Abstract
Introduction
Canine parvovirus (CPV) disease is one of the most threatening to domestic and wild dogs.
Material and Methods
A total of 132 clinical samples were isolated from domestic dogs with diarrhoea from Henan, Hubei, Jiangsu, and Anhui provinces from 2016 to 2017, and 56 were positive for CPV-2 by PCR. A phylogenetic tree was constructed for the isolate sequences incorporating 53 non-Chinese reference strains.
Results
VP2 sequences showed the strains mainly to be new CPV-2a/2b and CPV-2c genotypes. The Ala5Gly, Phe267Tyr, Ser297Ala, Tyr324Ile, Gln370Arg, Asn426Asp or Asn426Glu, and Thr440Ala sites in the VP2 protein antigenic region were found to have high mutation rates. The VP2 tertiary structural model shows that the change at these mutation points is a factor for the changes in the protein structure. Significant differences between the Central Chinese strains and others were found, indicating that evolution is geographically related and extended in major regions. The homology between the identified strains confirmed their relationship. Phylogenetic analysis indicated that the common genotypes in the same clusters differ slightly in homology and evolutionary history.
Conclusion
This epidemiological study enriches the available data and serves as an important reference for studies on the evolution of CPV and selection of vaccines in China.
Keywords: canine parvovirus, epidemiology, phylogenetic tree, sequence analysis, mutation analysis
Introduction
Canine parvovirus (CPV; Protoparvovirus genus, Parvoviridae family) (2) is a non-enveloped, single-stranded DNA virus (18). It is a highly contagious pathogen that usually causes severe diarrhoea and is distributed worldwide; further, it is associated with high morbidity and mortality, and it has severely impacted dog breeding (12, 17). Serological analyses revealed that CPV is closely related to feline panleukopenia virus (FPV) and mink enteritis virus. The CPV variant of FPV, designated as CPV type 2 (CPV-2), was first reported in dogs in the 1970s (9, 15). The genotype of CPV-2 has evolved continuously, and its major antigenic variants (CPV-2a, CPV-2b, and CPV-2c), which are clinically prevalent, have completely replaced the classic CPV-2 (11, 17). Dogs infected with CPV-2 mainly present acute haemorrhagic gastroenteritis and myocarditis. Dogs of any age can be infected; however, puppies aged 2–6 months have the highest infection rate with mortality of ≥70% (8, 15). Healthy dogs can be affected via physical contact, and sick dogs can continue to excrete faeces containing viral particles even after treatment (3).
The genomic DNA of CPV-2 contains two major open reading frames (ORFs): ORF1 and OFR2 (16). ORF1 encodes nonstructural proteins (NS1 and NS2) that regulate gene expression and are the first translated in the early stage of viral infection; ORF2 encodes capsid proteins (VP1 and VP2) that regulate viral tendency and antigenicity (7). VP2 constitutes the main antigenic determinant, which is strongly immunogenic and can stimulate the body to produce neutralising antibodies as well as be used to prepare subunits or DNA vaccines. In addition, VP2 can bind to transferrin receptors on the host cell membrane; this mediates the extent of infection of the virus particles, and is related to the haemagglutination of the virus (4). Therefore, amino acid substitution in the protein sequence of VP2 may lead to changes in host range, tissue tropism, and the genetic and antigenic properties of the virus.
In the early 1980s, the two new antigenic variants CPV-2a and CPV-2b acquired high virulence and pathogenicity, and they gradually replaced CPV-2 worldwide. As CPV-2a strains, the virus has regained its ability to infect cats and other canines (1). Compared with the VP2 gene of classic CPV-2, that of CPV-2a contains five amino acid mutations, i.e., at Met87Leu, Ile101Thr, Ala300Gly, Asp305Try, and Val555Ile. The main difference between CPV-2b and CPV-2a is that the VP2 of CPV-2b strains has only two amino acid mutations (Asn426Asp and Ile555Val). In recent years, the new CPV-2a/2b were considered the dominant epidemic strains. The difference between the CPV-2a/2b and new CPV-2a/2b is that VP2 has mutated Ser297Ala; this is because the immune pressure by the host on the virus causes the emergence of a new genotype of CPV-2c (Asp426Glu) (4). Changes in the main amino acids of the amino acid sequence of VP2 affect its antigenic characteristics, host range, and pathogenicity (20). In recent years, reports of CPV-2 in Central China have been limited. Therefore, the aim of this study is to clarify the evolution of CPV 2 isolated from Central China, analysing the variation and pathogenic characteristics of its strains and thereby providing a theoretical basis for the prevention and control of related diseases.
Material and Methods
Sample collection and DNA extraction. A total of 132 faecal samples were isolated from rectal swabs of dogs suspected to be affected by CPV disease and admitted to animal hospitals in Henan, Hubei, Anhui, and Jiangsu provinces in Central China from 2016 to 2017. The isolated samples were placed in an EP tube with 1 mL of 0.9% sterilised normal saline. After 30s whirlpool oscillation, the mixture was evenly mixed. High-speed centrifugation was performed for 5 min at a rate of 12,000 rpm/min. The supernatant was transferred into a new, sterile 1.5-mL EP tube and stored at −20°C. Genomic DNA was extracted from the supernatant using the commercial EasyPure® Viral DNA/RNA Kit (TransGen Biotech, China) according to the manufacturer’s instructions. The extracted genomic DNA samples were stored at −80°C.
