Abstract
Carefully maintained and precisely inherited chromosomal DNA provides long-term genetic stability, but eukaryotic cells facing environmental challenges can benefit from the accumulation of less stable DNA species. Circular DNA molecules lacking centromeres segregate randomly or asymmetrically during cell division, following non-Mendelian inheritance patterns that result in high copy number instability and massive heterogeneity across populations. Such circular DNA species, variously known as extrachromosomal circular DNA (eccDNA), microDNA, double minutes or extrachromosomal DNA (ecDNA), are becoming recognised as a major source of the genetic variation exploited by cancer cells and pathogenic eukaryotes to acquire drug resistance. In budding yeast, circular DNA molecules derived from the ribosomal DNA (ERCs) have been long known to accumulate with age, but it is now clear that aged yeast also accumulate other high-copy protein-coding circular DNAs acquired through both random and environmentally-stimulated recombination processes. Here, we argue that accumulation of circular DNA provides a reservoir of heterogeneous genetic material that can allow rapid adaptation of aged cells to environmental insults, but avoids the negative fitness impacts on normal growth of unsolicited gene amplification in the young population.
Keywords: Circular DNA, Extrachromosomal DNA, Double minutes, Extrachromosomal circular DNA, Ageing, Non-Mendelian inheritance
From a human perspective, the concept of mutation appears purely negative, associated only with degeneration and cancer. However, all living organisms must maintain an appropriate mutation rate to survive: too high a rate leads to genome instability and degradation of vital functions, but conversely too little mutation strangles evolution by suppressing genetic diversity, preventing adaptation to new environmental challenges and condemning an organism to be out-competed. Evolutionary studies in E. coli and asexual yeasts reveal that strains with high mutation rates outperform those with low mutation rates, showing that mutation provides an adaptive advantage (Arjan et al. 1999; Desai et al. 2007; Wielgoss et al. 2013). However, increasing the mutation rate only improves adaptability up to a critical “error threshold”: above this, further increases reduce adaptability because fitness-enhancing mutations only account for a very small proportion of total mutations, and the accumulation of more frequent deleterious mutations becomes limiting for fitness (Eyre-Walker and Keightley 2007; Gerrish et al. 2013; Lynch et al. 1993; Sprouffske et al. 2018).
It is well understood that prokaryotes supplement chromosomal genetic material with circular DNA plasmids that deviate from normal rules of Mendelian inheritance and accelerate adaptation. In contrast, the evolutionary significance of circular DNA in eukaryotes has until recently been largely ignored, despite evidence for circular DNA in eukaryotic nuclei dating back over half a century (Cox et al. 1965; Hotta and Bassel 1965). Here, we examine the potential of circular DNA to accelerate adaptation in eukaryotes in general and ask whether the accumulation of particular circular DNA species during ageing in yeast enhances the potential of those species to confer adaptive phenotypes, providing a beneficial outcome of ageing in simple eukaryotes.
Circular DNA ranges in size from a few hundred base pairs (100–1000 bp microDNA) to several megabases (1–5 Mb ecDNA also known as double minutes) (Paulsen et al. 2018; Shibata et al. 2012; Turner et al. 2017). Circular DNA can derive from sites with little or no sequence homology, however, highly repetitive genomic regions, such as the ribosomal DNA (rDNA), telomeres, transposon remnants, and tandemly repeated genes are the largest producers of circular DNA at least in yeast (Moller et al. 2015; Sinclair and Guarente 1997). Double-strand break repair is the primary source of circular DNA from these repetitive regions although other formation mechanisms have been proposed and may well apply to circular DNA arising from non-repetitive regions (Hull et al. 2019; Park et al. 1999; Paulsen et al. 2018). The existence of circular DNA appears to be common amongst eukaryotes and has so far been reported in fungi, trypanosomes, worms, flies, frogs, mammals and plants (Beverley 1991; Cohen et al. 1999; Hotta and Bassel 1965; Koo et al. 2018; Shoura et al. 2017; Stanfield and Helinski 1976). In healthy humans, circular DNA from many genomic regions are detected in somatic tissue and also form as a side-product of V(D)J recombination (Moller et al. 2018; Serana et al. 2013), but conversely circular DNA is detected in approximately half of human cancers, is more prevalent in malignant than benign tumours, and is associated with poor prognosis (Fan et al. 2011; Koche et al. 2020; Turner et al. 2017).
