Abstract
Porcine deltacoronavirus (PDCoV) has been recently identified as an emerging enteropathogenic coronavirus that mainly infects newborn piglets and causes enteritis, diarrhea and high mortality. Although coronavirus N proteins have multifarious activities, the subcellular localization of the PDCoV N protein is still unknown. Here, we produced mouse monoclonal antibodies against the PDCoV N protein. Experiments using anti-haemagglutinin antibodies and these monoclonal antibodies revealed that the PDCoV N protein is shuttled into the nucleolus in both ectopic PDCoV N-expressing cells and PDCoV-infected cells. The results of deletion mutagenesis experiments demonstrated that the predicted nucleolar localization signal at amino acids 295–318 is critical for nucleolar localization. Cumulatively, our study yielded a monoclonal antibody against the PDCoV N protein and revealed a mechanism by which the PDCoV N protein translocated into the nucleolus. The tolls and findings from this work will facilitate further investigations on the functions of the PDCoV N protein.
Keywords: Porcine deltacoronavirus, Nucleocapsid protein, mAb, Nucleolar localization
Introduction
Porcine deltacoronavirus (PDCoV), an emerging swine enteric pathogen, is a major causative agent of watery diarrhea, vomiting, and mortality in piglets, leading to significant losses in the pig industry [1, 2]. In addition to causing global outbreaks of diarrhea in pigs, PDCoV can cross the species barrier to infect calves, chickens, and even humans [3–5], thus posing a significant threat to public health. PDCoV is an enveloped RNA virus with a single-stranded, positive-sense genome. It is classified into the genus Deltacoronavirus, in the family of Coronaviridae and the order Nidovirale. PDCoV has the shortest genome (approximately 25.4 kb in length) among the known members in the family of Coronaviridae. Two-thirds of the genome at the 5ʹ ORF1a/1b encodes pp1a and pp1ab, which are two polymerase proteins that can be proteolytically cleaved into 15 mature non-structural proteins (nsps), while the remaining one-third of the genome at 3′ terminal region encodes four structural proteins and three newly identified accessory proteins, NS6, NS7, and NS7a. The four structural proteins are the spike (S), envelope (E), membrane (M) and nucleocapsid (N) proteins [6–9].
Among these proteins, N protein is structurally and functionally conserved within all the coronaviruses (CoVs), and it plays critical roles in packaging the viral genomic RNA into the virion for viral assembly [10, 11]. Additionally, the N protein is the most abundant and ubiquitous viral protein in the context of CoV infection or assembled virions, and has roles in multifarious activities throughout the life cycle of a CoV [12]. In addition to promoting viral genome transcription or replication, the N protein also modulates the processes of inflammatory cytokine productions, RNA interference and apoptosis, and counteracting the host innate immune defense [13–16]. Interestingly, based on studies of representative CoVs, a cytoplasmic nucleolar location pattern is common for the N proteins of several CoVs, including porcine epidemic diarrhea virus (PEDV), mouse hepatitis virus (MHV) and infectious bronchitis virus (IBV) [17–19]. Proteins imported to the nucleolus contain at least one nucleolar localization signal (NoLS) that specifies their nucleolar localization; however, how viral or cellular proteins traffic into the nucleolus is not clearly understood [17]. Meanwhile, the localization of the PDCoV N protein and its traffic mechanisms are yet unknown.
In this study, we produced monoclonal antibodies (mAbs) against the PDCoV N protein, and then used them to observe that the PDCoV N protein distribution pattern in the cytoplasm and nucleolus in both ectopic PDCoV N-expression cells and PDCoV-infected cells. We also performed deletion mutagenesis to identify potential NoLS. The findings from these experiments will provide new insights into the properties and functions of the PDCoV N protein.
Materials and methods
Ethics statement
With the approval of the Laboratory Animal Ethics Committee of Jiangxi Agricultural University and in accordance with The Guidelines for the Care of Laboratory Animals established by the Ministry of Agriculture of China, an animal use protocol (JXAULL-20190016) was used. The mice used to produce ascites for harvesting monoclonal antibodies against the N protein of PDCoV were euthanized. Ascites were harvested immediately on the day when abdominal enlargement was observed to avoid ascites accumulation, which may cause distress.
