SUMMARY
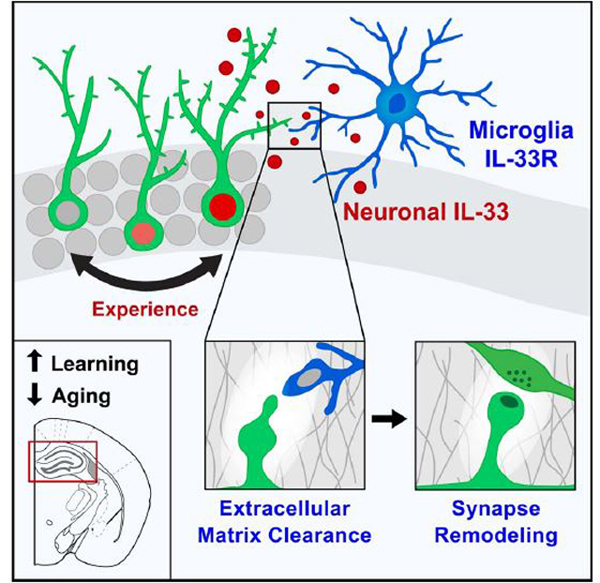
Synapse remodeling is essential to encode experiences into neuronal circuits. Here, we define a molecular interaction between neurons and microglia that drives experience-dependent synapse remodeling in the hippocampus. We find that the cytokine interleukin-33 (IL-33) is expressed by adult hippocampal neurons in an experience-dependent manner and defines a neuronal subset primed for synaptic plasticity. Loss of neuronal IL-33 or the microglial IL-33 receptor leads to impaired spine plasticity, reduced newborn neuron integration, and diminished precision of remote fear memories. Memory precision and neuronal IL-33 are decreased in aged mice, and IL-33 gain of function mitigates age-related decreases in spine plasticity. We find that neuronal IL-33 instructs microglial engulfment of the extracellular matrix (ECM) and that its loss leads to impaired ECM engulfment and a concomitant accumulation of ECM proteins in contact with synapses. These data define a cellular mechanism through which microglia regulate experience-dependent synapse remodeling and promote memory consolidation.
In Brief
A form of experience-dependent neuron-microglial communication is mediated by IL-33, which promotes hippocampal dendritic spine formation, synapse plasticity, and ECM engulfment and is required for memory consolidation.
Graphical Abstract
INTRODUCTION
Neural circuits undergo ongoing structural changes in synaptic connectivity in response to experience. Defining the mechanisms of these remodeling processes is critical for understanding how experiences become stably encoded in the brain. Components of the innate immune system regulate synapse formation and stabilization (Bennett and Molofsky, 2019; Kaneko et al., 2008; Lee et al., 2014; Stevens et al., 2007). Consistent with this, microglia, the predominant immune cells within the brain parenchyma, have been increasingly implicated in synapse remodeling. Microglia make ongoing and activity-dependent contact with dendritic spines (Eyo et al., 2014, 2015; Li et al., 2012; Madry et al., 2018; Tremblay et al., 2010; Wake et al., 2009). They engulf synapse components during early thalamic development and in the hippocampus (Paolicelli et al., 2011; Schafer et al., 2012; Vainchtein et al., 2018; Wang et al., 2020). However, microglia can also promote spine formation in an experience-dependent manner (Miyamoto et al., 2016; Parkhurst et al., 2013) and are required for some forms of cortical plasticity (Sipe et al., 2016).
However, the mechanisms by which these activity-dependent contacts between neurons and microglia lead to stable changes in synaptic connectivity are unknown. This is in part because molecules known to regulate microglial function are only beginning to be studied and have been challenging to examine with cell-type-specific manipulations. Our group recently identified the interleukin-1 (IL-1) family cytokine IL-33 as a novel regulator of microglial activation and function (Vainchtein et al., 2018). IL-33 is normally sequestered in the nucleus in an inactive state but, upon release into the extracellular space, is a potent activator of nuclear factor κF (NF-κB) signaling in target cells that express its obligate receptor, IL1RL1 (also known as ST2; Molofsky et al., 2015). We found that, during early postnatal development, IL-33 expressed by astrocytes in the thalamus and spinal cord drove microglial synapse engulfment and restricted excitatory synapse numbers.
Synapse remodeling continues throughout life in brain regions that change in response to experience, including the hippocampus, which is essential for learning and memory consolidation. Here we define a mechanism by which neuron-microglia signaling via IL-33 augments hippocampal plasticity in an experience-dependent manner. We found that, in the hippocampus, IL-33 was predominantly expressed by neurons, that its expression was experience dependent, and that this expression defined a molecularly distinct subset of neurons enriched for markers of synaptic plasticity. Conditional deletion of Il33 from neurons or its receptor from microglia led to fewer dendritic spines, decreased numbers of newborn neurons, and impaired precision of fear memory retrieval. Aging was associated with reduced neuronal IL-33 as well as deficits in memory precision, and IL-33 gain of function mitigated agerelated declines in spine plasticity and neuronal activation. Finally, we found that IL-33 was necessary and sufficient for extracellular matrix (ECM) engulfment by microglia. Loss of IL-33 led to accumulation of ECM proteins, particularly around synapses and dendritic spines, and exogenous enzymatic clearance of ECM restored dendritic spine numbers in IL-33-deficient animals. These studies reveal a molecular mechanism through which microglia remodel the ECM to promote synapse plasticity and reveal a requirement for this pathway in the precise consolidation of long-term memories.
RESULTS
Experience-Dependent Regulation of Il33 Expression in Adult Hippocampal Neurons
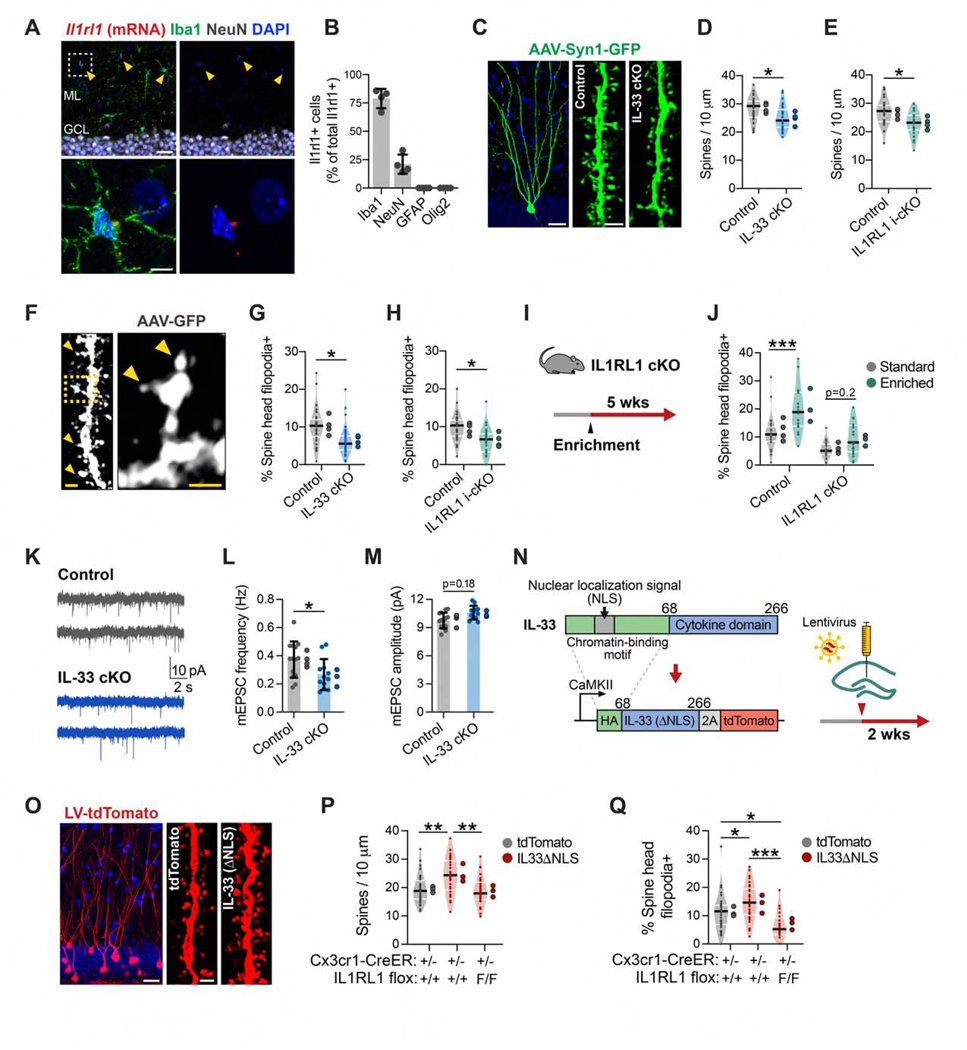
To identify the cellular sources of IL-33 in the hippocampus, we used a nucleus-localized knockin reporter line (Il33mCherry/+; Figure 1A). Unlike in developing subcortical regions, where Il33 is primarily expressed by astrocytes (Vainchtein et al., 2018), we found that most Il33-expressing cells in the hippocampus were neurons (Figures 1A and 1B; Figure S1A). Il33 has two isoforms that produce the same protein but are regulated by alternate promoters (Talabot-Ayer et al., 2012). We found that neurons expressed the Il33b isoform, whereas astrocytes produced Il33a, suggesting that neurons may have unique regulatory mechanisms for IL-33 production (Figure S1B). Il33 was expressed by ~74% of neurons in the dentate gyrus (DG) and ~24% of neurons in the CA1 subregion (Figures 1C and 1D). Within the DG, Il33 was expressed in Prox1 + granule cells, but not PV+ interneurons and rarely in Doublecortin+ immature granule cells (Figures S1C and S1D). We did not detect neuronal Il33 expression in the entorhinal cortex, amygdala, or thalamus, although some neurons in layer 2/3 of the prefrontal cortex also expressed Il33 (Figure S1E). These results were validated by immunostaining for IL-33 protein, which demonstrated neuronal expression that was abrogated by conditional deletion with a neuron-specific Syn1Cre recombinase line (Figure 1E; Figure S1F).
Figure 1. Experience-Dependent Regulation of IL-33 Expression in Adult Hippocampal Neurons.
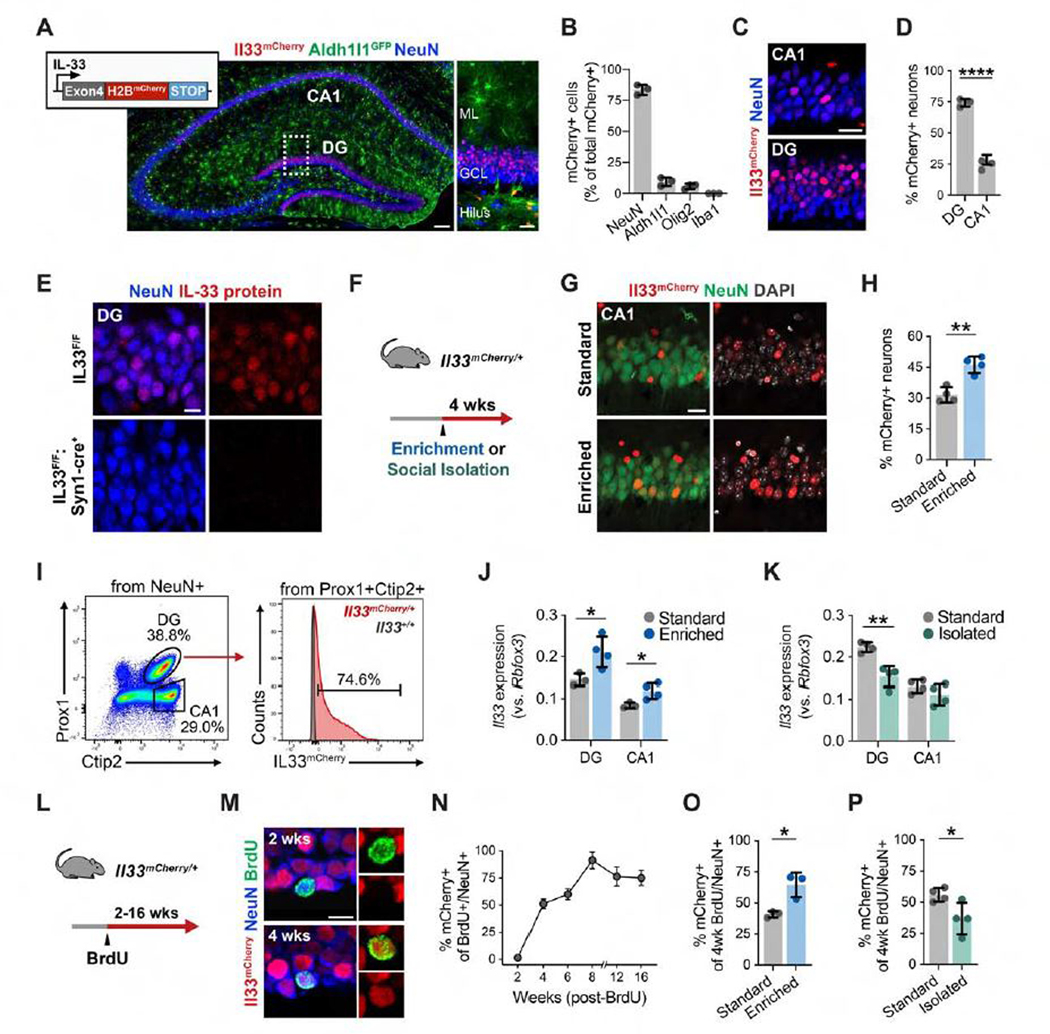
(A and B) Post-natal day 30 (P30) hippocampus showing Il33mCherry, Aldh1l1eGFP (astrocytes), and NeuN (neurons). Inset, DG. Also shown is quantification of Il33-mCherry+ cells labeled with cell-type-specific markers. Scale bars, 100 μm and 20 μm (inset). ML, molecular layer; GCL, granule cell layer.
(C and D) Il33-mCherry+ neurons in the DG and CA1. Scale bar, 20 μm.
(E) IL-33 protein in DG neurons in the control (Il33fl/f) or after neuron-specific excision of Il33 (Il33fl/fl: Synapsin1-cre). Scale bar, 10 μm.
(F) Schematic of environmental enrichment (EE) and social isolation (SI).
(G and H) Image and quantification of the CA1 from Il33mCherry/+ mice under standard or EE conditions. Scale bar, 20 μm.
(I) Fluorescence-activated cell sorting (FACS) gating of Il33-mCherry+ neurons from the DG and CA1.
(J and K) Effect of an EE or SI on Il33 expression by qPCR.
(L) Schematic of newborn neuron labeling.
(M and N) Image and percentage of Il33-mCherry+ newborn neurons at the indicated time points after BrdU injection (n = 3 mice/time point; >20 neurons/mouse). Injected at 4 weeks of age. Scale bar, 10 μm.
(O) Effect of an EE on the percentage of Il33-mCherry+ newborn neurons; BrdU injection and EE at 12 weeks, analysis at 16 weeks.
(P) Effect of SI on the percentage of Il33-mCherry+ newborn neurons; BrdU injection and SI at 4 weeks, analysis at 8 weeks.
Statistics: two-tailed unpaired t tests. Dots represent individual mice. Data are mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S1.
To determine whether Il33 expression is regulated by experience, 3month-old Il33mCherry/+ mice were placed in an enriched environment (EE), which promotes circuit remodeling and neurogenesis (Figure 1F; Bednarek and Caroni, 2011; Bergami et al., 2015; Kempermann et al., 1997). We observed that 4 weeks of EE led to a significant increase in the proportion of Il33+ CA1 neurons (Figures 1G and 1H). To examine this further, we isolated neuronal nuclei by flow cytometry from DG granule cells (Prox1+Ctip2+) and CA1 pyramidal neurons (Prox1+Ctip2−; Figure 1I; Jaeger et al., 2018). The EE increased Il33 mRNA expression in the DG and CA1 (Figure 1J) and increased the percentage of Il33+ neurons in the CA1 over a period of 14 days (Figures S1G–S1I). In contrast, social isolation (SI) reduces hippocampal remodeling and impairs neurogenesis (Dranovsky et al., 2011; Ibi et al., 2008). We found that 4 weeks of SI significantly reduced Il33 mRNA expression and the percentage of Il33+ neurons in the DG (Figure 1K; Figure S1J). In the CA2, ~5% of neurons were Il33+, and this increased with an EE (Figures S1K–S1L).
Neurogenesis and integration of newborn neurons in the DG are also modulated by experience (Alvarez et al., 2016; Bergami et al., 2015; Kempermann et al., 1997). We labeled newborn neurons by bromodeoxyuridine (BrdU) injection into 1-month-old Il33mCherry/+ mice and found that Il33 was induced in newborn neurons between 2–4 weeks of age (Figures 1L–1N). Additionally, an EE for adult mice increased the percentage of Il33+ newborn neurons (Figure 1O), whereas SI decreased it (Figure 1P). These results demonstrate that Il33 expression in hippocampal neurons is bidirectionally modulated by experience.