The vast majority of circular DNA molecules lack centromeres and, in the absence of other mechanisms, will segregate randomly in mitosis. Therefore, the copy number of a circular DNA is not necessarily the same in each daughter cell after mitosis and can be substantially higher or lower than that of the parental cell. Because of this non-Mendelian character, simulations and experiments show that individual circular DNA species can accumulate very rapidly (Nathanson et al. 2014; Turner et al. 2017), and evolution via circular DNA can be highly advantageous compared to chromosomal change under selection for increased gene dosage (deCarvalho et al. 2018; Ubeda et al. 2014). Dramatic copy number amplification of driving oncogenes and drug resistance factors are frequent in cancer (Beroukhim et al. 2010; Corcoran et al. 2010; Frei et al. 1984; Katoh 2008; Little et al. 2011), and it is perhaps unsurprising that these amplifications commonly occur through accumulation of circular DNA (deCarvalho et al. 2018; Koche et al. 2020; Storlazzi et al. 2010; Turner et al. 2017; Vogt et al. 2004; Von Hoff et al. 1990; Wu et al. 2019). Similarly, as circular DNA is often absent from one daughter after division, a sub-population is readily selected for decreased gene dosage (Gresham et al. 2010; Haber and Schimke 1981; Nathanson et al. 2014).
In budding yeast, circular DNA molecules derived from the ribosomal DNA (ERCs) are highly focused in aged cells and are suggested to drive premature ageing and shortened lifespan (Borghouts et al. 2004; Sinclair et al. 1997). Various mutants with exaggerated ERC accumulation have a reduced lifespan, for example, cells lacking the DNA helicase Sgs1 accumulate more ERCs than wild-types and display many age-associated phenotypes in addition to a shortened lifespan (Kaeberlein et al. 2004; Sinclair et al. 1997). Conversely, loss of the replication fork blocking protein Fob1 decreases the formation of ERCs and extends the lifespan of mother cells by 30–40% (Defossez et al. 1999). In addition to ERCs generated from the rDNA locus, sensitive sequencing methods have identified almost 1800 circular DNA species in young populations, covering 23% of the budding yeast genome (Moller et al. 2015). The vast majority of these circular DNAs contain at least a fragment of a protein-coding gene, but unlike ERCs these species are largely sub-stoichiometric, such that any given cell will only contain a few circular DNA molecules that increase the copy number of genes encoded on those circular DNAs by 1 copy. Although such changes can be adaptively useful, in most situations and for most genes, this degree of gene amplification is likely to have minimal effect. Indeed, fitness assays under different challenging environments show that genes present on low copy plasmids confer fitness effects rarely and of much lesser magnitude than those on multi-copy plasmids (Payen et al. 2016). Therefore, although young yeast populations contain a wide diversity of circular DNA, it is only with substantial accumulation of any given circular DNA that major phenotypic effects are likely to manifest.
Circular DNAs including ERCs accumulate in aged yeast through a highly asymmetric mitotic segregation process that is very different from the random segregation observed in mammalian cells (Murray and Szostak 1983). During cell division in budding yeast, mother cells retain ERCs and other molecules considered harmful, thereby promoting daughter cell fitness (Mortimer and Johnston 1959; Sinclair and Guarente 1997). ERC retention in mother cells is mediated by attachment via SAGA and TREX2 to nuclear pore complexes that are largely retained by the mother cell (Denoth-Lippuner et al. 2014; Shcheprova et al. 2008). However, asymmetric inheritance alone is insufficient to mediate circular DNA accumulation as additional copies of the circular DNA must be generated. ERCs and many other circular DNA species carry active replication origins and are, therefore, replicated during the cell cycle. Once replicated, both copies are retained in the mother cell and so the copy number doubles, accounting for the massive abundance of ERCs in aged cells which increase genome size by over 50% (Cruz et al. 2018). Surprisingly, we discovered that replication is not required for all circles, as CUP1 circular DNA does not replicate efficiently (despite carrying an annotated replication origin), and is instead generated from chromosomal DNA at such a high rate that simple asymmetric retention of the newly generated copies in the mother cell is sufficient for CUP1 accumulation (Hull et al. 2019). However, many circular DNA species do not form at a high rate, nor contain active replication origins and, therefore, the copy number of these species in mother cells will remain static or decrease with age. A curious outcome of these differences in formation speed, replication capacity and asymmetric retention is that the diversity of circular DNA species observed in young cells is high, but the copy number of each individual circle and, therefore, the phenotypic impact is low. As cells age, only a subset of circular DNA can accumulate but that subset reaches higher copy numbers that will have a greater phenotypic effect, beneficial or not (Fig. 1, steps 1 and 2). In other words, the diversity of circular DNA decreases with age in yeast, which has been observed experimentally (Prada-Luengo et al. 2020), but the effects of the remaining circular DNA species will progressively increase.
