Highlights
-
•
The cassette system enables automatic extraction, amplification and identification of target nucleic acids.
-
•
The closed-type cassette decreases the cross-contamination risk from amplicons.
-
•
The cassette system performs sample-in/answer-out nucleic acid test.
-
•
The cassette system can simultaneously detect 10 genotypes at 5 SNP sites using a 0.5 μL blood sample.
Keywords: Genetic test, Cassette, Point-of-care, Colorimetric detection, Magnetic separation
Abstract
Nucleic acid detection is important for clinical diagnostics; however, it is challenging to perform genetic testing at the point-of-care due to the tedious steps involved in DNA extraction and the risk of cross-contamination from amplicons. To achieve a fully-automated and contamination-free nucleic acid detection, we propose a closed-type cassette system which enables the following steps to be operated automatically and sequentially: sample preparation based on magnetic beads, target amplification using multiplex polymerase chain reaction, and colorimetric detection of amplicons using a serial invasive reaction coupled with the aggregation of gold nanoparticle probes. The cassette was designed to be round and closed, and 10 targets in a sample could be simultaneously detected by the naked eye or using a spectrophotometer in the system. In addition, a cassette-driven device was fabricated to transfer reagents between wells, to control the temperature of each reaction, and to sense the colour in the detection wells. The cassette system was sensitive enough to detect 10 genotypes at 5 single nucleotide polymorphism sites related to the anticoagulant’s usage, by using a 0.5 μL blood sample. The accuracy of the system was evaluated by detecting 12 whole blood samples, and the results obtained were consistent with those obtained using pyrosequencing. The cassette is airtight and the whole system is fully automatic; the only manual operation is the addition of the sample to the cassette, performing point-of-care genetic testing in a sample-in/answer-out way.
1. Introduction
Genetic testing is an important tool in disease diagnosis [1], pathogen detection [2], personalised medicine [3] and forensic medicine [4]. Along with the increasing number of genetic biomarkers discovered [5], genetic testing has become essential in clinical diagnosis. At the moment, many kinds of genetic testing methods have been developed, such as polymerase chain reaction (PCR)-based technologies [6] and isothermal amplification methods [7,8]. Most of them require the following steps: nucleic acid extraction, amplification and amplicon identification. Unlike an immunoassay for protein detection, each of these steps should be accomplished in an isolated room in a qualified biosafety lab and operated by a specially trained person. Therefore, regular genetic testing is time-consuming and labour-intensive, and thus, not suitable for a point-of-care-test (POCT) in a resource-limited environment.
In many cases, genetic testing results need to be quickly obtained at the point-of-care. For example, doctors need to know the cytochrome P450 2C19 (CYP2C19) genotype of a stroke patient as soon as possible to help choose the right anticoagulant and the corresponding dosage [9]. Therefore, the development of a fully automated POCT system for detecting nucleic acids is preferable in a situation where qualified staff and a biosafety lab are unavailable. The common POCT method integrates the sample preparation, target DNA amplification and the amplicon identification steps into one. Currently, many microfluidic chip-based POCT devices have been developed [[10], [11], [12], [13], [14], [15]], showing the potential to detect nucleic acids on site. However, these have some drawbacks, such as the small sample processing volume, the open operation system, the usual single-plex detection, the need of a sensitive sensing system for target identification; thus, their clinical application is very limited.
Nucleic acid detection in a qualified biosafety lab is the gold standard for molecular diagnosis. The best POCT strategy is to simulate all the lab-based operation steps necessary for DNA detection in a cassette. A cassette-based POCT is superior to a chip-based POCT in clinical diagnosis, due to the complete integration of all the detection processes (similar to that in a regular biolab) into an airtight cassette. Therefore, several cassette-based POCTs have been approved for clinical use by the Food and Drug Administration for pathogen detection in the USA, such as the GeneXpert® [[16], [17], [18]], iCubate® [19,20], FilmArray® [21,22], among others. However, they are mostly based on fluorescence for amplicon detection, leading to a complicated instrumentation since they require the use of laser irradiation and a light sensor system.
As the naked eye is a good enough sensor, colorimetric detection is an instrument-free type of detection, which is very suitable for POCT [[23], [24], [25]]. Lateral flow strip is a cheap and simple way for colorimetrically detecting amplicons, and has been used for POCT in no-airtight devices [26,27]. However, the transfer of amplicons into the strip is needed after amplification, which limits the use of the lateral flow strip in POCTs based on an airtight cassette. On the other hand, the lateral flow detection is amplicon-sensitive; thus, it is very hard to achieve single-base resolution. It is also difficult to couple this method with multiplexing amplification, which is needed in most clinical diagnosis (for example, in multiple pathogen detection and multiple single nucleotide polymorphism (SNP) typing) for simultaneously detecting multiple existing targets in a sample.
Previously, we have proposed a method of colorimetric detection of nucleic acids based on a serial invasive reaction coupled with the aggregation of gold nanoparticle probes (AuNPs) [28]. Benefitting from the high specificity of the invasive reaction and the unambiguous identification of the colour change due to the aggregation of the nanoparticle probes, amplicons from PCR or loop-mediated isothermal amplification could be identified with one-base resolution by the naked eye [28,29], showing a good performance in the detection of genotypes, circulating tumour DNAs, and pathogens [30,31]. Although this AuNPs-based colorimetric assay only needs a common PCR engine, an additional DNA extraction step is still required, which limits the application of the method to POCTs.
