Abstract
Background
Trifluridine (FTD) is an active cytotoxic component of the metastatic colorectal cancer (mCRC) drug TAS-102, and thymidine phosphorylase inhibitor (TPI) inhibits the rapid degradation of FTD. We tested whether single nucleotide polymorphisms (SNPs) in genes involved in FTD metabolism and TPI excretion could predict outcome in patients with mCRC treated with TAS-102.
Patients and methods
We investigated three different cohorts: a training cohort (n = 52) and a testing cohort (n = 129) both receiving TAS-102 and a control cohort (n = 52) receiving regorafenib. SNPs of TK1, ENT1, CNT1, MATE1, MATE2 and OCT2 were analysed by polymerase chain reaction-based direct DNA sequencing.
Results
In the training cohort, patients with any ENT1 rs760370 G allele had a significantly longer progression-free survival (PFS; 3.5 versus 2.1 months, respectively, hazard ratio [HR] 0.44, P = 0.004) and overall survival (OS; 8.7 versus 5.3 months, respectively, HR 0.27, P = 0.003) than the A/A genotype. These findings were validated in the testing cohort (P = 0.021 and 0.009 for PFS and OS, respectively). In addition, the combination of ENT1 rs760370, MATE1 rs2289669 and OCT2 rs316019 SNPs significantly stratified patients with the risk of PFS and OS in both cohorts (P < 0.001 for PFS and OS in the training cohort; P = 0.053 and 0.025 for PFS and OS, respectively, in the testing cohort). No significant differences were observed in the control group.
Conclusions
The combination of ENT1, MATE1 and OCT2 SNPs may serve as a predictive and prognostic marker in mCRC patients treated with TAS-102.
Keywords: TAS-102, Trifluridine, Thymidine phosphorylase inhibitor, Transporter, Metastatic colorectal cancer
1. Introduction
The metastatic colorectal cancer (mCRC) drug TAS-102 is an orally administered combination of the thymidine- based nucleoside analogue trifluridine (FTD) and the thymidine phosphorylase inhibitor (TPI) tipiracil hydrochloride [1]. Incorporation of tri-phosphorylated FTD into DNA confers its anti-tumour effect. FTD is rapidly degraded to inactive 5-trifluoromethyl-2,4(1H,3H)-pyr- imidinedione (FTY) by thymidine phosphorylase (TP); hence, TP inhibition by TPI is critical for maintaining increased FTD concentrations and enhanced TAS-102 cytotoxicity [2,3].
Nucleoside transporters (NTs) include human concentrative NTs (hCNTs) and human equilibrative NTs (hENTs). These cell membrane proteins mediate the uptake and release of nucleosides and nucleoside analogues such as FTD [4—6]. Human equilibrative NTs transport material bi-directionally depending on the nucleoside concentration gradient, whereas hCNTs transport purine nucleosides inwards against the concentration gradient [7]. Previous in vivo studies revealed that FTD was absorbed via CNT1 in rat intestinal lumens, indicating that FTD is a substrate for CNT1 [8]. Thymidine kinase 1 (TK1) subsequently converts FTD to FTD monophosphate and then FTD triphosphate causes DNA strand breaks [9,10]. NTs and TK1 are thought to be correlated not only with anticancer action but also FTD toxicity as potential biomarkers [11]. Recently, decreased hENT1 and TK1 expressions were suggested to decrease FTD nuclear intake and impair overall activity [12].
TPI lacks anti-tumour activity but inhibits FTD degradation and is potentially anti-angiogenic [2]. Most FTD is metabolised and excreted in urine after conversion to the inactive form FTY, whereas most TPI is not metabolised and mainly excreted in urine as an unchanged form [13]. TPI is also a substrate of organic cation transporter 2 (OCT2), which together with human multidrug and toxin extrusion 1 (MATE1) facilitates tubular reabsorption and drug secretion [14,15]. TPI is mainly excreted by OCT2 and MATE1 in the proximal tubular cell membrane as their substrate, whereas the role of glomerular filtration in renal TPI elimination is negligible.
MATE1 and OCT2 are important in TPI excretion and renal clearance (CLrenal), which might be responsible for TPI blood concentration equilibration. We hypothesised that circulating unchanged TPI in the blood inhibits TP in the liver, leading to diminished FTD degradation by TP, which might complement NTs in producing a synthetic anti-tumour effect of TAS-102 (Fig. 1). We therefore tested whether polymorphisms in genes involved in FTD and TPI pharmacokinetics, especially FTD absorption by NTs (CNT1 and ENT1), metabolism (TK1) and TPI excretion (OCT2 or MATE1), are associated with outcomes and toxicities in patients with refractory mCRC treated with TAS-102.
Fig. 1.
FTD metabolism and TPI excretion. FTD, trifluridine; TPI, thymidine phosphorylase inhibitor; MATE1, multidrug and toxin extrusion 1; OCT2, organic cation transporter 2; hCNT1, human concentrative nucleoside transporter 1; hENT1, human equilibrative nucleoside transporter; TK-1, thymidine kinase 1; TP, thymidine phosphorylase; FTY, 5-trifluoromethyl-2,4(1H,3H )-pyrimidinedione.
2. Materials and methods
2.1. Study design and patients
This retrospective exploratory study investigated three independent cohorts of patients with refractory mCRC: a training cohort receiving TAS-102 (n = 52), a testing cohort receiving TAS-102 (n = 129) and a control cohort treated with regorafenib (n = 52). The training cohort had been referred to the Cancer Institute Hospital (Tokyo, Japan); the testing cohort had been referred to Azienda Ospedaliero-Universitaria Pisana (Pisa, Italy), Istituto Oncologico Veneto (Padua, Italy) and Istituto Nazionale Tumori (Milan, Italy); and the control cohort had been referred to Azienda Ospedaliero-Universitaria Pisana. All patients were Japanese in the training cohort and Italian in the testing and control cohorts. We were fully compliant with the Reporting Recommendations for Tumour Marker Prognostic Studies guidelines. Analyses were approved by the Institutional Review Boards of each institute and conducted at the University of Southern California/ Norris Comprehensive Cancer Center in accordance with the Declaration of Helsinki and Good Clinical Practice Guidelines. Details of eligibility of the patients and dosage information of the treatment were described in Appendix A.
