Figure 2 a iv and Figure S4 show the swimming of MC3 in an ex vivo mouse oviduct channel (see details in the Supporting Information). When in the middle of the oviduct channel, the microcarrier swam as efficiently as without tissue, occasionally the micromotor was halted by protrusions or sticky parts of the tissue. Thanks to the precise response of the microcarrier to the external magnetic field, the navigation strategy was easily adjusted to adapt to the topography of the swimming path (Figure S4, and Video S8).
Figure 2.
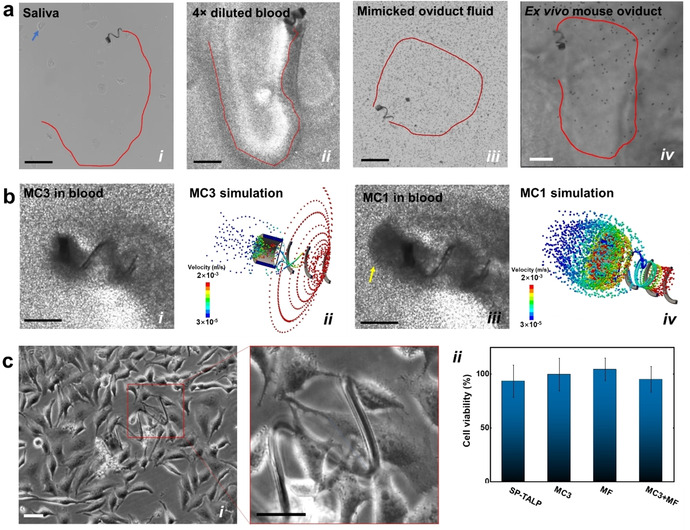
a) Track of micromotors swimming in different biological fluids (i) saliva, (ii) 4× diluted blood, and (iii) mimicked oviduct fluid. Blue arrow points at a squamous cell. Mimicked oviduct fluid was prepared with sperm medium (SP‐TALP) and 0.4 % methylcellulose containing 10 μm microparticles with a concentration of ≈3×105 particles per mL. (iv) Track of a microcarrier swimming inside an ex vivo mouse oviduct channel in a parafilm chip (Figure S4). Scale bars: 300 μm. b) Comparison of MC3 and MC1 swimming in 4× blood. Color bars indicate flow velocity (m s−1). Yellow arrow points at the spheroidal blood cell cluster accumulated in front of MC1′s front cavity. Scale bars: 100 μm. Parameters of the simulation can be found in the Experimental Section. c) MC biocompatibility assay, (i) exemplary optical images of HeLa cells co‐cultured with MC3 and microflakes. Scale bars: 50 μm, and (ii) cell viability analysis comparing different samples (SP‐TALP, MC3, MF and MC3+MF). (n=4).
To get a better understanding of the interaction between the micromotors and the surrounding cells during micromotor swimming, we implemented hydrodynamic simulations with particles of 5 μm diameter that simulate blood or epithelial cells (Figure 2 b). The surrounding cells in the vicinity of MC3 were sucked through its lumen and pushed backward, facilitating its forward motion. In contrast, the cells close to MC1 increased the fluid resistance when accumulated into a spheroidal cell cluster on the front cavity, slowing MC1 down. The agreement between the simulation and the experimental results highlight the importance of the hydrodynamics analysis for the design of microcarriers which can properly swim in blood or other cell‐containing solutions. As a conclusion, the proposed microcarrier (MC3) shows great swimming ability in both synthetic and ex vivo biological fluids, in in vitro and ex vivo tissues.
