Abstract
Bacteria have evolved defence mechanisms against bacteriophages. Restriction-modification systems provide innate immunity by degrading invading DNAs that lack proper methylation. CRISPR–Cas systems provide adaptive immunity by sampling the genome of past invaders and cutting the DNA of closely related DNA molecules. These barriers also restrict horizontal gene transfer mediated by conjugative plasmids. IncC conjugative plasmids are important contributors to the global dissemination of multidrug resistance among pathogenic bacteria infecting animals and humans. Here, we show that IncC conjugative plasmids are highly resilient to host defence systems during entry into a new host by conjugation. Using a TnSeq strategy, we uncover a conserved operon containing five genes (vcrx089–vcrx093) that confer a novel host defence evasion (hde) phenotype. We show that vcrx089–vcrx090 promote resistance against type I restriction-modification, whereas vcrx091–vcxr093 promote CRISPR–Cas evasion by repairing double-strand DNA breaks via recombination between short sequence repeats. vcrx091, vcrx092 and vcrx093 encode a single-strand binding protein, and a single-strand annealing recombinase and double-strand exonuclease related to Redβ and λExo of bacteriophage λ, respectively. Homologous genes of the integrative and conjugative element R391 also provide CRISPR–Cas evasion. Hence, the conserved hde operon considerably broadens the host range of large families of mobile elements spreading multidrug resistance.
INTRODUCTION
The global rise of multidrug-resistant bacterial pathogens is a serious public health issue and a growing economic burden. This ongoing antibiotic resistance crisis is mostly due to improper use and overuse in clinical settings and animal husbandry that is exacerbated by a dried-out antibiotic development pipeline (1–3). While a minority of bacterial pathogens have evolved multidrug resistance exclusively by chromosomal mutations, most have acquired resistance by horizontal gene transfer. Resistance genes are frequently associated with mobile genetic elements (MGEs), such as conjugative plasmids, integrative and conjugative elements (ICEs), and bacteriophages that act as vehicles for dissemination (4,5). Conjugative plasmids of the IncC group are large, broad-host range plasmids found in pathogenic species of Enterobacteriaceae, Vibrionaceae and Morganellaceae (6). IncC plasmids are commonly associated with multidrug resistance and drive the global dissemination of New Delhi metallo-β-lactamase genes (blaNDM) that confer resistance against most β-lactams including carbapenems (7,8). Most of the conserved core genes of IncC conjugative plasmids are also present and syntenic in ICEs of the SXT/R391 family that have played a key role in the emergence of multidrug resistance in seventh-pandemic Vibrio cholerae O1, the infectious agent of the diarrhoeal disease cholera (9).
Upon entry into a new host, these MGEs face multiple host-defence systems that protect the cell against invading DNA molecules (10). Restriction-modification systems (R-M) provide innate immunity against foreign DNA that lacks the proper modification at specific recognition sites, while the host genome is protected from restriction endonuclease activity by methylation of the same recognition sites (11). In contrast, Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated proteins (Cas) provide adaptive immunity based on an RNA-guided endonuclease that seeks for sequence complementarity in nucleic acids and cuts the targeted sequence (12). Guide RNAs are produced from spacer sequences in the CRISPR array that are acquired from foreign invading DNA molecules during previous infections by MGEs. Type I CRISPR–Cas systems use the Cas3 endonuclease to target double-strand DNA (dsDNA) and are frequently found in Enterobacteriaceae and Vibrionaceae (13–15). Given the vast distribution of host-defence systems and their important role as a barrier against horizontal genes transfer, many MGEs have evolved strategies to evade these defences. Bacteriophages and conjugative elements can produce anti-CRISPR proteins, restriction endonuclease inhibiting proteins, or subvert R-M activities by producing their own modification enzymes (16–20).
Here, we investigated the resilience of the multidrug resistance-conferring IncC conjugative plasmid pVCR94 against CRISPR–Cas immunity. We carried out a high-throughput, transposon insertion site sequencing analysis (TnSeq) on pVCR94 to identify genes that allowed resistance to Cas3 cleavage during conjugation in V. cholerae. Our analysis revealed a conserved cluster of five genes that provide resistance to DNA restriction and repair Cas3-mediated double-strand DNA breaks (DSB). Two proteins encoded by these genes (vcrx092 and vcrx093) are related to the single-strand annealing protein Redβ (synaptase) and exonuclease λExo of bacteriophage λ’s Red recombination system (21,22). Further investigation using the ICE R391 confirmed the role of its synaptase and exonuclease homologues in the repair of Cas3-mediated DSB during conjugation. Altogether, our work demonstrates that the widespread synaptase–exonuclease recombination systems allow conjugative MGEs to evade a new host's CRISPR–Cas immunity, and suggests a similar role for lambdoid bacteriophages during infection.
MATERIALS AND METHODS
Bacterial strains, plasmids and media
The bacterial strains and plasmids used in this study are described in Supplemental Table S1. Bacterial strains were grown at 37°C on Luria-Bertani (LB) agar or with agitation in LB broth. Bacterial strains were maintained at −75°C in LB broth containing 20% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; kanamycin (Kn), 50 μg/ml; spectinomycin (Sp) 50 μg/ml; rifampicin (Rf), 50 μg/ml for Escherichia coli and 100 μg/ml for V. cholerae; chloramphenicol (Cm), 20 μg/ml for E. coli and 2 μg/ml for V. cholerae and nalidixic acid (Nx), 40 μg/ml. For complementation assays, bacterial cultures were supplemented with l-arabinose (0.02%, wt/vol).
Conjugation assays
Bacteria were grown for 16 h in LB broth with the appropriate antibiotics. Mating assays were carried out by mixing 100 μl of the donor (containing conjugative plasmid pVCR94 or ICE R391) and the recipient cells. Cells were pelleted by centrifugation, then washed once in 1 volume of LB broth and resuspended in 1/20 volume of LB broth. Bacterial mixtures were incubated for 2.5 h on LB agar plates at 37°C to allow conjugation. For ICE R391, excision and conjugative transfer were enhanced by overexpressing setCD from pGG2B in the presence of 0.02% arabinose. Serial dilutions were then plated on selective LB-agar plates with appropriate antibiotics to discriminate between donor, recipient and transconjugant CFUs. The frequency of transfer was calculated by dividing the number of transconjugant CFUs by the number of donor CFUs. For each assay, 8 transconjugant colonies were randomly picked and stored at −75°C for whole-genome Illumina sequencing.
Statistical analyses for conjugation assays
One-way analysis of variance (ANOVA) or unpaired Student t-test was used to assess statistical differences for frequencies of transconjugant formation in conjugation assays. A P value < 0.05 was considered a statistically significant difference. Each conjugation assay was repeated at least three times.
Plasmids and strains construction
Plasmid DNA was prepared using the QIAprep Spin Miniprep kit (Qiagen), and genomic DNA was isolated with the QIAamp DNA mini kit (Qiagen) as recommended by the manufacturer. PCR products were purified using the PCR purification kit (Qiagen). All molecular biology manipulations were carried out by standard procedures in accordance with the Current Protocols in Molecular Biology (23). Electroporations were done using a Biorad Gene Pulser Xcell apparatus (BioRad) in 1 mm cuvettes under the following conditions: 200 Ω, 25 μF and either 18 kV/cm for E. coli or 10 kV/cm for V. cholerae.
The oligonucleotides used in this study are described in Supplemental Table S2. Gene deletions were achieved by using the one-step chromosomal gene inactivation technique (λ Red-mediated mutagenesis) with pKD3 and pKD4 as templates for antibiotic resistance cassettes (24). All deletions were confirmed by Sanger sequencing. Slight modifications were made to adapt λ Red-mediated mutagenesis in V. cholerae to obtain the Δcas, ΔhsdR and ΔrecA mutants based on previously reported procedures for V. cholerae (25). Briefly, ∼1000-bp fragments upstream and downstream of casA-E, hsdR or recA were PCR-amplified using Q5 high-fidelity polymerase (NEB) and cloned into pCR2.1, flanking the chloramphenicol resistance cassette of pKD3 to yield pCRcrispr, pCRhsdR and pCRrecA, respectively. The resulting plasmids were used as PCR templates for λ Red-mediated mutagenesis to obtain the three deletion mutants.
