Abstract
The genetic analysis of large numbers of Saccharomyces cerevisiae × S. uvarum (“cevarum”) and S. kudriavzevii × S. uvarum (“kudvarum”) hybrids in our previous studies revealed that these species are isolated by a postzygotic double-sterility barrier. We proposed a model in which the first barrier is due to the abruption of the meiotic process by the failure of the chromosomes of the subgenomes to pair (and recombine) in meiosis and the second barrier is assumed to be the result of the suppression of mating by allospecific MAT heterozygosity. While the former is analogous to the major mechanism of postzygotic reproductive isolation in plants and animals, the latter seems to be Saccharomyces specific. To bolster the assumed involvement of MAT in the second sterility barrier, we produced synthetic alloploid two-species cevarum and kudvarum hybrids with homo- and heterothallic backgrounds as well as three-species S. cerevisiae × S. kudvarum × S. uvarum (“cekudvarum”) hybrids by mass-mating and examined their MAT loci using species- and cassette-specific primer pairs. We found that the allospecific MAT heterozygosity repressed MAT switching and mating in the hybrids and in the viable but sterile spores produced by the cevarum hybrids that had increased (allotetraploid) genomes. The loss of heterozygosity by meiotic malsegregation of MAT-carrying chromosomes in the latter hybrids broke down the sterility barrier. The resulting spores nullisomic for the S. uvarum chromosome produced vegetative cells capable of MAT switching and conjugation, opening the way for GARMe (Genome Autoreduction in Meiosis), the process that leads to chimeric genomes.
Keywords: Interspecies hybridisation, Sterility barrier, Yeast, MAT, Mating, Sporulation
Introduction
Nature employs various ‘barriers’ to keep closely related species distinct from one another. One of these barriers is reproductive isolation that maintains the integrity of species over time by preventing genetic admixture (Coyne and Orr 2004). The reproductive isolation mechanisms can be prezygotic or postzygotic depending on whether they operate before or after fertilisation (Seehausen et al. 2014). The former prevents the formation of viable hybrid zygotes; while, the latter results in hybrids in which the incompatibility of the parental genomes causes aberrant development, low fitness, and/or sterility (Ouyang and Zhang 2013). Hybrid sterility is one of the earliest reproductive isolation mechanisms to evolve between two recently diverged species (Coyne and Orr 2004). The hybrids of such species are viable but incapable of sexual reproduction. Their sterility is mainly due to the failure of the chromosomes of the subgenomes to pair in meiosis (e.g. White 1978; Levin 2002; Li et al. 2009; Sigel 2016; Lavinscky et al. 2017; Forejt 1996) and/or to functional incompatibility of genes involved in gametogenesis (Johnson 2010). In the former case, the synteny and sequence differences prevent the (homeologous, allosyndetic) chromosomes of the subgenomes from efficient pairing in prophase-I of meiosis. This problem can be circumvented by genome duplication because within the duplicated subgenomes, each chromosome has a homologous (autosyndetic) partner to pair with, without interacting (allosyndetic pairing) with the chromosomes of the companion subgenome. As a result, each subgenome divides essentially autonomously and each gamete receives a copy of each chromosome of each subgenome. This process resulting in allodiploid gametes is referred to as autodiploidisation of the allotetraploid meiosis (Hutchinson et al. 1983). The allodiploid gametes can mate with other allodiploid gametes to produce allotetraploid progeny (Rieseberg and Willis 2007). If the subgenomes are functionally compatible, the allotetraploid hybrids (in contrast to the allodiploids) are capable of sexual propagation and can evolve into novel species distinct from the parental species. It is generally accepted that many plant species evolved in this way (by hybrid speciation) from interspecies hybrids (e.g. Soltis and Soltis 2009).
The species of the budding-yeast genus Saccharomyces are postzygotically isolated by a double-sterility barrier (for a recent review, see Sipiczki 2018). The Saccharomyces species form viable allodiploid hybrids in all combinations. The hybrid cells propagate asexually by mitotic divisions but cannot produce gametes (viable ascospores) by meiosis. Like in animal and plant allodiploid hybrids, the failure of viable gamete production (sporulation deficiency) in the Saccharomyces hybrids is also due to poor and promiscuous pairing between the chromosomes of the subgenomes. The inability of the allodiploid hybrids to produce viable ascospores is the first part of the double-sterility barrier. However, the spore viability problem can be circumvented by genome duplication. Like the allotetraploid plant hybrids, the allotetraploid Saccharomyces hybrids also produce viable gametes (ascospores) but, unlike the plant and animal allodiploid gametes, the allodiploid yeast ascospores cannot function as gametes because they cannot mate (fertilise). Thus, in the Saccharomyces hybrids, the restoration of spore viability does not imply that fertility is also restored. The mating incompetence of the allodiploid spores represents the second sterility barrier (Pfliegler et al. 2012) which has no equivalent in plants. This difference between the biological isolation of the plant species and the Saccharomyces species was proposed to be due to the different modes of the genetic determination of sexuality (Pfliegler et al. 2012). Being viable, the allodiploid spores germinate and produce clones of vegetatively propagating sterile cells (so-called spore clones or clones of “propagating gametes”) which is another difference from higher plants.
Our previous studies on “cevarum” (S. cerevisiae × S. uvarum) and “kudvarum” (S. kudriavzevii × S. uvarum) hybrids revealed that the allopolyploid hybrids can occasionally form fertile spores. These spores generate clones of cells which are able to mate with each other and the resulting “intraclonal F2 hybrids” also produce fertile spores. We noticed that these fertile spores had alloaneuploid genomes nullisomic for one or the other parental Chr III (Antunovics et al. 2005; Pfliegler et al. 2012; Karanyicz et al. 2017). As Chr III of S. cerevisiae and its counterparts in the other species carry the MAT locus, we proposed that the reactivation of the sexual processes (break-down of sterility barrier) is attributable to the loss of MAT heterozygosity (Pfliegler et al. 2012). All sterile spores had both parental types of Chr III; whereas, all fertile spores had only one type of Chr. III.
The “mating-type locus” MAT is the central regulator of the sexual processes (both gametogenesis/sporulation and fertilisation/mating) in S. cerevisiae (for reviews, see, e.g. Nasmyth 1982; Herskowitz 1988; Haber 2012; Thon et al. 2019). It has two idiomorphs (orthologous cassettes), MATa and MATalpha. In homothallic background (the HO gene is active), haploid cells can reversibly switch their mating types during vegetative propagation (by reversible replacement of the MAT cassettes in the MAT locus) and can conjugate with other haploid cells of opposite mating type but cannot sporulate. This is because both the MATa and the MATalpha alleles (cassettes) activate the mating-type switching machinery and the so-called haploid-specific genes whose activity is required for mating but both alleles repress the diploid-specific genes that are required for meiosis and sporulation. The mating of a MATa cell with a MATalpha cell results in a MATa/MATalpha zygote. The zygote produces vegetative cells of MATa/MATalpha genotype. In these cells, the proteins encoded by the different MAT cassettes interact to suppress mating-type switching and mating, but activate the diploid-specific genes that launch meiosis and sporulation in response to starvation signals. Thus, the haploid cells having single MAT loci (either MATa or MATalpha) are mating competent and sporulation incompetent; whereas, the diploid cells having both MATa and MATalpha cassettes are mating deficient and sporulation competent. Another difference between them is that the haploid cells can switch mating types; whereas, the MATa/MATalpha diploids cannot. As all Saccharomyces species have MAT loci which are interchangeable between the species (e.g. Pfliegler et al. 2012), the sexual processes are regulated much in the same way in the entire genus. Our model of the double-sterility barrier assumes that the allodiploid spores are sterile because they receive copies of different MAT cassettes from the subgenomes during the autodiploidised allotetraploid meiosis (Karanyicz et al. 2017) and the interactions of the cassettes block the mating process. The suppression of the mating activity which prevents the allodiploid spores from functioning as gametes is the second sterility barrier. The model also assumes that the subgenomes preserve the cassettes of the hybridising parental cells unchanged (no cassette switching takes place) during the propagation of the hybrid cells. The role of MAT in the sterility barrier may not be confined to Saccharomyces. MAT and MTL (MAT-like) loci were implicated in the sterility of alloploid Zygosaccharomyces strains isolated from natural substrates (Watanabe et al. 2017; Ortiz-Merino et al. 2017; Braun-Galleani et al. 2018; Bizzarri et al. 2019).
