Abstract
Background
Ethiopian sheep living in different climatic zones and having contrasting morphologies are a most promising subject of molecular-genetic research. Elucidating their genetic diversity and genetic structure is critical for designing appropriate breeding and conservation strategies.
Objective
The study was aimed to investigate genome-wide genetic diversity and population structure of eight Ethiopian sheep populations.
Methods
A total of 115 blood samples were collected from four Ethiopian sheep populations that include Washera, Farta and Wollo (short fat-tailed) and Horro (long fat-tailed). DNA was extracted using Quick-DNA™ Miniprep plus kit. All DNA samples were genotyped using Ovine 50 K SNP BeadChip. To infer genetic relationships of Ethiopian sheep at national, continental and global levels, genotype data on four Ethiopian sheep (Adilo, Arsi-Bale, Menz and Black Head Somali) and sheep from east, north, and south Africa, Middle East and Asia were included in the study as reference.
Results
Mean genetic diversity of Ethiopian sheep populations ranged from 0.352 ± 0.14 for Horro to 0.379 ± 0.14 for Arsi-Bale sheep. Population structure and principal component analyses of the eight Ethiopian indigenous sheep revealed four distinct genetic cluster groups according to their tail phenotype and geographical distribution. The short fat-tailed sheep did not represent one genetic cluster group. Ethiopian fat-rump sheep share a common genetic background with the Kenyan fat-tailed sheep.
Conclusion
The results of the present study revealed the principal component and population structure follows a clear pattern of tail morphology and phylogeography. There is clear signature of admixture among the study Ethiopian sheep populations
Electronic supplementary material
The online version of this article (10.1007/s13258-020-00984-y) contains supplementary material, which is available to authorized users.
Keywords: Fat-tail, Genetic diversity, Ovine 50 K SNP, Population structure
Introduction
Given its proximity to the Arabian Peninsula, Ethiopia is considered as a corridor for the introduction of livestock species including sheep to the African continent (Hanotte et al. 2002; Muigai and Hanotte 2013). Sheep and their products play a critical role in the livelihood of millions of farmers and pastoral communities in Ethiopia and are important for the national economy (Assefa et al. 2015). Ethiopia possesses highly diversified indigenous sheep populations adapted to highly diverse agro-ecologies and the populations are maintained by difference ethnic communities (Haile et al. 2002; Gizaw et al. 2008). There are about 29.33 million heads of sheep which are phenotypically identified into 14 populations (Gizaw et al. 2008; Leta and Mesele 2014). However, the local sheep populations with very low productivity dominate smallholder production systems, which are mainly confounded by lack of effective long-term sheep genetic improvement, multiplication and effective delivery systems, environmental as well as socio-economic factors (Tadele 2010; Gizaw et al. 2013). On the other hand, the demand for sheep and their products has increased from time to time due to a growing human population and urbanization. Therefore, it is an urgent need to improve their productivity in order to raise smallholder farmers’ incomes, to meet the demand of the growing human population as well as the national economy (FAO 2012).
The local sheep populations in Ethiopia are mainly named after the geographic location or ethnic group, based on phenotypic characteristics or agro-ecology (Solomon et al. 2011). This might have led to group genetically similar populations into phenotypically distinct groups. The previous studies indicated that Ethiopian indigenous sheep were clustered together based on their geographic distribution and tail phenotypes (Gizaw et al. 2007; Edea et al. 2017; Ahbara et al. 2019). Characterizing genetic diversity and understanding of population structure is a key aspect of developing sustainable breed improvement strategies (Groeneveld et al. 2010) and understanding adaptation to extreme environments (Ai et al. 2014). Molecular characterization were done on genetic diversity and population structure of Ethiopian sheep populations using microsatellite markers (Gizaw et al. 2007; Hellen 2015). In a recent study, Edea et al (2017) and Ahbara et al (2019) evaluated population structure of Ethiopian indigenous sheep populations (Kefis, Adane, Arabo, Gafera, Molale, Bonga, Gesses, Kido, Doyogena, ShubiGemo, Loya, Adilo, Menz, Arsi-Bale, Horro and Black Head Somali) and revealed high level of admixture among the populations using high and medium density SNP chip panel, respectively. Recently developed genome-wide ovine SNP array has provided a tool for investigating genetic diversity at high resolution, inferring population history, and mapping genomic regions subject to selection and adaptation (Kijas et al. 2009; Yang et al. 2016; Zhao et al. 2017).
However, there are still indigenous sheep populations in Ethiopia yet to be evaluated at genome-wide. These sheep populations include Farta, Wollo, Washera and Horro sheep which are adapted to diverse agro-ecologies and with different tail phenotype. Therefore, in this study we aimed to investigate genome-wide genetic diversity and population structure of the indigenous sheep populations in Ethiopian.
Material and methods
Sheep populations, sampling and SNP genotyping
Blood samples were collected from four Ethiopian sheep populations (n = 115) adapted to diverse agro-ecological environments. Detail description of the populations is summarized in Table 1. Farta (n = 26) and Wollo (n = 28) sheep are short fat-tailed coarse-wool sheep and adapted to sub-alpine environments (2000–3200 m a.s.l); Washera (n = 31) sheep is short fat-tailed hairy sheep and mainly inhabits wet, warmer mid-highlands (1600–2497 m a.s.l); Horro (n = 30) sheep is long fat-tailed hairy sheep and predominant in mid-to high-altitude environments (1400–2000 m a.s.l) (Gizaw et al. 2008). Geographic positioning system (GPS) coordinates were recorded for each sheep population and geographical distribution map was developed based on their GPS coordinates (Supplementary Fig. 1).
