Abstract
DNA topoisomerase II (TOP2) is a major DNA metabolic enzyme, with important roles in replication, transcription, chromosome segregation and spatial organisation of the genome. TOP2 is the target of a class of anticancer drugs that poison the DNA-TOP2 transient complex to generate TOP2-linked DNA double-strand breaks (DSBs). The accumulation of DSBs kills tumour cells but can also result in genome instability. The way in which topoisomerase activity contributes to transcription remains unclear. In this work we have investigated how transcription contributes to TOP2-dependent DSB formation, genome instability and cell death. Our results demonstrate that gene transcription is an important source of abortive TOP2 activity. However, transcription does not contribute significantly to apoptosis or cell death promoted by TOP2-induced DSBs. On the contrary: transcription-dependent breaks greatly contribute to deleterious mutations and translocations, and can promote oncogenic rearrangements. Importantly, we show that TOP2-induced genome instability is mediated by mutagenic canonical non-homologous end joining whereas homologous recombination protects cells against these insults. Collectively, these results uncover mechanisms behind deleterious effects of TOP2 abortive activity during transcription, with relevant implications for chemotherapy.
INTRODUCTION
The study of the DNA dynamics during gene expression is providing new insights into transcriptional regulation. In higher eukaryotes, the role of DNA torsion in gene expression is much more complex than previously thought. Key steps in transcriptional processes are not only coupled but coordinated with the generation and release of DNA supercoiling (1–3). The torsional state of the transcribed region is controlled by the action of DNA topoisomerases. It has been shown that DNA topoisomerase II (TOP2) has multiple direct roles in transcription: promoting the activation and repression of initiation by maintaining the structure of either active or inactive promoters, as well as releasing paused RNA polymerases and facilitating transcriptional elongation (4–6). At the same time, TOP2 is involved in many other processes of DNA metabolism including DNA replication, chromosome segregation and spatial organisation of the genome (2,7–9). Mammalian cells express two TOP2 isoforms, TOP2α and TOP2ß. Whereas TOP2ß is expressed thorough the cell cycle, TOP2α expression correlates with cellular proliferation and peaks at S and G2/M(10). TOP2α has a major role in replication and chromosome segregation although it has also been implicated in transcription. TOP2ß activity has been mainly associated to transcription (1–3,7).
DNA topoisomerases remove torsional stress by introducing transient breaks in DNA. TOP2 cleaves both strands of a DNA duplex to allow passage of another duplex through it. An intermediate, known as the ‘cleavage complex’ (TOP2cc), is created, within which the topoisomerase has cleaved both strands of DNA and is covalently linked to the 5′-terminus of the DNA via a phosphotyrosyl bond. The cleavage complex is normally transient, because the break is resealed at the end of the topoisomerase catalytic cycle. However, TOP2cc can, under uncertain circumstances, become ‘abortive’ resulting in a DNA double strand break (DSB) with the DNA 5′ termini ‘blocked’ by trapped protein adducts. Trapped TOP2 can be denatured and, at least partially, degraded by the proteasome. The remaining peptide can be then removed via the nuclease activity of the MRN complex (11) or by tyrosyl-DNA phosphodiesterase 2 (TDP2) (12,13). TDP2 cleaves the phosphotyrosyl bond between the topoisomerase peptide and the 5′ phosphate of the DNA, generating error-free ligatable ends that can be processed by the non-homologous end-joining (NHEJ) pathway (14,15).
Homologous recombination (HR) is largely an error-free DNA pathway that prevents genome instability during S and G2 phases of the cell cycle (14). In contrast, NHEJ is a rapid and efficient repair pathway that is active throughout the cell cycle, but can be considered error-prone as, under some circumstances, nucleases may modify the DNA to make it compatible for ligation. The canonical NHEJ pathway (cNHEJ) is required for cell survival following ionizing radiation-induced DNA breaks, and is essential for the lymphocyte maturation (16). In the absence of core cNHEJ factors, microhomology-mediated alternative NHEJ (altNHEJ) pathway may operate (16), although the physiological circumstances where these are favoured, and their consequences, remain obscure. In the case of the TOP2-dependent DSBs, the role of distinct NHEJ processes are poorly understood.
