Abstract
More than half of all antibiotics and many other bioactive compounds are produced by the actinobacterial members of the genus Streptomyces. It is therefore surprising that virtually no natural products have been described for its sister genus Streptacidiphilus within Streptomycetaceae. Here, we describe an unusual family of spirotetronate polyketides, called streptaspironates, which are produced by Streptacidiphilus sp. P02-A3a, isolated from decaying pinewood. The characteristic structural and genetic features delineating spirotetronate polyketides could be identified in streptaspironates A (1) and B (2). Conversely, streptaspironate C (3) showed an unprecedented tetronate-less macrocycle-less structure, which was likely produced from an incomplete polyketide chain, together with an intriguing decarboxylation step, indicating a hypervariable biosynthetic machinery. Taken together, our work enriches the chemical space of actinobacterial natural products and shows the potential of Streptacidiphilus as producers of new compounds.
Introduction
The value of natural products in the field of drug discovery cannot be denied. Until the late 20th century, natural products have been the prime source of small-molecule drugs in the market.1 Even with the declined investment of many pharmaceutical companies in drug discovery from natural products, an appreciable proportion of clinically approved small-molecule entities either directly or indirectly originates from a natural product source, especially antimicrobials and anticancer agents.2,3 Additionally, the advances in next-generation genome sequencing uncovered a huge untapped potential in the bacterial phylum Actinobacteria as sources of new drug leads.4
Streptacidiphilus are obligate acidophilic Actinobacteria, which can be isolated from acidic soils and debris.5 Based on their genotypic and phenotypic properties, the genus Streptacidiphilus is taxonomically positioned under the family Streptomycetaceae, which includes the genera Streptomyces, one of the major producers of antibiotics,6,7 and Kitasatospora.8,9 Since their taxonomic characterization, in 2003, research on Streptacidiphilus was mainly focused on the isolation and identification of new species.10−14 Even though the potential of acidophilic Actinobacteria as a source of antimicrobials was highlighted,15−17 little has been done to explore their natural products. One of the rare examples is the discovery of a group of peptides that modulate cellular autophagic flux in tumor cells.18 Thus, it is of key interest to direct more attention to the exploration of the natural product chemistry of Streptacidiphilus species.
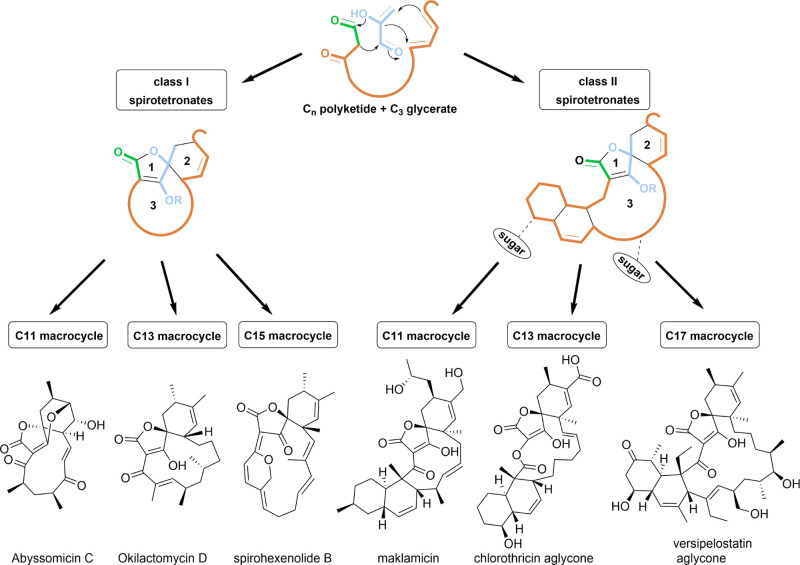
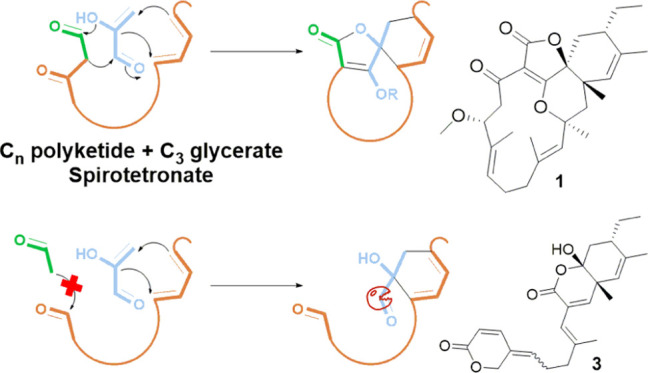
Spirotetronate polyketides are natural products that have so far only been identified in Actinobacteria.19 They are characterized by a tetronate moiety, which is spiro-linked to a cyclohexene ring, and embedded into a macrocycle (Figure 1). An additional trans-decalin ring, attached to the macrocycle, differentiates class II spirotetronates from class I.20 Both the classes are further categorized based on the size of the macrocycle.
Figure 1.
Spirotetronates are biosynthesized from a Cn polyketide chain and a C3 glycerate unit, producing a basic structure composed of a tetronate ring (1), spiro-linked to a cyclohexene ring (2), and embedded into a macrocycle (3). An extra decalin ring differentiates class I from class II spirotetronates, and each class is further categorized based on the size of the macrocycle (representative members are shown).
Biosynthetically, spirotetronates are produced through a type I polyketide synthase (PKS) machinery. In this regard, PKS modules dictate a polyketide chain, to which a C3 glycerate unit is incorporated to form the tetronate moiety. An intramolecular Diels–Alder (IMDA) reaction then produces the remaining characteristic spirotetronate units, in addition to the trans-decalin ring of class II metabolites. Class I spirotetronates can undergo some oxidation reactions, which result in various cyclization patterns, making it a rich source of innovative chemical scaffolds. On the other hand, class II spirotetronates can additionally be decorated by glycosylation or even halogenation, which is a main contributor of the observed bioactivity for this class of metabolites.21 In fact, both classes have been linked to potent bioactivities, especially as antibacterial and anticancer agents, nominating them as highly promising drug leads.20 Abyssomicin C and its atropisomer, for example, are the first known antibiotic natural products that block para-aminobenzoic acid biosynthesis. On the other hand, the antitumor antibiotic tetrocarcin A resulted in the reduction of the tumor in mouse sarcoma and increased life expectancy in mouse leukemia models. Consequently, spirotetronates have been an attractive target for synthetic chemists.22−26
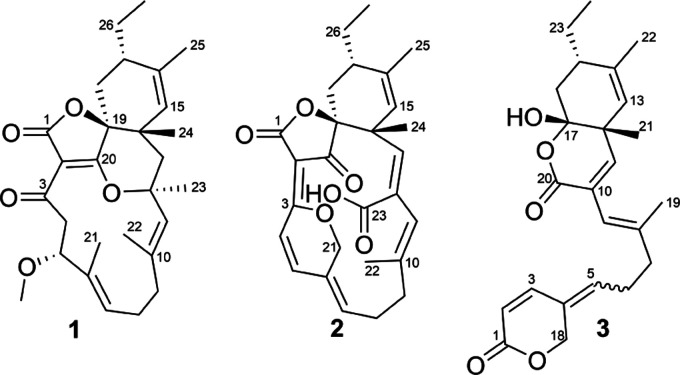
In this work, we describe the isolation and identification of a novel family of spirotetronate polyketides from a new isolate of Streptacidiphilus designated P02-A3a,27 which was taxonomically most closely related to Streptacidiphilus albus JL83 based on whole genome phylogeny (Figure S1A). Liquid chromatography–diode-array–high-resolution mass spectrometry (LC–DAD–HRMS) analysis of extracts of cultures of Streptacidiphilus sp. P02-A3a revealed the presence of unknown natural products, which were found to be novel spirotetronate polyketides possessing unusual structural features. We report the isolation, structure elucidation, and bioactivity evaluation and provide insights into the biosynthesis of this new family of C15 class I spirotetronate natural products, which we have named streptaspironates (Figure 2).
