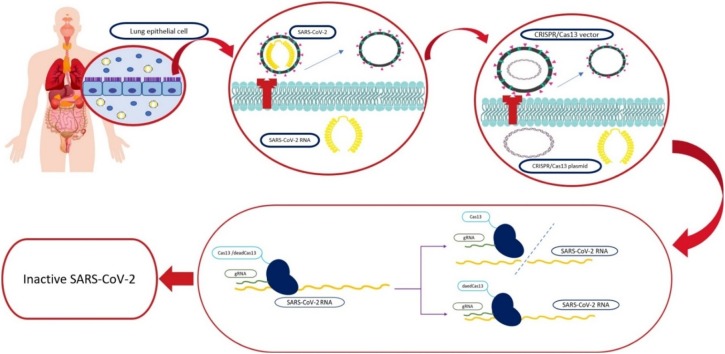
Graphical abstract
Keywords: CRISPR/Cas13, CRISPR/dead Cas13, COVID-19, SARS-CoV-2, Treatment
Abstract
The novel coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can be considered as the most important current global issue, as it has caused the novel coronavirus disease (COVID-19) pandemic, which has resulted in high mortality and morbidity rates all around the world. Although scientists are trying to discover novel therapies and develop and evaluate various previous treatments, at the time of writing this paper, there was no definite therapy and vaccine for COVID-19. So, as COVID-19 has called ideas for treatment, controlling, and diagnosis, we discussed the application of Clustered Regularly Interspaced Short Palindromic Repeats/Cas13 (CRISPR/Cas13) as a treatment of COVID-19, which received less attention compared with other potential therapeutic options.
1. Introduction
The novel coronavirus disease (COVID-19) outbreak, accompanying numerous pneumonia cases caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), emerged in Wuhan, a city in the Hubei province of China, late 2019. Although it was indicated that the patients infected with SARS-CoV-2 (formerly named 2019-nCoV [1]) in China might have used (as a source of food) or encountered infected animals in the seafood market, more inquiry displayed some patients with no record of visiting the seafood market. So, the person-to-person contagion of the virus through coughing, sneezing, and aerosols that could infiltrate the lungs via nose or mouth is inevitable [2,3].
Until now, seven coronaviruses have caused disease in humans [4,5]. Two strains with zoonotic origination containing severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) were connected to the emergence of severe respiratory diseases in humans in 2003 and 2012 subsequently [5], and now, SARS-CoV-2 as the seventh virus of the coronavirus family has caused severe respiratory diseases pandemic in humans. Like SARS and MERS, the symptoms of COVID-19 are commonly recognized from mild to critical as dry cough, sore throat, nasal congestion, malaise, headache, muscle pain, fever, shortness of breath, dyspnea, respiratory distress, tachypnea less than 30 breaths per minute, hypoxia less than 90 % of SpO2 on room air, conjunctivitis, diarrhea, confusion, chest pain, nausea, vomiting, anosmia, and dyspepsia [[6], [7], [8], [9], [10]]. Like other coronaviruses, SARS-CoV-2 is a positive-sense RNA virus, which infects the upper and lower respiratory tracks [11]. Moreover, its life cycle is probably related to SARS. As it is severely contagious, there is a rapid progression in its morbidity and mortality rates. Besides, there is no specific vaccine and treatment for that; so, the best solution for controlling the pandemic in addition to following preventive methods can be suggesting and finding probable effective therapies. In this way, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) has a potential in the development of diagnostic methods and therapeutic approaches. CRISPR is the new genome engineering technique produced of two components, consisting of a guide RNA (gRNA), which is particular to the target DNA or RNA sequence, and a non-specific CRISPR associated endonuclease protein or CRISPR- associated (Cas) protein.
In contrast to traditional vaccines and therapies, which rely on priming the human immune system to identify viral proteins and components and reduce viral entrance into cells [12], the CRISPR-based system has focused on identifying and degrading the intracellular viral genome and its resulting viral mRNAs. Thus, for using CRISPR as a therapeutic option, it is critical to identify the SARS-CoV-2 molecular characteristics. It must be taken in to account that Abbott et al., have expanded the application of CRISPR/Cas13 in addition to its diagnostic function for therapeutic goals by their Prophylactic Antiviral CRISPR in the huMAN cells (PAC-MAN) approach [13]. They found that the two highly-conserved regions in SARS-CoV-2 genome, which can be appropriate to be targeted by PAC-MAN as a potential pan-coronavirus inhibition strategy are respectively the RNA-dependent RNA polymerase (RdRP) gene in the open reading frame1a/b or ORF1a/b region, which maintains the proliferation of all coronaviruses, and the Nucleocapsid (N) gene at the 3′ end of the genome, which encodes the capsid protein for viral packaging [13]. Moreover, it has been mentioned in another study Nguyen et al., have designed 10,333 guide RNAs to specifically target ten peptide-coding regions of the SARS-CoV-2 virus RNA genome, including ORF1a/b and the spike (S) gene [14].
In this paper, we have discussed the molecular characteristic of SARS-CoV-2 and the potential of the CRISPR/Cas13 system as a treatment for COVID-19.