DNA topoisomerases are key targets of chemotherapeutic drugs. TOP2 ‘poisons’ such as etoposide are commonly used in the treatment of a broad range of tumours (17). These drugs stabilise TOP2cc, promoting abortive TOP2cc and DSB formation. Their efficacy relies on the proliferative status of tumour cells (18), since DNA replication accounts for the majority of cellular TOP2 activity. However, treatment with TOP2-targeting drugs can also result in chromosome translocations (in MLL, for example) that underlie secondary leukaemias (19,20). We have recently shown that TDP2 suppresses chromosomal translocations induced by TOP2 during transcription (21). The role of NHEJ in the formation or prevention of chromosomal translocation is still controversial. Whereas studies in mice lymphoblasts demonstrated that cNHEJ prevents genomic translocations that are mediated by altNHEJ (16), recent studies in human cells have shown that ionizing radiation and nuclease-induced translocations are dependent on cNHEJ (22–24). In the case of TOP2-induced translocations, this question remains unresolved (14).
Transcription has recently emerged as one of the main endogenous sources of TOP2 abortive cycles and DSBs (4,5,25,26). We recently showed that TOP2 activity during transcription is involved in the formation of chromosome translocations (21), but the mechanisms that mediate their formation are still unclear. The global contribution of transcription-induced breaks to topoisomerase-associated instability and cell death has not been studied in depth. Since replication-associated TOP2 activity is very high in cycling cells, discriminating transcription-dependent events has been challenging. Here, we have analysed the overall influence of transcription on the formation of abortive TOP2cc and DSBs. We show that gene transcription is associated with a high proportion of TOP2-derived DSBs in replicating and non-replicating cells, with little effect on cell survival, but making a key contribution to genomic instability. Moreover, we demonstrate that this genomic instability is mediated by cNHEJ, which promotes mutagenesis and facilitates the formation of genomic translocations. These data clarify the impact of TOP2 activity on gene transcription and the mutagenic role of NHEJ in human cells. Notably, our results have important implications for TOP2 activity during transcription, as well as for toxicity and long-term secondary effects of chemotherapy with TOP2 poisons.
MATERIALS AND METHODS
Cell lines and culture conditions
hTERT RPE-1 cells (originally purchased from ATCC) were propagated in DMEM/F12 medium supplemented with 10% fetal bovine serum (FBS) and with 1% penicillin and streptomycin. For serum starvation, cells were grown until confluency, washed twice with serum-free media and then cultured in 0% FBS for 3–6 days. RPE-1 control and LIG4−/− knockout cell lines were a generous gift from Professor SP Jackson's laboratory (27). KG1a cells (originally purchased from ATCC) were cultured in IMDM medium supplemented with 10% FBS and with 1% penicillin and streptomycin. LIG4 knockdown KG1a cell line was generated using LIG4 shRNA MISSION Lentiviral Transduction Particles (Sigma-Aldrich, NM_002312). A total of 1 × 105 KG1a cells were incubated with 8μg/ml of polybrene and lentiviral particles (MOI = 5) overnight at 37°C. Cells were selected in 5μg/ml Puromycin and LIG4 expression was checked regularly. LIG4 (CAAGAUGUUUACAGAAAGGAA) and Control (Luciferase CGUACGCGGAAUACUUCGA) siRNA was transfected using RNAi Max (Invitrogen) according to manufacturer instructions 48 h before assays.
All cell lines were grown at 37°C, 5% CO2 and were regularly tested for mycoplasma contamination.
Western blotting
Protein extracts were obtained by lysing cell pellets at 100°C for 10 min in 2× protein buffer (125 mM Tris, pH 6.8, 4% SDS, 0.02% bromophenol blue, 20% glycerol, 200 mM DTT). Extracts were then sonicated in a Bioruptor (Diagenode) for 1 min at high intensity. Primary antibodies were blocked in Tris buffered saline buffer 0.1% Tween20 5% BSA and employed as follows: TDP2 antibody ((28)) 1:5000, TOP2α (Santa Cruz, sc-5348) 1:500, TOP2ß (Santa Cruz, sc-13059) 1:500, LIG IV (Santa Cruz, sc-271299) 1:100, Vinculin (Santa Cruz, sc-25336) 1:1000, GAPDH (Santa Cruz, sc-47724) 1:1000, γH2AX (Millipore, 05-636) 1:1000, Cleaved caspase 3 (Cell Signaling, 966) 1:500 and Cyclin A (Santa Cruz, sc-751) 1:1000. Full blots are included in Supplementary Figures S13_1 and S13_2.
