Abstract
Nuclear factor erythroid 2-related factor 2 (NRF2) is a well-characterized transcription factor that protects cells against oxidative and electrophilic stresses. Emerging evidence has suggested that NRF2 protects cells against DNA damage by mechanisms other than antioxidation, yet the mechanism remains poorly understood. Here, we demonstrate that knockout of NRF2 in cells results in hypersensitivity to ionizing radiation (IR) in the presence or absence of reactive oxygen species (ROS). Under ROS scavenging conditions, induction of DNA double-strand breaks (DSBs) increases the NRF2 protein level and recruits NRF2 to DNA damage sites where it interacts with ATR, resulting in activation of the ATR–CHK1–CDC2 signaling pathway. In turn, this leads to G2 cell cycle arrest and the promotion of homologous recombination repair of DSBs, thereby preserving genome stability. The inhibition of NRF2 by brusatol increased the radiosensitivity of tumor cells in xenografts by perturbing ATR and CHK1 activation. Collectively, our results reveal a novel function of NRF2 as an ATR activator in the regulation of the cellular response to DSBs. This shift in perspective should help furnish a more complete understanding of the function of NRF2 and the DNA damage response.
INTRODUCTION
DNA double-strand breaks (DSBs) are highly toxic DNA lesions that are associated with various developmental, immunological, and neurological disorders as well as tumorigenesis (1). DSBs can be generated by exogenous agents, including ionizing radiation (IR) and radiomimetic chemicals, and endogenous factors, such as V(D)J recombination, meiosis, and replication fork stress (1). To preserve genome integrity, error-free homologous recombination (HR) competes and collaborates with error-prone nonhomologous end-joining (NHEJ) to repair DSBs (2). HR mainly functions in S/G2 phases, during which homologous sister chromatids are present and several critical HR proteins are activated (3).
Cell cycle checkpoint pathways are indispensable to cope with DNA damage and are traditionally defined as molecular signaling cascades that delay or arrest the cell cycle in response to DNA damage, thereby providing more time for DNA repair. Furthermore, the checkpoint machinery is integrated with activation of DNA repair, chromatin remodeling, modulation of transcription programs, and cell death (4,5). Phosphoinositide 3-kinase-related protein kinases, including ataxia-telangiectasia mutated (ATM) and ATM- and RAD3-related (ATR), are the master regulators of the DNA damage response (DDR) and act by controlling cell cycle transitions. ATM is recruited to chromatin where it phosphorylates lots of substrates in response to DSBs (6). The kinase CHK2 is a well-characterized substrate of ATM. CHK2 is phosphorylated at multiple sites by ATM, and mediates cell cycle arrest and apoptosis (7,8). Differently than ATM, ATR is thought to primarily deal with single-stranded DNA (ssDNA) breaks and tends to be recruited to replication protein A (RPA)-coated ssDNA (9,10). However, several findings indicate that ATR can also respond to DSBs caused by IR (11,12). Assembly of the ATR complex at DNA lesions activates signaling that coordinates the cell cycle, DNA repair and DNA replication. The CHK1–CDC2 pathway, which controls cell cycle transitions, is mainly dependent on activation of ATR (13,14). ATR is recruited to ssDNA via its partner ATR-interacting protein (ATRIP), and its optimal activation relies on its activators such as TopBP1 and ETAA1, which contain the ATR activation domain (AAD) (15–17). The identification of potential ATR regulators is important to elucidate the molecular mechanism by which ATR controls the DDR and DNA repair.
The transcription factor nuclear factor erythroid 2-related factor 2 (NRF2) is the master responder to oxidative and electrophilic stresses. NRF2 is usually maintained at a low basal protein level in unstressed condition by Keap1, which promotes the ubiquitination and proteasomal degradation of NRF2 (18). NRF2 escapes from this Keap1-dependent repression when cells are exposed to oxidative, electrophilic, or xenobiotic stress. Thereafter, NRF2 translocates into the nucleus and regulates transcription of genes that contain antioxidant response elements (19,20). Recent studies identified additional functions of NRF2 that extend beyond its redox-regulation capacity, such as functions in drug metabolism and excretion; energy, iron and amino acid metabolism; cell survival and proliferation; autophagy; proteasomal degradation; DNA repair and mitochondrial physiology (21,22). NRF2 may perform these additional functions by coordinating the transcription of genes involved in redox homeostasis, however, Jayakumar et al. recently showed that NRF2 regulated HR by influencing the mRNA level and foci formation of RAD51 in a reactive oxygen species (ROS)-independent manner (23,24). Despite this report, further investigation is required to characterize how NRF2 may regulate DDR and DNA repair by mechanisms other than antioxidation. Here, we report that the NRF2 protein level was increased in cells with DSBs and that NRF2 regulated radiosensitivity also in a ROS-independent manner. NRF2 accumulated in the nucleus and formed foci at DNA damage sites, thereby facilitating the DDR and DNA repair. The ATR–CHK1–CDC2 signaling cascade was activated by the interaction of NRF2 with ATR, and this was dependent on the AAD-like domain of NRF2. Ablation of NRF2 impaired activation of the ATR–CHK1 signaling pathway and G2 cell cycle arrest and decreased the HR efficiency in cells with DSBs. Brusatol, an NRF2 inhibitor, effectively decreased the NRF2 protein level in tumor xenografts and increased the radiosensitivity of tumor xenografts by compromising the ATR–CHK1 pathway.
MATERIALS AND METHODS
Cell culture
The human non-small cell lung cancer cell lines A549, H460 and H1299 were purchased from the American Type Culture Collection (ATCC; USA) and cultured in RPMI 1640 (HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, New Zealand). DR-GFP-U2OS cells were provided by Dr Xingzhi Xu (Shenzhen University, China) and cultured in DMEM supplemented with 10% FBS. HEK293T and U2OS cells were purchased from ATCC and cultured in DMEM supplemented with 10% FBS. A549-NRF2KO cells were generated using CRISPR-Cas9 by Shanghai Genechem Co., Ltd. (China). Briefly, cells were transfected with LV-sgCas9-P2A-puro containing guide RNAs targeting exon 1 of NRF2 (3′-CACCGACAGCTCATCATGATGGACT-5′) and selected with 1.5 μg/ml puromycin for three days before plating of individual clones. Deletion of the exon 1 splice junction was predicted to cause out-of-frame splicing and nonsense-mediated decay. Homozygous editing of the NRF2 locus was confirmed by PCR and sequencing. Furthermore, the protein level of NRF2 was confirmed by western blotting.
Antibodies and reagents
The following antibodies were used: anti-NRF2 (Abcam, ab62352 and Proteintech, 16396-1-AP), anti-KEAP1 (Proteintech, 10503-2-AP), anti-γH2AX (Ser139) (Abcam, ab26350), anti-ATR (GeneTex, GTX70109), anti-p-ATR (Ser428) (Cell Signaling Technology, #2853), anti-53BP1 (Abcam, ab36823), anti-p-53BP1 (Ser1778) (Cell Signaling Technology, #2675), anti-Cdk/CDC2 polyclonal (Cell Signaling Technology, #9116), anti-p-CDC2 (Tyr15) antibody (Cell Signaling Technology, #4539), anti-Cyclin A (Santa Cruz, sc-271682), anti-NQO1 (Abcam, ab28947), anti-BRCA1 (Santa Cruz, sc-6954 and GeneTex, GTX70111), anti-p-BRCA1 (Ser1524) (Cell Signaling Technology, #9009), anti-CHK1 (GeneTex, GTX100070), anti-p-CHK1 (Ser317) (Cell Signaling Technology, #12302), anti-p-CHK1 (Ser345) (Cell Signaling Technology, #2341), anti-p-ATM (Ser1981) (Cell Signaling Technology, #13050), anti-ATM (Cell Signaling Technology, #2873), anti-β-tubulin (Proteintech, 66240-1-Ig), anti-β-actin (Proteintech, 60008-1-Ig), anti-GAPDH (Proteintech, 60004-1-Ig), IgG Control Antibody (Proteintech, 30000-0-AP), HRP-conjugated Affinipure Goat Anti-Mouse IgG (H+L) (Proteintech, SA00001-1), HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (Proteintech, SA00001-2), Cy3-conjugated Affinipure Goat Anti-Mouse IgG (H+L) (Proteintech, SA00009-1), Cy3-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (Proteintech, SA00009-2), FITC-conjugated Affinipure Goat Anti-Mouse IgG (H+L) (Proteintech, SA00003-1) and FITC-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (Proteintech, SA00003-2).
The following reagents and kits were used: VE-821 (MedChemExpress, USA, HY-14731), KU-55933 (MedChemExpress, USA, HY-12016), brusatol (Tauto Biotech, Shanghai, China), propidium iodide (PI; Beijing Solarbio Life Science, C0080), N-acetyl-l-cysteine (NAC; Sigma-Aldrich, Germany, A9165), l-buthionine- sulfoximine (BSO; MedChemExpress, USA, HY-106376), a FITC-Annexin V Apoptosis Detection Kit (BD Biosciences, USA), a TUNEL Apoptosis Kit (Biotime Biotechnology, Shanghai, China), and a Pierce™ GST Protein Interaction Pull-down Kit (Thermo Fisher Scientific, USA, #21516), Total Reactive Oxygen Species (ROS) Assay Kit (Thermo Fisher Scientific, USA, 88-5930-74), Micro Reduced Glutathione (GSH) Assay Kit (Beijing Solarbio Science & Technology Co., Ltd., BC1175), formamidopyrimidine-DNA glycosylase (FPG enzyme; New England BioLabs, M0240S).
RNA interference assay
Small interfering RNAs (siRNAs) were synthesized by GenePharma Co., Ltd (Shanghai, China) and transfected using Lipofectamine RNAiMax (Invitrogen, USA) at a concentration of siRNAs of 20 nM. Cells were treated with reagents or radiation 48 h after transfection. The siRNA sequences were as follows: NRF2-targeting siRNA (siNRF2) sense sequence (5′-CCGGCAUUUCACUAAACACAA-3′), BRCA1-targeting siRNA (siBRCA1) sense sequence (5′-GGAACCUGUCUCCACAAAG-3′), KEAP1-targeting siRNA (siKEAP1#1) sense sequence (5′-ACAACAGUGUGGAGAGGUA-3′) and KEAP1-targeting siRNA (siKEAP1#2) sense sequence (5′- GAAACAGAGACGUGGACUU-3′).