We also evaluated the biocompatibility of the employed materials and carriers prior to sperm capture/transport experiments by co‐incubation with HeLa cells. Trypan blue was used as an indicator of dead cells and HeLa cell media was used as blank control. Cell viability was calculated as a ratio of live cell number in the specific group relative to the blank control. Magnets were first placed underneath the cell culture plates to attract the microcarriers down to the substrate. After 3 days co‐incubation, no significant difference was found on the live cell number among the evaluated samples (Figure 2 c i–ii). HeLa cells even started to grow on the microflake and the helical body of the microcarrier with several anchoring points. Both the BSA‐HA microflake and the TiO2‐coated MC enhanced cell adhesion. The latter was attributed to the rough surface and the oxide film, which are well‐known factors that improve cell affinity to TiO2.54
Different from immotile sperm or other passive cargoes, the trapping of motile ones, whose typical speeds are ca. 100 μm s−1[55] is very challenging. While the immotile sperm are efficiently trapped by the fluid vortex, motile sperm easily escape due to their strong flagella beating and large propulsion forces. To solve this problem, we synthesized a sperm trapping scaffold based on HA and BSA, which can be transported by the previously optimized magnetic carrier. Firstly, HA/BSA microbeads were synthesized by using a micro‐emulsion method (details in the Supporting Information). The BSA scaffold was created by heating (75 °C for 0.5 h), while the HA microparticles were cross‐linked in 1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodiimide (EDC) solution.56 The microbead size decreased with increasing concentration of the emulgator (see details in the Supporting Information). As mentioned above, HA was used to trap motile sperm with intact DNA39 while BSA served as a matrix to stabilize the HA microparticles and allow the sperm release after hydroxylation. As shown in Figure 3 a, the initially produced BSA‐HA microbeads have a smooth surface which did not allow the exposure of HA particles for proper sperm binding, thus sperm easily slipped away (Video S9). Hence, we gently milled the microbeads at 4 °C using a mortar (see Supporting Information) to roughen the surface and expose HA molecules to the swimming sperm cells (see Figure 3 b). Although the sperm capturing can be accelerated by increasing the sperm amount and motility, the final number of immobilized sperms was mainly dependent on the microflake size (as shown in Figure 3 c). The size of the micromotor‐microflake assembly was varied from ca. 30 to 80 μm (longer axis dimension), smaller than the smallest dimension of the oviduct channel.57 Figure 3 d and Video S10 show the sperm capturing on the microflake. The 80 μm microflake could capture ca. 50 sperm during 10 min co‐incubation. Despite the fact that the sperm capturing becomes more difficult over time (probably due to the repelling from the captured sperms), additional attachment could still occur even after 30 min of co‐incubation.
Figure 3.
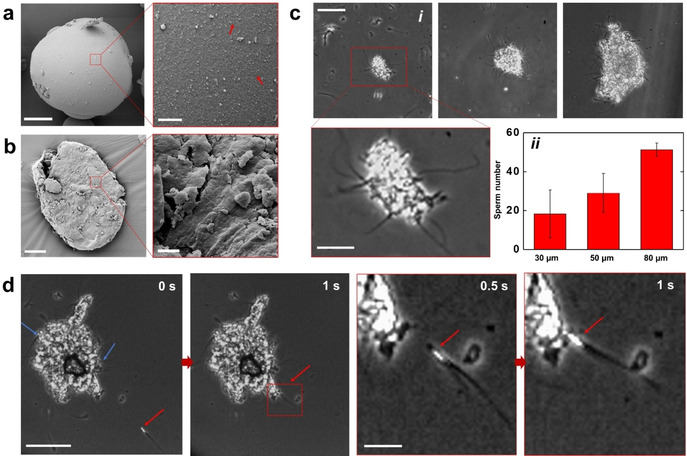
a) SEM image of a BSA‐HA microbead. Red arrows point at the HA particles (<1 μm size). Scale bar: 20 μm in the low magnification image and 2 μm in the magnified image. b) SEM image of the resulting microflake after milling the microbead of Figure a. Scale bar: 20 μm in the low magnification image and 2 μm in the magnified image. c) Sperm immobilization on microflakes of different sizes. (i) Optical microscopy images. Scale bar: 50 μm; 20 μm in the magnified image. (ii) Sperm number related to the microflake size (n=3). d) Image sequence showing the sperm capturing on a microflake. Red arrows point at one sperm before and after being captured. Blue arrows point at the already immobilized sperm. Scale bar: 50 μm.