Table 1.
Distinguishing physical features and agro-ecology of Ethiopian sheep populations
| Physical features and their ecology | Population | |||||||
|---|---|---|---|---|---|---|---|---|
| Washera | Farta | Horro | Wollo | BHS | Arsi-Bale | Menz | Adilo | |
| Fiber type | Short-haired | Coarse wool | Short-haired | Coarse wool | Short-haired | Hairy fiber | Coarse wool | Hairy fiber |
| Tail phenotype | Short fat-tail | Short fat-tail | Long fat-tail | Short fat-tail | Short fat-rump | Long fat-tail | Short fat-tail | Long fat-tail |
| Coat color | Brown, white, white and brown | White, brown, black | Brown, fawn | Black, white or brown | White body, black head and neck | Brown, red, black, gray, white | Black, Brown, white | Brown, white, gray |
| Horn | Polled | Male short horned, most females polled | Polled | Male short horned, most females polled | Polled | Males and most females are horned | Horned | Male short horned, most females polled |
| Altitude (m) | 1600–2497 | 2000–3142 | 1400–2000 | 2000–3200 | 500–1500 | 2492–2810 | 2500–3000 | 1618–2043 |
| Agro-ecology | Wet, warmer mid- highlands | Sub-alpine | Wet highlands | Sub-alpine | Arid lowland | Wet highland | Sub-alpine | Wet highland |
| Use | Meat | Meat & wool | Meat | Meat &wool | Meat | Meat | Meat &wool | Meat |
| Management | Mixed crop-livestock | Mixed crop-livestock | Mixed crop-livestock | Mixed crop-livestock | Pastoral/agro-pastoral | Mixed crop-livestock | Sheep-barley | Mixed crop-livestock |
| Community | Amhara and Agew | Amhara | Oromo | Amhara | Oromo/Somali | Oromo | Amhara | Wolaita/Hadiya |
The samples were collected from different households in different villages using 5 ml of vacutainer tube with 1 ml EDTA as anti-coagulant. A maximum number of 1–3 distinct breeding adult animals were sampled per household based on flock owners’ information and typical phenotypic characteristics that allow to avoid sampling of related animals. Genomic DNA was extracted using Quick-DNA™ Miniprep plus kit following the procedures of Biological Fluid and Tissue protocol (https://WWW.zymoresearch.com/m/D4068). All 115 genomic DNA samples were genotyped using Ovine 50 K SNP BeadChip (Illumina, San Diego, CA, USA) by GeneSeek/Neogen (Lincoln, NE, USA).
To infer genetic relationships of Ethiopian sheep populations at national level, Ovine 600 K SNP BeadChip genotype data of 44 animals representing four extra Ethiopian sheep populations (Adilo = 11; Arsi-Bale = 8; Blackhead Somali = 13; Menz = 12) were included in the study. Detail description of the extra four Ethiopian sheep populations is summarized in Table 1. The genotype data (600 K) were obtained from NRSP-8 Community File Sharing Platform (https://www.animalgenome.org/repository/pub/KORE2017.1122/).Ovine 50 K SNP BeadChip genotype data of 105 animals representing six breeds from east (Kenya: Red Maasai), north (Egyptian Barki; Moroccan sheep) and south (South Africa: Namaqua Afrikaner) Africa, Middle East (Iran: Afshari) and south-west Asia (India: Garole) were included in the study to investigate the genetic relationships in greater detail and infer population structure of Ethiopian sheep populations at continental and global levels. The genotype data (50 K) were obtained from Sheep HapMap and Animal Resources dataset (https://www.sheephapmap.org/hapmap.php). Therefore, a total of 14 sheep populations, including extra four Ethiopian sheep, are included in the study as reference.
Quality control and genetic diversity analyses
Genotypic data for 600 K and 50 K platforms were merged and common autosomal SNP markers (42,227) were obtained. These common autosomal SNPs with call rate less than 90% and minor allele frequency (MAF) less than 0.05 were filtered out using PLINK v1.9 (Purcell et al. 2007) and leaving 37,771 autosomal SNPs. Using the same software, this generated dataset were further subjected to linkage disequilibrium (LD) pruning to avoid the possible influence of clusters of SNPs on population relationships and structure analyses (Yuan et al. 2017). Following the LD pruning, 30,292 autosomal SNPs were retained for population structure analyses.