The proposed involvement of the MAT locus in the second sterility barrier relies upon the assumption that the MAT cassettes of the hybridising species remain active and functionally compatible with each other upon hybridisation. As this part of the model was largely hypothetical, we set out in this work to examine the MAT genotypes of hybrids and their viable spores. With this aim in view, we produced synthetic alloploid two-species cevarum (S. cerevisiae × S. uvarum) and kudvarum (S. kudriavzevii × S. uvarum) hybrids with homo- and heterothallic backgrounds as well as three-species cekudvarum (S. cerevisiae × S. kudriavzevii × S. uvarum) hybrids by crossing heterothallic and homothallic strains and examined their MAT loci. To be able to identify the MATa and MATalpha cassettes of each of the three species, we designed six pairs of species- and cassette-specific primers. Consistent with our model, the alloploid hybrids of the species and the sterile spores had complete alloploid karyotypes and stable MAT heterozygosity; whereas, the fertile spores were alloaneuploid, possessed only one parental Chr III, and formed clones in which the cells switched their mating types and mated with each other. The allotriploid cekudvarum hybrids were sterile and had three stable (non-switching) parental MAT cassettes.
Materials and methods
Strains and culture media
All strains used in this study are listed in Table 1. The strains involved in the tests of the MAT-specific primers and the parental strains used for hybridisation were maintained on YEA plates (yeast extract glucose agar) or in YEL broth (L stands for liquid). Mating tests were performed on YEA plates. Sporulation was tested on acetate SPA (sporulation agar). Hybrids were selected and maintained on MMA (minimal medium agar) or on MMA supplemented with uracil. The composition of the media was described in Sipiczki and Ferenczy (1977) and Antunovics et al. (2005).
Table 1.
List of strains
| Identification number | Strain | Genotype/phenotype | References |
|---|---|---|---|
| 10-170 | Saccharomyces cerevisiae X4005-11A | MATaSc hoSc leu2Sc | Antunovics et al. (2005) |
| 10-512 | Saccharomyces uvarum CBS 395 | MATaSu | CBS |
| 10-522 | Saccharomyces uvarum m9 | MATaSu, MATalphaSu HOSu ura3Su | Antunovics et al. (2005) |
| 10-642 | Saccharomyces cerevisiae ATCC 204508/S288c | MATalphaSc hoSc | ATCC |
| 10-643 | Saccharomyces kudriavzevii CBS 8840T | MATaSk, MATalphaSk HO | CBS |
| 10-1650 | Saccharomyces uvarum JRY9192 SSS110 | MATaSu ade2Su ura3Su hoSu | Scannel et al. (2011) |
| 10-1651 | Saccharomyces uvarum JRY9193 SSS111 | MATalphaSu ade2Su ura3Su hoSu | Scannel et al. (2011) |
| 10-1652 | Saccharomyces kudriavzevii FM1183 SSS410 | MATalphaSk trp1Sk ura3Sk hoSk | Scannel et al. (2011) |
| 10-1653 | Saccharomyces kudriavzevii FM1193 SSS411 | MATaSk trp1Sk ura3Sk hoSk | Scannel et al. (2011) |
| A2, A3, A27 | Two-species cevarum hybrids produced with mass-mating of 10-170 and 10-522 | MATaSc/MATalphaSu HOSu/hoSc LEU2Su/leu2Sc URA3Sc/ura3Su | This study |
| A3/1a, A3/1c | Spore clones of cevarun A3 | Prototrophic | This study |
| A3/1b, A3/1d | Spore clones of cevarun A3 | leu2Sc | This study |
| II/1, II/6 | Two-species kudvarum hybrids produced with mass-mating of 10-1651 and 10-1653 | MATaSk/MATalphaSu hoSk/hoSu ADE2Sk/ade2Su trp1Sk/TRP1Su ura3Sk/ura3Su | This study |
| II/6.1, II/6.2, II/6.3 | Three-species cekudvarum hybrids produced with mass-mating of 10-170 and II/6 | MATaSc/ MATaSk/MATalphaSu hoSc/hoSk/hoSu ADE2Sc/ADE2Sk/ade2Su TRP1Sc/trp1Sk/TRP1Su URA3Sc/ura3Sk/ura3Su leu2Sc/LEU2Sk/LEU2Su | This study |
CBS: CBS-KNAW Collections, Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands
ATCC: American Type Culture Collection, Manassas, VA 20108, USA
Hybridisation
Two types of synthetic two-species hybrids were produced. The cevarum hybrids were constructed by mass-mating of 10-170 S. cerevisiae and 10-522 S. uvarum cultures grown on the sporulation medium SPA in YEL. The sporulation step was included in the procedure to increase the efficiency of mating and ensure that preferentially haploid × haploid mating occurs. The cells of the homothallic 10-522 S. uvarum strain are diploid and cannot mate due to their MATa/MATalpha heterozygosity. Its spores are haploid of either a or alpha mating type. The latter can mate with the haploid cells of 10-170 S. cerevisiae in the complete medium YEL (Antunovics et al. 2005; Pfliegler et al. 2012). To obtain kudvarum hybrids, exponential-phase 10-1651 S. uvarum and 10-1653 S. kudriavzevii cultures were mass-mated in YEL. Conjugation was monitored microscopically in both cases and samples containing zygotes were spread on MMA plates (for the identification of cevarum hybrids) or MMA plates supplemented with uracil (for the identification of kudvarum hybrids). To produce three-species cekudvarum hybrids, cells of exponential-phase 10-170 S. cerevisiae and II/6 kudvarum cultures grown in YEL were mass-mated in YEL. Their hybrids formed prototrophic colonies on MMA plates. Individual colonies (as products of individual zygotes) were isolated from the plates and stored at − 80 °C to preserve the homogeneity of the hybrid cell populations as much as possible. We considered this precaution necessary because the hybrid genomes can segregate during mitotic propagation of the cells (for a review, see Sipiczki, 2018). We used this culture as inoculum in all experiments instead of streaking out samples and selecting a colony which might already be a segregant. The hybrid nature of the isolates was verified by comparing their electrophoretic karyotypes with those of the parental strains as described previously (Antunovics et al. 2005) and by PCR–RFLP analysis of the FUS1 gene (see in “Molecular methods”).
