Abstract
The CRISPR-Cas system offers a programmable platform for eukaryotic genome and epigenome editing. The ability to perform targeted genetic and epigenetic perturbations enables researchers to investigate questions in basic biology and potentially develop novel therapeutics for the treatment of disease. While CRISPR systems have been engineered to target DNA and RNA with increased precision, efficiency, and flexibility, assays to identify off-target editing are becoming more comprehensive and sensitive. Furthermore, techniques to perform high-throughput genome and epigenome editing can be paired with a variety of readouts and are uncovering important cellular functions and mechanisms. These technological advances drive and are driven by accompanying computational approaches. Here, we briefly present available CRISPR technologies and review key computational advances and considerations for various CRISPR applications. In particular, we focus on the analysis of on- and off-target editing and CRISPR pooled screen data.
The CRISPR-Cas system has accelerated the development of tools for genome and epigenome editing. Since the initial adaptation of Streptococcus pyogenes Cas9 (SpCas9) for eukaryotic genome editing, the CRISPR toolbox has rapidly expanded to include new CRISPR-Cas orthologs and variants with various target sequence requirements, fidelities, and perturbation mechanisms (Jiang and Doudna, 2017). CRISPR-Cas systems have also been repurposed in various ways for gene activation (CRISPR activation [CRISPRa]), gene repression (CRISPR interference [CRISPRi]), epigenome editing via fusion to epigenetic modifiers (Thakore et al., 2016), and DNA sequence alteration in the absence of a double strand break (DSB) (base editing and prime editing) (Anzalone et al., 2019; Rees and Liu, 2018).
Characterizing and quantifying products of genome editing is essential for the development of new tools and for bridging the knowledge gap between genome sequence and function. Biochemical assays for measuring editing events with simple—sometimes binary—readouts are being replaced by next-generation sequencing (NGS) approaches that improve accuracy and sensitivity, while also providing a more comprehensive view of genome editing outcomes. However, data from NGS-based experiments require several steps of downstream processing, which has led to the development of various computational tools for analysis.
In addition, it is well established that CRISPR-Cas editing may also occur at unintended genomic loci, also known as off-target sites. Locating and quantifying editing at these off-target sites is critical for interpretation of genome editing experiments as well as for assessing the safety of therapeutic genome editing programs. In silico methods for predicting off-targets have evolved from the simple enumeration of sites based on on-target sequence similarity to more advanced tools that incorporate common or personal genomic variants. At the same time, experimental techniques have also become more sophisticated, leveraging downstream computational analysis to identify sites of off-target editing with increased sensitivity.
The programmability of CRISPR-Cas systems has enabled the association of genomic perturbations to phenotypes at scale. High-throughput CRISPR perturbations can be performed on many cells in parallel using a pool of guide RNAs (gRNAs) targeting many genes or tiled across a region of interest. The perturbation response in each cell can be measured using a variety of readouts. This approach has enabled the dissection of critical functional elements within a region of interest and the identification of critical genes and gene networks associated with a phenotype of interest. An enormous amount of data is generated with each screen, and computational analysis methods have been developed to efficiently and properly interpret the experimental results.
In this review, we outline biological questions that can be investigated using current tools from the CRISPR-Cas toolbox and provide computational perspectives into the advantages, disadvantages, and analytical challenges of these technologies. Table 1 summarizes these technologies, associated computational methods, and key references with example applications and tools to help address these questions.
Table 1.
Summary of Key Technologies and Computational Strategies
| Purpose | Approach | Technology | Comments | Examples |
|---|---|---|---|---|
| Quantify/validate editing at a region | Mismatch assays | Heteroduplex cleavage assays A locus of interest is amplified from genomic DNA using PCR, followed by denaturation and annealing to form heteroduplex complexes between wildtype (non-edited) and nuclease-modified DNA strands. DNA mismatch enzymes cleave sites of sequence mismatch. The editing proportion is indicated by measuring the proportion of cleaved products. |
• Poor ability to reliably detect low-frequency mutations • Heterogeneous sensitivities for different variant types (e.g., limited detection of single nucleotide polymorphisms [SNPs]) • Under-estimation of editing frequencies including inability to detect high editing frequencies with an editing frequency of 30–40% possibly representing an upper limit that can be reliably detected (Mashal et al., 1995) • False positives in the setting of heterozygous germline mutations (Qiu et al., 2004), which can be falsely attributed to CRISPR-mediated mutagenesis if proper controls are not performed |
T7E1 (Mashal et al., 1995), Surveyor (Qiu et al., 2004) |
| Amplicon sequencing | Conventional chain termination (Sanger) sequencing Amplified DNA is sequenced using Conventional chain termination (Sanger) sequencing, producing a quantification of the proportion of each base at each position. |
• Inexpensive sequencing costs • Useful for screening clones or homogeneous populations • Deconvolution of alleles in heterogeneous populations may be inaccurate |
Brinkman et al., 2014, 2018; Hsiau et al., 2019 | |
| Next-generation sequencing Amplified DNA is sequenced using next-generation sequencing, producing the DNA sequence of alleles from different cells |
• Deep coverage at known loci, with the ability to detect and quantify rare editing events • Precise quantification of alleles and editing rates on heterogenous populations • Moderate sequencing costs |
Boel et al., 2016; Clement et al., 2019; Güell et al., 2014; Hwang et al., 2018; Lindsay et al., 2016; Park et al., 2017; Pinello et al., 2016 | ||
| Measure (single) guide specificity | In silico | Sequence homology to reference genome A reference genome sequence is scanned for similarity to a given guide sequence. Similarity is measured by the number of mismatches or gaps between the guide sequence and the match in the reference genome. |
• Quickly determine number and location of putative off-targets without any experimental labor • Useful in designing guides or selecting guides with few off-targets • Cellular conditions may affect CRISPR activity to prefer or avoid sites with few mismatches to the guide • Genomic sequence in target cells may vary from reference sequences |
Bae et al., 2014; Cancellieri et al., 2019; Haeussler et al., 2016; Heigwer et al., 2014; Klein et al., 2018; Lei et al., 2014; McKenna and Shendure, 2018; Montague et al., 2014; Xiao et al., 2014 |
| Machine learning approaches Machine learning models are trained on datasets of genome editing outcomes to predict the activity of untested guides or the activity of guides at untested regions. |
• Predicts activity rates at sites with sequence homology • Models are based on experimental data and may be biased based on cell type, guide sequence, or other variables |
Abadi et al., 2017; Allen et al., 2018; Lin and Wong, 2018; Listgarten et al., 2018; Peng et al., 2018 | ||
| Experimental |
In vitro
Genomic DNA is tested for the ability to be cleaved by a guide, outside of the cellular context, and not in the presence of native DSB repair machinery. |
• Edited genomic fragments can be selected, resulting in reduced sequencing costs • Double-stranded break repair mechanisms are not present in the in vitro context, so adaptor ligation or other biochemical tagging can be performed • Sensitivity of method may overestimate editing in cellular context • Some methods allow the detection of off-targets for individuals/samples for which limited genomic material is available |
SITE-seq (Cameron et al., 2017), Digenome-seq (Kim et al., 2015, 2016), CIRCLE-seq (Tsai et al., 2017) | |
| In cellula The occurrence of genome editing in cellular context is observed by observing events associated with DSB repair, or the introduction of molecular tags to edited regions. |
• Epigenetic context of the off-target is taken into consideration • Many cell types do not tolerate genomic manipulation required for readout |
Cas-ChIP (Duan et al., 2014; Kuscu et al., 2014; Wu et al., 2014), DISCOVER-seq (Wienert et al., 2019), IDLV insertion at DSB (Wang et al., 2015b), GUIDE-seq (Tsai et al., 2015), iGUIDE (Nobles et al., 2019), BLESS (Crosetto et al., 2013), BLISS (Yan et al., 2017) | ||
| Characterize gene function using a single perturbation | Single gene knockout | Single guide edit resulting in frameshift mutation or early stop codon A single guide is designed to target Cas9 or another editing enzyme to an early exon in the gene. The frameshift mutation resulting from the edit renders the gene non-functional. |
• Targeting region is well defined by exon boundaries • Genomic sequencing can validate success of edit • Edit knocks out genes and is propagated across cell divisions • Variety of CRISPR editing nucleases and editors can be used—Cas9, Cas12, BE, PE • Early exons may be skipped in edited cells (Mou et al., 2017) • Allelic diversity of edits may affect phenotype—can be overcome by growing and selecting edited clones |
Cong et al., 2013 |
| Non-coding perturbation | CRISPRi/a targeting promoter A single guide targets deactivated Cas9 to the promoter of a gene of interest. The deactivated Cas9 is tethered to an activating or inhibiting enzyme which activates or inhibits transcription of the gene. |
• Target is defined by a region of putative regulatory activity, e.g., an enhancer identified with epigenetic marks • Perturbations are transient (can be overcome by genomic integration of CRISPR system) • Requires annotated (single) promoter • Results in overall reduction in number of transcripts of the target gene • qPCR or RNA-seq (or ChIP of CRISPR binding at target) to validate changes in expression of target gene • Targeting boundaries are less well defined (as opposed to exon boundaries in genetic targeting) |
Gilbert et al., 2013; Qi et al., 2013 | |
| Disruption of binding site sequence A single guide targets Cas9 or another editing enzyme to a binding site that controls expression of a gene. After gene editing, the binding site is disrupted, and the gene is not transcribed. |
• Target sequence must be known (e.g., ChIP-seq or TF models) and well-described | Zhou et al., 2014a | ||
| Deletion of large element of DNA | Editing with a flanking pair of guides A pair of guides target a cleaving enzyme to the flanking sites of a region of interest. After cleavage, the region of interest is excluded upon DSB repair, resulting in a large deletion. |
• Genetic element is completely removed or disrupted • Inversions, NHEJ, or other complex outcomes are likely |
Chen et al., 2014; Mali et al., 2013 | |
| Discover putative genes associated with a phenotype | Pooled genome-wide genetic screen | Phenotype readout Many guides are designed to target genes across the genome. Cells are sorted by phenotype, and the guides producing high versus low phenotype are compared. |
• Measure selectable phenotypes • Potentially sequence alleles resulting in strongest phenotype |
Allen et al., 2019; Hart and Moffat, 2016; Li et al., 2014; Lindsay et al., 2016; Love et al., 2014; Shalem et al., 2014; Spahn et al., 2017; Wang et al., 2014, 2015a; Yu et al., 2016; Zhou et al., 2014b) |
| Single-cell RNA-seq readout Many guides are designed to target genes across the genome. Cells are treated with one guide per cell, and the transcription levels of all genes are read out using single-cell RNA-sequencing. |
• Granular results at single-gene expression level • Measure effect on genes or sets of genes • Readout of heterogeneity of response • Identify gene networks with similar response profiles • Limited number of cells profiled (per perturbation) • Requires high analysis resources |
Adamson et al., 2016; Datlinger et al., 2017; Dixit et al., 2016; Duan et al., 2019; Jaitin et al., 2016; Xie et al., 2017; Yang et al., 2020 | ||
| Pooled genome-wide epigenetic screen | CRISPRi/a targeting of gene promoters Many guides are designed to target genes across the genome with deactivated Cas and an enhancer/inhibitor. Phenotypic or transcriptomic changes associated with activation/repression of genes are measured. |
• CRISPRi/a must be targeted precisely to achieve consistent gene knockout, as compared to genetic mutations which can disable translation with a single mutation | Gilbert et al., 2014; Konermann et al., 2015 | |
| Discover critical elements within an annotated gene/enhancer | Tiling screen profiling a single locus | Genetic screen of coding region Many guides are designed to target a region of the genome with coding significance. Cas9 or another editing enzyme create genetic modifications to the coding region, altering the physical properties of the resulting protein. | • Can be used to discover functional domains of a protein | He et al., 2019; Neggers et al., 2018; Shi et al., 2015 |
| Genetic screen of noncoding region Many guides are designed to target a region of the genome with putative regulatory significance (e.g., enhancer). DNA edits by Cas9 or another editing enzyme affect the regulatory properties near the guide. Guides associated with a phenotype change indicate specific locations critical for function. |
• Effect of single-base-pair changes can be measured • Critical regions may be missed if not near perturbation window of guide • Low probability of creating activating mutations |
Canver et al., 2015; Sanjana et al., 2016 | ||
| Epigenetic screen of noncoding region Many guides are designed to target a region of the genome with putative regulatory significance. Epigenetic effectors are targeted using these guides, and guides associated with phenotype change indicate specific locations critical for function. |
• Larger regions can be interrogated using fewer guides • Can be used to identify silenced enhancers |
Fulco et al., 2016 |
CRISPR-Cas Perturbation Technologies
The CRISPR-Cas system is a versatile perturbation platform that can introduce different forms of genetic and epigenetic modifications. CRISPR-Cas enzymes were first discovered as components of an adaptive immunity system used by bacteria and archaea to counter foreign DNA and have been repurposed and engineered to perform genome editing in other organisms. The CRISPR-Cas-induced DSBs are repaired using endogenous cellular machinery involved in the non-homologous end joining (NHEJ), microhomology-mediated end joining (MMEJ), and homology-directed repair (HDR) pathways. Repair by NHEJ or MMEJ typically results in a spectrum of variable-length insertion or deletion (indel) mutations (Allen et al., 2018; Shen et al., 2018) introduced with relatively high frequencies. The induction of indels is particularly useful in introducing frameshift mutations or disrupting non-coding regulatory elements. By contrast, exogenous DNA templates can be provided to take advantage of HDR pathways to introduce precise genomic edits, albeit at lower efficiencies (though recent studies suggest that efficiency may be enhanced with small molecules [Riesenberg and Maricic, 2018; Song et al., 2016; Vartak and Raghavan, 2015]).
