Abstract
Background and Aim
Interferon‐stimulated gene 20 (ISG20) is an interferon‐inducible exonuclease that inhibits the replication of several RNA viruses. In patients with chronic hepatitis B, ISG20 expression is related to the interferon‐α treatment response. However, the molecular mechanism of ISG20‐mediated anti‐hepatitis B virus (HBV) activity is unclear.
Methods
We have investigated the effect of ISG20 on antiviral activity to address that. The life cycle of HBV was analyzed by the ectopic expression of ISG20 in HepG2 and HepG2‐NTCP cells. Finally, to provide physiological relevance of our study, the expression of ISG20 from chronic hepatitis B patients was examined.
Results
Interferon‐stimulated gene 20 was mainly induced by interferon‐β and dramatically inhibited HBV replication. In addition, ISG20 decreased HBV gene expression and transcription. Although ISG20 inhibited HBV replication by reducing viral enhancer activity, the expression of transcription factors that bind the HBV enhancer was not affected. Particularly, ISG20 suppressed HBV enhancer activity by binding to the enhancer II and core promoter (EnhII/Cp) region.
Conclusion
Our findings suggest that ISG20 exerts the anti‐HBV activity by acting as a putative repressor binding to the HBV EnhII/Cp region.
Keywords: Enhancer, Hepatitis B virus, Interferon‐stimulated gene 20, Interferon‐β
Introduction
Interferons (IFNs) are key molecules in the innate immune response against viral infection and bind to cell surface IFN receptors, leading to the induction of hundreds of IFN‐stimulated genes (ISGs).1 The well‐characterized classic ISGs RNA‐dependent protein kinase R, 2′‐5′ oligoadenylate synthetase/RNase L, and myxovirus resistance protein are essential for suppressing viral replication. Additionally, the tripartite motif protein family and IFN‐induced proteins with tetratricopeptides repeats (IFIT), ISG15, and ISG20 participate in IFN‐mediated antiviral activity by targeting various steps of the viral life cycle.2, 3
Interferon‐stimulated gene 20 is a 3′‐5′ exonuclease induced by both type I and II IFNs and is involved in antiviral functions against several RNA viruses.4, 5, 6 Ectopic expression of ISG20 inhibits the replication of RNA viruses such as hepatitis C virus, bovine viral diarrhea virus, yellow fever virus, hepatitis A virus, and human immunodeficiency virus type 1 (HIV‐1) through their exonuclease activities.6, 7, 8 In influenza A virus, ISG20 showed antiviral activity by interacting with nucleoproteins.9 Additionally, ISG20 suppressed the West Nile virus and dengue virus replication by disrupting the post‐transcription step.10 These studies suggest that ISG20 plays a key role in IFN‐mediated antiviral activity. In addition to the antiviral activity of ISG20, several studies using microarray data reported that ISG20 expression was increased by virus infection.11 Rabies virus infection induced ISG20 expression through the production of IFN‐β in neuron‐derivative NT2‐N cell lines.12 Epstein–Barr virus infection significantly upregulated ISG20 mRNA levels in B lymphocytes.13 Additionally, Vpr protein of HIV‐1 led to induction of ISG20 expression in monocyte‐derived macrophages.14
In hepatitis B virus (HBV) infection, ISG20 was found to be associated with HBV clearance in the liver of chimpanzees with acute HBV infection.15 Moreover, ISG20 expression has been implicated in the response to IFN‐α treatment in patients with chronic hepatitis B (CHB). ISG20 levels were reduced in the liver tissue of non‐responders before IFN‐α treatment16 and highly upregulated in responders to IFN‐α treatment.17 In addition to CHB patients, ISG20 was shown to have anti‐HBV activity in vivo and in vitro.18, 19 However, the detailed mechanism of the antiviral effect against HBV remains unclear. Therefore, we investigated the molecular target of ISG20‐mediated HBV suppression in the HBV life cycle and examined the role of ISG20 in the natural course of chronic infection with HBV.
Materials and methods
Cell culture and transfection
Human hepatoma HepG2, HepG2‐NTCP, and HepAD38 cells were described in our previous studies.20, 21 All cells were cultured at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Gibco, Grand Island, NY, USA). HepG2‐NTCP and HepAD38 cells were maintained in the presence of G418 and tetracycline, respectively. Plasmid DNA and small interfering RNA were transfected into HepG2 cells grown to ~50% confluence in 6‐well or 12‐well plates with Lipofectamine 2000 and RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol, respectively.
Plasmids and reagents
Plasmid HBV1.2 encoding 1.2 copies of the HBV genome (3.8 kb, genotype D), EnhI·II, Xp·EnhII, NRE·EnhII, EnhI·ΔEnhII, and EnhII/Cp were described in our previous studies.20, 21 ΔNRE·EnhII was cloned into the PGL3‐basic vector (Promega, Madison, WI, USA). ISG20 was cloned into pFLAG‐CMV1 at the HindIII and EcoRI sites. siISG20 (GE Healthcare, Buckinghamshire, UK) and IFNs (IFN‐α, IFN‐β, and IFN‐γ, PBL Assay Science, Piscataway Township, NJ, USA) were used in this study.
Measurement of HBeAg and HBsAg levels
Three days after HBV 1.2 transfection with or without ISG20 in HepG2 cells, the culture supernatants were collected to assess viral antigen levels. HBeAg and HBsAg secretion was measured using a commercial kit (Wantai Bio‐Pharm, Beijing, China) according to the manufacturer's protocol. To avoid signal saturation, the supernatants were diluted in phosphate‐buffered saline.
