SUMMARY
Given their sessile nature, land plants must use various mechanisms to manage dehydration under water‐deficit conditions. Osmostress‐induced activation of the SNF1‐related protein kinase 2 (SnRK2) family elicits physiological responses such as stomatal closure to protect plants during drought conditions. With the plant hormone ABA receptors [PYR (pyrabactin resistance)/PYL (pyrabactin resistance‐like)/RCAR (regulatory component of ABA receptors) proteins] and group A protein phosphatases, subclass III SnRK2 also constitutes a core signaling module for ABA, and osmostress triggers ABA accumulation. How SnRK2 is activated through ABA has been clarified, although its activation through osmostress remains unclear. Here, we show that Arabidopsis ABA and abiotic stress‐responsive Raf‐like kinases (AtARKs) of the B3 clade of the mitogen‐activated kinase kinase kinase (MAPKKK) family are crucial in SnRK2‐mediated osmostress responses. Disruption of AtARKs in Arabidopsis results in increased water loss from detached leaves because of impaired stomatal closure in response to osmostress. Our findings obtained in vitro and in planta have shown that AtARKs interact physically with SRK2E, a core factor for stomatal closure in response to drought. Furthermore, we show that AtARK phosphorylates S171 and S175 in the activation loop of SRK2E in vitro and that Atark mutants have defects in osmostress‐induced subclass III SnRK2 activity. Our findings identify a specific type of B3‐MAPKKKs as upstream kinases of subclass III SnRK2 in Arabidopsis. Taken together with earlier reports that ARK is an upstream kinase of SnRK2 in moss, an existing member of a basal land plant lineage, we propose that ARK/SnRK2 module is evolutionarily conserved across 400 million years of land plant evolution for conferring protection against drought.
Keywords: Raf‐like kinase, SnRK2, osmostress, ABA, Arabidopsis, B3‐MAPKKK
Significance Statement
Here, we show that Arabidopsis ABA and abiotic stress‐responsive Raf‐like kinases (AtARKs) belonging to the B3 clade of the mitogen‐activated kinase kinase kinase family play a crucial role in Arabidopsis drought responses, such as stomatal closure, through phosphorylation‐mediated activation of subclass III SNF1‐related protein kinase 2 (SnRK2) partly in an abscisic acid‐independent manner. Our findings indicate that ARK is a novel component of osmostress signaling that connects osmosensing with SnRK2 activation.
INTRODUCTION
Responding to osmostress was an important trait for land colonization by a common plant ancestor in the Paleozoic era. This feature allows plants to survive in an environment with limited water availability, although the mechanism of osmostress sensing and its signaling cascades remains largely unknown. Plant‐specific SNF1‐related protein kinase 2 (SnRK2) plays a central role in Arabidopsis drought tolerance (Fujii et al., 2011) and is categorized into three subclasses (I–III). Previous reports suggest that the precursor of the SnRK2 genes (subclass III‐type) arose before the evolution of land plants, with gene duplication and subfunctionalization (the emergence of subclasses I and II) occurring after the separation of bryophytes and vascular plants (Umezawa et al., 2010; Takezawa et al., 2011; Shinozawa et al., 2019) Osmostress activates subclass I‐III SnRK2s, whereas the stress hormone ABA specifically activates subclass III SnRK2 (Yoshida et al., 2002; Boudsocq et al., 2004; Kobayashi et al., 2004), which are an essential component in ABA signal transduction (Fujii and Zhu, 2009; Nakashima et al., 2009; Fujita et al., 2009). In Arabidopsis soluble ABA receptors [PYR (pyrabactin resistance)/PYL (pyrabactin resistance‐like)/RCAR (regulatory component of ABA receptors) proteins] (Park et al., 2009; Ma et al., 2009) and group A protein phosphatase 2Cs (PP2CAs) (Umezawa et al., 2009) regulate auto‐activation of Subclass III SnRK2 (Fujii et al., 2009). The mechanism of ABA‐associated activation of SnRK2 has been clarified, although its osmostress‐dependent activation is poorly understood (Boudsocq et al., 2007).
Previously, we showed that the subclass III SnRK2‐mediated signaling pathway exclusively contributes to ABA responses and osmostress tolerance in the moss Physcomitrella patens (Shinozawa et al., 2019). By contrast to the proposed model of ABA signaling in angiosperms, activation of SnRK2 in the moss depends on the upstream kinase ARK (ABA and abiotic stress‐responsive Raf‐like kinase) (Saruhashi et al., 2015), also known as ABA non‐responsive (Stevenson et al., 2016) or constitutive triple response 1‐like (Yasumura et al., 2015). ARK is a single‐copy gene in the moss genome and encodes a group B3 mitogen‐activated kinase kinase kinase (MAPKKK) (MAPK Group, 2002). This clade of MAPKKKs has been expanded in angiosperms, and the Arabidopsis genome encodes six B3‐MAPKKK genes (Stevenson et al., 2016), including constitutive triple response (CTR1) for ethylene signaling (Kieber et al., 1993) and enhanced disease resistance (EDR) for disease resistance (Frye et al., 2001). ARK was assumed to be a bryophyte‐specific factor that was lost in angiosperms during vascular plant evolution (Stevenson et al., 2016); however, our results from a cross‐species complementation assay indicate that Arabidopsis B3 MAPKKK can complement ARK function when expressed in an ark mutant of P. patens (Saruhashi et al., 2015). That analysis highlighted three genes, namely At1G18160 (AtARK1), At1G73660 {AtARK2; also known as SUGAR INSENSITIVE8 (SIS8)} (Huang et al., 2014) and At4G24480 (AtARK3), which all recover the ABA‐insensitivity of the ark mutant. Few studies are available describing the functional and downstream factors involved with these B3‐MAPKKKs (Huang et al., 2014). Here, we show that AtARKs play a crucial role in Arabidopsis drought responses, such as stomatal closure, through phosphorylation‐mediated activation of subclass III SnRK2s partly in an ABA‐independent manner. These results indicate that ARK is a component of osmostress signaling that connects osmosensing with SnRK2 activation. Our findings suggest that the ARK/SnRK2 module has been evolutionarily conserved to protect plants from drought across 400 million years of land plant evolution.
RESULTS AND DISCUSSION
ARK has been identified as the causal gene in an ABA‐insensitive AR7 mutant of P. patens, in which a point mutation effaces ARK function by causing a non‐synonymous substitution of Ser at 532 to Phe located in EDR domain (Saruhashi et al., 2015). The corresponding Ser is conserved in AtARK1/2/3, but not in other B3 MAPKKKs of Arabidopsis (Figure S1), consistent with functional complementation of ARK by these AtARKs in P. patens. Here, we investigated the expression patterns of AtARK1/2/3 using the Arabidopsis eFP browser (http://bar.utoronto.ca/efp2/Arabidopsis/Arabidopsis_eFPBrowser2.html) (Figures S2–S4). AtARKs are ubiquitously expressed, showing strong expression in stomata. The browser also indicated that ABA treatment enhances expression of AtARK3. Using a quantitative reverse transcriptase‐polymerase chain reaction (qRT‐PCR), we confirmed the ABA‐induced accumulation in seedlings of AtARK3 mRNA and, to a lesser extent, AtARK1 mRNA (Figure S5).