Il33 Expression Identifies a Neuronal Subset Primed for Synaptic Plasticity
Memory encoding and consolidation are circuit-specific and competitive processes (Josselyn and Tonegawa, 2020). We observed that DG neurons expressed heterogeneous levels of IL-33 (Figure 1E). To determine whether this heterogeneity had functional correlates, we performed single-nucleus RNA sequencing of hippocampal neuronal nuclei (Figures 2A–2F; Figures S2A–S2E). Unbiased clustering revealed cell subsets corresponding to DG granule cells (expressing Proxl, Stxbp6, and Dock10) as well as other major neuron classes (Figure 2A; Figures S2D and S2E). The DG population further clustered into two subsets, clusters 0 and 2 (Figure 2B). To identify Il33+ subsets, we also sequenced flow-sorted Il33mCherry+ neurons and included these metadata, revealing that the majority of mCherry+ neurons overlapped with DG cluster 2 (Figures 2C and 2D; Figures S2F and 2G; STAR Methods).
Figure 2. IL-33 Expression Identifies a Neuronal Subset Primed for Synaptic Plasticity.
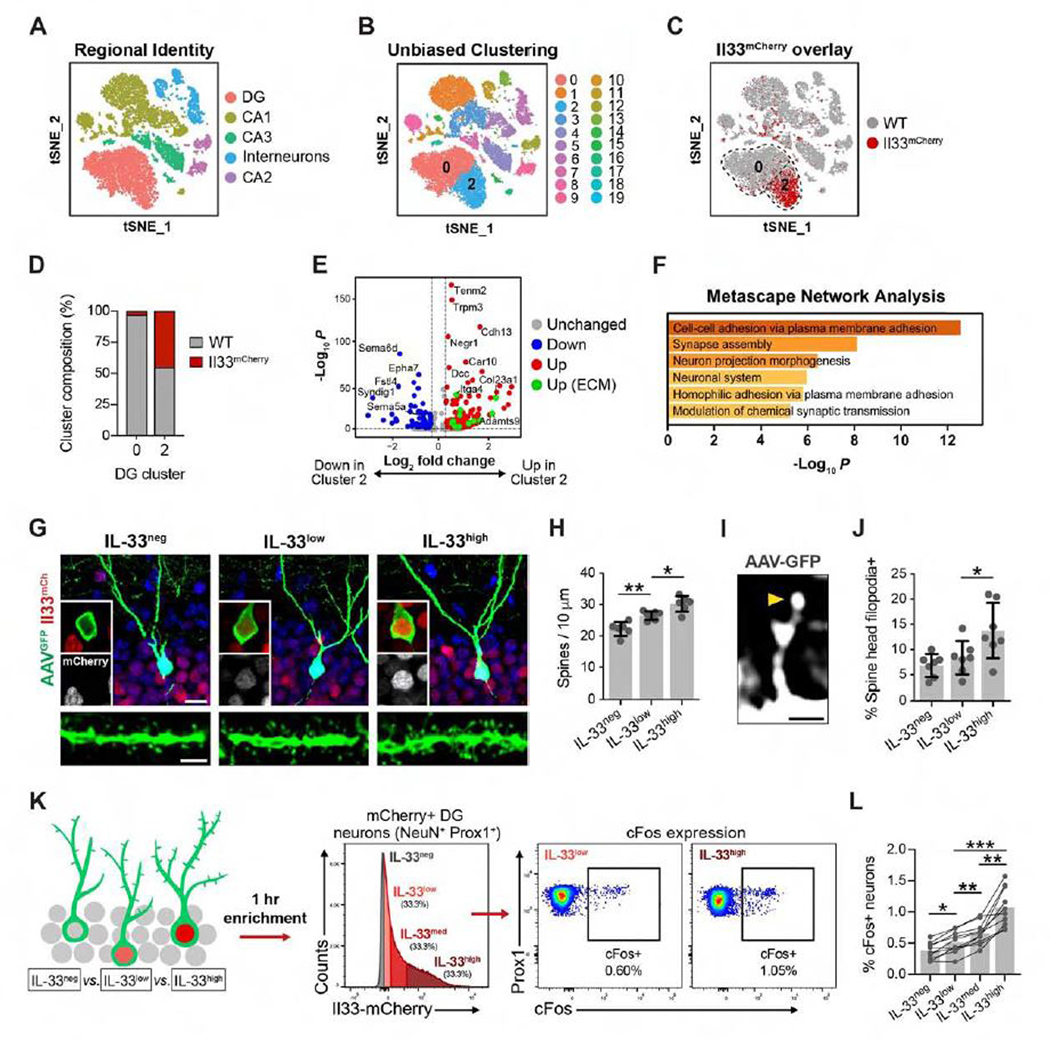
(A) t stochastic nearest neighbor embedding (tSNE) of 10,800 NeuN+ nuclei labeled by cell type.
(B) Unsupervised clustering of (A).
(C) Wild-type (WT) nuclei (gray) overlaid with Il33mCherry nuclei (red).
(D) WT and Il33mCherry composition in DG clusters 0 and 2.
(E) Volcano plot of differentially expressed genes between DG clusters 0 and 2 (p < 0.001). Green, ECM and ECM-associated genes (Naba et al., 2016).
(F) Top Gene Ontology terms for upregulated genes in (E).
(G) DG granule cells sparsely labeled with AAV9-Syn1-GFPfrom Il33mCherry/+ mice showing IL-33-negative, -low, and -high subsets. Scale bars, 10 μm (top) and 2 μm (bottom).
(H) Dendritic spine density in neuronal subsets (ANOVA, p < 0.0001, Tukey’s post hoc test, n = 6–7 neurons/group, 3 mice).
(I and J) Percentage of spines with spine head filopodia (ANOVA, p = 0.0096, Tukey’s post hoc test, n = 6–7 neurons/group, 3 mice). Arrowhead, spine head filopodia. Scale bar, 1 μm.
(K) Schematic of neuronal nucleus isolation by mCherry intensity.
(L) Percentage of cFos+ cells in each subset (repeated-measures ANOVA, p < 0.0001, Tukey’s post hoc test, n = 10 mice).
*p < 0.05, **p < 0.01, ***p < 0.001. Data in (G)–(L) are mean ± SD. See also Figure S2 and Table S1.
We next ranked differentially expressed genes between these subsets (Figures 2E and 2F; Table S1; Finak et al., 2015). Il33-enriched cluster 2 neurons expressed genes linked to cell adhesion, synapse assembly, and synaptic plasticity, including cadherins (Cdh10 and Cdh13; Basu et al., 2017; Duan et al., 2018). We also found enriched expression of genes related to assembly and remodeling of the ECM, including Adamts9, Itga4, Col23a1, and Col25a1. We crossreferenced our data with a curated dataset of matrix-associated genes (Naba et al., 2016; Figure 2E, green dots) and identified 18 genes encoding ECM-associated proteins. Thus, Il33 expression defines a molecular subtype enriched for markers of synapse formation and ECM remodeling.
We next investigated whether Il33 expression was associated with structural and functional correlates of synaptic plasticity. In the DG, granule cells recruited to encode a memory undergo a period of enhanced plasticity and spine growth that is required for proper memory consolidation (Kitamura et al., 2017; Roy et al., 2017; Ryan et al., 2015). We quantified dendritic spines in sparsely labeled mature DG granule cells in Il33mCherry/+ mice and compared neurons with different levels of Il33 expression (Figure 2G). We found that neurons with high mCherry had increased dendritic spine density compared with other subsets, as analyzed by binning (Figure 2H) or by Pearson correlation (Figure S2H). We also quantified spine head filopodia as structural markers of spine plasticity. These protrusions form prior to new spine formation or spine relocation and have been observed in the olfactory bulb and CA1 regions (Breton-Provencher et al., 2016; Orlando et al., 2012; Richards et al., 2005; Weinhard et al., 2018). We found that DG neurons with high mCherry levels had a significantly greater proportion of spines harboring spine head filopodia (Figures 2I and 2J; Figure S2I).
The expression of immediate-early genes such as Cfos and Arc identifies neuronal ensembles recruited during encoding of contextual memories (Denny et al., 2014; Liu et al., 2012). These neurons are allocated to a memory trace in a competitive process based on their relative excitability (Cai et al., 2016; Han et al., 2007;Park et al., 2016). To determine whether Il33-high neurons were more likely to be activated in response to learning, we placed animals in a novel environment for 1 h. We found that neuronal subsets with higher mCherry levels were more likely to express c-Fos than other subsets (Figures 2K and 2L), suggesting that these subsets may be functionally distinct. However, the total number of c-Fos+ granule cells did not differ in IL-33 cKO mice (Figure S2J), indicating that IL-33 does not control the size of an engram population. Rather, this could indicate that neurons with high levels of Il33 and spine remodeling represent a population more likely to engage in response to novel experiences. Altogether, our data suggest that Il33 expression is associated with a subset of neurons more likely to undergo synaptic remodeling.
Neuron-Microglial Signaling via IL-33 Drives Experience-Dependent Spine Remodeling
To identify cellular targets of IL-33 signaling, we quantified expression of Il1rl1, which encodes its obligate co-receptor (Molofsky et al., 2015; Palmer and Gabay, 2011). Microglia were the predominant cell type expressing Il1rl1 in the hippocampus, as assessed by qPCR of cell subsets isolated by flow cytometry (Figures S3A and S3B) and consistent with other regions of the CNS (Vainchtein et al., 2018). Quantitative in situ hybridization supported these results, although we also identified a small proportion of Illrll+NeuN+ cells in the hilus (Figures 3A and 3B). Genetic deletion of Il1rl1 with a myeloid-specific inducible Cre recombinase line (Il1rl1flox/flox:Cx3cr1CreER/+, “IL1RL1 i-cKO”) led to a 93% ± 6% reduction in Il1rl1 expression in microglia (Figures S3C and S3D). Thus, microglia are competent to respond to IL-33 signaling.
Figure 3. Neuron-Microglia Signaling via IL-33 Drives Experience-Dependent Spine Remodeling.
(A and B) ll1rl1 transcript co-labeled with antibodies to cell-type-specific markers in the DG (n = 4 mice). Scale bars, 25 μm and 5 μm (inset).
(C and D) Dendritic spine density in DG granule cells sparsely labeled with AAV9-Syn1-GFP in IL-33 cKO versus littermate controls (nested t test, n = 36 dendritic segments, 4 mice/genotype). Scale bars, 20 μm and 2 μm (spine inset).
(E) Spine density in IL1RL1 i-cKO versus littermate controls (nested t test, n = 27 dendritic segments, 4 mice/genotype).
(F-H) Percentage of spines with spine head filopodia in IL-33 cKO and IL1RL1 i-cKO versus littermate controls (arrowheads, spine head filopodia; statistics as in D and E). Scale bars, 1 μm and 0.5 μm (inset).
(I and J) Percentage of spines with spine head filopodia under standard or enriched conditions in IL1RL1 cKO mice versus littermate controls (2-way ANOVA, Tukey’s post hoc test, n = 18–24 dendritic segments, 3–4 mice/group).
(K-M) Frequency and amplitude of mEPSCs from IL-33 cKO mice and littermate controls (nested t test, n = 12–14 neurons, 3–4 mice/genotype).
(N) Schematic of IL-33ΔNLS viral gain-of-function strategy.
(O and P) Spine density after injection of control (tdTomato) or IL-33ΔNLSvirus into control orIL1RL1 i-cKO mice(one-way ANOVA, Tukey’s post hoc test, n = 29–33 dendritic segments, 3 mice/group). Scale bars, 20 μm and 1 μm (inset).
(Q) Percentage of spine head filopodia (sample sizes and statistics as in P).
*p < 0.05, **p < 0.01, ***p < 0.001. Data are mean ± SD (bar graphs) and median ± interquartile range (violin plots). Larger dots to the right of each plot indicate mean per individual mouse. See also Figure S3 and Table S2.
Given the strong expression of markers linked to synapse formation in Il33-enriched neurons, we next investigated whether neuronal IL-33 or microglial IL1RL1 affected dendritic spine numbers. We quantified dendritic morphology and spine density in mice with neuron-specific deletion of Il33 (Il33ffox/ffox:Syn1Cre, “IL-33 cKO”) using AAV9-Syn1-GFP to sparsely label hippocampal neurons (Figure 3C). We found that spine density was significantly decreased in IL-33 cKO in the DG and CA1 regions (Figure 3D; Figure S3E), although dendritic branching was largely preserved (Figure S3F). Conditional deletion of Il1rl1 in microglia using IL1RL1 i-cKO mice phenocopied this decreased spine density (Figure 3E). The numbers of microglia were not altered in these mice (Figure S3G), but constitutive deletion of Il1rl1 (Il1rl1 flox/flox:Cx3cr1 Cre/+, “IL1RL1 cKO”) led to altered microglial morphology and process coverage (Figure S3H), suggesting alterations in microglial function.
Microglial contact has been correlated with formation of new dendritic spines and induction of spine head filopodia (Miyamoto et al., 2016; Weinhard et al., 2018). We found that loss of neuronal IL-33 or microglial IL1RL1 resuted in fewer spines with spine head filopodia (Figures 3F–3H). Enrichment increased the number of spine head filopodia in control mice, whereas IL1RL1 cKO limited this increase (Figures 3I and 3J). To determine the effect of these spine alterations on synaptic function, we performed whole-cell patch-clamp recordings of DG granule cells from IL-33 cKO animals and littermate controls. We found a reduction in the frequency of miniature excitatory postsynaptic currents (mEPSCs) in IL-33 cKO mice and a trend toward increased amplitude of these mEPSCs (Figures 3K–3M; Table S2). These data are consistent with our finding of fewer dendritic spines. Taken together, these data suggest that neuron-microglia signaling via IL-33 promotes new spine formation and plasticity.
To determine whether IL-33 signaling through microglia is sufficient to drive spine formation, we next used a gain-of-function approach. To drive constitutive release of IL-33, we removed the N-terminal chromatin-binding domain containing the nuclear localization signal (NLS) and replaced it with a hemagglutinin (HA) epitope tag (Bessa et al., 2014). We then used lentiviral delivery to express this modified protein (IL-33ΔNLS-P2A-tdTomato) or a control (tdTomato) in DG neurons using a Ca2+/calmodulin-dependent protein kinase II (CaMKII) promoter (Figure 3N). We detected cytoplasmic and dendritic HAtagged IL-33 protein, indicating that the construct was functioning as expected (Figure S3I). We found that IL-33ΔNLS was sufficient to increase dendritic spine numbers and spine head filopodia and that these effects were abrogated in IL1RL1 i-cKO mice, indicating that microglial sensing of IL-33 was required (Figures 3O–3Q). These results suggest that IL-33 acts locally to increase dendritic spines, although non-cell-autonomous effects of IL-33 release on nearby neurons are also possible. Taken together, these data indicate that IL33 release from neurons is sufficient to promote dendritic spine formation and plasticity.
Neuron-Microglia Signaling through IL-33 Promotes Experience-Dependent Increases in Newborn Neurons and Is Required for Remote Memory Precision
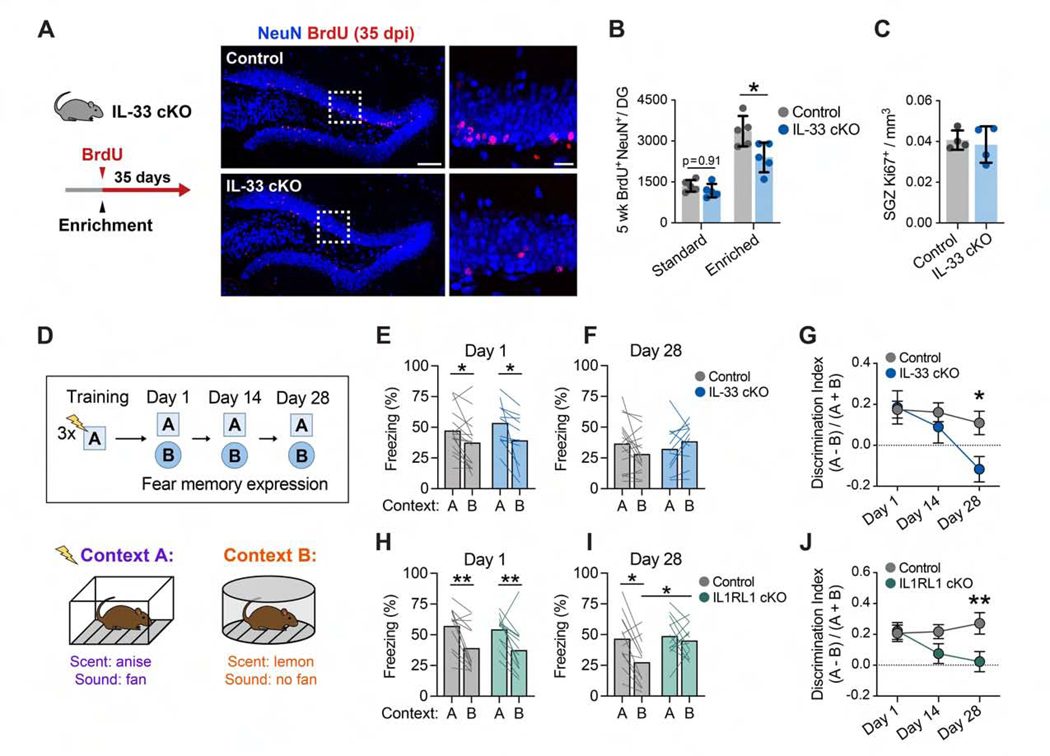
Integration of newborn neurons in the DG requires circuit remodeling and is increased in EEs, suggesting that this process is regulated by experience (Bergami et al., 2015; Kheirbek et al., 2012; Toni et al., 2007). We tested whether neuron-microglia signaling via IL-33 affects newborn neurons by pulse-chase labeling with BrdU and quantification of label-retaining NeuN+ neurons 5 weeks later (Figure 4A; Figure S4A). Although there was no difference in the number of newborn neurons in IL-33 cKO mice at baseline, mice housed in an EE for 5 weeks had significantly fewer newborn neurons compared with controls (Figure 4B). We did not detect differences in the number of Ki67+ proliferating cells in the subgranular zone (Figure 4C). Additionally, we found reduced dendritic spine density in newborn neurons of Il33−/− mice (Figure S4B), indicating that synaptic integration of newborn neurons might be delayed or impaired. These results suggest that neuronal IL-33 is required for experience-dependent increases in newborn neurons.
