SUMMARY
Terpenoid metabolism plays vital roles in stress defense and the environmental adaptation of monocot crops. Here, we describe the identification of the terpene synthase (TPS) gene family of the panicoid food and bioenergy model crop foxtail millet (Setaria italica). The diploid S. italica genome contains 32 TPS genes, 17 of which were biochemically characterized in this study. Unlike other thus far investigated grasses, S. italica contains TPSs producing all three ent‐, (+)‐ and syn‐copalyl pyrophosphate stereoisomers that naturally occur as central building blocks in the biosynthesis of distinct monocot diterpenoids. Conversion of these intermediates by the promiscuous TPS SiTPS8 yielded different diterpenoid scaffolds. Additionally, a cytochrome P450 monooxygenase (CYP99A17), which genomically clustered with SiTPS8, catalyzes the C19 hydroxylation of SiTPS8 products to generate the corresponding diterpene alcohols. The presence of syntenic orthologs to about 19% of the S. italica TPSs in related grasses supports a common ancestry of selected pathway branches. Among the identified enzyme products, abietadien‐19‐ol, syn‐pimara‐7,15‐dien‐19‐ol and germacrene‐d‐4‐ol were detectable in planta, and gene expression analysis of the biosynthetic TPSs showed distinct and, albeit moderately, inducible expression patterns in response to biotic and abiotic stress. In vitro growth‐inhibiting activity of abietadien‐19‐ol and syn‐pimara‐7,15‐dien‐19‐ol against Fusarium verticillioides and Fusarium subglutinans may indicate pathogen defensive functions, whereas the low antifungal efficacy of tested sesquiterpenoids supports other bioactivities. Together, these findings expand the known chemical space of monocot terpenoid metabolism to enable further investigations of terpenoid‐mediated stress resilience in these agriculturally important species.
Keywords: crop stress resilience, natural products, pathway discovery, plant specialized metabolism, Setaria italica, terpene synthases
Significance Statement
The diverse class of terpenoids serves important functions in inter‐organismal and environmental interactions in many plant species. The discovery of terpenoid metabolic pathways in the model crop foxtail millet (Setaria italica) provides foundational knowledge of the diversity of terpenoid metabolism in poaceous food and bioenergy crops for possible future application of these natural defense mechanisms for improving crop traits.
INTRODUCTION
Foxtail millet (Setaria italica) is a grain crop of the grass family (Panicoideae; Poaceae) that was domesticated from its wild relative green foxtail (Setaria viridis) in northern China over 8700 years ago (Lu et al., 2009). Valued for their nutritional benefits, climate resilience and low‐input cultivation, foxtail millet and related small millets are cultivated in many arid and semi‐arid regions of Asia, southern Europe, India, South America and North Africa (Goron and Raizada, 2015; Pant et al., 2016). In addition, their relatively short lifecycle and small diploid genomes (about 500 Mb) have made S. italica and S. viridis well‐suited as model species for related food and bioenergy crops (Doust et al., 2009; Bennetzen et al., 2012; Huang et al., 2016).
While the molecular mechanisms underlying environmental resilience in millets have remained largely unknown, related monocot crops, especially rice (Oryza sativa) and maize (Zea mays), deploy species‐specific mixtures of terpenoid metabolites as core modulators of both biotic and abiotic stress defenses (Schmelz et al., 2014; Murphy and Zerbe, 2020). Rice accumulates distinct groups of stress‐inducible diterpenoid phytoalexins with potent antimicrobial efficacy against major rice pathogens such as rice blast (Magnaporthe oryza) (Xu et al., 2012; Bagnaresi et al., 2012; Han et al., 2015; Lu et al., 2018). Among these bioactive diterpenoids, momilactones further demonstrate allelopathic functions (Kato‐Noguchi and Peters, 2013; Toyomasu et al., 2014). Species‐specific arrays of stress‐elicited volatile sesquiterpenoids and non‐volatile diterpenoids have also been identified in maize and shown to confer substantial resilience to various fungal pathogens, including species of Fusarium, Aspergillus and Colletotrichum, as well as against herbivore pests such as the European corn borer (Ostrinia nubilalis) and lepidopteran larvae (Harris et al., 2005; Schnee et al., 2006; Köllner et al., 2008; Dafoe et al., 2011; Huffaker et al., 2011; Schmelz et al., 2011; Köllner et al., 2013; Ding et al., 2017; Mafu et al., 2018; Ding et al., 2019). Beyond the functions of terpenoids in biotic stress responses, the accumulation of terpenoids in response to abiotic stressors has been reported in maize and rice (Kodama et al., 1988; Horie et al., 2015; Vaughan et al., 2015; Mafu et al., 2018). In addition, terpenoid‐deficient maize mutants show increased drought susceptibility, and high CO2 levels have a negative impact on terpenoid production and stress resistance, supporting the importance of terpenoids also in abiotic stress tolerance (Vaughan et al., 2014; Christensen et al., 2018).
Despite their structural diversity, all terpenoids are derived from only a few acyclic prenyl pyrophosphate precursors that differ by the number of condensed five‐carbon isoprenoid units (C10, C15, C20, etc.) (Chen et al., 2011). Downstream of this central precursor pool, the vast chemical space of species‐specific terpenoids is largely determined by terpene synthases (TPSs), cytochrome P450 monooxygenases (P450s) and other modifying enzyme classes (Banerjee and Hamberger, 2018; Karunanithi and Zerbe, 2019). Most commonly, TPSs catalyze the cyclization and rearrangement of their respective prenyl pyrophosphate substrates to generate a range of mono‐ (C10), sesqui‐ (C15) and di‐ (C20) terpenoid scaffolds (Chen et al., 2011). The majority of these enzymes represent class I TPSs that initiate substrate conversion through cleavage of the pyrophosphate leaving group. Uniquely, in angiosperms the formation of labdane diterpene scaffolds, which include kauranes, pimaranes, abietanes and related terpene groups, recruits pairs of class II and class I diTPSs that function sequentially to generate distinct scaffolds (Peters, 2010; Karunanithi and Zerbe, 2019). Here, the central diterpenoid precursor, geranylgeranyl pyrophosphate (GGPP), first undergoes protonation‐dependent cyclization by a class II diTPS to generate bicyclic prenyl pyrophosphate intermediates of different normal (+)‐, ent‐ or syn‐stereochemistry. Ionization‐dependent cyclization and rearrangement of the resulting intermediate by class I diTPSs then yields an array of distinct labdane scaffolds. Functional decoration of these terpene building blocks, typically initiated by oxygenation through the activity of cytochrome P450 monooxygenases (P450), ultimately generates the broad structural and functional diversity of plant terpenoids (Banerjee and Hamberger, 2018).
Enabled by the increasing availability of genomic resources, the TPS families and selected terpenoid metabolic members of the vast P450 families have been identified in several crop species of the Panicoideae, including rice, maize, wheat (Triticum aestivum) and switchgrass (Panicum virgatum) (reviewed in Schmelz et al., 2014; Murphy and Zerbe, 2020). Functional enzyme characterization has provided detailed insights into the expansion of the TPS and P450 families through repeated duplication and functional diversification of ancestral gibberellin (GA) biosynthetic genes that forms the basis for the evolution of species‐specific terpenoid networks as critical components of monocot stress resilience (Schmelz et al., 2014; Zi et al., 2014).
To deepen our understanding of the diversity of terpenoid metabolism in monocot crops, this study describes the genome‐wide discovery of the S. italica TPS family. Biochemical characterization of 17 TPSs and a diterpenoid‐metabolic P450 (CYP99A17) demonstrates the presence of both common and species‐specific terpenoid pathways. Terpenoid bioactivity assays combined with patterns of stress‐elicited terpenoid biosynthesis suggest possible roles for S. italica terpenoid metabolism in both biotic and abiotic stress responses.
RESULTS
Identification of TPS candidates
To identify the S. italica TPS gene family, we mined publicly available genome data (v.2.2; https://phytozome.jgi.doe.gov) using BLAST analysis against a curated protein database of known plant TPSs (Zerbe et al., 2013). A total of 39 genes with best matches to known TPSs were identified, of which 32 represented full‐length sequences (Table S1 in the online Supporting Information). Most TPS genes mapped to S. italica chromosomes 1, 6 and 7, whereas no TPS genes were located on chromosomes 4 and 5 (Table S2). Next, analysis of signature sequence motifs and phylogenetic comparison with TPSs from related monocot crops were used to functionally categorize the identified gene candidates. The presence of the catalytic DxDD motif and lack of a DDxxD motif characteristic for class I catalysis, combined with a close phylogenetic relationship to known TPSs of the TPS‐c clade, supported class II diTPS activity for SiTPS6, SiTPS9, SiTPS34 and SiTPS35 (Figures 1a, S1 and S2). Specifically, SiTPS34 and SiTPS35 clustered most closely with characterized ent‐copalyl pyrophosphate (CPP) synthases of general metabolism (e.g. Z. mays ANTHER EAR 1, ZmAn1) or specialized metabolism (e.g. Z. mays ANTHER EAR 2, ZmAn2), respectively, and contained the catalytic His–Asn dyad conserved among ent‐CPP synthases involved in GA biosynthesis (Harris et al., 2005; Köksal et al., 2014; Potter et al., 2014; Lemke et al., 2019), suggesting related functionalities (Figures 1a and S1). By contrast, SiTPS6 and SiTPS9 lacked the His–Asn motif, and phylogenetic analysis placed both enzymes with class II diTPSs of specialized metabolism, such as the maize 8,13‐CPP synthase ZmCPS4 and the wheat (+)‐CPP synthase TaCPS2 (Wu et al., 2012; Murphy et al., 2018) (Figures 1a and S1). The remaining 28 TPS candidates were designated as putative class I TPSs on account of the presence of only the conserved C‐terminal DDxxD motif (Figure S2). Of these TPSs, five clustered with diTPSs of the TPS‐e/f clade (Figure 1a). A close phylogenetic relationship of SiTPS28 and SiTPS29 to ZmTPS1, ZmKSL3 and ZmKSL5 indicated possible ent‐kaurene synthase activity for the encoded enzymes. This is further supported by the presence of a catalytic isoleucine residue previously shown to be conserved among ent‐kaurene synthases (Xu et al., 2007a; Jia and Peters, 2016) (Figure S2). By contrast, SiTPS5, SiTPS8 and SiTPS13 were most closely related to class I diTPSs of known functions in specialized metabolism (Figure 1a). Notably, SiTPS5 and SiTPS8 clustered on a separate branch distant from other specialized diTPSs. In addition, SiTPS8 was the only S. italica diTPS candidate featuring the βα‐bi‐domain architecture more commonly present in mono‐ and sesqui‐TPSs. Similar bi‐domain diTPS have also been discovered in a range of other plant species including wheat and switchgrass (Hillwig et al., 2011; Zhou et al., 2012; Pelot et al., 2018). However, SiTPS8 does not phylogenetically cluster with the respective bi‐domain diTPSs from wheat (TaKSL5; Hillwig et al ., 2011) and switchgrass (PvKS3/4; Pelot et al ., 2018) (Figure 1a). Outside the TPS‐e/f clade, SiTPS1 and SiTPS2 were placed with members of the TPS‐g clade that predominantly contains class I TPSs producing acyclic terpenes (Gao et al., 2018). The remaining 23 class I TPS candidates were assigned to the TPS‐a2 clade comprising monocot sesqui‐TPSs (Figure 1a). Putative functions of these TPSs in sesquiterpenoid metabolism were further supported by the absence of the N‐terminal RRx8W motif characteristic of mono‐TPSs and the lack of plastidial transit peptides in at least 19 of the identified TPS candidates. Close phylogenetic relationships of several S. italica TPS candidates with known sesqui‐TPSs from related monocot species, such as maize and switchgrass, suggested similar sesqui‐TPS activities (Schnee et al., 2002; Köllner et al., 2008a; Köllner et al., 2008b, Köllner et al., 2009; Muchlinski et al., 2019).
Figure 1.
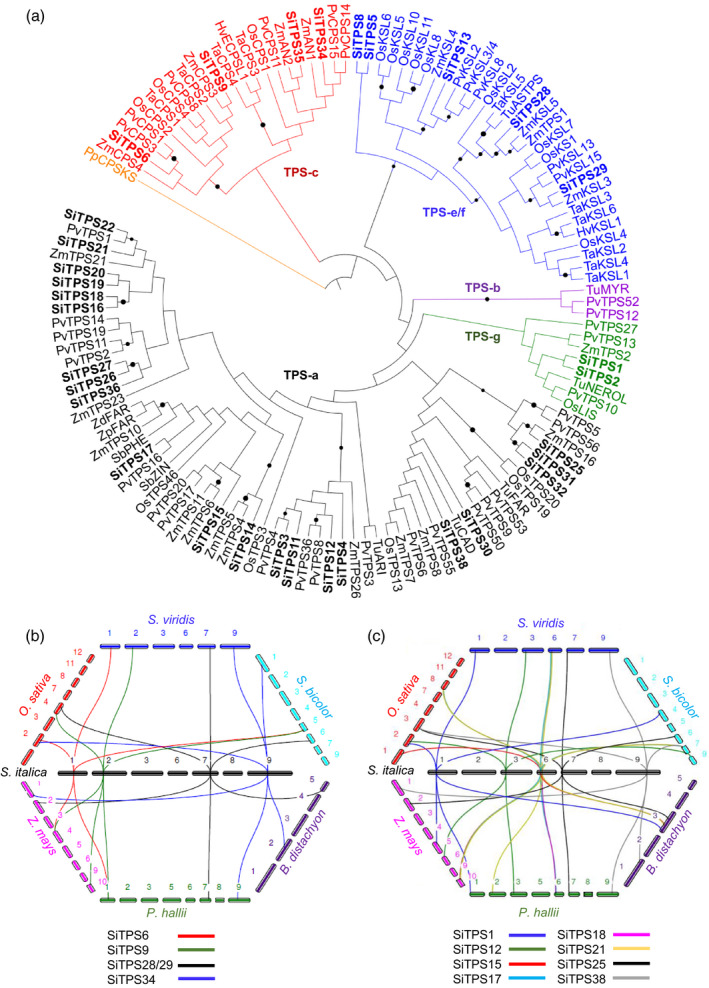
Phylogenetic and gene synteny analysis of Setaria italica terpene synthases (TPSs).
(a) Maximum likelihood phylogeny of S. italica TPSs (bold) and selected TPS‐c, TPS‐e/f, TPS‐g and TPS‐a2 clade TPSs from related poaceous crop species. Tree rooted with the ent‐CPP/ent‐kaurene synthase from Physcomitrella patens, PpCPS/KS. Black circles denote bootstrap support of > 80% (1000 repetitions). Accession numbers and protein sequences are given in Table S1.