To evaluate the levels of within-population genetic diversity, the expected (HE) and observed (Ho) heterozygosity and inbreeding coefficient (F) were estimated for each population using Arlequin v.3.5.2 (Excoffier and Lischer 2010). To partition genetic variation among groups, among population within groups and individuals within population, analysis of molecular variance (AMOVA) was performed following 10,000 permutations in Arlequin v.3.5.2 (Weir and Cockrham 1984). The analysis was done for the 14 global sheep populations grouped into major clusters based their geographical origin and community the populations are kept. This include East African (Ethiopia and Kenya), Arab region (Egypt, Morocco and Iran), India (Indian Garole sheep) and South African (Namaqua Afrikaner sheep). We further regrouped Ethiopian sheep populations separately according to their tail phenotype (Short fat-tailed: Wollo, Farta, Washera and Menz; Long fat-tailed: Arsi-Bale, Adilo, and Horro; Fat rumped: Blackhead Somali) and geographical location (Northern: Washera, Farta, Wollo and Menz; Southern: Adilo, Arsi-Bale and Horro; Eastern: Blackhead Somali).
Genetic population structure analyses
Principal components analysis (PCA) was performed using PLINK v1.9 (Purcell et al. 2007) to investigate the genetic structure and relationships among the studied populations based on genetic correlations between individuals. A graphical display of the first two principal components (PC1 and PC2) was generated using gglot2 package provided by R (Wickham 2009). Population structure analyses carried out in STRUCTURE v.2.3.4 (Pritchard et al. 2000) was used to investigate underlying genetic structure and estimate the proportion of shared genome ancestry between the studied populations. The STRUCTURE output was further analyzed in STRUCTURE HARVESTER (Earl 2012) to determine the optimal number of ancestral genomes (K) and proportions of genome ancestry shared among the studied populations using ΔK method (Evanno et al. 2005). To further evaluate the within and between Ethiopian and global sheep populations relationships, we used pairwise genetic differentiation estimates to construct the neighbor-network tree using SplitsTree4v.4.6 (Huson and Bryant 2005).
Results
Genetic diversity
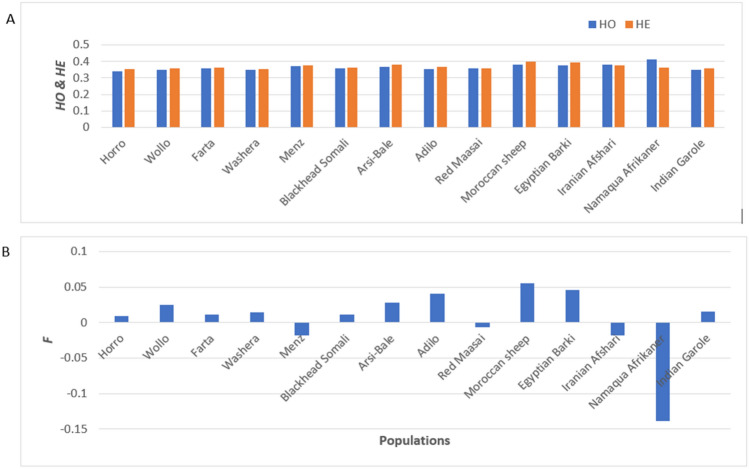
The mean values of HE, Ho and F, as measures of within-population genetic diversity, for 14 sheep populations are shown in Fig. 1 and supplementary Table S1. The lowest values of HE and Ho were observed in Horro sheep while the highest values were observed in Moroccan, Egyptian Barki, Arsi-Bale and Menz sheep populations, respectively. The highest and lowest inbreeding coefficient (F) was observed in Moroccan and Namaqua Afrikaner sheep populations, respectively. Analysis of molecular variance for the 14 global populations grouped according to geographical regions revealed that 9.67% (P < 0.0001) of the variance was among groups (East Africa; Arab region; India and South Africa) (Table 2). When an analysis was performed for Ethiopian sheep populations grouped based on their tail phenotype (Table 1), 3.14% (P < 0.001) of the variance was among groups (Table 2). We further ran AMOVA by grouping the Ethiopian sheep populations according to their geographical distribution (Table 1). Results indicated that 4.67% (P < 0.001) of the variation was among groups, 3.40% among populations within groups and 91.93% within populations (Table 2).
Fig. 1.
Distribution of the genetic diversity indices within each population. a observed heterozygosity (Ho); expected heterozygosity (HE) and b the inbreeding coefficient (F)
Table 2.