Sequencing of the VP2 fragment. According to the CPV VP2 gene sequence published by GenBank, one pair of primers CPV-F (5′-AGAGACAATCTTGCA CCAAT-3′) and CPV-R (5′-ATGTTAATATAATTT TCTAGGTGCT-3′) (from nucleotides 2761–4536 at MF805797) was designed using Primer Premier 5.0 software. DNA was added to a mix containing reaction buffer, GC enhancer, 6 pmol upstream/downstream primers, 0.4 mM dNTPs (3 μL), and Primer STAR HS DNA polymerase (TaKaRa Biotechnology Co., China) to obtain a total reaction volume of 20 μL. Sequence amplification was performed under the following cycling conditions: initial denaturation at 95°C for 3 min followed by 34 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1.48 min, with final extension at 72°C for 10 min. The VP2 gene sequences of CPV-2 isolates were cloned into the pMD19-T vector (commercial plasmid; TaKaRa Biotechnology Co.) and subjected to sequencing.
All 56 strains (length- 1,755 bp) were designated as follows: CN/HN1601–CN/HN1723, CN/HB1601–CN/ HB1715, CN/AH1601–CN/AH1710, and CN/JS1601– CN/HNJS1708. All complete genome sequences of the VP2 gene were simultaneously submitted to the NCBI GenBank database (http://www.ncbi.nlm.nih.gov) under accession numbers MK517966–MK518021. Detailed epidemiological data of these strains are presented in Table 1.
Table 1.
Epidemiological data of the 56 strains in the study and the reference strains for genotyping
| Strains | Accession nos. | Origin | Vaccination record | Age | CPV type | Substitution of amino acid residues in VP2 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5 | 267 | 297 | 324 | 370 | 426 | 440 | ||||||
| CPV-6.us.80 | EU659117 | the USA, 1980 | N.I.a | N.I. | CPV-2 | A | F | S | Y | Q | N | T |
| CPV-13.us.81 | EU659118 | the USA, 1981 | N.I. | N.I. | CPV-2a | A | F | S | Y | Q | N | A |
| CPV-411b.us.98 | EU659121 | the USA, 1998 | N.I. | N.I. | CPV-2b | A | F | A | Y | Q | N | A |
| CPV-HN1506 | MF467240 | Henan, 2017 | N.I. | N.I. | New-2a | A | Y | A | I | Q | N | A |
| RVC57 KNA | KY399053 | SNA, 2016 | N.I. | N.I. | New-2a | A | F | A | I | Q | N | A |
| BJ15-15 | KT162046 | Beijing, 2015 | N.I. | N.I. | New-2a | A | Y | A | I | Q | N | A |
| BJ15-11 | KT162026 | Beijing, 2015 | N.I. | N.I. | New-2b | A | Y | A | I | Q | D | A |
| VP2 | KR058183 | China, 2013 | N.I. | N.I. | New-2b | A | Y | A | I | Q | D | A |
| CPV-HN1506 | MF467239 | Henan, 2015 | N.I. | N.I. | New-2b | A | Y | A | I | Q | D | A |
| CPV dog HCM | LC216910 | Indonesia,2013 | N.I. | N.I. | CPV-2c | G | Y | A | I | R | E | T |
| CPV-HN1617 | MF467229 | Henan, 2016 | N.I. | N.I. | CPV-2c | G | Y | A | I | R | E | T |
| CPV-SH1516 | MG013488 | Shanghai, 2017 | N.I. | N.I. | CPV-2c | G | Y | A | I | R | E | T |
| CN/HN1601 | MK517966 | Henan, 2016 | N.I. | 3 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HN1602 | MK517967 | Henan, 2016 | 1 dose | 2 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HN1603 | MK517968 | Henan, 2016 | 2 doses | 5 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HN1604 | MK517969 | Henan, 2016 | 1 dose | 2 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HN1605 | MK517970 | Henan, 2016 | 1 dose | 2 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HN1606 | MK517971 | Henan, 2016 | 2 doses | 7 m | New-2a | A | Y | A | I | Q | N | A |
| CN/HN1607 | MK517972 | Henan, 2016 | 1 dose | 2 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HN1708 | MK517973 | Henan, 2017 | N.I. | 3 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HN1709 | MK517974 | Henan, 2017 | N.I. | 4 m | New-2a | A | Y | A | I | Q | N | A |
| CN/HN1710 | MK517975 | Henan, 2017 | 1 dose | 3 m | New -2a | A | Y | A | I | Q | N | A |
| CN/HN1711 | MK517976 | Henan, 2017 | 1 dose | 2 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HN1712 | MK517977 | Henan, 2017 | N.I. | 5 m | New-2a | A | Y | A | I | Q | N | A |
| CN/HN1713 | MK517978 | Henan, 2017 | 1 dose | 42 d | CPV-2c | G | Y | A | L | R | E | T |
| CN/HN1714 | MK517979 | Henan, 2017 | 1 dose | 2 m | CPV-2c | A | Y | A | Y | R | E | T |
| CN/HN1715 | MK517980 | Henan, 2017 | 1 dose | 5 m | New-2a | A | Y | A | I | Q | N | A |
| CN/HN1716 | MK517981 | Henan, 2017 | 2 doses | 4 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HN1717 | MK517982 | Henan, 2017 | N.I. | 2 m | New-2a | G | Y | A | I | Q | N | A |
| CN/HN1718 | MK517983 | Henan, 2017 | 1 dose | 37 d | New-2b | A | Y | A | I | Q | D | A |
| CN/HN1719 | MK517984 | Henan, 2017 | 1 dose | 2 m | New-2b | A | Y | A | I | Q | D | A |
| CN/HN1720 | MK517985 | Henan, 2017 | 2 doses | 4 m | New-2a | G | Y | A | I | Q | N | A |