(b), (c) Synteny and orthology of diterpene synthases (b) and sesquiterpene synthases (c) between S. italica, Setaria viridis and related diploid poaceous grasses. All chromosomes for Zea mays (magenta), Panicum hallii (green), Brachypodium distachyon (purple), Sorghum bicolor (cyan), S. viridis (blue) and Oryza sativa (red) are arranged in a polygon to emphasize synteny with S. italica (black). Chromosome IDs are given on each line segment. A syntenic network is depicted as a colored line specific to a TPS gene located on S. italica chromosomes (black) connecting to respective syntenic orthologs on chromosomes of other genomes (Table S3).
Having identified gene candidates for the S. italica TPS family, we next applied the same strategy to identify related TPS genes in the wild ancestor S. viridis. Consistent with the close evolutionary relationship between both species (Huang et al., 2016), the S. viridis genome (v.1.1; https://phytozome.jgi.doe.gov) contains 34 putative full‐length TPS genes. High protein sequence identities of 86–100% between these genes and their respective S. italica homologs. with the majority of amino acid variation occurring in regions outside the predicted active site domains, suggested identical or closely related catalytic activities of the TPSs in these two species (Figure S3). To further examine the evolutionary interrelations of the S. italica and S. viridis TPS families with TPSs in closely related diploid poaceous species, including maize, rice, sorghum, Panicum hallii (diploid relative of the allotetraploid Panicum virgatum) and Brachypodium distachyon, gene synteny studies were conducted (Figure 1b,c, Table S3). Comparison of the syntenic orthologs across the respective genomes demonstrated that, with the exception of SiTPS19, SiTPS26 and SiTPS31, all S. italica TPS loci had syntenic orthologs on S. viridis chromosomes. The degree of syntenic ortholog conservation for S. italica TPS candidates on other genomes varied, with 53% to P. hallii, 38% to B. distachyon, 59% to rice, 59% to sorghum and 50% to maize (Table S3). Of the 32 S. italica TPS candidates, 13 TPS syntenic networks were conserved across almost all genomes analyzed in this study (Figure 1b,c, Table S3). Of the five conserved diTPS syntenic networks, those originating from S. italica genes (SiTPS29 and SiTPS34) were observed among the genomes of all species, whereas SiTPS6 and SiTPS9 were common to all but the B. distachyon genome and SiTPS28 was common to all but the O. sativa genome (Figure 1b). Similarly, among the eight conserved S. italica class I TPS syntenic networks, SiTPS1, SiTPS21, SiTPS25 and SiTPS38 showed gene synteny across all investigated genomes, while SiTPS12 and SiTPS17 lacked syntenic orthologs only in the genome of B. distachyon, and SiTPS15 and SiTPS18 lacked syntenic orthologs only in the genome of Sorghum bicolor or Z. mays, respectively (Figure 1c). All 13 of these TPS syntenic networks were highly conserved among the genomes of S. italica and S. viridis, whereas 11 were conserved between the genomes of S. italica and P. hallii, consistent with their close evolutionary relatedness. Syntenic TPS networks on S. italica chromosomes 3, 6, 7 and 9 are mostly conserved on the respective homologous chromosomes 3, 6, 7 and 9 of S. viridis and P. hallii, consistent with the expected collinearity between genomes of closely related species (Table S3). Select syntenic network associations included genes of known function, including switchgrass 1,8‐cineole synthase (PvTPS3, chromosome 3), β‐bisabolene synthase (PvTPS17, chromosome 6), and a germacrene d‐synthase (PvTPS55, chromosome 9) (Muchlinski et al., 2019). Additionally, TPSs on S. italica chromosome 6 featured syntenic orthologs mapping to one chromosome in S. viridis (chromosome 6), P. hallii (chromosome 6), B. distachyon (chromosome 3), S. bicolor (chromosome 7) and Z. mays (chromosome 10), the latter including ZmTPS10 producing α‐bergamotene and β‐farnesene (Köllner et al., 2009). In contrast, TPSs on S. italica chromosome 7 have syntenic orthologs mapping to one chromosome in S. viridis (chromosome 6), S. bicolor (chromosome 6) and Z. mays (chromosome 2), but different chromosomes in P. hallii (chromosomes 2 and 7), B. distachyon (chromosomes 3 and 5) and O. sativa (chromosomes 3, 4 and 8), indicating diverse chromosomal origins for evolutionary hotspots for TPS genes across the Poaceae (Table S3). Interestingly, SiTPS34 (chromosome 9) with a predicted ent‐CPP synthase function, was placed in a syntenic network with known ent‐CPP synthases from rice (OsCPS1, chromosome 2), maize (ZmAN1, chromosome 1) and switchgrass (PvCPS14/15, chromosome 9) (Bensen et al., 1995; Toyomasu et al., 2015; Pelot et al., 2018). Similarly, the predicted ent‐ kaurene synthases SiTPS28/29 (chromosome 7) clustered in a syntenic network with ent‐kaurene synthases from rice (OsKS1, chromosome 4), maize (ZmKSL3/5, chromosome 2) and switchgrass (PvKSL13/15, chromosome 7). Additionally, S. italica class II diTPS candidates potentially involved in specialized metabolism, such as SiTPS6, were in syntenic networks with the ent‐CPP synthase of specialized metabolism in rice (OsCPS2, chromosome 2) and 8,13‐CPP synthases from maize (ZmCPS4, chromosome 4) and switchgrass (PvCPS3, chromosome 1) (Prisic et al., 2004; Murphy et al., 2018; Pelot et al., 2018). Likewise, SiTPS9 was placed in a syntenic network with ZmCPS4 and the maize (+)‐CPP synthase ZmCPS3 (chromosome 10) (Murphy et al., 2018), PvCPS3 (Pelot et al., 2018) and the rice syn‐CPP synthase OsCPS4 (chromosome4) (Xu et al., 2004; Otomo et al., 2004).
Functional characterization of class II diTPSs from S. italica
To biochemically analyze the predicted TPS activities we focused on the S. italica TPS family on account of the agricultural relevance of this species. Enzyme functional studies were conducted via in vivo combinatorial expression assays using Nicotiana benthamiana or Escherichia coli as platforms (Zerbe et al., 2013; Kitaoka et al., 2015a), dependent on the relative activity of individual TPS candidates in each host system. Consistent with their predicted activity as CPP synthases, expression of the class II diTPSs SiTPS9, SiTPS34 and SiTPS35 in N. benthamiana showed that these three enzymes produced CPP as compared to the authentic product of the maize ent‐CPP synthase, ZmAn2 (Harris et al., 2005) (Figure 2a). To verify the stereochemistry of the respective CPP products, each enzyme was further co‐expressed with class I diTPSs specific to CPP of ent‐ or normal (+)‐stereochemistry, represented by an ent‐kaurene synthase from Grindelia robusta (GrEKS) (Zerbe et al., 2013) converting ent‐CPP into ent‐kaurene and a class I diTPS from Marrubium vulgare, MvELS, that converts (+)‐CPP into miltiradiene (Zerbe et al., 2014). This modular co‐expression approach revealed that SiTPS34 and SiTPS35 showed coupled activity only with GrEKS, identifying both enzymes as ent‐CPP synthases (Figure 2b). Conversely, SiTPS9 showed activity only in the pairwise reaction with MvELS, verifying the SiTPS9 product as (+)‐CPP (Figure 2c). Small quantities of ent‐kaurene observed in the product profiles of SiTPS34 and SiTPS35 probably result from the activity of endogenous ent‐kaurene synthases in N. benthamiana (Zerbe et al., 2013). Expression of SiTPS6 in N. benthamiana did not yield detectable products. However, the use of E. coli co‐expression assays revealed that SiTPS6 produced syn‐CPP as verified by comparison with the product of the rice syn‐CPP synthase, OsCPS4, and characteristic product mass ions of m/z 192 and 177 (Xu et al., 2004) (Figure 2d). Mass spectra of enzyme products are given in Figure S4.
Figure 2.
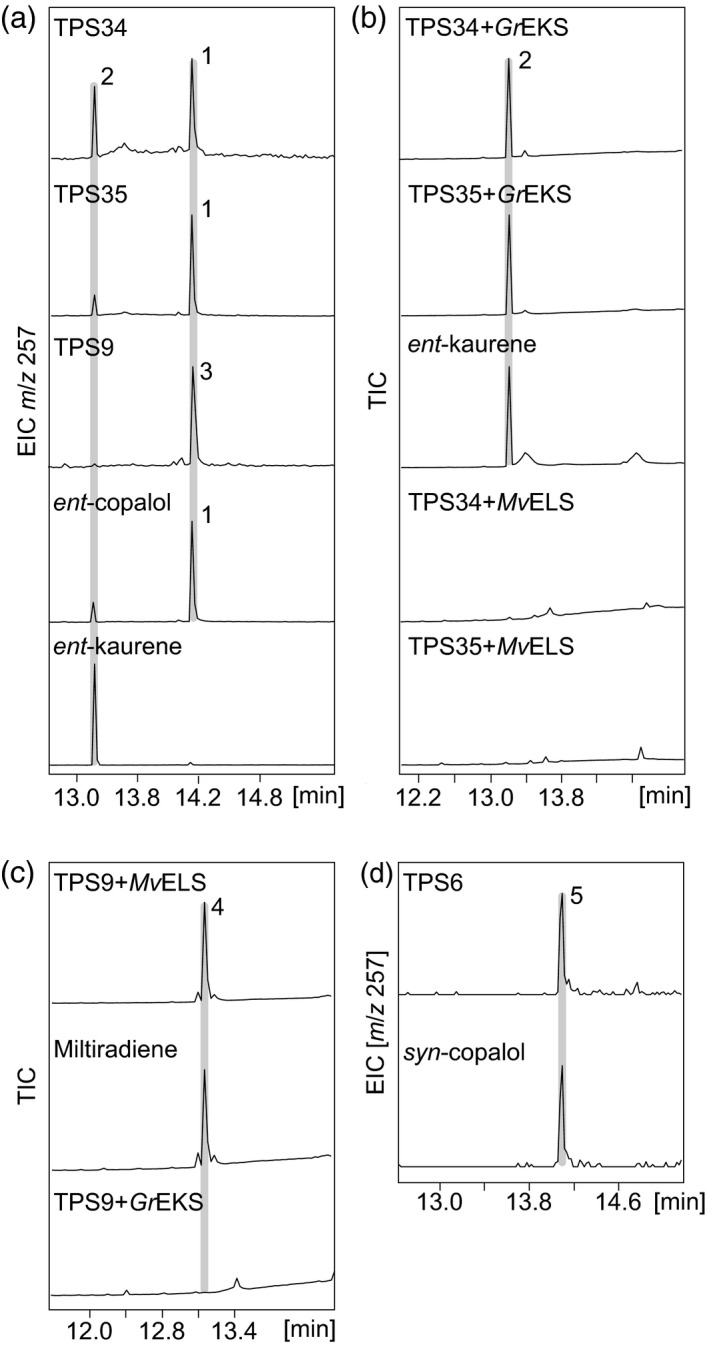
The GC‐MS analysis of Setaria italica class II diterpene synthase (diTPS) reaction products.
(a) The GC‐MS extracted ion chromatograms (EIC, m/z 257) of reaction products from Nicotiana benthamiana co‐expression assays of the class II diTPS candidates SiTPS9, SiTPS34 and SiTPS35. Enzyme products were identified by comparison with the product of the known ent‐copalyl pyrophosphate synthase (CPS), ZmAn2 (Harris et al., 2005).
(b) The GC‐MS total ion chromatograms (TIC) of reaction products from Nicotiana benthamiana co‐expression assays of SiTPS34 and SiTPS35 with the ent‐kaurene synthase from Grindelia robusta (GrEKS) (Zerbe et al., 2015) specific to converting ent‐copalyl pyrophosphate (ent‐CPP) into ent‐kaurene or a class I diTPS from Marrubium vulgare (MvELS) (Zerbe et al., 2014) specific to converting (+)‐CPP into miltiradiene verified the stereochemistry of the SiTPS34 and SiTPS35 products as ent‐CPP.
(c) The GC‐MS TICs of reaction products from N. benthamiana co‐expression assays of SiTPS9 with GrEKS or MvELS identify the SiTPS9 product as (+)‐CPP.
(d) The GC‐MS EICs (m/z 257) of reaction products from Escherichia coli co‐expression assays of SiTPS6 compared with the product of the rice (Oryza sativa) syn‐CPP synthase, OsCPS4, identified the SiTPS6 product as syn‐CPP (Xu et al., 2004). Note that all TPS products were analyzed in the form of their respective dephosphorylated derivatives to enable detection via GC‐MS analysis. Mass spectra for all compounds are given in Figure S2. 1, dephosphorylated ent‐CPP (ent‐copalol); 2, ent‐kaurene; 3, dephosphorylated (+)‐CPP ((+)‐copalol); 4, miltiradiene; 5, dephosphorylated syn‐CPP (syn‐copalol).
Functional characterization of class I diTPSs from S. italica
Having identified the ent‐, (+)‐ or syn‐CPP activity of the four class II diTPSs present in the S. italica genome, we set out to functionally analyze the identified S. italica class I TPSs using co‐expression in N. benthamiana or E. coli. Individual expression of the class I TPS candidates SiTPS1 and SiTPS2 (sharing 82% amino acid sequence identity) in N. benthamiana resulted in the formation of geranyllinalool and minor amounts of trans‐nerolidol, derived from GGPP and FPP, respectively, endogenously produced in N. benthamiana (Figures 3a and S3). Next, to verify the predicted ent‐kaurene synthase activity of the two class I diTPSs SiTPS28 and SiTPS29, co‐expression with the ent‐CPP synthase from maize (ZmAn2) (Harris et al., 2005) or a (+)‐CPP synthase IrTPS3 from Isodon rubescens (Pelot et al., 2017) (used here in place of SiTPS9 due to higher catalytic activity in N. benthamiana) were performed. Both SiTPS28 and SiTPS29 showed activity when combined with ent‐CPP synthases, whereas no class I diTPS products were detected with (+)‐CPP as a substrate, identifying these enzymes as ent‐kaurene synthases (Figures 3b and S5). Close phylogenetic relatedness of SiTPS28 to ZmTPS1 further indicated a possible dual diTPS and sesqui‐TPS activity, as previously demonstrated for ZmTPS1 (Fu et al., 2016). Corresponding E. coli co‐expression assays of SiTPS28 with the maize trans‐FPP synthase, ZmFPPS (Cervantes‐Cervantes et al., 2006), indeed resulted in the formation of trans‐nerolidol in addition to the ent‐kaurene synthase activity of SiTPS28 (Figure 3c).
Figure 3.