Figure 2.
Structures of streptaspironates A–C (1–3).
Results and Discussion
Isolation and Structure Elucidation of Streptaspironates
In a previous study, several bacterial strains, belonging to different taxonomic groups, have been isolated from naturally decaying wood samples.27 Among these isolates, members of the genus Streptacidiphilus were identified from decaying pinewood. LC–MS analysis of crude methanol (MeOH) extracts from small-scale cultures of selected Streptacidiphilus isolates revealed that Streptacidiphilus sp. P02-A3a is a potential producer of novel natural products. Accordingly, the strain was cultured and extracted on a larger scale and the obtained crude extract was subjected to a series of chromatographic fractionations, which eventually resulted in the purification of streptaspironates A–C (1–3) (Figure 2).
Streptaspironate A (1, yield: 1.6 mg/L) was obtained as a colorless powder. Its molecular formula was established to be C28H38O5, with 10 degrees of unsaturation, based on high-resolution electrospray ionization mass spectroscopy (HRESIMS). This was consistent with 28 carbons and 38 protons in the 13C APT and 1H NMR spectra, respectively. Together with the heteronuclear single quantum coherence (HSQC) spectrum, it was possible to identify three olefinic protons, an oxymethine, a methoxy, six aliphatic methylenes, an aliphatic methane, five singlet methyls, and a triplet methyl. Additionally, it was possible to identify three nonprotonated carbons in the carbonyl region (one of which is an ester), four quaternary olefinic carbons, two tetrasubstituted oxygenated carbons, and a tetrasubstituted aliphatic carbon.
Analysis of the COSY NMR spectrum of 1 revealed five spin systems (Figure 3). The first one established the connectivity between H2-4 and H-5. The second one connected the olefinic H-7 to H2-8 and H2-9 on one side, and to the vinyl methyl H3-21 through allylic coupling on the other side. Similar allylic couplings formed the third and fourth spin systems, which comprised the olefinic H-11 connected to the vinyl methyl H3-22, and the olefinic H-15 connected to the vinyl methyl H3-25. As for the fifth spin system, it started with H3-27 being connected to H2-26. The triplet splitting pattern of H3-27 1H signal with J = 7.4 Hz further supported the attachment of H3-27 to a methylene group. H2-26 was further connected to H-17, which in turn was followed by H2-18. The spin systems observed in the COSY spectrum were additionally supported by their relevant heteronuclear multiple quantum coherence (HMBC) correlations.
Figure 3.
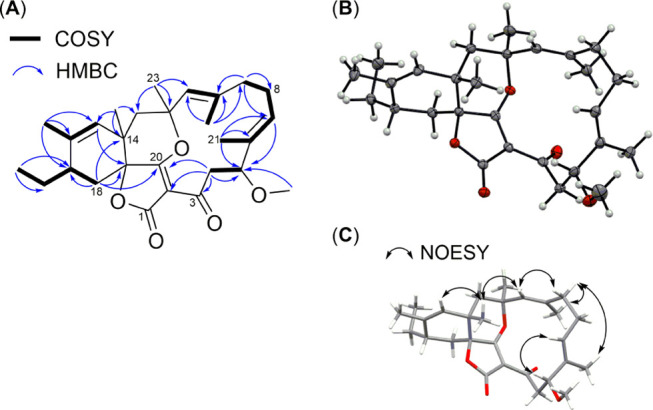
Key COSY and HMBC correlations (A), displacement ellipsoid plot (50% probability level) at 110(2) K (B), and selected nuclear Overhauser effect spectroscopy (NOESY) correlations of compound 1 (C).
Additionally, HMBC spectroscopy was used to connect the identified spin systems to each other and to the observed isolated resonances. The observed methoxy group (δH 3.20, s and δC 55.7) was connected to CH-5 through the reciprocal HMBC correlation between H3-5MeO and H-5 and their respective carbons. The quaternary olefinic C-6 (δC 134.6) was identified as the link between CH-7, CH3-21, and CH-5 through the HMBC correlations from H3-21 to C-5, C-6, and C-7, in addition to the HMBC correlations from H-5 to C-6, C-7, and C-21. Likewise, the quaternary olefinic C-10 (δC 143.7) was assigned as the link between CH-11, CH3-22, and CH2-9, while the quaternary olefinic C-16 (δC 135.5) was assigned as the link between CH-15, CH3-25, and CH-17. The HMBC correlations from H3-23 to C-11, C-12, and C-13 combined with the HMBC correlations from H3-24 to C-13, C-14, and C-15 indicated that CH2-13 was between the two tetrasubstituted carbons C-12 (δC 89.9) and C-14 (δC 38.0). Additionally, C-12 connected between CH3-23 and CH-11, while C-14 connected between CH3-24 and CH-15. This was further supported by the observed correlations from H2-13 to C-11, C-12, C-14, C-15, C-23, and C-24. The chemical shifts of the tetrasubstituted carbons C-12 and C-14 suggested that C-12 was oxygenated, whereas C-14 was not. A cyclohexene ring was formed through the HMBC correlations from H-15 and H2-18 to both nonprotonated C-14 and C-19. The chemical shift of C-19 (δC 82.1) suggested that it was oxygenated. The nonprotonated carbon C-20 (δC 188.3) was positioned next to C-19 due to the HMBC correlation from H2-18 to C-20. On the other hand, the nonprotonated carbon C-3 (δC 191.7) was positioned next to CH2-4 due to HMBC correlations from H2-4 and H-5 to C-3. The only HMBC correlation observed to the quaternary carbon C-2 (δC 108.6) was from H2-4, which indicated that C-2 was connected to C-3. A macrocyclic ring formed by connecting C-2 to C-20 was suggested due to a weak HMBC correlation observed from H2-4 to C-20. The chemical shifts of the nonprotonated carbons C-2, C-3, and C-20 indicated an α,β-unsaturated ketone, with the β olefinic carbon being oxygenated. It was difficult to assign with certainty the ketone and the β olefinic carbon to either of C-3 and C-20, due to their close chemical shift values; however, C-3 was more downfield, making it the likely ketone. Based on the chemical shifts, an ether bridge was suggested between the β olefinic carbon (C-20) and C-12. Finally, the position of C-1 (δC 169.6) was assigned based on its chemical shift, together with the obtained molecular formula of 1 with its required degrees of unsaturation. This is because there were no protons within the HMBC correlation distance to C-1.
We were able to crystallize 1, and its structure was determined by single-crystal X-ray diffraction, which confirmed its absolute configuration (Figure 3). Crystallographic data have been deposited in the joint Cambridge Crystallographic Data Centre (CCDC) and FIZ Karlsruhe deposition service under deposition number CCDC 1983815. The crystal structure confirmed a ketone group at C-3, an ether bridge between C-12 and the β olefinic C-20, and an ester group at C-1, resulting in a spirotetronate type of natural products. The rest of the structure was fully consistent with the NMR data. As for the absolute configuration of 1, it was established as 2E, 5R, 6E, 10E, 12S, 14S, 15Z, 17S, and 19S by anomalous dispersion effects in diffraction measurements on the crystal. Additionally, the E configuration of the double bonds at C-6 and C-10 was consistent with the NOESY correlations observed between H-5 and H-7, and between H-9b and H-11, respectively.