| CN/HN1721 | MK517986 | Henan, 2017 | 1 dose | 2 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HN1722 | MK517987 | Henan, 2017 | N.I. | 6 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HN1723 | MK517988 | Henan, 2017 | 1 dose | 5 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HB1601 | MK517989 | Hubei, 2016 | N.I. | 40 d | CPV-2c | G | Y | A | I | R | E | T |
| CN/HB1602 | MK517990 | Hubei, 2016 | 1 dose | 5 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HB1603 | MK517991 | Hubei, 2016 | 1 dose | 3 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HB1704 | MK517992 | Hubei, 2017 | 1 dose | 3 m | CPV-2c | G | Y | A | I | R | E | A |
| CN/HB1705 | MK517993 | Hubei, 2017 | 2 doses | 4 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HB1706 | MK517994 | Hubei, 2017 | 1 dose | 2 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HB1707 | MK517995 | Hubei, 2017 | N.I. | 3 m | New-2a | A | Y | A | I | Q | N | A |
| CN/HB1708 | MK517996 | Hubei, 2017 | 1 dose | 33 d | CPV-2c | A | Y | A | I | R | E | T |
| CN/HB1709 | MK517997 | Hubei, 2017 | 1 dose | 2 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HB1710 | MK517998 | Hubei, 2017 | 2 doses | 4 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/HB1711 | MK517999 | Hubei, 2017 | N.I. | 6 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/HB1712 | MK518000 | Hubei, 2017 | 1 dose | 39 d | New-2a | A | Y | A | I | Q | N | A |
| CN/HB1713 | MK518001 | Hubei, 2017 | 1 dose | 3 m | CPV-2c | G | Y | A | L | R | E | T |
| CN/HB1714 | MK518002 | Hubei, 2017 | N.I. | 5 m | CPV-2a | A | Y | S | Y | Q | N | T |
| CN/HB1715 | MK518003 | Hubei, 2017 | 1 dose | 2 m | New-2a | G | Y | A | I | R | E | T |
| CN/AH1601 | MK518004 | Anhui, 2016 | 2 doses | 6 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/AH1602 | MK518005 | Anhui, 2016 | N.I. | 4 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/AH1603 | MK518006 | Anhui, 2016 | 1 dose | 3 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/AH1604 | MK518007 | Anhui, 2016 | 1 dose | 5 m | New-2b | A | Y | A | I | R | D | T |
| CN/AH1705 | MK518008 | Anhui, 2017 | N.I. | 8 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/AH1706 | MK518009 | Anhui, 2017 | 1 dose | 3 m | New-2b | A | Y | A | I | Q | D | A |
| CN/AH1707 | MK518010 | Anhui, 2017 | N.I. | 2 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/AH1708 | MK518011 | Anhui, 2017 | N.I. | 3 m | New-2b | G | Y | A | I | Q | D | T |
| CN/AH1709 | MK518012 | Anhui, 2017 | 2 doses | 4 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/AH1710 | MK518013 | Anhui, 2017 | 1 dose | 5 m | New -2a | G | Y | A | I | Q | N | T |
| CN/JS1601 | MK518014 | Jiangsu, 2016 | N.I. | 3 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/JS1602 | MK518015 | Jiangsu, 2016 | 1 dose | 2 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/JS1603 | MK518016 | Jiangsu, 2016 | N.I. | 41 d | New-2a | A | Y | A | I | Q | N | A |
| CN/JS1704 | MK518017 | Jiangsu, 2017 | N.I. | 4 m | New-2a | G | Y | A | I | Q | N | A |
| CN/JS1705 | MK518018 | Jiangsu, 2017 | 1 dose | 4 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/JS1706 | MK518019 | Jiangsu, 2017 | N.I. | 2 m | CPV-2c | A | Y | A | I | R | E | T |
| CN/JS1707 | MK518020 | Jiangsu, 2017 | 2 doses | 5 m | CPV-2c | G | Y | A | I | R | E | T |
| CN/JS1708 | MK518021 | Jiangsu, 2017 | 1 dose | 3 m | CPV-2c | A | Y | A | I | R | E | T |
no information
Phylogenetic and epidemiological analysis of CPV-2. To analyse the genetic diversity of the CPV-2 strains identified in Central China, the VP2 gene of the full-length sequences of CPV-2a, CPV-2b, CPV-2c, new CPV-2a, and new CPV-2b strains isolated from different geographical locations within China and from foreign countries were retrieved from the NCBI nucleotide database to construct a phylogenetic tree (detailed information of each reference strain used for constructing the phylogenetic tree is shown in Supplementary Table 1). A phylogenetic tree was generated for all 56 CPV-2 strains and 53 reference strains using MEGA 7.0. The maximum likelihood method was used to construct the phylogenetic tree with the pound value of 1,000. Then, the mutation sites of VP2 of strains in this study were summarised, compared with the original CPV-2 strain (accession no. ABD03872), and structurally analysed in the SWISS-MODEL (https://swissmodel.expasy.org/interactive) application, and finally post-modelling Pdb files were constructed with the PyMOL software for collation and preservation.
