ABSTRACT
Objective.
To describe the resistance profile and the genetic characteristics of Escherichia coli isolates that harbor the mobilizable colistin resistance gene mcr-1 in Argentina.
Methods.
This was a retrospective study of 192 E. coli isolates positive for mcr-1 obtained from 69 hospitals of Buenos Aires City and 14 Argentinean provinces in 2012 – 2018. The antimicrobial susceptibility was performed by agar diffusion, broth macrodilution, and/or agar dilution. Standard polymerase chain reaction (PCR) was performed to detect resistance genes and incompatibility groups; specific PCR was applied to discriminate between blaCTX-M allelic groups and mcr-1.5 variant. The genetic relatedness among isolates was evaluated by XbaI-pulsed field gel electrophoresis and multilocus sequence typing in a subset of isolates.
Results.
All E. coli isolates showed minimal inhibitory concentrations to colistin ≥ 4μg/mL; nearly 50% were resistant to third-generation cephalosporins, with CTX-M-2 being the main extended-spectrum β-lactamase detected. Five E. coli were carbapenemase-producers (3 NDM, 2 KPC). The mcr-1.5 variant was detected in 13.5% of the isolates. No genetic relationship was observed among the mcr-1-positive E. coli clinical isolates, but a high proportion (164/192; 85.4%) of IncI2 plasmids was detected.
Conclusions.
The presence of IncI2 plasmids among highly diverse E. coli clones suggests that the mcr-1 gene’s wide distribution in Argentina may be driven by the horizontal transmission of IncI2 plasmids.
Keywords: Drug resistance, multiple; colistin; Enterobacteriaceae; Escherichia coli; Argentina
RESUMEN
Objetivo.
Describir el perfil de resistencia y las características genéticas de aislamientos clínicos de Escherichia coli que portan el gen movilizable de resistencia a colistina mcr-1 en Argentina.
Métodos.
Se realizó un estudio retrospectivo para analizar 192 aislamientos de E. coli mcr-1 positivo, obtenidos en 69 hospitales de la Ciudad de Buenos Aires y 14 provincias de Argentina entre 2012 y 2018. La sensibilidad a los antimicrobianos se analizó mediante los métodos de difusión en agar, macrodilución en caldo y/o dilución en agar. Se aplicó la técnica estándar de reacción en cadena de la polimerasa (PCR) para detectar genes de resistencia y grupos de incompatibilidad; se aplicó PCR específica para distinguir entre variantes alélicas del gen blaCTX-M y la variante mcr-1.5. La relación genética entre los aislamientos fue evaluada mediante la técnica de electroforesis en gel de campo pulsado usando la enzima Xbal y la tipificación por secuencias de múltiples locus en un subconjunto de aislamientos.
Resultados.
Todos los aislamientos de E. coli mostraron concentraciones inhibitorias mínimas de colistina ≥ 4μg/mL. Casi el 50% mostró resistencia a las cefalosporinas de tercera generación y CTX-M-2 fue la β-lactamasa de espectro extendido que más se detectó. Cinco aislamientos de E. coli mostraron ser productoras de carbapenemasas (3 NDM, 2 KPC). La variante mcr-1.5 se detectó en 13,5% de las cepas aisladas. No se observó relación genética entre los aislamientos clínicos estudiados de E. coli positivas para mcr-1, aunque sí se detectó una proporción elevada (164/192; 85,4%) de plásmidos Incl2.
Conclusiones.
La elevada ocurrencia de plásmidos IncI2 en un grupo altamente diverso de clones de E. coli podría indicar que la amplia difusión del gen mcr-1 en Argentina estaría asociada a la transmisión horizontal de plásmidos IncI2.
Palabras clave: Resistencia a múltiples medicamentos, colistina, Enterobacteriaceae, Escherichia coli, Argentina
RESUMO
Objetivo.
Descrever o perfil de resistência e as características genéticas de isolados clínicos de Escherichia coli que carregam o gene mobilizábel de resistência à colistina mcr-1 na Argentina.
Métodos.
Neste estudo retrospectivo, foram analizados 192 isolados de E. coli positivos para mcr-1 obtidos em 69 hospitais da Cidade de Buenos Aires e 14 províncias da Argentina, entre 2012 e 2018. A sensibilidade aos antimicrobianos foi examinada usando métodos de difusão em ágar, macrodiluição em caldo e/ou diluição em ágar. A técnica padrão de reação em cadeia da polimerase (PCR) foi aplicada para detectar genes de resistência e grupos de incompatibilidade; a PCR específica foi aplicada para discriminar entre variantes alélicas do gene blaCTX-M e a variante mcr-1.5. A relação genética entre os isolados foi avaliada por eletroforese em gel de campo pulsado usando a enzima XbaI e a tipagem por sequências de múltiplos lócus, em um subconjunto de isolados.
Resultados.
Todos os isolados de E. coli apresentaram concentrações inibitórias mínimas de colistina ≥4μg/mL. Quase 50% foram resistentes às cefalosporinas de terceira geração, e CTX-M-2 foi a β-lactamase de espectro estendido mais detectada. Cinco isolados de E. coli foram produtores de carbapenemase (3 NDM, 2 KPC). A variante mcr-1.5 foi detectada em 13,5% dos isolados. Não foi observada relação genética entre os isolados clínicos de E. coli positivos para mcr-1, mas foi detectada uma alta proporção (164/192; 85,4%) de plasmídeos IncI2.
Conclusões.
A alta ocorrência de plasmídeos IncI2 em um grupo altamente diverso de clones de E. coli sugere que a ampla distribuição do gene mcr-1 na Argentina estaria associada a transmissão horizontal de plasmídeos IncI2.
Palavras-chave: Resistência a múltiplos medicamentos, colistina, Enterobacteriaceae, Escherichia coli, Argentina
Polymyxins, including polymyxin B and colistin, are “last-line” treatment options against multidrug-resistant (MDR) gram-negative bacteria, such as carbapenem-resistant Enterobacterales. Until November 2015, the main colistin resistance mechanisms reported were chromosome-mediated mutations involving alterations in the PmrAB or PhoPQ, a two-component regulatory system (1). The situation changed with the report of mobile colistin resistance mediated by mcr-1 gene, revealing for the first time the horizontal spread of a colistin resistance determinant (2). This gene encodes a plasmid-borne phosphoethanolamine transferase that has been reported in Escherichia coli isolates from animal, food, environment, and human samples worldwide (3, 4). At this time, nine mcr genes have been described; but, mcr-1 is, by far, the most prevalent (3 - 5). Mcr-1 has been described in almost all countries in the Region of the Americas, while mcr-3 and mcr-5 genes have been sporadically described in only Brazil and Colombia, respectively (3, 5 – 7). Although detected in other Enterobacterales, the mcr-1 gene has been associated mainly with E. coli isolates, including Klebsiella pneumoniae and Salmonella spp. (5). Regardless of species, mcr-1 has been associated with a ~2609 bp DNA fragment containing the mcr-1 and pap2 genes, and has mobilized into an ISApI1-based composite transposon and different plasmid replicons, of which IncI2, IncX4, and IncHI2 are the most common incompatibility groups described so far (8 - 10).
Mcr-1-positive E. coli isolates causing bloodstream infections are still rare (1.0%), and generally, they remain susceptible to many antimicrobial agents (11). However, it is especially worrisome that mcr-1 gene acquisition by extended-spectrum β-lactamase- (ESBL) or carbapenemase-producing Enterobacterales, render extensively- or pan-drug resistant strains (3, 10 – 15). NDM has been the main carbapenemase reported in mcr-1-positive isolates, not only in hospitalized patients where it colonizes or causes infections, but also in healthy individuals within the community; sporadic cases of mcr-1-producers harboring KPC or OXA-48 carbapenemases have also been described (10 - 15).
