Abstract
Identification of glomerular lesions and structures is a key point for pathological diagnosis, treatment instructions, and prognosis evaluation in kidney diseases. These time‐consuming tasks require a more accurate and reproducible quantitative analysis method. We established derivation and validation cohorts composed of 400 Chinese patients with immunoglobulin A nephropathy (IgAN) retrospectively. Deep convolutional neural networks and biomedical image processing algorithms were implemented to locate glomeruli, identify glomerular lesions (global and segmental glomerular sclerosis, crescent, and none of the above), identify and quantify different intrinsic glomerular cells, and assess a network‐based mesangial hypercellularity score in periodic acid–Schiff (PAS)‐stained slides. Our framework achieved 93.1% average precision and 94.9% average recall for location of glomeruli, and a total Cohen's kappa of 0.912 [95% confidence interval (CI), 0.892–0.932] for glomerular lesion classification. The evaluation of global, segmental glomerular sclerosis, and crescents achieved Cohen's kappa values of 1.0, 0.776, 0.861, and 95% CI of (1.0, 1.0), (0.727, 0.825), (0.824, 0.898), respectively. The well‐designed neural network can identify three kinds of intrinsic glomerular cells with 92.2% accuracy, surpassing the about 5–11% average accuracy of junior pathologists. Statistical interpretation shows that there was a significant difference (P value < 0.0001) between this analytic renal pathology system (ARPS) and four junior pathologists for identifying mesangial and endothelial cells, while that for podocytes was similar, with P value = 0.0602. In addition, this study indicated that the ratio of mesangial cells, endothelial cells, and podocytes within glomeruli from IgAN was 0.41:0.36:0.23, and the performance of mesangial score assessment reached a Cohen's kappa of 0.42 and 95% CI (0.18, 0.69). The proposed computer‐aided diagnosis system has feasibility for quantitative analysis and auxiliary recognition of glomerular pathological features. © 2020 The Authors. The Journal of Pathology published by John Wiley & Sons Ltd on behalf of Pathological Society of Great Britain and Ireland.
Keywords: computational pathology, glomerular lesion classification, intrinsic glomerular cells identification, mesangial hypercellularity score assessment, IgAN
Introduction
Chronic kidney disease (CKD) has become a significant public health problem in the world [1, 2]. IgA nephropathy (IgAN) is the leading cause of CKD worldwide, especially in Asian regions, with up to 30–40% of patients developing end‐stage renal disease (ESRD) within 10–25 years [3, 4]. Renal pathology provides the most important reference for diagnosis, treatment instructions, and prognosis evaluation of kidney diseases. Patients with IgAN have varied clinical presentations and histological lesions, ranging from mild mesangial proliferation, endocapillary hypercellularity, crescentic glomerulonephritis, to global and segmental sclerosis. All of these should be described in pathology reports [5]. The 2016 Oxford Classification of IgAN considers mesangial hypercellularity (M), endocapillary hypercellularity (E), segmental glomerular sclerosis (S), interstitial fibrosis and tubular atrophy (T), and crescents (C) [6]. Several studies have indicated that the ‘MSTC’ score provides a prediction of prognosis, and guides management decisions in patients with IgAN [6, 7, 8, 9, 10, 11, 12, 13]. At present, these complex pathological features are mainly evaluated by manual quantitative evaluation, which is time‐consuming and laborious with subjective variations. Due to the heavy workload of pathologists, requirements for automatic identification and quantitative analysis have become more and more urgent.
In recent years, much effort has been made to locate glomeruli or glomerular structures (Table 1) [14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25]. Among these methods, deep convolutional neural networks (CNNs) have produced inspiring results, and are widely used for auxiliary diagnosis in whole‐slide images. Sarder et al addressed the glomerulus location task through image processing algorithms, and obtained 87.8% accuracy on 15 slides [22]. Studies based on handcrafted feature extraction techniques [15, 16] obtained an ~87% F1 score, while those based on CNN‐based methods mostly stand at more than 90%. Due to the diversity of stain qualities and pathological features, fixed handcrafted feature extractor and color threshold‐based methods cannot obtain superior results compared with strong, robust deep neural networks. Hermsen et al [25] obtained comprehensive results from renal tissue stained with periodic acid–Schiff (PAS) to segment glomeruli, tubules, and interstitium using five ensembling U‐Nets, which was the first attempt at multi‐class segmentation of PAS‐stained nephrectomy samples and transplant biopsies. Since the number of glomerular lesions provides diagnostic information, prognosis prediction, and guides management decisions in patients with CKD, it is crucial to develop models for glomerulus classification. To address this issue, researchers such as Ginley et al [23] developed CNN models with a long short‐term memory (LSTM) layer to classify five different types of glomeruli in diabetic nephropathy (DN), while others [21, 28, 29, 30] aimed to distinguish sclerotic glomeruli, normal glomeruli, and non‐glomeruli [31].
Table 1.
Previous work on location of glomeruli and the performance obtained.
| Source | Data/slide | Approach | Performance |
|---|---|---|---|
| Temerinac‐Ott et al [14] |
Train: 16 Test: 4 |
Mutual information + CNN | 68.94% F1 |
| Kato et al [15] |
Train: * Test: 20 |
S‐HOG + SVM | 86.6% F1 |
| Marée et al [16] | Total: 200 | Ellipsoidal shape + decision tree | 87% F1 |
| Gadermayr et al [17] | Total: 24 | U‐Net | 90% Dice |
| Gallego et al [18] | Total: 40 | AlexNet | 93.7% F1 (pixel‐wise) |
| Sheehan et al [19] | Total: 90 | AlexNet + SVM | 92% recall, 90% specificity |
| Bukowy et al [20] |
Train: 72 Test: 13 |
Faster RCNN | 96.9% precision, 96.8% recall |
| Bueno et al [21] |
Train: 38 Test: 9 |
SegNet‐VGG19 |
99.8% precision, 99.2% F1 (pixel‐wise) |
| Sarder et al [22] |
Train: * Test: 15 |
Gabor filters | 87.8% accuracy |
| Ginley et al [23] |
Train: 41 Test: 13 |
DeepLab v2 | 93% accuracy |
| Simon et al [24] |
Train: 25 Test: 9 |
Multi‐radial LBP + SVM | 90.4% precision, 76.7% recall |
| Hermsen et al [25] |
Train: 37 Test: 3 |
5 U‐Nets ensemble | 79% Dice |
| Comparison I |
Train: 360 Test: 40 |
U‐Net | 89.2% precision, 90.6% recall |
| Comparison II | U‐Net‐SSIM | 91.4% precision, 92.1% recall | |
| Comparison III | Mask R‐CNN [26] | 88.1% precision, 92.3% recall | |
| Comparison IV | FCOS [27] | 91.7% precision, 92.8% recall | |
| APRS | U‐Net‐SSIM + marked watershed |
93.1% precision, 94.0% F1 94.9% recall, 90.1% Dice |
Not mentioned. Dice, dice coefficient; F1, F1 score.