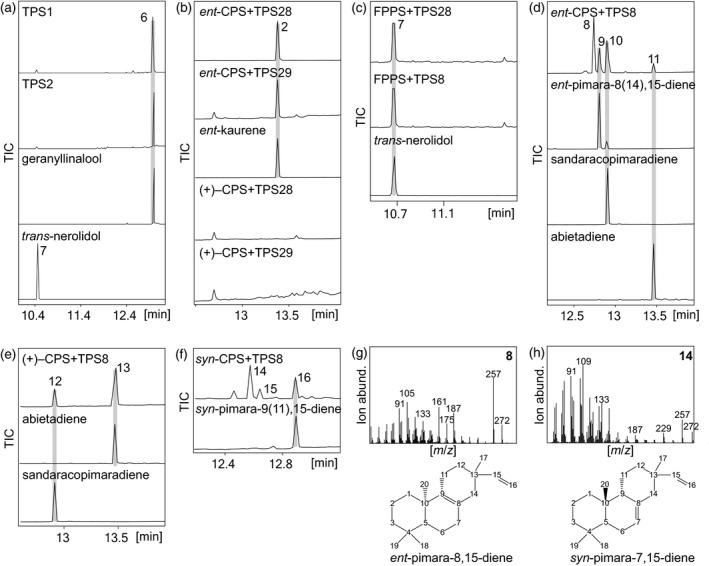
Functional characterization of the Setaria italica class I diterpene synthase (diTPS) candidates.
(a) GC‐MS traces of products resulting from expression of SiTPS1 or SiTPS2 in Nicotiana benthamiana identifying both enzymes as geranyllinalool/trans‐nerolidol synthases. (b) Co‐expression of SiTPS28 or SiTPS29 with the ent‐copalyl pyrophosphate (ent‐CPP) synthase (CPS), ZmAN2 (Harris et al., 2005) or the (+)‐CPP synthase, IrTPS3, from Isodon rubescens (Pelot et al., 2017). (c) The GC‐MS traces of products resulting from Escherichia coli co‐expression assays of SiTPS28 and SiTPS8 with E,E‐farnesyl pyrophosphate (FPP) produced by the maize FPP synthase ZmFPPS (Cervantes‐Cervantes et al., 2006). (d) The GC‐MS traces of products resulting from E. coli co‐expression assays of SiTPS8 with the ent‐CPP synthase ZmAn2. (e) The GC‐MS traces of products resulting from E. coli co‐expression assays of SiTPS8 with the grand fir (Abies grandis) abietadiene synthase variant D621A that produces (+)‐CPP (Cyr et al., 2007; Morrone et al., 2010). (f) The GC‐MS traces of products resulting from E. coli co‐expression of SiTPS8 with the syn‐CPP synthase OsCPS4 (Xu et al., 2004). (g) Mass spectrum of the major product of the coupled reaction of ZmAn2 and SiTPS8 (compound 8) and NMR‐based identification of the product as pimara‐8,15‐diene. (h) Mass spectrum of the primary product of the coupled reaction of OsCPS4 and SiTPS8 (compound 14) and NMR‐verified structure of the product as syn‐pimara‐7,15‐diene. Mass spectra for all detected TPS products are given in Figure S3. 2, ent‐kaurene; 6, geranyllinalool; 7, trans‐nerolidol; 8, ent‐pimara‐8,15‐diene; 9, ent‐pimara‐8(14),15‐diene; 10, ent‐sandaracopimaradiene; 11, ent‐abietadiene; 12, sandaracopimaradiene; 13, abietadiene; 14, syn‐pimara‐7,15‐diene; 15, unknown diterpene; 16, syn‐pimara‐9,11‐diene.
To examine the class I diTPSs with probable functions in specialized diterpenoid metabolism (SiTPS5, SiTPS8, SiTPS13), E. coli co‐expression assays were performed with ent‐, (+)‐ and syn‐CPP as substrates using, as partnering class II diTPSs, the ent‐CPP synthase ZmAn2, the rice syn‐CPP synthase OsCPS4 and a grand fir (Abies grandis) abietadiene synthase variant producing (+)‐CPP (Morrone et al., 2010). No class I diTPS products were detectable when co‐expressing SiTPS5 and SiTPS13 in E. coli with enzymes producing ent‐CPP, (+)‐CPP or syn‐CPP or in N. benthamiana expression assays with the (+)‐CPP synthase SiTPS9 or the ent‐CPP synthases SiTPS34/35 (Figure S6). In contrast, SiTPS8 showed catalytic promiscuity, converting all CPP stereoisomers (Figure 3d,f). The SiTPS8‐catalyzed conversion of ent‐CPP yielded multiple diterpene products, including ent‐pimara‐8(14),15‐diene, ent‐sandaracopimaradiene, ent‐abietadiene and a major product (product 8) that featured dominant mass ions of m/z 133, 161, 187, 257 and 272, indicating a related diterpene olefin (Figure 3d,g). To structurally identify product 8 via NMR analysis, in excess of 1 mg of the product was generated using large‐scale E. coli co‐expression assays and purified by silica chromatography and semi‐preparative HPLC (Murphy et al., 2019). The purified product was then subject to 1D (1H and 13C) and 2D NMR (HSQC, COSY, HMBC, H2BC) analyses that identified the compound as ent‐pimara‐8,15‐diene, tentatively assigning the same ent‐stereochemistry at C9 (13CH2 20.67 p.p.m.) and C10 (13CH3 19.43 p.p.m.) based on ent‐CPP as the initial substrate. Similarly, stereochemistry of methyl groups at C18 (CH3, 13C 21.68 p.p.m.) and C19 (CH3, 13C 33.25 p.p.m.) was tentatively assigned based on ent‐CPP as substrate and mutual HMBC correlations to each other as well as C3 (CH2, 13C 41.86 p.p.m.), C4 (C, 13C 33.32 p.p.m.) and C5 (CH, 13C 51.82 p.p.m.), whereas the defining double bond between quaternary carbons C8 (13C 124.15 p.p.m.) and C9 (13C 136.61 p.p.m.) was verified by HMBC correlations with the neighboring C14 (CH2, 13C 42.48 p.p.m.), as well as the double bond between C15 (CH, 13C 149.45 p.p.m.) and C16 (CH2, 13C 109.43 p.p.m.) with HMBC correlation to C17 (CH3, 13C 23.20 p.p.m.) (Figures 3g and S7). SiTPS8‐catalyzed conversion of (+)‐CPP yielded abietadiene and sandaracopimaradiene as a minor byproduct (Figure 3e). Additionally, co‐expression of SiTPS8 with a syn‐CPP synthase yielded syn‐pimara‐9(11),15‐diene and two unidentified diterpene products (Figure 3f). The major product (compound 14) featured characteristic labdane diterpene mass fragments of m/z 229, 257 and 272. The NMR analysis of the purified compound identified it as syn‐pimara‐7,15‐diene as based on a comparison with published NMR spectra (Mafu et al., 2016; Ye et al., 2017) (Figures 3h and S8). Beyond its diTPS activity, co‐expression of SiTPS8 with ZmFPPS resulted in trans‐nerolidol, highlighting the substrate promiscuity of this enzyme (Figure 3c). Mass spectra of all identified class I TPS products are given in Figure S5.
Functional characterization of selected class I sesquiterpene synthases from S. italica
Activity screening of selected S. italica sesqui‐TPS candidates using N. benthamiana and E. coli co‐expression with ZmFPPS showed several different TPS activities (Figure 4). Nicotiana benthamiana expression revealed SiTPS15 as an α‐bisabolol synthase, SiTPS20 as a β‐caryophyllene synthase, SiTPS19 and SiTPS27 as δ‐cadinene synthases and SiTPS26 as a germacrene d‐synthase compared with commercial standards, while SiTPS38 produced germacrene‐d‐4‐ol as verified by comparison with the product of a δ‐cadinene synthase variant (GhCAD:N403P/L405H) from cotton (Gossypium arboreum) (Yoshikuni et al., 2006) (Figures 4a,e and S5). Co‐expression of enzymes not showing activity in N. benthamiana with ZmFPPS in E. coli identified SiTPS21 as a cubebol synthase as validated by comparison with the product profile of cubebol synthase from Coniophora puteana (Mischko et al., 2018) (Figures 4e,f and S5). The primary product of SiTPS25 featured mass ions of m/z 161, 207, 222 and 240 (Figure 4g,h), which closely matched the fragmentation pattern of the sesquiterpene alcohol eudesme‐2,11‐diol, recently identified in maize (Liang et al., 2018). Additional 2D NMR analysis (H2BC and HMBC) of the purified compound verified the presence of two hydroxyl groups at quaternary carbons C2 (C, 13C 74.257 p.p.m.) and C11 (C, 13C 73.090 p.p.m.) and identified the product as eudesme‐2,11‐diol (Figures 4h and S9).
Figure 4.
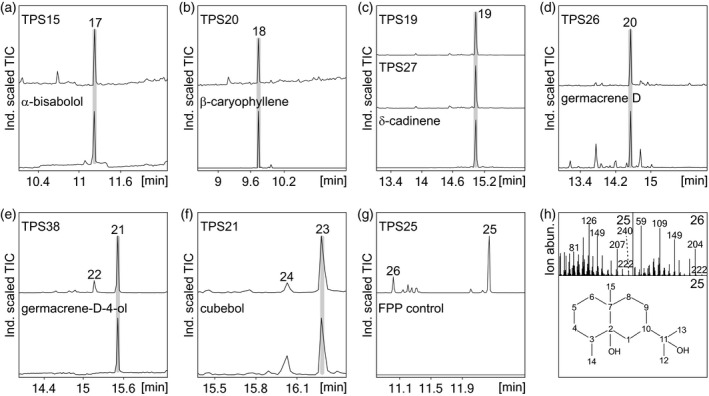
Functional characterization of the Setaria italica class I sesquiterpene synthases.
(a)–(e) The GC‐MS traces of reaction products resulting from Nicotiana benthamiana co‐expression assays of the focal S. italica sesquiterpene synthases with product identification by comparison with authentic standards identified SiTPS15 as an α‐bisabolol synthase (a), SiTPS20 as a β‐caryophyllene synthase (b), SiTPS19 and SiTPS27 as δ‐cadinene synthases (c), SiTPS26 as a germacrene d‐synthase (d) and SiTPS38 as a germacrene‐d‐4‐ol synthase (e). (f), (g) The GC‐MS traces of reaction products resulting from Escherichia coli co‐expression assays of the maize E,E‐farnesyl pyrophosphate (E,E‐FPP) synthase, ZmFPPS (Cervantes‐Cervantes et al., 2006), with focal S. italica sesquiterpene synthases identified SiTPS21 as a cubebol synthase (f), whereas SiTPS25 produced two distinct products, compound 25 and a minor byproduct, compound 26 (g). (h) Mass spectrum of compounds 25 and 26, and NMR‐verified structure of compound 25 identifying the SiTPS25 product as eudesme‐2,11‐diol. Mass spectra for all detected compounds are given in Figure S3. 17, α‐bisabolol; 18, β‐caryophyllene; 19, δ‐cadinene; 20, germacrene D; 21, germacrene‐D‐4‐ol; 22, unidentified terpene product; 23, cubebol; 24, unidentified terpene product; 25, eudesme‐2,11‐diol; 26, unidentified terpene product.
Identification of SiCYP99A17 as a P450 catalyzing C19‐hydroxylation of labdane scaffolds
Monocot diterpenoids are almost invariably functionally modified by P450 enzymes, with members of the CYP701, CYP71, CYP76 and CYP99 subfamilies having been shown to function in rice and maize diterpenoid metabolism (Swaminathan et al, 2009; Wang et al., 2011; Wu et al., 2011; Wang et al., 2012a; Wang et al., 2012b; Mao et al., 2016; Mafu et al., 2018; Ding et al., 2019). Previous rice studies further revealed patterns of gene co‐regulation and genomic clustering of diTPSs and P450s involved in the biosynthesis of diterpenoid phytoalexins, such as momilactones and phytocassanes (Wilderman et al., 2004; Prisic et al., 2004; Shimura et al., 2007; Swaminathan et al., 2009; Boutanaev et al., 2015). Based on these insights, we analyzed publicly available gene co‐expression data (https://phytozome.jgi.doe.gov) to identify similar diTPS/P450 co‐expression patterns in S. italica. Indeed, significant co‐expression was observed for SiTPS8, SiTPS9 and two P450 candidates, namely Seita.2G145800 and Seita.6G139100, henceforth designated CYP99A17 and CYP99A19, respectively (Figure 5a, Table S1). Both P450s showed best BLAST matches against rice CYP99A2 and CYP99A3 that catalyze the consecutive oxidation at C19 of syn‐pimara‐7,15‐diene in momilactone biosynthesis (Wang et al., 2011; Kitaoka et al., 2015b). Pearson‐correlated gene expression between SiTPS9 and SiTPS8 (0.88), CYP99A17 and SiTPS8 (0.89), CYP99A19 and SiTPS9 (0.87) and CYP99A19 and SiTPS8 (0.91) supported a functional relationship between these genes (Figure 5a). Further mining of the S. italica genome regions upstream and downstream of the identified TPS loci revealed that CYP99A17 was located proximal to SiTPS8 and SiTPS9 on chromosome 2 with a distance of about 550 kb from SiTPS9 and about 590 kb from SiTPS8 (Figure 5b). To biochemically characterize CYP99A17 and CYP99A19, the corresponding genes were synthesized as codon‐optimized and N‐terminally truncated (removal of the predicted endoplasmic reticulum membrane anchor; Table S1). Since SiTPS8 showed correlated gene expression with both CYP99A17 and CYP99A19, combinatorial activity assays were performed for both P450s via E. coli co‐expression with SiTPS8 and an ent‐, syn‐ or (+)‐CPP synthase, thus encompassing all specialized labdane diterpene scaffolds identified in this study. No P450 products were observed for CYP99A19 with any of the tested diTPS combinations (Figure S10), whereas co‐expression of CYP99A17 showed product formation with all diTPS combinations to form several new products with mass fragments of m/z 288, indicating hydroxylated labdane diterpenoid scaffolds (Figure 5c). Combining HSQC, HMBC and NOESY analyses identified the purified products of the coupled activity of (+)‐CPP synthase, SiTPS8 and CYP99A17 as abietadien‐19‐ol as compared with previously reported NMR data (Lee et al., 2001) (Figures 5d,g and S11), the presence of a hydroxyl group at C19 (CH2OH, 13C 64.99 p.p.m.) and NOESY correlation with C20 (CH3, 13C 14.69 p.p.m.). Low abundance of the product formed by SiTPS8 and CYP99A17 with ent‐CPP as the substrate prevented compound purification and NMR structural verification (Figure 5e). The product resulting from the co‐expression of SiTPS8 and CYP99A17 with a syn‐CPP synthase was tentatively identified via NMR analysis and comparison with reference spectra as syn‐pimara‐7,15‐dien‐19‐ol (Figure 5f,h and S12) (Wang et al ., 2011).
Figure 5.