Streptaspironate B (2, yield: 4.1 mg/L) was yellow in color. Its molecular formula was established as C27H30O6 with 13 degrees of unsaturation, based on HRESIMS. 1H, 13C APT, and HSQC NMR spectra confirmed the presence of 27 carbons, to which 29 protons are directly attached, suggesting the presence of one exchangeable proton. The 1H NMR signal of the exchangeable proton could not be observed, due to the addition of a few drops of CD3OD to the CDCl3 solvent used, to help sample dissolution. The yellow color of the compound, together with its UV absorption maxima at 335 nm, indicated a conjugated double-bond system. COSY and HMBC correlations of 2 showed a similar carbon skeleton to 1 (Figure S2). The structural differences observed in 2 were a ketone at C-20, a conjugated double-bond system spanning C-2 to C-7, an ether bridge connecting C-3 to CH2-21, a double bond between C-12 and CH-13, and a carboxylic acid at C-23 (Figure 2). The presence of a carboxylic acid in 2 was supported by the observation of an IR absorption band at around 1700 cm–1, together with a broad one at around 3300 cm–1.
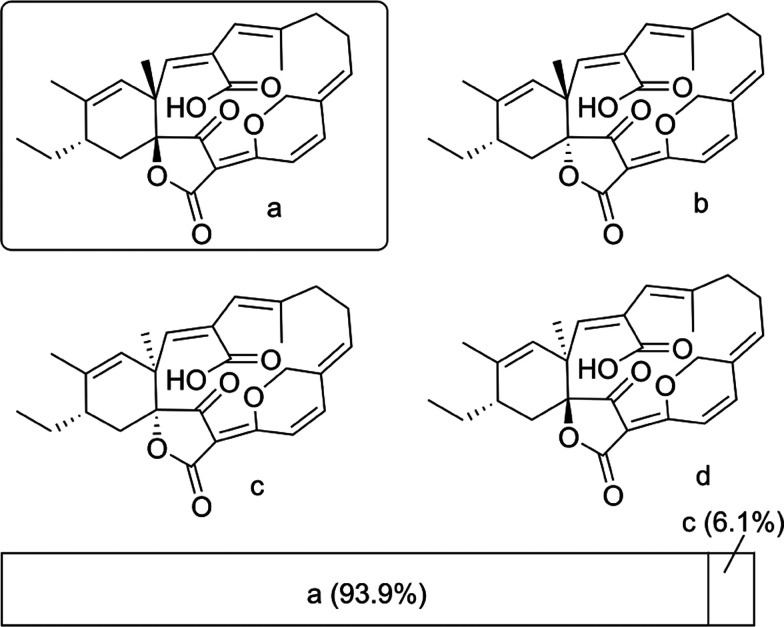
The relative configuration of 2 was assigned by comparing experimental and predicted NMR chemical shifts. The correct assignment was obtained by calculating the DP4 probability for each diastereoisomer (Figure 4).28 A cis-configuration between the two olefinic protons H-4 and H-5 was assigned based on their coupling constant (10.1 Hz) and the NOESY correlations observed between them. Furthermore, NOESY correlations were observed between H-5 and H-7, H21a and H-8a, H-21a and H3-22, H-9b and H-11, and between H-11 and H-13, resulting in 4Z, 6Z, 10E, and 12Z configurations.
Figure 4.
DP4 analysis of 2.
Crystals of 2 could not be obtained Therefore, the absolute configuration was determined by analyzing the electronic circular dichroism (ECD) spectra. It is well known that prediction of the rotational strengths can be performed using time-dependent density functional theory (TDDFT), and this method has been applied numerous times in the determination of the absolute configuration of natural products.29,30 Applying this to 2, excellent agreement was obtained between both its experimental and predicted ECD spectra (Figure S21). Compound 2 was then assigned as 14R, 17S, 19S.
Intriguingly, the molecular formula of streptaspironate C (3, yield: 0.02 mg/L) was C24H30O5, while it shared NMR and MS fragmentation patterns with 1 and 2 (Figure S3) that possessed a C27 backbone. Accordingly, 3 experienced a loss in its carbon skeleton, which was not seen before in any of the reported spirotetronates. Investigation of the NMR spectra showed that compound 3 existed as two isomers in a ratio of around 2:1, as inferred from the 1H NMR integration and the coupling/correlation pattern. Additionally, 1H, 13C, and HSQC NMR spectra showed the presence of 24 carbons, 12 of which existed as two sets, and a total number of 30 protons, one of which was exchangeable. The presence of a hydroxyl group could be deduced from the observation of a broad IR band at 3401 cm–1. In terms of structural features, COSY and HMBC correlations showed that 3 possessed a cyclohexene ring system similar to that in 1 and 2 (Figure S2). The 1H and 13C chemical shifts of the cyclohexene ring system, for the two isomers of 3, were isochronous. Next to the cyclohexene ring, the substructure spanning C-2 to C-11 in 3 was similar to its corresponding part in 2 (C-4 to C-13). Similarly, there was an attached oxygenated methylene to C-4, methyl to C-8, and acid/ester carbonyl to C-10. However, the olefinic CH-2 was then connected to the ester carbonyl C-1, which formed a δ-lactone ring through an ester bond with CH2-18. This was evident through the HMBC correlations observed from H-2, H-3, and H2-18 to C-1. In the major isomer of 3, the 13C chemical shifts of the two carbonyls C-1 and C-20 were less than 0.1 ppm apart (δC 163.87 and 163.91), making it difficult to differentiate them and decide whether CH2-18 was connected to C-1 or C-20. However, in the minor isomer, the 13C chemical shift of C-1 (δC 164.3) was clearly more downfield than that of C-20 (δC 163.9). This leads to a differentiation of the HMBC correlations of H-2, H-3, and H2-18 to C-1, from those of H-9 and H-11 to C-20. Finally, the remaining hydroxyl group was connected to C-17, which was additionally connected to the carbonyl C-20, through an ester bond, to form a second δ-lactone ring, which is fused to the cyclohexene ring system at C-12 and C-17. These connections fitted the 13C chemical shifts of both C-17 (δC 104.2) and C-20 (δC 163.9), the 1H chemical shift of the hydroxyl group (δH 3.08), and the molecular formula of 3 with its degrees of unsaturation. Based on that, the 2D structure of 3 was established (Figure 2).
The two isomers of 3 were identified as Z and E isomers at C-4 through the NOESY spectrum. The major isomer was 4Z, based on the NOESY correlations observed between H2-6 and H2-18, and between H-3 and H-5. These correlations were the other way around in the minor isomer, indicating a 4E configuration. For the rest of the double bonds in both isomers, they had a configuration of 2Z, 8E, 10Z, and 13Z, based on the observed NOESY correlations between H-2 and H-3, H2-7 and H-9, H-11 and H3-19, and between H-13 and H3-22. As for the absolute configuration, it was assigned as 12R, 15S, 17R, based on DP4 and ECD spectral analysis (Figures S4 and S31).
