Abstract
Effect size can differ as a function of the elapsed time since treatment or as a function of other key covariates, such as sex or age. In evidence synthesis, a better understanding of the precise conditions under which treatment does work or does not work well has been highly valued. With increasingly accessible individual patient or participant data (IPD), more precise and informative inference can be within our reach. However, simultaneously combining multiple related parameters across heterogeneous studies is challenging because each parameter from each study has a specific interpretation within the context of the study and other covariates in the model. This paper proposes a novel mapping method to combine study-specific estimates of multiple related parameters across heterogeneous studies, which ensures valid inference at all inference levels by combining sample-dependent functions known as Confidence Distributions (CD). We describe the “CD-based mapping method” and provide a data application example for a multivariate random-effects meta-analysis model. We estimated up to 13 study-specific regression parameters for each of 14 individual studies using IPD in the first step, and subsequently combined the study-specific vectors of parameters, yielding a full vector of hyperparameters in the second step of meta-analysis. Sensitivity analysis indicated that the CD-based mapping method is robust to model misspecification. This novel approach to multi-parameter synthesis provides a reasonable methodological solution when combining complex evidence using IPD.
Keywords: Multi-parameter synthesis, Multivariate random-effects meta-analysis, Mapping matrix, Combining confidence density functions, Individual patient data, Individual participant data
1. INTRODUCTION
Meta-analysis is a well-established statistical procedure for quantitatively synthesizing evidence from independent studies [43]. In recent years, meta-analysis has increasingly been discussed as an important research method to strengthen statistical inference [25]. Compared to traditional, standard meta-analysis of aggregate data (AD), meta-analysis of individual patient or participant data (IPD) in a one-step or two-step approach [49] or in a one-step, simultaneous “integrative data analysis” (IDA) of IPD [8, 24] represents a major innovation, which can expand the scope of evidence synthesis and produce clinically most meaningful results. When IPD are available, the same model can be applied across all studies, ensuring that the combined data have the same interpretation. In addition, it is possible to address more complex research questions with more appropriate and sophisticated models. Overall, meta-analysis of IPD provides unparalleled flexibility for analysts. At the same time, it is challenging to combine IPD from heterogeneous trials because they differ in key study features, including designs, populations, measures, or settings [20, 53]. For example, with respect to different measures, a commonly used screening equipment in medical settings may not always be available in other locations. Furthermore, longitudinal clinical trials included in a meta-analysis typically differ in their study duration and follow-up frequency and period [32, 57]. It is also not uncommon that binary or categorical covariates may not have any variability either by design (e.g., a study of all women) or naturally in a data set [11, 33]. Any one of these situations can pose significant estimation challenges for an IPD meta-analysis because they essentially represent study-level missing data. Typically, IPD meta-analysis applications have included a subset of studies that have all covariates or used a simpler model with a fewer number of covariates, either of which essentially deletes partially available data (i.e., listwise deletion) and represents an important loss of information, precision, power, and generalizability.
We propose a new information combination method that combines confidence distributions, hereon called the “CD-based approach” [59]. A confidence distribution (CD) is a sample-dependent distribution function that contains information about confidence intervals of a parameter of interest at all levels. It can be referred to as a confidence density if presented in a density function form [34]. This new method has been demonstrated as a powerful inference tool in connection with meta-analysis [6, 34, 60, 62]. The current study extends the CD-based approach to a multivariate random-effects meta-analysis model, which is based on more reasonable assumptions but computationally more challenging, compared with a multivariate fixed-effects meta-analysis model [34]. The current work, which explicitly accommodates heterogeneous designs and partial information, also provides a more general framework than the existing model [63] for random-effects meta-analysis models.
To implement the CD-based approach, we utilize a “two-step” IPD meta-analysis approach [54]. We proceed as follows. First, we identify an underlying “full” model for all studies included in a meta-analysis and conduct separate analyses for each study to obtain study-specific parameter estimates. Second, we identify appropriate connections between the expectation of study-specific vectors of parameters and the hyperparameters of the full model via appropriately identified “mapping” matrices. We subsequently estimate all hyperparameters of the full model in a multivariate random-effects meta-analysis model. Upon obtaining all hyperparameters, we can flexibly derive any relevant population-level inferences as needed (e.g., different treatment effect sizes for men vs. women).
The rest of this paper has four main sections. Section 2 discusses the data set that motivated the CD-based mapping method. Section 3 introduces the method in greater detail. Section 4 presents the findings, including those from sensitivity analyses. We conclude this paper with discussion in Section 5.
2. MOTIVATING DATA
The current study was motivated by Project INTEGRATE [37]. Project INTEGRATE obtained de-identified, item-level IPD from 24 trials through a network of interested collaborators. All trials tested the efficacy of brief motivational interventions (BMIs) to reduce excessive alcohol use and prevent harm among college students. Typical BMIs are brief, and provide personalized feedback on alcohol use and alcohol-related problems, as well as general educational information on alcohol.
2.1. Intervention and control groups
IPD for the current analysis come from 7,996 participants from 14 trials at baseline after excluding studies featuring a single intervention (i.e., no comparison group) or unique interventions, resulting in 10 stand-alone, personalized feedback intervention (“PF”) groups; eight in-person motivational intervention with personalized normative feedback profile (“MI + PF”) groups; and 13 no-treatment control (“Control”) groups. Note that studies 13 and 14 were originally independent trials but were subsequently collapsed (designated as study 13/14), given their similarities in study design characteristics and small samples. In addition, no systematic differences existed across intervention groups at baseline for studies 13 and 14. Tables 1 and 2, as well as Figure 1, provide an overview of all studies and descriptive statistics. With the exception of one study (study 1), all remaining 13 studies had a control group. Eleven of the 13 studies had an assessment-only control group, and the remaining two studies (studies 18 and 20) had a control group who received a single page information sheet containing very limited, generally-written information about alcohol use (e.g., alcohol has no nutritional value; space your drinks). Based on the quantitative content analysis of all intervention materials across groups, we determined that the exposure for the latter two groups was essentially the same as what other 11 control groups received [40, 45].
Table 1.
Baseline and follow-up assessment schedule by study
| Study | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | X | X | X | ||||||||||
| 2 | X | X | X | ||||||||||
| 8a | X | X | |||||||||||
| 8b | X | X | |||||||||||
| 8c | X | X | |||||||||||
| 9 | X | X | X | ||||||||||
| 10.1 | X | X | |||||||||||
| 11 | X | X | X | ||||||||||
| 12 | X | X | X | X | |||||||||
| 13/14 | X | X | X | X | |||||||||
| 18 | X | X | X | ||||||||||
| 20 | X | X | |||||||||||
| 21 | X | X | X | X | |||||||||
| 22 | X | X |
Notes. “X” indicates that baseline or follow-up outcome data exist at that time.
Table 2.
Means, standard deviations in parentheses, and percentages of the variables by study
| Variable | |||||
|---|---|---|---|---|---|
| Study | Man | White | First-year | PBS | AlcProb |
| 1 | 60% | 73% | 62% | 1.06(0.79) | 0.21(0.72) |
| 2 | 71% | 69% | 63% | 1.01(0.84) | −0.60(0.72) |
| 8a | 33% | 87% | 50% | 0.55(0.85) | −0.08(0.97) |
| 8b | 41% | 62% | 47% | 0.57(0.89) | −0.11(1.00) |
| 8c | 38% | 83% | 36% | 0.46(0.91) | 0.02(0.94) |
| 9 | 38% | 73% | 100% | 0.42(0.77) | 0.75(0.71) |
| 10.1 | 46% | 84% | 100% | - | 1.03(0.76) |
| 11 | 59% | 64% | 100% | - | −0.82(0.95) |
| 12 | 47% | 93% | 3% | −0.46(0.92) | 0.39(0.60) |
| 13/14 | 38% | 95% | 26% | - | 1.08(0.51) |
| 18 | 25% | 89% | 33% | 0.49(0.88) | −0.17(0.96) |
| 20 | 52% | 84% | 78% | - | 0.62(0.83) |
| 21 | 36% | 85% | 42% | 0.36(0.92) | 0.89(0.77) |
| 22 | 43% | 87% | 100% | 0.38(0.89) | −0.18(0.90) |
Notes. Man (coded 1; 0 = woman), White (coded 1; non-White = 0), First-year (coded 1; 0 = non first-year). PBS = Estimated latent trait (θ) scores at baseline for utilizing protective behavioral strategies. AlcProb = Estimated latent trait scores at baseline for alcohol-related problems. “−” indicate that the variable was not assessed. The presented descriptive statistics were obtained from the entire study sample.
Figure 1.