To introduce protospacers into pVCR94Kn, pVCR94Sp and R391 and its Δ(bet-exo) mutant, the PCR template for λ Red-mediated mutagenesis was constructed as follows. The cat chloramphenicol-resistance cassette flanked by FRT sites of pKD3 was digested by HindIII and ligated into pCR2.1. Protospacers V1, V1R, V1NT and V3 were synthesised by oligonucleotide annealing as follows. Complementary pairs of oligonucleotides were designed to include the specific protospacer adjacent motif (PAM) sequence AA in 3′ (or 5′ for V1NT) of spacer 1 (V1 and V1R, position 343 477–343 509) or spacer 20 (V3, position 344 636–344 668) of the CRISPR array of V. cholerae O395 (Genbank CP000627.1_1) and BamHI restriction sites at both extremities (26) (Figure 1a). V. cholerae O395 CRISPR spacer sequences length are typically 32–33 bp and are available in the CRISPRCasdb database at https://crisprcas.i2bc.paris-saclay.fr/. For annealing, oligonucleotides were resuspended at 1 mg/ml in annealing buffer (10 mM Tris, pH 7.5–8.0, 50 mM NaCl, 1 mM EDTA). 2 μg of each complementary oligonucleotides were mixed in 50 μl of nuclease-free water and heated at 95°C for 5 min in an aluminium heat block. The heat block was then removed from the heater unit and let to cool down for 60 min. 300 pmol of the mixture was then incubated with 10 units of T4 polynucleotides kinase (New England Biolabs) for 1 h at 37°C, and the enzyme was then inactivated at 65°C for 20 min. Annealed oligonucleotides were digested by BamHI and cloned into pCR2.1::cat to yield pCRV1, pCRV1R, pCRV1NT and pCRV3 that were used as PCR templates for λ Red-mediated mutagenesis after validation of each construction by Sanger sequencing.
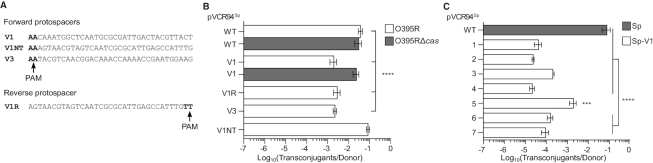
Figure 1.
Insertion of O395 protospacers in pVCR94 reduces its transfer to Cas+V. cholerae O395. (A) Sequence of the protospacers introduced into pVCR94Sp. (B) Conjugative transfer of pVCR94Sp derivatives bearing O395 protospacers from E. coli VB111 to V. cholerae O395R (Cas+) or O395RΔcas (Cas−). (C) Impact of protospacer insertion site on the transfer of pVCR94Sp to O395R. Positions of V1 protospacer insertions are indicated by orange arrowheads in Figure 2B. Bars are the mean of three independent experiments. Error bars are the standard error of the mean. Statistical significance: ****P< 0.0001 and ***P< 0.001.
The rifampicin-resistant mutants of V. cholerae O395 (O395R) was obtained by gradual selection of spontaneous resistant clones grown in LB broth supplemented with increasing concentration of rifampicin (0.1–100 μg/ml). The culture was serially diluted and plated onto LB agar to isolate Rf-resistant clones. Colonies were reisolated and sequenced to validate mutations in rpoB (27).
RNA extraction and cDNA synthesis
Briefly, RNA extractions were performed as follows. E. coli VB111 containing pVCR94Kn-V1 and V. cholerae O395R were grown at 37°C for 16 h in LB broth containing the appropriate antibiotics and a conjugation assay was performed as described above. After incubation, the conjugation mixture was resuspended in 1 ml of LB broth and this bacterial suspension was used for total RNA extraction using Direct-zol RNA extraction kit (Zymo Research) and TRI Reagent (Sigma-Aldrich) according to the manufacturer's instructions. Once purified, the RNA samples were treated using 2 units of DNase I (New England Biolabs) according to the manufacturer's instructions to eliminate any residual gDNA. cDNA was synthesised from 0.5 μg of RNA and 2 pmol of gene-specific primer vcrx094RT (Integrated DNA Technologies), using the reverse transcriptase SuperScript III (Invitrogen), according to the manufacturer's instructions. Control reactions without reverse transcriptase treatment (‘noRT’) were performed for each sample.
High-density Tn5 transposon mutagenesis
A conjugation-assisted random transposon mutagenesis experiment was performed on E. coli VB111 bearing pVCR94Kn-V1. The transposition system was composed of E. coli MFDpir+ carrying pFG036 (a plasmid coding for a thermosensitive cI transcriptional repressor) and pFG051 (a Pi-dependent RP4-mobilizable plasmid coding for the Tn5 transposition machinery repressed by cI, and carrying a mini-Tn5 (Sp) transposon). This diaminopimelate (DAP)-auxotrophic strain contains a chromosomal RP4 conjugative machinery and expresses the Pi protein required for pFG051 replication. The TnSeq experiment was performed in several successive steps. First, pFG051 was transferred by conjugation from MFDpir+ to VB111 bearing pVCR94Kn-V1 in a 2-h mating experiment at 30°C on LB agar plates supplemented with DAP in duplicates. Once in the recipient strain that lacks cI, the Tn5 machinery of pFG051 was constitutively expressed to mediate random mini-Tn5 (Sp) insertions in the genome. The mating mixture was then entirely spread onto 40 large LB agar plates (150 mm) supplemented with Cm, Kn, Nx and Sp. Plates were incubated until near confluence (10k to 40k CFUs) to select clones carrying mini-Tn5 (Sp) insertions. After overnight incubation at 37°C, Cm Nx Kn Sp-resistant colonies were collected using a cell scraper and resuspended in LB broth. The collected sample, designated as the ‘input library’ was washed, then resuspended in 4.5 ml of LB broth and cryopreserved. Total DNA of a 1.5 ml aliquot of the input library was extracted for sequencing. Another 1 ml aliquot of input library was used to inoculate 50 ml of LB broth supplemented with Cm, Kn, Nx and Sp, which was then grown overnight at 37°C. The resulting culture was used as the donor in a mating, where it was mixed in equal volumes with either V. cholerae O395R or O395RΔcas. After a 5 h incubation at 37°C, mating mixtures were spread onto 30 large LB agar plates (150 mm) supplemented with Rf, Kn and Sp to select for mini-Tn5 (Sp) insertions in pVCR94Kn-V1 that allowed transmission and replication of the plasmid into the recipients. After overnight incubation at 37°C, Rf, Kn and Sp-resistant colonies were collected and subsequently resuspended in LB broth, washed and resuspended in 4.5 ml of LB broth and cryopreserved. These samples were designated as ‘output libraries’. Total DNA of a 1.5 ml aliquot of the output libraries was extracted and used for sequencing.
Preparation of TnSeq sequencing libraries
For each input or output library, a 1.5 ml frozen stock aliquot was thawed on ice for 15 minutes. The aliquot was centrifuged, and cells were resuspended in 300 μl of cell lysis buffer from the Quick gDNA Miniprep kit (ZymoResearch). Total genomic DNA was then purified with AMPure DNA XP magnetic beads (Agencourt). Purified DNA was then fragmented and prepared for Nextera sequencing with the NEBNext Ultra II FS DNA Library Prep Kit (New England Biolabs) for Illumina. At the step of adapter ligation, we used our adapter for Nextera sequencing. Nextera adapter B was generated by annealing two oligonucleotides: 5′-PO4-CTGTCTCTTATACACATCTCCGAGCCCACGAGAC-InvdT-3′ and 5′- CAAGCAGAAGACGGCATACGAGATTCGCCTTAGTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGT-3′ together. Annealing was performed by heating of each oligonucleotide (40 μM final concentration) in annealing buffer (10 mM Tris NaCl pH 7.5, 50 mM NaCl) to 98°C and then slowly decreasing the temperature by 0.1°C every 10 s down to 4°C. DNA was purified again using DNA Ampure XP beads (Agencourt), and barcoding was performed in a qPCR machine using Veraseq 2.0 High-Fidelity DNA polymerase (Enzymatics). The amplification reaction was stopped at the end of the exponential phase. DNA was purified again and quantified using Quant-it PicoGreen dsDNA assay (Thermo Fisher). The quality and size distribution of the amplified mutant library was assessed on an Agilent 2100 Bioanalyzer instrument using a High Sensitivity DNA Chip. Mutant libraries were then pooled and sequenced by Illumina using the NextSeq® 500/550 High Output Kit v2 at the RNomics platform of the Laboratoire de Génomique Fonctionnelle de l’Université de Sherbrooke (https://rnomics.med.usherbrooke.ca) (Sherbrooke, QC, Canada). The transposon data analysis began by trimming the reads based on their quality and the presence of the Nextera Illumina adapters using Trimmomatic version 0.36 with the parameters SLIDINGWINDOW:4:20 MINLEN:36 ILLUMINACLIP:<adapters_file.fa>:2:30:15 (28). The quality of the reads was assessed with FastQC version 0.11.5 before and after trimming. The reads were aligned on the genome present in the respective strains with BWA MEM version 0.7.15 using default parameters. SAMtools version 1.3.1 was used to generate alignments statistics and to discard low quality alignments (MAPQ < 10) or multiple alignments (29). In order to get the exact positions of the transposon insertions, we made sam2sites.py, a python script that parses the alignments data and output all the insertions positions and read counts. We used the parameters—read_len_threshold 30 and—score_threshold 0. Note that we chose the middle base pair of the 9-bp Tn5 insertion site duplication to represent the insertion sites (30). The insertions sites files were then encoded in bigWig format using the Kent utilities (31). Visual inspection of the bigWig files was performed using an AssemblyHub on the UCSC Genome Browser (32). To get gene level insertion statistics, we used BEDTools intersect version 2.26.0 together with our python script sites2genes.py (33). The python scripts sam2sites.py and sites2genes.py are available at https://github.com/fredericQC/HDTM_tools.