To examine the role of AtARKs in ABA and osmostress responses of Arabidopsis, we generated ark disruptants using T‐DNA insertion lines obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA). In addition to the single disruptants of each AtARK gene, the double disruptant for AtARK1 and AtARK2 [double knockout (DKO)] and the triple disruptants for AtARK1, AtARK2 and AtARK3 [triple knockout (TKO)] were generated. We used RT‐PCR to confirm the null expression of each gene in the disruptants (Figure S6). No clear difference emerged between wild‐type (WT) and ark disruptants during germination under standard culture conditions or under ABA treatment (Figure S7a); however, ark disruptants showed more greening cotyledons than did WT after germination on ABA‐containing medium (Figure S7b,c), indicating that ark disruptants, especially ark1/2 DKO and ark1/2/3 TKO, are insensitive to ABA during the post‐germination stage. Compared to WT, the ark disruptants also showed reduced sensitivity to osmostress‐induced inhibition of cotyledon greening after germination (Figure S7b,d) in a pattern similar to that with ABA treatment. These results suggested that AtARKs act as positive regulators of ABA and osmostress signaling during the post‐germination growth of Arabidopsis.
Because AtARKs appeared to act redundantly in ABA and osmostress signaling, we focused on ark TKO plants for further analyses. In vegetative growth, ark TKO plants showed no significant difference from WT in ABA sensitivity, based on root growth on ABA‐containing medium (Figure S8), although mutants were more sensitive to osmostress at 300 mm or greater concentrations of sorbitol (Figure 1a,b). Osmostress treatment also was associated with significant effects on expression of several stress‐associated marker genes in ark TKO seedlings, although expression levels after ABA treatment varied (Figure 1c). To confirm this trend, we performed RNA‐seq analysis of ark TKO and WT plants after ABA or sorbitol treatment. This analysis also demonstrated that disruption of AtARK genes had a significant effect on osmostress‐regulated gene expression, but not on ABA‐regulated genes (Figure 1d; Figure S9, Table S1). These data suggest that AtARKs are also involved in vegetative osmostress responses of Arabidopsis, at least in part in an ABA‐independent manner. RNA‐seq analysis showed that ABA biosynthesis–related genes are expressed in ark TKO plants at a level comparable to that of WT except for NCED3, which is the limiting step of the ABA biosynthesis pathway (Sato et al., 2018). Although osmostress‐induced expression of NCED3 was significantly reduced compared to that of WT, it still showed considerable expression in response to osmostress (Figure S10a). We also compared stomatal closure of ark TKO plants with that of the ABA‐deficient mutant aba2‐1 (Leon‐Kloosterziel et al., 1996) using leaf disks (Figure S10b). Stomatal closure of aba2‐1 mutant plants did not occur after 3 h of sorbitol treatment, indicating that ABA accumulation is essential for osmostress‐induced stomatal closure in this condition. By contrast, stomata of ark TKO plants closed like WT in the same condition. These results suggest that ark TKO plants accumulate ABA to a level sufficient to close stomata in response to osmostress.
Figure 1.

Sensitivities of ark disruptants toward ABA and osmostress
(a) Seeds of wild‐type (WT) and ark1/2/3 triple knockout (TKO) were sown and germinated on a standard agar medium plate for 1 week, then transferred on agar medium plates with or without sorbitol. The photos were taken after 2 weeks of culture.
(b) Chlorophyll contents of WT and ark TKO plants after 2 weeks of osmostress treatment. Values are the mean ± SE (n = 60). *P < 0.05
(c) Quantitative reverse transcriptase‐polymerase chain reaction analysis of ABA‐ and stress‐associated marker gene expression in response to ABA or sorbitol. Two‐week‐old seedlings were treated with 50 µm ABA solution or 300 mm sorbitol solution for 1 or 3 h. Values are the mean ± SE (n = 3). ***P < 0.001.
(d) Numbers of genes more than two‐fold difference in mRNA accumulation in wild‐type (WT), and the numbers of genes affected in ark TKO plants after 3 h treatment by ABA or sorbitol (osmostress). Genes with more than 1.5‐fold difference in the expression level between ABA‐treated WT and ark TKO or sorbitol‐treated WT and ark TKO (false discovery rate (FDR) ≤0.05) were categorized as Decreased or Increased in TKO.
In Arabidopsis, four bZIP transcription factors (AREB1/ABF2, AREB2/ABF4, ABF3 and ABF1) play the dominant role in induction of osmostress‐regulated genes downstream of subclass III SnRK2 (Yoshida et al., 2014). To investigate whether AtARKs mediate the subclass III SnRK2/AREB transcription factor pathway in the regulation of osmostress signal, we compared osmostress‐ and AtARK‐regulated genes with AREB/ABF‐regulated genes (Yoshida et al., 2014). This comparison demonstrated that 60–70% of osmostress‐ and AREB/ABF‐regulated genes overlapped with osmostress‐ and AtARK‐regulated genes (Figure S11 and Table S2). This result suggested that, at least in part, AtARKs mediate the subclass III SnRK2–AREB pathway in regulating osmostress responses.
To further evaluate the role of AtARKs in osmostress responses of Arabidopsis, we measured water loss from detached leaves. On room atmosphere, detached rosette leaves of ark TKO plants lost water content significantly and shrunk more rapidly than WT, suggesting impaired stomata closure in the mutants in response to drought (Figure 2a,b). We then observed stomatal apertures after detachment under light microscopy. WT stomata started to close after 30 min of detachment and completely closed after 150 min, whereas the ark TKO stomatal aperture decreased only by 20% on average, even after 150 min (Figure 2c,d). It has been reported that leaves of Arabidopsis start to accumulate ABA 1–2 h after detachment (Ikegami et al., 2009). Therefore, it is likely that the phenotype of stomatal aperture in ark TKO mutants shown in Figure 2(d) occurred before ABA accumulation. To further investigate the response of stomata to osmostress, we isolated leaf disks from WT and ark TKO plants and treated them with 400 mm sorbitol solution (Figure 2e). The WT stomatal apertures decreased to approximately one‐quarter of the initial opening after 60 min of sorbitol treatment; however, more than one‐half of the initial aperture remained open in TKO stomata. After 3 h of treatment, stomata in both lines mostly closed, suggesting that ark TKO plants have defects in the initial steps of stomata closure in response to osmostress. These results clearly demonstrated that ark TKO plants are defective in stomatal closure in response to osmostress, suggesting an important role for AtARKs in drought‐induced stomatal closure. Because the stomata of ark TKO plants were less sensitive to ABA than were those of WT and the ABA‐deficient aba2‐1 mutant (Figure 2f), AtARKs may also have a role in mediating drought‐induced stomatal closure after ABA accumulation.
