Abstract
Background and Aims
The gap between patients on transplant waiting lists and available donor organs is steadily increasing. Human organoids derived from leucine‐rich repeat‐containing G protein‐coupled receptor 5 (LGR5)–positive adult stem cells represent an exciting new cell source for liver regeneration; however, culturing large numbers of organoids with current protocols is tedious and the level of hepatic differentiation is limited.
Approach and Results
Here, we established a method for the expansion of large quantities of human liver organoids in spinner flasks. Due to improved oxygenation in the spinner flasks, organoids rapidly proliferated and reached an average 40‐fold cell expansion after 2 weeks, compared with 6‐fold expansion in static cultures. The organoids repopulated decellularized liver discs and formed liver‐like tissue. After differentiation in spinner flasks, mature hepatocyte markers were highly up‐regulated compared with static organoid cultures, and cytochrome p450 activity reached levels equivalent to hepatocytes.
Conclusions
We established a highly efficient method for culturing large numbers of LGR5‐positive stem cells in the form of organoids, which paves the way for the application of organoids for tissue engineering and liver transplantation.
Abbreviations
- A1AT
alpha‐1‐antitrypsin
- ALB
albumin
- BMP‐7
bone morphogenetic protein 7
- CYP
cytochrome P450
- CYP3A4
cytochrome p450 3A4
- DM
differentiation medium
- ECM
extracellular matrix
- EM
expansion medium
- GLDH
glutamate dehydrogenase
- HE
hematoxylin and eosin
- K18
keratin 18
- K19
keratin 19
- LGR5
leucine‐rich repeat‐containing G protein‐coupled receptor 5
- MRP2
multidrug resistance–associated protein 2
- NSG
NOD.Cg‐Prkdc scid Il2rg tm1Wjl/SzJ
- RT‐PCR
reverse‐transcription PCR
- Rh123
rhodamine 123
- SOX9
SRY‐box 9
Liver disease and the subsequent loss of liver function is a major global health care challenge with poor long‐term clinical outcome. The only curative treatment for end‐stage liver failure is transplantation; however, available donor organs are limited, and the number of patients on the transplant wait list is steadily increasing. Approximately 13,000 patients are currently on the transplant wait list in the United States, which far exceeds the number of available donor organs. According to the Organ Procurement and Transplantation Network, only 8,250 livers were transplanted in 2018, and 20% of patients on the liver transplant wait list die or become too ill to be transplanted. This increasing shortage of donor organs demands the development of new techniques to replace damaged or severely diseased livers.1
Alternatives to liver transplantation include liver cell therapy and liver tissue engineering. Current approaches rely on primary hepatocytes. However, fully differentiated hepatocytes are difficult to work with, as they exhibit minimal growth in culture and rapidly dedifferentiate in vitro.2 This has led to the search for a renewable source of stem cells that can be expanded in vitro and subsequently differentiated into hepatocyte‐like cells.3 Several such cell types include human pluripotent stem cells such as embryonic stem cells4 or induced pluripotent stem (iPS) cells,5 which have been investigated extensively. With these sources, an unlimited supply of human hepatocytes could be envisioned; however, generation and maintenance of stable and mature adult liver cells ex vivo have not yet been achieved.6, 7
Another attractive alternative are organoids generated from leucine‐rich repeat‐containing G protein‐coupled receptor 5 (LGR5)–positive adult tissue stem cells. Organoids offer exciting new possibilities as an autologous cell source for tissue engineering or cell therapy,3, 8 as they can be obtained easily from a liver biopsy and expanded in culture for years while remaining genetically stable.9, 10 Unfortunately, reproducibility and upscaling of current organoid systems remain major bottlenecks in their clinical application.11 Establishing large numbers of organoids with current protocols is tedious, as organoids are cultured in droplets of Matrigel,9, 10 which hampers clinically relevant production times of organoids for tissue engineering and transplantations. It is estimated that the adult human liver contains about 240 billion (109) hepatocytes,12 and although transplantation studies show that replacing 10% of the liver mass results in clinical improvement,13 billions of cells are still required for clinical application.14
To enable mass production of viable cell cultures, bioreactors, including spinner flasks, have been used extensively for the large‐scale expansion and controlled differentiation of stem cells.15, 16 Bioreactors offer certain advantages including minimization of gradient formation (e.g., pH, nutrients, metabolites, dissolved oxygen), increased transport of oxygen and nutrients, and prevention of cell sedimentation, thus overcoming the intrinsic limitations of static culture systems.17 As an example, Lancaster et al. clearly demonstrated limited growth potential of brain organoids due to stationary diffusion of oxygen and nutrients in static Matrigel cultures, and the use of spinner flasks markedly improved organoid survival and development.18
In this study, we used spinner flasks for the large‐scale production of human adult stem cell–derived liver organoids. We then examined the differentiation potential of the organoids toward functional hepatocytes, and assessed their use for tissue engineering approaches.
Experimental Procedures
Organoid Culture and Differentiation in Spinner Flasks
Disposable 125‐mL spinner flasks (Corning, New York, NY) were inoculated with 2.5 million cells in 25 mL expansion medium (EM), including 10% vol/vol Matrigel. Due to single cell seeding, 10 mM Y‐27632 (Rho kinase‐inhibitor; Selleckchem, Munich, Germany) was added to the medium during the first week of culture. Rotation speed was set to 85 rpm. Every 2‐3 days, new medium was added to the spinner flasks.
For hepatic organoid differentiation in spinner flasks, organoids were primed for 2 days with the addition of bone morphogenetic protein 7 (BMP‐7; 25 ng/mL) to the EM. Subsequently, to separate organoids from the EM, the organoid suspension was filtered through a 70‐µm cell strainer. The organoids were transferred to a new spinner flask and differentiated for 12 days in human organoid differentiation medium (DM) supplemented with 10% Matrigel. The procedure is schematically outlined in Supporting Fig. S4.
Cryopreserved Hepatocyte Culture
LiverPool cryoplateable hepatocytes (Admixed from 10 donors, mixed gender) were purchased from BioreclamationIVT (Brussels, Belgium). The hepatocytes were brought into a collagen sandwich culture according to the manufacturer’s instructions, with the recommended InvitroGRO CP Medium (BioreclamationIVT). For mRNA sequencing, alpha‐1‐antitrypsin (A1AT) and glutamate dehydrogenase (GLDH) expression and albumin (ALB) secretion assays, hepatocytes, and medium were harvested after 4 hours of sandwich culture. For midazolam metabolism assays, the medium from hepatocytes cultured in collagen sandwich culture with 5 mM midazolam was collected after 24 hours.