Results
Genotyping of CPV-2. Among the 132 samples, 56 were positive for CPV-2. Fig. 1 shows the target band size of some samples which tested positive. Sequence comparison revealed 97.3%–99.9% and 95.2%–99.6% nucleotide and amino acid homology between the Central Chinese and foreign reference strains, respectively. Further, 98.1%–99.8% nucleotide homology and 98.4%–100% amino acid homology were revealed among the 23 strains isolated from Henan Province.
Fig. 1.
Detection of CPV-VP2 strains by electrophoresis. M – 2000 Maker; 1-9 – CN/HN1601-1607, CN/HN1708, and CN/HN1709; N – negative control
Supplementary Table 1.
Information about the reference strains in our study
| Strain | Accession no. | Genotype | Place of isolation | Submission date |
|---|---|---|---|---|
| nn171025 | MK332005 | CPV-2a | Guangxi | 2017 |
| nn17101 | MK332003 | CPV-2a | Guangxi | 2017 |
| nn1693 | MK332002 | CPV-2b | Guangxi | 2016 |
| nn1681 | MK331996 | CPV-2b | Guangxi | 2016 |
| nn171105 | MK332007 | CPV-2c | Guangxi | 2017 |
| nn171024 | MK332004 | CPV-2c | Guangxi | 2017 |
| CPV-411b.us.9 | EU659121 | CPV-2b | the USA | 1998 |
| CPV-13.us.81 | EU659118 | CPV-2a | the USA | 1981 |
| CPV-6.us.80 | EU659117 | CPV-2 | the USA | 1980 |
| Raccoon/ WI/ 37/ 10 | JN867618 | CPV-2a | the USA | 2010 |
| 110/ 07-27 | FJ005236 | CPV-2c | the USA | 2007 |
| 08-B | GU362934 | CPV-2a | Italy | 2008 |
| 260-00 | MF177231 | CPV-2a | Italy | 2000 |
| 140/ 05 | FJ005265 | CPV-2b | Italy | 2005 |
| CPV /IZSSI /25835/ 09 | KU508407 | CPV-2c | Italy | 2009 |
| 56/00 | FJ222821 | CPV-2c | Italy | 2000 |
| CPV/dog/HCM/20/2013 | LC216910 | CPV-2c | Indonesia | 2013 |
| Pome | EF599098 | CPV2c(a) | South Korea | 2007 |
| DH326 | EF599097 | CPV-2b | South Korea | 2007 |
| DH426 | EF599096 | CPV-2a | South Korea | 2007 |
| 16M130 | MH643886 | CPV-2 | South Korea | 2016 |
| 2670/CPV-2c/2010/Ind | KX425920 | CPV-2c | India | 2010 |
| CU267 | MH711901 | CPV-2c | Thailand | 2017 |
| TH011401 | KT364589 | CPV-2c | Thailand | 2014 |
| T37 | CPU72698 | CPV-2a | Taiwan | 1996 |
| T10 | CPU72696 | CPV-2b | Taiwan | 1996 |
| 2017090801 | MH127909 | CPV-2c | Taiwan | 2017 |
| Protein(VP2) | KU244254 | CPV-2c | Taiwan | 2015 |
| PV/PL/HeN02/08 | EU441280 | CPV-2a | Henan | 2008 |
| Henan42 | KJ438805 | CPV-2a | Henan | 2013 |
| CPV-HN1617 | MF467229 | CPV-2c | Henan | 2016 |
| CPV-zj18 | KM386948 | CPV-2b | Zhejiang | 2014 |
| CPV-zj7 | KM386937 | CPV-2a | Zhejiang | 2014 |
| Beijing | HQ883267 | CPV-2a | Beijing | 2010 |
| BJ-1 | MN101726 | CPV-2a | Beijing | 2018 |
| 2011-BJ-B43 | KF803527 | CPV-2b | Beijing | 2011 |
| 2011-BJ-B6 | KF803606 | CPV-2b | Beijing | 2011 |
| CPV-SH1516 | MG013488 | CPV-2c | Shanghai | 2017 |
| Shanghai/04g/2016 | KY937646 | CPV-2a | Shanghai | 2016 |
| ShangHai/3-2/2016 | KY937640 | CPV2a | Shanghai | 2016 |
| Shanghai/03g/2016 | KY937637 | CPV-2c | Shanghai | 2016 |
| CPVpf/2007(vaccine) | FJ197847 | CPV-2 | South Korea | 2007 |
| 29/97(vaccine) | FJ222823 | CPV-2b | N.I.a | 2008 |
| CPV-GX1581 | MF467242 | CPV-2c | Guangxi | 2015 |
| CPV-HN1506 | MF467240 | New-2a | Henan | 2017 |
| RVC57 KNA 2016 | KY399053 | New-2a | Saint Kitts and Nevis | 2016 |
| SY40 | KY625992 | New-2a | China | 2016 |
| PU4 | KC429669 | New-2a | India | 2011 |
| BJ15-15 | KT162046 | New-2a | Beijing | 2015 |
| SY38 | KY625998 | New-2b | China | 2016 |
| BJ15-11 | KT162026 | New-2b | Beijing | 2015 |
| VP2 | KR058183 | New-2b | China | 2013 |
| CPV-HN1506 | MF467239 | New-2b | Henan | 2015 |
no information
The corresponding ranges elsewhere were 97.4%– 99.9% nucleotide homology and 96.1%–99.8% amino acid homology among the 15 strains isolated from Hubei Province, 98.8%–99.8% nucleotide homology and 98.3%–99.8% amino acid homology among the 8 strains isolated from Jiangsu Province, and 98.9%–99.8% nucleotide homology and 98.8%–99.7% amino acid homology among the 10 strains isolated from Anhui Province.