Herein, we aimed to integrate the DNA extraction step, the amplification and the AuNPs-based colorimetric identification into a closed-type cassette to achieve a sample-in/answer-out POCT. The system was designed to be able to automatically transfer reagents between wells, and to sequentially run the DNA extraction, multiplex PCR amplification, serial invasive reaction and colour generation. The colour of the AuNPs in each detection well can be sensed by the naked eye or using a spectrophotometric scanning module in the system. The proposed cassette-based POCT enables the simultaneous detection of 10 targets with a single-base resolution.
2. Materials and methods
2.1. Design and fabrication of the cassette
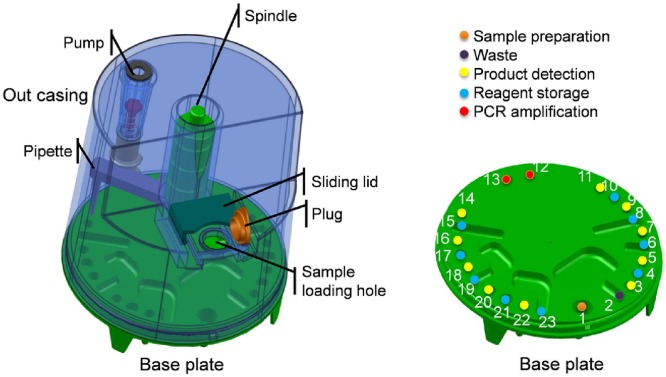
The cassette is an airtight and cylindrical cavity composed of a base plate and an outer case (Fig. 1 ).There are 23 wells on the base plate, including a sample preparation well, two PCR amplification wells, 10 product detection wells and 10 reagent storage wells. The reagents can be sealed in individual wells with aluminium foil in advance. The outer case of the cassette is equipped with a spindle that can rotate and move vertically, driving a pipette connected to the bottom of the spindle to transfer liquid between wells on the base. The pipetting of the reagents in each well is achieved by using a pump, which is on the outer case and is connected to the pipette by a soft silica tube. The outer case is designed to be tall on one side and low on the other. A sample loading hole is located on the low side, and the sample is added to the sample extraction well on the base plate through this hole. After adding a sample, the sample-loading hole is sealed with a plug, and subsequently covered with a sliding lid to fix the plug, this way ensuring that the cassette is airtightly sealed during the test. Moreover, sealing rings are used at the connection regions of each part to prevent air leaks. Except for the silica soft tube and the sealing rings, all parts of the cassette are manufactured using injection moulding. The base plate, the plug and the pipette are made of polypropylene, and the other parts are made of 3-aminopropyltriethoxysilane. The whole cassette is disposable, and a new cassette should be used for every sample.
Fig. 1.
3-D model of the cassette used for the POCT. The cassette consists of a base plate with 23 wells specified with different colours to illustrate their function (on the right), an outer case, a pipette, a pump, a spindle, a sliding lid, a plug and a sample loading hole.
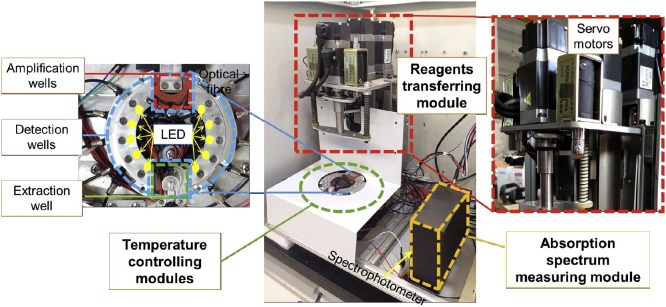
2.2. Design and fabrication of the device
The whole device mainly includes a reagents transferring module, temperature controlling modules and an absorption spectrum measuring module (Fig. 2 ). The reagents transferring module is composed of three servo motors and some connection parts to drive the spindle and the pump plunger of the cassette. Two of the three servo motors are connected to the spindle and in charge of the rotation and vertical movement of the spindle, which drives the pipette to move inside the cassette. By using a program to control the rotation angle and stroke of the two servo motors, it is possible to control the precise position of the pipette inside the cassette. The other servo motor is responsible for controlling the precise movement of the pump plunger, so that different volumes of reagents can be pipetted in each well. Three separate semiconductor-based temperature controlling modules are employed to precisely control the temperature required for nucleic acid extraction, amplification, and detection. The detection results are read out using a commercial spectrophotometer (Nanjing Wins Technology Company Limited, China) embedded in the instrument to monitor the colour produced by the AuNPs. A light emitting diode (LED) with an emission spectrum ranging from 420 nm to 660 nm was set on one side of the detection well near the centre of the temperature controlling module, and the other side was connected to the spectrophotometer through an optical fibre to determine the absorption spectrum. The detection well in the cassette is located between the LED and the optical fibre.
Fig. 2.
Image of the cassette-driven device, including the reagents transferring module, temperature controlling modules and absorption spectrum measuring module. The pipetting and the transferring of reagents were operated using the servo motors. The base plate containing the extraction wells, amplification wells and detection wells is heated and cooled down by the temperature controlling modules.
