Abstract
Background:
There is a lack of precision medicine in pancreatic ductal adenocarcinoma (PDA) and related cancers, and outcomes for patients with this diagnosis remain poor despite decades of research investigating this disease. Therefore, it is necessary to explore novel therapeutic options for these patients who may benefit from personalized therapies.
Objective:
Molecular profiling of hepatopancreaticobiliary malignancies at our institution, including but not limited to PDA, was initiated to assess the feasibility of incorporating molecular profiling results into patient oncological therapy planning.
Methods:
All eligible patients from Thomas Jefferson University (TJU) with hepatopancreaticobiliary tumors including PDA, who agreed to molecular testing profiling, were prospectively enrolled in a registry study from December 2014 to September 2017 and their tumor samples were tested to identify molecular markers that can be used to guide therapy options in the future. Next generation sequencing (NGS) and protein expression in tumor samples were tested at CLIA-certified laboratories. Prospective clinicopathologic data were extracted from medical records and compiled in a de-identified fashion.
Results:
Seventy eight (78) patients were enrolled in the study, which included 65/78 patients with PDA (local and metastatic) and out of that subset, 52/65 patients had surgically resected PDA. Therapy recommendations were generated based on molecular and clinicopathologic data for all enrolled patients. NGS uncovered actionable alterations in 25/52 surgically resected PDAs (48%) which could be used to guide therapy options in the future. High expression of three proteins, TS (p ¼ 0.005), ERCC1 (p = 0.001), and PD-1 (p = 0.04), was associated with reduced recurrence-free survival (RFS), while TP53 mutations were correlated with longer RFS (p = 0.01).
Conclusions:
The goal of this study was to implement a stepwise strategy to identify and profile resected PDAs at our institution. Consistent with previous studies, approximately half of patients with resected PDA harbor actionable mutations with possible targeted therapeutic implications. Ongoing studies will determine the clinical value of identifying these mutations in patients with resected PDA.
1. Introduction
In recent years, there has been great interest in the role that precision medicine might play in the treatment of patients with pancreatic ductal adenocarcinoma (PDA) who have an overall five year survival rate of nine percent [1]. After decades of research investigating this devastating disease, the main treatment options for advanced disease are a combination of chemotherapeutics [2,3] that offer only a limited survival benefit [4,5]. For example, ESPAC-4 showed a survival benefit of just over two months in patients with resected PDA receiving gemcitabine and capecitabine over gemcitabine alone [3]. The minimal survival benefit and known cytotoxicity of these available regimens make it imperative to explore novel therapeutic options for these patients. Thus, it is important to develop methods that can accurately select patients who may derive benefit from existing targeted therapies (i.e., a personalized therapeutic approach). One method to streamline this process is to consent a patient for molecular profiling at the time of surgical consent, and at the time of resection, send it directly for sequencing. Once processed and analyzed these results can be made available seamlessly into an electronic health record system for the provider [6].
The main goal of molecular profiling is to understand the specific biologic events driving a patient’s tumor. In the process of profiling, the hope is to identify actionable mutations that can be targeted with therapy [7]. The foundation of molecular profiling is next generation sequencing (NGS) which can identify mutations, gene copy number alterations, and rearrangements [8–10], and has been applied in several studies of PDA [11–14]. The multitude of potentially actionable mutations identified in PDA parallels other tumor types [7,15–17]. In fact, it was estimated in a recent study that roughly a quarter of all PDAs sequenced have an actionable mutation [6]. Personalized oncology has been applied to recent “basket studies” that categorize patients based on their actionable mutation for therapy as opposed to their pathology or tumor type [18–22].
Based on the aforementioned studies, we aimed to test the feasibility of consenting and profiling patients with PDA from a high volume single institution practice [23]. We focused primarily on resected PDA patients, with the ultimate goal to develop a personalized therapeutic approach that would complement surgical resection of this disease.
2. Methods
2.1. Patients
For this study, our inclusion criteria included any patient from Thomas Jefferson University (TJU) with hepatopancreaticobiliary cancers who agreed and gave consent for this study from December 2014 to September 2017. All patients had a tissue diagnosis of pancreatic ductal adenocarcinoma (PDA) cancer, either endoscopic or surgical, but did not have to be a surgical candidate necessarily. For the purposes of this study, all other non-PDA samples were excluded for this study analysis. Patients were enrolled during December 2014 to September 2017 and offered participation in the study. This study was performed with IRB-approval and an approved biobanking protocol.
2.2. Multi-omic profiling and therapy recommendation
To assess patient tumors for potentially actionable findings, resection or biopsy specimens were sent to Perthera, Inc. (McLean, VA) for a comprehensive multi-omic profile. Perthera, in collaboration with TJU, collected clinicopathologic data, including tumor characteristics and past treatment history for correlational analysis. Clinical variables included gender, age, smoking history, past medical history (diabetes, pancreatitis, hypertension, hyperlipidemia, arthritis, GERD, obesity), and family history of cancer. Pathologic variables assessed included stage at diagnosis, location of tumor (head vs. body/tail), tumor grade, tumor size, pathologic staging, lymph node positivity, presence of IPMN, lympho-vascular invasion, and perineural invasion. To see if there were any association between pathological variables and molecular variables, we performed statistical analysis on NGS samples with a cutoff of three or more genes mutated, or IHC samples with a cutoff of four or more gene alterations. This yielded 39 variables, for a total of 741 comparisons.
Tissue specimens collected were sent to CLIA-certified, CAP-accredited labs for two types of profiling: next generation sequencing (NGS) using the FoundationOne test from Foundation Medicine, Inc. (Cambridge, MA) and protein immunohistochemistry (IHC) from either Caris Life Sciences (Irving, TX) or NeoGenomics (Fort Myers, FL). The FoundationOne testing platform was used to compare patient sample genomics to 315 cancer-related genes plus introns from 28 genes often altered in cancer to identify these cancer driver genes [24]. The clinical and molecular data were integrated and processed in the Perthera Therapeutic Intelligence Engine to generate matching therapies and clinical trials.
2.3. Data infrastructure processing
To assure a streamline process, samples once collected were sent directly to Perthera and their collaborating companies for NGS processing and FoundationOne testing. Once the reports were generated, Perthera’s molecular tumor board reviewed the treatment recommendations individually and provided a summary to the referring physician. The referring physician would then review the findings and conclusions with the patients to discuss their therapy options. Further details on the Perthera workflow and method development have been previously published [6].
2.4. Statistical analysis
Correlations among molecular and clinicopathologic variables were assessed using Fisher’s exact test, implemented with the fisher.test function in the stats package of the R statistical programming language. Correlations between molecular and clinicopathologic variables and recurrence-free survival (RFS) and overall survival (OS) were assessed using a log-rank test of Kaplan-Meier survival estimates, implemented using the survdiff function in the survival package of R.