Hepatitis B virus enhancer activity
HepG2 cells were grown to 40–50% confluence in 12‐well plates and transfected with the plasmids of enhancer mutants indicated in Figure 3a. After 48 h, the cells were harvested and lysed to measure luciferase activity by using the Luciferase Assay System (Promega). β‐Galactosidase activity was measured by using a β‐galactosidase enzyme assay system (Promega) to normalize the transfection efficiency.
Figure 3.
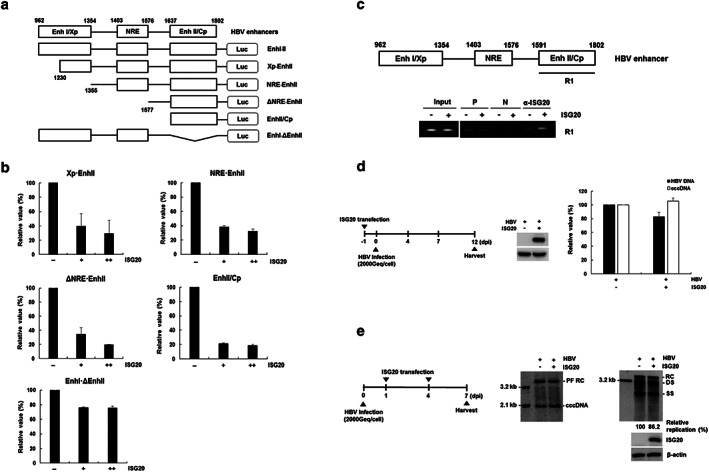
Interferon‐stimulated gene 20 (ISG20) decreases viral enhancer II activity via direct binding. (a) Schematic diagram of the reporter plasmids containing hepatitis B virus (HBV) enhancers of various lengths. (b) Relative luciferase activity of HBV enhancer mutants. Values represent the mean ± SD calculated from two independent experiments (each performed in duplicate). +, 0.5 μg; ++, 1 μg. [Correction added on 22 February 2020, after first online publication: figure image and caption have been amended.]
Hepatitis B virus infection
Hepatitis B virus particles were collected from the 100‐fold concentrated supernatant of HepAD38 cells using PEG Virus Precipitation Kit (BioVision, USA) and stored at −80 °C. HepG2‐NTCP cells (1 × 106 cells) were seeded onto six‐well plate coated with collagen I (Gibco) and infected with 2000 HBV genome equivalent per cells (GEq per cells) in DMEM containing 4% PEG 8000 (Sigma) and 2.5% dimethylsulfoxide (Sigma) for 16–20 h. Then cells were washed three times with DMEM, maintained in DMEM containing 2% dimethylsulfoxide, and harvested 7–12 days post‐infection (dpi).
Southern and northern blotting
Capsid‐associated HBV DNA and RNA were evaluated by southern and northern blotting as described in our previous reports.20, 21 For the detection of cccDNA, modified Hirt's extraction procedure was used. Briefly, the cell pellets were lysed in Tris–HCl (pH 7.4) buffer containing 10‐mM EDTA, 150‐mM NaCl, and 1% sodium dodecylsulfate and then treated with 2.5‐M KCl at 4 °C for overnight. A fraction of cccDNA was purified with a mixture of phenol/chloroform/isoamyl alcohol (25:24:1) (Sigma) and chloroform (Sigma) extraction. Purified cccDNA was treated with plasmid safe DNase I (PSD, Epicenter Technologies, USA) and inactivated by incubation for 30 min at 70 °C. PSD‐treated cccDNA was separated on a 0.8% agarose gel and transferred onto a Hybond‐N+ nylon membrane (GE Healthcare). Twenty micrograms of total RNA extracted using TRIzol reagent (Invitrogen) were separated on 1% formaldehyde‐agarose gel. HBV DNA and RNA were detected with highly pure randomly primed probes labeled with [α‐32P] dCTP (PerkinElmer, Waltham, MA, USA). As loading control, 28S and 18S RNA were detected for gel electrophoresis.
Western blot
Various proteins were detected by the following antibodies: ISG20 (Abcam, Cambridge, UK, 1:2000), HBV core protein (B0586, Dako, Glostrup, Denmark, 1:2000), HBsAg (ab9193, Abcam, or Dako, 1:2000), hepatocyte nuclear factor 1α (HNF1α) (sc‐393668, 1:1000, Santa Cruz Biotechnology, Dallas, TX, USA), FLAG‐M2 (A5316, 1:2000, Sigma), CEBPα (sc‐365318, 1:1000, Santa Cruz Biotechnology), HNF4α (sc‐374229, 1:2000, Santa Cruz Biotechnology), PGC‐1α (sc‐518025, 1:2000, Santa Cruz Biotechnology), and actin (A5316, 1:5000, Sigma).
