Abstract
Cervical cancer is the most common cancer affecting sub-Saharan African women and is prevalent among HIV positive (HIV+) patients. No comprehensive profiling of cancer genomes, transcriptomes or epigenomes has been performed in this population to date. We characterized 118 tumors from Ugandan patients, of which 72 were HIV+, and performed extended mutation analysis on an additional 89 cases. We detected human papillomavirus (HPV) clade-specific differences in tumor DNA methylation, promoter- and enhancer-associated histone marks, gene expression and pathway dysregulation. Histone modification changes at HPV integration events were correlated with upregulation of nearby genes and endogenous retroviruses.
Introduction
Persistent human papillomavirus (HPV) infection, in episomal or integrated forms, is necessary but not sufficient for the development of cervical cancer1. HPV16 and HPV18 are detected in at least 70% of cases2. HPV16 (clade A9) is common in both squamous cell carcinomas and adenocarcinomas, while HPV18 (clade A7) is associated with adenocarcinomas2 and inferior survival3–5.
Cervical cancer prevention strategies include HPV vaccination, screening, and treatment of high-grade precancer. Although effective6, vaccine use remains low in low- and middle-income countries7 where HIV is prevalent. Resource constraints similarly complicate screening, surgery8 and radiotherapy9, such that a 50% increase in cervical cancer mortality by 2040 is predicted10.
Genomic cervical cancer studies, primarily conducted in non-African patients11,12, identified APOBEC mutational signatures, copy number amplifications of CD274 (PD-L1) and PDCD1LG2 (PD-L2), somatic alterations affecting the PI3K–MAPK and TGFβR2 pathways11,12 and mutations in chromatin modifier genes11–13. Studies in HPV-infected head and neck squamous cell carcinomas linked HPV integration to histone modification14 and DNA methylation changes15, suggesting the potential for similar findings in cervical cancer.
As part of the National Cancer Institute’s (NCI) HIV+ Tumor Molecular Characterization Project (HTMCP), we characterized genomic, transcriptomic and epigenomic landscapes of cervical cancers from Ugandan patients. We identified previously uncharacterized differences in the epigenomes and transcriptomes of cervical tumors infected by different HPV clades, and note that these clades appear relevant to prognosis.
Results
Patient samples and clinical data
Our cohort of 212 cervical cancer patients received treatment at the Uganda Cancer Institute in Kampala. Of these, 118 comprised our discovery cohort and 89 comprised our extension cohort (Supplementary Tables 1 and 2, Methods). HIV+ patients (72/118, 61%) were 10 years younger, on average, than HIV-negative (HIV-) patients (mean, 42.9 vs. 52.4).
Genomic alterations in HIV+ and HIV- cervical cancers
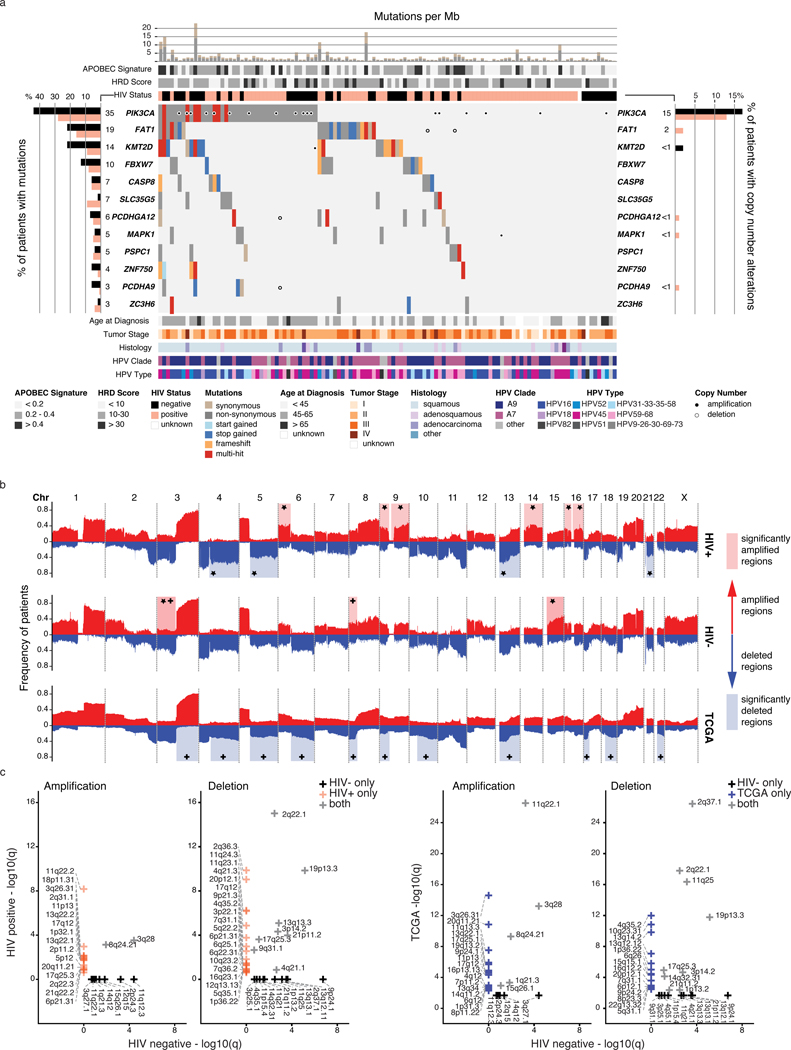
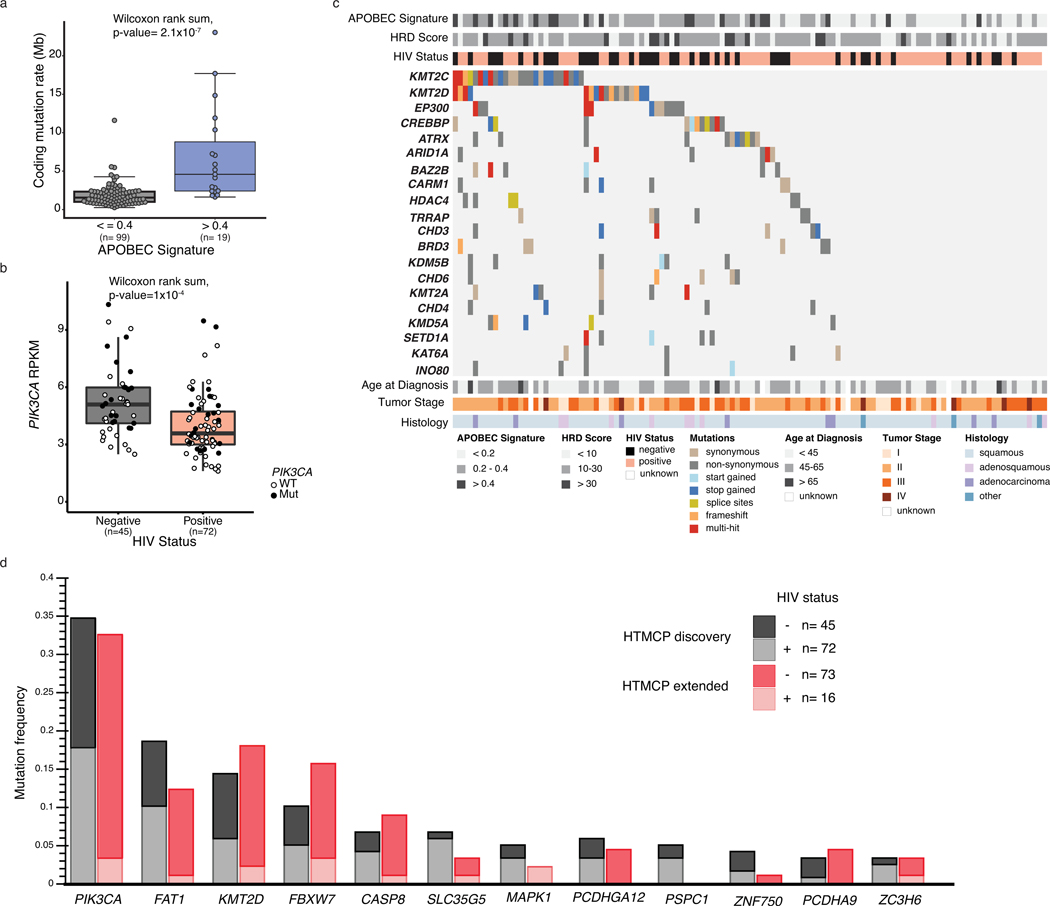
Whole genome sequencing (WGS) of samples from our discovery cohort identified an average of 22,942 somatic mutations (range 3,033 – 179,513) per sample, including 311 coding mutations (range 30–2,683, Figure 1a). We detected APOBEC mutation signatures 2 and 1316,17, confirming previous reports18, consistent with a mutational process driven by a cellular response to viral infections19 (Figure 1a). Tumors with high APOBEC signatures (proportion ≥0.4) exhibited significantly more coding mutations (3-fold increase per Mb, median, Wilcoxon, p-value=2.1×10–07) than those with lower proportions (Extended Data Figure 1a). Fifteen samples (13%) exhibited moderate to high homologous recombination (HR) deficiency scores (>30), indicative of a dysfunctional HR repair pathway20 (Figure 1a, Methods). There were no differences in mutation burden, mutation signatures or HRD scores between HIV+ and HIV- cases.
Figure 1: Mutational landscape of cervical cancers from Ugandan patients.
a. Mutation and copy number alterations for each tumor (n=118) ordered by frequency of alterations in significantly mutated genes (SMGs). Synonymous and non-synonymous mutation counts per megabase (Mb) are shown with the proportion of the APOBEC signatures (COSMIC 2 and 13) and homologous recombination deficiency (HRD) scores. HIV status, age at diagnosis, histology (”other” includes neuroendocrine and undifferentiated), tumor stage, HPV clade and type are annotated below the oncoprint. Left bar chart; Percentage of samples with mutations, by HIV status. Numbers to the right of the bar plot indicate the percentage of the entire cohort. Right bar chart; Proportion of samples with copy number alterations in SMGs, by HIV status. Numbers to the left of the bar plot indicate the percentage of the entire cohort. b. Broad somatic copy number alterations in our HIV+ and HIV- cohorts and TCGA cohort. * indicates the region is significantly amplified or deleted (determined by GISTIC, FDR <0.25, Methods) in only HIV+ or HIV- samples, and + indicates the region is significantly amplified or deleted in only TCGA or HIV- samples. c. Focal regions associated with significant copy number changes between HIV+ and HIV- samples, and between HIV- samples and TCGA. Number of tumor samples used to determine differences in b and c are: HIV+, n=72; HIV-, n=45; TCGA, n=178.
Of 12 significantly mutated genes (SMGs) in our cohort, PIK3CA was the most recurrent (Figure 1a, Supplementary Table 3), as reported in other studies11,12. A higher proportion of HIV- tumors (45%, 20/45) than HIV+ tumors (29%, 21/72) had PIK3CA mutations, and PIK3CA expression was 1.3 times higher in HIV- samples (Wilcoxon, p-value=1×10−04, Extended Data Figure 1b). Other SMGs included FAT1, KMT2D, FBXW7, CASP8, MAPK1 and ZNF750, all previously reported in cervical cancer11,12. Notably, 87% of the cohort (101/118) had at least one mutation in an annotated chromatin modifier gene20 (Supplementary Table 4; Extended Data Figure 1c).
We performed targeted sequencing of 2,735 selected genes in our extension cohort (HIV-, n=73; HIV+, n=16), confirming mutations in 11 of the 12 SMGs (Extended Data Figure 1d) and observing similar mutation frequencies between the discovery and extension cohorts.
Analysis of copy number landscapes showed that broad copy number alterations were comparable between HIV+ and HIV- samples, with shared amplifications of chromosomes 1, 8, 20 and arms 3q, 5p, 19q; and shared deletions of chromosome 11 and arms 3p, 4p, 19p, 21p (Figure 1b, top two). We found six amplified and four deleted chromosome arms unique to HIV+ samples and two amplified arms unique to HIV- samples (Figure 1b, Supplementary Table 5). HIV+ samples exhibited more unique focal amplifications and deletions compared to HIV- samples (Figure 1c, Supplementary Table 5).