In computer‐aided diagnosis, the amount of data is a crucial factor for model evaluation. Although some previous studies have used a moderate number of rat renal biopsy samples, research on human renal biopsy samples is still insufficient (shown in Tables 1 and 2). In this study, we aimed to establish an analytic renal pathology system (ARPS) to identify the basic lesions and pathological changes in IgAN by employing deep learning methods on a large number of whole‐slide images (WSIs). To address this issue, our work was divided into the following subtasks: (1) detect the glomeruli within the WSI; (2) distinguish glomeruli with global glomerular sclerosis (GS), segmental glomerular sclerosis (SS), crescents (C), and none of the above (NOA); (3) identify the intraglomerular structures including intrinsic glomerular cells (M: mesangial cells; E: endothelial cells; P: podocytes), mesangial area, and glomerulus parameters (glomerular diameter, proportion of the three types of intrinsic glomerular cells); and (4) apply ARPS to automatic mesangial score (M score) evaluation in IgAN. The convolutional neural networks implemented in this paper consist of U‐Net, DenseNet, LSTM‐GCNet, and 2D V‐Net. All of these architectures receive images with labels, and output the corresponding predictions. U‐Net [33], consists of several contracting paths to capture context and symmetric expanding paths which enable precise segmentation and location. Due to its outstanding performance, many researchers have employed this architecture to tackle the bio‐medical image segmentation task [34, 35]. Based on these approaches, we applied it with structural similarity loss to segment the glomeruli from the WSIs, followed by a glomerular lesion classifier. Densely connected convolutional neural network (DenseNet [36]) alleviates vanishing‐gradient problem and strengthens feature reuse and propagation, as well as reducing the quantity of parameters. Global context block and squeeze‐excitation block [37] were proposed in 2019 to assist the attention focusing mechanism in natural image classification tasks. Here, we introduce this approach with a long short‐term memory (LSTM) layer to tackle the bio‐medical image fine‐grained classification task, named LSTM‐GCNet. In the final position, a 2D V‐Net [38] with self‐attention module and center channel is presented so as to identify intrinsic glomerular cells, since the residual block can reduce the number of parameters while the center channel produces the center of each intrinsic cell. Moreover, image processing algorithms such as marked watershed were implemented to detect the glomerular structures.
Table 2.
Previous work on glomerulus classification and intrinsic cell recognition and performance obtained.
| Glomerulus classification | ||||
|---|---|---|---|---|
| Source | Approach | Data/glomeruli | Class | Performance |
| Chagas et al [31] | CNN + SVM | Total: 811 | 4 |
82.0% accuracy 81.9% F1 score |
| Ginley et al [23] | RNN | Total: 1989 | 5 | Cohen's kappa 0.55 |
| Barros et al [30] | LoG + kNN | Total: 811 | 3 | 88.3% accuracy |
| Jayapandian et al [32] | CNN | Total: 146 | 2 | 82.5% accuracy |
| Marsh et al [28] | CNN + FCN | Total: 3867 | 2 | 84.8% F1 score |
| Kannan et al [29] | Inception v3 | Total: 1496 | 3 | 92.4% accuracy |
| Bueno et al [21] | AlexNet | Total: 1245 | 3 |
98.2% accuracy 99.4% F1 score |
| Comparison I | ResNet50 |
Train: 10 935 Test: 1483 |
5 | 84.9% accuracy |
| Comparison II | Inception‐ResNet‐v2 | 85.9% accuracy | ||
| Comparison III | DenseNet | 87.1% accuracy | ||
| Comparison IV | ResNeXt‐50 | 84.9% accuracy | ||
| APRS |
DenseNet + LSTM‐GCNet |
92.8% accuracy Cohen's kappa 0.91 |
||
| Intrinsic glomerular cell recognition | ||||
| Source | Approach | Data/glomeruli | Class | Performance |
| Ginley et al [23] | DeepLab v2 |
Train: 616 Test: 22 |
1 (RC) |
94% sensitivity 93% specificity |
| Sarder et al [22] | Threshold + morphology |
Train: * Test: 50 |
1 (RC) | 92.3% accuracy |
| ARPS CI | 2D V‐Net |
Train: 300 Test: 160 |
3(M, E, P) | 89.4% accuracy |
| ARPS CII | 2D Attention V‐Net | 3(M, E, P) | 90.8% accuracy | |
| APRS | 2D Attention V‐Net‐center | 3(M, E, P) | 92.2% accuracy | |
Not mentioned. RC, regardless of category.
Materials and methods
An outline of our scheme is shown in Figure 1. Given an input WSI, the glomeruli are first segmented through the location model, followed by glomerular lesion identification and decomposition of glomeruli. The proposed method mainly consists of five parts, including preparation of specimens, annotation details, convolutional neural network design and training strategies, workflow demonstration, and evaluation metric illustration (shown in supplementary material, Supplementary materials and methods). The algorithm is developed on PyCharm Community v2019.3.4 and TensorFlow v1.14 (TensorFlow: Large‐scale machine learning on heterogeneous systems, 2015; software available from https://www.tensorflow.org).
Figure 1.
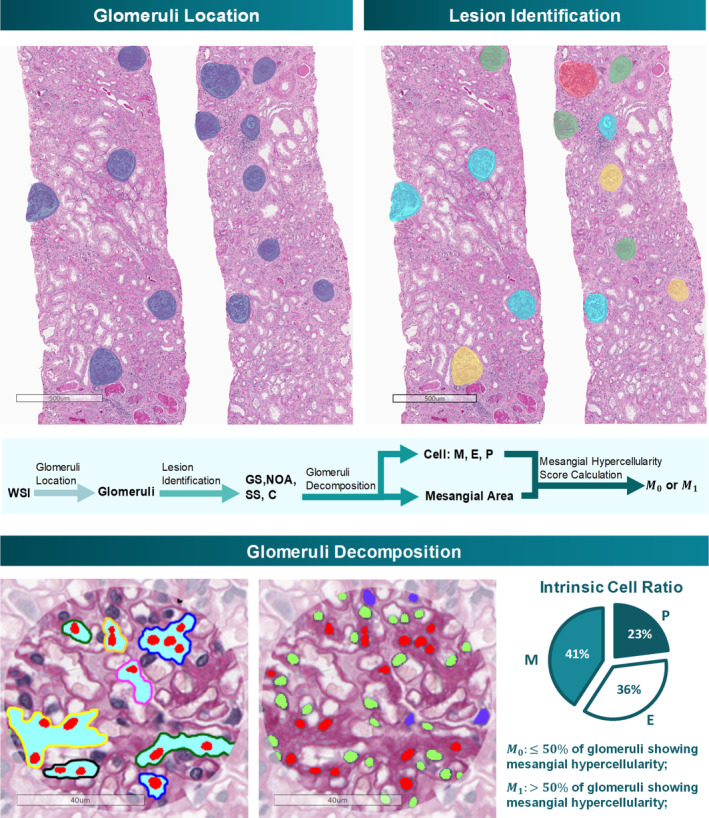
Workflow of the automatic detection model. The entire workflow is demonstrated in this figure, including glomerulus location, lesion identification, and glomerulus decomposition. Each glomerulus is divided into NOA, SS, GS, and C, followed by the NOA decomposition, which can identify intrinsic glomerular cells and evaluate the mesangial hypercellularity score automatically.