2.3. DNA extraction
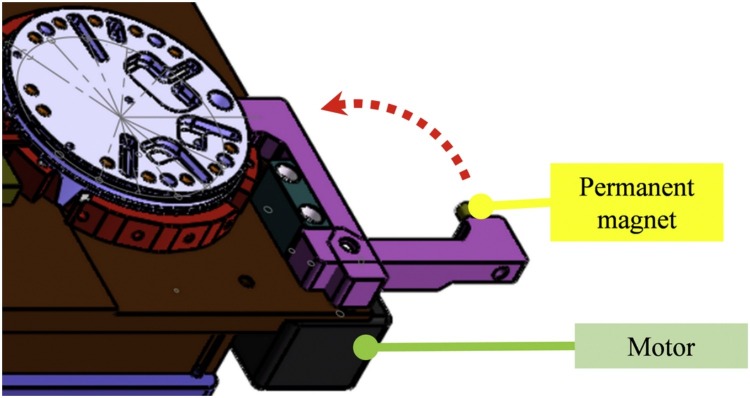
A whole blood DNA extraction kit (TianLong Science and Technology, Xi’an, China) was used for DNA extraction, including the sample lysis buffer, washing buffer Ⅰ, washing buffer Ⅱ, elution buffer, protease K (20 mg/mL) and magnetic beads, which were pre-stored in well-1, well-23, well-19, well-6, well-21 and well-17, respectively. After adding 50 μL of blood sample to well-1, the sample loading hole was closed, and the cassette was put into the device. The reagent transferring module of the device drove the pipette in the cassette for DNA extraction, and the temperature controlling module under the well-1 was in charge of controlling the sample incubation temperature. Firstly, 10 μL of protease K in well-21 and 8 μL of magnetic beads in well-17 were transferred to well-1 and mixed well by using the pipette. After incubation at 56℃ for 15 min, a permanent magnet was moved close to well-1 by using a motor (as shown in Fig. 3 ) and the magnetic beads were assembled in well-1. One minute later, the suspension was moved to the waste well (well-2) and 240 μL of washing buffer I from well-23 were added to well-1 by using the pipette. Then, the permanent magnet was moved away from well-1, and the magnetic beads were suspended in the washing buffer I by using the pipette. The permanent magnet was moved close to well-1 to assemble the beads again. One minute later, the suspension was moved to the waste well (well-2) and 240 μL of washing buffer II from well-19 were added to the well-1 by using the pipette. Then, the permanent magnet was moved away from well-1, and the magnetic beads were suspended in the washing buffer II by pipetting. The permanent magnet was moved close to well-1 to assemble the beads. After removing the suspension in well-1, 100 μL of elution buffer from well-6 were added to well-1, and the permanent magnet was moved away from well-1. After incubation at 65℃ for 5 min, the purified nucleic acid was eluted. A volume of 5 μL of eluted solution was respectively transferred to the amplification wells (well-12 and well-13) for PCR.
Fig. 3.
3-D model of the magnet control mode, consisting of a permanent magnet and a motor for driving. The motor controls the permanent magnet to approach or move away from the extraction well, along the red dotted line.
2.4. Multiplex PCR
Multiplex PCR was performed in well-12 and well-13. The PCR mixture contained 1× Master mix (QIAGEN Multiples PCR Kit, Qiagen, Germany), 0.25 μM each primer for amplifying CYP2C19*2, *3, cytochrome P450 2C9 (CYP2C9)*2, *3, and vitamin K epoxide reductase 1 (VKORC1) fragments (the sequences are shown in Table S1) and 20 μL of mineral oil was pre-stored in well-12 and well-13. After adding 5 μL of the eluted solution to the amplification wells (the total volume of the reaction mixture was 20 μL, excluding the mineral oil), PCR was performed using the following conditions: 95 °C for 5 min, 40 cycles of 94 °C for 30 s, 63 °C for 90 s, 72 °C for 60 s, followed by 72 °C for 5 min, and 98 °C for 5 min. After the PCR, 60 μL of H2O from well-8 were added to each amplification well and mixed well for the product detection step.
2.5. Serial invasive reactions
The PCR products were detected using a serial invasive reaction coupled with the aggregation of AuNPs. The serial invasive reactions were performed in well-3, well-5, well-7, well-9, well-11, well-14, well-16, well-18, well-20 and well-22 using 20 μL of the reaction mixture containing 1× reaction buffer (10 mM 3-morpholinopropane sulfonic acid, pH 7.5, 0.5% polyethylene glycol sorbitan monolaurate 20, 0.5% nonidet P-40, 10% bovine serum albumin, 30 mM MgCl2, 0.5 mM KCl, 10% polyethylene glycol 8000, 15 U of archaeoglobus fulgidus flap endonuclease 1 (FEN1) (prepared in our lab), 0.1 μM upstream probes, 0.1 μM downstream probes and 0.2 μM hairpin probe (the sequences of the probes are shown in Table S1). A volume of 5 μL of diluted PCR products was transferred to the 10 detection wells and the serial invasive reaction was performed at 85 °C for 1 min, followed by 63 °C for 16 min.
2.6. AuNPs hybridisation reaction
After the serial invasive reaction, 5 μL of NaCl (4 M) stored in well-10 and 6 μL of AuNPs stored in well-4 were transferred to each of the detection wells. The reaction was performed at 55 °C for 40 min. When the temperature of the detection wells reached 55 °C, the absorption spectrum of each detection well was measured using a spectrophotometer and used as a blank control. After the reaction, the absorption spectrum of each detection well was measured again. The detection results could be identified either by analysing the absorption spectrum of each detection well, or by observing the colour of each well based on the naked eye.