Mating and sporulation tests and generation of spore clones
Mating activity was tested in exponential-phase mixed cultures. Equal volumes of overnight cultures (~ 5 × 106 cells ml) of the strains grown in YEL were mixed, vortexed, centrifuged and then 10 µl of the wet pellet was dropped on YEA. After incubation at room temperature for 4–6 h, samples were taken from the mixed population of cells and examined microscopically. The testers of mating competences were S. cerevisiae 10-170 (MATa), S. cerevisiae 10-642 (MATalpha), S. kudriavzevii 10-1652 (MATalpha), S. kudriavzevii (MATa), S. uvarum 10-1650 (MATa) and S. uvarum 10-1651 (MATalpha).
Spore viability was examined by tetrad analysis. Cells of the hybrid to be tested were inoculated onto SPA plates and incubated for 5 days at room temperature. Four-spore asci were pulled out from samples of the sporulating culture transferred onto YEA plates, dissected by micromanipulation and their spores were separated. The viable spores formed colonies on the medium. As the colonies were formed by single spores, we considered them spore clones (clones of vegetative descendants of spores). The clones to be further examined were isolated and preserved at – 70 °C to prevent genomic changes by GARMi (Sipiczki 2018).
Molecular methods
For the amplification of nuclear sequences, genomic DNA was isolated from 50-ml overnight cultures grown in YEL at 25 °C as described in Antunovics et al. (2005). Segments of the FUS1 genes were amplified with two primer pairs. FUS1F (ACCGCAGCATATACTGACACC) and FUS1R (ACTTTTTCACCCAGCGAGAT) amplified a 870-bp-long fragment from FUS1Sc and FUS1Su but did not recognise FUS1Sk; whereas, FUS1kF (CGACAACAACTGTGATGACGAC) and FUS1kR (TGAAATATGTAGAACCTCTCAAGAACC) produced a 792-bp-long fragment from FUS1Sk. To distinguish the FUS1 markers of the species, the amplicons were digested with TaqI that generated unique restriction patterns for each species (S. cerevisiae: 342, 266, 262; S. kudriavzevii: 473, 276, 43; S. uvarum: 371, 336, 163). The MAT cassettes were amplified by primer pairs suitable for the specific amplification of regions of the three types of MATa and the three types of MATalpha cassettes. The sequences of the primers and the length of the amplicons are shown in Table 2. PCR reactions were performed with the following programme: 95 °C for 5 min, 30 × (94 °C for 1 min, Tm × °C for 1 min), 72 °C for 5 min. Tm was set to 47 °C for the amplification of the MATa segments of all species and MATalphaSu, 50 °C for the amplification of the rest of the MAT sequences, and 54 °C for FUS1. As certain MAT amplicons did not differ significantly in size, we did not apply multiplex PCR. All MAT amplifications were carried out with single primer pairs. The amplified DNA fragments were examined by electrophoresis in agarose gel. Electrophoretic karyotyping was performed with the Bio-Rad CHEF Mapper system as described previously (Antunovics et al. 2005).
Table 2.
Primers used for the identification of the MAT cassettes of the species
| Species | Primer | Sequence | Amplicon size (bp) | |
|---|---|---|---|---|
| S. cerevisiae | MATaSc | F: CCACATTAAAAAAGAGAAGAGC | MATaSc–Scout: | 509 |
| MATalphaSc | F: TAAAATCCAAATTCACAGGATAGCGTCT | MATalphaSc- Scout: | 672 | |
| Scout | R: TATGGTTAAGATAAGAACAAAGAATG | |||
| S. kudriavzevii | MATaSk | F: GTATGAAAAATCAAGCTAA | MATaSk- Skout: | 309 |
| MATalphaSk | F: GTAATGGCATAGTGAAACGAATAAGT | MATalphaSk- Skout: | 657 | |
| Skout | R: GTAAATACCTCAAAGGAATTATCA | |||
| S. uvarum | MATaSu | F: CAACGTGAATCAATCCTAA | MATaSu- Suout: | 439 |
| MATalphaSu | F: TCGAGAAAAGCATCAATAACAC | MATalphaSu- Suout: | 603 | |
| Suout | R: TCACCAAATACGAAAAGTAA | |||
Results
Hybridisation
For the examination of the involvement of the MAT alleles in the second sterility barrier, two-species cevarum and kudvarum hybrids and three-species cekudvarum hybrids were constructed. The hybrids were subjected to karyotyping and mating tests with the heterothallic parental strains. All hybrids had hybrid karyotypes (examples are shown in Fig. 1) and none of them conjugated (mated) with any of the testers. From these results, we inferred that the hybrids were alloploid and heterozygous at the MAT locus.
Fig. 1.
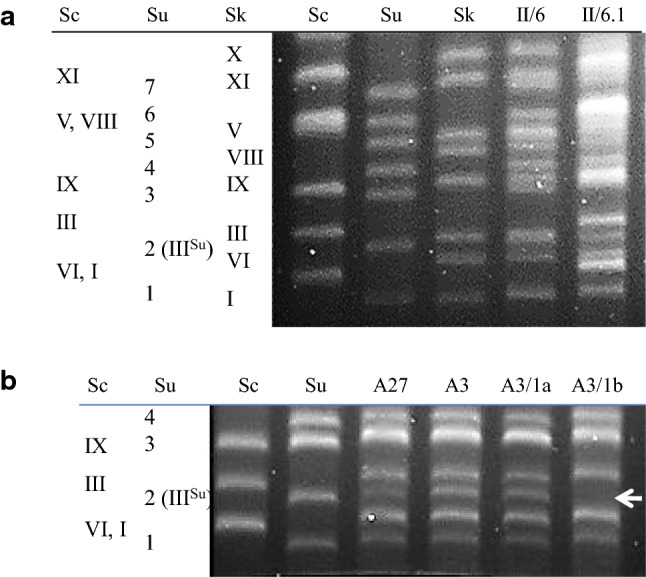
Karyotypes of parental strains, hybrids and spore clones. a Karyotypes of a two-species kudvarum hybrid, a three-species cekudvarum hybrid and the parental strains. Only the regions of smaller chromosomes are shown in which individual bands can be distinguished in the cekudvarum karyotype. b Karyotypes of two-species cevarum hybrids and spore clones. Only the regions containing the bands of the MAT-carrying chromosomes are shown. The columns on the left sides of the gel photographs show the conventional numbering of chromosomes in the S. cerevisiae, S. uvarum and S. kudriavzevii genomes. The S. uvarum chromosomes are numbered according to Nguyen et al (2000). Sc: S. cerevisiae 10-170; Sk: S. kudriavzevii 10-1653; Su: S. uvarum 10-522; II/6: kudvarum hybrid; II/6.1: cekudvarum hybrid; A27 and A3: cevarum hybrids, A3/1a: sterile spore clone of A3; A3/1b: fertile spore clone of A3. Arrowhead marks the position of the missing Chr IIISu (Chr 2) in the fertile spore clone A3/1b
The cevarum hybrids obtained from mass-mating of the heterothallic 10-170 S. cerevisiae and the homothallic 10-522 S. uvarum strains had inactive hoSc and functional HOSu genes. The mass-mating of the heterothallic 10-1653 S. kudriavzevii and 10-1651 S. uvarum cells resulted in kudvarum hybrids having inactive ho genes in both subgenomes. For further studies, we selected three HOSu/hoSc cevarum hybrids (A2, A3 and A27) and two hoSk/hoSu kudvarum (II/1 and II/6) hybrids.