Reverse transcription–polymerase chain reaction and real‐time polymerase chain reaction
Total RNA was extracted using the RNeasy Plus mini kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Two micrograms of total RNA were reverse‐transcribed into cDNA with the SuperScript III first‐strand synthesis kit for reverse transcription–polymerase chain reaction (PCR) (Invitrogen). Reverse transcription–PCR was performed using primers for ISG20 and GAPDH: ISG20 forward, 5′‐CGGCTACACAATCTAC GACA‐3′; reverse, 5′‐CTGTTCTG GATGCTCTTGTG‐3′ and GAPDH forward, 5′‐ATCATCCCTGCCTCTACTGG‐3′; reverse, 5′‐TGGGTGTCGCTGTTGAAGTC‐3′. For the analysis of HBV DNA and cccDNA, total genomic and viral DNA in HepG2‐NTCP cells were extracted using QIAamp DNA minikit (Qiagen) according to the manufacturer's protocol. To assess the cccDNA, 500 ng of total DNA were treated with PSD. To quantify HBV RNA, DNA, cccDNA, and GAPDH, real‐time PCR was performed using Power SYBR green PCR master mix (Applied Biosystems, Foster City, CA, USA) and amplified using QuantStudio 3.0 (Applied Biosystems) with the following primers: HBV RNA and DNA forward (nt 256 to 274), 5′‐CTCGTGGTG GACTTCTCTC‐3′; reverse (nt 404 to 421), 5′‐CTGCAGGAT GAAGAGGAA‐3′. HBV cccDNA forward (nt 1824 to 1843), 5′‐TTCACCTCTGCCTAATCATC‐3′; reverse (nt 2048 to 2068), 5′‐CCTGAGTGCTGTATGGTGAG‐3′. To normalize the cell number, the GAPDH gene was amplified.
Chromatin immunoprecipitation assay
The chromatin immunoprecipitation (ChIP) assay was performed using the Ez‐Magna ChIP™ A/G kit (Millipore, Billerica, MA, USA). Briefly, HepG2 cells were seeded into 10‐cm dishes and transfected with HBV 1.2 with or without ISG20. At 3 days post‐transfection, the cells were cross‐linked by formaldehyde, washed with phosphate‐buffered saline, and harvested. The lysates were sonicated by 3 pulses for 15 s at 30% power, and chromatin was precipitated with anti‐ISG20 (sc‐514979, 1 μg per sample, Santa Cruz Biotechnology). Normal mouse IgG and anti‐RNA polymerase II (CTD4H8) were used as negative and positive controls, respectively. Immunoprecipitated DNA fragments were amplified by PCR with EnhII/Cp primers, which had the following sequences: forward, 5′‐AGCTAGCTCTTGCCCAAGGTCTTA CAT‐3′; reverse, 5′‐TAGATCTACAGACCAATTTATGCCTA CAG‐3′.
Microarray data analysis
Microarray data of CHB patients were obtained from the Gene Expression omnibus (GEO) microarray data repositories.22 GSE65359 included 22 immune‐tolerant (IT), 50 immune activator (IA or immune clearance), and 11 inactive carrier (IC) samples.23 These phases are defined by serum alanine aminotransferase, serum HBV DNA levels, and HBeAg status.24, 25 The IA phase involves an elevated immune response to HBV, and the IC phase involves HBeAg seroconversion to reduce HBV DNA.24 The data were processed with the affy R package using robust multiarray average normalization. Statistical significance was determined by an unpaired t‐test. The P values **< 0.001 and ***< 0.0001 were considered to indicate significance.
Results
Interferon‐stimulated gene 20 inhibits hepatitis B virus replication
To investigate whether ISG20 exerts the antiviral activity against HBV, we constructed an expression plasmid for ISG20, and the anti‐HBV activity was examined after co‐transfection of HBV 1.2 into HepG2 cells. The expression of ISG20 was confirmed by western blotting (Fig. 1a). ISG20 reduced the secretion of viral antigens (HBeAg and HBsAg) into the media in a dose‐dependent and time‐dependent manner (Figs 1b and S1). Additionally, HBV replication was suppressed by ISG20 expression (Fig. 1c). Finally, to elucidate whether ISG20 is a cellular inhibitor of HBV infection, we examined the effect of endogenous ISG20 on the secretion of viral antigens. We co‐transfected HBV 1.2 with or without small interfering RNA to prevent ISG20 expression in HepG2 cells and measured the secretion of HBeAg and HBsAg. As shown in Figure 1d, HBV antigens were elevated by siISG20 treatment, indicating that endogenous ISG20 has anti‐HBV activity. These data demonstrate that ISG20 is a cellular inhibitor of HBV infection.
Figure 1.

Interferon‐stimulated gene 20 (ISG20) inhibits hepatitis B virus (HBV) replication in a dose‐dependent manner. (a) Ectopic expression of ISG20. ISG20 was detected by anti‐FLAG antibody. +, 1 μg; ++, 2 μg. Figure represented the representative data obtained from three independent experiments. (b) Effects of ISG20 on the secretion of  , HBeAg and
, HBeAg and  , HBsAg. Values represent the mean ± SD calculated from at least three independent experiments. +, 1 μg; ++, 2 μg. **
P < 0.005, ***
P < 0.0005. P values were determined using unpaired t‐test. (c) Effect of ISG20 on HBV replication. Values represent the mean ± SD calculated from at least three independent experiments. M, marker; +, 1 μg; ++, 2 μg; DS DNA, double‐stranded HBV DNA; SS DNA, single‐stranded HBV DNA. *
P = 0.01, **
P = 0.0025. P values were determined using unpaired t‐test. (d) Restoration of HBV replication by siISG20 (20 nM).