Specimen preparation
This study strictly followed the tenets of the Declaration of Helsinki, and was approved by the Ethics Committee of the Jinling Hospital (2019NZGKJ‐101). We selected in total 400 IgAN patients who underwent renal biopsy between 2016 and 2018 in the National Clinical Research Center of Kidney Diseases, Jinling Hospital. Renal biopsy tissues were processed using the standard techniques of light microscopy (LM) and were scanned by a Leica Aperio ScanScope AT Turbo (Leica, Wetzlar, Germany) under 400× magnification. We selected slides showing crescents from as many cases as possible for the training model. The clinicopathological data of 400 patients are shown in Table 3.
Table 3.
Clinicopathological features of patients with IgAN at time of biopsy.
| Variable | Value |
|---|---|
| Age (years) | 33 (27–42) |
| Male (%) | 43.98 |
| Ethnicity (% Asian) | 100 |
| BMI (kg/m2) | 23.0 ± 3.7 |
| SBP (mm Hg) | 126.9 ± 14.8 |
| DBP (mm Hg) | 78.2 ± 11.3 |
| MAP (mm Hg) | 94.2 ± 12.5 |
| Hypertension before biopsy (%) | 20.2 |
| Serum creatinine (mg/dl) | 0.96 (0.76–1.34) |
| eGFR (ml/min per 1.72m2) | 78.2 (57.5–107.0) |
| CKD stage (%) | |
| 1 | 50 |
| 2 | 28.3 |
| 3 | 17.3 |
| 4 | 3.5 |
| 5 | 0.9 |
| Proteinuria (g/day) | 1.1 (0.6–2.0) |
| Serum albumin (g/l) | 40.1 (36.6–43.0) |
| Serum cholesterol (mmol/l) | 4.6 (4.0–5.4) |
| Serum triglycerides (mmol/l) | 1.4 (1.0–2.0) |
| Serum uric acid (μmol/l) | 359 (303–428) |
| Oxford Classification (%) | |
| Ml | 29.9 |
| E1 | 22.5 |
| S1 | 86.1 |
| T1/T2 | 17.2/2.7 |
| C1/C2 | 87.3/3.0 |
BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure; MAP, mean arterial pressure; eGFR, estimated glomerular filtration rate (CKD‐EPI); CKD, chronic kidney disease; M, mesangial hypercellularity; E, endocapillary hypercellularity; S, segmental glomerular sclerosis; T, interstitial fibrosis and tubular atrophy; C, crescent.
Data are presented as the mean ± SD, the median with range or percentages.
Annotation details
Annotations of glomeruli were provided by three experienced junior pathologists through labeling along the outer margin of the glomerular capsule and their subcategories (global sclerosis, segmental sclerosis, crescent, etc.) using Aperio Image Scope software (Leica). These annotations were then checked by three senior pathologists. Glomerular sclerosis is defined as the obliteration of capillary lumen by increased extracellular collagenous matrix. Definitions of different classes can be described as global glomerular sclerosis (GS, sclerosis involving 100% of the tuft), segmental glomerular sclerosis (SS, sclerosis involving a portion of the glomerular tuft), crescent (C, containing more than two layers of cells within Bowman's space, often with fibrin and collagen deposition); a glomerulus with mild to moderate or severe mesangial proliferation without any sclerosis and crescent is defined as none of the above (NOA). Furthermore, each intrinsic glomerular cell was annotated by three senior pathologists who either have worked for more than 10 years, signing about 1600 renal biopsy reports per year and participating in the study of the Oxford Classification of IgAN and further research on crescents [6, 39]. The hypercellularity score was defined as when any mesangial area within the glomerulus had no fewer than four mesangial cells. Simplification of the mesangial hypercellularity score is defined as M1, when more than half of non‐sclerosing glomeruli within a slide appeared with mesangial hypercellularity, and M0 otherwise. All of the conflicted cases were carefully discussed by three senior pathologists to reach a consensus. Inter‐observer agreement evaluation was conducted for glomeruli and intrinsic cells. Assuming each senior pathologist would not miss a target without considering its types, we treated the coherence assessment of the detection task as that of a sequential classification task. The Kendall correlation coefficient (KCC) between three senior pathologists for intrinsic cells was high (Kendall's W = 0.891, Fleiss' kappa = 0.89, p < 0.0001) and that for glomerular lesions was comparable (Kendall's W = 0.768, Fleiss' kappa = 0.733, p < 0.0001).
Convolutional neural network design and training strategies
Details of our proposed neural network and train strategies are shown in supplementary material, Supplementary materials and methods.
Workflow demonstration
The workflow (shown in Figure 1) of ARPS includes four continuous components: (1) slide preprocessing, including ROI (region of interest) orientation and patch extraction; (2) location of glomeruli; (3) identification of glomerular lesions; and (4) decomposition of glomeruli, including recognition of intrinsic glomerular cells, mesangial region extraction, and M‐score assessment.
The whole‐slide pathological images range in size from 20 000 × 30 000 to 50 000 × 70 000 pixels, and cannot be trained directly due to hardware limitations. In total, 41 000 patches with a size of 1024 * 1024 and the corresponding semantic masks were extracted from 400 slide images, with 30 000 for training, 5000 for validation, and 5000 for test (patches in the training set, validation set, and test set are extracted from different slides). We then trained the attention U‐Net on these patches for location of glomeruli.
With accurate location of glomeruli, researchers tried to distinguish different glomerular lesions. Lesion identification can be treated as a fine‐grained classification problem, which is a challenging field in computer vision due to the subtle differences between subcategories. We selected 12 418 glomeruli from 400 slides, with 5361 NOA, 3416 GS, 2268 SS, 1373 C, and 5100 Neg (negative samples were regions excluding glomeruli), and allocated the training set, validation set, and test set in the ratio of 7:2:1. Following the location of glomeruli, those easily classified glomeruli such as GS and Neg were picked out by DenseNet. Then the glomerular boundary was utilized to remove redundant information outside the margin of the glomerular capsule, and the LSTM‐GCNet was trained to classify SS, NOA, and C through the internal glomerular features.