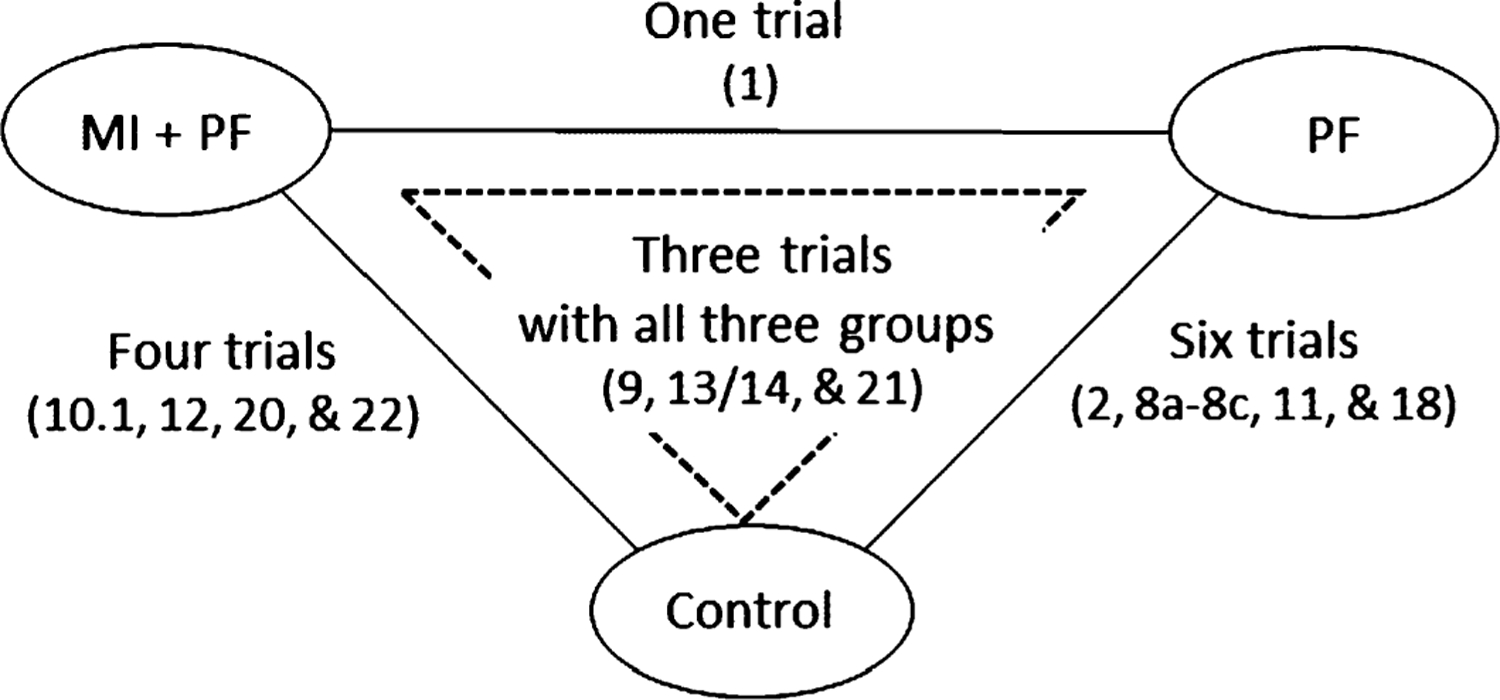
A diagram of the evidence network (numbers in parenthesis indicate studies).
2.2. Measures
We focus on alcohol-related problems (e.g., neglecting responsibilities; friends and relatives avoiding you) as the outcome variable of the current analysis. Because this outcome variable was assessed differently across studies (i.e., slightly differently worded items, different referent time frames, and response options), they could not be directly pooled. Therefore, we previously utilized a 2-parameter logistic item response theory (2-PL IRT) model to derive latent trait scores (called theta [θ] scores) in a separate hierarchical, multi-unidimensional IRT analysis using the Markov chain Monte Carlo algorithms that we specifically developed and validated for Project INTEGRATE [23]. Latent trait scores from IRT models can be interpreted with direct reference to item parameters, and are independent of which items that participants were tested on or who else was tested together [12]. IRT models are widely used in educational test settings to estimate latent trait (e.g., ability or severity) scores and increasingly utilized also for psychological and medical research [18]. The latent trait scores for one’s tendency to adopt protective behavioral strategies prior to, during, and after drinking to protect oneself from experiencing negative consequences from drinking [36], such as setting limits or alternating drinks, were estimated using a generalized partial credit IRT model [41] to accommodate polytomous responses [23, 37, 39]. The latent trait scores are estimated based on the assumption that the distribution follows a standard normal (i.e., an expected population mean of 0).
2.3. Motivating challenges
Example 1. Heterogeneity in study interventions. Figure 1 shows that there are up to three intervention arms in data. However, only three studies (studies 9, 13/14, and 21) have all three intervention groups, providing direct comparative evidence between three pairs of groups. The remaining 11 studies are two-arm trials, of which one did not have a control (study 1). The challenge that arises is how to draw valid inference regarding relative effectiveness of two competing interventions (i.e., “MI + PF” vs. “PF”) from the perspective of network meta-analysis [35], despite that we have (1) one missing treatment arm for 11 studies and (2) one study (i.e., study 1) without a control group. The latter condition results in several unique study-specific parameters, which require a “blueprint” directing how they can be related to study-specific parameters of other studies, and to their corresponding hyperparameters of the full model.
Example 2. Heterogeneity in the assessment of covariates. Studies 9, 10.1, 11, and 22 recruited exclusively first-year students. In these studies, study-specific parameters for first-year student status (coded 1 = first-year; 0 = other) are not estimable and cannot be linked directly to their counterpart from other studies or to the corresponding full model hyperparameter because the intercept parameters from these four studies would indicate the mean outcome level for their first-year students, assuming all study-specific covariates in the model are held constant (e.g., constrained to zero) within studies. In contrast, study-specific intercept parameters from the remaining 10 studies would reflect the mean outcome level of the students in 2nd year and above (i.e., referent demographic group) when all other covariates are held constant within studies. Therefore, the interpretation of the study-specific intercept parameters differs between studies due to the study design difference. Ignoring the study design difference would mean that the combined hyperparameter estimate for the intercept would be biased and/or not interpretable.
If one were to drop this covariate, the four studies with all first-year students would be retained in the analysis. However, first-year college students have higher levels of alcohol-related problems, compared with students in the second year and beyond. Due to their high risk status, the studies with only first-year students may have specific intervention content and different intervention effect sizes, compared with other studies that provide the same intervention for students across all years in college. If we exclude the studies that recruited exclusively first-year students, then resulting inferences would suffer from reduced power. Therefore, either option-excluding the covariate or the studies-can result in non-negligible loss of information and biased inference in a typical meta-analysis. Clearly, there is a need to properly separate the effect of interest from potential confounding effects when simultaneously combining multiple related parameters from heterogeneous studies.
Example 3. Availability of follow-up assessments. Some studies had a single post-intervention follow-up assessment, whereas others had at least two follow-up assessments within 12 months post intervention (see Table 1). Such between-study design differences can cause study-level missing data under certain full models. For example, if a true full model has a quadratic functional form, then it would require at least three data points over time to fit linear and quadratic terms in a longitudinal model. One may choose a simpler model that does not correctly reflect true change processes. Alternatively, one may limit the analysis to a subset of studies with a sufficient number of follow-up assessments. Neither option is optimal.
Note that the motivating challenges illustrated above do not represent a missing data problem within individual studies. However, when data from independently conducted studies are pooled in a meta-analysis, any between-study design heterogeneity, including differences in the number of treatment arms, comparison or control group, and lack of variability in covariates, can lead to a challenge in estimation and interpretation. In other words, study-specific parameters need to be made equivalent via “mapping” so that each study-specific parameter can be validly linked to the corresponding hyperparameter(s) of the full model. A multivariate CD-based approach provides a novel method, which incorporates mapping matrices in the estimation, for a multivariate fixed-effects or random-effects meta-analysis model.
3. METHODS
3.1. A mapping method with a CD-based meta-analysis approach
Consider k independent studies each with ni observations for i = 1, ⋯ , k. First, we formulate a full model as a generalized linear mixed model. We assume a random-intercept model with a total of p − 1 covariates across k studies:
where g(·) is the link function, E(·) denotes expectation, and βi = (β0i, β1i, β2i, … , β(p−1)i)T represents a study-specific parameter vector. yijt indicates the outcome for participant j in study i at time t, and xdijt is value of the dth covariate for participant j in study i at time t, where βdi indicates the coefficient associated with the dth covariate for study i with d = 0, ⋯ , p − 1. The term uij0 indicates a participant-specific random intercept effect. The link function is an identity link for a linear model with a continuous outcome. Other link functions can be specified depending on the distribution of outcomes [62].
The full model for the motivating example at the participant level is:
where the terms Man, White, First-year, PF, and MI + PF are binary indicator variables; PBS and Month are continuous variables; and the rest of the terms are either interaction terms between the aforementioned ones and/or a quadratic form. Table 2 shows the descriptive statistics of all covariates.
Throughout Section 3, we denote bi as the study-specific estimates of the corresponding Level-1 parameters βi for study i, E(βi) as the expectation of the study-specific vector of parameters at Level 1 (with dimension pi, where pi ≤ p), and β as the full vector of hyperparameters at Level 2 (with dimension p), respectively. We assume that a study-specific parameter vector βi follows a multivariate normal distribution with mean β and covariance matrix Σ for a multivariate random-effects meta-analysis model, an extension of its univariate counterpart [43]. As long as the the expectation of study-specific parameters has the same interpretation across studies, we can directly fit the full model (i.e., without any mapping) and obtain the full vector of hyperparameter estimates β and its covariance matrix Σ as follows:
where MVNpi stands for the multivariate normal distribution with dimension pi, Si is the observed covariance matrix for study i, and Σ is the unknown between-study covariance matrix that needs to be estimated. With a set of heterogeneous studies (study i = 1, 2, ⋯ , k) and covariate d = 1, 2, ⋯ , p − 1, we additionally identify and adopt an appropriate study-specific mapping matrix Mi for study i where Mi is a pi × p matrix, and obtain the full model as follows:
We first obtain bi for study-specific parameters βi for study i in the first step. Next, we identify an appropriate mapping matrix Mi for study i and link all study-specific parameters to their corresponding hyperparameters of the full model in the second step. Let Mi be the mapping function for study i that links βi to β: E(βi) ≡ Mi (β). In a linear model, the relationship E(βi) ≡ Mi (β) can typically be simplified to the following linear equation: E(βi) ≡ Miβ, where Mi is a pi × p matrix.
3.2. Mapping matrix to connect study-specific parameters to hyperparameters
As an illustration, we show how to determine an appropriate mapping matrix connecting the study-specific parameter vector βi to the hyperparameter vector β. Let us assume that we have a model of a continuous response variable y with two continuous covariates x1 and x2:
where subscripts i and j index study and participant, respectively. If study i has all the covariates required, then
In this case, study-specific parameters can be linked to the population hyperparameter vector directly in a standard multivariate random-effects meta-analysis model [26, 28]. However, consider a situation where β2i cannot be estimated for study i because x2i was not assessed by design. If we can assume that x2 has an expected zero mean, then its average influence on the outcome would be zero, which can be reflected in the following mapping matrix:
Consequently, the resulting reduced vector of estimable parameters in this situation for study i (Level-1 parameters) would be E(βi) = Miβ = (β0, β1)T.