Figure 2.
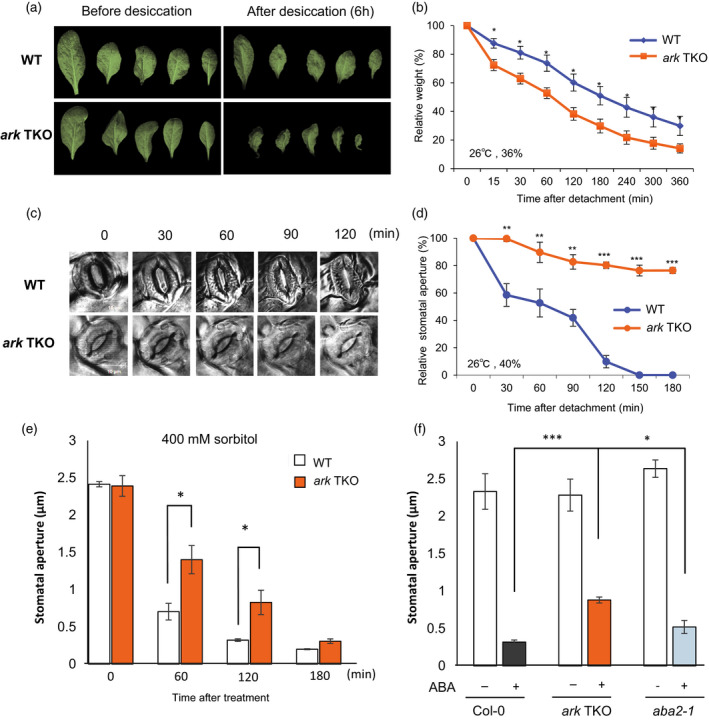
Stomatal closure in response to drought, osmostress, and ABA in ark troiple knockout (TKO) plants
(a) Photos of detached rosette leaves from 3‐week‐old wild‐type (WT) or ark TKO before and after 6 h of desiccation under laboratory room temperature.
(b) Detached leaves of WT and ark TKO were exposed to the atmosphere (26°C, 36% relative humidity) and the weights were measured at the indicated time points. Water loss was presented as relative to initial weights. Values are the mean ± SE (n = 12); **P < 0.01.
(c) Microscopic observation of stomata from WT and ark TKO detached leaves after exposure to laboratory room temperature.
(d) Stomatal apertures were calculated from the captured images. Values are the mean ± SE (n = 10); **P < 0.01, ***P < 0.001.
(e) Stomata of leafdisks from 4‐week‐old WT or ark TKO plants were treated with 400 mm sorbitol and observed under microscopy at 20°C (17% relative humidity), and then 15–40 images for each sample were captured at the indicated time points. Values are the mean ± SE (n = 4); *P < 0.05.
(f) Stomata of leafdisks from 4‐week‐old wild‐type (WT), ark TKO and aba2‐1 plants were treated with 20 µm ABA for 3 h and observed under microscopy at 20°C (17% relative humidity), and then 15–40 images for each sample were captured. Values are the mean ± SE (n = 3); *P < 0.05, ***P < 0.001.
ABA‐dependent stomatal closure of Arabidopsis relies on the activation of subclass III SnRK2 in guard cells (Mustilli et al., 2002). In P. patens, ARK directly phosphorylates serine residues in the activation loop of SnRK2 in vitro (Saruhashi et al., 2015), which is essential for activation of SnRK2 (Shinozawa et al., 2019). Therefore, we tested in vitro if AtARKs phosphorylate SRK2E/SnRK2.6, the causal gene in the Arabidopsis ost1 mutant that lacks ABA‐induced stomatal closure. The AtARKs and the kinase‐inactive mutant of SRK2E (SRK2EK50N) (Fujii et al., 2009) were fused to maltose‐binding protein (MBP) and bacterially synthesized, and then affinity‐purified recombinant proteins were used for the in vitro phosphorylation assay. The results indicated that AtARK1 and AtARK2, along with AtARK3 to a lesser extent, phosphorylate SRK2EK50N (Figure 3a; Figure S12a). We previously showed that, in the activation loop of PpSnRK2, the amino acid (aa) sequence between the two Ser residues (SQPKS) phosphorylated by ARK is evolutionarily conserved among land plant subclass III SnRK2s and is important for recognition by ARK (Figure S12b) (Shinozawa et al., 2019). Phosphorylation in these Ser residues plays an essential role in the activation of Arabidopsis SnRK2s (Belin et al., 2006). Mutation in both the conserved Ser171 and 175 to Ala (S171A and S175A) in SRK2E resulted in the loss of phosphorylation by AtARK1 (Figure 3b), indicating that conserved Ser residues in the activation loop of SRK2E are also target phosphorylation sites of AtARK. In addition, we investigated whether SRK2E phosphorylates AtARK in vitro. SRK2E showed only autophosphorylation activity and did not phosphorylate the N‐terminal region or kinase domain with the C‐terminal region of AtARK1 (Figure 3c), suggesting that SRK2E does not phosphorylate AtARK and acts downstream of AtARK. To examine the physical interaction of AtARKs and Arabidopsis SRK2E in vitro, we employed wheat cell‐free protein synthesis technology (Nomoto and Tada, 2018) coupled with AlphaScreen® (Amplified Luminescent Proximity Homogeneous Assay) (Ullman et al., 1994). The kinase domain with the C‐terminal region of AtARKs interacted with SRK2E, whereas the N‐terminal region did not (Figure 3d). We further confirmed the interaction between AtARK2 and SnRK2E using a biofluorescence complementation (BiFC) assay (Walter et al., 2004) in Nicotiana benthamiana epidermal cells. We observed a yellow fluorescent protein (YFP) signal between AtARK2 and SRK2E mainly in the cytosol and less frequently in the nucleus (Figure 3e), indicating the interaction between AtARK2 and SRK2E in planta. Taken together with the in vitro phosphorylation of SnRK2 by AtARKs, our experimental data suggest a role for AtARKs as upstream kinases of SnRK2 in Arabidopsis. Because the BiFC assay suggested a positive interaction of AtARK with subclass I and subclass II SnRK2s (Figure S13a), we investigated whether these subclasses are also the target of AtARK. In vitro, AtARK1 strongly phosphorylated subclass III SRK2E (K50N) and, to a lesser extent, subclass II SRK2C (K33N), although only slight phosphorylation of subclass I SRK2G (K33N) was observed (Figure S13b). We then tested whether this in vitro phosphorylation leads to activation of SnRK2 kinase activity in an in‐gel phosphorylation assay, using histone IIIS as the substrate (Figure 3f). The result clearly demonstrated that AtARK1 strongly activates subclass III SnRK2, but not subclass I SnRK2, suggesting that AtARK is involved in the activation of subclass III SnRK2 through phosphorylation.