The genotype of the CPV-2 strain and the amino acid mutation in the VP2 sequence are shown in Table 1. Among the four provinces in Central China, the genotypes of 23 strains in Henan were new CPV-2a (30.43%), new CPV-2b (8.70%), and CPV-2c (60.87%). Among the 15 strains in Hubei, the genotypes of CPV-2a comprised 6.67%, those of new CPV-2a 13.33%, and those of CPV-2c 80.00%. There were only two genotypes of new CPV-2a (25.00%) and CPV-2c (75.00%) in Jiangxi. The three genotypes in Anhui were new CPV-2a (10.00%), new CPV-2b (30.00%), and CPV-2c (60.00%). The geographical distribution and epidemiological survey results are presented in Fig. 2.
Fig. 2.
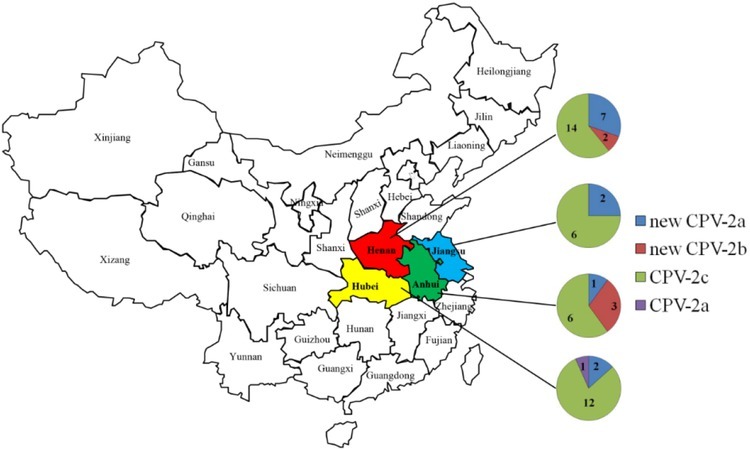
Geographical distribution and genotype of CPV isolates obtained in the study
Analysis of the mutation site in the VP2 protein. Based on the analysis of VP2 sequences, a total of five amino acid mutations were identified in the 56 CPV-2 strains. In the present study, residue 5 (Ala→Gly) was present in CPV-2c (44.64%), and all instances fit the mutation of the CPV-2c reference strain. Residue 267 (Phe→Tyr), residue 297 (Ser→Ala), and residue 324 (Tyr→Ile) were present in new CPV-2a, new CPV-2b, and CPV-2c at rates of 100%, 98.21%, and 92.86%, respectively. Regarding the identified CPV-2 strain, the mutation of residue 370 (Gln→Arg) requires further investigation in terms of its relationship with pathogenicity. In the present study, the mutation rate of this residue reached 69.64%, and it was specific for CPV-2c. Residue 426 was the main residue to distinguish CPV-2a, CPV-2b, and CPV-2c. 12 strains were Asn (21.43%) and a total of 5 strains had a mutation of residue 426 as Asn→Asp (8.93%); this mutation corresponds to that observed in CPV-2b. In the remaining 39 strains (69.64%), the mutation of residue 426 involved Asn→Glu. A total of 15 strains harboured the mutation of residue 440 (Thr→Ala) including 9, 3, 2, and 1 strain isolated from Henan, Hubei, Jiangsu, and Anhui provinces, respectively. This mutation was identical to those observed in the CPV-2a, CPV-2b, CPV-2c, new CPV-2a, and new CPV-2b reference strains. The distribution of relevant amino acid mutation residues in the VP2 tertiary structural model of canine parvovirus is presented in Fig. 3.
Fig. 3.
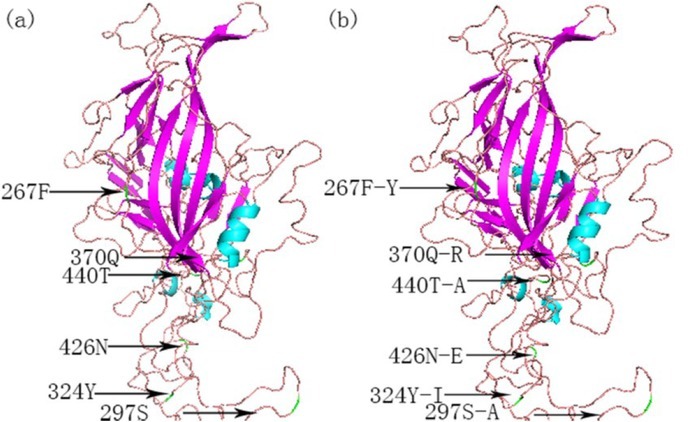
Tertiary structural model of canine parvovirus (CPV) capsid protein (VP2) and the distribution of the main amino acid mutation residues. (a) – conserved structure of VP2 and (b) – structure of the mutant VP2 protein
Phylogenetic analysis. Sequence analysis of 56 strains isolated from Central China and 53 isolated from foreign countries and regions in the database was conducted to obtain a phylogenetic tree (Fig. 4). The tree shows that CPV strains from four provinces in Central China are close to CPV-2c, new CPV-2a, and new CPV-2b. The evolutionary relationships of these strains indicate that they are distinct from the strains isolated from the United States, Italy, and South Korea. Similar to certain CPV-2c reference strains were 14 strains in Henan Province, 12 in Hubei Province, 6 in Jiangsu Province and 8 in Anhui Province. They were similar to CPV-2c reference strains, isolated from Thailand, Indonesia, Taiwan, Guangxi, Shanghai, and Henan Province. New CPV-2a in references was very close to the new Chinese CPV-2a isolates, and 12 strains isolated in this study were highly similar to them. The new CPV-2a strain from India was highly similar to the new CPV-2a strain isolated from Central China. In addition, CN/HB1714 was close to the CPV-2a reference strain from Henan Province. One strain from Anhui and two from Henan are similar to the new CPV-2b reference strain.