To demonstrate the overall potential of the MC‐MF based system for multiple sperm transport, we fabricated a microfluidic chip (Figure 4 a i) with a narrow constriction of 0.5×0.5 mm2 to mimic the oviduct, where the average narrowest part is ≈0.5 mm diameter.57 Figure 4 a ii illustrates the entire transport process: first, the microcarrier was slowly guided toward the sperm‐loaded microflake at a low rotating frequency. If the microcarrier approached the targeted microflake at a high speed, the docking was easily lost as the trapping force of the vortex declined rapidly outside the tube. Once the front opening of the microcarrier got close enough to the microflake, the rotating frequency was increased to over 20 Hz to generate a powerful vortex to firmly trap the microflake. After MC‐MF reached the target position, the cargo release was accomplished by simply unscrewing the MC by rotating the magnetic field in the opposite direction. The whole transport process can be seen in Video S11. Finally, the captured sperm were released by enzymatically hydrolyzing the BSA matrix, In general, protein hydrolysis occurs on the peptide linkages.58 During the preparation of the microflake, the aggregated proteins lose the solubility after heating due to an exposure of the hydrophobic groups.59 The aggregated proteins become more unstable, especially in the presence of proteases, as the peptide linkages are also exposed to water.59, 60 To demonstrate the enzymatic sperm release from microflake, we used trypsin as a model protease to mimic the function of the proteases in the oviduct. Trypsin was added into the microfluidic chip through inlet 2 at a final concentration of 100 μg mL−1 (see Figure 4 a). Figure 4 b and Video S12 show the release by enzymatic disintegration of the BSA‐HA sperm‐loaded microflake. At the beginning, the microflake was disintegrated very slowly in part due to the slow diffusion of trypsin from the channel opening to the location of the microflake as well as due to the microflake's hydrophobic nature. After about 4 min, the first sperm started to be released from the microflake. After that, the microflake was rapidly disintegrated, releasing the total amount of captured sperms after ca. 10 min (600 s) (see Figure 4 c). We also tested the sperm release in SP‐TALP without proteases. The immobilized sperm were well attached to the microflake in 1 h of co‐incubation at 37 °C, revealing a stable coupling. Moreover, the release was also influenced by the microflake composition. Video S13 shows the release from the microflake with a low 1:1 ratio of BSA/HA. As little amount of BSA did not form a dense scaffold to separate HA particles, the HA particles agglomerated in the presence of EDC. As a consequence, during the dissolution of the BSA scaffold, the captured sperm remained close to each other and stayed attached, affecting their motion performance after release. We therefore optimized the BSA/HA ratio to 4:1 to ensure a proper HA particle distribution, sperm immobilization and release. After being released, sperm conserved their motility, showing just a slightly decrease of ca. 20 % compared to the untreated sample (see Figure 4 d).
Figure 4.
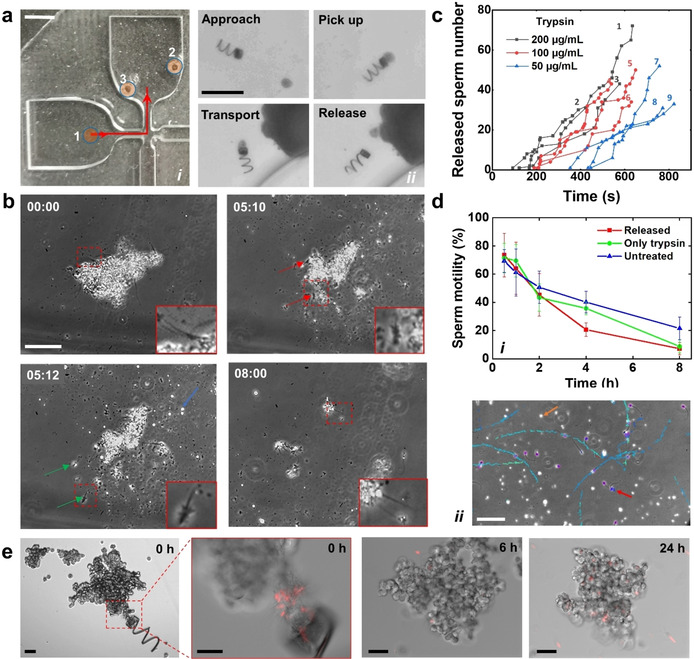