Recent technological advancements have led to newer forms of CRISPR-based genetic editors that modify DNA sequences without the requirement for DSBs. Base editors (BEs), a fusion of nickase Cas9 and a cytidine or adenosine deaminase enzyme, introduce targeted substitution mutations within a defined editing window at target DNA sequences (Rees and Liu, 2018). Prime editors (PEs), a fusion of nickase Cas9 and a reverse transcriptase, can be used with an associated RNA template contained on the prime editing guide RNA (pegRNA) to introduce a wide variety of genetic modifications ranging from single-base substitutions to short indel mutations (Anzalone et al., 2019). BEs and PEs are useful tools for sequence mutagenesis because they can introduce precise mutations and differ from CRISPR-Cas nucleases in that they significantly reduce the frequency of repair outcomes containing unwanted insertion or deletion mutations.
CRISPRa and CRISPRi technologies can be used to induce robust gene activation and repression, respectively (Gilbert et al., 2013; Kampmann, 2018; Qi et al., 2013; Thakore et al., 2016). For CRISPRa/i, the CRISPR-Cas enzyme is catalytically disabled and utilized to target effector domains such as transcriptional activators (e.g., p65, VPR), repressors (e.g., KRAB), or other epigenetic modifiers (e.g., DNMT3A) (Qi et al., 2013) to a specific loci and has also been used to block transcriptional initiation or elongation through steric hindrance. CRISPRa and CRISPRi have been useful in studying the effects of gene activation and knockdown in single gene or pooled settings (Adli, 2018; Thakore et al., 2016).
Characterization of DNA Editing at Defined Loci
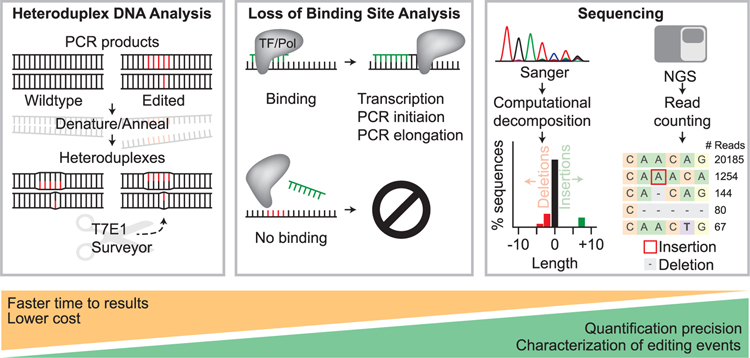
A variety of methods exist to quantify on- and off-target genome editing at individual loci (Figure 1 and Table 1). These methods include sequencing and non-sequencing-based techniques. In general, non-sequencing-based assays offer cost-effective and rapid solutions to semiquantitatively detect the presence of nuclease-mediated sequence modification, while sequencing-based methods offer the ability to accurately quantify editing frequencies and define mutation alleles induced by genome editing.
Figure 1. Overview of Strategies to Detect Editing at Known Loci Including Heteroduplex DNA Analysis, Loss of Binding Site Analysis, and Sequencing-Based Approaches.
Heteroduplex DNA (left panel) is formed by denaturing and annealing of PCR amplicons generated from a bulk population of edited cells. The mismatches in the heteroduplex are detected and cleaved by enzymes such as T7E1 or Surveyor. Loss of binding site analysis (middle panel) relies on the ability of a PCR primer (green DNA sequence) to bind based on Watson-Crick complementarity or a transcription factor or restriction enzyme to identify its recognition sequence. Editing can be identified by binding site modification that results in the loss of PCR primer binding or loss of restriction enzyme mediated cleavage. In sequencing-based assays (right panel), Sanger sequencing or Next Generation Sequencing (NGS) are used to analyze a given site(s).
Assessment of Editing at Defined Loci Using Non- Sequencing-Based Methods
Mismatch cleavage and heteroduplex mobility assays are examples of non-sequencing-based techniques for indel detection and rely on similar principles to detect editing frequencies within bulk-edited cell populations. Specifically, a locus of interest (e.g., an on-target or predicted off-target site) is amplified from genomic DNA using polymerase chain reaction (PCR), followed by denaturation and annealing steps to form heteroduplex complexes between wild-type (non-edited) and nuclease-edited DNA strands (Vouillot et al., 2015; Zhu et al., 2014). This heteroduplex DNA can be analyzed in several ways: heteroduplexes can be treated with DNA mismatch endonucleases, such as Surveyor or T7E1, that cleave at sites of sequence mismatch. The sizes and relative intensities of resulting cleavage products can be quantified by either gel or capillary electrophoresis, and these values can be used to estimate editing frequencies (Mashal et al., 1995; Qiu et al., 2004; Vouillot et al., 2015). Alternatively, heteroduplex DNA products can be resolved by polyacrylamide gel electrophoresis (PAGE), which relies on heteroduplex (edited) DNA running with slower mobility than homoduplex (non-edited) DNA (Yu et al., 2014; Zhu et al., 2014). Another analysis strategy is high-resolution melting analysis, which relies on differences in melting temperature (Tm) between homoduplex and heteroduplex DNA to identify nuclease-induced mutations (Thomas et al., 2014). Importantly, these non-sequencing-based methods resolve single nucleotide substitutions poorly and therefore have minimal utility for base and prime editing experiments yielding substitution edits.
Other approaches to detect genome editing include methods that measure disruption of a PCR primer binding site (Yu et al., 2014) or restriction endonuclease sites (Kim et al., 2014). Insertions and deletions as small as 1 base pair (bp) can be detected by fluorescent capillary electrophoresis (Cho et al., 2014; Ramlee et al., 2015; Yang et al., 2015), but these methods are unable to detect substitution edits (Yang et al., 2015). In addition, digital droplet PCR (ddPCR) offers a quantitative method to evaluate NHEJ/MMEJ and HDR outcomes. One strategy using ddPCR relies on the usage of two fluorescent probes with one probe at the predicted cleavage site and the other at a distant site. This and other ddPCR-based assays offer precise quantification of editing outcomes including the ability to distinguish mono-allelic and bi-allelic modifications. Of note, ddPCR cannot resolve sequence information from NHEJ- and MMEJ-mediated out- comes (Findlay et al., 2016).
The methods described above are predominantly focused on directly detecting DNA sequence alterations. However, surrogate outcomes can also be evaluated as functional readouts. For example, changes in gene expression or protein abundance can indicate that a genomic change has occurred to knock out a gene or otherwise influence the production of the protein of interest. While many approaches exist, quantitative reverse-transcription PCR (RT-qPCR) and western blotting are common methods for detection of RNA and protein expression changes, respectively. Surrogate outcomes are particularly useful in the context of CRISPR-based screens, which seek to identify candidate functional elements or regions through changes in gene expression, protein abundance, or cell viability.
Assessment of Editing at Defined Loci Using Sequencing-Based Methods
Analysis of genome editing outcomes based on sequencing can take several forms. First, Sanger sequencing trace decomposition can be performed by tools such as Tracking Indels by Decomposition (TIDE) or Inference of CRISPR Edits (ICE) to calculate editing and allelic frequencies from bulk edited cells. These tools can be used for the quantification of both NHEJ/ MMEJ and HDR outcomes and are able to infer individual allelic frequencies (Brinkman et al., 2014, 2018; Hsiau et al., 2019). Sanger trace decomposition offers a rapid and low-cost methodology to assess editing outcomes and is particularly useful in screening clones of edited cells. Additionally, PCR amplicons for a given locus can be cloned into a plasmid backbone (e.g., TA, TOPO, or blunt end cloning), transformed into bacteria, and sequenced by Sanger methods to identify specific alleles (Canver et al., 2014).
Next-generation sequencing (NGS) represents the gold standard for both determination of editing frequency and characterization of the resulting alleles. A recent comparison of T7E1, Indel Detection by Amplicon Analysis (IDAA) (Yang et al., 2015), and TIDE-based Sanger decomposition to NGS revealed that T7E1 exhibited poor performance when compared to NGS. In contrast, both TIDE and IDAA offered comparable results to NGS for indel identification and indel frequency calculation (assuming indels with a frequency of >5%); however, both TIDE and IDAA approaches overestimated the presence of wild-type alleles and were less sensitive compared to NGS (Sentmanat et al., 2018). Depending on access to NGS technology, NGS assays may be limited by high cost, laborious preparation, and time delay to the data. However, as sequencing costs continue to decline, the analysis of genetic editing by NGS is becoming increasingly common, facilitating the development of multiplexed NGS readouts and more sensitive assays for the detection of rare alleles.
Several computational tools have been developed to analyze NGS data from genetic editing experiments with CRISPR-Cas nucleases (Boel et al., 2016; Güell et al., 2014; Lindsay et al., 2016; Park et al., 2017; Pinello et al., 2016) and base editors (Clement et al., 2019; Hwang et al., 2018). A variety of alignment and analysis methods are used to separate sequencing errors from true genome edits. One example is the use of an “editing window,” whereby only mutations overlapping the predicted target of activity are quantified, and mutations distal to the predicted target of activity are ignored (Clement et al., 2019; Hwang et al., 2018). This editing window approach on simulated sequencing data demonstrated a significant reduction in the quantification of false-positive mutations (e.g., sequencing error at the ends of reads) (Pinello et al., 2016).
Technology Outlook
Current efforts to quantify on-target editing events are focused on expanding the ability to measure all types of genome editing events. For example, existing methods can measure short insertions and deletions, but large insertions or deletions (>100 bp) or translocations are not detectable with standard amplicon sequencing approaches. For example, these editing outcomes may not be discovered if one or both primer binding sites are deleted by a large deletion or if a large insertion occurs, since it may not be efficiently amplified (Cullot et al., 2019; Kosicki et al., 2018). More specialized strategies such as anchored multiplex PCR (AMP) (Zheng et al., 2014) and UDiTaS (Giannoukos et al., 2018) utilize single-anchor amplicon sequencing to detect translocations and insertions at on-target regions, but would also not discover deletions if the PCR primer site was deleted. Long-read sequencing may be a useful alternative to capture these larger-scale events (Dastidar et al., 2018; Gasperini et al., 2017; Kosicki et al., 2018).