Fresh Hepatocyte Isolation
Hepatocytes were obtained from healthy liver parenchyma from 2 patients with liver papilloma and adenoma, ages 40 and 41 years, respectively, undergoing liver resections. The hepatocytes were isolated by a modified two‐step collagenase perfusion technique, as described.30 The cells were snap‐frozen directly after isolation. The procedure was in accordance with the ethical standards of the institutional committee on human experimentation (protocol number 03/024) and the Helsinki Declaration of 1975. Informed consent in writing was obtained from each patient.
Whole Transcriptome Analysis
For mRNA sequencing, organoids, LiverPool cryoplateable hepatocytes, and freshly isolated hepatocytes were used. RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturers instructions. The polyadenylated mRNA fraction was isolated using Poly(A) Beads (NEXTflex; Bio Scientific, Austin, TX). The Rapid Directional RNA‐Seq Kit (NEXTflex) was used to prepare sequencing libraries. Illumina NextSeq500 sequencing produced single‐end 75 base long reads. RNA‐sequencing reads were mapped using STAR version 2.4.2a. Read groups were added to the BAM files with Picard’s AddOrReplaceReadGroups (v1.98) and sorted with Sambamba v0.4.5. Transcript abundances were quantified with HTSeq‐count version 0.6.1p1, using the union mode. The raw files were uploaded to Gene Expression Omnibus under the accession GSE123498. Differentially expressed genes were identified using the DESeq2 package with standard settings. Genes with a threshold fold change of four or more in all spinner flasks compared with their respective static controls were used for pathway analysis and visualization in heatmaps.
ToppFun was used for functional enrichment analysis based on functional annotations and protein interaction networks. Heatmaps were generated using edgeR.
Decellularized Liver Disc Preparation
Preparation of decellularized rat liver discs was performed as previously described22 and is outlined in the Supporting Experimental Procedures.
Recellularization of Liver Discs With Organoids
The seeding procedure was performed as previously described.22 Briefly, organoids were mechanically disrupted, and for each disc, organoid fragments corresponding to 2.5 × 105 cells were suspended in 10 μL EM supplemented with 25 ng/mL BMP‐7. The organoid suspension was slowly pipetted on top of each disc and incubated for 1 hour at 37°C to allow the cells to attach. Subsequently, 250 μL of additional EM with 25 ng/mL BMP‐7 was added to each well. Medium was changed twice per day. After 2 days of culture, the discs were carefully transferred to a 24‐well plate and cultured in DM for 5 days. DM was refreshed after 2 days.
RNA Isolation and Quantitative RT‐PCR
RNA was isolated from organoids, hepatocytes, and recellularized discs using the RNeasy Micro Kit according to the manufacturers instructions. For isolating RNA from the recellularized discs, we added the lysis buffer (RLT buffer from the RNeasy Micro Kit) to the discs, and after cell lysis, we transferred the lysate to a new tube and continued with the RNA isolation according to the RNeasy Micro Kit protocol. RNA was quantified by optical density. Complementary DNA (cDNA) was synthesized using the iScript cDNA synthesis kit (Bio‐Rad, Hercules, CA) as described by the manufacturer. Relative mRNA abundance of the selected genes was measured by real‐time PCR using validated primers (Supporting Table S5) with the SYBR Green method (Bio‐Rad). Messenger RNA abundances of the reference genes hypoxanthine‐guanine phosphoribosyltransferase (HPRT) and ribosomal protein L19 (RPL19) were used for normalization.
Microscopy, Immunohistochemistry, and Immunofluorescence Analysis
An overview of the used antibodies, dilutions, incubation times, and antigen retrieval method are listed in Supporting Table S6, and the procedure is extensively described in the Supporting Experimental Procedures.
ALB Secretion and Midazolam Metabolism Assays
To determine ALB secretion and midazolam metabolism, liver organoids were differentiated for 12 days as described. For spinner flask samples, organoids were harvested from spinner flasks at day 12 of differentiation, seeded in Matrigel droplets and cultured in DM for 24 hours, after which the culture medium was collected and concentrated using Amicon Ultra centrifugal filters (Amicon, Darmstadt, Germany). The concentration of ALB was then determined using a DxC‐600 Beckman chemistry analyzer (Beckman Coulter). Values were normalized for cell counts.
Midazolam metabolism was determined as a readout for cytochrome p450 3A4 (CYP3A4) functionality. Organoids were harvested from spinner flasks at day 12 of differentiation, seeded in Matrigel droplets and cultured in DM supplemented with 5 mM midazolam. After 24 hours, medium was harvested and the concentration of the midazolam metabolite 1‐hydroxymidazolam (1‐OH‐M) was determined using liquid chromatography–tandem mass spectrometry (LC/MS‐MS). Analysis was carried out at the Clinical Pharmaceutical and Toxicological Laboratory of the Department of Clinical Pharmacy of the University Medical Center Utrecht, the Netherlands. All experiments were performed on a Thermo Fisher Scientific (Waltham, MA) triple quadrupole Quantum Access LC/MS‐MS system with a Surveyor MS pump and a Surveyor Plus autosampler with an integrated column oven. Analytes were detected using tandem mass spectrometry, with an electrospray ionization interface in selected reaction monitoring mode, by their parent and product ions.
Rhodamine 123 Transport Assay
For rhodamine 123 (Rh123) transport assays, liver organoids were differentiated for 12 days as described. For spinner flask samples, organoids were harvested from spinner flasks at day 12 of differentiation and seeded in Matrigel droplets. Organoids were pretreated with DMSO or 10 μM Verapamil (Sigma‐Aldrich, St. Louis, MO) for 30 minutes. Organoids were then removed from Matrigel and incubated with 100 μM Rh123 (Sigma‐Aldrich) for 10 minutes. Fluorescence was visualized by an EVOS FL Cell Imaging System (Life Technologies, Carlsbad, CA).
GLDH and A1AT Expression
For the quantification of intracellular levels of GLDH and A1AT, liver organoids were differentiated for 12 days, as described. For spinner flask samples, organoids were harvested from spinner flasks at day 12 of differentiation, seeded in Matrigel droplets and cultured in DM for 24 hours. All wells were then trypsinized for cell counts. Subsequently, cells were lysed in MilliQ water. GLDH was measured in the lysates using a DxC‐600 Beckman chemistry analyzer (Beckman Coulter). A1AT was measured using an AssayMax A1AT enzyme‐linked immunosorbent assay kit (AssayPro, St. Charles, MO). Values were normalized to live cell numbers.