, HBsAg. Values represent the mean ± SD calculated from at least three independent experiments. +, 1 μg; ++, 2 μg. **
P < 0.005, ***
P < 0.0005. P values were determined using unpaired t‐test. (c) Effect of ISG20 on HBV replication. Values represent the mean ± SD calculated from at least three independent experiments. M, marker; +, 1 μg; ++, 2 μg; DS DNA, double‐stranded HBV DNA; SS DNA, single‐stranded HBV DNA. *
P = 0.01, **
P = 0.0025. P values were determined using unpaired t‐test. (d) Restoration of HBV replication by siISG20 (20 nM).  , HBeAg; □, H12BsAg. Values represent the mean ± SD calculated from three independent experiments. *
P < 0.05, ***
P < 0.01. P values were determined using unpaired t‐test. [Correction added on 22 February 2020, after first online publication: figure image and caption have been amended.]
, HBeAg; □, H12BsAg. Values represent the mean ± SD calculated from three independent experiments. *
P < 0.05, ***
P < 0.01. P values were determined using unpaired t‐test. [Correction added on 22 February 2020, after first online publication: figure image and caption have been amended.]
Interferon‐stimulated gene 20 suppresses hepatitis B virus transcription by targeting viral enhancer activity
To clarify the molecular mechanism of anti‐HBV activity by ISG20, we examined HBV gene expression and transcription. Overexpression of ISG20 significantly suppressed HBV proteins (particularly small surface and core) and RNA (Fig. 2a,b). These results indicate that ISG20 regulates HBV at the transcriptional level. HBV transcription is mainly regulated by two enhancers (EnhI and EnhII) containing transcription factors binding sites.26, 27 Thus, we first investigated the effect of ISG20 on viral enhancer activity. As shown in Figure 2c, ISG20 decreased HBV enhancer activity. Several studies reported that HBV enhancers are positively regulated by liver‐enriched transcription factors such as HNF1α, HNF4α, C/EBP1, and PGC1α.28, 29, 30, 31 Because ISG20 decreased viral enhancer activity, we next evaluated the expression of HNF1α, HNF4α, C/EBP1α, and PGC1α. Unexpectedly, ISG20 did not affect the expression of these transcription factors (Fig. 2d), suggesting that ISG20 directly affects HBV enhancer activity. Collectively, our data demonstrate that ISG20 inhibits HBV replication by suppressing viral enhancer activity without dysregulation of liver‐enriched transcription factors.
Figure 2.

Interferon‐stimulated gene 20 (ISG20) suppresses hepatitis B virus (HBV) gene transcription by decreasing viral enhancer activity. (a) Effect of ISG20 on HBV gene expression. L, M, and S, large, middle, and small envelop proteins, respectively; +, 1 μg; ++, 2 μg. (b) Effect of ISG20 on HBV transcription. HBV RNAs were analyzed by northern blot (left panel) and real‐time polymerase chain reaction (right panel). +, 1 μg; ++, 2 μg. Values represent the mean ± SD calculated from three independent experiments. * P < 0.05, *** P < 0.001. P values were determined using unpaired t‐test. (c) Relative luciferase activity of HBV enhancer following treatment with different doses of ISG20. Values represent the mean ± SD calculated from two independent experiments (each performed in duplicate). +, 0.5 μg; ++, 1 μg. (d) Effect of ISG20 on protein level of C/EBPα and hepatocyte nuclear factors (HNFs). +, 1 μg; ++, 2 μg. Figures of western (a, d) and northern blot (b) represented the representative data obtained from three independent experiments. [Correction added on 22 February 2020, after first online publication: figure image and caption have been amended.]
Interferon‐stimulated gene 20 suppresses the EnhII/Cp activity through its binding
To determine how ISG20 inhibited viral enhancer activity, we identified which region is targeted by ISG20. We constructed the several mutant plasmids with serial deletions of the HBV enhancer and determined the activity of the enhancer mutants (Fig. 3a). ISG20 decreased the activities of all deletion mutants by up to 80% except the EnhI·ΔEnhII construct (Fig. 3b), indicating that the EnhII/Cp region is a main target of ISG20‐mediated HBV inhibition.
Interferon‐stimulated gene 20 shows 3′ to 5′ exonuclease activity towards RNA and DNA in vitro,32 suggesting that interactions occur between ISG20 and HBV DNA. Accordingly, ChIP assay was performed to identify the interaction between ISG20 and EnhII/Cp region. As shown in Figure 3c, ISG20 bound to the EnhII/Cp region. In HBV infection system, we finally examined whether ISG20 implicates in the covalently close circular DNA (cccDNA) degradation. Before and after HBV infection, the transfected ISG20 slightly decreased the HBV DNA but the cccDNA level was not reduced (Fig. 3d,e). Taken together, these results indicate that ISG20 inhibits HBV replication by decreasing the transcription of HBV RNA from cccDNA through its binding.
Interferon‐stimulated gene 20 is involved in interferon‐β‐mediated suppression of hepatitis B virus
Several studies reported that ISG20 is induced by Type I and II IFNs.4, 5, 33 Consistently with previous studies, the mRNA and protein level of ISG20 were increased after treatment with Type I and II IFNs, particularly IFN‐β (Fig. 4a). To determine whether ISG20 mediates the antiviral effect of IFN‐β against HBV, we assessed the secretion of viral antigens following IFN‐β treatment with and without siISG20. The levels of viral antigens were restored by knockdown of ISG20 (Fig. 4b). Moreover, the expression of core protein was also rescued in HepG2‐NTCP‐shISG20 cells (Fig. S2C). These results indicate that endogenous ISG20 may be involved in IFN‐β‐mediated inhibition of HBV. To validate the physiological relevance of the anti‐HBV activity of ISG20, we examined the relationship between ISG20 expression and phases of CHB patients who are considered as IC, IA (also termed immune clearance), and IT (GSE65359). We observed that ISG20 expression was significantly upregulated in IC and IA patients compared with IT patients (P < 0.001 and P < 0.0001, Fig. 4c). This finding indicates that ISG20 expression is associated with HBV elimination following the natural history of chronic HBV infection.