Cell, virus and reagents
LLC-PK1 cells, a porcine kidney cell line, were purchased from ATCC and subsequently cultured in Dulbecco’s modified Eagle’s medium (Gibco) containing 10% heat-inactivated fetal bovine serum (PAN-biotech) and a combination of penicillin and streptomycin (Solarbio), in a humidified incubator with an atmosphere of 5% CO2 at 37 °C. PDCoV strain CH/JXJGS01/2016 (GenBank accession number KY293677.1) was isolated in our laboratory in 2016 from a newborn piglet with diarrhea. Mouse mAb against β-actin and hemagglutinin (HA) were bought from Medical and Biological Laboratories (Japan). Rabbit anti-B23 antibody was purchased from Proteintech (CHI, USA). Horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (H + L) was bought from antGene (China). FITC-conjugated goat anti-mouse IgG(H + L) and Alexa Fluor® 594 donkey anti-rabbit IgG(H + L) were bought from and TransGen Biotech (China). All enzymes used for the cloning procedures were purchased from Takara (Dalian, China). Freund’s complete/incomplete adjuvants, polyethylene glycol 1450 and HT/HAT medium were purchased from Sigma-Aldrich (MO, USA).
Plasmids
To construct the DNA expression vector pCAGGS-HA-PDCoV-N coding an HA-tagged PDCoV N protein, standard reverse transcription (RT)-PCR with primers N1-F and N3-R primers was used to amplify the PDCoV N gene. The RNA from LLC-PK1 cells infected with PDCoV strain CH/JXJGS01/2016 was extracted and used as a template for reverse transcription, generating cDNA for amplification. Mutants of the PDCoV N protein NR157A, N Δ 161–197, N Δ 295–318, NR157A Δ 161–197, NR157A Δ 295–318, N△161–197 Δ 295–318 and NR157A Δ 161–197 Δ 295–318 were constructed by truncation and/or site-directed mutagenesis (as described previously [20]) with overlap extending PCR employing the indicated templates and primers, listed in Tables 1 and 2. PCR products were digested with EcoR I and Cla I, followed by ligation into pCAGGS-HA. All constructs were verified by DNA sequencing. pCold-PDCoV-N that can express His-tagged N protein and recombinant PDCoV N protein that was expressed and purified from Escherichia coli were stored in our lab [21].
Table 1.
Primers and templets using in plasmid construction
| Fragment amplified | Primers used | Templates used |
|---|---|---|
| WT N (aa 1 to 342) | N1-F/N3-R | PDCoV |
| NR157A | N1-F/N157-R and N157-F/N3-R | WT N |
| N△161–197 | N1-F/N1-R and N2-F/N3-R | WT N |
| N△295–318 | N1-F/N2-R and N3-F/N3-R | WT N |
| NR157A△161–197 | N1-F/N157-R and N2-F/N3-R | NR157A |
| NR157A△295–318 | N1-F/N2-R and N3-F/N3-R | NR157A |
| N△161–197 △295–318 | N1-F/N2-R and N3-F/N3-R | N△161–197 |
| NR157A△161–197△295–318 | N1-F/N157-R and N157-F/N3-R | N△161–197 △295–318 |
Table 2.
Primer sequences for amplification of PDCoV N gene and mutants
| Name of primers | Sequence (5′ → 3′) |
|---|---|
| N1-F | TTTGAATTC ATGGCTGCACCAGTAGTCCCTA |
| N1-R | CTGAGAAATGGTTTTAGATTGAGATCTTGGGCC |
| N2-F | AGATCTCAATCTAAAACCATTTCTCAGGTATTTGGC |
| N2-R | TGCTGGCAGAGTTACTTTGGTGGGTGGCTC |
| N3-F | CCACCCACCAAAGTAACTCTGCCAGCAGACAAA |
| N3-R | TGGATCGATTTACTACGCTGCTGATTCCTGCTTTA |
| N157-F | TCTGGAGTTAACAGATTGAGATGCTGGGCC |
| N157-R | AGTGGCCCAGCATCTCAATCTGTTAACTCC |
The fonts-bold mean protective bases; the italics mean restriction enzyme cutting sites
Enzyme-linked immunosorbent assay (ELISA)
To establish an ELISA for screening hybridomas secreting mAbs specific to the N protein of PDCoV, a standard method was employed. Briefly, the recombinant PDCoV N protein (5 µg/ml) was coated onto 96-well ELISA plates. The coated plates were washed three times with 0.01 M PBS (pH 7.2) and then blocked with 5% skimmed milk. After three times washing, the plates were incubated with the sera of mice who had been immunized with the recombinant PDCoV N protein. Subsequently, the plates were incubated with HRP-conjugated goat anti-mouse IgG (1:2000, TransGen Biotech). The unbound secondary antibody was washed off with PBS. Signal reaction was activated utilizing 3,3′,5,5′-tetramethylbenzidine (TMB) substrate, and then stopped with 2 M H2SO4, after which the absorbance was read at OD450. Tested samples that gave an absorbance value greater than 0.080 were defined as positive.