Figure 4. Neuron-Microglia Signaling through IL-33 Promotes Experience-Dependent Increases in Newborn Neurons and Is Required for Remote Memory Precision.
(A and B) Newborn neuron quantification in IL-33 cKO mice versus controls in standard or enriched housing (n = 5 mice/group, 2-way ANOVA, Tukey’s post hoc test). Scale bars, 100 μm and 20 μm (inset).
(C) Ki67+ proliferating cells in the subgranular zone (SGZ).
(D) Schematic of the contextual fear discrimination assay.
(E-G) Freezing in the conditioned fear context A versus unconditioned context B in control and IL-33 cKO animals. (G) shows the same data quantified as a memory discrimination index and includes a 14-daytime point (n = 16 control and 11 IL-33 cKO mice, 2-way repeated measures [RM] ANOVA, Sidak’s post hoc tests; Figure S4C).
(H-J) Freezing in context A versus context B in control and IL1RL1 cKO animals (n = 12 mice/genotype, 2-way RM ANOVA, Sidak’s post hoc tests; Figure S4D).
*p < 0.05, **p < 0.01. Data are mean ± SD (B and C) and mean ± SEM (G and J). See also Figure S4.
The decreased number of newborn neurons and impaired spine plasticity prompted us to investigate whether the IL-33 pathway is involved in hippocampus-dependent memory. The DG is critical for pattern separation, a process that separates similar experiences into non-overlapping representations in the hippocampus (Miller and Sahay, 2019). Synaptic plasticity in mature granule cells plays an important role in this process (McHugh et al., 2007; Sun et al., 2020), as does newborn neuron function (Danielson et al., 2016; Kheirbek et al., 2012; Nakashiba et al., 2012; Sahay et al., 2011). We used a contextual fear discrimination assay to assess memory and pattern separation. Mice were conditioned to a context with 3 footshocks (context A) and then exposed to the conditioned context followed by an unconditioned but partially similar context (context B). We then quantified their ability to discriminate between the two contexts 1,14, and 28 days post-training (Figure 4D; McAvoy et al., 2016). We first tested animals with neuronal deletion of Il33, which preferentially targeted the hippocampus and prefrontal cortex, because neuronal IL-33 was not detected in other regions relevant to contextual fear memory, including the entorhinal cortex, amygdala, and nucleus reuniens (Figure S1E; Josselyn and Tonegawa, 2020; Ramanathan et al., 2018).
Immediately after training, IL-33 cKO animals had intact memory recall and context discrimination (Figure 4E). However, we observed a progressive decrement in the ability of IL-33 cKO mice to discriminate contexts emerging between 14 and 28 days post-training relative to controls (Figures 4F and 4G; Figure S4C). This was largely driven by an increase in freezing behavior in the unconditioned context, suggesting generalization of the fear memory. These deficits were phenocopied in animals with constitutive deletion of Il1rl1 in microglia (Figures 4H–4J; Figure S4D). We did not detect differences in anxiety-related behaviors in the open field or elevated plus maze tests in the IL-33 cKO or IL1RL1 cKO cohorts (Figures S4E–S4H). Collectively, our observations indicate that neuron-microglia communication through IL-33 is required for memory precision at remote time points, suggesting a role in memory consolidation.
IL-33 Expression Decreases in the Aged Hippocampus and Is Associated with Decreased Memory Precision and Spine Plasticity
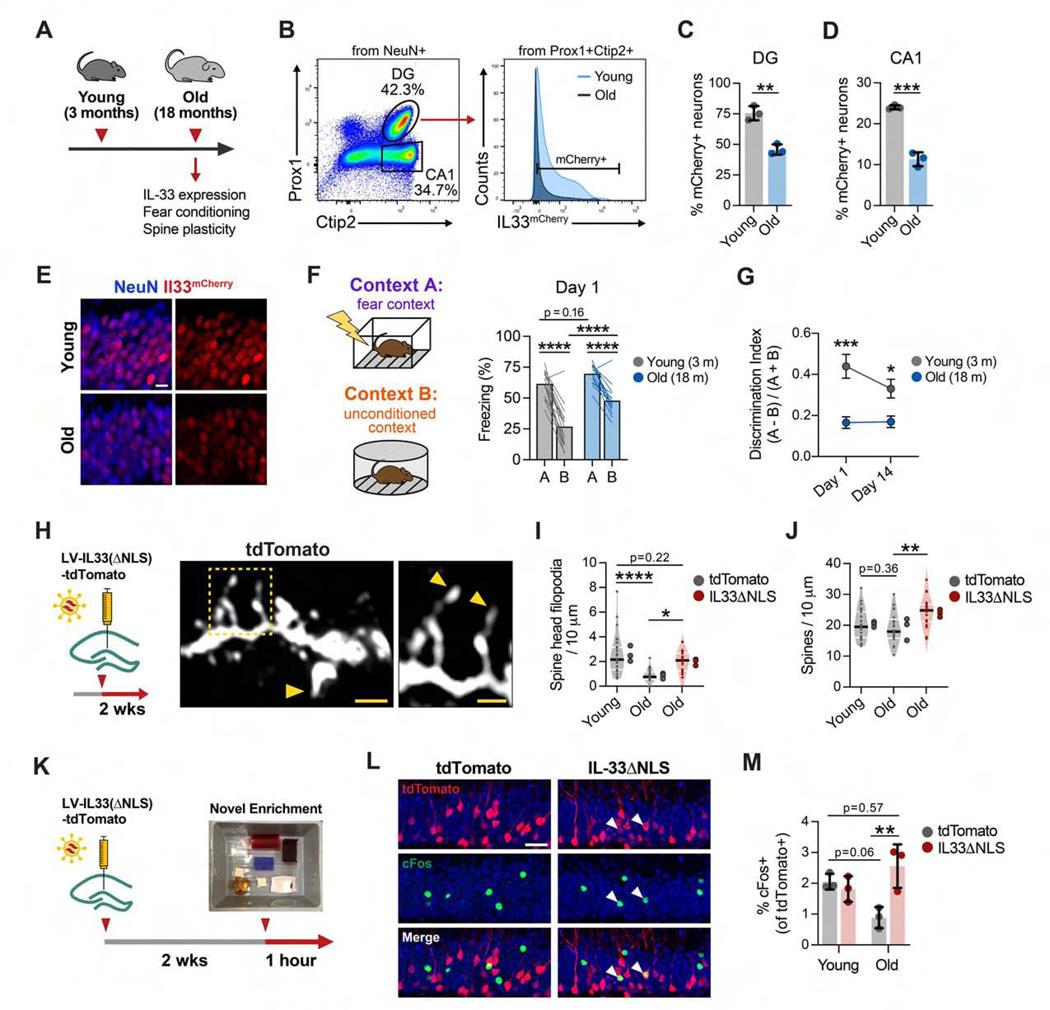
Decreased synaptic plasticity and memory impairment are among the phenotypes associated with physiological aging (Wyss-Coray, 2016), which led us to investigate whether this pathway might be modulated with age. We observed a significant decline in the number of Il33+ neurons in the DG and CA1 of 18-month-old mice (Figures 5A–5E; Figure S5A). To determine whether aging might affect memory precision, we quantified contextual fear learning and memory precision in young (3-month-old) versus aged (18-month-old) mice, using the same fear conditioning paradigm as in Figure 4. We found that, after training, aged mice recognized the conditioned context A similarly as young mice but had impaired context discrimination and overgeneralized to the unconditioned context B (Figures 5F and 5G; Figures S5B and S5C). The motor response to shock did not differ between young and old mice, suggesting that differences in freezing were not a result of altered shock sensitivity (Figure S5D). Thus, like IL33 cKO animals, aged mice showed reduced memory precision, although this deficit was already evident 1 day after training and persisted at a remote time point (14 days; Figure 5G). Context discrimination in this memory task tended to correlate with Il33 mRNA levels in DG granule cells (Figure S5E).
Figure 5. IL-33 Expression Decreases in the Aged Hippocampus and Is Associated with Decreased Memory Precision and Spine Plasticity.
(A) Overview schematic.
(B–D) Quantification of the percentage of mCherry+ neurons, using flow cytometry, in the DG and CA1 of young and old mice (t test, n = 3 mice/group).
(E) Representative image of Il33-mCherry in the DG of young and old Il33mCherry/+ mice. Scale bar, 10 μm.
(F) Contextual fear discrimination 1 day post-training in young and old mice(n = 16young and 14 old mice, 2-way RM ANOVA, Sidak’s post hoc tests; FigureS5C).
(G) Discrimination indices 1 and 14 days post-training (statistics as in F; Figure S5C).
(H–J) Number of spine head filopodia (I) and total spines (J) after injections with control or IL-33 gain-of-function virus(one-way ANOVA, Tukey’s post hoc test, n = 20–27 dendritic segments, 3 animals/group). Scale bars, 2 μm and 1 μm (inset).
(K) Schematic of the neuronal activation assay after IL-33 gain of function.
(L and M) Percentage of virally labeled DG neurons that were cFos+ after 1 h i n a novel environment (one-way ANOVA, Tukey’s post hoc test, 3 mice/group). Scale bar, 20 μm.
*p < 0.05, **p < 0.01, ****p < 0.0001. Data are mean ± SD (bar graphs) and median ± interquartile range (violin plots). Larger dots to the right of violin plots indicate mean per individual mouse. See also Figure S5.
These data suggested the possibility that at least some features of cognitive aging could be affected by augmenting IL-33 signaling. Increased dendritic spine plasticity and neuronal activity are associated with improved cognition in the aged brain (Cai et al., 2016; Villeda et al., 2014) and in mouse models of Alzheimer’s disease (Roy et al., 2016). To test the effect of IL-33 on dendritic spines in the aged brain, we used the lentiviral gain-of-function approach described in Figure 3 (IL-33ΔNLS). We observed that aging was associated with a substantial decrease in the number of spines with spine head filopodia and that IL-33ΔNLS in aged mice increased the number of spine head filopodia (Figures 5H and 5I). This increase was at least partly driven by a supraphysiological increase in total spines (Figure 5J), which were not significantly decreased with age, consistent with findings by other groups (Fan et al., 2017). Aging is also associated with decreased neuronal excitability (Oh et al., 2010), and age-related deficits in cognitive flexibility are ameliorated by increasing neuronal excitability and activation (Cai et al., 2016; Villeda etal., 2014). Consistent with prior studies, we found that neuronal activation decreased with age in the DG, as measured by the percentage of labeled neurons that were c-Fos+ after exposure to a novel environment (Figures 5K–5M). IL-33ΔNLS did not affect this metric in young mice but was sufficient to increase c-Fos+ neurons in old mice to the levels observed in young animals (Figure 5M). These data suggest that aging is a physiologic context associated with decreased Il33 expression and impaired memory precision and that increasing neuronal IL-33 release in individual neurons improves some structural and functional correlates of brain aging.
Neuronal IL-33 Drives Microglial Engulfment of the ECM
The deficient spine numbers after loss of IL-33-IL1RL1 signaling indicated a role of IL-33 in synapse formation or maintenance and a requirement for microglia in this process. To interrogate potential mechanisms by which IL-33 affects microglia, we transcriptionally profiled hippocampal microglia after 4 h of in vivo exposure to IL-33 (Figure 6A; Figure S6A). As expected, we identified a strong activation signature after IL-33 treatment, including Il1b, Tnf, and multiple chemokines (Figure 6B). Among the top upregulated genes were the class A scavenger receptor Marco, a regulator of filopodial morphogenesis and debris clearance (van der Laan et al., 1999; Pikkarainen et al., 1999), and the adaptor protein Gas7, which is necessary for phagocytic cup formation in macrophages (Hanawa-Suetsugu et al., 2019). These findings are consistent with IL-33’s role in promoting microglial phagocytosis (Fu et al., 2016; Vainchtein et al., 2018).
Figure 6. Neuronal IL-33 Drives Microglial Engulfment of the ECM.
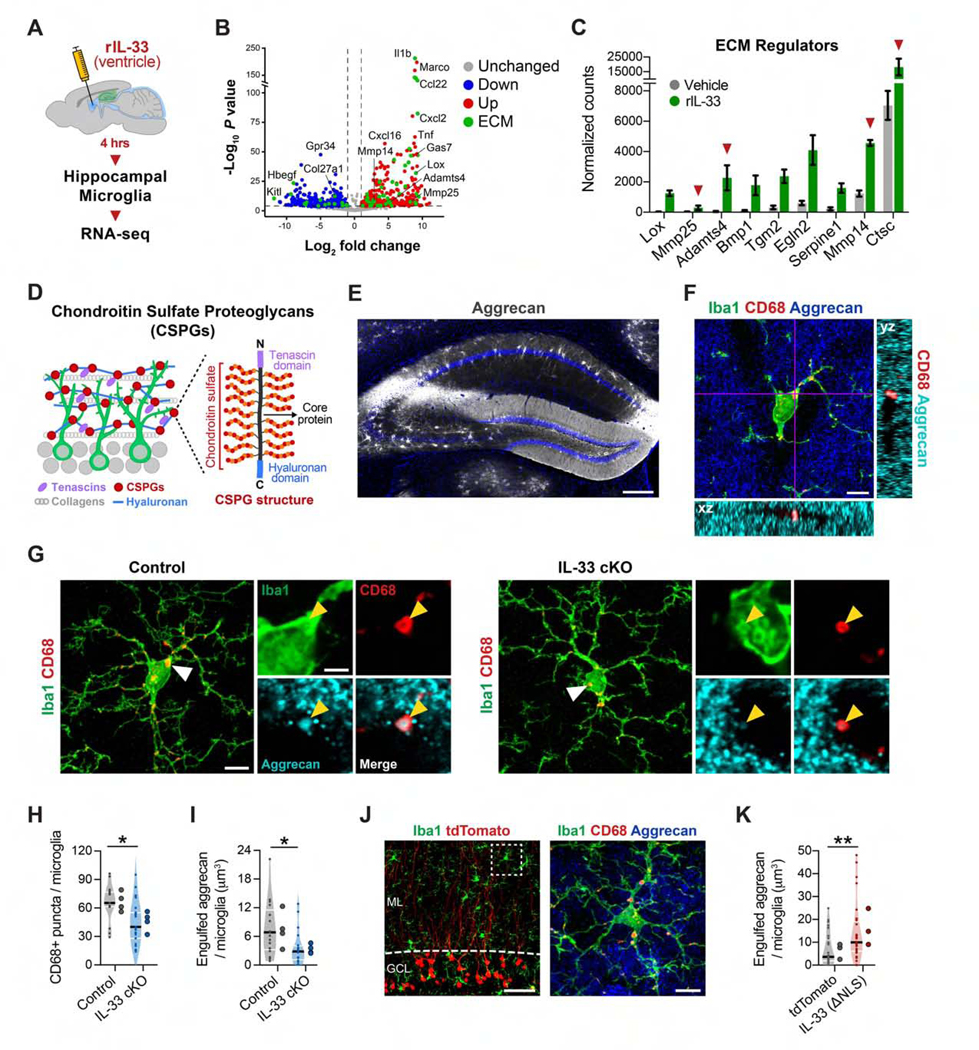
(A) Experimental overview of microglial transcriptomics.
(B) Volcano plot of differentially expressed genes after IL-33 or PBS treatment (PAdj < 0.01). Green, ECM and ECM-associated genes (Naba et al., 2016).
(C) Genes in (B) classified as ECM regulators. Red arrows, ECM proteases.
(D) Schematic of the ECM in the DG.
(E)Aggrecan staining in the hippocampus. Scale bar, 250 μm.
(F) z stack of Iba1+ microglia in the DG co-labeled with Aggrecan and CD68+ lysosomes. Scale bar, 5 μm.
(G-I) CD68+ lysosomes within microglia (H) and Aggrecan protein within microglial lysosomes (I) (nested t test, n = 17–20 microglia, 4 mice/genotype). Scale bat, 5 μm and 2 μm (inset).
(J and K) Aggrecan in microglial lysosomes 2 weeks after infection with a control or IL-33ΔNLS virus (t test, 28 microglia, 3 mice/group). Scale bars, 25 μm and 2 μm (inset).
*p < 0.05, **p < 0.01. Data are mean ± SEM (bar graphs) and median ± interquartile range (violin plots). Larger dots to the right of violin plots indicate mean per individual mouse. See also Figure S6 and Table S3.