Fig. 4.
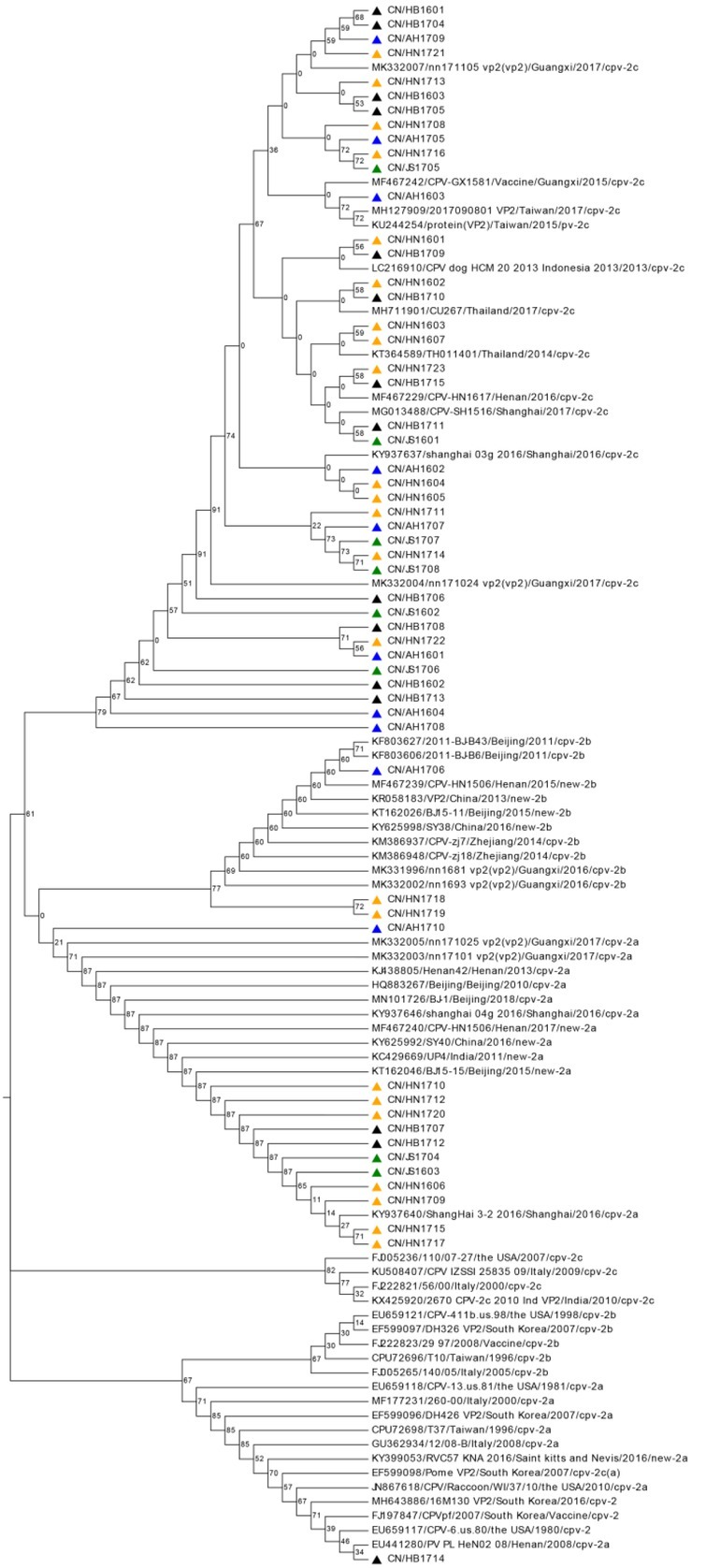
Phylogenetic tree based on VP2 gene sequences of canine parvovirus type 2 (CPV-2). Strains isolated from four central Chinese provinces in this study are marked with coloured triangles. The strains isolated from Hubei Province are indicated by black triangles; those from Anhui blue; those from Henan by orange; and those from Jiangsu green
Discussion
Since the first report on CPV-2 infection, the epidemic characteristics of CPV-2 have substantially varied among different regions (13). CPV isolates from the United States and some European countries mainly belong to the CPV-2b and CPV-2c subtypes, and the CPV-2a and CPV-2c subtypes are the most prevalent in Asian countries (5). In China, CPV-2 was the first isolated in 1982 and has developed into one of the most harmful canine pathogens (6). At present, vaccine-based immunisation is the major strategy to prevent and control CPV-2 in China; thus, understanding the latest epidemic situation and genetic variation of the circulating CPV-2 strains is necessary.