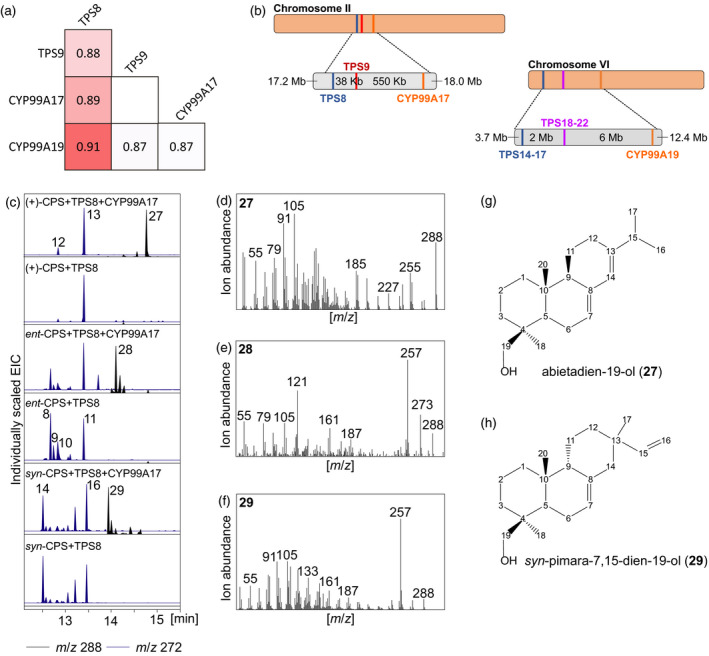
Identification of Setaria italica CYP99A17 as a diterpene monohydroxylase.
(a) Pearson correlation coefficients of SiTPS8, SiTPS9 and CYP99A17 (data obtained from Phytozome, https://phytozome.jgi.doe.gov). (b) Chromosomal location of SiTPS8, SiTPS9 and CYPP99A17 on S. italica chromosome 2. (c) The GC‐MS extracted ion chromatograms (EIC) for dominant mass ions of m/z 272 (blue) and 288 (black) of reaction products resulting from Escherichia coli co‐expression assays of SiTPS8 with (+)‐, ent‐ or syn‐copalyl pyrophosphate (CPP) synthases and CYP99A17. (d)–(f) Mass spectra of SiTPS8‐CYP99A17 products with (+)‐CPP (d), ent‐CPP (e) or syn‐CPP (f) as a substrate. (g) Structure of CYP99A17 product 27 as abietadien‐19‐ol as verified by NMR analysis. (h) Structure of CYP99A17 product 29 as syn‐pimara‐7,15‐dien‐19‐ol as verified by NMR analysis.
Abundance of terpene‐metabolic genes in S. italica
Numerous gene expression studies across poaceous crops have demonstrated stress‐inducible transcriptome accumulation of TPSs and P450s in response to biotic and/or abiotic stressors (reviewed in Schmelz et al., 2014), suggesting a related defensive response in S. italica. To test this hypothesis, we performed quantitative (q)PCR‐based gene expression studies of the identified TPS and P450 genes following different abiotic and biotic stress treatments. Abiotic stresses were applied to roots of 4‐week‐old S. italica plants in the form of oxidative stress by soil treatment with 10 mm CuSO4 for 2 days, in form of drought stress by lack of watering for seven consecutive days and by UV irradiation of leaves for 2 h. In addition, pathogen stress was approximated by treatment of stems of 10‐week‐old S. italica plants with mycoprotein derived from Fusarium venenatum (QuornTM) (Figures 6 and S13). In addition, publicly available data on S. italica tissue‐ and treatment‐specific gene expression (https://phytozome.jgi.doe.gov) were queried. Although comparison of these experimental and public data has to be viewed with caution given the use of, for example, different growth conditions and plant ages at harvest, some notable trends could be observed. SiTPS19 was the only transcript showing significantly induced expression in response to all stresses, except for UV irradiation (Figure 6), despite its apparently higher abundance in leaf and panicle (Figure S13). SiTPS15 was also most abundant in panicles and showed inducible expression patterns upon drought stress and mycoprotein treatment (Figures 6 and S13). All other TPS genes showed significant changes in gene expression predominantly under only one of the applied treatments. SiTPS26 showed the highest transcript changes in response to oxidative stress, although it was most abundant in leaves (Figures 6 and S13). Increases in gene expression in response to drought stress were highest for SiTPS19 and SiTPS35 (Figure 6). Ultraviolet irradiation caused increased transcript abundance most significantly for SiTPS1, SiTPS27 and SiTPS28, consistent with the primary occurrence of these genes in leaf and panicle tissue (Figures 6 and S13). SiTPS8, SiTPS9, SiTPS15, SiTPS19 and SiTPS20 were also most abundant in aerial S. italica organs and showed the most significantly induced gene expression in response to mycoprotein treatment, indicating a possible function in defense against above‐ground pathogens (Figures 6 and S13).
Figure 6.
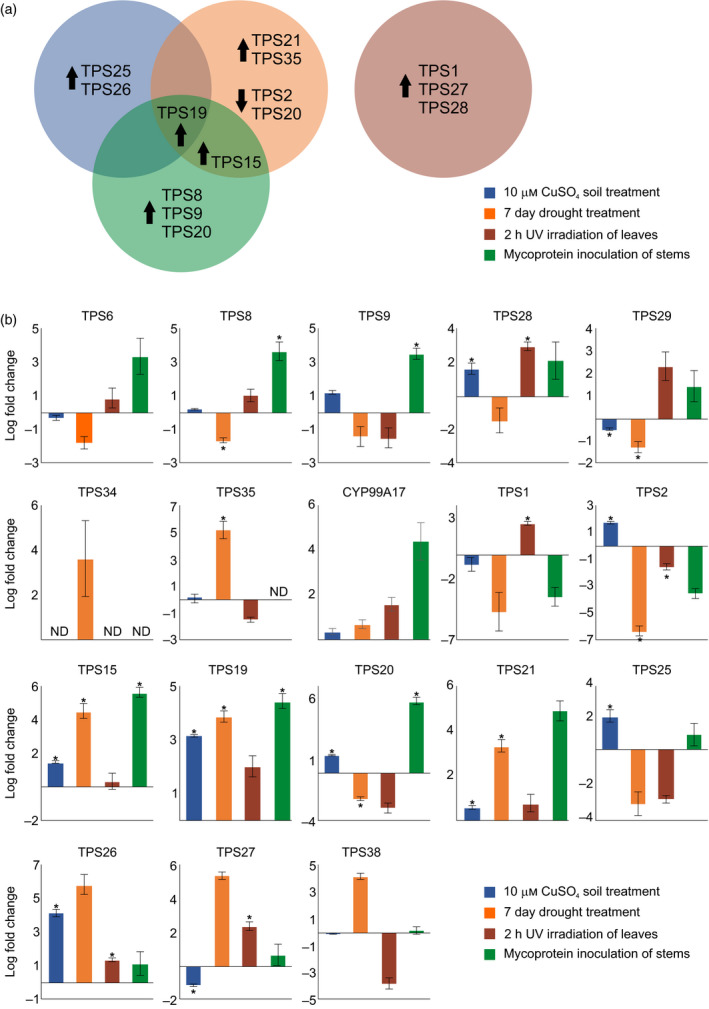
Abundance of terpene synthase genes and CYP99A17 in Setaria italica.
(a) Venn diagram representation of S. italica genes significantly up‐ or downregulated by two‐fold or more in organs. Treatments included root oxidative stress (soil treatment with 10 mm CuSO4 for 48 h; blue), drought stress (seven consecutive days with relative water content = 30%; orange), leaf exposure to UV irradiation (254 nm) for 2 h (purple) and pathogen stress (approximated by stem inoculation with QuornTM mycoprotein from Fusarium venenatum; green). Transcript levels were measured by qPCR gene expression analysis with normalization to the internal reference gene SiEF‐1α (Kumar et al., 2013) using log fold‐change (ΔΔCt) compared with untreated control samples (n = 3). Statistically significant differences between treated and control samples are based on the Welch two‐sample t‐test (P < 0.05; Figure S10).
(b) Heatmap and dendrogram showing gene expression of S. italica terpene synthases (TPSs) based on publicly available transcript data (https://phytozome.jgi.doe.gov/phytomine) with red indicating high expression and blue representing low expression levels.
Occurrence of TPS and P450 products in planta
To examine the presence of enzyme products of stress‐inducible TPSs and CYP99A17 in planta, control and stress‐treated S. italica leaf, stem and root samples used for gene expression studies were subjected to organic solvent extraction and targeted metabolite analysis using ultra performance liquid chromatography (UPLC)–hydrophilic interaction liquid chromatography (HILIC)–tandem mass spectrometry (MS/MS) against the purified enzyme products as authentic standards. Of the 20 identified TPS products, germacrene‐d‐4‐ol and the diterpene alcohols abietadien‐19‐ol and syn‐pimara‐7,15‐dien‐19‐ol were detected in the tested plant tissues under both controlled and stress‐treated conditions (Figure S14). Germacrene‐d‐4‐ol was detected exclusively in roots with product identification based on a dominant mass fragment m/z 205.195076 corresponding to the dehydrated protonated adduct ([M‐H2O + H]+, parent mass ion [M]+ = 222.1984) (Figure S14). By contrast, abietadien‐19‐ol and/or syn‐pimara‐7,15‐dien‐19‐ol (parent ion [M]+ m/z 288.2453) were present in leaves, stems and roots as verified by characteristic mass ions m/z 289.2525 and 271.2419 corresponding to the protonated adducts ([M + H]+) and the protonated dehydrated adducts ([M‐H2O + H]+), respectively (Figure S14). However, overlapping retention times and a nearly identical fragmentation pattern of abietadien‐19‐ol and syn‐pimara‐7,15‐dien‐19‐ol prevented an unambiguous verification if both or only one compound occurs in planta. Furthermore, abietadien‐19‐ol and/or syn‐pimara‐7,15‐dien‐19‐ol were detected in both stress‐exposed and control plants with no apparent difference in abundance.
Antifungal activity of terpenes from select S. italica TPSs
Fungal‐elicited transcript accumulation of SiTPS6, SiTPS8, SiTPS9, SiTPS15, SiTPS19, SiTPS20 and SiCYP99A17 suggested possible antifungal bioactivities of the corresponding enzyme products (Figure 6). To investigate possible antifungal activity of the corresponding terpenoids, in vitro fungal growth inhibition assays were performed using commercial standards of α‐bisabolol, β‐caryophyllene and δ‐cadinene, and purified enzyme products of abietadiene, syn‐pimara‐7,15‐diene, abietadien‐19‐ol and syn‐pimara‐7,15‐dien‐19‐ol. The purified SiTPS21 product cubebol proved to be unstable at the assay temperature and was excluded from further analysis. The fungal pathogens Fusarium verticillioides and Fusarium subglutinans were chosen for bioactivity assays, as these species have been shown to cause pathogenic symptoms in S. italica (Nwanma et al., 1991; Besharati fard et al., 2014; Li et al., 2015). Overall, at concentrations of 2 µg ml−1 and 10 µg ml−1 all tested metabolites showed no or moderate inhibitory activity on fungal growth within the 48 h assay duration (Figure 7). The strongest antifungal activity was observed for abietadien‐19‐ol, with a 40% reduction of F. verticillioides and F. subglutinans growth after 48 h at a metabolite concentration of 10 µg ml−1. Interestingly, the corresponding olefin precursor abietadiene exhibited a similar antifungal activity against F. subglutinans but was less effective in inhibiting the growth of F. verticillioides with only a 13% reduction in fungal growth at the same metabolite concentration (Figure 7). syn‐Pimara‐7,15‐dien‐19‐ol showed a comparable antifungal efficacy, inhibiting growth of F. subglutinans by about 40% at both tested metabolite concentrations, but showing less impact on F. verticillioides with only a 6% decrease in fungal growth after 48 h even at the higher metabolite concentration. Unlike abietadiene, the corresponding olefin precursor of syn‐pimara‐7,15‐dien‐19‐ol, syn‐pimara‐7,15‐diene, showed only minor antifungal activity. Regardless of the metabolite concentration tested, the targeted sesquiterpenoids, α‐bisabolol, β‐caryophyllene and δ‐cadinene, reduced fungal growth at lower levels of 10–13% compared with the diterpene alcohols.
Figure 7.
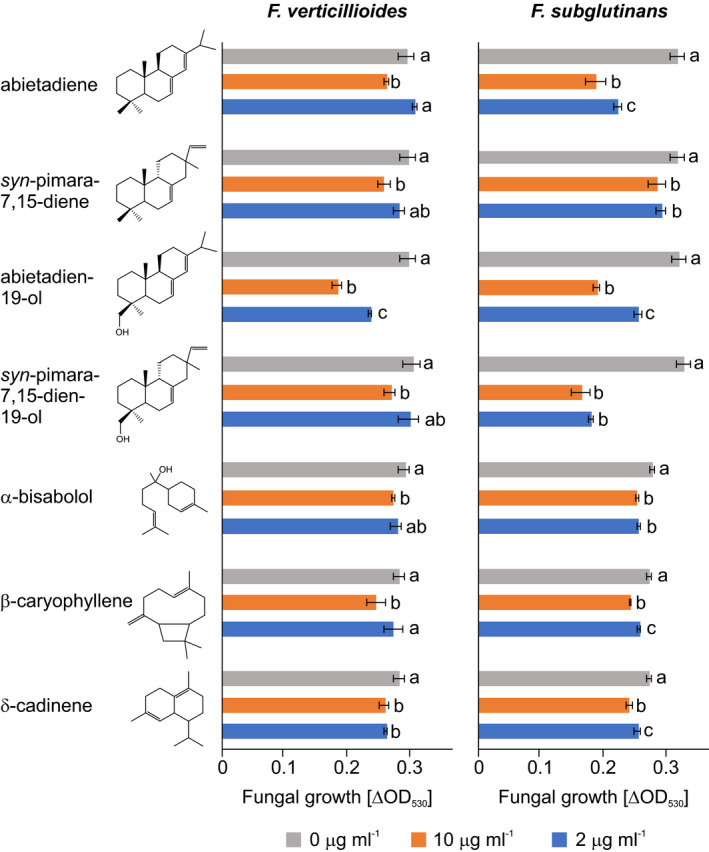
In vitro antifungal activity of selected terpenoid products.
Average growth (OD530) of Fusarium verticillioides and Fusarium subglutinans measured over a 48 h time course in defined minimal broth medium using a microtiter plate assay in the absence and presence of purified terpenoids at a concentration of 0 µg ml−1 (grey), 2 µg ml−1 (blue) and 10 µg ml−1 (orange). Error bars represent propagated SE values (n = 4) and letters represent significant differences at P < 0.05 as measured using analysis of variance and Tukey's honestly significant difference tests to correct for multiple comparisons between control and treatments.
DISCUSSION
Species‐specific terpenoid networks serve as important chemical defense mechanisms in monocot crops against both biotic and abiotic stressors. In particular, the potent pest‐ and disease‐protective functions of terpenoids in maize and rice have been well established (reviewed in Schmelz et al., 2014; Murphy and Zerbe, 2020). In addition, diverse terpenoid metabolic networks comprising both common and unique pathway branches and products have been described in wheat and switchgrass, suggesting similar physiological roles (Wu et al., 2012; Zhou et al., 2012; Pelot et al., 2018; Muchlinski et al., 2019). Continued investigation of the biosynthesis, diversity and function of terpenoid chemical defense mechanisms can enable new strategies for improving crop resilience in the face of projected increased harvest losses caused by shifting environmental pressures and associated biotic and abiotic stressors that can overwhelm the natural defense systems of plants (Chakraborty and Newton, 2011; de Sassi and Tylianakis, 2012; Leng and Hall, 2019). In this context, we report here the discovery and functional analysis of the TPS family of S. italica as a model food and bioenergy species and an important grain resource across Asia and Africa (Huang et al., 2016; Pant et al., 2016).