We next investigated potential pathways relevant to spine formation and plasticity. Outside of the brain, IL-33 promotes tissue remodeling, which includes turnover of the ECM (Bonnans et al., 2014; Molofsky et al., 2015). Live imaging studies suggest that the brain ECM is dynamic and responsive to synaptic cues (T0nnesen et al., 2018), and multiple studies have shown that ECM proteins can restrict synaptic plasticity and spine remodeling (Bolos et al., 2018; Frischknecht et al., 2009; Oray et al., 2004; Orlando et al., 2012; Pizzorusso et al., 2006; de Vivo et al., 2013). We found that 76 genes differentially expressed in microglia after IL-33 treatment were associated with the ECM ((Naba et al., 2016; Figure 6B, green dots). Of these, we identified 9 genes classified as ECM regulators that were significantly upregulated after IL-33 treatment (Figure 6C). These included several ECM proteases (Adamts4, Mmp14, Mmp25, and Ctsc). Interestingly, Adamts4 and Mmp14 encode membrane-localized metalloproteases that predominantly cleave chondroitin sulfate proteoglycans (CSPGs), which are a major class of ECM proteins in the brain (Figure 6D; Barbolina and Stack, 2008; Kelwick etal., 2015; Levy etal., 2014).
Aggrecan is a CSPG expressed prominently in perineuronal nets throughout the brain but is also highly enriched in the diffuse ECM of the DG (Figures 6D and 6E; Figures S6B and S6C; Bolos et al., 2018; Rowlands et al., 2018). We therefore investigated whether microglia can engulf Aggrecan. Using superresolution imaging and 3D reconstruction, we identified Aggrecan immunostaining within microglia and found that it localized to CD68+ lysosomes (Figure 6F). In microglia from IL-33 cKO mice, we found decreased numbers of CD68+ lysosomes compared with controls, indicating that IL-33 promotes a phagocytic phenotype (Figures 6G and 6H). Additionally, we also found a 54% reduction in the amount of engulfed Aggrecan in microglia from IL-33 cKO mice compared with controls (Figure 6I). Conversely, IL-33 gain of function was sufficient to increase microglial process coverage, the phagocytic index, and engulfment of Aggrecan (Figures 6J and 6K; Figures S6D–S6F). In summary, we show that neuron-derived IL-33 promotes microglial phagocytosis and engulfment of the CSPG Aggrecan.
IL-33 Deficiency Leads to Accumulation of Perisynaptic ECM
To determine whether microglial remodeling of the ECM could affect accumulation of ECM near synapses, we analyzed Aggrecan distribution in the hippocampus. Using superresolution imaging, we found that Aggrecan in the DG molecular layer formed punctate structures that colocalized with the postsynaptic density protein Homer1. IL-33 cKO mice had a significant increase in the density of Aggrecan puncta compared with littermate controls as well as increased colocalization of Aggrecan with Homer1 (Figures 7A–7D). Brevican is another CSPG prevalent in the adult brain that can restrict synaptic plasticity (Favuzzi et al., 2017; Frischknecht et al., 2009) and is proteolytically cleaved into a smaller 53-kDa fragment by ADAMTS4 (Mayer et al., 2005; Valenzuela et al., 2014); Figure 7E). We found that IL-33 cKO mice had significantly more total Brevican and a decrease in the levels of proteolyzed Brevican compared with controls (Figure 7F–7H).
Figure 7. IL-33 Deficiency Leads to Accumulation of Perisynaptic ECM.
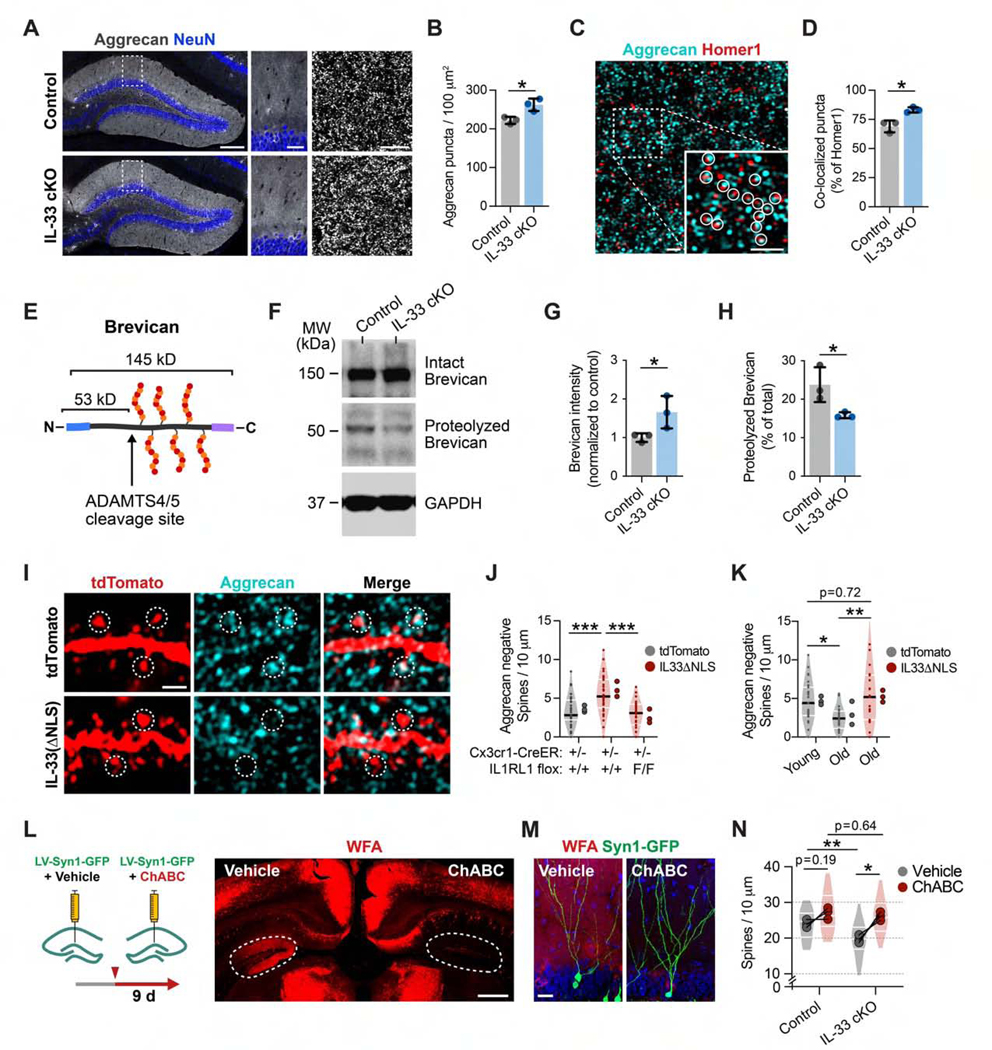
(A and B) Quantification of Aggrecan punctum density in control and IL-33 cKO mice. Insets, DG ML (t test, n = 3 mice/genotype). Scale bars, 200 μm (left), 40 μm (center), 5 μm (right).
(C and D) Aggrecan and Homerl immunostaining and co-localization in the ML (circles, co-localized puncta; t test, n = 3 mice/genotype). Scale bars, 1 μm and 1 μm (inset).
(E) Brevican CSPG and proteolytic cleavage site.
(F-H) Western blot of Brevican from hippocampal lysate in IL-33 cKO and littermate controls, including uncleaved (145 kD, G) and cleaved (53 kD, H) forms (t test, n = 3 mice/genotype).
(I and J) Aggrecan co-localization with dendritic spines in the DG injected with control (tdTomato) orIL-33 gain-of-function (IL-33ΔNLS) virus in control or IL1RL1 i-cKO mice (one-way ANOVA, Tukey’s post hoc test, n = 29–33 dendritic segments, 3 mice/group). Scale bar, 1 μm.
(K) Aggrecan co-localization with spines in young and old mice injected with a control or IL-33ΔNLS virus (one-way ANOVA, Tukey’s post hoc test, n = 19–27 dendritic segments, 3 mice/group).
(L and M) Schematic and representative images of ChABC digestion imaged with Wisteria floribunda agglutinin (WFA) staining (dashed circles, DG). Scale bars, 500 μm (L) and 20 μm (M).
(N) Dendritic spine density in IL-33 cKO or control mice injected with ChABC or vehicle (2-way RM ANOVA, Sidak’s post hoc test, n = 3 mice/genotype; dots, means per mouse; lines connect vehicle- and ChABC-injected hemispheres; violin plots show distribution of dendritic segments per group, 23–30 segments).
*p < 0.05, **p < 0.01, ***p < 0.001. Data are mean ± SD (bar graphs) and median ± interquartile range (violin plots). Larger dots to the right of violin plots indicate means per mouse.
We next used IL-33 gain of function to further examine the relationship between Aggrecan and dendritic spines. We found that IL33ΔNLS increased the number of spines devoid of Aggrecan contact and that this effect required microglial expression of the IL-33 receptor IL1RL1 (Figures 7I and 7J). In aged animals, we found a 2-fold decrease in the number of spines devoid of perisynaptic Aggrecan, consistent with studies that have found marked accumulation of ECM proteins with age (Végh et al., 2014a). IL-33ΔNLS increased the number of Aggrecan-negative spines to levels seen in young mice (Figure 7K). These data indicate that IL-33 promotes ECM remodeling and restricts ECM deposition near synapses.
Finally, we sought to determine the relationship between IL-33’s effect on ECM and dendritic spine maintenance. We injected the enzyme Chondroitinase ABC (ChABC) into the DG, which compromises ECM integrity by degrading the glycosaminoglycan side chains present on CSPGs (Figure 7L). We found that, 9 days after ChABC injection, spine density was modestly increased in controls but significantly increased in IL-33 cKO mice to the levels of ChABC-treated controls (Figures 7M and 7N). These data support the hypothesis that IL-33’s effects on spines are mediated at least in part through its effect on the ECM. Taken together, these findings directly implicate microglia as regulators of ECM clearance and spine formation.
DISCUSSION
This study defines a cytokine-mediated mechanism that acts via microglia to promote dendritic spine plasticity and precise retrieval of remote memories. We find that neurons determine microglial responses by experience-dependent regulation of IL-33, coupling neuronal state to microglial activation. Finally, we demonstrate that IL33 drives microglial engulfment of the ECM, revealing a molecular mechanism through which microglial activation leads to increases in synaptic plasticity.
Microglia Optimize Experience-Dependent Neural Circuit Plasticity
The instructive role of IL-33 in promoting microglial functions and the fact that IL-33 expression is experience-dependent suggest two levels at which this pathway may optimize encoding. First, we show that environmental inputs, including enrichment and SI, can alter the homeostatic set point of Il33 expression in neurons. These changes occur over days to weeks and suggest that this pathway could represent a mechanism whereby experiences such as stress and deprivation affect the capacity for information storage.
Second, the heterogeneous expression of IL-33 within the neuronal population suggests that not all DG neurons are equally poised for experience-dependent remodeling. Only a fraction of granule cells recruited to an engram are reactivated during memory recall, indicating that most neurons activated during learning eventually dissociate from the memory trace (Guo et al., 2018; Kitamura et al., 2017). Thus, it is possible that engram cells expressing high levels of IL-33 could gain a competitive edge by engaging microglia that promote spine plasticity. Such a mechanism could help optimize circuit connectivity and reduce memory interference, which we found to be increased in mice deficient in IL-33-IL1RL1 signaling.
Further insights into how this mechanism might underlie diversity within engram networks would require a better understanding of the time course of Il33 expression. For example, how does new IL-33 production compare with the time course of immediate-early genes such as Cfos or Arc? We were only able to detect experiencedependent changes in neuronal Il33 over days to weeks. However, the slow turnover kinetics of the reporter used in this study limit the conclusions that can be drawn. Defining the kinetics of Il33 expression with more temporal precision and at the level of single neurons as well as defining the regulation of IL-33 protein release, which is required for its downstream effects, are important areas for future study.
Microglia Remodel the Brain ECM
How do microglial interactions with synapses promote spine formation and plasticity? It has been proposed that memories may be stored in the “pattern of holes” in the ECM (Tsien, 2013). Live imaging has shown that microglia contact dendritic spines in an activity-dependent manner. Spine head filopodium formation correlates with perisynaptic microglia contact (Weinhard et al., 2018) and can be induced by local digestion of the ECM (Orlando et al., 2012). Our data raise the possibility that these frequent microglial contacts with spines could enable microglia to locally clear or modulate the ECM around individual synapses in support of activity-dependent structural plasticity.
Notably, the ECM is not only relevant to synapses. For example, oligodendrocytes produce ECM, which is required for proper assembly of nodes of Ranvier (Susuki et al., 2013). New myelin formation also promotes long-term memory precision (Pan et al., 2020; Steadman et al., 2020). This raises the question of whether microglia remodel myelin or other components of the extracellular space.
IL-33 Function in Physiology, Aging, and Pathology
Dysregulation of innate immune homeostatic mechanisms and of microglia are strongly implicated in the cognitive dysfunction that occurs in brain aging and disease (Bennett and Molofsky, 2019; Kelley et al., 2018). The progressive decline in neuronal IL-33 expression with age coincides with defects in memory precision and accumulation of perisynaptic ECM (Vegh et al., 2014a). Interestingly, clearance of ECM and exogenous recombinant IL-33 are beneficial in mouse models of Alzheimer’s disease (Fu et al., 2016; Vegh et al., 2014b). Thus, precisely defining the molecular mechanisms, such as IL33, that regulate microglial synaptic function in the healthy brain may lead to new tools and approaches for cognitive repair.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Anna Molofsky (anna.molofsky@ucsf.edu).
Materials Availability
Plasmids generated in this study will be deposited to Addgene.