2.2. Selection of single nucleotide polymorphisms
We selected eight candidate SNPs in genes involved in FTD metabolism and TPI excretion according to the following criteria: i) SNPs with statistical significance reported in the literature; ii) tagging SNPs from HapMap genotype data with r2 > 0.8(http://snpinfo.niehs.nih.gov/snpinfo/snptag.html); or iii) minor allele frequency with a cut-off of ≥10% in both Caucasians and East Asians (http://uswest.ensembl.org/index.html). Functional significance was predicted using the F-SNP database http://compbio.cs.queensu.ca/F-SNP/(Table A. 1).
2.3. DNA extraction and genotyping
Genomic DNA was extracted from peripheral whole blood using a QIAmp Kit (Qiagen, Valencia, CA) according to the manufacturer’s protocol. Candidate SNPs were genotyped using polymerase chain reaction (PCR)- based direct DNA sequencing analysis by an ABI 3100A Capillary Genetic Analyzer and Sequencing Scanner, version 1.0 (Applied Biosystems, Foster City, CA). PCR amplification was carried out using both forward and reverse primers for each single nucleotide polymorphism (SNP) for 35 cycles in PTC-100 or PTC-200 Thermal Cyclers (Marshall Scientific, Hampton, NH).
2.4. Development of a new signature based on OCT2—MATE1 interactions in TPI excretion
Given the reported variations in CLrenal and secretory clearance (CLsec) of metformin in healthy volunteers in a study investigating the association among CLrenal and OCT2 SNPs and the interaction of OCT2 and MATE1 SNPs [16], we established a CLsec cut-off of 25 L/h to determine whether individual CLsec from the nine assemblies composed of OCT2 rs316019 (CC, CA and AA) and MATE1 rs2252281 (TT, TC and CC) variants were above or below the cut-off. We divided the nine assemblies into ‘high clearance (HC)’ and ‘low clearance (LC)’, respectively (Fig. A. 1A). Meanwhile, the variations of OCT2 rs316019 and MATE1 rs2289669 on metformin pharmacokinetics were investigated in another study that showed consistent data with the aforementioned study [17]. Together with these previous reports, we adopted a gene—gene interaction-based classification for use with our candidate SNPs, for example, MATE1 rs2289669 and OCT2 rs316019. We replaced ‘LC’ and ‘HC’ with good clinical outcome (Good: longer progression-free survival [PFS] or overall survival [OS]) and poor clinical outcome (Poor: shorter PFS or OS) to more clearly represent the clinical value of the categories (Appendix A and Fig. A. 1B).
2.5. Statistical analysis
Details of statistical analysis were shown in Appendix A. The primary end-point in this study was PFS, and the secondary end-points were OS and disease control rate (DCR). Power analysis was performed to calculate statistical power for the cohorts. All analyses were carried out with SAS software, version 9.4 (SAS Institute, Cary, NC). All tests were two-sided at a significance level of 0.05.
3. Results
3.1. Baseline patients and tumour characteristics
Baseline characteristics of the three cohorts are summarised in Table A. 2. Patient characteristics were similar except for a higher number of males and lower number of chemotherapy lines received before TAS-102 in the testing cohort and a higher percentage of patients with the Eastern Cooperative Oncology Group performance status = 0 and without adjuvant treatment history in the control cohort compared with the other cohorts. The median follow-up time was 6.4 months (range 0—15.4 months), and the median PFS and OS were 2.6 and 8.0 months, respectively, in the training cohort. The median follow-up time was 5.3 months (range 0—8.9 months), and the median PFS and OS were 2.0 and 5.7 months in the testing cohort. All patients died in the control cohort; the median PFS and OS were 1.9 and 5.3 months, respectively. OCT2 rs316000 was not in Hardy-Weinberg equilibrium (HWE) in the training and testing cohorts (P < 0.050) and was thus excluded from further analysis. No high-linkage disequilibrium was observed between SNPs.
3.2. Association of clinical outcomes and FTD metabolism—related genetic variants in patients receiving TAS-102
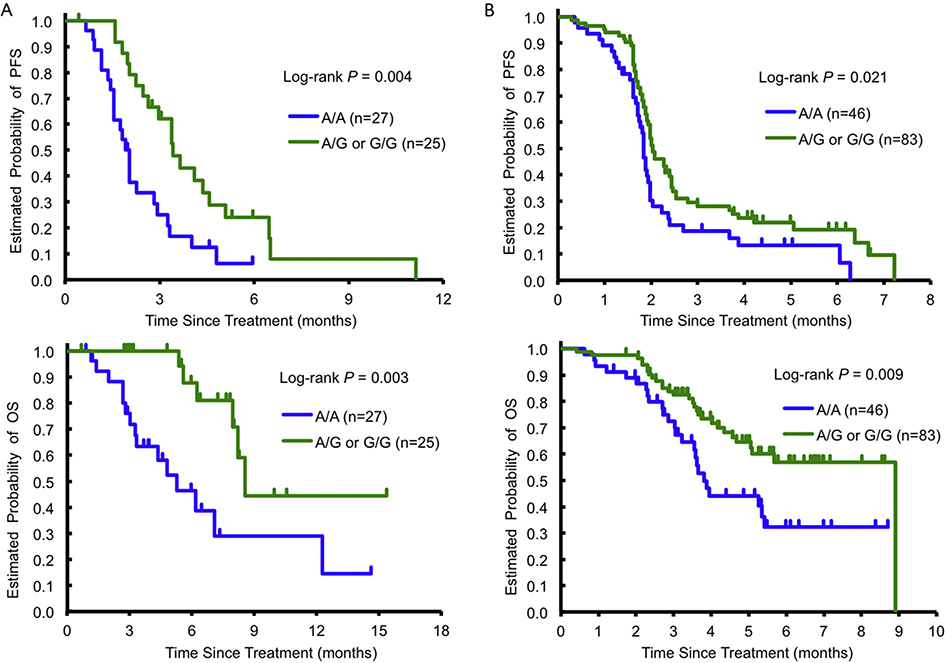
Univariate analysis of the training cohort showed that patients carrying any G allele in ENT1 rs760370 had a significantly longer PFS (hazard ratio [HR] 0.44, P = 0.004) and OS (HR 0.27, P = 0.003) than A/A variants (Fig. 2A). Patients with any T allele in ENT1 rs9394992 also had a significantly longer PFS and OS than C/C variants. Multivariable analysis revealed that both ENT1 rs760370 and ENT1 rs9394992 were significantly associated with PFS and OS. ENT1 rs760370 remained significant for PFS and OS following both univariate (PFS, 2.1 versus 1.9 months, respectively, HR 0.64, P = 0.021; OS, 9.0 versus 3.9 months, HR 0.50, P = 0.009; Fig. 2B) and multivariable (PFS, HR 0.65, P = 0.038; OS, HR 0.54, P = 0.027) analyses of the testing cohort. Although ENT1 rs760370 and ENT1 rs9394992 were both marginally significantly correlated with DCR in the training cohort, no association was confirmed in the testing cohort (Table 1, Table A. 3).