a) Transport of a sperm‐loaded microflake by a magnetic microcarrier. (i) Microchip for sperm transport. 1: inlet for sample introduction; 2: inlet for trypsin introduction; 3: outlet for pressure balance. Red arrow line indicates the transport path. Scale bar: 2 mm. (ii) Image sequence illustrating the transport and release of the microflake by MC3. Scale bar: 200 μm. b) Phase contrast images showing sperm release during microflake hydrolysis by trypsin. Blue arrow points at a HA aggregation. Red and green arrows point at the sperms before and after release, respectively. Time stamp: min:s. Scale bar: 50 μm. c) Influence of trypsin concentration on sperm release. Total number of initially captured sperms in the microflakes of gray color (1–3): 75, 37, 48 captured sperm; red color (4–6): 50, 55, 40 captured sperm; blue color (7–9): 56, 38, 39 captured sperm. d) Sperm motility under trypsinization. (i) Sperm motility over 8 h (n=4). (ii) Swimming track of sperm released from a microflake after 0.5 h of trypsinization. Purple circles point at sperm heads. Brown arrow points at a HA aggregation. Red arrow points at a re‐immobilized sperm. Scale bar: 50 μm. Error bars come from the standard deviation of sperm motility among 4 replicates (with 10 monitored fields of view). e) DOX‐HCl distribution in the cancer spheroid over time. A microflake containing drug‐loaded sperm was transported and delivered by MC3. Trypsin was terminated at 20 min after the microflake delivery. Scale bars: 50 μm.
To monitor sperm motility over time after sperm release, we first separated the captured sperm from the free ones. The microflakes were immobilized onto 3.5 cm diameter petri dishes in advance. After the immobilization, the petri dishes were washed with SP‐TALP to remove the uncoupled sperm, thus avoiding any artifacts caused by the free ones on the motility analysis. Trypsin was added to the solutions at a final concentration of 100 μg mL−1 to release the sperm. Figure 4 d i depicts the motility percentage over time under trypsinization. No difference was found between trypsin‐treated free sperm and the untreated ones, as trypsin cannot induce direct cell death despite the fact that it might induce membrane protein degradation over time in high concentrations.61 In the first 4 h after treatment, released sperm also showed no difference from the other groups, implying that the microflake‐based capturing and enzymatic release processes have no significant influence on sperm motility. As an example, swimming trajectories of the released sperm at 0.5 h were recorded (see Figure 4 d ii). Released sperm and HA particles were both found in the solution. While most sperm swam away, some of them remained on the residual HA parts after the microflake disintegration (see Video S14). Although we did not observe significant differences among the analyzed groups at the beginning, there was a more pronounced motility decrease after 4 h, probably due to the accumulation of pyrolysis products of BSA which might affect sperm metabolism.62 Another factor that affects release performance is the trypsin concentration. As depicted in Figure 4 c, the release rate declined as the trypsin concentration decreased. The release time can be further regulated by tuning the crosslinking degree of the BSA, considering the normal lifetime of sperm cells under physiological conditions (for bull sperms, it is about 8 h).35
To show the potential application of the proposed system as drug carriers, we evaluated its drug delivery capability on HeLa spheroids as tumor model. DOX‐HCl was used here as a model anticancer drug as well as fluorescence indicator due to its self‐fluorescence at 470 nm excitation wavelength. We first transported a single microflake containing drug‐loaded sperm toward a cancer spheroid in a trypsin‐enriched environment as shown in Figure 4 e and Video S15. Neither DOX‐HCl leakage nor sperm escape was found during the transport (<3 min) as shown in Figure 4 e. After 20 min, fetal bovine serum was introduced to terminate the trypsin effect. After 24 h culture, the drug was well distributed on the cancer spheroid (Figure 4 e). Given the fact that the sperm amount from a single microflake