3. Results and discussion
3.1. DNA extraction efficiency
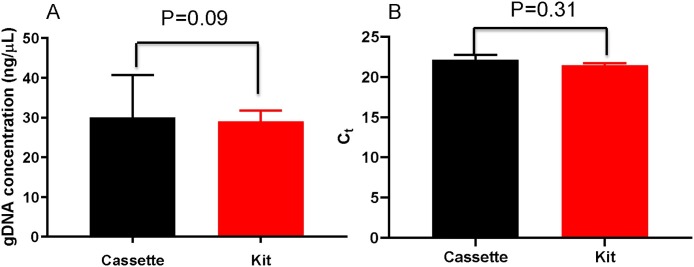
Sample preparation is a key step for a successful genetic test, because the quality and the quantity of an extracted DNA template may affect the detection results. Our system stores the reagents from the magnetic beads-based blood DNA extraction kit in the cassette for sample preparation. After loading a sample into the DNA extraction well from the loading hole, the system automatically performs all the extraction steps in the airtight cassette. To ensure the accuracy of the detection, the DNA extraction efficiency using the cassette should be as high as that achieved using the conventional manual operation in a conventional lab. This was evaluated by extracting the DNA from a 50 μL blood sample with the proposed cassette system and the suggested manual operation, respectively. The concentrations of the extracted DNA obtained using the two methods were compared by using an independent t-test (n = 3). As shown in Fig. 4 A, no significant concentration differences (calculated with the absorbance at 260 nm) of the extracted DNA using the 2 methods were observed. Moreover, no significant difference was found between the cycle threshold (Ct) values of quantitative PCR for amplifying the VKORC1 gene fragment in the DNA templates extracted using the two methods (Fig. 4B). Therefore, the DNA extraction efficiency by the cassette was similar to that using the suggested kit procedure.
Fig. 4.
The concentration (A) and Ct values (B) of the gDNA extracted automatically using the cassette and manually, using the kit. The VKORC1 gene fragment was amplified using quantitative PCR (n = 3).
Currently, there are various commercialised devices for an automatic DNA extraction, but most of them are not in an airtight format. Our proposed system achieved an automatic DNA extraction using an air-tight closed cassette; thus, the risk of cross contamination from amplicons was prevented. We believe that our airtight cassette system is very suitable for POCT.
3.2. Multiplex PCR in the cassette
Usually, the detection of multiple targets in a sample is needed; thus, multiplex PCR should be carried out for simultaneously amplifying fragments containing the target regions. The difficulty of multiplex PCR optimisation increased, along with the increase of the multiplex level. The cassette has 10 detection wells; thus, a multiplex PCR that detects 10 targets is needed. To simplify the optimisation process of the multiplex PCR, we designed two wells for PCR, for example, each for 5-plex PCR, if necessary. Regarding the genotyping, a 5-plex PCR was used for detecting 10 genotypes at 5 SNPs. A prerequisite for a successful multiplex PCR is the design of optimal primer pairs. The key for the design is to optimise the length and the GC content of the primers. In principle, the probability of a nonspecific binding is lower at higher annealing temperatures (for which the activity of Taq DNA polymerase is optimized); thus, here, longer primers with a higher melting temperature (64 - 68 °C) and an appropriate GC content (40 - 60%) were designed for 5-plex PCR. The optimal annealing temperature (63 °C) was determined by performing a gradient PCR. It is worth to emphasise that the templates with a high degree of secondary structure due to GC-rich regions may lead to an unsuccessful multiplex PCR; thus, the use of additives, such as Q-solution in the QIAGEN multiplex PCR kit, in multiplex PCR is recommended.
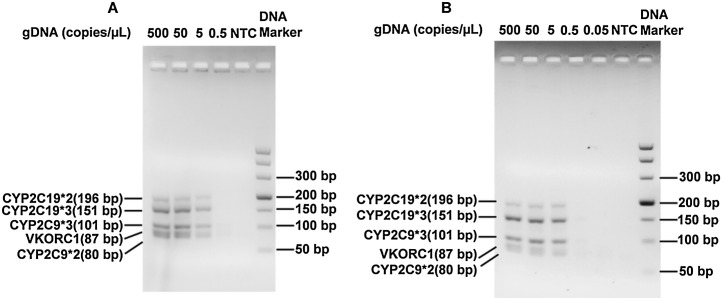
To verify the performance of multiplex PCR in the cassette, pre-extracted genomic DNA samples, which were serially diluted to different concentrations, were amplified using the cassette system and a conventional PCR engine. Amplicons from both instruments were analysed using electrophoresis (Fig. 5 ). It was observed that all five different target DNAs were successfully amplified in the cassette (Fig. 5A), even at a concentration of 5 copies genomic DNA/μL. This result is similar to that obtained using a conventional PCR engine (Fig. 5B), indicating that the temperature controlling module of our system could meet the multiplex PCR requirements.
Fig. 5.
Electropherograms of multiplex PCR amplicons using different concentrations of genomic DNA templates, using our cassette system (A) and a conventional PCR engine (B).
3.3. AuNPs aggregation-assisted serial invasive reaction for amplicon identification in the cassette
To identify each target in the amplicons from multiplex PCR, serial invasive reaction, which has a one-base resolution, was employed. Moreover, AuNPs were used as an indicator of colorimetric detection.