Figure 4.
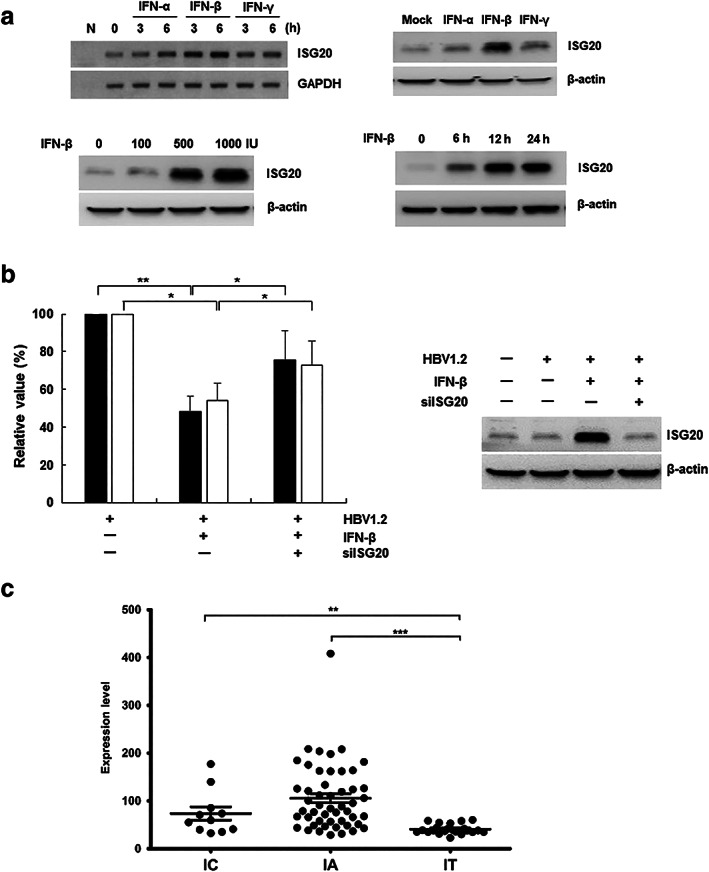
Interferon‐stimulated gene 20 (ISG20) is involved in interferon (IFN)‐β‐mediated suppression of hepatitis B virus (HBV). (a) The mRNA and protein level of ISG20 induced by IFNs. To detect the mRNA level of ISG20, HepG2 cells were treated with the indicated IFNs (100 IU) for 3 or 6 h (left upper panel). N, negative control. The indicated IFNs (500 IU) were treated for 24 h (right panel). In a dose‐dependent manner, IFN‐β was treated for 24 h. Figure represented the representative data obtained from three independent experiments. (b) Effect of siISG20 (20 nM) on IFN‐β‐mediated suppression of HBV.  , HBeAg; □, HBsAg. Values represent the mean ± SD calculated from at least three independent experiments. *
P < 0.05, **
P < 0.005. P values were determined using unpaired t‐test. (c) Expression level of ISG20 during natural course of chronic hepatitis B in patients. IA, immune activation; IC, inactive carrier; IT, immune tolerance. **
P < 0.001, ***
P < 0.0001. [Correction added on 22 February 2020, after first online publication: figure image and caption have been amended.]
, HBeAg; □, HBsAg. Values represent the mean ± SD calculated from at least three independent experiments. *
P < 0.05, **
P < 0.005. P values were determined using unpaired t‐test. (c) Expression level of ISG20 during natural course of chronic hepatitis B in patients. IA, immune activation; IC, inactive carrier; IT, immune tolerance. **
P < 0.001, ***
P < 0.0001. [Correction added on 22 February 2020, after first online publication: figure image and caption have been amended.]
Discussion
Interferons play an essential role in the innate immune response against viral infection. IFNs are induced by pathogen recognition receptors, which recognize viral RNA and show antiviral effects on their target during various stages of the viral life cycle.1, 34 In HBV infection, IFNs exert a broad spectrum of anti‐HBV activity. IFN‐γ participated in cytotoxic T lymphocyte‐mediated HBV clearance through a noncytopathic mechanism in transgenic mouse35, 36 and reduced cccDNA levels in HBV infected hepatocytes.37 IFN‐α regulates HBVs at various stages of the HBV life cycle such as by reducing cccDNA, viral transcription, and capsid assembly.38, 39, 40, 41 However, the details of IFN‐β‐mediated suppression of HBV are largely unknown. Although IFN‐β inhibited HBV replication by disrupting the capsid,40, 41 the precise mechanism and downstream molecules of IFN‐β inhibitory effect on HBV have not been identified. Here, we found that ISG20 was mainly induced IFN‐β in hepatoma cells (Fig. 4a) and strongly inhibited the HBV replication (Fig. 1c). Furthermore, IFN‐β‐mediated suppression of viral antigen secretion was restored by knockdown of endogenous ISG20 (Fig. 4b). These data suggest that ISG20 is one of the downstream molecules that participate in IFN‐β‐mediated antiviral effects on HBV infection.