was not enough to draw a statistic conclusion, we evaluated the cell killing efficiency by mimicking the transport of 10 microflakes (with ca. 50 drug‐loaded sperm on each) to tumor spheroids by manual pipetting (details are included in the Experimental Section and Figure S6). The results are in agreement with previously reported data on free swimming sperm,35, 36 and show that microflakes‐drug loaded sperm are efficient in killing cancer cells (see Figure S6c). Remarkably, a good cancer cell killing effect is also observed for sperm delivered without anti‐cancer drugs, which can be explained by the lethal impact of sperm tail beating on the cancer spheroids.35, 36
Conclusion
A helical micromotor actuated by an external magnetic field was used to actively transport and release a HA‐BSA microflake which was employed as scaffold to capture mature and motile sperm cells. The magnetic microcarrier was characterized in different biofluids (e.g., saliva, blood and synthetic oviduct fluid), and optimized to ensure efficient cargo transport. The release of motile sperm from the microflake relied on the enzymatic hydrolysis of the BSA scaffold which disintegrated over time (ca. 10 min). In the reproductive system, proteases including oviduct proteases, acrosin, and hyaluronidase facilitate the hydrolysis of foreign proteins,42, 43 thus being suitable for the disintegration of the BSA‐HA microflake when applying the proposed system in a living organism. The method to temporarily attach moving cargoes using a protein‐chemoattractant based scaffold opens up new possibilities for multiple sperm cargo‐delivery applications as well as for other motile living organisms or cells such as bacteria and macrophages whenever multiple of them are necessary for an effective treatment. However, there are remaining challenges to overcome before the proposed sperm carrier can be applied in the clinic either for assisted fertilization or drug delivery applications. Real‐time navigation in deep tissue is perhaps one of the most important ones. When conventional imaging strategies are either too slow or too coarse, the recent developed optoacoustic imaging system is one promising alternative to solve the problem of real‐time imaging of moving single micromotors in deep tissue (ca. 1 cm deep).63, 64, 65 Another hurdle that needs to be considered is the complex environment inside the reproductive system in vivo, from which there is very little known from the literature, for example the oviduct hydrodynamics in the presence of oocytes and micromotors or the exact fertilization timing. Fortunately, versatile micromotors not only enable various bio‐applications but can also be used as non‐invasive microtools to study cell/tissue biophysics66 or perform local sensing.63
Conflict of interest
The authors declare no conflict of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Acknowledgements
We thank Masterrind GmbH for kind donation of cryopreserved bovine semen. We thank F. Hebenstreit for the help with the medium preparation. We thank R. Engelhard, S. Nestler and C. Krien for clean room support. We thank L. Schwarz and F. Striggow for helpful discussions. We also thank to Dr. B. Ibarlucea for proof‐reading and M.M.‐S. for the TOC image. Haifeng Xu acknowledges the China Scholarship Council (CSC) for funding. O.G.S. acknowledges financial support by the Leibniz Program of the German Research Foundation (SCHM 1298/26‐1). This work was also supported by the German Research Foundation SPP 1726 “Microswimmers‐From Single Particle Motion to Collective Behavior” and by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (grants agreement No. 835268 and No. 853609). M.M.‐S. and O.G.S supervised the work.