As both 10-1653 and 10-1651 were ura3, the kudvarum hybrids were auxotrophic for uracil. To produce three-species cekudvarum hybrids with the method described in Sipiczki (2019), one of them (II/6) was mass-mated with the heterothallic leucine-auxotrophic S. cerevisiae strain 10-170. Although the kudvarum parent proved to be inactive in the mating tests, we presumed that their mating block was not absolute and mating-competent allodiploid cells might occasionally be produced during vegetative propagation. This assumption was based on the observations that heterozygous MATa/MATalpha S. cerevisiae cells converted at very low frequency to homozygous cells capable of mating (“rare mating”) by mitotic gene conversion (Gunge and Nakatomi 1972). As expected, a few prototrophic colonies appeared on the selective medium when larger samples of the mass-mated mixed population were spread on the plates. The karyotypes of these colonies had bands characteristic of the S. cerevisiae parent in addition to the kudvarum bands (Fig. 1a) and their cells did not conjugate with cells of the parental strains. To reinforce the karyotyping results, we also examined the FUS1 genes located on Chr III (between MAT and the centromere) in the genomes of the parental species (Scannel et al. 2011). The PCR–RFLP analysis detected all the three orthologues in the prototrophic colonies. The prototrophic phenotype, the presence of characteristic chromosomal bands of three parental strains and the presence of all parental versions of the FUS1 marker implied that these colonies were allotriploid cekudvarum hybrids. The FUS1 genotype and the lack of mating competence further indicated that they had heterozygous MAT genotypes.
Hybrid ploidy
Numerous previous studies found that allodiploid Saccharomyces hybrids possessing single sets of parental chromosomes were either defective in sporulation or produced viable spores at extremely low frequencies; whereas, the spores of the allopolyploids that had duplicated sets of chromosomes in the subgenomes were viable (for a review, see Sipiczki 2018). Thus, allodiploid hybrids can be distinguished from allopolyploid hybrids by testing their spores for viability. We found that the cevarum hybrid A27 formed no spores; whereas, A2 and A3 and the kudvarum hybrids II/1 and II/6 formed asci when being cultivated on the sporulation medium SPA. To examine the viability of the spores of the latter hybrids, 10 four-spored asci were dissected with micromanipulation from each sporulating culture and their spores were separated on the complete medium YEA. None of the spores of the kudvarum hybrids formed colonies; whereas, high proportions (32/40 and 37/40, respectively) of the spores of the cevarum hybrids A2 and A3 proved viable. From these results, we inferred that the cevarum hybrid A27 and the kudvarum hybrids were allodiploids; whereas, the ploidy of A2 and A3 was higher. The high level of spore viability indicates that the latter hybrids had allotetraploid genomes because triploids produce viable spores at much lower rates even when they have autoploid genomes (e.g. St Charles et al. 2010). Allotetraploids can arise from allodiploids by spontaneous genome duplication. Recent studies demonstrated that genome duplication by endoreduplication is a fairly common event in Saccharomyces (Harari et al. 2018). Besides, all spores of A2 and the majority of the spores of A3 formed prototrophic colonies. As the hybrids were heterozygous LEU2Su/leu2Sc URA3Sc/ura3Su, their prototrophy indicated that they were heterozygous allodiploid products of the tetraploid meiosis. A shown previously, Saccharomyces allotetraploids form allodiploid spores due to the autodiploidisation of the allotetraploid meiosis (Karanyicz et al. 2017). None of the spores of the three-species cekudvarum hybrid II/6.1 formed colonies, suggesting that they had single sets of parental chromosomes.
Meiotic segregation
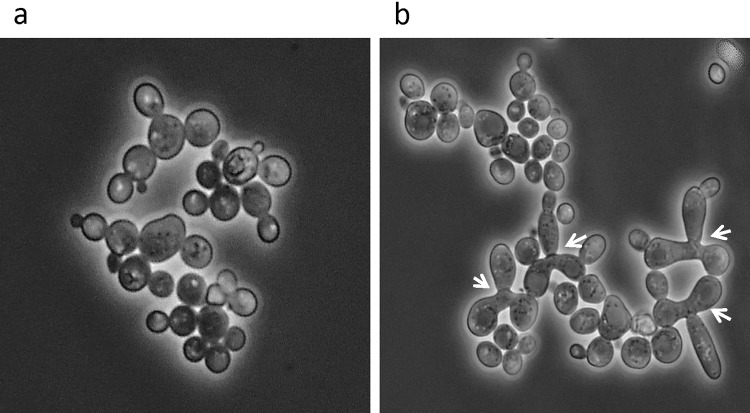
However, not all spores of the hybrid A3 were prototrophs. In one of the tetrads of A3, two spores (A3/1b and A3/1d) formed colonies auxotrophic for lucine. As A3 had LEU2Su/leu2Sc heterozygous genotype, these spores must have been aneuploid products of segregation bias against the S. uvarum chromosome carrying the wild-type LEU2Su gene. Occasional loss of LEU2Su/leu2Sc heterozygosity during cevarum allotetraploid meiosis was also observed in previous studies and found to correlate with the loss of Chr. IIISu that carries this gene, near its centromere (Antunovics et al. 2005; Pfliegler et al. 2012). Consistent with those observations, the karyotype of the leu− spore clones of A3 lacked the band corresponding to the S. uvarum Chr III (Fig. 1). As the MAT locus is also located on this chromosome, with the loss of Chr. IIISu, the spores also lost MATSu. Thus the leu2− spores were alloaneuploid, monosomic for Chr. IIISc and hemizygous for MATaSc. According to our model of the second sterility barrier, the loss of MAT heterozygosity relieves the mating block and restores mating competence and mating-type switching (Pfliegler et al. 2012). To verify the effect of the loss of MAT heterozygosity on the mating activity, we tested a leu+ (A3/1a) and a leu− (A3/1b) spore clone for conjugation with the parental strains. The cells of the prototrophic clones conjugated neither with each other nor with the testers; whereas, the cells of the leu− clones formed zygotes both within the clones (Fig. 2) and with both testers. This difference corroborates the notion that the loss of MAT heterozygosity breaks down the second sterility barrier and makes mating and mating-type switching possible.
Fig. 2.
Microscopic images of cultures tested for mating. a Cells of the sterile spore clone A3/1a. b Cells of the fertile spore clone A3/1b. Arrowheads point to germinating zygotes
Stable allospecific MAT heterozygosity in the hybrids
Since hybridisation took place between different species, the zygotes and the hybrid clones formed by them had allospecific combinations of MAT cassettes. To examine the postzygotic fate of the cassettes, we examined the MAT loci of the hybrids. Although the cassettes have conserved structures, the species differ in the sequences of certain segments. The primers proposed by Huxley et al. (1999) and widely used for the detection of the MATa and MATα cassettes in S. cerevisiae and the primers used by us in a previous study to amplify the entire MAT locus of S. cerevisiae (Pfliegler et al. 2012) are complementary to segments conserved in the three species involved in this study. Therefore, we set out to design primers suitable for the specific identification of the cassettes of the three species (Fig. 3 and Table 2). In each pair, the forward primer is complementary to a segment of the Ya or Yalpha region variable in the species; while, the reverse primer hybridises to an external chromosomal sequence adjacent to the Z2 region which is also different in each species. By choosing sequences located outside of the cassettes for reverse primers, we could prevent amplification from the silent HMR and HML cassettes. The amplification tests proved that each pair is species- and cassette specific (Fig. 4 and Table 2).