Analysis of molecular variance of the studied sheep populations from analysis of 37,771 SNPs
| Grouping | Source of variation | DF | Variance components | Percentage of variation |
|---|---|---|---|---|
| Geographical region (Inter-regional) | Among groups | 3 | 727.082 Va | 9.67 |
| Among population within groups | 301 | 332.156 Vb | 4.42 | |
| Within populations | 305 | 6462.026 Vc | 85.92 | |
| Total | 609 | 7521.264 | 100 | |
| Tail phenotype | Among groups | 2 | 213.379 Va | 3.14 |
| Among population within groups | 197 | 174.803 Vb | 2.58 | |
| Within populations | 200 | 6397.488 Vd | 94.28 | |
| Total | 399 | 6785.669 | 100 | |
| Geographical region (Inter-local) | Among groups | 2 | 721.083 Va | 4.67 |
| Among population within groups | 197 | 255.405 Vb | 3.40 | |
| Within populations | 200 | 6361.020 Vd | 91.93 | |
| Total | 399 | 7337.508 | 100 |
Bold values indicate degree of freedom within population
DF degree of freedom, Va, Vb, Vc, Vd explain null distributions of the variance; P < 0.0001; components in the respective sources of variance (Weir and Cockrham 1984)
Principal component analyses
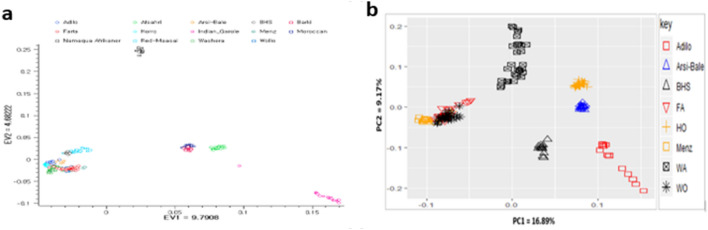
The principal component analysis (PCA) plot incorporating the global populations indicated that the 14 sheep populations are differentiated based on regions (east Africa: Kenya and Ethiopia; North Africa: Moroccan and Barki sheep populations; South Africa: Namaqua Afrikaner; Middle East: Afshari; India: Garole) (Fig. 2a). Moreover, the east Africa sheep populations differentiated based on their tail morphology. PC1 separates east Africa populations from South Africa, North Africa, Middle East and India sheep populations. Ethiopian (except BHS) and India populations are separated from east Africa, north Africa, South Africa and Middle East sheep populations by PC2. Among east Africa populations, Ethiopian fat-rumped sheep, BHS, and Kenyan fat-tailed sheep, Red Maasai, cluster together which is well supported by phylogenetic and structure analysis results (Figs. 3, 4, respectively).
Fig. 2.
Clustering of individual animals in the studied sheep populations a in a global and b Ethiopian indigenous sheep populations datasets
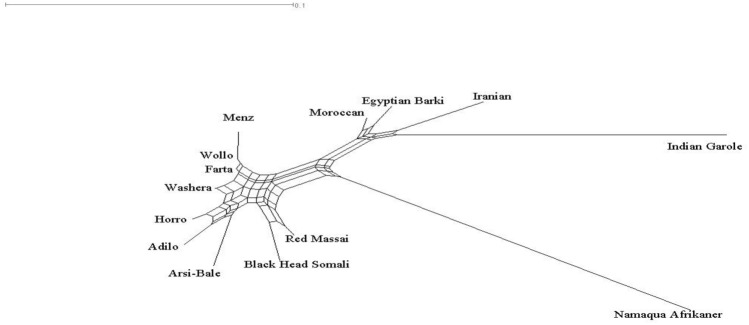
Fig. 3.
Neighbor network constructed in a global geographic context
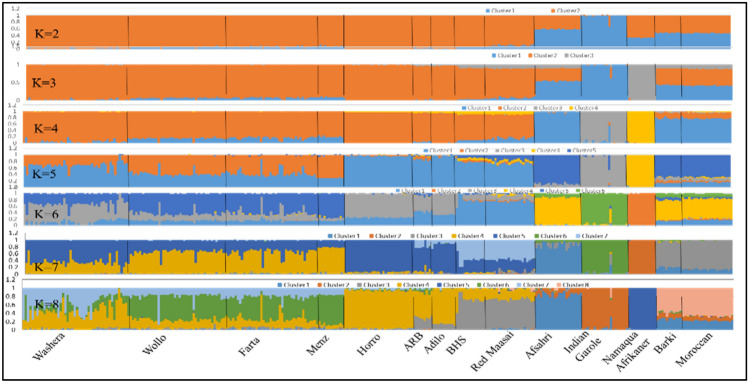
Fig. 4.
Population structure analyses of the studied populations in a global context. The eight genetic clusters are designated (Cluster 1) ~ (Cluster 8), respectively
To illustrate relationships within individuals and among Ethiopian sheep populations, PCA was performed (Fig. 2b). Principal components 1 and 2 accounted for 16.89% and 9.17% of the total variation, respectively and clustered the eight sheep populations according to their tail phenotypes and geographical distribution: long fat-tailed (Arsi-Bale, Horro, Adilo), short fat-tailed (Washera, Farta, Wollo, Menz), and fat-rumped (Black Head Somali). PC1 separate short fat-tailed sheep (FA: Farta, WO: Wollo, and Menz) from the rest of sheep populations including most individual animals of the other short fat-tailed sheep (WA: Washera). PC2 separate the short fat-tailed sheep (WA:Washera), the long fat-tailed sheep (Horro), few individuals of Farta and Arsi-Bale sheep from the rest of sheep populations.
Phylogenetic network analysis
A Neighbor-Net network constructed from pairwise comparison cluster the global dataset (14 sheep populations) according to their geographic region and tail phenotype as indicated in Fig. 3. The line in the phylogenetic network refers to population subdivision and the nodes refers to subpopulations. Among Ethiopian sheep, the short fat-tailed sheep (Farta, Wollo and Washera) were closely clustered. The two highland and long fat-tailed Ethiopian sheep (Horro and Adilo) were closely clustered. The Ethiopian fat-rumped (BHS) and the Kenyan fat- tailed (Red Maasai) sheep as well as the two North Africa sheep (Egyptian Barki and Moroccan sheep) were closely clustered. Long branches were noted for Namaqua Afrikaner (fat-tailed), Indian Garole (thin-tailed), and Iranian Afshari (thin-tailed) sheep possibly due to proximity in geographical region and their tail variation which could be associated to adaptation. The findings were further supported by global principal component and population STRUCTURE analyses (Figs. 2a, 4, respectively).
