Abstract
Mass spectrometry imaging (MSI) is an established analytical tool capable of defining and understanding complex tissues by determining the spatial distribution of biological molecules. Three-dimensional (3D) cell culture models mimic the pathophysiological environment of in vivo tumors and are rapidly emerging as a valuable research tool. Here, multimodal MSI techniques were employed to characterize a novel aggregated 3D lung adenocarcinoma model, developed by the group to mimic the in vivo tissue. Regions of tumor heterogeneity and the hypoxic microenvironment were observed based on the spatial distribution of a variety of endogenous molecules. Desorption electrospray ionization (DESI)-MSI defined regions of a hypoxic core and a proliferative outer layer from metabolite distribution. Targeted metabolites (e.g., lactate, glutamine, and citrate) were mapped to pathways of glycolysis and the TCA cycle demonstrating tumor metabolic behavior. The first application of imaging mass cytometry (IMC) with 3D cell culture enabled single-cell phenotyping at 1 μm spatial resolution. Protein markers of proliferation (Ki-67) and hypoxia (glucose transporter 1) defined metabolic signaling in the aggregoid model, which complemented the metabolite data. Laser ablation inductively coupled plasma (LA-ICP)-MSI analysis localized endogenous elements including magnesium and copper, further differentiating the hypoxia gradient and validating the protein expression. Obtaining a large amount of molecular information on a complementary nature enabled an in-depth understanding of the biological processes within the novel tumor model. Combining powerful imaging techniques to characterize the aggregated 3D culture highlighted a future methodology with potential applications in cancer research and drug development.
Mass spectrometry imaging (MSI) is a sophisticated technology capable of simultaneously mapping a variety of molecules within a biological sample. The benefits of conventional MSI techniques compared to other imaging modalities are the abilities to detect ionizable compounds in both targeted and untargeted methods without the use of specific labeling reagents.1 The spatial localization of a molecule can determine the interplay of biological functions and interactions within a tissue. This also enables a greater biological understanding of cellular phenotypes and their structural organization, in addition to the surrounding microenvironment. MSI has therefore had a major influence on cancer research and drug development by utilizing spatial localization of biomarkers, therapeutics, their active metabolites, and cellular responses.1
Depending on the biological sample and the molecules of interest, different MSI techniques can be employed for optimum analysis. Matrix-assisted laser desorption/ionization (MALDI) is the most widely used MSI technique across many applications due to its high spatial resolution and speed of acquisition. MALDI-MSI can detect a wide range of analytes in an untargeted manner including metabolites, lipids, peptides, and proteins.2−4 Desorption electrospray ionization (DESI)-MSI is also widely used due to the minimal sample preparation requirement of this ambient ionization methodology.5 The combination of DESI with Orbitrap and QTOF type mass spectrometers has generated images with high mass specificity for metabolites and small molecule therapeutics in tissue samples.6,7 The requirement for the study of trace elements or metal isotope distribution in tissues has also seen the development of laser ablation-inductively coupled plasma (LA-ICP)-MSI,8,9 a technique that has been applied to the analysis of metal-containing therapeutics such as cisplatin.10,11 Advancements of LA-ICP-MSI have evolved to imaging mass cytometry (IMC), a novel multiplex method capable of tissue phenotyping, and imaging biological processes at a high spatial resolution (<1 μm). Detection of proteins is achieved in this technique using metal-labeled antibodies specific to proteins and protein modifications. IMC has demonstrated high-dimensional single-cell analysis capabilities on numerous tissue types12,13 and directly visualized platinum-based therapeutics and the biological responses to treatment.14 Recent studies have demonstrated the benefits of the use of multimodal MSI for the same study for the extraction of complementary molecular information to enable a wider detection of a diverse range of analytes within corresponding samples.15,16
Parallel to the developments in MSI, aspects of preclinical therapeutic research are exploring more sustainable and cheaper biological models than the use of in vivo models. Three-dimensional (3D) cell cultures offer a biologically relevant model that can be used for early stage drug development and screening studies.17,18 The cellular complexity of 3D models mimics the biological microenvironment of tissues in a way that 2D cultures cannot. Due to recent societal issues regarding the use of animal models in science, the demand for 3D cultures has grown significantly. They offer a way in which the 3Rs principle, i.e., the reduction, replacement, and refinement of the use of animals in scientific research can be implemented in preclinical studies.19 Tumor spheroids are among the most common biological systems developed. These cellular spheres mirror the tumor microenvironment of proliferative, hypoxic, and necrotic regions through gradients of oxygen and nutrients. By recapitulating the cell–cell interactions and the growth and differentiation processes of tumors, spheroids are a valuable research tool to study realistic drug behavior.
Over the past decade, advancements in technology, especially the achievable spatial resolution, have enabled the analysis of tumor spheroids by MSI. Li and Hummon20 were the first to establish the combination of MALDI-MSI with spheroids for the characterization of protein distributions in a HCT116 colon carcinoma model. Proteins including Histone H4 and cytochrome c were localized across the spheroid, and a specific, unidentified peak (m/z 12 828) was localized within the necrotic core. More recently, Tucker et al.21 employed high spectral resolution Fourier transform-ion cyclotron resonance (FT-ICR) MALDI-MSI to determine metabolite distributions in MCF-7 breast cancer spheroids. The group mapped metabolites associated with hypoxia through biochemical processes including glycolysis and the hexosamine biosynthetic pathway. By improving the understanding of cellular functions in tumor spheroids, MSI techniques give opportunities to unravel a drug’s absorption, distribution, and metabolism. The Hummon group have reported numerous drug toxicity applications with MALDI-MSI, investigating the distribution of small molecule chemotherapeutics,22,23 combinational drugs,24 and immunotherapy25 in colon carcinoma spheroid cultures.
Recently, literature has debated whether tumor spheroids fully recapitulate the heterogeneous nature of an in vivo tumor.26 This is due to methods in which they are cultured. Multicellular tumor spheroids (MCTS) are the most common model formed by aggregation, rather than proliferation, via ultralow attachment techniques. Their homogeneity negatively impacts true biological behavior and therefore limits drug efficacy experiments. To overcome this limitation, our group has developed a novel 3D model which is formed via the aggregation of clonal spheroids.27 This 3D model, termed an “aggregoid” is cultured by isolating and aggregating tumor spheres generated from an alginate bead culture. Similar to the spheroid model, the aggregoid displays a gradient of oxygen and nutrients, whereby a depletion in the core produces a hypoxic environment. This gradient was observed by fluorescently staining the aggregoid culture, identifying a viable outer region and a necrotic core (Supplementary Figure 1). The asymmetric nature of the aggregoid model produces a heterogeneous tissue and therefore allows for a more morphological representation of an in vivo tumor. The aggregoid model can also be cultured to an approximate diameter of 1 mm, which is large enough for in-depth spatial distribution studies. MALDI-MSI analysis of this aggregoid model was demonstrated by Palubeckaite et al.27 for the determination of drug and endogenous metabolite distributions. Metabolites were localized in regions corresponding to the hypoxic gradient, including m/z 426.1 within the core, and m/z 281.3 located within the outer areas of an SAOS-2 osteosarcoma aggregoid. MALDI-MSI analysis was also employed to detect doxorubicin, a major chemotherapeutic, within the core of the SAOS-2 aggregoid and for mapping the metabolic responses to the treatment.
In this study, we have characterized a novel aggregoid tumor model created from HCC827 lung adenocarcinoma cells using multiple MSI modalities: DESI-MSI, IMC, and LA-ICP-MSI. We show how the aggregoid model displays similar phenotypical characteristics to tumor spheroid cultures, demonstrating its potential as an in vitro research tool. Additionally, we have identified specific molecular markers that define regions of hypoxia and key biological processes by the analysis of metabolites, proteins and protein modifications, and elemental compounds by their respective imaging platforms.