Fig. 2.
Progression-free survival (PFS) and overall survival (OS) according to ENT1 rs760370 A/A variant or any G allele in the training cohort (A) and the testing cohort (B) treated with TAS-102. ENT1, equilibrative nucleoside transporter 1.
Table 1.
Association between single nucleotide polymorphism and clinical outcome.
| |
Disease control |
Progression-free survival |
Overall survival |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | CR/PR/SD | PD | P-value* | Median, months (95% CI) | HR (95% CI)§ | P-value* | HR (95% CI)‡ | P-value* | Median, months (95% CI) | HR (95% CI)§ | P-value* | HR (95% CI)‡ | P-value* | |
| Training cohort | ||||||||||||||
| ENT1 rs760370 | 0.061 | 0.004 | 0.023 | 0.003 | 0.008 | |||||||||
| A/A | 27 | 4 (17%) | 19 (83%) | 2.1 (1.6, 2.9) | 1 (Reference) | 1 (Reference) | 5.3 (3.1, 12.4) | 1 (Reference) | 1 (Reference) | |||||
| A/Ga | 22 | 9 (45%) | 11 (55%) | 3.5 (2.5, 4.6) | 0.44 (0.23, 0.83) | 0.44 (0.22, 0.89) | 8.7 (8.1, 15.6+) | 0.27 (0.10, 0.70) | 0.17 (0.05, 0.63) | |||||
| G/Ga | 3 | 2 (67%) | 1 (33%) | |||||||||||
| ENT1 rs9394992 | 0.071 | 0.011 | 0.016 | <0.001 | 0.001 | |||||||||
| C/C | 21 | 3 (17%) | 15 (83%) | 1.9 (1.6, 2.7) | 1 (Reference) | 1 (Reference) | 4.4 (2.7, 7.2) | 1 (Reference) | 1 (Reference) | |||||
| C/Ta | 25 | 8 (36%) | 14 (64%) | 3.4 (2.3, 4.4) | 0.48 (0.25, 0.91) | 0.44 (0.23, 0.86) | 8.7 (8.1, 15.6+) | 0.21 (0.08, 0.51) | 0.20 (0.08, 0.51) | |||||
| T/Ta | 6 | 4 (67%) | 2 (33%) | |||||||||||
| MATE1 rs2289669 | 0.091 | 0.035 | 0.046 | 0.096 | 0.19 | |||||||||
| G/G | 12 | 1 (10%) | 9 (90%) | 2.0 (1.4, 3.4) | 1 (Reference) | 1 (Reference) | 8.1 (5.4, 10.7+) | 1 (Reference) | 1 (Reference) | |||||
| G/A | 28 | 12 (46%) | 14 (54%) | 3.4 (2.1, 4.4) | 0.44 (0.20, 0.93) | 0.41 (0.18, 0.94) | 12.4 (5.7, 15.6+) | 0.76 (0.26, 2.28) | 0.69 (0.23, 2.11) | |||||
| A/A | 12 | 2 (20%) | 8 (80%) | 2.3 (1.2, 3.3) | 0.94 (0.40, 2.18) | 0.92 (0.38, 2.24) | 4.9 (2.8, 6.5+) | 2.40 (0.69, 8.37) | 1.98 (0.56, 6.99) | |||||
| 0.13 | 0.086 | 0.10 | 0.96 | 0.88 | ||||||||||
| G/G | 12 | 1 (10%) | 9 (90%) | 2.0 (1.4, 3.4) | 1 (Reference) | 1 (Reference) | 8.1 (5.4, 10.7+) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 40 | 14 (39%) | 22 (61%) | 3.0 (2.1, 3.7) | 0.56 (0.28, 1.13) | 0.53 (0.25, 1.13) | 8.3 (5.3, 15.6+) | 1.03 (0.37, 2.83) | 0.93 (0.33, 2.59) | |||||
| OCT2 rs316019 | 0.16 | 0.40 | 0.50 | 0.93 | 0.35 | |||||||||
| C/C | 37 | 9 (27%) | 24 (73%) | 2.5 (2.1, 3.4) | 1 (Reference) | 1 (Reference) | 8.1 (5.7, 12.4) | 1 (Reference) | 1 (Reference) | |||||
| C/Aa | 11 | 3 (33%) | 6 (67%) | 3.0 (1.2, 6.0+) | 0.74 (0.36, 1.52) | 1.32 (0.58, 3.02) | 10.7+ (2.7, 10.7+) | 1.04 (0.38, 2.89) | 1.74 (0.55, 5.51) | |||||
| A/Aa | 4 | 3 (75%) | 1 (25%) | |||||||||||
| MATE1_OCT019 | 0.070 | 0.006 | 0.020 | 0.026 | 0.060 | |||||||||
| Poor | 15 | 1 (8%) | 11 (92%) | 2.3 (1.2, 2.9) | 1 (Reference) | 1 (Reference) | 4.9 (2.8, 10.7) | 1 (Reference) | 1 (Reference) | |||||
| Good | 37 | 14 (41%) | 20 (59%) | 3.4 (2.1, 4.1) | 0.44 (0.22, 0.85) | 0.45 (0.23, 0.88) | 8.3 (6.3, 15.6) | 0.39 (0.15, 0.98) | 0.41 (0.16, 1.04) | |||||
| ENT1370_ MATE/OCT | 0.044 | <0.001 | 0.016 | <0.001 | 0.009 | |||||||||
| Poor | 11 | 0 | 9 (100%) | 1.6 (0.9, 2.9) | 1 (Reference) | 1 (Reference) | 3.4 (2.7, 6.5) | 1 (Reference) | 1 (Reference) | |||||
| Fair | 16 | 4 (29%) | 10 (71%) | 2.1 (1.6, 4.1) | 0.42 (0.17, 1.03) | 0.48 (0.20, 1.16) | 7.2 (2.7, 14.8) | 0.37 (0.12, 1.16) | 0.37 (0.11, 1.23) | |||||
| Gooda | 4 | 1 (33%) | 2 (67%) | 3.5 (2.5, 4.6) | 0.26 (0.11, 0.59) | 0.28 (0.11, 0.66) | 8.7 (8.1, 15.6) | 0.14 (0.04, 0.48) | 0.08 (0.02, 0.41) | |||||
| Excellenta | 21 | 10 (50%) | 10 (50%) | |||||||||||