If x2 is a binary variable with a constant value (e.g., x2i = 1) for study i, then β2i cannot be estimated because there is no variability for that covariate. In this situation, a proper mapping matrix is
and
The first term on the right side of the equation above indicates that the expectation of the intercept parameter β0i cannot be directly linked to the corresponding hyperparameter β0 of the full model. Rather, the expectation of β0i, in the context of the full model, is linked to the sum of hyperparameters β0 and β2. In contrast, β1i can be linked directly to its corresponding hyperparameter β1. Since no variability exists for x2 for study i, there is no β2i.
Table 3 shows two mapping matrices for two different studies from the motivating data example. Table 4 shows covariate availability by study for the entire motivating data. Most studies have reduced sets of covariates, requiring mapping matrices. Let us consider a few specific mapping cases in the current study. The first motivating challenge example in Section 2.3 can be illustrated in this specific case (i.e., Mapping Pattern 1 in Table 3). Study 1 tested the efficacy of two BMIs (i.e., “MI + PF” and “PF”) without a no-treatment control group, whereas all other trials were two-arm or three-arm trials with a control group (see Figure 1).
Table 3.
Mapping matrix pattern. The pattern indices here correspond to the numbers shown in the last column in Table 4. Underlined elements indicate how the identity matrix was modified to link study-level parameters to hyperparameters
| Covariate | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pattern 1 (Study 1) | |||||||||||||
| 0. Intercept1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1. Man (= 1 vs. woman = 0) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2. White (= 1 vs. nonwhite = 0) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3. First-year (= 1 vs. other = 0) | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4. PBS at Baseline | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5. PF1(= 1 vs. control = 0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6. MI + PF1(= 1 vs. control = 0) | 0 | 0 | 0 | 0 | 0 | −1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7. LS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 8. QS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 9. LS × PF1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10. LS × (MI + PF)1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 1 | 0 | 0 |
| 11. QS × PF1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 12. QS × (MI + PF)1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | −1 | 1 |
| Pattern 5 (Study 11) | |||||||||||||
| 0. Intercept | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1. Man (= 1 vs. woman = 0) | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 2. White (= 1 vs. nonwhite = 0) | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 3. First-year (= 1 vs. other = 0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4. PBS at Baseline | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 5. PF (= 1 vs. control = 0) | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 6. MI + PF (= 1 vs. control = 0) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 7. LS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| 8. QS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| 9. LS × PF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 |
| 10. LS × (MI + PF) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 11. QS × PF | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| 12. QS × (MI + PF) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Note.
indicate that study 1 had two treatment groups.
Table 4.
Estimable covariates for the underlying full model by study and by mapping matrix pattern
| Study | N | Covariate position in the full model | Mapping matrix pattern | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |||
| 1+ | 348 | X | X | X | X | X | - | X | X | X | - | X | - | X | 1 |
| 2 | 230 | X | X | X | X | X | X | - | X | - | X | - | - | - | |
| 8a | 1486 | X | X | X | X | X | X | - | X | - | X | - | - | - | 2 |
| 8b | 2155 | X | X | X | X | X | X | - | X | - | X | - | - | - | |
| 8c | 600 | X | X | X | X | X | X | - | X | - | X | - | - | - | |
| 9 | 302 | X | X | X | - | X | X | X | X | X | X | X | X | X | 3 |
| 10.1 | 348 | X | X | X | - | - | - | X | X | - | - | X | - | - | 4 |
| 11 | 383 | X | X | X | - | - | X | - | X | X | X | - | X | - | 5 |
| 12 | 167 | X | X | X | X | X | - | X | X | X | - | X | - | X | 6 |
| 13/14 | 109 | X | X | X | X | - | X | X | X | X | X | X | - | X | 7 |
| 18 | 215 | X | X | X | X | X | X | - | X | X | X | - | X | - | 8 |
| 20 | 928 | X | X | X | X | - | - | X | X | - | - | X | - | - | 9 |
| 21 | 216 | X | X | X | X | X | X | X | X | X | X | X | X | X | 10 |
| 22 | 509 | X | X | X | - | X | - | X | X | - | - | X | - | - | 11 |
Notes. “X” indicates estimable parameters whereas “−” indicates inestimable parameters. Covariate in the full model are (0) Intercept; (1) Man (= 1 vs. woman = 0); (2) White (= 1 vs. non-white = 0); (3) First-year (= 1 vs. other = 0); (4) PBS (Estimated latent trait (θ) scores for utilizing protective behavioral strategies) at Baseline; (5) PF (stand-alone personalized feedback intervention) (= 1 vs. control = 0); (6) MI + PF (in-person motivational intervention with personalized normative feedback profile) (= 1 vs. control = 0); (7) LS (Linear slope of time in month); (8) QS (Quadratic slope of time in month); (9) LS × PF (vs. control); (10) LS × (MI + PF) (vs. control); (11) QS × PF (vs. control); (12) QS × (MI + PF) (vs. control). N represents sample size at baseline. += Study 1 did not have a control group, thus, PF served as a comparison group.
In the context of a network meta-analysis [5, 29], the relative intervention benefit between two BMIs − “MI + PF” and “PF” − can be seen as follows: “MI + PF” vs. “PF” = ((MI + PF) − control) − (PF − control) = ((MI + PF) − PF). Therefore, the study-specific parameter of the relative intervention effect from study 1 can provide valuable information as long as its expectation can properly be aligned to match up with the hyperparameters for intervention effects.
To include data from study 1 in our meta-analysis and draw valid inference, we need to identify an appropriate mapping matrix to link E(βi) to β. A mapping matrix for study 1 can be identified as follows. First, change the diagonal 1s from a 13 × 13 identity matrix into 0s in the 6th, 10th, and 12th rows (numbered as covariates 5, 9, and 11 in Table 3). These three rows correspond to the three hyperparameters comparing PF with control (i.e., PF vs. control; [PF vs. control] × linear growth slope; and [PF vs. control] × quadratic growth slope, respectively). These rows are then removed. Second, change the first row to indicate that the intercept parameter from study 1 describes the study-specific average outcome response of PF, rather than control, in the context of the full model. Therefore, the expectation of the intercept parameter β0i from study 1 corresponds to the sum of hyperparameters β0 and β5 at Level 2 and contributes to their estimation for the full model. Third, change the 8th and 9th rows (covariates 7 and 8 in Table 3) to indicate that the linear and quadratic slope parameters from study 1 represents the slope parameters for PF. Finally, contrast “MI + PF” against “PF” when applicable. The study-specific vector of Level-1 parameters for study 1 can be seen as
whose expectation can be linked to the full vector of hyperparameters β at Level 2 as follows:
To provide another motivating challenge example (Example 2 in Section 2.3 & Mapping Pattern 5 in Table 3), study 11 exclusively recruited first-year students, did not assess protective behavioral strategies at baseline, and tested the efficacy of PF against a control in a two-arm trial. Therefore, to map the expectation of estimable parameters from study 11 into the full hyperparameter vector, the rows of an identify matrix corresponding to these variables should be modified (Table 3). The first row of the mapping matrix are modified to indicate that the expectation of the intercept parameter from study 11 is the average outcome of first-year students in the context of the full model. The row for PBS can be removed because the estimated PBS trait scores follows a standard normal distribution with an expected population mean of 0. We expect that the omission of this row for missing PBS does not influence the mean model part of the full model, although the variation surrounding the mean may influence the error part of the model. The resulting vector of study-specific parameters for study 11 and its expectation E(βi) can be linked to β as follows:
A reduced model is analyzed separately and sequentially for each study. At this step, we save study-specific regression parameter estimates, bi and their covariance matrix, Si. For studies with partial information, one can obtain estimates of the pi length parameter vector βi for study i with pi ≤ p. All of the study-specific parameters are then linked to the full vector of hyperparameters β of the underlying full model via Mi.
3.3. Estimation of a multivariate random-effects meta-analysis model with mapping matrices
Once we obtain all study-specific parameter estimates and identify their appropriate mapping patterns to the full model, we need to estimate the full vector of hyperparameters, β ≡ (β0, β1, β2, ⋯ , βp−1)T. We denote bi as the study-specific estimates of the corresponding parameters βi for study i.
To estimate the hyperparameter vector β, one needs to estimate the between-study covariance matrix Σ. In the current study, we used the restricted maximum likelihood (REML) method while using the estimates from the method of moments [3] for starting values to achieve faster convergence. The estimation of Σ can be done by modifying the formula given by Jennrich and Schluchter [30] to incorporate mapping matrices:
where
and is the Moore-Penrose generalized inverse of Mi. Once is estimated, β can be estimated from a combined multivariate normal CD as follows.
First, to accommodate the multivariate nature of β (see [59] for the CD approach for univariate applications), we construct a multivariate normal CD function for β [55]. By definition, H (·) is a multivariate normal CD function for a p×1 vector β, if the projected distribution Hλ (·) on a p×1 vector λ, for any given , is a univariate normal CD for λT β.
Second, at the individual study level, assuming that Si and Σ are known,
is a corresponding multivariate CD function for study i, where θi = E(βi) = Miβ and Φu(·) is the cumulative distribution function for the standard multivariate normal distribution with u dimension, where u is any dimension. On the conditions that Mi is positive and semidefinite, and that all parameters can be linked via appropriate mapping matrices, a combined multivariate CD function for the population-level hyperparameter vector β has been shown as
assuming that Σ is known (see Yang et al. [62] for a formal definition). Here, in this definition, because Si and Σ are known and its covariance matrix , where .
If we plug in the consistent estimator of the variance matrix Σ, then β can be directly estimated from the asymptotic combined multivariate normal CD. In other words, from H(c) (β):
for the estimated mean vector and
for its covariance matrix, where is defined as
Therefore, and are CD estimators of β and Σc, respectively. Note that we use the sample covariance estimators Si and because the combined CD function H(c) (β) would be an asymptotic multivariate normal CD as long as these estimators are consistent. The CD-based approach yields estimates with several desirable properties (e.g., asymptotically efficient and robust against model misspecification). See the Appendix for more details.
