Abstract
CRISPR-associated Rossmann Fold (CARF) and SMODS-associated and fused to various effector domains (SAVED) are key components of cyclic oligonucleotide-based antiphage signaling systems (CBASS) that sense cyclic oligonucleotides and transmit the signal to an effector inducing cell dormancy or death. Most of the CARFs are components of a CBASS built into type III CRISPR–Cas systems, where the CARF domain binds cyclic oligoA (cOA) synthesized by Cas10 polymerase-cyclase and allosterically activates the effector, typically a promiscuous ribonuclease. Additionally, this signaling pathway includes a ring nuclease, often also a CARF domain (either the sensor itself or a specialized enzyme) that cleaves cOA and mitigates dormancy or death induction. We present a comprehensive census of CARF and SAVED domains in bacteria and archaea, and their sequence- and structure-based classification. There are 10 major families of CARF domains and multiple smaller groups that differ in structural features, association with distinct effectors, and presence or absence of the ring nuclease activity. By comparative genome analysis, we predict specific functions of CARF and SAVED domains and partition the CARF domains into those with both sensor and ring nuclease functions, and sensor-only ones. Several families of ring nucleases functionally associated with sensor-only CARF domains are also predicted.
INTRODUCTION
CRISPR–Cas (Clustered Regularly Interspaced Short Palindromic Repeats—CRISPR-associated genes) are adaptive immunity systems that are present in nearly all archaea and ∼40% of bacteria (1–3). The CRISPR–Cas machinery incorporates fragments of virus or plasmid DNA into CRISPR arrays, where they become spacers between repeats, and employs processed transcripts of these spacers (CRISPR(cr) RNAs) as guides to cleave the cognate foreign DNA or RNA. The CRISPR immune response consists of three stages, each mediated by a distinct subset of Cas proteins: (i) adaptation, i.e. spacer acquisition, (ii) crRNA maturation, (iii) interference, i.e. recognition and cleavage of the target DNA or RNA (2–6). The CRISPR–Cas systems split into two classes that differ with respect to the architecture of their effectors, i.e. Cas protein modules that are involved in crRNA maturation and target recognition and cleavage (1,7). Class 1 systems employ multisubunit effector complexes that consist of multiple Cas proteins, whereas in Class 2 systems, the effector is a single multidomain protein. Deeper classification of each CRISPR–Cas class into types and subtypes is based on the comparison of the compositions and genomic arrangements of core cas genes that encode proteins responsible for the key functions in each of the three stages of the CRISPR response.
In addition to the core cas genes, CRISPR–Cas systems are more loosely associated with numerous ancillary genes that are particularly abundant and diverse in type III systems. The most common among the ancillary genes, known as csm6/csx1-like genes, encode proteins containing the CARF (CRISPR–Cas Associated Rossmann Fold) domain (1,8,9). A distinctive feature of the CARF domain superfamily is the conserved (D/N)-X-(S/T)-X3-(R/K) motif, which is part of the ligand-binding surface of the Rossmann fold (8). Comparative analysis of the CARF superfamily proteins revealed numerous domain architectures, in most of which the CARF domain is fused to a nuclease, such as RNases of the HEPN, PIN and RelE families, PD-D/ExK endonucleases, and other, less common nucleases (8). Apart from their association with the CARF domain in CRISPR ancillary proteins, these nucleases are the toxin moieties of widespread toxin-antitoxin (TA) or abortive infection (ABI) systems that are activated by virus infection or other forms of stress, typically, resulting in dormancy or cell death (10). Based on the typical activities of the numerous, well-characterized Rossmann fold proteins, it has been predicted that CARF is a nucleotide-binding domain which, upon binding a nucleotide ligand, allosterically activates an effector domain that then functions analogously to the toxins in the TA and ABI systems (8).
The CARF-containing Csx1 protein of the subtype III-B CRISPR–Cas systems of the archaeon Sulfolobus islandicus has been shown to contribute to interference (11). In agreement with these findings, deletion of the csm6 gene that also encodes a CARF domain protein abrogates CRISPR immunity in Staphylococcus epidermidis type III-A systems (12). The first biochemical characterization of Csx1 from Pyrococcus furiosus has shown that the HEPN domain of this protein is an endoribonuclease that acts selectively on single-stranded RNA and cleaves specifically after adenosines (13). Subsequent comparative genomic analysis of nucleotide signaling systems led to the discovery of CARF domain-containing proteins that are encoded next to a protein denoted mCpol (minimal CRISPR polymerase) that shares the active cyclase domain with Cas10, the large subunit of the effector complexes of type III CRISPR–Cas systems (14). The mCpol protein is much smaller than Cas10, in particular, lacking the HD nuclease domain of the latter, and is not linked to CRISPR–Cas systems. The discovery of the CARF-mCpol association prompted a more specific hypothesis, according to which cyclases, such as mCpol and Cas10, produce cyclic nucleotide ligand that are bound by the associated CARF domains resulting in activation of the respective effectors (14). This prediction was validated independently by two laboratories that demonstrated that binding of the crRNA-containing effector complex of several bacterial and archaeal type III CRISPR–Cas to cognate target RNAs stimulates the cyclase activity of Cas10 resulting in the synthesis of cyclic oligoadenylates (cOAn, n = 2–6). The CARF domain of Csm6 binds cOA and allosterically activates the HEPN domain of Csm6 which degrades RNA non-specifically. Site-directed mutagenesis of the cyclase active site of Cas10, the nucleotide-binding loop of the CARF domain of Csm6, or the RNase active site of HEPN each abolished the RNA degradation. These findings demonstrate the existence of a distinct signaling pathway of type III CRISPR–Cas activation by virus infection (hereafter, the cOA pathway) (15–17).
The cOA pathway is a typical signaling system that includes a second messenger synthetase (Cas10), a sensor domain (CARF), and an effector domain (HEPN) (17). Most signaling systems also contain a fourth component, an enzyme that cleaves the messenger and halts the effector activation. Remarkably, such a component, indeed, has been identified shortly after the discovery of the built-in cOA pathway in type III CRISPR–Cas systems. It has been shown that several CARF domain-containing proteins in Sulfolobus solfataricus degrade cOA (ring) molecules (18). These enzymes were denoted ‘ring nucleases’ and the genes encoding them were named crn1. It has been further demonstrated that the CARF domain of the ring nucleases is responsible for the metal-independent cOA cleavage, and the catalytic residues have been identified. Many archaea and bacteria that possess type III systems encode a single CARF-domain protein in the entire genome suggesting that either the same CARF domain alternates between signal transduction and cOA cleavage or some other, unrelated proteins are responsible for the ‘off-switch’ step of the pathway. Strikingly, both of these alternative mechanisms have been discovered. In Thermococcus onnurineus, the CARF domain-containing protein has a dual function: it first triggers the RNase activity of the HEPN domain upon cOA4 binding, but then, starts to cleave the bound cOA molecule, thus, limiting the HEPN domain-mediated RNA cleavage (19). Similar observations have been made for the TTHB144 protein from Thermus thermophilus (20). Furthermore, yet another ring nuclease has been discovered that does not belong to the CARF superfamily, but rather, to the unrelated DUF1874 protein family that is most often represented in viruses, but is also encoded in several bacterial and archaeal genomes (21). This protein is a more potent ring nuclease than those identified previously and can neutralize type III CRISPR defense systems by depleting cOA. Thus, viruses apparently employ this ring nuclease as a type III-specific anti-CRISPR protein, and DUF1874 protein was, accordingly, denoted AcrIII-1 and Crn2 (21). Kinetic modelling of the antiviral signaling pathway demonstrated the importance of the controlled removal of the messenger molecule, implying that the ring nuclease is an essential component of type III CRISPR–Cas systems (22).
The diversity of the CARF superfamily continued to grow during the last few years. Several new families of CARFs and new architectures of CARF domain-containing proteins have been identified in the course of a systematic analysis of cas genes neighborhoods (23). Furthermore, several instances of the link between type III CRISPR–Cas systems and the SAVED (SMODS-associated and fused to various effector domains) domain have been reported as well (14,23). The SAVED domains are strongly associated with genes encoding SMODS (Second Messenger Oligonucleotide or Dinucleotide Synthetase) family proteins which include cyclic 2′-5′ GMP-AMP synthases and 2′-5′ oligoadenylate synthetases (14). A limited sequence similarity between SAVED and CARF domains has been detected, and it has been proposed that SAVED is a highly divergent version of the CARF domain (23). Recently, this prediction has been validated by structural analysis of the SAVED-containing Cap4 protein of Enterobacter cloacae (24). Similarly to CARF, SAVED domains are often fused to various effector domains, typically, nucleases.
However, many CARF domain-containing proteins are not associated with CRISPR–Cas systems. The largest and best characterized family among these is RtcR, a sigma54 transcriptional coactivator of the RNA repair system that also includes RtcA, an RNA 3′-terminal phosphate cyclase, and RtcB, a RNA ligase (25,26). The Rtc system is activated in a response to RNA damage and general stress (27,28). RtcR consists of CARF, AAA ATPase and helix-turn-helix (HTH) DNA-binding domains (29). The ligand(s) of RtcR has not been identified so far, but it appears likely to be 2′,3′-cyclic phosphate that is formed by RtcA at RNA termini (29,30).
Due to the extreme sequence divergence and the diversity of domain architectures of CARF domain-containing proteins, CARF domains are often overlooked or mis-annotated in genome analyses. Here we present a comprehensive census of the CARF and SAVED domains encoded in bacterial and archaeal genomes, together with a collection of subfamily-specific sequence profiles for the identification of CARF domains in sequence databases. We further describe the results of phylogenetic, contextual and comparative genomic analyses of CARF and SAVED domain-containing proteins, focusing, primarily, on those protein families that are associated with CRISPR–Cas systems. Based on this analysis, we propose a classification of CARF and SAVED proteins, predict several families of novel ring nucleases, and provide new insights on the organization of the cOA signaling pathway.
A CENSUS OF CARF AND SAVED DOMAIN-CONTAINING PROTEINS: CLASSIFICATION, DOMAIN ARCHITECTURE AND COMPARATIVE GENOMICS
To obtain a representative collection of the CARF and SAVED domain-containing proteins, we combined several sources. First, we collated manually curated sets of CARF and SAVED domain proteins from two previously published analyses (8,23). Next, we identified all CARF domain-containing proteins in Uniprot (UniRef90, 26 October 2019) and defined their CARF domain boundaries. This was achieved by using CARF domains with known 3D structure as a starting point and applying protocol similar to that described by Schaeffer and Grishin (31). The obtained domain sequences were clustered with CLANS resulting in 480 clusters (32). Sequence alignments of full-length proteins, corresponding to each cluster, were constructed and trimmed to retain only CARF or SAVED domains (Supplementary Data File 1). These alignments were assessed for specificity and selectivity, and optimal parameters were defined to search for CARF domains in large databases (See Supplementary Methods for details). These alignments and the corresponding search parameters were used to identify CARF domains in a database of 13 116 complete genomes (March 2019; see (1)).
Altogether, 7143 protein sequences containing the CARF or SAVED domain were identified and subjected to phylogenetic and domain architecture analyses (Supplementary Table S1). To delineate the relationships between CARF/SAVED proteins, two classification dendrograms were constructed (see Supplementary methods). The first dendrogram is based on CARF/SAVED domain sequences only (Figure 1A and Supplementary Data File 2). This dendrogram encompasses 7397 domains from 6925 proteins (some proteins contain two CARF domains; 219 (3%) proteins were excluded because the length of the mapped CARF domain in these sequences was <75% of the alignment length of the respective profile). The dendrogram consists of two major branches that encompass most of the CARF domains and most of the SAVED domains, with one smaller CARF domain family (cluster 4) appearing to be an outlier. The majority of the CARF domains belong to the vast cluster 1. An additional dendrogram was built using complete protein sequences to obtain a better resolution within the two largest clusters, CARF cluster 1 and SAVED cluster 1 (Supplementary Data File 3). This analysis identified 9 strongly supported major clades among the CARFs (CARF 1–9) and 13 minor clades (Figure 1B), and 7 clades among the SAVEDs (Figure 1C). Sigma 54 transcriptional regulators that are never associated with CRISPR–Cas systems form the largest clade within CARF cluster 1 (Figure 1A; designated RtcR, after the best-characterized member of this group). The RtcR clade can be further divided into three large branches, as discussed below.