3. Results
3.1. All samples: tissue collection rates for all patients included
During the study timeframe, Perthera delivered multi-omic profiling reports for a total of 78 TJU patients with hepatopancreaticobiliary cancers. The majority of these were pancreatic ductal adenocarcinoma cases (PDA, n = 65, Table 1), and the remaining 13 cases were non-PDA cancers and were excluded from this study analysis. Although tissue specimen quality is often a limiting factor in obtaining molecular test results, genomic and proteomic profiling had high success rates in this study. NGS results were obtained for all the patients, while IHC results were obtained for nearly all the patients (75/78, 96%). For the current analysis, we focused only on PDA samples collected for molecular testing (n = 65) for this study’s analysis.
Table 1.
Type of tumor and resection in this cohort of patients: Summary of all 65 patients included in the study with baseline demographics, diagnosis and status at tissue collection.
| Surgery | Biopsy | |
|---|---|---|
| Diagnosis | ||
| Pancreatic Adenocarcinoma | 52 | 13 |
| Age | ||
| <50 | 2 | |
| 50–59 | 11 | 1 |
| 60–69 | 20 | 7 |
| >70 | 19 | 5 |
| Gender | ||
| Male | 25 | 8 |
| Female | 27 | 5 |
| Status at Tissue Collection | ||
| Localized/Borderline Resectable | 51 | 1 |
| Metastatic | 0 | 13 |
3.2. Multi-omic profiles reveal therapeutic actionability in half of the patients with PDA
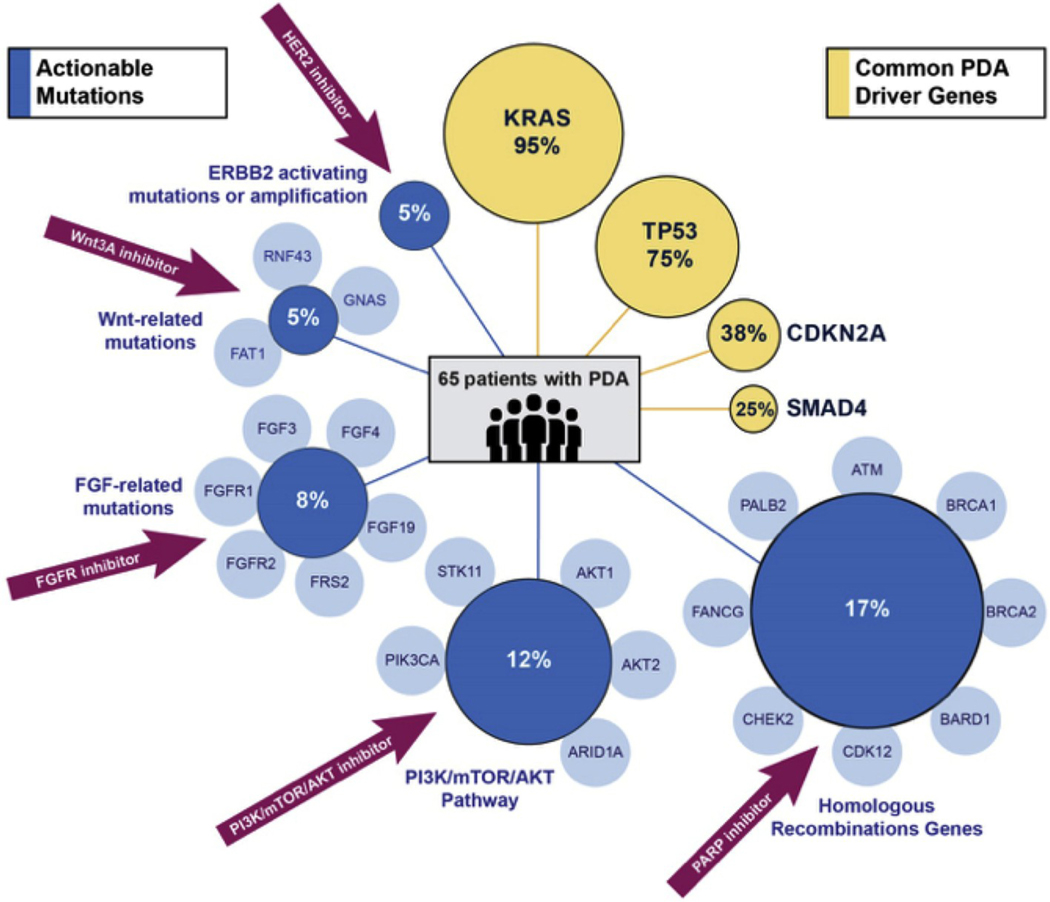
Actionable alterations are defined as mutations in the person’s genome that can be specifically targeted by molecular therapies against that mutation. In the 65 PDA cases, pathogenic mutations were detected in all tumor specimens, with a median of 3.5 pathogenic mutations or copy number alterations (CNA) per patient. Common PDA driver mutations were present at the expected frequencies (Fig. 1) [12]: KRAS mutations [25] were seen in 62/65 (95%) cases, TP53 mutations [25] found in 49/65 (75%), CDKN2A mutations or losses [25] found in 25/65 (38%), and SMAD4 mutations or losses [25] found in 16/65 (25%). Additionally, out of the 65 patients with PDA, 47% of patients had mutations in targetable genes, such as genes important for homologous recombination [26] (11/16, 17%), or AKT/PIK3CA amplification [27] and ARID1A/STK11 mutations [27] involving the PI3K/mTOR pathway (8/65, 12%)—these are actionable mutations that can be targeted with therapies that exploit these mutations and pathways.
Fig. 1. Driver and actionable mutations:
Summary of all mutations identified using NGS in the 65 patients with PDA. From those 65 patients, actionable mutations with specific therapeutic options are identified in blue, with common PDA driver genes identified in yellow.
As published in large sequencing studies [28,29], the most common actionable alterations in PDA patients were mutations in the DNA damage response pathway 8–10% [6,30,31] which can be targeted by platinum agents and PARP (poly ADP ribose polymerase) inhibitors. Other less common actionable abnormalities (and the potential associated therapies) included FGFR alterations (FGFR inhibitors; 5/65, 8%); HER2 amplifications [32] and/or ERRB2 activating mutations (HER2 inhibitors, 3/65, 5%), and RNF43/GNAS/FAT1 mutations important for the WNT pathway [33,34] that can be targeted with WNT inhibitors (3/65, 5%, Fig. 1).