Ammonium Elimination Assay
For ammonium elimination assays, liver organoids were differentiated for 12 days, as described. For spinner flask samples, organoids were harvested from spinner flasks at day 12 of differentiation and seeded in Matrigel droplets. All conditions were then incubated with DM supplemented with 1.5 mM NH4Cl for 24 hours. All medium samples were harvested after 24 hours of incubation and stored at −20°C. Subsequently, all wells were trypsinized, and the number of living cells per well was determined by counting with trypan blue. Ammonium concentrations in the medium were determined using the Urea/Ammonia Assay Kit (Megazyme; Bray, County Wicklow, Ireland). DM supplemented with 1.5 mM NH4Cl that was incubated for 24 hours without cells was used to determine the starting concentration. Ammonia elimination rates were normalized to live cell numbers.
Mouse Xenograft Studies
All mouse experiments have been regulated under the Erasmus MC license for experimental animals (AVD101002017867/17‐867‐19). The mice were housed in individually ventilated cages with standard bedding, enrichment with tissues and ad libitum pellets.
For subcutaneous grafts, organoid fragments equal to 1 and 5 million cells were suspended in 1:1 Matrigel:Advanced DMEM/F12 and were injected into four flank injection sites per female NOD.Cg‐Prkdc scid Il2rg tm1Wjl/SzJ (NSG) mice (Charles River, Wilmington, MA). Two mice were injected with organoids at the age of 10 weeks during the light cycle. As a positive control, human tumoroid liver organoids derived from a cholangiocarcinoma of a 76‐year old female donor were used.33 Visible tumors developed in approximately 8 weeks (tumoroid liver organoids), at which point the mice were euthanized and skin tissues were harvested to assess the growth and histology of the grafted cells. Samples were fixed in 4% paraformaldehyde overnight at room temperature, embedded in paraffin, sectioned at 4 μm and processed for immunofluorescent analysis as described previously.
Statistical Analyses
Quantitative RT‐PCR results (Fig. 4B), ALB secretion (Fig. 4E), A1AT expression (Fig. 4F), midazolam metabolism (Fig. 4G), GLDH expression (Fig. 4H), ammonium elimination (Fig. 4I) and mRNA sequencing data (Supporting Tables S2‐S4) were analyzed using a one‐tailed (unpaired) Mann‐Whitney U test. The P values are indicated in the corresponding figures and tables.
Figure 4.
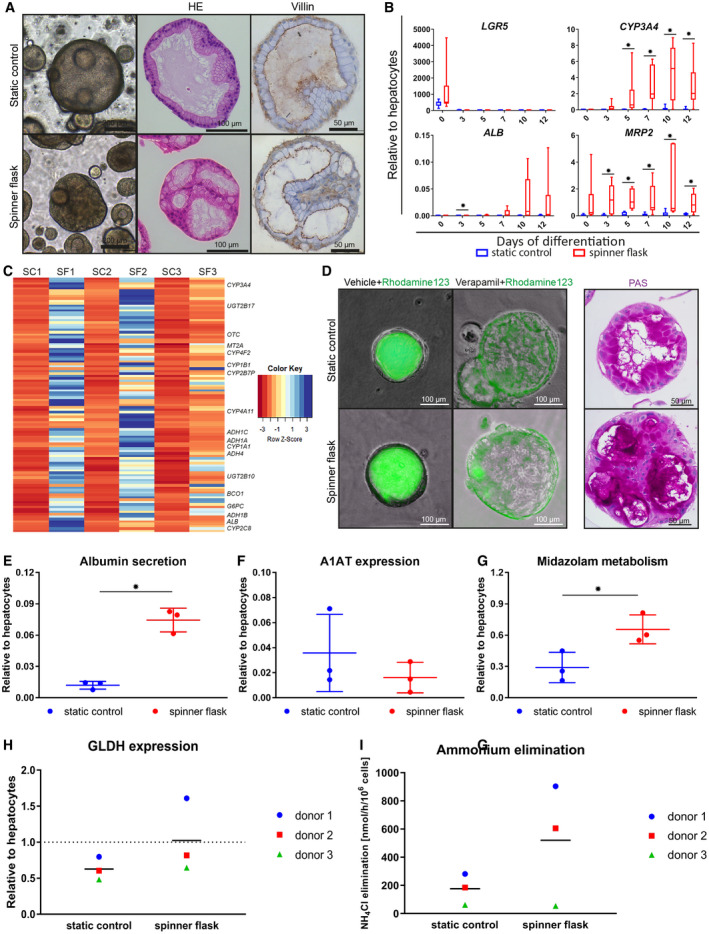
Organoid differentiation into functional hepatocytes in spinner flasks. Organoids were differentiated in spinner flasks or under static controls for 12 days. (A) Light microscopy images, HE stainings, and immunohistochemical analyses of paraffin‐embedded organoids for the canalicular marker Villin‐1. Nuclei were counterstained with hematoxylin. Expression of hepatocyte markers in differentiated organoids, determined by quantitative RT‐PCR (B) and mRNA‐sequencing (C). (B) Transcript levels of LGR5, CYP3A4, ALB, and MRP2. Graphs indicate five independent experiments for five different donors. Cryopreserved hepatocytes were used as positive control. (C) mRNA sequencing on organoids from three independent donors at day 12 of differentiation. Genes that were more than 4‐fold up‐regulated in the spinner flasks at day 12 of differentiation compared with the respective static controls are shown in a heatmap. Some well‐known hepatic genes are annotated. For a full list of genes and their liver‐related functions, see Supporting Table S4. (D) Rh123 transport was determined as readout for MDR1 activity, and PAS staining indicates glycogen storage in organoid cells. (E‐I) Hepatocyte functionality of spinner flask organoids was assessed. Three independent donors were analyzed in three independent experiments. Graphs indicate mean ± SD. Cryopreserved hepatocytes cultured for 24 hours in standard sandwich culture served as a positive control. ALB concentration in supernatant (E) and intracellular A1AT levels (F). Midazolam metabolism (G) was determined as readout for CYP3A4 functionality. Intracellular GLDH levels (H) and ammonium elimination (I) from the culture medium. *P ≤ 0.05. Abbreviations: ADH, alcohol dehydrogenase; BC01, beta‐carotene oxygenase 1; G6PC, glucose‐6‐phosphatase catalytic subunit; MT2A, metallothionein 2A; OTC, ornithine carbamoyltransferase; PAS, Periodic acid–Schiff; SC, static control; SF, spinner flask; UGT2B17, UDP glucuronosyltransferase family 2 member B17.