Finally, upon obtaining all estimates of the full model, flexible inference can be made using the combined full model. For example, to interpret decaying intervention effects over time, one can compare estimated outcomes at a given time across intervention groups. To estimate outcomes at specific values of the covariates, the estimated hyperparameters from the full model can be used to construct the estimated full model. We can then use the full model to obtain model-based mean ŷ0 and its variance by plugging in a set of in-sample covariate values x0 using the following formula:
and
4. DATA EXAMPLE
4.1. Underlying full model specification
Alcohol use trajectories among college students after various interventions typically show a sharp immediate decline, followed by a slow rebound to levels on par with or above the pre-intervention level over time [2, 58] because alcohol use and alcohol-related problems tend to peak during ages 18–24. Therefore, we chose a quadratic growth model and tested it using IPD from several individual studies separately, which supported the appropriateness of the model. We visually examined all available data, tested unconditional growth models, compared their fit indices (e.g., AIC, BIC), and examined growth coefficients and residual plots. To test intervention effects over time, we included interaction terms between time and intervention groups. We included gender, first-year student status, and race (white or otherwise) as demographic covariates. In addition, we conducted a separate analysis within individual studies to see if attrition at follow-ups could be explained by participant-level covariates. Based on this attrition analysis, we discovered that the tendency to use protective behavioral strategies prior to and while drinking, such as setting drinking limits, was related to greater chances for participants to drop out at follow-ups in some of the studies. We subsequently added this covariate to the full model.
All analyses were performed using R (version 3.4.4). The “nlme” R package [44] was used to fit a random-intercept growth model. We developed R codes to identify patterns of estimable covariates and to construct mapping matrices and used the “optimx” package [42] to obtain the REML estimates of the between-study covariance matrix.
4.2. Partial information and mapping matrices
Table 4 shows all 13 coefficients included in the current analysis and their availability by study. Some coefficients could not be estimated because (1) variables were not assessed by study design (e.g., PBS; studies 10.1, 11, 13/14, and 20); (2) the entire sample consisted of only first-year students (studies 9, 10.1, 11, and 22); (3) not all intervention groups were included in original studies (studies 1, 2, 8a, 8b, 8c, 10.1, 11, 12, 18, 20, and 22; see also Figure 1); and (4) only one follow-up assessment was available (i.e., only a linear slope term could be estimated; studies 2, 8a, 8b, 8c, 10.1, 20, and 22; see Table 1). A total of just three covariate coefficients were estimable across all studies (i.e., man vs. woman, white vs. non-white, and a linear slope of time), and only one study (study 21) had the necessary data to estimate all coefficients. There were a total of 11 different mapping matrix patterns. With the exception of study 21, all other studies required mapping matrices with reduced dimensions.
4.3. Estimation and interpretation
The full vector of hyperparameter estimates (see Table 5) and its corresponding covariance matrix were obtained by applying the estimation procedures described in Section 3. Table 6 shows that the correlation estimates of the regression coefficients derived in the current study were not boundary estimates (i.e., away from ±1), which usually suggests estimation difficulties in certain multivariate meta-analysis models [47]. The estimated full model for the data example at the participant level was:
Table 5.
Combined parameter estimates from the multivariate random-effects meta-analysis
| Covariate | Estimate | p value |
|---|---|---|
| 0. Intercept | 0.4449 | 0.0051 |
| 1. Man (=1 vs. woman=0) | 0.0172 | 0.7184 |
| 2. White (=1 vs. nonwhite=0) | 0.0564 | 0.1309 |
| 3. First-year (=1 vs. other=0) | 0.0403 | 0.1928 |
| 4. PBS at Baseline | −0.2825 | 0.0000 |
| 5. PF (=1 vs. control=0) | −0.0028 | 0.9223 |
| 6. MI + PF (=1 vs. control=0) | 0.0841 | 0.1270 |
| 7. LS | −0.0442 | 0.0178 |
| 8. QS | 0.0041 | 0.0276 |
| 9. LS × PF | 0.0006 | 0.8729 |
| 10. LS × (MI + PF) | −0.0311 | 0.0006 |
| 11. QS × PF | −0.0004 | 0.5774 |
| 12. QS × (MI + PF) | −0.0003 | 0.8314 |
Notes. PBS = Estimated latent trait (θ) scores for utilizing protective behavioral strategies; PF = stand-alone personalized feedback intervention; MI + PF = in-person motivational intervention with personalized normative feedback profile; LS = Linear slope (time in month); and QS = Quadratic slope (months squared).
Table 6.
Synthesized between-study correlation matrix of the combined parameter estimates
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.23 | −0.46 | −0.37 | −0.39 | −0.03 | −0.49 | 0.22 | −0.57 | 0.36 | 0.20 | 0.07 | 0.57 |
| 1 | −0.32 | −0.28 | 0.12 | −0.07 | −0.38 | 0.29 | −0.24 | 0.43 | 0.54 | −0.27 | 0.03 | |
| 2 | 0.32 | 0.28 | 0.12 | 0.07 | 0.38 | 0.29 | 0.24 | 0.43 | −0.03 | −0.10 | ||
| 3 | −0.06 | 0.26 | 0.47 | −0.28 | 0.30 | −0.42 | −0.41 | 0.13 | −0.13 | |||
| 4 | −0.02 | −0.11 | −0.61 | 0.71 | 0.01 | −0.21 | −0.15 | 0.05 | ||||
| 5 | 0.27 | −0.31 | 0.24 | −0.47 | −0.33 | 0.13 | 0.13 | |||||
| 6 | −0.13 | 0.22 | −0.42 | −0.51 | 0.13 | −0.24 | ||||||
| 7 | −0.87 | −0.29 | 0.72 | 0.14 | −0.33 | |||||||
| 8 | −0.31 | −0.56 | −0.13 | 0.00 | ||||||||
| 9 | 0.40 | −0.37 | 0.12 | |||||||||
| 10 | −0.25 | −0.40 | ||||||||||
| 11 | 0.26 |
Notes. 0 = Intercept, 1 = Man, 2 = White, 3 = First-year, 4 = PBS at baseline, 5 = PF, 6 = MI + PF, 7 = LS, 8 = QS, 9 = LS × PF, 10 = LS × (MI + PF), 11 = QS × PF, and 12 = QS × (MI + PF). PBS = Estimated latent trait (θ) scores for utilizing protective behavioral strategies; PF = stand-alone personalized feedback intervention; MI + PF = in-person motivational intervention with personalized normative feedback profile; LS = Linear slope (time in month); and QS = Quadratic slope (months squared).
Substantively, results indicated that there was a significant interaction between MI + PF and the linear slope of time. To interpret this interaction effect, we calculated model-implied outcome values for all groups based on the estimated full model. Namely, we used in-sample covariate values (i.e., first-year, male, white students, a mean PBS score) and obtained model-implied means for alcohol-related problems and their estimated variances at various time points post intervention. The estimates were derived based on the estimated full model.
Figure 2 shows the expected mean levels for all three groups, which shows a reduction in alcohol-related problems at 6 months, followed by a rebound at 12 months (top). To demonstrate the flexibility of this approach, we plotted monthly estimates of alcohol-related problems up to 12 months post intervention (bottom), which shows the linear and quadratic growth functions for the following three groups: MI + PF, PF, and control. MI + PF showed the best post-intervention trajectory over time, showing a clearer intervention benefit, which was better maintained over time, compared with PF. We further probed this by comparing estimated alcohol-related problems for PF vs. control and for MI + PF vs. control every three months (top) and every month (bottom) for up to 12 months post intervention (Figure 3). Figure 3 (bottom) shows that statistically significant group differences in alcohol-related problems emerged around 5–6 months post intervention (95% confidence intervals are below the dotted horizontal line).
Figure 2.