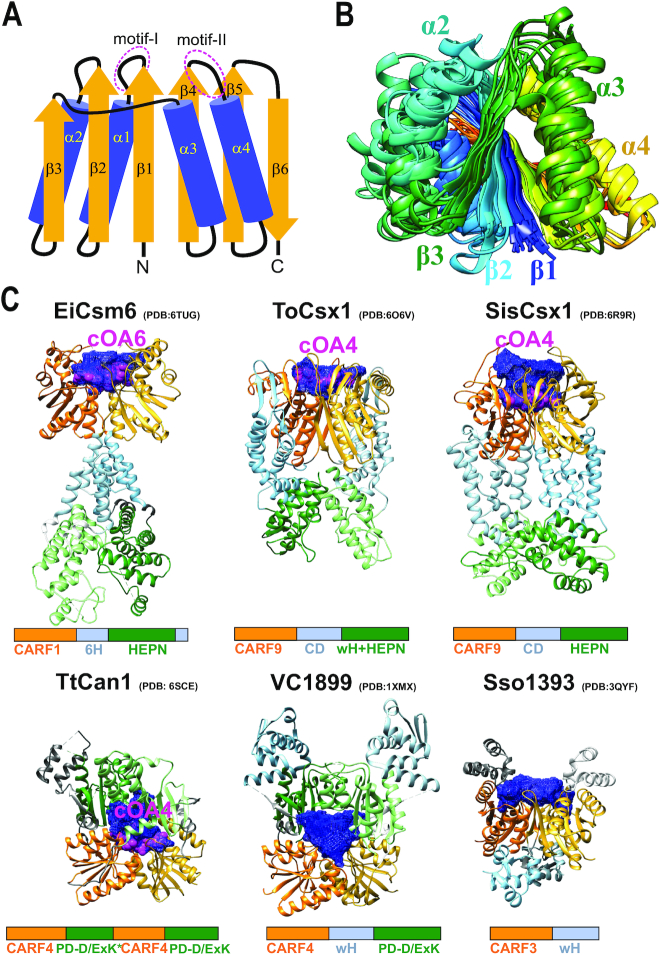
Figure 1.
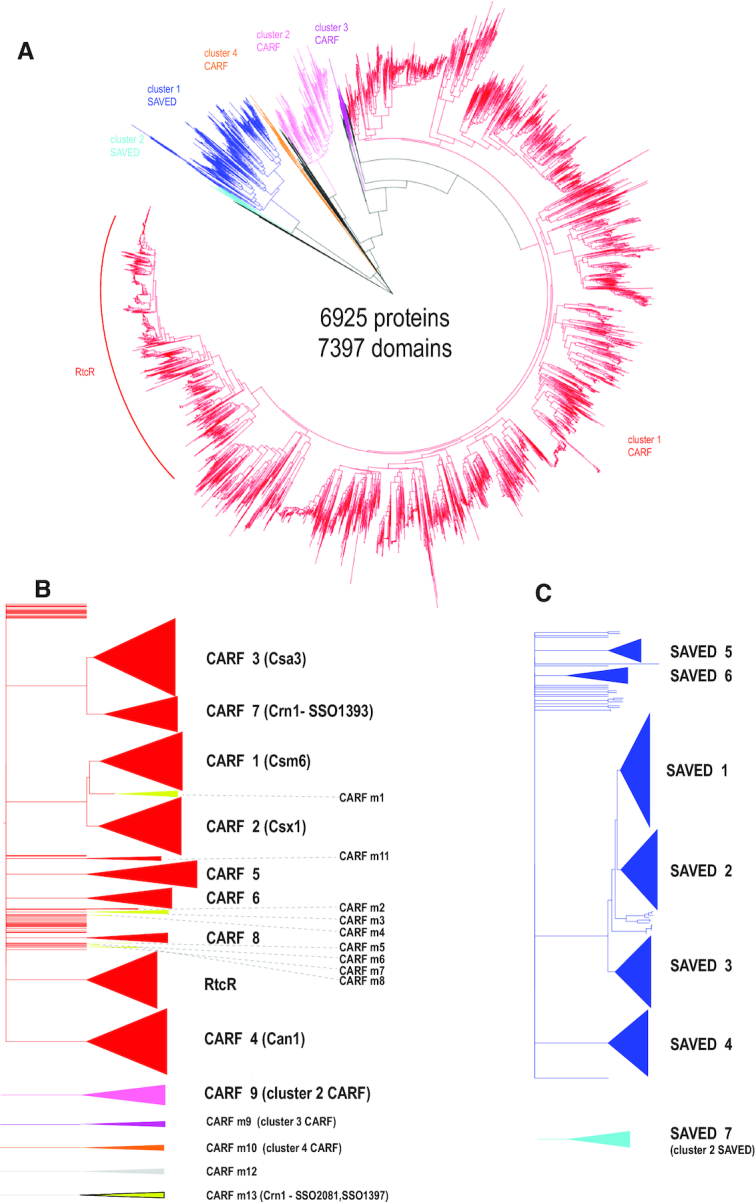
Relationships between CARF and SAVED domain-containing protein sequences. (A) Dendrogram built from the alignment of CARF and SAVED domain sequences only. The dendrogram was built using the ‘hybrid’ approach for sequence classification. Briefly, the FastTree program was used to infer relationships within alignable clusters, and the relationships between these clusters were inferred from HHalign pairwise scores using the matrix-based UPGMA method as described in detail previously (1). Distinct major alignable clusters are color coded. (B) Dendrogram built using alignment of complete amino acid sequences of CARF domain-containing proteins. Major and minor CARF clades corresponding to well-supported branches that include five or more sequences from diverse genomes are shown schematically on the right. The color coding is the same as in panel A. CARF_m13 group sequences are highly divergent and are included only in the second dendrogram. (C) Dendrogram built from the alignment of complete amino acid sequences of SAVED domain-containing proteins. Seven SAVED clades corresponding to well-supported branches that include 5 or more sequences from diverse genomes are shown schematically on the right. The color coding is the same as in panel A. The dendrograms in panels B and C were built using the same approach as the dendrogram in panel A. The subtrees including the sequences from the major cluster CARF1 (red) and SAVED (blue) were extracted from the tree built using complete protein sequences. Common names used in the literature are indicated in parentheses.
For all 7143 proteins in this set, we identified additional domains that are fused to the CARF or SAVED domain and also predicted transmembrane segments (Supplementary Table S1, Supplementary methods). Altogether, 60 distinct domain architectures of CARF domain proteins and 22 for SAVED domain proteins were identified (Supplementary Table S2). Of the 6665 identified CARF domain proteins, 2062 either contained no detectable domains other than CARF or contained only a fused HTH domain; of the 478 SAVED domain proteins, 176 contained no other detectable domains. The most common three enzymatic domains in CARF-containing proteins were HEPN, detected in 2019 proteins; PD-D/ExK family nuclease detected in 929 proteins; and AAA ATPase, mostly, represented in the RtcR family, in 796 proteins (Supplementary Table S2). Additionally, 488 CARF domain proteins were predicted to be membrane-associated. Most of the major clades of CARF and SAVED domains contain proteins with different domain architectures suggesting that domain shuffling is common in the evolution of these proteins. To facilitate the recognition of the domains associated with CARF and SAVED, we constructed 63 multiple alignment profiles many of which are not represented in the available profile databases (Supplementary Data File 1).
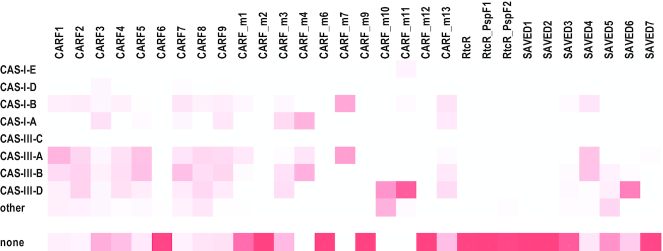
To uncover potential functional links of CARF domain proteins, we analyzed the loci, in which these proteins are encoded, in order to identify genes that are overrepresented in these genomic neighborhoods, separately for each major clade (Supplementary Table S3 and Supplementary Data File 4). For this purpose, we used the weighted ’guilt by association’ (33) method to search the database of 13 116 complete genomes of prokaryotes (see Supplementary methods and Supplementary Table S3 for more details). We also tabulated all the cases of association of CRISPR–Cas types and subtypes with different groups of CARF and SAVED proteins using the results of the recent comprehensive analysis of the CRISPR-cas loci (1) (Figure 2). From the same analysis, we extracted the data on the presence-absence of CRISPR–Cas systems in the genomes that encode CARF or SAVED domain proteins from our set (within or outside the CRIPSR-cas loci) (Supplementary Table S2) and the presence-absence of type III associated proteins. These results are discussed below for each CARF/SAVED clade separately.
Figure 2.
Association of different CARF and SAVED domain clades with CRISPR–Cas systems. The relative frequencies of CRISPR–Cas systems associated with distinct CARF and SAVED clades is shown.
CLADES OF CARF AND SAVED DOMAINS
CARF1
The CARF1 clade roughly corresponds to a combination of two PFAM families, PF09659 and PF09670 (Table 1). The proteins of this clade are encoded in both bacterial and archaeal genomes, and are strongly linked to subtype III-A CRISPR–Cas systems. The CARF domain of the Csm6 protein encoded in many subtype III-A operons belongs to this cluster (1,34). However, CARF1 clade genes are also found in some subtype III-B and subtype III-D loci, and even in type I loci (Figure 2, Supplementary Table S1). The main architectural distinction of this family is the fusion of CARF with the 6H (six helices) domain (8,35). A substantial majority of these proteins are also fused to the HEPN domain, a non-specific endoribonuclease (36,37), but some apparently have lost both the 6H and the HEPN domains (Supplementary Table S1). The structure of TTHB152 protein from T. thermophilus belonging to CARF1 clade has been solved, and the domains that have been originally identified in silico were validated (35). The Csm6 protein of the type III-A system from Staphylococcus epidermidis is essential for anti-plasmid interference although it is not an integral component of the Csm effector complex (12). Similar results have been reported for Csm6 from Lactococcuslactis and Streptococcus thermophilus (38). CARF1 domain in TTHB144 protein associated with type III-A system in T. thermophilus is a cOA4 sensor that can also cleave the signaling molecule (20). Finally, the Csm6 protein of the III-A system from Enterococcus italicus have been also shown to possess ring nuclease activity, in addition to the HEPN activation, so that, in the absence of dedicated ring nucleases, the CARF domain-mediated cOA6 cleavage provides an intrinsic off-switch to limit the period of activity of the HEPN RNase (39). CARF1 is the only clade in which some proteins have been shown to bind the cOA6 ligand.
Table 1.