2. SARS-CoV-2 features
Although the viral genome investigation revealed that SARS-CoV-2 is 88 % alike to bat-SL-CoVZC45, bat-SL-CoVZXC21 and 96.2 % identical to a bat CoV RaTG13 [15], recent studies proposed Malaysian smuggled pangolins to China as the probabilistic provenance of the virus. It must be mentioned that this approximate 4 % difference of SARS-CoV-2 from RaTG13 accounts for almost 20–50 years of evolution. Moreover, it is similar to RaTG13, but molecularly too far away to be a direct ancestor of SARS-CoV-2. Still, it is the most probable that bats were the origin of SARS-CoV-2, and the transmission to humans occurred via intermediate hosts; unless it was a direct transmission with a prolonged period of positive selection leading to optimization to human host receptor.
The most genomic encoded proteins of SARS-CoV-2 are 79.5 % and 51 % related to SARS-CoV and MERS-CoV, sequentially [1,16,17]. Furthermore, similar to SARS-CoV, it uses the angiotensin-converting enzyme 2 (ACE2) receptor [15] for cellular entry, which the high virus attraction to the ACE2 receptor is likely because of natural separation instead of premeditated manipulation [18].
Coronavirus spike (S) protein has been ascertained as a substantial determinative of virus insertion into host cells [19] as the receptor-binding domain (RBD) for binding to ACE2 receptor, and direct membrane fusion between the virus and plasma membrane [20] has been placed on the head of that. Furthermore, alongside the membrane fusion, the clathrin-dependent and -independent endocytosis mediates the SARS-CoV entry [21,22]. After admittance, the viral RNA genome is liberated into the cytoplasm and is translated into two polyproteins and structural proteins, after which the viral genome begins replication [23]. The newly formed envelope glycoproteins enter the endoplasmic reticulum or Golgi membrane, and the nucleocapsid is created by combining genomic RNA and nucleocapsid protein. The virus particles then germinate in the endoplasmic reticulum of the Golgi Network (ERGIC). Finally, the vesicles containing the virus particles are fused to the plasma membrane [19] to release the virus from the infected cell.
Novel coronavirus or 2019-nCoV is an enveloped, positive-sense, and single-stranded RNA beta-coronavirus [24,25], which is about 29.9 kb with a cap structure at its 5′ end and a poly-A tail at the 3′ end [26]. Like other coronaviruses, it has particular viral replicase genes in a variable number [[6], [7], [8], [9], [10], [11], [12], [13], [14]] open reading frame (ORF), which codifies vital proteins (27–29 ones) for viral replication, nucleocapsid, and spikes formation [5,[27], [28], [29]]. Two-thirds of viral RNA, principally placed in the 5′-terminal in the first ORF (ORF1a/b), translates two polyproteins (PP), pp1a and pp1b, which codifies 16 non-structural proteins (NSP) [30]. In contrast, the rest of ORFs encode at least eight accessory proteins (ORF3a, ORF6, ORF7a, ORF7b, ORF8, ORF9a, ORF9b, and ORF10) intervening with the host innate immune response [5,29]. It must be mentioned that there are intra-viral protein-protein intercommunications between most of these proteins, which make the situation more complicated than before.
2.1. Non-structural proteins (NSPs)
2.1.1. NSP1
It reduces the speed of the production of infected cells’ proteins, but it also enhances the production of SARS-CoV-2 essential proteins [31].
2.1.2. NSP2
Although its role has not been discovered yet, some evidence has shown its attachment to some protein, which helps in moving endosomes around the cell [31]. Furthermore, recent studies have offered that the mutation in NSP2 and NSP3 has a crucial function in the infectious potential and differentiation mechanism of SARS-CoV-2 [31,32]. It must be noticed that although there are no amino acid replacements in NSP7, NSP13, envelope, matrix, or accessory proteins p6 and 8b, there are some amino acid substitutions in spike protein (s), receptor-binding domain (RBD), underpinning subdomain, NSP2, and NSP3 [29].
2.1.3. NSP3
Generally, healthy cells label their obsolete proteins to be destroyed. However, one of the duties of NSP3 is to unlabeled them, which causes disorder and imbalance in cellular proteins. So, it weakens and disables the infected cell against the virus, and it can also prepare a suitable environment for viral proteins to do their tasks [31].
2.1.4. NSP4
It assists with other proteins like NSP3 and NSP6 in producing fluid-filled endosomes for composing various parts of virus new copies [31].
2.1.5. NSP5
Not only like human immunodeficiency virus (HIV), SARS-CoV-2 includes a potential cleavage site for furin proteases for actuating the polyproteins [33], but also it uses Chymotrypsin-Like protease (3CLpro) or Main protease (Mpro), Papain-Like protease (PLpro), and NSP5 to cleave the polyproteins into other NSPs [34]. So, PLP inhibitors, like diarylheptanoids [35], grazoprevir, telaprevir, boceprevir [36] and formoterol chloroquine [37] and 3CLpro inhibitors, like cinanserin [38], flavonoids [39], cobicistat [40], atazanavir, efavirenz, ritonavir, dolutegravir [41], and thymopentin, carfilzomib, saquinavir [42], ledipasvir, and velpatasvir [43].
2.1.6. NSP7
It helps with NSP8 and NSP12 in producing new copies of the virus RNA [31].
2.1.7. NSP9
It might affect the movement of molecules in and out of the infected cell nucleus by infiltrating narrow channels in that. It also works with NSP10 to inhibit NKRF (inhibits IL-8 and IL-6 induction) for accelerating the production of IL-8 and IL-6 in lung epithelial cells, which enhance neutrophil count that might mediate the consequences of acute respiratory distress syndrome (ARDS) in COVID-19 patients [44,45].