Previous studies reported that ISG20 inhibited the replication of several RNA viruses via its exonuclease activity. Positive‐strand RNA virus infection such as hepatitis C virus, bovine viral diarrhea virus, yellow fever virus, hepatitis A virus, and HIV‐1 were inhibited by overexpressed ISG20, which depended on its exonuclease activity.6, 7, 8 In West Nile virus and dengue virus infection, ISG20 inhibited replication steps in protein and RNA biosynthesis10 through its enzymatic activity. However, these studies only showed that antiviral activity was blocked by overexpression of mutant ISG20 lacking nuclease activity but did not reveal ISG20‐mediated viral RNA degradation. Interestingly, ISG20 inhibited influenza A virus replication by impairing polymerase activity through interactions with nucleoproteins.9 A recent study reported that ISG20 restricted chikungunya virus and Venezuelan equine encephalitis virus replication by inhibiting translation of infecting genomic RNA.42 Moreover, ISG20D94G, the enzymatic inactive form, showed anti‐HBV activity through binding to the epsilon structure of HBV pgRNA without RNA degradation.33 Our data also demonstrated that ISG20D94G decreased the enhancer activity by targeting the EnhII/Cp region (Fig. S3). In addition, ISG20 suppressed HBV transcription by interacting with the viral enhancer (Fig. 3c). These studies support that ISG20 has antiviral function without exonuclease function.
Interferon‐stimulated gene 20, a 3′‐5′ exonuclease, was first identified as novel ISG induced by Type I and II IFN in various cell lines.4 ISG20 mainly localizes in the nucleolus and Cajal bodies.42, 43 In crystal structure analysis, the active site of ISG20 was found to be very similar to the DEDDh group of DNase.44 Moreover, our data showed that ISG20 directly interacts with HBV EnhII/Cp region (Fig. 3c). These findings suggest that ISG20 may be involved in the cccDNA degradation binding to HBV DNA. Thus, we investigated cccDNA degradation by ISG20 in HepG2‐NTCP cells. Unexpectedly, ectopic expression of ISG20 did not affect the cccDNA level (Fig. 3d,e). However, recent studies reported that IFN‐α and its agonist reduced the cccDNA level through induction of IFN signaling molecules such as APOBEC3 deaminases and several ISGs,39, 45 suggesting that depletion of cccDNA by ISG20 could require for several host factors.
Several studies reported that various molecules are involved in cytokine‐mediated inhibition of HBV transcription. Interleukin‐32, induced by IFN‐γ and TNF‐α, controls HBV transcription by downregulating HNF1a and HNF4a in hepatoma cells and primary human hepatocytes.46 Hepatocystin contributes to IFN‐γ‐mediated anti‐HBV activity by decreasing viral enhancer activity by regulating HNF4a expression,47 and p22‐FLIP is involved in TNF‐α‐mediated suppression of HBV transcription through dysregulating HNF3β and HNF4α.20 Additionally, IFN‐α and IFN‐γ‐induced tripartite motif‐containing 22 impaired HBV enhancer activity, which depended on the nuclear‐located RING domain.48 In this context, our results demonstrate that ISG20 induced by IFN‐β suppressed HBV transcription by interacting with the EnhII/Cp region (Figs 3c and 4a). Our findings reveal a novel mechanism of cytokine‐mediated inhibition of HBV transcription.
Hepatitis B virus is a partially 3.2‐kb double‐stranded DNA virus that chronically infects hepatocytes.49 The natural history of chronic HBV infection is dependent on the interaction between the host immune response and HBV and divided into four phases, IT, IA (or immune clearance), IC, and reactivation, which are defined by serum alanine aminotransferase, serum HBV DNA levels, and HBeAg status.24, 25 Although these phases are defined based on clinical and not immunological measures, the IA phase involves an elevated immune response to HBV, and the IC phase involves HBeAg seroconversion to reduce HBV DNA.24 To search for the host factors involved in the phases of chronic HBV infection, previous researchers investigated immune response‐associated genes and signaling pathways in the liver and peripheral blood mononuclear cells from different phases in patients with CHB.22, 50 However, these studies focused on immune response signaling‐mediated hub genes or serum cytokines induced by leukocytes. Thus, we analyzed the level of ISG20 expression using a GEO dataset (GSE65359) to investigate the physiological relevance of the anti‐HBV activity of ISG20 and found that ISG20 was increased in IC and IA patients compared with IT patients (Fig. 4c), suggesting that ISG20 is responsible for HBV clearance in CHB patients.
In this study, we determined the antiviral effect of ISG20 on HBV replication. We showed that ISG20 is induced by IFN‐β in HepG2 cells and that it strongly inhibits HBV transcription and replication. Additionally, ISG20 was found to be involved in IFN‐β‐mediated suppression of HBV. Further studies revealed that ISG20 inhibits the HBV enhancer activity by directly binding to the EnhII/Cp region, leading to inhibition of viral transcription and replication. Finally, we found that ISG20 expression is associated with HBV elimination following the natural history of chronic HBV infection in CHB patients. Our findings reveal a novel mechanism by which IFN‐β suppresses HBV infection.
Supporting information
Figure S1. ISG20 inhibits HBV antigen secretion in a time‐dependent manner.
Figure S2. The knock‐down of ISG20 using shRNA restores the inhibition of HBV infection by IFN‐β.
Figure S3. ISG20D94G decreases viral enhancer activity by targeting Enh II/Cp region.