Classification of CARF and SAVED domains
| Group | Number of proteins* | Frequent fusions with enzymatic domains | CRISPR-CAS link | Taxonomic spread | Closest PFAM | CDD HHpred∧ | Comments |
|---|---|---|---|---|---|---|---|
| CARF1 | 329 | HEPN | CAS-III-A mostly | Mostly, bacteria | PF09659 | cd09699 | Known as Csm6 family |
| CARF2 | 239 | HEPN, mCpol | CAS-III | Archaea and bacteria | PF09455 | cd09732 | Known as Csx1 family; some lost HEPN |
| CARF3 | 436 | No | CAS-I mostly | Mostly, archaea and some bacteria | PF09002 | cd09655 | Known as Csa3 (or CasR) family |
| CARF4 | 400 | PD-D/ExK, csx16, AAA ATPase | partially | Mostly, bacteria | PF09002 | cd09723 | Some fused to other CARF4 domains; known as Can1 |
| CARF5 | 128 | ADA | CAS-III | Archaea and bacteria | PF09623 | cd09686 | |
| CARF6 | 167 | STK_AAA | No | Mostly bacteria, some Thaumarchaeota | PF06956 | cd09655 | Two fused CARF domains, often in defense islands, sometimes encoded within T7 transposon |
| CARF7 | 154 | RelE, CYTH, HD | CAS-III | Bacteria and some archaea, mostly, Crenarchaeota | PF09651 | cd09694 | Established ring nuclease Crn1 |
| CARF8 | 50 | csx16 | CAS-III | Archaea and bacteria | cd09747 | Membrane associated (2–4 segments) | |
| CARF9 | 183 | HEPN | CAS-III | Archaeal and bacterial thermophiles | PF09455 | cd09728 | Known as Csx1 family |
| SAVED1 | 176 | HNH or PD-D/ExK | No | Mostly, bacteria | PF18145 | Linked to 2′-5′ oligoA synthetase | |
| SAVED2 | 119 | peptidase M48, CHAT, nucleosidase | No | Bacteria only | PF18145 | 2TM or 3TM, linked to 2′-5′ oligoA synthetase | |
| SAVED3 | 122 | PD-D/ExK | Partially | Bacteria only | PF18179 | Some have 2TM (typically not fused with other domains); linked to 2′-5′ oligoA synthetase and often to ubiquitin system components: ubiquitin activating E1 and E2 family enzymes and JAB protease | |
| SAVED4 | 33 | LON | CAS-III | Mostly, bacteria | PF18145 | Mostly membrane, some don’t have LON domain | |
| SAVED5 | 27 | TIR, JAB | Partially | Mostly, bacteria | PF18145 | Some linked to 2′-5′ oligoA synthetase | |
| SAVED6 | 14 | No | Partially (CAS-III-D) | Actinobacteria | PF18145 | ||
| SAVED7 | 35 | No | No | Bacteria only | PF18145 | Mostly membrane | |
| RtcR | 1925 | ||||||
| RtcR | AAA | No | Proteobacteria only | PF06956 | cd09723 | Linked to RNA cyclase RtcA, RNA ligase RtcB, TROVE domain, stomatin-like proteins | |
| PspF1 | AAA | No | Bacteria only | PF06956 | cd09723 | Defense island context, often encoded within Tn7 transposon | |
| PspF2 | AAA | No | Bacteria only | PF06956 | cd09723 | Defense island context | |
| CARF_m1 | 54 | PIN | Partially | Archaea only (Crenarchaeota) | Only those with PIN domain are encoded in the loci with type III systems | ||
| CARF_m2 | 2 | LON | No | Mostly Planctomycetes | cd09747 | ||
| CARF_m3 | 47 | PIN | Partially type III | Archaea (Thermoprotei only) | cd09723 | ||
| CARF_m4 | 5 | No | CAS-III | Archaea (Thermoprotei only) | cd09694 | ||
| CARF_m5 | 5# | No | No | Asgard archaea | PF09002 | cd09723 | |
| CARF_m6 | 4 | unk_domain | No | Haloferax only | cd09655 | ||
| CARF_m7 | 4 | Nitrilase | CAS-III | Archaea (Methanosarcinales only) | cd09747 | ||
| CARF_m9 | 9 | No | No | Mostly, cyanobacteria | cd09723 | Membrane associated, several either fused or encoded next to linked to mCpol | |
| CARF_m10 | 3 | HEPN | CAS-III-D | Bacteria | cd09699 | ||
| CARF_m11 | 15 | HEPN, CorA | CAS-III-D | Actinobacteria only | cd09742 | ||
| CARF_m12 | 7 | PIN | No | Archaea (Desulfurococcales only) | cd09723 | Often co-occurred with type I-A CRISPR–Cas system | |
| CARF_m13 | 37 | No | Partially type III | Archaea (Thermoprotei only) | cd09723 | Established ring nuclease Crn1 |
Note: * – in prok1903 (redundant); # – five distinct CARF proteins from Asgard archaea, not represented among complete genomes; ∧ – best hit in HHpred with probability >90% (however, many homologous CDD profiles have very close probability values, so relationships are approximate).
CARF2
The CARF2 clade includes the majority of the PF09455 family proteins (Table 1) and confidently groups with the CARF1 clade in the tree (Figure 1). The CARF2 proteins are widespread in bacteria and archaea, and are often annotated as Csx1 or DxTHG family proteins. The rest of the PF09455 family belongs to the CARF9 clade that differs from CARF2 by the presence of several insertions in the CARF domain and deletions in the HEPN domain ((40) and see below). The great majority of CARF2 domains are fused to HEPN domains, but loss of the HEPN domain and fusions with other domains were also detected, in particular, with an additional CARF domain of the CARF1 cluster and the CRISPR–Cas ancillary proteins Csx3 and Csx16 (Supplementary Table S1). Csx3 shows no detectable sequence similarity with CARF domains but structural comparisons have been interpreted to suggest that this protein contains a highly derived CARF (41). However, more extensive structural analysis indicates that Csx3 instead might be related to the STAS domain (42), a structure distinct from the Rossmann fold. Our structural comparisons performed in the course of this work support the potential relationship between Csx3 and STAS domains (Supplementary Figure S1). Csx3 is a Mn-dependent deadenylating exoribonuclease that binds and cleaves tetraribonucleotides (43) and has been recently characterized as a ring nuclease that rapidly degrades cOA4, whereas the deadenylation activity is a much weaker side reaction (44). Csx16 (PF09652) has not been experimentally characterized but appears to be distantly related to DUF1874 (AcrIII-1) family proteins, which are potent ring nucleases (Supplementary Figure S1) (21,23). Thus, CARF2 domains, unlike at least some of the CARF1 domains (see above), appear to be unable to cleave oligoA and require distinct ring nucleases to control the RNase activity of HEPN domains. These ring nucleases could be either fused to CARF2 domains or encoded in the same locus, or even elsewhere in the genome (see the detailed discussion of ring nucleases below). CARF2 is the most abundant ancillary protein family associated with the III-A, B and D CRISPR–Cas systems in approximately equal proportions (Table 1, Figure 2).
CARF3
The CARF3 clade consists of proteins containing a fusion of CARF and HTH domains, but typically, no effector domains. These proteins are often annotated with the legacy name Csa3 because many of them are encoded in type I-A loci (34). Also, some of these proteins are referred to as CasR, CRISPR-associated transcriptional regulator. The structure of CARF3 protein from S. solfataricus (Sso1445) was among the first to be solved, but the ligand of the CARF3 domain remains unknown (45). Csa3 activates the expression of Cas1 and Cas4a (Csa1) in Sulfolobus islandicus (46). Additionally, this protein has been shown to activate several DNA repair genes (47). However, the regulatory functions of Csa3 seem to be more complex than transcription activation alone. In S. solfataricus, Csa3b binds to two palindromic repeat sites in the promoter region of the subtype I-A CRISPR array and facilitates binding of the Cascade complex to the promoter region, resulting in repression of the pre-crRNA expression. Upon virus infection, loading of Cascade complexes onto crRNA-matching protospacers relieves the transcriptional repression resulting in activation of the crRNA production (48).
Although CARF3 is found in both archaea and bacteria, the archaeal members of this group form several distinct clades in the tree (Supplementary Data File 4) suggesting that horizontal transfer of the respective genes between the two domains occurred on more than one occasion. This is the only CARF subfamily with the strongest affinity to type I-A CRISPR–Cas systems albeit, occasionally, found also in I-B and I-D as well as III-A, B and D loci (Table 1, Figure 2). Most of the genomes encoding CARF3 proteins lack Cas10 that would supply cOA. Moreover, in most Halobacteria and Methanobacteriales, CARF3 is encoded outside the CRISPR–Cas loci, and several of these genomes lack any CRISPR–Cas. Thus, CARF3 domains might bind ligands other than cOA and could regulate functions of cas genes without effector activation as well as functions of non-cas genes (Supplementary Table S1).
CARF4
The CARF4 clade roughly corresponds to the PF09002 family. The structure of a CARF4 protein from Vibrio cholerae (VC1899; PDB: 1XMX) was the first CARF-containing protein structure to be solved, revealing a Rossmann-like fold (hence the acronym CARF), an HTH domain and a PD-D/ExK nuclease domain (9). Most of the proteins in this group have the same domain architecture, but a subgroup represented in many bacteria and a few mesophilic archaea (Supplementary Table S1) also contains a second CARF4 domain. Recently, one of these proteins containing two CARF4 domains (T. thermophilus TTHB155) has been experimentally characterized and named Can1, CRISPR ancillary nuclease 1 (49). Can1 is a monomer, with both CARF domains contributing to cOA4 binding. The PD-D/ExK nuclease, when activated upon cOA4 binding, nicks supercoiled DNA which apparently slows down viral replication by collapsing replication forks (49). CARF4 is only weakly associated with CRISPR–Cas, with ∼30% of the CARF4 proteins encoded in subtype III-A, B or D loci, often, along with other CARF proteins (Figure 2, Supplementary Table S1). CARF4 proteins are encoded in many bacterial genomes that lack CRISPR–Cas systems. Given the absence of Cas10, the ligand of these CARF4 domains remains obscure. In several bacteria, CARF4 proteins are encoded within a type VII secretion system loci that are predicted to be involved in DNA-transfer and carry ter genes implicated in phage restriction (50). This genomic context implies that the majority of the CARF4 proteins that are not associated with CRISPR–Cas function as regulators of other defense mechanisms or stand-alone defense systems.
CARF5
The CARF5 clade roughly corresponds to the PF09623 family. None of this clade members have been experimentally characterized. The CARF5 proteins are sometimes annotated as Csx14 or Cas_NE0113 and, typically, have domain organization similar to that of CARF3, namely, a fusion of CARF with a wHTH domain. In some of these proteins, a Fe-S cluster binding subdomain is inserted between the CARF and wHTH domains. Several CARF5 proteins contain an additional adenine deaminase domain. The adenine deaminase is likely to be involved in defense functions because it is also found in association with other defense systems (51), and notably, A-to-I editing is implicated in the regulation of innate immunity in animals (52). The mechanisms through which the deaminase activity contributes to defense remain to be elucidated. CARF5 proteins are predominantly found in bacteria, but are encoded in several archaeal genomes, probably as a result of horizontal transfer from bacteria. The CARF5 genes are strongly linked to type III systems, especially, subtype A and B. In a few proteobacteria, where CARF5 is present, but there is no type III systems, the CARF5 genes are located in putative defense islands along with restriction-modification systems and other defense genes, e.g. WP_038868443.1, from Vibrio jasicida or WP_096041929.1 Pseudoalteromonas agarivorans (Supplementary Table S2), again implicating CARF in the regulation of defense functions other than CRISPR–Cas.
CARF6
This clade consists, mostly, of bacterial proteins with two fused CARF domains (Table 1) none of which have been experimentally characterized. The sequences of both domains are highly divergent, but the C-terminal portion shares significant similarity with CARF1, CARF3 and CARF4. Another domain architecture found in proteins of this group is the fusion with a serine-threonine protein kinase and an AAA ATPase (e.g. WP_012790223.1). These proteins are never found in CRISPR–Cas loci (Figure 2) but are mostly located in defense islands (Supplementary Table S1, Supplementary Data File 4). In these defense islands, many CARF6 genes appear to be cargo in Tn7-like transposons, judging from the presence of Tn7 genes, such as tniQ, tnsA and tniB, in the vicinity. Some of these CARF6 genes are linked to a diverged AAA ATPase from the same family with the ATPase fused to other CARF6 proteins. The latter loci often include also cpdA, the gene for the phosphodiesterase responsible for the degradation of the ubiquitous signaling molecule, cAMP (53). Thus, it appears likely that cAMP is the ligand for at least some of the CARF6 proteins.