We compared the copy number landscapes of our HIV- samples to the Cancer Genome Atlas (TCGA) cervical cancers (HIV-, Supplementary Table 6, Methods). TCGA samples exhibited a larger number of significantly deleted regions, affecting 11 chromosomes, while only 21p was lost in our cohort. In comparison to TCGA, our HIV- cohort exhibited three significantly amplified regions on chromosomes 3p, 8p, and 15q (Figure 1b). Five focal amplifications and nine deletions identified in the TCGA cohort were also detected in our HIV- cohort (Figure 1c). Focal deletions unique to TCGA or HIV- cases were comparable in number and more abundant than focal amplifications in either cohort (Supplementary Table 5). 11q22.1 and 22.2, containing YAP1, were amplified in HIV+, HIV- and TCGA samples. Six of the 12 SMGs were impacted by copy number alterations, with PIK3CA being the most frequently altered gene, by mutation or copy number (Figure 1a).
Recurrent non-coding mutations
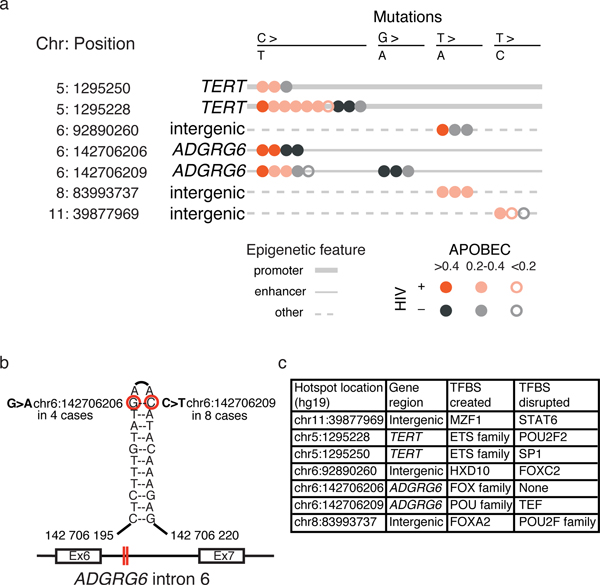
We leveraged WGS to identify seven high confidence non-coding “hotspots” (Methods; Figure 2a), including two in the TERT promoter first described in melanomas21,22, in 11% (13/118) of our discovery cohort samples. TERT transcript levels were not dysregulated in these samples. Two hotspots in a potential intronic enhancer (Methods) of ADGRG6 were observed in 9% (11/118) of samples. These non-coding mutations, at chr6:142,706,206 (G>A) and chr6:142,706,209 (C>T), resided within palindromic sequences predicted to form hairpin loops, accessible to APOBEC enzymes23 (Figure 2b). These hotspots have been reported in approximately 3% of breast cancers23 and 46% of bladder cancers24,25, and have been associated with increased ADGRG6 protein expression and angiogenesis. We did not observe dysregulated ADGRG6 mRNA expression in mutated cases. Three additional hotspots, on chromosomes 6, 8 and 11, were not associated with potential promoters or enhancers. All reported hotspots were present in HIV+ and HIV- cases and the samples with mutations (C>T, C>G) exhibited moderate APOBEC signatures (Figure 2a). Since TERT promoter mutations can create new transcription factor binding sites (TFBS) for c-ETS21,22, we investigated the potential for other non-coding hotspots to alter TFBS26 (Figure 2c), and noted that POU and FOX family binding sites were either created or destroyed by mutations in five of seven hotspots.
Figure 2: Recurrent non-coding mutations.
a. Schematic representation of the seven non-coding hotspots found in our cohort. Each dot represents a sample carrying the base substitution reported on the top of the plot. HIV status and strength of APOBEC signature in each sample with the mutation are indicated by the color and fill of the dots, and the line type represents the epigenetic characteristics surrounding the hotspot. b. Example of 2 hotspot mutations in ADGRG6 intron 6, found in 11 samples. Mutated nucleotides are circled in red and red lines in the diagram of the ADGRG6 gene highlight their location within intron 6. c. Predicted impact of the seven non-coding hotspot mutations on transcription factor binding sites (TFBS).
Distribution of HPV types
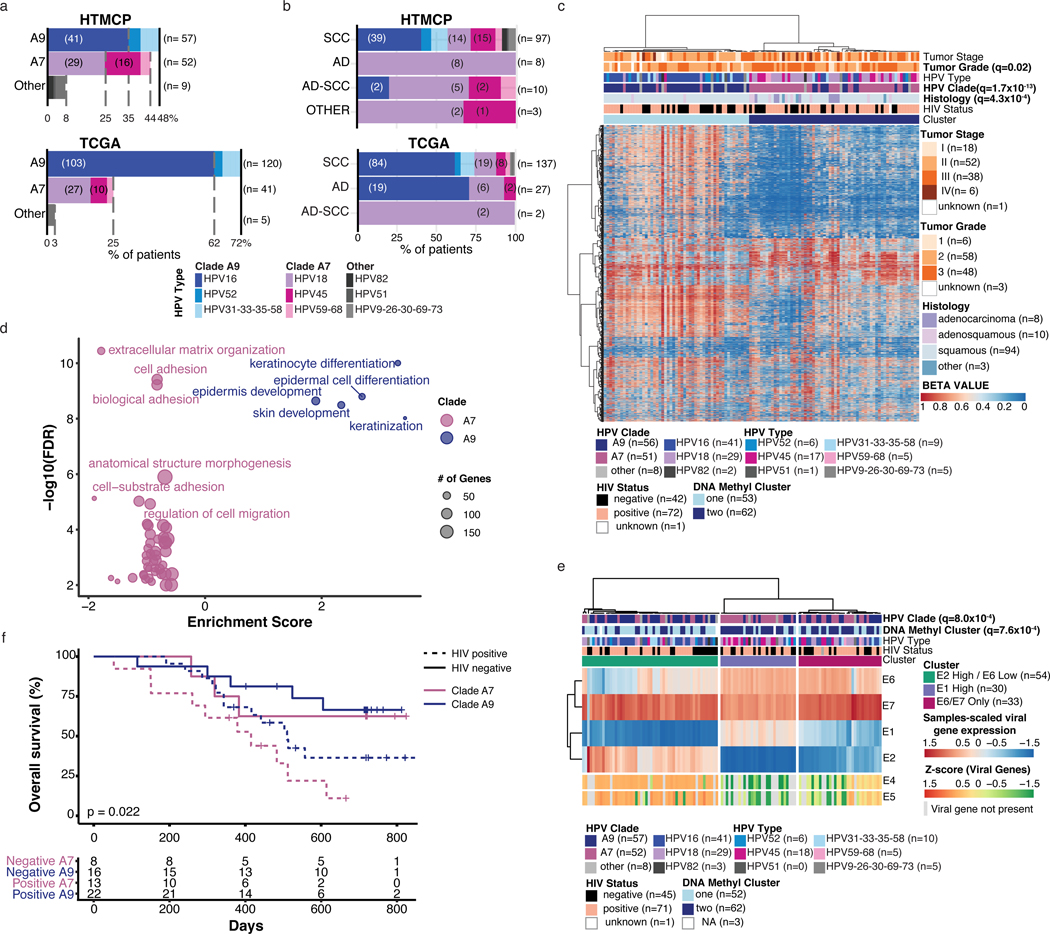
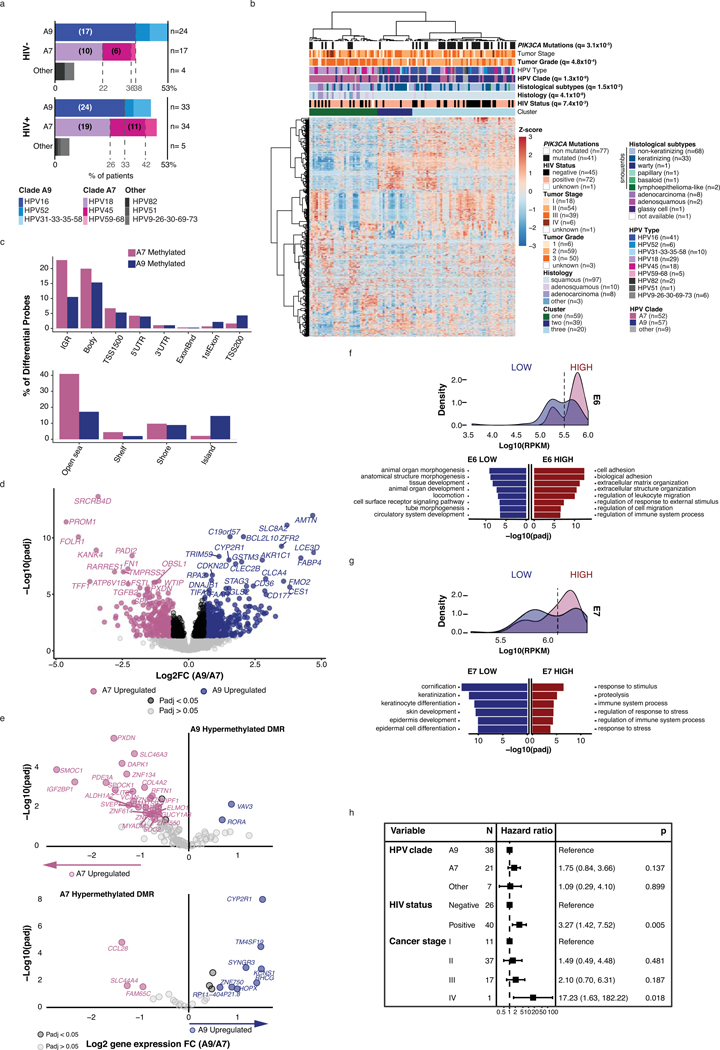
WGS detected 17 HPV types and their associated clades in our cohort (Methods)27,28. High-risk HPV16 (clade A9), 18 and 45 (clade A7) were the most abundant types (Figure 3a) and clade A7 was more prevalent in our cohort than TCGA, particularly among the squamous cell carcinomas (SCCs; Figure 3b). Unlike previous reports29,30, no difference in HPV types between HIV+ and HIV- tumors was found (Extended Data Figure 2a).
Figure 3: Clade specific molecular characteristics and prognosis.
a. Distribution of HPV types in our cohort (top, n=118 tumors) and TCGA (bottom, n=166 HPV+ tumors), and b. proportion of types split by histology. SCC= Squamous Cell Carcinoma, AD= Adenocarcinoma, AD-SCC= Adenosquamous Carcinoma, OTHER= Neuroendocrine (2 samples) and undifferentiated (1 sample). For a and b, the x-axis indicates the percentage of samples in that cohort infected by the indicated HPV type, and numbers in brackets indicate the number of samples. c. Unsupervised clustering analysis of DNA methylation for the top 8,000 variable probes in 115 samples. HIV status, HPV type and clade, histology, tumor grade and stage are annotated. d. Results from functional enrichment analysis of differentially expressed genes between clade A9- vs clade A7-infected samples (STRING, Methods). The size of the circles is proportional to the number of differentially expressed genes represented in each gene ontology. e. Unsupervised clustering analysis of sample-scaled HPV viral gene expression (n=117 samples). Z-scores for HPV genes not annotated in every HPV type are included below the clustering. HPV clade and type, DNA methylation clusters, and HIV status, are annotated. f. Overall survival of 59 patients stratified by the clade of infected HPV and HIV status. Kaplan-Meier overall survival statistics were determined using a log-rank test, and q-values for each variable on the heatmaps were determined using Benjamini-Hochberg corrected Fisher exact tests. All statistical tests were two-sided.
Expression and methylation profiles and HPV clades
We characterized expression and DNA methylation landscapes by performing unsupervised clustering on the most variably expressed genes (n=1,000) and methylation probes (n=8,000, 850k array), and correlated these with tumor features. We identified three gene expression clusters, enriched for adenocarcinomas (q-value=4.1×10−8; Cluster 1), non-keratinizing SCCs (Cluster 3), or keratinizing SCCs (q-value=0.015; Cluster 2), similar to those reported previously11 (Extended Data Figure 2b). Additionally, Cluster 1 was enriched for HPV clade A7 cases (q-value=1.3×10−9; Extended Data Figure 2b) and Cluster 3 for PIK3CA-mutated cases (q-value=3.1×10−5). Two DNA methylation clusters (Figure 3c) identified separation of clade A9-infected SCCs (Cluster 1) from clade A7-infected squamous and non-squamous carcinomas (Cluster 2, q-value=1.7×10−13). Cluster 2 was also enriched in cases exhibiting higher tumor grade (q-value=0.020).