Methods
3D Culture Growth
Epithelial HCC827 lung adenocarcinoma cell line (ATCC) was cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza Ltd., U.K.), supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (Lonza Ltd., U.K.). Aggregoids were generated based on the method of Palubeckaite et al.,27 as follows: Cells were maintained at 37 °C, 5% CO2 and grown to 80% confluence prior to use. To generate the initial tumor spheres, cells were suspended in 1.2% (w/v) alginic acid (Sigma-Aldrich, U.K.) in 0.15 M NaCl at 1 × 106 cells/mL and extruded out of a needle into 0.2 M CaCl2 to polymerize the alginate into beads. Beads were washed with 0.15 M NaCl before culturing in DMEM media for 14 days to yield spheroids ∼100 μm in diameter, and media was replaced every 72 h. Alginate beads were dissolved in an alginate buffer (55 mM sodium citrate, 30 mM EDTA, 0.15 M NaCl) to release spheroids into solution. Spheroids were washed with PBS (Lonza, Castleford, U.K.) before seeding spheroids into 1% agarose-coated 96-well plate in growth medium. Spheroids were cultured for 7 days to form aggregoids of an approximate 1 mm diameter before harvesting. Spheroid and aggregoid development were analyzed by fluorescent staining with Hoechst 33342 and propidium iodide staining (10 μg/mL each) for 30 min. Fluorescent images were obtained using the Olympus IX81 Microscope (Southend-on-Sea, U.K.) and images were captured using Cell^F Multifluorescence and Imaging Software (Europa Science Ltd., Cambridge, U.K.) (Supplementary Figure 1).
Tissue Preparation
Aggregoids were prepared for imaging analysis based on the tissue embedding protocol. Briefly, aggregoids were washed in PBS prior to embedding in media made of 7.5% hydroxypropyl-methylcellulose (HPMC) and 2.5% polyvinylpyrrolidone (PVP). Embedded tissues were flash frozen in liquid nitrogen and stored at −80 °C. Frozen aggregoids were sectioned at 10 μm thickness using a Leica CM3050 cryostat (Leica Microsystems, U.K.) operating at −18 °C. Sections were thaw-mounted onto polylysine glass slides followed by immediate desiccation with N2 and subsequent vacuum packing for storage at −80 °C.28
DESI-MSI
Small molecule imaging was performed using a Q-Exactive mass spectrometer (Thermo Fisher Scientific Inc., Germany) operated in negative mode. The mass spectrometer was equipped with a custom-built automated DESI ion source. The mass resolution was set to 70 000, and mass spectra were collected in the mass range m/z 80–900 at a spatial resolution 30 μm. The electrospray solvent was MeOH/water (95:5 v/v) set at a flow rate of 1 μL/min with nebulizing nitrogen used as gas at pressure of 2 bar. Imaging analysis was performed by combining individual horizontal line scans and converting into imzML format using the imzML converter V.1.1.4.5 (www.maldi-msi.org). The images were analyzed by SCiLS Lab MVS Premium 3D Version 2020a (Bruker Daltonics, Germany) employing root-mean-square (RMS) normalization.
Discriminatory Analysis
The aggregoid DESI-MSI data file was segregated into regional clusters by spatial segmentation processing by which the “core” and “outer” regions were identified (SCiLS, Bruker Daltonics). Discrimination between the two regions was achieved by automatically finding m/z values by employing the receiver operating characteristic (ROC) tool to calculate the area under the curve (AUC) value (Supplementary Table 1). The raw data file from the DESI-MSI was uploaded to METASPACE (https://metaspace2020.eu/) for metabolite identification of the discriminated m/z values by employing the Human Metabolome Database (HMDB) (tolerance <1 ppm). Metabolic pathways were assigned based on the KEGG database by importing identified m/z values into Pathos software (http://motif.gla.ac.uk/Pathos/). The ion abundances for the m/z values were generated into histograms for comparison between regions using GraphPad Prism software (La Jolla, CA).
IMC Staining
Tissues were fixed with 4% paraformaldehyde for 10 min at room temperature (RT). Prior to staining, tissues were permeabilized with 1× casein solution containing 0.1% Triton X-100 for 5 min at RT. Tissues were then incubated with blocking buffer (1× casein solution) for 30 min at RT. An antibody cocktail was made containing the appropriate dilutions for the antibodies. Tissues were incubated with the antibody cocktail overnight at 4 °C. DNA Ir-Intercalator (Fluidigm) was diluted 1:400 and applied to tissues for 30 min at RT. Washes with PBS were performed three times between each step, with the last step washed in deionized water for 30 s. Slides were left to air-dry until analysis.
IMC Analysis
Images were acquired using the Hyperion Imaging System (Fluidigm), rasterizing at 200 Hz and with the laser tuned to fully ablate the tissue without etching the glass. TIFF files of each acquisition were then exported for analysis in the HALO image analysis platform (Indica Laboratories). Using a random forest machine learning Tissue Classifier module, each image was segmented into the background and inner, core, and outer areas of each aggregoid. Using the Hiplex module, the DNA intercalator was used to first segment the nucleus of each cell, and a proxy for the cytoplasm of each cell defined in a 1 μm radius from the nucleus, before thresholds set to define positive cell staining for each marker. Percentage positivity of each cell was then defined within the inner and outer region of the aggregoid.
LA-ICP-MS Analysis
Experiments were conducted using a NexION 350X ICPMS (PerkinElmer) coupled to an UP-213 LA system (New Wave Research) with a frequency quintupled 213 nm Nd: YAG laser. Laser parameters were optimized to a 6 μm spot size with laser power 46%, 25 μm/s scan speed, 0.07 J cm–2 laser fluence, and 20 Hz repetition rate. The sample was ablated line by line with 6 μm raster spacing at 1.31 min acquisition time. For the ICP-MS instrument there was a direct flow with a rate of 1.4 L/min. The following settings were used in standard mode with an 18 L/min plasma gas flow, 1.2 L/min auxiliary gas flow at 1600 W RF power. Isotopes monitored included 24 Mg, 66Zn, and 63Cu, and the instrument was controlled using Syngistix software. Data analysis was achieved using Iolite Software on Igor Pro (WaveMetrics, USA).
Histological Staining
After DESI-MSI, aggregoid sections were stained using Mayer’s hematoxylin and eosin solutions. Sections were fixed in 4% paraformaldehyde for 10 min before staining with hematoxylin for 1 min. Tissues were rinsed in tap water before and after submerging in acid alcohol. Tissues were subsequently stained with eosin for 30 s prior to washing tap water, then subsequently washed 3 times with absolute ethanol for 1 min. Finally, tissues were submerged in xylene substitute for 1 min twice and mounted using DPX mountant. Stained tissues were imaged with an Aperio CS2 digital pathology scanner (Aperio Tech., Oxford, U.K.) at 40× and visualized with ImageScope software (Aperio Tech.).
Results and Discussion
Metabolite Imaging
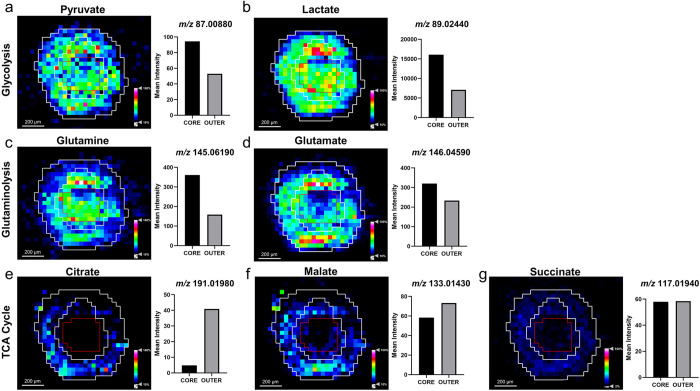
One major hallmark of cancer is an altered cellular metabolism to generate a sufficient energy source contributing to the initiation, growth, and maintenance of tumors.29 During tumor growth, a hypoxic microenvironment is developed due to the gradient of oxygen and nutrients. In the present study, the metabolic profile within the lung adenocarcinoma aggregoid model was investigated by employing a DESI Thermo Q-Exactive MSI to classify regions of a hypoxic core and a proliferative outer area. Initial processing of the aggregoid images was conducted to spatially segment the data. This is a process whereby the image is segregated into regions; pixels within proximity that share similar spectral characteristics are grouped together into a segment. These segments are then classified into regions which represent phenotypical features of a tissue. The aggregoid data was segmented into three main regions that depicted a gradient-like phenotype: a core, an annular zone, and an outer region (Figure 1b,c). From the 2- and 3-dimensional images, the clear discrimination of the regional clusters corresponded to the histology stain of the same section after MSI analysis (Figure 1a). From this, the spectra from each region were extracted to distinguish the distributions of key metabolites within the aggregoid. For the purpose of separating metabolites to distinct regions, the core and the outer zones were the main focus when observing the distribution of species, as the intermediate region was anticipated to be nondiscriminatory.