| Testing cohort | ||||||||||||||
| ENT1 rs760370 | 0.24 | 0.030 | 0.029 | 0.032 | 0.064 | |||||||||
| A/A | 46 | 9 (20%) | 36 (80%) | 1.9 (1.7, 2.0) | 1 (Reference) | 1 (Reference) | 3.9 (3.3, 5.5) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 64 | 22 (34%) | 42 (66%) | 2.1 (1.9, 2.5) | 0.59 (0.38, 0.90) | 0.58 (0.37, 0.89) | 8.7+ (4.7, 8.7+) | 0.48 (0.27, 0.85) | 0.50 (0.28, 0.89) | |||||
| G/G | 19 | 6 (32%) | 13 (68%) | 2.0 (1.6, 2.5) | 0.86 (0.48, 1.55) | 1.00 (0.55,1.80) | 9.0 (2.9, 9.0+) | 0.58 (0.24, 1.41) | 0.78 (0.31, 1.99) | |||||
| 0.15 | 0.021 | 0.038 | 0.009 | 0.027 | ||||||||||
| A/A | 46 | 9 (20%) | 36 (80%) | 1.9 (1.7, 2.0) | 1 (Reference) | 1 (Reference) | 3.9 (3.3, 5.5) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 83 | 28 (34%) | 55 (66%) | 2.1 (1.9, 2.4) | 0.64 (0.43, 0.95) | 0.65 (0.43, 0.98) | 9.0 (5.1, 9.0+) | 0.50 (0.29, 0.86) | 0.54 (0.31, 0.93) | |||||
| ENT1 rs9394992 | 1.00 | 0.90 | 0.56 | 0.51 | 0.76 | |||||||||
| C/C | 58 | 17 (29%) | 41 (71%) | 2.0 (1.9, 2.1) | 1 (Reference) | 1 (Reference) | 5.8 (4.7, 8.8+) | 1 (Reference) | 1 (Reference) | |||||
| C/T | 59 | 17 (29%) | 41 (71%) | 2.0 (1.8, 2.3) | 0.92 (0.61, 1.38) | 0.81 (0.54, 1.22) | 4.4 (3.6, 9.0+) | 1.38 (0.78, 2.43) | 1.23 (0.69, 2.18) | |||||
| T/T | 12 | 3 (25%) | 9 (75%) | 2.5 (0.4, 3.9) | 0.90 (0.45, 1.78) | 0.79 (0.39, 1.58) | 7.3+ (2.5, 7.3+) | 1.06 (0.40, 2.79) | 1.02 (0.38, 2.71) | |||||
| 1.00 | 0.65 | 0.28 | 0.32 | 0.54 | ||||||||||
| C/C | 58 | 17 (29%) | 41 (71%) | 2.0 (1.9, 2.1) | 1 (Reference) | 1 (Reference) | 5.8 (4.7, 8.8+) | 1 (Reference) | 1 (Reference) | |||||
| Any T | 71 | 20 (29%) | 50 (71%) | 2.0 (1.9, 2.4) | 0.92 (0.62, 1.35) | 0.81 (0.54, 1.19) | 5.2 (3.7, 9.0+) | 1.31 (0.76, 2.27) | 1.19 (0.69, 2.07) | |||||
| MATE1 rs2289669 | 0.90 | 0.98 | 0.83 | 0.34 | 0.32 | |||||||||
| G/G | 38 | 10 (27%) | 27 (73%) | 1.9 (1.9, 2.4) | 1 (Reference) | 1 (Reference) | 9.0 (4.4, 9.0+) | 1 (Reference) | 1 (Reference) | |||||
| G/A | 64 | 20 (31%) | 44 (69%) | 2.0 (1.8, 2.3) | 1.04 (0.66, 1.64) | 1.15 (0.73, 1.83) | 4.6 (3.7, 8.8+) | 1.55 (0.81, 2.96) | 1.67 (0.86, 3.25) | |||||
| A/A | 27 | 7 (26%) | 20 (74%) | 2.0 (1.7, 2.3) | 1.01 (0.58, 1.74) | 1.11 (0.63, 1.96) | 8.5+ (3.7, 8.5+) | 1.13 (0.50, 2.58) | 1.42 (0.61, 3.32) | |||||
| 0.83 | 0.87 | 0.55 | 0.25 | 0.15 | ||||||||||
| G/G | 38 | 10 (27%) | 27 (73%) | 1.9 (1.9, 2.4) | 1 (Reference) | 1 (Reference) | 9.0 (4.4, 9.0+) | 1 (Reference) | 1 (Reference) | |||||
| Any A | 91 | 27 (30%) | 64 (70%) | 2.0 (1.9, 2.1) | 1.03 (0.68, 1.58) | 1.14 (0.74, 1.75) | 5.3 (3.9, 8.8+) | 1.42 (0.76, 2.64) | 1.60 (0.85, 3.04) | |||||
| OCT2 rs316019 | 0.65 | 0.60 | 0.31 | 0.57 | 1.00 | |||||||||
| C/C | 98 | 27 (28%) | 70 (72%) | 2.0 (1.9, 2.3) | 1 (Reference) | 1 (Reference) | 5.8 (4.7, 9.0+) | 1 (Reference) | 1 (Reference) | |||||
| C/A | 31 | 10 (32%) | 21 (68%) | 1.9 (1.7, 2.4) | 1.12 (0.72, 1.76) | 1.28 (0.79, 2.07) | 4.6 (3.6, 7.1+) | 1.19 (0.64, 2.19) | 1.00 (0.53, 1.89) | |||||
| MATE1_OCT019 | 0.83 | 0.77 | 0.68 | 0.59 | 0.73 | |||||||||
| Poor | 38 | 10 (26%) | 28 (74%) | 2.0 (1.8, 2.1) | 1 (Reference) | 1 (Reference) | 8.5+ (3.6, 8.5+) | 1 (Reference) | 1 (Reference) | |||||
| Good | 91 | 27 (30%) | 63 (70%) | 2.0 (1.9, 2.3) | 0.94 (0.62, 1.43) | 0.91 (0.59, 1.42) | 5.4 (4.1, 9.0+) | 1.18 (0.64, 2.17) | 1.12 (0.59, 2.10) | |||||
| ENT1370_ MATE/OCT | 0.42 | 0.053 | 0.11 | 0.025 | 0.032 | |||||||||
| Poor | 14 | 3 (21%) | 11 (79%) | 2.0 (1.7, 2.4) | 1 (Reference) | 1 (Reference) | 3.6 (2.3, 8.5) | 1 (Reference) | 1 (Reference) | |||||
| Fair | 32 | 6 (19%) | 25 (81%) | 1.9 (1.6, 2.0) | 1.24 (0.64, 2.40) | 1.13 (0.57, 2.24) | 4.0 (3.1, 8.8) | 0.77 (0.34, 1.75) | 0.58 (0.25, 1.37) | |||||