Figure 3.
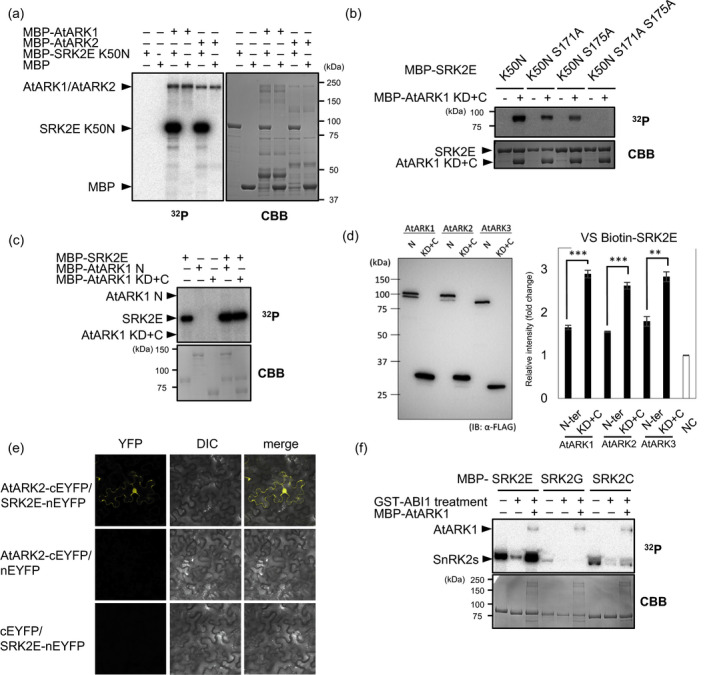
Role of Arabidopsis ARKs in phosphorylation and activation of SRK2E
(a) In vitro phosphorylation assay of recombinant maltose‐binding protein (MBP)‐SRK2EK50N by MBP‐AtARK1 and MBP‐AtARK2. Various combinations of proteins were reacted with γ‐[32P]ATP. The reaction mixtures were electrophoresed on a sodium dodecyl sulfate‐polyacrylamide gel, followed by autoradiography and Coomassie Brilliant Blue staining.
(b) Recombinant MBP‐SRK2EK50N proteins with or without alanine substitutions at S171 and S175 were used for in vitro phosphorylation by the AtARK1 kinase domain with the C‐terminal region (AtARK1 KD+C) fused to MBP.
(c) The recombinant AtARK N‐terminal region (AtARK1 N) and the kinase domain with the C‐terminal region (AtARK1 KD+C) fused to MBP were used for the in vitro phosphorylation assay by SRK2E.
(d) The C‐terminal FLAG ‐tagged AtARK truncated proteins were expressed using an in vitro transcription/translation system in wheat germ extract (WGE). The synthesized proteins were checked by western blotting with anti‐FLAG antibody. The biotinylated SRK2E and FLAG‐tagged AtARKs were incubated, and AlphaScreen® luminescence generated by the protein–protein interactions were detected by a multimode plate reader. WGE with no expressed proteins was employed as a negative control (NC) for estimation of the luminescence caused by endogenous wheat germ proteins.
(e) Biofluorescence complementation analysis to detect the physical interaction between SRK2D/SRK2E and AtARK2 in Nicotiana benthamiana epidermal cells.
(f) In vitro re‐activation assay of SnRK2s. Recombinant SRK2E (subclass III), SRK2G (Subclass I) and SRK2C (subclass II) fused to MBP, which were dephosphorylated by ABI1 protein phosphatase type C fused to GST, were then subjected to in vitro phosphorylation by AtARK1 fused to MBP. The protein mixtures were subjected to an in‐gel phosphorylation assay using histone IIIS as the substrate.
Next, we investigated the osmostress‐ or ABA‐activated kinase activity in ark TKO using an in‐gel phosphorylation assay. This assay detects osmostress‐ or ABA‐activated SnRK2 activity in Arabidopsis (Umezawa et al., 2009; Fujii et al., 2011). Two‐week‐old seedlings of WT and ark TKO plants were immersed in 300 mm sorbitol or 50 µm ABA solution for 60 or 90 min, and total proteins were extracted and subjected to the assay (Figure 4a). Kinase activity exceeding 40 kDa was strongly induced after osmostress or ABA treatment in WT; however, the kinase activity was not detected in srk2dei mutants, indicating that these activities are derived from subclass III SnRK2s (Umezawa et al., 2009). Of interest, these subclass III SnRK2 kinase activities were barely detected in ark TKO plants treated by osmostress during the time span examined. By contrast, ABA‐activated kinase activity of subclass III SnRK2s was still observed in ark TKO plants. To evaluate the role of each AtARK gene in the osmostress‐dependent activation of SnRK2, we performed an in‐gel phosphorylation assay using sorbitol‐treated ark single KO plants along with ark TKO and srkdei plants (Figure 4b). The results clearly indicated the importance of ARK2, along with AtARK1, to a lesser extent, for osmostress‐dependent activation of subclass III SnRK2 in a moderate osmostress condition. To examine whether AtARKs affect osmostress‐activated kinase activities of other SnRK2 subclasses, we performed an in‐gel phosphorylation assay of ark TKO plants treated under even greater osmostress (800 mm sorbitol), in which all three SnRK2 subclasses are activated (Figure S13c). In this greater osmostress condition, we detected osmostress‐activated kinase activity corresponding to all SnRK2 subclasses in both WT and ark TKO plants; however, the activity of subclass III SnRK2 of ark TKO plants was significantly lower than that of WT. Taken together, our results indicated that AtARK1 and AtARK2 mainly phosphorylate and activate subclass III SnRK2 in response to osmostress signals in Arabidopsis.
Figure 4.