After glomerular lesion classification, we employed V‐Net [38] architecture with center channel to obtain an accurate prediction for intrinsic glomerular cells. In total, 460 images of glomeruli (about 70 000 cells) were selected from NOA samples, with 240 for training, 60 for validation, and 160 for testing. Each intrinsic glomerular cell was annotated by senior pathologists. Since those adherent cells will affect the accuracy of the intrinsic glomerular cells' statistic, a digital image processing algorithm was applied to optimize the segmentation results. The concave region is calculated by extracting the convex hull of each contour and the interference points are carefully removed by area threshold. With the results of intrinsic glomerular cells, an ingenious method was designed to compute the mesangial region and mesangial hypercellularity score. We combined the segmentation results of intrinsic cells with the raw images, and developed a novel method to assess the M score, which is depicted as follows: (1) obtain the intrinsic cell and glomerulus segmentation results based on the above work; (2) remove the background outside of the glomeruli and Inpaint the mesangial cells' region with the value of their surrounding pixels; (3) extract the white regions based on the pixel value threshold; and (4) apply a marker‐controlled watershed segmentation algorithm, with the mesangial cells' region as the foreground, podocytes as white regions, and endothelial cells as the background (Figure 1).
Evaluation metrics of CNN performance
The evaluation metrics implemented in this study are described in supplementary material, Supplementary materials and methods.
Results
Location of glomeruli
Since identification of glomeruli is the basis for glomerular structure and lesion analysis, the first step is to locate and identify all glomeruli. We employed 360 slides (approximately 41 000 patches) with PAS stain from patients with IgAN for training and validation, and 40 slides (nearly 5000 patches) for testing. Performance of glomerulus location was evaluated through the dice coefficient (a common metric for segmentation), precision, and recall (a widely used metric for object detection). The results showed that our model could achieve 0.931 average precision and 0.949 average recall for all kinds of glomeruli (NOA, SS, GS, C) in IgAN on 40 slides, which indicates a superior performance in locating glomeruli. In addition, we also compared the performance of glomerulus localization with those of previous studies and architectures, and our model obtained an outstanding performance according to precision and recall (Table 1).
Glomerular lesion identification
To evaluate the utility of ARPS for identifying glomerular lesions, 1438 images of glomeruli (almost 300 images per category) were used for assessment. DenseNet‐121 [36] was applied in step 1 to classify each glomerulus into three categories: negative, GS, and others, followed by the LSTM‐SENet, classifying SS, C, and NOA. The ability of ARPS to identify the glomerular lesions is presented in Figure 2. As shown in Figure 2B, the glomerular lesion classification confusion matrix was calculated by combining the results of two stages, indicating that ARPS can achieve an extraordinary performance for classification of GS, and Neg, and achieve superior results for C, NOA, and SS identification. The combination of glomerular lesion classification in IgAN yielded an average accuracy of 92.8%, 0.912 Cohen's kappa, and 95% CI (0.892–0.923).
Figure 2.
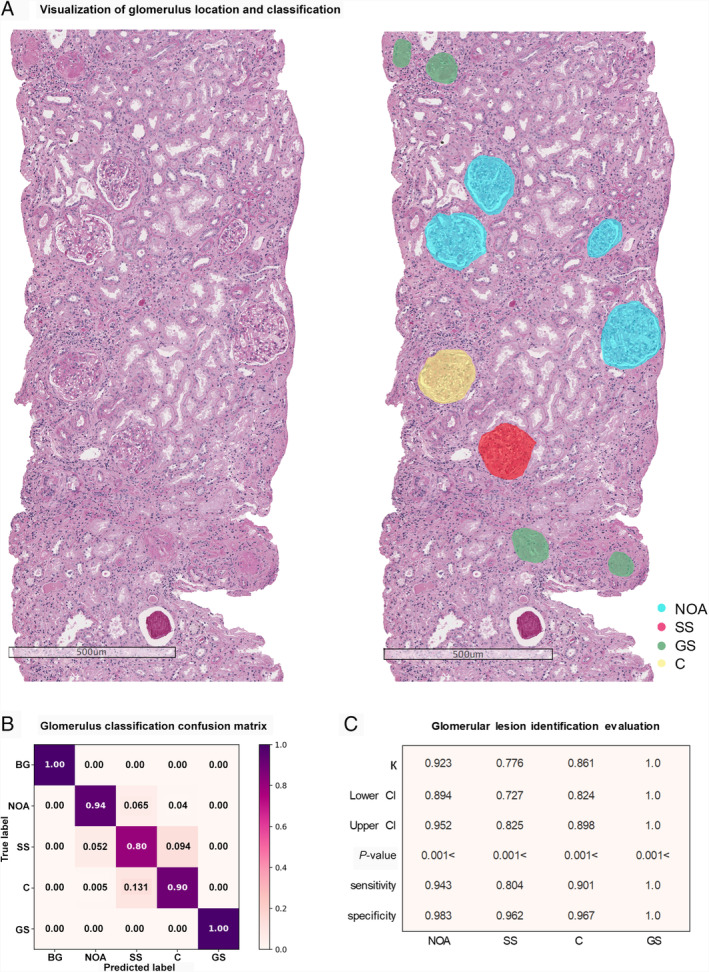
Assessment of glomerular lesion identification. (A) Visualization of location and classification of glomeruli with PAS stain. (B) Glomerular lesion classification confusion matrix, which provides the condition of how predictions are distributed over subcategories. (C) Metrics of the classification task, with Cohen's kappa, 95% confidence interval score, P value, sensitivity, and specificity.
Complex glomerular structure analysis
Intrinsic glomerular cell recognition
To analyze intraglomerular structures, we used 460 glomeruli (nearly 70 000 intrinsic glomerular cells) from NOA to train the 2D V‐Net. With sufficient training, our model could obtain 0.882 average precision and 0.879 average recall for intrinsic glomerular cell recognition (Table 4). Combined with glomerular parameters, APRS was able to acquire several types of information, such as the numbers of three kinds of intrinsic glomerular cells, identify mesangial regions, glomerulus diameter, and the area of these structures (Figure 3).
Table 4.
Quantitative results of intrinsic glomerular cell recognition.
| Metric | ARPS | Doctor 1 | Doctor 2 | Doctor 3 | Doctor 4 |
|---|---|---|---|---|---|
| Precision | 0.882 | 0.763 | 0.764 | 0.828 | 0.856 |
| Recall | 0.879 | 0.742 | 0.740 | 0.816 | 0.823 |
| Specificity | 0.931 | 0.852 | 0.857 | 0.903 | 0.918 |
| Accuracy | 0.922 | 0.810 | 0.814 | 0.874 | 0.882 |
| Time taken | 0.6 s/image | 43 ± 11 s/image | |||
Figure 3.