| Gooda | 24 | 7 (29%) | 17 (71%) | 2.1 (1.9, 2.4) | 0.73 (0.40, 1.34) | 0.71 (0.38, 1.30) | 9.0 (5.1, 9.0) | 0.42 (0.20, 0.90) | 0.37 (0.17, 0.80) | |||||
| Excellenta | 59 | 21 (36%) | 38 (64%) | |||||||||||
| Control cohort | ||||||||||||||
| ENT1 rs760370 | 0.93 | 0.71 | 0.68 | 0.77 | 0.95 | |||||||||
| A/A | 21 | 7 (35%) | 13 (65%) | 2.1 (1.8, 3.1) | 1 (Reference) | 1 (Reference) | 5.9 (4.5, 8.0) | 1 (Reference) | 1 (Reference) | |||||
| A/G | 19 | 7 (39%) | 11 (61%) | 1.9 (1.6, 3.9) | 1.26 (0.67, 2.37) | 1.08 (0.57, 2.08) | 5.7 (2.0, 8.9) | 1.25 (0.67, 2.35) | 0.98 (0.50, 1.90) | |||||
| G/G | 12 | 3 (27%) | 8 (73%) | 1.4 (0.4, 4.3) | 1.24 (0.59, 2.59) | 1.57 (0.57, 4.35) | 2.4 (1.2, 9.1) | 1.17 (0.55, 2.46) | 1.11 (0.48, 2.56) | |||||
| 1.00 | 0.41 | 0.67 | 0.48 | 0.97 | ||||||||||
| A/A | 21 | 7 (35%) | 13 (65%) | 2.1 (1.8, 3.1) | 1 (Reference) | 1 (Reference) | 5.9 (4.5, 8.0) | 1 (Reference) | 1 (Reference) | |||||
| Any G | 31 | 10 (34%) | 19 (66%) | 1.8 (1.5, 2.2) | 1.25 (0.71, 2.20) | 1.15 (0.61, 2.15) | 4.1 (2.2, 7.8) | 1.22 (0.70, 2.13) | 1.01 (0.55, 1.88) | |||||
| ENT rs9394992 | 0.30 | 0.29 | 0.62 | 0.039 | 0.22 | |||||||||
| C/C | 20 | 6 (30%) | 14 (70%) | 2.0 (1.2, 2.7) | 1 (Reference) | 1 (Reference) | 4.4 (2.6, 6.0) | 1 (Reference) | 1 (Reference) | |||||
| C/Ta | 25 | 7 (30%) | 16 (70%) | 1.9 (1.7, 3.9) | 0.75 (0.42, 1.32) | 0.85 (0.46, 1.59) | 7.7 (3.7, 8.9) | 0.57 (0.31, 1.05) | 0.64 (0.32, 1.30) | |||||
| T/Ta | 7 | 4 (67%) | 2 (33%) | |||||||||||
| MATE1 rs2289669 | 0.44 | 0.56 | 0.66 | 0.66 | 0.98 | |||||||||
| G/G | 19 | 7 (41%) | 10 (59%) | 2.1 (1.5, 3.1) | 1 (Reference) | 1 (Reference) | 5.7 (2.2, 7.8) | 1 (Reference) | 1 (Reference) | |||||
| G/Aa | 27 | 7 (27%) | 19 (73%) | 1.8 (1.7, 2.8) | 0.85 (0.48, 1.52) | 0.86 (0.43, 1.70) | 5.3 (3.7, 8.0) | 0.88 (0.50, 1.57) | 0.99 (0.52, 1.91) | |||||
| A/Aa | 6 | 3 (50%) | 3 (50%) | |||||||||||
| OCT2 rs316019 | 0.49 | 0.99 | 0.36 | 0.51 | 0.81 | |||||||||
| C/C | 44 | 16 (39%) | 25 (61%) | 2.1 (1.7, 3.1) | 1 (Reference) | 1 (Reference) | 5.3 (2.6, 7.9) | 1 (Reference) | 1 (Reference) | |||||
| C/Aa | 7 | 1 (14%) | 6 (86%) | 1.8 (1.5, 2.0) | 1.00 (0.44, 2.28) | 1.50 (0.62, 3.61) | 5.3 (3.6, 9.6) | 0.79 (0.35, 1.75) | 0.90 (0.39, 2.09) | |||||
| A/Aa | 1 | 0 | 1 (100%) | |||||||||||
CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; CI, confidence interval; HR, hazard ratio; ENT1, equilibrative nucleoside transporter 1; MATE1, multidrug and toxin extrusion 1; OCT2, organic cation transporter 2.
P-value based on Fisher’s exact test for response, log-rank test in univariate analysis (§) and Wald test in multivariable analysis (‡) within the Cox regression model adjusted for liver metastasis and adjuvant history in the training cohort; age group (<61 versus ≥61), liver metastasis, the Eastern Cooperative Oncology Group (ECOG) performance status, previous anti-EGFR in the testing cohort and ECOG performance status and number of metastases in the control cohort.
Estimates not yet reached.
Significant P-values are in bold.
Combined for estimates of HR.
3.3. A signature based on the OCT2 and MATE1 gene—gene interaction in TPI excretion
Univariate analysis of the combination of MATE1 rs2289669 and OCT2 rs316019 variants showed that patients in the Good category had a significantly longer PFS (3.4 versus 2.3 months, respectively, HR 0.44, P = 0.006) and OS (8.3 versus 4.9 months, respectively, HR 0.39, P = 0.026) than those in the Poor category of the training cohort. Multivariable analysis revealed that the association remained significant for PFS (HR 0.45, P = 0.020) and marginally significant for OS (HR 0.41, P = 0.060). No significance was observed in the testing cohort (Table 1).