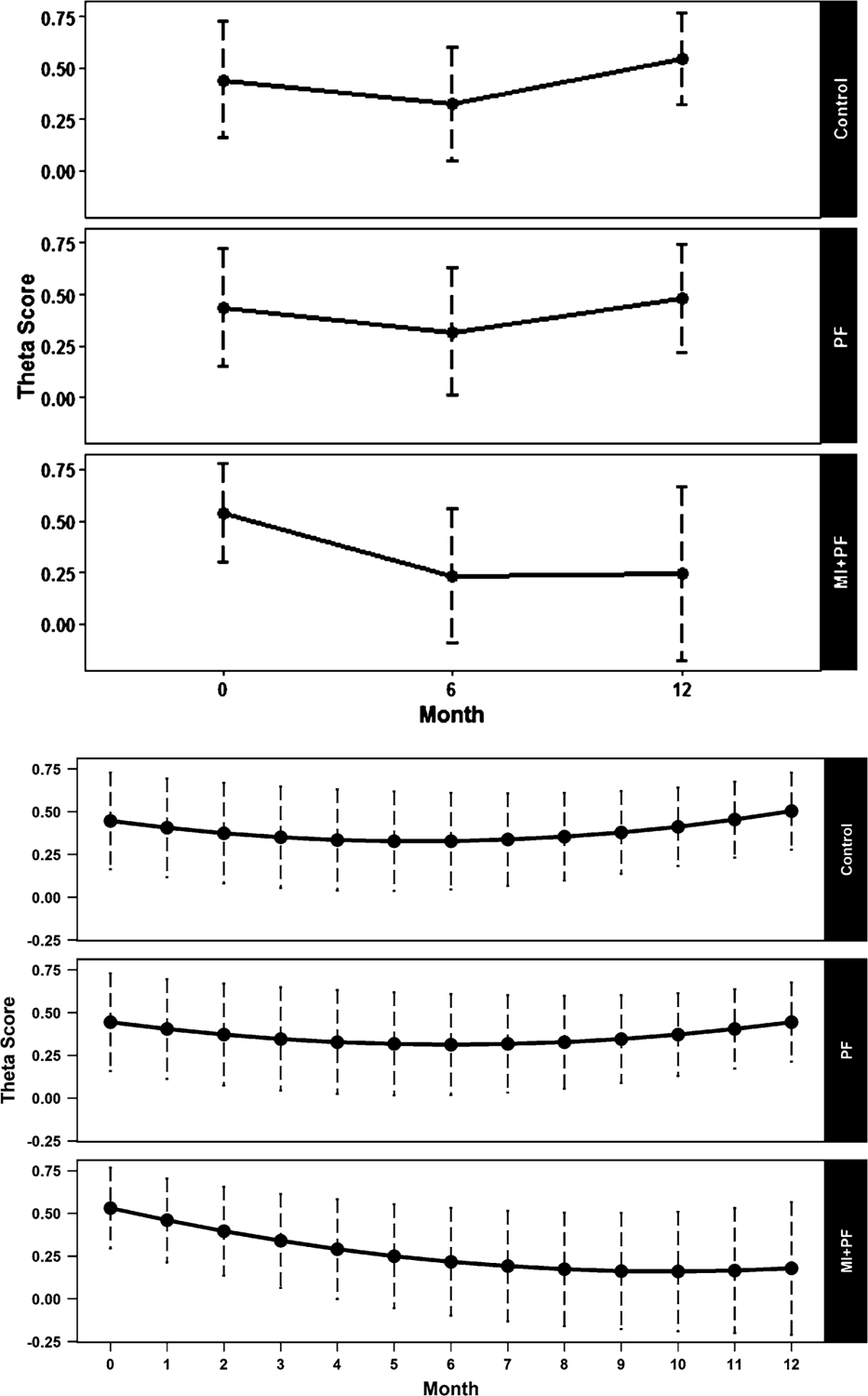
Model-based mean estimates for three different groups using the estimated full model shown in Table 5. The top figure shows estimates at baseline, and 6- and 12-month follow-ups. The bottom figure shows monthly estimates for 12 months post intervention. Theta score = latent trait severity score for alcohol-related problems. PF = stand-alone personalized feedback intervention; MI + PF = in-person motivational intervention with personalized normative feedback profile. Values for covariates were set for White, first-year, male students with a mean PBS score at baseline. Vertical dotted lines indicate 95% confidence intervals.
Figure 3.
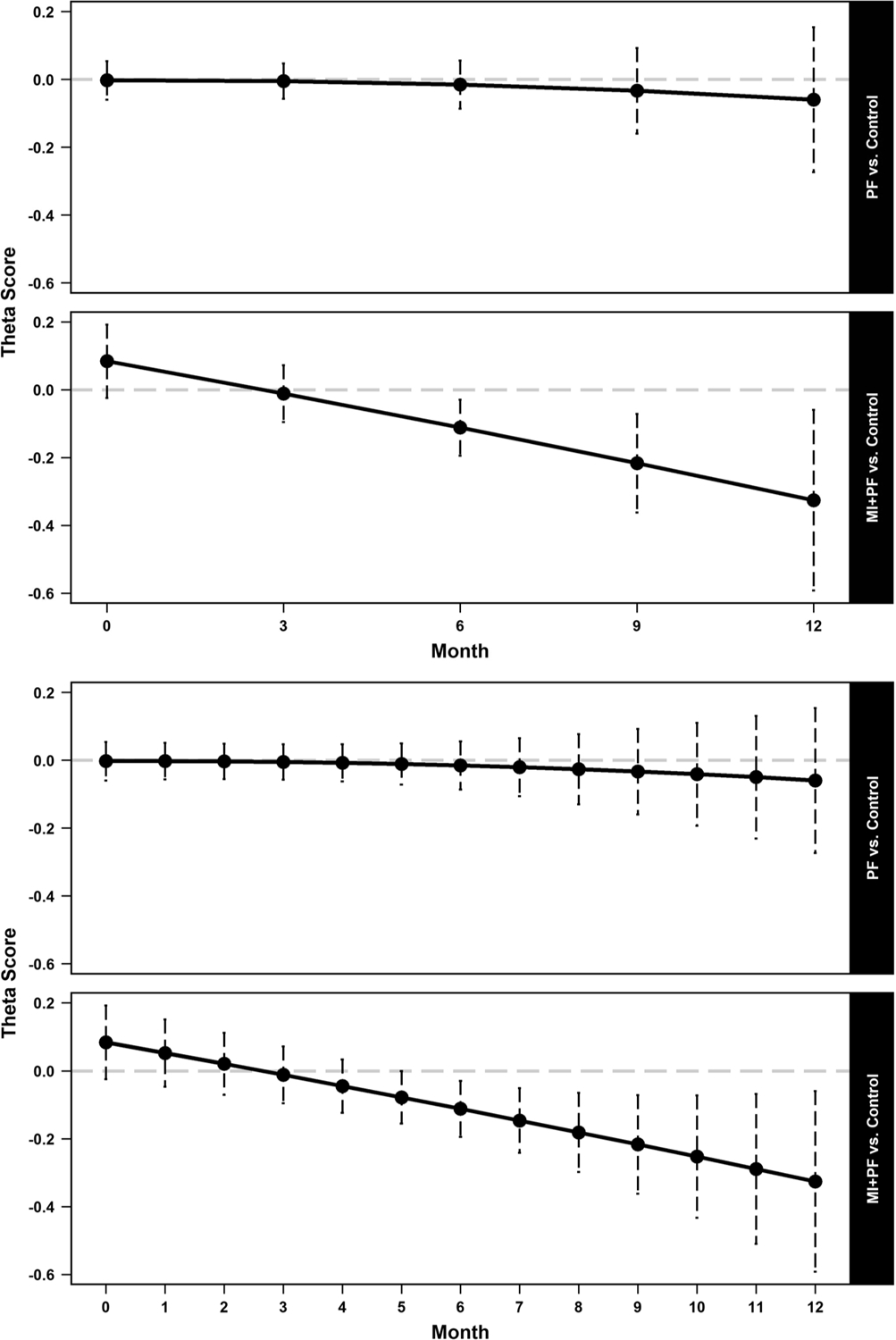
Model-based mean difference estimates of the two intervention groups, compared to control, in alcohol-related problems at baseline and every three months (top), and at baseline and each subsequent month post intervention (bottom). Theta score = latent trait severity score for alcohol-related problems. PF = stand-alone personalized feedback intervention; MI + PF = in-person motivational intervention with personalized normative feedback profile. Vertical dotted lines indicate 95% confidence intervals. A horizontal dashed line at zero indicates no group difference.
4.4. Sensitivity analyses
To examine if the reported results were overly influenced by outlying studies, we conducted a sensitivity analysis by excluding one study at a time and repeating the analysis. Results indicated that while individual regression parameters changed in magnitude to some extent, the overall findings remained largely the same (not shown). In addition, we sequentially removed two different covariates from the analysis at two different steps of the analysis (i.e., when estimating Level-1 parameters in the first step and when estimating Level-2 hyperparameters in the second step) and examined the impact of model misspecification on two key coefficients (i.e., PF × Linear Slope and (PF + MI) × Linear Slope).
Figure 4 shows the results when we removed PBS (top) and first-year student status (bottom) during the second step when hyperparameters were estimated. Figure 5 shows the results when we removed covariates during the first step of the analysis when each covariate was removed from the study-specific analysis (i.e., removed from both Level-1 and Level-2 analyses). Results from both sensitivity analyses suggest that the estimated hyperparameters (shown in filled diamond symbols in Figures 4 and 5) were fairly robust to model misspecification, and that an omitted covariate made little impact on the final estimates, regardless of whether it was a continuous or binary covariate. We also inspected other estimated coefficients and concluded that the derived hyperparameter estimates could be trusted.
Figure 4.
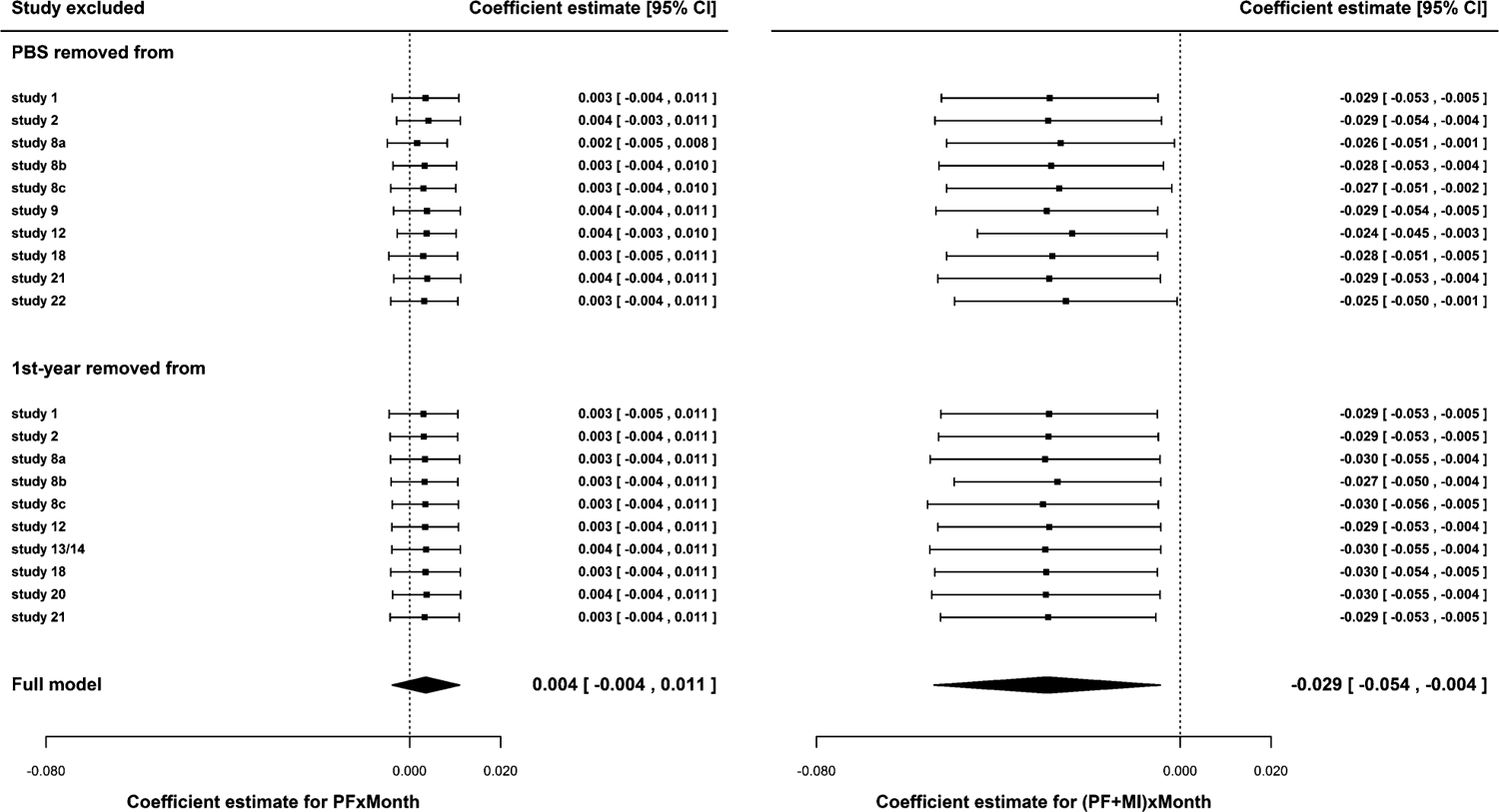
Results from sensitivity analyses where each covariate from each study was treated as systematically missing in the second step. The effects of the exclusion of a continuous covariate (PBS at baseline; top) and a binary covariate (first-year student status; bottom) on the combined estimates of PF × Linear Slope (left) and (MI + PF) × Linear Slope (right) are shown, respectively. Filled diamond symbols indicate the combined estimates from all 14 studies as reported in Table 5. The estimates from sensitivity analyses are shown in filled squares.