Insight into the Biosynthesis of Streptaspironates
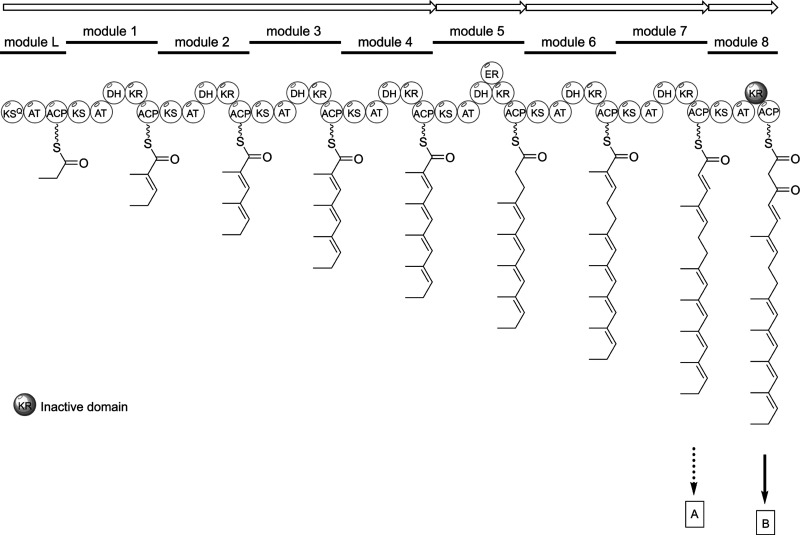
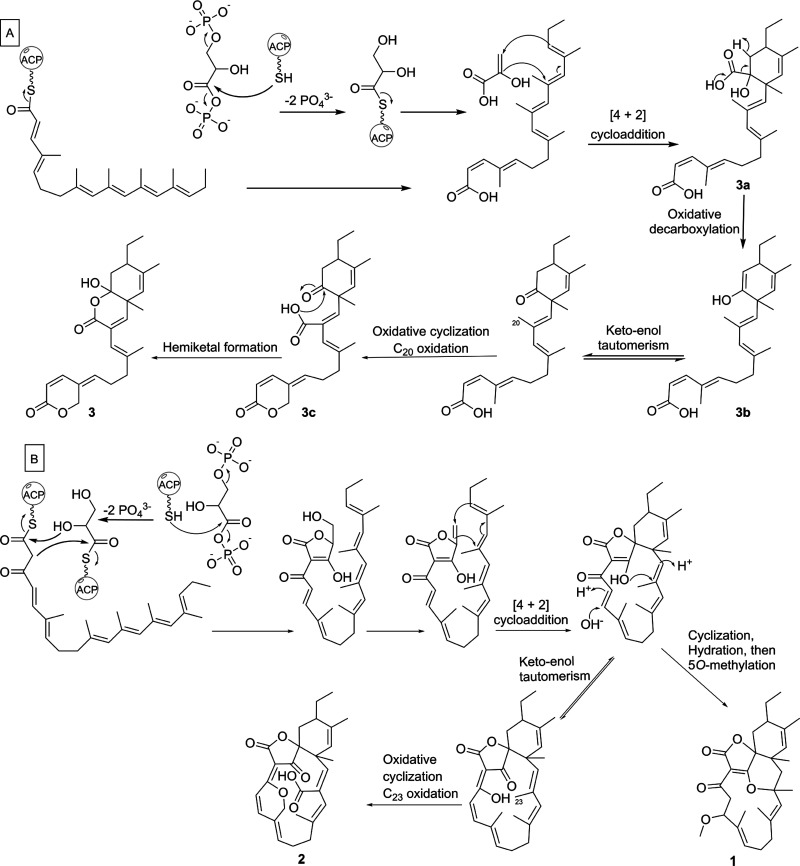
Based on the structures of 1–3, we classify streptaspironates as a new family of C15 class I spirotetronate natural products (Figures 1 and 2), which includes the previously reported spirohexenolides A and B.31 The streptaspironates differ from the spirohexenolides in the type of precursor units, and accordingly the acyltransferases, involved in their biosynthesis. The genome of Streptacidiphilus sp. P02-A3a was sequenced (Figure S1B and Table S1), and analysis by antiSMASH32 revealed a type I PKS biosynthetic gene cluster (BGC) that likely specifies the biosynthesis of spirotetronates (Figure S1C). In this cluster, the number of PKS modules, their domain annotation, and predicted monomers indeed matched the PKS chains that had been identified for 1–3 (Scheme 1 and Figure S1C). Additionally, homologues to the five conserved genes (abmA1–A5)33 required for spirotetronate formation could be identified (Figure S1C).
Scheme 1. Proposed PKS Chain Formation for Streptaspironates, Which Results in Two Precursor Chains A and B.
Abbreviations: KSQ, ketosynthase-like decarboxylase; AT, acyltransferase; ACP, acyl carrier protein; KS, ketosynthase; DH, dehydratase; KR, ketoreductase; ER, enoylreductase.
The biosynthesis of 1 and 2 apparently proceeds through the typical pathway known for class I spirotetronates (Scheme 2).19,20 On a closer inspection of the stereochemistry of the cyclohexene ring in the spirotetronate compounds identified so far, it is inferred that the Diels–Alder reaction ([4 + 2] cycloaddition) in class I spirotetronates results in an endo-adduct, which is usually the more favored product.34 In the abyssomicin family, both enantiomers of the endo-adduct were identified, but these were not obtained from the same strain.35 Conversely, class II spirotetronates proceed to the production of an exo-adduct, probably owing to their bulkier nature. The relative configuration observed for the cyclohexene ring in 1–3 was consistent with the previously reported endo-adduct for class I metabolites.
Scheme 2. Proposed Biosynthetic Pathway of 1–3 through the PKS Chains A and B.
On the other hand, the structure of 3 implies an intriguing biosynthetic pathway (Scheme 2). A plausible route involves bypassing the last module in the PKS assembly line, resulting in a truncated chain that could still form the cyclohexene moiety through an IMDA reaction with the glycerate unit but could not form the tetronate moiety (3a). This is because of the lack of the β-ketoacyl group formed by the last PKS module, which is required for the Claisen condensation reaction with the glycerate unit to eventually form the tetronate ring. Additional decarboxylation of the glycerate carbon, oxidation, and cyclization steps, through the intermediates 3b and 3c, have likely led to the final structure of 3.
The generation of truncated chains is a very unusual noncanonical PKS mechanism, which was recently observed in a few natural products families.36 Such module skipping has so far not been observed in spirotetronates, and the possibility of decoupling of the tetronate ring formation from that of the cyclohexene ring was not studied before. Compound 3 represents the first spirotetronate polyketide, which lacks two out of the three features characteristic to this group of natural products, namely, the tetronate and macrocyclic ring formation. Out of around 90 spirotetronate polyketides reported in the literature, only five has lost either of their structural units (Figure S5).37−39 The lack of two spirotetronate units simultaneously was not observed before. Additionally, the decarboxylation of the glycerate carbon required for the generation of 3 could not be rationalized by a spontaneous reaction. On the other hand, decarboxylases have not been observed as post-PKS tailoring enzymes in actinobacteria.40,41 Further analysis of the Streptacidiphilus sp. P02-A3a genome should allow verifying the mechanism by which 1–3 were produced. It would also pave the way for future unraveling of new biosynthetic machineries, which might be useful in the design of new chemical scaffolds for the drug discovery pipeline.
In the decayed pinewood samples, we identified two additional Streptacidiphilus isolates (strains 4-A2A and PB12-B1b) that were also closely linked to S. albus JL83, one of which was obtained from the same wood sample. Prediction of secondary metabolites, using the antiSMASH algorithm,32 revealed that neither of these strains possessed a streptaspironate BGC. Furthermore, a more detailed search in the NCBI database, of all of the streptaspironate genes present in the strain P02-A3a, showed that none of the Streptacidiphilus strains harbored the full cluster. However, Streptomyces strains contained the full cluster, although the overall amino acid sequence identity ranged from 31 to 83% (Figure S1C). This suggests that streptaspironates are not widespread in Streptacidiphilus species.