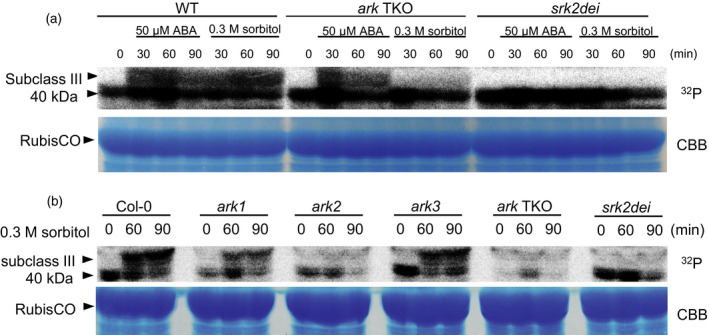
Role of Arabidopsis ARKs in the activation of subclass III SnRK2
(a) In‐gel phosphorylation assay of proteins extracted from 2‐week‐old seedlings of wild‐type (WT), ark triple knockout (TKO) and srk2dei plants treated with 50 μm ABA or 0.3 m sorbitol for the indicated periods using histone IIIS as the substrate. The position of kinase activities of subclass III SnRK2 induced by ABA and osmostress is indicated by an arrowhead. Bottom: Coomassie Brilliant Blue (CBB) staining of the protein samples to demonstrate equal loading.
(b) In‐gel phosphorylation assay of proteins extracted from 2‐week‐old seedlings of WT, each ark single disruptants, ark TKO and srk2dei plants treated with 0.3 m sorbitol for the indicated periods using histone IIIS as the substrate. The position of kinase activities of subclass III SnRK2 induced by ABA and osmostress is indicated by an arrowhead. Bottom: CBB staining of the protein samples to demonstrate equal loading.
A recent study, published when this article was in review, has also highlighted the importance of B3‐MAPKKKs in osmostress‐induced activation of subclass III SnRK2s (Takahashi et al., 2020). In that study, it was found that another Arabidopsis B3‐MAPKKK (At5g11850/M3Kδ1) also phosphorylates a critical Ser171 of SRK2E and that the Arabidopsis triple mutant for AtARK1/AtARK2/At5g11850 failed to activate subclass III SnRK2s in response to osmostress, which may explain why SnRK2 activation by 0.8 m sorbitol was not fully abolished in ark TKO plants (Figure S13c). The same study also showed weak ABA‐insensitive phenotypes in an ark1/ark2 double mutant, which is in agreement with our results (Figure 2f; Figure S2b,c). It is possible that AtARKs also play a role in ABA‐dependent activation of subclass III SnRK2s.
Although activation of SnRK2 has been suggested to involve upstream kinases(s) (Boudsocq et al., 2007), the identity of the kinase(s) has been unknown. Recently, the brassinosteroid (BR) signaling molecules glycogen synthase kinase 3‐like kinase (BIN2) (Cai et al., 2014) and BR1‐associated receptor kinase 1 (BAK1) (Shang et al., 2016) have been shown to play a role in modulation of ABA‐dependent activation of subclass III SnRK2 through its phosphorylation. BIN2 phosphorylates subclass III SnRK2 at serine or threonine residues located in the kinase and in the N‐terminal regulatory domain. These sites are different from those of AtARKs, which phosphorylate Ser residues in the activation loop and are essential for SnRK2 activation. The findings suggest that activation and function of subclass III SnRK2 are mediated through phosphorylation by multiple kinases. Here, we demonstrate that AtARKs are upstream kinases of subclass III SnRK2 in regulating stomatal closure in response to osmostress. By contrast to results with osmostress, ABA treatment activated subclass III SnRK2 in ark TKO plants and in WT. Therefore, we speculate that osmostress activation of SnRK2 kinase activity by AtARK is different from the PYL‐PP2C system. How AtARKs activate SnRK2 in response to osmostress remains to be clarified, although we note that ARK from P. patens interacts with ethylene receptors, which encode the sensor histidine kinases, orthologous to a bacterial two‐component system (Yasumura et al., 2015). The activity of CTR1, a member of Arabidopsis B3‐MAPKKKs, is regulated through physical interaction with ethylene receptors (Merchante et al., 2013). It is possible that AtARK activity is regulated through physical interaction with unidentified sensor histidine kinases at the endoplasmic reticulum membrane, which may perceive osmostress through conformational changes in endoplasmic reticulum membrane structure.
Experimental procedures
Plant materials and growth conditions
Arabidopsis seeds were sown on agar (0.8%, w/v) plates containing full‐strength Murashige and Skoog (MS) salts with a vitamin mixture (10 mg L−1 myoinositol, 200 μg L−1 glycine, 50 μg L−1 nicotinic acid, 50 μg L−1 pyridoxine hydrochloride, 10 μg L−1 thiamine hydrochloride, pH 5.7) and 1% sucrose. Plates were sealed with surgical tape; the seeds were stratified at 4°C for 4–7 days and then transferred to a growth chamber (80 μmol photons m2 sec−1; 16:8 h light/dark photocycle; 22°C) for germination and growth.
Seeds of the following Arabidopsis mutants were obtained from the Arabidopsis Biological Resource Center (Ohio State University): atark1 (SALK_082710), atark2 (SALK_004541) and atark3 (SALK_025685). To generate the atark1/2 double disruptant (DKO), the atark1 mutant was crossed with the atark2 mutant. To identify the homozygosity of each mutation, we genotyped F2 seedlings using genomic PCR (Figure 1 and Table S1). The F3 progeny of the DKO mutant was then crossed with the atark3 mutant to generate the atark1/2/3 triple disruptant (TKO).
ABA and osmostress treatment
For seed germination assay, seeds of WT and ark disruptants were sown on ABA or sorbitol‐containing media and incubated for 4–7 days at 4°C in the dark. Then, seeds were germinated under the above‐described conditions. Any seed with root emergence or green cotyledons was counted for germination rates and greening rates, respectively.
One‐week‐old seedlings of WT and ark disruptants were used for ABA and osmostress treatment. In the ABA sensitivity assay for root growth, seedlings were transferred with forceps onto MS agar plates with or without ABA supplement, and each root length was recorded. The plates were placed vertically in the growth cabinet and incubated under the conditions described above. Root length was measured after 2 weeks and the root growth was expressed relative to the initial root length. In the osmostress‐tolerant assay, seeds were sown on nylon mesh (990‐µm pore size)‐layered MS agar plates and grown for 1 week, then transferred with the nylon mesh onto MS agar plates with or without sorbitol supplement. After 2 weeks, chlorophyll content was measured as described below.
Measurement of chlorophyll content
The chlorophyll of the homogenized protonemal tissue was extracted with 10 ml of 80% (v/v) acetone. To remove cell debris, the samples were centrifuged for 5 min at 9000 g. Supernatants were measured at wavelengths of 645 and 663 nm for the chlorophyll quantification. After measurement, the supernatant was dried up in the initial tube at room temperature until a stable dry weight of each sample was obtained. The total chlorophyll was calculated as: mg chlorophyll per g dry weight 1⁄4 [(A663) (0.00802) + (A645) (0.0202)] × 50/dry weight.
qRT‐PCR analysis
Two‐week‐old WT and ark TKO seedlings were soaked in sterile distilled water for 24 h and then transferred to 50 µm ABA solution or 300 mm sorbitol solution for 0, 1 or 3 h. Total RNA was extracted using the RNeasy plant mini kit (Qiagen, Valencia, CA, USA). cDNA synthesis was carried out using ReverTra qPCR RT‐Master MIX (Toyobo, Osaka, Japan). qRT‐PCR was performed in LightCycler 96 (Roche, Basel, Switzerland) using THUNDERBIRD SYBR qPCR Mix (Toyobo). All qRT‐PCRs were performed using total RNA samples extracted from three independent biological replicates. The reaction mixture was incubated at one cycle of 10 sec at 95°C, 10 sec at 61°C and 10 sec at 72°C and then at 50 cycles of 10 sec at 95°C and 10 sec at 58°C. Actin2 (At3g18780) was used as an internal control. Primers used in the experiment are listed in Table S3.