We compared clade A7-infected samples to clade A9-infected samples (Supplementary Table 7) using differential methylation analysis31–33(Methods). We identified 107,685 differentially methylated probes (DMPs), with 46,639 DMPs in A9 tumors and 61,646 DMPs in A7 (FDR<0.05). The distribution of DMPs with respect to genomic features and proximity to CpG islands differed by clade. More common in clade A7 (79% vs. 45% in A9) were DMPs in ‘open sea’ and ‘shelf’ CpGs34,35(>2 kb from CpG islands), often in intergenic regions. More common in clade A9 (35% vs. 3.7% for clade A7) were DMPs in CpG islands, the majority of which resided in candidate transcriptional regulatory regions (Extended Data Figure 2c) (e.g. <1,500 bp from transcription start site (TSS), 5’UTR, 1st exon).
Motivated by the differences in DMP distribution between clades, and after accounting for histological differences, we detected 721 differentially expressed genes (Methods, Supplementary Table 8) between clades, with approximately equal proportions of genes upregulated in each (A7, n=363; A9, n=358, Extended Data Figure 2d). Functional enrichment analysis36 (Figure 3d, Supplementary Table 9) showed enrichment of keratin family genes, AMNT, LCE3D, and BCL2L10, and ontologies linked to keratinocyte and epithelial differentiation in clade A9 samples. The tightly regulated keratinocyte differentiation pathways are known to be exploited during HPV infection for active production of the virus, and later to direct uncontrolled cell growth in cervical epithelial cells37. Clade A7 samples had increased expression of PROM1, TGFB2, PXDN, and FN1, and genes were enriched for pathways linked to extracellular matrix organization, cell adhesion and migration, consistent with the more aggressive cancer grades that correlated with clade A7 tumors (Figure 3c).
To relate the effect of DNA methylation at promoter regions to changes in gene expression, we identified differentially methylated regions (DMRs; Methods, Supplementary Table 10), defined as nearby probes exhibiting consistent methylation changes, and associated these regions with gene expression. In agreement with the higher number of DMPs at CpG islands in A9-infected cases, we identified 558 methylated DMRs in these cases (490 overlapping with genes) and 53 in A7 cases (48 overlapping with genes). There were 26 upregulated genes in A7 cases that were associated with a methylated A9 DMR, compared to only 8 upregulated genes in A9 cases with a methylated A7 DMR (Extended Data Figure 2f). Thus, differential expression of genes between clades may result from differential methylation.
HPV viral gene expression influences host gene expression
HPV viral genes regulate epithelial cell differentiation and promote tumorigenesis5,38. To probe the impact of viral gene expression in our cohort, we performed unsupervised clustering of viral E1, E2, E6 and E7 transcripts, present in all HPV types in our cohort (n=117, Supplementary Table 11, Figure 3e, Methods), identifying three clusters. Cluster 1, enriched for A9-infected tumors (Fisher exact Test, q=0.0029), exhibited high E2 and low E6 expression, indicating dominant HPV episome transcription. Cluster 2, enriched for A7-infected cases (Fisher exact Test, q=0.0029), exhibited low E2 and high E1 expression. Cluster 3 contained cases infected with both clades and exhibited low expression of both E1 and E2. From the absence of E2, we inferred that Clusters 2 and 3 reflected viral expression originating from the integrated form of HPV. The expression distribution of E6 and E7 was bimodal (Extended Data Figure 2f,g), and was used to separate samples into high- and low-expressing tumors for each gene, on which we performed differential expression analysis. We identified 107 differentially expressed genes between E6 high/low tumors, and 60 between E7 high/low tumors. The E6-high group overlapped with clade A7-enriched pathways, while the E7-low group overlapped with clade A9-enriched pathways (Extended Data Figure 2f,g). Thus, clade-enriched tumor gene expression patterns may be influenced by the expression of HPV genes.
HPV clades are linked to prognosis
Consistent with our observation that A7-infected tumors displayed expression profiles indicative of a more aggressive phenotype, (Figure 3c,d), they also appeared to be more aggressive clinically, with A7-infected patients exhibiting inferior prognosis compared to A9-infected patients (hazard ratio (HR)=1.83, CI=1.02–3.30, p=0.04, log-rank test). This observation held true even after accounting for other covariates in our cohort, including HIV status (Figure 3f), stage and histology (Methods, Extended Data Figure 2h). Despite the relatively small number of patients available for analysis, the impact of clade (HR=1.75, p=0.14) on overall survival was similar to that of disease stage (HR=2.10, p=0.19), the latter being an established prognostic factor39. Similar observations have been reported previously40.
Histone modification profiles associated with HPV clades
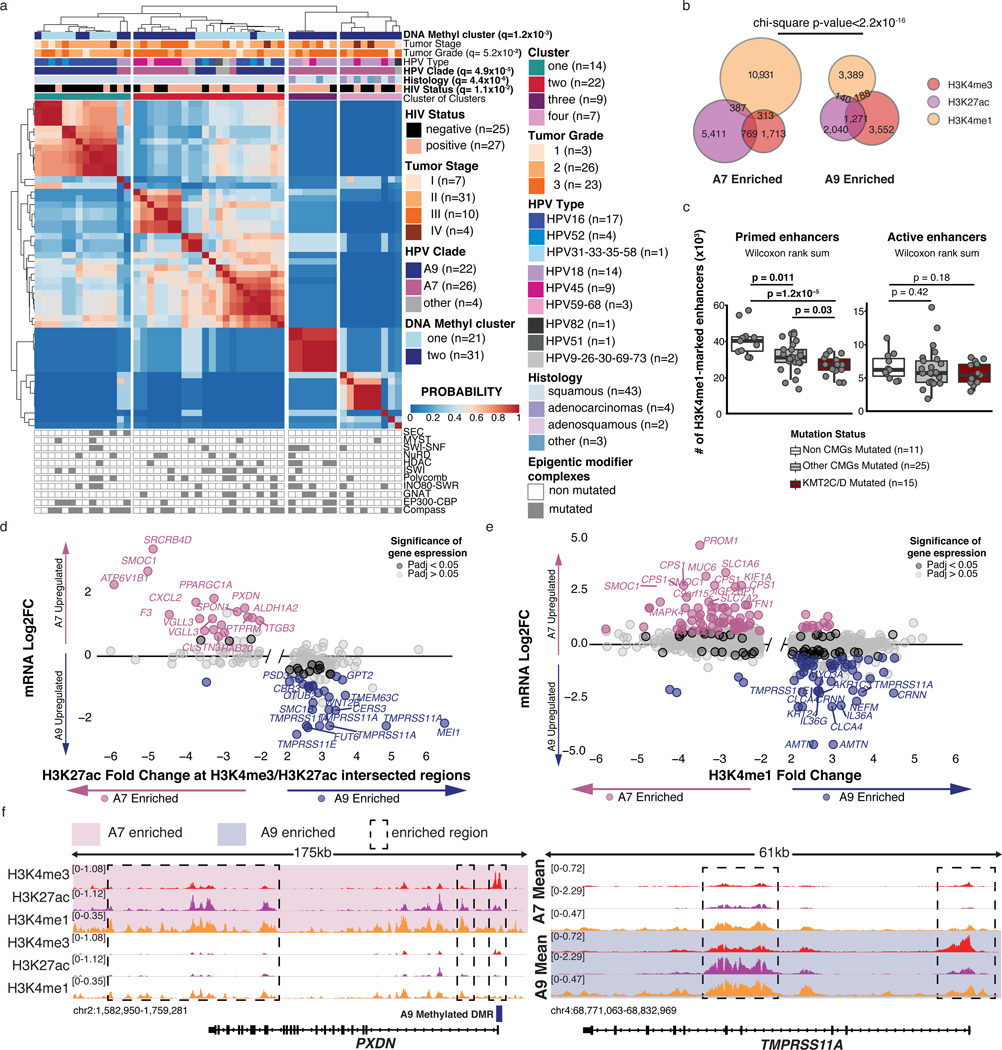
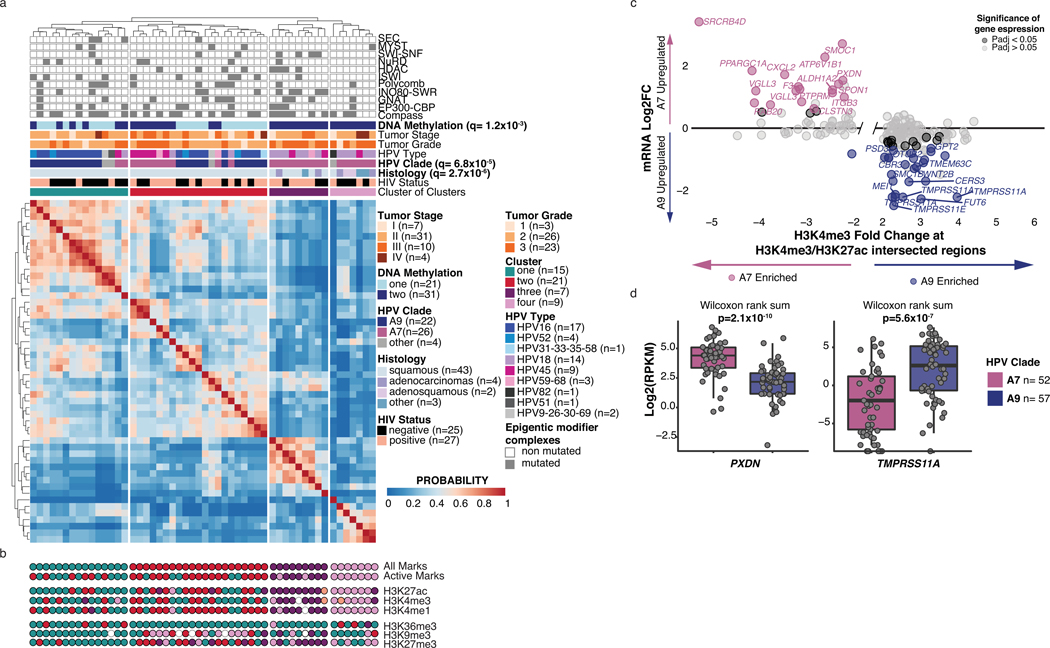
Motivated by our DNA methylation results (Figure 3c) and the preponderance of somatic mutations in chromatin modifying genes, we investigated whether histone modifications also exhibited clade-specific differences. Using ChIP-seq, we profiled four histone modification marks associated with transcriptional activation (H3K4me1, H3K4me3, H3K27ac, H3K36me3) and two marks associated with repression (H3K9me3 and H3K27me3) in 52 cases. Cluster of clusters analysis41 of these marks identified a clustering solution that resembled the individual solutions for the promoter- and enhancer-associated marks H3K4me1/3 and H3K27ac, but not H3K36me3, H3K9me3 and H3K27me3 (Extended Data Figure 3a, b). We thus re-performed the analysis using only these three active marks, which identified four clusters (Methods; Figure 4a). Cluster 1 was enriched in clade A9-infected tumors (q-value=4.9×10−5), while Cluster 2 included an equal number of tumors infected by clades A7 (mostly HPV45) and A9. The remaining clade A7-infected tumors were found in Clusters 3 and 4, which was enriched for non-SCC tumors (Fisher exact test, p-value=4.4×10−6). None of the clusters were enriched for somatic mutations in chromatin modifying complex members, although Cluster 4 conspicuously lacked alterations in SEC, NURD, HDAC and ISWI complex members (Figure 4a and Extended Data Figure 3a).