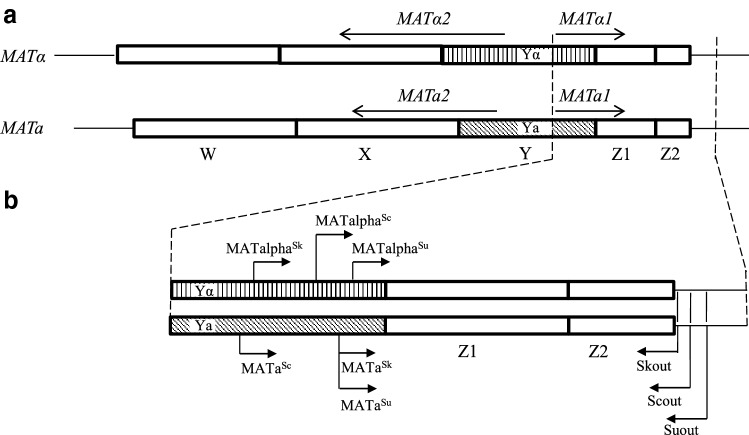
Fig. 3.
Selection of binding segments for primers that can be used to amplify species-specific sections of the MAT loci. a Structures of the MATalpha and MATa cassettes of S. cerevisiae distinguished by their Ya or Yalpha regions differing from each other both in length and in nucleotide sequence. W, X, Z1 and Z2 are conserved regions present both in the MATalpha and MATa cassettes (Haber 1998). b Locations of primers used for the amplification of species-specific segments of the MAT alleles
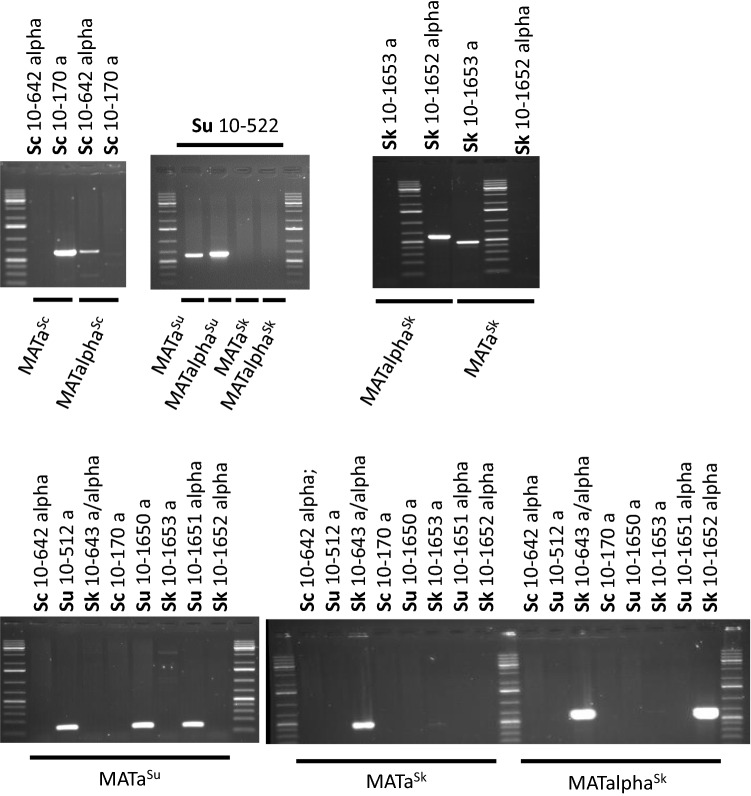
Fig. 4.
Verification of the specificity of primers. Amplification of MAT segments of homo- and heterothallic strains of S. cerevisiae (10-170, 10-642), S. kudriavzevii (10-643, 10-1652, 10-1653) and S. uvarum (10-512, 10-522, 10-1650, 10-1651) with primer pairs specific for MATaSc, MATalphaSc, MATaSk, MATalphaSk, MATaSu and MATalphaSu. Sc: S. cerevisiae; Sk: S. kudriavzevii; Su: S. uvarum
Using the species- and cassette-specific primers, PCR reactions were performed with genomic DNAs extracted from the hybrids. From the cevarum hybrids, MATaSc and MATalphaSu could be amplified; whereas, the MATalphaSc and MATaSu primers generated no bands. The lack of the latter cassettes implies that these hybrids arose from conjugation of MATa S. cerevisiae cells with MATalpha S. uvarum spores and no cassette replacement (mating-type switching) took place during the vegetative propagation of the hybrid cells in spite of the presence of the wild-type HOSu gene in their genomes. In the kudvarum hybrids II/1 and II/6, the PCR reactions detected MATaSk and MATalphaSu cassettes which was consistent with their hoSk/hoSu genotype. The cekudvarum hybrid produced by “rare mating” of the MATaSk/MATalphaSu hoSk/hoSu kudvarum hybrid with the MATaSc hoSc S. cerevisiae strain possessed all parental MAT cassettes. No cassette replacement (MAT switching) was detected in this hybrid either (Table 3).
Table 3.
Amplification of MAT cassettes
| Strain | Amplification with specific primer pairs | |||||
|---|---|---|---|---|---|---|
| S. cerevisiae | S. kudriavzevii | S. uvarum | ||||
| a | alpha | a | alpha | a | alpha | |
| MATaSc Scout | MATalphaSc Scout | MATaSk Skout | MATalphaSk Skout | MATaSu Suout | MATalphaSu Suout | |
| S. cerevisiae 10-170 a | + | − | − | − | − | − |
| S. cerevisiae 10-642 alpha | − | + | − | − | − | − |
| S. kudriavzevii 10-643 a/alpha | − | − | + | + | − | − |
| S. kudriavzevii 10-1652 alpha | − | − | − | + | − | − |
| S. kudriavzevii 10-1653 a | − | − | + | − | − | − |
| S. uvarum 10-512 a | − | − | − | − | + | − |
| S. uvarum 10-522 a/alpha | − | − | − | − | + | + |
| S. uvarum 10-1651 alpha | − | − | − | − | − | + |
| Cevarum hybrids A2, A3 and A27 | + | − | − | − | − | + |
| Cevarum spore clones A3/1a and A3/1c | + | − | − | − | − | + |
| Cevarum spore clones A3/1b and A3/1d | + | + | − | − | − | − |
| Kudvarum hybrids II/1 and II/6 | − | − | + | − | − | + |
| Cekudvarum hybrid II/6.1 | + | − | + | − | − | + |
Reactivated MAT switching upon the loss of allospecific MAT heterozygosity
The ability of the cells of the alloaneuploid spore clones A3/1b and A3/1d to produce zygotes within the clones and with both mating-type testers indicated that these clones were heterogeneous in terms of the mating activities of their cells. As the cells had MAT-carrying chromosome(s) only in the S. cerevisiae subgenome, heterogeneity could only be attributed to cassette replacements (MAT switching) in the S. cerevisiae MAT locus. To bolster this conclusion with experimental results, we amplified MAT cassettes from the cells of these clones (Fig. 5). As expected, we detected both S. cerevisiae-type cassettes (MATaSc and MATalphaSc) but neither S. uvarum-type cassette. To verify that the cells of these clones were MATaSc/ MATalphaSc heterozygotes, samples of their cultures were plated out on YEA and individual colonies were isolated. Three colonies for each clone were tested for MAT. All were heterozygotes. In contrast, the non-conjugating allodiploid spore clones A3/1a, A3/1c preserved the MATaSc/MATalphaSu genotype of the A3 hybrid. Table 4 summarises all the relevant properties of the spore clones of the cevarum tetrad A3/1. We also tested the MAT genotype in four prototrophic and one additional leu− clone randomly selected from other tetrads. The prototrophs had allospecific MAT heterozygosity; whereas, the leu− clone was heterozygous for the S. cerevisiae MAT cassettes.