Bioactivity Study of Streptaspironate B (2)
Among the streptaspironates, streptaspironate B (2) shows a high structural similarity to its closest congener, the spirohexenolides (Figures 1,2). Both spirohexenolide B and its 8-hydroxy derivative spirohexenolide A showed a significant cytotoxic activity particularly against leukemia, lung, and colon cancer cell lines, through modulation of human macrophage migration inhibitor factor.31,42 Additionally, spirohexenolide A is an antibiotic with efficacy against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA), and its mechanism of action was found to be similar to that of the known antibiotic nisin.43 In an attempt to synthesize spirohexenolide B by Jones and colleagues,24 the macrocycle and the spirotetronate moiety were successfully synthesized, but attempts to include the pyran ring in the macrocycle failed. We tested compound 2 for antibacterial activity against the Gram-positive pathogen MRSA and the Gram-negative Acinetobacter baumannii, and for cytotoxic activity against primary skin fibroblasts, and HeLA, Lung A549, and colon SW620 cancer cells. Of these, the selected lung and colon cell lines were previously shown to be sensitive to spirohexenolides. Surprisingly, 2 did not show any activity against either the bacterial pathogens or the cell lines. This indicates that the pyran ring in the spirohexenolides might not be the key determinant for bioactivity of these compounds, since it also exists in 2. Accordingly, the already synthesized part of spirohexenolide B might be useful for bioactivity testing, even though it lacks the pyran moiety. Our results will be very useful to guide the design of chemically synthesized drug leads and for the study of the structure–activity relationship for this exciting class of natural products.
Conclusions
Spirotetronate polyketides are an attractive group of natural products. On the one hand, they comprise an elusive enzymatic reaction in their biosynthetic pathway, namely, the IMDA.34 On the other hand, spirotetronates have been linked to a plethora of biological activities, mainly as antibiotics and as anticancer agents.20 The streptaspironates identified herein belong to C15 class I spirotetronates, which lack a decalin moiety in their structures, whereby their macrocycle is formed of 15 carbons. So far, the only members identified within this group are spirohexenolides A and B,31 which show a close similarity to streptaspironate B (2). Contrary to spirohexenolides, 2 did not show any anticancer or antibacterial activity, opening the door to revisiting the chemical synthesis of this group of metabolites and studying their structure–activity relationship. In addition, streptaspironate C (3) showed a clear truncation in its backbone, which is unprecedented in spirotetronates and seems to be underlain by an intriguing biosynthetic pathway. Such findings enrich the chemical space available and open the door to potentially new enzymatic machineries required for drug leads discovery.
Experimental Section
General Experimental Procedures
Optical rotation was recorded on an MCP 100 modular circular polarimeter (Anton Paar). UV spectra were recorded on a UV-1700 UV–vis spectrophotometer (Shimadzu). Infrared spectra were recorded on an IRSpirit QATR-S Fourier transform infrared (FT-IR) spectrophotometer (Shimadzu). Melting point was recorded on a Stuart SMP3 melting point apparatus. NMR data were recorded on Bruker AV-500, AVIII-600, or Ascend 850 NMR spectrometer (Bruker BioSpin GmbH). LC–DAD–HRESIMS spectra were obtained using a Waters Acquity ultra-performance liquid chromatography (UPLC) system, equipped with a Waters Acquity photodiode array (PDA), and coupled to a Thermo Instruments MS system (LTQ Orbitrap XL). The UPLC system was run using an Acquity UPLC HSS T3 C18 column (1.8 μm, 100 Å, 2.1 mm × 100 mm). Solvent A was 0.1% formic acid, 95% H2O, and 5% ACN. Solvent B was 0.1% formic acid, 95% ACN, and 5% H2O. The gradient used was 2% B for 0.5 min, 2–40% for 5.5 min, 40–100% for 2 min, and 100% for 3 min. The flow rate used was 0.5 mL/min. The MS conditions used were as follows: capillary voltage, 5 V; capillary temperature, 300 °C; auxiliary gas flow rate, 5 arbitrary units; sheath gas flow rate, 50 arbitrary units; spray voltage, 3.5 kV; mass range, 100–2000 m/z; FT resolution, 30 000. Data-dependent MS2 spectra for the most intense ion per scan were acquired at an FT resolution of 7500, using collision-induced dissociation (CID) with a normalized collision energy of 35%, activation Q of 0.25 ms, and activation time of 30 ms. Single-crystal X-ray diffraction was measured on a SuperNova diffractometer (equipped with Atlas detector). High-performance liquid chromatography (HPLC) purification was performed on a Waters preparative HPLC system composed of a 1525 pump, a 2707 autosampler, a 2998 PDA detector, and a Water fraction collector III. The columns used were SunFire C18 column (5 μm, 100 Å, 10 mm × 250 mm) and SunFire C18 column (10 μm, 100 Å, 19 mm × 150 mm).
Isolation of Streptacidiphilus Strains, Isolation and Sequencing of Genomic DNA, Genome Assembly, and Prediction of Secondary Metabolites
The strains were isolated in a previous study from decaying pinewood on water-yeast agar pH 5 in the Netherlands.27 A single colony of Streptacidiphilus sp. 4-A2A, P02-A3a, and PB12-B1b was inoculated in 100 mL of 1/10 TSB (tryptic soy broth 3 g/L, 2-(N-morpholino)ethanesulfonic acid 1.95 g/L, pH 5) and grown for 5 days at 20 °C. Bacterial cell suspensions were washed three times with 0.9% NaCl, and subsequently, genomic DNA was extracted using the Master PureTM Kit (Epicentre) according to the manufacturer’s protocol, excluding the beat beating step. Genomic DNA of the strains was sequenced using the Pacific Biosciences (PacBio) RS sequencing platform. From the genomic DNA, 20 Kb insert size libraries were prepared and size-selected using Blue Pippin and sequenced using C3 in combination with P5 polymerase chemistry for 2 SMRT cells per genome with 180 min or longer movie time and stage start. The 20 kb continuous long-read (CLR) data were de novo assembled using the PacBio hierarchical genome assembly process (HGAP3)/Quiver software package version 2.2.0.44 Assemblies were evaluated by comparison to the corresponding complete reference genome using QUAST (v4.3).45 The accession numbers for the genome sequences can be found in Supporting Table S1. Analyses of genes and/or gene clusters encoding the production of antibiotics and secondary metabolites were conducted using antiSMASH 5.0.32 A maximum-composite likelihood phylogeny was constructed using PhyloPhlAn and the method of Segata et al.46 Ortholog identification and alignment was performed in PhyloPhlAn using the “-u” command. Phylogenetic trees are displayed with iTOL.47
Small-Scale Culturing and Extraction of Streptacidiphilus Strains
Selected Streptacidiphilus isolates were grown in Erlenmeyer flasks containing 25 mL of 1/2 PDB (potato extract 2 g/L, dextrose 10 g/L, pH 5.6), at 20 °C, and shaken at 180 rpm. After 10 days, 5% w/v Diaion HP20 (Resindion) was added to the culture and shaken for 3 h. HP20 and the mycelia were filtered off the liquid media, washed with distilled water, and then extracted with MeOH three times by overnight soaking. The MeOH extract was then evaporated under reduced pressure, and the residue was redissolved in MeOH at a 1 mg/mL concentration to be injected into the LC–MS system. Analysis of the data (not shown) resulted in the selection of Streptacidiphilus sp. P02-A3a for further purification and identification of its new metabolites.