Data and Code Availability
Single nuclei RNA sequencing and microglia bulk RNA sequencing data were deposited in NCBI’s Gene Expression Omnibus (Barrett et al., 2013) under accession number GSE150714.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice
All mouse strains were maintained in the University of California San Francisco specific pathogen-free animal facility, and all animal protocols were approved by and in accordance with the guidelines established by the Institutional Animal Care and Use Committee and Laboratory Animal Resource Center. Littermate controls were used for all experiments when feasible, and all mice were back-crossed >10 generations on a C57BL/6 background unless otherwise indicated. The mouse strains used are described in the Key Resources Table and as referenced in the text. The following strains were used: IL-33 reporter line Il33H2B-mCherry (Vainchtein et al., 2018); Astrocyte reporter line Aldh1l1eGFP (Gong et al., 2003; Molofsky et al., 2014); Synapsin-Cre transgenic mice for neuron-specific excision of IL-33, cre expression was maintained in the female line for breeding (Knoferle et al., 2014; Rempe et al., 2006; Zhu et al., 2001); Il1rl1flox/flox and Il33fl/fl(Chen et al., 2015); Cx3cr1creER (used in studies referring to ‘IL1RL1 i-cKO’; (Yona et al., 2013)Cx3cxr1cre (used for studies referring to ‘IL1RL1 cKO’; (Yona etal., 2013)). All experiments, including behavioral analyses incorporated animals of both sexes in approximately equal numbers. All experiments of n > 5 were analyzed for sex-specific trends, and none were evident unless specifically noted.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| rat anti-mCherry 1:1000 | Thermo Fisher | Cat# M11217; RRID:AB_2536611 |
| rabbit anti-dsRed 1:1000 | Takara Bio | Cat#632496; RRID:AB_10013483 |
| chicken anti-GFP 1:1000 | Aves Labs | Cat#GFP-1020; RRID:AB_10000240 |
| mouse anti-NeuN 1:500 | Millipore | Cat# MAB377; RRID:AB_2298772 |
| chicken anti-NeuN 1:400 | Millipore | Cat#ABN91; RRID:AB_11205760 |
| rabbit anti-Iba1 1:1000 | Wako Chemicals | Cat#019-19741; RRID:AB_839504 |
| mouse anti-Iba1 1:1000 | Wako Chemicals | Cat#016-26721; RRID:AB_2811160 |
| rat anti-GFAP 1:1000 | Thermo Fisher | Cat#13-0300; RRID:AB_2532994 |
| rabbit anti-Olig2 1:500 | Thermo Fisher | Cat#P21954; RRID:AB_2539836 |
| mouse anti-Parvalbumin 1:1000 | Sigma-Aldrich | Cat#P3088; RRID:AB_477329 |
| rabbit anti-Prox1 1:1000 | BioLegend | Cat#925201; RRID:AB_2749865 |
| rat anti-Ctip2 1:500 | BioLegend | Cat#650603; RRID:AB_2686936 |
| goat anti-Doublecortin 1:1000 | Santa Cruz Biotech | Cat# sc-8066; RRID:AB_2088494 |
| rat anti-BrdU 1:500 | Abcam | Cat# ab6326; RRID:AB_305426 |
| rabbit anti-Aggrecan 1:500 | Millipore | Cat# AB1031 ; RRID:AB_90460 |
| goat anti-IL-33 1:500 | R&D Systems | Cat# AF3626; RRID:AB_884269 |
| goat anti-cFos 1:500 | Santa Cruz Biotech | Cat# sc-52-G; RRID:AB_2629503 |
| rabbit anti-cFos 1:1000 | Cell Signaling Technologies | Cat#2250S; RRID:AB_2247211 |
| rabbit anti-Hemagglutinin (HA) 1:500 | Cell Signaling Technology | Cat#3724S; RRID:AB_1549585 |
| rat anti-CD68 1:1000 | Bio-Rad | Cat#MCA1957; RRID:AB_2074849 |
| Biotin Lectin from Wisteria floribunda (WFA) | Millipore | Cat#L1516 |
| Hoechst 33342 solution 1:5000 | Thermo Fisher | Cat#62249 |
| TruStain rat anti-CD16/32 1:100 | BioLegend | Cat#101319; RRID:AB_1574973 |
| APC rat anti-CD11b 1:100 | BioLegend | Cat#101212; RRID:AB_312795 |
| PE rat anti-CD11b 1:100 | Thermo Fisher | Cat#12-0112-81; RRID:AB_465546 |
| FITC rat anti-CD45 1:100 | BioLegend | Cat#103108; RRID:AB_312973 |
| Alexa Fluor 488 mouse anti-NeuN 1:1500 | Millipore | Cat# MAB377X; RRID:AB_2149209 |
| Alexa Fluor 647 mouse anti-GFAP 1:500 | BD Biosciences | Cat#561470; RRID:AB_10646037 |
| rabbit anti-Olig2 1:100 | Thermo Fisher | Cat# P21954; RRID:AB_2539836 |
| rabbit anti-Prox1 1:1500 | BioLegend | Cat#925201; RRID:AB_2749865 |
| rat anti-Ctip2 1:600 | BioLegend | Cat#650603; RRID:AB_2686936 |
| goat anti-cFos 1:500 | Santa Cruz Biotech | Cat# sc-52-G; RRID:AB_2629503 |
| mouse anti-Brevican | BioLegend | Cat# MMS-5127; RRID:AB_2564837 |
| Bacterial and Virus Strains | ||
| AAV9-hSyn1-eGFP | Addgene/UPENN vector core | Addgene 105539-AAV9 |
| LV-CaMKII-tdTomato | Generated in house | Addgene deposit pending |
| LV-CaMKII-Il33(ΔNLS)-2A-tdTomato) | Generated in house | Addgene deposit pending |
| LV-Synapsin1-GFP | SignaGen Laboratories | SL100271 |
| MMLV-CAG-GFP | Addgene | Addgene16664 |
| Critical Commercial Assays | ||
| RNAscope Multiplex Fluorescent Reagent Kit v2 assay, Mouse Il1rl1 RNAscope Probe | ACD Bio | Cat# 440661 |
| Chromium single cell gene expression platform, version 2 | 10x Genomics | Library and gel bead kit - V2, 120267: Chip A kit: 1000009 |
| Recombinant mouse IL-33 | R&D Systems | Cat# 3626-ML-010/CF |
| Chondroitinase ABC from Proteus vulgaris | Sigma | Cat# C3667 |
| TaqMan gene expression assays for qPCR of Il33 and Il1rl1 | Thermo Fisher | Mm00505403 Mm00516117 |
| TaqMan gene expression assays for Actb and Rplp0 | IDT Technologies | Mm.PT.39a.22214843.g Mm.PT.58.43894205 |
| Deposited Data | ||
| Single nucleus RNA-sequencing of adult hippocampal neurons from WT and II33mCherry/+ mice | Gene Expression Omnibus | GSE150714 |
| Bulk RNA-sequencing of microglia after intracerebroventricular injections of rIL-33 or PBS | Gene Expression Omnibus | GSE150714 |
| Experimental Models: Mouse strains | ||
| IL-33 reporter line II33H2B-mCherry was derived from an insertional mutation of a gene trap cassette into the intron upstream of exon 5 of the II33 gene. | Vainchtein et al., 2018 | Courtesy of Mucosal Immunology Study Team (MIST) |
| Astrocyte reporter line Aldh1l1eGFP is a BAC transgenic generated by the GENSAT project | Gong et al., 2003; Molofsky et al., 2014 | MGI:3843271 |
| Il33flox/flox mice | R.T. Lee (Chen et al., 2015) | |
| Synapsin-Cre transgenic mice for neuron-specific excision of IL-33, cre expression was maintained in the female line for breeding | Knoferle et al., 2014; Rempe et al., 2006; Zhu et al., 2001 | JAX: 003966; RRID:IMSR_JAX:003966 |
| Il1rl1fiox/fiox | Chen et al., 2015 | |
| Il33flox/flox | Chen et al., 2015 | |
| Cx3cr1creER (used in studies referring to ‘IL1RL1 i-cKO’) | Yona et al., 2013 | JAX: 020940; RRID:IMSR_JAX:020940 |
| Cx3cxr1cre (used for studies referring to ‘IL1RL1 cKO’) | Yona et al., 2013 | JAX: 025524; RRID:IMSR_JAX:025524 |
| Oligonucleotides | ||
| Il33a primers, Sybr green assay | In house | F: GCTGCAGAAGGGAGAAATCACG; R:GGAGTTGGAATACTTCATTCTAGGTCTCAT |
| Il33b primers, Sybr green assay | In house | F: GGCTCACTGCAGGAAAGTACAGCA; R:GGAGTTGGAATACTTCATTCTAGGTCTCAT |
| Software and Algorithms | ||
| Seurat (version 2.3.4) | Satija et al., 2015 | https://satijalab.org/seurat/ |
| STAR (version 2.5.4b) | Dobin et al., 2013 | https://github.com/alexdobin/STAR/releases |
| DESeq2 package (version 1.24.0) | Love et al., 2014 | http://www.bioconductor.org/packages/release/bioc/html/DESeq2.html |
Environmental enrichment consisted of housing 2–3-month old mice in large ventilated rat cages (210 sq. in, Lab Products) with standard nesting, rodent foraging toys, and one running wheel per cage (https://larc.ucsf.edu/rodent-enrichment). Social isolation conditions consisted of singly housing 4-week old mice immediately after weaning in standard mouse cages devoid of enrichment toys.
Viral Constructs
For sparse labeling of hippocampal neurons, adeno-associated virus (AAVg-hSyn1-eGFP) was packaged and supplied by the Addgene/UPENN vector core (Addgene viral prep # 105539-AAV9) at titers of ~1×1013 vg/mL. For IL-33 gain of function experiments, lentiviral vectors (LV-CaMKII-tdTomato and LV-CaMKII-IL33(ΔNLS)-2A-tdTomato) were designed in house and packaged by the UCSF virus core at titers of ~1×107 vg/mL. For labeling adultborn neurons, MMLV retrovirus vector (CAG-GFP) was supplied by Addgene (Plasmid #16664) and packaged into retrovirus by the UCSF virus core at titers of ~1×106 vg/mL. LV-Synapsin-GFP lentivirus was purchased from SignaGen Laboratories.
METHOD DETAILS
Tamoxifen and BrdU injections
Tamoxifen (Sigma, T5648) was diluted in Corn Oil (Sigma-Aldrich) at 37°C overnight and administered by oral gavage at a concentration of 200 mg/kg four times every other day beginning at 4 weeks of age. For all BrdU labeling experiments, BrdU (Sigma-Aldrich) was diluted in a 10 mg/mL stock solution in sterile 1X PBS and injected intraperitoneally at a concentration of 100 mg/kg daily for 3 days.
Stereotaxic injections
All brain injections were performed with a Kopf stereotaxic apparatus (David Kopf, Tujunga, CA) with a microdispensing pump (World Precision Instruments) holding a beveled glass needle with ~50 μm outer diameter. For perinatal injections to sparsely label hippocampal neurons, P2–3 mice were anesthetized by hypothermia, headfixed with a custom clay mold, and 0.5 μL of AAVg-hSyn1-eGFP was injected at a rate of 15 nL/sec into the right ventricle using in house coordinates (1.5 mm AP from lambda, 0.8 mm ML, −1.6 mm DV). A dilution of 1:250 in sterile saline was used for experiments measuring dendritic spine density and a 1:750 dilution was used for experiments requiring single neuron morphology. Mice were allowed to recover on a heat pad before being returned to their cage.
For injections into adult hippocampus, mice were anesthetized with 1.5% isoflurane at an oxygen flow rate of 1L/min, headfixed with a stereotaxic frame, and treated with ophthalmic eye ointment. Fur was shaved and the incision site was sterilized with 70% ethanol and Betadine prior to surgical procedures. A hole was drilled in the skull and 1.5 uL of LV-CaMKII-tdTomato or LV-CaMKII-IL33(ΔNLS)-2A-tdTomato was injected at a 1:25 dilution in sterile saline (−2.1 mm AP, 1.375 mm ML, x −1.7 mm DV) at a rate of 3 nL/sec. For injections with Chondroitinase ABC (Sigma), 40 U/mL were injected with LV-Synapsin-GFP (SignaGen Laboratories) at a dilution of 1:150. Body temperature was maintained throughout surgery using a heating pad. Post-surgery, Buprenorphine (Henry Schein Animal Health) was administered as needed by intraperitoneal injection at a concentration of 0.1 mg/kg.
Immunohistochemistry and Confocal Microscopy
For all immunohistochemistry experiments, mice were perfused transcardially with ~10 mL of ice-cold 1X PBS followed by ~10 mL of 4% (weight/volume) paraformaldehyde diluted in PBS. Brains were post-fixed in 4% PFA for a minimum of 4 hours and then transferred to a 30% sucrose solution for a minimum of 2 days. Brains were then flash frozen and sliced in 40 um thick coronal sections on a HM 440E freezing microtome (GMI Instruments).
For immunohistochemistry, brain sections were incubated in a blocking solution consisting of 10% normal goat or donkey serum (Thermo Fisher) and 0.4% Triton (Sigma-Aldrich) diluted in 1X PBS. Primary antibodies were diluted in 3% normal goat or donkey serum in 0.4% Triton and tissue was incubated on a shaker overnight at 4°C. Secondary antibodies were diluted in 3% normal goat or donkey serum and tissue was incubated on a shaker for 2 hours at room temperature with Hoechst 33342 (Thermo Fisher) added at 1:5000 during the last 5 minutes of incubation. Brain sections were mounted on coverslips with ProLong Glass without DAPI (Thermo Fisher) for high resolution imaging and ProLong Gold (Thermo Fisher) for all other experiments.
For IL-33 staining, brain sections were heated in a 1X Sodium Citrate solution (Fisher Scientific) at 95°C for 10 minutes and then washed 3x with PBS prior to blocking. For BrdU staining, brain sections were stained with non-BrdU primary antibodies and secondary antibodies as described above and then incubated with 4% PFA for 10 minutes at room temperature. Brain sections were incubated in 2N HCl (Sigma-Aldrich) for 30 minutes at 37°C and then quenched with Boric Acid buffer (0.31 g/50 mL in H2O, pH 8.5) for 10 minutes at room temperature before being carefully washed four times with 1X PBS. Tissue was incubated in a blocking solution of 10% normal goat serum and 0.4% Triton on a shaker for 1 hour at room temperature and then incubated with an anti-BrdU antibody (Abcam) diluted in 3% normal goat serum and 0.4% Triton on a shaker overnight at 4°C. Secondary staining and mounting were performed as described above.
Histology slides were imaged on an LSM 880 confocal microscope with AiryScan (Zeiss) on Superresolution mode using a 63x objective for high resolution imaging and an LSM 700 confocal microscope (Zeiss) using 20x and 63x objectives for all other imaging experiments.
Fluorescent In Situ Hybridization (FISH)
FISH experiments were performed using the RNAscope Multiplex Fluorescent Reagent Kit v2 assay (ACD Bio) as described by the manufacturer for fixed-frozen tissue, except tissue was not baked for 30 minutes at 60°C prior to tissue dehydration. Brains were embedded in OCT following 30% sucrose treatment and frozen at −80°Cfor a minimum of 1 day and then sliced in 15 um thick coronal sections on a CryoStar NX70 Cryostat (Thermo Fisher) before being mounted on coverslips for downstream RNAscope processing. Mouse Il1rl1 RNAscope Probe (ACD Bio, 440661) and TSA Plus Cyanine 3 reagent (Perkin Elmer) were used to detect Il1rl1 transcript. For immunohistochemical labeling with antibodies following the RNAscope assay, tissues were incubated with blocking and antibody solutions as described above immediately after developing the HRP-TSA Plus signal and carefully washing four times.
Flow cytometry
For all flow cytometry experiments, animals were perfused transcardially with ice-cold 1X PBS and hippocampi were isolated under a dissecting microscope where meninges and excess myelin were removed. Antibodies are outlined in the Key Resource Table. Gating was based on fluorescence minus one (FMO) negative staining controls. All data analysis was performed using FlowJo™ software.
For sorting neuronal nuclei, cells were isolated using a protocol modified from Jaegaret al., 2018. Hippocampi were isolated under a dissecting microscope and mechanically dissociated using a 2 mL glass tissue homogenizer (Kontes Glassware) on ice using five strokes of the loose pestle and ten strokes of the tight pestle in nuclei isolation media (250 mM sucrose, 25 mM KCl, 5 mM MgCl2, 10 mM Tris pH 8.0, and nuclease free H2O). Dissociated nuclei were pelleted in an eppendorf tube at 1000 g for 5 minutes at 4°C and then resuspended in nuclei storage buffer (166.5 mM sucrose, 5 mM MgCl2,10 mM Tris pH 8.0, and nuclease free H2O) using a P1000 tip. Nuclei were again pelleted at 1000 g for 5 minutes at 4°C and resuspended in blocking buffer consisting of 1% (weight/volume) RNase-free BSA and 1X PBS. Cells were sorted on a BD Facs Aria III and gated on forward/side scatter, nuclei by Hoechst, NeuN (Millipore) to identify neuronal nuclei, and Prox1 (BioLegend) and Ctip2 (BioLegend) to identify nuclei from dentate granule cells and CA1 pyramidal neurons. For select experiments, cFos (Santa Cruz Biotech) was used to identify activated neurons. For experiments with downstream qPCR and single-nuclei RNA sequencing, all reagents were treated with 0.2 U/uL of SUPERasein RNase inhibitor (Thermo Fisher) and instruments and working surfaces treated with RNase Away surface decontaminant (Thermo Fisher). For experiments stratifying neuronal nuclei by Il33-mCherry expression, the mCherry positive population was gated using an mCherry negative control and then divided into 3 equal populations such that each gate comprised of 33.3% of the mCherry positive population.
For sorting microglia for downstream RNA-sequencing, cells were isolated as described (Galatro et al., 2017). Briefly, hippocampi were mechanically dissociated using a glass tissue homogenizer in isolation medium (HBSS, 15 mM HEPES, 0.6% glucose, 1 mM EDTApH 8.0). Cells were filtered and then pelleted at 300 g for 5 minutes at 4°C before being resuspended in 22% Percoll (GE Health-care) and centrifuged at 900 g for 20 minutes with acceleration set to 4 and deceleration set to 1 in order to remove cellular debris. Pelleted microglia were then washed with isolation medium and incubated in block solution consisting of anti-mouse CD16/32 antibody (Biolegend). Cells were sorted on a BD FACS Aria III and gated on forward/side scatter, live cells by Hoechst, CD11b-PE (eBioscience), and CD45-FITC (BioLegend) to isolate microglia.
For sorting multiple CNS cell-types in parallel for comparative qPCR, cells were isolated and fixed as described (Srinivasan et al., 2016). Hippocampi were roughly minced with a sterile razor blade, incubated in 1X Accutase (Millipore) at 4°C for 1 hour, pelleted at 2000 g for 1 minute, and resuspended in Hibernate A Low Fluorescence medium (BrainBits). Cells were then lightly pelleted using a benchtop microcentrifuge and supernatant filtered through a 70 μm cell strainer into a FACS tube. 1 mL of Hibernate A media was then added to the cell pellet and slowly triturated with a P1000 tip. Cells were again lightly pelleted and supernatant was filtered into FACS tubes. This process was repeated 2 additional times. To remove excess myelin, cells were then resuspended in 22% Percoll diluted with Hibernate A and centrifuged at 900 g for 20 minutes with acceleration set to 4 and deceleration set to 1. To fix and permeabilize cells, cell pellets were resuspended in 500 uLof Hibernate A and 500 uL of ice cold 100% ethanol was slowly added. Cells were incubated on ice for 15 minutes, washed twice with Hibernate A, and then incubated in block solution consisting of anti-mouse CD16/32 antibody (Biolegend). Cells were sorted on a BD FACS Aria III and gated on forward/side scatter, intact cells by Hoechst, CD11b (eBioscience) to identify microglia, GFAP (Millipore) to identify astrocytes, Olig2 (Thermo Fisher) to identify oligolineage cells, and NeuN (Millipore) to isolate neurons.
Single nuclei RNA-sequencing
Neuronal nuclei were isolated from hippocampus, stained with Alexa Fluor 488 mouse anti-NeuN 1:1500 (Millipore), and sorted on an Aria III (BD) flow cytometer. To account for “dropouts,” we also isolated NeuN+ neuronal nuclei from Il33mCherry/+ mice and sorted the top 30% of mCherry+ neurons by intensity and sequenced them as metadata. FACS purified neuronal nuclei were sequenced using the Chromium single cell gene expression platform (10x Genomics, version 2). Approximately 15,000 nuclei from each sample were loaded into each well of Chromium Chip A according to the manufacturer instructions and combined into droplets with barcoded beads using the Chromium controller. Libraries were prepared by the Gladstone Institutes Genomics Core following the instructions in the Chromium Single Cell 3′ Reagent Kits version 2 user guide. The samples were sequenced to an average depth of 40,000–60,000 reads on an Illumina HiSeq 4000 sequencer.