3.3. Correlations among molecular and clinical variables
Molecular and clinicopathologic data were systematically analyzed for correlations in the 52/65 surgically resected PDA specimens. Clinical variables included gender, age, smoking history, other medical conditions and family history of cancer. Testing for associations using Fisher’s exact test revealed that 36 of these comparisons were significant at a threshold of p < 0.05 (Table 2).
Table 2.
Correlations among molecular and clinicopathologic features: For the patients with PDA, mutated NGS markers (>3) are listed in red text, while altered IHC markers (>4) are listed in orange text. Using Fisher’s exact test association testing was performed to identify significant variables that correlated between the clinical and molecular variables in patients with PDA. These significant variables are shown on the table. LVI: lympho-vascular invasion, GERD: gastroesophageal reflux disease.
| Variable 1 | Variable 2 | p value | Odds ratio |
|---|---|---|---|
| LVI present | Positive lymph node, N1 | 6.50E-07 | 73.9 |
| Hyperlipidemia | Hypertension | 3.60E-04 | 10.9 |
| IHC: ERCC1 | IHC: TS | 5.90E-04 | 14.4 |
| NGS: CDKN2A | NGS: CDKN2B | 9.10E-04 | Infinity |
| GERD | Hyperlipidemia | 0.0023 | 8.1 |
| IHC: PTEN | NGS: CDKN2A | 0.0027 | 0 |
| Hypertension | NGS: DNMT3A | 0.0033 | Infinity |
| NGS: CDKN2A | NGS: CDKN2ARF | 0.0033 | Infinity |
| GERD | Hypertension | 0.0035 | 6.9 |
| Metastatic presentation | NGS: BRCA2 | 0.0067 | 64 |
| Gender, male | IHC: TS | 0.0097 | 0.2 |
| NGS: CDKN2ARF | NGS: SMAD4 | 0.011 | 15.7 |
| Hypertension | Obesity | 0.011 | Infinity |
| Family cancer history | Size, > 3 cm | 0.014 | 0.2 |
| NGS_DNMT3A | Size, > 3 cm | 0.018 | Infinity |
| Grade 3/4 | IHC: MET | 0.019 | 0 |
| Age >66 | Recent smoker | 0.019 | 0.2 |
| Age >66 | NGS: CDKN2ARF | 0.02 | Infinity |
| NGS: BRCA2 | Location, pancreatic head | 0.021 | 0 |
| IHC: TS | Obesity | 0.024 | Infinity |
| Location, pancreatic head | Pancreatitis | 0.027 | 6 |
| Family cancer history | IHC: ERCC1 | 0.028 | 8 |
| Diabetes | Hypertension | 0.028 | 4.2 |
| NGS: CDKN2A | NGS: TP53 | 0.031 | 5.4 |
| Diabetes | IHC: PTEN | 0.031 | 0.1 |
| Arthritis | Obesity | 0.034 | 12.1 |
| Hyperlipidemia | NGS: DNMT3A | 0.034 | 9.9 |
| Family cancer history | Metastatic presentation | 0.036 | 18.4 |
| Age >66 | IHC: PD1 | 0.039 | 0.1 |
| Arthritis | Hyperlipidemia | 0.042 | 4.1 |
| NGS: TP53 | Location, pancreatic head | 0.044 | 4 |
| Grade 3/4 | NGS: DNT3A | 0.047 | 8.7 |
| IHC: PD1 | NGS: CDKN2B | 0.048 | 11.2 |
| Age >66 | IHC: PTEN | 0.048 | Infinity |
| IHC: TUBB3 | NGS: ARID1A | 0.049 | 0 |
| Grade 3/4 | LVI present | 0.05 | 5.2 |
Multiple significant correlations (p < 0.05) between common PDA driver genes were observed. CDKN2A mutations or copy number losses were more frequent when TP53 was mutated (p = 0.031). SMAD4 mutations were more common in tumors with mutations at the p14 ARF locus of CDKN2A (p = 0.011). Several correlations were found between the four common PDA drivers (TP53, SMAD4, CDKN2A, PTEN) and certain pathological and molecular features. For example, all tumors with PTEN protein loss had CDKN2A mutations/loss (p = 0.0027), and high protein expression of PD-1 was more frequently observed in patients with loss of CDKN2B (p = 0.048), as seen in Table 2 (for diagnosis, complete NGS sequencing, and proteomic results for each patient see Table 3).
Table 3.
A summary of each patient’s sex, pathologic diagnosis, all relevant sequence coding region sequence changes, and proteomic findings.
| SEX | DIAGNOSIS | GENOMIC FINDINGS |
|---|---|---|
| Male | Pancreatic adenocarcinoma | HER2 S310F, KRAS G12D, p53 R282W, CSF1R T37M, GATA6 Amplification, RBM10 Y508* |
| Male | Pancreatic adenocarcinoma | HER2 Amplification, KRAS G12R, p53 H193P, MYC Amplification |
| Male | Pancreatic adenocarcinoma | CHEK2 T367fs*15, KRAS G12D, NF1 Truncation exon 35, DNMT3A C818* |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 R248W, CDKN2A Loss, SMAD4 V112fs*8 |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 Y220C, CDKN2A R80*, SMAD4 R361C |
| Female | Pancreatic adenocarcinoma | BRCA2 T3085fs*19 |
| Male | Pancreatic adenocarcinoma | KRAS G12V, p53 P153fs*28, CDKN2A Loss, CCNE1 Amplification, SOX2 Amplification, MAP2K2 Amplification, PIK3CA Amplification, PRKCI Amplification, SNCAIP R499W, TERC Amplification |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 I195F, CDKN2A Loss, CCND1 Amplification, EMSY Amplification, GATA6 Amplification, FGF19 Amplification, FGF3 Amplification, FGF4 Amplification, LRP1B deletion exon 4–16 |
| Male | Pancreatic adenocarcinoma | ATM K2589fs*8, KRAS Q61R, p53 L43fs*9, CDKN2A Loss, SMAD4 V163fs*3, GATA6 Amplification, MAGI2 M593V, SLIT2 A276T |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 Y163C, PRKN Rearrangement |
| Male | Pancreatic adenocarcinoma | BRCA2 C1200fs*1, HER2 H878Y, KRAS G12R, SMAD4 D493H, SMAD4 V335fs*48, BRCA2 R2336H |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 K319_K320insKKPLDGEYFT*, AKT2 Amplification, CCNE1 Amplification |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 M237_N239del2 |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 C176Y, CDKN2A R80*, CDKN2A Truncation, intron 1, AKT2 Amplification, MYC Amplification |
| Male | Pancreatic adenocarcinoma | KRAS G12V, p53 A159fs*21, ARID1A Q575*, ARID1A S1828*, SLIT2 N775S |
| Female | Pancreatic adenocarcinoma | FGFR1 Amplification, KRAS G12V, p53 H214R, CDKN2A Loss |
| Male | Pancreatic adenocarcinoma | KRAS G12V, p53 G266E |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 P278L, CDKN2A R22_G23del |
| Female | Pancreatic adenocarcinoma | KRAS G12V |