2.1.8. NSP10
In addition to its duty mentioned before, it works with NSP16 to camouflage the virus’s genes for inhibiting antiviral proteins of human cells that have to find and attack them. Besides, it is a critical co-factor for NSP14 as well [31,34].
2.1.9. NSP11
2.1.10. NSP12
It is RNA-dependent RNA polymerase (RdRp), which helps with NSP7 and NSP8 to congregate genetic letters into the new SARS-CoV-2 genome. So, it is essential for replication and transcription of the viral genome [47]. Moreover, remdesivir has been suggested to interfere with NSP12.
2.1.11. NSP13
It is a helicase that unwinds the SARS-CoV-2 RNA to prepare it for other proteins to read its sequence and produce new copies [34,48].
2.1.12. NSP14
Like NSP12, it is essential for viral replication and transcription. Moreover, it is a viral proofreading. When NSP12 duplicates the SARS-CoV-2 genome, it seldom adds the wrong letter to the new copy. At this time, NSP14 (the N-terminal exoribonuclease (ExoN) domain cuts out the wrong letter and substitutes it with the correct letter to prevent lethal mutagenesis [34]. Also, its C-terminal domain acts as a guanine-N7 methyltransferase (N7-MTase) for mRNA capping [34].
2.1.13. NSP15
It is also named Uridylate-specific endoribonuclease, which can inhibit the infected cells’ antiviral mechanisms by hiding the leftover virus RNA by chopping them up [49].
2.1.14. NSP16
It is an S-Adenosyl Methionine (SAM) dependent nucleoside-2′-O methyltransferase, that it is only activated by NSP10 to hide the virus’s genome from proteins cleave viral RNA [34,50].
2.2. Accessory proteins
2.2.1. ORF3a
It facilitates the new viruses escaping from infected cells by making some holes in their membranes. Besides, it induces inflammation as well. However, it is not proved that SARS-CoV-2 applies to this gene.
2.2.2. ORF6
Not only it works as a signal blocker which inhibits the signals of the infected cell send out to induce immune system responses, but also it inhibits some of the cell’s proteins that combat against the virus.
2.2.3. ORF7
There are two types of that, including ORF7a, ORF7b [45]. It facilitates the produced new viruses escaping through reducing tetherin proteins, which are responsible for snaring new escaping viruses. It has also been reported that ORF7a can induce suicide to infected cells, which leads to lung damage.
2.2.4. ORF8
It is different in 2019-nCoV [31].
2.2.5. ORF9b
It inhibits interferon induction and signaling, which is the human innate immune response against SARS-CoV-2. However, it is not evident whether it uses this protein or not.
2.2.6. ORF10
It is also like ORF8, a mysterious protein that SARS-CoV-2 probably does not have its encoding gene.
2.3. Structural proteins
2.3.1. Spike glycoprotein
It is composed of three S1- S2 heterodimers. The receptor-binding domain (RBD) located on the head of S1 begins the membrane fusion of SARS-CoV-2 by binding it to the cellular receptor angiotensin-converting enzyme 2 (ACE2). Plus, S2 works as a post-fusion of crown-like spike proteins. It must be mentioned that due to mutation (insertion of 12 genetic letters (31)) of spike protein in SARS-CoV-2, there is a high affinity of the virus to ACE2 receptors [34].
2.3.2. Membrane protein (M protein)
It contributes along with other structural proteins to produce the SARS-CoV-2 coat. Moreover, it is identified as a protective factor against humoral responses [26].
2.3.3. Envelope (E) protein
It is also called the E channel as it makes pentamer and acts as an ion channel, which helps form the oily bubble of the virus [34].
2.3.4. Nucleocapsid (N) protein
It covers the viral genome and has various roles in the virus replication cycle [34].
3. CRISPR/Cas13
The Clustered Regularly-Interspaced Short Palindromic Repeats (CRISPR) has adapted from the prokaryotic adaptive immune system CRISPR-associated (Cas) system, to be used as a novel and specific genome editing tool for other organisms. CRISPR–Cas systems are vastly divided into two major classes and are more subdivided into six main types (I–VI) and over 19 subtypes [51].
CRISPR/Cas13 has appeared as a robust eukaryotic gene expression repression approaches at the posttranscriptional level whereby encoded information in the genome is modified at the level of the RNA that is the Class 2 type VI CRISPR-associated RNA-guided ribonuclease. Four protein families have been recognized, including Cas13a (initially referred as Class 2 candidate 2 (C2c2)), Cas13b, Cas13c, and Cas13d (named CasRx) [[52], [53], [54]]. Despite Cas9, Cas13 lacks homology with any known DNA nuclease domain. However, it has two higher prokaryotes and eukaryotes nucleotide-binding (HEPN) domains, which together form the ribonuclease-active site, let it act as an RNA-guided RNA-targeting CRISPR effector [55,56]. It has composed of the programmable single-effector RNA-guided RNase Cas13 and a 64–66-nt CRISPR RNA (crRNA), which only identifies a 24–30-nt sequence on the target RNA through the protospacer-flanking site [57]. The gRNA binds to the complementary target RNA sequence, then Cas13 cleaves, and knocks down the intended cellular RNA targets. If the targeting components are present in the appropriate cellular compartments at adequate doses, it can exert remarkable posttranscriptional suppression of gene expression in almost 24 h [58]. So, as it can cleave any complementary sequence to gRNA, and it has the ability to efficiently target and cleave RNA in several model systems, including mammalian cells [59,60], it can cleave the specific sequence of SARS-CoV-2 RNA and inactivate the virus at the posttranscriptional level in mammalian cells.