Figure 1.
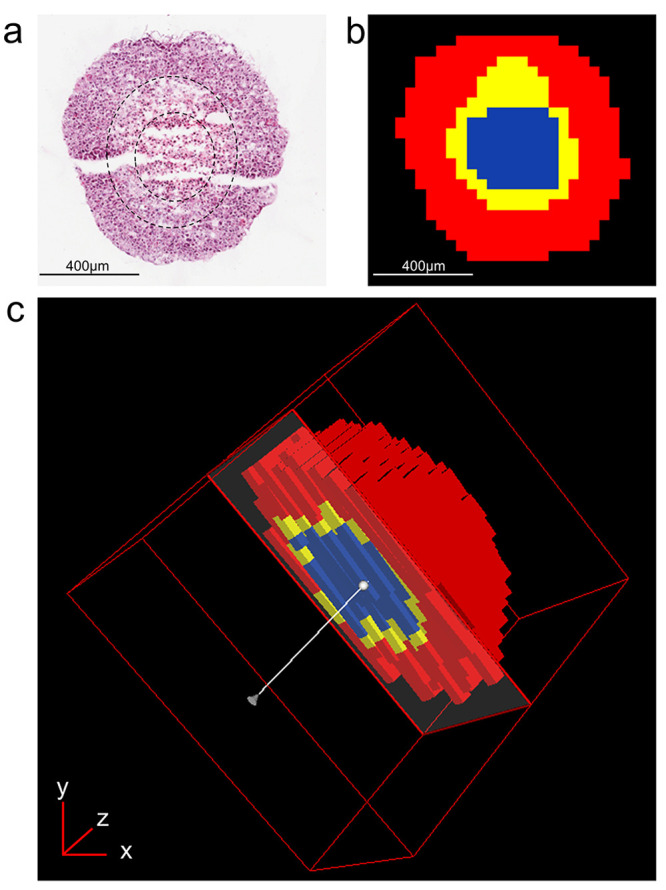
Spatial segmentation of HCC827 aggregoid model from metabolite data by DESI-MSI. (a) H&E stain of central aggregoid section showing three separate regions within the tissue. Slight fissures can be observed in the tissue which formed during sectioning. Scale bar 400 μm. (b) Spatial segmentation of central aggregoid section identified three clustering regions that correspond to the hypoxia gradient: necrotic core (blue), annular quiescent region (yellow), and proliferative outer region (red). Scale bar 400 μm. (c) Realigned 3D construct of aggregoid displaying segmentation pattern throughout the model.
Within the aggregoid model, key metabolites involved in cancer metabolism were identified with a mass error of <0.5 ppm (Supplementary Table 1). A major metabolic substrate that is regulated by the tumor microenvironment is lactate. Within the aggregoid, a high intensity of lactate (m/z 89.02440) was distributed throughout indicating the presence of metabolic activity (Figure 2b). An increase in the expression of lactate converted from glucose via the glycolysis reaction is thought to be the predominant pathway to promote tumor survival and growth rather than following oxidative metabolism, otherwise known as the Warburg effect.30 An elevated expression of lactate in the core of the aggregoid also implies the presence of hypoxia. In anaerobic conditions, the rate of glycolysis increases due to insufficient oxygen levels to promote tumor survival. A similar distribution of a glycolysis intermediate, pyruvate (m/z 87.00880) was also observed (Figure 2a). The localization of pyruvate across the aggregoid, with elevated levels in the core validates the assumption of an increased rate of glycolysis in response to hypoxia. A key molecule associated with increased lactate production is the expression of hypoxia-inducible factor alpha (HIF-1α), which is stabilized in a hypoxic environment due to the lack of oxygen and therefore a direct marker of hypoxia. HIF-1α is responsible for regulating the expression of numerous genes under hypoxic conditions. Specifically, HIF-1α promotes the transportation of glucose into the cell by increasing the expression of the glucose transporter 1 (Glut1).31 Additionally, HIF-1α promotes a high glycolysis rate by inducing both pyruvate dehydrogenase kinase (PDK) and lactate dehydrogenase A (LDH-A) to prevent the metabolism of pyruvate into acetyl-CoA to feed the tricarboxylic acid (TCA) cycle and rather by favoring the conversion of lactate.32,33
Figure 2.
Distribution of metabolites regulating cancer growth and survival within the HCC827 aggregoid central section by DESI-MSI. Ion density maps of metabolites outlining the core and the outer area on the image. Mean intensity plotted on bar graph against the core and outer regions. Scale bar 200 μm. Intermediates of the glycolysis reaction: (a) pyruvate, m/z 87.00880 and (b) lactate, m/z 89.02440. Glutaminolysis reaction: (c) glutamine, m/z 145.06190 and (d) glutamate, m/z 146.04590. TCA cycle: (e) citrate, m/z 191.01980; (f) malate, m/z 133.01430; and (g) succinate, m/z 117.01940.
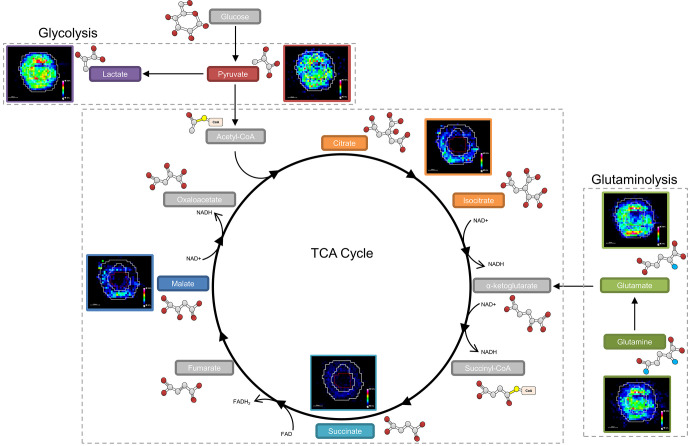
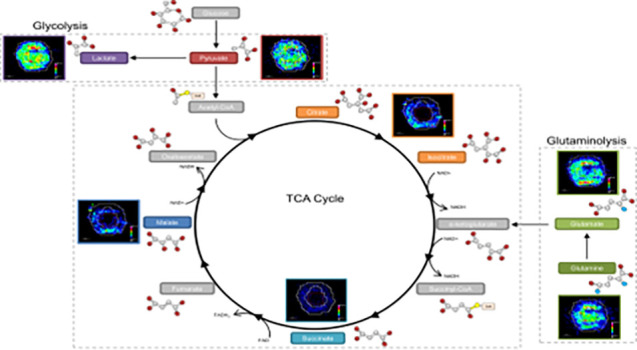
The image analysis identified an increased distribution of glutamine (m/z 145.06190) within the core of the aggregoid (Figure 2c). Glutamine is considered a major bioenergetic substrate that sources the TCA cycle by its metabolism to the intermediate α-ketoglutarate achieved by the anaplerotic pathway, glutaminolysis.34 The TCA cycle is described as the epicenter of cell metabolism due to the extensive supply of metabolic substrates that are utilized for energy production.35 The localization of glutamine within the core suggests the cells within the hypoxic environment are substituting for the lack of pyruvate sourcing the TCA cycle. Interestingly, glutamate (m/z 146.04590), an intermediate of glutaminolysis, is distributed toward the outer region of the aggregoid (Figure 2d). The TCA cycle is heavily utilized by proliferating cells for growth, the suppression of glutamine conversion to glutamate in the core therefore implies the presence of necrosis. Several spheroid studies have reported that the increase in diameter decreases the cell viability due to the reduced levels of oxygen and nutrients, thus the spheroid eventually develops an inner necrotic core.36,37 By examining the gene expression profiles, Daster et al.38 reported the development of a necrotic region in multicellular spheroids larger than 500 μm. Since the diameter of the aggregoid model is approximately 1 mm, the presence of an inner necrotic core is highly likely. Fluorescent staining of the aggregoid with propidium iodide validates this necrotic region (Supplementary Figure 1) and additionally shows the large asymmetric hypoxic area opposed to the simple radial gradient in a typical spheroid model, which could explain the asymmetric metabolite distribution in Figure 2a–d. In contrast, the distributions of the major TCA cycle intermediates citrate (m/z 191.01980), malate (m/z 133.01430), and succinate (m/z 117.01940) were observed with more annular features within the outer proliferative region of the aggregoid (Figure 2e–g), implying a surplus of oxygen and nutrients surrounding the aggregoid and the absence of cell proliferation in the core. By identifying the significant metabolites that drive cancer metabolism, it was possible to map the ion density images onto their corresponding pathways to associate the metabolic activity with specific regions of the aggregoid (Figure 3).