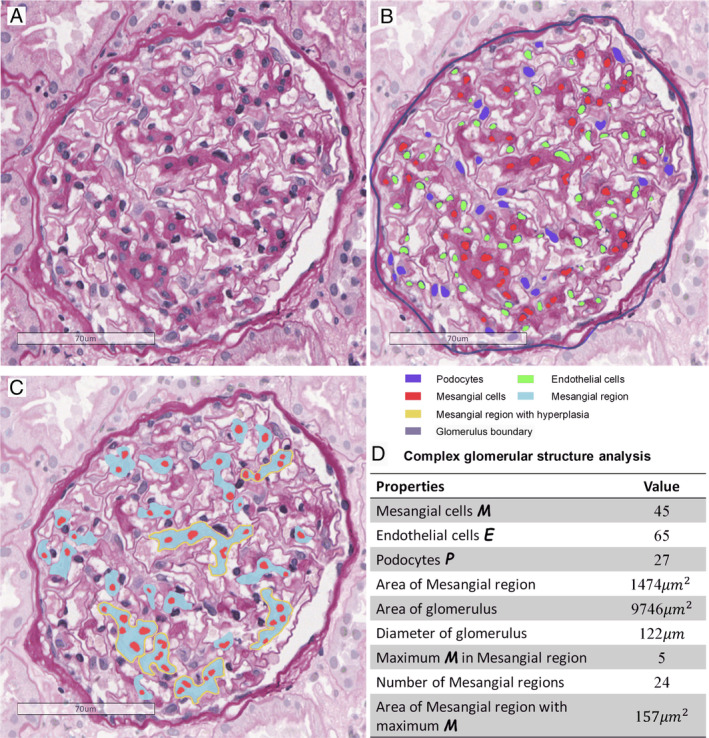
Assessment of internal glomerular structures. (A) Original glomerulus image with PAS stain. (B) Intrinsic glomerular cell segmentation, with detected mesangial cells (M), podocytes (P), endothelial cells (E), and glomerulus boundary. (C) Assessment of mesangial regions, including mesangial regions and mesangial cells. (D) Measurement results of the glomerulus in C.
To better evaluate the performance of cell recognition by ARPS, we plotted the residual between ground truth and the prediction given by junior pathologists (doctors 1–4) and APRS, shown in Figure 4. Experimental results showed that ARPS had the minimum gap with senior pathologists for identifying podocytes, followed by endothelial cells and mesangial cells. The residual of ARPS was significantly less than that of junior pathologists, especially in mesangial and endothelial cells, indicating the approving performance of ARPS (Figure 4). Overall, ARPS performed better with a mean accuracy of 92.2%, surpassing by about 5–11% that of the junior pathologists (Table 4). It took only 0.6 s to estimate the number of intrinsic cells per glomerulus, which was about 1/50 to 1/100 times that of the junior pathologists. In the final step, we employed t‐tests to evaluate the difference between ARPS and the average of four junior pathologists. The result shows that there was a significant difference (p < 0.0001) between ARPS and junior pathologists for identifying mesangial and endothelial cells, with t‐statistic of 10.859 and 11.644, respectively, but not for identifying podocytes (p = 0.0602; t‐statistic 1.896).
Figure 4.
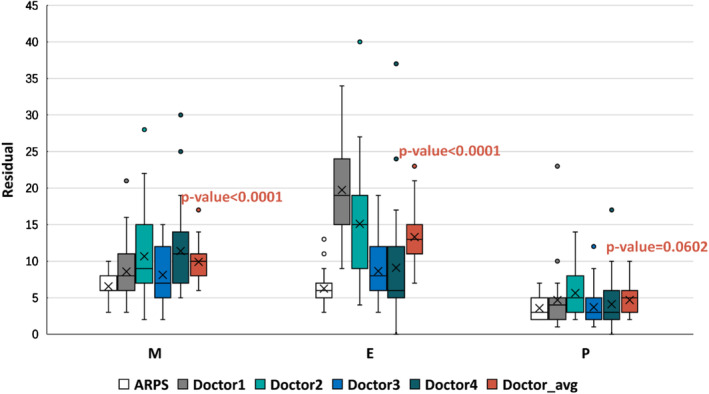
Boxplot of the residual map in the intrinsic glomerular cell recognition task. The horizontal axis represents the ground truth, while the vertical axis indicates the residual between the competitors (ARPS and four doctors) and ground truth (senior pathologists). The horizontal lines from top to bottom in each boxplot represent the maximum, third quartile, median, first quartile, and minimum, respectively, and the cross represents the mean value of residual.
Intrinsic glomerular cell distribution
The first step in evaluating the pattern of glomerular injury is to determine whether glomeruli are hypercellular or not. To better study the hypercellularity of intrinsic glomerular cells, ARPS was employed to measure the quantification of the intrinsic cells including mesangial cells (Figure 5B), endothelial cells (Figure 5C), and podocytes (Figure 5D) based on 3592 qualified NOA samples. Since we can only observe one cross‐section from a WSI, calculating the intrinsic cell ratio can be better suited for analyzing the association between intrinsic cells and a lesion rather than the number of cells. As shown in Figure 5A, each tri‐fold line represents the distribution of intrinsic cells in a single glomerulus. The distribution histogram (Figure 5B–D) describes the ratio distribution of each kind of intrinsic glomerular cell across 3592 glomeruli, while the kernel density estimation (KDE, green line) represents the real distribution. The red line indicates the normal distribution with mean and variance from real data. Since each diagram in Figure 5B–D appears to follow a normal distribution, the mean value of sample distribution is treated as the mean value of overall distribution, and the confidence interval can be calculated from supplementary material, Supplementary materials and methods, equation (5). The average proportion of M, E, and P of each glomerulus in IgAN was 0.41:0.36:0.23 and their margin of error calculated from the expectation and sample capacity was 0.0082, 0.0080, and 0.0070, with 95% CI (0.39–0.42), (0.35–0.38), and (0.22–0.25), respectively. The correlation between M cells' ratio in ARPS and M score in the Oxford Classification was also calculated, with Pearson's r = 0.368. In addition, we conducted cell distribution analysis on biopsies from patients with Henoch–Schönlein purpura nephritis (80 slides, 758 glomeruli); the results showed that the average proportion of M, E, and P of each glomerulus in Henoch–Schönlein purpura nephritis was 0.36:0.37:0.27.
Figure 5.
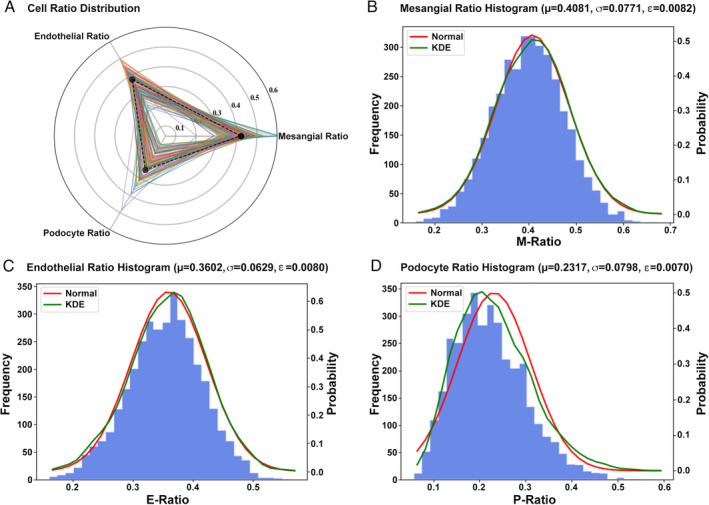
Distribution of intrinsic glomerular cells. (A) Distribution map of three kinds of intrinsic glomerular cells. (B–D) Histogram of mesangial cells, endothelial cells, and podocytes, respectively. The kernel density estimation (KDE, green line) represents the real distribution, while the red line indicates the normal distribution.