To illustrate this method, the simultaneous genotyping of multiple SNPs was employed as an example. As shown in Fig. S1, an upstream probe (UP) and an SNP-specific downstream probe (DP-W/DP-M) forms a three-base overlapping structure at the SNP site if the amplicons completely match the probes. FEN1 recognises the structure and cleaves the DP to release the 5′-flap fragment, which is captured by a hairpin probe to form another three-base overlapping structure, causing the cleavage of the hairpin probe by FEN1. Then, AuNPs are added into the detection wells using the pipette in the cassette to generate the signals. The cleaved hairpin probe cannot bridge two gold nanoparticle probes, so that no aggregation occurs; thus, a red colour appears in the detection well. On the contrary, if the amplicons do not match the probes, no cleavage occurs, and the AuNPs aggregate, leading to a colourless solution in the detection well. Therefore, the genotyping results could be readily obtained by observing the solution colour in each of the detection wells by the naked eye or automatically, using a spectrophotometer in the cassette system.
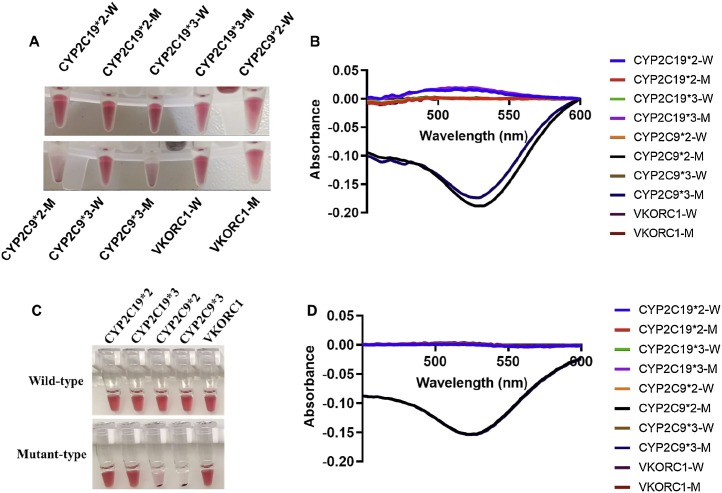
The demonstration of multiplex PCR combined with AuNPs aggregation-assisted serial invasive reaction in the cassette was carried out by detecting 10 genotypes at 5 SNPs. The genotypes of a purified genomic DNA sample with the concentration of 5 copies/μL were automatically detected in the cassette. As shown in Fig. 6 A, B, CYP2C19*2, *3 and VKORC1 were heterozygotes, and CYP2C9*2 and *3 were homozygotes of the wild-type, consistent with the results from the same reactions manually performed using a conventional PCR engine (Fig. 6C, D). Therefore, the cassette system could achieve an accurate target identification using multiplex PCR combined with a serial invasive reaction and AuNPs aggregation.
Fig. 6.
The genotyping results of a genomic DNA sample at 5 copies/μL using multiplex PCR combined with AuNPs aggregation assisted the serial invasive reaction using the cassette (A, B) and a traditional PCR engine (C, D). A: images of detection wells in the cassette; B: absorption spectra of the corresponding detection wells in the cassette; C: images of the tubes from the genotyping reactions performed using a traditional PCR engine. D: absorption spectra of the corresponding tubes in C.
3.4. The detection limit of the cassette system
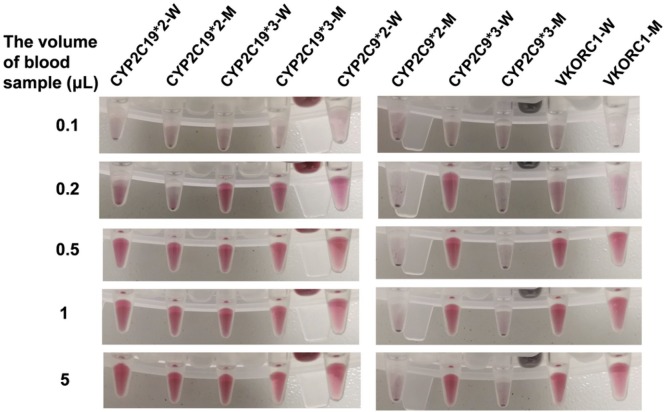
The cassette system could accurately detect the genotypes of the purified genomic DNA sample with a concentration of 5 copies/μL, but the detection limit of the cassette for running the whole steps should be verified from the starting material, the blood. Blood samples with a volume of 0.1, 0.2, 0.5, 1 and 5 μL were dispensed into the cassettes, and all the steps from the genomic DNA extraction to target identification were automatically and sequentially ran. As shown in Fig. 7 and Fig. S2, our proposed cassette system is sensitive enough to get accurate genotyping results of all 5 SNPs using 0.5 μL of a blood sample, while a 0.1 μL blood sample could not yield any positive results. Although we found a weak signal in some detection wells (VKORC1-W, VKORC1-M, CYP2C19*2-W and CYP2C19*2-M in Fig. 7) when the blood sample volume used was 0.2 μL, to obtain accurate and reproducible results using the cassette system, a volume of 0.5 μL of whole blood, at least, is required, similar to that using manual operation (Fig. S3). Therefore, it is possible to use fingertip blood for on-site genotyping using the cassette system.
Fig. 7.