The close clustering of East African sheep populations and distinct separation from their Northern counterparts was well demonstrated by our phylogenetic and global PCA analysis results (Figs. 3, 2a, respectively).
Genetic population structure analyses
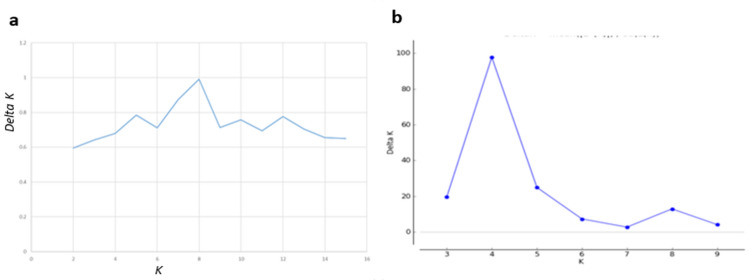
Genetic population structure analysis on the global dataset (14 sheep populations) was carried out using hypothetical ancestral clusters (K) ranging from 2 to 15 and cluster the studied populations according to their geographic region and tail phenotype (Fig. 4). The highest ∆K value registered at K = 8 suggesting this to be the most optimal number of clusters explaining the variation in the dataset (Fig. 5a). The proportion of each ancestral cluster in each population at K = 8 is shown in Fig. 4 and Supplementary Table S2. Indian Garole and Namaqua Afrikaner sheep separated from the rest of sheep populations at K = 2 and K = 3, respectively. East Africa (Ethiopia and Kenya) sheep populations separated from the rest of the sheep populations at K = 4.
Fig. 5.
Graph of cross validation (CV) error generated for: a global and b Ethiopian indigenous sheep populations datasets
At optimal K (K = 8), Ethiopian (Blackhead Somali) and Kenyan (Red Maasai) sheep share up to 94% a single common genetic background (Cluster 3). The Asian thin-tailed sheep, Indian Garole, formed an independent cluster (Cluster 2) with high proportion (~ 95%). The South Africa fat-tailed sheep, Namaqua Afrikaner, formed a clear separate cluster (Cluster 5) with the highest proportion (~ 100%) from the rest of the populations. The two North African representatives shared one genetic background (Cluster 8), and both shared 22–26% common genetic background with Afshari of Iranian sheep (Cluster 1). The Asian thin-tailed sheep showed distinct clustering from east Africa and South African fat-tailed and from east Africa (Ethiopia) fat-rumped sheep.
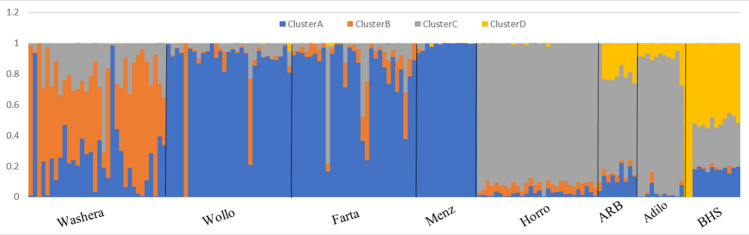
Three to nine hypothetical ancestral clusters (K) were tested with structure on the Ethiopian dataset (national dataset). The highest ∆K value suggested K = 4 as most optimal number of ancestral clusters present in the national dataset (Fig. 5b). The proportion of each ancestral cluster in each population at K = 4 is shown in Fig. 6 and Supplementary Table S3. Menz (highest proportion ~ 100%), Wollo, Farta and few individuals of Washera sheep share one genetic cluster group (Cluster A). Cluster B observed in Washera (proportion ~ 55%) and few individuals of Wollo, and Farta sheep. Horro, Arsi-Bale (ARB) and Adilo sheep form Cluster C whereas Cluster D observed in Blackhead Somali (BHS) and some individuals of Arsi-Bale (ARB) and Adilo sheep. Ethiopian short fat-tailed sheep (Washera, Wollo, Farta, and Menz) did not represent one genetic cluster group rather they form two different genetic cluster groups (Cluster A and B) which is supported by admixture analysis for other Ethiopian short fat-tailed sheep populations (Ahbara et al. 2019). Farta, Wollo and Menz sheep formed one genetic cluster group (Cluster A) while Washera and few individuals of Farta and Wollo sheep formed another genetic cluster group (Cluster B). Close clustering was observed among Ethiopian long fat-tailed sheep (Horro, Adilo, and Arsi-Bale) (Cluster C). The findings are further supported by national principal component and global sheep genetic population structure analyses results (Figs. 2b, 4, respectively).
Fig. 6.