Lentiviral transduction
Lentiviruses and plasmids were produced by Shanghai Genechem Co., Ltd. The short hairpin RNA (shRNA) sequences were as follows: NRF2-targeting shRNA (shNRF2 #1):
5′-CCGGGCTCCTACTGTGATGTGAAATCTCGAGATTTCACATCACAGTAGGAGCTTTTT-3′ and shNRF2 #2: 5′-CCGGCCGGCATTTCACTAAACACAACTCGAGTTGTGTTTAGTGAAATGCCGGTTTTT-3′. For lentiviral infection, cells were plated one day prior to infection and cultured overnight to reach 70–80% confluency. The spent culture medium was then aspirated, and fresh medium was added containing concentrated lentiviruses harboring an empty vector (as a control) or shRNA. Cells and viruses were incubated for 24–48 h in the presence of polybrene 10 μg/ml, and then cells were selected by incubation in the presence of 1.5 μg/ml puromycin for 3–7 days. The mRNA and protein levels of NRF2 were assessed by RT-PCR and western blotting, respectively. Cells were infected with a lentivirus harboring Flag-NRF2 using the same procedure. Wild-type (WT) NRF2 and NRF2 mutant proteins (aa1–338, Δ180–230, and W188A) were cloned into GV366 vector (Shanghai Genechem Co., Ltd). Cells in six-well plates were transfected with 4 μg/well of plasmids using 8 μl/well of Lipofectamine 2000 and collected or treated 24–48 h post-transfection.
Immunofluorescence and quantitative analysis of individual cells
Cells were grown on chamber slides, washed once with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 20 min, washed once with PBS, treated with 0.3% Triton X-100 diluted in PBS, blocked with 5% FBS diluted in PBS for 1 h at room temperature (RT), and washed three times with PBS. Samples were then incubated with primary antibodies diluted in 1% bovine serum albumin at 4°C overnight, washed three times with PBS, incubated with FITC- or Cy3-conjugated secondary antibodies diluted in PBS for 1–2 h at RT, and washed three times with PBS. Finally, samples were mounted using VECTASHIELD® mounting medium with DAPI (Vector Laboratories, CA, USA) and analyzed by fluorescence microscopy at RT. Images were acquired using a 40× objective and processed using Photoshop CS6. The numbers of foci in individual cells were counted, and the data were plotted using GraphPad Prism 5 software.
Whole-cell lysate collection and western blotting
To collect whole-cell lysates, cells were harvested, washed with PBS, resuspended in pre-chilled RIPA lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, EDTA, protease inhibitors, and phosphatase inhibitors), and incubated on ice for 30 min. Lysates were supplemented with 4× loading buffer and boiled for 10 min. The protein concentration was determined using a BCA protein assay kit (Beyotime, Shanghai, China, P0010). Proteins in whole-cell lysates were separated on 8% or 10% SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride membranes. Membranes were blocked with 1% bovine serum albumin at RT for 1 h, incubated with primary antibodies at 4°C overnight, and then incubated with secondary antibodies at RT for 1 h. Immunoreactive bands were detected using Thermo Scientific™ SuperSignal™ West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, 34580) and a ChemiDoc™ MP Imaging System (Bio-Rad, Shanghai, China).
Co-immunoprecipitation
Cells were collected by trypsinization, washed once with PBS, and lysed on ice in 600 μl of NP-40 lysis buffer (Beyotime, P0013F) containing a cocktail of phosphatase inhibitors (Roche, Switzerland) and PMSF (Sigma). Lysates were centrifuged at 12 000 rpm for 10 min at 4°C. Proteins in the lysates were incubated overnight at 4°C with protein A/G magnetic beads (Bimake, USA, B23202) bound to an anti-NRF2 antibody, an anti-ATR antibody, or normal IgG. To bind antibodies to the magnetic beads, 40 μl volume of magnetic beads was washed twice with wash buffer (50 mM Tris–HCl, 150 mM NaCl and 0.5% Triton X-100, pH 7.5), incubated with 200 μl of the antibody solution for 1 h, and then collected using a magnetic stand. The mixture of proteins and beads was washed twice with wash buffer and collected using a magnetic stand. Proteins were eluted by boiling the beads in 2× loading buffer and subjecting the samples to SDS-PAGE and immunoblotting analysis.
Comet assay/ single-cell gel electrophoresis (SCGE) assay
Cells were pretreated with 5 mM NAC or 10 μM BSO for 2 h. The media was then removed, and cells were digested with trypsin and collected into EP tubes. Next, the cells were washed with 500 ml of PBS and were suspended in culture medium at a density of 4–5 × 105 cells/mL. Cells were exposed to 8 Gy Cs137 γ-rays and then returned to the incubator for 1 h for later use. The comet slides were coated with 500 μl of 0.75% normal-melting-point agarose/PBS and coverslips were quickly applied. Once the first layer of normal-melting-point agarose was coagulated, the coverslips were removed. Next a mixture of 70 μl of 0.75% low-melting-point agarose/PBS and 30 μl of cell suspensions were applied as the second layer, and the comet slides were incubated at 4°C for 10 min. The comet slides were then immersed in cold fresh lysis solution (2.5 M NaCl, 10 mM Tris base, 1% N-sodium lauryl sarcosinate, 30 mM Na2EDTA, 10% DMSO and 1% Triton X-100) for 2.5 h at 4°C. The 1× enzyme reaction buffer (40 mM HEPES, 0.1 M KCl, 0.5 mM EDTA, 0.2 mg/ml BSA, pH 8.0 with KOH) was prepared, and the comet slides were washed in a staining jar with this enzyme reaction buffer three times, 5 min each wash. Gel surfaces were covered with 50 μl of the enzyme solution (1:103 dilution of FPG enzyme). The comet slides were put in a moist box and incubated at 37°C for 30 min. Next, the comet slides were washed with PBS and immersed in TBE buffer for 20 min at 4°C in a horizontal electrophoresis tank. Electrophoresis was performed at 30 V for 20 min in TBE buffer, followed by neutralization for 20 min. The slides were then washed with PBS and stained with ethidium bromide. Finally, the comet slides were viewed with a fluorescence microscope and data were collected with a digital imaging system and analyzed with CASP software (Wroclaw, Poland).
Micronucleus formation experiments
Cells were pretreated with or without 5 mM NAC for 2 h, exposed to 8 Gy Cs137 γ-rays (Atomic Energy of Canada Ltd.), collected 24 h post-irradiation, washed twice with PBS, fixed in a 3:1 mixture of methanol: glacial acetic acid, spread onto slides, air-dried, and stained with Giemsa. Micronuclei were observed using a 40× objective. More than 500 cells were counted per group.
HR repair assay
DR-GFP-U2OS cells were used to assay the HR efficiency (25). Cells were transfected with a plasmid expressing I-SceI for 48 h. GFP-positive cells were identified and quantified by flow cytometry. The HR repair efficiency was scored as the percentage of GFP-positive cells. To examine the role of NRF2 in DSB repair, cells were treated with siNRF2 24 h prior to transfection of I-SceI, and siBRCA1 was used as a positive control.
Flow cytometric analysis
To analyze the cell cycle distribution, cells were exposed to 8 Gy IR for 6, 12 and 24 h or treated with camptothecin (CPT) for 4 h, released for 10 h, and then collected. Cells were washed twice with PBS, fixed in 75% ice-cold ethanol overnight, incubated with staining buffer (10 μg/ml RNase A, 50 μg/mL PI, and 4 mM sodium citrate) at 37°C for 15 min in the dark, and then assessed by flow cytometry. Cell cycle profiles were analyzed using FlowJo 7.6 software.
Apoptosis was determined using an FITC-Annexin V/PI Apoptosis Detection Kit (BD Biosciences) according to the manufacturer's instructions. Briefly, cells were exposed to 8 Gy IR, cultured for 24 h, collected, washed once with PBS, gently suspended in Annexin V binding buffer, and incubated with 20 μg/mL FITC-Annexin V and 20 μg/ml PI for 15 min in the dark. Flow cytometric analysis was performed using FlowJo 7.6 software.
Measurement of ROS level
Intracellular ROS levels were measured using the total reactive oxygen species (ROS) Assay Kit. ROS inhibition was implemented by treating cells with 5 mM NAC for 1 h before IR. After 1 h of IR, cells were treated with ROS assay stain solution for 60 min in a 37°C incubator with 5% CO2 and then washed with PBS before trypsinization. After detaching with trypsin, the cells were collected, washed twice, and resuspended in PBS. Fluorescence was measured by FACS based on signal at 488 nm (blue laser) in the FITC channel.
Reduced glutathione (GSH) assay
GSH was measured with the micro reduced glutathione (GSH) assay kit. Briefly, at least 106 cells were treated with trypsin, washed twice with PBS, and resuspended in reagent 1 at three times the volume of the cells. The cell suspensions were frozen and thawed in liquid nitrogen and 37°C water bath three times. The cells were then centrifuged at 8000g for 10 min to collect supernatants and the supernatants were collected and stored at 4°C for later use. Reagent 2 was warmed in a 37°C water bath for 30 min. Next, 20 μL ddH2O or samples, 140 μl warmed reagent 2 and 40 μl reagent 3 were added to wells of a 96-well plate. The mixed samples were allowed to stand for 2 min and then the absorbance at 412 nm was measured by Multiscan Spectrum (Bio-tek Instruments). GSH contents were calculated according to the formula specified in the kit.
GST pull-down assay
Recombinant GST and GST-NRF2 proteins (GST-NRF2 (1–338), GST-NRF2 (1–338, Δ180–230), GST-NRF2 (1–338, W188A) were expressed in BL21 cells. Flag-ATR was overexpressed in HEK293T cells. The GST pull-down assay was performed using a Pierce™ GST Protein Interaction Pull-down Kit. The obtained proteins were analyzed by western blotting.
Animal experiments
Athymic nude mice were purchased from Beijing HFK Bioscience Co., Ltd., and A549 cells were injected into the thighs of 4–6-week-old mice. Once tumors reached ∼50 mm3, mice were randomly allocated into four groups and locally treated with saline (groups 1 and 2) or 2 mg/kg brusatol (groups 3 and 4). After 24 h, mice in groups 2 and 4 were locally exposed to 8 Gy Cs137 γ-rays.