Single nuclei data analysis
Sequenced samples were processed using the Cell Ranger 2.1 pipeline and aligned to the GRCm38 (mm10) mouse reference genome. In order to count the numerous intronic reads aligned from our nuclear dataset, a custom pre-mRNA GTF file was made using cellranger by converting the “transcript” annotation in the reference genome to “exon,” enabling CellRanger to count pre-mRNAs. Clustering and differential expression analysis were conducted using Seurat version 2.3.4 (Butler et al., 2018; Satija et al., 2015). Cells with fewer than 500 detected genes/cell or over 3000 genes/cell, comprising the lower and upper 5% of cells respectively, or more than 1% mitochondrial DNA were excluded during quality control steps before identification of variable genes in the dataset, cell centering, and scaling. Additionally, 3000 genes were eliminated that were expressed by fewer than 5 cells in the dataset.
After log normalization and scaling the data with a scale factor of 10,000, approximately 1500 of the most variable genes were identified and used for calculating nearest neighbors, PCA, and t-SNE plots. The most variable genes were identified using a LogVMR function from the Seurat package with parameters of expression mean between 0.125 and 5, and dispersion > 0.5. 40 PCs were calculated using these genes and the first 15 principle components were used for subsequent calculations. Clustering and t-SNE plots were computed using Seurat (Butler et al., 2018; Satija et al., 2015). A resolution of 0.5 was used to generate clusters, and the clusters were named based on expression of canonical marker genes for each region of the hippocampus. For differential expression analysis of the dentate gyrus, cells from the two clusters identified in the dentate gyrus (0,2) were extracted from the dataset, renormalized, and scaled. The top 3000 most variable genes were calculated using the LogVMR method in Seurat; only genes with mean expression between 0.1 and 5 and dispersion over 0.1 were included in subsequent calculations. The cells were then clustered as described for the full dataset. Cells were identified as “female” or “male” based on their expression of the gene Xist; any cells expressing one or more counts of Xist were labeled female, while all others were labeled male. Female cells from sample 3 were known to express IL-33 based on IL-33mCherry fluorescence during sorting prior to sequencing. Differentially expressed genes were identified using the “FindMarkers” function in Seurat, comparing the cluster enriched in sorted IL-33 expressing cells with the other dentate gyrus cell cluster. Only genes expressed in 25% of the cells in a given cluster that pasted a log fold change threshold of 0.25 were included in the differentially expressed gene list. GO analysis was conducted using the Metascape webpage (https://www.metascape.org).
Bulk RNA sequencing
Microglia were isolated from hippocampus, stained with CD45-FITC 1:100 (BioLegend) and CD11b-APC 1:100 (BioLegend), and sorted on an Aria III (BD) flow cytometer into RLT plus buffer (QIAGEN). RNA was extracted and purified using an RNeasy Mini Kit (QIAGEN). RNA quality was measured using the Agilent RNA 6000 Pico kit on an Agilent Bioanalyzer. All samples had RNA Integrity Number (RIN) > 7. For each sample, 10 ng of RNA was used as input for cDNA amplification and library construction using the Quant-Seq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen) following manufacturer’s instructions. Library quality and quantity was assessed using the Agilent High Sensitivity DNA kit on an Agilent Bioanalyzer and Quant-iT dsDNA Assay Kit on a Qubit Fluorometer (Thermo Fisher Scientific). Single-end (65-bp reads) sequencing was performed using an Illumina HiSeq 4000 yielding 30–45 million reads per sample.
Bulk RNA sequencing analysis
Quality of reads was evaluated using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), all samples passed quality control, and reads were aligned to mm10 (GRCm38; retrieved from Ensembl, version November 2019) using STAR (version 2.5.4b) (Dobin et al., 2013) with ‘outFilterMultimapNmax 1’ to only keep reads that map one time to the reference genome. Mapped reads were counted using HTSeq (version 0.9.0)(Anders et al., 2015) and DESeq2 package (version 1.24.0) (Love et al., 2014) was used to normalize the raw counts and perform differential gene expression analysis (using theapeglm method (Zhu et al., 2019) for effect size shrinkage). Volcano plot was made using genes from the comparison IL-33 versus PBS with adjusted p value < 0.01 with the EnhancedVolcano package (version 1.2.0).
Behavioral Assays
Conditioned fear was elicited by administering three mild footshocks (0.75 mA) following a five-minute exposure to an array of contextual cues (conditioning chamber, chamber lights, white noise, scent of anise extract). Retrieval of the fear memory was assessed by a three-minute re-exposure of the animal to the conditioning context in the absence of shock and freezing (the cessation of all movement outside of respiration) was interpreted as expression of fear memory. Video recordings were acquired and scored automatically in FreezeFrame (Actimetrics). Mice were habituated to transport and holding in a separate room for at least one hour prior to all conditioning or retrieval sessions and subjected to three days of handling prior to all behavioral tests. For assessment of fear discrimination, freezing was measured in a context similarto the conditioning context two hours later, but with the following variations: the chamber fan was turned off, the scent of anise extract was swapped for lemon extract, and a plastic divider was inserted to make the chamber walls circular and opaque. Freezing in the similar context was tested two hours following retrieval testing in the original conditioning context, and animals were rested in a holding room between sessions.
Anxiety-like behaviors were assessed via ten-minute exposures to the open field test (OFT) and five minute exposures to the elevated plus maze (EPM). Video recordings were acquired in Ethovision (Noldus) and time spent in the periphery of the OFT and in the closed arms of the EPM were scored through automated object detection and interpreted as anxiety-like behavior. Freezing and anxiety-like behaviors were both scored via an automated, unbiased process and all behavioral testing and analysis was performed blinded to genotype.
Slice Preparation and Patch-Clamp Electrophysiology
After euthanizing mice with 4% isoflurane and removing brains, 300 mm-thick transverse slices were prepared in ice-cold sucrose cutting solution (234 mM sucrose, 2.5 mM KCl, 1.25 mM NaH2PO4, 10 mM MgSO4, 0.5 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose, equilibrated with 95% O2 and 5% CO2, pH 7.4) using a Leica VT1200 microtome (Leica Microsystems). Hippocampal sections were incubated for an hour at 32–34°C and then at room temperate in artificial cerebrospinal fluid (aCSF; 126 mM NaCl, 2.5 mM KCl, 1.25 mM NaH2PO4, 1 mM MgCl2, 2 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose, equilibrated with 95% O2 and 5% CO2, pH 7.4).
Recording electrodes were made from borosilicate glass with a resistance 4–5.5 MΩ when filled with intracellular solution (115 mM potassium gluconate, 11 mM KCl, 1 mM MgCl2, 1 mM CaCl2,10 mM HEPES, 11 mM EGTA, and 2 mM K2ATP, pH adjusted to 7.35 with KOH; 286 mOsm). Series resistance was monitored in all recordings, and the recordings were excluded from analysis if the series resistance was > 33 MΩ or varied by more than 10%. Recordings were obtained using a MultiClamp 700B microelectrode amplifier (Molecular Devices, Sunnyvale, CA), digitized using Digidata 1550B (Molecular Devices), and acquired at 20 kHz using pClamp 10 software (Molecular Devices). Recordings were performed in voltage-clamp mode at a holding potential of −70 mV and obtained from visually identified mature granule cell neurons in the mid and outer granule cell layer of the dentate gyrus, which were confirmed by morphology and intrinsic membrane properties. In the presence of aCSF supplemented with 0.5 μm tetrodotoxin (TTX) and 50 μm picrotoxin, miniature excitatory post-synaptic currents (mEPSCs) were isolated and recorded for 5 minutes. mESPCs were analyzed using ClampFit (Molecular Devices) and MiniAnalysis (Synaptosoft) software. Experimenter was blinded to genotype.
qPCR
To extract RNA from cells isolated by FACS, freshly sorted cells were pelleted at 500 g for 10 minutes at 4° and then resuspended in RLT Plus buffer (QIAGEN). Cells were vortexed and frozen for at least one day at −80° before being thawed on ice and processed for RNA using an RNeasy Mini Kit (QIAGEN). Purified mRNA was converted tocDNA with the High Capacity cDNA Reverse Transcription kit (Life Technologies) and amplified using eitherthe Fast SYBR Green Master Mix (Thermo Fisher) orTaqMan Gene Expression Master Mix (Thermo Fisher) and a 7900HT Fast Real-Time PCR System (Applied Biosystems).
Western blotting
Hippocampi were dissected from freshly perfused mouse brains and excess meninges and myelin removed under a dissecting microscope. Tissues were then flash frozen on dry ice and sonicated for 20 s in lysis buffer (50 mM tris-HCl, 1 mM EDTA, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate and 0.1% SDS). The sample was centrifuged for 10 minutes at 15,000 rpm at 4°C and the pellet was discarded. Samples were run on a denaturing gel and transferred to PVDF membrane, blocked with 5% milk in TBST for1 hour at room temperature, incubated with Brevican antibody (BioLegend, MMS-5127) at 1:200 overnight at 4°C and secondary at room temperature for one hour, and developed with ECL plus.
BrdU quantification
BrdU+ NeuN+ cells in the granule cell layer in the DG were counted in every sixth coronal hemibrain section and the total from six sections were multiplied by six to estimate the number of adultborn neurons.
Dendritic Spine and Spine Head Filopodia quantification
Images were acquired with an LSM 880 Confocal Microscope with AiryScan (Zeiss) on Superresolution mode using a 63x objective (NA 1.4) with 5x optical zoom (0.03 um pixel size). Z stacks were acquired at a step-size set at half the optical section and laser power and gain were consistent across all experiments. AiryScan processing was performed in Zen software (Zeiss) at a setting of 6 (“optimal” setting). Images were taken in the molecular layer of the DG. Dendritic spine density was quantified using ImageJ by measuring a distance of at least 15 μm along a dendritic branch and then counting dendritic spines. The experimenter was blinded to genotype for all quantifications. For experiments comparing IL-33 cKO, IL1RL1 cKO, and IL1RL1 i-cKO mice to controls, AAV9-Syn1-GFP virus was injected into P2–3 pups intracerebroventricularly at a dilution of 1:250 in sterile saline and mice were analyzed at 2–4 months of age.
For experiments stratifying dendritic spines and spine head filopodia by Il33-mCherry expression, the soma of sparsely labeled granule cells were imaged using a 20x objective with 16-bit depth and the position of each neuron was recorded. Dendritic spines for each neuron were imaged at 63x as described above. Mean fluorescent intensity for mCherry was then quantified in the nucleus using ImageJ and mCherry negative neurons were identified manually. For mCherry+ neurons, Il33-mCherry low and high neurons were designated as the bottom and top half of neurons by mCherry intensity, respectively. For these experiments, AAV9-Syn1-GFP virus was injected into P2–3 pups intracerebroventricularly at a dilution of 1:750 in sterile saline and mice were analyzed at 2 months of age.
Scholl analysis
Z stacks were acquired from 60 μm thick coronal sections containing hippocampus with an LSM 700 Confocal microscope (Zeiss) using a 20x objective. Sholl analysis was performed in ImageJ using the simple neurite tracer plugin (https://imagej.net/SNT).
Aggrecan and Homer1 puncta quantification
Images were acquired with an LSM 880 Confocal Microscope with AiryScan on Superresolution mode using the same specifications as described above for dendritic spines. Aggrecan puncta were quantified using the Puncta Analyzer plugin (Ippolito and Eroglu, 2010) in ImageJ and thresholding was consistent for every image. For colocalization of Aggrecan and Homer1 puncta, images were analyzed with the Puncta Analyzer plugin with a minimum pixel specification of 4. Three images in the molecular layer were averaged per mouse for analysis.
Aggrecan engulfment
Z stacks encompassing entire microglia were collected on an LSM 880 Confocal Microscope with AiryScan (Zeiss) on Superresolution mode using a 63x objective (NA 1.4) with 5x optical zoom (0.03 um pixel size); laser power and gain were consistent across all experiments. Images were analyzed using Imaris software (Bitplane) by creating a 3D surface rendering of individual microglia, which were thresholded to ensure microglia processes were accurately reconstructed and maintained consistent thereafter. The microglia rendering was used to mask the CD68 channel within that microglia, which was then 3D rendered, and the CD68 volume was subsequently used to mask the Aggrecan channel. Aggrecan engulfment per microglia was then calculated as the total volume of Aggrecan within the masked CD68 volume.
Microglia coverage
Images of Iba1 immunostaining were acquired on an LSM 700 Confocal microscope (Zeiss) using a 20x objective with 16-bit depth. Microglia coverage was quantified in ImageJ by first thresholding the Iba1 channel and then measuring the area of Iba1+ pixels and normalizing that value by the total area per field of view in the DG molecular layer. Thresholding was held consistent for each image.
QUANTIFICATION AND STATISTICAL ANALYSIS
Graphpad Prism 8.3.0 was used for most statistical analyses. Statistical tests are as described in text and figure legends. Violin plots were used for data with n ≥ 20 to better visualize the distribution of individual data points. RNA-sequencing data was analyzed in R as described in the methods section above.
Supplementary Material
(A) Il33 expression (Il33-mCherry) in hippocampal astrocytes (Aldh1l1-GFP) and oligolineage cells (Olig2). Scale bar: 10 μm.
(B) Representative gating and expression of Il33 transcript isoforms from qPCR of flow sorted hippocampal astrocytes and dentate granule cells (t test, n = 4 mice).
(C) DG neurons with immunolabeling for Il33-mCherry, the granule cell marker Proxl, and interneuron marker parvalbumin (PV). Scale bar: 20 μm.
(D) DG neurons with immunolabeling for Il33-mCherry, NeuN, and the progenitor/immature neuron marker Doublecortin (Dcx). Scale bar: 10 μm.
(E) Expression of Il33-mCherry with the astrocyte reporter Aldh1l1-eGFP and NeuN across brain regions in adult mice. Scale bar: 50 μm. 3V: third ventricle.
(F) Immunostaining for IL-33 protein and neuronal labeling with AAV9-Syn1-GFP. Scale bar: 5 μm.
(G and H) Schematic and time course of the percentage of Il33-mCherry+ neurons in the CA1 region after indicated time periods of environmental enrichment quantified by flow cytometry (2-way ANOVA, Tukey’s post hoc test, n = 3-4 mice/group).
(I) Proportion of Il33-mCherry+ neurons in DG and CA1 after 4 weeks of enrichment versus standard housing quantified by flow cytometry (t test; n = 4 mice).
(J) Proportion of Il33-mCherry+ neurons after 4 weeks of social isolation versus standard housing quantified by flow cytometry (t test; n = 4 mice).
(K and L) Percentage of NeuN+ neurons that are Il33-mCherry+ after4weeks of enrichment versus standard housing in the CA2 region, as demarcated by RGS14 staining, and CA3 (t test, n = 3 mice/condition). Scale bar: 200 μm.
*p < 0.05, **p < 0.01, ***p < 0.001. Data = mean ± SD.
(A and B) Quality control metrics for single nuclei RNA sequencing. (A) Number of genes recovered per cell. (B) Mitochondrial RNA content per cell. Lines show thresholds used for cell selection prior to all other analyses.
(C) t-SNE plot showing clustering of the independent wild-type replicates (coral and turquoise).
(D and E) Violin and t-SNE plots of markers used to assign subtype identities (Figure 2B) showing genes for interneurons (Gad2), CA3 pyramidal neurons (Socs2), DG granule cells (Prox1), CA1 pyramidal neurons (Satb2), and CA2/CA region pyramidal neurons (Sv2b).
(F) Percentage of cells per cluster originating from WT (blue) and Il33-mCherry (red) samples.
(G) Gating strategy to isolate neuronal nuclei and Il33-mCherry nuclei for single-cell RNA sequencing.
(H and I) Correlation between mCherry levels and dendritic spine or spine head filopodia numbers (Pearson correlation, n = 20 neurons, > 110 spines per neuron, 3 mice). Different colors correspond to neurons from individual mice.
(J) Representative image and quantification of the number of cFos+ cells in the DG of IL-33 cKO and control mice (t test, n = 4 mice/genotype). Scale bar: 200 μm.
(A) Gating strategy for isolation of hippocampal microglia (CD11b), neurons (NeuN), astrocytes (GFAP), and oligodendrocytes (Olig2) by FACS and expression of cell-type specific markers by qPCR (n = 4 mice).
(B) Expression of II1rI1 transcript in neurons and glia by qPCR of flow sorted hippocampal cell types (n = 3 mice).
(C) Schematic and representative image of Il1rl1 excision specificity in the hippocampus using the Cx3cr1creER allele fate mapped with the inducible Rosa26-tdTomato reporter (Ai14) and immunostaining with Iba1 and NeuN (n = 3 mice). Scale bar: 50 μm.
(D) Sensitivity of ll1rl1 excision by qPCR of flow sorted microglia using IL1RL1 i-cKO mice (Cr3cr1CreER/+:Il1rl1flox/flox; one-way ANOVA; Tukey’s post hoc test, n = 3 mice/group).
(E) Representative imageofCA1 pyramidal neuron and quantification of dendritic spine density after loss of neuronal IL-33 (t test, n = 30-34 dendritic segments, 3 mice/genotype). Scale bar: 20 μm.
(F) Scholl analysis of dendritic branching in DG granule cells after loss of neuronal IL-33 (t test, Holm-Sidak multiple comparisons; n = 12-16 neurons, 3 mice). Scale bar: 20 μm.