Figure 4: Clade-specific histone mark landscapes.
a. Cluster of clusters analysis for 27 consensus clustering solutions for 3 active marks on 52 samples (k=2–10 for each mark). HIV status, histology, HPV type, clade, tumor grade, stage, DNA methylation cluster and mutation status of genes in epigenetic modifier complexes are annotated. q-values for each variable were determined using Benjamini-Hochberg corrected Fisher exact tests. b. Overlap of H3K27ac, H3K4me3 and H3K4me1 peaks significantly enriched in each clade. c. Number of H3K4me1-marked enhancers at primed (H3K4me1-only) regions (left) or active (H3K4me1/H4K27ac) regions (right). The samples are divided by mutation status of chromatin modifiers genes. Boxplots represent the median, upper and lower quartiles of the distribution and whiskers represent the limits of the distribution (1.5-times interquartile range). P-values were calculated using Wilcoxon rank sum tests. d and e. Fold changes of histone mark abundance and gene expression between clades associated with TSS of genes found between −5/+20kb of intersecting H3K4me3 and H3K27ac peaks and (e) between −20/+20kb from differential H3K4me1 peaks. Sample Ns used for differential analyses (and derivation of adjusted p-values) were: expression A7=52, A9=57; H3K4me3, H3K27ac, and H3K4me1 A7=25, A9=22. Genes with BH-adjusted p-values<0.05 (DESeq2, Methods) are highlighted. f. Example of differential active histone marks (H3K4me3, H3K27ac and H3K4me1) near PXDN (left) and TMPRSS11A (right), differentially expressed between clades. All statistical tests were two-sided.
With HPV clade-specific differences observed, we assessed whether clades were associated with differential abundance42 of histone marks at regulatory regions, including active promoters (H3K27ac and H3K4me3) and enhancers (H3K4me1). We identified differential abundance of 15,245 H3K4me1 peaks, 9,902 H3K27ac peaks and 7,736 H3K4me3 peaks (adjusted p-value<0.01, fold change>2) between clades, after normalizing for histology differences (Methods, Figure 4b, Supplementary Table 12).
Clade A7-infected samples had three times more H3K4me1 enriched regions (11,530 clade A7 vs. 3,715 clade A9) and approximately double the number of H3K27ac enriched regions compared to A9-infected samples (6,405 clade A7 vs. 3,497 clade A9), suggesting an enrichment for enhancers. In contrast, clade A9-infected tumors had almost twice the number of differential H3K4me3 peaks (4,997 clade A9 vs. 2,739 clade A7) and a quarter of them (1,271 peaks) overlapped with H3K27ac enriched regions (chi-square p-value<2.2×10−16, Figure 4b), indicating enrichment for promoter marks in clade A9.
The two most frequently mutated chromatin modifying genes (CMGs) in our cohort, KMT2C/D, deposit H3K4me1 at enhancer regions43–46. We therefore sought to investigate the impact of KMT2C/D loss-of-function (LOF) mutations on the number of H3K4me1-marked regions. We observed that, for the 15 samples with KMT2C/D LOF mutations, the number of primed enhancer regions (H3K4me1 only) was lower than in tumors with no CMG mutations or with other CMG mutations, while there was no difference in the number of active enhancer regions (H3K4me1 and H3K27ac) (Methods, Figure 4c). Despite different peak enrichments between clades (Figure 4b), the effect of KMT2C/D mutations on primed enhancers was not clade-specific.
To relate mark profiles to differential expression, we mapped the H3K27ac and H3K4me3 differential peak regions to nearby genes (Methods; Supplementary Table 13). This identified 769 A7-enriched differential H3K27ac/H3K4me3 regions, of which 18 were near the TSS of clade A7-upregulated genes, including the invasion and extracellular matrix genes SRCRB4D, PXDN and CXCL2 (Figure 4d and Extended Data Figure 3c). We also identified 1,271 A9-enriched differential H3K27ac/H3K4me3 regions, 25 of which were near the TSS of clade A9-upregulated genes (Figure 4d and Extended Data Figure 3c) including TMPRSS11A, WNT2B, and MEI1. We similarly observed clade-specific correlations between differential H3K4me1 marked regions and expression of the nearest gene (Figure 4e). Relationships between histone modification and gene expression differences between clades are displayed in Figure 4f and Extended Data Figure 3d, using the genes PXDN and TMPRSS11A as examples.
Our results thus indicate that DNA methylation (Figure 3c), and epigenetic modification patterns attributed to H3K27ac, H3K4me3 and H3K4me1 (Figure 4b) are altered in an HPV clade-specific manner in our cohort.
Altered RNA and histone profiles at HPV integration sites
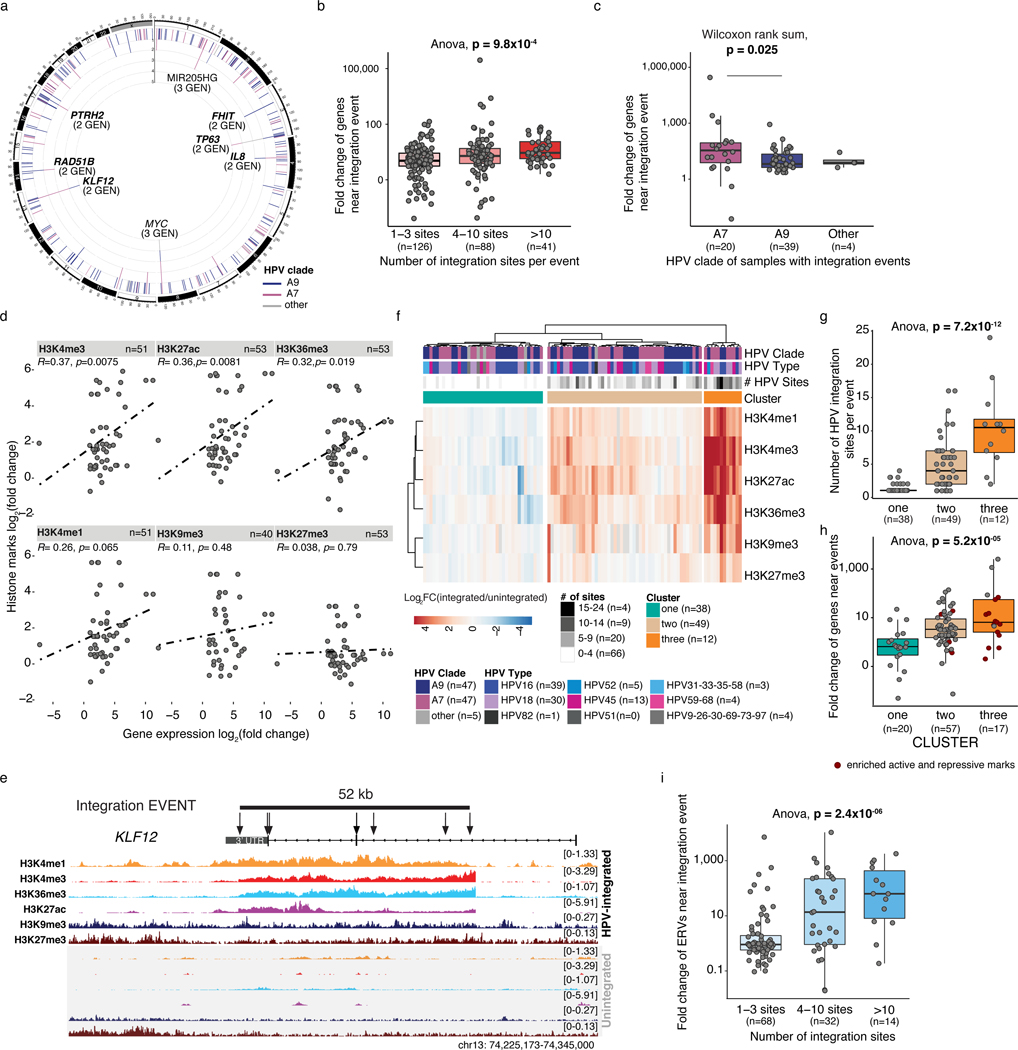
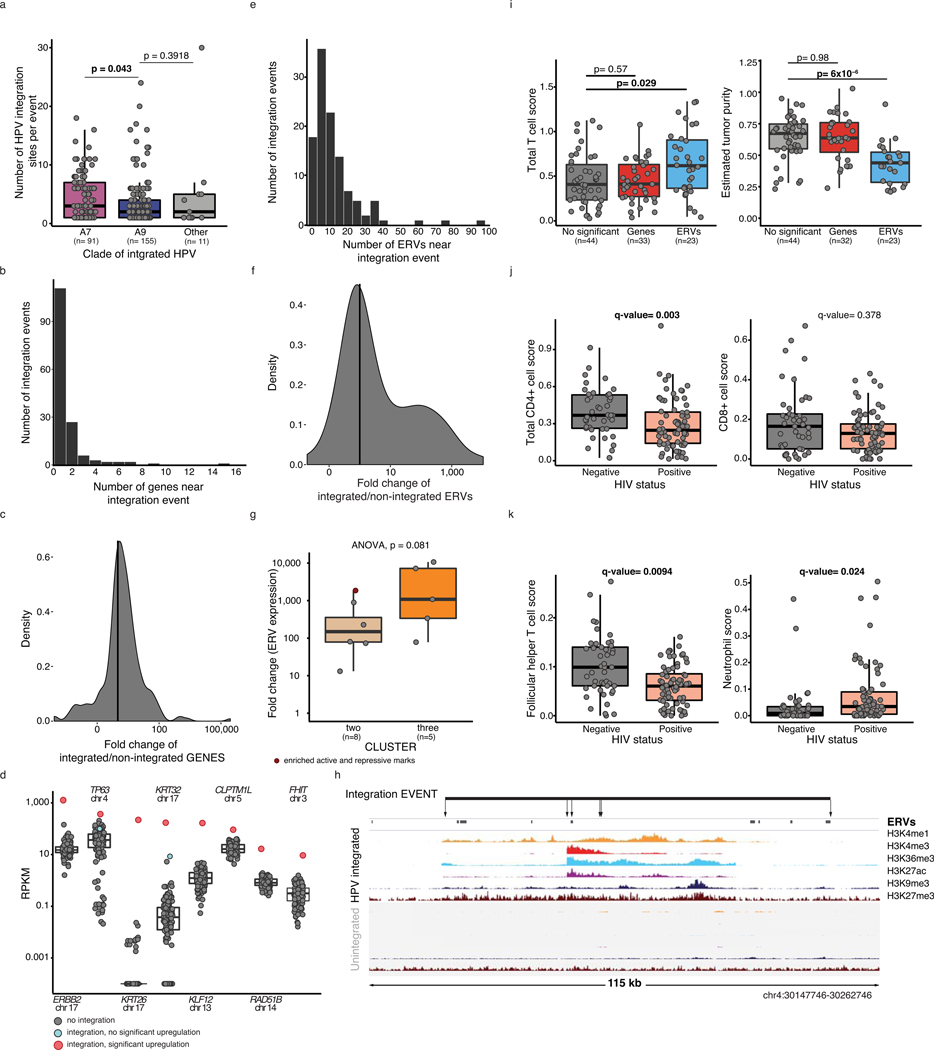
We studied the genomic impacts of 1,010 unique HPV integration sites in 109 of the 118 tumors (Supplementary Table 2). Grouping of integration sites near one another (<500 kb) within samples resulted in the identification of 257 “integration events” (median length: 2.6 kb, range: 1 bp - 409 kb; (Methods, Supplementary Table 14). Clade A7 integration events contained more integration sites per event than clade A9 events (Wilcoxon, p-value=0.043, Extended Data Figure 4a).
Of these events, 155 (60%) were within 10 kb of one or more genes (Methods, Extended Data Figure 4b). KLF12, TP63, RAD51B and MYC were among 16 genes that were the closest in proximity to an integration event in multiple samples, as previously reported12,47 (Figure 5a and Supplementary Table 15). Of the genes nearby integration events, 61 (from 45 events) displayed significantly higher expression in samples with integration (fold change ≥2 adjusted p-value≤ 0.05, Extended Data Figure 4c, Supplementary Table 15, Methods). Furthermore, the events containing a higher number of integration sites were associated with increased expression fold change (ANOVA, p-value=9.8×10−4; Figure 5b). Clade A7 integration events appeared to have a more pronounced effect on expression than clade A9 events (Wilcoxon, p-value=0.025, Figure 5c), which may result from the higher number of integration sites per event in this clade. Of the 15 genes identified in multiple samples near integration events (Figure 5a), eight were significantly upregulated in integrated samples (Extended Data Figure 4d), including the oncogenes ERBB2 (69-fold, adjusted p-value=0.033) and TP63 (8.3-fold, adjusted p-value=0.033).