Generation of monoclonal antibodies
The monoclonal antibodies were produced based on the protocol described in “Antibodies”, A laboratory manual edited by Ed Harlow and David Lane [22]. Briefly, BALB/c mice were subcutaneously immunized with 100 µg of recombinant PDCoV N protein mixed with Freund’s complete adjuvant (Sigma-Aldrich), followed by two boosts with 100 µg of recombinant PDCoV N protein mixed with Freund’s incomplete adjuvant (Sigma-Aldrich), at 2 weeks intervals. Serum samples collected on the tenth day after the third immunization were tested. The titer of serum samples was detected using the established ELISA described above; when the titer was higher than 1:10,000, 100 µg of recombinant PDCoV N protein were intraperitoneally (i.p.) injected into the mouse for the final boost. The splenocytes from immunized mice were collected and fused with sp2/0 cells to generate hybridomas. The resulting hybridomas secreting PDCoV N-specific mAbs were selected and continuously subcloned at least three times to verify their clonality. To acquire mAb in ascitic fluid, the hybridomas were transplanted intraperitoneally into mice, and once the abdomens of the mice swelled, the resulting ascitic fluid was collected and stored at − 20 °C.
Western blot
After being infected with PDCoV or transfected with PDCoV wildtype N-protein-expressing plasmids and/or mutants with lip2000, LLC-PK1 cells were lysed with lysis buffer (containing 3% DTT, 0.065 mol/L Tris–HCl [PH 6.8], 4% SDS and 30% glycerol) supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF) and then subjected to western blot. Briefly, the cell lysate samples were boiled in 1 × Laemmli buffer, separated by SDS-PAGE, and electroblotted onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad). The membrane was then blocked with PBST (PBS with 0.05% Tween-20) containing 5% milk, incubated with mouse anti-HA or anti-β-actin antibody at 1:2500, and subsequently incubated with HRP-conjugated goat anti-mouse IgG at 1:2500 before being developed with the ECL western blot substrate.
Indirect immunofluorescence assay (IFA)
To examine the subcellular localization of wild type and artificial mutant PDCoV N proteins. LLC-PK1 cells were transfected with 2 µg of the indicated plasmids or with negative vectors or infected with PDCoV when cells gown on microscope coverslips in 24-well plates reached approximately 80% confluence. The cells were subsequently harvested for fixation with 4% paraformaldehyde for 10 min, followed by permeabilization with 0.1% Triton X-100 for 10 min, and blocking with PBS containing 3% bovine serum albumin for 1 h. After washed three times with 0.01 M pH 7.0 PBS, the cells were then incubated separately with mouse mAb against the HA tag or PDCoV N (1:200) for 1 h, followed by FITC-conjugated goat anti-mouse IgG antibody for 1 h, and finally 4,6-diamidino-2-phenylindole (DAPI) for 15 min to detect nuclear DNA. The cells were observed with a confocal laser scanning fluorescent microscope (Olympus Fluoviewer. 3.1, Tokyo, Japan) after washed three times with PBS.
Results
mAbs produced can react with the PDCoV N protein
To generate a monoclonal antibody against the N protein, the pCold-PDCoV-N vector was constructed, and the resulting recombinant protein, which was expressed and purified from Escherichia coli as described previously [21]. The purified N protein was used to immunize female BALB/c mice. One positive clone, designated as 2G12, was obtained through ELISA screening. Indirect ELISAs were then performed to determine the mAb titers. They revealed that the antibody titers of ascites and hybridoma cell culture supernatants were over 1,000,000 and 400,000, respectively (Fig. 1a). Western blot analysis showed that mAb 2G12 specifically reacted with the E. coli-produced N protein, N protein expressing cells and PDCoV-infected cells (Fig. 1b). IFAs further confirmed the specificity and reactiveness of mAb 2G12 against the N protein in the context of PDCoV infection (Fig. 1c). These results demonstrate that mAb 2G12 recognizes the PDCoV N protein.