3.4. A novel classification composed of ENT1 and OCT2/ MATE1 SNPs with clinical outcome
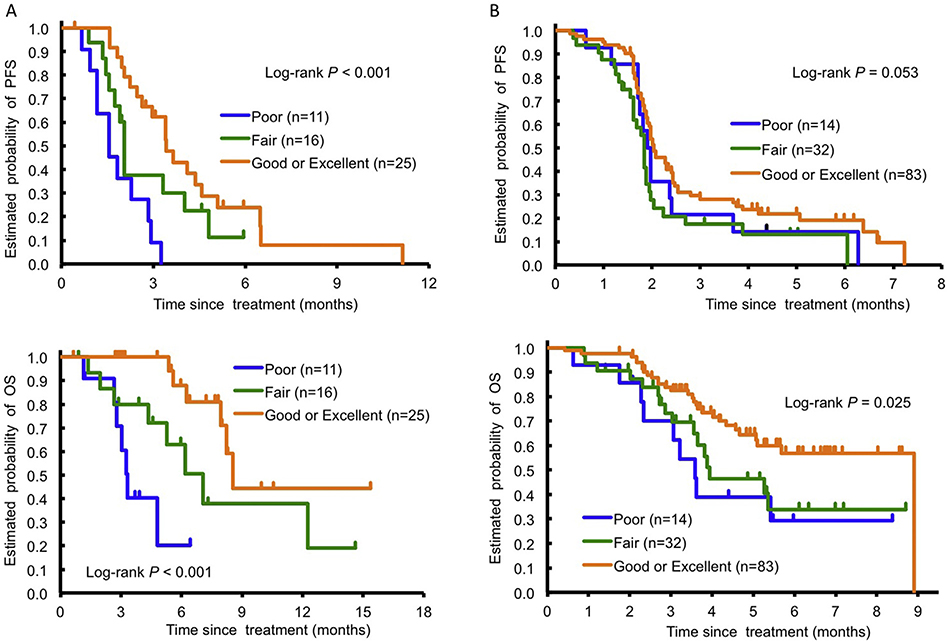
Next, we tested a combination of ENT1 rs760370 and the aforementioned signature based on the OCT2 and MATE1 gene—gene interaction for prediction of overall TAS-102 efficacy (Table 1). We newly defined four categories related to a combination of these genes: ENT1/OCT2—MATE1: Excellent (n = 21), any G allele/Good; Good (n = 4), any G allele/Poor; Fair (n = 16), A/A variant/Good and Poor (n = 11), A/A variant/Poor. We integrated Good and Excellent into one category to increase the sample size (Fig. 4A). PFS, OS and frequency of grade 2+ neutropenia were included in each category. Univariate analysis of the training cohort recognised the benefit on PFS and OS among the three categories (Poor versus Fair versus Good or Excellent: PFS, 1.6 versus 2.1 versus 3.5 months, P < 0.001; OS, 3.4 versus 7.2 versus 8.7, P < 0.001; Fig. 3A). This significance remained in multivariate analysis for PFS (P = 0.016) and OS (P = 0.009). In the testing cohort, these findings were confirmed for OS (3.6 versus 4.0 versus 9.0 months, respectively, P = 0.025) and PFS (2.0 versus 1.9 versus 2.1 months, respectively, P = 0.053). In multivariate analysis, the significance remained for OS (P = 0.032) (Fig. 3B and Fig. A. 2). Univariate and multivariate analyses for all candidate SNP genotypes in the control cohort receiving regorafenib with no previous TAS-102 treatment showed no significant differences among the SNPs in PFS or OS (Table 1).
Fig. 4.
(A) Novel classification of ENT1 rs760370 and OCT2 rs316019/MATE1 rs2289669 based on the association of SNPs and clinical outcome in the training cohort. Excellent and Good groups were combined to increase the sample size. (B) Potential treatment algorithm for TAS-102 in patients with metastatic colorectal cancer. PFS, progression-free survival; OS, overall survival; MATE1, multidrug and toxin extrusion 1; OCT2, organic cation transporter 2; ENT1, equilibrative nucleoside transporter 1; NEU, neutropenia; Gr, grade. * OCT2 rs316019/MATE1 rs2289669: CC/AA, CA/GG or AA and AA/GG. ** OCT2 rs316019/MATE1 rs2289669: CC/GG or GA, CA/ GA and AA/GA or AA.
Fig. 3.
Progression-free survival (PFS) and overall survival (OS) by combination of ENT1 rs760370 and OCT2 rs316019/MATE1 rs2289669 in the training cohort (A) and the testing cohort (B) treated with TAS-102: Poor, Fair and Good or Excellent. OCT2, organic cation transporter 2; ENT1, equilibrative nucleoside transporter 1; MATE1, multidrug and toxin extrusion 1.
3.5. SNPs and toxicity with clinical outcomes
Toxicities were analysed for association with clinical outcomes. In the training cohort, grade 3+ neutropenia (n = 20) was marginally associated with longer PFS and OS compared with grade 3— (n = 32). These findings were confirmed in the testing cohort for PFS (2.3 versus 1.9 months, respectively, HR 0.50, P < 0.001) and OS (8.8 versus 3.7 months, respectively, HR 0.21, P < 0.001) (Table A. 4). No significant association between SNPs and grade 3+ neutropenia was observed in both cohorts. In addition, grade 2+ neutropenia (n = 32) was significantly associated with longer OS compared with grade 2— neutropenia (8.3 versus 4.4 months, respectively, HR 0.41, 95% confidence interval: 0.17—0.98, P = 0.028) in the training cohort, whereas grade 2+ neutropenia was more frequent in patients with any G allele (n = 20/25, 80%) at ENT1 rs760370 compared with A/A variants (n = 12/27, 44%) (Table A. 5).
4. Discussion
We provide the first evidence that the NT SNP ENT1 rs760370 involved in the cellular uptake of FTD confers clinical outcomes in refractory mCRC patients treated with TAS-102. The fact that TPI also participates in the enhancement of FTD anti-tumour activity is particularly important because TPI demonstrates low efficacy and toxicity.