Acknowledgments
This work was supported by the intramural fund (grant numbers 2017‐NI51002‐00 and 2019‐NI065‐00) from Korea National Institutes of Health.
Park, Y. K. , Lee, S. Y. , Lee, A. R. , Kim, K.‐C. , Kim, K. , Kim, K.‐H. , and Choi, B.‐S. (2020) Antiviral activity of interferon‐stimulated gene 20, as a putative repressor binding to hepatitis B virus enhancer II and core promoter. Journal of Gastroenterology and Hepatology, 35: 1426–1436. 10.1111/jgh.14986.
Declaration of conflict of interest: The authors who have taken part in this study declared that they do not have any conflict of interest with respect to this manuscript.
References
- 1. Sen GC. Viruses and interferons. Annu. Rev. Microbiol. 2001; 55: 255–281. [DOI] [PubMed] [Google Scholar]
- 2. Schneider WM, Chevillotte MD, Rice CM. Interferon‐stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 2014; 32: 513–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zheng Z, Wang L, Pan J. Interferon‐stimulated gene 20‐kDa protein (ISG20) in infection and disease: review and outlook. Intractable Rare Dis Res. 2017; 6: 35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gongora C, David G, Pintard L et al Molecular cloning of a new interferon‐induced PML nuclear body‐associated protein. J. Biol. Chem. 1997; 272: 19457–19463. [DOI] [PubMed] [Google Scholar]
- 5. Espert L, Degols G, Gongora C et al ISG20, a new interferon‐induced RNase specific for single‐stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 2003; 278: 16151–16158. [DOI] [PubMed] [Google Scholar]
- 6. Zhou Z, Wang N, Woodson SE et al Antiviral activities of ISG20 in positive‐strand RNA virus infections. Virology 2011; 409: 175–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang D, Guo H, Xu C et al Identification of three interferon‐inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 2008; 82: 1665–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Espert L, Degols G, Lin YL, Vincent T, Benkirane M, Mechti N. Interferon‐induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 2005; 86: 2221–2229. [DOI] [PubMed] [Google Scholar]
- 9. Qu H, Li J, Yang L, Sun L, Liu W, He H. Influenza A virus‐induced expression of ISG20 inhibits viral replication by interacting with nucleoprotein. Virus Genes 2016; 52: 759–767. [DOI] [PubMed] [Google Scholar]
- 10. Jiang D, Weidner JM, Qing M et al Identification of five interferon‐induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 2010; 84: 8332–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Degols G, Eldin P, Mechti N. ISG20, an actor of the innate immune response. Biochimie 2007; 89: 831–835. [DOI] [PubMed] [Google Scholar]
- 12. Prehaud C, Megret F, Lafage M, Lafon M. Virus infection switches TLR‐3‐positive human neurons to become strong producers of beta interferon. J. Virol. 2005; 79: 12893–12904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yuan J, Cahir‐McFarland E, Zhao B, Kieff E. Virus and cell RNAs expressed during Epstein‐Barr virus replication. J. Virol. 2006; 80: 2548–2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zahoor MA, Xue G, Sato H, Murakami T, Takeshima SN, Aida Y. HIV‐1 Vpr induces interferon‐stimulated genes in human monocyte‐derived macrophages. PLoS ONE 2014; 9: e106418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc. Natl. Acad. Sci. U. S. A. 2004; 101: 6669–6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xiao C, Qin B, Chen L, Liu H, Zhu Y, Lu X. Preactivation of the interferon signalling in liver is correlated with nonresponse to interferon alpha therapy in patients chronically infected with hepatitis B virus. J. Viral Hepat. 2012; 19: e1–e10. [DOI] [PubMed] [Google Scholar]
- 17. Lu X, Qin B, Ma Q, Yang C, Gong XY, Chen LM. Differential expression of ISG20 in chronic hepatitis B patients and relation to interferon‐alpha therapy response. J. Med. Virol. 2013; 85: 1506–1512. [DOI] [PubMed] [Google Scholar]
- 18. Leong CR, Funami K, Oshiumi H et al Interferon‐stimulated gene of 20 kDa protein (ISG20) degrades RNA of hepatitis B virus to impede the replication of HBV in vitro and in vivo. Oncotarget 2016; 7: 68179–68193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Michailidis E, Pabon J, Xiang K et al A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci. Rep. 2017; 7: 16616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park YK, Park ES, Kim DH et al Cleaved c‐FLIP mediates the antiviral effect of TNF‐α against hepatitis B virus by dysregulating hepatocyte nuclear factors. J. Hepatol. 2016; 64: 268–277. [DOI] [PubMed] [Google Scholar]
- 21. Lee AR, Lim KH, Park ES et al Multiple functions of cellular FLIP are essential for replication of hepatitis B virus. J. Virol. 2018; 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu H, Li F, Zhang X et al Differentially expressed intrahepatic genes contribute to control of hepatitis B virus replication in the inactive carrier phase. J Infect Dis 2018; 217: 1044–1054. [DOI] [PubMed] [Google Scholar]
- 24. Croagh CM, Lubel JS. Natural history of chronic hepatitis B: phases in a complex relationship. World J. Gastroenterol. 2014; 20: 10395–10404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Trepo C, Chan HL, Lok A. Hepatitis B virus infection. Lancet 2014; 384: 2053–2063. [DOI] [PubMed] [Google Scholar]
- 26. Moolla N, Kew M, Arbuthnot P. Regulatory elements of hepatitis B virus transcription. J. Viral Hepat. 2002; 9: 323–331. [DOI] [PubMed] [Google Scholar]