Figure 3.
Mapping metabolites to biological pathways defined areas of tumor metabolism. The glycolysis reaction is highly expressed across the whole aggregoid section demonstrating the Warburg effect. Conversion of glutamine to glutamate is showing reduced expression in the core. The TCA intermediates present within the proliferative outer region. Metabolite images obtained by DESI-MSI analysis. Intermediates acetyl-CoA, α-ketoglutarate, succinyl-CoA, fumarate, and oxaloacetate were not observed.
Proliferating cancer cells utilize fatty acids as they have essential roles as structural components of the membrane matrix, secondary messengers for signaling pathways, and sources for energy production.39 Here fatty acid distribution was imaged within the aggregoid by DESI-MSI and identified with a mass error ≤0.7 ppm (Supplementary Table 1). The image analysis demonstrated the presence of two polyunsaturated fatty acids, FA (18:2), e.g., linoleic acid at m/z 279.23280, and FA (20:4), e.g., arachidonic acid at m/z 303.23300, within the proliferative region (Figure 4a,b). The metabolite glutathione (GSH) (m/z 306.07650) displayed a similar localization to the fatty acids described with elevated levels surrounding the hotspot within the outer region (Figure 4c). GSH protects cells against reactive oxygen species (ROS), a normal product from cellular metabolism, through the oxidation of its sulfhydryl group to form glutathione disulfide (GSSG).40 The colocalization of GSH with the fatty acids suggests an area of high metabolic activity. Interestingly, there was a lack of GSH in the core of the aggregoid, which is heavily associated with oxidative stress (Figure 4c). In hypoxia, the expression of antioxidant genes including GSH metabolic genes are induced to allow cells to regulate ROS. However, it has been shown that in the presence of excess ROS, GSH is depleted which leads to the activation of apoptosis.41 The presence of GSH (or the lack of) can therefore be a potential measure of oxidative stress within the aggregoid model. The importance of defining heterogeneity within the in vitro aggregoid model allows a further understanding of a realistic tumor microenvironment and the true metabolic behavior of an in vivo cancer. This information can then be utilized in applications of drug development.
Figure 4.
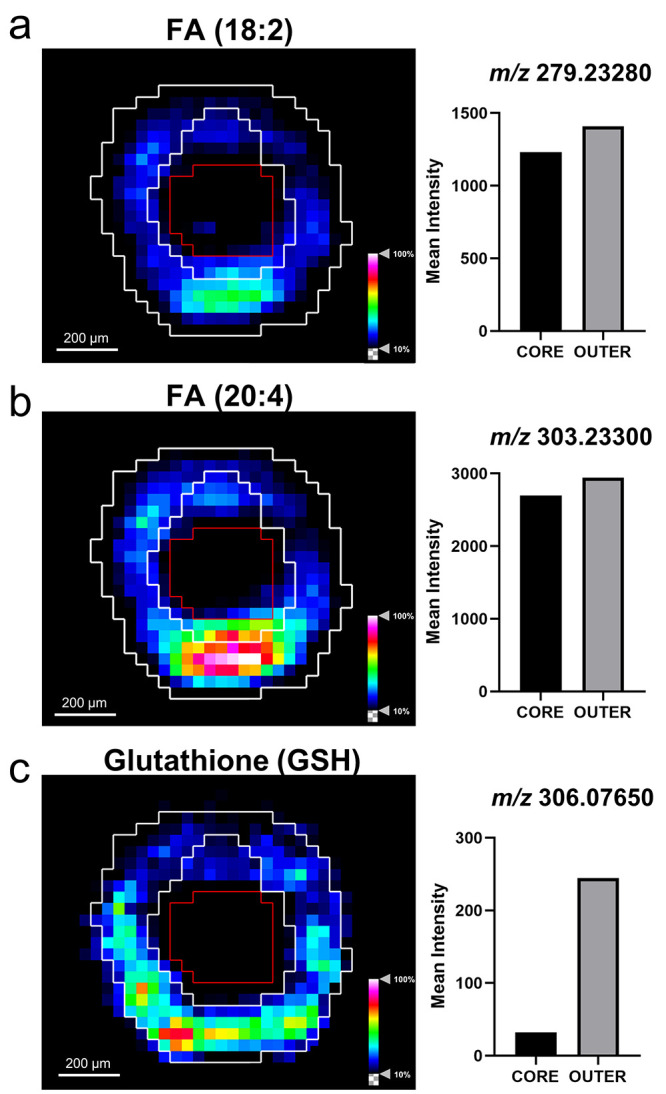
Fatty acid species observed in proliferative outer region by DESI-MSI. Ion density maps of metabolites outlining the core and the outer area on the image. Mean intensity plotted on bar graph against the core and outer regions. Scale bar 200 μm. (a) FA (18:2), m/z 279.23280; (b) FA (20:4), m/z 303.23300; (c) GSH, m/z 306.07650.
Single-Cell Tumor Characterization
IMC is a novel, multiplex imaging platform capable of high-dimensional tissue phenotyping and the detection of signaling activities by the analysis of protein and protein modification markers at single-cell resolution (1 μm). The analysis of proteins within tissues can define essential cellular functions such as proliferation, metabolism, gene expression, organization, and apoptosis.42 Modifications to such proteins can manipulate their spatial distribution, composition, and their function,43 which can contribute to tumor progression. In the present study, IMC was employed for single-cell phenotyping of the HCC827 aggregoid model for an in-depth characterization of the tumor microenvironment. Proteomic markers relevant to lung adenocarcinoma were selected to identify key components of cellular organizations, functions, and signaling.
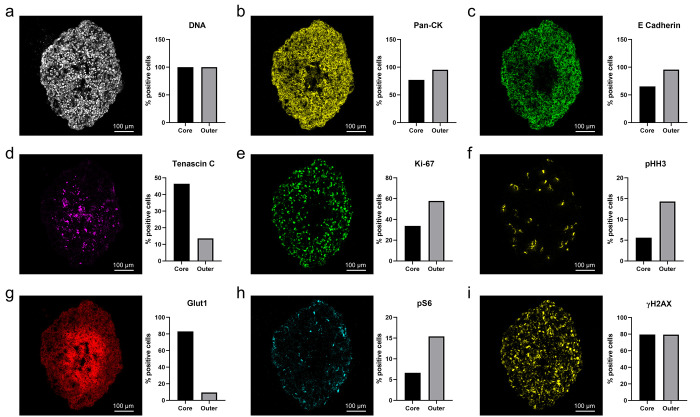
Due to the complex heterogeneity of cancer tissues, morphological and structural components provide a navigational aid to determine the initial tissue organization.13 In this study, such cellular elements included DNA and epithelial tumor markers. The DNA intercalator selected was a generic marker, selected to identify the size and shape of the nucleus in individual cells within the aggregoid (Figure 5a). This data was used to spatially segment the image to calculate the percentage positive cells for each marker (HALO, Indica Laboratories) (Supplementary Figure 3).
Figure 5.
Representative IMC images of biological processes at subcellular detail in the HCC827 aggregoid model. Scale bar, 100 μm. Percentage positive cells plotted on bar graph against the core and outer regions. (a) DNA intercalator identified individual cells within the aggregoid section. Epithelial tumor markers: (b) Pan-CK, (c) E-Cadherin, and (d) Tenascin C (TNC). Proliferation markers: (e) Ki-67 and (f) pHH3. Hypoxia influenced markers: (g) pHH3 and (h) pS6. DNA damage marker: (i) γH2AX.