Cas13a enzymes can be a suitable tool for nucleic acid detection [61], and there is the protocol for using it as a pre-clinical SARS-CoV-2 detective method [62,63]. There is a sensitive CRISPR-based detective technique called Specific High-sensitivity Enzymatic Reporter unlocking (SHERLOCK). The latest SHERLOCK technique is known as SHERLOCKv2, which uses multiple Cas13 enzymes and other enzymes, like an enzyme known as Csm6, which exhibits RNase activity when activated by some of the Cas13 nucleases products [64]. SHERLOCK is used for diagnostic and detective goals [65]. It must be mentioned that food and drug administration (FDA) approved the first CRISPR-based detective kit of COVID-19, providing results in approximately one hour [66].
Moreover, CRISPR/Cas13 might also have efficacy in the medical treatment of COVID-19 patients. During the past two decades, several variants of coronavirus like SARS-CoV-2, SARS, and MERS, emerged from animal sources and infected humans, each time caused remarkable disease and deaths. So, there was a considerable need for designing a strategy that could vastly prohibit and stop the viruses especially all coronavirus strains that currently reside in animals. In this way, through analysis of all known coronavirus genomes (3,051) to identify all possible crRNAs that can target each of those genomes, which ended up in 6 crRNAs, Abbott et al., discovered a CRISPR-based therapy called Prophylactic Antiviral CRISPR in the huMAN cells (PAC-MAN-T6) method using Cas13, which has vigorous target cleavage activity [13,67]. They applied the class 2 type VI-D CRISPR (CRISPR/Cas13d) system derived from Ruminococcus flavefaciens XPD3002 to inhibit RNA viruses in human cells [13,68].
The Cas13d was used due to its small size (967 amino acids), which makes it proper for an ‘all-in-one’ adeno-associated virus (AAV) delivery with a guide-RNA array, high catalytic activity in mammalian cells for targeting SARS-CoV-2 genome, and high specificity [13,14,67].
There are several reasons why CRISPR/Cas13d can be practical as a therapeutic option of COVID-19 in infected persons. The known Cas13 effectors can efficiently cut complementary target single-stranded RNA (ssRNA), which represent a likely more reliable and protected alternative to Cas9, as it provokes loss-of-function phenotypes without genomic loss of the targeted gene [69]. CRISPR/Cas13d system is flexible in designing guide RNA to apply any sequence in the virus genome as Cas13 does not have a specific need of Cas9 to an adjacent protospacer motif (PAM) like the NGG at the editing site [70]. So, it meets the need for the rapid designing of gRNAs to target different virus variants that evolve and may escape traditional drugs. Also, in contrast to Cas13, as it cleaves RNA, there are some problems with using Cas9, which cleaves DNA. Mismatches or off-targets (cleaving the host DNA at a random location that has not been programmed for that) [71] are one of those problems that can induce accidental mutations, which might result in cancer [72,73]. Moreover, Cas9 often cleaves the DNA at the right location, which has been programmed for that; but the cellular repairing system does not do what we want. There are two repairing ways after cleavage, including non-homologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ, which is active in replicating and post-mitotic cells, might make impressive gene disruption through composing a mixture of insertion or deletion (indel) mutations, which could result in frame-shifts in protein-coding genes. However, indels production are under control in post-mitotic cells and second precise gene editing through template-dependent homology-directed repair (HDR), which is highly restricted to replicating cells and is mediated through host pieces of machinery [74].
Although the appropriate size of guide RNA is 20 bp [75], longer Cas13 guide RNAs than 20 bp (has low effectivity) can be used in sufficient concentration for gaining the best result. Therefore, CRISPR/Cas13 can be directly used in tissues affected by the SARS-CoV-2 as a probable effective therapeutic option against it. Moreover, Cas13 can process its CRISPR array and release individual CRISPR RNA (crRNAs), so it can be used for multiplexed targeting applications [76]. It is feasible to assess its effectiveness in vitro through cultivating affected tissue cells in the appropriate culture media.
One obstacle in the way of using CRISPR/Cas13 as a therapeutic option against COVID-19 is the delivery strategy. For breaking or inhibiting the RNA sequences of 2019-nCoV before they have any time to create any enzyme through using Cas13 enzymes along with gRNA sequences targeting different locations of SARS-CoV-2 genome, there are several methods for transferring the CRISPR system into the target cells like lipofection, electroporation, nucleofection, microinjection, and viral vectors [77].
Due to the small size of the Cas13d effector, one of the best options is AAV, which up to three guide RNAs targeting different peptide-encoding regions of the SARS-CoV-2 RNA genome can be packaged into one AAV. Moreover, AAV has serotypes highly specific to the lung, the central organ infected by SARS-CoV-2, and can be employed for targeted transmission of the CRISPR system [14]. However, as it is possible to cultivate SARS-CoV-2 in primary monkey cells and cell lines like Vero and LLCMK2 [78], we can make a SARS-CoV-2 vector-only composed of structural proteins like the S and N proteins due to high affinity of ACE2 as a capable viral vector for transfecting the CRISPR/Cas13 components with simultaneously using of immunosuppressors.