Data Availability Statement
All of the transcriptome data sets generated and analyzed during the current study are available in the Gene Expression Omnibus repository under the accession GSE123498. Other data sets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Results
Human Liver Organoids Are Highly Proliferative in Stirred Suspension Culture
To establish a suspension culture for efficient organoid expansion, we inoculated spinner flasks with 105 human liver organoid cells per milliliter of organoid EM10 supplemented with 10% (vol/vol) Matrigel, cultured the cells for 2 weeks, and compared the proliferation with that of cells conventionally cultured in Matrigel droplets (static controls; 105 cells/100 µL Matrigel). Light microscopy showed that the single cells grew out to form organoids within the first 4 days of culture in both static controls and spinner flasks (Fig. 1A). From culture day 7 onward, the diameter of the organoids in the spinner flasks increased and the outer cell layers appeared thicker compared with those in static controls. This was confirmed in histological sections, where spinner flask organoids often consisted of a multilayer of cells, whereas organoids in static controls consisted of a single epithelial layer (Fig. 1C). On day 14, organoid sizes were heterogeneous, with those in the spinner flasks clearly larger compared with those in static controls. The biggest organoids reached a diameter of about 5 mm in the spinner flasks and 1 mm in the static controls (Fig. 1A).
Figure 1.
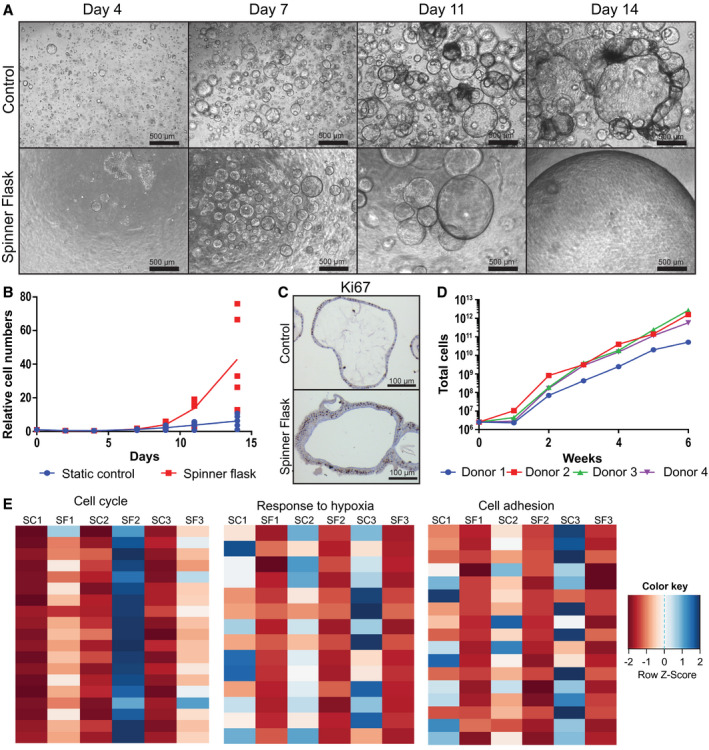
Enhanced liver organoid expansion in spinner flasks. Spinner flasks were inoculated with 2.5 × 106 single organoid cells (105 cells/mL) at day 0 and cultured in human organoid EM supplemented with the Rho kinase inhibitor Y27632 and 10% vol/vol Matrigel; 4‐5 different donors were analyzed in independent experiments. As controls, single cells were seeded in Matrigel droplets (105 cells/100 µL Matrigel) and cultured in EM supplemented with Y27632. (A) Light microscopy images of organoids grown from single cells in spinner flasks or in static control. (B) In vitro growth curves. An aliquot of cells was counted every 2‐3 days, and cell numbers relative to day 0 were calculated. Each red dot represents a different donor. Blue dots represent the corresponding donors cultured in static control. Lines represent the mean of five different donors. (C) Ki67 stainings confirmed that organoids were highly proliferative at day 14 after seeding in spinner flasks and controls. (D) Long‐term in vitro growth curves. Organoids from four different donors were cultured for 6 weeks in spinner flasks. Cultures were split every 2 weeks, and new spinner flasks were inoculated with organoid fragments corresponding to 2.5 × 106 cells. An aliquot of cells was counted every week, and theoretical total cell numbers were calculated. (E) mRNA sequencing data from organoids from three different donors that were cultured in spinner flasks or in static control for 14 days. Heatmaps show the three major pathways that were differentially regulated in spinner flask organoids compared with static controls. Abbreviations: SC, static control; SF, spinner flask.
To quantify the cells, we harvested an aliquot from the spinner flasks every 2 to 3 days, and trypsinized and counted the single cells. Expansion rates varied between donors, but were significantly increased in spinner flasks compared with static controls for all five donors that were analyzed in independent experiments (Fig. 1B). The average cell expansion after 2 weeks was 43‐fold in spinner flasks and 6‐fold in static controls (Fig. 1B). Relative cell numbers for each donor are provided in Supporting Table S1. Immunohistochemical staining for Ki67 confirmed that almost every cell in the spinner flasks was proliferative (Fig. 1C), and quantitative RT‐PCRs showed comparable expression of the stem cell marker LGR5 in spinner flasks and static controls (Supporting Fig. S1A). We next analyzed whether the stem cell phenotype and the high proliferation of the organoids were sustainable over several weeks, to obtain billions of cells as required for transplantations or tissue engineering. We inoculated four independent spinner flasks with single cells from four different donors and passaged the organoids every 2 weeks. After passaging, one new spinner flask per donor was inoculated with organoid fragments equal to a density of 105 cells/mL and expanded in EM. Quantitative RT‐PCR showed that the expression of LGR5 in spinner flasks samples remained stable over the course of 6 weeks (Fig. S1B). For quantification of the cells, an aliquot from the spinner flasks was trypsinized and single cells were counted every week. We observed an exponential cell expansion during the 6 weeks of culture for organoids from all four donors. The calculation of the theoretical number of cells after 6 weeks of culture revealed a total amount of cells ranging between 5.1 × 1010 and 2.7 × 1012, depending on the donor (Fig. 1D). Therefore, with a starting number of 2.5 million cells, our method allowed us to produce sufficient numbers of cells to restore 17%‐900% of the liver mass of an adult person in only 6 weeks.
To assess what caused the high proliferation rate in the spinner flasks, we performed mRNA sequencing on organoids from three different donors that were cultured in spinner flasks or in Matrigel droplets for 14 days. We selected genes that were down‐regulated or up‐regulated more than 4‐fold in the spinner flasks compared with their respective static controls, and obtained a list of 69 annotated down‐regulated genes (Supporting Table S2) and 62 annotated up‐regulated genes (Supporting Table S3) that were overlapping for organoids from all three donors. Gene ontology analysis of those genes revealed that three major pathways were differentially regulated in spinner flask organoids compared with static controls: cell cycle genes (n = 19) were significantly up‐regulated; and genes related to cell adhesion (n = 17) and genes related to hypoxia (n = 14) were significantly down‐regulated in the spinner flask organoids (Fig. 1E and Supporting Tables S2 and S3), suggesting that the increased expansion rate in the spinner flasks may be associated with an improved oxygen supply.