Fig. 1.
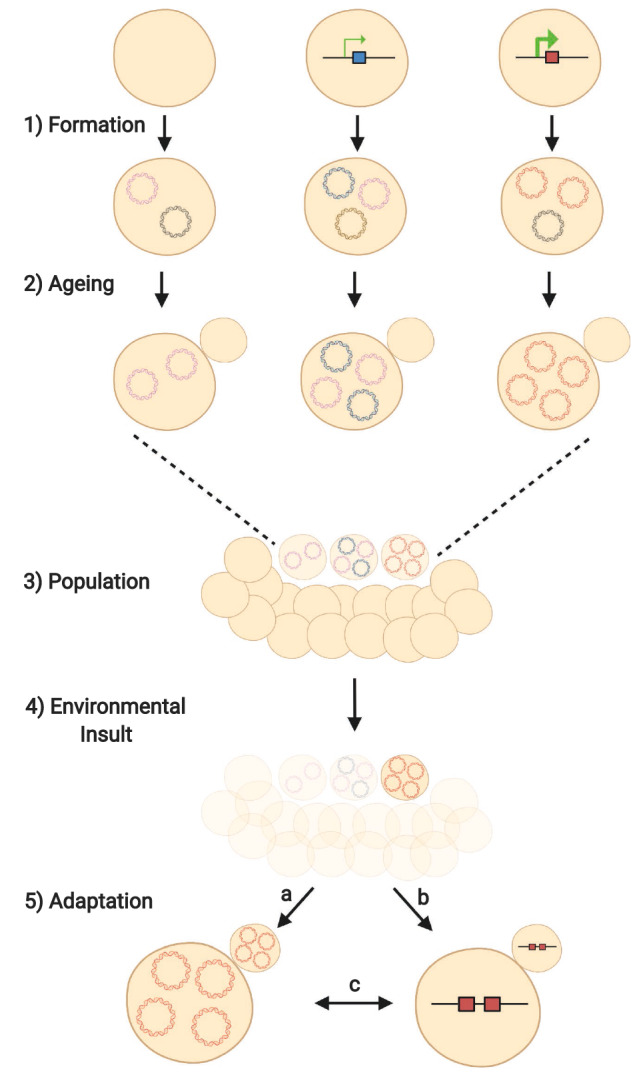
Rapid adaptation of aged cells to environmental insults. (1) Circular DNA molecules are formed from the linear chromosomes by both random (purple, black, and brown circles) and transcriptionally-stimulated (blue, and red circles) recombination and repair processes, producing a high diversity of low-copy circles in young cells. Highly-transcribed inducible genes (red box) produce more circles (red) than lowly-transcribed inducible genes (blue box and blue circles). (2) During ageing some circles (black and brown) are lost, whilst other circles (blue, purple and red) accumulate in mother cells by asymmetric segregation, creating a less-diverse but higher-copy circular DNA repertoire. (3) Circles are retained in the aged sub-population, which displays a fitness deficit, enabling the rest of the population of pre-dominantly young cells to grow unimpeded. (4) High-copy circles enhance adaptability in aged cells, increasing the likelihood of an aged cell having the necessary copy number mutation to survive an environmental insult. (5) The adaptive mutation is then propagated through the population by: a relaxing the asymmetric segregation of circular DNA and enabling donations to daughter cells; b re-integrating the beneficial circular DNA into the chromosome as a heritable duplication; c a combination of strategies a and b. Figure created with BioRender.com
The ageing yeast sub-population is, therefore, enriched for high-copy circular DNA containing protein-coding genes, and there are many examples of gene copy number accumulation providing adaptive advantages (Fig. 1, steps 3 and 4). For example, yeast can adapt to sulphate, nitrogen and glucose limitation, and to environmental toxins, through gene amplification (Beaupere and Labunskyy 2019; Brown et al. 1998; Fogel and Welch 1982; Gresham et al. 2008, 2010). Furthermore, a recent study in cancer cells revealed that circular DNA has an unusually open chromatin structure which would amplify the phenotypic effects of genes encoded on circular DNA relative to their chromosomal counterparts (Wu et al. 2019). Therefore, advantages exist in accumulating multiple copies of circular DNA encoding specific genes in certain environments (Fig. 1, step 4). However, there would be a fitness cost because unbalanced copy number amplification of most genes will impair gene regulatory networks and protein homeostasis to some extent. In other words, accumulation of multiple copies of random genes on circular DNA is more likely to be neutral at best or detrimental at worst, and it is not unreasonable to suggest that accumulation of genic circular DNA species may contribute to the age-related fitness decline currently attributed to ERCs.