| Female | Pancreatic adenocarcinoma | PALB2 P1152fs*9, FGFR2 P253R, PALB2 S804fs*10, PRKCI Amplification, SLIT2 A276T, TERC Amplification |
| Male | Pancreatic adenocarcinoma | KRAS Q61H, ARID1A R1335*2 |
| Female | Pancreatic adenocarcinoma | ATM K1066fs*6, KRAS G12D, MDM2 Amplification, ARID1A Y216*, FRS2 Amplification, ATM W1710*, KMT2D P601fs*329 |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 G266E |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 M133K, CDKN2A H83D, SMAD4 Loss, MUTYH Y165C |
| Female | Pancreatic adenocarcinoma | BRCA1 Rearrangement intron 2, KRAS G12V, p53 R249S, CDKN2A Loss, MYC Amplification, PIK3CA Amplification, PRKCI Amplification, TERC Amplification |
| Male | Pancreatic adenocarcinoma | KRAS G12D |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 C124*, CDKN2A Loss |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 R213*, CDKN2A Loss, RICTOR Amplification, STAG2 X435_splice |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 R175H, GNAS R201H, RNF43 E43* |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 C176F, CDKN2A Loss, RNF43 E777fs*10+, GATA6 Amplification |
| Female | Pancreatic adenocarcinoma | BARD1 S551*, KRAS G12R, p53 Q136*, p53 S366A, CDK12 Truncation, CDK12 Truncation exon 10, MYC Amplification, DNMT3A R729W, GRM3 D280N, MYC Amplification equivocal, RUNX1T1 R520H |
| Male | Pancreatic adenocarcinoma | ABL2 Rearrangement, KRAS Q61H |
| Female | Pancreatic adenocarcinoma | KRAS G12R, p53 W91*, SMAD4 R445* |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 V157F, CDKN2A Loss |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 A76fs*55, SMARCA4 T910M |
| Female | Pancreatic adenocarcinoma | BRCA2 L557*, KRAS G12V, p53 R248W, CDKN2A X51_splice, CDKN2A X0_splice, DNMT3A R882H, FAT1 Y4540fs*8, HGF Amplification, EPHB1 R79W, HGF Amplification equivocal |
| Male | Pancreatic adenocarcinoma | KRAS Q61H, p53 R213*, CDKN2A A17fs*9 |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 R248Q |
| Male | Pancreatic adenocarcinoma | KRAS G12D, CDKN2A H83R |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 G334V, BRSK1 Loss exon 2–17 |
| Male | Pancreatic adenocarcinoma | KRAS Q61H, p53 Q192*, CDKN2A R87fs*21, SMAD4 W302*, KDM6A E226*, PBRM1 R710*, HGF E199K, LRP1B D2702N |
| Female | Pancreatic adenocarcinoma | KRAS G12R, p53 X307_splice, SMAD4 R361H, SMAD4 R445*, CCND3 Amplification, VEGFA Amplification, APC I2615fs*1, TMB Intermediate |
| Male | Pancreatic adenocarcinoma | KRAS G12V, p53 V157F, NOTCH2 Amplification |
| Male | Pancreatic adenocarcinoma | KRAS Q61R, p53 R337C, SMAD4 G286fs*50, MYCL Amplification, CCNE1 Amplification, MYCL Amplification equivocal |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 R213W, CDKN2A H83Y, SMAD4 C523*, ARID1A D322fs*40, TMB Intermediate |
| Male | Pancreatic adenocarcinoma | KRAS G12R, p53 Y220C, CDKN2A Loss, TGFBR2 R537C |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 R282W, CDKN2A Loss, SMAD4 Truncation intron 4, SMAD4 Truncation, NOTCH3 deletion exon 7–31, NOTCH3 deletion |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 D49fs*76, RBM10 L195fs*71 |
| Female | Pancreatic adenocarcinoma | KRAS G12D |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 R175H, NTRK3 K732T |
| Male | Pancreatic adenocarcinoma | KRAS G12V, p53 R196*, CDKN2A L94P |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 R282W |
| Female | Pancreatic adenocarcinoma | KRAS G12D, SMAD4 S474* |
| Male | Pancreatic adenocarcinoma | p53 R282W |
| Male | Pancreatic adenocarcinoma | KRAS G12C, p53 G245D, MYCL R330*, VEGFA Amplification, SF3B1 K666R, VEGFA Amplification equivocal |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 V218E, CDKN2A A100fs*46, NF1 X244_splice |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 D281fs*31, KDM6A A516fs*9, SETD2 X2037_splice, DNMT3A K844*, TMB Intermediate, SPTA1 T681fs*76 |
| Male | Pancreatic adenocarcinoma | KRAS G12D, p53 C141fs*8 |
| Female | Pancreatic adenocarcinoma | KRAS G12D, p53 P322fs*23, ARID2 Loss, ARID2 Loss exon 17–21 |
| Male | Pancreatic adenocarcinoma | FANCG W599fs*49, KRAS G12V |
| Female | Pancreatic adenocarcinoma | KRAS G12D, DNMT3A R771* |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 H193R, SMAD4 W99*, CUL3 R709Q |
| Female | Pancreatic adenocarcinoma | KRAS G12V, p53 V173L, CDKN2A deletion exon 2, CDKN2A deletion exon 2 - intron 2, SMAD4 Q116*, FRS2 Amplification |
| Female | Pancreatic adenocarcinoma | ATM Q1084fs*9, KRAS G12V, CDKN2A M54del, SMAD4 Q28* |
| Male | Pancreatic adenocarcinoma | KRAS G12V, SMAD4 Truncation intron 8 |
| Female | Pancreas neuroendocrine carcinoma | MEN1 L89R |
| Female | Extrahepatic cholangiocarcinoma | STK11 R304W, MDM2 Amplification, FRS2 Amplification, U2AF1 S34F |
| Male | Extrahepatic cholangiocarcinoma | p53 W91*, CDKN2A R107fs*37, AKT1 Amplification, NCOR2 G781fs*15 |
| Male | Extrahepatic cholangiocarcinoma | KRAS Amplification, KRAS G12V, p53 G293fs*13, SF3B1 K700E |
| Female | Extrahepatic cholangiocarcinoma | SPEN M2790V, KRAS G12V, CTNNB1 S45F, CD36 C243* |
| Male | Duodenal adenocarcinoma | KRAS G12D, p53 R213*, ERBB3 G284R |
| Male | Colon adenocarcinoma | KRAS G12V, GNAS R201H |
| Male | Ampullary adenocarcinoma | KRAS G12D, APC K1543fs*2, FH V435M, MAP2K4 S251N |
| Male | Ampullary adenocarcinoma | BARD1 Y404fs*1, CIC G797fs*114 |
| Male | Ampullary adenocarcinoma | BRCA2 E1518fs*25 |
| Female | Ampullary adenocarcinoma | HER2 D769Y, p53 G245V |
| Female | Solid pseudopapillary neoplasm of the pancreas | CTNNB1 S37A |
| Female | Solid pseudopapillary neoplasm of the pancreas | CTNNB1 I35_G38del |
TMB was either low or not reported unless otherwise specified; Microsatellite instability was not detected in any of these cases.