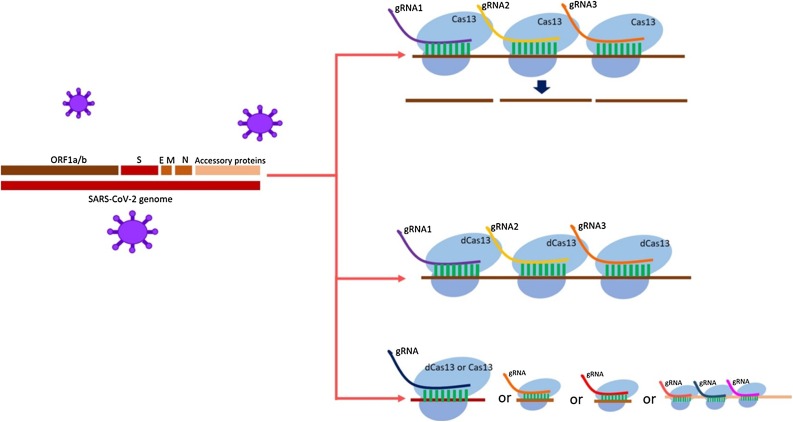
Although all the protein-coding genes mentioned before (NSP1-NSP16, and accessory proteins) individually and independently can be considered as a therapeutic target of CRISPR/Cas13, ORF1a/b (scientists have designed multiple guide RNAs to specifically target peptide-coding regions of ORF1a/b [13,14]), which encodes polyproteins and is essential for SARS-CoV-2 pathogenesis can be an appropriate target for active and catalytically inactive Cas13 (dead Cas13) for inhibiting the production and activation of polyproteins. Through applying multiple complementary guide RNAs of various sequences in ORF1a/b (the innate capability of Cas13 to process pre-crRNA into single crRNAs could simplify simultaneous targeting of multiple mRNAs contributed in one or several pathways [79]), we can inactivate it by cutting its RNA or by using dead Cas13 [80] and numerous guide RNAs instead of cleaving it we can just inactivate it by making some hurdles in the way of its translation (producing proteins) and transcription (Fig. 1 ). Another potential therapeutic method can be inactivating the viral main proteinase 3CLpro or Mpro and NSP5 through dead Cas13 or Cas13 or cleaving the cleavage site for furin proteases by Cas13, which all of them can inhibit the activation of polyproteins.
Fig. 1.
Various appropriate site of SARS-CoV-2 genome, which can be targeted by CRISPR Cas13 or CRISPR deadCas13.
Through methods negotiated above, it is possible to inactive SARS-CoV-2 in infected mammalian cells and to prevent its transmission to remaining healthy cells in early stages and inhibiting the development of COVID-19 to severe stages and prepare the situation for effective immune system responses. Moreover, scientists concluded in several studies that CRISPR/Cas13 is a beneficial, flexible, and rapid novel method for treating and preventing RNA virus infections, especially COVID-19 [13,14]. Further, multiplexed therapy of SARS-CoV-2 by combining CRISPR-based treatment with other therapeutic options might be impressive. It must be mentioned that according to several launched clinical trials for appraising therapeutic and detective options against COVID-19, scientists are working hard to develop suitable preventive and therapeutic approaches comprising vaccines, monoclonal antibodies, peptides, interferon therapies, and small-molecule drugs to overcome SARS-CoV-2. In the meanwhile, as we are not the only creature battling viruses there are also bacteria, it deserves to evaluate the effectiveness of CRISPR/Cas13 or dead Cas13 derived from bacteria against SARS-CoV-2 as it is the novel, the most cost-effective and practical genome editing agent for RNA up until now [77]. Although CRISPR/Cas13 system has collateral cleavage activity, there are minimal off-target effects on the host transcriptome in mammalian cells [68,81], which may lead to unwanted and unpredicted mutations.
Moreover, it is a novel gene engineering tool, which its different aspects and side effects should be studied and its delivery strategy [82,83] in mammalian cells is a challenge. Thus, may be due to those reasons despite its enormous potential clinical applications CRISPR/Cas13 has received less attention as a therapeutic option against SARS-CoV-2 in clinic otherwise it is easy to use, more cost-effective, sensitive, and specific than other diagnostic and therapeutic methods. It must be mentioned that using dead Cas13 is safer than Cas13 as it does not cut the RNA. Moreover, there are anti-CRISPR proteins, which can be used for deactivating the Cas13, whenever there are side effects [[84], [85], [86]]. Thus, if Cas13 can be programmed to efficiently target and cleave a wide variety of mammalian viruses’ RNA genome especially SARS-CoV-2 genome by investigating the prevalence of target sites in ssRNA viral genomes, it has the potential as an antiviral therapy, which has fewer off-targets. However, if there are side effects, deadCas13 and anti-CRISPR proteins can be used. So, although several studies have investigated the effectiveness of CRISPR/Cas13d against SARS-CoV-2 in vitro, further studies in animal models must be done for evaluating its safety to develop it to be used in the clinic. Besides, the effectiveness of dead Cas13 against COVID-19 should be appraised.