Excitingly we have observed that the rate of copy number variation events including circular DNA formation is not completely random, which may improve this cost–benefit balance for aged cells (Hull et al. 2017, 2019). Formation of CUP1 circular DNA, which encodes the copper resistance protein Cup1, is highly dependent on the transcription of the CUP1 locus, which in turn is tightly regulated based on copper in the environment (Hull et al. 2019). Therefore, yeast ageing in the presence of copper transcribe CUP1 and so produce many more copies of the CUP1 circular DNA, which pre-adapts these cells to further increases in copper concentration. A link between transcriptional activity and circular DNA content has also been observed by the Regenberg lab, who found that per megabase of DNA, gene-rich chromosomes contribute more to the total level of circular DNA in healthy human tissues (Moller et al. 2018). Furthermore, TTN (titin), the most transcribed protein-coding gene in muscle tissue, is also the largest producer of circular DNA per gene (Moller et al. 2018). Formalising this idea, we suggest that genes which have evolved to be induced in response to particular environmental conditions are excellent candidates for adaptive amplification, and that simply connecting circular DNA formation to transcriptional induction is a clever means by which cells could gain the maximum chance of accumulating useful circular DNA, rather than unhelpful or negative species.
By itself, an adaptive phenotype in an individual aged cell is of little use if the causal circular DNA is selfishly retained in the mother cell, as would be the case if asymmetric segregation is maintained, and we must consider how circular DNA accumulation is translated into a heritable advantage. First, once circular DNA has accumulated, segregation can be relaxed under stress allowing circles with replication origins to propagate at high copy number in the population (Fig. 1, step 5a). This release of the asymmetric segregation system under heat stress has been observed and represents a general response to signalling from the cell wall integrity pathway (Baldi et al. 2017). Secondly, accumulation of high levels of circular DNA increases the chances of chromosome re-integration and, therefore, restoration of normal heritability for the amplified allele (Fig. 1, 5b). Such adaptive chromosomal re-integration events have been repeatedly observed, although it is unclear whether they happened in aged cells (Beverley et al. 1984; Brewer et al. 2015; Demeke et al. 2015; Durkin et al. 2012; Galeote et al. 2011; Koche et al. 2020; Lauer et al. 2018; Vogt et al. 2004).
The idea that a sub-population trades short-term growth for adaptive capacity is formalised in bet-hedging (concisely reviewed in (Levy et al. 2012)), and facets of ageing that fit with a bet-hedging model have been demonstrated experimentally including that aged yeast can be more stress resistant and more adaptable to nutrient transitions than young cells (Frenk et al. 2017; Levy et al. 2012). Cell-to-cell heterogeneity in the expression of metabolic enzymes and stress resistance factors also increases with age suggesting general phenotypic diversification of the ageing population (Radzinski and Reichmann 2019). These studies invoke transcriptional mechanisms linked to ageing to mediate switching to an adaptive state, whereas circular DNA excision represents a reversible genetic mechanism for phenotypic switching (Moller et al. 2013). The exciting feature of age-linked circular DNA accumulation in bet-hedging scenarios is that it forms a system that accrues diversity without impacting the genetic integrity of chromosomal DNA, but has the potential to be fixed rapidly in evolutionary timescales by chromosomal re-integration. Circular DNA, therefore, provides cells with the ability to explore the benefits of a high mutation rate, without the associated risk of generating deleterious chromosomal mutations, and with the possibility to make beneficial mutations permanent.
Acknowledgements
We would like to thank Birgitte Regenberg and Henrik Møller for inspiring our move into the circular DNA field. RH is funded by the Swedish Foundations’ Starting Grant (Ragnar Söderberg Foundation), JH is funded by the Wellcome Trust [110216] and Biological Sciences Research Council BBSRC [BI Epigenetics ISP: BBS/E/B/000C0423].