Mesangial hypercellularity score evaluation
The mesangial hypercellularity score is the indispensable parameter for predicting clinical outcome. In this study, in total 100 slides were used for M‐score assessment, and each of the slides had a visual score of mesangial hypercellularity made by senior pathologists. Regions of indeterminate mesangial cellularity were excluded through the Oxford Classification, including global sclerosis/advanced segmental sclerosis, global endocapillary hypercellularity, retracted glomerular tuft, incomplete mesangial area, and crescent‐only glomeruli. After carefully scoring and counting each glomerulus and calculating the mesangial score, ARPS achieved 0.42 Cohen's kappa and 95% CI (0.18–0.69).
Discussion
Accurate renal pathological diagnosis relies on the recognition of pathological patterns within glomeruli, tubules, and vascular compartments under light microscopy, combined with immunofluorescence, electron microscopy, and clinical information [5]. Due to the complexity and diversity of renal pathology, computer‐aided diagnosis for renal pathology demands the recognition of all kinds of glomeruli and their pathological features. Since the change of intrinsic glomerular cells is highly correlated with the basic lesion in renal disease, we designed ARPS, employing deep learning methods to automatically identify three types of intrinsic glomerular cell, which has been rarely reported in previous literature. We also applied ARPS to calculate the mesangial hypercellularity score automatically in IgAN, although the performance still needs to be improved.
In this study, in total 100 human slides (60 for validation and 40 for testing) were used for evaluation to obtain convincing results for localization of glomeruli. Deep neural learning is a data‐driven method. In addition to neural architectures, the network's performance relies greatly on the quality and quantity of datasets, since the more data, the more precise features that can be extracted by networks. Most existing studies (Table 1) were implemented on a middle‐sized dataset (10–90) with limited test images. In this paper, we adopted in total 400 whole‐slide images for developing ARPS, and designed ablation experiments to prove the proposed method. Use of the original U‐Net could only achieve 89.2% precision and 90.6% recall, which is close to the Mask R‐CNN‐based method. When aided by SSIM loss function, the performance could be increased to the FCOS level (~91% precision and 92% recall). Furthermore, the marked watershed algorithm aimed at adherent glomeruli led to a 2% and 3% improvement of precision and recall, respectively. The adequate training data and the attention U‐Net [40] trained with the instance similarity loss enabled our work to yield an average recall of 94.9% in the localization of glomeruli task.
For classification of glomeruli, previous studies have shown that computational techniques can be used to identify global sclerosis and non‐sclerosis glomeruli [28, 29, 30, 32]. Due to the diversity and co‐existence of pathological changes in IgAN (such as the co‐existence of segmental sclerosis and crescents), doctors are more interested in understanding the types of lesions in the non‐sclerosis glomeruli. Thus, computational tools for IgAN require a more detailed sub‐division of glomerular lesions. To classify these fine‐grained glomerular lesions, we applied the LSTM‐GCNet, combining the LSTM layer [41] with the GCNet [37] to extract more useful features, and focused more on regional information instead of local message. The experimental results demonstrated that identification of global sclerosis and crescents gave satisfactory results with 1.0 and 0.861 Cohen's kappa, respectively, and that identification of segmental sclerosis still needs to be improved (Cohen's kappa 0.776). It was also proved that obtaining satisfactory agreement for segmental sclerosis recognition was difficult, even by expert pathologists (intra‐class correlation coefficients of GS, C, and SS were 0.9, 0.66, and 0.46, assessed by three to five pathologists in the Oxford Classification in IgAN [39]).
Intrinsic glomerular cell proliferation is a manifestation of certain glomerular diseases, including IgAN, lupus nephritis, etc., and is closely related to the diagnosis, severity, and prognosis. Different from Barros et al [30], who applied the Gaussian method to extract the features and could only segment intrinsic cells without category information, we carefully designed a deep neural network to extract pathological features and segment intrinsic cells. The convincing results showed that ARPS can distinguish M, E, and P, with none of the previous work accomplishing this task, to our knowledge. Sufficient experiments show that ARPS can surpass junior pathologists in the identification of M and E (significant difference between ARPS and the average of four doctors, with p < 0.0001), while the performance of podocytes is similar, with p = 0.0602 (due to the relative isolated location and morphological features). We also found that junior pathologists were more likely to misclassify M and E, which was also mentioned in the previous study [39] [the ICC (intra‐class correlation coefficient) of the endothelial proliferation achieved only 0.36] and can be alleviated with the application of ARPS.
In addition, inspired by Steffes et al [42], who analyzed intrinsic cell distribution from normal kidney donors (proportion of M, E, and P was 0.37:0.37:0.26), we employed ARPS in calculating the distribution of intrinsic cells from patients with IgAN at an extremely low cost. Experiments showed that the average proportion of M, E, and P within each glomerulus from patients with IgAN was 0.41:0.36:0.23, illustrating that IgAN was a disease with the proliferation of mesangial cells. The relatively weak correlation (Pearson's r 0.368) between the M cells' ratio in ARPS and M score in the Oxford Classification is probably because of the different definitions of these two parameters. The Oxford Classification M score reflects the absolute mesangial cellularity, whilst the M cells' ratio reflects the proportion of glomerular cells that are mesangial cells, and is therefore affected by the number of endothelial, epithelial, and inflammatory cells. Meanwhile, we conducted cell distribution analysis on Henoch–Schönlein purpura nephritis and the results showed that the average proportion of M, E, and P of each glomerulus in this disease was 0.36:0.37:0.27. In the future, we propose to investigate the relationship between the ratio variation of M, E, P and clinical data, including clinical parameters, pathological classification, and the prognosis of different kidney diseases using more cases.
Mesangial score is the crucial parameter in the Oxford Classification, which has great significance to the prognosis of IgAN. Evaluation of the mesangial score is time‐consuming and of great variability for pathologists, with a consistency of 0.64 ICC [39]. Here we applied ARPS to automatically assess mesangial score for IgAN patients according to the simplified mesangial score. Due to the disturbance of the mesangial area adjacent to the vascular stalk, our model only reached a Cohen's kappa of 0.42, indicating moderate consistency with senior doctors. We found that we obtained this imperfect Cohen's kappa even with a high accuracy of recognition of intrinsic glomerular cells, which could be related to the following: (1) the central mesangial regions are not well excluded, which leads to error in M‐score assessment; and (2) there exists variability on M‐score assessment among several studies as the ICC of the median mesangial score varied from 0.37 to 0.80 [10, 13, 39]. The consistency of ARPS may be further improved by accurate segmentation of mesangial regions and intrinsic cells. The model in this study could also be helpful for assisting Oxford Classification score and proposing a new mesangial‐related indicator to predict prognosis, which can be verified through many more IgAN cases.