Recently, epidemiological investigation in Central China has only been reported in Henan Province. In the present study, dogs suspected of CPV-2 infections in Henan, Hubei, Anhui, and Jiangsu provinces in Central China were investigated, and a total of 56 dogs were identified to be infected with it. The VP2 gene of the 56 CPV-2 strains was amplified and sequenced. Analysis showed that the main subtype of these strains was CPV-2c followed by new CPV-2a; new CPV-2b was not identified in Hubei or Jiangsu provinces. The survey results can only be used as a reference to judge the current situation of the viral epidemics in Central China; thus, a large-scale survey with more samples and analysis data is needed.
In recent years, new CPV-2a has been emerging as the predominant epidemic strain of CPV-2 in some Chinese provinces (7, 19). To the best of our knowledge, a wide distribution of CPV-2c and new CPV-2a variants has not been reported in Central China before this study.
To summarise the current reports on residue analysis, the seven reported mutation residues related to virulence strength and host range were analysed in this study and compared with mutation residues in the reference strains. The amino acid mutation Thr440Ala in VP2 is associated with the evolution of antigenic variants. All new CPV-2a strains described in this study harboured a mutation of residue 440 (Thr→Ala). The mutation of residue 426 as Asn→Asp serves as a marker for distinguishing CPV-2b, whereas that of residue 426 as Asn→Glu helps determine CPV-2c genotype. Regarding the identified CPV-2c, the mutation rate for residue 370 (Gln→Arg) reached 69.64%, which is suspected to be caused by long-term antibody pressure. The mutation of residue 324 (Tyr→Ile) displayed in the 56 strains and all reference strains has spread worldwide since its first report, which indicated that the change at residue 324 has been a common mutation in CPV-2c in Asian countries. Furthermore, the amino acids at position 324 are present on the outer surface of the virions, and mutation at this position may cause changes in the tertiary structural model structure of the protein, leading to the enhancement of the binding ability of the viral receptor and a change in the host range (10, 14). In this study, mutations of Ser297Ala (98.21%) and Phe267Tyr (100%) were found in CPV-2c and new CPV-2a/2b; a change at these points causes a change in the antigenicity of CPV-2 and is, therefore, used as a marker to distinguish new CPV-2a/2b strains (14). It can be clearly seen from the tertiary structural model of the protein that change at an amino acid mutation site will affect the protein structure, which may change the pathogenicity of the virus. During virus transmission, the risk of enhanced virulence or increased clinical severity is extremely high. All these mutation sites may be affected by changes in the fine structure of the virus, thereby weakening the binding ability of specific antibodies to viral antigens. Therefore, the immune response of immunised animals should be strengthened, and protective measures should be taken for the clinical treatment and prevention of CPV disease. The biological function of all site mutations identified in the present study needs to be confirmed with animal regression studies, and extensive epidemiological investigations are needed. According to the currently reported information on site mutations, timely monitoring of mutations at each site as well as adjusting CPV-2 treatment and prevention measures are necessary.
In the present study, the type and geographical distribution of the reference strains in the phylogenetic tree were analysed, and the results were found to be highly comprehensive and complete. Evolutionary analysis of the classical strains isolated from the United States and Italy compared with those isolated from four provinces showed the degree of difference. Since the discovery of CPV-2, its evolution has been rapid. New CPV-2a and new CPV-2b were highly similar to CPV-2a and CPV-2b, respectively, which indicated that these two new genotypes mutated from their respective ancestors. The trend of the VP2 phylogenetic tree was consistent with that reported in a previous study (7). Meanwhile, the CN/HN/1714 strain belonged to CPV-2a, which was closely related to the Japanese reference strains.
In conclusion, the present study revealed new epidemiological data of CPV-2 in Central China, including the co-circulation of new CPV-2a and new CPV-2c with high variation. In addition, the molecular characterisation of the strains isolated from Central China increased the understanding of the epidemic characteristics of CPV-2 strains worldwide. This study may be a scientific reference for CPV vaccine-related research and development.
Footnotes
Conflict of Interest
Conflict of Interests Statement: The authors declare that there is no conflict of interests regarding the publication of this article.
Financial Disclosure Statement: This study was supported by the National Natural Science Foundation of China (grants no. 31802185 and 31870917), the Scientific and Technological Project of Henan Province (grant no. 182107000040), the programme for Innovative Research Team of Science and Technology in the University of Henan Province (grant no. 20IRTSTHN024), the Key Scientific and Technological Project of the Education Department of Henan Province (grant no. 18A230012), and Technological Project of Nanyang Normal University (grants no. 18046 and 2019QN009).
Animal Rights Statement: None required.