(G) Number of Iba1 + microglia in the DG of IL-33 cKO and control mice (t test, n = 3-4 mice). Scale bar: 200 μm.
(H) Representative images of Iba1 + microglia from control and IL1RL1 cKO mice housed under standard or enriched conditions (4 weeks). Quantification of the percentage of Iba1 + pixels (microglia coverage) in the molecular layer of the DG (one-way ANOVA, Tukey’s post hoc test, n = 3-4 mice/group). Scale bar: 20 μm.
(I) Representative image of HA epitope tag immunostaining after viral transduction with control (tdTomato) or IL-33 gain-of-function (IL-33ΔNLS) constructs. Scale bar: 20 μm.
*p < 0.05, **p < 0.01, ***p < 0.001. Data = mean ± standard deviation (bar graphs) and median ± interquartile range (violin plots).
(A) Representative image of BrdU labeled neurons35 days post-injection and quantification ofthe percentage of BrdU+ label-retaining cells that are NeuN+ in IL-33 cKO and control mice (t test, n = 5 mice/genotype). Scale bar: 20 μm.
(B) Dendritic spine density in 4-weekold newborn neurons labeled with MMLV-GFP retrovirus, which selectively targets proliferating cells, in IL-33 KO and control animals (t test, n = 34-53 dendritic segments, 3 mice). Scale bar: 20 μm, 2 μm (inset).
(C) Statistics for contextual fear discrimination assay in IL-33 cKO mice versus controls (related to Figures 4E–4G)
(D) Statistics for contextual fear discrimination assay in IL1RL1 cKO mice versus controls (related to Figures 4H–4J)
(E and F) Open field and Elevated plus maze testing of IL-33 cKO mice versus controls (t test, n = 16 control mice, 11 IL-33 cKO mice).
(G and H) Open field and Elevated plus maze testing of IL1RL1 cKO mice (t test, n = 12 mice/genotype).
*p < 0.05. Data = mean ± SD.
(A) Representative gating strategy to isolate neuronal nuclei from Prox1+Ctip2+ dentate granule cells and Prox1−Ctip2+ pyramidal neurons with fluorescence minus one (FMO) negative staining controls.
(B and C) Quantification of freezing in context A versus context B fourteen days post-training in young and old mice(n = 16 young mice and 14 old mice, 2-wayRM ANOVA, Sidak’s post hoc tests). Statistics for contextual fear discrimination assay in young versus old mice (also related to Figures 5F and 5G).
(D) Quantification of distance traveled in young and old mice during the first 2 s footshock (t test, n = 16 young and 14 old mice. Mean ± SD).
(E) Correlation between day 14 discrimination index and expression of Il33 mRNA in flow sorted dentate granule cells in individual old mice (Pearson correlation, n = 11 mice
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
(A) Representative gating strategy to isolate live microglia (CD11b+CD45+).
(B) Representative hemisection from adult (P90) mouse showing Aggrecan immunostaining. Inset = hippocampus. Scale bar: 500 μm, 250 μm (inset).
(C) Aggrecan colocalization with WFA+ perineuronal nets around Parvalbumin+ interneurons in CA1. Scale bar: 50 μm.
(D and E) Representative image and quantification of microglial process coverage index calculated as the percentage of Iba1+ pixels in afield of view normalized to the percentage of tdTomato+ pixels after viral transduction with control or IL-33ΔNLS constructs (t test, n = 3 mice). Scale bar: 20 μm.
(F) Quantification of number of CD68+ lysosomes in microglia after viral transduction with control or IL-33ΔNLS constructs (t test, 28 microglia, 3 mice/group).
*p < 0.05. Data = mean ± standard deviation (bar graph) and median ± interquartile range (violin plot).
Highlights.
IL-33 is expressed by subsets of hippocampal neurons and is modulated by experience
Microglia drive dendritic spine plasticity and memory precision via neuronal IL-33
IL-33 gain of function mitigates some age-related decreases in spine plasticity
Neuronal IL-33 induces microglial remodeling of the extracellular matrix
ACKNOWLEDGMENTS
We are grateful to Drs. E. Huang, S. Villeda, and members of the Molofsky Lab for helpful comments on the manuscript. We also thank the Mucosal Immunology Studies Team (MIST) for Il33H2B-mCherry, the UCSF Laboratory for Cell Analysis, and the ChanZuckerberg Biohub for sequencing support. A.V.M is supported by the Pew Charitable Trusts, NIMH (R01MH119349 and DP2MH116507), the Brain and Behavior Research Foundation, and the Burroughs Wellcome Fund. P.T.N. is supported by a graduate research fellowship from the National Science Foundation. M.A.K. is supported by NIMH (R01 MH108623, R01 MH111754, and R01 MH117961), a One Mind Rising Star Award, the Human Frontier Science Program, the Pew Charitable Trusts, the Esther A. and Joseph Klingenstein Fund, and the McKnight Memory and Cognitive Disorders Award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2020.05.050.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alvarez DD, Giacomini D, Yang SM, Trinchero MF, Temprana SG, Büttner KA, Beltramone N, and Schinder AF (2016). A disynaptic feed-back network activated by experience promotes the integration of new granule cells. Science 354, 459–465. [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, and Huber W (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbolina MV, and Stack MS (2008). Membrane type 1-matrix metalloproteinase: substrate diversity in pericellular proteolysis. Semin. Cell Dev. Biol. 19, 24–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, et al. (2013). NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res. 41,D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu R, Duan X, Taylor MR, Martin EA, Muralidhar S, Wang Y, Gangi-Wellman L, Das SC, Yamagata M, West PJ, et al. (2017). Heterophilic Type II Cadherins Are Required for High-Magnitude Synaptic Potentiation in the Hippocampus. Neuron 96, 160–176.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarek E, and Caroni P (2011). β-Adducin is required for stable assembly of new synapses and improved memory upon environmental enrichment. Neuron 69, 11321146. [DOI] [PubMed] [Google Scholar]
- Bennett FC, and Molofsky AV (2019). The immune system and psychiatric disease: a basic science perspective. Clin. Exp. Immunol. 197, 294–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergami M, Masserdotti G, Temprana SG, Motori E, Eriksson TM, Göbel J, Yang SM, Conzelmann K-K, Schinder AF, Götz M, and Berninger B (2015). A critical period for experience-dependent remodeling of adult-born neuron connectivity. Neuron 85, 710–717. [DOI] [PubMed] [Google Scholar]
- Bessa J, Meyer CA, deVera Mudry MC, Schlicht S, Smith SH, Iglesias A , and Cote-Sierra J (2014). Altered subcellular localization of IL-33 leads to nonresolving lethal inflammation. J. Autoimmun. 55, 33–41. [DOI] [PubMed] [Google Scholar]
- Bolós M, Perea JR, Terreros-Roncal J, Pallas-Bazarra N, Jurado-Arjona J, Ávila J, and Llorens-Martín M (2018). Absence of microglial CX3CR1 impairs the synaptic integration of adult-born hippocampal granule neurons. Brain Behav. Immun. 68, 76–89. [DOI] [PubMed] [Google Scholar]
- Bonnans C, Chou J, and Werb Z (2014). Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 15, 786–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton-Provencher V, Bakhshetyan K, Hardy D, Bammann RR, Cavarretta F, Snapyan M, Côté D, Migliore M, and Saghatelyan A (2016). Principal cell activity induces spine relocation of adult-born interneurons in the olfactory bulb. Nat. Commun. 7, 12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai DJ, Aharoni D, Shuman T, Shobe J, Biane J, Song W, Wei B, Veshkini M, La-Vu M, Lou J, et al. (2016). A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W-Y, Hong J, Gannon J, Kakkar R, and Lee RT (2015). Myocardial pressure overload induces systemic inflammation through endothelial cell IL-33. Proc. Natl. Acad. Sci. USA 112, 7249–7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielson NBB, Kaifosh P, Zaremba JDD, Lovett-Barron M, Tsai J, Denny CA, Balough EM, Goldberg AR, Drew LJ, Hen R, et al. (2016). Distinct Contribution of Adult-Born Hippocampal Granule Cells to Context Encoding. Neuron 90, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deVivo L, Landi S, Panniello M, Baroncelli L, Chierzi S, Mariotti L, Spolidoro M, Pizzorusso T, Maffei L, and Ratto GM (2013). Extracellular matrix inhibits structural and functional plasticity of dendritic spines in the adult visual cortex. Nat. Commun. 4, 1484. [DOI] [PubMed] [Google Scholar]
- Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laugh-man KB, Tomm NK, Turi GF, Losonczy A, and Hen R (2014). Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron 83, 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranovsky A, Picchini AM, Moadel T, Sisti AC, Yamada A, Kimura S, Leonardo ED, and Hen R (2011). Experience dictates stem cell fate in the adult hippocampus. Neuron 70, 908–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, Laboulaye MA, Liu J, Peng YR, Yamagata M, Toma K, and Sanes JR (2018). Cadherin Combinations Recruit Dendrites of Distinct Retinal Neurons to a Shared Interneuronal Scaffold. Neuron 99, 1145–1154.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Peng J, Swiatkowski P, Mukherjee A, Bispo A, and Wu L-J (2014). Neuronal hyperactivity recruits microglial processes via neuronal NMDA receptors and microglial P2Y12 receptors after status epilepticus. J. Neurosci. 34, 10528–10540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyo UB, Gu N, De S, Dong H, Richardson JR, and Wu LJ (2015). Modulation of microglial process convergence toward neuronal dendrites by extracellular calcium. J. Neurosci. 35, 2417–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favuzzi E, Marques-Smith A, Deogracias R, Winterflood CM, Sánchez-Aguilera A, Mantoan L, Maeso P, Fernandes C, Ewers H, and Rico B (2017). Activity-Dependent Gating of Parvalbumin Interneuron Function by the Perineuronal Net Protein Brevican. Neuron 95, 639–655.e10. [DOI] [PubMed] [Google Scholar]
- Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht R, Heine M, Perrais D, Seidenbecher CI, Choquet D, and Gundelfinger ED (2009). Brain extracellular matrix affects AMPA receptor lateral mobility and short-term synaptic plasticity. Nat. Neurosci. 12, 897–904. [DOI] [PubMed] [Google Scholar]
- Fu AKYY, Hung K-WW, Yuen MYFF, Zhou X, Mak DSYY, Chan ICWW, Cheung TH, Zhang B, Fu W-YY, Liew FY, and Ip NY (2016). IL-33 ameliorates Alzheimer’s disease-like pathology and cognitive decline. Proc. Natl. Acad. Sci. USA 113, E2705–E2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galatro TF, Vainchtein ID, Brouwer N, Boddeke EWGM, and Eggen BJL (2017). Isolation of Microglia and Immune Infiltrates from Mouse and Primate Central Nervous System. Methods Mol. Biol. 1559, 333–342. [DOI] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, and Heintz N (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Guo N, Soden ME, Herber C, Kim MT, Besnard A, Lin P, Ma X, Cepko CL, Zweifel LS, and Sahay A (2018). Dentate granule cell recruitment of feedforward inhibition governs engram maintenance and remote memory generalization. Nat. Med. 24, 438–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J-H, Kushner SA, Yiu AP, Cole CJ, Matynia A, Brown RA, Neve RL, Guzowski JF, Silva AJ, and Josselyn SA (2007). Neuronal Competition and Selection During Memory Formation. Science 316, 457–460. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu K, Itoh Y, Ab Fatah M, Nishimura T, Takemura K, Takeshita K, Kubota S, Miyazaki N, Wan Mohamad Noor WNI, Inaba T, et al. (2019). Phagocytosis is mediated by two-dimensional assemblies of the F-BAR protein GAS7. Nat. Commun. 10, 4763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibi D, Takuma K, Koike H, Mizoguchi H, Tsuritani K, Kuwahara Y, Kamei H, Nagai T, Yoneda Y, Nabeshima T, and Yamada K (2008). Social isolation rearinginduced impairment of the hippocampal neurogenesis is associated with deficits in spatial memory and emotion-related behaviors in juvenile mice. J. Neurochem. 105, 921–932. [DOI] [PubMed] [Google Scholar]
- Ippolito DM, and Eroglu C (2010). Quantifying synapses: an immunocyto-chemistrybased assay to quantify synapse number. J. Vis. Exp. (45), 2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeger BN, Linker SB, Parylak SL, Barron JJ, Gallina IS, Saavedra CD, Fitzpatrick C, Lim CK, Schafer ST, Lacar B, et al. (2018). A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun. 9, 3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josselyn SA, and Tonegawa S (2020). Memory engrams: Recalling the past and imagining the future. Science 367, eaaw4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M, Stellwagen D, Malenka RC, and Stryker MP (2008). Tumor necrosis factor-α mediates one component of competitive, experience-dependent plasticity in developing visual cortex. Neuron 58, 673–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley KW, Nakao-Inoue H, Molofsky AV, and Oldham MC (2018). Variation among intact tissue samples reveals the core transcriptional features of human CNS cell classes. Nat. Neurosci. 21, 1171–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelwick R, Desanlis I, Wheeler GN, and Edwards DR (2015). The ADAMTS (A Disintegrin and Metalloproteinase with Thrombospondin motifs) family. Genome Biol. 16, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempermann G, Kuhn HG, and Gage FH (1997). More hippocampal neurons in adult mice living in an enriched environment. Nature 386, 493–495. [DOI] [PubMed] [Google Scholar]
- Kheirbek MA, Tannenholz L, and Hen R (2012). NR2B-dependent plasticity of adult-born granule cells is necessary for context discrimination. J. Neurosci. 32, 8696–8702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Ogawa SK, Roy DS, Okuyama T, Morrissey MD, Smith LM, Redondo RL, and Tonegawa S (2017). Engrams and circuits crucial for systems consolidation of a memory. Science 356, 73–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoferle J, Yoon SY, Walker D, Leung L, Gillespie AK, Tong LM, Bien-Ly N, and Huang Y (2014). Apolipoprotein E4 produced in GABAergic interneurons causes learning and memory deficits in mice. J. Neurosci. 34, 14069–14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Brott BK, Kirkby LA, Adelson JD, Cheng S, Feller MB, Datwani A, and Shatz CJ (2014). Synapse elimination and learning rules co-regulated by MHC class I H2-Db. Nature 509, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy AD, Omar MH, and Koleske AJ (2014). Extracellular matrix control of dendritic spine and synapse structure and plasticity in adulthood. Front. Neuroanat. 8, 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Du X-FF, Liu C-SS, Wen Z-LL, and Du J-LL (2012). Reciprocal regulation between resting microglial dynamics and neuronal activity in vivo. Dev. Cell 23, 1189–1202. [DOI] [PubMed] [Google Scholar]
- Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, and Tonegawa S (2012). Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature 484, 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Kyrargyri V, Arancibia-Carcamo IL, Jolivet R, Kohsaka S, Bryan RM, and Attwell D (2018). Microglial Ramification, Surveillance, and Interleukin-1β Release Are Regulated by the Two-Pore Domain K+ Channel THIK-1. Neuron 97, 299312.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Hamel MG, and Gottschall PE (2005). Evidence for proteolytic cleavage of brevican by the ADAMTSs in the dentate gyrus after excitotoxic lesion of the mouse entorhinal cortex. BMC Neurosci. 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh TJ, Jones MW, Quinn JJ, Balthasar N, Coppari R, Elmquist JK, Lowell BB, Fanselow MS, Wilson MA, and Tonegawa S (2007). Dentate gyrus NMDA receptors mediate rapid pattern separation in the hippocampal network. Science 317, 94–99. [DOI] [PubMed] [Google Scholar]
- Miller SM, and Sahay A (2019). Functions of adult-born neurons in hippocampal memory interference and indexing. Nat. Neurosci. 22, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto A, Wake H, Ishikawa AW, Eto K, Shibata K, Murakoshi H, Koizumi S, Moorhouse AJ, Yoshimura Y, and Nabekura J (2016). Microglia contact induces synapse formation in developing somatosensory cortex. Nat. Commun. 7, 12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AV, Kelley KW, Tsai H-H, Redmond SA, Chang SM, Madireddy L, Chan JR, Baranzini SE, Ullian EM, and Rowitch DH (2014). Astrocyte-encoded positional cues maintain sensorimotor circuit integrity. Nature 509, 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molofsky AB, Savage AK, and Locksley RM (2015). Interleukin-33 in Tissue Homeostasis, Injury, and Inflammation. Immunity 42, 1005–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, and Hynes RO (2016). The extracellular matrix: Tools and insights for the “omics” era. Matrix Biol. 49, 10–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashiba T, Cushman JD, Pelkey KA, Renaudineau S, Buhl DL, McHugh TJ, Rodriguez Barrera V, Chittajallu R, Iwamoto KS, McBain CJ, et al. (2012). Young dentate granule cells mediate pattern separation, whereas old granule cells facilitate pattern completion. Cell 149, 188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MM, Oliveira FA, and Disterhoft JF (2010). Learning and aging related changes in intrinsic neuronal excitability. Front. Aging Neurosci. 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oray S, Majewska A, and Sur M (2004). Dendritic spine dynamics are regulated by monocular deprivation and extracellular matrix degradation. Neuron 44, 1021–1030. [DOI] [PubMed] [Google Scholar]