In the future, with a much better performance than junior pathologists in kidney pathology, ARPS could be employed in training and improving the level of pathologists in glomerular lesion identification. We can extract the quantified pathological features of a glomerulus through ARPS, and combine them with clinical data, prognostic information, genomics, transcriptome, proteomics, metabolomics, and other multi‐group data to find an individual's pathogenesis and predict the treatment and prognosis of patients.
Our study has several limitations. It was initially based on assessment of PAS stains and cannot identify the crescent subcategories due to insufficient cases with different types of crescents. Furthermore, we did not study endocapillary hypercellularity, as the incidence of endocapillary hypercellularity is 11–42% [9, 13, 43], which is relatively low compared with other pathological features, and we did not have enough cases for training our models. In addition, we did not obtain comprehensive assessment about the MESTC score based on present research. Finally, the performance of mesangial score evaluation still needs to be improved. In our work, only partial glomerular lesions were evaluated by AI technology, and much work remains to be done before the entire renal biopsy pathology can be evaluated and generate a pathological report automatically.
In conclusion, this paper presents ARPS, which provides a digital indicator for renal pathology. Our system can provide a computational approach for the identification of glomerular lesions and recognition of intrinsic glomerular cells, and provides a mesangial hypercellularity score, which could be used directly for assisting the pathological diagnosis of kidney diseases.
Author contributions statement
ZCH, NY and XF had full access to all of the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. LZH, ZCH, NY and LCF were responsible for study concept and design. LZH, ZCH, NY, XF, HXS and LFY acquired, analyzed, or interpreted data. NY, XF, CTY, LQJ, LFY, HXS, ZCH and LZH drafted the manuscript. NY, LFY and HXS carried out statistical analysis. LZH obtained funding and supervised the study. XGT, LCF, LX, ZCH and LZH provided administrative, technical, or material support. All the authors critically revised the manuscript for important intellectual content.
References 44 ‐ 50 are cited only in the supplementary material.
Supporting information
Supplementary materials and methods
Acknowledgements
We thank Tao Jingli, Wang Huichao, ZouYun, Liu Li, Li He, Song Yashan, Sun Jing, Tang Wen, Jiang Linglin, and Li Li for pathological annotation work. This research was funded by the National Key Research and Development Program of China (2016YFC0901202), the National Natural Science Foundation of China (81570644), and the Project of Invigorating Health Care through Science, Technology and Education of Jiangsu Province Medical Key Talent (ZDRCA2016098).
No conflicts of interest were declared.
References
- 1. Zhang L, Wang F, Wang L, Wang W, et al. Prevalence of chronic kidney disease in China: a cross‐sectional survey. The Lancet 2012; 379 : 815–822. [DOI] [PubMed] [Google Scholar]
- 2. Eckhardt K‐U, Coresh J, Devuyst O, et al Evolving importance of kidney disease: from subspecialty to global health burden. Lancet 2013; 382 : 158–169. [DOI] [PubMed] [Google Scholar]
- 3. Magistroni R, D'Agati VD, Appel GB, et al New developments in the genetics, pathogenesis, and therapy of IgA nephropathy. Kidney Int 2015; 88 : 974–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wyatt RJ, Julian BA. IgA nephropathy. N Engl J Med 2013; 368 : 2402–2414. [DOI] [PubMed] [Google Scholar]
- 5. Sethi S, Haas M, Markowitz GS, et al Mayo Clinic/Renal Pathology Society consensus report on pathologic classification, diagnosis, and reporting of GN. J Am Soc Nephrol 2016; 27 : 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Haas M, Verhave JC, Liu Z‐H, et al A multicenter study of the predictive value of crescents in IgA nephropathy. J Am Soc Nephrol 2017; 28 : 691–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alamartine E, Sauron C, Laurent B, et al The use of the Oxford classification of IgA nephropathy to predict renal survival. Clin J Am Soc Nephrol 2011; 6 : 2384–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bellur SS, Lepeytre F, Vorobyeva O, et al Evidence from the Oxford Classification cohort supports the clinical value of subclassification of focal segmental glomerulosclerosis in IgA nephropathy. Kidney Int 2017; 91 : 235–243. [DOI] [PubMed] [Google Scholar]
- 9. Cattran DC, Rosanna C, Terence HC, et al The Oxford classification of IgA nephropathy: rationale, clinicopathological correlations, and classification. Kidney Int 2009; 76 : 534–545. [DOI] [PubMed] [Google Scholar]
- 10. Herzenberg AM, Fogo AB, Reich HN, et al Validation of the Oxford classification of IgA nephropathy. Kidney Int 2011; 80 : 310–317. [DOI] [PubMed] [Google Scholar]
- 11. Shi S‐F, Wang S‐X, Jiang L, et al Pathologic predictors of renal outcome and therapeutic efficacy in IgA nephropathy: validation of the Oxford classification. Clin J Am Soc Nephrol 2011; 6 : 2175–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Trimarchi H, Barratt J, Cattran DC, et al Oxford Classification of IgA nephropathy 2016: an update from the IgA Nephropathy Classification Working Group. Kidney Int 2017; 91 : 1014–1021. [DOI] [PubMed] [Google Scholar]
- 13. Zeng C‐H, Le W, Ni Z, et al A multicenter application and evaluation of the Oxford classification of IgA nephropathy in adult Chinese patients. Am J Kidney Dis 2012; 60 : 812–820. [DOI] [PubMed] [Google Scholar]
- 14. Temerinac‐Ott M, Forestier G, Schmitz J, et al Detection of glomeruli in renal pathology by mutual comparison of multiple staining modalities In Proceedings of the 10th International Symposium on Image and Signal Processing and Analysis. IEEE: Ljubljana, 2017; 19–24. [Google Scholar]
- 15. Kato T, Relator R, Ngouv H, et al Segmental HOG: new descriptor for glomerulus detection in kidney microscopy image. BMC Bioinformatics 2015; 16 : 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marée R, Dallongeville S, Olivo‐Marin J‐C, et al An approach for detection of glomeruli in multisite digital pathology In 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). IEEE: Prague, 2016; 1033–1036. [Google Scholar]
- 17. Gadermayr M, Dombrowski A, Klinkhammer BM, et al CNN cascades for segmenting sparse objects in gigapixel whole slide images. Comput Med Imaging Graph 2019; 71 : 40–48. [DOI] [PubMed] [Google Scholar]
- 18. Gallego J, Pedraza A, Lopez S, et al Glomerulus classification and detection based on convolutional neural networks. J Imaging 2018; 4 : 20. [Google Scholar]
- 19. Sheehan S, Mawe S, Cianciolo RE, et al Detection and classification of novel renal histologic phenotypes using deep neural networks. Am J Pathol 2019; 189 : 1786–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bukowy JD, Dayton A, Cloutier D, et al Region‐based convolutional neural nets for localization of glomeruli in trichrome‐stained whole kidney sections. J Am Soc Nephrol 2018; 29 : 2081–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bueno G, Fernandez‐Carrobles MM, Gonzalez‐Lopez L, et al Glomerulosclerosis identification in whole slide images using semantic segmentation. Comput Methods Programs Biomed 2020; 184 : 105273. [DOI] [PubMed] [Google Scholar]
- 22. Sarder P, Ginley B, Tomaszewski JE. Automated renal histopathology: digital extraction and quantification of renal pathology In Medical Imaging 2016: Digital Pathology. International Society for Optics and Photonics, 2016; 97910F. [Google Scholar]
- 23. Ginley B, Lutnick B, Jen K‐Y, et al Computational segmentation and classification of diabetic glomerulosclerosis. J Am Soc Nephrol 2019; 30 : 1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Simon O, Yacoub R, Jain S, et al Multi‐radial LBP features as a tool for rapid glomerular detection and assessment in whole slide histopathology images. Sci Rep 2018; 8 : 2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hermsen M, de Bel T, den Boer M, et al Deep learning‐based histopathologic assessment of kidney tissue. J Am Soc Nephrol 2019; 30 : 1968–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. He K, Gkioxari G, Dollár P, et al Mask R‐CNN. 2017 IEEE International Conference on Computer Vision (ICCV), Venice, 2017; 2980–2988, DOI: 10.1109/ICCV.2017.322.