Tumor markers observed within the HCC827 aggregoid were epithelial cadherin (E-cadherin) and pan-cytokeratin (Pan-CK). The image analysis of both markers identified similar distributions with elevated expression levels within the outer proliferative region of the aggregoid (Figure 5b,c). In epithelial cells, E-cadherin and cytokeratin are responsible for mediating cell–cell adhesion and mechanical support via intermediate filaments, respectively. The absence of these markers within the core is possibly due to the breakdown of cell interactions as a result of necrosis. Simiantonaki et al.44 reported a similar correlation with cellular necrosis and a lack of E-cadherin distribution in the core of HT-29 colorectal carcinoma spheroids via immunohistochemistry. Interestingly, the expression of both E-cadherin and cytokeratin can determine epithelial-mesenchymal transition (EMT), a process which promotes tumor progression and metastasis. In EMT, both epithelial markers are either downregulated or lost coupled with a gain of mesenchymal markers, N-cadherin and vimentin.45 Studies have demonstrated that EMT signaling can be induced by HIF-1α in tumor spheroids.46,47 Without analyzing markers of the mesenchymal phenotype, this process cannot be confirmed. However, as necrosis and EMT have a crucial part in tumor progression, future aggregoid analysis with IMC has potential for applications in drug development and resistance.
Alternatively, Tenascin C (TNC) is an extracellular matrix (ECM) marker considered an active component of cancer. Relatively high expression of the marker was localized within the necrotic core of the aggregoid (Figure 5d). TNC is thought to promote survival and invasion by regulating the expression of proangiogenic factors such as vascular endothelial growth factor (VEGF) modulated by HIF-1α.48 Additionally, TNC has been associated with inducing EMT changes with the downregulation of E-cadherin.49 Thus, the TNC marker correlates with the distribution of the E-cadherin marker (Figure 6b).
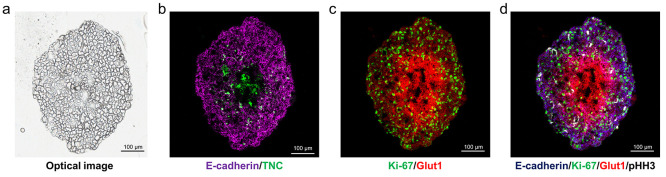
Figure 6.
Structural organization of biological processes for in-depth phenotyping of HCC827 aggregoid model by IMC. (a) Optical image of aggregoid prior to staining with antibodies and image analysis. Scale bar, 200 μm. Overlay of IMC markers displays representative images of (b) epithelial tumor markers: Ecadherin, TNC; (c) proliferation and hypoxia, Ki-67 and Glut1; (d) overlay image combining markers of epithelial tumor, proliferation, hypoxia, and mitosis: E-cadherin, Ki-67, Glut1, and pHH3, respectively. Scale bar, 100 μm.
To distinguish regions of the tumor microenvironment and to complement the findings from the metabolite distributions, specific markers of proliferation and hypoxia were included. Ki-67 is a cellular marker, present in all stages of the cell cycle except for early G1 and G0 quiescent phases. The high expression of Ki-67 present within the outer region of the aggregoid therefore implies an active proliferative zone (Figure 5e). In addition, phosphorylated Histone H3 (pHH3) marker was identified in only a few specific cells, yet still located primarily in the outer region of the aggregoid (Figure 5f). HH3 is a nuclear core protein, and when phosphorylated at serine-10, is specifically involved in mitotic chromatin condensation.50 Hence, the expression of pHH3 can identify cells undergoing mitosis. It can be concluded that the cells within the outer region of the aggregoid are highly proliferative implying a nonhypoxic area compared to the cells within the core, thus tightly corresponding to the distributions of the TCA cycle intermediates from the DESI-MSI analysis.
On the other hand, Glut1 (glucose transporter 1) is a proxy hypoxia marker. Elevated levels of the marker were observed within the necrotic core of the aggregoid (Figure 5g). Glut1 is a hypoxia responsive gene, which is upregulated by HIF-1α to maintain an adequate energy supply in response to reduced oxidative phosphorylation.51 High levels of Glut1 complement the high lactate expression from the metabolite analysis, implying an increase in glucose transport into cells for lactate production via glycolysis. From the overlay image analysis, an inverse distribution of Ki-67 and Glut1 can distinguish the two major regions of the tumor microenvironment: proliferative outer and hypoxic core (Figure 6c). Phosphorylated S6 ribosomal protein (pS6), an active marker for mTOR signaling for growth and metabolism, is also regulated by hypoxia. In contrast, the expression of pS6 was observed primarily within the outer region of the aggregoid, with high levels within specific cells (Figure 5h). In hypoxic conditions, the activity of the mTOR pathway is reduced, negatively impacting on the pS6 expression.52 Both Glut1 and pS6 markers therefore identified metabolic signaling within the aggregoid that is affected by hypoxia.
Alternatively, phosphorylated Histone H2AX (γH2AX) is a marker for DNA damage and stress and can be indicative of cellular apoptosis.53 This therefore explains the accumulation of γH2AX within the hypoxic core of the aggregoid (Figure 5i). From the image analysis, however, high expression levels of γH2AX was also observed within the proliferative outer region (Figure 5i). Due to oxidative stress, induced by natural ROS from metabolic activity, proliferative cells are subjected to constant DNA damage.54 Therefore, the distribution of γH2AX throughout the aggregoid is supported. In the future, γH2AX marker has potential to be used for the detection of cellular stress within the aggregoid, with elevated levels when subject to therapeutic treatment.
This is the first report on the analysis of 3D cell culture models with IMC. The usage of IMC to characterize the HCC827 aggregoid model based on morphological and structural markers specific for an epithelial tumor, growth and proliferation, and the hypoxia gradient of the tumor microenvironment has been demonstrated (Figure 6d). Furthermore, with the single-cellular resolution capabilities of IMC, it was possible to distinguish individual cells and the matrix surrounding based on the cellular localization of such protein markers. Therefore, IMC demonstrates promise for high-dimensional phenotyping of 3D cell culture models at a greater spatial resolution than is currently possible with the other MSI modalities employed in this study.
Endogenous Elemental Analysis
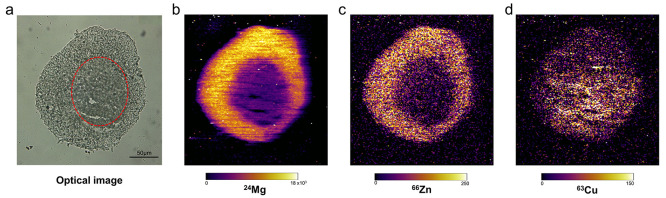
Deficiencies, defects, and accumulation of metal compounds within cells are known to be a hallmark of cancer and disease. Within tissues, metals have a heterogeneous distribution whereby high concentrations can be associated with high metabolic activity.9 Therefore, visualizing the metal composition within a tissue can provide essential information to understanding their key functions in different environments, such as hypoxia or nutrient rich areas. The composition of abundant metal isotopes 24 Mg, 66Zn, and 63Cu were selected to analyze within the HCC827 aggregoid model. To measure the abundance of low mass range metal ions at high sensitivity, LA-ICP-MSI was employed.
In the cell, Mg and Zn are essential components to drive cell growth, division, and proliferation.55,56 Observations from the LA-ICP-MSI analysis localized both elements of high expression solely within the outer proliferative region of the aggregoid (Figure 7b,c). Similar to Ki-67, Mg plays a key role in the cell cycle except for early G1 and G0 quiescent phases.57 Thus, the absence of Mg is indicative of a nonproliferative region or necrotic core. Zn, on the other hand, has been directly linked to the degradation of HIF-1α under normoxic conditions.58 Under hypoxic conditions, however, this process is downregulated to enable stabilization of HIF-1α. It is therefore possible that the absence of Zn within the aggregoid core is associated with the activation of HIF-1α in hypoxia; whereby Zn is possibly exported to the proliferative zone where high levels are required for metabolic activity.
Figure 7.
Elemental distributions within HCC827 aggregoid sections obtained using LA-ICP-MS. (a) Optical image taken before acquisition; necrotic region outlined by red dotted line. Scale bar 50 μm. Elemental maps of (b) 24Mg, (c) 66Zn, and (d) 63Cu within the section of aggregoid.