Statistical analysis
All statistical analyses were performed using GraphPad Prism v.5.01 or SPSS software. Values of P ≤ 0.05 were considered statistically significant in the two-tailed Student's t test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
RESULTS
NRF2 affects radiosensitivity by modulating DNA repair independently of ROS
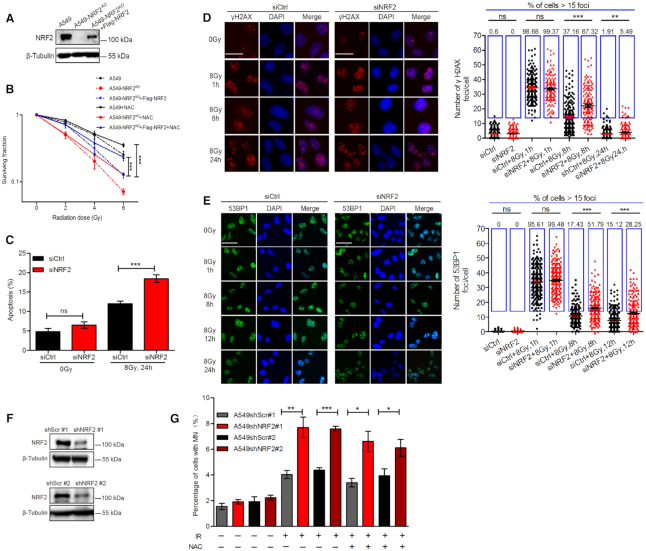
The transcription factor NRF2 mainly regulates expression of a wide array of genes that encode antioxidants and other proteins responsible for detoxification of xenobiotics and ROS (26). Gain of NRF2 function protects cells against IR toxicity, an activity that has been attributed to its antioxidant properties (27). However, other mechanisms may also be involved. Jayakumar et al. reported a novel ROS-independent function of NRF2 in DNA repair (23). To confirm that NRF2 protects cells against IR via a ROS-independent mechanism, we generated NRF2-knockout cells (A549-NRF2KO cells) using the CRISPR-Cas9 system and A549-NRF2KO cells that express wild-type (WT) NRF2 (A549-NRF2KO+Flag-NRF2 cells) (Figure 1A and Supplementary Figure S1). We scavenged ROS by treating cells with 5 mM N-acetyl-L-cysteine (NAC), a commonly used ROS scavenger (28). NAC effectively increased the levels of glutathione (GSH) while l-buthionine-sulfoximine (BSO), a GSH synthesis inhibitor, significantly reduced GSH levels in cells exposed to IR (Supplementary Figures S2A–C). ROS levels were also measured in A549-NRF2KO cells. IR significantly increased the levels of ROS and NAC effectively inhibited the increase of ROS (Supplementary Figures S2D and E). To determine whether NAC reduced oxidative DNA damage, we performed a comet assay using formamidopyrimidine-DNA glycosylase (FPG) to detect oxidative DNA damage (29). We found that NAC significantly reduced oxidative DNA damage while BSO significantly increased oxidative DNA damage in cells exposed to irradiation (Supplementary Figure S2F), highlighting the ability of NAC to effectively scavenge ROS.
Figure 1.
NRF2 affects radiosensitivity by modulating DNA repair independently of ROS. (A) Western blotting of NRF2 in A549, A549-NRF2KO and A549-NRF2KO+Flag-NRF2 cells. (B) Colony-formation assay with A549, A549-NRF2KO and A549-NRF2KO+Flag-NRF2 cells. A549-NRF2KO cells were generated using CRISPR-Cas9 and transfected with a Flag-NRF2 expression vector to generate A549-NRF2KO+Flag-NRF2 cells. Cells were pretreated with 5 mM NAC for 1 h and then exposed to 0, 2, 4 or 6 Gy IR. Data are means ± S.E.M. (n = 3). ***P ≤ 0.001. (C) A549 cells transfected with siCtrl or siNRF2 were pretreated with NAC for 1 h and exposed to 8 Gy Cs137 γ-rays. At 24 h post-irradiation, the percentage of apoptotic cells was determined by co-staining with FITC-Annexin V and PI. Data are means ± S.E.M. (n ≥ 3). ***P ≤ 0.001, ns: no significance. (D) A549 cells transfected with siNRF2 or siCtrl were pretreated with NAC for 1 h and exposed to 8 Gy Cs137 γ-rays. γH2AX foci were counted at 1, 8, and 24 h post-irradiation. Scale bars, 25 μm. Data are means ± S.E.M., with >150 cells were counted in three independent experiments. **P ≤ 0.01, ***P ≤ 0.001. (E) A549 cells transfected with siCtrl or siNRF2 were pretreated with NAC for 1 h and exposed to 8 Gy Cs137 γ-rays. 53BP1 foci were counted at 1, 8, 12 and 24 h post-irradiation. Scale bars, 50 μm. Data are means ± S.E.M., with more than 150 cells were counted in three independent experiments. ***P ≤ 0.001. (F) Western blotting of NRF2 in A549 cells treated with shScr#1, shNRF2#1, shScr#2 and shNRF2#2. (G) A549 cells treated with shScr#1, shNRF2#1, shScr#2 or shNRF2#2 were pretreated with or without 5 mM NAC and exposed to 8 Gy Cs137 γ-rays. The percentage of cells with micronuclei (MN) was determined at 24 h post-irradiation. Data are means ± S.E.M. (n = 3). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001.
We next examined radiosensitivity by colony-formation assay. In agreement with the result reported previously, A549-NRF2KO cells were hypersensitive to IR while expression of WT NRF2 in A549-NRF2KO cells rescued this defect (Figure 1B and Supplementary Figure S3A). Although NAC did increase radio-resistance of all cells, A549-NRF2KO cells were still more sensitive to IR than A549-NRF2KO+Flag-NRF2 cells after scavenging ROS (Figure 1B). As an additional way to eliminate the influence of ROS, we induced DNA damage using 30 nM camptothecin (CPT), a topoisomerase I inhibitor, which interferes with DNA replication and eventually induces formation of DSBs (30). Consistently, knockout of NRF2 led to decreased cell survival and expression of Flag-NRF2 in A549-NRF2KO cells restored cell survival upon CPT treatment (Supplementary Figure S3B), suggesting that NRF2 protects cells from DNA damage also in a ROS-independent manner.
Ionizing radiation can induce cellular apoptosis. So we next depleted NRF2 using small interfering RNA (siRNA) in A549 cells and tested how loss of NRF2 affected the percentage of apoptotic cells upon IR exposure. Apoptosis of A549 cells transfected with control siRNA (siCtrl) or siNRF2 was investigated in cells pretreated with NAC and then exposed to IR. NRF2-depleted cells showed significantly higher levels of apoptosis than control cells (Figure 1C and Supplementary Figure S4). IR was previously shown to induce upregulation and nuclear accumulation of NRF2 (31). Western blotting showed that the NRF2 protein level markedly increased upon exposure to IR, regardless of NAC pretreatment (Supplementary Figure S5), which suggests that IR-induced NRF2 expression may be independent of ROS. To further investigate the role of NRF2 in the DDR or DNA repair, we used an immunofluorescence assay to longitudinally monitor γH2AX and 53BP1 foci as indicators of DNA damage. There were significantly lower numbers of γH2AX foci (Figure 1D) and 53BP1 foci (Figure 1E) in siCtrl-transfected cells than in siNRF2-transfected cells after exposure to 8Gy of IR, indicating repair of IR-damaged DNA was defective in NRF2 knockdown cells compared to control cells. Insufficient DNA repair usually leads to the formation of micronuclei. Therefore, we generated NRF2-knockdown cells via infection of lentiviruses harboring shNRF2 (Figure 1F) and determined the percentage of cells containing micronuclei after exposure to IR (Figure 1G). There was a significantly higher level of micronuclei in NRF2-knockdown cells regardless of whether cells were pretreated with NAC, indicating that NRF2 might play a role in preventing genetic instability. Thus, we conclude that NRF2 affects radiosensitivity and preserves genomic integrity also by regulating DNA repair independently of ROS.
NRF2 promotes HR during DNA repair
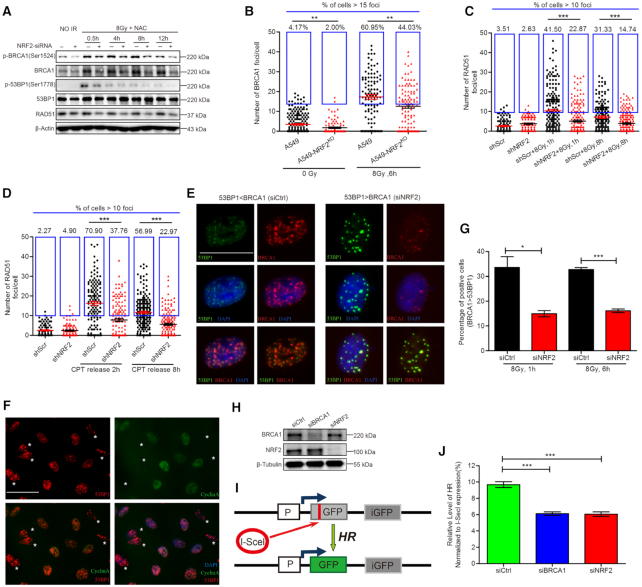
NHEJ and HR are the two major pathways for repair of DSBs. BRCA1 antagonizes 53BP1-dependent NHEJ to promote HR (2,5,32). Recent studies showed that NRF2 can simultaneously regulate BRCA1 and 53BP1 expression (33,34). To determine how NRF2 coordinates functions of these two proteins during DNA repair, we investigated the active phosphorylated forms of BRCA1 and 53BP1. The phosphorylated BRCA1 (p-BRCA1) and total BRCA1 protein levels were markedly lower in siNRF2-transfected cells than in siCtrl-transfected cells after exposure to IR. However, depletion of NRF2 did not obviously affect the total protein level of 53BP1. The 53BP1 phosphorylation level was greatly reduced in siNRF2-transfected cells at 0.5 h post-irradiation, but dephosphorylation of 53BP1 occurred subsequently at 4, 8 and 12 h after IR exposure (Figure 2A). Consistent with the finding of Jayakumar et al. that NRF2 influences mRNA level of RAD51, depletion of NRF2 decreased the protein level of RAD51, a key protein in HR. Therefore, we hypothesized that NRF2 might promote HR during repair of DSBs. To investigate this, we analyzed BRCA1 and RAD51 foci as indicators of the HR efficiency. As expected, there were lower percentages of BRCA1 and RAD51 foci-positive cells in NRF2-depleted cells than in control cells, regardless of whether cells were treated with IR or CPT (Figure 2B–D). Co-staining with anti-BRCA1 and anti-53BP1 antibodies revealed that BRCA1 and 53BP1 exhibited mutually exclusive foci formation. The BRCA1 foci were larger and more obvious than 53BP1 foci in some cells and vice versa in other cells (Figure 2E). Co-staining for the S/G2 phase marker Cyclin A further demonstrated that cells with large and obvious 53BP1 foci were mainly in G1 phase (Figure 2F). Feng et al. reported a similar phenomenon and demonstrated that BRCA1 antagonizes 53BP1 signaling to ensure HR in S/G2 phases (35). We counted cells in which BRCA1 foci were stronger than 53BP1 foci because HR tended to occur in such cells following exposure to IR. The percentage of these cells was significantly higher for control cells than for NRF2-depleted cells (Figure 2G). These results demonstrated that NRF2 facilitated HR in cells with DNA damage resulting from IR or CPT treatment. To quantify HR efficiency in NRF2-depleted and control cells, we used DR-GFP-U2OS cells, which harbor a HR efficiency reporter system (36). BRCA1 was depleted as a positive control. As expected, depletion of NRF2 significantly decreased the HR efficiency (Figure 2H–J), suggesting that NRF2 could promote HR during repair of DSBs.