Spinner Flask Organoids Do Not Form Tumors In Vivo
Because the spinner flask organoids were highly proliferative and genes related to cell cycle progression were highly expressed, we wondered how the organoids would behave in vivo, particularly with regard to tumorigenic potential. We therefore performed xenografts in immune‐deficient NSG mice. Organoids from two independent donors were harvested after 8 weeks of culture in spinner flasks. Subsequently, 1 and 5 million organoid cells from each donor were injected subcutaneously into the flanks of two NSG mice. As a positive control, we injected human liver cancer organoids derived from a cholangiocarcinoma. After approximately 8 weeks, the site that was injected with the tumoroid liver organoids had a visible tumor and the mice were euthanized. We found many keratin 19 (K19)‐positive liver structures in the tumor organoid–injected skin; only one K19 positive organoid was found back in skin transplanted with organoids derived from a single spinner flask donor (Fig. 2). None of the other injection sites showed any K19‐positive structures. The tumor organoids stained positive for Ki67, showing proliferation in vivo. In contrast, no Ki67 staining was seen in the one spinner flask organoid that was found back in the skin tissue, indicating that the surviving cells were quiescent, and not proliferative (Fig. 2).
Figure 2.
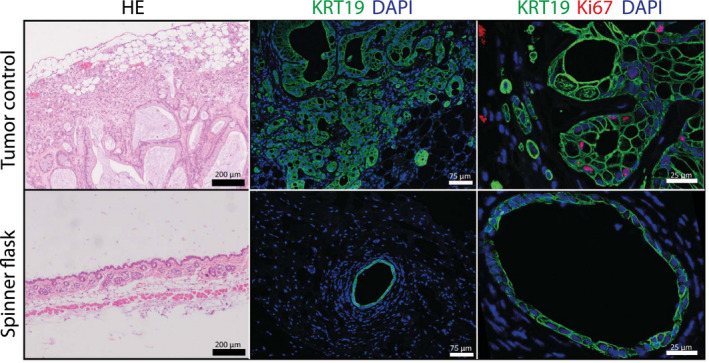
Spinner flask organoids do not form tumors in vivo. Organoids from two independent donors were harvested after 8 weeks of culture in spinner flasks. Subsequently, organoids equal to 1 and 5 million cells from each donor were injected subcutaneously into NSG mice, as were human liver cancer organoids derived from a cholangiocarcinoma as a positive control. HE stainings and immunofluorescent analysis of paraffin‐embedded injection sites are shown. A single K19‐positive organoid was found back in the skin transplanted with spinner flask organoids, but no Ki67‐positive nuclei were seen. All other injection sites did not show any K19‐positive structures. Abbreviation: DAPI, 4′,6‐diamidino‐2‐phenylindole.
Spinner Flask Organoids Repopulate Decellularized Liver Discs
Because mRNA sequencing analysis revealed that a group of genes related to cell adhesion was significantly down‐regulated in the spinner flask organoids compared with the static controls (Fig. 1E), we investigated whether the spinner flask organoids were able to engraft, a crucial requirement for clinical application of organoids. We therefore seeded the spinner flask organoids on decellularized rat liver discs. We and others had previously shown that primary hepatocytes,19 human liver stem/progenitor cells,20, 21, 22 and induced hepatocyte‐like cells23 can repopulate decellularized liver tissue. We harvested organoids after 14 days of expansion in spinner flasks, mechanically dissociated them and seeded organoid fragments onto decellularized rat liver discs. After 1 week of culture in DM, the organoid cells had repopulated the discs, were viable and had adapted a columnar shape, with the nuclei on the extracellular matrix (ECM) side (Fig. 3A). Analysis of the discs by immunofluorescent staining revealed that we had different cell populations on the discs, which we quantified: Most of the cells (56%) showed a K19+/ keratin 18 (K18−) cholangiocyte‐like phenotype (Fig. 3B), and 28% were K19/SRY‐box 9 (SOX9) double positive (Fig. 3C), 22% of cells had an intermediate K19+/K18+ phenotype (Fig. 3B), and only a minority of the cells (10%) were K18+/hepatocyte nuclear factor‐4‐alpha hepatocyte‐like cells (Fig. 3D). This indicates that organoids seeded on decellularized discs clearly interact with the ECM, adapt a columnar shape, and can obtain either a cholangiocyte‐like or a hepatocyte‐like fate, as such proving the true bipotential nature of these organoids and their great potential for tissue engineering approaches.
Figure 3.
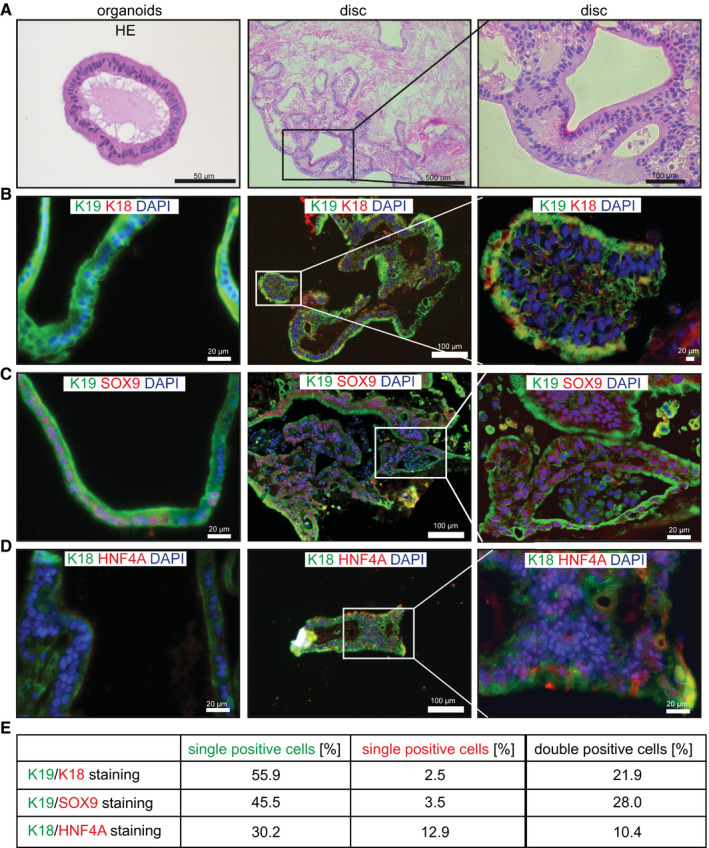
Tissue formation of spinner flask organoids on liver ECM. (A‐D) Organoids were expanded in spinner flasks for 14 days and then seeded on decellularized rat liver discs. Reseeded discs were cultured for 2 days in EM supplemented with BMP‐7 and then differentiated for 5 days in human organoid DM. Four different donors (2‐3 discs per donor) were analyzed in independent experiments. HE stainings (A) and immunofluorescent analysis (B‐D) of paraffin‐embedded organoids and liver discs are shown. Note that K18+ and HNF4a+ cells were not present in the organoids at the time of seeding on decellularized discs. (E) Quantification of the different cell populations. Abbreviation: HNF4a+, hepatocyte nuclear factor‐4‐alpha.