Figure 5: HPV integration alters local histone modifications and expression.
a. HPV integration events in 109 samples collapsed into frequent regions within 500 kb of one another. The number of integrations, colored by clade, are presented radially. The number of unique genes closest to integration events are labeled (GEN), and upregulated genes are highlighted (bold). b and c. Fold change of genes nearby integration events, by the number of integration sites per event (b) and clade (c). d. Fold change of local gene expression and histone mark coverage at events. Statistics are determined using Spearman tests. e. Histone marks coverage of KLF12 3’ region. Arrows indicate individual integration sites within the event (line). Top tracks show a sample with an event in this region, and bottom tracks show a sample without. f. Unsupervised clustering of fold changes of histone mark coverage at integration events (n=99). g. Number of integration sites per event in each cluster (f). h. Fold change of genes near events by clusters (f). i. Fold change of ERVs near events by the number of sites within the event. All fold-changes refer to the integrated sample (n=1) vs. the cohort (n=117 expression, n=46 H3K9me3, n=50 H3K4me1/3, n=51 H3K27ac/me3 and H3K36me3). Boxplots in b, c, g, h and i represent the median, upper and lower quartiles of the distribution and whiskers represent the limits of the distribution (1.5-times interquartile range). Where relevant, all statistical tests were two-sided.
To explore possible epigenomic mechanisms of altered gene expression at HPV integration events, we examined the fold change in histone mark enrichment within integration events (Methods). Fold changes in histone mark enrichment of H3K27ac, H3K4me3, H3K4me1 and H3K36me3 were positively correlated with gene expression changes (Figure 5d). In our unsupervised clustering analyses, we noted that increased H3K36me3, typically associated with transcription48,49, was associated with local transcriptional dysregulation at integration events but not global dysregulation (Extended Data Figure 3b). The KLF12 3’-region provides a visual example, highlighting the relationship between an HPV integration event, altered histone modifications (mean log2 fold change= 2.8 for H3K27ac, H3K4me3, H3K4me1, H3K36me3, Figure 5e) and increased gene expression (fold change=112, adjusted p-value=0.033, Methods, Extended Data Figure 4d).
Unsupervised clustering of the histone modification fold changes near integration events identified three clusters with varying levels of histone mark enrichment (Figure 5f, Supplementary Table 16, 17). Cluster 3 contained 12 events with increased coverage of H3K27ac, H3K36me3, H3K4me3 and H3K4me1. Seven of these also had significant enrichment of the repressive modifications H3K9me3 and H3K27me3. Cluster 2 included 49 events with a lower degree of enrichment of all active histone modifications than Cluster 3 (14/49 vs. 12/12 events), while Cluster 1 included events with no enrichment of all active histone modifications (0/38 events). The average number of HPV integration sites per event was significantly different between the three clusters, with Cluster 3 having the highest (ANOVA, p-value=7.2×10−12, Figure 5g). Consistent with our observation that a higher number of HPV integration sites was associated with increased expression of nearby genes (Figure 5b), events in Cluster 3 were associated with genes exhibiting the highest increases in expression (ANOVA, p-value=5.2×10−5, Figure 5h). Events enriched for active and repressive marks appeared to have a dampened upregulation of local genes compared to those without increased repressive marks (Wilcoxon, p-value=0.04, Figure 5h, n=18 active and repressive, n=76 active only).
Thirty-two percent (32/99) of integration events in samples with available ChIP data were not within 10 kb of a protein coding gene, but were associated with locally altered histone modification patterns (clusters 2 and 3). We thus sought other genomic features potentially influenced by these events, identifying endogenous retroviral sequences (ERVs) near 114/257 (44%) of them. ERVs are epigenetically silenced in the human genome and their reactivation is associated with induction of antiviral pathways, such as double stranded RNA (dsRNA) response signaling50. We analysed the expression of ERVs near integration events and observed that their expression was significantly higher in the integrated samples and was also positively correlated with the number of HPV insertions within the event (Figure 5i, Supplementary Table 18). As with genes, upregulation of ERVs near integration events was associated with histone modification changes (ANOVA, p-value=0.081, Extended Data Figure 4g, h).
To explore the tumor microenvironments and to determine whether increased ERV expression was associated with increased immune cell presence, as described previously50, we inferred immune expression scores using RNA-Seq51(Methods). Samples with upregulated ERVs at integration events had higher total T-cell scores (Extended Data figure 4i, left) but, due to the lower estimated tumor content for these cases (Methods, Extended Data Figure 4i, right) and the inverse correlation of tumor content and total immune scores in our samples, we could not confidently assess the relationship between ERV upregulation and immune cell abundance. We also examined the expression of genes in pathways involved in ERV recognition, including type I interferon signalling (GO:0060337) and dsRNA sensing pathways (GO:0043330), but did not observe increased expression of such genes in samples with an ERV integration event, nor did we observe evidence of immune escape through point mutation, deletion or methylation of these genes.
As HIV infection targets CD4+ T cells, we compared CD4+ T cell scores between samples from HIV+ and HIV- patients. Total CD4+ T cell scores were lower in HIV+ than HIV- samples (Wilcoxon, p-value=0.0030, Extended Data Figure 4j), particularly for follicular helper T-cell scores (Wilcoxon, Benjamini-Hochberg corrected p-value=0.0094, Extended Data Figure 4k, left), which is consistent with HIV infection primarily affecting these cells52. The only immune score found to be higher in HIV+ samples was of neutrophils (Wilcoxon, Benjamini-Hochberg corrected p-value=0.024) (Extended Data Figure 4k, right), the role of which is unclear in the mucosal environment of the genital tract53.
These observations indicate that HPV integration sites in tumor genomes are associated with local histone modification changes that correlate with altered expression of genes and ERVs, including known oncogenes such as ERBB2, which may contribute to tumor progression.
Discussion
We characterized the genomic, transcriptomic and epigenomic landscapes of 118 cervical cancers from an understudied population of HIV+ and HIV- Ugandan patients and identified HPV clade-associated dysregulation. Large-scale genomics studies like this are important, particularly in underrepresented ethnicities, to understand molecular phenotypes of these cancers, which can lead to improved treatment options.
The composition of this cohort, including comparable representation of clade A9 and clade A7-infected samples, allowed us to describe molecular characteristics associated with clades. Clade A7-infected samples exhibited distinct gene expression patterns converging on pathways linked to the extracellular matrix and to cell adhesion and migration, indicating a more aggressive phenotype. While inferior prognosis associated with clade A7 has been previously reported in invasive cervical cancer40, our study provides insight into the cellular pathways that may promote the aggressive phenotype in these tumors. Genes upregulated in clade A7 samples, such as PXDN, are upregulated in cancers that have more potential to progress through the epithelial-mesenchymal transition54,55. DNA methylation, for which we also observed clade-specific patterns, is tightly regulated through cell differentiation56. It is therefore reasonable to hypothesize that these two HPV clades may push epithelial cells to replicate at the two ends of the epithelial differentiation spectrum, with clade A7 driving a less differentiated phenotype.
We related distinct patterns of viral gene expression to HPV clades and linked these to dysregulated genes in the tumor. The absence of E2 expression in the A7-enriched cluster supports the current understanding that HPV18-infected tumors (clade A7) are always associated with integration, which leads to loss of E2 expression57. Conversely, only about 76% of HPV16-infected cervical tumors (clade A9) show evidence of HPV integration11, supporting the presumed presence of episomal HPV DNA due to the persistent expression of the E2 gene observed in our samples. The presence of episomal HPV is associated with epithelial differentiation and active HPV infection58. This higher expression of episomal HPV genes in clade A9-infected samples further supports the hypothesis that molecular characteristics associated with clade A9 indicate more epithelial differentiation than clade A7-infected samples, and may have more active HPV infection.
We found that HPV clades exhibit distinct histone modification profiles. HPV viral proteins have been reported to interact with different epigenetic modifiers including CREBBP, CHD4, KAT2B, EP300, SNW159, however clade-specific interactions that may explain our epigenomic changes at distinct genomic regions remain unexplored. The high frequency of samples with mutations in chromatin modifiers in our cohort (87%) may suggest a mechanism beyond simple transcriptome dysregulation, perhaps encompassing variation in chromatin accessibility or three-dimensional chromatin structure that could promote HPV infection or cancer progression14. Such ideas await future studies.
Enrichment of active histone marks in close proximity to HPV integration events was associated with increased expression of nearby genes/ERVs. In our study, we cannot distinguish between the possibility that we are observing several distinct HPV integrations in similar regions in different cells within the same tumor (type 1 integrations57,60), or multiple HPV integrations in tandem within single cells (type 2 integration57,60). However, the increased upregulation of genes/ERVs in tumors with a higher number of integration sites per event suggests that it may be the latter. Local alterations associated with HPV integration events may also result from focal amplification of the integrated viral genome and neighboring regions in the human genome61. These amplifications may support the enhanced recruitment of chromatin modifiers to viral regulatory regions61 contributing to increased histone modifications in the regions surrounding HPV integration.
Online Methods
Ethical compliance, consent and cohort enrolment
The study was approved by Fred Hutchinson Cancer Research Center Institutional review board (#7662) and complies with ethical regulation. Accrual received institutional and governmental approval, and informed consent was obtained from all patients. Approval was obtained from BC Cancer Research Ethics Board (UBC BC Cancer REB H16–02279) for molecular characterization. 212 patients were enrolled initially and split into discovery (n=123) and extension (n=89) groups before further exclusions.
Pathology and molecular review
Formalin-fixed, paraffin-embedded (FFPE) tumor blocks or unstained sections were submitted for histopathological review by the Uganda Cancer Institute (Kampala) to the University of California, San Francisco (San Francisco, CA). Hematoxylin and Eosin (H&E) and p16 immunohistochemistry (IHC) slides sent to Nationwide Children’s Hospital (Columbus, OH) were imaged at 40X using an Aperio scanner, and assigned to 3 pathologists (M.H.S, T.C.W, and T.M.D) for consensus review. Tumors were evaluated for histological type, subtype, grade, and p16 immunoreactivity.
Gene expression analysis flagged 17 cases for re-review that appeared discordant with initial H&E diagnosis of p16 positive poorly differentiated SCC. Twelve cases were re-queried using IHC stains for p63 and p40 (SCC), and also BER-EP4, MOC 31, and B72.3 (adenocarcinoma). For two of these, neuroendocrine markers (synaptophysin and chromogranin) were also performed, leading to a revised histological classification of 2 samples as adenocarcinoma, 7 as adenosquamous carcinoma, 2 as neuroendocrine carcinoma, and 1 as an undifferentiated carcinoma. Of the re-reviewed cases, five, negative for high-risk HPV by BBT27, were excluded from the final discovery cohort (n=118). Of these, four were uterine primaries and one an equivocal cervical/uterine primary.
A manual of Standard Operating Procedures for NCI’s Office of Cancer Genomics Cancer Genome Characterization Initiative are provided: https://ocg.cancer.gov/sites/default/files/HTMCP_SOP_manual.pdf
Clinical data and survival analyses
Clinical data, including overall survival was obtained from https://cgci-data.nci.nih.gov/PreRelease/HTMCP-CC in January 2020. To mitigate clinical bias, we excluded patients that had not received a therapeutic intervention (n=83), as these had more advanced disease (p=0.01 chi squared), poorer prognosis (p=0.00082, log-rank test) and would not have been followed with curative intent. To account for prognostic differences between histologies, the analyses described only assess survival differences in a subset of these patients with SSCs (n=66). Hazard ratios and p-values were determined using log-rank tests.
Whole genome sequencing library construction
PCR-free whole genome sequencing libraries were constructed using the TruSeq DNA PCR-free kit (E6875–6877B-GSC, New England Biolabs), automated on a Microlab NIMBUS liquid handling robot (Hamilton), as previously described62. Libraries were purified using paramagnetic (Aline Biosciences) beads and prior to sequencing, concentrations were quantified using a qPCR Library Quantification kit (KAPA, KK4824).