CARF7
The CARF7 clade roughly corresponds to the PF09651 family that is represented in both bacteria and archaea. These proteins are often annotated as Cas_APE2256. The structure has been solved for SSO1393 from S. solfataricus (3QYF) which contains CARF and wHTH. Proteins of this group have two typical domains organizations. One is the same as in SSO1393, whereas the other one is the fusion with RelE domain, a non-specific RNase, a well-characterized toxin in numerous toxin-antitoxin systems (54). In several thermophiles, mostly, bacteria of the order Thermotogales, there are additional fusions, CARF-HTH-HD and CYTH-CARF-HTH-HD, where HD is a predicted DNA endonuclease, and CYTH is a triphosphate tunnel metalloenzyme (TTM). The TTM family includes, in particular, class IV adenylyl cyclase CyaB that produces cAMP (55–57). In a few bacteria, these proteins contain another CARF domain of the CARF2 group (Supplementary Data File 4, Supplementary Table S1). Recently, it has been shown that SSO1393 is cOA4 specific ring nuclease in which the CARF domain is the active moiety (18). The CARF7 genes are strongly linked to type III systems, especially, to subtype III-B (Figure 2).
CARF8
CARF8 is a relatively small clade of experimentally uncharacterized, membrane-associated CARFs. Typically, these proteins contain 2 transmembrane segments, but proteins with different numbers of transmembrane segments have been identified as well. The only observed fusion is with the Csx16 domain (Supplementary Table S2). The CARF8 genes are present in both bacteria and archaea, and are strongly associated with type III-A, B and D systems, in roughly equal proportions (Figure 2).
CARF9
The CARF9 proteins possibly represent a divergent version of CARF2, with several distinct insertions in the HEPN domain that are implicated in the hexamerization of some of these proteins (40). Similarly to CARF2, in these proteins, the CARF domain is fused with a HEPN domain, a non-specific RNase. These proteins are mostly found in archaea and in some thermophilic bacteria (Supplementary Table S1). Structures have been solved for 4 proteins: TON_0898 from T. onnurineus (PDB:6O6Y), PF1127 from P. furiosus (PDB:4EOG), SSO1389 from S. solfataricus (PDB:2I71), and SisCsx1 from Sulfolobus islandicus (PDB: 6R9R). All these proteins activate the HEPN RNase upon cOA4 binding. Reports on ring nuclease activity are mixed: in TON_0898 CARF domain functions as a ring nuclease, in PF1127 (4EOG), the HEPN domain has been reported to possess this activity, whereas neither of the Sulfolobus proteins appears to be a ring nuclease (19,40,58). As with the CARF2 group, CARF9 proteins are strongly associated with type III-B, III-A and III-D systems (Figure 2).
RtcR
The RtcR protein received its name because of its association with the Rtc (RNA terminal phosphate cyclase) system and involvement in the regulation of the Rtc expression in a sigma54-dependent manner (25). Rtc is an RNA repair system that consists of two enzyme-encoding genes: the cyclase rtcA and the RNA ligase rtcB (26,27,29). The Rtc system has been studied in some detail, but the ligand of the CARF domain of RtcR remains unknown although the product of RtcA, the 2′,3′-cyclic phosphate at the modified RNA terminus, seems to be a strong candidate (8). Like most of the sigma 54-dependent enhancers, RtcR contains an AAA+ ATPase and HTH domains, in addition to the N-terminal sensory CARF domain (59). Although functionally distinct, the CARF domain of RtcR is confidently assigned to the largest cluster 1 of CARF domains (Figure 1). In addition to the RtcR proper, the RtcR clade includes at least two additional, distinct branches which we provisionally denoted PspF1 and PspF2, after their closest homolog PspF (phage shock protein F) that consists of an AAA+ ATPase and an HTH domain but lacks the CARF domain (Supplementary Figure S1, Supplementary Data File 3). Unlike RtcR, PspF1 and PspF2 are not associated with the Rtc system, and instead, are typically encoded in defense islands, most often, next to R-M genes (Supplementary Figure S2, Supplementary Data File 4). PspF1 and PspF2 proteins have not been characterized experimentally, but PspF is known to be involved in the activation of the phage shock membrane-associated complex in response to phage infection and other stress factors (60,61). A similar function can be inferred for PspF1–2, with the addition of sensing nucleotide signals via the CARF domain. None of the RtcR clade genes are associated with CRISPR–Cas systems. Despite the diversity of the proteins in the RtcR clade, it is so far represented only in Proteobacteria and Bacteroidetes. The variation of the Rtc system has been recently addressed in detail (29), so we skip further discussion of this family here.
EMERGING DIVERSITY OF THE CARF DOMAINS: MINOR CLADES
We discussed above the 10 most abundant clades of CARF proteins. In addition to these, there are many smaller groups of CARFs, often lineage-specific and/or with unique domain organization. Many of such groups are found only in archaea (Figure 1, Table 1, Supplementary Table S1). Among these, three distinct clades, namely, Thermococcales-specific CARF_m1, and CARF_m3 and CARF_m12, both limited to Crenarchaeota, include CARF domains fused with or encoded next to a PIN domain nuclease. These genes are not stably associated with type III CRISPR-cas loci (Supplementary Table S1).
The Pyrobaculum-specific CARF_m4 and Methanosarcina-specific CARF_m7 are strongly linked to type III-A or type III-B systems, whereas CARF_m5 specific to the Asgard archaea and CARF_m6 specific to Haloferacales are not CRISPR-associated (Table 1). The CARF_m8 proteins that are found specifically in Bathyarchaeota have unusual domain architecture, with fusions to the lipoprotein release LolE-like protein and, in some case, additionally, to beta-galactosidase, suggesting potential involvement in lipoprotein turnover and/or glycosylation (Supplementary Table S1). Finally, the archaea-specific clade CARF_m13 includes two ring nucleases that have been experimentally characterized in S. solfataricus (18). These genes are rarely encoded in CRISPR–cas loci but are typically present in genomes along with type III systems encoded elsewhere (Supplementary Table S1).
The few remaining bacteria-specific groups seem to link CARF to membrane processes. The CARF_m11 proteins are strongly associated with type III-D systems. They typically contain a C-terminal divergent HEPN domain, but in a few cases, the CARF domain is instead fused with the membrane protein CorA, a recently identified ancillary CRISPR–Cas component (23) (Supplementary Table S1). In several Planctomycetales species that encode CARF_m2 proteins, the CARF domain is fused to a Lon family protease, another ancillary protein (23). Another clade, CARF_m9, is present in diverse bacteria and consists of proteins with a CorA-like transmembrane domain. In several cyanobacteria, they also contain the mCpol domain, or alternatively, mCpol is encoded next to these genes in a putative two-gene operon (Supplementary Table S1). These domain configurations strongly suggest that mCpol synthesizes the ligand recognized by the CARF_m9 domains.
Several even smaller groups of CARFs (all distinct branches within Cluster 1 in the CARF tree) possess other unique features. Among these, a CARF-mCpol-HEPN fusion found in several cyanobacteria is of particular interest because it represents a putative complete signal transduction pathway analogous to the cOA pathway in type III CRISPR–Cas, in which mCpol would synthesize a ligand, whereas CARF would bind the produced ligand and activate the HEPN RNase. This configuration could be ancestral to the type III CRISPR–Cas effector modules (1). Other catalytic domains fused with CARFs of these minor groups include 3′,5′-cyclic AMP phosphodiesterase CpdA, 2OG-Fe(II) oxygenase family domain (62) that possibly functions in oxidative dealkylation during RNA and DNA repair (63,64) and several other domains (Supplementary Table S1).
THE SAVED DOMAIN SUPERFAMILY
The SAVED domain was originally described in association with cyclic 2′-5′ GMP-AMP synthase and 2′-5′ oligoA synthetase in bacteria and some archaea, and was hypothesized to function as a sensor for 2′-5′ GMP-AMP (cGAMP) and, possibly, also, 2′-5′OA ((14) (Supplementary Table S2). A weaker link of SAVED domains with CRISPR–Cas systems has been noticed before, as well as a detailed description of SAVED domain fusions with various effector domains available at the time of the analysis was given (14). The SAVED domain was subsequently rediscovered in a larger set of complete genomes, but this time, starting from the analysis of the context of type III CRISPR-cas loci (23). A limited sequence similarity between the SAVED and CARF domains has been detected, and it has been proposed that SAVED is a highly divergent homolog of CARF (23). Indeed, the region of similarity between SAVED and CARF sequences detected by HHpred is located within the most conserved region of the CARF domain, and the two families have similar (predicted) secondary structures, which is suggestive of homology (Supplementary Figure S1). Furthermore, both families have clearly analogous functions, and both can be associated with type III CRISPR–Cas systems. Analysis of the recently solved crystal structure of the antivirus proteins Cap4 from Enterobacter cloacae that consists of a SAVED and a restriction endonuclease domains confirms the structural similarity and, by inference, common origin of the SAVED and CARF domains (24). More specifically, the SAVED domain of Cap4 consists of two CARF-like domains (that is, corresponds to a CARF domain dimer) and, as predicted, has been shown to specifically bind 2'-5′OA (24).
Here we provide information on all SAVED proteins, but mostly, focus on the SAVED families that are found in association with CRISPR–Cas systems (Supplementary Table S2). Phylogenetic analysis divides the SAVED domains into 7 clades (Figure 1; Supplementary Table S2). Only three of these, clades 4, 5 and 6, are often found in type III CRISPR–Cas loci (Figure 2). Members of the SAVED4 and SAVED6 groups, with the strongest link to type III systems, typically, are membrane-associated proteins. In addition to transmembrane segments, many SAVED4 proteins are fused to a Lon-like protease domain (23). Both SAVED4 and SAVED6 clades have been described in detail previously (denoted derived CARF domains at the time) (23). The SAVED5 clade consists of predicted intracellular proteins in which SAVED is, typically, fused to a TIR domain, and in some case, also, to a JAB domain. TIR domains are components of diverse immune systems, and some are enzymes that cleave NAD+ and, potentially, other signaling molecules (14,65,66). However, specific experimental evidence on the roles of TIR domains in immunity remains scarce. The JAB domain is a metal-dependent de-ubiquitinating isopeptidase of the JAB1/MPN/MOV34 family (67).
STRUCTURAL INSIGHTS INTO THE FUNCTIONS AND EVOLUTION OF CARF DOMAIN PROTEINS
The core of the CARF domain is a 6-stranded Rossmann-like fold, with the core strand-5 and strand-6 forming a β-hairpin (Figure 3). As indicated above, sequence conservation is mostly associated with strand-1 and strand-4: strand-1 often ends with a hydroxyl side chain residue (S/T), whereas the characteristic motif with the [DN]x[ST]xxx[RK] signature is located immediately after strand 4 (8).
Figure 3.
Structures of the CARF domain containing proteins. (A) Schematic representation of the conserved core of the CARF fold. Motifs I and II (corresponding to β1-α1 and β4-α4 junctions, respectively) are involved in cyclic oligoadenylate binding/cleavage activities. (B) Superimposed structures of 11 CARF domains colored according to chain progression from N-terminus (blue) to C-terminus (red). Non-conserved loops/insertions were removed for clarity. (C) Selected structures of CARF proteins with different domain architectures. Domain homodimers (different chains) or heterodimers (single chain as in TtCan1) are represented by different shades of the same color. All CARF domain dimers have a cleft (blue mesh) in the corresponding structural regions. This cleft (pocket) is where a cyclic oligoadenylate (shown in pink and labeled) binds. CARF domains, orange; domains in topologically equivalent positions following CARF (Csx1 connector domain, Csm6 6H domain and wHTH domain), light blue; toxin domains (HEPN, wHTH-HEPN and PD-D/ExK), green.
Functional forms of CARF domains
So far, CARF domains in all solved structures have been found to form dimers, most often, homodimers (Supplementary Table S4). Some proteins form higher order assemblies as exemplified by the hexameric structure of SisCsx1 (CARF9 clade), which is a trimer of dimers. The trimer formation is mediated by a unique insertion region in the HEPN domain of SisCsx1 (40). This interaction appears to be biologically relevant because it contributes to the cOA4-dependent cooperativity of the HEPN ribonuclease (40). In some proteins, CARF dimers are part of the same polypeptide chain and form a pseudosymmetric structure as exemplified by Can1, of the CARF4 clade (49). Can1 contains a duplication of the [CARF]-[PD-D/ExK] module, with one of the two nuclease domains inactivated, and is most similar to the uncharacterized V. cholerae VC1899 protein (PDB: 1xmx), which has the CARF-wHTH-PD-D/ExK domain architecture; the CARF domains in these proteins form closely similar dimeric structures (49).