Figure 2.
NRF2 promotes HR during DNA repair. (A) Western blotting of RAD51, p-BRCA1, BRCA1, p-53BP1 and 53BP1 in A549 cells transfected with siCtrl or siNRF2, pretreated with 5 mM NAC for 1 h, and exposed to 8 Gy Cs137 γ-rays. (B) BRCA1 foci in A549-NRF2KO and A549 cells pretreated with NAC for 1 h and exposed to 0 or 8 Gy Cs137 γ-rays. BRCA1 foci were counted at 6 h post-irradiation. Cells containing >15 BRCA1 foci were considered BRCA1 foci-positive cells. Data are means ± S.E.M., and >150 cells were counted in three independent experiments. **P ≤ 0.01. (C) RAD51 foci in A549 cells treated with shNRF2 or shScr, pretreated with NAC for 1 h, and exposed to 8 Gy Cs137 γ-rays. RAD51 foci were counted at 1 and 8 h post-irradiation. Data are means ± S.E.M., and >150 cells were counted in three independent experiments. ***P ≤ 0.001. (D) RAD51 foci in A549 cells treated with shNRF2 or shScr, pretreated with 30 nM CPT for 6 h, and released for 2 or 8 h. Cells containing more than 10 RAD51 foci were counted as RAD51 foci-positive cells. Data are means ± S.E.M., with >150 cells were counted in three independent experiments. ***P ≤ 0.001. (E) A549 cells were transfected with siCtrl or siNRF2. 24 h later, cells were exposed to 8 Gy Cs137 γ-rays and subjected to immunofluorescence analysis of BRCA1 (red), 53BP1 (green), and DAPI (blue) at 6 h post-irradiation. Scale bars, 20 μm. (F) A549 cells were transfected with siCtrl or siNRF2. 24 h later, cells were exposed to 8 Gy Cs137 γ-rays and subjected to immunofluorescence analysis of 53BP1 (red), Cyclin A (green), and DAPI (blue) at 6 h post-irradiation. Scale bars, 50 μm. Asterisks indicate Cyclin A-negative cells. (G) A549 cells were transfected with siCtrl or siNRF2. 24 h later, cells were exposed to 8 Gy Cs137 γ-rays and subjected to immunofluorescence analysis of BRCA1 (red), 53BP1 (green) and DAPI (blue) at 1 h or 6 h post-irradiation. Percentage of cells in which BRCA1 foci were stronger than 53BP1 foci. Data are means ± S.E.M. (n = 3). *P ≤ 0.05, ***P ≤ 0.001. (H) Western blotting of BRCA1 and NRF2 in DR-GFP-U2OS cells transfected with siCtrl, siNRF2, or siBRCA1. (I) Schematic of the DR-GFP reporter used to monitor HR. (J) Flow cytometric analysis of the HR efficiency in DR-GFP-U2OS cells transfected with siCtrl, siNRF2 or siBRCA1. Data are means ± S.E.M. (n ≥ 3). ***P ≤ 0.001.
NRF2 is a vital regulator of G2 cell cycle arrest during DNA repair
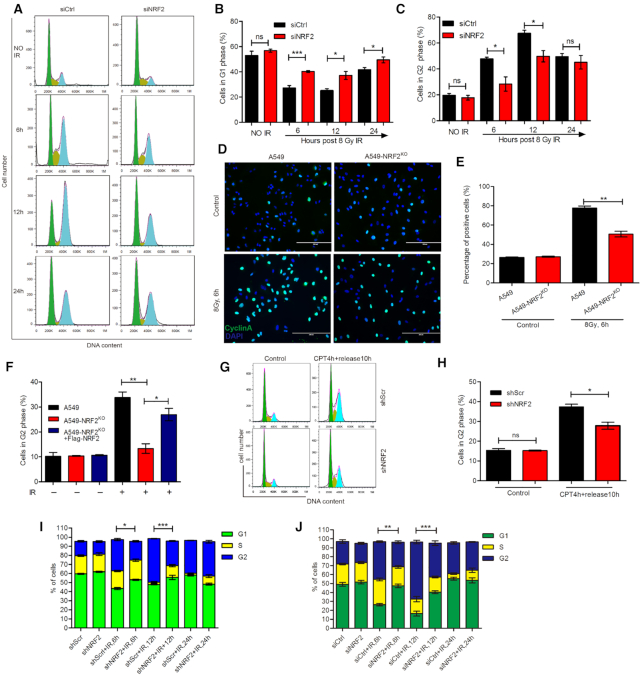
As HR occurs only in S/G2 phase, the decreased HR efficiency after the depletion of NRF2 may be related to the change of cell cycle. Cell cycle arrest upon DNA damage is important for preserving genomic integrity (37). We hypothesized that NRF2 might regulate HR not only by affecting BRCA1 and RAD51 but also by affecting cell cycle arrest. To determine how NRF2 influences cell cycle arrest during DNA repair, we analyzed the cell cycle distribution. About 50% and 70% of control cells were in G2 phase, but only 30% and 50% of NRF2-depleted cells were in G2 phase at 6 h and 12 h, respectively, after exposure to 8 Gy of IR. This result indicates that control cells were arrested in G2 phase after exposure to IR, but there was significantly attenuated G2 cell cycle arrest following exposure to IR in NRF2-depleted cells (Figure 3A–C). As observed following IR exposure, induction of G2 cell cycle arrest by CPT was also significantly reduced in NRF2-knockdown cells (Figure 3G and H). In addition, we assessed G2 cell cycle arrest in H460 and H1299 cells. We found that ablation of NRF2 also perturbed G2 cell cycle arrest in these two cell lines (Figure 3I, J and Supplementary Figures S6A–S6D). The above results all indicate that depletion of NRF2 impairs G2 cell cycle arrest. To further confirm the effect of NRF2 on G2 cell cycle arrest, we examined the cell cycle distribution of A549, A549-NRF2KO and A549-NRF2KO+Flag-NRF2 cells after IR. G2 cell cycle arrest was compromised in A549-NRF2KO cells, but this defect was rescued in A549-NRF2KO+Flag-NRF2 cells (Figure 3F). Additionally, a significantly higher percentage of A549 cells were positively stained for Cyclin A after irradiation compared to the percentage of A549-NRF2KO cells (Figure 3D and E). Western blotting showed that depletion of NRF2 markedly inhibited the S/G2 phase-specific accumulation of Cyclin A in cells exposed to IR (Figure 4B and D). These results demonstrate that NRF2 is required for G2 cell cycle arrest upon formation of DSBs.
Figure 3.
NRF2 is a vital regulator of G2 cell cycle arrest during DNA repair. (A–C) A549 cells transfected with siCtrl or siNRF2 were exposed to 8 Gy Cs137 γ-rays. The distribution of cells in G1/S/G2 phases was determined by PI staining and flow cytometry at 6, 12, and 24 h post-irradiation (A). The percentages of cells in G1 (B) and G2 (C) phases were determined. Data are means ± S.E.M. (n = 4). *P ≤ 0.05, ***P ≤ 0.001, ns: no significance. (D, E) Immunofluorescence analysis of CyclinA (green) and DAPI (blue) in A549 and A549-NRF2KO cells exposed to 8 Gy Cs137 γ-rays for 6 h. Scale bars, 200 μm (D). The percentage of positively stained cells was determined (E). Data are means ± S.E.M. (n = 3). **P ≤ 0.01. (F) A549, A549-NRF2KO and A549-NRF2KO+Flag-NRF2 cells were treated with 8 Gy Cs137 γ-rays. Cells in G2 phase were detected by PI staining and flow cytometry at 6 h post-irradiation. Data are means ± S.E.M. (n = 3). *P ≤ 0.05, **P ≤ 0.01. (G, H) G2 cell cycle arrest in A549 cells treated with shScr or shNRF2, followed by CPT treatment to induce DNA damage. Cells were treated with 30 nM CPT for 4 h and released for 10 h. Cells in G2 phase were detected by PI staining and flow cytometry (F). The percentage of cells in G2 phase was determined (G). Data are means ± S.E.M. (n = 3). *P ≤ 0.05, ns: no significance. (I) H460 cells treated with shScr or shNRF2 were exposed to 8 Gy Cs137 γ-rays. The distribution of cells in G1/S/G2 phases was determined by PI staining and flow cytometry at 6, 12 and 24 h post-irradiation. Data are means ± S.E.M. (n = 3). *P ≤ 0.05, ***P ≤ 0.001. (J) H1299 cells transfected with siCtrl or siNRF2 were exposed to 8 Gy Cs137 γ-rays. The distribution of cells in G1/S/G2 phases was determined by PI staining and flow cytometry at 6, 12 and 24 h post-irradiation. Data are means ± S.E.M. (n ≥ 3). **P ≤ 0.01, ***P ≤ 0.001.
Figure 4.