4. Conclusion
In conclusion, although several studies have evaluated the potential of CRISPR/Cas13 as a novel therapeutic option of COVID-19, among other various therapeutic options of COVID-19, CRISPR/Cas13 has received less attention. However, it has the potential to be successful against SARS-CoV-2. Contrary to traditional therapies, which have concentrated on triggering the human immune system to distinguish viral proteins and diminish viral entry into cells, CRISPR/Cas13 system has focused on cleaving the viral RNA genome inside the infected cells or preventing the expression of protein-coding genes of the virus. Thus, CRISPR/Cas13-based therapy might be more sensitive and specific than traditional treatments. There are several target sites for it to cleave and inhibit the SARS-CoV-2 RNA genome like NSP1-NSP16, accessory proteins, and ORF1-10, which scientists have reported that ORF1a/b can be the best target. Although in contrast to CRISPR/Cas9, CRISPR/Cas13 does not have the mismatch problem and the unpredicted mutations (made by repairing system (NHEJ) after cleavage) on its way of the development to clinical use further studies must be done for apprising its safety and effectivity against COVID-19 and other viral infections. Moreover, although scientists have used the AAV vector as a vehicle for transferring the CRISPR/Cas13d system into the infected cells, other transferring methods must be evaluated as AAV might be ineffective in transferring the CRISPR/Cas13d system into the infected cells due to several reasons.
Authors’ contributions
All authors contributed to the study conception and design. Melika Lotfi had the idea for the article and contributed to the data collection, writing the initial draft of the manuscript, and designing the figure. Nima Rezaei contributed to scientific and structural editing, critical revising, and verifying the manuscript before submission. All authors commented on the last versions of the manuscript, and all of them read and approved the final manuscript.
Funding
We declare that we received no funding for the publication of this review study.
Availability of data and material
Not applicable.
Code availability
Not applicable.
Declaration of Competing Interest
The authors report no declarations of interest.
References
- 1.Guo Y.-R., Cao Q.-D., Hong Z.-S., Tan Y.-Y., Chen S.-D., Jin H.-J., et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak–an update on the status. Mil. Med. Res. 2020;7(1):1–10. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riou J., Althaus C.L. Pattern of early human-to-human transmission of Wuhan 2019 novel coronavirus (2019-nCoV), December 2019 to January 2020. Eurosurveillance. 2020;25(4) doi: 10.2807/1560-7917.ES.2020.25.4.2000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parry J. British Medical Journal Publishing Group; 2020. China Coronavirus: Cases Surge as Official Admits Human to Human Transmission. [DOI] [PubMed] [Google Scholar]
- 4.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;38(2):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui J., Li F., Shi Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hui D.S., Azhar I.E., Madani T.A., Ntoumi F., Kock R., Dar O., et al. The continuing 2019-nCoV epidemic threat of novel coronaviruses to global health—the latest 2019 novel coronavirus outbreak in Wuhan, China. Int. J. Infect. Dis. 2020;91:264–266. doi: 10.1016/j.ijid.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J-PO Li, Lam D.S.C., Chen Y., Ting D.S.W. BMJ Publishing Group Ltd; 2020. Novel Coronavirus Disease 2019 (COVID-19): The Importance of Recognising Possible Early Ocular Manifestation and Using Protective Eyewear. [DOI] [PubMed] [Google Scholar]
- 8.Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cascella M., Rajnik M., Cuomo A., Dulebohn S.C., Di Napoli R. Statpearls [internet]: StatPearls Publishing; 2020. Features, Evaluation and Treatment Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 10.Lotfi M., Rezaei N. SARS‐CoV‐2: a comprehensive review from pathogenicity of the virus to clinical consequences. J. Med. Virol. 2020;92(10):1864–1874. doi: 10.1002/jmv.26123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., et al. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS‐CoV, MERS‐CoV, and 2019‐nCoV. J. Med. Virol. 2020;92(5):491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rappuoli R. Glycoconjugate vaccines: principles and mechanisms. Sci. Transl. Med. 2018;10(456):eaat4615. doi: 10.1126/scitranslmed.aat4615. [DOI] [PubMed] [Google Scholar]
- 13.Abbott T.R., Dhamdhere G., Liu Y., Lin X., Goudy L., Zeng L., et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–876. doi: 10.1016/j.cell.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen T.M., Zhang Y., Pandolfi P.P. Nature Publishing Group; 2020. Virus against Virus: A Potential Treatment for 2019-nCov (SARS-CoV-2) and Other RNA Viruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P., Yang X., Wang X., Hu B., Zhang L., Zhang W., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;2020 doi: 10.1038/s41586-020-2012-7. Published online February 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paraskevis D., Kostaki E.G., Magiorkinis G., Panayiotakopoulos G., Sourvinos G., Tsiodras S. Full-genome evolutionary analysis of the novel corona virus (2019-nCoV) rejects the hypothesis of emergence as a result of a recent recombination event. Infect. Genet. Evol. 2020;79 doi: 10.1016/j.meegid.2020.104212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banerjee A., Kulcsar K., Misra V., Frieman M., Mossman K. Bats and coronaviruses. Viruses. 2019;11(1):41. doi: 10.3390/v11010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020:1–3. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmons G., Reeves J.D., Rennekamp A.J., Amberg S.M., Piefer A.J., Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc. Natl. Acad. Sci. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., et al. SARS coronavirus entry into host cells through a novel clathrin-and caveolae-independent endocytic pathway. Cell Res. 2008;18(2):290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuba K., Imai Y., Ohto-Nakanishi T., Penninger J.M. Trilogy of ACE2: A peptidase in the renin–angiotensin system, a SARS receptor, and a partner for amino acid transporters. Pharmacol. Ther. 2010;128(1):119–128. doi: 10.1016/j.pharmthera.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7(6):439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., et al. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. Nature. 2020;57(9):270–273. [Google Scholar]