After the description of mcr-1 in November 2015, the National Reference Laboratory on Antimicrobial Resistance in Argentina (NRL) set national and regional alerts for detection of mcr-1 gene in E. coli clinical isolates (16). Through December 2018, a total of 192 E. coli clinical isolates were confirmed as positive for mcr-1 at the NRL. The present study aims to describe the resistance profiles and the genetic characteristics of mcr-1-producing E. coli clinical isolates in Argentina.
MATERIALS AND METHODS
This retrospective study comprised the entire NRL collection: 192 E. coli clinical isolates (one per patient) previously confirmed as mcr-1 positive, submitted by 69 hospitals from 14 of Argentina’s provinces and Buenos Aires City (Figure 1). Isolates were recovered mainly from urine (n = 117; 60.9%), blood (n = 26; 13.6%), and other samples (n = 49; 25.5%). The first 10 isolates were recovered in July 2012 – January 2016, as previously described by Rapoport and colleagues (17) and Martino and colleagues (18). The remaining 182 isolates were submitted from February 2016 – December 2018, with minimal inhibitory concentration (MIC) to colistin > 2µg/ml and/or a positive growth on Mueller-Hinton screening agar plates containing 3 µg/mL colistin (19).
FIGURE 1. Geographical distribution of 192 mcr-1 positive Escherichia coli clinical isolates, by province and capital city, Argentina, 2012 – 2018.
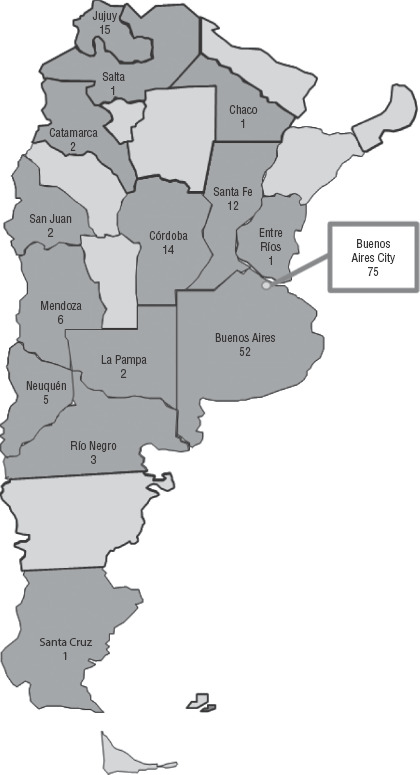
Source: Prepared by the authors from the study results.
Data on sample type and patient age and sex were obtained from the clinical laboratory documentation that accompanied each isolate. Patient data were anonymized to preserve the patient’s identity.
Susceptibility profiles were determined by the agar diffusion method, with the exception of colistin which was tested by broth macrodilution and/or agar dilution according to the Clinical and Laboratory Standards Institute guidelines (CLSI; 20). CLSI criteria were used to interpret all results, except for colistin and tigecycline, for which the 2018 European Committee on Antimicrobial Susceptibility Testing guidelines (21) were used. ESBL- or plasmidic-AmpC-phenotype were defined as resistant to third-generation cephalosporins with clavulanic acid or phenyl boronic acid inhibition, respectively.
PCR was performed to detect mcr-1, plasmidic-AmpC, broad-spectrum and ESBL, and carbapenemases using the primers and conditions described in Table 1. Briefly, DNA templates were prepared by boiling for 10 min a suspension of one or two colonies of each isolate in 100 µL of Milli-Q water; 2.5µL were used for the PCR reactions. A final volume of 25µL containing 10 pmol of each primer, 25 µM of each dNTP, 1.5 mM MgCl2, 1X Taq buffer, and 2.5 U of Taq polymerase (Invitrogen,TM ThermoFisher Scientific Inc., Waltham, MA, United States) was used. Amplifications were performed using a 2720 Thermal Cycler™ (Applied Biosystems, ThermoFisher Scientific Inc., Waltham, MA) following a standard program: pre-denaturation for 5 min at 94°C; 35 cycles of 94°C for 30 sec, T°C annealing (Table 1) for 30 sec, 72°C for 30 sec, and final extension at 72°C for 5 min. If the expected amplicon was ≥ 700 bp, the elongation step of cycling was increased to 1 min and the final elongation to 10 min. PCR products were run on 1% agarose gel for 60 min and stained with ethidium bromide. PCR was also used to discriminate between blaCTX-M-2, blaCTX-M-1/15, blaCTX-M-8/25, and blaCTX-M-9/14 groups (Table 1). Identification of incompatibility groups IncI2, IncX4, and IncHI2 was analyzed by PCR using previously described conditions (22, 23). To identify the mcr-1.5 variant, we developed an allele specific PCR that detected the C1354T modification (H452Y). A degenerated primer, MCR-1.5-F, with a mismatch at the third base from the 3’extreme was designed to increase the PCR specificity (Table 1).
TABLE 1. Primers for PCR analysis of antimicrobial resistance mechanisms.
Target |
Primer |
Oligonucleotide sequence |
Amplicon size (bp) |
T °C annealing |
|---|---|---|---|---|
mcr-1 |
CLR5-F CLR5-R |
5’ CGGTCAGTCCGTTTGTTC 3’ 5’ CTTGGTCGGTCTGTAGGG 3’ |
309 |
45 |
mcr-1.5 |
MCR-1.5-F MCR-1 Full-R |
5’ TCCAGTGGCTGCAGAAGT 3’ 5’ TCAGCGGATGAATGCGGT 3’ |
288 |
62 |
blaNDM |
NDM-F NDM-R |
5’ AGCACACTTCCTATCTCGAC 3’ 5’ GGCGTAGTGCTCAGTGTC 3’ |
512 |
50 |
blaIMP |
IMP-UF1 IMP-UR1 |
5’ GGYGTTTWTGTTCATACWTCKTTYGA 3’ 5’ GGYARCCAAACCACTASGTTATCT 3’ |
404 |
50 |
blaVIM |
VIM-F VIM-R |
5’ AGTGGTGAGTATCCGACAG 3’ 5’ ATGAAAGTGCGTGGAGAC 3’ |
261 |
50 |
blaCTX-M |
CTX-MU1 CTX-MU2 |
5’ ATGTGCAGYACCAGTAARGT 3’ 5’ TGGGTRAARTARGTSACCAGA 3’ |
593 |
52 |
CTX-M-G2 |
CTXM2G-F CTXM2G-R |
5’ GCCGCTCAATGTTAACGGTGA 3’ 5’ ACCGTGGGTTACGATTTTCGC 3’ |
851 |
55 |
CTX-M-G9 |
CTXM9G-F CTXM9G-R |
5’ ATGGTGACAAAGAGAGTGCAACG 3’ 5’ GCGGCTGGGTAAAATAGGTCACC 3’ |
808 |
56 |
CTX-M-G1/15 |
CTXM1/15G-F CTXM1/15G-R |
5’ CAGTTCACGCTGATGGCGACG 3’ 5’ CGGCGCACGATCTTTTGGCCA 3’ |
756 |
60 |
CTX-M-G8/25 |
CTXM8/25G-F CTXM8/25G-R |
5’ CTGGAGAAAAGCAGCGGGGG 3’ 5’ CGCTGCCGGTTTTATCCCCGAC 3’ |
604 |
58 |
blaPER |
PER-U-Fw PER-U-Rv |
5’ GTGTGGGAGCCTGACGATCT 3’ 5’ CTSTGGTCCTGTGGTGGTTTC 3’ |
524 |
59 |
blaTEM |
OT-1 OT-2 |
5’ TTGGGTGCACGAGTGGGTTA 3’ 5’ TAATTGTTGCCGGGAAGCTA 3’ |
504 |
55 |
blaSHV |
OS1 OS2 |
5’ TCGGGCCGCGTAGGCATGAT 3’ 5’ AGCAGGGCGACAATCCCGCG 3’ |
626 |
59 |
blaCMY |
CITMF CITMR |
5’ TGGCCAGAACTGACAGGCAAA 3’ 5’ TTTCTCCTGAACGTGGCTGGC 3’ |
462 |
64 |
Source: Prepared by the authors from the study results.