- 27. Su H, Yee JK. Regulation of hepatitis B virus gene expression by its two enhancers. Proc. Natl. Acad. Sci. U. S. A. 1992; 89: 2708–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lopez‐Cabrera M, Letovsky J, Hu KQ, Siddiqui A. Multiple liver‐specific factors bind to the hepatitis B virus core/pregenomic promoter: trans‐activation and repression by CCAAT/enhancer binding protein. Proc. Natl. Acad. Sci. U. S. A. 1990; 87: 5069–5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shlomai A, Paran N, Shaul Y. PGC‐1α controls hepatitis B virus through nutritional signals. Proc. Natl. Acad. Sci. U. S. A. 2006; 103: 16003–16008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. U. S. A. 2001; 98: 1841–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zheng Y, Li J, Ou JH. Regulation of hepatitis B virus core promoter by transcription factors HNF1 and HNF4 and the viral X protein. J. Virol. 2004; 78: 6908–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen LH, Espert L, Mechti N, Wilson DM 3rd. The human interferon‐ and estrogen‐regulated ISG20/HEM45 gene product degrades single‐stranded RNA and DNA in vitro. Biochemistry 2001; 40: 7174–7179. [DOI] [PubMed] [Google Scholar]
- 33. Liu Y, Nie H, Mao R et al Interferon‐inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem‐loop structure of viral RNA. PLoS Pathog. 2017; 13: e1006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schulz KS, Mossman KL. Viral evasion strategies in type I IFN signaling—a summary of recent developments. Front. Immunol. 2016; 7: 498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guidotti LG, Ando K, Hobbs MV et al Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 1994; 91: 3764–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guidotti LG, Ishikawa T, Hobbs MV, Matzke B, Schreiber R, Chisari FV. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 1996; 4: 25–36. [DOI] [PubMed] [Google Scholar]
- 37. Xia Y, Stadler D, Lucifora J et al Interferon‐γ and tumor necrosis factor‐α produced by T cells reduce the HBV persistence form, cccDNA, without cytolysis. Gastroenterology 2016; 150: 194–205. [DOI] [PubMed] [Google Scholar]
- 38. Belloni L, Allweiss L, Guerrieri F et al IFN‐α inhibits HBV transcription and replication in cell culture and in humanized mice by targeting the epigenetic regulation of the nuclear cccDNA minichromosome. J. Clin. Invest. 2012; 122: 529–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lucifora J, Xia Y, Reisinger F et al Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science 2014; 343: 1221–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pasquetto V, Wieland SF, Uprichard SL, Tripodi M, Chisari FV. Cytokine‐sensitive replication of hepatitis B virus in immortalized mouse hepatocyte cultures. J. Virol. 2002; 76: 5646–5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wieland SF, Guidotti LG, Chisari FV. Intrahepatic induction of alpha/beta interferon eliminates viral RNA‐containing capsids in hepatitis B virus transgenic mice. J. Virol. 2000; 74: 4165–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weiss CM, Trobaugh DW, Sun C et al The interferon‐induced exonuclease ISG20 exerts antiviral activity through upregulation of type I interferon response proteins. mSphere 2018; 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Espert L, Eldin P, Gongora C et al The exonuclease ISG20 mainly localizes in the nucleolus and the Cajal (Coiled) bodies and is associated with nuclear SMN protein‐containing complexes. J. Cell. Biochem. 2006; 98: 1320–1333. [DOI] [PubMed] [Google Scholar]
- 44. Horio T, Murai M, Inoue T, Hamasaki T, Tanaka T, Ohgi T. Crystal structure of human ISG20, an interferon‐induced antiviral ribonuclease. FEBS Lett. 2004; 577: 111–116. [DOI] [PubMed] [Google Scholar]
- 45. Furutani Y, Toguchi M, Shiozaki‐Sato Y et al An interferon‐like small chemical compound CDM‐3008 suppresses hepatitis B virus through induction of interferon‐stimulated genes. PLoS ONE 2019; 14: e0216139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kim DH, Park ES, Lee AR et al Intracellular interleukin‐32γ mediates antiviral activity of cytokines against hepatitis B virus. Nat. Commun. 2018; 9: 3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shin GC, Ahn SH, Choi HS et al Hepatocystin contributes to interferon‐mediated antiviral response to hepatitis B virus by regulating hepatocyte nuclear factor 4α. Biochim. Biophys. Acta 1842; 2014: 1648–1657. [DOI] [PubMed] [Google Scholar]
- 48. Gao B, Duan Z, Xu W, Xiong S. Tripartite motif‐containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear‐located RING domain. Hepatology (Baltimore, Md). 2009; 50: 424–433. [DOI] [PubMed] [Google Scholar]
- 49. Seeger C, Mason WS. Molecular biology of hepatitis B virus infection. Virology 2015; 479‐480: 672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vanwolleghem T, Hou J, van Oord G et al Re‐evaluation of hepatitis B virus clinical phases by systems biology identifies unappreciated roles for the innate immune response and B cells. Hepatology (Baltimore, Md). 2015; 62: 87–100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. ISG20 inhibits HBV antigen secretion in a time‐dependent manner.
Figure S2. The knock‐down of ISG20 using shRNA restores the inhibition of HBV infection by IFN‐β.
Figure S3. ISG20D94G decreases viral enhancer activity by targeting Enh II/Cp region.