Fig. 1.
mAb against PDCoV N protein generated. a Titers of anti-PDCoV N protein mAb from Ascites and cell culture supernatants from a monoclonal hybridoma cell line were analyzed by ELISA. Serum from a mock vector immunized mouse acted as a control. The coating concentration of PDCoV N protein was 5 µg/ml. The optical density (OD) values of each point represent the mean value and standard deviation from three determinations (n = 3). b Purified recombinant PDCoV-N protein samples or lysates of LLC-PK1 cells that had been transfected with pCAGGS-HA-PDCoV N or infected with PDCoV were subjected to western blotting with anti-PDCoV N protein mAb. c LLC-PK1 cells were infected with PDCoV, then fixed with paraformaldehyde for an IFA. The green and blue signals indicate PDCoV N and DAPI, respectively. Fluorescent cells were imaged using confocal laser scanning microscopy
N protein localizes in the nucleolus of both cells ectopically expressing N protein and cells infected with PDCoV
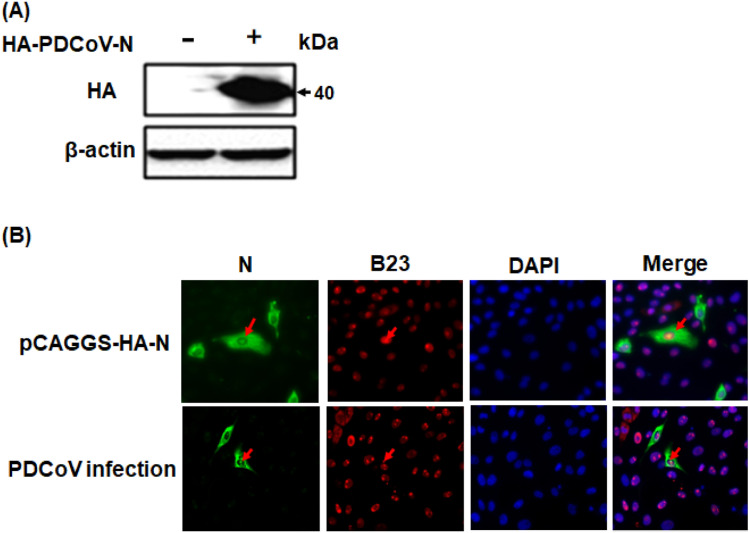
To determine the subcellular distribution of the PDCoV N protein, the pCAGGS-HA-PDCoV-N vector was constructed and transfected into LLC-PK1 cells. The results of a western blot analysis showed that the PDCoV N protein was expressed normally (Fig. 2a). Next, the results of an IFA analysis of LLC-PK1 cells transfected with pCAGGS-HA-PDCoV-N demonstrated that although the fluorescence indicating the presence of the N protein was predominantly observed in the cytoplasm, some fluorescence was also observed in the nucleolus for a low percentage in ectopic N-expressing cells. The nucleolus location was confirmed by B23, which is a nucleolar biomarker. A similar result was observed in the context of PDCoV infection (Fig. 2b), indicating that PDCoV N protein localizes in cytoplasm and can also be imported into nucleolus.
Fig. 2.
PDCoV N protein distributes in both the nucleolus and cytoplasm. a LLC-PK1 cells transfected with pCAGGS-HA-PDCoV-N plasmids were subjected to western blotting using anti-HA antibodies or β-actin antibodies. b LLC-PK1 cells were transfected with pCAGGS-HA-PDCoV-N vectors or infected with PDCoV, then fixed with Triton X-100. After incubation with mouse monoclonal antibodies against HA or PDCoV-N protein and rabbit polyclonal antibody against B23, and then cells were stained with DAPI. The cells were observed and photographed using a confocal laser scanning microscope. The N protein is colored green, and B23 is colored red
At least one of the predicted NoLS is critical for nucleolar localization
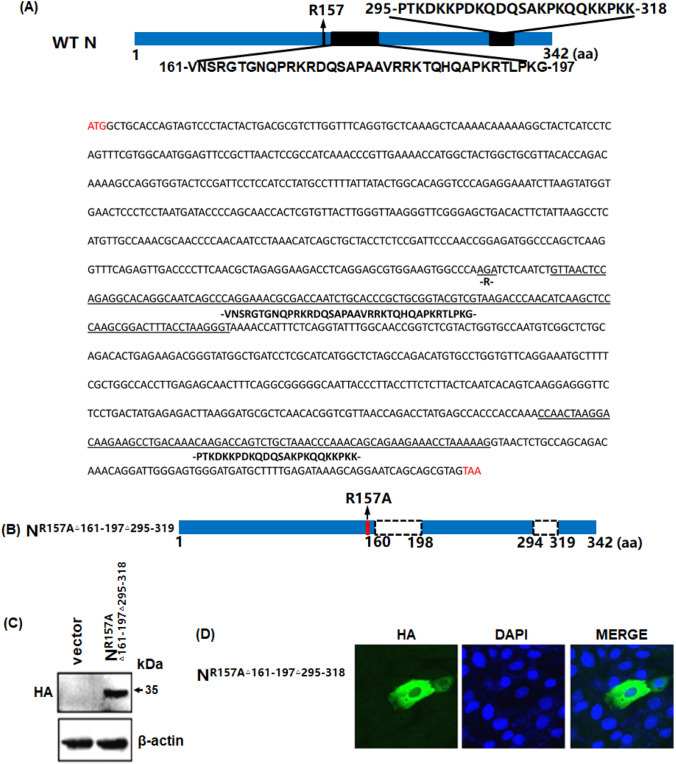