To increase the quantification accuracy of amplicon sequencing, the use of unique molecular identifiers (UMIs) has been proposed to reduce the impact of PCR amplification bias or other artifacts that may skew quantification of paired (Kennedy et al., 2014; Kinde et al., 2011) or single-anchor amplicon sequencing (Tsai et al., 2015). These strategies introduce a unique DNA barcode during library preparation or in the first PCR cycle that can be read out via NGS. Reads with the same barcode likely originate from the same molecule and can be considered as duplicates in the downstream analysis. Computational frameworks for incorporating UMIs into genome editing quantification will be useful to increase quantification accuracy (Clement et al., 2018).
Another challenge facing the field is lowering the limit of detection for rare editing events. Currently, the limit of detection is bounded by PCR amplification and sequencing error rates—with current NGS and amplification technologies, it can be difficult to determine whether a deviation from the expected reference sequence is due to sequencing error, PCR error, or a rare genome editing event. One solution is to use UMIs coupled with high sequencing coverage, so that reads from each UMI are sequenced several times and can be used to error-correct sequencing and/or PCR errors. Several additional experimental and computational strategies could lower this detection limit, including machine learning approaches to distinguish sequencing errors from genome edits (Poplin et al., 2018).
Assessing CRISPR-Cas Targeting Specificity
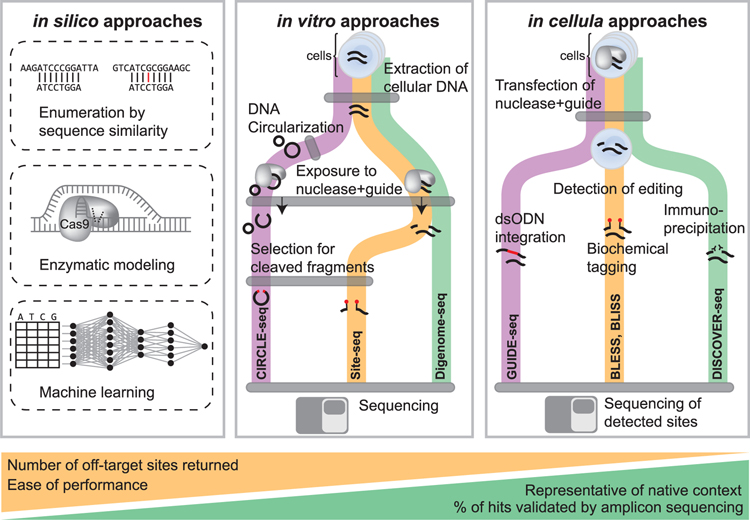
Many techniques have been developed to identify sites of potential off-target editing activity, i.e., editing at unwanted locations that may confound the interpretation of on-target perturbations or potentially complicate therapeutic applications (Figure 2; Table 1). The identification of off-target editing represents an important technical challenge for all genome editing platforms, including CRISPR. Thus, it is of great importance and interest to the scientific community to reliably detect and accurately quantify CRISPR off-target editing.
Figure 2. Overview of In Silico, In Vitro, and In Cellula Strategies to Nominate Off-Target Editing at Known/Unknown Loci.
In silico approaches (left panel) utilize sequence homology to identify genomic loci (top strand) with similarity to the gRNA sequence (bottom strand) up to a particular number of mismatches, 2019In silico approaches (left panel) utilize sequence homology to identify genomic loci (top strand) with similarity to the gRNA sequence (bottom strand) up to a particular number of mismatches. Other approaches use enzymatic modeling to predict gRNA binding specificity at putative off-target sites. Machine learning approaches have also been developed to identify off-target sites. In vitro approaches (middle panel) first extract DNA from cells, and in CIRCLE-seq, DNA is circularized. Next DNA is exposed to editing reagents and cleaved fragments can be selected and sequenced to identify off-target cleavage or subjected to whole genome re-sequencing with identification of cleavage sites by identifying reads that start or end at the same base position. In cellula strategies (right panel) introduce CRISPR reagents into cells in the native cellular context. Cleavage events can be detected through a variety of methods such as ligation of known sequences to double strand breaks, biochemical tagging with biotinylated primers, or immunoprecipitation of DNA repair factors recruited to sites of cleavage.
A two-step approach of nomination followed by validation has been widely used to identify off-targets. First, a superset of potential off-target editing sites are nominated using one or more in silico, in vitro, or in cellula approaches. Second, these sites are individually validated in the target cell type using the assays for characterizing editing at a single locus, with a strong preference for NGS given its greater sensitivity. Together, these steps have been used to comprehensively and sensitively identify off-targets in vivo (Akcakaya et al., 2018).
In Silico Nomination of Sites with On-Target Homology
Off-target genomic cleavage known to occur at certain sequences with high similarity to the on-target site (Cho et al., 2014; Pattanayak et al., 2013). However, the rules governing whether or not certain high-homology sequences are cleaved remain incompletely understood (Tycko et al., 2016). Given the sequence-dependence of off-target activity, there have been extensive efforts to computationally predict off-target sites at the gRNA design stage (Hanna and Doench, 2020). These efforts have focused on the identification of sites with sequence homology up to a specified number of mismatches and/or RNA/DNA bulges; some of these methods also attempt to incorporate predictions about the effects of mismatches on cleavage activities (Doench et al., 2014, 2016b; Sanson et al., 2018).
Putative off-target sites with high homology to the on-target site can be identified using standard sequence aligners. Ontarget sequence alignment to the reference genome of interest is performed, and all genomic loci up to a desired number of mismatches are reported (e.g., Blast [CRISPR-P (Lei et al., 2014)], Bowtie [Langmead et al., 2009] [CHOPCHOP (Montague et al., 2014), GT-Scan (O’Brien and Bailey, 2014), Bowtie2 (Langmead and Salzberg, 2012) (E-CRISP [Heigwer et al., 2014]), or BWA (Li and Durbin, 2009) (CRISPOR [Haeussler et al., 2016])]). Sequence aligners offer rapid and scalable in silico enumeration of closely matched sites but are limited because they do not perform an exhaustive genomic search, which can oftentimes lead to an incomplete list of such sites (Naito et al., 2004).
Newer search algorithms have been developed to overcome limitations of using general sequence aligners for finding potential off-target sites. Off-target sites can be enumerated rapidly if only mismatches to the on-target sequence are considered (McKenna and Shendure, 2018). Other search algorithms further include the ability to incorporate biological information about sequence requirements for different PAM sites (Xiao et al., 2014; Zhu et al., 2014), to identify sites with RNA and/or DNA bulges (Bae et al., 2014; Cancellieri et al., 2019), and to perform biochemical modeling of the enzymatic binding and cleaving process (Klein et al., 2018). Genetic sequence variants can also alter the off-target profiles of a genetic editor (Canver et al., 2018b; Scott and Zhang, 2017), which requires additional capabilities for identifying sites affected by these variants (Cancellieri et al., 2019). Of note, many of these search algorithms offer user-friendly web interfaces with a variety of customizable parameters to facilitate user-specific analysis (Bae et al., 2014; Heigwer et al., 2014; Montague et al., 2014; Naito et al., 2015). After enumerating sites from in silico analysis, the potential editing activity at these sites can also be predicted (Doench et al., 2016a; Hsu et al., 2013; Singh et al., 2015; Stemmer et al., 2015; Xu et al., 2017). The gRNAs can be chosen to minimize the overall number of closely matched sites in a genome of interest and to avoid off-targets that lie in annotated functional genomic regions such as coding sequences, promoters, putative enhancers, or insulators (Cancellieri et al., 2019).
In general, the presence of off-target editing decreases with an increasing number of mismatches (Cho et al., 2014; Haeussler et al., 2016). However, various experiments have shown that off-target mutations can occur in sites with as many as six mismatches relative to the on-target site (Tsai et al., 2015), but why some more closely matched sites are not mutated while other less closely matched sites are mutated remains poorly understood. (Tsai et al., 2015). As a result, the number of mismatches that should be tolerated during in silico off-target site enumeration remains an open question. Further insights may be derived from the increasing availability of larger experimental datasets cataloguing genome editing outcomes. Models have been trained on experimental data using a variety of computational approaches including logistic models (Allen et al., 2018), two-layer regression models (Elevation [Listgarten et al., 2018]), neural nets (Lin and Wong, 2018), or random forest regression models (CRISTA [Abadi et al., 2017]). However, generating sufficient data for model training has been a challenge. For example, previous work primarily utilized a published dataset of only 30 gRNAs for model training (Lin and Wong, 2018). In addition, these models were trained only for the CRISPR-Cas9 nuclease and are not directly applicable to other editors (e.g., Cas12a nuclease or base editors).
In Vitro Off-Target Nomination Assays
In vitro approaches measure the biochemical cleavage activities of CRISPR-Cas nucleases on DNA substrates in a cell-free or in vitro environment (as compared to in cellula assays in which the CRISPR-Cas nucleases are exposed to genomic DNA in a cellular context, see below). Multiple different in vitro off-target cleavage assays that were originally developed for zinc finger nucleases and TAL effector nucleases have been adapted for assessment of CRISPR-mediated off-target cleavage (Pattanayak et al., 2011). For example, one such strategy uses the generation of concatemeric DNA oligonucleotide libraries containing all possible variants with up to 8 mismatches relative to a gRNA sequence of interest (~1012 distinct variations). After exposure to a CRISPR-Cas nuclease, flanking adapters are ligated at DSB positions to allow for PCR amplification and NGS to identify the cleaved sites (Pattanayak et al., 2013).
Whole genome sequencing (WGS)-based strategies have also been used to identify off-target cleavage events. The Digenome-seq method detects cleavage activity by fragmenting genomic DNA, treating the sample with a given CRISPR-Cas nuclease and gRNA of interest, and then performing WGS on the sample. Nuclease-mediated cleavage events are then identified by a pileup of reads that consistently terminate at a particular base position. Digenome-seq can be used to analyze multiple gRNAs in a single sequencing run, although reduced sequencing depth can lead to lower sensitivity for identifying low-frequency off-target events (Kim et al., 2015, 2016). WGS-based approaches are also limited by high cost, limited access to specialized high-throughput sequencing platforms (HiSeq x10 or NovaSeq), and highly inefficient yield of information due to the lack of enrichment for reads cleaved by the nuclease.
One strategy for improving the yield of information from an in vitro off-target identification strategy is to perform an enrichment for those sequencing reads that are informative for nuclease-mediated cleavage. The SITE-seq assay accomplishes this by ligating biotinylated adapters at DSB sites in genomic DNA, which can then be selectively enriched prior to sequencing (Cameron et al., 2017). Similarly, the CIRCLE-seq method accomplishes enrichment for desired cleavage events by first circularizing sheared genomic DNA fragments and then treating with a nuclease of interest. Following nuclease cleavage, the resulting free DNA ends can then be substrates for sequencing adaptor ligation followed by NGS. This strategy greatly enriches for nuclease cleavage events and enables NGS to be more efficient, requiring the use of only a MiSeq run to detect rare off-target events. In addition, CIRCLE-seq can be performed without the need for a reference genome and therefore can be utilized in cases where the reference genome sequence is unknown or when the genomic sequence of interest deviates significantly from the reference (Tsai et al., 2017).