The genetic relatedness between the isolates was evaluated by XbaI-digested pulsed-field gel electrophoresis (PFGE) using a Chef-DR® III System (Bio-RadTM, Hercules, CA, United States) as previously reported (24). DNA fragments were resolved in 1% agarose gel applying a switch time of 2.4 to 54.2 seconds during 20 hr at 14°C. Those PFGE patterns showing > 6 bands of difference were considered to be non-genetically related. Selected isolates were also genotyped by multilocus sequence typing (MLST) and the sequence types (STs) were analyzed according to the E. coli MLST (available from http://enterobase.warwick.ac.uk/species/index/ecoli).
RESULTS
All 192 strains from the E. coli clinical isolates harboring mcr-1 strains grew on Mueller-Hinton screening agar plates containing 3 µg/mL colistin and showed MIC to colistin ≥ 4μg/mL. According to Figure 2, the percentage of resistance to other antimicrobials was as follows: ampicillin (85.9%), ciprofloxacin (74.9%), tetracycline (65.2%), trimethoprim-sulfamethoxazole (52.7%), third-generation cephalosporins (cefotaxime and/or ceftazidime; 48.9%), minocycline (39.7%), fosfomycin (23.9%), gentamicin (17.2%), carbapenems (ertapenem and/or imipenem; 3.6%), nitrofurantoin (5.0%), amikacin (2.6%), and tigecycline (2.3%). Nine isolates were resistant only to colistin, while 80.2% showed an MDR phenotype (resistance to > 2 families of antimicrobials).
FIGURE 2. Resistance profile of 192 mcr-1-positive Escherichia coli clinical isolates in Argentina, 2012 – 2018.
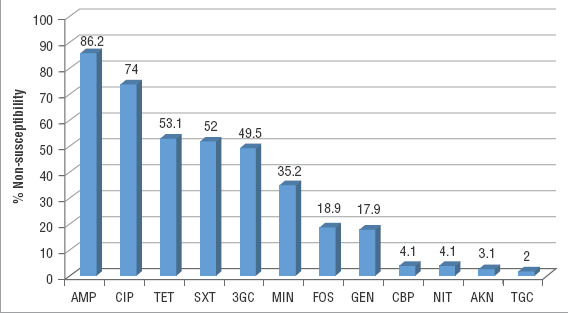
Note: AMP = ampicillin; CIP = ciprofloxacin; TET = tetracycline; SXT = trimethoprim-sulfamethoxazole; 3GC = third-generation cephalosporins (cefotaxime and/or ceftazidime); MIN = minocycline; FOS = fosfomycin; GEN = gentamicin; CBP = carbapenem (ertapenem and/or imipenem); NIT = nitrofurantoin; AKN = amikacin; TGC = tigecycline.
Source: Prepared by the authors from the study results.
Nearly one-half of the isolates (n = 94; 48.9%) showed resistance to third-generation cephalosporins, with ESBLs being the main mechanism (n = 77; 81.9%), followed by AmpC (n = 12; 12.8%) and carbapenemases (n = 5; 5.3%). The ESBLs detected were CTX-M (n = 73; 94.8%), SHV (n = 3; 3.9%), and PER-2 (n = 1; 1.3%). BlaCTX-M genes were grouped by sequence-similarity-based PCR as follow: CTX-M-2 (n = 37; 50.7%); CTX-M-9/14 (n = 21; 28.8%); CTX-M-8/25 (n = 11; 15.0%), and CTX-M-1/15 (n = 4; 5.5%). Nine of 12 isolates showing AmpC-phenotype were positive for blaCMY-2 plasmidic-AmpC gene. Carbapenemases were detected in 5 isolates from five hospitals in four cities that had been recovered from screening (n = 2), blood (n = 2), and urine samples (n = 1). As shown in Table 2, these carbapenemases were characterized as blaNDM-1 (n = 3) and blaKPC-2 (n = 2). Those isolates harboring blaNDM-1 were also positive for blaCMY-6 variant and rmtC genes. The presence of the mcr-1.5 variant was confirmed in 26 isolates (13.5%). Other mcr-1 variants or mcr-genes (mcr-2 to mcr-9) were not evaluated.
TABLE 2. Epidemiological data of five carbapenemase-producing mcr-1 positive Enterobacterales clinical isolates.
INEI ID |
Species |
Date |
Sample |
Hosp. |
Province |
Genes |
CIM COL |
Resistance profile |
ST |
|---|---|---|---|---|---|---|---|---|---|
E. coli |
May 2014 |
Blood |
A |
CABA |
blaNDM-1; blaCMY-6; mcr-1 |
≥ 4μg/mL |
3GC, CBP, GEN, AMK, COL |
10 |
|
E. coli |
September 2015 |
Blood |
B |
Córdoba |
blaKPC-2; mcr-1 |
≥ 4μg/mL |
3GC, CBP, CIP, MIN, SXT, COL |
156 |
|
E. coli |
April 2016 |
Screening |
C |
Santa Cruz |
blaKPC-2; mcr-1 |
≥ 4μg/mL |
3GC, CBP, TET, MIN, COL |
5208-like |
|
E. coli |
March 2018 |
Screening |
D |
CABA |
blaNDM-1; blaCMY-6; mcr-1 |
≥ 4μg/mL |
3GC, CBP, CIP, TET, MIN, GEN, AMK, SXT, NIT, COL |
354 |
|
E. coli |
May 2018 |
Urine |
E |
Entre Ríos |
blaNDM-1; blaCMY-6; mcr-1 |
≥ 4μg/mL |
3GC, CBP, CIP, GEN, AMK, COL |
8492 |
Note: Hosp = hospital; CABA = Ciudad Autonoma de Buenos Aires; 3GC = third generation cephalosporins (cefotaxime and/or ceftazidime); CBP = carbapenem (ertapenem and/or imipenem); GEN = gentamicin; AKN = amikacin; COL = colistin; CIP = ciprofloxacin; MIN, minocycline; SXT = trimethoprim-sulfamethoxazole; TET = tetracycline; NIT = nitrofurantoin; NA = not applicable; ST = sequence type. ST5208-like is a SLV of ST5208 (adk: 10; fum: 7; gyr: 265; icd: 8; mdh: 12; purA: 8; recA: 194).
Source: Prepared by the authors from the study results.
Among 110 E. coli analyzed by PFGE (57.3%), a high genetic diversity was observed defining 103 pulsotypes, while 7 isolates were repeatedly non-typeable. MLST was analyzed in the 5 carbapenemase-producing E. coli isolates identifying 5 unrelated STs: ST10, ST156, ST354, ST8492, and a SLV-ST5208 (adk: 10; fum: 7; gyr: 265; icd: 8; mdh: 12; purA: 8; recA: 194).