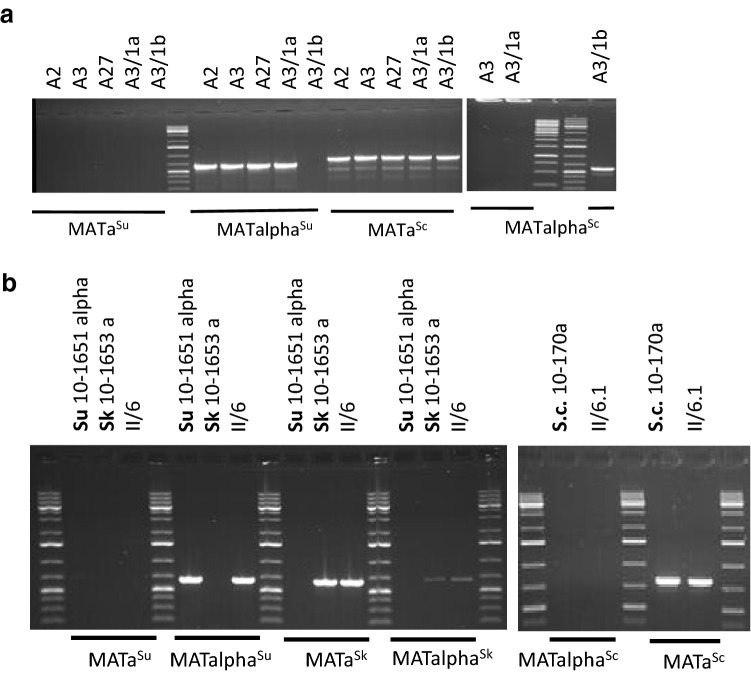
Fig. 5.
Examination of the MAT genotypes of hybrids and spore clones with species-specific primers. a HO/ho cevarum hybrids (A2, A3 and A27), the sterile spore clone A3/1a and the fertile spore clone A3/1b. All hybrids and the sterile spore clone have MATaSc/MATalphaSu genotypes. The fertile spore clone lacks MATalphaSu but has MATalphaSc. b The ho/ho kudvarum hybrid II/6 and the ho/ho/ho cekudvarum hybrid II/6.1. The kudvarum hybrid has MATaSk/MATalphaSu genotype whereas the cekudvarum hybrid also has a MATaSc cassette
Table 4.
Properties of the spore clones of the cevarum tetrad A3/1
| Spore clone | Phenotype | ChrIII | HO | MAT | Viable spores |
|---|---|---|---|---|---|
| A3/1a | Prototrophic | ChrIIISc, ChrIIISu | HOSu/hoSc | MATaSc/MATalphaSu | − |
| A3/1b | Leu− | ChrIIISc | HOSu/hoSc | MATaSc/MATalphaSc | + |
| A3/1c | Prototrophic | ChrIIISc, ChrIIISu | HOSu/hoSc | MATaSc/MATalphaSu | − |
| A3/1d | Leu− | ChrIIISc | HOSu/hoSc | MATaSc/MATalphaSc | + |
Discussion
In previous analyses of alloploid Saccharomyces hybrids, we noticed that the species of the genus are reproductively isolated by a pair of postzygotic sterility barriers which ensure that neither the allodiploid (first barrier) nor the allopolyploid (second barrier) hybrids produce functional gametes of euploid genomes (for a review, see Sipiczki 2018). We proposed a model in which the first barrier is due to the abruption of the meiotic process by the failure of the chromosomes of the subgenomes to pair (and recombine) in the prophase-I of meiosis and the second barrier is attributed to MAT heterozygosity. While the former is analogous to the major mechanism underlying the reproductive isolation in plants and animals (Sigel 2016; Lavinscky et al. 2017; Forejt 1996), the latter seems to be Saccharomyces specific. In the model, the MAT system contributes to the sterility by the interactions of the MATa and MATalpha loci of the subgenomes that repress the activity of the mating-specific genes (required for fertilisation) and MAT switching (reciprocal cassette replacements in the MAT loci). The current study contributes to the experimental validation of the model by investigating the MAT loci of interspecies hybrids and their meiotic derivatives.
The results of the analysis of the MAT loci of the two- and three-species hybrids and the viable but sterile allodiploid spores of an allotetraploid two-species hybrid carried out in this study corroborate the proposed interaction of the allospecific MAT cassettes to silence the mating functions. Neither the two-species cevarum (MATaSc/MATalphaSu) and kudvarum (MATaSk/MATalphaSu) hybrids nor the three-species cekudvarum (MATaSc/ MATaSk/MATalphaSu) hybrids conjugated with the tester heterothallic strains. The non-mating allodiploid gametes (ascospores) of the cevarum hybrid producing viable spores were also MATaSc/MATalphaSu. No MATaSu and MATalphaSu cassettes were detected in their cells. The correlation of the inability of the alloploid cells of these spore clones to mate and the allospecific combinations of MATa and MATalpha cassettes in their genomes bolsters the notion that MAT heterozygosity silences their mating-specific genes and, hence, prevents them from acting as gametes.
Our double-sterility model further assumes that the interaction of the allospecific MAT cassettes also represses MAT switching. Consistent with this assumption, we found that both the two-species allodiploid and the three-species allotriploid hybrids preserved the cassettes of their parental strains unchanged. The parental cassettes did not change in the MAT loci of the sporulating tetraploid hybrid and its non-mating allodiploid spore clones either.
It was also proposed in the model that the allospecific MATa and MATalpha cassettes can cooperatively launch meiosis. Consistent with this proposal, two of the cevarum hybrids and the kudvarum hybrids formed spores. However, only the cevarum spores were viable. As the meiotic pairing and segregation of chromosomes require two sets of chromosomes in both subgenomes (autodiploidisation of meiosis, Karanyicz et al. 2017), these hybrids must have undergone genome duplication. We also observed frequent genome-size increase in our previous studies of cevarum and kudvarum hybrids (Pfliegler et al. 2012; Karanyicz et al. 2017). The alloploid karyotypes of their spores indicated that these hybrids had at least allotetraploid genomes. As neither subgenome had two different MAT loci, the meiosis was launched in these hybrids by the interaction of their complementary MAT loci (MATaSc and MATalphaSu). This interaction can also be functional in allodiploids, and allotriploids as demonstrated by the sporulation proficiency of the kudvarum hybrids II/1 and II/6 as well as the cekudvarum hybrids but cannot result in viable spores because of the abruption of the meiotic process by the lack of homologous partners for chromosome pairing within the subgenomes.