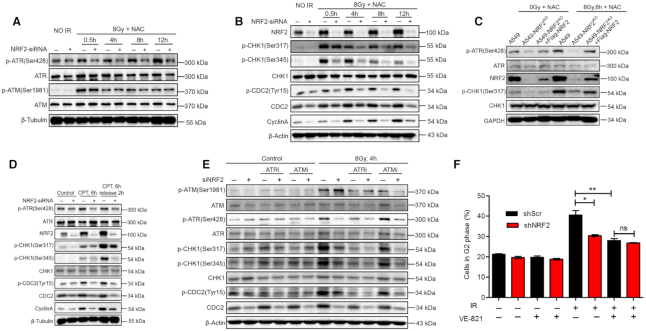
NRF2 facilitates G2 cell cycle arrest by activating the ATR-CHK1-CDC2 signaling pathway. (A) Western blotting of p-ATR, ATR, p-ATM and ATM in A549 cells transfected with siCtrl or siNRF2, pretreated with 5 mM NAC for 1 h, and exposed to 8 Gy Cs137 γ-rays. (B) Western blotting of NRF2, p-CHK1, CHK1, p-CDC2, CDC2 and CyclinA in A549 cells transfected with siCtrl or siNRF2, pretreated with 5 mM NAC for 1 h, and exposed to 8 Gy Cs137 γ-rays. (C) Western blotting of NRF2, CHK1, p-CHK1, ATR and p-ATR in A549, A549-NRF2KO and A549-NRF2KO+Flag-NRF2 cells pretreated with 5 mM NAC for 1 h and exposed to 0 or 8 Gy Cs137 γ-rays. (D) Western blotting of NRF2, p-ATR, ATR, p-CHK1, CHK1, p-CDC2, CDC2 and Cyclin A in A549 cells transfected with siCtrl or siNRF2, treated with 30 nM CPT for 6 h, and released for 2 h. (E) Western blotting of p-ATR, ATR, p-ATM, ATM, p-CHK1, CHK1 and p-CDC2 in A549 cells treated with siCtrl or siNRF2, pretreated with 10 μM VE-821 or 20 μM KU-55933 for 2 h, 5 mM NAC for 1 h, and exposed to 8 Gy Cs137 γ-rays. (F) Flow cytometric analysis of G2 cell cycle arrest in H1299 cells treated with shScr or shNRF2, pretreated with or without 10 μM VE-821 and 5 mM NAC for 1 h, and exposed to or not exposed to 8 Gy Cs137 γ-rays. Data are means ± S.E.M. (n = 3). *P ≤ 0.05, **P ≤ 0.01, ns: no significance.
NRF2 facilitates G2 cell cycle arrest by activating the ATR-CHK1-CDC2 signaling pathway
We next explored the molecular mechanism explaining the requirement for NRF2 in G2 cell cycle arrest in cells with DNA damage. In vertebrate cells, the DDR is mainly controlled by three related kinases: ATM, ATR and DNA-PK (38). Among these three proteins, ATR and ATM collaborate to regulate cell cycle arrest in cells with DNA damage (12,39). Therefore, we first investigated whether NRF2 regulates activation of these kinases. Phosphorylation of ATR was obviously reduced in NRF2-knockdown cells exposed to IR, while there was no marked difference in the phosphorylation of ATM between control and NRF2-knockdown cells (Figure 4A). This result is consistent with our above findings and with previous reports that ATR mainly regulates G2 cell cycle arrest by phosphorylating CHK1, while ATM mainly regulates G1 cell cycle arrest by phosphorylating CHK2 (38). Next, we concentrated on ATR and explored its signaling cascade. Similar to activation of ATR, there was markedly lower phosphorylation of CHK1 and CDC2 in NRF2-knockdown cells compared to control cells (Figure 4B). ATR and CHK1 were not phosphorylated in A549-NRF2KO cells after exposure to IR, but this defect was rescued in A549-NRF2KO+Flag-NRF2 cells (Figure 4C). We also monitored phosphorylation of ATR, CHK1 and CDC2 in CPT-treated cells, and found that activation of the ATR-CHK1-CDC2 signaling cascade was similarly dependent on NRF2 (Figure 4D). To determine whether elevated endogenous NRF2 level affects the activation of the ATR-CHK1 pathway, we used siRNAs (siKEAP1#1 and siKEAP1#2) to knock down KEAP1, a natural inhibitor of NRF2 that promotes the ubiquitination and degradation of NRF2. When the KEAP1 level decreased, the NRF2 level increased significantly in U2OS cells. The levels of p-ATR (Ser428), p-CHK1 (Ser317) and p-CHK1 (Ser345) in the KEAP1 knockdown cells were significantly higher than those in the control cells (Supplementary Figure S7). These results suggest that NRF2 is a potential activator of the ATR-CHK1-CDC2 signaling cascade in cells with DSBs. To confirm that NRF2 facilitated G2 cell cycle arrest by targeting ATR, we used the ATR inhibitor VE-821 to treat cells. Western blotting showed that IR induced phosphorylation of ATR, CHK1 and CDC2 in A549 cells treated with siCtrl, but the phosphorylation levels of ATR, CHK1 and CDC2 were obviously lower in cells treated with siNRF2, and VE-821 treatment attenuated the phosphorylation of ATR, CHK1 and CDC2 in IR-treated cells (Figure 4E). We also used the ATM inhibitor KU-55933, which effectively inhibited the activation of ATM, but had no obvious effect on the function of NRF2 and the ATR-CHK1 pathway (Figure 4E). Flow cytometry also demonstrated little difference in G2 cell cycle arrest following IR exposure between cells treated with shScr and those treated with shNRF2 upon VE-821 treatment (Figure 4F). Thus, we conclude that the NRF2 activity to promote G2 cell cycle arrest is dependent on ATR.
BRCA1 helps regulate the G2 cell cycle arrest following DNA damage (40) and NRF2 regulates the basal transcription activity of the BRCA1 gene (41). Although knockdown of NRF2 reduced the BRCA1 protein level in cells exposed to IR as expected (Figure 2A), depletion of BRCA1 using siBRCA1 did not abrogate the effect of NRF2 on G2 cell cycle arrest (Supplementary Figures S8A and B), implying the regulation by NRF2 of cell cycle arrest may be independent of BRCA1. To test whether ROS affect NRF2 and ATR-CHK1, we treated A549, A549-NRF2KO and A549-NRF2KO+F/G-NRF2 (A549-NRF2KO cells expressing Flag-GFP-NRF2) with H2O2. The levels of intracellular ROS were significantly increased in A549 and A549-NRF2KO cells, however, there was no obvious change in A549-NRF2KO+F/G-NRF2 (Supplementary Figure S8C). The western blotting results showed that the increase of ROS significantly increased the NRF2 level, but had no significant effect on p-ATR (Ser428) (Supplementary Figure S8D). Although ROS induced markedly increased phosphorylation levels of CHK1, the phosphorylation of CHK1 may occur through a p-ATR (Ser428)-independent pathway. Under this condition, NRF2 had no obvious effect on phosphorylation of CHK1, suggesting that NRF2–ATR–CHK1 activation may occur only for DNA damage that is caused by certain factors, with no obvious effect of ROS on this pathway. Thus, we conclude that NRF2 facilitates G2 cell cycle arrest by directly promoting ATR phosphorylation and thereby activating the ATR–CHK1–CDC2 signaling pathway.
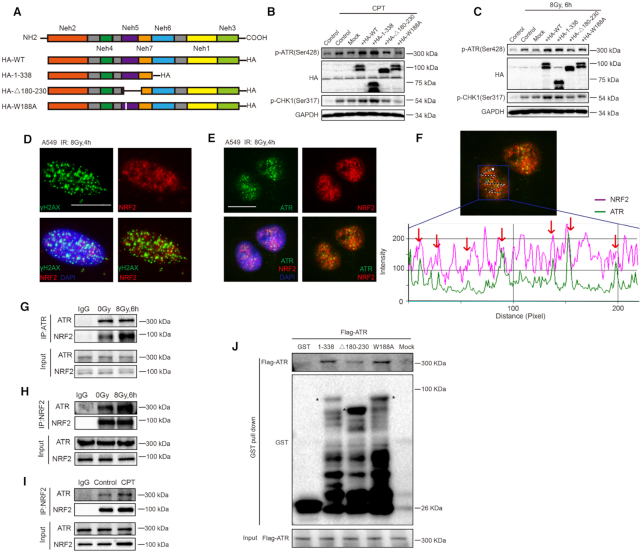
NRF2 activates the ATR-CHK1-CDC2 signaling pathway via its AAD-like domain and interacts with ATR at DNA damage sites
Based on the observations above, we hypothesized that NRF2 might directly activate ATR after induction of DSBs. TopBP1 and ETAA1 are only two proteins found in higher eukaryotes that directly stimulate ATR kinase activity and harbor an ATR-activating domain (AAD) (15–17). The unstructured AAD contains a high abundance of hydrophobic residues, the essential aromatic amino acid Trp, and consecutive acidic patches (42). Protein sequence alignment and hydrophobicity analysis show that NRF2 also contains a Trp residue at position 188 and desultory hydrophobic fragments, similar to TopBP1 and ETAA1 (Supplementary Figure S9A-D). Furthermore, sequence analysis also demonstrated that this region of NRF2 is highly evolutionarily conserved (Supplementary Figure S9E). To investigate the effect of this AAD-like domain in NRF2, we constructed plasmids encoding WT NRF2 (HA-WT) and various mutants of NRF2, namely, amino acids 1-338 of NRF2 (HA-1–338), NRF2 lacking amino acids 180–230 (HA-Δ180–230), and NRF2 in which the Trp (W) amino acid at position 188 was replaced by Ala (A) (HA-W188A). Thus, HA-WT and HA-1–338 contain the complete AAD-like domain and HA-Δ180–230 and HA-W188A do not. We transfected HEK293T cells with these NRF2 constructs (Figure 5A). When cells were treated with IR or CPT, expression of HA-WT and HA-1–338 significantly promoted ATR and CHK1 phosphorylation, whereas expression of HA-Δ180–230 and HA-W188A did not (Figure 5B and C). These results demonstrated the importance of the AAD-like domain in NRF2 for activation of the ATR–CHK1–CDC2 signaling pathway. We previously observed nuclear transposition of NRF2 in cells exposed to IR (43). We further carefully analyzed the nuclear distribution of NRF2, and found that NRF2 formed nuclear foci, some of which colocalized with γH2AX foci, which represent DNA damage sites (Figure 5D). The same observation was made in H1299 cells (Supplementary Figure S10A). Thus, we hypothesized that NRF2 interacts with ATR at DNA damage sites and promotes ATR activation. To test this, we examined NRF2 and ATR nuclear foci in cells exposed to 8 Gy of IR. Surprisingly, most ATR foci colocalized with NRF2 foci (Figure 5E and F). Next, we used co-immunoprecipitation to test potential interaction of NRF2 with ATR. NRF2 or ATR was detected after ATR or NRF2 was immunoprecipitated from A549 cell extracts, respectively. The interaction between ATR and NRF2 was enhanced in cells treated with IR or CPT (Figure 5G–I). This interaction was also detected in H1299 cells by co-immunoprecipitation (Supplementary Figures S10B and C). To determine whether NRF2 indeed interacts with ATR, we performed a GST pull-down assay using constructs that expressed GST-tagged proteins encoding 1–338 amino acids of WT NRF2 (1–338), NRF2 fragment (1–338) lacking amino acids 180–230 (Δ180–230) or a NRF2 mutant (1–338) in which the W amino acid at position 188 was replaced by A (W188A). As shown in Figure 5J, the WT NRF2 fragment (1-338) associated with purified Flag-ATR in vitro and the NRF2 mutant (W188A) also bound to Flag-ATR, but NRF2 mutant (Δ180–230) pulled down Flag-ATR much less efficiently than the other two proteins. These results suggest that the interaction of NRF2 and ATR may depend on the complete structure of AAD-like domain of NRF2 and the single Trp (W) at position 188 may be not sufficient to mediate this interaction. Altogether, these results suggest that NRF2 interacts with ATR at DNA damage sites and promotes activation of ATR via its AAD-like domain.