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arjan JA, Visser M, Zeyl CW, Gerrish PJ, Blanchard JL, Lenski RE. Diminishing returns from mutation supply rate in asexual populations. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- Baldi S, Bolognesi A, Meinema AC, Barral Y. Heat stress promotes longevity in budding yeast by relaxing the confinement of age-promoting factors in the mother cell. eLife 6. 2017 doi: 10.7554/eLife.28329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaupere C, Labunskyy VM. (Un)folding mechanisms of adaptation to ER stress: lessons from aneuploidy. Curr Genet. 2019;65:467–471. doi: 10.1007/s00294-018-0914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, Mc Henry KT, Pinchback RM, Ligon AH, Cho YJ, Haery L, Greulich H, Reich M, Winckler W, Lawrence MS, Weir BA, Tanaka KE, Chiang DY, Bass AJ, Loo A, Hoffman C, Prensner J, Liefeld T, Gao Q, Yecies D, Signoretti S, Maher E, Kaye FJ, Sasaki H, Tepper JE, Fletcher JA, Tabernero J, Baselga J, Tsao MS, Demichelis F, Rubin MA, Janne PA, Daly MJ, Nucera C, Levine RL, Ebert BL, Gabriel S, Rustgi AK, Antonescu CR, Ladanyi M, Letai A, Garraway LA, Loda M, Beer DG, True LD, Okamoto A, Pomeroy SL, Singer S, Golub TR, Lander ES, Getz G, Sellers WR, Meyerson M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beverley SM. Gene amplification in Leishmania. Annu Rev Microbiol. 1991;45:417–444. doi: 10.1146/annurev.mi.45.100191.002221. [DOI] [PubMed] [Google Scholar]
- Beverley SM, Coderre JA, Santi DV, Schimke RT. Unstable DNA amplifications in methotrexate-resistant Leishmania consist of extrachromosomal circles which relocalize during stabilization. Cell. 1984;38:431–439. doi: 10.1016/0092-8674(84)90498-7. [DOI] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J, Jazwinski SM. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics. 2004;166:765–777. doi: 10.1534/genetics.166.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Payen C, Di Rienzi SC, Higgins MM, Ong G, Dunham MJ, Raghuraman MK. Origin-dependent inverted-repeat amplification: tests of a model for inverted DNA amplification. PLoS Genet. 2015;11:e1005699. doi: 10.1371/journal.pgen.1005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evolut. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Cohen S, Menut S, Mechali M. Regulated formation of extrachromosomal circular DNA molecules during development in Xenopus laevis. Mol Cell Biol. 1999;19:6682–6689. doi: 10.1128/mcb.19.10.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Dias-Santagata D, Bergethon K, Iafrate AJ, Settleman J, Engelman JA. BRAF gene amplification can promote acquired resistance to MEK inhibitors in cancer cells harboring the BRAF V600E mutation. Sci Signal. 2010;3:ra84. doi: 10.1126/scisignal.2001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox D, Yuncken C, Spriggs AI. Minute chromatin bodies in malignant tumours of childhood. Lancet. 1965;1:55–58. doi: 10.1016/s0140-6736(65)90131-5. [DOI] [PubMed] [Google Scholar]
- Cruz C, Della Rosa M, Krueger C, Gao Q, Horkai D, King M, Field L, Houseley J. Tri-methylation of histone H3 lysine 4 facilitates gene expression in ageing cells. eLife. 2018;7:e34081. doi: 10.7554/eLife.34081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deCarvalho AC, Kim H, Poisson LM, Winn ME, Mueller C, Cherba D, Koeman J, Seth S, Protopopov A, Felicella M, Zheng S, Multani A, Jiang Y, Zhang J, Nam DH, Petricoin EF, Chin L, Mikkelsen T, Verhaak RGW. Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma. Nat Genet. 2018;50:708–717. doi: 10.1038/s41588-018-0105-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, Silver PA, Keil RL, Guarente L. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/S1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Demeke MM, Foulquie-Moreno MR, Dumortier F, Thevelein JM. Rapid evolution of recombinant Saccharomyces cerevisiae for Xylose fermentation through formation of extra-chromosomal circular DNA. PLoS Genet. 2015;11:e1005010. doi: 10.1371/journal.pgen.1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denoth-Lippuner A, Krzyzanowski MK, Stober C, Barral Y. Role of SAGA in the asymmetric segregation of DNA circles during yeast ageing. eLife. 