These results corroborate the tenet of the double-sterility barrier model that the second sterility barrier is attributable to allospecific MAT heterozygosity causing mating (fertilisation) incompetence. In the same time, this heterozygosity can launch meiosis which, however, gets stuck because of the failure of chromosome pairing. In principle, MAT heterozygosity can also confer spore sterility in autotetraploids. Even if it happens, it affects only a small fraction of spores (Pomper et al. 1954) because in an autotetraploid nucleus each chromosome can freely pair with any of its three homologues regardless of their origin.
Further proof bolstering the role of the MAT loci in the second sterility barrier is provided by the cevarum spore clones that lack Chr IIISu. By losing this chromosome, the spore also loses the MAT locus of the S. uvarum subgenome. We found that these spores nullisomic for Chr IIISu formed clones of vegetative cells capable of mating with each other within the clones. Their mating competence implies that the loss of MAT heterozygosity relieves the switching machinery and the mating programme from repression and, thus, inactivates the second sterility barrier. In accordance with this, we also detected the MATalphaSc cassette in the cultures of these clones. Obviously, MAT switching took place in the S. cerevisiae chromosome despite the absence of an active HOSc gene. This finding indicates that the wild-type HOSu gene of the S. uvarum subgenome can functionally substitute its S. cerevisiae orthologue. Thus, not only the MAT cassettes but also the HO genes are interchangeable between S. cerevisiae and S. uvarum in spite of the difference between their sequences (de Melo Pereira et al. 2010). Due to the reactivation of the mating competence and the mating-type switching mechanisms, the cells of the leu− spore clones can mate with each other and form zygotes. As they are nullisomic for Chr IIISu, the resulting zygotes and their vegetative progeny will also be nullisomic for this chromosome but disomic for Chr IIISc and heterozygous for the S. cerevisiae MAT cassettes (MATaSc/MATalphaSc). The MAT heterozygosity prevents them from further mating but allows sporulation because the a1p/alpha2p heterodimer protein complex activates IME1 under starvation conditions that codes for a central regulator of many meiotic genes (for a review of the regulation of meiosis in S. cerevisiae, see Mitchell 1994).
The break-down of the second sterility barrier represents an escape route from the reproductive isolation (Pfliegler et al. 2012; Karanyicz et al. 2017). The cells of the spore clones nullisomic for one or the other parental Chr III can mate with each other and with mating-competent cells of other strains. In the former case, they form zygotes whose vegetative progeny produce fertile gametes which then generate additional generations of fertile zygotes. However, the loss of Chr. III destabilises the genome. Additional chromosomes can be lost during meiotic divisions in the successive generations of spore clones. This gradual genome reduction is referred to as GARMe (Karanyicz et al. 2017). Figure 6 depicts the correlation between the sexual behaviour and the MAT genotype in the alloploid hybrids, their sterile and fertile spores (gametes).
Fig. 6.
A general scheme showing the experimental strategy and the detected correlation between MAT genotypes, alloploid sterility and the break-down of the sterility barrier. Hybrids and viable spore clones are grey. ↔ : MAT switching. GARMe genome autoreduction by meiosis (Karanyicz et al. 2017)
Recently MAT genes have been implicated in the evolution and sterility of certain natural Zygosaccharomyces strains whose genome structures indicated hybrid origin (Watanabe et al. 2017; Ortiz-Merino et al. 2017; Braun-Galleani et al. 2018; Bizzarri et al. 2019). These strains have complex and diverse repertoires of MAT and MAT-like (MTL) idiomorphs inherited from the partners of the ancient hybridisation events and modified later by intragenomic chimerisation events. Some of them were found to participate in the regulation of the sexual processes in ways somewhat different from the situation in Saccharomyces. For examples, the sterile allodiploid strain ATCC42981 is of MATa/MATalpha genotype but the deletion of the MATalpha locus did not restore fertility probably because of incomplete silencing at the chimeric HMLalpha cassette (Bizzarri et al. 2019). In S. cerevisiae, the HMR and HML loci contain completely silent MATa and MATalpha cassettes (reviewed in Haber 2012). In contrast to ATCC42981, the fertile natural interspecies hybrid ATCC60483 is assumed to have regained fertility as a consequence of irreparable damage to one of the two homeologous MAT loci that must have occurred some time ago during the evolution of the strain (Ortiz-Merino et al. 2017). The MAT genotypes of the natural Zygosaccharomyces hybrids indicate that a mechanism similar to the second sterility barrier of Saccharomyces species may also exist in Zygosaccharomyces. Genetic analysis of synthetic alloploid Zygosaccharomyces hybrids will reveal to what extent the mechanisms underlying the reproductive isolation of species are similar in these genera.
Acknowledgements
Open access funding provided by University of Debrecen (DE). We thank Anita Kovacs for skilful technical assistance. This study was supported by the Grant K-124417 provided by the National Research, Development and Innovation Office of Hungary.
Author contributions
Conceptualization: MS; methodology: MS, ZA, AS; hybrid construction: ZA, AS; rest of experimental work: MS; data analysis: MS; writing: MS; funding aquisition: MS.
Funding
This study was funded by the National Research, Development and Innovation Office of Hungary (Grant number K-124417).