Population structure analyses of Ethiopian sheep
Discussion
The genotype data generated by Ovine 50 K SNP BeadChip were used to investigate genetic diversity and population structure of Ethiopian indigenous sheep populations. To assess the genetic diversity and relationships in greater detail and infer population structure of Ethiopian sheep populations at the continental and global levels, populations from other regions of the African continent and the world were included. The findings revealed that the estimated within Ethiopian sheep population diversity is in line with previous studies on indigenous sheep populations in Ethiopia using microsatellite markers and SNP chip (Gizaw et al. 2008; Edea et al. 2017; Ahbara et al. 2019). The presence of high genetic diversity within-population is congruent with the high variability observed in phenotypic characters, particularly in coat color within the sub-alpine sheep populations (Gizaw et al. 2008). High degree of genetic diversity within-population is a characteristic of large traditional populations that have not been subjected to strong selection indicating the need to conserve such traditional populations (Lauvergne et al. 2000), and thus can be exploited through implementing appropriate breeding strategies.
Ethiopian indigenous sheep showed slightly lower levels of genetic diversity (HE = 0.366) than the presumed ancestral populations in the Middle East (Afshari; HE = 0.376) and North Africa (HE = 0.401). Populations found near to domestication center are expected to retain higher allelic diversity than those that migrated farther away (Peter et al. 2007). The higher level of diversity estimates in North Africa as compared to east Africa populations can be further explained by the fact that North Africa populations reflect a high degree of admixture between fat-tailed and thin-tailed sheep as demonstrated in the STRUCTURE analyses result (Fig. 4 and Supplementary Table S2). Given its proximity to the Near East and Mediterranean Sea, North Africa served as a gateway for early livestock introduction to the African continent and is considered as a secondary hotspot of genetic variation (Gautier 2002).
Although they are defined by the same tail phenotype, the four Ethiopian short fat-tailed sheep (Wollo, Menz, Farta and Washera) did not cluster together. Adaptation to different eco-climates that can impede gene flow, may have shaped this genetic sub-structuring (Madrigal et al. 2001; Gizaw et al. 2007). The close clustering of the three short fat-tailed sheep (Wollo, Farta and Menz) is consistent with adaptation to similar eco-climates (sub-alpine), similar in tail phenotype and they are maintained by the same ethnic group or community which may act as barriers to gene flow that shape population genetic structure (Madrigal et al. 2001). The distinct clustering of the warmer mid-highland short fat-tailed sheep (Washera) from the three sub-alpine short fat-tailed sheep (Wollo, Farta and Menz) could be associated with adaptation to different eco-climates and face different selection pressure, which may have shaped their genomes in different manner. Likewise, The close association observed among the three long fat-tailed sheep populations (Horro, Arsi-Bale, and Adilo) could be due to they are predominantly maintained by the same ethnic group or community, have same tail phenotype, and inhabit similar eco-climates (highland) and face common selective pressures, which may have shaped their genomes in a similar manner. The chances of animal exchange are greater within the same ethnic group or community than between any two different ethnic groups or communities (Gizaw et al. 2007).
The distant association observed between the short fat-tailed sheep, Menz, and the fat-rumped sheep, Black Head Somali, could be due to selection for ecological adaptation, geographical isolation and differences in tail phenotype (Gizaw et al. 2008; Edea et al. 2017). This finding is not in line with the report of Ahbara et al. (2019). The author indicated that the short fat-tailed, Molale-Menz, clustered together with the Ethiopian fat-rumped sheep which could be close proximity of sampling sites for the two different sheep populations since they are distinct in tail phenotype, adapted to different eco-climates, and maintained by different ethnic group or community which may have shaped their genomes differently (Gizaw et al. 2008).
The close association between Ethiopian fat-rumped sheep, BHS, and the Kenyan fat-tailed sheep, Red Maasai, could be because of the two sheep populations are reared under mobile pastoral and agro-pastoral systems and there is a high chance of exchanging animal across the border (Wilson 2011). They are also adapted to similar ecological environments (lowland) and face common selective pressures (Wilson 2011). The close clustering of Ethiopian fat-rump sheep (BHS) and Kenyan fat-tailed sheep (Red Maasai) suggests the dependent introduction and dispersion histories of Africa fat-tailed and fat-rumped sheep into the continent. This finding is agree with previous report that indigenous African sheep genetic resources have been classified into two main groups with a largely non-overlapping distribution: thin-tailed and fat-tailed (including fat-rump) sheep (Wilson 1991) but not in line with the report of Ryder (1984) which indicated that the fat-tailed sheep were introduced into Africa during the third wave of migration following thin-tailed hair sheep and thin-tailed wool sheep, fat-rumped sheep much later.
The close association of the two phenotypically different (variation in tail type) North African sheep populations could be due to gene flow between the two populations since they have close geographical proximity. Previous studies revealed that north Africa is mostly populated by fat-tailed sheep (Muigai and Hanotte 2013), but our STRUCTURE analysis indicated there is high signatures of admixture in the genomes of north Africa populations as compared to their east and South African populations and they shared 22–26% its genome with Middle East thin-tailed sheep (Iranian Afshari). The influence of Middle Eastern thin-tailed sheep detected in north Africa sheep can be explained by the historical introduction of sheep into Africa and their dispersion across the continent through the Nile Valley; for instance, thin-tailed sheep spread into the Western Sahara via Northern Africa (Muigai and Hanotte 2013), which may have left its genomic footprint in the current north Africa sheep populations.