Spinner Flask Organoids Show Increased Hepatocyte Differentiation and Function
We previously showed that human liver organoids can be differentiated toward hepatocyte‐like cells following change of medium conditions to DM, in which growth stimuli R‐spondin and forskolin are removed, and N‐[(3,5‐difluorophenyl)acetyl]‐l‐alanyl‐2‐phenyl]glycine‐1,1‐dimethylethyl ester, fibroblast growth factor 19, dexamethasone, and BMP‐7 are added.10 However, the expression of differentiation markers in DM organoids was still significantly lower compared with hepatocytes. We reasoned that differentiation might be improved in our spinner flask organoids due to a more homogenous supply of oxygen and growth factors. Therefore, after 11 of days of culture in EM (to which BMP‐7 was added for the last 2 days), we recovered the organoids in a cell strainer and transferred them to a new spinner flask containing DM. Light microscopy and hematoxylin and eosin (HE) staining of the differentiated organoids at day 5 revealed that the spinner flask organoids had a more folded morphology compared with control organoids (Fig. 4A). Immunohistological analysis for Villin‐1 demonstrated that the spinner flask organoids in general had the same polarization as the control organoids, with the Villin‐positive canalicular membrane facing the inside of the organoids, and the Villin‐negative sinusoidal membrane facing the outside (Fig. 4A). However, polarization in the spinner flask organoids was less defined compared with control organoids (Fig. 4A); in particular, some of the larger and highly folded organoids displayed mixed polarization (Supporting Fig. S2), putatively due to less stable adjacent polarizing Matrigel concentrations when compared with static cultures in Matrigel droplets. Analysis of the organoids by quantitative RT‐PCR at different timepoints of differentiation confirmed that the stem cell marker LGR5 was down‐regulated in all samples (Fig. 4B). In contrast, several hepatocyte markers such as CYP3A4, ALB, and multidrug resistance‐associated protein 2 (MRP2) were up‐regulated, and were significantly higher expressed in the spinner flasks compared with static controls (Fig. 4B). Importantly, ALB transcript levels reached on average about 3%‐4% of hepatocyte transcript levels, and those of CYP3A4 and MRP2 were higher in the spinner flask organoids than in hepatocytes. As a comparison, our previously published protocol for organoid differentiation in Matrigel droplets resulted in ALB levels of about 0.1% and CYP3A4 levels of about 10% of liver tissue.10 To obtain a more general overview of the expression patterns of our spinner flask organoids, we then performed mRNA sequencing on organoids from three independent donors at days 5 and 12 of differentiation. Principal component analysis for all samples showed that the spinner flask samples clustered together and were closer to primary hepatocytes than the static control samples (Supporting Fig. S3). We then selected genes that were up‐regulated more than 4‐fold in the spinner flasks at day 12 of differentiation compared with their respective static controls, and obtained a list of 105 genes (99 annotated genes) that overlapped across all three donors. Gene ontology analysis revealed that more than 30% of those genes were directly related to liver functions, such as the cytochrome p450 (CYP) pathway and lipid, drug, hormone, and alcohol metabolism (Fig. 4C and Supporting Table S4).
Finally, we analyzed the functionality of the differentiated spinner flask organoids using several assays. First, we incubated organoids with Rh123, a fluorescent chemical compound that can be actively secreted from hepatocytes by MDR1. We observed fluorescence accumulation inside the lumen of the organoids for both static controls and spinner flasks (Fig. 4D). In contrast, Rh123 was retained in the cytoplasm of the cells when organoids had been pretreated with the competitive MDR1 inhibitor Verapamil, confirming the MDR1‐specific transport of Rh123. Another characteristic of hepatocytes is glycogen storage. During a feeding state, glucose enters the hepatocytes and is synthesized to glycogen, which can then be hydrolyzed to generate glucose during a fasting state. Periodic acid–Schiff staining for glycogen confirmed that differentiated organoids from static conditions and spinner flasks showed large intracellular accumulations of glycogen (Fig. 4D). Another important function of hepatocytes is the production of serum proteins; therefore, we determined ALB secretion and the expression of A1AT of the organoids at day 12 of differentiation. Compared with static controls, we observed a significantly higher concentration of ALB in the spinner flask organoid medium, which reached about 8% of the concentration in the medium of cultured hepatocytes (Fig. 4E). A1AT levels did not differ between spinner flask organoids and static controls (Fig. 4F). Another key function of hepatocytes is drug metabolism, with CYP enzymes as important players. To measure CYP activity in the organoids, we exposed differentiated organoids to 5 mM midazolam for 24 hours, and then determined the concentration of the midazolam metabolite 1‐OH midazolam in the culture medium. In spinner flask organoids, we observed that midazolam metabolism was induced to about 70% of the levels of hepatocytes (Fig. 4G). Finally, we assessed ammonium elimination from the medium by spinner flask organoids and static controls. Hepatocytes eliminate ammonia from the medium by urea cycle activity or fixation into amino acids. Ammonia reacts with α‐ketoglutaric acid to l‐glutamate. This reaction is catalyzed by l‐ GLDH. The expression of GLDH showed a higher trend in the spinner flask organoids compared with static controls (Fig. 4H), and accordingly, ammonia elimination from the medium appeared to be more efficient in spinner flask organoids compared with static controls (Fig. 4I).
Discussion
The shortage of available donor organs for patients on transplant wait lists highlights the need for alternatives to liver transplantation. Liver organoids from LGR5‐positive adult tissue stem cells offer exciting new possibilities for tissue engineering or cell transplantations.3, 8 However, the lack of reliable and reproducible methods for the expansion of liver organoids hampers clinical application in which billions of cells are needed. To restore 10%‐20% of the hepatocyte liver mass of an adult, it is essential to culture an estimated 20‐50 × 109 cells.14
In this study, we describe a robust spinner flask–based method for large‐scale culture of human LGR5‐positive adult stem cell–derived liver organoids. This method allows high proliferation levels, repopulation potential, and maturation toward cholangiocyte‐like and hepatocyte‐like cells. These cells do not induce tumor formation and can be serially passaged and maintained for at least 6 weeks (Fig. 1A‐D). This greatly improves upon current techniques for the expansion of large numbers of liver stem cells, and therefore approaches to liver regeneration.