H. Xu, M. Medina-Sánchez, O. G. Schmidt, Angew. Chem. Int. Ed. 2020, 59, 15029.
References
- 1. Medina-Sánchez M., Schmidt O. G., Nature 2017, 545, 406–508. [DOI] [PubMed] [Google Scholar]
- 2. Medina-Sánchez M., Xu H., Schmidt O. G., Ther. Delivery 2018, 9, 303–316. [DOI] [PubMed] [Google Scholar]
- 3. Wang J., Nanomachines: Fundamentals and Applications, Wiley, Hoboken, 2013, pp. 119–138. [Google Scholar]
- 4. Gao W., de Ávila B. E.-F., Zhang L., Wang J., Adv. Drug Delivery Rev. 2018, 125, 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peng F., Tu Y., Wilson D. A., Chem. Soc. Rev. 2017, 46, 5289–5310. [DOI] [PubMed] [Google Scholar]
- 6. Jurado-Sánchez B., Pacheco M., Rojo J., Escarpa A., Angew. Chem. Int. Ed. 2017, 56, 6957–6961; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2017, 129, 7061–7065. [Google Scholar]
- 7. Pacheco M., López M. Á., Jurado-Sánchez B., Escarpa A., Anal. Bioanal. Chem. 2019, 411, 6561–6573. [DOI] [PubMed] [Google Scholar]
- 8. Xu D., Wang Y., Liang C., You Y., Sanchez S., Ma X., Small 2020, 16, 1902464. [DOI] [PubMed] [Google Scholar]
- 9. Novotný F., Wang H., Pumera M., Chem 2020, 6, 1032. [Google Scholar]
- 10. Lin X., Wu Z., Wu Y., Xuan M., He Q., Adv. Mater. 2016, 28, 1060–1072. [DOI] [PubMed] [Google Scholar]
- 11. Felfoul O., Mohammadi M., Taherkhani S., De Lanauze D., Xu Y. Z., Loghin D., Essa S., Jancik S., Houle D., Lafleur M., Gaboury L., Tabrizian M., Kaou N., Atkin M., Vuong T., Batist G., Beauchemin N., Radzioch D., Martel S., Nat. Nanotechnol. 2016, 11, 941–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang H., Li Z., Wu Z., He Q., Adv. Ther. 2019, 2, 1900096. [Google Scholar]
- 13. Chen X.-Z., Hoop M., Mushtaq F., Siringil E., Hu C., Nelson B. J., Pané S., Appl. Mater. Today 2017, 9, 37–48. [Google Scholar]
- 14. Kangarlu A., Burgess R. E., Zhu H., Nakayama T., Hamlin R. L., Abduljalil A. M., Robitaille P. M. L., Magn. Reson. Imaging 1999, 17, 1407–1416. [DOI] [PubMed] [Google Scholar]
- 15. Tottori S., Zhang L., Qiu F., Krawczyk K. K., Franco-Obregón A., Nelson B. J., Adv. Mater. 2012, 24, 811–816. [DOI] [PubMed] [Google Scholar]
- 16. Dreyfus R., Baudry J., Roper M. L., Fermigier M., Stone H. A., Bibette J., Nature 2005, 437, 862–865. [DOI] [PubMed] [Google Scholar]
- 17. Giltinan J., Diller E., Sitti M., Lab Chip 2016, 16, 4445–4457. [DOI] [PubMed] [Google Scholar]
- 18. Wu Z., Troll J., Jeong H.-H., Wei Q., Stang M., Ziemssen F., Wang Z., Dong M., Schnichels S., Qiu T., Fischer P., Sci. Adv. 2018, 4, eaat4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Go G., Jeong S.-G., Yoo A., Han J., Kang B., Kim S., Nguyen K. T., Jin Z., Kim C.-S., Seo Y. R., Kang J. Y., Na J. Y., Song E. K., Jeong Y., Seon J. K., Park J.-O., Choi E., Sci. Robot. 2020, 5, eaay6626. [DOI] [PubMed] [Google Scholar]
- 20. Venugopalan P. L., Sai R., Chandorkar Y., Basu B., Shivashankar S., Ghosh A., Nano Lett. 2014, 14, 1968–1975. [DOI] [PubMed] [Google Scholar]
- 21.D. J. Bell, S. Leutenegger, K. M. Hammar, L. X. Dong, B. J. Nelson, Proc. 2007 IEEE Int. Conf. Robot. Autom 2007, pp. 1128–1133.