To identify potential localization signals in the PDCoV N protein, putative NoLSs were predicted using the online algorithm NLStradamus, (https://www.moseslab.csb.utoronto.ca/NLStradamus/) [23]. An analysis showed that three regions, i.e., the including 157th arginine residue, amino acid residues 161–197, and amino acid residues 295–318, might play a role in N protein localization as a NoLS (Fig. 3a). To investigate whether these predicted signals contribute to the subcellular trafficking of the N protein, the 157th arginine was mutated to an alanine with amino acid residues 161–197 and 295–319 deletions in an N protein mutant named NR157AΔ161−197Δ295−318 (Fig. 3b). Western blotting confirmed that NR157AΔ161−197Δ295−318 can be expressed normally in LLC-PK1 cells (Fig. 3c), and an IFA showed that this mutant was only located in the cytoplasm (Fig. 3d), indicating that the 157th arginine residue, and/or amino acid residues 161–197, and/or amino acid residues 295–318 are indispensable for the N nucleolar localization.
Fig. 3.
Putative NoLSs are critical for PDCoV N nucleolar localization. a and b A schematic diagram of PDCoV N protein (a) and PDCoV N mutant NR157AΔ161−197 Δ295−318 (b) is shown. Below the diagram is the nucleotide sequence of PDCoV N. Red words in the 5′ and 3′ are start and stop codons. Words underlined are mutated or deleted, and indicated amino acids are shown below. The amino acid sequence is listed above the black boxes or bars. The dashed box represents an internal deletion; the red bar represents a mutant in which the amino acid site 157 Arg was replaced by Ala; and the numbers indicate the amino acid position. c A western blotting analysis of LLC-PK1 cell lysates expressing mutant NR157AΔ161−197 Δ295−318 was performed using anti-HA antibodies. (D) LLC- PK1 cells were transfected with the mutant NR157A△161–197△295–318, then fixed with paraformaldehyde for an IFA. The recombinant proteins are colored green and the nucleus is colored blue. Images were examined as described in Fig. 1c
Deletion of the N protein amino acids at 295–318 abrogates the nucleolar distribution of the N protein
To map the residues responsible for the N protein nucleolar localization, N protein mutants with an alanine as amino acid 157 and/or a deletion of amino acids at the position 161–197 and/or a deletion of amino acids at 295–319 were constructed and transfected into LLC-PK1 cells, which were then subjected to immunoblotting. All mutants were expressed normally (Fig. 4a, b). IFA and confocal microscopy revealed that all the N protein mutants with deleted amino acids 295–319 (NΔ161−197 Δ295−319, NR157AΔ295−319, and NΔ295−319) had abrogated nucleolar localization compared with wildtype N protein. The other N protein mutants that contained amino acids 295–319 (NR157AΔ161−197, NR157A, and NΔ161−197) were still able to distribute in both the cytoplasm and nucleolus. This result indicates that amino acids 295–319 are responsible for the nucleolar localization of the N protein of PDCoV.
Fig. 4.