In contrast, the Cu levels in the aggregoid were elevated within the necrotic core (Figure 7d). Increasing evidence has linked Cu with HIF-1α via the hypoxia signaling pathway as a response to oxidative stress to regulate Cu-dependent genes.59,60 These include BNIP3, a cell death factor that induces necrosis,61 and VEGF, which stimulates angiogenesis.62 Both are stimulating factors in hypoxia. In agreement with this, VEGF is also known to be regulated by TNC, which from the IMC analysis was also localized within the core of the aggregoid (Figure 5d). In addition, HIF-1α accordingly promotes the upregulation of the Cu-efflux transporter, ATP7A,63 which tightly regulates levels of free Cu ions to prevent the formation of ROS. Thus, elevated Cu concentrations could imply an active export of free Cu ions into the ECM, accumulating in a less dense area of the aggregoid. As necrotic cells are unregulated, the core therefore becomes the source of metabolic debris.
At present, there is only a limited amount of literature on the study of endogenous elemental compounds in 3D cultures by LA-ICP-MSI. Yet the analysis of tumor spheroids with this technique has had some interest regarding the localization of platinum-based therapeutics and hypoxia-responsive drugs.10,64 Theiner et al.11 differentiated morphological characteristics of a necrotic core, quiescent zone, and proliferative outer region through the analysis of platinum accumulation within HCT116 colon cancer spheroids. However, the elemental compositions in this study are consistent with literature reported in studies employing X-ray fluorescence microscopy (XFM), an alternative analytical technique capable of elemental analysis at high sensitivity. Zhang et al.65 reported similar distributions of Zn and Cu within DLD-1 colon carcinoma spheroids implying the accumulation of such compounds highlight regions of a proliferative outer zone and a necrotic core, respectively.
Conclusion
We have applied advanced molecular imaging techniques for an in-depth phenotyping of a novel aggregated tumor model. This is the first example of an IMC application with a 3D cell culture model. Combining the IMC data with molecular information from DESI-MSI and LA-ICP-MSI, a detailed characterization of the tumor microenvironment within the aggregoid was possible. Distinct regions of a necrotic core and a proliferative outer was distinguished by each method. The localization of metabolites including lactate, glutamine, and citrate within the aggregoid highlighted the metabolic activity in relation to hypoxia. Mapping the ion density images onto the central biological pathways enabled a clearer understanding of the metabolite behavior within the tumor microenvironment. IMC enabled single-cell phenotyping of protein signaling activity. The protein expression complimented the metabolite data including the expression of the Glut1 with elevated lactate levels in the core. In addition, the endogenous elemental compositions of Mg, Zn, and Cu corresponded to the protein information and further validated the presence of a hypoxia gradient. This study improved our understanding of the molecular activity within a 3D cell culture tumor microenvironment. Therefore, MSI analysis of tumor aggregoids highlights a potential methodology for in vitro applications of biomedical research and pharmaceutical development.
Acknowledgments
Lucy Flint is supported by a Ph.D. studentship sponsored by the Biomolecular Sciences Research Centre, Sheffield Hallam University. AstraZeneca are gratefully acknowledged for the continued support and their collaboration during this project. AstraZeneca would like to acknowledge Cancer Research UK Grand Challenge Consortium for discussions in support of their research. Sheffield Hallam University would like to acknowledge that the aggregated tumour model was originally developed in work supported by National Centre for the Replacement Reduction and Refinement of Animals in Research (NC3Rs) Strategic Imaging Technology for the 3Rs award NC/L001896/1.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c02389.
Fluorescent images of 3D cell culture; table of metabolite assignments, ppm error, and discriminatory analysis; average mean spectra of core, intermediate, and outer regions for metabolite data; and IMC classification and segmentation (PDF)
Video S1, 3D video of aggregoid segmentation (AVI)
Video S2, 3D video of glutamine and GSH distribution within whole aggregoid (AVI)
Author Contributions
L.E.F. collected the data and wrote the manuscript with feedback from all authors. G.H. contributed to the DESI-MSI analysis. S.L. contributed to the IMC analysis. J.D.R. and C.J.D. assisted with the LA-ICP-MSI analysis. M.R.C., N.A.C., D.P.S., R.J.A.G., and L.M.C. designed the project. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Swales J. G.; Hamm G.; Clench M. R.; Goodwin R. J. A. Mass spectrometry imaging and its application in pharmaceutical research and development: A concise review. Int. J. Mass Spectrom. 2019, 437, 99–112. 10.1016/j.ijms.2018.02.007. [DOI] [Google Scholar]
- Buchberger A. R.; DeLaney K.; Johnson J.; Li L. Mass Spectrometry Imaging: A Review of Emerging Advancements and Future Insights. Anal. Chem. 2018, 90, 240–265. 10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckli M.; Chaurand P.; Hallahan D. E.; Caprioli R. M. Imaging mass spectrometry: A new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 2001, 7, 493–496. 10.1038/86573. [DOI] [PubMed] [Google Scholar]
- Weaver E. M.; Hummon A. B. Imaging mass spectrometry: From tissue sections to cell cultures. Advanced Drug Delivery Reviews. Adv. Drug Delivery Rev. 2013, 65, 1039–1055. 10.1016/j.addr.2013.03.006. [DOI] [PubMed] [Google Scholar]
- Takats Z.; Wiseman J. M.; Gologan B.; Cooks R. G. Mass Spectrometry Sampling Under Ambient Conditions with Desorption Electrospray Ionization. Science 2004, 306, 471–473. 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- Chen H.; Talaty N. N.; Takáts Z.; Cooks R. G. Desorption Electrospray Ionization Mass Spectrometry for High-Throughput Analysis of Pharmaceutical Samples in the Ambient Environment. Anal. Chem. 2005, 77, 6915–6927. 10.1021/ac050989d. [DOI] [PubMed] [Google Scholar]
- Guenther S.; Muirhead L. J.; Speller A. V. M.; Golf O.; Strittmatter N.; Ramakrishnan R.; Goldin R. D.; Jones E.; Veselkov K.; Nicholson J.; Darzi A.; Takats Z. Spatially Resolved Metabolic Phenotyping of Breast Cancer by Desorption Electrospray Ionization Mass Spectrometry. Cancer Res. 2015, 75, 1828–1837. 10.1158/0008-5472.CAN-14-2258. [DOI] [PubMed] [Google Scholar]
- Pozebon D.; Scheffler G. L.; Dressler V. L.; Nunes M. A. G. Review of the applications of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to the analysis of biological samples. J. Anal. At. Spectrom. 2014, 29, 2204–2228. 10.1039/C4JA00250D. [DOI] [Google Scholar]
- Hare D. J.; New E. J.; de Jonge M. D.; McColl G. Imaging metals in biology: balancing sensitivity, selectivity, and spatial resolution. Chem. Soc. Rev. 2015, 44, 5941–5958. 10.1039/C5CS00055F. [DOI] [PubMed] [Google Scholar]