References
- 1.Battilani M., Balboni A., Ustulin M., Giunti M., Scagliarini A., Prosperi S.. Genetic complexity and multiple infections with more Parvovirus species in naturally infected cats. Vet Res. 2011;42:43. doi: 10.1186/1297-9716-42-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cotmore S.F., Agbandje-McKenna M., Chiorini J.A., Mukha D.V., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A.J.. The family Parvoviridae. Arch Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decaro N., Desario C., Campolo M., Elia G., Martella V., Ricci D., Lorusso E., Buonavoglia C.. Clinical and virological findings in pups naturally infected by canine parvovirus type 2 Glu-426 mutant. J Vet Diagn Invest. 2005;17:133–138. doi: 10.1177/104063870501700206. [DOI] [PubMed] [Google Scholar]
- 4.Decaro N., Desario C., Parisi A., Martella V., Lorusso A., Miccolupo A., Mari V., Colaianni M.L., Cavalli A., Di Trani L., Buonavoglia C.. Genetic analysis of canine parvovirus type 2c. Virology. 2009;385:5–10. doi: 10.1016/j.virol.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Dogonyaro B.B., Bosman A.M., Sibeko K.P., Venter E.H., van Vuuren M.. Genetic analysis of the VP2-encoding gene of canine parvovirus strains from Africa. Vet Microbiol. 2013;165:460–465. doi: 10.1016/j.vetmic.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Geng Y., Guo D., Li C., Wang E., Wei S., Wang Z., Yao S., Zhao X., Su M., Wang X., Wang J., Wu R., Feng L., Sun D.. Co-Circulation of the Rare CPV-2c with Unique Gln370Arg Substitution, New CPV-2b with Unique Thr440Ala Substitution, and New CPV-2a with High Prevalence and Variation in Heilongjiang Province, Northeast China. PLoS One. 2015;10:e0137288. doi: 10.1371/journal.pone.0137288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han S.C., Guo H.C., Sun S.Q., Shu L., Wei Y.Q., Sun D.H., Cao S.Z., Peng G.N., Liu X.T.. Full-length genomic characterizations of two canine parvoviruses prevalent in Northwest China. Arch Microbiol. 2015;197:621–626. doi: 10.1007/s00203-015-1093-4. [DOI] [PubMed] [Google Scholar]
- 8.Horiuchi M., Goto H., Ishiguro N., Shinagawa M.. Mapping of determinants of the host range for canine cells in the genome of canine parvovirus using canine parvovirus/mink enteritis virus chimeric viruses. J Gen Virol. 1994;75:1319–1328. doi: 10.1099/0022-1317-75-6-1319. [DOI] [PubMed] [Google Scholar]
- 9.Horiuchi M., Yamaguchi Y., Gojobori T., Mochizuki M., Nagasawa H., Toyoda Y., Ishiguro N., Shinagawa M.. Differences in the evolutionary pattern of feline panleukopenia virus and canine parvovirus. Virology. 1998;249:440–452. doi: 10.1006/viro.1998.9335. [DOI] [PubMed] [Google Scholar]
- 10.Hueffer K., Parker J.S., Weichert W.S., Geisel R.E., Sgro J.Y., Parrish C.R.. The natural host range shift and subsequent evolution of canine parvovirus resulted from virus-specific binding to the canine transferrin receptor. J Virol. 2003;77:1718–1726. doi: 10.1128/JVI.77.3.1718-1726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miranda C., Thompson G.. Canine parvovirus: the worldwide occurrence of antigenic variants. J Gen Virol. 2016;97:2043–2057. doi: 10.1099/jgv.0.000540. [DOI] [PubMed] [Google Scholar]
- 12.Paradiso P.R., Rhode S.L. 3rd, Singer I.I.. Canine parvovirus: a biochemical and ultrastructural characterization. J Gen Virol. 1982;62:113–125. doi: 10.1099/0022-1317-62-1-113. [DOI] [PubMed] [Google Scholar]
- 13.Parrish C.R., Have P., Foreyt W.J., Evermann J.F., Senda M., Carmichael L.E.. The global spread and replacement of canine parvovirus strains. J Gen Virol. 1988;69:1111–1116. doi: 10.1099/0022-1317-69-5-1111. [DOI] [PubMed] [Google Scholar]
- 14.Perez R., Calleros L., Marandino A., Sarute N., Iraola G., Grecco S., Blanc H., Vignuzzi M., Isakov O., Shomron N., Carrau L., Hernandez M., Francia L., Sosa K., Tomas G., Panzera Y.. Phylogenetic and genome-wide deep-sequencing analyses of canine parvovirus reveal co-infection with field variants and emergence of a recent recombinant strain. PLoS One. 2014;9:e111779. doi: 10.1371/journal.pone.0111779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pletcher J.M., Toft J.D. 2nd, Frey R.M., Casey H.W.. Histopathologic evidence for parvovirus infection in dogs. J Am Vet Med Assoc. 1979;175:825–828. [PubMed] [Google Scholar]
- 16.Reed A.P., Jones E.V., Miller T.J.. Nucleotide sequence and genome organization of canine parvovirus. J Virol. 1988;62:266–276. doi: 10.1128/jvi.62.1.266-276.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Truyen U.. Emergence and recent evolution of canine parvovirus. Vet Microbiol. 1999;69:47–50. doi: 10.1016/s0378-1135(99)00086-3. [DOI] [PubMed] [Google Scholar]
- 18.Wang D., Yuan W., Davis I., Parrish C.R.. Nonstructural protein-2 and the replication of canine parvovirus. Virology. 1998;240:273–281. doi: 10.1006/viro.1997.8946. [DOI] [PubMed] [Google Scholar]
- 19.Xu J., Guo H.C., Wei Y.Q., Shu L., Wang J., Li J.S., Cao S.Z., Sun S.Q.. Phylogenetic analysis of canine parvovirus isolates from Sichuan and Gansu provinces of China in 2011. Transbound Emerg Dis. 2015;62:91–95. doi: 10.1111/tbed.12078. [DOI] [PubMed] [Google Scholar]
- 20.Zhou P., Zeng W., Zhang X., Li S.. The genetic evolution of canine parvovirus - A new perspective. PLoS One. 2017;12:e0175035. doi: 10.1371/journal.pone.0175035. [DOI] [PMC free article] [PubMed] [Google Scholar]