The S. italica TPS family represents a medium‐sized gene family of 32 members similar to those of related diploid grasses, including maize, rice and wheat (Schmelz et al., 2014; Murphy and Zerbe, 2020). Diterpenoid biosynthesis in S. italica is controlled by four class II diTPSs, two of which, SiTPS34 and SiTPS35, represent catalytically redundant ent‐CPP synthases (Figure 8). Similar ent‐CPP synthase pairs are also present in maize (Bensen et al., 1995; Harris et al., 2005), rice (Prisic et al., 2004) and switchgrass (Pelot et al., 2018), and genetic studies showed these enzyme pairs to be functionally separate, with roles in GA biosynthesis and specialized metabolism, respectively (Bensen et al., 1995; Prisic et al., 2004; Sakamoto et al., 2004; Vaughan et al., 2015). Inducible gene expression of SiTPS35, but not SiTPS34, under drought stress and phylogenetic clustering of SiTPS34 with the generalized GA‐biosynthetic maize proteins ZmAn1 and SiTPS35 with the specialized ZmAn2 indicate a similar functional separation in S. italica. Syntenic conservation of SiTPS34 and the ent‐kaurene synthase SiTPS29 with known or predicted ent‐CPP synthase/ent‐kaurene synthase pairs in all seven analyzed Poaceae genomes support this hypothesis. In contrast, only weak gene synteny was observed for the additional ent‐CPP synthase/ent‐kaurene synthase pair in S. italica, SiTPS35 and SiTPS28, where SiTPS35 lacks syntenic orthologs in multiple species and SiTPS28 lacks a syntenic ortholog in rice. Although elucidation of SiTPS34 and SiTPS35 will require genetic analyses beyond the scope of this study, these combined results suggest that SiTPS34 and SiTPS29 function in GA biosynthesis, whereas SiTPS35 and SiTPS28 are more likely to be involved in specialized diterpenoid metabolism. Dual activity of SiTPS28 in forming ent‐kaurene and converting FPP into trans‐nerolidol also supports this conclusion.
Figure 8.
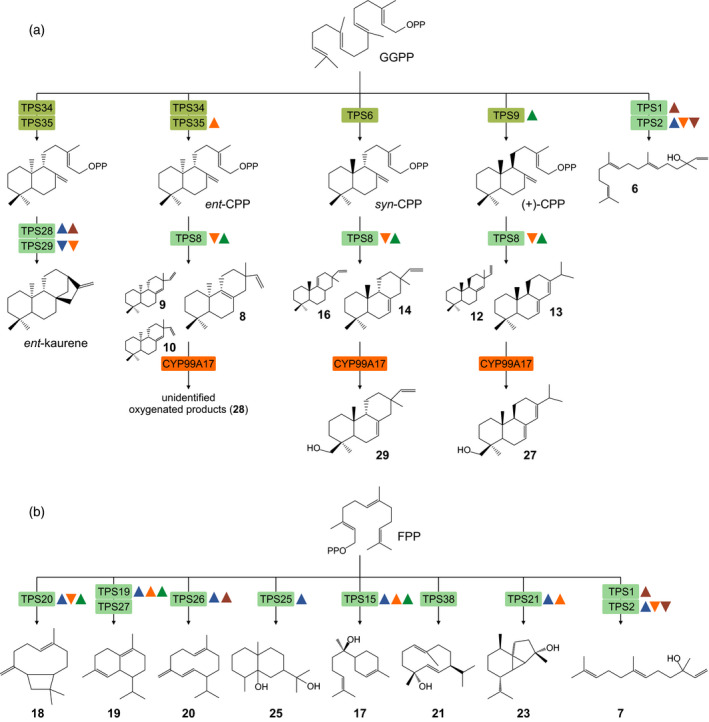
The Setaria italica terpenoid metabolic network.
(a) Scheme of identified diterpenoid biosynthetic pathways in S. italica, involving monofunctional class II and class I diterpene synthases (diTPSs), as well as CYP99A17 that convert the central precursor C20 E,E,E‐geranylgeranyl pyrophosphate (GGPP) into a range of diterpenoid metabolites.
(b) Scheme of sesquiterpenoid biosynthesis in S. italica, involving distinct class I TPSs converting the C15 precursor E,E‐farnesyl pyrophosphate (FPP) into distinct sesquiterpenoid metabolites. Triangles depict genes showing significant stress‐elicited expression in response to root oxidative stress with CuSO4 (blue), root drought stress (orange), leaf UV irradiation (brown), and stem inoculation with Fusarium venenatum‐derived mycoprotein (Quorn, green). 6, geranyllinalool; 7, trans‐nerolidol; 8, ent‐pimara‐8,15‐diene; 9, ent‐pimara‐8(14),15‐diene; 10, ent‐sandaracopimaradiene; 12, abietadiene; 13, sandaracopimaradiene; 14, syn‐pimara‐7,15‐diene; 16, syn‐pimara‐9(11),15‐diene; 17, α‐bisabolol; 18, β‐caryophyllene; 19, δ‐cadinene; 20, germacrene D; 21, germacrene‐D‐4‐ol; 23, cubebol; 25, eudesme‐2,11‐diol; 27, abietadien‐19‐ol; 29, syn‐pimara‐7,15‐dien‐19‐ol.
The additional presence of a syn‐CPP synthase (SiTPS6) and a (+)‐CPP synthase (SiTPS9) sets the catalytic range of class II diTPSs in S. italica apart from all other poaceous crop species investigated so far, which, according to current knowledge, feature enzymes for producing only syn‐CPP (e.g. rice, switchgrass) or (+)‐CPP (e.g. maize, wheat) in addition to the ubiquitously occurring ent‐CPP (Schmelz et al., 2014; Murphy et al., 2018; Pelot et al., 2018). Although syntenic orthologs for SiTPS6 and SiTPS9 were present in all analyzed genomes (except for B. distachyon), the respective maize, rice and switchgrass orthologs did not show functional conservation. For example, the SiTPS6 orthologs of rice (OsCPS2) and maize (ZmCPS4) encode for an ent‐CPP synthase and 8,13‐CPP synthase, respectively, rather than syn‐CPP synthases (Prisic et al., 2004; Murphy et al., 2018). Likewise, of the identified maize and rice orthologs to the (+)‐CPP synthase SiTPS9, only ZmCPS3 encodes for a (+)‐CPP synthase, whereas rice OsCPS4 encodes a syn‐CPP synthase and maize ZmCPS4 forms 8,13‐CPP (Xu et al., 2004; Otomo et al., 2004; Murphy et al., 2018). Close phylogenetic interrelations of SiTPS6 with ZmCPS4 and of SiTPS9 with ZmCPS3 and OsCPS4 are consistent with this functional divergence. Indeed, structure‐guided mutagenesis studies in several poaceous species highlight the ease with which neo‐functionalization of class II diTPSs may occur. For example, single active site residue substitutions are sufficient to alter the product specificity of the switchgrass 8,13‐CPP synthase PvCPS3 to the form (+)‐CPP, and similar active site mutations lead to a re‐programming of the rice OsCPS4 from syn‐CPP to 5,13‐halimadienyl pyrophosphate (HDP) as a major product (Potter et al., 2016; Pelot et al., 2018).
Next to the pair of ent‐kaurene synthases (SiTPS28 and SiTPS29), SiTPS8 showed expansive substrate promiscuity, converting all S. italica CPP stereoisomers into distinct pimaradiene, sandaracopimaradiene and abietadiene scaffolds (Figure 8). Several ent‐CPP‐ and syn‐CPP‐derived SiTPS8 products, including syn‐pimara‐7,15‐diene, ent‐pimara‐8,15‐diene, ent‐sandaracopimaradiene and (+)‐abietadiene, also occur in rice, switchgrass and presumably wheat (Xu et al., 2007b; Zhou et al., 2012; Pelot et al., 2018). Additionally, SiTPS8 formed the previously unrecognized pimarane double bond isomer, ent‐pimara‐8,15‐diene, with the most closely related function described for the wheat class I diTPS, TaKSL4, that produces the corresponding (+)‐stereoisomer (Zhou et al., 2012). Notably, in rice, ent‐sandaracopimaradiene and syn‐pimara‐7,15‐diene serve as precursors to oryzalexins and momilactones, respectively, with have demonstrated functions in pathogen defense (Atawong et al., 2002; Hasegawa et al., 2010; Toyomasu et al., 2014). Abundance in above‐ground tissues and elicitation of SiTPS8 gene expression in response to mycoprotein treatment may point toward similar functions in S. italica. However, unlike the upregulation of momilactone production in rice plants exposed to UV irradiation, SiTPS8 expression was not elicited by UV light, indicating species‐specific differences. Consistent with this hypothesis, SiTPS8 seems to have evolved independently, since it is only distantly related to specialized βα‐domain enzymes of the TPS‐e/f clade (Morrone et al., 2011; Zerbe and Bohlmann, 2015; Pelot et al., 2018), and showed no gene synteny across the analyzed Poaceae genomes.
Downstream of diterpene scaffold formation, functional modifications typically initiated by P450 monooxygenases are key to the various bioactivities of the compound group (Banerjee and Hamberger, 2018). In rice, formation of momilactones from syn‐pimara‐7,15‐diene is catalyzed by members of the CYP99A family (Shimura et al., 2007; Wang et al., 2011). With CYP99A17, we also identified a functional CYP99 enzyme in S. italica, which is capable of converting several SiTPS8 products of ent‐, syn‐ and (+)‐stereochemistry (Figure 8). In addition to its more expansive substrate range, CYP99A17 differs from rice CYP99 enzymes in its catalytic sequence. Unlike rice CYP99A2/3, which oxygenate their respective substrates at C19 to generate the corresponding diterpene acids (Wang et al., 2011), NMR analysis of selected CYP99A17 products showed that the enzyme catalyzed C19 hydroxylation reactions, as exemplified by the diterpene alcohol products syn‐pimara‐7,15‐dien‐19‐ol and abietadien‐19‐ol verified here. Mycoprotein‐elicited gene expression of CYP99A17 in S. italica stems and, albeit moderate as compared with, for example, maize diterpenoids (Schmelz et al., 2011; Mafu et al., 2018), in vitro antifungal activity of abietadien‐19‐ol and syn‐pimara‐7,15‐dien‐19‐ol against F. verticillioides and F. subglutinans support possible roles of these pathway branches in disease resistance, although structurally distinct from those observed in rice. Considering the expansive functional decoration of many antimicrobial diterpenoids, it appears likely that the observed diterpene alcohols do not represent pathway end products but are further modified. The low abundance of abietadien‐19‐ol and/or syn‐pimara‐7,15‐dien‐19‐ol in S. italica tissues is consistent with this hypothesis.
Next to the diversity of diterpenoid products, several TPS enzymes were characterized that produce a range of sesquiterpenoids (Figure 8). Among these, the catalytically redundant SiTPS1 and SiTPS2 of the TPS‐g clade showed dual activity, converting FPP into trans‐nerolidol and GGPP into geranyllinalool. Related TPS‐g enzymes identified, for example, in Arabidopsis, switchgrass, tomato (Solanum lycopersicum) and maize function in forming the geranyllinalool‐derived 4,8,12‐trimethyltrideca‐1,3,7,11‐tetraene (TMTT) with antifeedant activity (Ament et al., 2004; Herde et al., 2008; Richter et al., 2016; Muchlinski et al., 2019). SiTPS1 and SiTPS2 are co‐located on chromosome 1 and show 82% protein sequence identity. Yet, unlike SiTPS1, SiTPS2 lacks syntenic orthologs in several analyzed genomes and appears to be devoid of a plastidial transit peptide, supporting the emergence of SiTPS2 through a more recent gene duplication event and possible alteration of compartmentalization and associated substrate restriction as also proposed for functionally redundant acyclic TPSs in switchgrass and snapdragon (Antirrhinum majus) (Nagegowda et al., 2008; Muchlinski et al., 2019). Similarly, two δ‐cadinene synthases, SiTPS19 and SiTPS27, showed catalytic redundancy, yet distinct patterns of pathway regulation. Here, SiTPS19 expression was elicited in response to several stresses (except for UV irradiation) in roots and stems, whereas SiTPS27 expression was most significantly increased in UV‐stressed leaves. The remaining characterized sesquiterpene synthases appear to occur as single‐copy genes in S. italica. Among these, identification of SiTPS20 as a β‐caryophyllene synthase is consistent with β‐caryophyllene production in rice and maize, where it is associated with volatile above‐ground herbivore defenses (Cheng et al., 2007; Köllner et al., 2008a). Beyond these commonly occurring sesquiterpenoids in monocot crops, several S. italica TPSs formed more unusual sesquiterpene alcohols, including germacrene‐d‐4‐ol, α‐bisabolol, cubebol and eudesme‐2,11‐diol, with as yet unknown biological functions. Production of cubebol and α‐bisabolol is broadly distributed across angiosperm and gymnosperm species, but this is less well described in Poaceous crops (Asadollahi et al., 2010; Kamatou and Viljoen, 2010; Son et al., 2014; Muchlinski et al., 2019). Increased transcript abundance of the α‐bisabolol synthase, SiTPS15, and the cubebol synthase SiTPS21 in response to drought stress and mycoprotein treatment may indicate functions in both biotic and abiotic stress responses. Similarly, detection of germacrene‐d‐4‐ol in extracts of S. italica roots (although at low abundance) and inducible expression of the corresponding biosynthetic gene, SiTPS38, in roots exposed to drought suggest a possible function in below‐ground stress responses. Biosynthesis of the dihydroxylated sesquiterpenoid eudesme‐2,11‐diol has only recently been described in monocots as a function of the maize ZmTPS16 (Liang et al., 2018). The S. italica eudesme‐2,11‐diol synthase, SiTPS25, represents a syntenic ortholog of ZmTPS16, thus indicating a common ancestry. However, contrasting the pathogen‐inducible accumulation of eudesme‐2,11‐diol in maize roots, eudesme‐2,11‐diol was not detected in S. italica tissue extracts and no substantial changes in gene expression were observed under the tested stressors. Interestingly, although germacrene‐d‐4‐ol, α‐bisabolol, cubebol and eudesme‐2,11‐diol are less commonly observed in Poaceous species, the genomic regions encoding the corresponding TPS enzymes are syntenically conserved across the Poaceae species tested here (with the exception of SiTPS15 that lacks an ortholog in S. bicolor). It can be speculated that species‐specific variation in the presence of these sesquiterpenoids is due to functional divergence of several such orthologs. Indeed, the switchgrass syntenic orthologs for SiTPS15 and SiTPS38 encode TPSs that produce structurally related terpenes to their S. italica counterparts, β‐bisabolene and germacrene d, respectively (Muchlinski et al., 2019).