2014 doi: 10.7554/eLife.03790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai MM, Fisher DS, Murray AW. The speed of evolution and maintenance of variation in asexual populations. Curr Biol. 2007;17:385–394. doi: 10.1016/j.cub.2007.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin K, Coppieters W, Drogemuller C, Ahariz N, Cambisano N, Druet T, Fasquelle C, Haile A, Horin P, Huang L, Kamatani Y, Karim L, Lathrop M, Moser S, Oldenbroek K, Rieder S, Sartelet A, Solkner J, Stalhammar H, Zelenika D, Zhang Z, Leeb T, Georges M, Charlier C. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature. 2012;482:81–84. doi: 10.1038/nature10757. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8:610–618. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- Fan Y, Mao R, Lv H, Xu J, Yan L, Liu Y, Shi M, Ji G, Yu Y, Bai J, Jin Y, Fu S. Frequency of double minute chromosomes and combined cytogenetic abnormalities and their characteristics. J Appl Genet. 2011;52:53–59. doi: 10.1007/s13353-010-0007-z. [DOI] [PubMed] [Google Scholar]
- Fogel S, Welch JW. Tandem gene amplification mediates copper resistance in yeast. PNAS. 1982;79:5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frei E, 3rd, Rosowsky A, Wright JE, Cucchi CA, Lippke JA, Ervin TJ, Jolivet J, Haseltine WA. Development of methotrexate resistance in a human squamous cell carcinoma of the head and neck in culture. PNAS. 1984;81:2873–2877. doi: 10.1073/pnas.81.9.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenk S, Pizza G, Walker RV, Houseley J. Aging yeast gain a competitive advantage on non-optimal carbon sources. Aging Cell. 2017;16:602–604. doi: 10.1111/acel.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galeote V, Bigey F, Beyne E, Novo M, Legras JL, Casaregola S, Dequin S. Amplification of a Zygosaccharomyces bailii DNA segment in wine yeast genomes by extrachromosomal circular DNA formation. PLoS ONE. 2011;6:e17872. doi: 10.1371/journal.pone.0017872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish PJ, Colato A, Sniegowski PD. Genomic mutation rates that neutralize adaptive evolution and natural selection. J R Soc Interface. 2013;10:20130329. doi: 10.1098/rsif.2013.0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, DeSevo CG, Botstein D, Dunham MJ. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. PNAS. 2010;107:18551–18556. doi: 10.1073/pnas.1014023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber DA, Schimke RT. Unstable amplification of an altered dihydrofolate reductase gene associated with double-minute chromosomes. Cell. 1981;26:355–362. doi: 10.1016/0092-8674(81)90204-x. [DOI] [PubMed] [Google Scholar]
- Von Hoff DD, Forseth B, Clare CN, Hansen KL, VanDevanter D. Double minutes arise from circular extrachromosomal DNA intermediates which integrate into chromosomal sites in human HL-60 leukemia cells. J Clin Invest. 1990;85:1887–1895. doi: 10.1172/JCI114650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta Y, Bassel A. Molecular size and circularity of DNA in cells of mammals and higher plants. PNAS. 1965;53:356–362. doi: 10.1073/pnas.53.2.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull RM, Cruz C, Jack CV, Houseley J. Environmental change drives accelerated adaptation through stimulated copy number variation. PLoS Biol. 2017;15:e2001333. doi: 10.1371/journal.pbio.2001333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull RM, King M, Pizza G, Krueger F, Vergara X, Houseley J. Transcription-induced formation of extrachromosomal DNA during yeast ageing. PLoS Biol. 2019;17:e3000471. doi: 10.1371/journal.pbio.3000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol. 2004;2:E296. doi: 10.1371/journal.pbio.0020296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M. Cancer genomics and genetics of FGFR2 (Review) Int J Oncol. 2008;33:233–237. [PubMed] [Google Scholar]