All CARF dimers contain a preformed cleft in equivalent structural regions (Figure 3), and in all known cases, the cognate ligands bind within these clefts. The size and the depth of the clefts vary, suggesting substantial variability of the ligand-binding specificity and affinity among CARF domains.
Structural similarity among the CARF domains
Although sharing the same overall fold, CARF domain structures vary considerably, in particular, due to additional structural elements that are often inserted into the common core fold. The currently solved CARF structures can be divided into two major groups by structural similarity, Csx1 and the rest (Supplementary Table S5). This subdivision is consistent with the sequence similarity dendrogram of the CARF domains because Csx1 of P. furiosus (PDB: 4eog) belongs to the CARF cluster 2 (CARF9), whereas all other CARFs belong to the largest CARF cluster 1 (Figure 1, Supplementary Data File 2). The Csx1/CARF9 clade includes four CARF proteins with solved structures, P. furiosus Csx1 (PDB: 4eog) being the typical representative. The CARF cluster 1 includes 6 structurally characterized proteins with various architectures that are best approximated by the basic CARF structure of VC1899 (PDB: 1xmx). Notably, similarity of the cognate ligands is not necessarily a good indicator of the structural similarity between the respective CARF domains. For example, each of the two CARF domains of Can1, which binds cOA4, is more structurally similar to the CARF domain of Enteroccocus italicus Csm6 (EiCsm6), which binds cA6, than to those in any of the cOA4-binding Csx1 proteins (Supplementary Table S5).
Ligand binding and signal transduction
So far, most of the functionally characterized CARF proteins have been found to bind either cyclic or linear An (n = 4 or n = 6) ligands, and upon ligand binding, allosterically activate the fused effector domains. Once several structures with the cognate ligands bound were solved, it became clear that conserved motifs located at the ends of β-strands 1 and 4 are directly involved in ligand binding (19,39,40,49). Structures of two proteins from the CARF9 clade (ToCsx1/Csm6 and SisCsx1) were solved both with and without the bound cyclic oligoadenylate (19,40). Unexpectedly, these structures revealed that the differences between the bound and unbound states are local and small, indicating that the allosteric regulation of HEPN domains in these proteins does not involve major structural rearrangements. The same behavior might be expected for other CARF proteins that feature a combination of CARF and HEPN domains. How the signal is transferred from the ligand-binding pocket in CARF domains to the HEPN active site, remains unclear because the two sites are ∼60–70 Å apart from each other (Figure 3). By contrast, Can1 that contains two CARF domains in a single chain undergoes a major structural rearrangement upon binding the cognate cOA4 ligand (49). Notably, unlike the HEPN domains in Csx1/Csm6 that are positioned away from the CARF domains, the nuclease and nuclease-like domains of Can1 interact with the ligand-binding site of the CARF domain. A closely similar arrangement of the PD-D/ExK modules was observed in a related protein (VC1889; PDB: 1xmx), suggesting that it also undergoes substantial conformation changes upon ligand binding to the CARF cleft.
Insights from the sequences and structures of CARF ring nucleases
The CARF ring nucleases identified so far are metal-independent ribonucleases. The degradation of a cyclic oligoadenylate proceeds by nucleophilic attack of the ribose 2′-OH group onto the scissile phosphate bond, producing a 2′,3′-cyclic phosphate and a 5′-OH (18). It has been proposed that CARF domains primarily contribute to the hydrolysis of cOAs by sterically positioning the ribose 2′-OH group for inline nucleophilic attack. Indeed, from the enzymology standpoint, CARF ring nucleases are extremely inefficient enzymes, that is, they degrade cOAs very slowly (Supplementary Table S6). Detailed enzyme kinetics studies have been performed for only some of the identified ring nucleases, but clearly, even the most active CARF ring nucleases, such as Sso2081 (Crn1), are orders of magnitude less efficient than the more recently discovered viral anti-CRISPR ring nucleases with a distinct fold unrelated to CARF (21). Conceivably, the low enzymatic efficiency of the CARF domain ring nucleases stems from the fact that, similarly to other Rossmann fold domains, the primary function of CARF is (oligo)nucleotide binding, whereas the nuclease activity is a secondary adaptation that seems to have evolved independently in different lineages of CARFs.
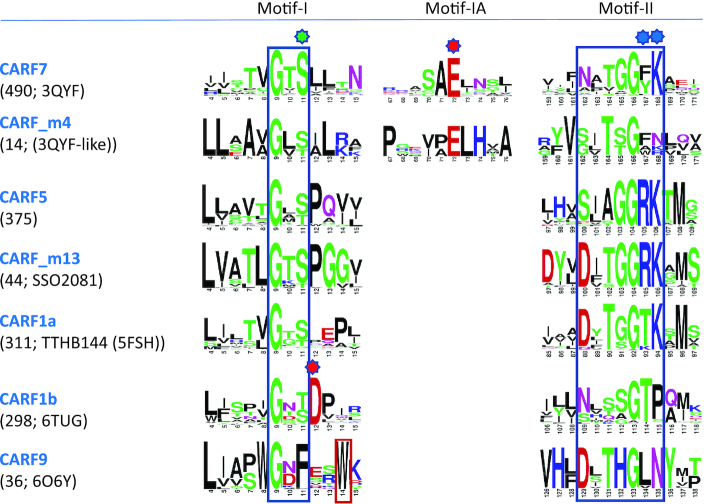
Consistent with the catalytic inefficiency of the CARF ring nucleases, their active sites remain poorly defined. Initially, two motifs adjacent to β-strands 1 and 4 of the core fold (motif-I and II in Figure 3), predicted as ligand-binding regions in the original CARF domain analysis (8), were implicated in the ring nuclease activity (18). However, a subsequent study found that a conserved Lys residue in motif-II is critical for ligand binding (20) (Figure 4). These findings suggest that motif-II might be primarily involved in cOA binding, whereas motif-I could be largely responsible for the catalytic activity. Despite uncertainties regarding the exact roles of the two motifs, biochemical experiments complemented with a growing number of CARF structures co-crystallized with cognate ligands provide for some inferences.
Figure 4.
Sequence motifs of ring nucleases. Conserved motifs in different groups of CARF ring nucleases are represented as sequence logos. For each CARF group, a number of sequences in the group and known representatives are indicated in parentheses. Motifs I and II are framed. Positions of residues in motif-I and motif-II known to be important for binding/catalysis of cOA are indicated by green and blue stars, respectively. Additional conserved Glu (motif-IA) in CARF7 and CARF_m4 groups as well as structurally analogous conserved Asp (motif-I) in CARF1b predicted to be important for ring nuclease activity are indicated with a red star. CARF9 is represented by close homologs of ToCsx1 protein, in which Trp14 was identified as a catalytic residue (enclosed in red frame).
There appears to be at least two distinct signatures of motif-I that is associated with the ring nuclease activity. The first signature is characteristic of the Csx1 group, in which only one representative (ToCsx1/Csm6) has been shown to harbor a ring nuclease activity in its CARF domain (19). In this case, a single conserved tryptophan (Trp 14) that forms a hydrogen bond with the scissile phosphate was implicated as an active site residue (Figure 4). The key role of this Trp residue seems to be supported by the observation that two other Csx1 proteins, SsoCsx1 (2i71) and SisCsx1 (6r9r), that have Tyr in the corresponding positions, were unable to degrade cOA4 (18,40). However, a recent study of PfuCsx1 (4eog) has shown that, although there is a conserved Trp (Trp 16) in the corresponding position, the CARF domain of PfuCsx1 does not function as a ring nuclease (58). Thus, in CARF9 proteins, the presence of conserved Trp in motif-I seems to be a necessary but not a sufficient requirement for CARF domain to possess the ring nuclease activity. The second motif-I signature can be defined as GX(S/T), and the residue in the third position (S/T) has been shown to participate in catalysis (18,20,39). This signature motif corresponds to a tight turn interacting with the sugar-phosphate backbone of cOA. Some ring nucleases (Sso1393, EiCsm6) contain an insert immediately next to the GX(S/T) motif. In such cases, there is an additional, highly conserved acidic residue (Asp12 in EiCsm6 and Glu72 in Sso1393), which interacts with Gly of the GX(S/T) (Figure 4). Although the effects of mutating either of these two conserved acidic residues have not been studied, our structural analysis suggests that they might play an important role in catalysis by keeping motif-I positioned adjacent to the scissile phosphate. The GX(S/T) signature in motif-I is present in a number of experimentally uncharacterized clades, such as CARF5, CARF_m1, CARF_m4, suggesting that they could also be ring nucleases (Figure 4, Supplementary Figure S2, Supplementary Table S1). The variation of the catalytic residues in the active sites of CARF domains implies that there are multiple modes of positioning and activation of 2′-OH group of the ribose for nucleophilic attack that results in the phosphodiester bond cleavage during cyclic oligoadenylate (cOA4 and cOA6) degradation.
THE ROLES OF DIFFERENT CARFS AND ASSOCIATED PROTEINS IN THE cOA SIGNALING PATHWAY
The core of the cOA signaling system consists of three key components: (i) signal molecule synthetase, (ii) sensor and (iii) effector. In type III CRISPR–Cas systems, the first role is performed by the cyclase/polymerase domain of Cas10 that is activated by RNA target binding to the effector complex and synthesizes a signaling molecule, cOAn (n = 4 or n = 6). The sensor domain, CARF, binds this molecule and undergoes a conformational change to activate the effector domain, which is most often a non-specific RNase, in particular, HEPN, but in some cases, a more specific effector, such as a PD-D/ExK DNA endonuclease cleaving supercoiled plasmid DNA. This dual response, combining the highly specific crRNA-guided RNA and DNA cleavage with a less specific, abortive infection-like mechanism, is the key feature of most type III systems containing an active cyclase/polymerase domain. In more general terms, this is a mechanism of coupling active immunity with dormancy induction or programmed cell death (68). Recently, the fourth important component of the cOA signaling cascade was discovered, the ring nucleases that cleave cOA, tuning down the response and thus avoiding excessive cell toxicity. Surprisingly, the first ring nucleases to be discovered were CARF proteins. Moreover, two CARF ring nucleases (SSO2081, SSO1393) were identified in one genome, S. solfataricus (18). None of these CARF domains is fused to an enzymatic effector domain, and so, they appear to be dedicated ring nucleases. In other genomes, however, no CARF proteins without a fused effector domain were identified, raising the question of the identity of the ring nuclease (if any) in these organisms. Subsequently, it has been shown that, at least, in some of these proteins, the sensor CARF domain itself doubles as a ring nuclease (20). Independently, it has been shown that proteins of the DUF1874 family, unrelated to CARF, are even more potent ring nucleases that are encoded, mostly, by viruses and function as anti-CRISPR proteins (21).
As more CARF domain containing proteins were tested for ring nuclease activity, it became clear that, in different CARF families, distinct sets of amino acids are important for the catalytic mechanism (Figure 4), which makes it difficult to predict this activity from amino acid conservation only. The original function of CARF might have been (oligo)nucleotide-binding, with the catalytic activity evolving subsequently, on several independent occasions. Presently, however, it is difficult to rule out that the dual function of the CARF domain is ancestral to type III CRISPR–Cas systems but was lost in some variants. Notably, a similar phenomenon was observed for the Cas ribonucleases involved in pre-crRNA processing in Class 1 CRISPR–Cas systems (Cas6 and, in some cases, other members of the RAMP protein superfamily). Generally, RAMPs are RNA-binding proteins, but some have evolved the capacity for catalysis of RNA cleavage, likely, on several independent occasions (69).
Even more puzzling are the findings that, among the proteins of the CARF9 clade, some are capable of cleaving cOA4 (TON_0898), whereas others are not (SisCsx1) (20,40). In some of the CARF9 proteins with a non-enzymatic CARF domain (PF1127), it is the HEPN domain that appears to be responsible for cOA4 cleavage (58). Notably, S. islandicus encodes several other CARF proteins, in addition to SisCsx1, including a representative of the CARF7 clade that includes experimentally validated ring nucleases, whereas in P. furiosus and T. onnurineus, no members of the known ring nuclease families were identified. Furthermore, a CARF1 clade protein from E. italicus, the only type III-associated CARF protein encoded in the genome, has been shown to possess ring nuclease activity (39). Thus, it has been hypothesized that, if there is no dedicated ring nuclease encoded in a genome, the CARF domain protein associated with type III locus performs this role (39). Moreover, as mentioned above, kinetic modelling suggests a crucial role of the ring nucleases for CRISPR–Cas systems that induce dormancy or cell death via the activity of cOA-activated toxins (22).