EXPERIMENTAL PROCEDURES
Plant material
Seeds of S. italica (Yugu1) were provided by Dr Katrien Devos (Department of Plant Biology, University of Georgia, USA). Seeds were germinated in Conviron TCR120 growth chambers (https://www.conviron.com/) under a photoperiod of 16 h, 60% relative humidity, 100 μmol m−2sec−1 light intensity and a day/night temperature cycle of 21ºC /18ºC. After 2 weeks, S. italica plants were either maintained in growth chambers for an additional 2 weeks (for drought, UV or CuSO4 treatments) or were grown in a greenhouse using nutrient water solution at an output of about 300 ml day−1 for 10 weeks under an ambient photoperiod and 22°C/17°C day/night temperatures (for mycoprotein assays). Nicotiana benthamiana plants were cultivated in growth chambers for a total of 5 weeks before use for Agrobacterium‐mediated co‐expression studies using A. tumefaciens GV3101.
Gene discovery, synthesis and cloning
The TPS and P450 genes reported in this study were identified by querying the publicly available genomes of S. italica (v.2.2) and S. viridis (v.1.1) (https://phytozome.jgi.doe.gov/pz/portal.html) using Blast search against a manually curated plant TPS database (E‐value threshold of ≤ 1 × 10–30) (Zerbe et al., 2013). For phylogenetic analysis, protein sequence alignments were generated using ClustalW2, curated with Gblocks, and maximum‐likelihood phylogenetic analyses were performed using PhyML‐aBayes v.3.0.1 beta with four rate substitution categories, the LG substitution model, a BIONJ starting tree and 1000 bootstrap repetitions (http://phylogeny.lirmm.fr). The phylogenetic tree was visualized using Interactive Tree of Life (https://itol.embl.de/).
Targeted TPS genes were synthesized as follows. For functional characterization of the encoded enzymes in N. benthamiana, full‐length native sequences were synthesized and inserted into the PacI site of the pLIFE (pCAMBIA130035Su) expression vector. For co‐expression in E. coli N‐terminally truncated (lacking the predicted plastidial transit peptide) and codon‐optimized genes were generated and cloned into the pET‐DUET1 or pET28b(+) vectors (Novagen, https://www.emdmillipore.com/; Sigma, https://www.sigmaaldrich.com/). Gene synthesis was conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, under contract no. De‐AC02‐05CH11231. The P450 genes CYP99A17 and CYP99A19, were synthesized as codon‐optimized and N‐terminally modified (removal of the endoplasmic reticulum membrane anchor) genes and cloned into the pET‐DUET1 vector also containing the Z. mays cytochrome P450 reductase ZmCPR2 for expression in E. coli. (Table S1) (Wang et al., 2011; Mafu et al., 2018). Additional genes used for producing authentic standards, namely G. arboreum germacrene‐d‐4‐ol/δ‐cadinene synthase (NM_001330020) variant N403P/L405H and C. puteana cubebol synthase, Copu3 (XP_007765978), were synthesized for expression in E. coli and cloned into pET‐DUET1 according to published literature (Yoshikuni et al., 2006; Mischko et al., 2018).
Synteny analysis
The latest coding sequence and gene annotations (GFF) for each species were obtained from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html). The GFF files were converted to BED files using the Python package JCVI (https://pypi.org/project/jcvi/). Candidate TPS genes were aligned to the candidate species using the MCscan workflow contained within JCVI. Genes were aligned using a ‘cscore’, the ratio of BLAST hit to the best BLAST hits to either the query or hit with a cutoff at 0.8. The best alignment for each TPS gene was considered a potential ortholog. If a tie occurred, the alignments of the 25 genes both upstream and downstream of the candidate gene were inspected to determine blocks of synteny, which were used to break the tie. All candidate TPS gene alignments were plotted using the graphics library of JCVI utility libraries | Zenodo (https://zenodo.org/record/31631#.XVX8_uhKjct.; accessed 3 August, 2019).
Enzyme characterization in E. coli
For microbial expression assays, individual TPS constructs were transformed into E. coli BL21DE3‐C41 along with plasmids encoding for key genes of the MEP pathway, and the A. grandis GDPP synthase for diterpene production or a Z. mays FPP synthase (ZmFPPS) for sesquiterpene production, as described previously (Cyr et al., 2007; Morrone et al., 2010). Additionally, for coupled SiTPS8 and CYP99A17 (pET‐DUET1:CYP99A17/ZmCPR2C) co‐expression assays in E. coli we used, the ent‐CPP synthase from Z. mays (ZmAn2), the rice syn‐CPP synthase (OsCPS4) or the A. grandis abietadiene synthase variant D621A that produces CPP of (+)‐stereochemistry (Cyr et al., 2007; Morrone et al., 2010; Mafu et al., 2018). Cultures were grown at 37°C and 180 r.p.m. in 50 ml of Terrific Broth (TB) medium to an OD600 of about 0.6 before cooling to 16°C and induction with 1 mm isopropyl‐β‐d‐1‐thiogalactopyranoside for 72 h with the addition of 25 mm sodium pyruvate. For co‐expression assays of P450s, 75 mg L−1 aminolevulinic acid and 5 mg L−1 riboflavin were added during the 72 h incubation. Products were extracted with 50 ml of hexane, air dried, and resuspended in 1 ml of n‐hexane for GC‐MS analysis.
Transient co‐expression in N. benthamiana
The TPS candidates were expressed in N. benthamiana using the pLIFE (pCAMBIA130035Su) vector system in conjunction with the RNA‐silencing suppressor gene p19 (Zerbe et al., 2013). The TPS genes cloned into individual pLIFE expression vectors were transformed separately into A. tumefaciens strain GV3101. Bacterial cultures were grown at 28°C in Luria‐Bertani (LB) medium supplemented with 50 mg L−1 kanamycin, pelleted and resuspended to a final OD600 of 1.0 in 10 mm MES buffer with 10 mm MgCl2. Following incubation for 2 hours at 22°C and gentle shaking, cell suspensions were mixed and syringe‐infiltrated into the underside of the leaves of 5‐week‐old N. benthamiana plants. Transfected plants were maintained for 4 days before metabolites were extracted with 1.5 ml of hexane from a single transformed leaf and analyzed via GC‐MS.
The GC‐MS analysis of enzyme products
Metabolite extracts from plant material or microbial cultures were freed from residual water by the addition of anhydrous Na2SO4, dried under a N2 stream and re‐dissolved in 1 ml of hexane. Extracts from microbial co‐expression assays with CYP99A17 and CYP99A19 were further derivatized using tetramethylsilane (TMS)‐diazomethane (Millipore, https://www.emdmillipore.com/; Sigma). A GC‐MS analysis was performed on an Agilent 7890B GC (https://www.agilent.com/) interfaced with a 5977 Extractor XL MS detector at 70 eV and 1.2 ml min−1 He flow, using a HP5‐ms column (30 m, 250 µm i.d., 0.25 µm film) and the following GC parameters for detection of diterpene products and sesquiterpene products of SiTPS15 and SiTPS20: 50°C for 1 min, 20°C min−1 to 300°C, hold 3 min with pulsed splitless injection at 250°C and 50°C oven temperature. The GC‐MS parameters for detection of sesquiterpene products of SiTPS19, SiTPS21, SiTPS26, SiTPS27 and SiTPS38 were: 50°C for 1 min, 10°C min−1 to 300°C, hold for 3 min with pulsed splitless injection at 250°C and 50°C oven temperature. The MS data from a 90–600 mass‐to‐charge ratio (m/z) were collected after a 9 min solvent delay. Products were identified through comparison of GC‐MS retention time and mass spectra with commercial standards, enzymatically produced authentic compounds or NMR analysis, as stated.
Metabolite profiling via LC‐MS analysis
Plant tissue was ground under liquid N2 and extracted with hexane:ethyl acetate (70:30 v/v) for 12 h. Extracts were dried‐down and resuspended in methanol before UPLC‐HILIC‐MS/MS analysis. All measurements were carried out on a Q Exactive HF mass spectrometer coupled with a Vanquish LC system (Thermo Scientific, https://www.thermofisher.com/). A total injection volume of 2 μl was separated on a Waters Acquity UPLC BEH Amide column (150 × 2.1 mm; 1.7 μm) coupled to an Acquity UPLC BEH Amide VanGuard pre‐column (5 × 2.1 mm; 1.7 μm). The column was maintained at 45°C at a flow rate of 0.4 ml min−1. The mobile phases consisted of: (A) water with 10 mm ammonium formate and 0.125% formic acid and (B) acetonitrile:water (95:5, v/v) with 10 mm ammonium formate and 0.125% formic acid. A 17 min separation was conducted under the following gradient: 0 min 15% B; 0–2 min 100% B; 2–7.7 min 70% B; 7.7–9.5 min 40% B; 9.5–10.25 min 30% B; 10.25–12.75 min 100% B; 12.75–16.75 min 100% B. The Orbitrap MS instrument was operated in positive electrospray ionization (ESI) mode with the following parameters: mass range 60–900 m/z; spray voltage +3.6 kV; sheath gas (nitrogen) flow rate 60 units; auxiliary gas (nitrogen) flow rate 25 units; capillary temperature 320°C; full scan MS1 mass resolving power 30,000; data‐dependent MSMS (dd‐MSMS) three scans per cycle; dd‐MSMS mass resolving power 15,000. Thermo Xcalibur 4.0.27.19 was used for data acquisition and analysis. MS‐DIAL v.3.70 was used for batch data processing.
Product purification and NMR analysis
For NMR analysis of enzyme products >1 mg of target compounds was extracted from 12 L E. coli cultures co‐expressing the desired enzyme combinations. Products were extracted using hexane and purified by silica chromatography and semi‐preparative HPLC as described previously (Murphy et al., 2019). Purified compounds were resuspended in chloroform‐D spiked with 0.03% TMS as internal standard and used to collect 1D (1H, 13C) and 2D (COSY, HSQC, HMBC, selective HSQC, selective HMBC and NOESY) spectra on a Brucker Avance 800 MHz spectrometer (https://www.bruker.com/) equipped with a 5 mm CPTCI cryoprobe. The NMR reports were generated using MestReNova and ACDLabs.
Elicitation of stress responses in S. italica tissues
Abiotic stress treatments were conducted using 4‐week‐old S. italica plants. To mimic below‐ground oxidative stress in S. italica, 10 mm CuSO4 in 300 ml of nutrient solution was added to the soil over a period of 48 h. Control plants were treated with nutrient solution only. For drought stress, watering was withheld for seven consecutive days, resulting in 30% relative water content at the time of harvest. Control plants were well watered to ≥ 80% relative water content at time of harvest. For UV irradiation, detached leaf blades were placed on wet paper towels, irradiated for 2 h under a 254 nm UV lamp situated 20 cm from the leaf surface, and then kept in the dark in a high‐humidity incubator at 30°C until sample collection. As a negative control, leaf blades were handled in the same manner, but without UV treatment. Fungal elicitor assays were performed using mycoprotein commercially produced from F. venenatum by QuornTM (https://www.quorn.us/mycoprotein). Here, stems of 10‐week‐old S. italica plants were cut lengthwise and incubated by wrapping a mycoprotein‐soaked cheesecloth around the incision, covering the incision with tape to retain moisture and incubating for 48 h (Schmelz et al., 2011). Appropriate controls were treated in the same fashion, but with the cheesecloth soaked with water only.
Gene expression analysis
The heatmaps and dendograms were generated using R‐statistical software with pheatmap 1.0.12 from CRAN (https://CRAN.R‐project.org/package=pheatmap) with gene expression data obtained from Phytomine (https://phytozome.jgi.doe.gov/phytomine). Quantitative real‐time PCR was performed with total RNA extracted from 100 mg of S. italica plant material used in stress assays (see above) using the Plant RNA Purification Kit (Qiagen, https://www.qiagen.com/). Equal amounts of RNA were used for cDNA synthesis with the iScript cDNA Synthesis Kit (Bio‐Rad, https://www.bio‐rad.com/) and oligo(dT) primers. The subsequent qPCR reaction was performed on a Bio‐Rad CFX96 real‐time system using the SsoFast kit and target‐specific oligonucleotides (Table S4). The log fold‐change in gene expression was calculated using the ΔΔCt method with S. italica elongation factor 1α (SiEF1α) (XM_004984777) (Kumar et al., 2013) as the reference gene and triplicate measurements with three biological replicates. Target specificity was confirmed by sequence verification of representative amplicons. Statistical significance for gene expression was analyzed using the Welch two‐sample t‐test (P‐value < 0.05).
Fungal growth‐inhibition assays
Fungal stock cultures of F. verticillioides (GL1093) and F. subglutinans (M2236) were grown on potato‐dextrose agar plates for 2 weeks under constant light at room temperature (23ºC) before use in bioactivity assays. The antimicrobial efficacy of purified terpene products was assessed following the Clinical and Laboratory Standards Institute M38‐A2 guidelines, as described previously (Mafu et al., 2018). Briefly, fungal growth at 30°C in RPMI 1640 (ThermoFisher, https://www.thermofisher.com/) medium was monitored using a SPECTRAmax 340 Microplate Reader (Molecular Devices, https://www.moleculardevices.com/) through periodic measurements of changes in OD530 over 48 h. Each well contained 200 µl of initial fungal inoculum (2.5 × 104 conidia ml−1) with 0.4 µl of either pure DMSO or DMSO containing dilutions of HPLC‐purified terpene products, as mentioned above, to yield final concentrations of 2 µg ml−1 and 10 µg ml−1 per well. The statistical significance for difference in OD530 measurements between 0 and 48 h was analyzed using analysis of variance and Tukey honestly significant difference tests (n = 4, P‐value < 0.05).
Accession numbers
Accession numbers for all TPS candidates described in this study are given in Table S1.
Author contributions
PZ conceived the original research and oversaw data analysis; PSK performed most of the experiments; DB assisted with qRT‐PCR; SW assisted with in vitro antifungal assays; JD and JM performed synteny analysis; TS and OF assisted with LC‐MS analyses; PSK and PZ wrote the article with contributions from all authors. All authors read and approved the manuscript.
Conflict of interest
The authors declare that they have no conflicts of interest in accordance with the journal policy.
Supporting information
Figure S1. Protein sequence alignment of select class II diterpene synthases.
Figure S2. Protein sequence alignment of select class I diterpene synthases.
Figure S3. Sequence similarity matrix of terpene synthase candidates from Setaria italica and Setaria viridis.
Figure S4. Mass spectra of class II diterpene synthase products identified in this study.
Figure S5. Mass spectra of class I terpene synthase products identified in this study.
Figure S6. Mass spectra of products resulting from co‐expression assays of SiTPS5 and SiTPS13.
Figure S7. The NMR analysis of ent‐pimara‐8,15‐diene.
Figure S8. The NMR analysis of syn‐pimara‐7,15‐diene.
Figure S9. The NMR analysis of eudesme‐2,11‐diol.
Figure S10. Functional analysis of CYP99A17 and CYP99A19.