Availability of data and material
All data generated or analysed during this study are included in this published article.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Antunovics Z, Nguyen HV, Gaillardin C, Sipiczki M. Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 2005;5:1141–1150. doi: 10.1016/j.femsyr.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Bizzarri M, Cassanelli S, Bartolini L, Pryszcz LP, Dušková M, Sychrová H, Solieri L. Interplay of chimeric mating-type loci impairs fertility rescue and accounts for intra-strain variability in Zygosaccharomyces rouxii interspecies hybrid ATCC42981. Front Genet. 2019;10:137. doi: 10.3389/fgene.2019.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun-Galleani S, Ortiz-Merino RA, Wu Q, Xu Y, Wolfe KH. Zygosaccharomyces pseudobailii, another yeast interspecies hybrid that regained fertility by damaging one of its MAT loci. FEMS Yeast Res. 2018;18:79. doi: 10.1093/femsyr/foy079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne J, Orr H. Speciation. Sunderland, MA: Sinauer Associates; 2004. [Google Scholar]
- de Melo Pereira GV, Ramos CL, Galvão C, Souza Dias E, Schwan RF. Use of specific PCR primers to identify three important industrial species of Saccharomyces genus: Saccharomyces cerevisiae, Saccharomyces bayanus and Saccharomyces pastorianus. Lett Appl Microbiol. 2010;51:131–137. doi: 10.1111/j.1472-765X.2010.02868.x. [DOI] [PubMed] [Google Scholar]
- Forejt J. Hybrid sterility in the mouse. Trends Genet. 1966;12:412–417. doi: 10.1016/0168-9525(96)10040-8. [DOI] [PubMed] [Google Scholar]
- Gorter de Vries AR, Pronk JT, Daran JG (2019) Lager-brewing yeasts in the era of modern genetics. FEMS Yeast Res 19:pii: foz063 [DOI] [PMC free article] [PubMed]
- Gunge N, Nakatomi Y. Genetic mechanisms of rare matings of the yeast Saccharomyces cerevisiae heterozygous for mating type. Genetics. 1972;70:41–58. doi: 10.1093/genetics/70.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber JE. Mating-type gene switching in Saccharomyces cerevisiae. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harari Y, Ram Y, Kupiec M. Frequent ploidy changes in growing yeast cultures. Curr Genet. 2018;64:1001–1004. doi: 10.1007/s00294-018-0823-y. [DOI] [PubMed] [Google Scholar]
- Herskowitz I. Life cycle of the budding yeast Saccharomyces cerevisiae. Microbiol Rev. 1988;52:536–553. doi: 10.1128/MMBR.52.4.536-553.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson ES, Price SC, Kahier AL, Morris MI, Allard RW. An experimental verification of segregation theory in a diploidized tetraploid: esterase loci in Avena barbata. J Hered. 1983;74:381–383. doi: 10.1093/oxfordjournals.jhered.a109816. [DOI] [Google Scholar]
- Huxley C, Green ED, Dunham I. Rapid assessment of Saccharomyces cerevisiae mating type by PCR. Trends Genet. 1990;6:236. doi: 10.1016/0168-9525(90)90190-H. [DOI] [PubMed] [Google Scholar]
- Johnson NA. Hybrid incompatibility genes: remnants of a genomic battlefield? Trends Genet. 2010;26:317–325. doi: 10.1016/j.tig.2010.04.005. [DOI] [PubMed] [Google Scholar]
- Karanyicz E, Antunovics Z, Kallai Z, Sipiczki M. Non-introgressive genome chimerisation by malsegregation in autodiploidised allotetraploids during meiosis of Saccharomyces kudriavzevii x Saccharomyces uvarum hybrids. Appl Microbiol Biotechnol. 2017;101:4617–4633. doi: 10.1007/s00253-017-8274-9. [DOI] [PubMed] [Google Scholar]
- Lavinscky MP, Souza MM, Silva GS, Melo CAF. Contributions of classical and molecular cytogenetic in meiotic analysis and pollen viability for plant breeding. Genet Mol Res. 2017 doi: 10.4238/gmr16039582. [DOI] [PubMed] [Google Scholar]
- Levin DA. The role of chromosomal change in plant evolution. New York: Oxford University Press; 2002. [Google Scholar]
- Li XC, Barringer BC, Barbash DA. The pachytene checkpoint and its relationship to evolutionary patterns of polyploidization and hybrid sterility. Heredity. 2009;102:24–30. doi: 10.1038/hdy.2008.84. [DOI] [PubMed] [Google Scholar]
- Mitchell AP. Control of meiotic gene expression in Saccharomyces cerevisiae. Microbiol Rev. 1994;58:56–70. doi: 10.1128/MMBR.58.1.56-70.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth KA. Molecular genetics of yeast mating type. Annu Rev Genet. 1982;16:439–500. doi: 10.1146/annurev.ge.16.120182.002255. [DOI] [PubMed] [Google Scholar]
- Nguyen HV, Boekhout T. Characterization of Saccharomycesuvarum (Beijerinck, 1898) and related hybrids: assessment of molecular markers that predict the parent and hybrid genomes and a proposal to name yeast hybrids. FEMS Yeast Res. 2017 doi: 10.1093/femsyr/fox014. [DOI] [PubMed] [Google Scholar]
- Nguyen HV, Lepingle A, Gaillardin CA. Molecular typing demonstrates homogeneity of Saccharomyces uvarum strains and reveals the existence of hybrids between S. uvarum and S. cerevisiae, including the S. bayanus type strain CBS 380. Syst Appl Microbiol. 2000;23:71–85. doi: 10.1016/S0723-2020(00)80048-X. [DOI] [PubMed] [Google Scholar]
- Ortiz-Merino RA, Kuanyshev N, Braun-Galleani S, Byrne KP, Porro D, Branduardi P, Wolfe KH. Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch. PLoS Biol. 2017;15:e2002128. doi: 10.1371/journal.pbio.2002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Zhang Q. Understanding reproductive isolation based on the rice model. Annu Rev Plant Biol. 2013;64:111–135. doi: 10.1146/annurev-arplant-050312-120205. [DOI] [PubMed] [Google Scholar]
- Pfliegler WP, Antunovics Z, Sipiczki M. Double sterility barrier between Saccharomyces species and its breakdown in allopolyploid hybrids by chromosome loss. FEMS Yeast Res. 2012;12:703–718. doi: 10.1111/j.1567-1364.2012.00820.x. [DOI] [PubMed] [Google Scholar]
- Pomper S, Daniels KM, McKee DW. Genetic analysis of polyploid yeast. Genetics. 1954;39:343–355. doi: 10.1093/genetics/39.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT (2011) The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for Saccharomyces sensu stricto genus. G3 (Bethesda) 1:11–25 [DOI] [PMC free article] [PubMed]
- Seehausen O, Butlin RK, Keller I, Wagner CE, Boughman JW, Hohenlohe PA, Peichel CL, Saetre GP, Bank C, Brännström A, Brelsford A, Clarkson CS, Eroukhmanoff F, Feder JL, Fischer MC, Foote AD, Franchini P, Jiggins CD, Jones FC, Lindholm AK, Lucek K, Maan ME, Marques DA, Martin SH, Matthews B, Meier JI, Möst M, Nachman MW, Nonaka E, Rennison DJ, Schwarzer J, Watson ET, Westram AM, Widmer A. Genomics and the origin of species. Nat Rev Genet. 2014;15:176–192. doi: 10.1038/nrg3644. [DOI] [PubMed] [Google Scholar]
- Sigel EM. Genetic and genomic aspects of hybridization in ferns. J System Evol. 2016;54:638–655. doi: 10.1111/jse.12226. [DOI] [Google Scholar]
- Sipiczki M. Interspecies hybridisation and genome chimerisation in Saccharomyces: combining of gene pools of species and its biotechnological perspectives. Front Microbiol. 2018;9:3071. doi: 10.3389/fmicb.2018.03071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M. Yeast two- and three-species hybrids and high-sugar fermentation. Microb Biotechnol. 2019;12:1101–1108. doi: 10.1111/1751-7915.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M, Ferenczy L. Protoplast fusion of Schizosaccharomyces pombe auxotrophic mutants of identical mating-type. Mol Gen Genet. 1977;151:77–81. doi: 10.1007/BF00446915. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. The role of hybridization in plant speciation. Annu Rev Plant Biol. 2009;60:561–588. doi: 10.1146/annurev.arplant.043008.092039. [DOI] [PubMed] [Google Scholar]
- St Charles J, Hamilton ML, Petes TD. Meiotic chromosome segregation in triploid strains of Saccharomycescerevisiae. Genetics. 2010;186:537–550. doi: 10.1534/genetics.110.121533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thon G, Maki T, Haber JE, Iwasaki H. Mating-type switching by homology-directed recombinational repair: a matter of choice. Curr Genet. 2019;65:351–362. doi: 10.1007/s00294-018-0900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe J, Uehara K, Mogi Y, Tsukioka Y. Mechanism for restoration of fertility in hybrid Zygosaccharomyces rouxii generated by interspecies hybridization. Appl Environ Microbiol. 2017;83(21):e01187–e1217. doi: 10.1128/AEM.01187-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJD. Modes of speciation. Freeman and Company, San Francisco, CA: W. H; 1978. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analysed during this study are included in this published article.