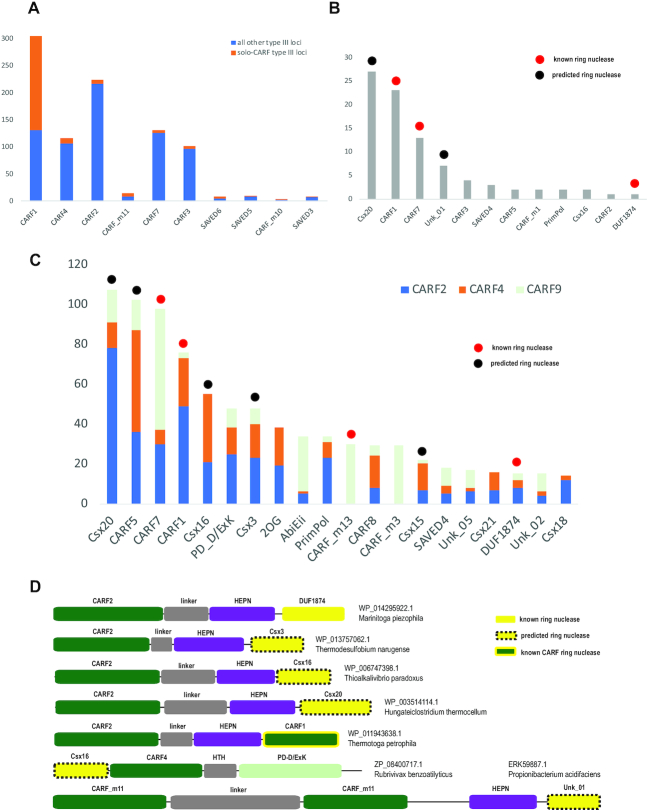
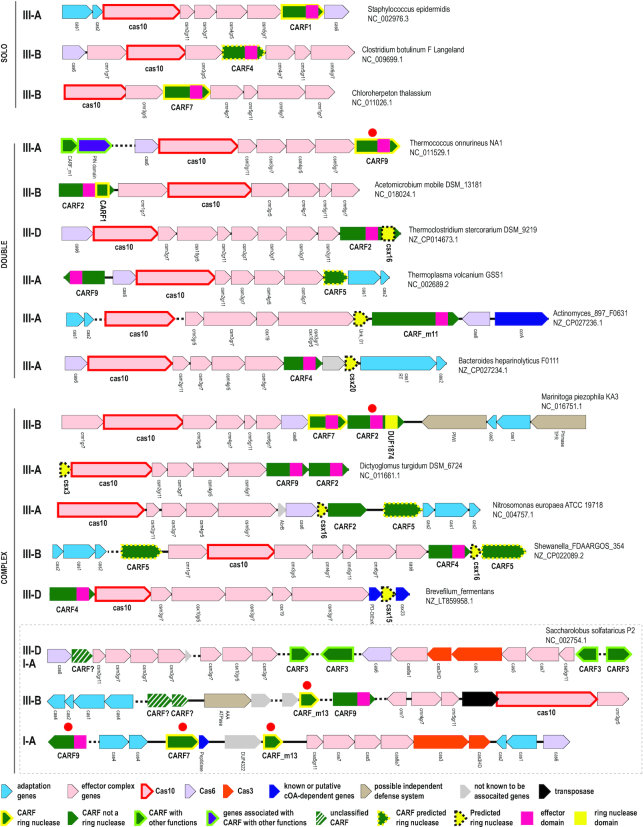
We then sought to identify type III loci containing a single CARF domain protein (hereafter, solo-CARF loci) in complete genomes, making sure that no other CARF proteins or type III associated genes that could be yet uncharacterized ring nucleases are found in the respective genomes. We identified 793 type III loci containing, altogether, 1271 genes encoding CARF or SAVED domain proteins. Only 213 of these (12%) of the CARF or SAVED genes are located in solo-CARF loci, suggesting that the majority of the type III systems containing an active cOA synthetase require a helper ring nuclease, assuming that this component of the cOA pathway is essential (Figure 5A). Most of the solo-CARF loci (174 loci, 82%) contain genes from the CARF1 clade, which is compatible with the experimental demonstrations of the intrinsic ring nuclease activity of CARF1 domains (20,39). CARF2, CARF4 and CARF9, the three largest clades associated with type III systems and fused to effector, (potentially) toxic domains, are typically found along with other CARFs or type III associated proteins, suggesting that the majority of these require a helper ring nuclease to tune down the activity of the effector domain. This is also compatible with experimental results showing that CARF domains of these groups are unable to cleave cOA (40,49,58,70).
Figure 5.
Analysis of the components of the cOA signaling pathway (A) Representation of different CARF groups in type III CRISPR-cas loci. Vertical axis, number of type III loci that encode the where respective CARF. Orange bars show the loci with a single CARF domain containing gene with additional requirement that no previously reported type III associated non-CARF genes (1) or CARF proteins from other groups are present in the respective genome (solo-CARF loci). Blue bars show the remaining loci. (B) Co-occurrence of different families of CARFs and type III associated proteins in the genome encoding only one such protein, in addition to CARF2, CARF4 or CARF9 (‘double’ CARF loci). The vertical axis is the number of ‘double’ CARF loci. (C) Co-occurrence of different families of CARFs and type III associated genes with three major groups of CARFs predicted to lack ring nuclease activity (CARF2, CARF4 and CARF9). The vertical axis is the number of CARF loci. (D) Domain organizations of selected proteins containing known or predicted ring nuclease domains. The domains are shown approximately to scale. The domain family name is indicated above each bar.
To predict helper ring nucleases among the protein families associated with type III CRISPR–Cas systems and other CARF domain groups, we examined the genomes with type III loci containing CARF2, CARF4 and CARF9 genes (Figure 5B and C, Supplementary Table S7). First, we analyzed genomes in which, in addition to one or more CARF proteins from these 3 groups, a single CARF protein from another group or a single type III-associated protein was identified, suggesting that this additional protein could be the helper ring nuclease (Figure 5B). Among 90 genomes that meet this criterion, uncharacterized type III CRISPR-associated csx20 genes are most frequent, followed by the genes encoding CARF1 and CARF7 group proteins, for which ring nuclease activity has been demonstrated, another, uncharacterized family (Unk-01), and a few other, rare protein families.
Next, we calculated the co-occurrence of CARF2, CARF4 and CARF9 with CARFs from other clades and type III-associated proteins for the remaining 886 CARF genes that occur in more complex genomic contexts (Figure 5C). Here, again, the top 5 families co-occurring with the CRISPR-associated CARF2, CARF4 and CARF9 include Csx20, CARF1, CARF7, and two additional uncharacterized families, CARF5 and Csx16. All these families appear to be viable candidates for the ring nuclease role. Some of the remaining families down the list also might be ring nucleases, considering that two of them are CARF_m13 and DUF1874, both experimentally characterized ring nucleases. Furthermore, a CARF2 family protein from Marinitoga piezophila fused to DUF1874 has been recently experimentally characterized and it has been shown that the DUF1874 domain is responsible for the ring nuclease activity (70). Thus, this protein combines three components of the cOA pathway, sensor, effector and an off-switch, within the same polypeptide (Figure 5D). Notably, we also identified CARF domain fusions with Csx3, Csx20, Csx16 and Unk_01 domains (Figure 5D), supporting these as solid candidates for ring nuclease activity. Indeed, the ring nuclease activity of Csx3 has been demonstrated (70). Csx20, Csx16 and Unk_01 are all small domains containing combinations of conserved polar residues, such as histidine, arginine, aspartate or glutamate, which is compatible with an enzymatic, and in particular, nuclease activity (Supplementary Figure S1). Furthermore, limited sequence similarity was detected between Csx20 and Csx16, and DUF1874 and Csx16, respectively, suggesting that all these proteins might belong to the same, highly diverged family of enzymes and strengthening the argument for their ring nuclease activity (Supplementary Figure S1). Among other CARF-associated protein families, Csx15 shows the same features in the alignment and shares limited sequence similarity to both SAVED and CARF domains (HHpred probability 58 and 39, respectively), suggesting that it could be a distinct variety of the same type of Rossmann fold with a ring nuclease activity (Supplementary Figure S1).
Among the uncharacterized CARF families that co-occur with CARF2, CARF4 and CARF9, CARF5 is the strongest candidate for ring nuclease activity. Examination of the CARF5 clade alignment revealed conserved threonine or serine and lysine residues as in the characterized ring nucleases of the CARF7 clade (Figure 4). Furthermore, in the dendrogram based on complete protein sequences, CARF5 groups with CARF_m13, an experimentally characterized ring nuclease (Supplementary Data File 3).
Several protein families that are often encoded in the same loci with CARF2, CARF4 and CARF9 are unlikely to possess ring nuclease activity. These include AbiEii-like AAA ATPase and predicted aspartic protease PEPT_D, especially, considering that none of them are detected in the ‘double’ loci (those that encompass CARF2, CARF4 or CARF9 along with a single CARF from another clade or a single non-CARF ancillary gene), and neither were they found as fusions with these CARF domains (Figure 5B). These and several other associated families could be components of additional defense systems that might be activated by cOA or by linear OA. One of such proteins that has been recently experimentally characterized is a PD-D/ExK family nuclease, NucC, that is associated with both 2′5′-OA synthetases and many type III systems (subtypes A,B and D), is activated by cOA3 and functions via an abortive infection mechanism (71).
Based on the above observations and inferences, the roles for most CRISPR-associated proteins found in type III loci can be tentatively assigned as follows:
In the solo-CARF loci (i.e. in the absence of any known or predicted ring nucleases encoded in the respective genome), either the CARF domain itself or the associated effector domain likely functions as the ring nuclease that tunes down the non-specific response to infection by cleaving cOA. The caveat is that we cannot rule out the presence of genes coding for novel ring nucleases encoded outside the CRISPR-cas loci in the respective genomes.
In most cases, when a non-enzymatic CARF domain is fused to an effector domain that lacks the ring nuclease activity, the function of controlling the cOA-dependent response is relegated to a helper ring nuclease that can reside either in an additional CARF domain protein or in an unrelated protein from one of the several CRISPR-associated families discussed above that are often encoded within type III loci.
Most of the remaining proteins in the type III loci are probably cOA-dependent or cOA-independent innate immunity modules that have been co-opted by type III systems to facilitate and enhance the immune response. The NucC family can be considered a typical example of a cOA-dependent abortive infection mechanism whereas the CRISPR-associated Argonaute nucleases present a case of cOA-independent mechanism (72).
Finally, cOA-dependent and cOA-independent transcriptional regulators are the least studied components of the type III systems. In type III CRISPR–Cas systems, numerous CARFs that lack effector domains are fused to HTH domains and thus are predicted to function as cOA-dependent transcription regulators. Other CRISPR-associated HTH-containing proteins are likely to regulate expression of type III systems genes in a cOA-independent manner.
Figure 6 shows how these considerations could be applied for selected type III loci, and a scheme of the general organization of cOA signaling is shown in the Figure 7.
Figure 6.
Functional organization of selected CARF domain-encoding loci. For each locus, species name and genome accession number are indicated. Genes are shown by arrows roughly to scale. Dashed line between arrows indicated that the loci encoded far from each other. Arrows are color-coded according to the scheme below. The gene names largely follow the nomenclature from (1), but the RAMP proteins of groups 5 and 7 and small subunits are denoted gr5, gr7 and gr11, respectively. The CRISPR–Cas system subtype is indicated on the left. Experimentally characterized ring nucleases are denoted by small red circle above the respective arrows.