Figure S11. The NMR analysis of abietadien‐19‐ol.
Figure S12. The NMR analysis of syn‐pimara‐7,15‐dien‐19‐ol.
Figure S13. Gene expression analysis of characterized Setaria italica terpene synthase genes.
Figure S14. Occurrence of terpene synthase and CYP99A17 products in Setaria italica.
Table S1. Accession numbers and cloned constructs for terpene synthases used in this study.
Table S2. Chromosomal location of Setaria italica terpene synthase candidate genes.
Table S3. Gene synteny analysis of Setaria italica terpene synthase candidates.
Table S4. Oligonucleotides used for gene expression analysis in this study.
Acknowledgements
Financial support for this work, by generous support from the US Department of Energy (DOE) Joint Genome Institute (JGI) DNA Synthesis Science Program (grant no. 2568, to PZ), is gratefully acknowledged. The gene synthesis work conducted by the JGI, a DOE Office of Science User Facility, is supported under contract no. DE‐AC02‐05CH11231. We thank Dr David Nelson (University of Tennessee) for assistance with the annotation of CYP99A17 and CYP99A19, Dr Reuben Peters (Iowa State University) and Dr Jörg Bohlmann (University of British Columbia) for sharing diterpenoid standards, Dr Thomas Gordon (UC Davis) for providing fungal stock cultures and Dr Bennett J. Addison (National Renewable Energy Laboratory) for assistance with NMR structural assignments. We further thank Ms. Puja Dhanota (Encoded Therapeutics Inc., Burlingame, CA United States) for excellent support with project and laboratory support contributing to this work.
References
- Ament, K. , Kant, M.R. , Sabelis, M.W. , Haring, M.A. and Schuurink, R.C. (2004) Jasmonic acid is a key regulator of spider mite‐induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol. 135, 2025–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asadollahi, M.A. , Maury, J. , Schalk, M. , Clark, A. and Nielsen, J. (2010) Enhancement of farnesyl diphosphate pool as direct precursor of sesquiterpenes through metabolic engineering of the mevalonate pathway in Saccharomyces cerevisiae . Biotechnol. Bioeng. 106, 86–96. [DOI] [PubMed] [Google Scholar]
- Atawong, A. , Hasegawa, M. and Kodama, O. (2002) Biosynthesis of rice phytoalexin: enzymatic conversion of 3beta‐hydroxy‐9beta‐pimara‐7,15‐dien‐19,6beta‐olide to momilactone A. Biosci. Biotechnol. Biochem. 66, 566–70. [DOI] [PubMed] [Google Scholar]
- Bagnaresi, P. , Biselli, C. , Orru, L. , Urso, S. , Crispino, L. , Abbruscato, P. , Piffanelli, P. , Lupotto, E. , Cattivelli, L. and Vale, G. (2012) Comparative transcriptome profiling of the early response to Magnaporthe oryzae in durable resistant vs susceptible rice (Oryza sativa L.) genotypes. PLoS ONE, 7, e51609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee, A. and Hamberger, B. (2018) P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem. Rev. 17, 81–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen, J.L. , Schmutz, J. , Wang, H. et al . (2012) Reference genome sequence of the model plant Setaria . Nat. Biotechnol. 30, 555–561. [DOI] [PubMed] [Google Scholar]
- Bensen, R.J. , Johal, G.S. , Crane, V.C. , Tossberg, J.T. , Schnable, P.S. , Meeley, R.B. and Briggs, S.P. (1995) Cloning and characterization of the maize An1 gene. Plant Cell, 7, 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besharati fard, M. , Mohammadi, A. and Darvishnia, M. (2014) Fusarium species associated with foxtail millet (Setaria Italica) in Iran. Intl. J. Adv. Biol. Biomed. Res. 2, 2549–2552. [Google Scholar]
- Boutanaev, A.M. , Moses, T. , Zi, J. , Nelson, D.R. , Mugford, S.T. , Peters, R.J. and Osbourn, A. (2015) Investigation of terpene diversification across multiple sequenced plant genomes. Proc. Natl. Acad. Sci. USA, 11, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes‐Cervantes, M. , Gallagher, C.E. , Zhu, C. and Wurtzel, E.T. (2006) Maize cDNAs expressed in endosperm encode functional farnesyl diphosphate synthase with geranylgeranyl diphosphate synthase activity. Plant Physiol. 141, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty, S. and Newton, A.C. (2011) Climate change, plant diseases and food security: an overview. Plant Pathol. 60, 2–14. [Google Scholar]
- Chen, F. , Tholl, D. , Bohlmann, J. and Pichersky, E. (2011) The family of terpene synthases in plants: a mid‐size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 66, 212–229. [DOI] [PubMed] [Google Scholar]
- Cheng, A.X. , Xiang, C.Y. , Li, J.X. , Yang, C.Q. , Hu, W.L. , Wang, L.J. , Lou, Y.G. and Chen, X.Y. (2007) The rice (E)‐beta‐caryophyllene synthase (OsTPS3) accounts for the major inducible volatile sesquiterpenes. Phytochemistry, 68, 1632–1641. [DOI] [PubMed] [Google Scholar]
- Christensen, S.A. , Huffaker, A. , Sims, J. et al . (2018) Fungal and herbivore elicitation of the novel maize sesquiterpenoid, zealexin A4, is attenuated by elevated CO2 . Planta, 247, 863–873. [DOI] [PubMed] [Google Scholar]
- Cyr, A. , Wilderman, P.R. , Determan, M. and Peters, R.J. (2007) A modular approach for facile biosynthesis of labdane‐related diterpenes. J. Am. Chem. Soc. 129, 6684–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dafoe, N.J. , Huffaker, A. , Vaughan, M.M. , Duehl, A.J. , Teal, P.E. and Schmelz, E.A. (2011) Rapidly induced chemical defenses in maize stems and their effects on short‐term growth of Ostrinia nubilalis . J. Chem. Ecol. 37, 984–991. [DOI] [PubMed] [Google Scholar]
- Ding, Y. , Huffaker, A. , Kollner, T.G. , Weckwerth, P. , Robert, C.A.M. , Spencer, J.L. , Lipka, A.E. and Schmelz, E.A. (2017) Selinene volatiles are essential precursors for maize defense promoting fungal pathogen resistance. Plant Physiol. 175, 1455–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Y. , Murphy, K.M. , Poretsky, E. et al . (2019) Multiple genes recruited from hormone pathways partition maize diterpenoid defences. Nat. Plants, 5, 1043–1056. [DOI] [PubMed] [Google Scholar]
- Doust, A.N. , Kellogg, E.A. , Devos, K.M. and Bennetzen, J.L. (2009) Foxtail millet: a sequence‐driven grass model system. Plant Physiol. 149, 137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, J. , Ren, F. , Lu, X. , Mao, H. , Xu, M. , Degenhardt, J. , Peters, R.J. and Wang, Q. (2016) A tandem array of ent‐Kaurene synthases in maize with roles in gibberellin and more specialized metabolism. Plant Physiol. 170, 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, F. , Liu, B. , Li, M. , Gao, X. , Fang, Q. , Liu, C. , Ding, H. , Wang, L. and Gao, X. (2018) Identification and characterization of terpene synthase genes accounting for volatile terpene emissions in flowers of Freesia x hybrida . J. Exp. Bot. 69, 4249–4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goron, T.L. and Raizada, M.N. (2015) Genetic diversity and genomic resources available for the small millet crops to accelerate a New Green Revolution. Front. Plant Sci. 6, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, Y.Q. , Zhang, K. , Yang, J. , Zhang, N. , Fang, A.F. , Zhang, Y. , Liu, Y.F. , Chen, Z.Y. , Hsiang, T. and Sun, W.X. (2015) Differential expression profiling of the early response to Ustilaginoidea virens between false smut resistant and susceptible rice varieties. BMC Genom. 16, 955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, L.J. , Saparno, A. , Johnston, A. , Prisic, S. , Xu, M. , Allard, S. , Kathiresan, A. , Ouellet, T. and Peters, R.J. (2005) The maize An2 gene is induced by Fusarium attack and encodes an ent‐copalyl diphosphate synthase. Plant Mol. Biol. 59, 881–994. [DOI] [PubMed] [Google Scholar]
- Hasegawa, M. , Mitsuhara, I. , Seo, S. , Imai, T. , Koga, J. , Okada, K. , Yamane, H. and Ohashi, Y. (2010) Phytoalexin accumulation in the interaction between rice and the blast fungus. Mol. Plant Microbe Interact. 23, 1000–1011. [DOI] [PubMed] [Google Scholar]
- Herde, M. , Gartner, K. , Kollner, T.G. , Fode, B. , Boland, W. , Gershenzon, J. , Gatz, C. and Tholl, D. (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect‐induced volatile C16‐homoterpene TMTT. Plant Cell, 20, 1152–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillwig, M.L. , Xu, M. , Toyomasu, T. , Tiernan, M.S. , Wei, G. , Cui, G. , Huang, L. and Peters, R.J. (2011) Domain loss has independently occurred multiple times in plant terpene synthase evolution. Plant J. 68, 1051–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie, K. , Inoue, Y. , Sakai, M. , Yao, Q. , Tanimoto, Y. , Koga, J. , Toshima, H. and Hasegawa, M. (2015) Identification of UV‐Induced Diterpenes Including a New Diterpene Phytoalexin, Phytocassane F, from Rice Leaves by Complementary GC/MS and LC/MS Approaches. J. Agric. Food Chem. 63, 4050–4059. [DOI] [PubMed] [Google Scholar]
- Huang, P. , Shyu, C. , Coelho, C.P. , Cao, Y. and Brutnell, T.P. (2016) Setaria viridis as a model system to advance millet genetics and genomics. Front. Plant Sci. 7, 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker, A. , Kaplan, F. , Vaughan, M.M. , Dafoe, N.J. , Ni, X. , Rocca, J.R. , Alborn, H.T. , Teal, P.E. and Schmelz, E.A. (2011) Novel acidic sesquiterpenoids constitute a dominant class of pathogen‐induced phytoalexins in maize. Plant Physiol. 156, 2082–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, M. and Peters, R.J. (2016) Extending a single residue switch for abbreviating catalysis in plant ent‐Kaurene synthases. Front Plant Sci. 7, 1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamatou, G.P.P. and Viljoen, A.M. (2010) A review of the application and pharmacological properties of alpha‐Bisabolol and alpha‐Bisabolol‐rich oils. J. Am. Oil Chem. Soc. 87, 1–7. [Google Scholar]
- Karunanithi, P.S. and Zerbe, P. (2019) Terpene synthases as metabolic gatekeepers in the evolution of plant terpenoid chemical diversity. Front. Plant Sci. 10, 1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato‐Noguchi, H. and Peters, R.J. (2013) The role of momilactones in rice allelopathy. J. Chem. Ecol. 39, 175–85. [DOI] [PubMed] [Google Scholar]
- Kitaoka, N. , Lu, X. , Yang, B. and Peters, R.J. (2015a) The application of synthetic biology to elucidation of plant mono‐, sesqui‐, and diterpenoid metabolism. Mol. Plant, 8, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaoka, N. , Wu, Y. , Xu, M. and Peters, R.J. (2015b) Optimization of recombinant expression enables discovery of novel cytochrome P450 activity in rice diterpenoid biosynthesis. Appl. Microbiol. Biotechnol. 99, 7549–7558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama, O. , Suzuki, T. , Miyakawa, J. and Akatsuka, T. (1988) Ultraviolet‐Induced Accumulation of Phytoalexins in Rice Leaves. Agr. Biol. Chem. Tokyo, 52, 2469–2473. [Google Scholar]
- Köksal, M. , Potter, K. , Peters, R.J. and Christianson, D.W. (2014) 1.55A‐resolution structure of ent‐copalyl diphosphate synthase and exploration of general acid function by site‐directed mutagenesis. Biochim. Biophys. Acta, 1840, 184–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner, T.G. , Held, M. , Lenk, C. , Hiltpold, I. , Turlings, T.C. , Gershenzon, J. and Degenhardt, J. (2008a) A maize (E)‐beta‐caryophyllene synthase implicated in indirect defense responses against herbivores is not expressed in most American maize varieties. Plant Cell, 20, 482–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner, T.G. , Schnee, C. , Li, S. , Svatos, A. , Schneider, B. , Gershenzon, J. and Degenhardt, J. (2008b) Protonation of a neutral (S)‐beta‐bisabolene intermediate is involved in (S)‐beta‐macrocarpene formation by the maize sesquiterpene synthases TPS6 and TPS11. J. Biol. Chem. 283, 20779–20788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner, T.G. , Gershenzon, J. and Degenhardt, J. (2009) Molecular and biochemical evolution of maize terpene synthase 10, an enzyme of indirect defense. Phytochemistry, 70, 1139–1145. [DOI] [PubMed] [Google Scholar]
- Köllner, T.G. , Lenk, C. , Schnee, C. , Köpke, S. , Lindemann, P. , Gershenzon, J. and Degenhardt, J. (2013) Localization of sesquiterpene formation and emission in maize leaves after herbivore damage. BMC Plant Biol. 13, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, K. , Muthamilarasan, M. and Prasad, M. (2013) Reference genes for quantitative real‐time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tiss. Org. 115, 13–22. [Google Scholar]
- Lee, H.‐J. , Ravn, M.M. and Coates, R.M. (2001) Synthesis and characterization of abietadiene, levopimaradiene, palustradiene, and neoabietadiene: hydrocarbon precursors of the abietane diterpene resin acids. Tetrahedron, 57, 6155–6167. [Google Scholar]
- Lemke, C. , Potter, K.C. , Schulte, S. and Peters, R.J. (2019) Conserved bases for the initial cyclase in gibberellin biosynthesis: from bacteria to plants. Biochem J. 476, 2607–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, G. and Hall, J. (2019) Crop yield sensitivity of global major agricultural countries to droughts and the projected changes in the future. Sci. Total Environ. 654, 811–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.Y. , Wang, N. , Dong, Z.P. , Dong, L. , Bai, H. , Quan, J.Z. and Liu, L. (2015) First report of Fusarium graminearum causing ear rot of foxtail millet in China. Plant Dis. 99, 1856. [Google Scholar]
- Liang, J. , Liu, J. , Brown, R. , Jia, M. , Zhou, K. , Peters, R.J. and Wang, Q. (2018) Direct production of dihydroxylated sesquiterpenoids by a maize terpene synthase. Plant J. 94, 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. , Zhang, J. , Liu, K.B. et al (2009) Earliest domestication of common millet (Panicum miliaceum) in East Asia extended to 10,000 years ago. Proc. Natl. Acad. Sci. USA, 106, 7367–7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, X. , Zhang, J. , Brown, B. et al . (2018) Inferring roles in defense from metabolic allocation of rice diterpenoids. Plant Cell, 30, 1119–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafu, S. , Jia, M. , Zi, J. , Morrone, D. , Wu, Y. , Xu, M. , Hillwig, M.L. and Peters, R.J. (2016) Probing the promiscuity of ent‐kaurene oxidases via combinatorial biosynthesis. Proc. Natl. Acad. Sci. USA, 113, 2526–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mafu, S. , Ding, Y. , Murphy, K.M. et al . (2018) Discovery, biosynthesis and stress‐related accumulation of dolabradiene‐derived defenses in maize. Plant Physiol. 176, 2677–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao, H. , Liu, J. , Ren, F. , Peters, R.J. and Wang, Q. (2016) Characterization of CYP71Z18 indicates a role in maize zealexin biosynthesis. Phytochemistry, 121, 4–10. [DOI] [PubMed] [Google Scholar]