Genome library construction for custom capture
DNA (500 ng) was sonicated (Covaris) to 250–350 bp, purified using PCRClean DX magnetic beads (Aline Biosciences), end-repaired, phosphorylated and bead purified before A-tailing using a custom NEB Paired-End Sample Prep Premix Kit. Illumina sequencing adapters were ligated overnight at 16oC, bead purified and enriched with 6 cycles of PCR using indexed primers enabling library pooling and sequenced using paired-end 125 base reads in a single flowcell lane.
PolyA RNA library construction
Polyadenylated (PolyA) mRNA was purified from total RNA and cDNA was synthesized as previously described62.
Native chromatin immunoprecipitation (ChIP) sequencing
Fifty-two tumor samples were lysed in 0.1% Triton X-100, 0.1% Deoxycholate buffer plus protease inhibitors (PI). Extracted chromatin was digested with micrococcal nuclease (MNase) enzyme (NEB) and the reaction quenched using 250 μM of EDTA. 1% Triton X-100 and 1% Deoxycholate were mixed and added to the samples on ice. 4% of digested chromatin was used as input control, the remaining was pre-cleared with Protein A/G Dynabeads (Invitrogen) in IP buffer (20 mM Tris-HCl [pH7.5], 2 mM EDTA, 150 mM NaCl, 0.1% Triton X-100, 0.1% Deoxycholate, PI) at 4oC for 1.5 hours. Supernatants were transferred to a 96-well plate containing the antibody-bead complex, and incubated overnight at 4oC with agitation. Immunoprecipitated samples were washed twice with low salt buffer (20 mM Tris-HCl [pH 8.0], 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl) and twice with high salt buffer (same, but with 500 mM NaCl). DNA-antibody complexes were eluted in Elution Buffer (100 mM NaHCO3, 1% SDS), at 65°C for 1.5 hours with mixing (1350 rpm). Qiagen Protease was used to digest protein in the eluted DNA at 50°C for 30 minutes with mixing (600 rpm). ChIP DNA was purified using Sera-Mag beads (Fisher Scientific) with 30% PEG before library construction as described for custom capture.
Amplified libraries were purified as described above (ALINE Biosciences) and the DNA quality and quantity determined using Caliper LabChip GX DNA High Sensitivity assay (PerkinElmer) and the Quant-iT dsDNA high sensitivity assay (ThermoFisher Scientific).
Whole genome, transcriptome and ChIP sequencing
Tumor genomes were sequenced to a target depth of 80X coverage, and normal blood samples to 40X coverage using 125bp reads. Transcriptomes were sequenced using 75bp paired-end reads. ChIP libraries were normalized and pooled before sequencing. All sequencing was performed on an Illumina HiSeq2500.
Estimation of tumor content
Tumor purity and ploidy were estimated using Ploidetect (https://github.com/lculibrk/ploidetect). Tumor reads and heterozygous SNP allele frequencies in non-overlapping bins (~100 kb and equal coverage in the matched normal samples) were computed for each case. Read counts were modelled using Gaussian mixture models (GMM), modified to restrict component means as a fixed depth apart and component variances to be equal to one another. Allele frequencies were modelled using a separate GMM, incorporating priors from the first. Models were generated for each possible value of tumor purity and scored based on the mean likelihood of both the depth and the allele frequency GMMs. All results were verified by review of GMM parameters and their fit to the data. Estimates were congruent with observed copy number data in 104 out of 118 samples. In the remaining cases, purity and ploidy were determined by review of alternate models.
Somatic alteration detection
Tumor and normal sequencing reads were aligned to the human reference genome (hg19) using BWA-MEM v0.7.6a63). Read duplicates were marked using sambamba64(v0.5.5). Somatic single nucleotide variants (SNVs) were identified using Strelka (v1.0.6)65. A panel of 2,735 genes including mutated oncogenes, tumor suppressors, epigenetic modifiers, splicing factors, or other genes recurrently mutated (≥3 cases) in this cohort, were selected for targeting sequencing in the extended cohort. The coding mutation rate was reported for each tumor as the number of coding SNVs (low, moderate or high SNPeff annotation66) per Mb.
Custom capture validation of SNVs
DNA from the 89 extension libraries were pooled prior to hybridization capture of 2,735 target genes using SureSelect XT custom probes (Agilent) and RNA probes at 65°C for 24 hours. Streptavidin-coated magnetic beads (Dynal, MyOne) were used for custom capture, followed by purification on MinElute columns (Qiagen) and enrichment with 10 PCR cycles using primers that maintain library-specific indices. Pooled libraries were sequenced generating 125bp paired-end reads. To capture the KMT2D gene and non-coding hotspots, 544 120bp xGen Lockdown probes were designed and synthesized (Integrated DNA Technologies) for targeted capture sequencing as above.
Significantly mutated genes (SMGs)
Significantly mutated genes were identified using MutSig2CV (https://software.broadinstitute.org/cancer/cga/mutsig) as previously described62.
Expression profiling
RNA-Seq reads were aligned to the human reference genome (hg19) and converted to RPKMs (reads per kilobase per million mapped reads) as described previously67.
ChIP-Seq alignment and peak calling
ChIP sequence reads (75nt) were aligned to the human reference genome (hg19) with BWA-MEM63 (v0.7.6a, parameters: -M). Read duplicates were marked using sambamba64 (v0.5.5). Forty-seven samples had all 6 histone marks (4 broad: H3K4me1, H3K9me3, H3K27me3, H3K36me3 and 2 narrow: H3K4me3 and H3K27ac), and 5 had a subset of these.
Peaks were called using MACS2 (v2.1.1)68 with default parameters, comparing each mark to its control. Bedgraph output files were converted to the library size-normalized bigWig format for manual inspection using the UCSC and IGV genome browsers69,70.
ChIP-seq data quality was assessed using Encode guidelines71. Samples had a minimum of 50 million sequenced reads for narrow marks and 100 million for broad marks. The percentage of uniquely mapped reads was above 70%, and the percentage of duplicated reads varied between 1 and 10%. The non-redundant fraction, fraction of reads in peaks (FRIP) and sequencing saturation using preseq v2.0.2 (https://github.com/smithlabcode/preseq) were also assessed.
HPV typing and expression
Microbial detection, HPV typing and HPV integration detection were performed using BioBloom tools (BBT, v2.0.11b)27. Where 2 or more HPV types were integrated (n=3), the dominant type was determined by E6/E7 expression. Where no integration was found (n=9), the dominant HPV was determined by the type with the most read evidence.
To determine expression of HPV genes, fasta genome references and gff annotation files were downloaded from NCBI for 16 HPV strains. HPV51 did not have a gff file so one infected sample was excluded (samples with HPV expression n=117). Samples were aligned to their HPV strain using BWA-mem v0.7.6a Sambamba. The fraction of reads with sequencing quality greater than Q10 within gene boundaries were counted and normalized to reads per kilobase of exons per million reads mapped to HPV (RPKM).
Mutation signatures and HRD score
SNVs were categorized into 96 mutation classes based on 6 variant types and 16 trinucleotide contexts. For each sample, values of the 96 classes were used to compute a non-negative least squares deconvolution based on 30 previously described mutational signatures (COSMIC)16,17. The APOBEC signature reported for each sample is the max exposure value of signature 2 or 13.
HRD scores were computed using the HRDtools (v0.0.0.9, R), as previously described72.
Copy ratio landscape comparisons
Copy number alterations (CNAs) between cohorts were called and analysed using GATK473 (v4.0.9, https://gatkforums.broadinstitute.org/gatk/discussion/9143/how-to-call-somatic-copy-number-variants-using-gatk4-cnv) and GISTIC2.074 (v2.0.17). Genomic intervals were prepared by dividing the reference genome into equally sized bins (1000bp). A panel of normals was generated to median sample reference counts. Allele counts were collected independently for the tumor and matching normal. Continuous segments were modelled with both the allelic ratios and copy ratios.
Germline CNAs previously identified in the TCGA cervical cancer (CESC) study11 were filtered out to remove any potential germline CNVs in this cohort. Segments were excluded if 75% or more of the segment overlapped with these.
Somatic copy number alterations in TCGA CESC tumors were determined previously using SNP 6.0 arrays11 and these were downloaded from the Broad GDAC website. The 178 samples in TCGA CESC core set were used for comparison to our HIV- samples.
To determine regions of CNA variance between cohorts (HIV- vs. HIV+, HIV- vs. TCGA), analyses were performed on each cohort separately according to the GISTIC2 (v2.0.22) documentation (http://software.broadinstitute.org/cancer/software/genepattern/modules/docs/GISTIC_2.0) with the parameters -qvt 0.25, -genegistic 1, -broad 1, -brlen 0.5, -conf 0.99, -armpeel 1, -savegene 1, -gcm extreme, -maxseg 3000. The genome was binned into 1kb segments and the fraction of patients having a copy gain (>0.1) or loss (<−0.1) was calculated based on the mean segment values for each cohort. Significantly amplified and deleted chromosome arms were identified using an FDR threshold < 0.25. Unique or shared arms and cytobands were identified as those significant in one cohort, and not the other.
Specific copy number alterations in samples
Regions of CNA in individual samples were identified as previously described62.
Non-coding mutation hotspots
Non-coding variants annotated by SNPeff66 as 5’ Flank, 3’ Flank, IGR, 3’ UTR, Intron, 5’UTR, RNA, Splice_Site, Translation_Start_Site were used as input to Rainstorm75, with all parameters set at default values (k=4).
Of the 3,094 hotspot regions identified, we focused on 3,539 potential point mutation hotspots, present in 3 or more samples. These were filtered for those called by both Strelka and MutationSeq76 and did not reside in centromeric regions. Further filtering removed any variant called in a normal sample, reducing the potential non-coding hotspots to 404, of which 7 (high confidence) were confirmed by manual review.
Hotspots were annotated as ‘potential promoter’, ‘potential enhancer’, or ‘intergenic’ using ChIP-seq data (enhancer= intersect of H3K4me1 and H3K27ac peaks, promoter= H3K4me3).
We assessed the potential for other non-coding hotspots to alter transcription factor binding using motifBreakR26.
DNA methylation analysis
Human DNA methylation using the EPIC array (Illumina) was performed by The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada. The DNA methylation beta-values for 115 samples were binarized as unmethylated (beta<=0.25) and methylated (beta>0.25). The 8,000 most variable probes were clustered using the ConsensusClusterPlus77 (v1.38.0, R) with a ‘binary’ distance and ‘ward.D2’ clustering method with 1,000 iterations.
Differentially methylated probes (DMP) and differentially methylated regions (DMR) between clade A7- and A9-infected samples were determined using CHAMP31,32(v2.10.2, R) (q<0.05); DMRs used the ‘bumphunter’ method. For the DMPs, associated gene, genomic feature, and CpG island features of these probes came from CHAMP31,32. DMRs were intersected with protein-coding genes (hg19 Ensembl (v75), n=20,232) using bedtools (v2.27.1)78.
Human and viral gene expression and gene ontology enrichment analyses
Clustering analysis was performed with ConsenusClusterPlus77(v1.38.0, R) using log10(RPKM) values using the ‘Pearson’ method and ‘ward.D2’ linkage with 1000 iterations. Human genes used included the top 1,000 most variable genes (RPKM > 5 in at least one patient). All 118 samples were included in human gene clustering and 117 in viral gene clustering (no gff file was available for HPV51).
Differential gene expression between groups (A7 vs. A9; E6 and E7 high vs. low) was performed using the DESeq2 (v1.14.1, R)33. Genes were filtered using an adjusted p-value <0.05, >1.5-fold change in mean expression, and a baseMean expression >1000. For the A7 vs. A9 comparison, the differential analysis was normalized for histology using a multifactorial approach. Results from the normalized analysis were compared to those using only squamous A7 and A9 samples to ensure histology correction was only removing expression differences attributed to histologies (89% concordance).
Functional enrichment of the significantly differentially expressed genes in the A7 vs. A9 comparison was performed using STRING (v11.0)36. For visualization, enrichment scores for A7-enriched ontologies were set to negative values. Functional enrichment of the significantly differentially expressed upregulated and downregulated gene lists for the E6 and E7 analysis was performed using HOMER (v4.10.3)79.