- Koche RP, Rodriguez-Fos E, Helmsauer K, Burkert M, MacArthur IC, Maag J, Chamorro R, Munoz-Perez N, Puiggros M, Dorado Garcia H, Bei Y, Roefzaad C, Bardinet V, Szymansky A, Winkler A, Thole T, Timme N, Kasack K, Fuchs S, Klironomos F, Thiessen N, Blanc E, Schmelz K, Kunkele A, Hundsdorfer P, Rosswog C, Theissen J, Beule D, Deubzer H, Sauer S, Toedling J, Fischer M, Hertwig F, Schwarz RF, Eggert A, Torrents D, Schulte JH, Henssen AG. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat Genet. 2020;52:29–34. doi: 10.1038/s41588-019-0547-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo DH, Molin WT, Saski CA, Jiang J, Putta K, Jugulam M, Friebe B, Gill BS. Extrachromosomal circular DNA-based amplification and transmission of herbicide resistance in crop weed Amaranthus palmeri. PNAS. 2018;115:3332–3337. doi: 10.1073/pnas.1719354115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S, Avecilla G, Spealman P, Sethia G, Brandt N, Levy SF, Gresham D. Single-cell copy number variant detection reveals the dynamics and diversity of adaptation. PLoS Biol. 2018;16:e3000069. doi: 10.1371/journal.pbio.3000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy SF, Ziv N, Siegal ML. Bet hedging in yeast by heterogeneous, age-correlated expression of a stress protectant. PLoS Biol. 2012;10:e1001325. doi: 10.1371/journal.pbio.1001325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little AS, Balmanno K, Sale MJ, Newman S, Dry JR, Hampson M, Edwards PA, Smith PD, Cook SJ. Amplification of the driving oncogene, KRAS or BRAF, underpins acquired resistance to MEK1/2 inhibitors in colorectal cancer cells. Sci Signal. 2011;4:ra17. doi: 10.1126/scisignal.2001752. [DOI] [PubMed] [Google Scholar]
- Lynch M, Burger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84:339–344. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- Moller HD, Andersen KS, Regenberg B. A model for generating several adaptive phenotypes from a single genetic event: Saccharomyces cerevisiae GAP1 as a potential bet-hedging switch. Commun Integr Biol. 2013;6:e23933. doi: 10.4161/cib.23933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller HD, Parsons L, Jorgensen TS, Botstein D, Regenberg B. Extrachromosomal circular DNA is common in yeast. PNAS. 2015 doi: 10.1073/pnas.1508825112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller HD, Mohiyuddin M, Prada-Luengo I, Sailani MR, Halling JF, Plomgaard P, Maretty L, Hansen AJ, Snyder MP, Pilegaard H, Lam HYK, Regenberg B. Circular DNA elements of chromosomal origin are common in healthy human somatic tissue. Nat Commun. 2018;9:1069. doi: 10.1038/s41467-018-03369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Murray AW, Szostak JW. Pedigree analysis of plasmid segregation in yeast. Cell. 1983;34:961–970. doi: 10.1016/0092-8674(83)90553-6. [DOI] [PubMed] [Google Scholar]
- Nathanson DA, Gini B, Mottahedeh J, Visnyei K, Koga T, Gomez G, Eskin A, Hwang K, Wang J, Masui K, Paucar A, Yang H, Ohashi M, Zhu S, Wykosky J, Reed R, Nelson SF, Cloughesy TF, James CD, Rao PN, Kornblum HI, Heath JR, Cavenee WK, Furnari FB, Mischel PS. Targeted therapy resistance mediated by dynamic regulation of extrachromosomal mutant EGFR DNA. Science. 2014;343:72–76. doi: 10.1126/science.1241328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park PU, Defossez PA, Guarente L. Effects of mutations in DNA repair genes on formation of ribosomal DNA circles and life span in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:3848–3856. doi: 10.1128/mcb.19.5.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen T, Kumar P, Koseoglu MM, Dutta A. Discoveries of extrachromosomal circles of DNA in normal and tumor cells. Trends Genet. 2018;34:270–278. doi: 10.1016/j.tig.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payen C, Sunshine AB, Ong GT, Pogachar JL, Zhao W, Dunham MJ. High-throughput identification of adaptive mutations in experimentally evolved yeast populations. PLoS Genet. 2016;12:e1006339. doi: 10.1371/journal.pgen.1006339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada-Luengo I, Møller HD, Henriksen RA, Gao Q, Larsen CE, Alizadeh S, Maretty L, Houseley J, Regenberg B. Replicative aging is associated with loss of genetic heterogeneity from extrachromosomal circular DNA in Saccharomyces cerevisiae. bioRxiv. 2020 doi: 10.1101/2020.02.11.943357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzinski M, Reichmann D. Variety is the spice of life: how to explore a redox-dependent heterogeneity in genomically identical cellular populations. Curr Genet. 2019;65:301–306. doi: 10.1007/s00294-018-0878-9. [DOI] [PubMed] [Google Scholar]