Figure 5.
NRF2 promotes activation of the ATR–CHK1–CDC2 signaling pathway via its AAD-like domain and interacts with ATR at DNA damage sites. (A) Schematic illustration of the HA-tagged NRF2 mutants. (B) Western blotting of HA, p-ATR, and p-CHK1 in HEK293T cells treated with 60 nM CPT for 6 h and released for 2 h. (C) Western blotting of HA, p-ATR and p-CHK1 in HEK293T cells pretreated with 5 mM NAC for 1 h and exposed to 8 Gy Cs137 γ-rays. (D) Immunofluorescence analysis of NRF2 (red), γH2AX (green), and DAPI (blue) in A549 cells exposed to 8 Gy Cs137 γ-rays. Scale bars, 20 μm. (E) Immunofluorescence analysis of NRF2 (red), ATR (green), and DAPI (blue) in A549 cells exposed to 8 Gy Cs137 γ-rays. Scale bars, 20 μm. (F) The co-localization of red (NRF2) and green (ATR) foci was analyzed by Image-Pro Plus 6.0. (G) Extracts of A549 cells exposed to 0 or 8 Gy Cs137 γ-rays were subjected to immunoprecipitation with an anti-ATR antibody or control IgG. Samples were immunoblotted with the indicated antibodies. (H) Extracts of A549 cells exposed to 0 or 8 Gy Cs137 γ-rays were subjected to immunoprecipitation with an anti-NRF2 antibody or control IgG. Samples were immunoblotted with the indicated antibodies. (I) Extracts of A549 cells treated with or without 30 nM CPT were subjected to immunoprecipitation with an anti-NRF2 antibody or control IgG. Samples were immunoblotted with the indicated antibodies. (J) GST pull-down assay following incubation of (1–338), (1–338) lacking amino acids 180–230 (Δ180–230), (1–338) in which the W amino acid at position 188 was replaced by A (W188A) or GST produced in E. coli with Flag-ATR produced in HEK293T cells.
Brusatol inhibits NRF2 in tumor xenografts and increases radiosensitivity
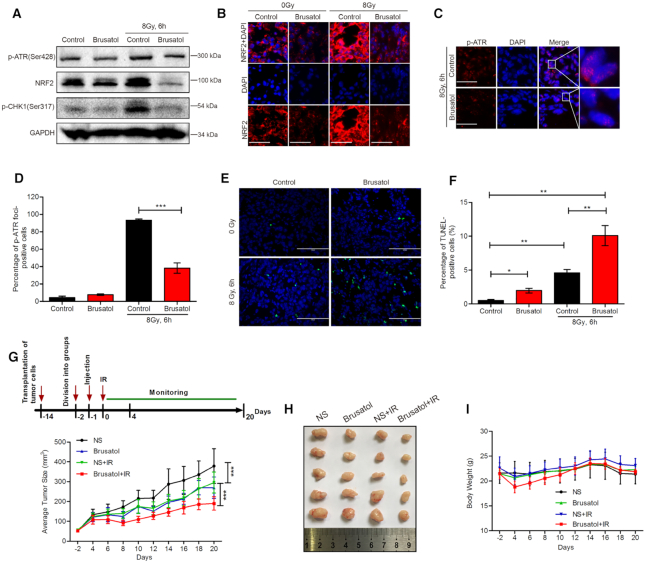
To explore whether NRF2 activates the ATR–CHK1 signaling pathway and regulates radiosensitivity in vivo, we transplanted A549 and A549-NRF2KO cells into nude mice. Unexpectedly, only A549 cells developed into tumor xenografts while A549-NRF2KO cells did not form stable tumor xenografts (data not shown). To overcome this complication, we used brusatol, an NRF2 inhibitor that increases cell radiosensitivity in vitro (43), to inhibit NRF2 in tumor xenografts formed by A549 cells. Consistent with the results obtained in vitro, brusatol effectively decreased the NRF2 protein level at 24 h after being injected around tumors and also attenuated upregulation of the NRF2 protein level upon exposure to IR (Figure 6A and B). We examined phosphorylation of CHK1 and ATR by western blotting and formation of p-ATR foci by immunofluorescence. Brusatol effectively reduced phosphorylation of CHK1 and ATR and formation of p-ATR foci (Figure 6A, C, and D). Apoptotic cells in tumor xenografts were detected by TUNEL assay after exposure to 8 Gy of γ-rays for 6 h. The percentage of apoptotic cells was 10.09% after treatment with brusatol and IR, but only 4.58% upon treatment with IR alone (Figure 6E and F). We also used this mouse model to assess the anti-tumor effect of brusatol upon exposure to IR. We found tumor size was reduced in the group treated with brusatol and IR (Figure 6G and H), while body weight did not significantly differ between the groups (Figure 6I). These results demonstrate that inhibition of NRF2 effectively increases tumor radiosensitivity in vivo, potentially through NRF2 activation of the ATR–CHK1 signaling pathway.
Figure 6.
Brusatol inhibits NRF2 in tumor xenografts and increases radiosensitivity. (A) Western blotting of NRF2 and p-CHK1 in tumor xenografts. Brusatol (2 mg/kg) was injected around tumors. After 24 h, tumors were exposed to 8 Gy Cs137 γ-rays for 6 h and whole-cell lysates of tumors were collected. (B) Immunofluorescence analysis of p-ATR foci in tumor xenografts treated with brusatol (2 mg/kg) for 24 h and exposed to 8 Gy Cs137 γ-rays for 6 h. Scale bars, 50 μm. (C, D) Immunofluorescence analysis of p-ATR foci in tumor xenografts treated with brusatol (2 mg/kg) for 24 h and exposed to 8 Gy Cs137 γ-rays for 6 h. Scale bars, 50 μm (C). Cells with >10 p-ATR foci were counted (D). Data are means ± S.E.M. (n ≥ 3). ***P ≤ 0.001. (E, F) TUNEL of apoptotic cells in tumor xenografts treated with brusatol (2 mg/kg) for 24 h and exposed to 8 Gy Cs137 γ-rays for 6 h. Scale bars, 100 μm (E). The percentage of TUNEL-positive cells was determined (F). Data are means ± S.E.M. (n ≥ 3). *P ≤ 0.05, **P ≤ 0.01. (G–I) Brusatol sensitizes xenografts to IR. Once tumors reached 50 mm3, mice (more than six per group) were treated with normal saline (NS) or 2 mg/kg brusatol and then exposed to 8 Gy Cs137 γ-rays 24 h later. Tumor size (G, H) and body weight (I) were measured every 2 days. Mice were dissected on day 20. Data are means ± SD. (n ≥ 6). ***P ≤ 0.001.
DISCUSSION
NRF2 is a transcription factor that protects cells against oxidative, electrophilic, and other stresses. To maintain redox homeostasis in cells exposed to radiation or drugs, NRF2 escapes from KEAP1-dependent repression and accumulates in the nucleus (44). Recent reports showed that not all NRF2 activity is ROS-dependent (21). Jayakumar et al. reported that the influence of NRF2 on radiosensitivity and DNA repair is linked to its regulation of the mRNA level of RAD51 and foci formation, and this function is independent of its antioxidant activity (23). However, the detailed mechanism by which NRF2 regulates the DNA repair process was not understood. We first confirmed that loss of NRF2 made cells hypersensitive to IR by measuring colony formation and apoptosis in the presence of the ROS scavenger NAC (Figure 1). This is probably a consequence of defective DNA repair caused by loss of NRF2, which is supported by our results of γH2AX and 53BP1 foci (Figure 1). We further demonstrated that NRF2 promoted HR during the repair of DSBs (Figure 2). BRCA1 and RAD51 are important proteins for the regulation of HR (45,46), and their mRNA levels are regulated by NRF2 (23,34,41). Consistent with previous reports (23,33), NRF2 influenced the protein levels of BRCA1 (Figure 4) and RAD51 (Figure 2). NRF2 enhanced phosphorylation of BRCA1 and increased formation of BRCA1 and RAD51 foci (Figure 2), and depletion of NRF2 significantly decreased the HR efficiency in an HR efficiency reporter system, suggesting that NRF2 could promote HR. HR competes with NHEJ, especially in S/G2 phases, but NHEJ is the major repair pathway and about 80% of DSBs are repaired by this process, irrespective of whether cells are in G1 or S/G2 phase (47). Here we investigated the influence of NRF2 on HR but did not assess the potential effect of NRF2 on NHEJ. The observed increases in the percentage of apoptotic cells and the percentage of cells with micronuclei among NRF2-depleted cells exposed to IR (Figure 1) may be only partially explained by a decrease in HR efficiency. Although NRF2 did not obviously influence the total protein level of 53BP1 (Figure 4), the effect on 53BP1 foci formation (Figure 2) is consistent with previous reports that NRF2 promotes the rapid disappearance of 53BP1-positive cells (34), implying a potential link between NRF2 and NHEJ to be explored.