Images of detection wells in the cassettes for genotyping 5 SNPs using blood samples with the following volumes: 0.1, 0.2, 0.5, 1 and 5 μL.
3.5. Feasibility of the detection of clinical samples
Although the detection limit of the cassette system was sufficient for the genotyping of 5 SNPs using 0.5 μL of whole blood, further verification of the system using more clinical samples was necessary. A total of 12 blood samples were tested using the cassette system, and the results were compared with those using pyrosequencing. As shown in Table 1 , the genotypes of 12 samples detected automatically using the cassette were consistent with those using pyrosequencing, indicating that the cassette system could accurately detect the genotypes of the clinical samples. Different from pyrosequencing, which requires four physically separated regions for reagent preparation, sample extraction, PCR amplification, and amplicons sequencing, the cassette system can automatically and sequentially run all the steps, from DNA extraction to target identification. Therefore, the cassette system has a potential to be used as a POCT of DNA.
Table 1.
The comparison of the genotyping results of 12 clinical samples detected using the cassette system and pyrosequencing.
| Genes | Methods | Genotyping results | ||||
|---|---|---|---|---|---|---|
| CYP2C19*2 | Pyrosequencing | Cassette system | ||||
| GG | GA | AA | Total | |||
| GG | 3 | 0 | 0 | 3 | ||
| GA | 0 | 9 | 0 | 9 | ||
| AA | 0 | 0 | 0 | 0 | ||
| Total | 3 | 9 | 0 | 12 | ||
| CYP2C19*3 | Pyrosequencing | Cassette system | ||||
| GG | GA | AA | Total | |||
| GG | 11 | 0 | 0 | 11 | ||
| GA | 0 | 1 | 0 | 1 | ||
| AA | 0 | 0 | 0 | 0 | ||
| Total | 11 | 1 | 0 | 12 | ||
| CYP2C9*2 | Pyrosequencing | Cassette system | ||||
| CC | CT | TT | Total | |||
| CC | 12 | 0 | 0 | 12 | ||
| CT | 0 | 0 | 0 | 0 | ||
| TT | 0 | 0 | 0 | 0 | ||
| Total | 12 | 0 | 0 | 12 | ||
| CYP2C9*3 | Pyrosequencing | Cassette system | ||||
| AA | AC | CC | Total | |||
| AA | 12 | 0 | 0 | 12 | ||
| AC | 0 | 0 | 0 | 0 | ||
| CC | 0 | 0 | 0 | 0 | ||
| Total | 12 | 0 | 0 | 12 | ||
| VKORC1 | Pyrosequencing | Cassette system | ||||
| GG | GA | AA | Total | |||
| GG | 0 | 0 | 9 | 9 | ||
| GA | 0 | 3 | 0 | 3 | ||
| AA | 0 | 0 | 0 | 0 | ||
| Total | 0 | 3 | 9 | 12 | ||
4. Conclusions
We have developed a cassette-based POCT, which is able to automatically and sequentially perform the whole genetic testing process in a closed-type cassette. The only manual operation required during the test is the addition of a sample to the sample well in the cassette; thus, no professionally trained personnel nor a qualified lab are needed for testing. In addition, the airtight cassette minimises the cross-contamination risk from the amplicon’s aerogel. Our fully automatic POCT system has a sensitivity similar to that obtained using manual operation in a lab, and on-site genotyping using fingertip blood is possible. This system is very suitable for genotype-guided medication in an emergency room, and resource-limited medical regions, such as the clinic and community medical centres.
Most importantly, the sequence identification and signal generation are universal, and could be principally used to identify any DNA targets in the cassette. During the development of the method, the only time-consuming step is the optimisation of multiplex PCR. By using commercialised multiplex PCR kits, we believe that it is not difficult to amplify 10 targets with two sets of multiplex PCRs in 2 wells. Further studies using the cassette-based POCT for the detection of multiple pathogens are in progress. We believe that our system could be an effective tool for rapidly detecting pathogens of infectious diseases (such as COVID-19) on-site.
CRediT authorship contribution statement
Tianhui Dong: Conceptualization, Software, Formal analysis, Validation, Investigation. Xueping Ma: Data curation, Investigation, Methodology. Nan Sheng: Methodology, Visualization. Xiemin Qi: Investigation, Validation. Yanan Chu: Investigation, Resources. Qinxin Song: Resources, Funding acquisition. Bingjie Zou: Conceptualization, Investigation, Project administration, Methodology, Writing - original draft. Guohua Zhou: Funding acquisition, Supervision, Project administration, Resources, Writing - original draft.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (61871403, 81673390), Jiangsu Provincial Science Fund for Distinguished Young Scholars (BK20180005), Jiangsu Provincial Medical Youth Talent Program (No. QNRC2016889).
Biographies
Tianhui Dong is a Ph.D. candidate in School of Life Science and Technology, China Pharmaceutical University. His current research is focused on the biosensors and analytical microsystems.
Xueping Ma is a Research Fellow in Department of Pharmacology, Jinling Hospital, Medical School of Nanjing University. Her current research is focused on genetic testing related to pharmacogenomics.
Nan Sheng received her Ph.D. degree at China Pharmaceutical University. Her current research is focused on the method development for molecular diagnosis related to personalized medicine.
Xiemin Qi is a Research Fellow in Department of Pharmacology, Jinling Hospital, Medical School of Nanjing University. She has been working on the development of analytical methods for personalized medicine and microbial diversity.