Preparation of whole-genome sequencing libraries
Whole-genome DNA library preparation was prepared as described for TnSeq libraries with the following modifications. For each sample, total DNA was extracted from 500 μl of overnight culture, purified and fragmented as specified for TnSeq procedures. For the adapter ligation step, NEBnext Adaptor for Illumina (New England Biolabs) was used. All the subsequent steps were done as described for TnSeq library preparation above. Different variants of rescued pVCR94Sp were sequenced to characterise scars. The assembly workflow started by trimming the reads based on their quality and the presence of the TruSeq Illumina adapters using Trimmomatic version 0.36 with the parameters SLIDINGWINDOW:4:20 MINLEN:36 ILLUMINACLIP:<adapters_file.fa>:2:30:15 (28). The quality of the reads was assessed with FastQC version 0.11.5 before and after trimming. De novo assemblies were then generated using SPAdes version 3.11.1 with the parameters -k 21 33 55 77 –careful –only-assembler (34). QUAST version 5.0.2 was used to evaluate the quality of the assemblies (35). Full description of sequenced strains and assembly statistics are available in Supplemental Table S3.
Bioinformatic analyses
The sequences corresponding to each identified mobile element were downloaded from Genbank Protein homologues of Vcrx089, Bet and Exo were aligned using MUSCLE (36). Pairwise similarity matrices were calculated using UGene 1.32 (37) and plotted as heatmaps using the Heatmapper server (http://www.heatmapper.ca/pairwise). Repeat sequences in Supplemental Figure S1 were aligned using ClustalW alignment software (38). mRNA structure predictions were performed using RibEx (39). mRNA structure folding was carried out using the RNAfold web server at http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi.
RESULTS
CRISPR–Cas3 of V. cholerae O395 impedes the transfer of a targeted pVCR94 mutant
To investigate the sensitivity of pVCR94 to CRISPR–Cas activity and identify new anti-CRISPR proteins encoded by IncC plasmids, we took advantage of V. cholerae O395, a strain that naturally expresses CRISPR–Cas3 immunity (26). Since the CRISPR array of O395 is devoid of spacers targeting pVCR94, protospacers corresponding to spacers 1 (V1) and 20 (V3) of O395 CRISPR array were inserted downstream of vcrx093 in the spectinomycin-resistant derivative pVCR94Sp (Figures 1A and 2B, orange arrowhead 5 on pVCR94Sp map).
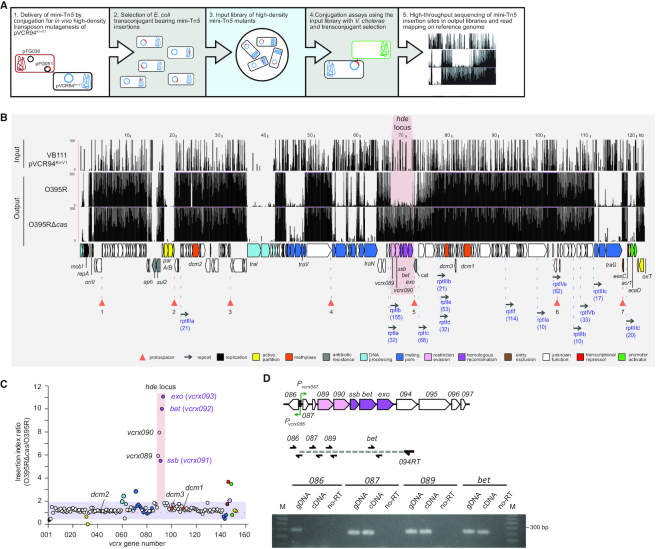
Figure 2.
Identification of CRISPR and restriction evasion functions by TnSeq. (A) Overview of the TnSeq workflow for the identification of anti-CRISPR functions in the IncC plasmid pVCR94. (B) A cluster of five genes is essential for the transfer of pVCR94Kn-V1 from E. coli VB111 to V. cholerae O395R (Cas+) but not O395RΔcas (Cas−). The three first tracks plot the number of mini-Tn5 reads mapped as a function of the position in pVCR94Kn-V1 (black bars). (C) Plot of mini-Tn5 insertion index for each gene (vcrx) of pVCR94Kn-V1. Insertion indexes were calculated as the ratios of insertion counts between O395RΔcas and O395R. Genes are color-coded as in panel b. (D) hde genes are part of a single operon. A 2% agarose gel from an assay to amplify genes of the hde locus on the vcrx094-derived reverse transcripted RNA. Genomic DNA of VB111 carrying pVCR94Sp (gDNA) and reverse transcription samples in the absence of reverse transcriptase (‘noRT’) were used as positive and negative PCR controls, respectively. M, 100 bp Plus II DNA ladder (Civic Bioscience).
The resulting plasmids were donated by conjugation in mating assays using E. coli VB111 (Nx) as the donor strain, and a rifampicin-resistant derivative of V. cholerae O395 (O395R) or a mutant lacking casA-E (O395RΔcas) as the recipient strains. Protospacer V1 reduced plasmid transfer to the Cas+ strain by 19-fold compared to the Cas− strain (Figure 1B). V1 in reverse orientation (V1R) and V3 produced a similar effect, whereas Non-Target V1 (identified as V1NT) with the protospacer adjacent motif (PAM) located at the 5′ end produced no interference, thereby confirming the specific yet partial immunity of O395R against pVCR94Sp-V1 (Figure 1A and B).
Next, to evaluate whether any positional effect resulted from the insertion site, protospacer V1 was also inserted at six alternative intergenic loci across pVCR94Sp (Figure 2B, orange arrowheads 1–7 on pVCR94 map) to compare with the phenotype obtained with V1 inserted downstream of vcrx093 at position 5. Again, we observed a partial immunity of O395R for all positions. However, the frequency of transfer of the resulting plasmids was differentially impacted by the position of V1, spanning an 80-fold difference between position 5 and positions 2 or 4 (Figure 1C). Notably, pVCR94Sp-V1 rescue was optimal with V1 inserted at position 5. Although reasons for the observed positional effect will be explored below, insertion of V1 at position 5 was used for all further experiments.
Identification by TnSeq of a conserved host defence evasion (hde) locus
Since the strain O395R failed to abolish the entry and persistence to pVCR94Sp-V1 (partial immunity), we hypothesized that this plasmid encodes a CRISPR–Cas evasion system. To identify the gene(s) responsible for this phenotype, we devised a TnSeq strategy aimed at targeting critical genes that allow evasion of the O395R CRISPR–Cas system during conjugation (Figure 2A). We constructed a high-density Tn5 insertion library in E. coli VB111 bearing the kanamycin-resistant variant pVCR94Kn-V1. This pVCR94 variant lacks acr2, the main repressor of IncC plasmid transfer (40,41), to increase conjugative transfer and prevent an insertion bias in this negative regulator gene. This set of pVCR94Kn-V1 mutants represents the ‘input library’ enriched in Tn5 insertions enabling plasmid replication and maintenance. The input library was used in mating assays to donate the set of mutated plasmids to V. cholerae O395R and O395RΔcas. The two sets of transconjugant colonies correspond to ‘output libraries’. Insertions in pVCR94Kn-V1 that are depleted in the output libraries compared to the input library were expected to reveal plasmid genes or sequences that are important for conjugation (with O395RΔcas) and involved in CRISPR–Cas evasion or DNA repair (with O395R).
Mean coverages of 289×, 3608× and 3537× were obtained for the input library, and the O395R and O395RΔcas output libraries, respectively. These results suggested sufficient saturation of the libraries to enable identification of genes essential for replication, maintenance, transfer, and repair. O395R and O395RΔcas outputs exhibited strikingly similar Tn5 insertion patterns (Figure 2B). Transfer-associated genes (e.g. traN and traG), plasmid replication and maintenance genes (e.g. repA and parAB) had virtually identical Tn5 insertion depletions in both conditions (Supplemental Table S4), except for a discrete ∼4.7-kb region, hereafter referred to as hde for Host Defence Evasion, exhibiting rare Tn5 insertions in O395R. Indeed, the five genes vcrx089, vcrx090, vcrx091, vcrx092 and vcrx093 located in the hde locus underwent a 5- to 12-fold depletion of Tn5 insertions in the Cas+ strain (Figure 2C and Supplemental Table S4).