Cell cycle arrest is important for essential DNA repair and preservation of genomic integrity. Activation of checkpoint cascades allows cell cycle arrest, chromatin remodeling, modulation of transcriptional programs, and activation of DNA repair (4,48). Arrest in G2 phase, when homologous sister chromatids are present and there are increased levels of critical HR proteins, preferentially induces HR, which allows more accurate repair of DSBs than NHEJ (5). Given the importance of NRF2 in HR, we next investigated whether NRF2 affects cell cycle arrest. We revealed that NRF2 deficiency perturbed G2 cell cycle arrest induced by DSBs (Figure 3), suggesting NRF2 functions as a cell cycle regulator independently of its roles as an antioxidant and a nuclear transcription factor. To further understand how NRF2 affects G2 cell cycle arrest, we examined the activation of ATR and ATM, two key kinases in the DDR, in the presence or absence of NRF2. We found depletion of NRF2 greatly reduced the active forms of ATR, CHK1 and CDC2 in response to DSBs (Figure 4), implying ATR could be mediated the NRF2 function in cell cycle arrest.
ATR is a master regulator of the DDR in mammary cells. In cells with DSBs, crosslinks, and those under replication stress, RPA coats ssDNA at the ends of DNA damage. ATR recognizes this RPA-coated ssDNA with its partner protein ATRIP (49,50). Recruitment of ATR/ATRIP to RPA-coated ssDNA is insufficient for its optimal activation, which requires activator proteins. Two activator proteins, TopBP1 and ETAA1, have been reported (16,17,51). Here, we showed that NRF2 is another potential activator of ATR and contains an AAD-like domain like TopBP1 and ETAA1 (Figures 4 and 5). High expression of ETAA1-AAD is sufficient to activate ATR (17). Consistently, overexpression of NRF2 (1–338 aa), which contained the AAD-like domain of NRF2, could activate ATR in cells with DSBs, while the mutated AAD-like domain of NRF2 could not activate ATR effectively (Figure 5), indicating the AAD-like domain of NRF2 may be critical for activation of ATR. Importantly, we also show the interaction and colocalization of NRF2 with ATR (Figure 5), suggesting NRF2 may be another activator of ATR. But the details of interaction between NRF2 and ATR need to be further clarified. Although both TopBP1 and ETAA1 are activators of ATR, only TopBP1 is necessary to activate the CHK1 signaling pathway and ETAA1 does not significantly affect CHK1 activation (16). Here, we found that NRF2 also activated the ATR–CHK1–CDC2 signaling pathway like TopBP1 (Figure 4). The relationship of NRF2 and TopBP1 is unclear and further work should determine whether NRF2 affects TopBP1 in DNA damage response and repair process.
NRF2 is known as a nuclear transcription factor, and previous reports have focused on its role in regulating transcription of target genes. However, NRF2 is widely distributed in cells, and can bind to other proteins to have other cellular functions. P21 was reported to bind directly to NRF2 by recognizing the DLG/ETGE motifs of NRF2 interfering with KEAP1 dependent NRF2 ubiquitination (52). Recent data indicate that NRF2 indirectly promotes activation of the AKT/p21 pathway by prompting SP1 recruitment and platelet-derived growth factor A-(PDGFA-) transcription (53). Studies also show that NRF2 binds to BRCA1 and Poly [ADP-ribose] polymerase 1 (PARP1), important proteins that regulate DNA damage repair (54,55), which is further consistent with a role of NRF2 in DNA damage repair. We observed interaction of NRF2 with ATR at DNA damage sites and activation of the ATR–CHK1–CDC2 signaling pathway, confirming a new function of NRF2 in DNA damage repair.
NRF2 is generally considered to attenuate toxicities of many carcinogenic factors by inducing the expression of a series of detoxifying and antioxidative stress genes. NRF2 accumulation is crucial for the prevention of chemicals-induced carcinogenesis (56–58). However, NRF2 overexpression in tumor cells is often responsible for radio- and chemo-resistance (27,59). Its double-sided role in tumor prevention and treatment remains to be explored and the mechanisms remain ambiguous. To explore the role of NRF2 in tumorigenesis and radiotherapy, we transplanted A549 and A549-NRF2KO cells into nude mice. However, A549-NRF2KO cells did not form stable tumor xenografts. This may be related to the role of NRF2 in driving cancer progression and metastasis. Ren et al. found that brusatol, a unique inhibitor of the Nrf2 pathway, enhances the efficacy of chemotherapeutic drugs (60). Consistent with this, we previously reported that brusatol increases the radiosensitivity of cells in vitro (43). Thus, we treated A549 tumor xenografts with brusatol to inhibit NRF2 and investigated the subsequent response to DNA damage induced by IR. Brusatol treatment increased radiosensitivity in tumor xenografts due to insufficient activation of the ATR–CHK1 pathway (Figure 6). Knockdown or inhibition of NRF2 was previously reported to be an effective strategy for increasing the radiosensitivity of cells by blocking NRF2-dependent antioxidant responses (27,31). Here, we found that brusatol increases the radiosensitivity of tumor xenografts by blocking the ATR–CHK1 signaling pathway. Cancer cells are particularly reliant on the ATR–CHK1 pathway. Specific inhibitors of this pathway were recently developed not only as chemosensitizers and radiosensitizers but also as single agents that can exploit the specific pathologies of tumor cells (61). Our results demonstrate the effectiveness of using brusatol to inhibit both NRF2 and ATR and to combat radioresistance, suggesting that NRF2 may be an effective tumor treatment target.
In conclusion, we uncovered a new function of NRF2 as an ATR activator to protect cells against DSBs in a manner that is independent of its role in regulating transcription and antioxidant defense. NRF2 deficiency resulted in the impairment of G2 cell cycle arrest and homologous recombination repair, the enhanced levels of apoptosis and micronuclei, and hypersensitivity of cells and xenografts to IR, implying NRF2 may be a new regulator for preserving genomic integrity.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Xingzhi Xu (Shenzhen University, China) for kindly providing DR-GFP-U2OS cells and the I-SceI plasmid.
Contributor Information
Xiaohui Sun, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Yan Wang, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Kaihua Ji, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Yang Liu, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Yangyang Kong, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Shasha Nie, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Na Li, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Jianxiu Hao, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Yi Xie, Institute of Modern Physics, Chinese Academy of Sciences, Lanzhou, China.
Chang Xu, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Liqing Du, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
Qiang Liu, Institute of Radiation Medicine, Chinese Academy of Medical Sciences & Peking Union Medical College, Tianjin Key Laboratory of Radiation Medicine and Molecular Nuclear Medicine, Tianjin, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31670859, 81772243, 81803172, 81803167, 31800703]; CAMS Innovation Fund for Medical Science [2017-I2M-1-016, 2019-I2M-2-006]; China Postdoctoral Science Foundation [2018M630106]; Natural Science Foundation of Tianjin [18JCYBJC26800, 18JCQNJC12300, 17JCYBJC42700]; Fundamental Research Funds for the Central Universities [10023201601602, 3332019159]; Non-profit Central Research Institute Fund of the Chinese Academy of Medical Sciences [2017-1001-08, 2018RC310020]. Funding for open access charge: CAMS Innovation Fund for Medical Science [2017-I2M-1-016, 2019-I2M-2-006].
Conflict of interest statement. None declared.
REFERENCES
- 1. Jackson S.P., Bartek J.. The DNA-damage response in human biology and disease. Nature. 2009; 461:1071–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chapman J.R., Taylor M.R., Boulton S.J.. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012; 47:497–510. [DOI] [PubMed] [Google Scholar]
- 3. Shrivastav M., De Haro L.P., Nickoloff J.A.. Regulation of DNA double-strand break repair pathway choice. Cell Res. 2008; 18:134–147. [DOI] [PubMed] [Google Scholar]
- 4. Bartek J., Lukas C., Lukas J.. Checking on DNA damage in S phase. Nat. Rev. Mol. Cell Biol. 2004; 5:792–804. [DOI] [PubMed] [Google Scholar]
- 5. Shibata A., Conrad S., Birraux J., Geuting V., Barton O., Ismail A., Kakarougkas A., Meek K., Taucher-Scholz G., Löbrich M. et al.. Factors determining DNA double-strand break repair pathway choice in G2 phase. EMBO J. 2011; 30:1079–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Simon B.J., Claudia L., Risa K., Fredrik M., Kastan M.B., Jiri B., Jiri L.. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 2006; 173:195–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ahn J.Y., Schwarz J.K., Piwnicaworms H., Canman C.E.. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res. 2000; 60:5934–5936. [PubMed] [Google Scholar]
- 8. Hirao A., Cheung A., Duncan G., Girard P.M., Elia A.J., Wakeham A., Okada H., Sarkissian T., Wong J.A., Sakai T.. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol. Cell. Biol. 2002; 22:6521–6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zou L., Elledge S.J.. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 10. Zou L., Liu D., Elledge S.J.. Replication protein A-mediated recruitment and activation of Rad17 complexes. PNAS. 2003; 100:13827–13832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cuadrado M. ATM regulates ATR chromatin loading in response to DNA double-strand breaks. J. Exp. Med. 2006; 203:297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Adams K.E., Medhurst A.L., Dart D.A., Lakin N.D.. Recruitment of ATR to sites of ionising radiation-induced DNA damage requires ATM and components of the MRN protein complex. Oncogene. 2006; 25:3894–3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smits V.A., Reaper P.M., Jackson S.P.. Rapid PIKK-dependent release of Chk1 from chromatin promotes the DNA-damage checkpoint response. Curr. Biol. 2006; 16:150–159. [DOI] [PubMed] [Google Scholar]
- 14. Boutros R., Dozier C., Ducommun B.. The when and wheres of CDC25 phosphatases. Curr. Opin. Cell Biol. 2006; 18:185–191. [DOI] [PubMed] [Google Scholar]