- Theiner S.; Schreiber-Brynzak E.; Jakupec M. A.; Galanski M.; Koellensperger G.; Keppler B. K. LA-ICP-MS imaging in multicellular tumor spheroids - a novel tool in the preclinical development of metal-based anticancer drugs. Metallomics. 2016, 8, 398–42. 10.1039/C5MT00276A. [DOI] [PubMed] [Google Scholar]
- Theiner S.; Van Malderen S. J. M.; Van Acker T.; Legin A.; Keppler B. K.; Vanhaecke F.; Koellensperger G. Ablation-Inductively Coupled Plasma Mass Spectrometry Imaging of the Distribution of Platinum-Based Anticancer Compounds in Multicellular Tumor Spheroids. Analytical chemistry. Anal. Chem. 2017, 89, 12641–12645. 10.1021/acs.analchem.7b02681. [DOI] [PubMed] [Google Scholar]
- Giesen C.; Wang H. A O; Schapiro D.; Zivanovic N.; Jacobs A.; Hattendorf B.; Schuffler P. J; Grolimund D.; Buhmann J. M; Brandt S.; Varga Z.; Wild P. J; Gunther D.; Bodenmiller B. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 2014, 11, 417–422. 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- Chang Q.; Ornatsky O. I.; Siddiqui I.; Loboda A.; Baranov V. I.; Hedley D. W. Imaging Mass Cytometry. Cytometry, Part A 2017, 91, 160–169. 10.1002/cyto.a.23053. [DOI] [PubMed] [Google Scholar]
- Chang Q.; Ornatsky O. I.; Siddiqui I.; Straus R.; Baranov V. I.; Hedley D. W. Biodistribution of cisplatin revealed by imaging mass cytometry identifies extensive collagen binding in tumor and normal tissues. Sci. Rep. 2016, 6, 36641. 10.1038/srep36641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chughtai S.; Chughtai K.; Cillero-Pastor B.; Kiss A.; Agrawal P.; MacAleese L.; Heeren R. M. A. A multimodal mass spectrometry imaging approach for the study of musculoskeletal tissues. Int. J. Mass Spectrom. 2012, 325–327, 150–160. 10.1016/j.ijms.2012.07.008. [DOI] [Google Scholar]
- Veličković D.; Chu R. K.; Carrell A. A.; Thomas M.; Paša-Tolić L.; Weston D. J.; Anderton C. R. Multimodal MSI in Conjunction with Broad Coverage Spatially Resolved MS2 Increases Confidence in Both Molecular Identification and Localization. Anal. Chem. 2018, 90, 702–707. 10.1021/acs.analchem.7b04319. [DOI] [PubMed] [Google Scholar]
- Yoshii Y.; Furukawa T.; Waki A.; Okuyama H.; Inoue M.; Itoh M.; Zhang M.; Wakizaka H.; Sogawa C.; Kiyono Y.; Yoshii H.; Fujibayashi Y.; Saga T. High-throughput screening with nanoimprinting 3D culture for efficient drug development by mimicking the tumor environment. Biomaterials 2015, 51, 278–289. 10.1016/j.biomaterials.2015.02.008. [DOI] [PubMed] [Google Scholar]
- Rimann M.; Laternser S.; Gvozdenovic A.; Muff R.; Fuchs B.; Kelm J. M.; Graf-Hausner U. An in vitro osteosarcoma 3D microtissue model for drug development. J. Biotechnol. 2014, 189, 129–135. 10.1016/j.jbiotec.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Russo C.; Lewis E. E. L.; Flint L.; Clench M. R. Mass Spectrometry Imaging of 3D Tissue Models. Proteomics 2018, 18, 1700462 10.1002/pmic.201700462. [DOI] [PubMed] [Google Scholar]
- Li H.; Hummon A. B. Imaging Mass Spectrometry of Three-Dimensional Cell Culture Systems. Anal. Chem. 2011, 83, 8794. 10.1021/ac202356g. [DOI] [PubMed] [Google Scholar]
- Tucker L. H.; Hamm G. R.; Sargeant R. J. E.; Goodwin R. J. A.; Mackay C. L.; Campbell C. J.; Clarke D. J. Untargeted Metabolite Mapping in 3D Cell Culture Models Using High Spectral Resolution FT-ICR Mass Spectrometry Imaging. Anal. Chem. 2019, 91, 9522–9529. 10.1021/acs.analchem.9b00661. [DOI] [PubMed] [Google Scholar]
- Liu X.; Weaver E. M.; Hummon A. B. Evaluation of Therapeutics in Three-Dimensional Cell Culture Systems by MALDI Imaging Mass Spectrometry. Anal. Chem. 2013, 85, 6295. 10.1021/ac400519c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowski J. K.; Weaver E. M.; Hummon A. B. Analyzing Liposomal Drug Delivery Systems in Three-Dimensional Cell Culture Models Using MALDI Imaging Mass Spectrometry. Anal. Chem. 2017, 89, 8453–8458. 10.1021/acs.analchem.7b02006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBonia G. J.; Ludwig K. R.; Mousseau C. B.; Hummon A. B. iTRAQ Quantitative Proteomic Profiling and MALDI–MSI of Colon Cancer Spheroids Treated with Combination Chemotherapies in a 3D Printed Fluidic Device. Anal. Chem. 2018, 90, 1423. 10.1021/acs.analchem.7b04969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Lukowski J. K.; Flinders C.; Kim S.; Georgiadis R. A.; Mumenthaler S. M.; Hummon A. B. MALDI-MSI of Immunotherapy: Mapping the EGFR-Targeting Antibody Cetuximab in 3D Colon-Cancer Cell Cultures. Anal. Chem. 2018, 90, 14156–14164. 10.1021/acs.analchem.8b02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Flinders C.; Mumenthaler S. M.; Hummon A. B. MALDI Mass Spectrometry Imaging for Evaluation of Therapeutics in Colorectal Tumor Organoids. J. Am. Soc. Mass Spectrom. 2018, 29, 516–526. 10.1007/s13361-017-1851-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palubeckaitė I.; Crooks L.; Smith D. P.; Cole L. M.; Bram H.; Le Maitre C.; Clench M. R.; Cross N. A. Mass spectrometry imaging of endogenous metabolites in response to doxorubicin in a novel 3D osteosarcoma cell culture model. J. Mass Spectrom. 2020, 55, e4461 10.1002/jms.4461. [DOI] [PubMed] [Google Scholar]
- Swales J. G.; Dexter A.; Hamm G.; Nilsson A.; Strittmatter N.; Michopoulos F.; Hardy C.; Morentin-Gutierrez P.; Mellor M.; Andren P. E.; Clench M. R.; Bunch J.; Critchlow S. E.; Goodwin R. J. A. Quantitation of Endogenous Metabolites in Mouse Tumors Using Mass-Spectrometry Imaging. Anal. Chem. 2018, 90, 6051–6058. 10.1021/acs.analchem.7b05239. [DOI] [PubMed] [Google Scholar]
- Hanahan D.; Weinberg R. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science 1956, 123, 309–314. 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Hayashi M.; Sakata M.; Takeda T.; Yamamoto T.; Okamoto Y.; Sawada K.; Kimura A.; Minekawa R.; Tahara M.; Tasaka K.; Murata Y. Induction of glucose transporter 1 expression through hypoxia-inducible factor 1alpha under hypoxic conditions in trophoblast-derived cells. J. Endocrinol. 2004, 183, 145. 10.1677/joe.1.05599. [DOI] [PubMed] [Google Scholar]
- Kim J.; Tchernyshyov I.; Semenza G. L.; Dang C. V. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006, 3, 177–185. 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Cui X.; Han Z.; He S.; Wu X.; Chen T.; Shao C.; Chen D.; Su N.; Chen Y.; Wang T.; Wang J.; Song D.; Yan W.; Yang X.; Liu T.; Wei H.; Xiao J. HIF1/2α mediates hypoxia-induced LDHA expression in human pancreatic cancer cells. Oncotarget. 2017, 8, 24840–24852. 10.18632/oncotarget.15266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J.; Kamphorst J. J.; Mathew R.; Chung M. K.; White E.; Shlomi T.; Rabinowitz J. D. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol. Syst. Biol. 2013, 9, 712. 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Reyes I.; Chandel N. S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102–111. 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio E.; Salerno S.; Barbieri G.; De Bartolo L.; Drioli E.; Bader A. Mass transfer and metabolic reactions in hepatocyte spheroids cultured in rotating wall gas-permeable membrane system. Biomaterials 2007, 28, 5487–5497. 10.1016/j.biomaterials.2007.08.033. [DOI] [PubMed] [Google Scholar]