3.4. Patient outcomes and therapy selection
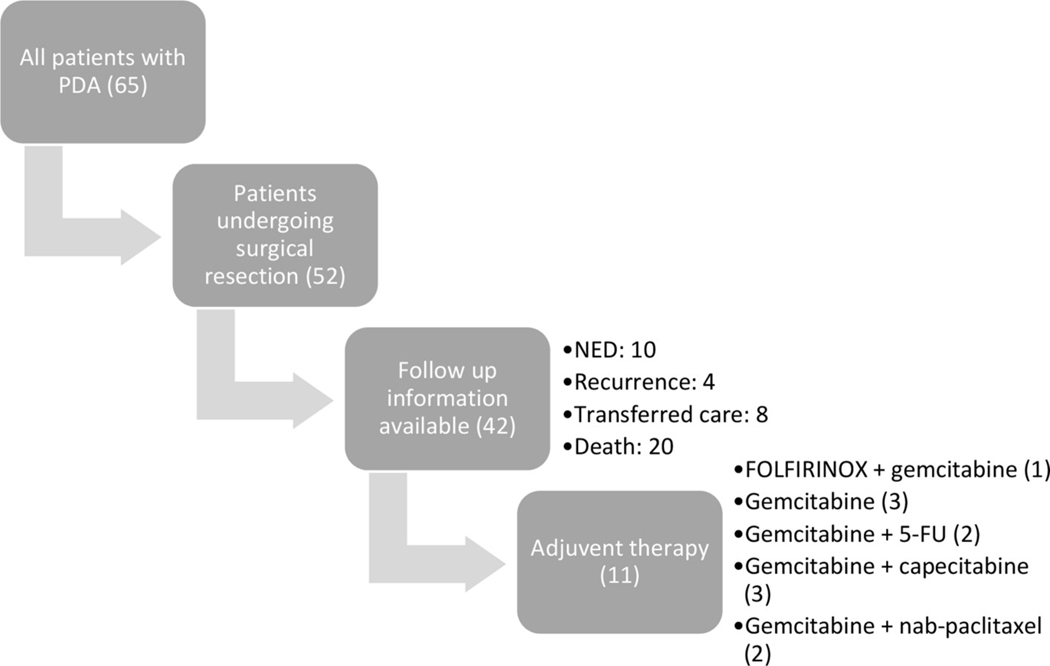
Of the 65 patients with PDA, 52 patients underwent surgical resection and outcomes data were collected on patients who continued receiving care at TJU. Median overall follow-up time was 459 days from time of initial diagnosis. Of the surgically resected patients with available outcomes, 14/42 (33.3%) had no evidence of disease or had recently developed disease recurrence as of last follow-up (Fig. 2). Eleven of these patients were on standard of care adjuvant chemotherapy, with initial therapies listed in Fig. 2. One of these followed a molecularly matched therapy recommendation: a patient with a BRCA2 mutation enrolled in a trial of a PARP inhibitor in combination with FOLFOX (folinic acid, 5- fluorouracil, oxaliplatin).
Fig. 2. Outcomes collection:
Summary of overall outcome data available from patients with PDA enrolled in the study. Outcome data for patients who had long term follow-up in TJU system is represented as of 2018 when the charts were last reviewed with details on any treatment ongoing. NED: no evidence of disease, 5-FU: 5-fluorouracil.
3.5. Survival analysis
Of the 52 patients with resected PDA, we screened the 39 molecular and clinicopathologic features (see Methods for details) against survival data to evaluate correlations with recurrence-free and overall survival. Median follow-up time was 459 days from initial diagnosis. Further analysis of the data showed that almost half of the patients (23/52) had developed disease recurrence, with a Kaplan-Meier estimate of median RFS of 12.4 months. In the patients who did develop disease recurrence, most recurrences were in the liver (17 patients), lung (1 patient), brain (1 patient), bone (1 patient), omentum (1 patient), lymph node (1 patient), and one patient had a locally recurrent tumor. Molecular profiling was not repeated after recurrence had occurred in these patients. Fewer than half of the patients (20/52) were deceased, with a Kaplan-Meier estimate of median OS of 21.7 months.
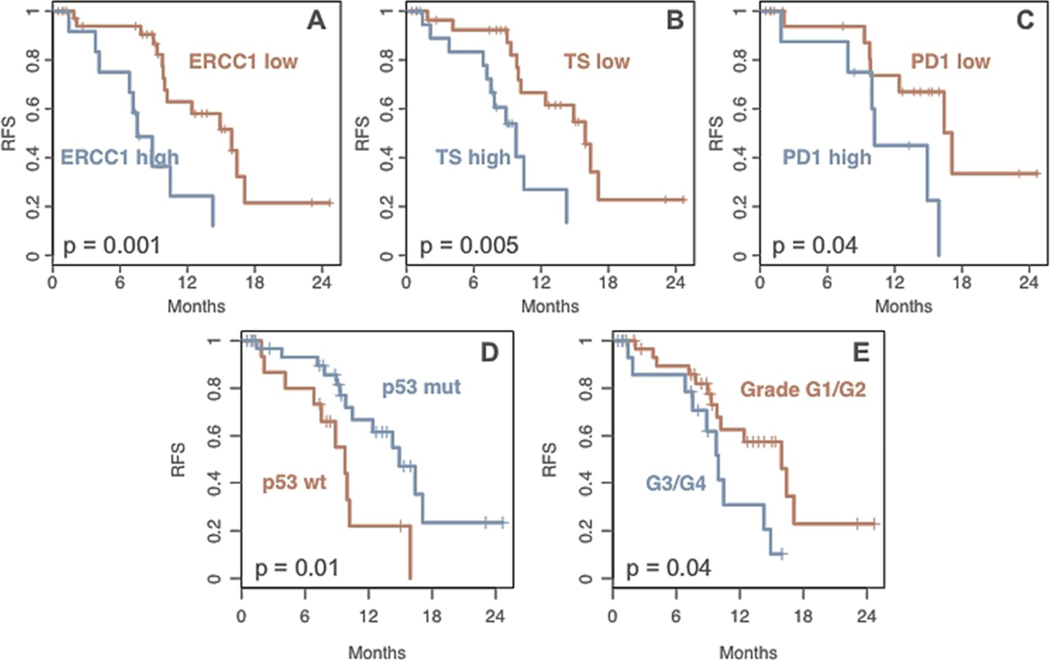
Five variables were correlated with RFS. High expression of three proteins, ERCC1 (p = 0.001), TS (p = 0.005), and PD-1 (p = 0.04), were associated with lower RFS (Fig. 3). Interestingly, TP53 mutations were correlated with longer RFS (p = 0.01). High tumor grade was negatively correlated with RFS (p = 0.04). No molecular variables were correlated with OS, but two pathologic features were associated with shorter OS: tumor size over 3 cm (p = 0.002) and high histologic grade (p = 0.02).