Figure 5.
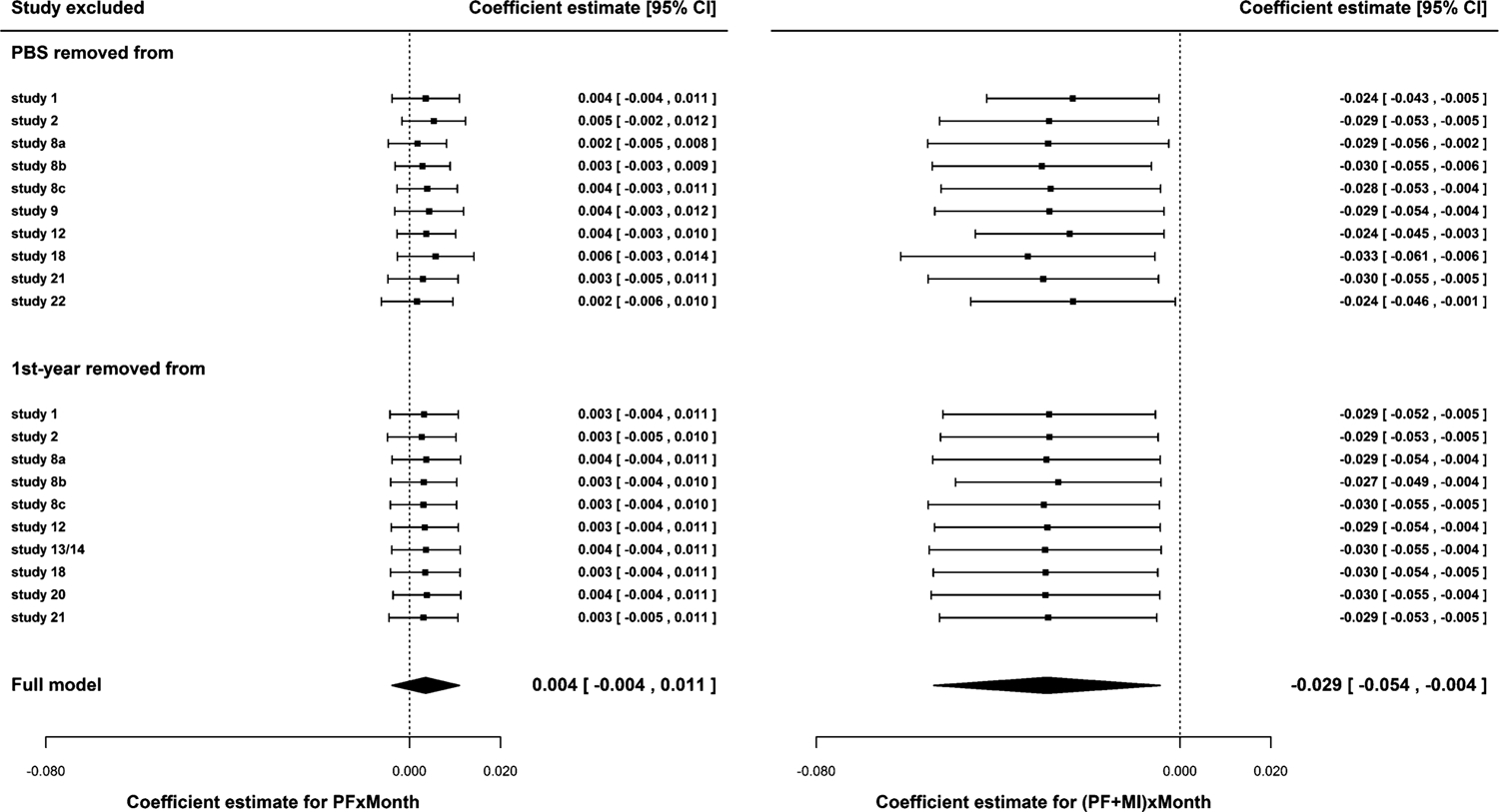
Results from sensitivity analyses where a covariate from each study was sequentially removed throughout the entire analysis. The effects of the exclusion of a continuous covariate (PBS at baseline; top) and a binary covariate (first-year student status; bottom) on the combined estimates of PF × Linear Slope (left) and (MI + PF) × Linear Slope (right) are shown, respectively. Filled diamond symbols indicate the combined estimates from all 14 studies as reported in Table 5. The estimates from sensitivity analyses are shown in filled squares.
5. DISCUSSION
The current study extended the CD-based mapping method to a multivariate random-effects meta-analysis model from the multi-parameter synthesis perspective [1, 16, 17]. We showed that data from heterogeneous trials can be validly accommodated by utilizing the CD concept [59, 60]. The two-step CD-based mapping method differs from the existing methods in the sense that it is aimed at combining the entire full model, which is subsequently used to derive model-based estimates for flexible inference. Broadly speaking, the two-step CD-based mapping method shares some features in common with Bayesian and meta-analytic structural equation modeling (MASEM) [4] approaches in the sense that the CD-based method does not focus on deriving isolated point estimates. Rather, the CD-based mapping method utilizes all available evidence that exist within studies to link to, and estimate, the underlying full model. This method makes fewer assumptions and can be broadly applicable, providing a more general synthesis framework (Table 7).
Table 7.
The premises, assumptions, and challenges of the CD-based mapping method
| Premises |
|
| Assumptions |
|
| Challenges |
|
In the present study, the combined full model had 13 hyperparameter estimates across 11 different estimable patterns of coefficients for 14 trials. To accommodate missing or inestimable covariates and covariates with different meanings across studies, we identified appropriate connections between the expectation of the study-specific parameters and the full model hyperparameters and subsequently, derived the joint distribution of hyperparameters using the multivariate CD-based approach to meta-analysis. This new method may provide the field with a methodological alternative to consider, in connection with methods of aggregating published prediction models [9, 10], dealing with systematic missing data [13, 31, 46, 52], exploring subgroups that may respond differently to an intervention [14, 50], and combining multiple parameter estimates from either AD or IPD [4, 17].
Multivariate meta-analysis, despite its well established rationale and premise, has resulted in a rather small improvement in the statistical properties of individual estimates [26, 56]. In typical multivariate meta-analysis applications, the dimension of combined coefficients has been rather limited, which may help explain the small gain thus far in the literature. The method illustrated in the current paper allows us to make more specific inference based on the full model, which may be helpful for the development of personalized treatment approaches using clinical trial data (i.e., the Precision Medicine Initiative [7]).
In addition, the CD-based mapping method for multivariate meta-analysis explicitly accommodates between-study differences. Consequently, the derived estimates are not confounded with between-study differences. This CD-based method may be helpful when estimating treatment effects for subgroups in data situations where no within-study estimates exist for some of the studies included in a meta-analysis. Riley et al. [48] explored a multivariate meta-analysis extension application, in which different treatment effects were examined separately for each subgroup. However, this approach, as Riley et al. discussed, can result in a confounded treatment effect estimate when one cannot reasonably assume that within-study and between-study covariate interactions are the same. Riley et al. discussed the advantages (e.g., power) and disadvantages (e.g., ecological inference bias and study-level confounding) of combining within-study estimates with between-study estimates of the approach. The method we illustrated in the current study may offer a more favorable solution for this challenge.
Table 7 shows a summarized list of the premises, assumptions, and challenges of the CD-based approach to multivariate meta-analysis. It is flexible and not computationally intensive. Most of the assumptions involved in this CD-based approach are assumed for the existing meta-analysis methods. For example, we assume that the designated full model is a true model for all studies. For a meta-analysis of clinical trials, a true full model is reasonable to assume. We also assume that the pattern of omitted covariates at the study level meets the MAR assumption, which may be quite reasonable for randomized clinical trials [48]. Other assumptions, such as common scales, can be explicitly checked and analyzed when item-level IPD (as opposed to scale-level IPD) are available. In addition, as long as mapping matrices can be identified, this CD-based approach can be used more generally without any restrictions on the distributions of outcome variables or the types of coefficients being combined for a full model. In the current study, we simultaneously combined three different types of related coefficients: the relative intervention benefits (network), informative covariates (regression), and repeated follow-up outcome data (longitudinal).