Immunofluorescence and FISH
For immunofluorescence (IF), cells were grown on coverslips for 2 days (for cycling cultures) or 4–8 days (for serum-starved and confluency-arrested cell cultures) and then treated as indicated. Cells were fixed (10 min in PBS–4% paraformaldehyde), permeabilized (5 min in PBS–0.2% Triton X-100), blocked (30 min in PBS–5% BSA), and incubated with the indicated primary antibodies for 1–3 h in PBS–1% BSA. Cells were then washed (3 × 5 min in PBS–0.1% Tween20), incubated for 30 min with the corresponding AlexaFluor-conjugated secondary antibody (1:1000 dilution in 1% BSA–PBS) and washed again as described above. Finally, cells were counterstained with DAPI (Sigma) and mounted in Vectashield (Vector Labs). 53BP1 foci were manually counted (double-blind) in 20–40 cells per data point per experiment. Primary anti-γH2AX (Millipore, 05-636) were employed at 1:1000 and anti-53BP1 (Abcam, ab32042) at 1:2500. Whole chromosome FISH was performed according to manufaturer's protocol (CytoCell, aquarius FISH probes). Click chemistry reaction was performed before DAPI staining by incubating (30 min at room temperature) with 1 mM AlexaFluor-conjugated azide (Invitrogen) in reaction cocktail (100 mM Tris–HCl pH 8.5, 1 mM CuSO4, 100 mM ascorbic acid).
Metaphase spreads
For metaphase spreads, cells were incubated with demecolcine (Sigma) at 0.2 μg/ml for 4–12 h and then harvested. Cells were collected using standard cytogenetic techniques and fixed in 3:1 methanol:acetic acid. Fixed cells were dropped onto acetic acid-humidified slides before dehydration and FISH.
RNA extraction and RT-qPCR
Total RNA from cultured cells was extracted with RNA Isolation Kit (Qiagen), according to manufacturer instructions. DNA was eliminated with RQ1 DNAse (Promega). For RT-qPCR, cDNA was obtained using M-MLV RT (H-) Point Mutant (Promega), according to manufacturer instructions. qPCR was performed using TB Green Premix Ex Taq II (Takara) on a Roche LightCycler 480 II machine. DNA oligonucleotides used in this experiment are: MLL_F (5′ CAAGAGAGGATCCTGCCCCAAA 3′), MLL_R (5′ GTGGTGGCCTGTTTGGATTCAG 3′), 45S_F (5′ CCGTGCCGAGTCGTGAC 3′), 45S_R (5′ CGCTTTCCCAGGGCCA 3′), 18S_F (5′ GTTGGTGGAGCGATTTGTCTGG 3′), 18S_R (5′ CTGAACGCCACTTGTCCCTCTA 3′).
Clonogenic survival assays
For quiescent RPE-1, cells were plated in 10 mm plates. After 4 days of serum-starvation, cells were treated with indicated concentrations of etoposide for 3 h and/or pre-incubated with DRB (100 μM, 1 h). Dishes were rinsed twice with PBS and trypsinized. A total of 500 cells were re-cultured in serum containing media for 10 days, and then fixed and stained in 70% ethanol/1% methylene blue. For asynchronous RPE-1, 2000 cells were split in 100 mm dishes, or 300 cells in 60 mm dishes. After 6 hours cells were treated with the indicated concentration of etoposide for 3 h, washed with PBS and growth in fresh new media for 10 days, and then fixed and stained in 70% ethanol/1% methylene blue. For KG1a, cells were treated with indicated concentrations of etoposide for 3 h. When indicated cells were pre-incubated with DRB (100 μM, 1 h). After that, cells were washed twice with PBS and cultured in semisolid medium (1% methylcellulose IMDM) for 15 days. Colonies were counted either manually (RPE-1), or using the image processing program ImageJ (29) (KG1a). The surviving fraction at each dose was calculated by dividing the average number of colonies in treated dishes by the average number in untreated dishes. In all biological replicates cells were split in duplicate for each experimental condition.