In cellula Off-Target Nomination Assays
Cell-based off-target nomination assays take the endogenous chromatin and cell-type DNA repair preferences into account for the identification of off-target CRISPR-Cas nuclease activity (Iyama and Wilson, 2013; Riesenberg and Maricic, 2018; Sun et al., 2019).This can be an advantage if the assay can be performed in the actual cell type of interest. However, it can also be a disadvantage if one needs to perform the assay in a different cell type because cell-type-specific effects can lead to different off-target profiles. Initial in cellula nomination assays for CRISPR-Cas nucleases used chromatin immunoprecipitation-sequencing (ChIP-seq) to identify genomic loci bound by a catalytically inactive Cas9 (dCas9) as a surrogate for off-target cleavage sites (Duan et al., 2014; Kuscu et al., 2014; Wu et al., 2014). However, little correlation was found between dCas9 binding and Cas9 cleavage with many false positive sites bound by dCas9 but not cleaved by Cas9 (Tsai et al., 2015). DISCOVER-seq offers a ChIP-seq-based in cellula (and in vivo) detection method that identifies DNA associated with the MRE11 DNA repair protein, which is recruited to genomic loci that have DSBs (Wienert et al., 2019). However, the recruitment of MRE11 to DSB sites may be unrelated to nuclease activity, the method may introduce false positives intrinsic to the use of ChIP (e.g., antibody specificity, non-specific binding), and the assay exhibits lower sensitivity overall compared to other in cellula methods.
Another general strategy for in cellula nomination of off-target activity leverages NHEJ-mediated insertion of known sequences into sites following DSBs. For example, capture of integration-deficient lentiviral vectors (IDLVs) (Wang et al., 2015b), protected double-stranded oligonucleotides (GUIDE-seq) (Nobles et al., 2019; Tsai et al., 2015), or AAV genomes (Hanlon et al., 2019) have been used to identify off-target sites. With these strategies, the sites of insertion are selectively amplified by using a primer that is designed to be complementary to the inserted sequence, followed by NGS and read mapping to a reference genome. An alternative approach called BLESS labels the two ends of a DSB with a biotinylated linker followed by streptavidin capture (Crosetto et al., 2013). Other methods blunt DSBs and then ligate an adaptor that harbors sequences such as barcodes, UMIs, sequencing adapters, and/or a T7 promoter (for T7-mediated in vitro transcription as demonstrated by BLISS) (Yan et al., 2017). Yet another nomination strategy, high-throughput genome-wide translocation sequencing (HTGTS), exploits the formation of translocations between off-target DSBs and ontarget DSBs. HTGTS uses a sequencing primer targeting one side of the on-target editing location to sequence across the predicted cleavage position to unbiasedly identify off-target loci following translocation events (Frock et al., 2015; Hu et al., 2016).
Validation of Nominated Sites
Candidate off-target sites nominated by in silico, in vitro, or cell-based assays can be validated in cellula or in vivo using targeted amplicon sequencing, AMP, or UDiTaS. The latter two approaches have the advantage of being able to capture events other than simple indels (e.g., larger deletions, inversions, trans-locations). A major limitation of all existing validation approaches is their inability to detect indel mutations at frequencies lower than the error rate of NGS (typically 0.1 to 0.01%). In addition, because no gold standard currently exists for identifying off-targets, the use of sensitive nomination assays to identify sites for validation is strongly recommended (Akcakaya et al., 2018).
Technological Outlook
One of the key challenges for improving off-target nomination assays is to increase their specificity without sacrificing sensitivity to detect rare off-targets. Recent experimental approaches to improve double-stranded oligonucleotide (dsODN)-based integration methods for the detection of in cellula off-targets have incorporated additional sequences into the dsODN tag to reduce mis-priming events during amplification (Nobles et al., 2019). Computational frameworks have shown progress in modeling editing at on-targets and could be extended to predict whether editing will occur at off-targets as well (Allen et al., 2018; Leenay et al., 2019; Shen et al., 2018). Models could also potentially incorporate epigenetic information related to off-target sites, which has been shown to alter cleavage efficiency (Verkuijl and Rots, 2019).
WGS has been proposed as an alternate method to identify off-target editing activity by comparing the whole genome sequence of an edited sample against an unedited sample (Iyer et al., 2015; Smith et al., 2014; Veres et al., 2014). Multiple technological challenges exist with this type of approach, but one major challenge is finding a suitable control sample so that one can distinguish editing events from pre-existing genetic variation, DNA replication errors, or other non-editing sources of mutation (Lareau et al., 2018; Lescarbeau et al., 2018; Nutter et al., 2018; Wilson et al., 2018). Genetic heterogeneity may be overcome to some degree by performing CRISPR editing in one cell of an organism at the 2-cell state (Zuo et al., 2019).
Another challenge in the field of off-target nomination assays is identifying potential off-target activity of non-cleaving enzymes such as base editors. WGS has been used with some success (Jin et al., 2019; Zuo et al., 2019), but the method is very low-throughput and expensive because most of the DNA fragments sequenced do not contain useful information about off-target editing. Methods such as GUIDE-seq (Tsai et al., 2015) that select for edited DNA using integration of known sequence tags at DSBs cannot be used to identify off-targets of non-cleaving enzymes. Some methods have been suggested to selectively measure off-target editing of base-editors (Doman et al., 2020), but as novel CRISPR technologies emerge, new assays and methods will be required to assess their specificity. This challenge is compounded by the fact that unintended editing may affect other cellular components such as RNA (Grünewald et al., 2019).
Through the further development of off-target assays, the field can better compare specificity across different editing enzymes, more sensitively identify accurate gRNAs, and drive the development of highly specific editing tools to approach safe and effective therapeutic editing.
Characterization of DNA Elements Using a Single Perturbation
CRISPR-Cas systems can be used to target a perturbation to a single locus (Bauer et al., 2015). For example, it is possible to characterize the function of a gene using a CRISPR-Cas nuclease whereby a gRNA is targeted to a gene exon (usually one of the first exons) to perform gene knockout. Phenotypic measurements such as cell viability, protein staining, or gene expression can be used to determine the perturbation effect. When using a CRISPR-Cas nuclease (Cebrian-Serrano and Davies, 2017), genetic editing can introduce frameshift mutations or premature stop codons, resulting in nonsense-mediated decay of the transcript or nonfunctional protein. Alternately, base editors can be used to introduce a novel stop codon in the coding sequence (Billon et al., 2017). The non-coding region controlling gene expression can also be targeted using CRISPR technology to disrupt—for example—a transcription factor binding site. Notably, genetic changes are heritable across cell division and edits can easily be verified using approaches described above.
Epigenetic editing can be used to characterize the function of genes or regulatory elements. CRISPRa uses dCas9 fused to an activating domain (e.g., VP64) which can increase gene expression when targeted to promoter regions. When CRISPRa is targeted to inactive enhancers, these enhancers can become activated and result in an increase in target gene expression. CRISPRi can be used to repress active regulatory elements and silence target genes in a similar manner (Kampmann, 2018; Simeonov et al., 2017; Thakore et al., 2016).
Large deletions can be mediated using a pair of gRNAs flanking the region of interest (Chen et al., 2014; Mali et al., 2013). This approach can be useful for deleting entire enhancers or even genes (Moorthy and Mitchell, 2016). The robust downstream functional changes resulting from enhancer or gene deletion can be read out using a variety of phenotypic assays. However, deletion rates may be low, and inversion or translocation events and incomplete editing are also possible outcomes when using this approach (Canver et al., 2014).
Technology Outlook
Prediction of editing effects by Cas9 has been approached with some success using computational frameworks (Allen et al., 2018; Doench et al., 2016b; Leenay et al., 2019; Listgarten et al., 2018; Moreno-Mateos et al., 2015; Shen et al., 2018). Forecasting the functional effect of mutations has also shown promise in other contexts (Gallion et al., 2017; Lyon and Wang, 2012; Ng and Henikoff, 2001), and combining predicted phenotype effects with predicted genome editing mutations could aid in gRNA design. However, modeling the phenotypic effect of epigenome editors remains an important and unsolved problem, especially in the context of the endogenous epigenetic environment of the intended target.
Discovery of Functional DNA Elements Using Pooled Screens
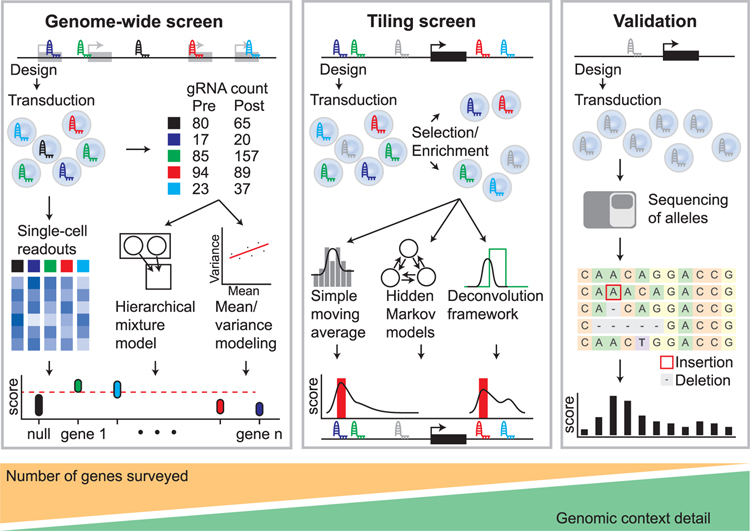
The utility of the CRISPR-Cas system at a single locus can be applied to multiple loci using a pooled screen design where individual cells are edited using one or more gRNAs to enable high-throughput functional interrogation of the genome (Doench, 2018) (Figure 3; Table 1). Broadly, pooled screens introduce a single gRNA into each cell within a large pool of cells. Each gRNA targets a particular element (e.g., gene, non-coding sequence), and the element function is assessed following a CRISPR perturbation (genetic or epigenetic). Cells can be sorted or selected based on phenotypes of interest, and gRNAs associated with the phenotypic changes can be read out to discover associated genes. Alternately, single-cell assays can be used to characterize the effect of each perturbation in each cell.
Figure 3.
Overview of Analysis Strategies for Tiling and Gene-Targeted Pooled Screens followed by Screen Validation Genome-wide screening approaches (left panel) utilize gene-targeted gRNA libraries in viral vectors. gRNA abundance is determined before and after phenotypic selection or enrichment. Scores are generated by comparing the relative gRNA abundance in pre-and post-selection populations, and identification of critical genes is performed using mean/variance modeling to address overdispersion or hierarchical mixture models to accommodate gRNA- and gene-specific variation. Alternately, single-cell readouts such as scRNA-seq can be applied to populations of cells to link phenotypes to specific perturbations. Tiling screens (middle panel) are performed by targeting gRNAs across a genomic interval. gRNA abundance is determined before and after phenotypic selection/enrichment. Scores are generated by comparing the relative gRNA abundance in pre-and post-selection populations. The effect of each gRNA can be computed using simple moving averages, hidden Markov models, or deconvolution frameworks. Pooled screen validation (right panel) often involves re-testing gRNAs in an arrayed format in bulk cell populations or individual clones using the techniques for measuring editing at known loci. For example, next-generation sequencing of an individual gRNA target site can be followed by computational analysis to identify generated alleles and calculate a per-base activity score.
Pooled genome-wide screens allow for flexibility in design and can be adapted to study many different phenotypes of interest. After the design of an gRNA library, gRNAs can be synthesized on a large-scale to construct pooled libraries with up to hundreds of thousands of unique library members, typically involving the use of viral vectors such as lentiviruses or adeno-associated viruses. Pooled libraries are transduced at low multiplicity of infection (MOI) into a population of cells, whereby individual cells receive less than one gRNA, on average. An antibiotic selection can then be applied to select for cells that were successfully transduced. The subpopulation of cells that receive a given gRNA represent an individual experiment assessing the functional effects of that gRNA (Canver et al., 2018a).