Yanan Chu is a Research Fellow in Department of Pharmacology, Jinling Hospital, Medical School of Nanjing University. Her current research is focused on genetic testing related to pharmacogenomics.
Qinxin Song is a Professor of the School of Pharmacy in China Pharmaceutical University. Her research is mainly focused on clinical molecular diagnosis, pharmacogenomics and pharmaceutical analysis.
Bingjie Zou is a Senior Research Fellow in the Department of Pharmacology, Jinling Hospital, Medical School of Nanjing University. He has devoted himself to the development of companion diagnostic techniques for personalized medicine.
Guohua Zhou is the Head of the Pharmacology Department in Jinling Hospital, Medical School of Nanjing University, and acquired the special government allowance in China. He is a tenured Professor in State Key Laboratory of Analytical Chemistry for Life Science in Nanjing University, and the Chairman of Pharmacogenomics Committee in Jiangsu Province. He has been developing various tools for highly sensitive and highly specific DNA detection in related to personalized medicine and infectious diseases.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.snb.2020.128919.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Eccles D.M. Uses of genetic testing for cancer prevention. Ann. Oncol. 2019;30:vi4. [Google Scholar]
- 2.Niemz A., Ferguson T.M., Boyle D.S. Point-of-care nucleic acid testing for infectious diseases. Trends Biotechnol. 2011;29:240–250. doi: 10.1016/j.tibtech.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans J.P., Watson M.S. Genetic Testing and FDA Regulation Overregulation Threatens the Emergence of Genomic Medicine. J. Am. Med. Assoc. 2015;313:669–670. doi: 10.1001/jama.2014.18145. [DOI] [PubMed] [Google Scholar]
- 4.McCord B.R., Gauthier Q., Cho S., Roig M.N., Gibson-Daw G.C., Young B. Forensic DNA Analysis. Anal. Chem. 2019;91:673–688. doi: 10.1021/acs.analchem.8b05318. [DOI] [PubMed] [Google Scholar]
- 5.Yang Y.H., Wu L., Shu X., Lu Y.C., Shu X.O., Cai Q.Y. Genetic Data from Nearly 63,000 Women of European Descent Predicts DNA Methylation Biomarkers and Epithelial Ovarian Cancer Risk. Cancer Res. 2019;79:505–517. doi: 10.1158/0008-5472.CAN-18-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang H.M., Subramaniam M., Targ S., Nguyen M., Maliskova L., McCarthy E. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 2018;36:89. doi: 10.1038/nbt.4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding X., Xu Z.H., Yin K., Sfeir M., Liu C.C. Dual-Priming Isothermal Amplification (DAMP) for Highly Sensitive and Specific Molecular Detection with Ultralow Nonspecific Signals. Anal. Chem. 2019;91:12852–12858. doi: 10.1021/acs.analchem.9b02582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varona M., Anderson J.L. Visual Detection of Single-Nucleotide Polymorphisms Using Molecular Beacon Loop-Mediated Isothermal Amplification with Centrifuge-Free DNA Extraction. Anal. Chem. 2019;91:6991–6995. doi: 10.1021/acs.analchem.9b01762. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka T., Yamagami H., Ihara M., Miyata T., Miyata S., Hamasaki T. Association of CYP2C19 Polymorphisms With Clopidogrel Reactivity and Clinical Outcomes in Chronic Ischemic Stroke. Circ. J. 2019;83:1385–1393. doi: 10.1253/circj.CJ-18-1386. [DOI] [PubMed] [Google Scholar]
- 10.Chinnadayyala S.R., Park J., Le H.T.N., Santhosh M., Kadam A.N., Cho S. Recent advances in microfluidic paper-based electrochemiluminescence analytical devices for point-of-care testing applications. Biosens. Bioelectron. 2019;126:68–81. doi: 10.1016/j.bios.2018.10.038. [DOI] [PubMed] [Google Scholar]
- 11.Liang C., Liu Y.C., Niu A.Y., Liu C., Li J.M., Ning D.X. Smartphone-app based point-of-care testing for myocardial infarction biomarker cTnI using an autonomous capillary microfluidic chip with self-aligned on-chip focusing (SOF) lenses. Lab Chip. 2019;19:1797–1807. doi: 10.1039/c9lc00259f. [DOI] [PubMed] [Google Scholar]
- 12.Nasseri B., Soleimani N., Rabiee N., Kalbasi A., Karimi M., Hamblin M.R. Point-of-care microfluidic devices for pathogen detection. Biosens. Bioelectron. 2018;117:112–128. doi: 10.1016/j.bios.2018.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu D.D., Huang X.W., Guo J.H., Ma X. Automatic smartphone-based microfluidic biosensor system at the point of care. Biosens. Bioelectron. 2018;110:78–88. doi: 10.1016/j.bios.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 14.Ye X., Li L., Li J., Wu X.D., Fang X.E., Kong J. Microfluidic-CFPA Chip for the Point-of-Care Detection of African Swine Fever Virus with a Median Time to Threshold in about 10 min. Acs Sensors. 2019;4:3066–3071. doi: 10.1021/acssensors.9b01731. [DOI] [PubMed] [Google Scholar]