Amino acid residues 295–318 of the PDCoV N protein are indispensable for N nucleolar localization. a A schematic diagram of PDCoV N mutants, represented as described above. b LLC-PK1 cells were transfected with expression vectors for the mutants NΔ161−197 Δ295−318, NR157AΔ161−197, NR157AΔ295−318, NR157A, NΔ161−197 and NΔ295−318 and then lysed for immunoblotting with anti-HA antibodies and anti-β-actin antibodies. c LLC-PK1 cells were transfected with the mutants NΔ161−197 Δ295−318, NR157AΔ161−197, NR157AΔ295−318, NR157A, NΔ161−197 and NΔ295−318, then subjected to an IFA. Mutant proteins are colored green, and the nucleus is colored blue. Fluorescent images were acquired as described in Fig. 1c
Discussion
Coronavirus N proteins show low amino acid homology, but they share several conserved functions, including immune regulation. Like the N proteins from α-coronavirus or β-coronavirus, the PDCoV N protein also possesses an IFN-inhibition function via interfering dsRNA and PACT binding to RIG-I or RIG-I K63-linked polyubiquitination [14, 24–26]. Little research into the function of PDCoV N protein has been conducted, but a recent study showed that PDCoV N could upregulate two HSP70 family members, glucose-regulated protein 78 (GRP78) and heat shock cognate 70-kDa protein (HSC70), which may facilitate virus infection. Furthermore, that study also observed that the PDCoV N protein can localize in the both nucleolus and the cytoplasm in PDCoV N protein expressing-cell lines [27], which is consistent with our findings here, where we generated mAb against the PDCoV N protein and confirmed that the N protein is distributed in either the cytoplasm alone, or in both the nucleolus and cytoplasm. Moreover, in the present study, we identified a functional NoLS that is critical for the nucleolus localization of the N protein of PDCoV.
The N proteins of Coronaviridae family members, including PEDV, avian IBV and severe acute respiratory syndrome coronavirus (SARS-CoV), have been reported to localize in the nucleolus [17, 28, 29]. The subcellular localization of the PDCoV N protein differs slightly from those of the N proteins of other CoVs. The N protein of PDCoV can localize in the cytoplasm alone or in both the cytoplasm and nucleolus, similarly to the N proteins of PEDV, a member in the genus Alphacoronavirus and IBV, a member in the genus Gammacoronavirus. The N-terminal domains of PEDV and IBV N proteins are both necessary and sufficient for nucleolar retention [17, 30], whereas the PDCoV N protein NoLS is located in the C-terminal domain (CTD). In contrast, the N protein from SARS-CoV is mostly distributed to the cytoplasm, and even a putative NLS is recognized in its the CTD [20, 29].
The nucleolus is a phase-separated cell condensate, which comprised three subcompartments: fibrillar centers (FCs), the dense fibrillar component (DFC), and the granular component (GC) [31]. The nucleolus is plurifunctional; it was primarily identified as a site of ribosome biogenesis [32], but later was found to participate in many biological processes including tRNA and mRNA processing, cell cycle regulation and cellular aging [33]. The replication, transcription, and virion assembly of most RNA viruses occur in the cytoplasm of an infected cell. Although the reason(s) why proteins from cytoplasmic RNA viruses are imported into the nucleolus are still unknown, it has been proven that the association of viral proteins with components in the nucleolus notably contributes to efficient viral replication. The capsid protein of West Nile virus (WNV) is a convincing example of this phenomenon, and it was demonstrated to interact with the nucleolar RNA helicase DDX56 and relocate it from the nucleoli to virus assembly sites for WNV particle assembly [34–36]. Additionally, default in the nucleolar localization of the Japanese Encephalitis Virus (JEV) capsid protein impaired the virus replication and pathogenesis of encephalitis induced by JEV [37]. Our data demonstrate that the N protein of PDCoV is found in the nucleolus. However, the nucleolar components with which PDCoV N combines and the functions in which they are involved need to be investigated in further studies.
Acknowledgements
This work was supported by grants from The National Natural Science Foundation of China (Grant No. 31860704), Jiangxi Province Project (Grant Nos. GJJ170261, 20192BAB214023) and Program for Jiangxi Agriculture Research System (Grant No. JXARS-01). We thank Katie Oakley, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author contributions
ZD, YT, ND, and JZ conceived and designed the experiments; SL, WG, JC, and JC carried out the experiments; ZD, TW, YY, and DS analyzed the data; ZD, SL, LK, and ND prepared the manuscript draft; and LW, YT, JZ, and ZD reviewed and edited the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Animal experiments (JXAULL-20190016) were conducted under the guidelines of the ethics committee of Jiangxi Agricultural University, China and The Care and Use Guidelines of Experimental Animals established by the Ministry of Agriculture of China.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jinghua Zhang, Email: zhangjh1122@jxau.edu.cn.
Yuxing Tang, Email: tang53ster@gmail.com.