The close clustering of east Africa sheep populations and distinct separation from their north counterparts was well demonstrated by our phylogenetic and PCA analyses (Figs. 3, 2a, respectively). This result coincides with the evidence that fat-tailed sheep were introduced into Africa via two independent routes: one via the north-east Africa and the Mediterranean Sea coastline, and the other via the Horn of Africa crossing through the strait of Babel-Mandeb (Ryder 1984).
The distinct clustering of Asian thin-tailed sheep from fat-tailed east and South Africa as well as from fat-rumped east Africa (Ethiopia) sheep suggests its independent introduction into the continent. It is in line with the distinct histories and non-overlapping geographic distributions of the African thin-tailed with fat-tailed and fat-rumped sheep (Hanotte et al. 2002; Muigai 2003b), and support the predominance of fat-tailed sheep in the east and South parts of Africa (Muigai and Hanotte 2013). Moreover, analyses of autosomal markers and the Y chromosome have revealed the distinct evolutionary histories of thin- and fat-tailed African sheep populations (Aswani 2007).
Conclusions
Our genome-wide SNP analyses revealed that there is clear signature of admixture among Ethiopian sheep populations which could be accounted by some level of current admixture which results in low variation among the sheep populations but large within population variation. The principal component (PCA) and population structure analyses of Ethiopian sheep revealed four distinct genetic cluster groups according to their tail phenotype and geographical distribution. The short fat-tailed sheep populations did not represent one genetic cluster group which require further investigation. Our population structure analyses of Ethiopian sheep population demonstrated a clear pattern of the tail morphology and their phylogeography. Further investigation is required on morphometric basis of tail morphology variation in indigenous Ethiopian sheep populations to confirm the genetic basis of tail morphology variation investigated (Ahbara et al. 2019). Principal component and population structure analyses of global sheep population suggests the independent introduction of thin-tailed sheep into the continent but the dependent introduction and dispersion histories of fat-tailed and fat-rumped sheep. Principal component and phylogenetic analysis of global sheep population results coincide with the evidence that fat-tailed sheep were introduced into Africa via two independent routes: Horn of Africa, via the strait of Bab-el-Mandeb and Northern Africa, via the Isthmus of Suez from the Middle East (Ryder 1984).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The field data collection was fully financed by the Amhara Regional Agricultural Research Institute (ARARI-Andassa Livestock Research Center) in Ethiopia. The laboratory work including genotyping was supported by a Grant from Biosciences eastern and central Africa-International Livestock Research Institute (BecA-ILRI) Hub through the Africa Biosciences Challenge Fund (ABCF) program, Nairobi, Kenya. We would like to acknowledge the sheep flock owners and the district agriculture and rural development offices in the study areas for their volunteer and support during sampling. The authors would also like to appreciate Mr. Mehari Ayalew and Mulugeta Meseret for their kind support in data collection.
Author contributions
AA, GMT, GB, YN, OH and TD designed the research and edited the manuscript. Material preparation and data collection were performed by AA. Data analysis were performed by AA, GMT and JB. GMT provide a critical input in writing the manuscript and supervised the first author throughout the work. MK and YN provided a critical support and supervision in the DNA extraction and facilitating genotyping of the samples. TG, AK guided the first author while sampling. ZE sharing genotype data of some reference sheep populations. The first draft of the manuscript was written by AA and all authors commented on previous versions of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical approval
The research was conducted in the absence of any ethical issue on animal research.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Agraw Amane, Email: agraw29@yahoo.com.
Gurja Belay, Email: gurja.belay@aau.edu.et.
Getinet Mekuriaw Tarekegn, Email: yafetgetinet@gmail.com.
References
- Ahbara A, Bahbahani H, Almathen F, et al. Genome-wide variation, candidate regions and genes associated with fat deposition and tail morphology in Ethiopian indigenous sheep. Front Genet. 2019;9:699. doi: 10.3389/fgene.2018.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai H, Yang B, Li J, Xie X, Chen H, Ren J. Population history and genomic signatures for high-altitude adaptation in Tibetan pigs. BMC Genom. 2014;15:834. doi: 10.1186/1471-2164-15-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assefa F, Tigistu T, Lambebo A. Assessment of the production system, major constraints and opportunities of sheep production in Doyogena Woreda, Kembata Tambaro zone, southern Ethiopia. J Biol Agric Healthc. 2015;5:37–41. [Google Scholar]
- Aswani O. B (2007) Genetic diversity and relationships of African sheep: AY chromosome perspective. Dissertation, University of Egerton.