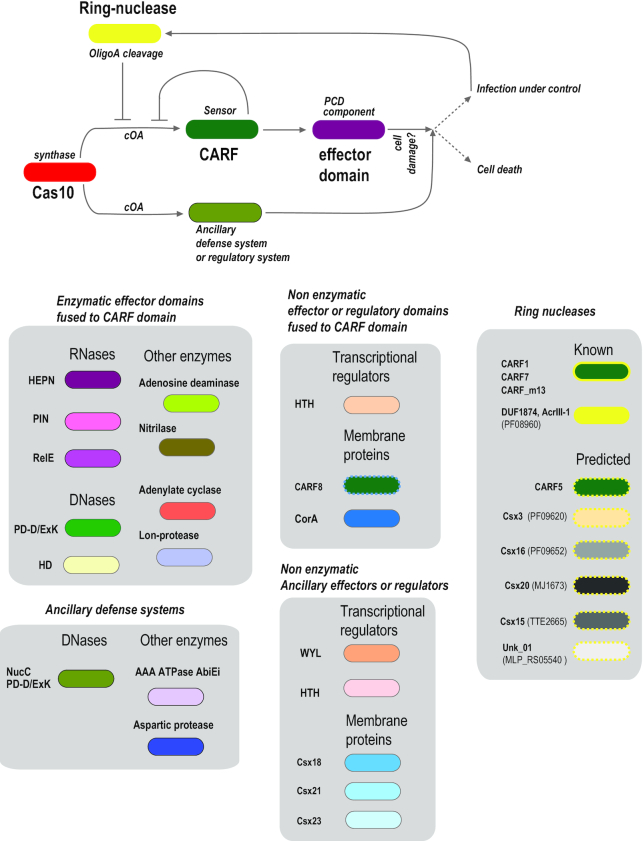
Figure 7.
Updated scheme of the cOA signaling pathway. The general scheme of cOA signaling pathway is shown on top, followed by experimentally characterized and predicted components of the pathway classified into five functional categories within gray shapes. Distinct proteins families are shown by oval shapes. CARF families are shown by dark green, and other proteins are shown by oval shapes of different colors, including shades of blue for uncharacterized membrane proteins and shades of gray for protein families without similarity to any characterized proteins. Experimentally characterized ring nucleases are shown by thick yellow outline, and predicted ring nucleases are shown by thick dashed yellow outline in panel A. Components are denoted by general family name if known, CRISPR–Cas ancillary protein names are indicated according to the current nomenclature of cas genes (1). Abbreviations: cOA, cyclic oligoadenylates; CARF, CRISPR-associated Rossmann Fold; WYL, predicted ligand-binding domain associated with many CRISPR–Cas systems (named after the respective amino acids that are partly conserved in the family); HEPN, PIN, RelE, ribonucleases of the respective families; HTH, helix-turn-helix DNA-binding domain; HD, PD-D/ExK, nuclease (or phosphatases) of the respective superfamilies; CorA, divalent cation channel or its homolog.
CONCLUDING REMARKS
Recent comparative-genomic and functional studies have shown that bacteria and archaea possess a broad spectrum of cyclic oligonucleotide-based anti-phage signaling system (CBASS) (73,74). Most if not all of the CBASS function via an Abi-type mechanism, whereby virus infection induces the synthesis of a cyclic oligonucleotide signaling molecule that is recognized by the sensor component and, through a conformation change in the sensor, activates the effector component, most often, a nuclease. The effector nuclease cleaves host RNA (or, in some cases, DNA) indiscriminately, to induce dormancy or PCD.
Type III CRISPR–Cas systems contain a built-in CBASS machinery that couples, via the cOA pathway, the CRISPR-mediated adaptive immune response with dormancy or PCD induction. The cOA subsystem of type III CRISPR–Cas consists of four key functional components: (i) polymerase-cyclase Cas10 (that also performs an essential structural role as the large subunit of the CRISPR–Cas effector complex) producing cOA in response to the crRNA binding to the target RNA, (ii–iii) two-domain protein that consists of a sensor CARF or SAVED domain and an effector, typically, a nuclease, (iv) a ring nuclease that cleaves cOA and thus dampens the non-specific immune reaction to prevent cell death (Figure 7).
The sensor component of the cOA pathway is a dedicated Rossmann fold domain, CARF, or in a minority of cases, the SAVED domain. The SAVED-containing effectors have been shown not only to discriminate between 2′-5′- and 3′-5′-linked cyclic oligonucleotides, apparently, the two main products synthetized by SMODs enzymes, but can also specifically bind hundreds of different nucleotide second messenger species (24). In addition to their function as sensors, some of the CARF domains also possess the ring nuclease activity, either as a secondary function of a sensor domain, or as a dedicated nuclease. There are thousands of CARF domains encoded in bacterial and archaeal genomes, and the majority are components of type III CRISPR–Cas. Of these, only a minority appear to combine the signal transduction role with the ring nuclease function. Some notable families of CARF domains are not CRISPR-associated, in particular, the mCpol-CARF-HEPN module that is predicted to function as a CBASS and is the likely evolutionary ancestor of the type III CRISPR–Cass effector modules (1). The CARF domains of RtcR protein, for which the ligand identity remains unknown, conversely, seem to be derivatives of CRISPR-associated CARFs that function as regulators of RNA repair systems.
Here we present a comprehensive census of CARF and SAVED domains in bacterial and archaeal genomes as well as their classification based on sequence and structure comparisons. The CARF domains are classified into 10 major families and many additional, smaller groups that differ in their structural features, association with distinct effector domains, and the presence or absence of the ring nuclease activity. Notably, most of the minor CARF groups were identified in archaea, emphasizing the importance of the cOA-dependent mechanisms in archaeal antivirus defense. The cOA pathway is likely to have evolved in archaea, along with the type III CRISPR–Cas systems themselves, as previously proposed (23,75).
CARF and SAVED domains as well as ring nucleases are central to an emerging major theme in microbial biology, the coupling between immunity and dormancy induction or programmed cell death (76,77). In addition to the comprehensive classification of the CARF and SAVED domains, we employ comparative genome analysis to predict their functions, and in particular, to differentiate those CARFs that possess both the sensor and the ring nuclease activity from non-enzymatic, sensor-only ones. We then predict ‘helper’ ring nucleases associated with the sensor-only CARFs, whether or not the nucleases themselves contain a CARF domain. As a result, we predict several families of previously unknown ring nucleases with different protein folds. In addition to the classification itself, we provide a collection of sequence profiles for multiple families of CARF and SAVED domains as well as effector domains and the predicted ring nucleases that will substantially facilitate identification and analysis of defense and signaling proteins encoded in microbial genomes.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Malcolm F. White for critical reading of the manuscript and helpful comments.
Author contributions: K.S.M. and E.V.K. initiated the project; K.S.M., A.B.G., Y.I.W., A.T. and Č.V. performed data analysis; all authors analyzed and interpreted the results; K.S.M., Č.V. and E.V.K. wrote the manuscript that was edited and approved by all authors.
Contributor Information
Kira S Makarova, National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD 20894, USA.
Albertas Timinskas, Institute of Biotechnology, Life Sciences Center, Vilnius University, Vilnius, Lithuania.
Yuri I Wolf, National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD 20894, USA.
Ayal B Gussow, National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD 20894, USA.
Virginijus Siksnys, Institute of Biotechnology, Life Sciences Center, Vilnius University, Vilnius, Lithuania.
Česlovas Venclovas, Institute of Biotechnology, Life Sciences Center, Vilnius University, Vilnius, Lithuania.
Eugene V Koonin, National Center for Biotechnology Information, National Library of Medicine, Bethesda, MD 20894, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
K.S.M., Y.I.W., A.B.G. and E.V.K. are supported by the funds of the Intramural Research Program of the National Library of Medicine, NIH (US Department of Health and Human Services); A.T. and Č.V. were supported by European Social Fund [09.3.3-LMT-K-712-01-0080] under grant agreement with the Research Council of Lithuania (LMTLT). The open access publication charge for this paper has been waived by Oxford University Press – NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
REFERENCES
- 1. Makarova K.S., Wolf Y.I., Iranzo J., Shmakov S.A., Alkhnbashi O.S., Brouns S.J.J., Charpentier E., Cheng D., Haft D.H., Horvath P. et al.. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020; 18:67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mohanraju P., Makarova K.S., Zetsche B., Zhang F., Koonin E.V., van der Oost J.. Diverse evolutionary roots and mechanistic variations of the CRISPR–Cas systems. Science. 2016; 353:aad5147. [DOI] [PubMed] [Google Scholar]
- 3. Hille F., Richter H., Wong S.P., Bratovic M., Ressel S., Charpentier E.. The Biology of CRISPR–Cas: backward and Forward. Cell. 2018; 172:1239–1259. [DOI] [PubMed] [Google Scholar]
- 4. Amitai G., Sorek R.. CRISPR–Cas adaptation: insights into the mechanism of action. Nat. Rev. Microbiol. 2016; 14:67–76. [DOI] [PubMed] [Google Scholar]
- 5. Jackson S.A., McKenzie R.E., Fagerlund R.D., Kieper S.N., Fineran P.C., Brouns S.J.. CRISPR–Cas: adapting to change. Science. 2017; 356:eaal5056. [DOI] [PubMed] [Google Scholar]
- 6. Barrangou R., Horvath P.. A decade of discovery: CRISPR functions and applications. Nat Microbiol. 2017; 2:17092. [DOI] [PubMed] [Google Scholar]
- 7. Makarova K.S., Wolf Y.I., Alkhnbashi O.S., Costa F., Shah S.A., Saunders S.J., Barrangou R., Brouns S.J., Charpentier E., Haft D.H. et al.. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015; 13:722–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Makarova K.S., Anantharaman V., Grishin N.V., Koonin E.V., Aravind L.. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front Genet. 2014; 5:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makarova K.S., Grishin N.V., Shabalina S.A., Wolf Y.I., Koonin E.V.. A putative RNA-interference-based immune system in prokaryotes: computational analysis of the predicted enzymatic machinery, functional analogies with eukaryotic RNAi, and hypothetical mechanisms of action. Biol. Direct. 2006; 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Makarova K.S., Wolf Y.I., Koonin E.V.. Comparative genomics of defense systems in archaea and bacteria. Nucleic. Acids. Res. 2013; 41:4360–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Deng L., Garrett R.A., Shah S.A., Peng X., She Q.. A novel interference mechanism by a type IIIB CRISPR-Cmr module in Sulfolobus. Mol. Microbiol. 2013; 87:1088–1099. [DOI] [PubMed] [Google Scholar]
- 12. Hatoum-Aslan A., Maniv I., Samai P., Marraffini L.A.. Genetic characterization of antiplasmid immunity through a type III-A CRISPR–Cas system. J. Bacteriol. 2014; 196:310–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sheppard N.F., Glover C.V. 3rd, Terns R.M., Terns M.P.. The CRISPR-associated Csx1 protein of Pyrococcus furiosus is an adenosine-specific endoribonuclease. RNA. 2016; 22:216–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burroughs A.M., Zhang D., Schaffer D.E., Iyer L.M., Aravind L.. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic. Acids. Res. 2015; 43:10633–10654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Niewoehner O., Garcia-Doval C., Rostol J.T., Berk C., Schwede F., Bigler L., Hall J., Marraffini L.A., Jinek M.. Type III CRISPR–Cas systems produce cyclic oligoadenylate second messengers. Nature. 2017; 548:543–548. [DOI] [PubMed] [Google Scholar]
- 16. Kazlauskiene M., Kostiuk G., Venclovas C., Tamulaitis G., Siksnys V.. A cyclic oligonucleotide signaling pathway in type III CRISPR–Cas systems. Science. 2017; 357:605–609. [DOI] [PubMed] [Google Scholar]
- 17. Koonin E.V., Makarova K.S.. Discovery of oligonucleotide signaling mediated by CRISPR-associated polymerases solves two puzzles but leaves an enigma. ACS Chem. Biol. 2018; 13:309–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Athukoralage J.S., Rouillon C., Graham S., Gruschow S., White M.F.. Ring nucleases deactivate type III CRISPR ribonucleases by degrading cyclic oligoadenylate. Nature. 2018; 562:277–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jia N., Jones R., Yang G., Ouerfelli O., Patel D.J.. CRISPR–Cas III-A Csm6 CARF domain is a ring nuclease triggering stepwise cA4 cleavage with ApA>p formation terminating RNase activity. Mol. Cell. 2019; 75:944–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Athukoralage J.S., Graham S., Gruschow S., Rouillon C., White M.F.. A Type III CRISPR ancillary ribonuclease degrades its cyclic oligoadenylate activator. J. Mol. Biol. 2019; 431:2894–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Athukoralage J.S., McMahon S.A., Zhang C., Gruschow S., Graham S., Krupovic M., Whitaker R.J., Gloster T.M., White M.F.. An anti-CRISPR viral ring nuclease subverts type III CRISPR immunity. Nature. 2020; 577:572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Athukoralage J.S., Graham S., Rouillon C., Gruschow S., Czekster C.M., White M.F.. The dynamic interplay of host and viral enzymes in type III CRISPR-mediated cyclic nucleotide signalling. Elife. 2020; 9:e55852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shmakov S.A., Makarova K.S., Wolf Y.I., Severinov K.V., Koonin E.V.. Systematic prediction of genes functionally linked to CRISPR–Cas systems by gene neighborhood analysis. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E5307–E5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lowey B., Whiteley A.T., Keszei A.F.A., Morehouse B.R., Mathews I.T., Antine S.P., Cabrera V.J., Kashin D., Niemann P., Jain M. et al.. CBASS immunity uses CARF-Related effectors to Sense 3′-5′- and 2′-5′-Linked cyclic oligonucleotide signals and protect bacteria from phage infection. Cell. 2020; 182:38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Genschik P., Drabikowski K., Filipowicz W.. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its sigma54-regulated operon. J. Biol. Chem. 1998; 273:25516–25526. [DOI] [PubMed] [Google Scholar]