Mcr-1-bearing plasmids previously characterized from Argentina belonged to the IncI2 incompatibility group with ca. 60 Kb in size (26 - 28). Considering these previous results, a PCR to detect the IncI2 group was performed on all 192 isolates. A high prevalence (164/192; 85.4%) of IncI2 group was observed among the strains. In the 28 IncI2-negative isolates, the presence of IncX4 and IncHI2 incompatibility groups was evaluated, with IncX4 being detected in 18 isolates (9.4%). The remaining 10 isolates (5.2%) were negative for IncI2, IncX4, and IncHI2 incompatibility groups.
DISCUSSION
The global distribution of mcr-1 gene shows that most belong to E. coli and only a few belong to other bacterial species (7, 25). This low frequency of mcr-1 in non-E. coli species is an epidemiological characteristic of the mcr-1 gene dissemination (5, 7, 25). The prevalence of mcr-1 gene in E. coli and K. pneumoniae recovered from bloodstream infections is still low, ~1.0% and ~0.2%, respectively; nevertheless, the clinical impact of this mechanism is not fully understood (10, 26). However, in two recent global surveillance studies, a high proportion of mcr-1 gene was detected among colistin-resistant E. coli clinical isolates, ranging from 32.2% – 42.2% (3, 6). Analyzing data collected in 2012 – 2018 through the National Antimicrobial Resistance Surveillance Network WHONET-Argentina (91 hospitals), among Enterobacterales clinical isolates (excluding community onset infections), an incremental 4.3-fold in colistin resistance was observed in E. coli (0.3% to 1.3%; P < 0.0001) and 2.7-fold in K. pneumoniae (3.3% to 8.9%; P < 0.0001); but no significant difference was observed for E. cloacae (2.4% to 2.1%; P = 1). The rise of colistin resistance in hospital settings may be associated with its increased use in treating human infections caused by MDR or extensively-drug resistant Enterobacterales, particularly KPC-producing K. pneumoniae, and carbapenem-resistant Pseudomonas aeruginosa or Acinetobacter baumannii.
The present study observed a wide distribution of mcr-1 E. coli clinical isolates, most recovered from urine samples, in several provinces and Buenos Aires City. A high proportion of the isolates showed an MDR profile, and nearly one-half were resistant to third-generation cephalosporins, which reduces therapeutic options for systemic infections mainly to carbapenems, aminoglycosides, or combined therapies. The main mechanism of resistance to third-generation cephalosporins was mediated by ESBLs, with CTX-M being the more relevant, as observed by Wise and colleagues in a global analysis (3).
Infections caused by carbapenemase-producing bacteria have high morbidity and mortality rates, so colistin has become a last-resort option for treatment. In Argentina, blaKPC-2 is the main carbapenemase among Enterobacterales; however, during recent years, an increase in detection of blaNDM-1 has been observed, especially in Providencia spp. (14, 18, 24, 27). In the present collection, 5 carbapenemase-producing mcr-1-positive E. coli isolates were detected, yielding an extensively drug-resistance phenotype for which the unique therapeutic option was tigecycline. These 5 carbapenemase-producing E. coli isolates were assigned to unrelated STs; nevertheless, one of them was ST10, the predicted founder of clonal complex 10 (CC10). The CC10 lineage has been defined as an epidemic clone, presents intrinsic ability to acquire antimicrobial resistance genes (including mcr-1), and has been detected in human and animal samples (8). In a previous study, we reported the case of a pediatric patient infected or colonized with 5 NDM-1-producing Enterobacterales, including E. coli M17386 mcr-1 isolate (Table 2). This study provides evidence of intra-patient dissemination of a blaNDM-1 harboring plasmid among 5 Enterobacterales species (18). Additionally, a C. amalonaticus clinical isolate—a species rarely reported to cause human infections, harboring 16 resistance genes including blaNDM-1 and mcr-1 determinants borne on different plasmids—was recently described in our country (14). Worldwide, detection of carbapenem- and mcr-1 colistin-resistant Enterobacterales clinical isolates has also been reported, with NDM, KPC, and OXA-48 being the main enzymes described (10 - 15).
The mcr-1.5 variant was previously described only in three countries: Japan, Bolivia, and Argentina (13, 28, 29). In the present collection, 13.5% of the isolates were positive for this variant. Moreover, the mcr-1.5 variant was detected in 8 of 10 plasmids from E. coli isolates recovered from healthy chickens on commercial farms, and in 4 of 12 E. coli from diarrheic piglets and healthy fattening pigs in Argentina (30). Therefore, tracking the distribution of mcr-1.5 among E. coli isolates from food-producing animals and from humans may be a clue to understanding the dissemination of this gene among different sources.
High clonal diversity among mcr-1 E. coli isolates was observed, as it has been by other authors (2, 3), suggesting that clonal expansion is not involved in the spread of this mechanism. To date, all mcr-1 plasmids from human isolates in Argentina were characterized as a ca. 60kb IncI2 plasmid (14, 28, 31). Similarly, in the present study, a high proportion (85.4%) of IncI2 plasmids was detected. Additionally, the same IncI2 plasmids have been reported in mcr-1 E. coli isolates recovered from gulls, chicken, dogs, and pigs in Argentina (30, 32, 33). The high proportion of IncI2 plasmids among genetically diverse E. coli isolates suggests that in Argentina, these plasmids might be the main vehicle for horizontal dissemination of mcr-1 among human and animal isolates. However, finding IncX4 plasmids in 9.4% of the isolates may indicate recent changes in the genetic platforms involved in mcr-1 dissemination in our country. A different epidemiological scenario seems to occur in Brazil, where IncX4 has been the major incompatibility group reported among mcr-1 harboring plasmids, while IncA/C2 and IncHI2 are rarely reported (7, 8). Moreover, IncX4 and IncI2 were the main incompatibility groups detected in other Latin American countries as well (5, 8). Interestingly, both IncX4 and IncI2 plasmids carry mcr-1 gene as the unique determinant of resistance, while IncHI2 generally harbor multiple resistance determinants (7, 8).
The One Health approach is directed to design and implement programs, policies, legislation, and research to combat antibiotic resistance among multiple sectors, including human, animal, and environmental health. Colistin is known to be used widely to prevent infection and promote growth in food-producing animals (1, 2, 34). The use of colistin in food animals is believed to be responsible for the emergence and transmission of mcr-genes (1, 2). It has been suggested that mcr-carrying plasmids move from animals to humans, since mcr-genes are prevalent in animal food production, which is where the most colistin is consumed (1, 4, 34). According to the National Antimicrobial Resistance Surveillance (35), within the National Service for Safety and Quality of Food and Agriculture of Argentina, high levels of colistin resistance were observed in poultry (31.5%), cattle (16.5%), and pigs (15.0%). In Argentina, mcr-1 gene has been reported not only in E. coli isolates recovered from chicken and swine, but also from pets and wild birds (32, 33). This suggests that this gene is successfully circulating among different environments. Given the situation and to reserve this drug for treating human infections, in January 2019 the Ministry of Agriculture of Argentina banned the use of colistin for veterinary purposes (available from http://servicios.infoleg.gob.ar/infolegInternet/anexos/315000-319999/318811/norma.htm). This initiative aims to intensify stewardship efforts for this last-resort antibiotic.
Limitations
This study’s main limitation was that the presence of other mcr-genes (mcr-2 to mcr-9) was not evaluated, nor were mcr-genes among species other than E. coli. Even though this collection of mcr-1 E. coli isolates represents the largest and most diverse in Argentina, local and/or regional epidemiological differences are probable and expected.