- Mischko, W. , Hirte, M. , Fuchs, M. , Mehlmer, N. and Bruck, T.B. (2018) Identification of sesquiterpene synthases from the Basidiomycota Coniophora puteana for the efficient and highly selective beta‐copaene and cubebol production in E. coli . Microb. Cell Fact. 17, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone, D. , Lowry, L. , Determan, M.K. , Hershey, D.M. , Xu, M. and Peters, R.J. (2010) Increasing diterpene yield with a modular metabolic engineering system in E. coli: comparison of MEV and MEP isoprenoid precursor pathway engineering. Appl. Microbiol. Biotechnol. 85, 1893–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrone, D. , Hillwig, M.L. , Mead, M.E. , Lowry, L. , Fulton, D.B. and Peters, R.J. (2011) Evident and latent plasticity across the rice diterpene synthase family with potential implications for the evolution of diterpenoid metabolism in the cereals. Biochem J. 435, 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchlinski, A. , Chen, X. , Lovell, J.T. et al . (2019) Biosynthesis and emission of stress‐induced volatile terpenes in roots and leaves of switchgrass. Front. Plant Sci. 10, 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K.M. and Zerbe, P. (2020) Specialized diterpenoid metabolism in monocot crops: biosynthesis and chemical diversity. Phytochemistry, 172, 112289. [DOI] [PubMed] [Google Scholar]
- Murphy, K.M. , Ma, L.T. , Ding, Y. , Schmelz, E.A. and Zerbe, P. (2018) Functional characterization of two class II diterpene synthases indicates additional specialized diterpenoid pathways in maize (Zea mays). Front. Plant Sci. 9, 1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, K.M. , Chung, S. , Fogla, S. , Minsky, H.B. , Zhu, K.Y. and Zerbe, P. (2019) A customizable approach for the enzymatic production and purification of diterpenoid natural products. J. Vis. Exp. 152, e59992 10.3791/59992. [DOI] [PubMed] [Google Scholar]
- Nagegowda, D.A. , Gutensohn, M. , Wilkerson, C.G. and Dudareva, N. (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J. 55, 224–239. [DOI] [PubMed] [Google Scholar]
- Nwanma, B.N.O. , Nelson, P.E. and Marasas, W.F.O. (1991) Fusarium species associated with millet grain from Nigeria, Lesotho, and Zimbabwe. Mycologia, 83, 708–712. [Google Scholar]
- Otomo, K. , Kanno, Y. , Motegi, A. et al . (2004) Diterpene cyclases responsible for the biosynthesis of phytoalexins, momilactones A, B, and oryzalexins A‐F in rice. Biosci. Biotechnol. Biochem. 68, 2001–6. [DOI] [PubMed] [Google Scholar]
- Pant, S.R. , Irigoyen, S. , Doust, A.N. , Scholthof, K.B. and Mandadi, K.K. (2016) Setaria: a food crop and translational research model for C4 grasses. Front. Plant Sci. 7, 1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelot, K.A. , Hagelthorn, D.M. , Addison, J.B. and Zerbe, P. (2017) Biosynthesis of the oxygenated diterpene nezukol in the medicinal plant Isodon rubescens is catalyzed by a pair of diterpene synthases. PLoS ONE, 12, e0176507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelot, K.A. , Chen, R. , Hagelthorn, D.M. , Young, C.A. , Addison, J.B. , Muchlinski, A. , Tholl, D. and Zerbe, P. (2018) Functional diversity of diterpene synthases in the biofuel crop switchgrass. Plant Physiol. 178, 54–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, R.J. (2010) Two rings in them all: the labdane‐related diterpenoids. Nat. Prod. Rep. 27, 1521–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, K. , Criswell, J. , Zi, J. , Stubbs, A. and Peters, R.J. (2014) Novel product chemistry from mechanistic analysis of ent‐copalyl diphosphate synthases from plant hormone biosynthesis. Angew. Chem. Int. Ed. Engl. 53, 7198–7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter, K.C. , Jia, M. , Hong, Y.J. , Tantillo, D. and Peters, R.J. (2016) Product rearrangement from altering a single residue in the rice syn‐Copalyl diphosphate synthase. Org. Lett. 18, 1060–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prisic, S. , Xu, M. , Wilderman, P.R. and Peters, R.J. (2004) Rice contains two disparate ent‐copalyl diphosphate synthases with distinct metabolic functions. Plant Physiol. 136, 4228–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, A. , Schaff, C. , Zhang, Z. et al . (2016) Characterization of Biosynthetic Pathways for the Production of the Volatile Homoterpenes DMNT and TMTT in Zea mays . Plant Cell, 28, 2651–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto, T. , Miura, K. , Itoh, H. et al . (2004) An overview of gibberellin metabolism enzyme genes and their related mutants in rice. Plant Physiol. 134, 1642–1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Sassi, C. and Tylianakis, J.M. (2012) Climate change disproportionately increases herbivore over plant or parasitoid biomass. PLoS ONE, 7, e40557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz, E.A. , Kaplan, F. , Huffaker, A. , Dafoe, N.J. , Vaughan, M.M. , Ni, X. , Rocca, J.R. , Alborn, H.T. and Teal, P.E. (2011) Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA, 108, 5455–5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz, E.A. , Huffaker, A. , Sims, J.W. , Christensen, S.A. , Lu, X. , Okada, K. and Peters, R.J. (2014) Biosynthesis, elicitation and roles of monocot terpenoid phytoalexins. Plant J. 79, 659–678. [DOI] [PubMed] [Google Scholar]
- Schnee, C. , Köllner, T.G. , Gershenzon, J. and Degenhardt, J. (2002) The maize gene terpene synthase 1 encodes a sesquiterpene synthase catalyzing the formation of (E)‐beta‐farnesene, (E)‐nerolidol, and (E, E)‐farnesol after herbivore damage. Plant Physiol. 130, 2049–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnee, C. , Köllner, T.G. , Held, M. , Turlings, T.C. , Gershenzon, J. and Degenhardt, J. (2006) The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. acad. Sci. USA, 103, 1129–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura, K. , Okada, A. , Okada, K. et al . (2007) Identification of a biosynthetic gene cluster in rice for momilactones. J. Biol. Chem. 282, 34013–34018. [DOI] [PubMed] [Google Scholar]
- Son, Y.J. , Kwon, M. , Ro, D.K. and Kim, S.U. (2014) Enantioselective microbial synthesis of the indigenous natural product (‐)‐alpha‐bisabolol by a sesquiterpene synthase from chamomile (Matricaria recutita). Biochem. J. 463, 239–248. [DOI] [PubMed] [Google Scholar]
- Swaminathan, S. , Morrone, D. , Wang, Q. , Fulton, D.B. and Peters, R.J. (2009) CYP76M7 is an ent‐cassadiene C11alpha‐hydroxylase defining a second multifunctional diterpenoid biosynthetic gene cluster in rice. Plant Cell, 21, 3315–3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyomasu, T. , Usui, M. , Sugawara, C. et al . (2014) Reverse‐genetic approach to verify physiological roles of rice phytoalexins: characterization of a knockdown mutant of OsCPS4 phytoalexin biosynthetic gene in rice. Physiol. Plant. 150, 55–62. [DOI] [PubMed] [Google Scholar]
- Toyomasu, T. , Usui, M. , Sugawara, C. et al . (2015) Transcripts of two ent‐copalyl diphosphate synthase genes differentially localize in rice plants according to their distinct biological roles. J. Exp. Bot. 66, 369–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, M.M. , Huffaker, A. , Schmelz, E.A. et al . (2014) Effects of elevated [CO2] on maize defence against mycotoxigenic Fusarium verticillioides . Plant Cell Environ. 37, 2691–2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, M.M. , Christensen, S. , Schmelz, E.A. , Huffaker, A. , McAuslane, H.J. , Alborn, H.T. , Romero, M. , Allen, L.H. and Teal, P.E. (2015) Accumulation of terpenoid phytoalexins in maize roots is associated with drought tolerance. Plant Cell Environ. 38, 2195–2207. [DOI] [PubMed] [Google Scholar]
- Wang, Q. , Hillwig, M.L. and Peters, R.J. (2011) CYP99A3: functional identification of a diterpene oxidase from the momilactone biosynthetic gene cluster in rice. Plant J. 65, 87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Hillwig, M.L. , Okada, K. , Yamazaki, K. , Wu, Y. , Swaminathan, S. , Yamane, H. and Peters, R.J. (2012a) Characterization of CYP76M5‐8 indicates metabolic plasticity within a plant biosynthetic gene cluster. J. Biol. Chem. 287, 6159–6168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q. , Hillwig, M.L. , Wu, Y. and Peters, R.J. (2012b) CYP701A8: a rice ent‐kaurene oxidase paralog diverted to more specialized diterpenoid metabolism. Plant Physiol. 158, 1418–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilderman, P.R. , Xu, M. , Jin, Y. , Coates, R.M. and Peters, R.J. (2004) Identification of syn‐pimara‐7,15‐diene synthase reveals functional clustering of terpene synthases involved in rice phytoalexin/allelochemical biosynthesis. Plant Physiol. 135, 2098–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Hillwig, M.L. , Wang, Q. and Peters, R.J. (2011) Parsing a multifunctional biosynthetic gene cluster from rice: Biochemical characterization of CYP71Z6 & 7. FEBS Lett. 585, 3446–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Y. , Zhou, K. , Toyomasu, T. et al . (2012) Functional characterization of wheat copalyl diphosphate synthases sheds light on the early evolution of labdane‐related diterpenoid metabolism in the cereals. Phytochemistry, 84, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Hillwig, M.L. , Prisic, S. , Coates, R.M. and Peters, R.J. (2004) Functional identification of rice syn‐copalyl diphosphate synthase and its role in initiating biosynthesis of diterpenoid phytoalexin/allelopathic natural products. Plant J. 39, 309–318. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Wilderman, P.R. and Peters, R.J. (2007a) Following evolution's lead to a single residue switch for diterpene synthase product outcome. Proc. Natl. Acad. Sci. USA, 104, 7397–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, M. , Wilderman, P.R. , Morrone, D. , Xu, J. , Roy, A. , Margis‐Pinheiro, M. , Upadhyaya, N.M. , Coates, R.M. and Peters, R.J. (2007b) Functional characterization of the rice kaurene synthase‐like gene family. Phytochemistry, 68, 312–326. [DOI] [PubMed] [Google Scholar]
- Xu, M. , Galhano, R. , Wiemann, P. et al . (2012) Genetic evidence for natural product mediated plant–plant allelopathy in rice. New Phytol. 193, 570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z. , Nakagawa, K. , Natsume, M. , Nojiri, H. , Kawaide, H. and Okada, K. (2017) Biochemical synthesis of uniformly 13C‐labeled diterpene hydrocarbons and their bioconversion to diterpenoid phytoalexins in planta. Biosci. Biotechnol. Biochem. 6, 1176–1184. [DOI] [PubMed] [Google Scholar]
- Yoshikuni, Y. , Martin, V.J. , Ferrin, T.E. and Keasling, J.D. (2006) Engineering cotton (+)‐delta‐cadinene synthase to an altered function: germacrene D‐4‐ol synthase. Chem. Biol. 13, 91–98. [DOI] [PubMed] [Google Scholar]
- Zerbe, P. and Bohlmann, J. (2015) Plant diterpene synthases: exploring modularity and metabolic diversity for bioengineering. Trends Biotechnol. 33, 419–428. [DOI] [PubMed] [Google Scholar]
- Zerbe, P. , Hamberger, B. , Yuen, M.M. , Chiang, A. , Sandhu, H.K. , Madilao, L.L. , Nguyen, A. , Hamberger, B. , Bach, S.S. and Bohlmann, J. (2013) Gene discovery of modular diterpene metabolism in nonmodel systems. Plant Physiol. 162, 1073–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbe, P. , Chiang, A. , Dullat, H. , O'Neil‐Johnson, M. , Starks, C. , Hamberger, B. and Bohlmann, J. (2014) Diterpene synthases of the biosynthetic system of medicinally active diterpenoids in Marrubium vulgare . Plant J. 79, 914–927. [DOI] [PubMed] [Google Scholar]
- Zerbe, P. , Rodriguez, S.M. , Mafu, S. , Chiang, A. , Sandhu, H.K. , O'Neil‐Johnson, M. , Starks, C.M. and Bohlmann, J. (2015) Exploring diterpene metabolism in non‐model species: transcriptome‐enabled discovery and functional characterization of labda‐7,13E‐dienyl diphosphate synthase from Grindelia robusta . Plant J. 83, 783–793. [DOI] [PubMed] [Google Scholar]
- Zhou, K. , Xu, M. , Tiernan, M. et al . (2012) Functional characterization of wheat ent‐kaurene(‐like) synthases indicates continuing evolution of labdane‐related diterpenoid metabolism in the cereals. Phytochemistry, 84, 47–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zi, J. , Mafu, S. and Peters, R.J. (2014) To gibberellins and beyond! Surveying the evolution of (di)terpenoid metabolism. Annu. Rev. Plant Biol. 65, 259–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Protein sequence alignment of select class II diterpene synthases.
Figure S2. Protein sequence alignment of select class I diterpene synthases.
Figure S3. Sequence similarity matrix of terpene synthase candidates from Setaria italica and Setaria viridis.
Figure S4. Mass spectra of class II diterpene synthase products identified in this study.
Figure S5. Mass spectra of class I terpene synthase products identified in this study.
Figure S6. Mass spectra of products resulting from co‐expression assays of SiTPS5 and SiTPS13.
Figure S7. The NMR analysis of ent‐pimara‐8,15‐diene.
Figure S8. The NMR analysis of syn‐pimara‐7,15‐diene.
Figure S9. The NMR analysis of eudesme‐2,11‐diol.
Figure S10. Functional analysis of CYP99A17 and CYP99A19.
Figure S11. The NMR analysis of abietadien‐19‐ol.
Figure S12. The NMR analysis of syn‐pimara‐7,15‐dien‐19‐ol.
Figure S13. Gene expression analysis of characterized Setaria italica terpene synthase genes.
Figure S14. Occurrence of terpene synthase and CYP99A17 products in Setaria italica.
Table S1. Accession numbers and cloned constructs for terpene synthases used in this study.
Table S2. Chromosomal location of Setaria italica terpene synthase candidate genes.
Table S3. Gene synteny analysis of Setaria italica terpene synthase candidates.
Table S4. Oligonucleotides used for gene expression analysis in this study.