- 26. Das U., Shuman S.. 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA. 2013; 19:1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Temmel H., Muller C., Sauert M., Vesper O., Reiss A., Popow J., Martinez J., Moll I.. The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res. 2017; 45:4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kurasz J.E., Hartman C.E., Samuels D.J., Mohanty B.K., Deleveaux A., Mrazek J., Karls A.C.. Genotoxic, metabolic, and oxidative stresses regulate the RNA repair operon of salmonella enterica serovar typhimurium. J. Bacteriol. 2018; 200:e00476-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burroughs A.M., Aravind L.. RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Res. 2016; 44:8525–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shigematsu M., Kawamura T., Kirino Y.. Generation of 2′,3′-cyclic phosphate-containing RNAs as a hidden layer of the transcriptome. Front Genet. 2018; 9:562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schaeffer D., Grishin N.V.. Identification of protein homologs and domain boundaries by iterative sequence alignment. Methods Mol. Biol. 2019; 1851:277–286. [DOI] [PubMed] [Google Scholar]
- 32. Frickey T., Lupas A.. CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 2004; 20:3702–3704. [DOI] [PubMed] [Google Scholar]
- 33. Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000; 10:1074–1077. [DOI] [PubMed] [Google Scholar]
- 34. Haft D.H., Selengut J., Mongodin E.F., Nelson K.E.. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput. Biol. 2005; 1:e60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Niewoehner O., Jinek M.. Structural basis for the endoribonuclease activity of the type III-A CRISPR-associated protein Csm6. RNA. 2016; 22:318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Anantharaman V., Makarova K.S., Burroughs A.M., Koonin E.V., Aravind L.. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013; 8:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gruschow S., Athukoralage J.S., Graham S., Hoogeboom T., White M.F.. Cyclic oligoadenylate signalling mediates Mycobacterium tuberculosis CRISPR defence. Nucleic Acids Res. 2019; 47:9259–9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Foster K., Kalter J., Woodside W., Terns R.M., Terns M.P.. The ribonuclease activity of Csm6 is required for anti-plasmid immunity by Type III-A CRISPR–Cas systems. RNA Biol. 2019; 16:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Garcia-Doval C., Schwede F., Berk C., Rostol J.T., Niewoehner O., Tejero O., Hall J., Marraffini L.A., Jinek M.. Activation and self-inactivation mechanisms of the cyclic oligoadenylate-dependent CRISPR ribonuclease Csm6. Nat. Commun. 2020; 11:1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Molina R., Stella S., Feng M., Sofos N., Jauniskis V., Pozdnyakova I., Lopez-Mendez B., She Q., Montoya G.. Structure of Csx1-cOA4 complex reveals the basis of RNA decay in Type III-B CRISPR–Cas. Nat. Commun. 2019; 10:4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Topuzlu E., Lawrence C.M.. Recognition of a pseudo-symmetric RNA tetranucleotide by Csx3, a new member of the CRISPR associated Rossmann fold superfamily. RNA Biol. 2016; 13:254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaur G., Burroughs A.M., Iyer L.M., Aravind L.. Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity. Elife. 2020; 9:e52696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yan X., Guo W., Yuan Y.A.. Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease. RNA Biol. 2015; 12:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Athukoralage J.S., McQuarrie S., Grüschow S., Graham S., Gloster T.M., White M.F.. Tetramerisation of the CRISPR ring nuclease Csx3 facilitates cyclic oligoadenylate cleavage. Elife. 2020; 9:e57627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lintner N.G., Frankel K.A., Tsutakawa S.E., Alsbury D.L., Copie V., Young M.J., Tainer J.A., Lawrence C.M.. The structure of the CRISPR-associated protein Csa3 provides insight into the regulation of the CRISPR/Cas system. J. Mol. Biol. 2011; 405:939–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu T., Li Y., Wang X., Ye Q., Li H., Liang Y., She Q., Peng N.. Transcriptional regulator-mediated activation of adaptation genes triggers CRISPR de novo spacer acquisition. Nucleic Acids Res. 2015; 43:1044–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu T., Liu Z., Ye Q., Pan S., Wang X., Li Y., Peng W., Liang Y., She Q., Peng N.. Coupling transcriptional activation of CRISPR–Cas system and DNA repair genes by Csa3a in Sulfolobus islandicus. Nucleic Acids Res. 2017; 45:8978–8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. He F., Vestergaard G., Peng W., She Q., Peng X.. CRISPR–Cas type I-A cascade complex couples viral infection surveillance to host transcriptional regulation in the dependence of Csa3b. Nucleic Acids Res. 2017; 45:1902–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McMahon S.A., Zhu W., Graham S., Rambo R., White M.F., Gloster T.M.. Structure and mechanism of a Type III CRISPR defence DNA nuclease activated by cyclic oligoadenylate. Nat. Commun. 2020; 11:500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anantharaman V., Iyer L.M., Aravind L.. Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol. Biosyst. 2012; 8:3142–3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Makarova K.S., Gao L., Zhang F., Koonin E.V.. Unexpected connections between type VI-B CRISPR–Cas systems, bacterial natural competence, ubiquitin signaling network and DNA modification through a distinct family of membrane proteins. FEMS Microbiol. Lett. 2019; 366:fnz088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liddicoat B.J., Chalk A.M., Walkley C.R.. ADAR1, inosine and the immune sensing system: distinguishing self from non-self. Wiley Interdiscip Rev RNA. 2016; 7:157–172. [DOI] [PubMed] [Google Scholar]
- 53. Kim H.S., Kim S.M., Lee H.J., Park S.J., Lee K.H.. Expression of the cpdA gene, encoding a 3′,5′-cyclic AMP (cAMP) phosphodiesterase, is positively regulated by the cAMP-cAMP receptor protein complex. J. Bacteriol. 2009; 191:922–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Griffin M.A., Davis J.H., Strobel S.A.. Bacterial toxin RelE: a highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues. Biochemistry. 2013; 52:8633–8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Iyer L.M., Aravind L.. The catalytic domains of thiamine triphosphatase and CyaB-like adenylyl cyclase define a novel superfamily of domains that bind organic phosphates. BMC Genomics. 2002; 3:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gallagher D.T., Smith N.N., Kim S.K., Heroux A., Robinson H., Reddy P.T.. Structure of the class IV adenylyl cyclase reveals a novel fold. J. Mol. Biol. 2006; 362:114–122. [DOI] [PubMed] [Google Scholar]
- 57. Gong C., Smith P., Shuman S.. Structure-function analysis of Plasmodium RNA triphosphatase and description of a triphosphate tunnel metalloenzyme superfamily that includes Cet1-like RNA triphosphatases and CYTH proteins. RNA. 2006; 12:1468–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Foster K., Gruschow S., Bailey S., White M.F., Terns M.P.. Regulation of the RNA and DNA nuclease activities required for Pyrococcus furiosus Type III-B CRISPR–Cas immunity. Nucleic Acids Res. 2020; 48:4418–4434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morett E., Segovia L.. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 1993; 175:6067–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Flores-Kim J., Darwin A.J.. The phage shock protein response. Annu. Rev. Microbiol. 2016; 70:83–101. [DOI] [PubMed] [Google Scholar]
- 61. Joly N., Engl C., Jovanovic G., Huvet M., Toni T., Sheng X., Stumpf M.P., Buck M.. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 2010; 34:797–827. [DOI] [PubMed] [Google Scholar]
- 62. Imamura R., Yamanaka K., Ogura T., Hiraga S., Fujita N., Ishihama A., Niki H.. Identification of the cpdA gene encoding cyclic 3′,5′-adenosine monophosphate phosphodiesterase in Escherichia coli. J. Biol. Chem. 1996; 271:25423–25429. [DOI] [PubMed] [Google Scholar]
- 63. Aravind L., Koonin E.V.. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001; 2:RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fedeles B.I., Singh V., Delaney J.C., Li D., Essigmann J.M.. The AlkB family of Fe(II)/alpha-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J. Biol. Chem. 2015; 290:20734–20742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nanson J.D., Kobe B., Ve T.. Death, TIR, and RHIM: self-assembling domains involved in innate immunity and cell-death signaling. J. Leukoc. Biol. 2019; 105:363–375. [DOI] [PubMed] [Google Scholar]
- 66. Horsefield S., Burdett H., Zhang X., Manik M.K., Shi Y., Chen J., Qi T., Gilley J., Lai J.S., Rank M.X. et al.. NAD(+) cleavage activity by animal and plant TIR domains in cell death pathways. Science. 2019; 365:793–799. [DOI] [PubMed] [Google Scholar]
- 67. Maupin-Furlow J.A. Prokaryotic ubiquitin-like protein modification. Annu. Rev. Microbiol. 2014; 68:155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Koonin E.V., Krupovic M.. Origin of programmed cell death from antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 2019; 116:16167–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Makarova K.S., Aravind L., Wolf Y.I., Koonin E.V.. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR–Cas systems. Biol. Direct. 2011; 6:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Samolygo A., Athukoralage J.S., Graham S., White M.F.. Fuse to defuse: a self-limiting ribonuclease-ring nuclease fusion for type III CRISPR defence. Nucleic Acids Res. 2020; 48:6149–6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lau R.K., Ye Q., Birkholz E.A., Berg K.R., Patel L., Mathews I.T., Watrous J.D., Ego K., Whiteley A.T., Lowey B. et al.. Structure and mechanism of a cyclic trinucleotide-activated bacterial endonuclease mediating bacteriophage immunity. Mol. Cell. 2020; 77:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kaya E., Doxzen K.W., Knoll K.R., Wilson R.C., Strutt S.C., Kranzusch P.J., Doudna J.A.. A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cohen D., Melamed S., Millman A., Shulman G., Oppenheimer-Shaanan Y., Kacen A., Doron S., Amitai G., Sorek R.. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature. 2019; 574:691–695. [DOI] [PubMed] [Google Scholar]
- 74. Zaver S.A., Woodward J.J.. Cyclic dinucleotides at the forefront of innate immunity. Curr. Opin. Cell Biol. 2020; 63:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Koonin E.V., Makarova K.S.. Origins and evolution of CRISPR–Cas systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2019; 374:20180087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Makarova K.S., Anantharaman V., Aravind L., Koonin E.V.. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct. 2012; 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Koonin E.V., Makarova K.S., Wolf Y.I.. Evolutionary genomics of defense systems in archaea and bacteria. Annu. Rev. Microbiol. 2017; 71:233–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.