Higher levels of ENT1 mRNA were previously reported in cancers including colorectal, breast, lung and stomach compared with normal tissues [18]. Spratlin et al. [19] first investigated the association of hENT1 protein expression with a nucleoside analogue for gemcitabine efficacy, revealing that tumour cells expressing high hENT1 were associated with longer survival compared with those without hENT1 expression in advanced pancreatic adenocarcinoma patients. Thus, the clinical evaluation of hENT1 protein expression has been considered a predictive marker for gemcitabine. A recent in vitro study identified hENT mRNA down- regulation as a possible mechanism of FTD resistance in human colorectal cancer [12]. Interestingly, low levels of hENT RNA were associated with low intracellular FTD in resistant cells compared with FTD-sensitive cells. No specific correlation between hCNT mRNA expression and FTD accumulation was observed, suggesting a critical role for hENT1, but not hCNT, in FTD uptake and sensitivity. Nevertheless, a previous in vivo study demonstrated that FTD was absorbed via rCNT1 in rat intestinal lumens [7].
Tanaka et al. [20] demonstrated that the ENT1 rs760370 G/G variant was associated with poor tumour response and that ENT1 rs9394992 with any T allele was associated with increased neutropenia in gemcitabine- based therapy for advanced pancreatic cancer patients. Gemcitabine and FTD intracellular uptake is mainly mediated by hENT1 and hCNT1, respectively, and hENT1 and hCNT1 act as bi-directional and inward transporters, respectively [8,20]. The difference may infer different roles of ENT1 SNPs between gemcitabine and FTD for efficacy, for example, the ENT1 rs760370 G allele and rs9394992 T allele show opposite anti-tumour effects between gemcitabine and FTD. Further studies are required to clarify this interesting issue.
Testing of ENT1 SNPs for 5-fluorouracil (5-FU) may be of interest to researchers as a predictive marker, although there has been no clear evidence to date. A recent study that comprehensively investigated the transcript levels of NTs in colorectal cancer (CRC) tissue showed that low hENT1 expression was correlated with more sensitive response to 5-FU [21]. Meanwhile, only ENT1 SNPs were individually revealed to be strong predictors in the present study, suggesting that these SNPs may be specific for TAS-102. Furthermore, considering that thymidylate synthase inhibition is the main anti-tumour action of 5-FU, whereas DNA incorporation mainly confers the anti-tumour action of FTD, a comprehensive genetic analysis of transporters and metabolic enzymes is required, especially for 5-FU.
Interactions between OCT2 and MATE1 SNPs were previously reported to determine the SNP association with metformin CLrenal [16,22,23]. However, to our knowledge, we are the first to suggest a novel categorisation based on estimated transporter gene—gene interactions to identify survival benefit. A further categorisation of ENT1 rs360370 with OCT2 rs316019 and MATE1 rs2289669 interactions demonstrated the survival benefit more clearly than individual SNPs. These results indicate that NTs for FTD uptake and renal tubule transporters for TPI excretion may complement each other during TAS-102 administration in patients, conferring efficacy as well as drug-related neutropenia. Furthermore, the mechanism that non- excreted circulating TPI re-induces the inhibition of FTD degradation by TP in the liver was previously reported [24]. However, further studies are necessary to confirm the presence of gene—gene interactions in association with CLrenal for TPI excretion and the collaboration of these transporters.
Our study is limited by its retrospective design, absence of correction for multiple testing and lack of evidence regarding the role of SNPs, including TPI CLsec categorised by the OCT2 and MATE1 SNP interaction. However, its strengths include the control group of patients with comparable clinical characteristics and disease stage and the presence of a large testing cohort of patients with comparable clinical characteristics receiving the same treatment, albeit the training and control cohorts lacked the information about extended RAS or BRAF status. We also clarified the role of specific transporters involved in FTD and TPI pharmacokinetics or pharmacodynamics. Nevertheless, we need a further study to confirm whether the combined SNPs of ENT1, OCT2 and MATE1 will be not only a prognostic but also a predictive factor, albeit ENT1 was revealed as both predictive and prognostic factor alone in patients receiving TAS-102. In addition, cost-effective analysis is also warranted to evaluate clinical utility of a test kit for our candidate SNPs as shown in UGT1A1 polymorphisms [25]. However, regarding easiness in the procedure and non-invasiveness to the patients, SNPs testing seems to be acceptable and one of the ideal modalities to identify specific population who benefit from therapeutic agents.
In conclusion, genetic variants in the TAS-102 pharmacokinetic pathway, ENT1 germline SNPs and combination variants of OCT2 and MATE1 may serve as predictive and prognostic markers in refractory mCRC patients receiving TAS-102. We suggest a potential treatment algorithm for TAS-102 in mCRC patients in terms of efficacy and toxicity (Fig. 4B): Group 3 patients might benefit greatly from TAS-102 with careful monitoring of the onset of neutropenia; TAS-102 might be also available for Group 2 patients but they should be carefully considered and monitored; Group 1 patients might be less sensitive to TAS-102; hence, early evaluation of tumour response are recommended during treatment.
Supplementary Material
Acknowledgements
Mitsukuni Suenaga is the recipient of a Takashi Tsuruo Memorial Fund; Martin D. Berger received grants from the Swiss Cancer League (BIL KLS-3334-02-2014) and the Werner and Hedy Berger-Janser Foundation of Cancer Research; Yuji Miyamoto received a grant from the Japan Society for the Promotion of Science (S2606).
Funding
This work was partially supported by the National Institutes of Health (P30CA014089-27S1), the Gloria Borges Wunderglo Project, the Dhont Family Foundation, the Dave Butler Research Fund and the Call to Cure Research Fund.
Footnotes
Conflict of interest statement
None declared.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ejca.2017.08.033.