While no Pfam domain was found for the translation product of vcrx090, vcrx089 is predicted to code for a protein containing a CbbQ_C domain (Pfam PF08406) and an ATPase AAA_5 domain (Pfam PF07728). vcrx091, vcrx092 and vcrx093 code for a single-strand binding protein (Pfam PF00436: SSB), a single-strand annealing protein (Pfam PF03837: RecT) and a double-strand specific 5′ to 3′ exonuclease (Pfam PF09588: YqaJ). The translation products of vcrx092, and vcrx093, hereafter named Bet and Exo, share 25% and 24% identity with Redβ and λExo encoded by bacteriophage λ. Together with gam, bet and exo form the λ Red system that is widely used as a chromosomal gene deletion and replacement tool due to its ability to facilitate RecA-independent recombination between identical sequences as short as 36 bp (24).
Based on their location, we hypothesized that the five genes of the hde locus are part of a polycistronic transcript. A reverse transcription experiment was performed using a primer located at the 3′ end of vcrx094 and total RNA extracted from VB111 cells bearing pVCR94Sp. PCR amplification using the generated cDNA revealed that vcrx087, vcrx089 and bet but not vcrx086 are part of the same mRNA transcript, likely initiated at Pvcrx087 (Figure 2D). Therefore, all five hde genes are part of the same operon.
vcrx089-090 is involved in restriction evasion
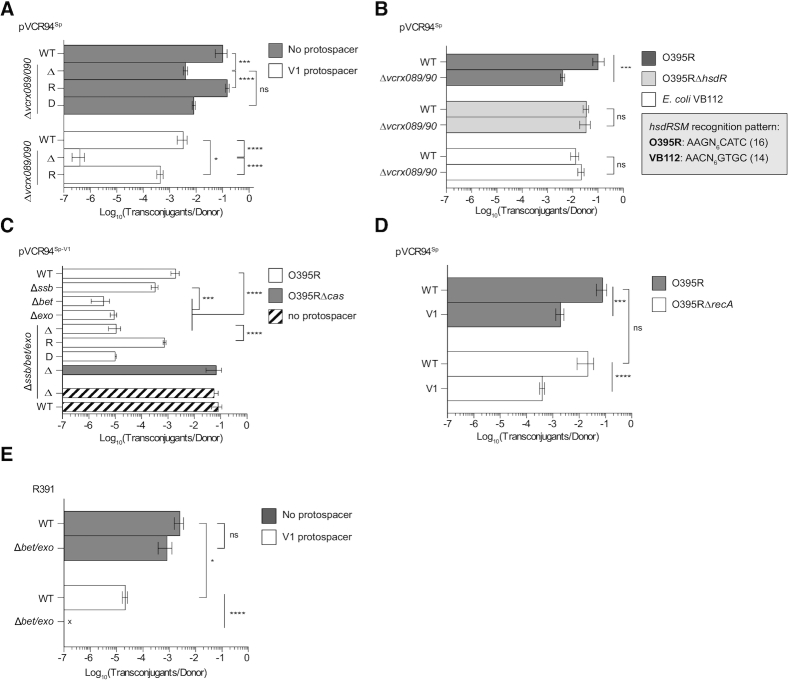
To investigate the role of the hde genes in the CRISPR–Cas evasion phenotype observed above, we constructed single and combined deletion mutants both in the presence and absence of protospacer V1 at position 5. In the absence of V1, deletion of both vcrx089 and vcrx090 led to a 25-fold reduction of conjugative transfer of pVCR94Sp to O395R (Figure 3A). Transfer was restored to wild-type level upon ectopic expression of both genes in the recipient, but not in the donor cells, suggesting that vcrx089-090 acts upon entry into the new hosts. The same experiment conducted using E. coli VB112 as the recipient strain showed that pVCR94Sp and its Δ(vcrx089-90) mutant transferred at the same rate (Figure 3B). E. coli VB111 and VB112 are virtually isogenic strains with identical EcoKI (AACN6GTGC) methylation patterns. In contrast V. cholerae O395 codes for a type I R-M enzyme predicted by REBASE (42) to recognizes the pattern AAGN6CATC. To test whether the transfer deficiency of pVCR94Sp Δ(vcrx089-90) resulted from restriction by the recipient strain, mating assays were carried out using a ΔhsdR mutant of O395R that is unable to produce the endonuclease subunit of the type I R-M enzyme (locus tag VC395_1879; Genbank CP001235.1). pVCR94Sp and its Δ(vcrx089-90) mutant transferred at the same rate to O395RΔhsdR, thereby suggesting that vcrx089-90 encodes an anti-restriction mechanism (Figure 3B).
Figure 3.
hde genes impact the resilience of translocated DNA against DNA cleavage in recipient cells during conjugation. (A) Effect of vcrx089/090 on transfer of pVCR94Sp with or without protospacer V1 using O395R as the recipient. Complementation of the Δ(vcrx089-090) mutation was carried out using pVcrx089-90 in either donor (D) or recipient (R) cells. (B) Effect of vcrx089/090 on transfer of pVCR94Sp devoid of protospacer using O395R and it restriction deficient ΔhsdR mutant or VB112 as the recipient. The pattern recognized by the type I R-M enzyme of each recipient is indicated. The corresponding number of sites in pVCR94Sp is shown in parenthesis. (C) Effect of ssb, bet and exo on transfer of pVCR94Sp with or without protospacer V1 using O395R or its Δcas mutant as the recipient. Complementation of the Δ(ssb-exo) mutation was carried out using pSsbBetExo in either donor (D) or recipient (R) cells. (D) Effect of bet and exo on the transfer of R391 with or without protospacer V1 using O395R as the recipient. (E) Effect of recA in recipient cells on transfer pVCR94Sp with or without protospacer V1. For all mating assays, E. coli VB111 containing pVCR94Sp variants was used as the donor strain and transconjugants were selected as Rf Sp-resistant colonies. Bars are the mean of three independent experiments. Errors bars are the standard error of the mean. An ‘x’ indicates that the frequency of transconjugant formation was below the detection limit (<10−7). Statistical significance: ****P< 0.0001; ***P< 0.001; *P< 0.05; ns, not statistically significant.
In the presence of protospacer V1, we observed a 5000-fold reduction of transfer of the Δ(vcrx089-90) mutant to O395R, suggesting a key role of vcrx089-090 in CRISPR–Cas evasion. However, complementation assays failed to restore the transfer of this mutant to the level of pVCR94Sp-V1 (Figure 3A). Hence, the strong transfer deficiency of pVCR94Sp-V1 Δ(vcrx089-90) likely results from both a sensitivity to restriction and a probable polar effect of the mutation on the expression of the downstream genes ssb, bet and exo. While the cause of this phenotype is not clearly understood, deletion of vcrx089-090 preserved a predicted RNA secondary structure that likely sequesters the ribosome binding site of vcrx089 (Supplemental Figure S2). The deletion places this structure 212 bp upstream of the start codon of ssb, leaving a large untranslated region that could negatively impact the translation initiation of ssb, bet and exo or the stability of the altered hde mRNA. Hence, in the context of Cas-mediated DSB, vcrx089-090 restriction evasion function seems to be at least in part dependent upon the downstream ssb/bet/exo genes.
ssb/bet/exo allows CRISPR–Cas evasion of IncC plasmid during conjugation
To test this hypothesis, ssb, bet and exo deletion mutants of pVCR94Sp or pVCR94Sp-V1 were used in mating experiments. The Δ(ssb-exo) mutation had no impact on the transfer of pVCR94Sp to O395R (Figure 3C). In contrast, deletion of any of the three genes resulted in a 6- to >200-fold reduction of pVCR94Sp-V1 transfer, thereby suggesting an enhanced sensitivity of these mutants to Cas3-mediated cleavage (Figure 3C). This phenotype was reversed when ssb-bet-exo was ectopically expressed in the recipient but not in the donor cells or when O395RΔcas was used as the recipient (Figure 3C). Altogether, these results confirm that ssb, bet and exo, while not required for transfer, provide an evasion mechanism against CRISPR–Cas immunity during entry by conjugation into a Cas+ strain.
Bet-Exo-mediated DNA repair is RecA-independent
λ Red mediates homologous recombination by single-strand annealing (SSA) in a RecA-independent fashion (43). Likewise, Bet and Exo of SXT/R391 ICEs have been shown to promote ICE diversity independently of RecA (21). To test whether RecA is involved in IncC plasmid rescue, a ΔrecA mutant of O395R was constructed and used as the recipient strain in mating assays. Transfer of pVCR94Sp-V1 remained unaffected by the recA mutation (Figure 3D), confirming that the DNA repair process mediated by Bet-Exo recombination occurs independently of RecA.