ChIP clustering analyses
The union of peaks for each histone mark was found by concatenating peak files and merging overlapping regions using bedtools v2.27.178. The normalized coverage of each sample in the peak union was counted using deeptools (v3.0.2)80. For each mark, the top 1% most variable peaks were clustered using the ConsensusClusterPlus (v1.38.0, R) using the ‘pearson’ distance and ‘complete’ clustering method with 1000 iterations for k=2–10 clusters. The 54 consensus clustering solutions (6 marks x 9 solutions) were then analysed using a Cluster of Clusters Analysis (COCA)41. For active marks, pairwise probabilities were generated for 27 solutions (3 marks x 9 solutions). For marks in which some samples had missing data, pairwise comparisons were normalized to exclude samples in those marks. Matrices of probabilities (54×52, 27×52) were clustered using pheatmap (v1.0.10, R) with the ‘pearson’ distance and ‘complete’ clustering method.
H3K4me3, H3K27ac and H3K4me1 peaks differentially present between HPV clades (A7 vs. A9) were determined using DiffBind with DESeq2 method (v2.2.12, R, FDR<0.01, fold-change>2)42. Coverage at peaks was counted 500bp around the centre of the peak, and a multifactorial experimental design was performed to normalize histology differences (referred to as blocking factor). Significantly differential peaks were intersected using bedtools (v2.27.1)78. Associated genes were identified using the nearest transcription start site (TSS) to the differential H3K4me1 and intersected H3K4me3/H3K27ac regions, identified using bedtools78 (v2.27.1) to RefSeq’s hg19 annotation.
H3K4me1-marked enhancer regions
H3K4me1-marked enhancer regions were selected from the union of H3K4me1 peaks81. This union was overlapped in each sample with H3K4me1 and H3K27ac to identify primed (H3K4me1) and active (H3K4me1+H3K27ac) regions, excluding those overlapping with H3K27me3. To eliminate regions marking promoters, they were filtered for a median H3K4me1:H3K4me3 ratio coverage >1 and >2000 bp from a TSS (n=324,447 regions).
HPV integration events and ChIP
HPV integration sites were determined using chimeric reads mapping to both human and HPV genomes. Within each sample, integration sites were merged into a single integration event (n=257) if they were <500 kb apart. HPV integration hotspots were determined by counting the number of events that fell within a 500 kb bin across the genome.
ChIP-seq alterations at HPV events were clustered using the log2(fold-change) of normalized coverage (RPM) of the integrated sample versus the mean RPM of the unintegrated samples using the ‘pheatmap’ (v1.0.10, R) with a ‘ward.D2’ clustering method. Events <20 kb were extended to 20 kb to obtain adequate coverage of the region.
For each mark per event (6 modifications in 99 events), a control peakset was made by randomly selecting 1,000 peak regions of the same mark on the same chromosome as the event, and extending the peaks from the center to the same size as the event. Normalized ChIP-seq coverage of the histone modification at these 1,000 random peaks was counted in the 52 samples, and the log2(fold change) of coverage was calculated for the integrated sample. A p-value was calculated for the fold change of the integration event based on the distribution of fold changes in these control peaks. Benjamini-Hochberg adjusted p-values<0.05 was regarded as significant.
HPV integration events and expression
For each integration event (n=257), we identified all protein-coding genes (hg19 Ensembl (v75), n=20,232) that fell within the event +/−10 kb37, which revealed 255 genes near integration events. Fold changes of integrated samples were calculated based on the mean expression of all samples lacking events, and p-values were derived from the distribution of expression of the gene across all samples. Oncogenes were identified by OncoKB82. The same method was applied to identify ERVs upregulated at HPV integration events (n=34 events). Samples were labelled as having a statistically significant integration event if they had a fold change≥2 and Benjamini-Hochberg adjusted p-value≤0.05 for genes, and fold change≥10 and Benjamini-Hochberg adjusted p-value≤0.05 for ERVs based on the distribution of fold changes for each respectively (Extended Data Figure 4). Samples with significant events were correlated with T cell infiltration scores from CIBERSORT51, and genes from the gene ontologies for dsRNA sensing pathways (GO:0043330) and type I interferon signalling (GO:0060337).
ERV quantification
5,467,457 repeat elements and hg19 coordinates (chromosomes 1–22, X) were downloaded from RepeatMasker Open v.4.0.5 (http://www.repeatmasker.org/faq.htm). To minimize read count bias from nearby expressed protein-coding genes, we filtered for ERVs >10 kb away from their nearest gene. Raw expression values were calculated by counting the number of reads that mapped unambiguously (mates mapped within 10 kb) to each region and were normalized for sequencing depth and length by conversion to RPKMs.
Estimation of immune cell content
CIBERSORT (v1.0.4)51 was used to quantify leukocyte expression signatures on the expression RPKMs as previously described62. Total CD4+ T-cell content is the sum of the following cells; naive, memory resting, memory activated, follicular helper and regulatory T cells.
Visualization
All heatmaps were visualized using pheatmap (v1.0.10, R).
Statistical analyses
No sample sizes were predetermined. Unless otherwise stated all statistical tests reflect two-sided tests. P-value methods and multiple test correction is reported in the text. Wilcoxon in the text refers to the Wilcoxon rank sum test.
Reporting Summary
Further information on research design and methods is available in the Life Science Reporting Summary linked to this article.
Data Availability
All molecular and clinical data used in this publication can be found on the National Cancer Institute’s Genome Data Commons Publication Page https://gdc.cancer.gov/about-data/publications/CGCI-HTMCP-CC-2020 . Data from this publication is publicly available for download through dbGaP (phs000528), as part of the NCI Cancer Genome Characterization initiative (CGCI, phs000235). Sample metadata is reported in Supplementary Table 2. Source data for all Figures and Extended Data Figures are presented with the paper.
TCGA cervical cancer data (file name: CESC.snp__genome_wide_snp_6__broad_mit_edu__Level_3__segmented_scna_minus_germline_cnv_hg19__seg.seg.txt) was obtained from http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/CESC/20160128/.
Code availability
Bioinformatics analyses in this study were conducted using open-source software, with the exception of tumor purity and ploidy estimation using Ploidetect (https://github.com/lculibrk/ploidetect).
Extended Data
Extended Data Fig. 1. Additional characteristics of the HTMCP discovery and extension cohorts.
a. Coding mutations per Mb in samples exhibiting low (≤ 0.4) and high (> 0.4) APOBEC signatures. b. Difference in PIK3CA expression by HIV status. c. Mutations in the top 20 most mutated epigenetic modifiers, ordered by frequency of alterations for the cohort (n=118). APOBEC signature proportion and homologous recombination deficiency (HRD) scores are reported above. HIV status, age at diagnosis, tumor histology (”other” includes neuroendocrine and undifferentiated) and stage are also annotated. d. Comparison of mutation frequencies of the 12 SMGs in the discovery vs. extension cohorts. Boxplots in a and b represent the median, upper and lower quartiles of the distribution and whiskers represent the limits of the distribution (1.5-times interquartile range), and statistics were determined using two-sided Wilcoxon rank sum tests.
Extended Data Fig. 2. Additional characteristics of the HTMCP discovery and extension cohorts.
a. HPV types in our cohort separated by HIV status (n=72 positive samples, n=45 negative), and clade. The x axis indicates the percentage of samples in that cohort infected by the indicated HPV type, and in brackets is the number of samples. b. Unsupervised clustering of the top 1,000 most variable genes across our cohort (n=118 samples). q-values were determined using Benjamini-Hochberg (BH) corrected Fisher exact tests. c. Percentage of differentially methylated probes between clades (A7=51 samples, A9=56 samples) at different genomic features, by HPV clade. d. Log2 fold change and adjusted (BH) p-value of differentially expressed genes between clade A7- (n=52) and A9-infected (n=57) samples. e. Volcano plots showing the log2 fold change and adjusted p-value (BH) of differentially expressed genes between clade A7- (n=52) and A9-infected (n=57) samples associated with A9 hypermethylated (top), and A7 hypermethylated (bottom) differentially methylated regions (DMRs). f, g. top: Kernel density of E6 (f) and E7 (g) expression in the HTMCP cohort separates samples into high- and low expressing cases. bottom: gene ontologies enriched in differentially expressed genes in samples with low / high E6 (n=68 / n=48) (f) and E7 (n=58 / n=59) (g). h. Multivariate cox proportional hazards model for HPV clade, HIV status and disease stage for 66 patients. Hazard ratios and p-values reported for each variable were determined using log-rank tests. Where relevant, all statistical tests were two-sided.
Extended Data Fig. 3. Correlations between histone modifications and gene expression.
a. Cluster of clusters analysis for 54 consensus clustering solutions for all histone marks on 52 samples (solutions with k = 2 to 10 for each mark). The heatmap color indicates the sample probabilities in the consensus matrix. q-values for each variable were determined using Benjamini-Hochberg corrected Fisher exact tests. b. Schematic showing the cluster of clusters solution (k=5 for H3K27ac and H3K4me3 and k=4 for the other marks) for all histone marks and for the 3 active marks. Each dot represents a sample and dot color represents the cluster membership of the sample. Hollow circles indicate no available ChIP data for that sample. c. Fold change of H3K4me3 abundance and gene expression between clades associated with TSS of genes (−5/+20 kb) found at intersecting H3K4me3 and H3K27ac peaks. Sample Ns used for differential analyses (and derivation of adjusted p-values) were: expression A7=52, A9=57; H3K4me3 and H3K27ac A7=25, A9=22. Genes with BH-adjusted p-values <0.05 (DESeq, Methods) are highlighted. d. Expression of the genes reported in Figure 4.f separated by HPV clade. Boxplots represent the median, upper and lower quartiles of the distribution and whiskers represent the limits of the distribution (1.5-times interquartile range), and p-values were calculated by Wilcoxon rank sum tests. Where relevant, all statistical tests were two-sided.
Extended Data Fig. 4. HPV integration events and tumor microenvironments.
a. Number of HPV integration sites per event separated by HPV clade. b, c, e, f. Distribution of the number (b, e) and fold change in integrated samples (c, f) of genes (b, c) and ERVs (e, f) near integration events. d. Expression (RPKM) of selected genes near HPV integration events in each sample (n=118). g. Fold change of ERVs nearby integration events separated based on the clusters identified in figure 5.f. h. Histone mark coverage of a 115 kb genomic region containing ERVs. The line represents an integration event, and arrows indicate individual integration sites. Top tracks refer to a case with integration, and the bottom to a control case without integration. i. Total T-cell scores and estimated tumor content of samples with HPV integration events that are associated with significant changes in expression of ERVs or genes, and those that are not. j, k. CIBERSORT scores for all CD4+ T-cells (sum) and CD8+ T-cells (j), Follicular helper T-cells and neutrophils (k) separated by HIV status (HIV+ n=72, HIV- n=45). Boxplots in a, d, g, i, j and k represent the median, upper and lower quartiles of the distribution and whiskers represent the limits of the distribution (1.5-times interquartile range). All p-values were determined by Wilcoxon tests unless otherwise stated, and q-values were corrected using the Benjamini-Hochberg method. Where relevant, all statistical tests were two-sided.
Supplementary Material
Acknowledgements
This project has been funded in whole or in part with U.S. Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E and HHSN261201500003. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
We gratefully acknowledge the Fred Hutchinson Cancer Research Center and the Uganda Cancer Institute for overseeing sample and data collection in Uganda.
We are grateful for contributions from the other members of the HTMCP Cervical Cancer Working Group at Department of Epidemiology, University of Alabama at Birmingham, Pancreas Centre BC and various groups at Canada’s Michael Smith Genome Sciences Centre including those from the Biospecimen, Library Construction, Sequencing, Bioinformatics, Technology development, Quality Assurance, LIMS, Purchasing and Project Management teams. We thank the AIDS and Cancer Specimen Resource for logistical coordination and support of this project through NIH grants U01CA066535, U01CA096230 and UM1CA181255. L.C. and V.L.P. are the recipients of the CIHR Frederick Banting and Charles Best Canada Graduate Scholarship GSD-164207 and GSD-152374, respectively. S.J.M.J is the recipient of the Canada Research Chair in Computational Genomics.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute (R.Y.). C.C. is supported by NIH Grant Number P30 AI027757. G.B.M. is supported by NCI grants U01 CA217842 and P50 CA098258. M.A.M is the recipient of the Canada Research Chair in Genome Science. This work was supported in part by funding provided by the Canadian Institutes for Health Research (CIHR Award #FDN-143288) to M.A.M.