There are challenges and caveats of the CD-based approach. First, the implementation of this approach using IPD requires considerable time and efforts by a research team with a wide range of complementary expertise and skills. In particular, the necessary identification of an appropriate full model and subsequent mapping matrices can be difficult especially when data dimensions (covariates × study) increase. For randomized controlled trials, the identification of an appropriate full model is more straightforward than observational studies. Nonetheless, it would be beneficial to include domain experts to help identify important factors that may affect outcomes so that full models can be reasonably identified. Despite the best effort, however, if a full model is incorrect or if mapping matrices are incorrect, how robust the method is to model misspecification remains to be more thoroughly studied.
Second, the studies included in a meta-analysis should be sufficiently similar in terms of their methodological and clinical characteristics to justify that data can be combined. Although this assumption is generally required for meta-analysis, it may not be reasonable or desirable in some data applications, depending on its goals. If a comprehensively inclusive meta-analysis is the goal, then creating a subgroup of studies sharing the same full model within the subgroup and developing multiple full models to accommodate other subgroups of studies may be needed.
Third, not all constructs can be directly observed or may share the same interpretation across studies. For example, a death is directly observable and has the same meaning regardless of study membership. Mental health outcomes and correlates, however, are not directly observable but derived, and measured in a number of different ways. In the case of depression, there are more than 280 different depression scales with little item overlap across scales [15]. If there is sufficient overlap in items across studies, items may be linked across studies via shared items and a commensurate metric may be established [38, 23].
Fourth, when the number of studies is small or when individual studies have small samples, it may be necessary to accommodate uncertainty surrounding covariance estimators Si and . In future meta-analysis studies, this uncertainty may be reflected, for example, by inflating confidence intervals for the REML method [27]. Finally, our previous measurement work to make IPD comparable across studies involved an additional set of assumptions and constraints, which need to be considered when developing appropriate mapping matrices.
Substantively, we found the positive effect of the MI + PF intervention on alcohol-related problems, which is consistent with the previously reported findings [21], despite using a different methodological approach to IPD meta-analysis. Huh et al. estimated a Bayesian three-level model in a “one-step” meta-analysis of IPD [19] (see also Huh et al. [22]). In addition, Huh et al. used a different full model, which specified alcohol-related problem scores at baseline as a covariate. Intervention effects were estimated within studies using posterior distributions. Consequently, studies without their own control group were excluded in the analysis.
Based on the comparison of the one-step approach with the current two-step CD-based approach, albeit indirectly, we reach two conclusions. First, the convergent findings from the two studies increase our confidence in the substantive conclusion that MI + PF has an advantage over PF in terms of reducing alcohol-related problems for college students. Second, the two-step CD-based mapping method approach to IPD multivariate meta-analysis may be better suited to build a “scalable” evidence base, compared to the one-step approach. The term “scalable” applies not only to the number of studies but also to the number of informative covariates that can be examined in a meta-analysis. Instead of imputing study-level missing data, the CD-based approach utilizes information from all available data across studies to provide inference. Given that with increasing data dimensions, between-study heterogeneity increases, it is critical that missing or inestimable data are appropriately handled in IPD meta-analysis. A recent study [51] discussed that accurate imputations of study-level missing data across 19 depression trials were not obtained, which is a critical step needed for conducting one-step IPD meta-analysis or IDA. This has been a major barrier to harmonizing and synthesizing IPD across studies. The CD-based method that we showcased in the present study may be an important new tool for the field of complex research synthesis.
In conclusion, to provide answers to complex questions from available large-scale data, it is critical to account for between-study differences and accommodate study-level missing data. The CD-based method is a promising new approach aimed at promoting large-scale, complex evidence synthesis of IPD for multivariate meta-analysis models.
ACKNOWLEDGEMENTS
We would like to thank the following contributors to Project INTEGRATE in alphabetical order: John S. Baer, Department of Psychology, The University of Washington, and Veterans’ Affairs Puget Sound Health Care System; Nancy P. Barnett, Center for Alcohol and Addiction Studies, Brown University; M. Dolores Cimini, University Counseling Center, The University at Albany, State University of New York; William R. Corbin, Department of Psychology, Arizona State University; Kim Fromme, Department of Psychology, The University of Texas, Austin; Joseph W. LaBrie, Department of Psychology, Loyola Marymount University; Mary E. Larimer, Department of Psychiatry and Behavioral Sciences, The University of Washington; Matthew P. Martens, Department of Educational, School, and Counseling Psychology, The University of Missouri; James G. Murphy, Department of Psychology, The University of Memphis; Scott T. Walters, Department of Health Behavior and Health Systems, The University of North Texas Health Science Center; Helene R. White, Center of Alcohol Studies, Rutgers, The State University of New Jersey; and the late Mark D. Wood, Department of Psychology, The University of Rhode Island.
FUNDING
The project described was supported by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) [R01 AA019511 to EYM] and in part by the National Science Foundation (NSF) [DMS1513483 and DMS1737857 to MX]. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAAA, the National Institutes of Health, or the NSF.
APPENDIX
Proof of the claims in Section 3.3. First, let us assume that Σ is known. At the individual study level, , where bi is a point estimator of θi = E(βi) = Miβ. By Singh et al. [55], is a corresponding multivariate CD function for θi, for i = 1, … , k.
Following Xie et al. [60] and Yang et al. [61], we know that combining normal CDs of individual studies can be achieved by a linear combination of normal CDs with weight Wi. In particular, when Σ is known, we have
Since , the combined CD function for the regression parameter of interest β is . When Σ is not known, we then replace it with a consistent estimator (as k → ∞). This leads to the following asymptotic combined CD: as described in Section 3.3.
Contributor Information
Yang Jiao, Department of Statistics, Rutgers, The State University of New Jersey, Piscataway, NJ, USA.
Eun-Young Mun, Department of Health Behavior and Health Systems, University of North Texas Health Science Center, Fort Worth, TX, USA.
Thomas A. Trikalinos, Department of Health Services, Policy and Practice, Brown University, Providence, RI, USA
Minge Xie, Department of Statistics, Rutgers, The State University of New Jersey, Piscataway, NJ, USA.
REFERENCES
- [1].Ades AE and Sutton AJ (2006). Multiparameter evidence synthesis in epidemiology and medical decision-making: Current approaches. Journal of the Royal Statistical Society: Series A (Statistics in Society) 169(1) 5–35. [Google Scholar]
- [2].Carey KB, Carey MP, Henson JM, Maisto SA, and DeMartini KS (2011). Brief alcohol interventions for mandated college students: Comparison of face-to-face counseling and computer-delivered interventions. Addiction 106(3) 528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen H, Manning AK, and Dupuis J (2012). A method of moments estimator for random effect multivariate meta-analysis. Biometrics 68(4) 1278–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cheung MWL and Hafdahl AR (2016). Special issue on meta-analytic structural equation modeling: Introduction from the guest editors. Research Synthesis Methods 7(2) 112–120. [DOI] [PubMed] [Google Scholar]
- [5].Cipriani A, Higgins JPT, Geddes JR, and Salanti G (2013). Conceptual and technical challenges in network meta-analysis. Annals of Internal Medicine 159(2) 130–137. [DOI] [PubMed] [Google Scholar]
- [6].Claggett B, Xie M, and Tian L (2014). Meta-analysis with fixed, unknown, study-specific parameters. Journal of the Amercan Statistical Association 109(508) 1660–1671. [Google Scholar]
- [7].Collins FS and Varmus H (2015). A new initiative on precision medicine. New England Journal of Medicine 372(9) 793–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Curran PJ and Hussong AM (2009). Integrative data analysis: The simultaneous analysis of multiple data sets. Psychological Methods 14(2) 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Debray TPA, Koffijberg H, Nieboer D, Vergouwe Y, Steyerberg EW, and Moons KGM (2014). Meta-analysis and aggregation of multiple published prediction models. Statistics in Medicine 33(14) 2341–2362. [DOI] [PubMed] [Google Scholar]
- [10].Debray TPA, Koffijberg H, Vergouwe Y, Moons KG, and Steyerberg EW (2012). Aggregating published prediction models with individual participant data: A comparison of different approaches. Statistics in Medicine 31(23) 2697–2712. [DOI] [PubMed] [Google Scholar]
- [11].Debray TPA, Moons KGM, Abo-Zaid GMA, Koffijberg H, and Riley RD (2013). Individual participant data meta-analysis for a binary outcome: One-stage or two-stage? PLoS ONE 8(4) e60650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Embretson SE (2006). The continued search for nonarbitrary metrics in psychology. American Psychologist 61(1) 50–55; discussion 62–71. [DOI] [PubMed] [Google Scholar]
- [13].Fibrinogen Studies Collaboration (2009). Correcting for multivariate measurement error by regression calibration in meta-analyses of epidemiological studies. Statistics in Medicine 28(7) 1067–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Fisher DJ, Copas AJ, Tierney JF, and Parmar MKB (2011). A critical review of methods for the assessment of patient-level interactions in individual participant data meta-analysis of randomized trials, and guidance for practitioners. Journal of Clinical Epidemiology 64(9) 949–967. [DOI] [PubMed] [Google Scholar]
- [15].Fried EI and Flake JK (2018). Measurement matters. Observer 31(3) 29–31. [Google Scholar]