Bet/Exo repairs Cas-generated DSBs by annealing conserved short direct repeats
Since the position of protospacer insertions influences the efficiency of DSB repair (Figure 1C), rescued plasmids that were recovered from O395R transconjugants were further investigated by PCR amplification and sequencing of scar sequences. Only a handful of transconjugants were obtained using pVCR94Sp with V1 inserted at positions 1, 2 and 4 (Table 1). Analysis of these rescued plasmids revealed an intact protospacer and surrounding region, suggesting Cas3 loss-of-function, a phenotype also reported for Cas9 endonuclease at a similar rate (44). Surprisingly, standard PCR assays failed to amplify regions adjacent to all other V1 insertion loci, suggesting large deletions or insertions at scar sites. Therefore, whole-genome Illumina sequencing was carried out to determine the structure of the repaired plasmids. Since fragments of the O395R recipient genome could have been used as templates for Bet/Exo-mediated repair, the rescued plasmids were first retransferred into E. coli VB111 to facilitate the identification of possible insertions of chromosomal DNA originating from O395R. Out of 85 selected transconjugants carrying rescued IncC plasmids, 78 could donate their plasmid to VB111. Seven plasmids partially or totally lost the ability to conjugate. Consequently, these plasmids were sequenced in O395R. De novo assemblies yielded a 61x mean coverage (Supplemental Table S3).
Table 1.
Summary of the scar patterns found in rescued pVCR94Sp carrying V1 protospacer at diverse positions
| Protospacer position | Number of sequenced clonesa | Type of scarsb (distribution) | Conjugation properties | Repeat ID used for repairc | Deletion size (bp)d |
|---|---|---|---|---|---|
| 5 | 73/85 | IA (43/85) | Transmissible | rptIb-rptIf | 23 702 |
| IB (1/85) | Transmissible | rptIb-rptIf | 23 702 | ||
| IC (7/85) | Transmissible | rptIb-rptIe | 14 076 | ||
| ID (1/85) | Transmissible | rptIb-rptIe | 14 076 | ||
| IE (1/85) | Transmissible | rptIb-rptId | 14 003 | ||
| IF (4/85) | Transmissible | rptIb-rptIc | 10 310 | ||
| IG (16/85) | Transmissible | rptIa-rptIc | 9268 | ||
| 1 | 0/85 | No scar | NA | NA | |
| 7 | 1/85 | III (1/85) | Deficient | rptIIIc-rptIIId | 6728 |
| 3 | 2/85 | IV (2/85) | Deficient | rptIIIa-rptIIIb | 59 432 |
| 6 | 9/85 | VA (1/85) | Transmissible | rptIIa-rptIIb | 5138 |
| VB (8/85) | Transmissible | rptIVa-rptIVb | 4936 | ||
| 2 | 0/85 | No scar | NA | NA | |
| 4 | 0/85 | No scar | NA | NA |
a0 values, no repaired transconjugants was obtained.
bPartial sequences of characterized scars are available in Supplemental Figure S2.
dExact deletion size from pVCR94Sp-V1 variants
Analysis of the scar sites revealed different repair patterns, even when DSBs occurred at the same position. Eleven different scar patterns were observed for all protospacer positions (Table 1 and Supplemental Table S3). All repairs led to deletions; no insertions were found. With a set of 73 representatives, rescued pVCR94Sp-V1 plasmids were the most frequently sequenced since the protospacer at position 5 yielded the highest rate of transconjugant formation (Figure 1C). Within this set, seven different deletion patterns spanning from positions 9268 to 23 702 bp were identified. No key transfer genes were deleted within this set, allowing the repaired plasmids to remain self-transmissible (Table 1) while being protected from O395R Cas3 activity. Rescued plasmids with protospacer at positions 3, 6 and 7 lost DNA fragments spanning from positions 4936 to 59 432 bp.
All repairs occurred by homologous recombination between repeated sequences distributed across pVCR94 (all repeats found in pVCR94 are listed in Supplemental Table S5). Four distinct repeated sequences (rptI-IV) were used for recombination depending on DSB location and resulted in different scar patterns (Table 1). Except for rptII that is located within two genes of unknown function, all repeats were found to be located within intergenic sequences. rptId allowed recombination between only seven identical nucleotides. The longest repeat, rptIb, was 155 bp and common to all repairs occurring at position 5 except for type IG scars (Table 1 and Supplemental Figure S3). All other type I repeats (rptIa, rptIc-f) were found to be partial repeats of rptIb (Table 2 and Supplemental Figure S1). The use of the same two repeats could yield slightly different scars depending on where homologous recombination took place within the repeats (Table 1 and Supplemental Figure S3), as observed for IA-IB and IC-ID. rptIb and rptIf shared the longest identity stretches (Supplemental Figure S1) and generated the majority of characterised scars (Table 1) when DSB occurred at position 5.
Table 2.
Repeated sequence motifs recombined in repaired plasmids

|
NCBI blastn search of rptI (rptIb), rptIII and rptIV sequences revealed a strong conservation among diverse species of Gammaproteobacteria. rptI was found in most IncC conjugative plasmids of Enterobacteriaceae and Vibrionaceae, such as multidrug resistance plasmid pMG252 from E. coli and pVPS129 from Vibrio parahaemolyticus, and many other IncC plasmids from Klebsiella, Salmonella, Providencia, Escherichia, Vibrio, Citrobacter and Shewanella. Most type 2 IncC plasmids shared repeats highly similar to rptI (>98% identity), whereas type 1a and 1b rptI-like repeats were more divergent (>92% identity). Interestingly, a 32-bp rptIb segment was also found (100% identity) in Salmonella genomic island 1 (SGI1), a multidrug resistance island that requires IncC plasmids for its own dissemination (45). The rptIII and rptIV repeats were also strictly conserved (100% identity) in most IncC plasmids.
ssb/bet/exo allows CRISPR–Cas evasion of ICE R391 during conjugation
The translation products of vcrx089, vcrx092, and vcrx093 share 53%, 59% and 45% with Orf70, Bet and Exo encoded by SXT/R391 ICEs, respectively, and homologues are found in several other mobile genetic elements including IncA, IncT and IncP-7 conjugative plasmids (Supplemental Figure S4a and S4b). Bet and Exo of SXT/R391 ICEs have been shown to promote hybrid ICE formation by RecA-independent homologous recombination between SXT and R391 integrated in a tandem fashion in the chromosome (21). Like IncC plasmids, SXT/R391 ICEs lack λ gam, which encodes an inhibitor of E. coli SbcCD and RecBCD exonuclease, but carry a ssb gene (46–48). Since SXT/R391 ICEs share homologous bet and exo genes (46), the role of this system was also investigated using R391. Insertion of protospacer V1 in R391 resulted in CRISPR–Cas interference (76-fold reduction of transfer) (Figure 3e). Deletion of bet and exo completely abolished the transfer of R391V1, confirming the key role of bet and exo in CRISPR–Cas evasion during ICE transfer.
DISCUSSION
Although many transfer and maintenance-related genes have been characterised for IncC conjugative plasmids and SXT/R391 ICEs, many genes remain to be characterised (6,8,22,49). Here, we sought genes that could be instrumental for these globally prevalent MGEs to evade CRISPR–Cas immunity deployed by bacteria as a barrier against invading DNA molecules during conjugation, the main mechanism of multidrug resistance dissemination. Several anti-CRISPR proteins are known to directly inhibit type I system Cas3 endonucleases such as AcrIE and AcrIF (16,50). Most of these anti-CRISPR proteins are produced by bacteriophages, and are challenging to predict due to their small size and the lack of conserved sequence motif (16,20,50).
In contrast, our TnSeq approach allowed us to identify host defence evasion (hde) functions that are conserved among conjugative plasmids belonging to several incompatibility groups as well as ICEs of the SXT/R391 family. While seeking anti-CRISPR functions, we serendipitously identified the hde genes vcrx089-090 as genes that protect IncC plasmids against type I R-M systems. De novo expression of these genes was required in the recipient but not the donor strain for effective protection against the recipient's type I R-M endonuclease activity. Phages and conjugative elements deploy diverse anti-restriction strategies. Besides evolving genome sequences depleted for restriction sites, these MGEs often code for active protection mechanisms. For instance, IncW plasmids produce ArdC that occludes restriction sites (51). Ral of phage λ stimulates the methylation activity of EcoKI to expedite the protection of its own genome (52). Phage T3 encodes a hydrolase that depletes the pool of S-adenosyl methionine, an essential cofactor for type I R-M endonuclease activity (53). ArdA proteins encoded by diverse conjugative elements such as IncN plasmids and Tn916 ICE mimic the DNA structure to bind and block type I R-M enzymes (54). IncC plasmids code for three conserved cytosine-specific methylation enzymes (dcm1 [vcrx109, PF00145], dcm2 [vcrx042, PF00145] and dcm3 [vcrx100, PF06044]) that were shown to change the host's methylation pattern and help the IncC plasmid pVC211 to cross the R-M barrier of V. cholerae O1 El Tor C6706 (17). However, none of the three dcm genes came out as essential for escaping restriction in our assays using V. cholerae O395R as the recipient (Figure 2B and C), suggesting possible functional redundancy. Neither Vcrx089 nor Vcrx090 share any homology with known anti-restriction factors. CbbQ_C domain proteins are part of CbbQ/NirQ/NorQ family of proteins which play a role in the post-translational activation of Rubisco. The biological role and origin of such proteins in bacteria is unknown (55). ATPase_AAA_5 domain proteins are part of AAA+ superfamily of ATPases that is largely distributed among bacteria and participate in a wide range of cellular processes such as membrane fusion, proteolysis and DNA replication (56). The presence of predicted CbbQ_C and ATPase AAA_5 domains in Vcrx089 suggests a novel anti-restriction mechanism that is currently being characterized.