A.I.O. was supported in part by the Endlichhofer Trust (OCCC #3120957) and V Foundation grant (DVP2018–007).
Footnotes
Competing interests
G.B.M. reports the following potentially competing interests: SAB/Consultant: AstraZeneca, Chrysallis Biotechnology, ImmunoMET, Ionis, Lilly, PDX Pharmaceuticals, Signalchem Lifesciences, Symphogen, Tarveda, Zentalis. Stock/ Options/Financial: Catena Pharmaceuticals, ImmunoMet, SignalChem, Tarveda Licensed Technology HRD assay to Myriad Genetics, DSP patents with Nanostring Sponsored research Nanostring Center of Excellence, Ionis (Provision of tool compounds). R.Y. reports the following potentially competing interests: research support from a CRADA with Celgene/BMS. T.C.W. reports the following potentially competing interests: consultant to Roche, BD, and Inovio with respect to HPV diagnostic tests and therapeutic vaccines.
References
- 1.Bodily J & Laimins LA Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 19, 33–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Sanjose S et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. 11, 1048–1056 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Wright JD et al. Human papillomavirus type and tobacco use as predictors of survival in early stage cervical carcinoma. Gynecol. Oncol. 98, 84–91 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Yang S-H, Kong S-K, Lee S-H, Lim S-Y & Park C-Y Human papillomavirus 18 as a poor prognostic factor in stage I-IIA cervical cancer following primary surgical treatment. Obstet. Gynecol. Sci. 57, 492–500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai C-H et al. Role of Human Papillomavirus Genotype in Prognosis of Early-Stage Cervical Cancer Undergoing Primary Surgery. J. Clin. Oncol. 25, 3628–3634 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Garland SM et al. Impact and Effectiveness of the Quadrivalent Human Papillomavirus Vaccine: A Systematic Review of 10 Years of Real-world Experience. Clin. Infect. Dis. 63, 519–527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruni L et al. Global estimates of human papillomavirus vaccination coverage by region and income level: a pooled analysis. Lancet Glob. Health 4, e453–e463 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Nakisige C, Schwartz M & Ndira AO Cervical cancer screening and treatment in Uganda. Gynecol. Oncol. Rep. 20, 37–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zubizarreta EH, Fidarova E, Healy B & Rosenblatt E Need for radiotherapy in low and middle income countries – the silent crisis continues. Clin. Oncol. R. Coll. Radiol. G. B. 27, 107–114 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Ferlay J et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 144, 1941–1953 (2019). [DOI] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research Network et al. Integrated genomic and molecular characterization of cervical cancer. Nature 543, 378–384 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ojesina AI et al. Landscape of genomic alterations in cervical carcinomas. Nature 506, 371–375 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X Emerging role of mutations in epigenetic regulators including MLL2 derived from The Cancer Genome Atlas for cervical cancer. BMC Cancer 17, 252 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley DZ et al. Integrated Analysis of Whole-Genome ChIP-Seq and RNA-Seq Data of Primary Head and Neck Tumor Samples Associates HPV Integration Sites with Open Chromatin Marks. Cancer Res. 77, 6538–6550 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lleras RA et al. Unique DNA methylation loci distinguish anatomic site and HPV status in head and neck squamous cell carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 19, 5444–5455 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ & Stratton MR Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 3, 246–259 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosenthal R, McGranahan N, Herrero J, Taylor BS & Swanton C DeconstructSigs: delineating mutational processes in single tumors distinguishes DNA repair deficiencies and patterns of carcinoma evolution. Genome Biol. 17, 31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Henderson S, Chakravarthy A, Su X, Boshoff C & Fenton TR APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 7, 1833–1841 (2014). [DOI] [PubMed] [Google Scholar]
- 19.Wallace NA & Münger K The curious case of APOBEC3 activation by cancer-associated human papillomaviruses. PLoS Pathog. 14, e1006717 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H-M et al. AnimalTFDB 2.0: a resource for expression, prediction and functional study of animal transcription factors. Nucleic Acids Res. 43, D76–81 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang FW et al. Highly recurrent TERT promoter mutations in human melanoma. Science 339, 957–959 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Horn S et al. TERT promoter mutations in familial and sporadic melanoma. Science 339, 959–961 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Nik-Zainal S et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature 534, 47–54 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garinet S et al. High Prevalence of a Hotspot of Noncoding Somatic Mutations in Intron 6 of GPR126 in Bladder Cancer. Mol. Cancer Res. MCR 17, 469–475 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Wu S et al. Whole-genome sequencing identifies ADGRG6 enhancer mutations and FRS2 duplications as angiogenesis-related drivers in bladder cancer. Nat. Commun. 10, 720 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coetzee SG, Coetzee GA & Hazelett DJ motifbreakR: an R/Bioconductor package for predicting variant effects at transcription factor binding sites. Bioinforma. Oxf. Engl. 31, 3847–3849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu J et al. BioBloom tools: fast, accurate and memory-efficient host species sequence screening using bloom filters. Bioinforma. Oxf. Engl. 30, 3402–3404 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schiffman M, Clifford G & Buonaguro FM Classification of weakly carcinogenic human papillomavirus types: addressing the limits of epidemiology at the borderline. Infect. Agent. Cancer 4, 8 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maranga IO et al. HIV Infection Alters the Spectrum of HPV Subtypes Found in Cervical Smears and Carcinomas from Kenyan Women. Open Virol. J. 7, 19–27 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clifford GM et al. Effect of HIV Infection on Human Papillomavirus Types Causing Invasive Cervical Cancer in Africa. J. Acquir. Immune Defic. Syndr. 1999 73, 332–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morris TJ et al. ChAMP: 450k Chip Analysis Methylation Pipeline. Bioinforma. Oxf. Engl. 30, 428–430 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y et al. ChAMP: updated methylation analysis pipeline for Illumina BeadChips. Bioinforma. Oxf. Engl. 33, 3982–3984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Love MI, Huber W & Anders S Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sandoval J et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 6, 692–702 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Shen J et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics 8, 34–43 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Szklarczyk D et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doolittle-Hall JM, Cunningham Glasspoole DL, Seaman WT & Webster-Cyriaque J Meta-Analysis of DNA Tumor-Viral Integration Site Selection Indicates a Role for Repeats, Gene Expression and Epigenetics. Cancers 7, 2217–2235 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moody CA & Laimins LA Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10, 550–560 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Monk BJ, Tian C, Rose PG & Lanciano R Which clinical/pathologic factors matter in the era of chemoradiation as treatment for locally advanced cervical carcinoma? Analysis of two Gynecologic Oncology Group (GOG) trials. Gynecol. Oncol. 105, 427–433 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rader JS et al. Genetic variations in human papillomavirus and cervical cancer outcomes. Int. J. Cancer 144, 2206–2214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoadley KA et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell 158, 929–944 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stark R & Brown G DiffBind: Differential binding analysis of ChIP-Seq peak data. 33. [Google Scholar]
- 43.Lin-Shiao E et al. KMT2D regulates p63 target enhancers to coordinate epithelial homeostasis. Genes Dev. 32, 181–193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herz H-M et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 26, 2604–2620 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu D et al. The MLL3/MLL4 Branches of the COMPASS Family Function as Major Histone H3K4 Monomethylases at Enhancers. Mol. Cell. Biol. 33, 4745–4754 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee J-E et al. H3K4 mono- and di-methyltransferase MLL4 is required for enhancer activation during cell differentiation. eLife 2, e01503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hu Z et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat Genet 47, 158–163 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Pokholok DK et al. Genome-wide Map of Nucleosome Acetylation and Methylation in Yeast. Cell 122, 517–527 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Gates LA, Foulds CE & O’Malley BW Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem. Sci. 42, 977–989 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hurst TP & Magiorkinis G Activation of the innate immune response by endogenous retroviruses. J. Gen. Virol. 96, 1207–1218 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Newman AM et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okoye AA & Picker LJ CD4(+) T-cell depletion in HIV infection: mechanisms of immunological failure. Immunol. Rev. 254, 54–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hensley-McBain T & Klatt NR The Dual Role of Neutrophils in HIV Infection. Curr. HIV/AIDS Rep. 15, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sitole BN & Mavri-Damelin D Peroxidasin is regulated by the epithelial-mesenchymal transition master transcription factor Snai1. Gene 646, 195–202 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Zheng Y-Z & Liang L High expression of PXDN is associated with poor prognosis and promotes proliferation, invasion as well as migration in ovarian cancer. Ann. Diagn. Pathol. 34, 161–165 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Gifford CA et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 153, 1149–1163 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McBride AA & Warburton A The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 13, e1006211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kajitani N, Satsuka A, Kawate A & Sakai H Productive Lifecycle of Human Papillomaviruses that Depends Upon Squamous Epithelial Differentiation. Front. Microbiol. 3, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ou HD, May AP & O’Shea CC The critical protein interactions and structures that elicit growth deregulation in cancer and viral replication. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 48–73 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeon S, Allen-Hoffmann BL & Lambert PF Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. J. Virol. 69, 2989–2997 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Groves IJ, Knight ELA, Ang QY, Scarpini CG & Coleman N HPV16 oncogene expression levels during early cervical carcinogenesis are determined by the balance of epigenetic chromatin modifications at the integrated virus genome. Oncogene 35, 4773–4786 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pleasance E et al. Pan-cancer analysis of advanced patient tumors reveals interactions between therapy and genomic landscapes. Nat. Cancer 1, 452–468 (2020). [DOI] [PubMed] [Google Scholar]
- 63.Li H & Durbin R Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinforma. Oxf. Engl. 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saunders CT et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinforma. Oxf. Engl. 28, 1811–1817 (2012). [DOI] [PubMed] [Google Scholar]
- 66.Cingolani P et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly (Austin) 6, 80–92 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chun H-JE et al. Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer Cell 29, 394–406 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang Y et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kent WJ et al. The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kent WJ, Zweig AS, Barber G, Hinrichs AS & Karolchik D BigWig and BigBed: enabling browsing of large distributed datasets. Bioinforma. Oxf. Engl. 26, 2204–2207 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Landt SG et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 22, 1813–1831 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhao EY et al. Homologous Recombination Deficiency and Platinum-Based Therapy Outcomes in Advanced Breast Cancer. Clin. Cancer Res. 23, 7521–7530 (2017). [DOI] [PubMed] [Google Scholar]
- 73.McKenna A et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mermel CH et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12, R41 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arthur SE et al. Genome-wide discovery of somatic regulatory variants in diffuse large B-cell lymphoma. Nat. Commun. 9, 4001 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding J et al. Feature-based classifiers for somatic mutation detection in tumour-normal paired sequencing data. Bioinforma. Oxf. Engl. 28, 167–175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wilkerson MD & Hayes DN ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinforma. Oxf. Engl. 26, 1572–1573 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quinlan AR BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinforma. 47, 11.12.1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Heinz S et al. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramírez F, Dündar F, Diehl S, Grüning BA & Manke T deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 42, W187–191 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pellacani D et al. Analysis of Normal Human Mammary Epigenomes Reveals Cell-Specific Active Enhancer States and Associated Transcription Factor Networks. Cell Rep. 17, 2060–2074 (2016). [DOI] [PubMed] [Google Scholar]
- 82.Chakravarty D et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis. Oncol. 2017, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All molecular and clinical data used in this publication can be found on the National Cancer Institute’s Genome Data Commons Publication Page https://gdc.cancer.gov/about-data/publications/CGCI-HTMCP-CC-2020 . Data from this publication is publicly available for download through dbGaP (phs000528), as part of the NCI Cancer Genome Characterization initiative (CGCI, phs000235). Sample metadata is reported in Supplementary Table 2. Source data for all Figures and Extended Data Figures are presented with the paper.
TCGA cervical cancer data (file name: CESC.snp__genome_wide_snp_6__broad_mit_edu__Level_3__segmented_scna_minus_germline_cnv_hg19__seg.seg.txt) was obtained from http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/CESC/20160128/.