Conclusions
The study findings show that mcr-1 E. coli is circulating among several provinces in Argentina. Mcr-1 E. coli is associated with MDR, with CTX-M being the main ESBL. Although infrequently, mcr-1 E. coli co-producing NDM or KPC carbapenemases have emerged. The presence of IncI2 plasmids among highly diverse E. coli clones indicates that they have driven wide distribution of the mcr-1 gene among clinical isolates through horizontal transmission.
This study provides a basic framework for understanding the molecular epidemiology of mcr-1-positive E. coli in Argentina. For comprehensive picture from the perspective of One Health, further studies are essential to understanding the dissemination of mcr-1 gene through the environment.
Funding.
This work was supported by the regular federal budget of the National Ministry of Health of Argentina and the Préstamo BID-PICT-2016-3154 to D.F. from ANPCYT. The funders had no role in the study design, data collection or analysis, decision to publish, or preparation of the manuscript.
Disclaimer.
Authors hold sole responsibility for the views expressed in the manuscript, which may not necessarily reflect the opinion or policy of the RPSP/PAJPH and/or PAHO.
Acknowledgments
Members of the MCR Group: Isabel Mirayes (Hospital Carlos Durand, Buenos Aires City); Adriana De Paulis (Instituto Lanari, Buenos Aires City); Adriana Ernst (Hospital de Niños V.J. Vilela, Santa Fe); Amalia Noemi Farias (Hospital Transito Cáceres de Allende, Córdoba); Ana Berejnoi (Hospital Público Materno Infantil, Salta); Ana Maria Togneri (Hospital Evita de Lanus, Buenos Aires); Ana Patricia Meo (Hospital Ramos Mejia, Buenos Aires City); Ana Sangoy (Hospital Antonio Centrangolo, Buenos Aires); Beatriz Cristina Garcia (Hospital Pediátrico Dr. Humberto Notti, Mendoza); Bernardo Lagos (Hospital Dr. R. Larcade, Buenos Aires); Cecilia Vescina (Hospital de Niños Sor Maria Ludovica, Buenos Aires); Claudia Fernandez (Hospital de Pediatría S.A.M.I.C. Pr. Dr. Juan Garrahan, Buenos Aires City); Claudia Fontan (Hospital 4 De Junio “Dr. Ramon Carrillo,” Buenos Aires); Dina Pedersen (Hospital Municipal de Agudos Dr. Leonidas Lucero, Buenos Aires); Echegaray Marina and Pace Julio (Trinidad Ramos Mejía, Buenos Aires); Mariela Schijman (Hospital Alvarez - Laboratorio Central, Buenos Aires City); Eduardo Gregorini and Laura Colombo (Hospital Escuela Eva Perón, Santa Fe); Estefania Biondi (Hospital de Niños “Dr. Ricardo Gutierrez,” Buenos Aires City); Flavia Amalfa (Hospital General de Agudos Parmenio Piñero, Buenos Aires City); Geni Bruni (Hospital El Carmen OSEP, Mendoza); Gladis Pino de Muñoz (Hospital Nuevo San Roque, Córdoba); Hebe Gullo (HIGA Vicente Lopez y Planes, Buenos Aires); Herman Sauer (Hospital Heller, Neuquén); Jimena Minoli (Hospital Córdoba, Córdoba); Josefina M. Villegas and Guillermo Garcia (Hospital Distrital Caleta Olivia, Santa Cruz); Laura Errecalde (Hospital Juan A. Fernandez, Buenos Aires City); Leonardo Papantoniou and Marcela Gonzalez (Laboratorio Manlab, Buenos Aires City); Liliana Lorena Gonzalez (Hospital Infantil Municipal de Córdoba, Córdoba); Lucia Velasco (Clínica Santa Isabel, Buenos Aires City); Silvana Manganello (Hospital Velez Sarsfield, Buenos Aires City); Marcelo Casabona (Hospital Lucio Molas, La Pampa); Marcelo Toffoli (Hospital de Niños Dr. Hector Quintana, Jujuy); Maria Gabriela Rivollier (Hospital Artemides Zatti, Rio Negro); Maria Rosa Baroni (Hospital De Niños “Dr. Orlando Alassia,” Santa Fe); Maria Rosa Nuñez (Hospital Provincial Neuquen “Dr. Castro Rendon,” Neuquén); Mariana Boleas (Hospital San Martin de Paraná, Entre Ríos); Mariana Carol Rey (Hospital Dr. Julio Perrando, Chaco); Marina Bottiglieri (Clínica Universitaria Reina Fabiola, Córdoba); Marta Beatriz Giovanakis (Hospital Británico de Buenos Aires, Buenos Aires City); Monica Machain (Hospital Interzonal de Junín-HIGA Dr. Abraham Piñeyro, Buenos Aires); Monica Millara (Hospital General de Agudos J. M. Penna, Buenos Aires City); Monica Patricia Borgo (Maternidad Martin-CEMAR-DSLAC, Santa Fe); Mriam Mortarini (Hospital F.J. Muñiz, Buenos Aires City); Myrian Figueroa (Hospital Nuestra Señora de la Misericordia, Córdoba); Nancy Ruth Vega (Hospital Marcial Quiroga, San Juan); Patricia Marchiaro (Hospital Centenario, Santa Fe); Patricia Valdez and Marta Ferres (Hospital Interzonal de Niños Eva Perón, Catamarca); Rosana Padlog Mais (HIGA San Martin, Buenos Aires); Sabrina De Bunder (Hospital Zonal Bariloche, Río Negro); Sandra Valle (Instituto Alexander Fleming, Buenos Aires City); María Rosa Sarrouf (Hospital Luis Lagomaggiore, Mendoza); Sonia Del Valle Medieta (Hospital Pablo Soria, Jujuy); Ana Julia Tanco (Hospital Español, Buenos Aires City); Teresa Nilda Lopez (Hospital Guillermo Rawson, Córdoba); Viviana Del Valle David (Hospital Interzonal San Juan Bautista, Catamarca); Viviana Vilches (Hospital Universitario Austral, Buenos Aires); Graciela Greco (Hospital Italiano de Buenos Aires, Buenos Aires City); Andrea Vila and Hugo Pagella (Hospital Italiano de Mendoza, Mendoza); Daniela Chianaino (HIGA “Dr. Oscar E. Alende,” Buenos Aires); Mónica Vallejo (Hospital Privado de Comunidad, Buenos Aires).
Footnotes
Author contributions.
DF and AC conceived the original idea. DF, MR, EA, FP, and AC planned the experiments. DF, MR, EA, FC, JDM, DDB, CL, SG, DD, and MG collected, contributed, and analyzed the data. DF, MR, EA, CL, SG, FP, and AC interpreted the results. DF and AC wrote the paper. All authors reviewed and approved the final version.
Conflicts of interest.
None declared.