CRISPR–Cas systems can be acquired by horizontal gene transfer and are widely distributed in Enterobacteriaceae and Vibrionaceae (13). Considering that as adaptive bacterial immune systems, CRISPR arrays collect spacers from past invading DNA molecules, acquisition of IncC plasmid-specific spacer by natural strains is a possible event. For instance, the CRISPR array of Shewanella putrefaciens W3-18-1 (Genbank CP000503.1) contains three spacers, 26, 29 and 41, that seem to target the conjugal transfer traC gene of IncC plasmids (data not shown). Hence, mitigation of CRISPR–Cas immunity via dsDNA break repair likely broadens the host range of IncC and related plasmids. We have identified Bet (Vcrx092) and Exo (Vcrx093) as key actors in the evasion of IncC plasmids from the host's CRISPR–Cas immunity (Figure 2). Consistent with this function, the deletion of bet and exo in the distantly related ICE R391 increased its sensitivity to CRISPR–Cas-mediated cleavage. Together with vcrx089-090 and ssb (vcrx091), bet and exo are part of a large operon that is conserved in IncC plasmids and SXT/R391 ICEs (Supplemental Figure S4a) (46). The proteins encoded by bet and exo are distantly related to the Red system of bacteriophage λ (Supplemental Figure S4b) (46,57). While λ Red is widely used for bacterial mutagenesis and synthetic biology (24), its exact biological role remains ambiguous despite its involvement in λ replication (58). Likewise, the products of bet and exo of SXT/R391 ICEs have been shown to promote recombination between ICEs integrated in the chromosome in a tandem fashion, increasing ICE plasticity and diversity (21). However, ICE tandem arrays were not found in nature, and were shown to be unstable and short-lived in laboratory conditions, suggesting that generation of ICE diversity is likely an accessory consequence of another key function mediated by bet and exo (46). Moreover, IncC plasmids do not naturally integrate into the chromosome in a site-specific manner, are mutually incompatible, and exclude each other's entry into the same host (59,60), thereby limiting the opportunities for two different IncC plasmids to coexist together within the same cell and recombine. Hence, bet and exo genes are unlikely dedicated catalysts of plasmid diversity through recombination. Rather, generation of diversity is likely a by-product of the repair function that depends on the availability of compatible repair templates. Indeed, our results strongly support the notion that the products of bet and exo of IncC plasmids and SXT/R391 ICEs repair DSBs mediated by Cas endonucleases upon entry into an immune host cell. Notably, successful complementation of the ssb, bet and exo null mutants occurred exclusively when the genes were expressed in recipient cells, confirming that CRISPR–Cas evasion does result from pre-existing diversity generated in donor cells prior to conjugation.
RecA-mediated homologous recombination and repair of DSBs by double-strand invasion requires long stretches of identical complementary DNA (61). IncC plasmids are large, low-copy number, entities that are likely maintained in a single copy per cell, providing little DNA substrate for such repair mechanism upon entry into a new host. Scars characterisation of rescued plasmids revealed that Bet/Exo repaired DSBs by recombining identical repeated sequences as short as 7 bp compared to 9 bp for λ Red (62). These repeats are conserved among IncC plasmids. Bet/Exo-mediated repairs resulted in different deletions (Table 1) consistent with the SSA homologous repair mechanism. Bet/Exo failed to repair DSBs occurring at positions 2 and 4 (Table 1). Several factors may affect the efficiency of DSB repair, including the proximity of essential genes at these two positions, such as partition system parAB, replication gene repA, antibiotic resistance genes aph and sul2 used for selection, or toxin/antitoxin addiction systems (49). The lack of available repeats near the cleavage sites is another factor that could prevent DSB repair. Bet/Exo-mediated repair of DSB induced by CRISPR–Cas is non-conservative since the region containing the protospacer was eliminated, thereby immunizing the rescued plasmids against the new host. As expected, several rescued plasmids lost their conjugative properties due to the deletion of conjugation-related genes (Table 1). Although our experimental design led to isolation of deletion mutant only, we cannot rule out that DSB repair facilitates DNA insertion. With the proper selective pressure and DNA template, SSA-mediated DSB repair could eventually led to the incorporation of antibiotic resistance genes. Such event could explain the existence of hotspots for the insertion of variable DNA in SXT/R391 ICEs (21,22).
The Ssb protein encoded by vcrx091 was important but not essential for DSB repair, suggesting an accessory role that facilitates DSB repair. The lack of an ssb homologue in the hde loci of IncA plasmid pRA1 and untyped plasmid pAsa4C is consistent with this observation (Supplemental Figure S4a). Redβ was recently shown to bind E. coli Ssb to facilitate annealing of the 3′-overhang to Ssb-coated ssDNA at the replication fork (63). Whether Bet encoded by IncC conjugative plasmids and SXT/R391 ICEs can bind their host-encoded Ssb protein is unclear. However, since DSB repair of the Δssb mutant of pVCR94Sp-V1 was impaired, we hypothesize that the plasmid-encoded Ssb protein can coat the 3′-overhangs generated by Exo and facilitate the loading of Bet on these 3′-overhangs. Alternatively, Bet could be directly loaded on the 3′-overhangs via an Exo-Bet protein-protein interaction, and then interacts with Ssb-coated ssDNA of the lagging strand at the replication fork of the same or another copy of the replicating plasmid.
Here, we report that the Bet/Exo system encoded by IncC plasmids and SXT/R391 ICEs protects these MGEs against the host-encoded defence mechanisms, by recombining direct repeats in a RecA-independent fashion to repair DSBs caused by CRISPR–Cas (58,64). Bacteriophage λ can mediate recombination by a RecA-independent SSA mechanism between λExo-processed complementary DNA sequences. By analogy, we proposed that, following CRISPR–Cas-mediated DSB, Exo resects the 5′-end of duplex DNA ends to generate a 3′-overhang (Supplemental Figure S5). Specific patterns such as palindromic sequences found in the repeats (Table 2) could pause or slow down the degradation mediated by Exo, similar to Chi sequences that regulate RecBCD nuclease activities in the RecA-dependent DNA break repair mechanism (65). Ssb binds to the resulting 3′-overhang, protects it from host nuclease and helps recruiting Bet to anneal to a complementary ssDNA strand in a RecA-independent manner. The host ligase, with the collaboration of Bet and Exo, likely seals the resulting nick upon cleavage of 3′ ssDNA regions excluded from the duplex (58,64,66). Consistent with our results and this SSA repair model, one copy of the repeat and the region within these repeats are lost, giving rise to different scars, and contributing to plasmid plasticity.
This novel CRISPR evasion mechanism differs from anti-CRISPR proteins as it does not prevent DSB but rescues plasmids by SSA repair after Cas-mediated cleavage (16). Hence, synaptase-exonuclease may protect MGEs against several types of CRISPR–Cas system considering the large distribution of these two-component recombination systems in MGEs. Besides bacteriophage λ and related phages, homologues of Bet and Exo are encoded by IncC, IncA, IncT and IncP-7 conjugative plasmids, SXT/R391 ICEs, as well as many other uncharacterized mobile elements in distantly related bacteria such as Bacillus subtilis and Borrelia hermsii (46,67,68). Interestingly, most IncC plasmids but not SXT/R391 ICEs share repeats I and III, thus suggesting that distinct Bet/Exo systems may have evolved together with specific sets of repeated motifs. The presence of a portion of rptIb in SGI variants may help explain the evolution of these genomic islands that are widespread in S. enterica and highly reliant on IncC plasmids for their propagation (69). The involvement of Bet (RecT) has been suggested in the elevated recombination rate that occurs in SGI1 in the presence of an IncC helper plasmid and leads to the generation of SGI1 deletion mutants (69). However, the involvement of rptIb has not been demonstrated.
Therefore, synaptase-exonuclease recombination systems, together with the Vcrx089-90 anti-restriction system, play a crucial role in the acquisition, persistence, and dissemination of antibiotic resistance genes in bacterial populations by helping the conjugative elements that carry these genes to evade defence systems deployed by bacteria as barriers against invading DNA molecules. By broadening the host range of highly prevalent MGEs, hde functions could hinder efforts to develop CRISPR–Cas targeting systems, such as engineered conjugative plasmids or bacteriophages, aimed at controlling the dissemination of multidrug-resistant bacteria (70–72).