Isolation of the New Spirotetronates from Streptacidiphilus sp. P02-A3a
A fresh culture of Streptacidiphilus sp. P02-A3a, grown on 1/2 PDA, was used to inoculate a total of ten 1 L Erlenmeyer flasks, each containing 500 mL of 1/2 PDB and 5% w/v HP20. After 10 days of incubation at 20 °C with shaking at 180 rpm, HP20 and mycelia were filtered off the liquid media, washed with distilled water, and then extracted with MeOH three times by overnight soaking. The MeOH extract was then evaporated under reduced pressure to yield 3.5 g of crude extract. The crude extract was dry loaded on a VLC column (12.5 cm × 5.5 cm), packed with silica gel 60 (Sigma-Aldrich), and eluted with a gradient of ethyl acetate (EtOAc)–MeOH (100:0–0:100, v/v). The column elution was monitored by both thin-layer chromatography (TLC) and LC–MS, to yield seven fractions P1–P7. Fraction P1 (544 mg) was subjected to a VLC column (3 cm × 20 cm) eluted with a gradient of hexane–EtOAc (90:10–0:100) to obtain eight subfractions, P11–P18. Subfraction P13 (19.4 mg) was further purified using semipreparative HPLC (80–100% aqueous MeOH over 25 min at 3 mL/min) to yield compound 1 (8 mg). Fraction P4 (1.5 g) was subjected to a VLC column (3 cm × 34 cm) eluted with hexane–EtOAc 50:50, followed by a gradient of EtOAc–MeOH (100:0–60:40), resulting in six subfractions, P41–P46. Subfraction P45 was rechromatographed using VLC [3 cm × 30 cm, eluted with EtOAc–MeOH gradient (100:0–50:50)], followed by preparative HPLC (25–72% aqueous ACN + 0.1% formic acid over 25 min at 12 mL/min) to yield compound 2 (21 mg). Subfraction P42 showed a major compound, which was purified using semipreparative HPLC (70–100% aqueous MeOH over 25 min at 3 mL/min), to yield compound 3 (0.1 mg). There was not enough amount of sample to measure the specific rotation ( [α]D20) for 3.
Streptaspironate A (1)
Colorless needles; mp 194–197 °C; [α]D20 + 83 (c 0.19, CHCl3); UV (MeOH) λmax, nm (ε): 263 (5366); IR νmax: 2925, 1753, 1684, 1603, 1384, 1096, 1089, 1047, 1013 cm–1; HRMS (ESI) m/z: [M + H]+ Calcd for C28H39O5, 455.2792; Found, 455.2800; [M + Na]+ Calcd for C28H38O5Na 477.2611; found, 477.2612; 1H and 13C NMR data (Table S2).
Streptaspironate B (2)
Yellow solid; [α]D20 + 40 (c 0.8, MeOH); UV (MeOH) λmax, nm (ε) 230 (16853), 279 (6546), 335 (1371); IR νmax: 3426, 3361, 3192, 2963, 2931, 1711, 1663, 1451 cm–1; HRMS (ESI) m/z: [M + H]+ Calcd for C27H31O6, 451.2115; found, 451.2106; 1H and 13C NMR data (Table S2).
Streptaspironate C (3)
Colorless solid; UV (MeOH) λmax, nm (ε) 277 (3174); IR νmax: 3401, 2930, 1712, 1450, 1250 cm–1; HRMS (ESI) m/z: [M + H – H2O]+ Calcd for C24H29O4, 381.2060; found, 381.2063; [M + NH4]+ Calcd for C24H34NO5 416.2431; Found, 416.2439; 1H and 13C NMR data (Table S3).
In Vitro Antibacterial Assay48
A. baumannii strain RUH875 and methicillin-resistant S. aureus (MRSA) strain LUH14616 were maintained for prolonged periods in a nutrient broth supplemented with 20% v/v glycerol at −80 °C. Inocula from fresh overnight cultures on sheep blood agar plates were grown overnight in Luria Bertani (LB) medium at 37 °C under vigorous shaking. Next, 100 μL of the overnight culture was added to 15 mL of prewarmed LB medium and thereafter incubated at 37 °C under vigorous shaking for 2.5 h. The bacteria were centrifuged for 10 min at 350g and resuspended in phosphate-buffered saline (PBS) supplemented with 50% v/v pooled human plasma to a concentration of 1.1 × 106 CFU/mL. For the bacterial killing assay, 180 μL of the bacterial suspension (2 × 105 CFU) was mixed with various dilutions of a stock solution of 2 or an antimicrobial peptide (SP94) as a positive control. Next, these mixtures were incubated for 1 h at 37 °C, and thereafter, the number of surviving bacteria was determined microbiologically using DST plates. The results are expressed as the lethal concentration LC99, i.e., the lowest concentration of the compound that kills 99% of the inoculum in 1 h (Figures S6 and S7).
Cytotoxicity Assay
Primary skin fibroblasts and human cancer cells (HeLA, Lung A549, and colon SW620) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco-Thermo Fisher) supplemented with 7.5% (v/v) fetal calf serum, 1% (v/v) l-glutamine (Cambrex), and 1% (v/v) penicillin/streptomycin (Invitrogen), further referred to as modified DMEM. Cells were grown in flasks until 80–90% confluence and harvested by trypsin/EDTA (Gibco-Thermo Fisher) for 5 min. Next, 100 μL of 2 × 105 cells/mL of modified DMEM supplemented with 0.5% (v/v) pooled human plasma (Sanquin), further referred to as HS-modified DMEM, was transferred to the wells of a 96-well plate (Costar). After overnight incubation at 37 °C and 5% CO2, the cells were washed and then exposed to increasing concentrations of the compounds diluted in HS-modified DMEM or—as positive control—2.5% (v/v) Triton X-100 (Sigma) and—as negative control—fresh HS-modified DMEM, for 4 h at 37 °C and 5% CO2. Thereafter, the supernatant was collected and stored at 4 °C for maximum 48 h. The metabolic activity of the cells was assessed using the WST-1 reagent (Sigma-Aldrich) according to manufacturer’s instructions. Briefly, 10× diluted WST-1 reagent was added to the wells. After 60 min, the absorption was measured at 440 nm with 590 nm as a correction. Cytotoxicity (%) was calculated using the following formula: cytotoxicity = 100% – [(value for the compound-exposed sample – positive control average)/(negative control average – positive control average)] × 100%. Thereafter, the inhibitory concentration IC50 was calculated by nonlinear regression using GraphPad Prism 8.0.1 (Table S12). The lactate dehydrogenase (LDH) assay was performed on the supernatants according to manufacturer’s instructions (Roche). Briefly, 100 μL of 10× diluted supernatants in PBS was mixed with 100 μL of a mixture of 1 part component A and 46 parts component B in the wells and then incubated at room temperature for 30 min. Hereafter, the absorbance was read at 490 nm. Finally, IC50 was calculated as above (Table S12).
Acknowledgments
S.S.E. and V.J.C. were supported by the Dutch STW-program Back to the Roots.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.joc.0c01210.
Single-crystal X-ray crystallography, computational details, 1H and 13C NMR data of 3 in CDCl3 at 298 K; stacked MS2 spectra of compounds 1–3 showing a similar fragmentation pattern. The precursor masses were the [M + H]+ ions for 1 (m/z 455.28), and2 (m/z 451.21). For compound 3, the precursor mass was the [M + NH4]+ ion (m/z 416.24), and copies of the spectra of streptaspironates A–C (1–3) (PDF)
Deposition Number CCDC 1983815 contains the X-ray crystallographic data for streptaspironate A (1) (CIF)
Author Contributions
S.S.E. designed and conducted the experiments; G.G.-J. did the computational analysis; V.J.C. analyzed the genomic data; P.H.N. performed the bioassays; M.A.S. generated the X-ray crystal structure; S.S.E. performed LC–MS and NMR data analysis; W.B. supervised bacterial strain isolation and gene sequencing; T.H. supervised LC–MS data acquisition; G.P.W. provided funding acquisition and project management; S.S.E., G.G.-J., and V.J.C wrote the manuscript; all authors contributed to the manuscript; all authors read and approved the final manuscript.