Screen Design
Genome-wide, gene-targeted screens typically include 4–10 gRNAs per gene to reduce gRNA-specific effects and increase confidence in screen hits (Sanson et al., 2018). Several pre-designed genome-wide gene-targeted libraries exist with gRNAs that have been selected based on low off-target potential and high on-target potency (Doench et al., 2016b; Horlbeck et al., 2016; Sanson et al., 2018). While genome-wide gene-targeted pooled screens are most common, genome-wide transcription factor binding sites or other regulatory regions can be targeted as well (Fei et al., 2019; Seruggia et al., 2019; Zhou et al., 2014b).
Phenotype Readouts
Pooled gRNA libraries can be analyzed at the population or single-cell level. The gRNA incorporated into each cell is typically read out using amplicon sequencing. Population-level readouts require a selective enrichment step (e.g., cell sorting) for a phenotype of interest (e.g., cell viability, gene expression, differentiation). The functional effects of gRNAs are then inferred based on comparing gRNA frequencies between pre- and post-enrichment populations or across multiple sorted populations (Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014b). Quantification of the perturbation effect for each guide can be improved with the use of UMIs (Michlits et al., 2017).
Single-Cell Readouts
Pooled genome-wide screens have been complemented by the development of single-cell assays that can be used to measure the effects of gene knockout on complex cellular phenotypes. Single cell RNA sequencing (scRNA-seq) has been shown to be effective in measuring transcriptomic changes resulting from CRISPR-based genetic and epigenetic perturbations (Adamson et al., 2016; Datlinger et al., 2017; Dixit et al., 2016; Genga et al., 2019; Jaitin et al., 2016; Mimitou et al., 2019; Replogle et al., 2020; Xie et al., 2017) and can be used to probe regulatory circuits and identify gene interactions. These screens are designed such that the gRNA sequence (or a barcode that can be linked to the gRNA sequence) are read out in the single-cell RNA-sequencing and link the global transcriptional changes to the gRNA present in that cell.
The epigenomic changes resulting from gene knockout can also be uncovered by combining single-cell ATAC-seq with targeted amplification of the gRNAs after the pooled screen (Rubin et al., 2019). Spatially resolved cellular characteristics such as protein localization can be linked to genetic perturbations in single cells using pooled screens that combine an optical readout of a given phenotype with in situ sequencing of the gRNA present in each cell (Feldman et al., 2019).
Analysis
Several analytical approaches applicable to CRISPR genome-wide knockout screens have been developed previously in other contexts, particularly in the field of RNA interference (RNAi). RNAi uses synthetic anti-sense oligonucleotides to degrade mRNA transcripts in a targeted manner, leading to gene silencing. Pooled RNAi screens in which small-interfering RNA (siRNA) or short-hairpin (shRNA) libraries targeting many genes are applied to a pool of cells to identify critical genes for a given phenotype (Boettcher and Hoheisel, 2010; König et al., 2007; Luo et al., 2008). Both RNAi and CRISPR approaches take into account the differences in knockdown/knockout efficiencies of different library members, so certain methods originally developed for RNAi analysis have been adapted for genome-wide CRISPR knockout screens (König et al., 2007; Luo et al., 2008). The following sections introduce the key analytical considerations in analyzing genome-wide pooled screens.
Fold Change Analysis
One challenge in analyzing pooled screens is that the number of gRNAs targeting each gene is not necessarily uniform in the pre-enrichment gRNA pool, causing some gRNAs to be apparently more or less abundant in the post-enrichment readout. This can be addressed by using the fold-change analysis method, which is performed by comparing the abundance of each gRNA in the post-enrichment pool to the abundance in the pre-enrichment pool. This simple processing step mitigates the effect of nonuniform distribution of gRNA counts and is employed as a first step in most processing pipelines.
Modeling gRNA Effects
Next-generation sequencing of gRNAs relies on several rounds of PCR to create sequencing libraries. Because of this, gRNAs that are highly represented in the population tend to have greater variability in NGS readouts as compared to gRNAs that are lowly represented, a phenomenon called overdispersion. For this reason, many analysis tools account for over-dispersion in pooled screen gRNA counts using negative binomial distributions (e.g., PinAPL-Py [Spahn et al., 2017], RSA [König et al., 2007], RIGER [Luo et al., 2008]) or beta binomial distributions (e.g., CRISPRBetaBinomial [Jeong et al., 2019]). Using these distributions, p values can be calculated for each gRNA. Permutation-based non-parametric analysis (PBNPA) permutes gRNA labels to assign p values to genes without assumptions on the underlying distributions (Jia et al., 2017).
Aggregating gRNAs
For pooled screens in which multiple gRNAs target each gene, p values representing the effect of each gRNA are aggregated to derive a gene-level statistic. Each gRNA may have varying effects on its target due to induced allelic diversity, perturbation range, or cleavage efficiency and may even have unintended off-target effects that may affect the phenotype readout. MAGeCK ranks gRNAs by p value, then calculates gene-level p values using modified Robust Ranking Aggregation (Li et al., 2014). Maximum likelihood estimation methods (e.g., MAGeCK-MLE [Li et al., 2015]) and hierarchical mixture models (e.g., CRISPhieRmix [Daley et al., 2018], ScreenBEAM [Yu et al., 2016]) that account for variable gRNA efficiencies have been proposed to calculate gene-level statistics. If multiple experiments are performed using the same gRNA library, the software package Jacks can model individual gRNA effects using a Bayesian approach (Allen, Behan et al., 2019).
Quality Control and Data Visualization
Assessing data quality and visualizing results from CRISPR genome-wide screens is addressed by several methods. The MAGeCK algorithm is supported by MAGeCK-VISPR and MAGeCKFlute, which provide comprehensive quality control (QC) analysis and visualizations when working with the MAGeCK pipeline (Li et al., 2015; Wang et al., 2019). Other analysis platforms such as caRpools (R package), CRISPRcloud (cloudbased platform), and CRISPRBetaBinomial (R package) provide user-friendly environments to explore CRISPR genome-wide screen data (Jeong et al., 2017, 2019; Winter et al., 2016). These tools have extensive QC metrics, intuitive data visualizations, and a wide selection of popular statistical methods (including several described above) to analyze CRISPR screen data. QC and other data visualization techniques can help ensure appropriate analysis of screen data by investigators.
Single-Cell Analysis
Computational analysis of scRNA-seq readouts can be performed to measure transcriptional effects of perturbations in a pool of cells. In order to overcome the sparsity of scRNA-seq data, cells with poor transcriptional signal are discarded, or imputation of missing values is applied in some cases. scMAGeCK (Yang et al., 2020) and MIMOSCA (Dixit et al., 2016) model the effect of gRNAs on gene expression using a regularized linear model. MUSIC (Duan et al., 2019) uses topic modeling to discover biological functions induced by perturbations. It should be noted that the methods discussed in this section are far from comprehensive, and this is still a rapidly developing field.
Technological Outlook
Technical developments in the area of pooled genome screens include innovations in characterizing multiplexed perturbation effects as well as perturbation readouts. Screens in which multiple elements (e.g., genes) are perturbed in the same cell in a controlled manner can give additional insights into gene networks and interactions, including the identification of synthetic lethal or buffering gene interactions (Du et al., 2017; Han et al., 2017; Horlbeck et al., 2018; Najm et al., 2018). These gene-interaction screens may also incorporate single-cell readouts to discover complex responses to gene network perturbation (Norman et al., 2019; Replogle et al., 2020).
Pooled screens can be coupled with many single-cell analyses to read the perturbation effects to understand complex phenotypic consequences of gene perturbation. Multi-omic single-cell approaches may yield richer data to more comprehensively describe perturbation responses. However, interpreting the results of gene perturbations remains a computational problem because the data collected in a single experiment is very large and because many single-cell assays still exhibit low resolution and sensitivity (e.g., the transcription of only a subset of genes can be reliably measured using single-cell RNA-sequencing).
Discovery of Critical Elements within an Annotated Region of Interest Using Pooled Screens
CRISPR tiling screens allow for scanning of genomic regions to uncover functional sequences related to a phenotype of interest. Typically, gRNAs are designed in an unbiased fashion to target a genomic interval, which can include an entire locus or specific annotated elements within a given locus. Unlike genome-wide screens where targets are annotated, tiling screens are performed on coding or noncoding sequences to discover functional protein domains and critical regulatory elements, respectively (Canver et al., 2015, 2017; Fulco et al., 2016; Klann et al., 2017; Sanjana et al., 2016; Schoonenberg et al., 2018; Shi et al., 2015; Simeonov et al., 2017).
The analysis of CRISPR tiling screens depend on the type of CRISPR perturbation introduced, largely due to the differences in how these perturbations affect endogenous DNA. While epigenome editors (CRISPRa/i) remodel chromatin across hundreds of bp, CRISPR-Cas nucleases typically introduce narrow indels (oftentimes 1–10 bp). In general, simple moving averages have been used for CRISPRa and CRISPRi tiling screens (Fulco et al., 2016; Simeonov et al., 2017), whereas hidden Markov models (HMMs) and deconvolution frameworks have been used for CRISPR-Cas nuclease tiling screens (Canver et al., 2015, 2017; Hsu et al., 2018). The following sections introduce commonly adopted methods to analyze tiling pooled screens.
Fold Change Analysis
Similar to data from a genome-wide pooled screen, the data generated from a tiling screen is represented as gRNA counts in a pre- and post-enrichment population. Fold change analysis between the pre- and post-enrichment populations generally measures the gRNA effect sizes reasonably if the gRNA library is sampled sufficiently. The gRNA fold change values serve as input for most tiling screen analyses described below.
Simple Moving Averages
Simple moving averages (SMAs) smooth signal from CRISPR tiling screens by averaging across gRNA fold change values within fixed windows. The number of gRNAs that go into each “averaging window” is the only parameter for this strategy. SMAs have been successfully used in the analysis of CRISPRa/i tiling screens primarily because of the extent of shared information between neighboring gRNAs, meaning that the CRISPRa/i effect produced by two closely neighboring gRNAs will likely be similar (Fulco et al., 2016; Simeonov et al., 2017). Although smoothing is desirable to reduce noise of individual gRNAs, the averaging window size must be carefully selected or else short functional elements may be missed. Furthermore, the gRNA spacing in a tiling screen is not uniform, which can present problems in using a constant-length averaging window, especially if both densely and sparsely targeted regions exist within a screen. Following SMA analysis, statistical tests (e.g., t test) can be used to identify significant regions.
Hidden Markov Models
Hidden Markov models (HMMs) infer underlying DNA regulatory states (i.e., neutral, active, and repressive) from observations in the form of gRNA fold change values derived from CRISPR tiling screen data. The HMM uses perturbation effects of gRNAs to predict the underlying regulatory state at each position across the perturbation locus. The HMM has been successfully applied to CRISPR-Cas9 tiling screens to uncover critical regulatory DNA sequences within enhancer elements (Canver et al., 2015, 2017).
Deconvolution Framework
Deconvolution frameworks have recently been proposed for the analysis of CRISPR tiling screen data. This framework models the observed gRNA fold change values by means of a convolution operation between an underlying genomic regulatory signal and a CRISPR perturbation profile. The CRISPR perturbation profile takes the form of a parameterized Gaussian window or is constructed based on empirical data. This type of framework attempts to leverage biological knowledge of how different CRISPR technologies perturb endogenous DNA and models shared information between neighboring gRNAs based on this knowledge. Importantly, the deconvolution framework explicitly parameterizes the exact targeting coordinates of all tiled gRNAs and only models shared information between neighboring gRNAs if perturbation profiles overlap. In contrast, SMAs and HMMs force information to be shared based on the qualitative ordering of gRNAs and do not account for the details of non-uniform gRNA spacing. The deconvolution framework has been successfully applied to CRISPRa/i and CRISPR-Cas nuclease tiling screens and implemented in a software package called CRISPR-SURF (Hsu et al., 2018).