Water loss measurement
Rosette leaves detached from 3‐week‐old WT and ark TKO seedlings grown in a soil pot were placed on a paper and left on the laboratory bench (24–25°C, relative humidity36%–45%). Loss in fresh weight was monitored at the indicated times.
Stomatal aperture measurements
For drought‐induced stomatal closure measurement, leaves of WT and ark TKO seedlings were detached and placed between a slide glass and a cover slip and left on the laboratory bench (24–26°C, relative humidity 34%–53%) under room light. For ABA‐induced stomatal closure measurement, leaves of WT and ark TKO seedlings were detached and incubated in dimethylsulfoxide with or without 50 µm ABA for 3 h at room temperature under room light, then placed between a slide glass and a cover slip. Guard cell images were captured using a laser scanning microscope (LSM710; Carl Zeiss, Oberkochen, Germany). We measured the width and length of each stomatal pore in the image.
In‐gel phosphorylation assay
SnRK2 activity was detected using an in‐gel phosphorylation assay as described before. Two‐week‐old WT and ark TKO seedlings were soaked in sterile distilled water for 24 h, then transferred to 50 µm ABA solution or 300 mm sorbitol solution for 0, 30 or 90 min. The treated seedlings were crushed and powdered in liquid nitrogen by a mortar and pestle and homogenized in extraction buffer [50 mm HEPES KOH (pH 7.5), 5 mm ethylenediaminetetraacetic acid, 5 mm ethylene glycol tetraacetic acid, 25 mm NaF, 1 mm o‐ranadate, 50 mm β‐glycerophosphate, 2 µg ml−1 leupeptin, 2 μg ml−1 pepstatin A, 20% glycerol, 2 mm dithiothreitol (DTT), 2 mm phenylmethylsulfonyl fluoride]. The crude extracts containing 30 µg total protein were separated by 12% sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE) with histone IIIS as a substrate. After renaturation, the gel was incubated with [γ‐32P]ATP for 1 h, then exposed to the imaging plate and analyzed using BAS ‐2000 (Fujifilm, Tokyo, Japan).
In vitro phosphorylation assay
For preparation of recombinant proteins, DNA fragments of SRK2E, SRK2C, SRK2G, AtARK1, AtARK2, AtARK1N (1–714 aa), AtARK1KD+C (715–992 aa) and AtARK3KD+C (669–951 aa) were amplified from cDNA and fused in‐frame to the pMAL‐c5X vector (New England Biolabs, Ipswich, MA, USA). SRK2EK50N, SRK2EK50N S171A, SRK2EK50N S175A, SRK2EK50N S171A S175A, SRK2CK33N and SRK2GK33N were generated by site‐directed mutagenesis. The maltose binding protein (MBP)‐fusion proteins were expressed and purified from Escherichia coli BL21 (DE3) using amylose resin (New England Biolabs) in accordance with the manufacturer’s instructions, and further purified with a Nanosep® 30‐kDa size‐exclusion column (Pall Corporation, Port Washington, NY, USA). In vitro phosphorylation assays were performed as described previously, with some modifications (Umezawa et al., 2009). The recombinant proteins of SnRK2s and AtARKs (2 µg for each) were mixed and incubated in 50 mm Tris‐HCl (pH 7.5), 5 mm MgCl2, 50 µm ATP and 0.037 MBq of [γ‐32P] ATP (PerkinElmer, Waltham, MA, USA) at 30°C for 1.5 h. Samples were subsequently separated by SDS‐PAGE and phosphorylation levels were detected by autoradiography using BAS‐2500 (Fujifilm).
Reactivation assay
Bacterial recombinant GST‐ABI1 protein was prepared as previously described (Umezawa et al., 2009). MBP‐SRK2E, MBP‐SRK2G or MBP‐SRK2C was bound to amylose resin beads (New England Biolabs) and incubated with 5 µg of GST‐ABI1 for 30 min at 30°C in 50 mm Tris‐HCl (pH 7.5), 5 mm MgCl2 and 5 mm MnCl2. The beads were washed four times with 50 mm Tris‐HCl (pH 7.5), 100 mm NaCl and 1 mm DTT, and SnRK2 proteins were eluted with 50 mm Tris‐HCl (pH 7.5), 100 mm NaCl, 1 mm DTT and 10 mm maltose. A total of 2 µg of each SnRK2 protein was incubated with 1 µg of MBP‐AtARK1 in reaction buffer [50 mm Tris‐HCl (pH 7.5), 5 mm MgCl2, 50 µm cold ATP] at 30°C for 30 min. These proteins were separated by SDS‐PAGE and SnRK2 protein kinase activity was detected using an in‐gel kinase assay with histone IIIS as substrate.
AlphaScreen®
The N‐terminal biotinylated SRK2E (biotin‐SRK2E) proteins and C‐terminal FLAG (DYKDDDDK)‐tagged AtARK truncated proteins, such as AtARK1N (1‐714 aa), AtARK1KD+C (715‐992 aa), AtARK2N (1‐747 aa), AtARK2KD+C (748‐1030 aa), AtARK3N (1‐668 aa) and AtARK3KD+C (669‐951 aa), were expressed using an in vitro transcription/translation system in wheat germ extract (WGE) as described (Nomoto and Tada, 2018). The synthesized proteins were checked by western blotting with streptavidin or anti‐FLAG antibody, respectively. To detect protein–protein interactions, the AlphaScreen® (Amplified Luminescent Proximity Homogeneous Assay) was performed using an AlphaScreen® FLAG® (M2) Detection Kit (PerkinElmer). The biotinylated SRK2E and FLAG‐tagged AtARKs were incubated at 21°C for 12 h in control buffer containing acceptor beads coated with anti‐FLAG antibody, donor beads coated with streptavidin, 0.01% Tween‐20 and 0.1% BSA. The AlphaScreen® luminescence was detected with the multimode plate reader infinite® M1000 Pro (Tecan, Männedorf, Switzerland). WGE with no expressed proteins was employed as negative control for estimation of the luminescence caused by endogenous wheat germ proteins.