REFERENCES
- 1.Sun J, Zhang H, Liu YH, Feng Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018;26(9):794–808. doi: 10.1016/j.tim.2018.02.006. [DOI] [PubMed] [Google Scholar]; 1. Sun J, Zhang H, Liu Y-H, Feng Y. Towards Understanding MCR-like Colistin Resistance. Trends Microbiol. 2018;26(9):794-808. [DOI] [PubMed]
- 2.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]; 2. Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16: (2):161–168. 2016. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed]
- 3.Wise MG, Estabrook MA, Sahm DF, Stone GG, Kazmierczak KM. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS ONE. 2018;13(4):e0195281. doi: 10.1371/journal.pone.0195281. [DOI] [PMC free article] [PubMed] [Google Scholar]; 3. Wise MG, Estabrook MA, Sahm DF, Stone GG, Kazmierczak KM. Prevalence of mcr-type genes among colistin-resistant Enterobacteriaceae collected in 2014-2016 as part of the INFORM global surveillance program. PLoS ONE. 2018. 13(4):e0195281. [DOI] [PMC free article] [PubMed]
- 4.Al-Tawfiq JA, Laxminarayan R, Mendelson M. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int J Infect Dis. 2017;54:77–84. doi: 10.1016/j.ijid.2016.11.415. [DOI] [PubMed] [Google Scholar]; 4. Al-Tawfiq JA, Laxminarayan R, Mendelson M. How should we respond to the emergence of plasmid-mediated colistin resistance in humans and animals? Int J Infect Dis. 2017;54:77–84. doi: 10.1016/j.ijid.2016.11.415. [DOI] [PubMed]
- 5.Mendes Oliverira VR, Paiva MC, Lima WG. Plasmid-mediated colistin resistance in Latin America and Caribbean: A systematic review. Travel Med Infect Dis. 2019;20:101459. doi: 10.1016/j.tmaid.2019.07.01. [DOI] [PubMed] [Google Scholar]; 5. Mendes Oliveira VR, Paiva MC, Lima WG. Plasmid-mediated colistin resistance in Latin America and Caribbean: A systematic review. Travel Med Infect Dis. 2019;20:101459. doi: 10.1016/j.tmaid.2019.07.01 [DOI] [PubMed]
- 6.Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RM, Flamm RK. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother. 2016;60(9):5623–5624. doi: 10.1128/AAC.01267-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; 6. Castanheira M, Griffin MA, Deshpande LM, Mendes RE, Jones RN, Flamm RK. Detection of mcr-1 among Escherichia coli clinical isolates collected worldwide as part of the SENTRY Antimicrobial Surveillance Program in 2014 and 2015. Antimicrob Agents Chemother. 2016;60(9):5623-4. doi: 10.1128/AAC.01267-16. [DOI] [PMC free article] [PubMed]
- 7.Wang R, Van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, Jin L, Zhang Q, Liu Y, Rieux A, Dorai-Schneiders T, Weinert LA, Iqbal Z, Didelot X, Wang H, Balloux F. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018;9(1):1179. doi: 10.1038/s41467-018-03205-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; 7. Wang R, van Dorp L, Shaw LP, Bradley P, Wang Q, Wang X, Jin L, Zhang Q, Liu Y, Rieux A, Dorai-Schneiders T, Weinert LA, Iqbal Z, Didelot X, Wang H, Balloux F. The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat Commun. 2018; 9(1):1179. [DOI] [PMC free article] [PubMed]
- 8.Matamoros S, van Hattem JM, Arcilla MS, Willemse N, Melles DC, Penders J, Vinh TN, Thi Hoa N, consortium COMBAT, de Jong MD, Schultsz C. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7(1):15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; 8. Matamoros S, van Hattem JM, Arcilla MS, Willemse N, Melles DC, Penders J, Vinh TN, Thi Hoa N, COMBAT consortium, de Jong MD, Schultsz C. Global phylogenetic analysis of Escherichia coli and plasmids carrying the mcr-1 gene indicates bacterial diversity but plasmid restriction. Sci Rep. 2017;7(1):15364. doi: 10.1038/s41598-017-15539-7. [DOI] [PMC free article] [PubMed]
- 9.Snesrud E, McGann P, Chandler M. The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: the Vehicle for Transferable Colistin Resistance. MBio. 2018;9(1):e02381–e02417. doi: 10.1128/mBio.02381-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; 9. Snesrud E, McGann P, Chandler M. The Birth and Demise of the ISApl1-mcr-1-ISApl1 Composite Transposon: the Vehicle for Transferable Colistin Resistance. MBio. 2018; 9(1). pii: e02381-17. [DOI] [PMC free article] [PubMed]
- 10.Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M, Yu Y. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400–410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed] [Google Scholar]; 10. Quan J, Li X, Chen Y, Jiang Y, Zhou Z, Zhang H, Sun L, Ruan Z, Feng Y, Akova M, Yu Y. Prevalence of mcr-1 in Escherichia coli and Klebsiella pneumoniae recovered from bloodstream infections in China: a multicentre longitudinal study. Lancet Infect Dis. 2017;17(4):400-410. doi: 10.1016/S1473-3099(16)30528-X. [DOI] [PubMed]
- 11.Delgado-Blas JF, Ovejero CM, Abadia-Patiño L, Gonzalez-Zorn B. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother. 2016;60(10):6356–6358. doi: 10.1128/AAC.01319-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; 11. Delgado-Blas JF, Ovejero CM, Abadia-Patiño L, Gonzalez-Zorn B. Coexistence of mcr-1 and blaNDM-1 in Escherichia coli from Venezuela. Antimicrob Agents Chemother. 2016;60(10):6356-8. doi: 10.1128/AAC.01319-16. [DOI] [PMC free article] [PubMed]
- 12.Dalmolin TV, Wink PL, de Lima-Morales D, Barth AL. Low prevalence of the mcr-1 gene among carbapenemase-producing clinical isolates of Enterobacterales. Infect Control Hosp Epidemiol. 2018;40(2):263–264. doi: 10.1017/ice.2018.301. [DOI] [PubMed] [Google Scholar]; 12. Dalmolin TV, Wink PL, de Lima-Morales D, Barth AL. Low prevalence of the mcr-1 gene among carbapenemase-producing clinical isolates of Enterobacterales. Infect Control Hosp Epidemiol. 2018;40(2):263-264. doi: 10.1017/ice.2018.301. [DOI] [PubMed]
- 13.Giani T, Sennati S, Antonelli A, Di Pilato V, di Maggio T, Mantella A, Niccolai C, Spinicci M, Monasterio J, Castellanos P, Martinez M, Contreras F, Balderrama Villaroel D, Damiani E, Maury S, Rocabado R, Pallecchi L, Bartoloni A, Rossolini GM. High prevalence of carriage of mcr-1-positive enteric bacteria among healthy children from rural communities in the Chaco region, Bolivia, September to October 2016. Euro Surveill. 2018;23(45):1800115. doi: 10.2807/1560-7917.ES.2018.23.45.1800115. [DOI] [PMC free article] [PubMed] [Google Scholar]; 13. Giani T, Sennati S, Antonelli A, Di Pilato V, di Maggio T, Mantella A, Niccolai C, Spinicci M, Monasterio J, Castellanos P, Martinez M, Contreras F, Balderrama Villaroel D, Damiani E, Maury S, Rocabado R, Pallecchi L, Bartoloni A, Rossolini GM. High prevalence of carriage of mcr-1-positive enteric bacteria among healthy children from rural communities in the Chaco region, Bolivia, September to October 2016. Euro Surveill. 2018;23(45):1800115. doi:10.2807/1560-7917.ES.2018.23.45.1800115. [DOI] [PMC free article] [PubMed]
- 14.Faccone D, Albornoz E, Tijet N, Biondi E, Gomez S, Pasterán F, Zaquez M, Melano RG, Corso A. Characterization of a multidrug resistant Citrobacter amalonaticus clinical isolate harboring blaNDM-1 and mcr-1.5 genes. Infect Genet Evol. 2019;67:51–54. doi: 10.1016/j.meegid.2018.10.020. [DOI] [PubMed] [Google Scholar]; 14. Faccone D, Albornoz E, Tijet N, Biondi, E, Gomez S, Pasterán F, Vazquez M, Melano RG, Corso A. Characterization of a multidrug resistant Citrobacter amalonaticus clinical isolate harboring blaNDM-1 and mcr-1.5 genes. Infect Genet Evol. 2019;67:51–54. doi:10.1016/j.meegid.2018.10.020 [DOI] [PubMed]
- 15.Lin YC, Kuroda M, Suzuki S, Mua JJ. Emergence of an Escherichia coli strain co-harboring mcr-1 and blaNDM-9 from a urinary tract infection in Taiwan. J Global Antimicrob Resist. 2019;16:286–290. doi: 10.1016/j.jgar.2018.10.003. [DOI] [PubMed] [Google Scholar]; 15. Lin YC, Kuroda M, Suzuki S, Mua JJ. Emergence of an Escherichia coli strain co-harboring mcr-1 and blaNDM-9 from a urinary tract infection in Taiwan. J Global Antimicrob Resist. 2019;16:286–290. doi:10.1016/j.jgar.2018.10.003. [DOI] [PubMed]
- 16.Servicio Antimicrobianos, INEI-ANLIS “Dr. Carlos G. Malbrán” Alerta epidemiológico: Emergencia de resistencia plasmídica (transferible) a colistina/polimixina B, mcr-1 en Argentina. Boletin informativo No. Feb 3, 2016. [Accessed on 10 March 20202]. Available from : http://antimicrobianos.com.ar/ATB/wp-content/uploads/2016/02/Alerta-epidemiol%C3%B3gico.pdf.Spanish.; 16. Servicio Antimicrobianos, INEI-ANLIS “Dr. Carlos G. Malbrán”. Alerta epidemiológico: Emergencia de resistencia plasmídica (transferible) a colistina/polimixina B, mcr-1 en Argentina. Boletin informativo No. 3 – Feb. 2016. Available from : http://antimicrobianos.com.ar/ATB/wp-content/uploads/2016/02/Alerta-epidemiol%C3%B3gico.pdf.Spanish. Accessed on 10 March 20202.