References
- 1.Jung K, Hu H, Saif LJ. Porcine deltacoronavirus infection: etiology, cell culture for virus isolation and propagation, molecular epidemiology and pathogenesis. Virus Res. 2016;226:50–59. doi: 10.1016/j.virusres.2016.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J. Porcine deltacoronavirus: overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016;226:71–84. doi: 10.1016/j.virusres.2016.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jung K, Hu H, Saif LJ. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch Virol. 2017;162:2357–2362. doi: 10.1007/s00705-017-3351-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang Q, Zhang H, Li B, Ding Q, Wang Y, Gao W, Guo D, Wei Z, Hu H. Susceptibility of chickens to porcine deltacoronavirus infection. Viruses. 2019;11(6):573. doi: 10.3390/v11060573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Hulswit RJG, Kenney SP, Widjaja I, Jung K, Alhamo MA, van Dieren B, van Kuppeveld FJM, Saif LJ, Bosch BJ. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc Natl Acad Sci USA. 2018;115:E5135–e5143. doi: 10.1073/pnas.1802879115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song D, Zhou X, Peng Q, Chen Y, Zhang F, Huang T, Zhang T, Li A, Huang D, Wu Q, He H, Tang Y. Newly emerged porcine deltacoronavirus associated with diarrhoea in swine in China: identification, prevalence and full-length genome sequence analysis. Transbound Emerg Dis. 2015;62:575–580. doi: 10.1111/tbed.12399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang YW, Yue H, Fang W, Huang YW. Complete genome sequence of porcine deltacoronavirus strain CH/Sichuan/S27/2012 from Mainland China. Genome Announc. 2015 doi: 10.1128/genomeA.00945-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fang P, Fang L, Liu X, Hong Y, Wang Y, Dong N, Ma P, Bi J, Wang D, Xiao S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology. 2016;499:170–177. doi: 10.1016/j.virol.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fang P, Fang L, Hong Y, Liu X, Dong N, Ma P, Bi J, Wang D, Xiao S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J Gen Virol. 2017;98:173–178. doi: 10.1099/jgv.0.000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Masters PS, Sturman LS. Background paper. Functions of the coronavirus nucleocapsid protein. Adv Exp Med Biol. 1990;276:235–238. doi: 10.1007/978-1-4684-5823-7_32. [DOI] [PubMed] [Google Scholar]
- 11.Masters PS, Parker MM, Ricard CS, Duchala C, Frana MF, Holmes KV, Sturman LS. Structure and function studies of the nucleocapsid protein of mouse hepatitis virus. Adv Exp Med Biol. 1990;276:239–246. doi: 10.1007/978-1-4684-5823-7_33. [DOI] [PubMed] [Google Scholar]
- 12.Chang CK, Lo SC, Wang YS, Hou MH. Recent insights into the development of therapeutics against coronavirus diseases by targeting N protein. Drug Discov Today. 2016;21:562–572. doi: 10.1016/j.drudis.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Likai J, Shasha L, Wenxian Z, Jingjiao M, Jianhe S, Hengan W, Yaxian Y. Porcine deltacoronavirus nucleocapsid protein suppressed IFN-beta production by interfering porcine RIG-I dsRNA-binding and K63-linked polyubiquitination. Front Immunol. 2019;10:1024. doi: 10.3389/fimmu.2019.01024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Fang P, Wang M, Peng Q, Ren J, Wang D, Peng G, Fang L, Xiao S, Ding Z. Porcine deltacoronavirus nucleocapsid protein antagonizes IFN-beta production by impairing dsRNA and PACT binding to RIG-I. Virus Genes. 2019 doi: 10.1007/s11262-019-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan YW, Fang S, Fan H, Lescar J, Liu DX. Amino acid residues critical for RNA-binding in the N-terminal domain of the nucleocapsid protein are essential determinants for the infectivity of coronavirus in cultured cells. Nucleic Acids Res. 2006;34:4816–4825. doi: 10.1093/nar/gkl650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emmott E, Munday D, Bickerton E, Britton P, Rodgers MA, Whitehouse A, Zhou EM, Hiscox JA. The cellular interactome of the coronavirus infectious bronchitis virus nucleocapsid protein and functional implications for virus biology. J Virol. 2013;87:9486–9500. doi: 10.1128/JVI.00321-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi D, Lv M, Chen J, Shi H, Zhang S, Zhang X, Feng L. Molecular characterizations of subcellular localization signals in the nucleocapsid protein of porcine epidemic diarrhea virus. Viruses. 2014;6:1253–1273. doi: 10.3390/v6031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hiscox JA, Wurm T, Wilson L, Britton P, Cavanagh D, Brooks G. The coronavirus infectious bronchitis virus nucleoprotein localizes to the nucleolus. J Virol. 