Fig. 3. Recurrence-free survival (RFS) is correlated with molecular and pathologic features:
From the patients with PDA who had long term follow-up at TJU, Kaplan Meier graphs were generated to assess RFS. High expression of ERCC1 (A), TS (B), and PD-1 (C) were correlated with lower RFS. Additionally, mutations in TP53 were correlated with higher RFS(D), while high grade tumors had lower RFS (E). All graphs shown are statistically significant with p values indicated on the graphs.
4. Discussion
There is a pressing need in today’s health care setting for a facile system wherein patient consenting is seamlessly tied to molecular analytic workflow and tracked throughout the course of a patient’s treatment. Integrated, structured data are critical for applying statistical and artificial intelligence-based or heuristic algorithms that will be necessary to detect complex relationships between patient data, treatment data, molecular data and treatment response. In this study, we sought to take a first-step to address these data infrastructure-related challenges and demonstrate that a comprehensive precision medicine program can be integrated into cancer care as part of a large clinical practice focused on pancreatic cancer.
Recent studies have demonstrated that precision medicine can suggest alternative therapeutic strategies in roughly 50% of pancreatic cancer patients [6,35,36]. Of the 52 surgically resected PDA patients in this study, 48% harbored at least one actionable alteration, similar to some other studies published [6,30,36]. While DNA repair genes and PI3K/mTOR pathway genes had actionable mutations in a relatively high number of patients, no single therapeutic target dominated the actionable alterations (Fig. 1). This suggests that profiling of a broad panel of genes is important in PDA in order to identify all potential targets. We note that the frequencies of actionable alterations, in addition to the four common PDA driver alterations, did not differ greatly from published datasets [28,37–39] or from the Know Your Tumor dataset of over 600 patients [6].
One major limitation of this study, and indeed of all similar profiling studies that have been published [30], is that a limited number of patients that had actionable molecular alterations have gone on to receive matched targeted therapies, especially in a clinical trial. Therefore, it is impossible to draw conclusions about the overall clinical effectiveness of this approach. Moreover, overall follow-up, to date, is not yet long enough to draw any meaningful conclusions about overall impact of the platform on patient care (to this point, in this study 76% of the surgically resected patients have not yet received treatment for recurrent or metastatic disease).
Although we could not perform a statistical analysis correlating molecular and clinicopathologic features with treatment data, we were able to find correlations with the time interval of recurrence-free survival (RFS). The high number of significant correlations with RFS (Fig. 3) in this relatively small dataset suggests that as the number of patients analyzed using this platform grows, treatment-specific correlations should become readily detectable. Of the five biomarkers that were significantly correlated with RFS, three were protein-based: ERCC1, TS and PD-1. These markers are putative predictive markers for chemotherapeutic (5-FU) efficacy [40], platinum based therapy resistance [41], and potentially future immunotherapeutic strategies. This work supports the notion that proteomic-based testing may also provide additional information for survival and response rates. Interestingly, in our current patient subset we also found that there was an increase in RFS in patients with TP53 mutations compared to TP53 wildtype, which contradicts what was previously published [42]. There have been some inconsistencies in the literature over TP53 mutations and prognostic significance, with some articles considering it a negative prognostic indicator [43] and some articles showing no significance [44, 45].
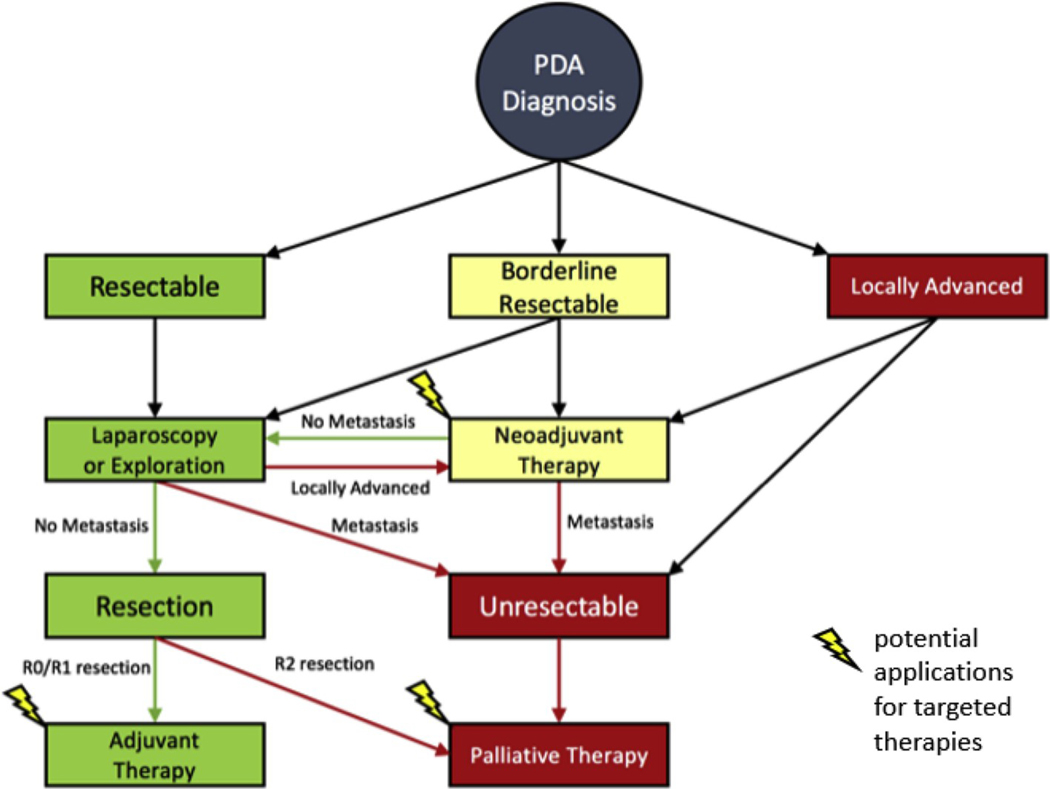
Our study establishes that profiling surgically resected primary pancreatic tumors yields a comparable amount of actionable alterations to profiling studies of metastatic sites. This is consistent with studies that showed actionable NGS alteration frequencies did not differ between primary and metastatic PDA [46,47]. Performing molecular profiling on surgical tissue specimens has multiple important benefits for the timeline of patient care. First, it allows for enough time to plan for therapy if recurrence occurs. This may facilitate enrollment in molecularly targeted clinical trials in the first line of therapy setting, when there is a greater chance of deriving benefit. For example, one patient in a separate study that had a rare IDH1 mutation received an IDH1 inhibitor as third line therapy after developing resistance to FOLFIRINOX (folinic acid, 5- fluorouracil, irinotecan, oxaliplatin) and gemcitabine with nab-paclitaxel [48]. This patient did not respond to the IDH1 inhibitor, but it is possible that earlier molecular profiling of her tumor could have led to earlier initiation of targeted therapy and a better response [48, 49]. Second, profiling surgical specimens allows time to study the tumor’s biology and propagate ex vivo models for various drug sensitivity assays (e.g., organoid model) [29,50]. Third, profiling a tumor after the administration of neoadjuvant therapy, would allow for a more accurate assessment of the tumor after it has been exposed to potent DNA damaging agents. Finally, this work lays the groundwork for the potential of a personalized adjuvant therapy for patients with lymph node positivity and R1/R2 resections. Based on these theories highlighting the utility of molecular profiling in an oncological setting, there are a couple of potential points of therapeutic intervention (Fig. 4). With the use of NGS and molecular profiling we can use this additional information to guide oncological therapy in the neoadjuvant and adjuvant settings to allow for the administration of a more unique and personalized therapy for the patient.