Flow cytometry apoptosis assay
Apoptosis assay was performed with Annexin V-FITC and propidium iodide (PI) kit (SantaCruz, sc4252-ak) according to the manufacturer instructions. After treatment, cells were rinsed twice with PBS and culture in drug-free media for indicated periods. After staining, percentage of apoptotic cells was analysed by flow cytometry. Graph shows early (Annexin V+ PI-, lower right quadrant in plot) and late apoptosis (Annexin V+ PI+, upper right quadrant) percentage. 20 000–30 000 cells were counted for each sample.
Cell cycle analysis
Cells were incubated with 10μM BrdU (Sigma, B5002) during 15 min. Cells were washed twice with PBS and fixed with 70% ethanol overnight. DNA was denatured with 2 N HCl/Triton X-100. Cells were incubated with anti-BrdU (SantaCRuz, sc-32323) at 1:1000 overnight at 4°C. After that, AlexaFluor-conjugated secondary antibody (Invitrogen) was added at 1:1000 during 1 h. Finally, before flow cytometry, cells were incubated with 100 μg/ml PI and 100 μg/ml RNAse A for 30 min.
Sister chromatid exchanges (SCEs)
Cells were cultured for 48 h in the presence of 10 μM BrdU. At 40 h, cultures were incubated with indicated concentration of etoposide or vehicle 30 min. After treatment, cells were rinsed twice with PBS and cultured in drug-free BrdU-containing media. Metaphase spreads were done according to the previous protocol. After dropping, slides were incubated in 10 μg/ml bisbenzimide solution (Hoescht) for 20 min and incubated in 20× SSC buffer for 5 min. Slides were irradiated in a 365 nm UV-lamp for 45 min. Next, slides were incubated in 20× SSC at 60°C for 60 min. Finally, slides were stained with 5% Giemsa for 20 min.
ICE assay
ICE assay was performed as previously described (30) with minor modifications. Briefly, a total of 2 × 106 cells were treated as indicated and lysed with 3 ml 1% Sarkosyl. The lysate was passed through a 25G5/8 gauge ten times. 2 ml of CsCl solution (1.5 g/ml) was layered into an ultracentrifugation tube. The volume of lysate was layered on top of the gradient. Samples were centrifuged in a Beckman VTI-65 rotor at 25 °C, 37 200 rpm for 20 h. Pellet was resuspended in TE 1× and DNA concentration was measured in a Nanodrop. 2 μg of DNA was loaded in nitrocellulose membrane preincubated in 25 mM NaPO4 pH 6,5 for 15 min using a slot-blot apparatus.
Statistical analysis
Statistical analysis is included in figure legends. In all cases, comparison tests were performed using GraphPad Prism version 6.0c for macOS. Statistical significance is indicated in plots (*P < 0.05, **P < 0.01, ***P < 0.001, NS, not significant).
RESULTS
TOP2ß activity is associated to gene transcription
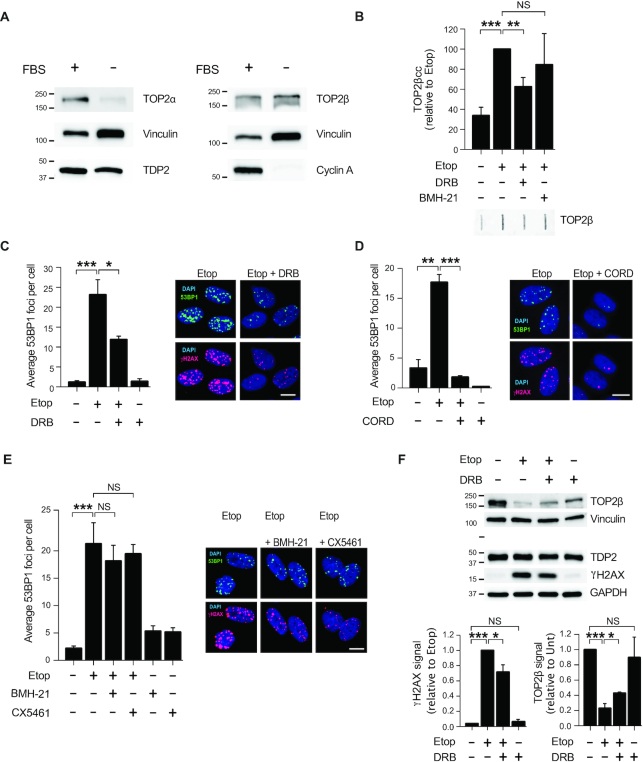
Our first goal was to measure TOP2 activity during transcription in human cells. To avoid the interference of replication-associated TOP2 activity we synchronised diploid human RPE-1 hTERT cells in G0/G1 (see Materials and Methods). As a result, over 97% of cells were arrested in G0/G1 and no replicating cells were detected (Supplementary Figure S1). In accordance with previous reports (31), quiescent cells strongly downregulated TOP2α, whereas TOP2ß levels remained unaltered (Figure 1A). We next studied the activity of TOP2-associated RNA polymerases (RNAP) using RNAP I and II specific inhibitors (Figure 1B and Supplementary Figure S2). We measured active TOP2ß by the isolation of TOP2ß cleavage complexes (ICE assay (30)) after inhibition of transcription. TOP2ßcc were not significantly altered by addition of the RNAPI inhibitor BMH-21 (32), but reduced by 40% in the presence of RNAP II elongation inhibitor 5,6-dichlorobenzimidazone-1-β-d-ribofuranoside (DRB) (Figure 1B). These results suggest that TOP2ß activity is associated with RNAP II-dependent transcription in quiescent cells.
Figure 1.
Transcription by RNAP II promotes TOP2β-induced DSBs in non-cycling cells. (A) Protein blots of the indicated proteins in proliferating (FBS+) and serum-starved (FBS–) RPE-1 cells. Molecular weight markers are in kDa. (B) Analysis of TOP2β cleavage-complexes (TOP2βcc) by ICE assay. Serum-starved RPE-1 cells were treated with 50 μM etoposide for 1 h. Where indicated, cells were pre-incubated with DRB (100 μM) or BMH21 (2 μM) for 1 h prior to etoposide treatment. Data are the mean (± s.e.m.) of five independent experiments. Statistical significance was determined by t-test (**P < 0.01, ***P < 0.001, NS, not significant). (C) 53BP1 foci in serum-starved RPE-1 cells following following treatment with 20 μM etoposide for 1 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 3 h prior to etoposide treatment. Representative images of 53BP1 foci (green), γH2AX foci (red) and DAPI counterstain (blue) are shown. Scale bar, 10 μm. In all cases, data are the mean (± s.e.m.) of at least three independent experiments. Statistical significance was determined by t-test (*P< 0.05, **P< 0.01, ***P< 0.001, NS, not significant). (D) 53BP1 foci in serum-starved RPE-1 cells following etoposide treatment. Where indicated, cells were pre-incubated with 100 μM cordycepin (CORD) for 2 h prior to etoposide treatment. Other details as in (C). (E) 53BP1 foci in serum-starved RPE-1 cells following etoposide treatment. Where indicated, cells were pre-incubated with BMH21 (2 μM) for 3 h or CX5461 (20 μM) for 3 h prior to etoposide treatment. Other details as in (C). (F) Top, protein blots of the indicated proteins in serum-starved RPE-1 cells following treatment with 100 μM etoposide for 6 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 3 h prior to etoposide treatment. Molecular weight markers are in kDa. Bottom, quantification of TOP2β and γH2AX signal normalized with respect to GAPDH. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by t-test (*P< 0.05, ***P< 0.001, NS, not significant).
Gene transcription induces TOP2 breaks
Next, we investigated the influence of transcription in DSBs generated by TOP2 activity. The TOP2 poison etoposide rapidly induced DSBs in quiescent cells, as measured using 53BP1 and H2AX serine 139 phosphorylation (hereafter γH2AX) immunofoci as surrogate markers of DSBs (33) (Figure 1C). More importantly, RNAP II elongation inhibition before TOP2 poisoning resulted in a 50% reduction in DSBs (Figure 1C). The addition of cordycepin, a functionally different RNAP II inhibitor, also resulted in a significant decrease in DSBs, indicating that the DSB reduction was not an artefact caused by DRB (Figure 1D). In contrast, in accordance with no substantial influence of RNAP I on TOP2 activity, inhibition of RNAP I caused no significant reduction in DSBs (Figure 1E).