- 15.Yeh E.C., Fu C.C., Hu L., Thakur R., Feng J., Lee L.P. Self-powered integrated microfluidic point-of-care low-cost enabling (SIMPLE) chip. Sci. Adv. 2017;3 doi: 10.1126/sciadv.1501645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hai H.T., Vinh D.N., Thu D.D.A., Hanh N.T., Phu N.H., Srinivasan V. Comparison of the Mycobacterium tuberculosis molecular bacterial load assay, microscopy and GeneXpert versus liquid culture for viable bacterial load quantification before and after starting pulmonary tuberculosis treatment. Tuberculosis. 2019;119 doi: 10.1016/j.tube.2019.101864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kost C.B., Rogers B., Oberste M.S., Robinson C., Eaves B.L., Leos K. Multicenter beta trial of the GeneXpert enterovirus assay. J. Clin. Microbiol. 2007;45:1081–1086. doi: 10.1128/JCM.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rindi L., Ali G., Fabiani B., Fontanini G., Garzelli C. Detection of Mycobacterium tuberculosis from paraffin-embedded tissues by GeneXpert MTB/RIF. Tuberculosis. 2017;106:53–55. doi: 10.1016/j.tube.2017.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Conover M.S., Heflin K., Liu H., Wagner L., Wells C. Detection of Gram-Negative Bacteria and Antimicrobial Resistance Markers Using the iCubate iC-GN Assay. J. Mol. Diagn. 2017;19:976–977. [Google Scholar]
- 20.Granato P.A., Unz M.M., Widen R.H., Silbert S., Young S., Heflin K.L. Clinical Evaluation of the iCubate iC-GPC Assay for Detection of Gram-Positive Bacteria and Resistance Markers from Positive Blood Cultures. J. Clin. Microbiol. 2018;56:e00485–18. doi: 10.1128/JCM.00485-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buss S.N., Leber A., Chapin K., Fey P.D., Bankowski M.J., Jones M.K. Multicenter Evaluation of the BioFire FilmArray Gastrointestinal Panel for Etiologic Diagnosis of Infectious Gastroenteritis. J. Clin. Microbiol. 2015;53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leber A.L., Everhart K., Daly J.A., Hopper A., Harrington A., Schreckenberger P. Multicenter Evaluation of BioFire FilmArray Respiratory Panel 2 for Detection of Viruses and Bacteria in Nasopharyngeal Swab Samples. J. Clin. Microbiol. 2018;56:e01945–17. doi: 10.1128/JCM.01945-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen H.V., Nguyen V.D., Nguyen H.Q., Chau T.H.T., Lee E.Y., Seo T.S. Nucleic acid diagnostics on the total integrated lab-on-a-disc for point-of-care testing. Biosens. Bioelectron. 2019;141 doi: 10.1016/j.bios.2019.111466. [DOI] [PubMed] [Google Scholar]
- 24.Van Nguyen H., Nguyen V.D., Lee E.Y., Seo T.S. Point-of-care genetic analysis for multiplex pathogenic bacteria on a fully integrated centrifugal microdevice with a large-volume sample. Biosens. Bioelectron. 2019;136:132–139. doi: 10.1016/j.bios.2019.04.035. [DOI] [PubMed] [Google Scholar]
- 25.Yin K., Pandian V., Kadimisetty K., Ruiz C., Cooper K., You J.X. Synergistically enhanced colorimetric molecular detection using smart cup: a case for instrument-free HPV-associated cancer screening. Theranostics. 2019;9:2637–2645. doi: 10.7150/thno.32224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim T.H., Park J., Kim C.J., Cho Y.K. Fully Integrated Lab-on-a-Disc for Nucleic Acid Analysis of Food-Borne Pathogens. Anal. Chem. 2014;86:3841–3848. doi: 10.1021/ac403971h. [DOI] [PubMed] [Google Scholar]
- 27.Roskos K., Hickerson A.I., Lu H.W., Ferguson T.M., Shinde D.N., Klaue Y. Simple System for Isothermal DNA Amplification Coupled to Lateral Flow Detection. Plos One. 2013;8 doi: 10.1371/journal.pone.0069355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu Y., Ma X.P., Wang J.P., Sheng N., Dong T.H., Song Q.X. Visualized detection of single-base difference in multiplexed loop-mediated isothermal amplification amplicons by invasive reaction coupled with oligonucleotide probe-modified gold nanoparticles. Biosens. Bioelectron. 2017;90:388–393. doi: 10.1016/j.bios.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 29.Ge Y.Y., Zhou Q., Zhao K.C., Chi Y., Liu B., Min X.Y. Detection of influenza viruses by coupling multiplex reverse-transcription loop-mediated isothermal amplification with cascade invasive reaction using nanoparticles as a sensor. Int. J. Nanomed. 2017;12:2645–2656. doi: 10.2147/IJN.S132670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J.P., Zou B.J., Ma Y.J., Ma X.P., Sheng N., Rui J.Z. Closed-Tube PCR with Nested Serial Invasion Probe Visualization Using Gold Nanoparticles. Clin. Chem. 2017;63:852–860. doi: 10.1373/clinchem.2016.263996. [DOI] [PubMed] [Google Scholar]
- 31.Pang S.Y., Sheng N., Wang J.P., Zou B.J., Song Q.X., Huang X.H. A Closed-Tube Colorimetric PCR Based on Serial Invasive Reaction Assisted Gold Nanoparticle Assembling for IL28B Genotyping. Nanosci. Nanotech. Let. 2018;10:32–38. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.