BiFC assay
We amplified the full‐length open‐reading frame without a stop codon for AtARK2 or SRK2E by PCR and subcloned it into the 5' upstream of C‐terminal half of EYFP gene (cEYFP) or N‐terminal half of EYFP (nEYFP) to generate translational fusion proteins using pRI201‐AN vector (Takara Bio Inc., Otsu, Japan). Agrobacterium tumefaciens GV3101 cultures harboring each plasmid were used for transformation of Nicotiana benthamiana epidermal cells via infiltration with a 1‐ml syringe. Plants were cultured for 3 days and the epidermal layers were subjected to microscopic observation using a laser scanning microscope (LSM710; Carl Zeiss).
Immunoblot analysis
A total of 6 μg each of total proteins used in the in‐gel phosphorylation assay was separated on 10% SDS‐PAGE and transferred onto a nylon membrane. Phosphorylation of serine residues in the SnRK2 activation loop was detected using an antibody recognizing phosphorylated serine residues in this region. The antibody‐reacted proteins were visualized using the chemiluminescent ECL Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Little Chalfont, UK) and detected with ChemiDoc XRS Plus (Bio‐Rad Laboratories, Hercules, CA, USA). The anti‐phosphorylated serine residues in the activation loop of SnRK2 and the anti‐rabbit IgG (H + L) (#170‐6515, Bio‐Rad) were used at dilutions of 1:25 and 1:50 000, respectively.
RNA sequencing analysis
Two‐week‐old seedlings of WT and ark TKO plans were treated with 300 mm sorbitol or 50 µm ABA solution for 1 or 3 h, and total RNA was extracted using the RNeasy plant mini kit (Qiagen). Total RNA quality was confirmed with a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). RNA libraries were prepared according to the protocol accompanying the NEBNext Ultra II RNA Library Prep Kit for Illumina (New England Biolabs, Beverly, MA, USA). The average length of the library measured with the Bioanalyzer was 350 bp. The concentration of each library was confirmed with a Kapa Library Quantification Kit (NIPPON Genetics, Tokyo, Japan) and the concentration was adjusted to 10 nm. The library was sequenced with the Hiseq 2500 system (Illumina) and single reads from each library were processed using bcl2fastq, version 2.18.0.12 (Illumina) in FASTQ format. The reference genome of Arabidopsis thaliana (TAIR10) was obtained from the TAIR website (https://www.arabidopsis.org). Mapping of reads to the reference genome of A. thaliana and extraction of differentially expressed genes (DEGs) were performed using clc genomics workbench, version 12 (Qiagen). The DEGs satisfy a false discovery rate of less than 0.05 and a fold change greater than 2. Genes with a fold change greater than 1.5 compared to WT were defined as AtARK‐regulated genes.
Statistical analysis
Statistical significance between two samples was analyzed by a two‐tailed Student’s t‐test. The numbers of biological repeats are indicated as and where appropriate. Experiments were repeated at least three times on different occasions to confirm the reproducibility, and representative results of experiments are presented.
Accession Numbers
The raw single reads of RNA sequencing in the present study are available as DDBJ Sequence Read Archive accession number DRA009196.
Author Contributions
SK, GM and HB generated the multiple combinations of Arabidopsis ark transgenic plants and performed physiological experiments. AS performed RNA sequencing, qRT‐PCR and the in‐gel phosphorylation assay. YK, TU and DT performed the in vitro phosphorylation assay and protein___protein interaction experiments. IY, TT and YS designed the experiments and wrote the manuscript.
Conflict of Interest
The authors declare no conflict of interest.
Supporting information
Figure S1. Comparison of the EDR domain of Arabidopsis B3 MAPKKK proteins with that of P. patens ARK.
Figure S2. Digital expression pattern of AtARK1 by the eFP browser.
Figure S3. Digital expression pattern of AtARK2 by the eFP browser.
Figure S4. Digital expression pattern of AtARK3 by the eFP browser.
Figure S5. Expression analysis of AtARK genes by qRT‐PCR.
Figure S6. Confirmation of null expression for AtARK genes in atark mutants by RT‐PCR.
Figure S7. ABA sensitivity in seed germination of WT and atark plants.
Figure S8. ABA sensitivity in root growth of WT and atark1/2/3 TKO plants.
Figure S9. Transcriptomic analysis of AtARK downstream factors.
Figure S10. Effect of ARK disruption on ABA accumulation.
Figure S11. Comparison of ABF‐regulated genes and AtARK‐regulated genes.
Figure S12. Phosphorylation of SRK2E by AtARK.
Figure S13. Analysis of SnRK2 activation by AtARKs.
Table S1. List of ABA and osmostress responsive genes and differentially expressed genes in ark TKO plants.
Table S2. Overlap of ‘ARK‐regulated’ and ABF‐regulated genes in response to ABA.
Table S3. Primer sequences used in the present study.
Acknowledgements
We thank Dr Yuki Hayashi for technical assistance. This study was supported in part by Grants‐in‐Aid for Scientific Research (No. 18H04774 to D.T., No. 15547992 and 19H03240 to T.U. and 19K06713 to Y.S.) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (MEXT), as well as by Precursory Research for Embryonic Science and Technology (PRESTO) from Japan Science and Technology Agency (T.U.).