- Grimes D. R.; Kelly C.; Bloch K.; Partridge M. A method for estimating the oxygen consumption rate in multicellular tumour spheroids. J. R. Soc., Interface 2014, 11, 20131124. 10.1098/rsif.2013.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Däster S.; Amatruda N.; Calabrese D.; Ivanek R.; Turrini E.; Droeser R. A.; Zajac P.; Fimognari C.; Spagnoli G. C.; Iezzi G.; Mele V.; Muraro M. G. Induction of hypoxia and necrosis in multicellular tumor spheroids is associated with resistance to chemotherapy treatment. Oncotarget. 2017, 8, 1725–1736. 10.18632/oncotarget.13857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beloribi-Djefaflia S.; Vasseur S.; Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis 2016, 5, e189 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong J. S.; Steinauer K. K.; Hornung B.; Irish J. M.; Lecane P.; Birrell G. W.; Peehl D. M.; Knox S. J. Role of glutathione depletion and reactive oxygen species generation in apoptotic signaling in a human B lymphoma cell line. Cell Death Differ. 2002, 9, 252–263. 10.1038/sj.cdd.4400959. [DOI] [PubMed] [Google Scholar]
- Kipp A. P.; Deubel S.; Arnér E. S. J.; Johansson K. Time- and cell-resolved dynamics of redox-sensitive Nrf2, HIF and NF-kB activities in 3D spheroids enriched for cancer stem cells. Redox Biol. 2017, 12, 403–409. 10.1016/j.redox.2017.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. The ins and outs of signalling. Nature 2001, 411, 759–762. 10.1038/35081138. [DOI] [PubMed] [Google Scholar]
- Hoffman M. D.; Sniatynski M. J.; Kast J. Current approaches for global post-translational modification discovery and mass spectrometric analysis. Anal. Chim. Acta 2008, 627, 50–61. 10.1016/j.aca.2008.03.032. [DOI] [PubMed] [Google Scholar]
- Simiantonaki N.; Kurzik-Dumke U.; Karyofylli G.; Jayasinghe C.; Kirkpatrick C. Loss of E-cadherin in the vicinity of necrosis in colorectal carcinomas: Association with NFκB expression. Int. J. Oncol. 2007, 31, 269–275. 10.3892/ijo.31.2.269. [DOI] [PubMed] [Google Scholar]
- Lamouille S.; Xu J.; Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essid N.; Chambard J. C.; Elgaaïed A. B. Induction of epithelial-mesenchymal transition (EMT) and Gli1 expression in head and neck squamous cell carcinoma (HNSCC) spheroid cultures. Bosnian J. Basic Med. Sci. 2018, 18, 336–346. 10.17305/bjbms.2018.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S.; te Boekhorst V.; Odenthal J.; Bianchi R.; van Helvert S.; Ikenberg K.; Ilina O.; Stoma S.; Xandry J.; Jiang L.; Grenman R.; Rudin M.; Friedl P. Hypoxia Induces a HIF-1-Dependent Transition from Collective-to-Amoeboid Dissemination in Epithelial Cancer Cells. Curr. Biol. 2017, 27, 392–400. 10.1016/j.cub.2016.11.057. [DOI] [PubMed] [Google Scholar]
- Wang Z.; Wei Q.; Han L.; Cao K.; Lan T.; Xu Z.; Wang Y.; Gao Y.; Xue J.; Shan F.; Feng J.; Xie X. Tenascin-c renders a proangiogenic phenotype in macrophage via annexin II. J. Cell. Mol. Med. 2018, 22, 429–438. 10.1111/jcmm.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y.; Sawada G.; Kurashige J.; Matsumura T.; Uchi R.; Ueo H.; Ishibashi M.; Takano Y.; Akiyoshi S.; Iwaya T.; Eguchi H.; Sudo T.; Sugimachi K.; Yamamoto H.; Doki Y.; Mori M.; Mimori K. Tumor-derived tenascin-C promotes the epithelial-mesenchymal transition in colorectal cancer cells. Anticancer Res. 2013, 33, 1927–1934. [PubMed] [Google Scholar]
- Kim J.; Jeong H. S.; Chung T.; Kim M.; Lee J. H.; Jung W. H.; Koo J. S. The value of phosphohistone H3 as a proliferation marker for evaluating invasive breast cancers: A comparative study with Ki67. Oncotarget. 2017, 8, 65064–65076. 10.18632/oncotarget.17775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouiddir A.; Planes C.; Fernandes I.; VanHesse A.; Clerici C. Hypoxia Upregulates Activity and Expression of the Glucose Transporter GLUT1 in Alveolar Epithelial Cells. Am. J. Respir. Cell Mol. Biol. 1999, 21, 710–718. 10.1165/ajrcmb.21.6.3751. [DOI] [PubMed] [Google Scholar]
- Schneider A.; Younis R. H.; Gutkind J. S. Hypoxia-induced Energy Stress Inhibits the mTOR Pathway by Activating an AMPK/REDD1 Signaling Axis in Head and Neck Squamous Cell Carcinoma. Neoplasia. 2008, 10, 1295–1302. 10.1593/neo.08586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E. P.; Nieves-Neira W.; Boon C.; Pommier Y.; Bonner W. M. Initiation of DNA fragmentation during apoptosis induces phoysphorylation of H2AX histone at serine 139. J. Biol. Chem. 2000, 275, 9390–9395. 10.1074/jbc.275.13.9390. [DOI] [PubMed] [Google Scholar]
- Tanaka T.; Halicka H. D.; Huang X.; Traganos F.; Darzynkiewicz Z. Constitutive Histone H2AX Phosphorylation and ATM Activation, the Reporters of DNA Damage by Endogenous Oxidants. Cell Cycle 2006, 5, 1940–1945. 10.4161/cc.5.17.3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernon W. B. The role of magnesium in nucleic-acid and protein metabolism. Magnesium 1988, 7, 234–248. [PubMed] [Google Scholar]
- MacDonald R. S. The Role of Zinc in Growth and Cell Proliferation. J. Nutr. 2000, 130, 1500S–1508S. 10.1093/jn/130.5.1500S. [DOI] [PubMed] [Google Scholar]
- Walker G. M. Magnesium and cell cycle control: an update. Magnesium 1986, 5, 9–23. [PubMed] [Google Scholar]
- Nardinocchi L.; Puca R.; Givol D.; D’Orazi G. HIPK2--A therapeutical target to be (re)activated for tumor suppression: Role in p53 activation and HIF-1α inhibition. Cell Cycle 2010, 9, 1270–1275. 10.4161/cc.9.7.11125. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Zhang W.; Kang Y. J. Copper affects the binding of HIF-1α to the critical motifs of its target genes. Metallomics. 2019, 11, 429–438. 10.1039/C8MT00280K. [DOI] [PubMed] [Google Scholar]
- Feng W.; Ye F.; Xue W.; Zhou Z.; Kang Y. J. Copper Regulation of Hypoxia-Inducible Factor-1 Activity. Mol. Pharmacol. 2009, 75, 174–182. 10.1124/mol.108.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowter H. M.; Ratcliffe P. J.; Watson P.; Greenberg A. H.; Harris A. L. HIF-1-dependent Regulation of Hypoxic Induction of the Cell Death Factors BNIP3 and NIX in Human Tumors. Cancer Res. 2001, 61, 6669–6673. [PubMed] [Google Scholar]
- Büchler P.; Reber H. A.; Büchler M.; Shrinkante S.; Büchler M. W.; Friess H.; Semenza G. L.; Hines O. J. Hypoxia-Inducible Factor 1 Regulates Vascular Endothelial Growth Factor Expression in Human Pancreatic Cancer. Pancreas 2003, 26, 56–64. 10.1097/00006676-200301000-00010. [DOI] [PubMed] [Google Scholar]
- Zimnicka A. M.; Tang H.; Guo Q.; Kuhr F. K.; Oh M.; Wan J.; Chen J.; Smith K. A.; Fraidenburg D. R.; Choudhury M. S. R.; Levitan I.; Machado R. F.; Kaplan J. H.; Yuan J. X. Upregulated Copper Transporters in Hypoxia-Induced Pulmonary Hypertension. PLoS One 2014, 9, e90544 10.1371/journal.pone.0090544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill E. S.; Kaur A.; Bishop D. P.; Shishmarev D.; Kuchel P. W.; Grieve S. M.; Figtree G. A.; Renfrew A. K.; Bonnitcha P. D.; New E. J. Hypoxia-Responsive Cobalt Complexes in Tumor Spheroids: Laser Ablation Inductively Coupled Plasma Mass Spectrometry and Magnetic Resonance Imaging Studies. Inorg. Chem. 2017, 56, 9860–9868. 10.1021/acs.inorgchem.7b01368. [DOI] [PubMed] [Google Scholar]
- Zhang J. Z.; Bryce N. S.; Siegele R.; Carter E. A.; Paterson D.; de Jonge M. D.; Howard D. L.; Ryan C. G.; Hambley T. W. The use of spectroscopic imaging and mapping techniques in the characterisation and study of DLD-1 cell spheroid tumour models. Integr. Biol. (Camb). 2012, 4, 1072–1080. 10.1039/c2ib20121f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.