Our mRNA sequencing data suggest that the enhanced proliferation may be caused by an improved oxygen supply (and also probably growth factors) in spinner flasks (Fig. 1E). This hypothesis is strengthened by the observation that organoids at the edges of a Matrigel droplet usually grow larger compared with organoids in the middle of the droplet, where oxygen and growth factor supply may be restricted.
Importantly, Huch et al. have extensively analyzed the genetic stability of cultured organoids in vitro and showed that their genetic integrity is preserved over several months.14 Because proliferation of our organoids in spinner flasks was significantly increased compared with conventional Matrigel culture, we assessed their behavior in vivo, and the lack of tumor formation suggests their promise for clinical application (Fig. 2). This is in strong contrast to hepatocyte‐like cells derived from iPS cells, where genetic instability raises concerns regarding their safety in a clinical setting.24
We observed that expansion of organoids in spinner flasks led to a significant down‐regulation of adhesion‐related genes in the organoids (Fig. 1E), raising doubts about their capacity for engraftment in vivo, which is mediated by integrin‐ECM interactions.25 We therefore seeded the spinner flask organoids on decellularized liver discs,22 and observed clear interaction with the ECM as well as a columnar cell shape and selective differentiation toward both a cholangiocyte‐like and a hepatocyte‐like fate, indicating their bipotential nature and great potential for tissue engineering approaches. Of note, only a minority of the cells obtained a K19−/K18+ hepatocyte phenotype, whereas the vast majority had a K19+/Sox9+ cholangiocyte phenotype or a K19+/K18+ intermediate phenotype (Fig. 3). This inefficient hepatocyte differentiation may partly be explained by the origin of the decellularized liver discs used in this study, which were derived from rats and not from humans. Additionally, we foresee that the hepatocyte maturation potential of the reseeded discs may be improved if organoids are seeded on human ECM and cultured under shaking conditions, for enhanced oxygenation and medium perfusion.26 Although the organoids engrafted well in vitro, unfortunately we did not observe any in vivo engraftment of the spinner flask organoids after xenotransplantation in Fah−/−/Rag2−/−/Il2rg−/y (FRG) mice (Supporting Fig. S5). This might be explained by the fact that even though spinner flask differentiation led to a more mature hepatocyte‐like phenotype compared with conventional Matrigel controls (Fig. 4), the organoids did not reach a fully mature hepatocyte phenotype. For example, ALB secretion reached only 8% of the concentration in the medium of cultured hepatocytes (Fig. 4E). A low ALB expression compared with other more mature hepatocyte markers is consistent with earlier observations of organoid differentiation10 and suggests that organoids follow a sequence of differentiation events that is different from the stages of fetal to mature hepatocyte development.27 Remarkably, the late hepatocyte marker CYP3A4 reached expression levels equivalent to mature hepatocytes (Fig. 4B), intracellular GLDH levels were comparable to hepatocytes (Fig. 4H), and midazolam metabolism was induced to about 70% of levels found in mature hepatocytes (Fig. 4G).
In conclusion, we have developed a method for highly efficient and safe expansion of LGR5‐positive liver stem cells, enabling subsequent differentiation toward highly mature and functional hepatocyte‐like cells. Interestingly, during the writing of this manuscript, culture conditions for the in vitro expansion of primary human hepatocytes in 2D28 and primary mouse and human fetal hepatocytes as 3D organoids29, 30 have been established. Those in vitro expanded hepatocytes are a more efficient source for transplantations than the cholangiocyte‐derived organoids described here, as hepatocyte maturation generally correlates with transplantation efficiency. We expect that the spinner flask method could also be applied to those hepatocyte organoids and other types of adult stem cell–derived organoids, which may benefit from an improved supply of oxygen and growth factors.31 As a next step, we should seek to replace Matrigel in the spinner flasks by a defined and clinically suitable hydrogel, following the lead by Gjorevski et al., who recently published a major breakthrough by replacing Matrigel with a modified PEG gel for the expansion and differentiation of intestinal organoids.32 The major advantage of the spinner flask culture method is that we can reproducibly culture functional liver organoid cells in a time‐, work‐, and cost‐efficient way, and the combination of a clinically suitable hydrogel and the spinner flask method for the culture of adult stem cell–derived organoids will establish an avenue for clinical applications of organoids in the near future.
Author Contributions
K.S., P.M.B. and B.S. were responsible for the study’s conceptualization. K.S., N.S.‐R., S.Y., L.A.O., F.G.v.S., I.P.P., M.E.v.W., G.v.T., R.L., H.C.‐V., and I.S. were responsible for the study’s methodology. K.S., B.S., S.Y., and F.G.v.S. were responsible for the study investigation. K.S. was responsible for drafting the original manuscript. C.C., B.S., M.M.A.V., P.M.B., N.S.‐R., S.A.F., L.C.P., R.H., and J.D.K. were responsible for the drafting, reviewing, and editing of the manuscript. K.S., S.Y., P.M.B., L.C.P., and B.S. were responsible for the funding acquisition. P.M.B., R.H., M.M.A.V., L.J.W.v.d.L., and S.A.F were responsible for the study’s resources. L.C.P., L.J.W.v.d.L., P.M.B., H.C., and B.S. were responsible for the study supervision.
Supporting information
Acknowledgment
We thank Hans Vernooij for his help with the statistical analysis and Dr. Rupert Ecker from TissueGnostics GmbH, Austria, for his kind help on the quantification of the positive cell populations in the immunofluorescence presented in Fig. 3 of this manuscript.