- Serana F, Chiarini M, Zanotti C, Sottini A, Bertoli D, Bosio A, Caimi L, Imberti L. Use of V(D)J recombination excision circles to identify T and B cell defects and to monitor the treatment in primary and acquired immunodeficiencies. J Transl Med. 2013;11:119. doi: 10.1186/1479-5876-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcheprova Z, Baldi S, Frei SB, Gonnet G, Barral Y. A mechanism for asymmetric segregation of age during yeast budding. Nature. 2008;454:728–734. doi: 10.1038/nature07212. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Kumar P, Layer R, Willcox S, Gagan JR, Griffith JD, Dutta A. Extrachromosomal microDNAs and chromosomal microdeletions in normal tissues. Science. 2012;336:82–86. doi: 10.1126/science.1213307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoura MJ, Gabdank I, Hansen L, Merker J, Gotlib J, Levene SD, Fire AZ. Intricate and cell type-specific populations of endogenous circular DNA (eccDNA) in Caenorhabditis elegans and Homo sapiens. G3 (Bethesda) 2017;7:3295–3303. doi: 10.1534/g3.117.300141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—a cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/S0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Mills K, Guarente L. Accelerated aging and nucleolar fragmentation in yeast sgs1 mutants. Science. 1997;277:1313–1316. doi: 10.1126/science.277.5330.1313. [DOI] [PubMed] [Google Scholar]
- Sprouffske K, Aguilar-Rodriguez J, Sniegowski P, Wagner A. High mutation rates limit evolutionary adaptation in Escherichia coli. PLoS Genet. 2018;14:e1007324. doi: 10.1371/journal.pgen.1007324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield S, Helinski DR. Small circular DNA in Drosophila melanogaster. Cell. 1976;9:333–345. doi: 10.1016/0092-8674(76)90123-9. [DOI] [PubMed] [Google Scholar]
- Storlazzi CT, Lonoce A, Guastadisegni MC, Trombetta D, D'Addabbo P, Daniele G, L'Abbate A, Macchia G, Surace C, Kok K, Ullmann R, Purgato S, Palumbo O, Carella M, Ambros PF, Rocchi M. Gene amplification as double minutes or homogeneously staining regions in solid tumors: origin and structure. Genome Res. 2010;20:1198–1206. doi: 10.1101/gr.106252.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner KM, Deshpande V, Beyter D, Koga T, Rusert J, Lee C, Li B, Arden K, Ren B, Nathanson DA, Kornblum HI, Taylor MD, Kaushal S, Cavenee WK, Wechsler-Reya R, Furnari FB, Vandenberg SR, Rao PN, Wahl GM, Bafna V, Mischel PS. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017;543:122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda JM, Raymond F, Mukherjee A, Plourde M, Gingras H, Roy G, Lapointe A, Leprohon P, Papadopoulou B, Corbeil J, Ouellette M. Genome-wide stochastic adaptive DNA amplification at direct and inverted DNA repeats in the parasite Leishmania. PLoS Biol. 2014;12:e1001868. doi: 10.1371/journal.pbio.1001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt N, Lefevre SH, Apiou F, Dutrillaux AM, Cor A, Leuraud P, Poupon MF, Dutrillaux B, Debatisse M, Malfoy B. Molecular structure of double-minute chromosomes bearing amplified copies of the epidermal growth factor receptor gene in gliomas. PNAS. 2004;101:11368–11373. doi: 10.1073/pnas.0402979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielgoss S, Barrick JE, Tenaillon O, Wiser MJ, Dittmar WJ, Cruveiller S, Chane-Woon-Ming B, Medigue C, Lenski RE, Schneider D. Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. PNAS. 2013;110:222–227. doi: 10.1073/pnas.1219574110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Turner KM, Nguyen N, Raviram R, Erb M, Santini J, Luebeck J, Rajkumar U, Diao Y, Li B, Zhang W, Jameson N, Corces MR, Granja JM, Chen X, Coruh C, Abnousi A, Houston J, Ye Z, Hu R, Yu M, Kim H, Law JA, Verhaak RGW, Hu M, Furnari FB, Chang HY, Ren B, Bafna V, Mischel PS. Circular ecDNA promotes accessible chromatin and high oncogene expression. Nature. 2019;575:699–703. doi: 10.1038/s41586-019-1763-5. [DOI] [PMC free article] [PubMed] [Google Scholar]