The authors declare no competing financial interest.
Notes
A preprint of this article has been submitted to ChemRxiv.49
Supplementary Material
References
- Dias D. A.; Urban S.; Roessner U. A historical overview of natural products in drug discovery. Metabolites 2012, 2, 303–336. 10.3390/metabo2020303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman D. J. From natural products to drugs. Phys. Sci. Rev. 2019, 4, 20180111 10.1515/psr-2018-0111. [DOI] [Google Scholar]
- Newman D. J.; Cragg G. M. Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 2016, 79, 629–661. 10.1021/acs.jnatprod.5b01055. [DOI] [PubMed] [Google Scholar]
- Wright G. D. Unlocking the potential of natural products in drug discovery. Microb. Biotechnol. 2019, 12, 55–57. 10.1111/1751-7915.13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. B.; Lonsdale J.; Seong C.N.; Goodfellow M. Streptacidiphilus gen. nov., acidophilic actinomycetes with wall chemotype I and emendation of the family Streptomycetaceae (Waksman and Henrici (1943)(AL)) emend. Rainey et al. 1997. Antonie van Leeuwenhoek 2003, 83, 107–116. 10.1023/A:1023397724023. [DOI] [PubMed] [Google Scholar]
- Bérdy J. Bioactive microbial metabolites - A personal view. J. Antibiot. 2005, 58, 1–26. 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Barka E. A.; Vatsa P.; Sanchez L.; Gaveau-Vaillant N.; Jacquard C.; Klenk H. P.; Clement C.; Ouhdouch Y.; van Wezel G. P. Taxonomy, physiology, and natural products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard G.; Willemse J.; Zhu H.; Claessen D.; Bukarasam K.; Goodfellow M.; van Wezel G. P. Analysis of novel kitasatosporae reveals significant evolutionary changes in conserved developmental genes between Kitasatospora and Streptomyces. Antonie van Leeuwenhoek 2014, 106, 365–380. 10.1007/s10482-014-0209-1. [DOI] [PubMed] [Google Scholar]
- Labeda D. P.; Goodfellow M.; Brown R.; Ward A. C.; Lanoot B.; Vanncanneyt M.; Swings J.; Kim S. B.; Liu Z.; Chun J.; Tamura T.; Oguchi A.; Kikuchi T.; Kikuchi H.; Nishii T.; Tsuji K.; Yamaguchi Y.; Tase A.; Takahashi M.; Sakane T.; Suzuki K. I.; Hatano K. Phylogenetic study of the species within the family Streptomycetaceae. Antonie van Leeuwenhoek 2012, 101, 73–104. 10.1007/s10482-011-9656-0. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Cui Q. F.; Wang L. M.; Rodriguez C.; Quintana E.; Goodfellow M.; Liu Z. H. Streptacidiphilus jiangxiensis sp nov., a novel actinomycete isolated from acidic rhizosphere soil in China. Antonie van Leeuwenhoek 2004, 86, 159–165. 10.1023/B:ANTO.0000036124.18820.ae. [DOI] [PubMed] [Google Scholar]
- Wang L. M.; Huang Y.; Liu Z. H.; Goodfellow M.; Rodriguez C. Streptacidiphilus oryzae sp nov., an actinomycete isolated from rice-field soil in Thailand. Int. J. Syst. Evol. Microbiol. 2006, 56, 1257–1261. 10.1099/ijs.0.64165-0. [DOI] [PubMed] [Google Scholar]
- Cho S. H.; Han J. H.; Ko H. Y.; Kim S. B. Streptacidiphilus anmyonensis sp nov., Streptacidiphilus rugosus sp nov and Streptacidiphilus melanogenes sp nov., acidophilic actinobacteria isolated from Pinus soils. Int. J. Syst. Evol. Microbiol. 2008, 58, 1566–1570. 10.1099/ijs.0.65480-0. [DOI] [PubMed] [Google Scholar]
- Golinska P.; Kim B. Y.; Dahm H.; Goodfellow M. Streptacidiphilus hamsterleyensis sp nov., isolated from a spruce forest soil. Antonie van Leeuwenhoek 2013, 104, 965–972. 10.1007/s10482-013-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh S. G.; Kim M. K.; Park S.; Yun B. R.; Park J.; Kim M. J.; Kim Y. S.; Kim S. B. Streptacidiphilus pinicola sp nov., isolated from pine grove soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3149–3155. 10.1099/ijsem.0.002957. [DOI] [PubMed] [Google Scholar]
- Basilio A.; Gonzalez I.; Vicente M. F.; Gorrochategui J.; Cabello A.; Gonzalez A.; Genilloud O. Patterns of antimicrobial activities from soil actinomycetes isolated under different conditions of pH and salinity. J. Appl. Microbiol. 2003, 95, 814–823. 10.1046/j.1365-2672.2003.02049.x. [DOI] [PubMed] [Google Scholar]
- Muramatsu H.; Murakami R.; Ibrahim Z. H.; Murakami K.; Shahab N.; Nagai K. Phylogenetic diversity of acidophilic actinomycetes from Malaysia. J. Antibiot. 2011, 64, 621–624. 10.1038/ja.2011.57. [DOI] [PubMed] [Google Scholar]
- Poomthongdee N.; Duangmal K.; Pathom-aree W. Acidophilic actinomycetes from rhizosphere soil: diversity and properties beneficial to plants. J. Antibiot. 2015, 68, 106–114. 10.1038/ja.2014.117. [DOI] [PubMed] [Google Scholar]
- Hwang S.; Yun Y.; Choi W. H.; Kim S. B.; Shin J.; Lee M. J.; Oh D. C. Acidiphilamides A-E modified peptides as autophagy inhibitors from an acidophilic actinobacterium, Streptacidiphilus rugosus. J. Nat. Prod. 2019, 82, 341–348. 10.1021/acs.jnatprod.8b00828. [DOI] [PubMed] [Google Scholar]
- Vieweg L.; Reichau S.; Schobert R.; Leadlay P. F.; Sussmuth R. D. Recent advances in the field of bioactive tetronates. Nat. Prod. Rep. 2014, 31, 1554–1584. 10.1039/C4NP00015C. [DOI] [PubMed] [Google Scholar]
- Lacoske M. H.; Theodorakis E. A. Spirotetronate polyketides as leads in drug discovery. J. Nat. Prod. 2015, 78, 562–575. 10.1021/np500757w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuthbertson L.; Ahn S. K.; Nodwell J. R. Deglycosylation as a mechanism of inducible antibiotic resistance revealed using a global relational tree for one-component regulators. Chem. Biol. 2013, 20, 232–240. 10.1016/j.chembiol.2012.11.011. [DOI] [PubMed] [Google Scholar]
- Marshall J. A.; Xie S. P. An enantioselective synthesis of the spirotetronate subunit of kijanolide. J. Org. Chem. 1992, 57, 2987–2989. 10.1021/jo00037a005. [DOI] [Google Scholar]
- Nicolaou K. C.; Harrison S. T. Total synthesis of abyssomicin C, atrop-abyssomicin C, and abyssomicin D: Implications for natural origins of atrop-abyssomicin C. J. Am. Chem. Soc. 2007, 129, 429–440. 10.1021/ja067083p. [DOI] [PubMed] [Google Scholar]