Fig. 4. Chronology of patients’ medical course from diagnosis with PDA to therapeutic intervention:
Flowchart representation a typical medical course of a patient diagnosed with PDA and the potential therapeutic options depending on PDA stage. Lightning bolts emphasize where potential applications of molecularly targeted therapies could intervene in each patient’s treatment strategy in the neoadjuvant, adjuvant or palliative setting.
In the future, molecular profiling combined with focused drug screens in ex vivo cultures, may inform a personalized approach to treating pancreatic and other cancers. Future randomized controlled trials, along with the optimization of targeted approaches, will determine whether and when molecular profiling will have a role in the treatment of patients with resectable disease (Fig. 4). This type of precision medicine platform may allow large hospital systems, such as ours, and cooperative groups to facilitate next generation molecular tumor boards in an effort to scale precision medicine for the benefit of large numbers of patients.
Supplementary Material
Acknowledgments
Funding
This study was performed with grant support from the Concetta Greenberg Fund in memory of Marvin S. Greenberg, M.D. at Thomas Jefferson University Department of Surgery; Perthera, Inc.; PanCAN-AACR Research Acceleration Network (RAN) Grant (JRB, JMW and MJP); Thomas Jefferson University Hospital; and T32 training grant NIH/NIGMS T32GM008562 (T. Dhir).
Footnotes
Declaration of competing interest
Joseph Bender, PhD, Talar Tatarian, MD, Subha Madhavan, PhD, Emanuel F Petricoin 3rd, PhD, Michael J Pishvaian, MD, PhD, and Jonathan R Brody, PhD have affiliations with Perthera, Inc.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.suronc.2020.02.003.
References
- [1].Siegel RL, Miller KD, Jemal A, Cancer statistics, CA A Cancer J. Clin. 67 (1) (2017) 7–30, 2017. [DOI] [PubMed] [Google Scholar]
- [2].Chapman BC, et al. , Perioperative and survival outcomes following neoadjuvant FOLFIRINOX versus gemcitabine abraxane in patients with pancreatic adenocarcinoma, JOP 19 (2) (2018) 75–85. [PMC free article] [PubMed] [Google Scholar]
- [3].Neoptolemos JP, et al. , Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): a multicentre, open-label, randomised, phase 3 trial, Lancet 389 (10073) (2017) 1011–1024. [DOI] [PubMed] [Google Scholar]
- [4].Conroy T, et al. , FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer, N. Engl. J. Med. 364 (19) (2011) 1817–1825. [DOI] [PubMed] [Google Scholar]
- [5].Von Hoff DD, et al. , Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine, N. Engl. J. Med. 369 (18) (2013) 1691–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Pishvaian MJ, et al. , Molecular profiling of patients with pancreatic cancer: initial results from the Know Your tumor initiative, Clin. Canc. Res. 24 (20) (2018) 5018–5027. [DOI] [PubMed] [Google Scholar]
- [7].Carr TH, et al. , Defining actionable mutations for oncology therapeutic development, Nat. Rev. Canc. 16 (5) (2016) 319–329. [DOI] [PubMed] [Google Scholar]
- [8].Heather JM, Chain B, The sequence of sequencers: the history of sequencing DNA, Genomics 107 (1) (2016) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hollingsworth SJ, Precision medicine in oncology drug development: a pharma perspective, Drug Discov. Today 20 (12) (2015) 1455–1463. [DOI] [PubMed] [Google Scholar]
- [10].Koboldt DC, et al. , The next-generation sequencing revolution and its impact on genomics, Cell 155 (1) (2013) 27–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Dreyer SB, et al. , Pancreatic cancer genomes: implications for clinical management and therapeutic development, Clin. Canc. Res. 23 (7) (2017) 1638–1646. [DOI] [PubMed] [Google Scholar]
- [12].Pishvaian MJ, Brody JR, Therapeutic implications of molecular subtyping for pancreatic cancer, Oncology (Williston Park) 31 (3) (2017). [PubMed] [Google Scholar]
- [13].Sibinga Mulder BG, et al. , Targeted next-generation sequencing of FNA-derived DNA in pancreatic cancer, J. Clin. Pathol. 70 (2) (2017) 174–178. [DOI] [PubMed] [Google Scholar]
- [14].Guan Z, et al. , Individualized drug screening based on next generation sequencing and patient derived xenograft model for pancreatic cancer with bone metastasis, Mol. Med. Rep. 16 (4) (2017) 4784–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sleijfer S, Bogaerts J, Siu LL, Designing transformative clinical trials in the cancer genome era, J. Clin. Oncol. 31 (15) (2013) 1834–1841. [DOI] [PubMed] [Google Scholar]
- [16].Simon R, Roychowdhury S, Implementing personalized cancer genomics in clinical trials, Nat. Rev. Drug Discov. 12 (5) (2013) 358–369. [DOI] [PubMed] [Google Scholar]
- [17].Andre F, et al. , Prioritizing targets for precision cancer medicine, Ann. Oncol. 25 (12) (2014) 2295–2303. [DOI] [PubMed] [Google Scholar]
- [18].N.C. Institute, Molecular Profiling-Based Targeted Therapy in Treating Patients with Advanced Solid Tumors, 2017. (USA). [Google Scholar]
- [19].N.C. Institute, NCI-MATCH: Targeted Therapy Directed by Genetic Testing in Treating Patients with Advanced Refractory Solid Tumors, Lymphomas, or Multiple Myeloma, 2017. (USA). [Google Scholar]
- [20].A.S.o.C. Oncology, TAPUR: Testing the Use of Food and Drug Administration (FDA) Approved Drugs that Target a Specific Abnormality in a Tumor Gene in People with Advanced Stage Cancer, TAPUR, USA, 2017. [Google Scholar]
- [21].Pancreatic cancer action network precision promise [Webpage], [cited 2017; Available from: https://www.pancan.org/research/precision-promise/, 2017.