- 22. Zhang L., Abbott J. J., Dong L., Kratochvil B. E., Bell D., Nelson B. J., Appl. Phys. Lett. 2009, 94, 064107. [Google Scholar]
- 23. Mark A. G., Gibbs J. G., Lee T.-C., Fischer P., Nat. Mater. 2013, 12, 802–807. [DOI] [PubMed] [Google Scholar]
- 24. Gao W., Sattayasamitsathit S., Manesh K. M., Weihs D., Wang J., J. Am. Chem. Soc. 2010, 132, 14403–14405. [DOI] [PubMed] [Google Scholar]
- 25. Xie H., Sun M., Fan X., Lin Z., Chen W., Wang L., Dong L., He Q., Sci. Robot. 2019, 4, eaav8006. [DOI] [PubMed] [Google Scholar]
- 26. Frutiger D. R., Vollmers K., Kratochvil B. E., Nelson B. J., Int. J. Rob. Res. 2010, 29, 613–636. [Google Scholar]
- 27. Xin C., Yang L., Li J., Hu Y., Qian D., Fan S., Hu K., Cai Z., Wu H., Wang D., Wu D., Chu J., Adv. Mater. 2019, 31, 1808226. [DOI] [PubMed] [Google Scholar]
- 28. Bozuyuk U., Yasa O., Yasa I. C., Ceylan H., Kizilel S., Sitti M., ACS Nano 2018, 12, 9617–9625. [DOI] [PubMed] [Google Scholar]
- 29. Medina-Sánchez M., Schwarz L., Meyer A. K., Hebenstreit F., Schmidt O. G., Nano Lett. 2016, 16, 555–561. [DOI] [PubMed] [Google Scholar]
- 30. Huang T. Y., Qiu F., Tung H. W., Chen X. B., Nelson B. J., Sakar M. S., Appl. Phys. Lett. 2014, 105, 114102. [Google Scholar]
- 31. Mortimer S. T., Hum. Reprod. Update 1997, 3, 403–439. [DOI] [PubMed] [Google Scholar]
- 32. Cohen J., Tyler K. R., J. Reprod. Fertil. 1980, 60, 213–218. [DOI] [PubMed] [Google Scholar]
- 33. Magdanz V., Sanchez S., Schmidt O. G., Adv. Mater. 2013, 25, 6581–6588. [DOI] [PubMed] [Google Scholar]
- 34. Magdanz V., Medina-Sánchez M., Schwarz L., Xu H., Elgeti J., Schmidt O. G., Adv. Mater. 2017, 29, 1606301. [DOI] [PubMed] [Google Scholar]
- 35. Xu H., Medina-Sanchez M., Magdanz V., Schwarz L., Hebenstreit F., Schmidt O. G., ACS Nano 2018, 12, 327–337. [DOI] [PubMed] [Google Scholar]
- 36.H. Xu, M. Medina-sánchez, D. R. Brison, R. J. Edmondson, S. S. Taylor, L. Nelson, K. Zeng, S. Bagley, C. Ribeiro, P. R. Lina, E. Lucena, C. K. Schmidt, O. G. Schmidt, arXiv:1904.12684 2019.
- 37. Chen C., Chang X., Angsantikul P., Li J., Esteban-Fernández de Ávila B., Karshalev E., Liu W., Mou F., He S., Castillo R., Liang Y., Guan J., Zhang L., Wang J., Adv. Biosyst. 2018, 2, 1700160. [Google Scholar]
- 38. Huszar G., Jakab A., Sakkas D., Ozenci C. C., Cayli S., Delpiano E., Ozkavukcu S., Reprod. Biomed. Online 2007, 14, 650–663. [DOI] [PubMed] [Google Scholar]
- 39. Huszar G., Ozenci C. C., Cayli S., Zavaczki Z., Hansch E., Vigue L., Fertil. Steril. 2003, 79, 1616–1624. [DOI] [PubMed] [Google Scholar]
- 40. Rawlings N. D., Barrett A. J., Biochem. J. 1993, 290, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kan F. W. K., Esperanzate P. W., Mol. Reprod. Dev. 2006, 766, 8–9. [DOI] [PubMed] [Google Scholar]
- 42. Hiyoshi M., Takamune K., Mita K., Kubo H., Sugimoto Y., Katagiri C., Dev. Biol. 2002, 243, 176–184. [DOI] [PubMed] [Google Scholar]
- 43. Adham I. M., Nayernia K., Engel W., Mol. Reprod. Dev. Inc. Gamete Res. 1997, 46, 370–376. [DOI] [PubMed] [Google Scholar]