- 27. Tian Z, Shen C, Chen H, et al FCOS: fully convolutional one‐stage object detection In International Conference on Computer Vision, 2019; 9627–9636; arXiv:1904.01355. [Google Scholar]
- 28. Marsh JN, Matlock M, Kudose S, et al Deep learning global glomerulosclerosis in transplant kidney frozen sections. IEEE Trans Med Imaging 2018; 37 : 2718–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kannan S, Morgan LA, Liang B, et al Segmentation of glomeruli within trichrome images using deep learning. Kidney Int Rep 2019; 4 : 955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barros GO, Navarro B, Duarte A, et al PathoSpotter‐K: a computational tool for the automatic identification of glomerular lesions in histological images of kidneys. Sci Rep 2017; 7 : 46769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chagas P, Souza L, Araujo I, et al Classification of glomerular hypercellularity using convolutional features and support vector machine. Artif Intell Med 2020; 103 : 101808. [DOI] [PubMed] [Google Scholar]
- 32. Jayapandian C, Chen Y, Janowczyk A, et al Deep learning based detection of normal and globally sclerotic glomeruli on whole slide images from the NEPTUNE renal biopsies with HE, PAS, trichrome and silver staining. In Lab Invest 2019; 32(Suppl 2): 16–17. [Google Scholar]
- 33. Ronneberger O, Fischer P, Brox T. U‐Net: convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer‐Assisted Intervention – MICCAI 2015, Navab N, Hornegger J, Wells W, et al. (eds). MICCAI 2015. Lecture Notes in Computer Science, vol 9351. Springer: Cham; 2015.
- 34. Norman B, Pedoia V, Majumdar S. Use of 2D U‐net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 2018; 288 : 177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Venhuizen FG, van Ginneken B, Liefers B, et al Robust total retina thickness segmentation in optical coherence tomography images using convolutional neural networks. Biomed Opt Express 2017; 8 : 3292–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang G, Liu Z, Van Der Maaten L, et al Densely connected convolutional networks In The IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2017; 4700–4708. [Google Scholar]
- 37. Cao Y, Xu J, Lin S, et al GCNet: non‐local networks meet squeeze‐excitation networks and beyond. arXiv 2019; arXiv:1904.11492. [Google Scholar]
- 38. Milletari F, Navab N, Ahmadi S‐A. V‐net: fully convolutional neural networks for volumetric medical image segmentation In 2016 Fourth International Conference on 3D Vision (3DV). IEEE, 2016; 565–571. 10.1109/3DV.2016.79. [DOI] [Google Scholar]
- 39. Working Group of the International IgA Nephropathy Network and the Renal Pathology Society , Roberts ISD, Cook HT, et al The Oxford classification of IgA nephropathy: pathology definitions, correlations, and reproducibility. Kidney Int 2009; 76 : 546–556. [DOI] [PubMed] [Google Scholar]
- 40. Oktay O, Schlemper J, Folgoc LL, et al Attention U‐net: learning where to look for the pancreas. arXiv 2018; arXiv:1804.03999. [Google Scholar]
- 41. Hochreiter S, Schmidhuber J. Long short‐term memory. Neural Comput 1997; 9 : 1735–1780. [DOI] [PubMed] [Google Scholar]
- 42. Steffes MW, Schmidt D, McCrery R, et al Glomerular cell number in normal subjects and in type 1 diabetic patients. Kidney Int 2001; 59 : 2104–2113. [DOI] [PubMed] [Google Scholar]
- 43. Coppo R, Troyanov S, Bellur S, et al Validation of the Oxford classification of IgA nephropathy in cohorts with different presentations and treatments. Kidney Int 2014; 86 : 828–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Z, Bovik AC, Sheikh HR, et al Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process 2004; 13 : 600–612. [DOI] [PubMed] [Google Scholar]
- 45. Huang G, Liu Z, Weinberger KQ, et al Densely connected convolutional networks. arXiv 2016; arXiv:1608.06993. [Google Scholar]
- 46. Ginley B, Tomaszewski JE, Sarder P. Automatic computational labeling of glomerular textural boundaries In Proc SPIE 10140, Medical Imaging 2017: Digital Pathology, 2017; 101400G; 10.1117/12.2254517. [DOI] [Google Scholar]
- 47. Hu J, Shen L, Sun G. Squeeze‐and‐excitation networks In The IEEE Conference on Computer Vision and Pattern Recognition, 2018; 7132–7141. [Google Scholar]
- 48. Lian S, Luo Z, Zhong Z, et al Attention guided U‐Net for accurate iris segmentation. J Vis Commun Image Represent 2018; 56 : 296–304. [Google Scholar]
- 49. Glorot X, Bengio Y. Understanding the difficulty of training deep feedforward neural networks In Proceedings of the 13th International Conference on Artificial Intelligence and Statistics, 2010; PMLR 9; 249–256. [Google Scholar]
- 50. Kingma DP, Ba J. Adam: a method for stochastic optimization. arXiv 2014; arXiv:14126980. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary materials and methods