- 25.Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fehr A.R., Perlman S. Coronaviruses. Springer; 2015. Coronaviruses: an overview of their replication and pathogenesis; pp. 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Boheemen S., de Graaf M., Lauber C., Bestebroer T.M., Raj V.S., Zaki A.M., et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3(6):e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Z., Xu Y., Bao L., Zhang L., Yu P., Qu Y., et al. From SARS to MERS, thrusting coronaviruses into the spotlight. Viruses. 2019;11(1):59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zumla A., Chan J.F., Azhar E.I., Hui D.S., Yuen K.-Y. Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15(5):327. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.nytimes . 2020. Bad News Wrapped in Protein: Inside the Coronavirus Genome 2020 April 3.https://www.nytimes.com/interactive/2020/04/03/science/coronavirus-genome-bad-news-wrapped-in-protein.html Available from: [Google Scholar]
- 32.Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID‐2019: The role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92(6):584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang T., Wu Q., Zhang Z. Probable pangolin origin of SARS-CoV-2 associated with the COVID-19 outbreak. Curr. Biol. 2020;30(7):1346–1351. doi: 10.1016/j.cub.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kong R., Yang G., Xue R., Liu M., Wang F., Hu J., et al. 2020. COVID-19 Docking Server: An Interactive Server for Docking Small Molecules, Peptides and Antibodies Against Potential Targets of COVID-19. arXiv preprint arXiv:200300163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J.-Y., Jeong H.J., Kim J.H., Kim Y.M., Park S.-J., Kim D., et al. Diarylheptanoids from Alnus japonica inhibit papain-like protease of severe acute respiratory syndrome coronavirus. Biol. Pharm. Bull. 2012:b12–00623. doi: 10.1248/bpb.b12-00623. [DOI] [PubMed] [Google Scholar]
- 36.Elfiky A.A., Ibrahim N.S. 2020. Anti-SARS and Anti-HCV Drugs Repurposing Against the Papain-like Protease of the Newly Emerged Coronavirus (2019-nCoV) [Google Scholar]
- 37.Arya R., Das A., Prashar V., Kumar M. 2020. Potential Inhibitors Against Papain-like Protease of Novel Coronavirus (SARS-CoV-2) from FDA Approved Drugs. [Google Scholar]
- 38.Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., et al. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79(11):7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jo S., Kim S., Shin D.H., Kim M.-S. Inhibition of SARS-CoV 3CL protease by flavonoids. J. Enzyme Inhib. Med. Chem. 2020;35(1):145–151. doi: 10.1080/14756366.2019.1690480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rismanbaf A. Potential treatments for COVID-19; a literature review. Arch. Acad. Emerg. Med. 2020;8(1):29. [PMC free article] [PubMed] [Google Scholar]
- 41.Beck B.R., Shin B., Choi Y., Park S., Kang K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (2019-nCoV), Wuhan, China through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020;18:784–790. doi: 10.1016/j.csbj.2020.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q., Zhao Y., Chen X., Hong A. 2020. Virtual Screening of Approved Clinic Drugs with Main Protease (3CLpro) Reveals Potential Inhibitory Effects on SARS-CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y.W., Yiu C.-P., Wong K.-Y. Prediction of the 2019-nCoV 3C-like protease (3CLpro) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nourbakhsh M., Kälble S., Dörrie A., Hauser H., Resch K., Kracht M. The NF-κB repressing factor is involved in basal repression and interleukin (IL)-1-induced activation of IL-8 transcription by binding to a conserved NF-κB-flanking sequence element. J. Biol. Chem. 2001;276(6):4501–4508. doi: 10.1074/jbc.M007532200. [DOI] [PubMed] [Google Scholar]
- 45.Liang Q., Li J., Guo M., Tian X., Liu C., Wang X., et al. Virus-host interactome and proteomic survey of PMBCs from COVID-19 patients reveal potential virulence factors influencing SARS-CoV-2 pathogenesis. Med. (N.Y.) 2020;(Jul (21)) doi: 10.1016/j.medj.2020.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y., Jedrzejczak R., Maltseva N.I., Endres M., Godzik A., Michalska K., et al. Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2. Protein Science. 2020;29(7):1596–1605. doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jia Z., Yan L., Ren Z., Wu L., Wang J., Guo J., et al. Delicate structural coordination of the severe acute respiratory syndrome coronavirus Nsp13 upon ATP hydrolysis. Nucleic Acids Res. 2019;47(12):6538–6550. doi: 10.1093/nar/gkz409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhardwaj K., Palaninathan S., Alcantara J.M.O., Yi L.L., Guarino L., Sacchettini J.C., et al. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J. Biol. Chem. 2008;283(6):3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., et al. Crystal structure and functional analysis of the SARS-coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 2011;7(5) doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., et al. An updated evolutionary classification of CRISPR-Cas systems. Nat. Rev. Microbiol. 2015;13(11):722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shmakov S., Smargon A., Scott D., Cox D., Pyzocha N., Yan W., et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017;15(3):169. doi: 10.1038/nrmicro.