- 44.D. Allan, T. Caswell, N. Keim, C. van der Wel, 2015, 10.5281/zendo.34028. [DOI]
- 45. Orozco J., Jurado-Sánchez B., Wagner G., Gao W., Vazquez-Duhalt R., Sattayasamitsathit S., Galarnyk M., Cortés A., Saintillan D., Wang J., Langmuir 2014, 30, 5082–5087. [DOI] [PubMed] [Google Scholar]
- 46. Rojas D., Jurado-Sánchez B., Escarpa A., Anal. Chem. 2016, 88, 4153–4160. [DOI] [PubMed] [Google Scholar]
- 47. Zhang L., Abbott J. J., Dong L., Peyer K. E., Kratochvil B. E., Zhang H., Bergeles C., Nelson B. J., Nano Lett. 2009, 9, 3663–3667. [DOI] [PubMed] [Google Scholar]
- 48. Kasemo B., J. Prosthet. Dent. 1983, 49, 832–837. [DOI] [PubMed] [Google Scholar]
- 49. Hatton M. N., Levine M. J., Margarone J. E., Aguirre A., J. Oral Maxillofac. Surg. 1987, 45, 496–499. [DOI] [PubMed] [Google Scholar]
- 50. Dintenfass L., Blood Viscosity, Springer Science & Business Media, Cham, 1985. [Google Scholar]
- 51. Romero-Aguirregomezcorta J., Sugrue E., Martínez-Fresneda L., Newport D., Fair S., Sci. Rep. 2018, 8, 16897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Theda C., Hwang S. H., Czajko A., Loke Y. J., Leong P., Craig J. M., Sci. Rep. 2018, 8, 4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neutra M., Leblond C. P., J. Cell Biol. 1966, 30, 137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou W., Zhong X., Wu X., Yuan L., Zhao Z., Wang H., Xia Y., Feng Y., He J., Chen W., Surf. Coat. Technol. 2006, 200, 6155–6160. [Google Scholar]
- 55. Parrish J. J., Foote R. H., J. Androl. 1987, 8, 259–266. [DOI] [PubMed] [Google Scholar]
- 56. Zhou Z., Yang Z., Kong L., Liu L., Liu Q., Zhao Y., Zeng W., Yi Q., Cao D., J. Macromol. Sci. Part B 2012, 51, 2392–2400. [Google Scholar]
- 57. Eddy C. A., Pauerstein C. J., Clin. Obstet. Gynecol. 1980, 23, 1177–1193. [DOI] [PubMed] [Google Scholar]
- 58. Adler-Nissen J., Enzymic Hydrolysis of Food Proteins, Elsevier Applied Science Publishers, Amsterdam, 1986. [Google Scholar]
- 59. Voutsinas L. P., Cheung E., Nakai S., Food Sci. 1983, 48, 26–32. [Google Scholar]
- 60. Murphy K. P., Privalov P. L., Gill S. J., Science 1990, 247, 559–561. [DOI] [PubMed] [Google Scholar]
- 61. Skog M., Sivlér P., Steinvall I., Aili D., Sjöberg F., Elmasry M., Cell Transplant. 2019, 28, 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fu J., Li Y., Wang L., Zhen L., Yang Q., Li P., Li X., Theriogenology 2017, 102, 87–97. [DOI] [PubMed] [Google Scholar]
- 63. Xie L., Pang X., Yan X., Dai Q., Lin H., Ye J., Cheng Y., Zhao Q., Ma X., Zhang X., Liu G., Chen X., ACS Nano 2020, 14, 2880–2893. [DOI] [PubMed] [Google Scholar]
- 64. Wu Z., Li L., Yang Y., Hu P., Li Y., Yang S.-Y., Wang L. V., Gao W., Sci. Robot. 2019, 4, eaax0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Aziz A., Medina-Sánchez M., Claussen J., Schmidt O. G., Nano Lett. 2019, 19, 6612–6620. [DOI] [PubMed] [Google Scholar]
- 66.P. L. Venugopalan, A. Ghosh, 2015 IEEE 15th Int. Conf. Nanotechnol 2015, pp. 428–431.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary
Supplementary