DATA AVAILABILITY
Raw sequencing data were submitted to Genbank under Bioproject accession number PRJNA602297 with the following Biosample accession numbers: for TnSeq with O395R and O395RΔcas as the recipient strains, SAMN13887834 and SAMN13887835, respectively; for TnSeq with VB111 bearing pVCR94Kn-V1 as the donor strain (input), SAMN13910572; for whole-genome sequencing of transconjugants bearing repaired IncC plasmids, SAMN13887703 to SAMN13887713. The python scripts sam2sites.py and sites2genes.py are available at https://github.com/fredericQC/HDTM_tools.
Complete data from aligned reads for TnSeq experiments can also be visualized using the UCSC genome browser at http://genome.ucsc.edu/cgi-bin/hgTracks?genome=pVCR94dX3dacr2spacer1&hubUrl=https://datahub-101-cw2.p.genap.ca/roy_2020/hub.txt.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sébastien Rodrigue (Université de Sherbrooke, Canada) for the kind gift of pFG036 and pFG051, and Lisa Craig (Simon Fraser University, Canada) for the kind gift of V. cholerae O395. We are grateful to Alain Lavigueur for his insightful comments on the manuscript and to Abigaëlle Bolduc for her technical assistance.
Contributor Information
David Roy, Département de biologie, Université de Sherbrooke, Sherbrooke J1K 2R1, Québec, Canada.
Kevin T Huguet, Département de biologie, Université de Sherbrooke, Sherbrooke J1K 2R1, Québec, Canada.
Frédéric Grenier, Département de biologie, Université de Sherbrooke, Sherbrooke J1K 2R1, Québec, Canada.
Vincent Burrus, Département de biologie, Université de Sherbrooke, Sherbrooke J1K 2R1, Québec, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Discovery grant [2016-04365] from the Natural Sciences and Engineering Research Council of Canada (NSERC); Project grant [PJT-153071 to V.B.] from the Canadian Institutes of Health Research (CIHR); D.R. is the recipient of a Fonds de recherche du Québec-Nature et Technologies (FRQ-NT) Postdoctoral fellowship; NSERC Postdoctoral fellowship; K.T.H. was supported by a postdoctoral fellowship [SPE20170336797] from the Fondation de la Recherche Médicale (FRM, France). Funding for open access charge: Discovery grant [2016-04365] from the Natural Sciences and Engineering Research Council of Canada (NSERC); Project grant [PJT-153071] from the Canadian Institutes of Health Research (CIHR).
Conflict of interest statement. None declared.
REFERENCES
- 1. Munita J.M., Arias C.A.. Mechanisms of antibiotic resistance. Microbiol. Spectr. 2016; 4:VMBF-0016-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Stokes H.W., Gillings M.R.. Gene flow, mobile genetic elements and the recruitment of antibiotic resistance genes into Gram-negative pathogens. FEMS Microbiol. Rev. 2011; 35:790–819. [DOI] [PubMed] [Google Scholar]
- 3. McEwen S.A., Collignon P.J.. Antimicrobial resistance: a one health perspective. Microbiol. Spectr. 2018; 6:ARBA-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Partridge S.R., Kwong S.M., Firth N., Jensen S.O.. Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 2018; 31:e00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker S., Thomson N., Weill F.X., Holt K.E.. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science. 2018; 360:733–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harmer C.J., Hall R.M.. The A to Z of A/C plasmids. Plasmid. 2015; 80:63–82. [DOI] [PubMed] [Google Scholar]
- 7. Wu W., Feng Y., Tang G., Qiao F., McNally A., Zong Z.. NDM metallo-beta-lactamases and their bacterial producers in health care settings. Clin. Microbiol. Rev. 2019; 32:e00115-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ambrose S.J., Harmer C.J., Hall R.M.. Evolution and typing of IncC plasmids contributing to antibiotic resistance in Gram-negative bacteria. Plasmid. 2018; 99:40–55. [DOI] [PubMed] [Google Scholar]
- 9. Weill F.X., Domman D., Njamkepo E., Tarr C., Rauzier J., Fawal N., Keddy K.H., Salje H., Moore S., Mukhopadhyay A.K. et al.. Genomic history of the seventh pandemic of cholera in Africa. Science. 2017; 358:785–789. [DOI] [PubMed] [Google Scholar]
- 10. Abedon S.T. Bacterial ‘immunity’ against bacteriophages. Bacteriophage. 2012; 2:50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vasu K., Nagaraja V.. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol. Mol. Biol. Rev. 2013; 77:53–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rath D., Amlinger L., Rath A., Lundgren M.. The CRISPR–Cas immune system: biology, mechanisms and applications. Biochimie. 2015; 117:119–128. [DOI] [PubMed] [Google Scholar]
- 13. McDonald N.D., Regmi A., Morreale D.P., Borowski J.D., Boyd E.F.. CRISPR–Cas systems are present predominantly on mobile genetic elements in Vibrio species. BMC genomics. 2019; 20:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Medina-Aparicio L., Davila S., Rebollar-Flores J.E., Calva E., Hernandez-Lucas I.. The CRISPR–Cas system in Enterobacteriaceae. Pathog. Dis. 2018; 76:fty002. [DOI] [PubMed] [Google Scholar]
- 15. Makarova K.S., Koonin E.V.. Annotation and classification of CRISPR–Cas systems. Methods Mol. Biol. 2015; 1311:47–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bondy-Denomy J., Garcia B., Strum S., Du M., Rollins M.F., Hidalgo-Reyes Y., Wiedenheft B., Maxwell K.L., Davidson A.R.. Multiple mechanisms for CRISPR–Cas inhibition by anti-CRISPR proteins. Nature. 2015; 526:136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wang R., Lou J., Li J.. A mobile restriction modification system consisting of methylases on the IncA/C plasmid. Mobile DNA. 2019; 10:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Melkina O.E., Goryanin I.I., Zavilgelsky G.B.. The DNA-mimic antirestriction proteins ArdA ColIB-P9, Arn T4, and Ocr T7 as activators of H-NS-dependent gene transcription. Microbiol. Res. 2016; 192:283–291. [DOI] [PubMed] [Google Scholar]
- 19. Hynes A.P., Rousseau G.M., Lemay M.L., Horvath P., Romero D.A., Fremaux C., Moineau S.. An anti-CRISPR from a virulent streptococcal phage inhibits Streptococcuspyogenes Cas9. Nat. Microbiol. 2017; 2:1374–1380. [DOI] [PubMed] [Google Scholar]
- 20. Hynes A.P., Rousseau G.M., Agudelo D., Goulet A., Amigues B., Loehr J., Romero D.A., Fremaux C., Horvath P., Doyon Y. et al.. Widespread anti-CRISPR proteins in virulent bacteriophages inhibit a range of Cas9 proteins. Nat. Commun. 2018; 9:2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garriss G., Waldor M.K., Burrus V.. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 2009; 5:e1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wozniak R.A., Fouts D.E., Spagnoletti M., Colombo M.M., Ceccarelli D., Garriss G., Dery C., Burrus V., Waldor M.K.. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009; 5:e1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K.. Current Protocols in Molecular Biology. 2003; Hoboken: John Wiley & Sons. [Google Scholar]
- 24. Datsenko K.A., Wanner B.L.. One-step inactivation of chromosomal genes in Escherichiacoli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yamamoto S., Izumiya H., Morita M., Arakawa E., Watanabe H.. Application of lambda Red recombination system to Vibriocholerae genetics: simple methods for inactivation and modification of chromosomal genes. Gene. 2009; 438:57–64. [DOI] [PubMed] [Google Scholar]
- 26. Box A.M., McGuffie M.J., O’Hara B.J., Seed K.D.. Functional analysis of bacteriophage immunity through a type I-E CRISPR–Cas system in Vibriocholerae and its application in bacteriophage genome engineering. J. Bacteriol. 2016; 198:578–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mariam D.H., Mengistu Y., Hoffner S.E., Andersson D.I.. Effect of rpoB mutations conferring rifampin resistance on fitness of Mycobacteriumtuberculosis. Antimicrob. Agents CH. 2004; 48:1289–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. . The sequence alignment/map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goryshin I.Y., Miller J.A., Kil Y.V., Lanzov V.A., Reznikoff W.S.. Tn5/IS50 target recognition. Proc. Natl. Acad. Sci. U.S.A. 1998; 95:10716–10721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rhead B., Karolchik D., Kuhn R.M., Hinrichs A.S., Zweig A.S., Fujita P.A., Diekhans M., Smith K.E., Rosenbloom K.R., Raney B.J. et al.. The UCSC genome browser database: update 2010. Nucleic Acids Res. 2010; 38:D613–D619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Casper J., Zweig A.S., Villarreal C., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Karolchik D. et al.. The UCSC genome browser database: 2018 update. Nucleic Acids Res. 2018; 46:D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quinlan A.R., Hall I.M.. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010; 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. et al.. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012; 19:455–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gurevich A., Saveliev V., Vyahhi N., Tesler G.. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013; 29:1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Edgar R.C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004; 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Okonechnikov K., Golosova O., Fursov M., team U.. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012; 28:1166–1167. [DOI] [PubMed] [Google Scholar]