References
- Belin, C. , de Franco, P.‐O. , Bourbousse, C. , Chaignepain, S. , Schmitter, J.‐M. , Vavasseur, A. , Giraudat, J. , Barbier‐Brygoo, H. and Thomine, S. (2006) Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 141, 1316–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq, M. , Barbier‐Brygoo, H. and Laurière, C. (2004) Identification of nine sucrose nonfermenting 1‐related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J. Biol. Chem. 279, 41758–41766. [DOI] [PubMed] [Google Scholar]
- Boudsocq, M. , Droillard, M.‐J. , Barbier‐Brygoo, H. and Laurière, C. (2007) Different phosphorylation mechanisms are involved in the activation of sucrose non‐fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol. Biol. 63, 491–503. [DOI] [PubMed] [Google Scholar]
- Cai, Z. , Liu, J. , Wang, H. et al . (2014) GSK3‐like kinases positively modulate abscisic acid signaling through phosphorylating subgroup III SnRK2s in Arabidopsis. Proc. Natl Acad. Sci. USA, 111, 9651–9656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye, C.A. , Tang, D. and Innes, R.W. (2001) Negative regulation of defense responses in plants by a conserved MAPKK kinase. Proc. Natl Acad. Sci. USA, 98, 373–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. and Zhu, J.‐K. (2009) Arabidopsis mutant deficient in 3 abscisic acid‐activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl Acad. Sci. USA, 106, 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. , Chinnusamy, V. , Rodrigues, A. et al . (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature, 462, 660–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, H. , Verslues, P.E. and Zhu, J.‐K. (2011) Arabidopsis decuple mutant reveals the importance of SnRK2 kinases in osmotic stress responses in vivo. Proc. Natl Acad. Sci. USA, 108, 1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, Y. , Nakashima, K. , Yoshida, T. et al . (2009) Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Li, C.Y. , Qi, Y. , Park, S. and Gibson, S.I. (2014) SIS8, a putative mitogen‐activated protein kinase kinase kinase, regulates sugar‐resistant seedling development in Arabidopsis. Plant J. 77, 577–588. [DOI] [PubMed] [Google Scholar]
- Ikegami, K. , Okamoto, M. , Seo, M. and Koshiba, T. (2009) Activation of abscisic acid biosynthesis in the leaves of Arabidopsis thaliana in response to water deficit. J. Plant Res. 122, 235–243. [DOI] [PubMed] [Google Scholar]
- Kieber, J.J. , Rothenberg, M. , Roman, G. , Feldmann, K.A. and Ecker, J.R. (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell, 72, 427–441. [DOI] [PubMed] [Google Scholar]
- Kobayashi, Y. , Yamamoto, S. , Minami, H. , Kagaya, Y. and Hattori, T. (2004) Differential activation of the rice sucrose nonfermenting1‐related protein kinase2 family by hyperosmotic stress and abscisic acid. Plant Cell, 16, 1163–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leon‐Kloosterziel, K.M. , Gil, M.A. , Ruijs, G.J. , Jacobsen, S.E. , Olszewski, N.E. , Schwartz, S.H. , Zeevaart, J.A. and Koornneef, M. (1996) Isolation and characterization of abscisic acid‐deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. and Grill, E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- MAPK Group (2002) Mitogen‐activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Merchante, C. , Alonso, J.M. and Stepanova, A.N. (2013) Ethylene signaling: simple ligand, complex regulation. Curr. Opin. Plant Biol. 16, 554–560. [DOI] [PubMed] [Google Scholar]
- Mustilli, A.‐C. , Merlot, S. , Vavasseur, A. , Fenzi, F. and Giraudat, J. (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell, 14, 3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima, K. , Fujita, Y. , Kanamori, N. et al . (2009) Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. [DOI] [PubMed] [Google Scholar]
- Nomoto, M. and Tada, Y. (2018) Cell‐free protein synthesis of plant transcription factors. Methods Mol. Biol. 1830, 337–349. [DOI] [PubMed] [Google Scholar]
- Park, S.‐Y. , Fung, P. , Nishimura, N. et al . (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saruhashi, M. , Kumar Ghosh, T. , Arai, K. et al . (2015) Plant Raf‐like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1‐related protein kinase2. Proc. Natl Acad. Sci. USA, 112, E6388–E6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato, H. , Takasaki, H. , Takahashi, F. et al . (2018) Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl Acad. Sci. USA, 115, E11178–E11187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, Y. , Dai, C. , Lee, M.M. , Kwak, J.M. and Nam, K.H. (2016) BRI1‐associated receptor kinase 1 regulates guard cell ABA signaling mediated by open stomata 1 in Arabidopsis. Mol. Plant, 9, 447–460. [DOI] [PubMed] [Google Scholar]
- Shinozawa, A. , Otake, R. , Takezawa, D. et al . (2019) SnRK2 protein kinases represent an ancient system in plants for adaptation to a terrestrial environment. Commun. Biol. 2, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson, S.R. , Kamisugi, Y. , Trinh, C.H. et al . (2016) Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON‐RESPONSIVE (ANR), a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell, 28, 1310–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, Y. , Zhang, J. , Hsu, P.‐K. et al . (2020) MAP3Kinase‐dependent SnRK2‐kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takezawa, D. , Komatsu, K. and Sakata, Y. (2011) ABA in bryophytes: how a universal growth regulator in life became a plant hormone? J. Plant Res. 124, 437–453. [DOI] [PubMed] [Google Scholar]
- Ullman, E.F. , Kirakossian, H. , Singh, S. et al . (1994) Luminescent oxygen channeling immunoassay: measurement of particle binding kinetics by chemiluminescence. Proc. Natl Acad. Sci. USA, 91, 5426–5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa, T. , Sugiyama, N. , Mizoguchi, M. , Hayashi, S. , Myouga, F. , Yamaguchi‐Shinozaki, K. , Ishihama, Y. , Hirayama, T. and Shinozaki, K. (2009) Type 2C protein phosphatases directly regulate abscisic acid‐activated protein kinases in Arabidopsis. Proc. Natl Acad. Sci. USA, 106, 17588–17593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umezawa, T. , Nakashima, K. , Miyakawa, T. , Kuromori, T. , Tanokura, M. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2010) Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M. , Chaban, C. , Schütze, K. et al . (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Yasumura, Y. , Pierik, R. , Kelly, S. , Sakuta, M. , Voesenek, L.A.C.J. and Harberd, N.P. (2015) An ancestral role for constitutive triple response 1 (CTR1) proteins in both ethylene and abscisic acid signaling. Plant Physiol. 169, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida, R. , Hobo, T. , Ichimura, K. , Mizoguchi, T. , Takahashi, F. , Aronso, J. , Ecker, J.R. and Shinozaki, K. (2002) ABA‐activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483. [DOI] [PubMed] [Google Scholar]
- Yoshida, T. , Fujita, Y. , Maruyama, K. , Mogami, J. , Todaka, D. , Shinozaki, K. and Yamaguchi‐Shinozaki, K. (2014) Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of the EDR domain of Arabidopsis B3 MAPKKK proteins with that of P. patens ARK.
Figure S2. Digital expression pattern of AtARK1 by the eFP browser.
Figure S3. Digital expression pattern of AtARK2 by the eFP browser.
Figure S4. Digital expression pattern of AtARK3 by the eFP browser.
Figure S5. Expression analysis of AtARK genes by qRT‐PCR.
Figure S6. Confirmation of null expression for AtARK genes in atark mutants by RT‐PCR.
Figure S7. ABA sensitivity in seed germination of WT and atark plants.
Figure S8. ABA sensitivity in root growth of WT and atark1/2/3 TKO plants.
Figure S9. Transcriptomic analysis of AtARK downstream factors.
Figure S10. Effect of ARK disruption on ABA accumulation.
Figure S11. Comparison of ABF‐regulated genes and AtARK‐regulated genes.
Figure S12. Phosphorylation of SRK2E by AtARK.
Figure S13. Analysis of SnRK2 activation by AtARKs.
Table S1. List of ABA and osmostress responsive genes and differentially expressed genes in ark TKO plants.
Table S2. Overlap of ‘ARK‐regulated’ and ABF‐regulated genes in response to ABA.
Table S3. Primer sequences used in the present study.