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smargon A.A., Cox D.B., Pyzocha N.K., Zheng K., Slaymaker I.M., Gootenberg J.S., et al. Cas13b is a type VI-B CRISPR-associated RNA-guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol. Cell. 2017;65(4):618–630. doi: 10.1016/j.molcel.2016.12.023. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang C., Konermann S., Brideau N.J., Lotfy P., Wu X., Novick S.J., et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell. 2018;175(1):212–223. doi: 10.1016/j.cell.2018.09.001. e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murovec J., Pirc Ž, Yang B. New variants of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol. J. 2017;15(8):917–926. doi: 10.1111/pbi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Granados-Riveron J.T., Aquino-Jarquin G. CRISPR-Cas13 precision transcriptome engineering in cancer. Cancer Res. 2018;78(15):4107–4113. doi: 10.1158/0008-5472.CAN-18-0785. [DOI] [PubMed] [Google Scholar]
- 58.Unniyampurath U., Pilankatta R., Krishnan M.N. RNA interference in the age of CRISPR: Will CRISPR Interfere with RNAi? Int. J. Mol. Sci. 2016;17(3):291. doi: 10.3390/ijms17030291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Freije C.A., Myhrvold C., Boehm C.K., Lin A.E., Welch N.L., Carter A., et al. Programmable inhibition and detection of RNA viruses using Cas13. Mol. Cell. 2019;76(5):826–837. doi: 10.1016/j.molcel.2019.09.013. e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bawage S.S., Tiwari P.M., Santangelo P.J. Synthetic mRNA expressed Cas13a mitigates RNA virus infections. bioRxiv. 2018 [Google Scholar]
- 61.Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang F., Abudayyeh O.O., Jonathan S.G. 2020. A Protocol for Detection of COVID-19 using CRISPR Diagnostics. [Google Scholar]
- 63.Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F., Gootenberg J.S. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–444. doi: 10.1126/science.aaq0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.asia B. 2020. FDA Approves First CRISPR Test for Coronavirus Detection.https://www.biospectrumasiaB.com/news/83/15927/fda-approves-first-crispr-test-for-coronavirus-detection.html 13 May, Available from: [Google Scholar]
- 67.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., et al. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70(2):327–339. doi: 10.1016/j.molcel.2018.02.028. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Konermann S., Lotfy P., Brideau N.J., Oki J., Shokhirev M.N., Hsu P.D. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–676. doi: 10.1016/j.cell.2018.02.033. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Granados-Riveron J.T., Aquino-Jarquin G. CRISPR–Cas13 precision transcriptome engineering in cancer. Cancer Res. 2018;78(15):4107–4113. doi: 10.1158/0008-5472.CAN-18-0785. [DOI] [PubMed] [Google Scholar]
- 70.Kim Y.B., Komor A.C., Levy J.M., Packer M.S., Zhao K.T., Liu D.R. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat. Biotechnol. 2017;35(4):371. doi: 10.1038/nbt.3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ali Z., Mahas A., Mahfouz M. CRISPR/Cas13 as a tool for RNA interference. Trends Plant Sci. 2018;23(5):374–378. doi: 10.1016/j.tplants.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 72.Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018;24(7):939–946. doi: 10.1038/s41591-018-0050-6. [DOI] [PubMed] [Google Scholar]
- 73.Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018;24(7):927–930. doi: 10.1038/s41591-018-0049-z. [DOI] [PubMed] [Google Scholar]
- 74.Kim H., Kim J.-S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014;15(5):321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 75.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8(11):2281. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60(3):385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.infobioquimica CRISPR 101 eBook: Your Guide to Understanding CRISPR 2017 June 23. https://www.infobioquimica.com/new/2017/06/23/crispr-101-ebook-your-guide-to-understanding-crispr/ Available from:
- 78.Loeffelholz M.J., Tang Y.-W. Laboratory diagnosis of emerging human coronavirus infections—the state of the art. Emerg. Microbes Infect. 2020:1–26. doi: 10.1080/22221751.2020.1745095. (just-accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.East-Seletsky A., O’Connell M.R., Knight S.C., Burstein D., Cate J.H., Tjian R., et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cox D.B., Gootenberg J.S., Abudayyeh O.O., Franklin B., Kellner M.J., Joung J., et al. RNA editing with CRISPR-Cas13. Science. 2017;358(6366):1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abudayyeh O.O., Gootenberg J.S., Essletzbichler P., Han S., Joung J., Belanto J.J., et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550(7675):280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Oude Blenke E., Evers M.J., Mastrobattista E., van der Oost J. CRISPR-Cas9 gene editing: delivery aspects and therapeutic potential. J. Control. Release. 2016;244(Pt B):139–148. doi: 10.1016/j.jconrel.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 83.Gori J.L., Hsu P.D., Maeder M.L., Shen S., Welstead G.G., Bumcrot D. Delivery and specificity of CRISPR-Cas9 genome editing technologies for human gene therapy. Hum. Gene Ther. 2015;26(7):443–451. doi: 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- 84.Scherz P. The mechanism and applications of CRISPR-Cas9. Natl. Cathol. Bioeth. Q. 2017;17(1):29–36. [Google Scholar]
- 85.Lin P., Qin S., Pu Q., Wang Z., Wu Q., Gao P., et al. CRISPR-Cas13 inhibitors block RNA editing in bacteria and mammalian cells. Mol. Cell. 2020;78(5):850–861. doi: 10.1016/j.molcel.2020.03.033. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marino N.D., Pinilla-Redondo R., Csörgő B., Bondy-Denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat. Methods. 2020;17(5):471–479. doi: 10.1038/s41592-020-0771-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.