- 15. Kumagai A., Lee J., Yoo H.Y., Dunphy W.G.. TopBP1 activates the ATR-ATRIP complex. Cell. 2006; 124:943–955. [DOI] [PubMed] [Google Scholar]
- 16. Bass T.E., Luzwick J.W., Kavanaugh G., Carroll C., Dungrawala H., Glick G.G., Feldkamp M.D., Putney R., Chazin W.J., Cortez D.. ETAA1 acts at stalled replication forks to maintain genome integrity. Nat. Cell Biol. 2016; 18:1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Haahr P., Hoffmann S., Tollenaere M.A.X., Ho T., Toledo L.I., Mann M., Bekkerjensen S., Räschle M., Mailand N.. Activation of the ATR kinase by the RPA-binding protein ETAA1. Nat. Cell Biol. 2016; 18:1196–1207. [DOI] [PubMed] [Google Scholar]
- 18. Tebay L.E., Robertson H., Durant S.T., Vitale S.R., Penning T.M., Dinkovakostova A.T., Hayes J.D.. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic. Biol. Med. 2015; 88:108–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kobayashi A., Kang M.I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M.. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-Based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 2004; 24:7130–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang H.C., Nguyen T., Pickett C.B.. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 2002; 277:42769. [DOI] [PubMed] [Google Scholar]
- 21. Rojo M.D.L.V., Chapman E., Zhang D.D.. NRF2 and the hallmarks of cancer. Cancer Cell. 2018; 34:21–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee S.B., Sellers B.N., Denicola G.M.. The regulation of NRF2 by nutrient-responsive signaling and its role in anabolic cancer metabolism. Antioxid. Redox Signal. 2017; 29:1774–1791. [DOI] [PubMed] [Google Scholar]
- 23. Jayakumar S., Pal D., Sandur S.K.. Nrf2 facilitates repair of radiation induced DNA damage through homologous recombination repair pathway in a ROS independent manner in cancer cells. Mutat. Res. 2015; 779:33–45. [DOI] [PubMed] [Google Scholar]
- 24. Sekhar K.R., Freeman M.L.. NRF2 promotes survival following exposure to ionizing radiation. Free Radic. Biol. Med. 2015; 88:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bing X., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., Livingston D.M.. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 2006; 22:719–729. [DOI] [PubMed] [Google Scholar]
- 26. Xiang M., Namani A., Wu S., Wang X.. Nrf2: bane or blessing in cancer. J. Cancer Res. Clin. Oncol. 2014; 140:1251–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Singh A., Bodas M., Wakabayashi N., Bunz F., Biswal S.. Gain of Nrf2 function in non-small-cell lung cancer cells confers radioresistance. Antioxid Redox Signal. 2010; 13:1627–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Moon-Taek P., Min-Jung K., Young-Hee K., Soon-Young C., Jae-Hoon L., Jung-A C., Chang-Mo K., Chul-Koo C., Seongman K., Sangwoo B.. Phytosphingosine in combination with ionizing radiation enhances apoptotic cell death in radiation-resistant cancer cells through ROS-dependent and -independent AIF release. Blood. 2005; 105:1724. [DOI] [PubMed] [Google Scholar]
- 29. Czene S., Harms-Ringdahl M.. Detection of single-strand breaks and formamidopyrimidine-DNA glycosylase-sensitive sites in DNA of cultured human fibroblasts. Mutat. Res. 1995; 336:235–242. [DOI] [PubMed] [Google Scholar]
- 30. Luciana F., Kmiec E.B.. Camptothecin enhances the frequency of oligonucleotide-directed gene repair in mammalian cells by inducing DNA damage and activating homologous recombination. Nucleic Acids Res. 2004; 32:5239–5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee S., Lim M.J., Kim M.H., Yu C.H., Yun Y.S., Ahn J., Song J.Y.. An effective strategy for increasing the radiosensitivity of human lung cancer cells by blocking Nrf2-dependent antioxidant responses. Free Radic. Biol. Med. 2012; 53:807–816. [DOI] [PubMed] [Google Scholar]
- 32. Ceccaldi R., Rondinelli B., D’Andrea A.D.. Repair pathway choices and consequences at the double-Strand break. Trends Cell Biol. 2016; 26:52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Q., Li J., Yang X., Sun H., Gao S., Zhu H., Wu J., Jin W.. Nrf2 is associated with the regulation of basal transcription activity of the BRCA1 gene. Acta Biochim. Biophys. Sin. (Shanghai). 2013; 45:179–187. [DOI] [PubMed] [Google Scholar]
- 34. Kim S.B., Pandita R.K., Eskiocak U., Ly P., Kaisani A., Kumar R., Cornelius C., Wright W.E., Pandita T.K., Shay J.W.. Targeting of Nrf2 induces DNA damage signaling and protects colonic epithelial cells from ionizing radiation. PNAS. 2012; 109:E2949–E2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Feng L., Li N., Li Y., Wang J., Gao M., Wang W., Chen J.. Cell cycle-dependent inhibition of 53BP1 signaling by BRCA1. Cell Discovery. 2015; 1:15019–15019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li Z., Li Y., Tang M., Peng B., Lu X., Yang Q., Zhu Q., Hou T., Li M., Liu C. et al.. Destabilization of linker histone H1.2 is essential for ATM activation and DNA damage repair. Cell Res. 2018; 28:756–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Branzei D., Foiani M.. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008; 9:297. [DOI] [PubMed] [Google Scholar]
- 38. Blackford A.N., Jackson S.P.. ATM, ATR, and DNA-PK: the trinity at the heart of the DNA damage response. Mol. Cell. 2017; 66:801–817. [DOI] [PubMed] [Google Scholar]
- 39. Stiff T., Walker S.A., Cerosaletti K., Goodarzi A.A., Petermann E., Concannon P., O’Driscoll M., Jeggo P.A.. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006; 25:5775–5782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yan Y., Spieker R.M., Stoeger S.M., Cowan K.H.. BRCA1-mediated G2/M cell cycle arrest requires ERK1/2 kinase activation. Oncogene. 2005; 24:3285–3296. [DOI] [PubMed] [Google Scholar]
- 41. Chiara G., Baniasadi P.S., Harris I.S., Jennifer S., Satoshi I., Bryan S., Joshi P.A., Andrew W., Molyneux S.D., Bernard M.. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J. Exp. Med. 2013; 210:1529–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang X., Ran T., Zhang X., Xin J., Zhang Z., Wu T., Wang W., Cai G.. 3.9 ? structure of the yeast Mec1-Ddc2 complex, a homolog of human ATR-ATRIP. Science. 2017; 358:1206. [DOI] [PubMed] [Google Scholar]
- 43. Sun X., Qin W., Yan W., Du L., Chang X., Qiang L.. Brusatol enhances the radiosensitivity of A549 cells by promoting ROS production and enhancing DNA damage. Int. J. Mol. Sci. 2016; 17:997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Michael B.S., Karen T.L.. NRF2 and cancer: the good, the bad and the importance of context. Nat. Rev. Cancer. 2012; 12:564–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Escribano-Díaz C., Orthwein A., Fradet-Turcotte A., Xing M., Young J.T.F., Tkáč J., Cook M.A., Rosebrock A.P., Munro M., Canny M.D. et al.. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell. 2013; 49:872–883. [DOI] [PubMed] [Google Scholar]
- 46. Ma C.J., Gibb B., Kwon Y., Sung P., Greene E.C.. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res. 2016; 45:749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Andrea B., Julie B., Leopoldine T., Atsushi S., Sandro C., Goodarzi A.A., Andrea K., Jeggo P.A., Markus L.B.. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009; 28:3413–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhou B.B., Elledge S.J.. The DNA damage response: putting checkpoints in perspective. Nature. 2000; 408:433–439. [DOI] [PubMed] [Google Scholar]
- 49. Cimprich K., Cortez D.. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008; 9:616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ball H.L., Ehrhardt M.R., Mordes D.A., Glick G.G., Chazin W.J., David C.. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol. Cell Biol. 2007; 27:3367–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhou Z.W., Liu C., Li T.L., Bruhn C., Krueger A., Min W.K., Wang Z.Q., Carr A.M.. An essential function for the ATR-Activation-Domain (AAD) of TopBP1 in mouse development and cellular senescence. PLos Genet. 2013; 9:e1003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen W., Sun Z., Wang X.-J., Jiang T., Huang Z., Fang D., Zhang D.D.. Direct interaction between Nrf2 and p21Cip1/WAF1 upregulates the Nrf2-Mediated antioxidant response. Mol. Cell. 2009; 34:663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu D., Zhang Y., Wei Y., Liu G., Zhang N.. Activation of AKT pathway by Nrf2/PDGFA feedback loop contributes to HCC progression. Oncotarget. 2014; 7:65389–65402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bae I., Fan S., Meng Q., Rih J.K., Kim H.J., Kang H.J., Xu J., Goldberg I.D., Jaiswal A.K., Rosen E.M.. BRCA1 induces antioxidant gene expression and resistance to oxidative stress. Cancer Res. 2004; 64:7893. [DOI] [PubMed] [Google Scholar]
- 55. Wu T., Wang X.-J., Tian W., Jaramillo M.C., Lau A., Zhang D.D.. Poly(ADP-ribose) polymerase-1 modulates Nrf2-dependent transcription. Free Radic. Biol. Med. 2014; 67:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Paonessa J.D., Ding Y., Randall K.L., Munday R., Argoti D., Vouros P., Zhang Y.. Identification of an unintended consequence of Nrf2-directed cytoprotection against a key tobacco carcinogen plus a counteracting chemopreventive intervention. Cancer Res. 2011; 71:3904–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. auf dem Keller U., Huber M., Beyer T.A., Kümin A., Siemes C., Braun S., Bugnon P., Mitropoulos V., Johnson D.A., Johnson J.A. et al.. Nrf transcription factors in keratinocytes are essential for skin tumor prevention but not for wound healing. Mol. Cell. Biol. 2006; 26:3773–3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Satoh H., Moriguchi T., Takai J., Ebina M., Yamamoto M.. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res. 2013; 73:4158–4168. [DOI] [PubMed] [Google Scholar]
- 59. Arlt A., Sebens S., Krebs S., Geismann C., Grossmann M., Kruse M.L., Schreiber S., Schäfer H.. Inhibition of the Nrf2 transcription factor by the alkaloid trigonelline renders pancreatic cancer cells more susceptible to apoptosis through decreased proteasomal gene expression and proteasome activity. Oncogene. 2013; 32:4825–4835. [DOI] [PubMed] [Google Scholar]
- 60. Ren D., Villeneuve N.F., Jiang T., Wu T., Lau A., Toppin H.A., Zhang D.D.. Brusatol enhances the efficacy of chemotherapy by inhibiting the Nrf2-mediated defense mechanism. PNAS. 2011; 108:1433–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rundle S., Bradbury A., Drew Y., Curtin N.J.. Targeting the ATR-CHK1 axis in cancer therapy. Cancers. 2017; 9:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.