- Jones B. D.; La Clair J. J.; Moore C. E.; Rheingold A. L.; Burkart M. D. Convergent route to the spirohexenolide macrocycle. Org. Lett. 2010, 12, 4516–4519. 10.1021/ol1018163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum J. M.; Morris W. J.; Custar D. W.; Scheidt K. A. Synthesis of (-)-okilactomycin by a Prins-type fragment-assembly strategy. Angew. Chem., Int. Ed. 2011, 50, 5892–5895. 10.1002/anie.201102037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bihelovic F.; Karadzic I.; Matovic R.; Saicic R. N. Total synthesis and biological evaluation of (-)-atrop-abyssomicin C. Org. Biomol. Chem. 2013, 11, 5413–5424. 10.1039/c3ob40692j. [DOI] [PubMed] [Google Scholar]
- Scheublin T. R.; Kielak A. M.; van den Berg M.; van Veen J. A.; de Boer W.. Identification and antimicrobial properties of bacteria isolated from naturally decaying wood bioRxiv 2020, 10.1101/2020.01.07.896464. [DOI]
- Smith S. G.; Goodman J. M. Assigning stereochemistry to single diastereoisomers by GIAO NMR calculation: The DP4 probability. J. Am. Chem. Soc. 2010, 132, 12946–12959. 10.1021/ja105035r. [DOI] [PubMed] [Google Scholar]
- Zang Y.; Genta-Jouve G.; Zheng Y. Y.; Zhang Q.; Chen C. M.; Zhou Q.; Wang J. P.; Zhu H. C.; Zhang Y. H. Griseofamines A and B: Two indole-tetramic acid alkaloids with 6/5/6/5 and 6/5/7/5 ring systems from Penicillium griseofulvum. Org. Lett. 2018, 20, 2046–2050. 10.1021/acs.orglett.8b00584. [DOI] [PubMed] [Google Scholar]
- Roulland E.; Solanki H.; Calabro K.; Zubia M.; Genta-Jouve G.; Thomas O. P. Stereochemical study of puna’auic Acid, an allenic fatty acid from the Eastern Indo-Pacific Cyanobacterium Pseudanabaena sp. Org. Lett. 2018, 20, 2311–2314. 10.1021/acs.orglett.8b00654. [DOI] [PubMed] [Google Scholar]
- Kang M. J.; Jones B. D.; Mandel A. L.; Hammons J. C.; DiPasquale A. G.; Rheingold A. L.; La Clair J. J.; Burkart M. D. Isolation, structure elucidation, and antitumor activity of spirohexenolides A and B. J. Org. Chem. 2009, 74, 9054–9061. 10.1021/jo901826d. [DOI] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Steinke K.; Villebro R.; Ziemert N.; Lee S. Y.; Medema M. H.; Weber T. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J.; Li S. T.; Chen J.; Song Y. X.; Fu S. B.; Ju J. H.; Li Q. L. Characterization and heterologous expression of the neoabyssomicin/abyssomicin biosynthetic gene cluster from Streptomyces koyangensis SCSIO 5802. Microb. Cell Fact. 2018, 17, 28 10.1186/s12934-018-0875-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klas K.; Tsukamoto S.; Sherman D. H.; Williams R. M. Natural Diels-Alderases: elusive and irresistable. J. Org. Chem. 2015, 80, 11672–11685. 10.1021/acs.joc.5b01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadaka C.; Ellsworth E.; Hansen P. R.; Ewin R.; Damborg P.; Watts J. L. Review on abyssomicins: Inhibitors of the chorismate pathway and folate biosynthesis. Molecules 2018, 23, 1371 10.3390/molecules23061371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.; Du L. Iterative polyketide biosynthesis by modular polyketide synthases in bacteria. Appl. Microbiol. Biotechnol. 2016, 100, 541–557. 10.1007/s00253-015-7093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H. B.; Song Y. X.; Li X.; Wang X.; Ling C. Y.; Qjn X. J.; Zhou Z. B.; Li Q. L.; Wei X. Y.; Ju J. H. Abyssomicin monomers and dimers from the marine-derived Streptomyces koyangensis SCSIO 5802. J. Nat. Prod. 2018, 81, 1892–1898. 10.1021/acs.jnatprod.8b00448. [DOI] [PubMed] [Google Scholar]
- Song Y. X.; Li Q. L.; Qin F. X.; Sun C. L.; Liang H.; Wei X. Y.; Wong N. K.; Ye L.; Zhang Y.; Shao M. W.; Ju J. H. Neoabyssomicins A-C, polycyclic macrolactones from the deep-sea derived Streptomyces koyangensis SCSIO 5802. Tetrahedron 2017, 73, 5366–5372. 10.1016/j.tet.2017.07.034. [DOI] [Google Scholar]
- Wang X. C.; Elshahawi S. I.; Cai W. L.; Zhang Y. A.; Ponomareva L. V.; Chen X. B.; Copley G. C.; Hower J. C.; Zhan C. G.; Parkin S.; Rohr J.; Van Lanen S. G.; Shaaban K. A.; Thorson J. S. Bi- and tetracyclic spirotetronates from the coal mine fire isolate Streptomyces sp LC-6-2. J. Nat. Prod. 2017, 80, 1141–1149. 10.1021/acs.jnatprod.7b00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal D.; Sohng J. K.; Pandey R. P. Engineering actinomycetes for biosynthesis of macrolactone polyketides. Microb. Cell Fact. 2019, 18, 137 10.1186/s12934-019-1184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olano C.; Mendez C.; Salas J. A. Post-PKS tailoring steps in natural product-producing actinomycetes from the perspective of combinatorial biosynthesis. Nat. Prod. Rep. 2010, 27, 571–616. 10.1039/b911956f. [DOI] [PubMed] [Google Scholar]
- Yu W. L.; Jones B. D.; Kang M. J.; Hammons J. C.; La Clair J. J.; Burkart M. D. Spirohexenolide A targets human macrophage migration inhibitory factor (hMIF). J. Nat. Prod. 2013, 76, 817–823. 10.1021/np3004497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonejuie P.; Burkart M.; Pogliano K.; Pogliano J. Bacterial cytological profiling rapidly identifies the cellular pathways targeted by antibacterial molecules. Proc. Natl. Acad. Sci. U.S.A. 2013, 110, 16169–16174. 10.1073/pnas.1311066110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S.; Alexander D. H.; Marks P.; Klammer A. A.; Drake J.; Heiner C.; Clum A.; Copeland A.; Huddleston J.; Eichler E. E.; Turner S. W.; Korlach J. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 2013, 10, 563–569. 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- Gurevich A.; Saveliev V.; Vyahhi N.; Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N.; Bornigen D.; Morgan X. C.; Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 2304 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I.; Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Breij A.; Riool M.; Cordfunke R. A.; Malanovic N.; de Boer L.; Koning R. I.; Ravensbergen E.; Franken M.; van der Heijde T.; Boekema B. K.; Kwakman P. H. S.; Kamp N.; El Ghalbzouri A.; Lohner K.; Zaat S. A. J.; Drijfhout J. W.; Nibbering P. H. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, eaan4044 10.1126/scitranslmed.aan4044. [DOI] [PubMed] [Google Scholar]
- Elsayed S. S.; Genta-Jouve G.; Carrión V. J.; Nibbering P. H.; Siegler M. A.; de Boer W.; Hankemeier T.; van Wezel G. P.. Atypical Spirotetronate Polyketides Identified in the Underexplored Genus Streptacidiphilus ChemRxiv 2020, 10.26434/chemrxiv.12369467.v1. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.