2001;75:506–512. doi: 10.1128/JVI.75.1.506-512.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J Virol. 2001;75:9345–9356. doi: 10.1128/JVI.75.19.9345-9356.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timani KA, Liao Q, Ye L, Zeng Y, Liu J, Zheng Y, Ye L, Yang X, Lingbao K, Gao J, Zhu Y. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005;114:23–34. doi: 10.1016/j.virusres.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F, Song D, Guo N, Ye Y, Zhou X, Li A, Zhang M, Peng Q, Chen Y, Huang D, Tang Y. Establishment of a recombinant nucleoprotein-based ELISA for detection of antibodies against newly emerged porcine deltacoronavirus. Chin J Prev Vet Med. 2016;388:795–799. [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: a laboratory manual. New York: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 23.Nguyen Ba AN, Pogoutse A, Provart N, Moses AM. NLStradamus: a simple Hidden Markov Model for nuclear localization signal prediction. BMC Bioinform. 2009;10:202. doi: 10.1186/1471-2105-10-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding Z, Fang L, Jing H, Zeng S, Wang D, Liu L, Zhang H, Luo R, Chen H, Xiao S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J Virol. 2014;88:8936–8945. doi: 10.1128/JVI.00700-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y, Li W, Gao T, Cui Y, Jin Y, Li P, Ma Q, Liu X, Cao C. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits Type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol. 2017 doi: 10.1128/JVI.02143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding Z, Fang L, Yuan S, Zhao L, Wang X, Long S, Wang M, Wang D, Foda MF, Xiao S. The nucleocapsid proteins of mouse hepatitis virus and severe acute respiratory syndrome coronavirus share the same IFN-beta antagonizing mechanism: attenuation of PACT-mediated RIG-I/ MDA5 activation. Oncotarget. 2017;8:49655–49670. doi: 10.18632/oncotarget.17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S, Lee C. Functional characterization and proteomic analysis of the nucleocapsid protein of porcine deltacoronavirus. Virus Res. 2015;208:136–145. doi: 10.1016/j.virusres.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed ML, Dove BK, Jackson RM, Collins R, Brooks G, Hiscox JA. Delineation and modelling of a nucleolar retention signal in the coronavirus nucleocapsid protein. Traffic. 2006;7:833–848. doi: 10.1111/j.1600-0854.2006.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.You J, Dove BK, Enjuanes L, DeDiego ML, Alvarez E, Howell G, Heinen P, Zambon M, Hiscox JA. Subcellular localization of the severe acute respiratory syndrome coronavirus nucleocapsid protein. J Gen Virol. 2005;86:3303–3310. doi: 10.1099/vir.0.81076-0. [DOI] [PubMed] [Google Scholar]
- 30.Ng LF, Liu DX. Further characterization of the coronavirus infectious bronchitis virus 3C-like proteinase and determination of a new cleavage site. Virology. 2000;272:27–39. doi: 10.1006/viro.2000.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grob A, McStay B. Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division. Cell Cycle. 2014;13:2501–2508. doi: 10.4161/15384101.2014.949124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L, Ren XM, Xing JJ, Zheng AC. The nucleolus and viral infection. Virol Sin. 2010;25:151–157. doi: 10.1007/s12250-010-3093-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsekrekou M, Stratigi K, Chatzinikolaou G. The nucleolus: in genome maintenance and repair. Int J Mol Sci. 2017;18:1411. doi: 10.3390/ijms18071411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Z, Anderson R, Hobman TC. The capsid-binding nucleolar helicase DDX56 is important for infectivity of West Nile virus. J Virol. 2011;85:5571–5580. doi: 10.1128/JVI.01933-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z, Hobman TC. The helicase activity of DDX56 is required for its role in assembly of infectious West Nile virus particles. Virology. 2012;433:226–235. doi: 10.1016/j.virol.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reid CR, Hobman TC. The nucleolar helicase DDX56 redistributes to West Nile virus assembly sites. Virology. 2017;500:169–177. doi: 10.1016/j.virol.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 37.Mori Y, Okabayashi T, Yamashita T, Zhao Z, Wakita T, Yasui K, Hasebe F, Tadano M, Konishi E, Moriishi K, Matsuura Y. Nuclear localization of Japanese encephalitis virus core protein enhances viral replication. J Virol. 2005;79:3448–3458. doi: 10.1128/JVI.79.6.3448-3458.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]