- Orlando C, Ster J, Gerber U, Fawcett JW, and Raineteau O (2012). Perisynaptic chondroitin sulfate proteoglycans restrict structural plasticity in an integrin-dependent manner. J. Neurosci. 32, 18009–18017, 18017a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer G, and Gabay C (2011). Interleukin-33 biology with potential insights into human diseases. Nat. Rev. Rheumatol. 7, 321–329. [DOI] [PubMed] [Google Scholar]
- Pan S, Mayoral SR, Choi HS, Chan JR, and Kheirbek MA (2020). Preservation of a remote fear memory requires new myelin formation. Nat. Neurosci. 23, 487–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, et al. (2011). Synaptic pruning by microglia is necessary for normal brain development. Science 333,1456–1458. [DOI] [PubMed] [Google Scholar]
- Park S, Kramer EE, Mercaldo V, Rashid AJ, Insel N, Frankland PW, and Josselyn SA (2016). Neuronal Allocation to a Hippocampal Engram. Neuropsychopharmacology 41, 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhurst CN, Yang G, Ninan I, Savas JN, Yates JR 3rd, Lafaille JJ, Hempstead BL, Littman DR, and Gan W-B (2013). Microglia promote learningdependent synapse formation through brain-derived neurotrophic factor. Cell 155, 15961609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikkarainen T, Brännström A, and Tryggvason K (1999). Expression of macrophage MARCO receptor induces formation of dendritic plasma membrane processes. J. Biol. Chem. 274, 10975–10982. [DOI] [PubMed] [Google Scholar]
- Pizzorusso T, Medini P, Landi S, Baldini S, Berardi N, and Maffei L (2006). Structural and functional recovery from early monocular deprivation in adult rats. Proc. Natl. Acad. Sci. USA 103, 8517–8522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan KR, Ressler RL, Jin J, and Maren S (2018). Nucleus reuniens is required for encoding and retrieving precise, hippocampal-dependent contextual fear memories in rats. J. Neurosci. 38, 9925–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rempe D, Vangeison G, Hamilton J, Li Y, Jepson M, and Federoff HJ (2006). Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis 44, 44–49. [DOI] [PubMed] [Google Scholar]
- Richards DA, Mateos JM, Hugel S, de Paola V, Caroni P, Gähwiler BH, and McKinney RA (2005). Glutamate induces the rapid formation of spine head protrusions in hippocampal slice cultures. Proc. Natl. Acad. Sci. USA 102,6166–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands D, Lensj0 KK Dinh T, Yang S, Andrews MR, Hafting T, Fyhn M, Fawcett JW, and Dick G (2018). Aggrecan directs extracellular matrix-mediated neuronal plasticity. J. Neurosci. 38, 10102–10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DS, Arons A, Mitchell TI, Pignatelli M, Ryan TJ, and Tonegawa S (2016). Memory retrieval by activating engram cells in mouse models of early Alzheimer’s disease. Nature 531, 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy DS, Muralidhar S, Smith LM, and Tonegawa S (2017). Silent memory engrams as the basis for retrograde amnesia. Proc. Natl. Acad. Sci. USA 114, E9972E9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan TJ, Roy DS, Pignatelli M, Arons A, and Tonegawa S (2015). Memory. Engram cells retain memory under retrograde amnesia. Science 348, 1007–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahay A, Scobie KN, Hill AS, O’Carroll CM, Kheirbek MA, Burghardt NS, Fenton AA, Dranovsky A, and Hen R (2011). Increasing adult hippocampal neurogenesis is sufficient to improve pattern separation. Nature 472, 466–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DP, Lehrman EK, Kautzman AG, Koyama R, Mardinly AR, Yamasaki R, Ransohoff RM, Greenberg ME, Barres BA, and Stevens B (2012). Microglia sculpt postnatal neural circuits in an activity and complement-dependent manner. Neuron 74, 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipe GO, Lowery RL, Tremblay ME, Kelly EA, Lamantia CE, and Majewska AK (2016). Microglial P2Y12 is necessary for synaptic plasticity in mouse visual cortex. Nat. Commun. 7, 10905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, Borneo J, Poon C, Ho T, Cai F, et al. (2016). Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 7, 11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadman PE, Xia F, Ahmed M, Mocle AJ, Penning ARA, Geraghty AC, Steenland HW, Monje M, Josselyn SA, and Frankland PW (2020). Disruption of Oligodendrogenesis Impairs Memory Consolidation in Adult Mice. Neuron 105, 150–164.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. (2007). The classical complement cascade mediates CNS synapse elimination. Cell 131, 1164–1178. [DOI] [PubMed] [Google Scholar]
- Sun X, Bernstein MJ, Meng M, Rao S, S0rensen AT, Yao L, Zhang X, Anikeeva PO, and Lin Y (2020). Functionally Distinct Neuronal Ensembles within the Memory Engram. Cell 181, 410–423.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susuki K, Chang K-J, Zollinger DR, Liu Y, Ogawa Y, Eshed-Eisenbach Y, Dours-Zimmermann MT, Oses-Prieto JA, Burlingame AL, Seidenbecher CI, et al. (2013). Three mechanisms assemble central nervous system nodes of Ranvier. Neuron 78, 469–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talabot-Ayer D, Calo N, Vigne S, Lamacchia C, Gabay C, and Palmer G (2012). The mouse interleukin (Il)33 gene is expressed in a cell type- and stimulus-dependent manner from two alternative promoters. J. Leukoc. Biol. 91, 119–125. [DOI] [PubMed] [Google Scholar]
- Toni N, Teng EM, Bushong EA, Aimone JB, Zhao C, Consiglio A, van Praag H, Martone ME, Ellisman MH, and Gage FH (2007). Synapse formation on neurons born in the adult hippocampus. Nat. Neurosci. 10,727–734. [DOI] [PubMed] [Google Scholar]
- T0nnesen J, Inavalli VVGK, and Nagerl UV (2018). Super-Resolution Imaging of the Extracellular Space in Living Brain Tissue. Cell 172, 1108–1121.e15. [DOI] [PubMed] [Google Scholar]
- Tremblay M-È, Lowery RL, and Majewska AK (2010). Microglial interactions with synapses are modulated by visual experience. PLoS Biol. 8, e1000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsien RY (2013). Very long-term memories may be stored in the pattern of holes in the perineuronal net. Proc. Natl. Acad. Sci. USA 110, 12456–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainchtein ID, Chin G, Cho FS, Kelley KW, Miller JG, Chien EC, Liddelow SA, Nguyen PT, Nakao-Inoue H, Dorman LC, et al. (2018). Astrocyte-derived interleukin-33 promotes microglial synapse engulfment and neural circuit development. Science 359, 1269–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela JC, Heise C, Franken G, Singh J, Schweitzer B, Seidenbecher CI, and Frischknecht R (2014). Hyaluronan-based extracellular matrix under conditions of homeostatic plasticity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan LJ, Dopp EA, Haworth R, Pikkarainen T, Kangas M, Elomaa O, Dijkstra CD, Gordon S, Tryggvason K, and Kraal G (1999). Regulation and functional involvement of macrophage scavenger receptor MARCO in clearance of bacteria in vivo. J. Immunol. 162, 939–947. [PubMed] [Google Scholar]
- Vegh MJ, Rausell A, Loos M, Heldring CM, Jurkowski W, van Nierop P, Paliukhovich I, Li KW, del Sol A, Smit AB, et al. (2014a). Hippocampal extracellular matrix levels and stochasticity in synaptic protein expression increase with age and are associated with age-dependent cognitive decline. Mol. Cell. Proteomics 13, 2975–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegh MJ, Heldring CM, Kamphuis W, Hijazi S, Timmerman AJ, Li KW, van Nierop P, Mansvelder HD, Hol EM, Smit AB, and van Kesteren RE (2014b). Reducing hippocampal extracellular matrix reverses early memory deficits in a mouse model of Alzheimer’s disease. Acta Neuropathol. Commun. 2, 76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeda SA, Plambeck KE, Middeldorp J, Castellano JM, Mosher KI, Luo J, Smith LK, Bieri G, Lin K, Berdnik D, et al. (2014). Young blood reverses agerelated impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 20, 659–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake H, Moorhouse AJ, Jinno S, Kohsaka S, and Nabekura J (2009). Resting microglia directly monitor the functional state of synapses in vivo and determine the fate of ischemic terminals. J. Neurosci. 29, 3974–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Yue H, Hu Z, Shen Y, Ma J, Li J, Wang XD, Wang L, Sun B, Shi P, et al. (2020). Microglia mediate forgetting via complement-dependent synaptic elimination. Science 367, 688–694. [DOI] [PubMed] [Google Scholar]
- Weinhard L, di Bartolomei G, Bolasco G, Machado P, Schieber NL, Neniskyte U, Exiga M, Vadisiute A, Raggioli A, Schertel A, et al. (2018). Microglia remodel synapses by presynaptic trogocytosis and spine head filopodia induction. Nat. Commun. 9, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyss-Coray T (2016). Ageing, neurodegeneration and brain rejuvenation. Nature 539, 180–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yona S, Kim K-W, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, and Parada LF (2001). Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 15, 859–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu A, Ibrahim JG, and Love MI (2019). Heavy-tailed prior distributions for sequence count data: removing the noise and preserving large differences. Bioinformatics 35, 2084–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Il33 expression (Il33-mCherry) in hippocampal astrocytes (Aldh1l1-GFP) and oligolineage cells (Olig2). Scale bar: 10 μm.
(B) Representative gating and expression of Il33 transcript isoforms from qPCR of flow sorted hippocampal astrocytes and dentate granule cells (t test, n = 4 mice).
(C) DG neurons with immunolabeling for Il33-mCherry, the granule cell marker Proxl, and interneuron marker parvalbumin (PV). Scale bar: 20 μm.
(D) DG neurons with immunolabeling for Il33-mCherry, NeuN, and the progenitor/immature neuron marker Doublecortin (Dcx). Scale bar: 10 μm.
(E) Expression of Il33-mCherry with the astrocyte reporter Aldh1l1-eGFP and NeuN across brain regions in adult mice. Scale bar: 50 μm. 3V: third ventricle.
(F) Immunostaining for IL-33 protein and neuronal labeling with AAV9-Syn1-GFP. Scale bar: 5 μm.
(G and H) Schematic and time course of the percentage of Il33-mCherry+ neurons in the CA1 region after indicated time periods of environmental enrichment quantified by flow cytometry (2-way ANOVA, Tukey’s post hoc test, n = 3-4 mice/group).
(I) Proportion of Il33-mCherry+ neurons in DG and CA1 after 4 weeks of enrichment versus standard housing quantified by flow cytometry (t test; n = 4 mice).
(J) Proportion of Il33-mCherry+ neurons after 4 weeks of social isolation versus standard housing quantified by flow cytometry (t test; n = 4 mice).
(K and L) Percentage of NeuN+ neurons that are Il33-mCherry+ after4weeks of enrichment versus standard housing in the CA2 region, as demarcated by RGS14 staining, and CA3 (t test, n = 3 mice/condition). Scale bar: 200 μm.
*p < 0.05, **p < 0.01, ***p < 0.001. Data = mean ± SD.
(A and B) Quality control metrics for single nuclei RNA sequencing. (A) Number of genes recovered per cell. (B) Mitochondrial RNA content per cell. Lines show thresholds used for cell selection prior to all other analyses.
(C) t-SNE plot showing clustering of the independent wild-type replicates (coral and turquoise).
(D and E) Violin and t-SNE plots of markers used to assign subtype identities (Figure 2B) showing genes for interneurons (Gad2), CA3 pyramidal neurons (Socs2), DG granule cells (Prox1), CA1 pyramidal neurons (Satb2), and CA2/CA region pyramidal neurons (Sv2b).
(F) Percentage of cells per cluster originating from WT (blue) and Il33-mCherry (red) samples.
(G) Gating strategy to isolate neuronal nuclei and Il33-mCherry nuclei for single-cell RNA sequencing.
(H and I) Correlation between mCherry levels and dendritic spine or spine head filopodia numbers (Pearson correlation, n = 20 neurons, > 110 spines per neuron, 3 mice). Different colors correspond to neurons from individual mice.
(J) Representative image and quantification of the number of cFos+ cells in the DG of IL-33 cKO and control mice (t test, n = 4 mice/genotype). Scale bar: 200 μm.
(A) Gating strategy for isolation of hippocampal microglia (CD11b), neurons (NeuN), astrocytes (GFAP), and oligodendrocytes (Olig2) by FACS and expression of cell-type specific markers by qPCR (n = 4 mice).
(B) Expression of II1rI1 transcript in neurons and glia by qPCR of flow sorted hippocampal cell types (n = 3 mice).
(C) Schematic and representative image of Il1rl1 excision specificity in the hippocampus using the Cx3cr1creER allele fate mapped with the inducible Rosa26-tdTomato reporter (Ai14) and immunostaining with Iba1 and NeuN (n = 3 mice). Scale bar: 50 μm.
(D) Sensitivity of ll1rl1 excision by qPCR of flow sorted microglia using IL1RL1 i-cKO mice (Cr3cr1CreER/+:Il1rl1flox/flox; one-way ANOVA; Tukey’s post hoc test, n = 3 mice/group).
(E) Representative imageofCA1 pyramidal neuron and quantification of dendritic spine density after loss of neuronal IL-33 (t test, n = 30-34 dendritic segments, 3 mice/genotype). Scale bar: 20 μm.
(F) Scholl analysis of dendritic branching in DG granule cells after loss of neuronal IL-33 (t test, Holm-Sidak multiple comparisons; n = 12-16 neurons, 3 mice). Scale bar: 20 μm.
(G) Number of Iba1 + microglia in the DG of IL-33 cKO and control mice (t test, n = 3-4 mice). Scale bar: 200 μm.
(H) Representative images of Iba1 + microglia from control and IL1RL1 cKO mice housed under standard or enriched conditions (4 weeks). Quantification of the percentage of Iba1 + pixels (microglia coverage) in the molecular layer of the DG (one-way ANOVA, Tukey’s post hoc test, n = 3-4 mice/group). Scale bar: 20 μm.
(I) Representative image of HA epitope tag immunostaining after viral transduction with control (tdTomato) or IL-33 gain-of-function (IL-33ΔNLS) constructs. Scale bar: 20 μm.
*p < 0.05, **p < 0.01, ***p < 0.001. Data = mean ± standard deviation (bar graphs) and median ± interquartile range (violin plots).
(A) Representative image of BrdU labeled neurons35 days post-injection and quantification ofthe percentage of BrdU+ label-retaining cells that are NeuN+ in IL-33 cKO and control mice (t test, n = 5 mice/genotype). Scale bar: 20 μm.
(B) Dendritic spine density in 4-weekold newborn neurons labeled with MMLV-GFP retrovirus, which selectively targets proliferating cells, in IL-33 KO and control animals (t test, n = 34-53 dendritic segments, 3 mice). Scale bar: 20 μm, 2 μm (inset).
(C) Statistics for contextual fear discrimination assay in IL-33 cKO mice versus controls (related to Figures 4E–4G)
(D) Statistics for contextual fear discrimination assay in IL1RL1 cKO mice versus controls (related to Figures 4H–4J)
(E and F) Open field and Elevated plus maze testing of IL-33 cKO mice versus controls (t test, n = 16 control mice, 11 IL-33 cKO mice).
(G and H) Open field and Elevated plus maze testing of IL1RL1 cKO mice (t test, n = 12 mice/genotype).
*p < 0.05. Data = mean ± SD.
(A) Representative gating strategy to isolate neuronal nuclei from Prox1+Ctip2+ dentate granule cells and Prox1−Ctip2+ pyramidal neurons with fluorescence minus one (FMO) negative staining controls.
(B and C) Quantification of freezing in context A versus context B fourteen days post-training in young and old mice(n = 16 young mice and 14 old mice, 2-wayRM ANOVA, Sidak’s post hoc tests). Statistics for contextual fear discrimination assay in young versus old mice (also related to Figures 5F and 5G).
(D) Quantification of distance traveled in young and old mice during the first 2 s footshock (t test, n = 16 young and 14 old mice. Mean ± SD).
(E) Correlation between day 14 discrimination index and expression of Il33 mRNA in flow sorted dentate granule cells in individual old mice (Pearson correlation, n = 11 mice
*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
(A) Representative gating strategy to isolate live microglia (CD11b+CD45+).
(B) Representative hemisection from adult (P90) mouse showing Aggrecan immunostaining. Inset = hippocampus. Scale bar: 500 μm, 250 μm (inset).
(C) Aggrecan colocalization with WFA+ perineuronal nets around Parvalbumin+ interneurons in CA1. Scale bar: 50 μm.
(D and E) Representative image and quantification of microglial process coverage index calculated as the percentage of Iba1+ pixels in afield of view normalized to the percentage of tdTomato+ pixels after viral transduction with control or IL-33ΔNLS constructs (t test, n = 3 mice). Scale bar: 20 μm.
(F) Quantification of number of CD68+ lysosomes in microglia after viral transduction with control or IL-33ΔNLS constructs (t test, 28 microglia, 3 mice/group).
*p < 0.05. Data = mean ± standard deviation (bar graph) and median ± interquartile range (violin plot).
Data Availability Statement
Single nuclei RNA sequencing and microglia bulk RNA sequencing data were deposited in NCBI’s Gene Expression Omnibus (Barrett et al., 2013) under accession number GSE150714.