- 17.Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, Corso A, on behalf of the MCR Group First Description of mcr-1-Mediated Colistin Resistance in Human Infections Caused by Escherichia coli in Latin America. Antimicrob Agents Chemother. 2016;60(7):4412–4413. doi: 10.1128/AAC.00573-16. [DOI] [PMC free article] [PubMed] [Google Scholar]; 17. Rapoport M, Faccone D, Pasteran F, Ceriana P, Albornoz E, Petroni A, on behalf of the MCR Group, Corso A. First Description of mcr-1-Mediated Colistin Resistance in Human Infections Caused by Escherichia coli in Latin America. Antimicrob Agents Chemother. 2016;60(7):4412–4413. doi:10.1128/AAC.00573-16 [DOI] [PMC free article] [PubMed]
- 18.Martino F, Tijet N, Melano R, Petroni A, Heinz E, De Belder D, Faccone D, Rapoport M, Biondi E, Rodrigo V, Vazquez M, Pasteran F, Thomson NR, Corso A, Gomez SA. Isolation of five Enterobacteriaceae species harbouring blaNDM-1 and mcr-1 plasmids from a single paediatric patient. PLoS ONE. 2019;14(9):e0221960. doi: 10.1371/journal.pone.0221960. [DOI] [PMC free article] [PubMed] [Google Scholar]; 18. Martino F, Tijet N, Melano R, Petroni A, Heinz E, De Belder D, Faccone D, Rapoport M, Biondi E, Rodrigo V, Vazquez M, Pasteran F, Thomson NR, Corso A, Gomez SA. Isolation of five Enterobacteriaceae species harbouring blaNDM-1 and mcr-1 plasmids from a single paediatric patient. PLoS ONE. 2019;14(9):e0221960. doi:10.1371/journal.pone.0221960 [DOI] [PMC free article] [PubMed]
- 19.Pasteran F, Danze D, Canrera C, Lucero C, Menocal A, Albornoz E, Castillo I, Rapoport M, Ceriana P, Gagetti P, Corso A. European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Abstract O0952; Madrid, Spain: 2018. Development and validation of simple tests (agar spot, colistin drop, 1ml-broth disk elution MIC and tablet pre-diffusion) as an alternative to improve accuracy in screening chromosomal and plasmid-mediated colistin resistance in Gram-negative bacilli F. [Google Scholar]; 19. Pasteran F, Danze D, Cabrera C, Lucero C, Menocal A, Albornoz E, Castillo I, Rapoport M, Ceriana P, Gagetti P, Corso A. Development and validation of simple tests (agar spot, colistin drop, 1ml-broth disk elution MIC and tablet pre-diffusion) as an alternative to improve accuracy in screening chromosomal and plasmid-mediated colistin resistance in Gram-negative bacilli F. European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) 2018. Abstract O0952, Madrid, Spain.
- 20.Wayne PAS Ed 2019, p Suppl M100-S11. 2019. NCLSI National committee for clinical laboratory standards: performance standards for antimicrobial testing NCCLS. [Google Scholar]; 20. NCLSI National committee for clinical laboratory standards: performance standards for antimicrobial testing NCCLS (2019), Wayne PAS Ed 2019, p Suppl M100-S11.
- 21.EUCAST European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters (Version 9.0) 2019. [Accessed on 10 March 2020]. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf.; 21. EUCAST European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters (Version 9.0). 2019. Available from: http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_9.0_Breakpoint_Tables.pdf Accessed on 10 March 2020.