Validation
Validation is typically required after computational analysis of gene-targeted or tiling pooled screens (Figure 3). At present, screen analysis is reliant on gRNA enumeration or single-cell phenotype readouts (e.g, scRNA-seq) as opposed to direct assessment of perturbations (i.e., DNA sequence or epigenetic modifications). Therefore, validation often involves re-testing gRNAs in an arrayed format in bulk cell populations or individual clones to confidently connect gRNAs or specific genetic alterations to the phenotype of interest. Upon re-testing of gRNAs implicated by a given screen, analysis can be performed using the full spectrum of techniques described above for defined loci to characterize resulting DNA modifications as well as assess changes in gene expression, protein expression, or epigenetic marks.
Technological Outlook
Current analysis approaches for CRISPR-based pooled screens measure the change in gRNA abundances pre- and post- selection or their effects on gene expression through scRNA-seq. However, the activity of gRNAs and the resulting mutational spectrum from CRISPR-Cas targeting is lost in this analysis approach. Efforts to directly observe the introduced perturbations may enable more sensitive and higher-resolution screening capabilities. Screens targeting coding sequences may benefit from the in silico separation of frameshift and in-frame mutations to enable more accurate identification of critical residues within the target protein (Shi et al., 2015). Screens targeting non-coding sequences could also benefit from the identification of the precise alleles that result in a given phenotype of interest, with the potential to map functional transcription factor binding sites with higher resolution. Similarly, barcodes or gRNAs themselves have been used as surrogate readouts for targeted deletion screens (Gasperini et al., 2017; Zhu et al., 2016).
New CRISPR-based editing mechanisms provide new avenues for pooled screens moving forward. Base editing introduces specific substitutions with low indel rates and may provide an alternative method to study coding sequences without introducing frameshift mutations (Rees and Liu, 2018). Prime editing offers the unique ability to introduce a wide range of mutations at a target site, ranging from all substitution mutations to small insertion and deletion mutations. Though still in its technological infancy, prime editing opens the possibility of introducing precise genomic perturbations to study DNA sequence at single-base resolution (Anzalone et al., 2019).
Conclusions
The rapid innovations in genome editing technologies and their applications have been substantially fueled by both experimental and computational efforts. For the assessment of ontarget editing, methods that were primarily experimental with simple binary or semiquantitative readouts have transitioned toward NGS and tailored computational analyses that are able to comprehensively characterize genome editing outcomes at the allelic level. Advances in assays for detecting and quantifying off-targets have enabled researchers to build nucleases with improved genome-wide specificities. At the same time, specialized alignment algorithms and machine learning models are being leveraged to better design gRNAs and predict their off-target sites, which may accelerate the development of safer and more predictable editing tools. In the applications of using CRISPR technology to study different cellular phenotypes, initial studies focused on characterizing gene function using a single perturbation and have expanded into genome-wide or tiling pooled screens with a variety of single-cell readouts, creating a rich, comprehensive, and high- throughput view of many perturbation effects. The availability of these tools will further enable the elucidation of the biological underpinnings, regulatory structures, and non-linear interactions of gene networks through the use of big data approaches. Moving forward, both experimental and computational efforts will continue to foster the improvement and creation of accurate and precise CRISPR technologies for a variety of research and clinical applications.
ACKNOWLEDGMENTS
L.P. is supported by NHGRI (R00HG008399 and R35HG010717), DARPA (HR0011-17-2-0042), and the Centers for Excellence in Genomic Science of the National Institutes of Health under award number RM1HG009490 through a New Collaborator Grant sub-award. J.K.J. is supported by a DARPA Safe Genes contract (HR0011-17-2-0042), an NIH Maximizing Investigators’ Research Award (MIRA) (R35 GM118158), and an NIH Centers of Excellence in Genomic Science award (RM1 HG009490). J.K.J. is also supported by the Desmond and Ann Heathwood MGH Research Scholar award and the Robert B. Colvin, M.D. Endowed Chair in Pathology. We are thankful for the input of reviewers and editors who have contributed useful feedback and suggestions.
Footnotes
DECLARATION OF INTERESTS
L.P. has financial interests in Edilytics, Inc. K.C. is an employee, shareholder, and officer of Edilytics, Inc. J.K.J. has financial interests in Beam Therapeutics, Editas Medicine, Excelsior Genomics, Pairwise Plants, Poseida Therapeutics, Transposagen Biopharmaceuticals, and Verve Therapeutics (f/k/a Endcadia). The interests of L.P., K.C., and J.K.J. were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. is a member of the Board of Directors of the American Society of Gene and Cell Therapy. J.K.J. is a co-inventor on various patents and patent applications that describe gene editing and epigenetic editing technologies.
REFERENCES
- Abadi S, Yan WX, Amar D, and Mayrose I (2017). A machine learning approach for predicting CRISPR-Cas9 cleavage efficiencies and patterns underlying its mechanism of action. PLoS Comput. Biol 13, e1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, et al. (2016). A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell 167, 1867–1882.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adli M (2018). The CRISPR tool kit for genome editing and beyond. Nat. Commun 9, 1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, Fellows MD, Porritt MJ, Firth MA, Carreras A, et al. (2018). In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature 561, 416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Páleńıkova P, Khodak A, Kiselev V, Kosicki M, et al. (2018). Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol 37, 64–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen F, Behan F, Khodak A, Iorio F, Yusa K, Garnett M, and Parts L (2019). JACKS: joint analysis of CRISPR/Cas9 knockout screens. Genome Res 29, 464–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, and Liu DR (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae S, Park J, and Kim J-S (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Canver MC, and Orkin SH (2015). Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J. Vis. Exp 95, e52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billon P, Bryant EE, Joseph SA, Nambiar TS, Hayward SB, Rothstein R, and Ciccia A (2017). CRISPR-Mediated Base Editing Enables Efficient Disruption of Eukaryotic Genes through Induction of STOP Codons. Mol. Cell 67, 1068–1079.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel A, Steyaert W, De Rocker N, Menten B, Callewaert B, De Paepe A, Coucke P, and Willaert A (2016). BATCH-GE: Batch analysis of Next-Generation Sequencing data for genome editing assessment. Sci. Rep 6, 30330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher M, and Hoheisel JD (2010). Pooled RNAi Screens - Technical and Biological Aspects. Curr. Genomics 11, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Chen T, Amendola M, and van Steensel B (2014). Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic Acids Res 42, e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkman EK, Kousholt AN, Harmsen T, Leemans C, Chen T, Jonkers J, and van Steensel B (2018). Easy quantification of template-directed CRISPR/Cas9 editing. Nucleic Acids Res 46, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, et al. (2017). Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat. Methods 14, 600–606. [DOI] [PubMed] [Google Scholar]
- Cancellieri S, Canver MC, Bombieri N, Giugno R, and Pinello L (2019). CRISPRitz: rapid, high-throughput and variant-aware in silico off-target site identification for CRISPR genome editing. Bioinformatics 36, 2001–2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Bauer DE, Dass A, Yien YY, Chung J, Masuda T, Maeda T, Paw BH, and Orkin SH (2014). Characterization of genomic deletion efficiency mediated by clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem 289, 21312–21324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. (2015). BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature 527, 192–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Lessard S, Pinello L, Wu Y, Ilboudo Y, Stern EN, Needleman AJ, Galactéros F, Brugnara C, Kutlar A, et al. (2017). Variant-aware saturating mutagenesis using multiple Cas9 nucleases identifies regulatory elements at trait-associated loci. Nat. Genet 49, 625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Haeussler M, Bauer DE, Orkin SH, Sanjana NE, Shalem O, Yuan G-C, Zhang F, Concordet J-P, and Pinello L (2018a). Integrated design, execution, and analysis of arrayed and pooled CRISPR genome-editing experiments. Nat. Protoc 13, 946–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canver MC, Joung JK, and Pinello L (2018b). Impact of Genetic Variation on CRISPR-Cas Targeting. CRISPR J 1, 159–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian-Serrano A, and Davies B (2017). CRISPR-Cas orthologues and variants: optimizing the repertoire, specificity and delivery of genome engineering tools. Mamm. Genome 28, 247–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Xu F, Zhu C, Ji J, Zhou X, Feng X, and Guang S (2014). Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci. Rep 4, 7581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim Y, Kweon J, Kim HS, Bae S, and Kim JS (2014). Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res 24, 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Farouni R, Bauer DE, and Pinello L (2018). AmpUMI: design and analysis of unique molecular identifiers for deep amplicon sequencing. Bioinformatics 34, i202–i210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, and Pinello L (2019). CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol 37, 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosetto N, Mitra A, Silva MJ, Bienko M, Dojer N, Wang Q, Karaca E, Chiarle R, Skrzypczak M, Ginalski K, et al. (2013). Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods 10, 361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullot G, Boutin J, Toutain J, Prat F, Pennamen P, Rooryck C, Teichmann M, Rousseau E, Lamrissi-Garcia I, Guyonnet-Duperat V, et al. (2019). CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun 10, 1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daley TP, Lin Z, Lin X, Liu Y, Wong WH, and Qi LS (2018). CRISPhieR-mix: a hierarchical mixture model for CRISPR pooled screens. Genome Biol 19, 159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dastidar S, Ardui S, Singh K, Majumdar D, Nair N, Fu Y, Reyon D, Samara E, Gerli MFM, Klein AF, et al. (2018). Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells. Nucleic Acids Res 46, 8275–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klughammer J, Schuster LC, Kuchler A, Alpar D, and Bock C (2017). Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit A, Parnas O, Li B, Chen J, Fulco CP, Jerby-Arnon L, Marjanovic ND, Dionne D, Burks T, Raychowdhury R, et al. (2016). Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG (2018). Am I ready for CRISPR? A user’s guide to genetic screens. Nat. Rev. Genet 19, 67–80. [DOI] [PubMed] [Google Scholar]
- Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, and Root DE (2014). Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol 32, 1262–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016a). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016b). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doman JL, Raguram A, Newby GA, and Liu DR (2020). Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base. Nat. Biotechnol 38, 620–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Roguev A, Gordon DE, Chen M, Chen SH, Shales M, Shen JP, Ideker T, Mali P, Qi LS, and Krogan NJ (2017). Genetic interaction mapping in mammalian cells using CRISPR interference. Nat. Methods 14, 577–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Lu G, Xie Z, Lou M, Luo J, Guo L, and Zhang Y (2014). Genome-wide identification of CRISPR/Cas9 off-targets in human genome. Cell Res 24, 1009–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan B, Zhou C, Zhu C, Yu Y, Li G, Zhang S, Zhang C, Ye X, Ma H, Qu S, et al. (2019). Model-based understanding of single-cell CRISPR screening. Nat. Commun 10, 2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei T, Li W, Peng J, Xiao T, Chen CH, Wu A, Huang J, Zang C, Liu XS, and Brown M (2019). Deciphering essential cistromes using genome-wide CRISPR screens. Proc. Natl. Acad. Sci. USA 116, 25186–25195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman D, Singh A, Schmid-Burgk JL, Carlson RJ, Mezger A, Garrity AJ, Zhang F, and Blainey PC (2019). Optical Pooled Screens in Human Cells. Cell 179, 787–799.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay SD, Vincent KM, Berman JR, and Postovit LM (2016). A digital pcr-based method for efficient and highly specific screening of genome edited cells. PLoS ONE 11, e0153901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frock RL, Hu J, Meyers RM, Ho Y-J, Kii E, and Alt FW (2015). Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol 33, 179–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, and Engreitz JM (2016). Systematic mapping of functional enhancer-promoter connections with CRISPR interference. Science 354, 769–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallion J, Koire A, Katsonis P, Schoenegge AM, Bouvier M, and Lichtarge O (2017). Predicting phenotype from genotype: Improving accuracy through more robust experimental and computational modeling. Hum. Mutat 38, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperini M, Findlay GM, McKenna A, Milbank JH, Lee C, Zhang MD, Cusanovich DA, and Shendure J (2017). CRISPR/Cas9-Mediated Scanning for Regulatory Elements Required for HPRT1 Expression via Thousands of Large, Programmed Genomic Deletions. Am. J. Hum. Genet 101, 192–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genga RMJ, Kernfeld EM, Parsi KM, Parsons TJ, Ziller MJ, and Maehr R (2019). Single-Cell RNA-Sequencing-Based CRISPRi Screening Resolves Molecular Drivers of Early Human Endoderm Development. Cell Rep 27, 708–718.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoukos G, Ciulla DM, Marco E, Abdulkerim HS, Barrera LA, Bothmer A, Dhanapal V, Gloskowski SW, Jayaram H, Maeder ML, et al. (2018). UDiTaS™, a genome editing detection method for indels and genome rearrangements. BMC Genomics 19, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC, et al. (2014). Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 159, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünewald J, Zhou R, Garcia SP, Iyer S, Lareau CA, Aryee MJ, and Joung JK (2019). Transcriptome-wide off-target RNA editing induced by CRISPR-guided DNA base. Nature 569, 433–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güell M, Yang L, and Church GM (2014). Genome editing assessment using CRISPR Genome Analyzer (CRISPR-GA). Bioinformatics 30, 2968–2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, and Bassik MC (2017). Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol 35, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanlon KS, Kleinstiver BP, Garcia SP, Zaborowski MP, Volak A, Spirig SE, Muller A, Sousa AA, Tsai SQ, Bengtsson NE, et al. (2019). High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat. Commun 10, 4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna RE, and Doench JG (2020). Design and analysis of CRISPR-Cas experiments. Nat. Biotechnol [DOI] [PubMed]
- Hart T, and Moffat J (2016). BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics 17, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Zhang L, Villarreal OD, Fu R, Bedford E, Dou J, Patel AY, Bedford MT, Shi X, Chen T, et al. (2019). De novo identification of essential protein domains from CRISPR-Cas9 tiling-sgRNA knockout screens. Nat. Commun 10, 4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heigwer F, Kerr G, and Boutros M (2014). E-CRISP: fast CRISPR target site identification. Nat. Methods 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M, and Weissman JS (2016). Compact and highly active next-generation libraries for CRISPR-mediated gene repression and activation. eLife 5, e19760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlbeck MA, Xu A, Wang M, Bennett NK, Park CY, Bogdanoff D, Adamson B, Chow ED, Kampmann M, Peterson TR, et al. (2018). Mapping the Genetic Landscape of Human Cells. Cell 174, 953–967.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiau T, Maures T, Waite K, Yang J, Kelso R, Holden K, and Stoner R (2019). Inference of CRISPR Edits from Sanger Trace Data. CRISPR J 2, 223–229. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JY, Fulco CP, Cole MA, Canver MC, Pellin D, Sher F, Farouni R, Clement K, Guo JA, Biasco L, et al. (2018). CRISPR-SURF: discovering regulatory elements by deconvolution of CRISPR tiling screen data. Nat. Methods 15, 992–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Meyers RM, Dong J, Panchakshari RA, Alt FW, and Frock RL (2016). Detecting DNA double-stranded breaks in mammalian genomes by linear amplification-mediated high-throughput genome-wide translocation sequencing. Nat. Protoc 11, 853–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang GH, Park J, Lim K, Kim S, Yu J, Yu E, Kim ST, Eils R, Kim JS, and Bae S (2018). Web-based design and analysis tools for CRISPR base editing. BMC Bioinformatics 19, 542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyama T, and Wilson DM 3rd (2013). DNA repair mechanisms in dividing and non-dividing cells. DNA Repair (Amst.) 12, 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer V, Shen B, Zhang W, Hodgkins A, Keane T, Huang X, and Skarnes WC (2015). Off-target mutations are rare in Cas9-modified mice. Nat. Methods 12, 479. [DOI] [PubMed] [Google Scholar]
- Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, Salame TM, Tanay A, van Oudenaarden A, and Amit I (2016). Dissecting Immune Circuits by Linking CRISPR-Pooled Screens with Single-Cell RNA- Seq. Cell 167, 1883–1896.e15. [DOI] [PubMed] [Google Scholar]
- Jeong HH, Kim SY, Rousseaux MWC, Zoghbi HY, and Liu Z (2017). CRISPRcloud: a secure cloud-based pipeline for CRISPR pooled screen deconvolution. Bioinformatics 33, 2963–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong HH, Kim SY, Rousseaux MWC, Zoghbi HY, and Liu Z (2019). Beta-binomial modeling of CRISPR pooled screen data identifies target genes with greater sensitivity and fewer false negatives. Genome Res 29, 999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Wang X, and Xiao G (2017). A permutation-based non-parametric analysis of CRISPR screen data. BMC Genomics 18, 545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu JL, Zhang F, and Gao C (2019). Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364, 292–295. [DOI] [PubMed] [Google Scholar]
- Kampmann M (2018). CRISPRi and CRISPRa Screens in Mammalian Cells for Precision Biology and Medicine. ACS Chem. Biol 13, 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy SR, Schmitt MW, Fox EJ, Kohrn BF, Salk JJ, Ahn EH, Prindle MJ, Kuong KJ, Shen JC, Risques RA, and Loeb LA (2014). Detecting ultralow-frequency mutations by Duplex Sequencing. Nat. Protoc 9, 2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, Kim D, Kim S, and Kim J-S (2014). Genotyping with CRISPR-Cas-derived RNA-guided endonucleases. Nat. Commun 5, 3157. [DOI] [PubMed] [Google Scholar]
- Kim D, Bae S, Park J, Kim E, Kim S, Yu HR, Hwang J, Kim J-I, and Kim J-S (2015). Digenome-seq: genome-wide profiling of CRISPR-Cas9 off- target effects in human cells. Nat. Methods 12, 237–243, 1, 243. [DOI] [PubMed] [Google Scholar]
- Kim D, Kim S, Kim S, Park J, and Kim J-S (2016). Genome-wide target specificities of CRISPR-Cas9 nucleases revealed by multiplex Digenome-seq. Genome Res 26, 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinde I, Wu J, Papadopoulos N, Kinzler KW, and Vogelstein B (2011). Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 108, 9530–9535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, Crawford GE, Reddy TE, and Gersbach CA (2017). CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol 35, 561–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein M, Eslami-Mossallam B, Arroyo DG, and Depken M (2018). Hybridization Kinetics Explains CRISPR-Cas Off-Targeting Rules. Cell Rep 22, 1413–1423. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R, Chiang CY, Tu BP, Yan SF, DeJesus PD, Romero A, Bergauer T, Orth A, Krueger U, Zhou Y, and Chanda SK (2007). A probability-based approach for the analysis of large-scale RNAi screens. Nat. Methods 4, 847–849. [DOI] [PubMed] [Google Scholar]
- Kosicki M, Tomberg K, and Bradley A (2018). Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol 36, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, and Adli M (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol 32, 677–683. [DOI] [PubMed] [Google Scholar]
- Langmead B, and Salzberg SL (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau CA, Clement K, Hsu JY, Pattanayak V, Keith Joung J, Aryee MJ, and Pinello L (2018). Response to “unexpected mutations after CRISPR-Cas9 editing in vivo.”. Nat. Methods 15, 238–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenay RT, Aghazadeh A, Hiatt J, Tse D, Roth TL, Apathy R, Shifrut E, Hultquist JF, Krogan N, Wu Z, et al. (2019). Large dataset enables prediction of repair after CRISPR-Cas9 editing in primary T cells. Nat. Biotechnol 37, 1034–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, and Chen LL (2014). CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant 7, 1494–1496. [DOI] [PubMed] [Google Scholar]
- Lescarbeau RM, Murray B, Barnes TM, and Bermingham N (2018). Response to “unexpected mutations after CRISPR-Cas9 editing in vivo. Nat. Methods 15, 237. [DOI] [PubMed] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Köster J, Xu H, Chen CH, Xiao T, Liu JS, Brown M, and Liu XS (2015). Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol 16, 281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, and Wong KC (2018). Off-target predictions in CRISPR-Cas9 gene editing using deep learning. Bioinformatics 34, i656–i663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H, Burger A, Biyong B, Felker A, Hess C, Zaugg J, Chiavacci E, Anders C, Jinek M, Mosimann C, and Robinson MD (2016). Crisp-RVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol 34, 701–702. [DOI] [PubMed] [Google Scholar]
- Listgarten J, Weinstein M, Kleinstiver BP, Sousa AA, Joung JK, Crawford J, Gao K, Hoang L, Elibol M, Doench JG, and Fusi N (2018). Prediction of off-target activities for the end-to-end design of CRISPR guide RNAs. Nat Biomed Eng 2, 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B, Cheung HW, Subramanian A, Sharifnia T, Okamoto M, Yang X, Hinkle G, Boehm JS, Beroukhim R, Weir BA, et al. (2008). Highly parallel identification of essential genes in cancer cells. Proceedings of the National Academy of Sciences of the United States of America, 20380–20385. [DOI] [PMC free article] [PubMed]
- Lyon GJ, and Wang K (2012). Identifying disease mutations in genomic medicine settings: current challenges and how to accelerate progress. Genome Med 4, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, and Church GM (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashal RD, Koontz J, and Sklar J (1995). Detection of mutations by cleavage of DNA heteroduplexes with bacteriophage resolvases. Nat. Genet 9, 177–183. [DOI] [PubMed] [Google Scholar]
- McKenna A, and Shendure J (2018). FlashFry: a fast and flexible tool for large-scale CRISPR target design. BMC Biol 16, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michlits G, Hubmann M, Wu SH, Vainorius G, Budusan E, Zhuk S, Burkard TR, Novatchkova M, Aichinger M, Lu Y, et al. (2017). CRISPRUMI: single-cell lineage tracing of pooled CRISPR-Cas9 screens. Nat. Methods 14, 1191–1197. [DOI] [PubMed] [Google Scholar]
- Mimitou EP, Cheng A, Montalbano A, Hao S, Stoeckius M, Legut M, Roush T, Herrera A, Papalexi E, Ouyang Z, et al. (2019). Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, and Valen E (2014). CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42 (Web Server issue, W1), W401–W407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy SD, and Mitchell JA (2016). Generating CRISPR/Cas9 mediated monoallelic deletions to study enhancer function in mouse embryonic stem cells. J. Vis. Exp (110), e53552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, and Giraldez AJ (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Smith JL, Peng L, Yin H, Moore J, Zhang X-O, Song C-Q, Sheel A, Wu Q, Ozata DM, et al. (2017). CRISPR/Cas9-mediated genome editing induces exon skipping by alternative splicing or exon deletion. Genome Biol 18, 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Yamada T, Ui-Tei K, Morishita S, and Saigo K (2004). siDirect: highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res 32, W124–W129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y, Hino K, Bono H, and Ui-Tei K (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najm FJ, Strand C, Donovan KF, Hegde M, Sanson KR, Vaimberg EW, Sullender ME, Hartenian E, Kalani Z, Fusi N, et al. (2018). Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol 36, 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neggers JE, Kwanten B, Dierckx T, Noguchi H, Voet A, Bral L, Minner K, Massant B, Kint N, Delforge M, et al. (2018). Target identification of small molecules using large-scale CRISPR-Cas mutagenesis scanning of essential genes. Nat. Commun 9, 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, and Henikoff S (2001). Predicting deleterious amino acid substitutions. Genome Res 11, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobles CL, Reddy S, Salas-McKee J, Liu X, June CH, Melenhorst JJ, Davis MM, Zhao Y, and Bushman FD (2019). iGUIDE: an improved pipeline for analyzing CRISPR cleavage specificity. Genome Biol 20, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman TM, Horlbeck MA, Replogle JM, Ge AY, Xu A, Jost M, Gilbert LA, and Weissman JS (2019). Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutter LMJ, Heaney JD, Lloyd KCK, Murray SA, Seavitt JR, Skarnes WC, Teboul L, Brown SDM, and Moore M (2018). Response to “Unexpected mutations after CRISPR-Cas9 editing in vivo”. Nat. Methods 15, 235–236. [DOI] [PubMed] [Google Scholar]