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1. Atala A. Regenerative medicine strategies. J Pediatr Surg 2012;47:17‐28. [DOI] [PubMed] [Google Scholar]
- 2. Fraczek J, Bolleyn J, Vanhaecke T, Rogiers V, Vinken M. Primary hepatocyte cultures for pharmaco‐toxicological studies: at the busy crossroad of various anti‐dedifferentiation strategies. Arch Toxicol 2013;87:577‐610. [DOI] [PubMed] [Google Scholar]
- 3. Forbes SJ. Organoid cultures boost human liver cell expansion. Hepatology 2015;62:1635‐1637. [DOI] [PubMed] [Google Scholar]
- 4. Hay DC, Zhao D, Fletcher J, Hewitt ZA, McLean D, Urruticoechea‐Uriguen A, et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 2008;26:894‐902. [DOI] [PubMed] [Google Scholar]
- 5. Sullivan GJ, Hay DC, Park IH, Fletcher J, Hannoun Z, Payne CM, et al. Generation of functional human hepatic endoderm from human induced pluripotent stem cells. Hepatology 2010;51:329‐335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen C, Soto‐Gutierrez A, Baptista PM, Spee B. Biotechnology challenges to in vitro maturation of hepatic stem cells. Gastroenterology 2018;154:1258‐1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baxter M, Withey S, Harrison S, Segeritz CP, Zhang F, Atkinson‐Dell R, et al. Phenotypic and functional analyses show stem cell‐derived hepatocyte‐like cells better mimic fetal rather than adult hepatocytes. J Hepatol 2015;62:581‐589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huch M. Regenerative biology: the versatile and plastic liver. Nature 2015;517:155‐156. [DOI] [PubMed] [Google Scholar]
- 9. Huch M, Dorrell C, Boj SF, van Es JH, Li VS, van de Wetering M, et al. In vitro expansion of single Lgr5+ liver stem cells induced by Wnt‐driven regeneration. Nature 2013;494:247‐250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huch M, Gehart H, van Boxtel R, Hamer K, Blokzijl F, Verstegen MM, et al. Long‐term culture of genome‐stable bipotent stem cells from adult human liver. Cell 2015;160:299‐312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huch M, Knoblich JA, Lutolf MP, Martinez‐Arias A. The hope and the hype of organoid research. Development 2017;144:938‐941. [DOI] [PubMed] [Google Scholar]
- 12. Bianconi E, Piovesan A, Facchin F, Beraudi A, Casadei R, Frabetti F, et al. An estimation of the number of cells in the human body. Ann Hum Biol 2013;40:463‐471. [DOI] [PubMed] [Google Scholar]
- 13. Dhawan A, Puppi J, Hughes RD, Mitry RR. Human hepatocyte transplantation: current experience and future challenges. Nat Rev Gastroenterol Hepatol 2010;7:288‐298. [DOI] [PubMed] [Google Scholar]
- 14. Willemse J, Lieshout R, van der Laan LJW, Verstegen MMA. From organoids to organs: bioengineering liver grafts from hepatic stem cells and matrix. Best Pract Res Clin Gastroenterol 2017;31:151‐159. [DOI] [PubMed] [Google Scholar]
- 15. dos Santos FF, Andrade PZ, da Silva CL, Cabral JM. Bioreactor design for clinical‐grade expansion of stem cells. Biotechnol J 2013;8:644‐654. [DOI] [PubMed] [Google Scholar]
- 16. King JA, Miller WM. Bioreactor development for stem cell expansion and controlled differentiation. Curr Opin Chem Biol 2007;11:394‐398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Massai D, Isu G, Madeddu D, Cerino G, Falco A, Frati C, et al. A versatile bioreactor for dynamic suspension cell culture. Application to the culture of cancer cell spheroids. PLoS One 2016;11:e0154610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lancaster MA, Knoblich JA. Generation of cerebral organoids from human pluripotent stem cells. Nat Protoc 2014;9:2329‐2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Soto‐Gutierrez A, Zhang L, Medberry C, Fukumitsu K, Faulk D, Jiang H, et al. A whole‐organ regenerative medicine approach for liver replacement. Tissue Eng Part C Methods 2011;17:677‐686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang Y, Cui CB, Yamauchi M, Miguez P, Roach M, Malavarca R, et al. Lineage restriction of human hepatic stem cells to mature fates is made efficient by tissue‐specific biomatrix scaffolds. Hepatology 2011;53:293‐305. [DOI] [PubMed] [Google Scholar]
- 21. Baptista PM, Siddiqui MM, Lozier G, Rodriguez SR, Atala A, Soker S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 2011;53:604‐617. [DOI] [PubMed] [Google Scholar]
- 22. Vyas D, Baptista PM, Brovold M, Moran E, Gaston B, Booth C, et al. Self‐assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 2017;67:750‐761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen C, Pla‐Palacin I, Baptista PM, Shang P, Oosterhoff LA, van Wolferen ME, et al. Hepatocyte‐like cells generated by direct reprogramming from murine somatic cells can repopulate decellularized livers. Biotechnol Bioeng 2018;115:2807‐2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bayart E, Cohen‐Haguenauer O. Technological overview of iPS induction from human adult somatic cells. Curr Gene Ther 2013;13:73‐92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kumaran V, Joseph B, Benten D, Gupta S. Integrin and extracellular matrix interactions regulate engraftment of transplanted hepatocytes in the rat liver. Gastroenterology 2005;129:1643‐1653. [DOI] [PubMed] [Google Scholar]
- 26. Adam AAA, van der Mark VA, Donkers JM, Wildenberg ME, Oude Elferink RPJ, Chamuleau RAFM, et al. A practice‐changing culture method relying on shaking substantially increases mitochondrial energy metabolism and functionality of human liver cell lines. PLoS One 2018;13:e0193664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kheolamai P, Dickson AJ. Liver‐enriched transcription factors are critical for the expression of hepatocyte marker genes in mES‐derived hepatocyte‐lineage cells. BMC Mol Biol 2019;10:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhang K, Zhang L, Liu W, Ma X, Cen J, Sun Z, et al. In vitro expansion of primary human hepatocytes with efficient liver repopulation capacity. Cell Stem Cell 2018;23:806‐819. [DOI] [PubMed] [Google Scholar]
- 29. Peng WC, Logan CY, Fish M, Anbarchian T, Aguisanda F, Álvarez‐Varela A, et al. Inflammatory cytokine TNFα promotes the long‐term expansion of primary hepatocytes in 3D culture. Cell 2018;175:1607‐1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hu H, Gehart H, Artegiani B, Löpez‐Iglesias C, Dekkers F, Basak O, et al. Long‐term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 2018;175:1591‐1606. [DOI] [PubMed] [Google Scholar]
- 31. Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordonez‐Moran P, et al. Designer matrices for intestinal stem cell and organoid culture. Nature 2016;539:560‐564. [DOI] [PubMed] [Google Scholar]
- 32. Hoekstra R, Deurholt T, ten Bloemendaal L, Desille M, van Wijk AC, Clement B, et al. Assessment of in vitro applicability of reversibly immortalized NKNT‐3 cells and clonal derivatives. Cell Transplant 2006;15:423‐433. [PubMed] [Google Scholar]
- 33. Broutier L, Mastrogiovanni G, Verstegen MM, Francies HE, Gavarro LM, Bradshaw CR, et al. Human primary liver cancer‐derived organoid cultures for disease modeling and drug screening. Nat Med 2017;23:1424‐1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the transcriptome data sets generated and analyzed during the current study are available in the Gene Expression Omnibus repository under the accession GSE123498. Other data sets generated and analyzed during the current study are available from the corresponding author on reasonable request.