- 22.Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50. doi: 10.1016/j.plasmid.2012.03.001. [DOI] [PubMed] [Google Scholar]; 22. Johnson TJ, Bielak EM, Fortini D, Hansen LH, Hasman H, Debroy C, Nolan LK, Carattoli A. Expansion of the IncX plasmid family for improved identification and typing of novel plasmids in drug-resistant Enterobacteriaceae. Plasmid. 2012;68(1):43–50. doi:10.1016/j.plasmid.2012.03.001. [DOI] [PubMed]
- 23.Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother. 2013;57(10):5019–5025. doi: 10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed] [Google Scholar]; 23. Chen L, Chavda KD, Al Laham N, Melano RG, Jacobs MR, Bonomo RA, Kreiswirth BN. Complete nucleotide sequence of a blaKPC-harboring IncI2 plasmid and its dissemination in New Jersey and New York hospitals. Antimicrob Agents Chemother. 2013;57(10):5019–5025. doi:10.1128/AAC.01397-13. [DOI] [PMC free article] [PubMed]
- 24.De Belder D, Lucero C, Rapoport M, Rosato A, Faccone D, Petroni A, Pasteran F, Albornoz E, Corso A, Gomez SA. Genetic Diversity of KPC-Producing Escherichia coli, Klebsiella oxytoca, Serratia marcescens, and Citrobacter freundii Isolates from Argentina. Microb Drug Resist. 2018;24(7):958–965. doi: 10.1089/mdr.2017.0213. [DOI] [PubMed] [Google Scholar]; 24. De Belder D, Lucero C, Rapoport M, Rosato A, Faccone D, Petroni A, Pasteran F, Albornoz E, Corso A, Gomez SA. Genetic Diversity of KPC-Producing Escherichia coli, Klebsiella oxytoca, Serratia marcescens, and Citrobacter freundii Isolates from Argentina. Microb Drug Resist. 2018;24(7):958–965. doi:10.1089/mdr.2017.0213 [DOI] [PubMed]
- 25.Elbediwi M, Li Y, Paudyal N, Pan H, Li X, Xie S, Rajkovic A, Feng Y, Fang W, Rankin SC, Yue M. Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980–2018) Microorganisms. 2019;7(10):461. doi: 10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed] [Google Scholar]; 25. Elbediwi M, Li Y, Paudyal N, Pan H, Li X, Xie S, Rajkovic A, Feng Y, Fang W, Rankin SC, Yue M. Global Burden of Colistin-Resistant Bacteria: Mobilized Colistin Resistance Genes Study (1980–2018). Microorganisms. 2019;7(10):461. doi:10.3390/microorganisms7100461. [DOI] [PMC free article] [PubMed]
- 26.Zheng B, Xu H, Yu X, Jiang X, Zhang J, Chen Y, Huang J, Huang C, Xiao Y. Low prevalence of MCR-1-producing Klebsiella pneumoniae in bloodstream infections in China. Clin Microbiol Infect. 2018;24(2):205–206. doi: 10.1016/j.cmi.2017.08.004. [DOI] [PubMed] [Google Scholar]; 26. Zheng B, Xu H, Yu X, Jiang X, Zhang J, Chen Y, Huang J, Huang C, Xiao Y. Low prevalence of MCR-1-producing Klebsiella pneumoniae in bloodstream infections in China. Clin Microbiol Infect. 2018;24(2):205–206. doi:10.1016/j.cmi.2017.08.004. [DOI] [PubMed]
- 27.Faccone D, Pasteran F, Albornoz E, Ceriana P, Gomez S, Lucero C, Rapoport M, Corso A, NDM Argentina Group . 26th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) Amsterdam; Netherlands: 2016. Molecular Epidemiology of Providencia spp. Harbouring NDM Carbapenemase in Argentina. Abstract P0293. [Google Scholar]; 27. Faccone D, Pasteran F, Albornoz E, Ceriana P, Gomez S, Lucero C, Rapoport M, NDM Argentina Group, Corso A. Molecular Epidemiology of Providencia spp. Harbouring NDM Carbapenemase in Argentina. 26th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) 2016. Abstract P0293, Amsterdam, Netherlands.
- 28.Tijet N, Faccone D, Rapoport M, Seah C, Pasteran F, Ceriana P, Albornoz E, Corso A, Petroni A, Melano RG. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS ONE. 2017;12:e0180347. doi: 10.1371/journal.pone.0180347. [DOI] [PMC free article] [PubMed] [Google Scholar]; 28. Tijet N, Faccone D, Rapoport M, Seah C, Pasteran F, Ceriana P, Albornoz E, Corso A, Petroni A, Melano RG. Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS ONE. 2017;12: e0180347. [DOI] [PMC free article] [PubMed]
- 29.Ishii Y, Aoki K, Endo S, Kiyota H, Aoyagi T, Kaku M, Bonomo RA, Tateda K. Spread of mcr-1.5 in the community: an emerging threat. Int J Antimicrob Agents. 2018;51(1):161–162. doi: 10.1016/j.ijantimicag.2017.10.015. [DOI] [PubMed] [Google Scholar]; 29. Ishii Y, Aoki K, Endo S, Kiyota H, Aoyagi T, Kaku M, Bonomo RA, Tateda K. Spread of mcr-1.5 in the community: an emerging threat. Int J Antimicrob Agents. 2018;51(1):161–162. doi:10.1016/j.ijantimicag.2017.10.015. [DOI] [PubMed]
- 30.Dominguez JE, Faccone D, Tijet N, Gomez S, Corso A, Fernández-Miyakawa ME, Melano RG. Characterization of Escherichia coli carrying mcr-1-plasmids recovered from food animals from Argentina. Front Cell Infect Microbiol. 2019;9:41. doi: 10.3389/fcimb.2019.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]; 30. Dominguez JE, Faccone D, Tijet N, Gomez S, Corso A, Fernández-Miyakawa ME, Melano RG. Characterization of Escherichia coli carrying mcr-1-plasmids recovered from food animals from Argentina. Front Cell Infect Microbiol. 2019;9:41. doi:10.3389/fcimb.2019.00041. [DOI] [PMC free article] [PubMed]
- 31.Elena A, Cejas D, Magariños F, Jewtuchowicz V, Facente A, Gutkind G, Di Conza J, Radice M. Spread of clonally related Escherichia coli strains harboring an IncA/C1 plasmid encoding IMP-8 and its recruitment into an unrelated MCR-1-containing isolate. Antimicrob Agents Chemother. 2018;62(6):e02414–e02417. doi: 10.1128/AAC.02414-17. [DOI] [PMC free article] [PubMed] [Google Scholar]; 31. Elena A, Cejas D, Magariños F, Jewtuchowicz V, Facente A, Gutkind G, Di Conza J, Radice M. Spread of clonally related Escherichia coli strains harboring an IncA/C1 plasmid encoding IMP-8 and its recruitment into an unrelated MCR-1-containing isolate. Antimicrob Agents Chemother. 2018;62(6):e02414-17. doi:10.1128/AAC.02414-17. [DOI] [PMC free article] [PubMed]
- 32.Liakopoulos A, Mevius DJ, Olsen B, Bonnedahl J. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother. 2016;71(8):2335–2336. doi: 10.1093/jac/dkw262. [DOI] [PubMed] [Google Scholar]; 32. Liakopoulos A, Mevius DJ, Olsen B, Bonnedahl J. The colistin resistance mcr-1 gene is going wild. J Antimicrob Chemother. 2016;71(8):2335–2336. doi:10.1093/jac/dkw262 [DOI] [PubMed]
- 33.Rumi MV, Mas J, Elena A, Cerdeira L, Muñoz ME, Lincopan N, Gentilini ER, Di Conza J, Gutkind G. Co-occurrence of clinically relevant β-lactamases and MCR-1 encoding genes in Escherichia coli from companion animals in Argentina. Vet Microbiol. 2019;230:228–234. doi: 10.1016/j.vetmic.2019.02.006. [DOI] [PubMed] [Google Scholar]; 33. Rumi MV, Mas J, Elena A, Cerdeira L, Muñoz ME, Lincopan N, Gentilini ER, Di Conza J, Gutkind G. Co-occurrence of clinically relevant β-lactamases and MCR-1 encoding genes in Escherichia coli from companion animals in Argentina. Vet Microbiol. 2019;230:228–234. doi:10.1016/j.vetmic.2019.02.006. [DOI] [PubMed]
- 34.Rhouma M, Beaudry F, Thériault W, Letellier A. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front Microbiol. 2016;7:1789. doi: 10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed] [Google Scholar]; 34. Rhouma M, Beaudry F, Thériault W, Letellier A. Colistin in pig production: chemistry, mechanism of antibacterial action, microbial resistance emergence, and One Health perspectives. Front Microbiol. 2016;7:1789. doi:10.3389/fmicb.2016.01789. [DOI] [PMC free article] [PubMed]
- 35.Comisión Nacional para el Control de la Resistencia a los Antimicrobianos. Resistencia Antimicrobiana: Estado Actual República Argentina. 2019. [Accessed on 10 March 2020]. Available from__ http://cvpba.org/wp-content/uploads/2019/10/folletoRAM2019Final.pdf.; 35. Comisión Nacional para el Control de la Resistencia a los Antimicrobianos. Resistencia Antimicrobiana: Estado Actual República Argentina. 2019. Available from__http://cvpba.org/wp-content/uploads/2019/10/folletoRAM2019Final.pdf Accessed on 10 March 2020.