- [22].Redig AJ, Janne PA, Basket trials and the evolution of clinical trial design in an era of genomic medicine, J. Clin. Oncol. 33 (9) (2015) 975–977. [DOI] [PubMed] [Google Scholar]
- [23].Paulson AS, et al. , Therapeutic advances in pancreatic cancer, Gastroenterology 144 (6) (2013) 1316–1326. [DOI] [PubMed] [Google Scholar]
- [24].Medicine FA, World-leading molecular insights company, Available from: https://www.foundationmedicine.com/, 2019.
- [25].Cicenas J, et al. , KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer, Cancers 9 (5) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tangutoori S, Baldwin P, Sridhar S, PARP inhibitors: a new era of targeted therapy, Maturitas 81 (1) (2015) 5–9. [DOI] [PubMed] [Google Scholar]
- [27].Yang L, et al. , Inhibition of PI3K/AKT signaling pathway radiosensitizes pancreatic cancer cells with ARID1A deficiency in vitro, J. Canc. 9 (5) (2018) 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jones S, et al. , Core signaling pathways in human pancreatic cancers revealed by global genomic analyses, Science 321 (5897) (2008) 1801–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tiriac H, et al. , Organoid profiling identifies common responders to chemotherapy in pancreatic cancer, Canc. Discov. 8 (9) (2018) 1112–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lowery MA, et al. , Real-time genomic profiling of pancreatic ductal adenocarcinoma: potential actionability and correlation with clinical phenotype, Clin. Canc. Res. 23 (20) (2017) 6094–6100. [DOI] [PubMed] [Google Scholar]
- [31].Pishvaian MJ, Brody JR, Therapeutic implications of molecular subtyping for pancreatic cancer, Oncology (Williston Park) 31 (3) (2017) 159–166, 168. [PubMed] [Google Scholar]
- [32].Chou A, et al. , Clinical and molecular characterization of HER2 amplified-pancreatic cancer, Genome Med. 5 (8) (2013) 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jiang X, et al. , Inactivating mutations of RNF43 confer Wnt dependency in pancreatic ductal adenocarcinoma, Proc. Natl. Acad. Sci. U. S. A. 110 (31) (2013) 12649–12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Katoh M, Function and cancer genomics of FAT family genes (review), Int. J. Oncol. 41 (6) (2012) 1913–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yamamoto KN, et al. , Personalized management of pancreatic ductal adenocarcinoma patients through computational modeling, Canc. Res. 77 (12) (2017) 3325–3335. [DOI] [PubMed] [Google Scholar]
- [36].Hayashi H, et al. , Genomic testing for pancreatic cancer in clinical practice as real- world evidence, Pancreatology 18 (6) (2018) 647–654. [DOI] [PubMed] [Google Scholar]
- [37].Waddell N, et al. , Whole genomes redefine the mutational landscape of pancreatic cancer, Nature 518 (7540) (2015) 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sausen M, et al. , Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients, Nat. Commun. 6 (2015) 7686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Biankin AV, et al. , Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes, Nature 491 (7424) (2012) 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Grimminger PP, et al. , TS and ERCC-1 mRNA expressions and clinical outcome in patients with metastatic colon cancer in CONFIRM-1 and −2 clinical trials, Pharmacogenomics J. 12 (5) (2012) 404–411. [DOI] [PubMed] [Google Scholar]
- [41].Tezuka S, et al. , Predictive value of ERCC1, ERCC2, ERCC4, and glutathione S- Transferase Pi expression for the efficacy and safety of FOLFIRINOX in patients with unresectable pancreatic cancer, Am. J. Canc. Res. 8 (10) (2018) 2096–2105. [PMC free article] [PubMed] [Google Scholar]
- [42].Ormanns S, et al. , pERK, pAKT and p53 as tissue biomarkers in erlotinib-treated patients with advanced pancreatic cancer: a translational subgroup analysis from AIO-PK0104, BMC Canc. 14 (2014) 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Liu SZ, et al. , Prognostic impact of cyclin D1, cyclin E and P53 on gastroenteropancreatic neuroendocrine tumours, Asian Pac. J. Cancer Prev. APJCP 14 (1) (2013) 419–422. [DOI] [PubMed] [Google Scholar]
- [44].Ansari D, et al. , Systematic review of immunohistochemical biomarkers to identify prognostic subgroups of patients with pancreatic cancer, Br. J. Surg. 98 (8) (2011) 1041–1055. [DOI] [PubMed] [Google Scholar]
- [45].Oshima M, et al. , Immunohistochemically detected expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4) strongly predicts survival in patients with resectable pancreatic cancer, Ann. Surg. 258 (2) (2013) 336–346. [DOI] [PubMed] [Google Scholar]
- [46].Yachida S, Iacobuzio-Donahue CA, Evolution and dynamics of pancreatic cancer progression, Oncogene 32 (45) (2013) 5253–5260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yachida S, et al. , Distant metastasis occurs late during the genetic evolution of pancreatic cancer, Nature 467 (7319) (2010) 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Brody JR, et al. , Identification of a novel metabolic-related mutation (IDH1) in metastatic pancreatic cancer, Canc. Biol. Ther. 19 (4) (2018) 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Brody JR, et al. , Identification of a novel metabolic-related mutation (IDH1) in metastatic pancreatic cancer, Canc. Biol. Ther. (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Boj SF, et al. , Organoid models of human and mouse ductal pancreatic cancer, Cell 160 (1–2) (2015) 324–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.