References
- [1].Hong DS, Abbruzzese JL, Bogaard K, Lassere Y, Fukushima M, Mita A, et al. Phase I study to determine the safety and pharmacokinetics of oral administration of TAS-102 in patients with solid tumors. Cancer 2006;107:1383–90. [DOI] [PubMed] [Google Scholar]
- [2].Temmink OH, Emura T, de Bruin M, Fukushima M, Peters GJ. Therapeutic potential of the dual-targeted TAS-102 formulation in the treatment of gastrointestinal malignancies. Cancer Sci 2007; 98:779–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bijnsdorp IV, Peters GJ, Temmink OH, Fukushima M, Kruyt FA. Differential activation of cell death and autophagy results in an increased cytotoxic potential for trifluorothymidine compared to 5-fluorouracil in colon cancer cells. Int J Cancer 2010;126:2457–68. [DOI] [PubMed] [Google Scholar]
- [4].Bonate PL, Arthaud L, Cantrell WR Jr, Stephenson K, Secrist JA 3rd, Weitman S. Discovery and development of clo- farabine: a nucleoside analogue for treating cancer. Nat Rev Drug Discov 2006;5:855–63. [DOI] [PubMed] [Google Scholar]
- [5].Jordheim LP, Durantel D, Zoulim F, Dumontet C. Advances in the development of nucleoside and nucleotide analogues for cancer and viral diseases. Nat Rev Drug Discov 2013;12:447–64. [DOI] [PubMed] [Google Scholar]
- [6].Smith KM, Slugoski MD, Loewen SK, Ng AM, Yao SY, Chen XZ, et al. The broadly selective human Na+/nucleoside cotransporter (hCNT3) exhibits novel cation-coupled nucleoside transport characteristics. J Biol Chem 2005;280:25436–49. [DOI] [PubMed] [Google Scholar]
- [7].Spratlin JL, Mackey JR. Human equilibrative nucleoside transporter 1 (hENT1) in pancreatic adenocarcinoma: towards individualized treatment decisions. Cancers (Basel) 2010;2:2044–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Okayama T, Yoshisue K, Kuwata K, Komuro M, Ohta S, Nagayama S. Involvement of concentrative nucleoside transporter 1 in intestinal absorption of trifluorothymidine, a novel antitumor nucleoside, in rats. J Pharmacol Exp Ther 2012;340: 457–62. [DOI] [PubMed] [Google Scholar]
- [9].Emura T, Suzuki N, Yamaguchi M, Ohshimo H, Fukushima M. A novel combination antimetabolite, TAS-102, exhibits antitumor activity in FU-resistant human cancer cells through a mechanism involving FTD incorporation in DNA. Int J Oncol 2004;25: 571–8. [PubMed] [Google Scholar]
- [10].Suzuki N, Nakagawa F, Nukatsuka M, Fukushima M. Tri- fluorothymidine exhibits potent antitumor activity via the induction of DNA double-strand breaks. Exp Ther Med 2011;2:393–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sakamoto K, Yokogawa T, Ueno H, Oguchi K, Kazuno H, Ishida K, et al. Crucial roles of thymidine kinase 1 and deoxy- UTPase in incorporating the antineoplastic nucleosides tri- fluridine and 2’-deoxy-5-fluorouridine into DNA. Int J Oncol 2015;46:2327–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Temmink OH, Bijnsdorp IV, Prins HJ, Losekoot N, Adema AD, Smid K, et al. Trifluorothymidine resistance is associated with decreased thymidine kinase and equilibrative nucleoside transporter expression or increased secretory phospholipase A2. Mol Cancer Ther 2010;9:1047–57. [DOI] [PubMed] [Google Scholar]
- [13].Doi T, Ohtsu A, Yoshino T, Boku N, Onozawa Y, Fukutomi A, et al. Phase I study of TAS-102 treatment in Japanese patients with advanced solid tumours. Br J Cancer 2012;107:429–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Yoshisue K, Takahashi K, Okayama T, Yamashita F, Chiba M. Investigation of transporters that play an important role in urinary secretion of thymidine phosphorylase inhibitor combined with a novel anti-cancer agent of TAS-102. Drug Metab Rev 2015;47:263. [Google Scholar]
- [15].Strobel J, Muller F, Zolk O, EndreB B, Konig J, Fromm MF, et al. Transport of asymmetric dimethylarginine (ADMA) by cationic amino acid transporter 2 (CAT2), organic cation transporter 2 (OCT2) and multidrug and toxin extrusion protein 1 (MATE1). Amino Acids 2013;45:989–1002. [DOI] [PubMed] [Google Scholar]
- [16].Christensen MM, Pedersen RS, Stage TB, Brasch-Andersen C, Nielsen F, Damkier P, et al. A gene-gene interaction between polymorphisms in the OCT2 and MATE1 genes influences the renal clearance of metformin. Pharmacogenet Genomics 2013;23: 526–34. [DOI] [PubMed] [Google Scholar]
- [17].Grün B, Kiessling MK, Burhenne J, Riedel KD, Weiss J, Rauch G, et al. Trimethoprim-metformin interaction and its genetic modulation by OCT2 and MATE1 transporters. Br J Clin Pharmacol 2013;76:787–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Damaraju VL, Damaraju S, Young JD, Baldwin SA, Mackey J, Sawyer MB, et al. Nucleoside anticancer drugs: the role of nucleoside transporters in resistance to cancer chemotherapy. Oncogene 2003;22:7524–36. [DOI] [PubMed] [Google Scholar]
- [19].Spratlin J, Sangha R, Glubrecht D, Dabbagh L, Young JD, Dumontet C, et al. The absence of human equilibrative nucleoside transporter 1 is associated with reduced survival in patients with gemcitabine-treated pancreas adenocarcinoma. Clin Cancer Res 2004;10:6956–61. [DOI] [PubMed] [Google Scholar]
- [20].Tanaka M, Javle M, Dong X, Eng C, Abbruzzese JL, Li D. Gemcitabine metabolic and transporter gene polymorphisms are associated with drug toxicity and efficacy in patients with locally advanced pancreatic cancer. Cancer 2010;116:5325–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Phua LC, Mal M, Koh PK, Cheah PY, Cheah PY, Chan EC, et al. Investigating the role of nucleoside transporters in the resistance of colorectal cancer to 5-fluorouracil therapy. Cancer Chemother Pharmacol 2013;71:817–23. [DOI] [PubMed] [Google Scholar]
- [22].Otsuka M, Matsumoto T, Morimoto R, Arioka S, Omote H, Moriyama Y. A human transporter protein that mediates the final excretion step for toxic organic cations. Proc Natl Acad Sci U S A 2005;102:17923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Meyer zu Schwabedissen HE, Verstuyft C, Kroemer HK, Becquemont L, Kim RB. Human multidrug and toxin extrusion 1 (MATE1/SLC47A1) transporter: functional characterization, interaction with OCT2 (SLC22A2), and single nucleotide polymorphisms. Am J Physiol Ren Physiol 2010;298:F997–1005. [DOI] [PubMed] [Google Scholar]
- [24].Lee JJ, Seraj J, Yoshida K, Mizuguchi H, Strychor S, Fiejdasz J, et al. Human mass balance study of TAS-102 using (14)C analyzed by accelerator mass spectrometry. Cancer Chemother Pharmacol 2016;77:515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gold HT, Hall MJ, Blinder V, Schackman BR. Cost effectiveness of pharmacogenetic testing for uridine diphosphate glucur- onosyltransferase 1A1 before irinotecan administration for metastatic colorectal cancer. Cancer 2009;115:3858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.