- 38. Thompson J.D., Higgins D.G., Gibson T.J.. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994; 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abreu-Goodger C., Merino E.. RibEx: a web server for locating riboswitches and other conserved bacterial regulatory elements. Nucleic Acids Res. 2005; 33:W690–W692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lang K.S., Johnson T.J.. Characterization of Acr2, an H-NS-like protein encoded on A/C2-type plasmids. Plasmid. 2016; 87–88:17–27. [DOI] [PubMed] [Google Scholar]
- 41. Carraro N., Matteau D., Luo P., Rodrigue S., Burrus V.. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014; 10:e1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Roberts R.J., Vincze T., Posfai J., Macelis D.. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015; 43:D298–D299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stahl M.M., Thomason L., Poteete A.R., Tarkowski T., Kuzminov A., Stahl F.W.. Annealing vs. invasion in phage lambda recombination. Genetics. 1997; 147:961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Forsberg K.J., Bhatt I.V., Schmidtke D.T., Javanmardi K., Dillard K.E., Stoddard B.L., Finkelstein I.J., Kaiser B.K., Malik H.S.. Functional metagenomics-guided discovery of potent Cas9 inhibitors in the human microbiome. eLife. 2019; 8:e46540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Carraro N., Durand R., Rivard N., Anquetil C., Barrette C., Humbert M., Burrus V.. Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet. 2017; 13:e1006705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garriss G., Poulin-Laprade D., Burrus V.. DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R391 family. J. Bacteriol. 2013; 195:1991–2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilkinson M., Troman L., Wan Nur Ismah W.A., Chaban Y., Avison M.B., Dillingham M.S., Wigley D.B.. Structural basis for the inhibition of RecBCD by Gam and its synergistic antibacterial effect with quinolones. eLife. 2016; 5:e22963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kulkarni S.K., Stahl F.W.. Interaction between the sbcC gene of Escherichiacoli and the gam gene of phage lambda. Genetics. 1989; 123:249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hancock S.J., Phan M.D., Peters K.M., Forde B.M., Chong T.M., Yin W.F., Chan K.G., Paterson D.L., Walsh T.R., Beatson S.A. et al.. Identification of IncA/C plasmid replication and maintenance genes and development of a plasmid multilocus sequence typing scheme. Antimicrob. Agents CH. 2017; 61:e01740-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Pawluk A., Staals R.H., Taylor C., Watson B.N., Saha S., Fineran P.C., Maxwell K.L., Davidson A.R.. Inactivation of CRISPR–Cas systems by anti-CRISPR proteins in diverse bacterial species. Nat. Microbiol. 2016; 1:16085. [DOI] [PubMed] [Google Scholar]
- 51. Belogurov A.A., Delver E.P., Agafonova O.V., Belogurova N.G., Lee L.Y., Kado C.I.. Antirestriction protein Ard (Type C) encoded by IncW plasmid pSa has a high similarity to the ‘protein transport’ domain of TraC1 primase of promiscuous plasmid RP4. J. Mol. Biol. 2000; 296:969–977. [DOI] [PubMed] [Google Scholar]
- 52. Debrouwere L., Zabeau M., Van Montagu M., Schell J.. The ral gene of phage lambda. II. Isolation and characterization of ral deficient mutants. Mol. Gen. Genet. 1980; 179:75–80. [DOI] [PubMed] [Google Scholar]
- 53. Eustaquio A.S., Harle J., Noel J.P., Moore B.S.. S-Adenosyl-L-methionine hydrolase (adenosine-forming), a conserved bacterial and archaeal protein related to SAM-dependent halogenases. Chembiochem. 2008; 9:2215–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chen K., Reuter M., Sanghvi B., Roberts G.A., Cooper L.P., Tilling M., Blakely G.W., Dryden D.T.. ArdA proteins from different mobile genetic elements can bind to the EcoKI Type I DNA methyltransferase of E.coli K12. Biochim. Biophys. Acta. 2014; 1844:505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hayashi N.R., Arai H., Kodama T., Igarashi Y.. The cbbQ genes, located downstream of the formI and formII RubisCO genes, affect the activity of both RubisCOs. Biochem. Biophys. Res. Commun. 1999; 265:177–183. [DOI] [PubMed] [Google Scholar]
- 56. Ogura T., Wilkinson A.J.. AAA+ superfamily ATPases: common structure–diverse function. Genes Cells. 2001; 6:575–597. [DOI] [PubMed] [Google Scholar]
- 57. Mosberg J.A., Lajoie M.J., Church G.M.. Lambda red recombineering in Escherichiacoli occurs through a fully single-stranded intermediate. Genetics. 2010; 186:791–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Murphy K.C. Lambda recombination and recombineering. EcoSal Plus. 2016; 7:doi:10.1128/ecosalplus.ESP-0011-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ambrose S.J., Harmer C.J., Hall R.M.. Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid. 2018; 96–97:7–12. [DOI] [PubMed] [Google Scholar]
- 60. Humbert M., Huguet K.T., Coulombe F., Burrus V.. Entry exclusion of conjugative plasmids of the IncA, IncC, and related untyped incompatibility groups. J. Bacteriol. 2019; 201:e00731-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bi X., Liu L.F.. recA-independent and recA-dependent intramolecular plasmid recombination. Differential homology requirement and distance effect. J. Mol. Biol. 1994; 235:414–423. [DOI] [PubMed] [Google Scholar]
- 62. Fujimoto R., Osakabe T., Saito M., Kurosawa N., Isobe M.. Minimum length of homology arms required for effective Red/ET recombination. Biosci. Biotech. Bioch. 2009; 73:2783–2786. [DOI] [PubMed] [Google Scholar]
- 63. Caldwell B.J., Zakharova E., Filsinger G.T., Wannier T.M., Hempfling J.P., Chun-Der L., Pei D., Church G.M., Bell C.E.. Crystal structure of the Redbeta C-terminal domain in complex with lambda Exonuclease reveals an unexpected homology with lambda Orf and an interaction with Escherichiacoli single stranded DNA binding protein. Nucleic Acids Res. 2019; 47:1950–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Caldwell B.J., Bell C.E.. Structure and mechanism of the Red recombination system of bacteriophage lambda. Prog. Biophys Mol. Biol. 2019; 147:33–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Smith G.R. How RecBCD enzyme and Chi promote DNA break repair and recombination: a molecular biologist's view. Microbiol Mol. Biol. Rev. 2012; 76:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cassuto E., Lash T., Sriprakash K.S., Radding C.M.. Role of exonuclease and beta protein of phage lambda in genetic recombination. V. Recombination of lambda DNA in vitro. Proc. Natl. Acad. Sci. U.S.A. 1971; 68:1639–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Iyer L.M., Koonin E.V., Aravind L.. Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC Genomics. 2002; 3:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Vellani T.S., Myers R.S.. Bacteriophage SPP1 Chu is an alkaline exonuclease in the SynExo family of viral two-component recombinases. J. Bacteriol. 2003; 185:2465–2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kiss J., Nagy B., Olasz F.. Stability, entrapment and variant formation of Salmonella genomic island 1. PLoS One. 2012; 7:e32497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rodrigues M., McBride S.W., Hullahalli K., Palmer K.L., Duerkop B.A.. Conjugative delivery of CRISPR–Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob. Agents CH. 2019; 63:e01454-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Park J.Y., Moon B.Y., Park J.W., Thornton J.A., Park Y.H., Seo K.S.. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcusaureus. Sci. Rep. 2017; 7:44929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Bikard D., Barrangou R.. Using CRISPR–Cas systems as antimicrobials. Curr. Opin. Microbiol. 2017; 37:155–160. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data were submitted to Genbank under Bioproject accession number PRJNA602297 with the following Biosample accession numbers: for TnSeq with O395R and O395RΔcas as the recipient strains, SAMN13887834 and SAMN13887835, respectively; for TnSeq with VB111 bearing pVCR94Kn-V1 as the donor strain (input), SAMN13910572; for whole-genome sequencing of transconjugants bearing repaired IncC plasmids, SAMN13887703 to SAMN13887713. The python scripts sam2sites.py and sites2genes.py are available at https://github.com/fredericQC/HDTM_tools.
Complete data from aligned reads for TnSeq experiments can also be visualized using the UCSC genome browser at http://genome.ucsc.edu/cgi-bin/hgTracks?genome=pVCR94dX3dacr2spacer1&hubUrl=https://datahub-101-cw2.p.genap.ca/roy_2020/hub.txt.