- Earl DA. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4:359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Edea Z, Dessie T, Dadi H, Do K-T, Kim K-S. Genetic diversity and population structure of ethiopian sheep populations revealed by high-density SNP markers. Front Genet. 2017;8:218. doi: 10.3389/fgene.2017.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HE. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10(3):564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Fao F (2012) Agriculture Organization of the United Nations. FAO statistical yearbook
- Gautier A (2002) The evidence for the earliest livestock in North Africa: or adventures with large bovids, ovicaprids, dogs and pigs. In: Droughts, Food and Culture, Springer, Boston, MA, pp 195–207
- Gizaw S, Abegaz S, Rischkowsky B, Haile A, Mwai AO, Dessie T (2013) Review of Sheep Research and Development Projects in Ethiopia. Nairobi: International Livestock Research Institute (ILRI)
- Gizaw S, Komen H, Hanotte O, Van Arendonk J. Indigenous sheep resources of Ethiopia: types, production systems and farmers preferences. Anim Genet Resour. 2008;43:25–39. doi: 10.1017/S1014233900002704. [DOI] [Google Scholar]
- Gizaw S, Getachew T, Edea Z et al (2013) Characterization of indigenous population strategies of the sheep farming communities of Ethiopia: a basis for designing community-based population programs
- Groeneveld L, Lenstra J, Eding H, Toro M, Scherf B, Pilling D, et al. Genetic diversity in farm animals–a review. Anim Genet. 2010;41:6–31. doi: 10.1111/j.1365-2052.2010.02038.x. [DOI] [PubMed] [Google Scholar]
- Haile A, Tembely S, Anindo D, et al. Effects of breed and dietary protein supplementation on the responses to gastrointestinal nematode infections in Ethiopian sheep. Small Rumin Res. 2002;44:247–261. doi: 10.1016/S0921-4488(02)00080-9. [DOI] [Google Scholar]
- Hanotte O, Bradley DG, Ochieng JW, Verjee Y, Hill EW, Rege JEO. African pastoralism: genetic imprints of origins and migrations. Science. 2002;296(5566):336–339. doi: 10.1126/science.1069878. [DOI] [PubMed] [Google Scholar]
- Helen N (2015) Phenotypic and genetic characterization of indigenous sheep breeds of eastern ethiopia A (Doctoral dissertation, Harmaya University)
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2005;23(2):254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Kijas JW, Townley D, Dalrymple BP, et al. A genome wide survey of SNP variation reveals the genetic structure of sheep populations. PLoS ONE. 2009;4(3):e4668. doi: 10.1371/journal.pone.0004668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauvergne JJ, Bourzat D, Minvielle F. Using morphometric indices to map goat resources. In: Blench RM, MacDonald KC, editors. The origins and development of African livestock: archaeology, genetics, linguistics and ethnography. London: Univ College London Press; 2000. pp. 290–301. [Google Scholar]
- Leta S, Mesele F. Spatial analysis of cattle and shoat population in Ethiopia: growth trend, distribution and market access. Springer Plus. 2014;3(1):31. doi: 10.1186/2193-1801-3-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal L, Ware B, Miller R, Saenz G, Chavez M, Dykes D. Ethnicity, gene flow, and population subdivision in Limon, Costa Rica. Am J Phys Anthropol Off Publ Am Assoc Phys Anthropol. 2001;114(2):99–108. doi: 10.1002/1096-8644(200102)114:2<99::AID-AJPA1010>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Mirkena T (2010) Identifying breeding objectives of smallholders/pastoralists and optimizing community-based breeding programs for adapted sheep breeds in Ethiopia. (Doctoral dissertation, University of Natural Resources and Life Sciences, Vienna)
- Muigai A. Characterization and conservation of indigenous animal genetic resources: genetic diversity and relationships of fat-tailed and thin-tailed sheep of Africa. Juja: Jomo Kenyatta University of Agriculture and Technology; 2003. [Google Scholar]
- Muigai AW, Hanotte O. The origin of African sheep: archaeological and genetic perspectives. Afr Archaeol Rev. 2013;30(1):39–50. doi: 10.1007/s10437-013-9129-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter C, Bruford M, Perez T, Dalamitra S, Hewitt G, Erhardt G, Econogene Consortium Genetic diversity and subdivision of 57 European and Middle Eastern sheep populations. Anim Genet. 2007;38(1):37–44. doi: 10.1111/j.1365-2052.2007.01561.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder ML (1984) Sheep. In: Mason IL (ed) Evolution of domestic animals Longman, London, pp 63–85
- Solomon A, Hegde B, Mengistie T (2011) Growth and physical body characteristics of Gumuz sheep under traditional management systems in Amhara Regional State, Ethiopia. Livestock Research for Rural Development 23(5)
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38(6):1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wickham H (2009) ggplot2: Elegant Graphics for Data Analysis (Use R!). New York, Springer. ISBN 0387981403
- Wilson RT (1991) Small ruminant production and the small ruminant genetic resource in tropical Africa, Food & Agriculture Org. Rome, Italy, Springer. ISBN 0387981403
- Wilson RT. Populations and production of fat-tailed and fat-rumped sheep in the Horn of Africa. Trop Anim Health Prod. 2011;43(7):1419–1425. doi: 10.1007/s11250-011-9870-9. [DOI] [PubMed] [Google Scholar]
- Yang J, Li WR, Lv FH, et al. Whole-genome sequencing of native sheep provides insights into rapid adaptations to extreme environments. Mol Biol Evol. 2016;33(10):2576–2592. doi: 10.1093/molbev/msw129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z, Liu E, Liu Z, Kijas JW, et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim Genet. 2017;48:55–66. doi: 10.1111/age.12477. [DOI] [PubMed] [Google Scholar]
- Zhao YX, Yang J, et al. Genomic reconstruction of the history of native sheep reveals the peopling patterns of nomads and the expansion of early pastoralism in East Asia. Mol Biol Evol. 2017;34(9):2380–2395. doi: 10.1093/molbev/msx181. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.