- [16].Gasparrini A and Armstrong B (2011). Multivariate meta-analysis: A method to summarize non-linear associations. Statistics in Medicine 30(20) 2504–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gasparrini A, Armstrong B, and Kenward MG (2012). Multivariate meta-analysis for non-linear and other multi-parameter associations. Statistics in Medicine 31(29) 3821–3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gibbons RD, Weiss DJ, Frank E, and Kupfer D (2016). Computerized adaptive diagnosis and testing of mental health disorders. In Annual Review of Clinical Psychology, Volume 12, pp. 83–104. [DOI] [PubMed] [Google Scholar]
- [19].Hadfield JD (2010). MCMC methods for multi-response generalized linear mixed models: The MCMCglmm R package. 2010 33(2) 22. [Google Scholar]
- [20].Hofer SM and Piccinin AM (2009). Integrative data analysis through coordination of measurement and analysis protocol across independent longitudinal studies. Psychological Methods 14(2) 150–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huh D, Mun E-Y, Larimer ME, White HR, Ray AE, Rhew IC, Kim S-Y, Jiao Y, and Atkins DC (2015). Brief motivational interventions for college student drinking may not be as powerful as we think: An individual participant-level data meta-analysis. Alcoholism: Clinical and Experimental Research 39(5) 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huh D, Mun E-Y, Walters ST, Zhou Z, and Atkins DC (2019). A tutorial on individual participant data meta-analysis using bayesian multilevel modeling to estimate alcohol intervention effects across heterogeneous studies. Addictive Behaviors 94 162–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huo Y, de la Torre J, Mun E-Y, Kim SY, Ray AE, Jiao Y, and White HR (2015). A hierarchical multi-unidimensional IRT approach for analyzing sparse, multi-group data for integrative data analysis. Psychometrika 80(3) 834–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hussong AM, Curran PJ, and Bauer DJ (2013). Integrative data analysis in clinical psychology research. The Annual Review of Clinical Psychology 9(1) 61–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ioannidis JPA (2005). Why most published research findings are false. PLoS Med 2(8) e124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jackson D, Riley R, and White IR (2011). Multivariate meta-analysis: Potential and promise. Statistics in Medicine 30(20) 2481–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jackson D and Riley RD (2014). A refined method for multivariate meta-analysis and meta-regression. Statistics in Medicine 33(4) 541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jackson D, White IR, and Thompson SG (2010). Extending DerSimonian and Laird’s methodology to perform multivariate random effects meta-analyses. Statistics in Medicine 29(12) 1282–1297. [DOI] [PubMed] [Google Scholar]
- [29].Jansen JP, Fleurence R, Devine B, Itzler R, Barrett A, Hawkins N, Lee K, Boersma C, Annemans L, and Cappelleri JC (2011). Interpreting indirect treatment comparisons and network meta-analysis for health-care decision making: Report of the ISPOR Task Force on indirect treatment comparisons good research practices: Part 1. Value in Health 14(4) 417–428. [DOI] [PubMed] [Google Scholar]
- [30].Jennrich RI and Schluchter MD (1986). Unbalanced repeated-measures models with structured covariance matrices. Biometrics 42(4) 805–820. [PubMed] [Google Scholar]
- [31].Jolani S, Debray TPA, Koffijberg H, van Buuren S, and Moons KGM (2015). Imputation of systematically missing predictors in an individual participant data meta-analysis: A generalized approach using MICE. Statistics in Medicine 34(11) 1841–1863. [DOI] [PubMed] [Google Scholar]
- [32].Jones AP, Riley RD, Williamson PR, and Whitehead A (2009). Meta-analysis of individual patient data versus aggregate data from longitudinal clinical trials. Clinical Trials 6(1) 16–27. [DOI] [PubMed] [Google Scholar]
- [33].Kim S-Y, Mun E-Y, and Smith S (2014). Using mixture models with known class membership to address incomplete covariance structures in multiple-group growth models. British Journal of Mathematical and Statistical Psychology 67(1) 94–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Liu DG, Liu RY, and Xie MG (2015). Multivariate meta-analysis of heterogeneous studies using only summary statistics: Efficiency and robustness. Journal of the American Statistical Association 110(509) 326–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lumley T (2002). Network meta-analysis for indirect treatment comparisons. Statistics in Medicine 21(16) 2313–2324. [DOI] [PubMed] [Google Scholar]
- [36].Martens MP, Ferrier AG, Sheehy MJ, Corbett K, Anderson DA, and Simmons A (2005). Development of the Protective Behavioral Strategies Survey. Journal of Studies on Alcohol 66(5) 698–705. [DOI] [PubMed] [Google Scholar]
- [37].Mun E-Y, de la Torre J, Atkins DC, White HR, Ray AE, Kim S-Y, Jiao Y, Clarke N, Huo Y Larimer ME, and Huh D (2015). Project INTEGRATE: An integrative study of brief alcohol interventions for college students. Psychology of Addictive Behaviors 29(1) 34–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mun E-Y, Huo Y, White HR, Suzuki S, and de la Torre J (2019). Multivariate higher-order IRT model and MCMC algorithm for linking individual participant data from multiple studies. Frontiers in Psychology 10 1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mun E-Y, Jiao Y, and Xie M (2016). Integrative data analysis for research in developmental psychopathology In Cicchetti D (Ed.), Developmental psychopathology: Theory and method (3 ed.), Volume 1, Chapter 23, pp. 1042–1087. Wiley. [Google Scholar]
- [40].Mun E-Y and Ray AE (2018). Integrative data analysis from a unifying research synthesis perspective In Fitzgerald HE and Puttler LI (Eds.), Alcohol use disorders: A developmental science approach to etiology, pp. 341–353. New York: Oxford University Press. [Google Scholar]
- [41].Muraki E (1992). A generalized partial credit model: Application of an EM algorithm. Applied Psychological Measurement 16(2) 159–176. [Google Scholar]
- [42].Nash JC and Varadhan R (2011). Unifying optimization algorithms to aid software system users: Optimx for R. Journal of Statistical Software 43(9) 14. [Google Scholar]
- [43].Normand S-LT (1999). Meta-analysis: Formulating, evaluating, combining, and reporting. Statistics in Medicine 18(3) 321–359. [DOI] [PubMed] [Google Scholar]
- [44].Pinheiro J, Bates D, DebRoy S, Sarkar D, and R Core Team (2019). nlme: Linear and nonlinear mixed effects models. R package version 3.1–141. [Google Scholar]
- [45].Ray AE, Kim S-Y, White HR, Larimer ME, Mun E-Y, Clarke N, Jiao Y, Atkins DC, and Huh D (2014). When less is more and more is mess in brief motivational interventions: Characteristics of intervention content and their associations with drinking outcomes. Psychology of Addictive Behaviors 15(4) 1026–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Resche-Rigon M, White IR, Bartlett J, Peters SAE, Thompson SG, and PROG-IMT Study Group (2013). Multiple imputation for handling systematically missing confounders in meta-analysis of individual participant data. Statistics in Medicine 32(28) 4890–4905. [DOI] [PubMed] [Google Scholar]
- [47].Riley R, Abrams K, Sutton A, Lambert P, and Thompson J (2007). Bivariate random-effects meta-analysis and the estimation of between-study correlation. BMC Medical Research Methodology 7(1) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Riley RD, Elia EG, Malin G, Hemming K, and Price MP (2015). Multivariate meta-analysis of prognostic factor studies with multiple cut-points and/or methods of measurement. Statistics in Medicine 34(17) 2481–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Riley RD, Lambert PC, and Abo-Zaid G (2010). Meta-analysis of individual participant data: Rationale, conduct, and reporting. BMJ 340 c221. [DOI] [PubMed] [Google Scholar]
- [50].Riley RD, Price MJ, Jackson D, Wardle M, Gueyffier F, Wang J, Staessen JA, and White IR (2015). Multivariate meta-analysis using individual participant data. Research Synthesis Methods 6(2) 157–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Siddique J, de Chavez PJ, Howe G, Cruden G, and Brown CH (2018). Limitations in using multiple imputation to harmonize individual participant data for meta-analysis. Prevention Science 19(1) 95–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Siddique J, Reiter JP, Brincks A, Gibbons RD, Crespi CM, and Brown CH (2015). Multiple imputation for harmonizing longitudinal non-commensurate measures in individual participant data meta-analysis. Statistics in Medicine 34(26) 3399–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Simmonds M and Higgins JPT (2007). Covariate heterogeneity in meta-analysis: Criteria for deciding between meta-regression and individual patient data. Statistics in Medicine 26(15) 2982–2999. [DOI] [PubMed] [Google Scholar]
- [54].Simmonds M, Stewart G, and Stewart L (2015). A decade of individual participant data meta-analyses: A review of current practice. Contemporary Clinical Trials 45, Part A, 76–83. [DOI] [PubMed] [Google Scholar]
- [55].Singh K, Xie M, and Strawderman WE (2007). Confidence distribution (CD): Distribution estimator of a parameter. Lecture Notes-Monograph Series 54 132–150. [Google Scholar]
- [56].Trikalinos TA, Hoaglin DC, and Schmid CH (2014). An empirical comparison of univariate and multivariate meta-analyses for categorical outcomes. Statistics in Medicine 33(9) 1441–1459. [DOI] [PubMed] [Google Scholar]
- [57].Trikalinos TA and Olkin I (2012). Meta-analysis of effect sizes reported at multiple time points: A multivariate approach. Clinical Trials 9(5) 610–620. [DOI] [PubMed] [Google Scholar]
- [58].White HR, Mun E-Y, Pugh L, and Morgan TJ (2007). Long-term effects of brief substance use interventions for mandated college students: Sleeper effects of an in-person personal feedback intervention. Alcoholism: Clinical and Experimental Research 31(8) 1380–1391. [DOI] [PubMed] [Google Scholar]
- [59].Xie M and Singh K (2013). Confidence distribution, the frequentist distribution estimator of a parameter: A review. International Statistical Review 81(1) 3–39. [Google Scholar]
- [60].Xie M, Singh K, and Strawderman WE (2011). Confidence distributions and a unifying framework for meta-analysis. Journal of the American Statistical Association 106(493) 320–333. [Google Scholar]
- [61].Yang G, Liu D, Liu RY, Xie M, and Hoaglin DC (2014). Efficient network meta-analysis: A confidence distribution approach. Statistical Methodology 20 105–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Yang G, Liu D, and Xie M (2014). Combining multivariate confidence distributions and its application to meta-analysis. Thesis. [Google Scholar]
- [63].Zeng D and Lin DY (2015). On random-effects meta-analysis. Biometrika 102(2) 281–294. [DOI] [PMC free article] [PubMed] [Google Scholar]