- O’Brien A, and Bailey TL (2014). GT-Scan: identifying unique genomic targets. Bioinformatics 30, 2673–2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lim K, Kim JS, and Bae S (2017). Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33, 286–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, and Liu DR (2011). Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 8, 765–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, and Liu DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol 31, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Zheng Y, Zhao Z, Liu T, and Li J (2018). Recognition of CRISPR/ Cas9 off-target sites through ensemble learning of uneven mismatch distributions. Bioinformatics 34, i757–i765. [DOI] [PubMed] [Google Scholar]
- Pinello L, Canver MC, Hoban MD, Orkin SH, Kohn DB, Bauer DE, and Yuan GC (2016). Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol 34, 695–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poplin R, Chang PC, Alexander D, Schwartz S, Colthurst T, Ku A, Newburger D, Dijamco J, Nguyen N, Afshar PT, et al. (2018). A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol 36, 983–987. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, and Lim WA (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P, Shandilya H, D’Alessio JM, O’Connor K, Durocher J, and Gerard GF (2004). Mutation detection using Surveyor nuclease. Biotechniques 36, 702–707. [DOI] [PubMed] [Google Scholar]
- Ramlee MK, Yan T, Cheung AMS, Chuah CTH, and Li S (2015). High-throughput genotyping of CRISPR/Cas9-mediated mutants using fluorescent PCR-capillary gel electrophoresis. Sci. Rep 5, 15587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees HA, and Liu DR (2018). Base editing: precision chemistry on the genome and transcriptome of living cells. Nat. Rev. Genet 19, 770–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle JM, Norman TM, Xu A, Hussmann JA, Chen J, Cogan JZ, Meer EJ, Terry JM, Riordan DP, Srinivas N, et al. (2020). Combinatorial single-cell CRISPR screens by direct guide RNA capture and targeted sequencing. Nat. Biotechnol [DOI] [PMC free article] [PubMed]
- Riesenberg S, and Maricic T (2018). Targeting repair pathways with small molecules increases precise genome editing in pluripotent stem cells. Nat. Commun 9, 2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin AJ, Parker KR, Satpathy AT, Qi Y, Wu B, Ong AJ, Mumbach MR, Ji AL, Kim DS, Cho SW, et al. (2019). Coupled Single-Cell CRISPR Screening and Epigenomic Profiling Reveals Causal Gene Regulatory Networks. Cell 176, 361–376.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Wright J, Zheng K, Shalem O, Fontanillas P, Joung J, Cheng C, Regev A, and Zhang F (2016). High-resolution interrogation of functional elements in the noncoding genome. Science 353, 1545–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanson KR, Hanna RE, Hegde M, Donovan KF, Strand C, Sullender ME, Vaimberg EW, Goodale A, Root DE, Piccioni F, and Doench JG (2018). Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nat. Commun 9, 5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonenberg VAC, Cole MA, Yao Q, Macias-Treviño C, Sher F, Schupp PG, Canver MC, Maeda T, Pinello L, and Bauer DE (2018). CRISPRO: identification of functional protein coding sequences based on genome editing dense mutagenesis. Genome Biol 19, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DA, and Zhang F (2017). Implications of human genetic variation in CRISPR-based therapeutic genome editing. Nat. Med 23, 1095–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentmanat MF, Peters ST, Florian CP, Connelly JP, and Pruett-Miller SM (2018). A Survey of Validation Strategies for CRISPR-Cas9 Editing. Sci. Rep 8, 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seruggia D, Oti M, Tripathi P, Canver MC, LeBlanc L, Di Giammartino DC, Bullen MJ, Nefzger CM, Sun YBY, Farouni R, et al. (2019). TAF5L and TAF6L Maintain Self-Renewal of Embryonic Stem Cells via the MYC Regulatory Network. Mol. Cell 74, 1148–1163.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelson T, Heckl D, Ebert BL, Root DE, Doench JG, and Zhang F (2014). Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MW, Arbab M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa CA, Liu DR, Gifford DK, and Sherwood RI (2018). Predictable and precise template-free CRISPR editing of pathogenic variants. Nature 563, 646–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Wang E, Milazzo JP, Wang Z, Kinney JB, and Vakoc CR (2015). Discovery of cancer drug targets by CRISPR-Cas9 screening of protein domains. Nat. Biotechnol 33, 661–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonov DR, Gowen BG, Boontanrart M, Roth TL, Gagnon JD, Mumbach MR, Satpathy AT, Lee Y, Bray NL, Chan AY, et al. (2017). Discovery of stimulation-responsive immune enhancers with CRISPR activation. Nature 549, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Kuscu C, Quinlan A, Qi Y, and Adli M (2015). Cas9-chromatin binding information enables more accurate CRISPR off-target prediction. Nucleic Acids Res 43, e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C, Gore A, Yan W, Abalde-Atristain L, Li Z, He C, Wang Y, Brodsky RA, Zhang K, Cheng L, and Ye Z (2014). Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 15, 12–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Yang D, Xu J, Zhu T, Chen YE, and Zhang J (2016). RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun 7, 10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn PN, Bath T, Weiss RJ, Kim J, Esko JD, Lewis NE, and Harismendy O (2017). PinAPL-Py: A comprehensive web-application for the analysis of CRISPR/Cas9 screens. Sci. Rep 7, 15854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, and Mateo JL (2015). CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS ONE 10, e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Osterman MD, and Li M (2019). Tissue specificity of DNA damage response and tumorigenesis. Cancer Biol. Med 16, 396–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakore PI, Black JB, Hilton IB, and Gersbach CA (2016). Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat. Methods 13, 127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas HR, Percival SM, Yoder BK, and Parant JM (2014). High-throughput genome editing and phenotyping facilitated by high resolution melting curve analysis. PLoS ONE 9, e114632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, and Joung JK (2017). CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat. Methods 14, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycko J, Myer VE, and Hsu PD (2016). Methods for Optimizing CRISPR-Cas9 Genome Editing Specificity. Mol. Cell 63, 355–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartak SV, and Raghavan SC (2015). Inhibition of nonhomologous end joining to increase the specificity of CRISPR/Cas9 genome editing. FEBS J 282, 4289–4294. [DOI] [PubMed] [Google Scholar]
- Veres A, Gosis BS, Ding Q, Collins R, Ragavendran A, Brand H, Erdin S, Cowan CA, Talkowski ME, and Musunuru K (2014). Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 15, 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkuijl SAN, and Rots MG (2019). The influence of eukaryotic chromatin state on CRISPR-Cas9 editing efficiencies. Curr. Opin. Biotechnol 55, 68–73. [DOI] [PubMed] [Google Scholar]
- Vouillot L, Thélie A, and Pollet N (2015). Comparison of T7E1 and surveyor mismatch cleavage assays to detect mutations triggered by engineered nucleases. G3 (Bethesda) 5, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, and Lander ES (2014). Genetic screens in human cells using the CRISPR-Cas9 system. Science 343, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, and Sabatini DM (2015a). Identification and characterization of essential genes in the human genome. Science 350, 1096–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wang Y, Wu X, Wang J, Wang Y, Qiu Z, Chang T, Huang H, Lin R-J, and Yee J-K (2015b). Unbiased detection of off-target cleavage by CRISPR-Cas9 and TALENs using integrase-defective lentiviral vectors. Nat. Biotechnol 33, 175–178. [DOI] [PubMed] [Google Scholar]
- Wang B, Wang M, Zhang W, Xiao T, Chen CH, Wu A, Wu F, Traugh N, Wang X, Li Z, et al. (2019). Integrative analysis of pooled CRISPR genetic screens using MAGeCKFlute. Nat. Protoc 14, 756–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienert B, Wyman SK, Richardson CD, Yeh CD, Akcakaya P, Porritt MJ, Morlock M, Vu JT, Kazane KR, Watry HL, et al. (2019). Unbiased detection of CRISPR off-targets in vivo using DISCOVER-Seq. Science 364, 286–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CJ, Fennell T, Bothmer A, Maeder ML, Reyon D, Cotta-Ramusino C, Fernandez CA, Marco E, Barrera LA, Jayaram H, et al. (2018). Response to “Unexpected mutations after CRISPR-Cas9 editing in vivo”. Nat. Methods 15, 236–237. [DOI] [PubMed] [Google Scholar]
- Winter J, Breinig M, Heigwer F, Bruügemann D, Leible S, Pelz O, Zhan T, and Boutros M (2016). caRpools: an R package for exploratory data analysis and documentation of pooled CRISPR/Cas9 screens. Bioinformatics 32, 632–634. [DOI] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol 32, 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A, Cheng Z, Kong L, Zhu Z, Lin S, Gao G, and Zhang B (2014). CasOT: a genome-wide Cas9/gRNA off-target searching tool. Bioinformatics 30, 1180–1182. [DOI] [PubMed] [Google Scholar]
- Xie S, Duan J, Li B, Zhou P, and Hon GC (2017). Multiplexed Engineering and Analysis of Combinatorial Enhancer Activity in Single Cells. Mol. Cell 66, 285–299.e5. [DOI] [PubMed] [Google Scholar]
- Xu X, Duan D, and Chen SJ (2017). CRISPR-Cas9 cleavage efficiency correlates strongly with target-sgRNA folding stability: from physical mechanism to off-target assessment. Sci. Rep 7, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Mirzazadeh R, Garnerone S, Scott D, Schneider MW, Kallas T, Custodio J, Wernersson E, Li Y, Gao L, et al. (2017). BLISS is a versatile and quantitative method for genome-wide profiling of DNA double-strand breaks. Nat. Commun 8, 15058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Steentoft C, Hauge C, Hansen L, Thomsen AL, Niola F, Vester-Christensen MB, Frödin M, Clausen H, Wandall HH, and Bennett EP (2015). Fast and sensitive detection of indels induced by precise gene targeting. Nucleic Acids Res 43, e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Zhu Y, Yu H, Cheng X, Chen S, Chu Y, Huang H, Zhang J, and Li W (2020). scMAGeCK links genotypes with multiple phenotypes in single-cell CRISPR screens. Genome Biol 21, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Zhang Y, Yao S, and Wei Y (2014). A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS ONE 9, e98282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Silva J, and Califano A (2016). ScreenBEAM: a novel meta-analysis algorithm for functional genomics screens via Bayesian hierarchical modeling. Bioinformatics 32, 260–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, Chen J, Robinson HE, Shim HS, Chmielecki J, et al. (2014). Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med 20, 1479–1484. [DOI] [PubMed] [Google Scholar]
- Zhou HY, Katsman Y, Dhaliwal NK, Davidson S, Macpherson NN, Sakthidevi M, Collura F, and Mitchell JA (2014a). A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev 28, 2699–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, and Wei W (2014b). High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature 509, 487–491. [DOI] [PubMed] [Google Scholar]
- Zhu X, Xu Y, Yu S, Lu L, Ding M, Cheng J, Song G, Gao X, Yao L, Fan D, et al. (2014). An efficient genotyping method for genome-modified animals and human cells generated with CRISPR/Cas9 system. Sci. Rep 4, 6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Li W, Liu J, Chen CH, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J, et al. (2016). Genome-scale deletion screening of human long non-coding RNAs using a paired-guide RNA CRISPR-Cas9 library. Nat. Biotechnol 34, 1279–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo E, Sun Y, Wei W, Yuan T, Ying W, Sun H, Yuan L, Steinmetz LM, Li Y, and Yang H (2019). Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos. Science 364, 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]