In line with our previous result, protein blots showed that DRB prevented ≃30% of γH2AX accumulation induced by etoposide (Figure 1F). As previously reported (34), etoposide triggered partial TOP2ß loss, reflecting proteasomal-mediated degradation of abortive TOP2cc (Figure 1F). Strikingly, inhibition of RNAP II before TOP2 poisoning slightly but significantly prevented this degradation (Figure 1F), confirming the reduction in abortive TOP2ßcc when RNAP II transcription is blocked and suggesting that transcription-associated breaks are subsequently processed and repaired. Although a minor contribution of remaining TOP2α is possible, these results demonstrate that TOP2ß activity during RNAP II transcription can be a prominent source of abortive TOP2 activity in non-cycling cells.
Gene transcription induces replication-independent TOP2 breaks in proliferating cells
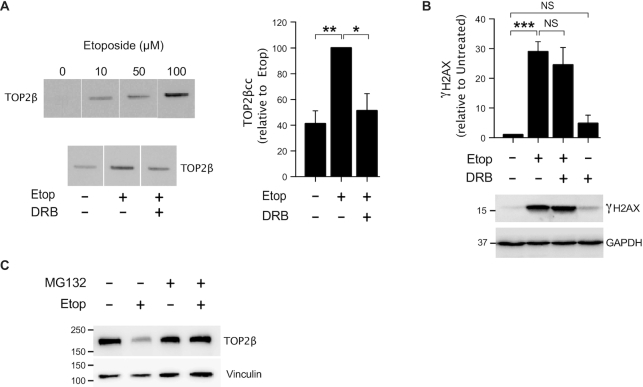
We then assessed the contribution of transcription-associated TOP2 activity to overall TOP2 activity in proliferating cells. DRB also promoted a decrease of TOP2ß activity in human myeloid KG1a cells (Figure 2A), suggesting that TOP2ß activity is linked to transcription in proliferating cells. However, RNAP II inhibition (Supplementary Figure S3) did not significantly decrease etoposide-induced γH2AX accumulation in proliferating myeloblasts (Figure 2B, Supplementary Figure S4A and B). Since TOP2ß activity was associated with transcription in these cells (Figure 2A), this result indicates that there is a high level of transcription-independent TOP2 activity in proliferating cells, probably related to other processes such as DNA replication. Nevertheless, TOP2 poisoning induced proteasome-dependent degradation of TOP2ß in KG1a cells, indicating that abortive TOP2 breaks are processed for repair in these cells (Figure 2C). It is important to note that changes are independent of cell cycle status since conditions used did not significantly alter cell cycle other than some cell death induced by DRB (Supplementary Figure S5).
Figure 2.
Transcription by RNAP II promotes TOP2β-induced DSBs in cycling cells. (A) Analysis of TOP2β cleavage-complexes (TOP2βcc) by ICE assay. KG1a cells were treated with indicated concentrations of etoposide for 1 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 1 h prior to etoposide treatment. Left, representative slot blot images and right, data (mean ± s.e.m.) of four independent experiments. Statistical significance was determined by t-test (*P< 0.05, **P< 0.01). (B) Protein blots of γH2AX and GAPDH in KG1a cells treated with 40 μM etoposide for 3 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 1 h prior to etoposide treatment. Data are the mean (± s.e.m) of three independent experiments. γH2AX protein level was normalized with respect to GAPDH. Statistical significance was determined by t-test (***P< 0.001, NS, not significant). Molecular weight markers are in kDa. (C) Proteins blots of indicated proteins. Cells were treated with 40 μM etoposide for 1 h. Where indicated, cells were pre-incubated with the proteasome inhibitor MG132 (10 μM) for 2 h prior to etoposide treatment. Vinculin was used as a loading control. Molecular weight markers are in kDa.
These results suggest that, while TOP2ß activity is associated with transcription in proliferating cells, the majority of TOP2 activity is transcription-independent. In the presence of TOP2 poisons, these processes promote the formation of a much higher proportion of DSBs to than transcription-related processes.
Gene transcription does not contribute to rapid etoposide-induced cell death in proliferating cells
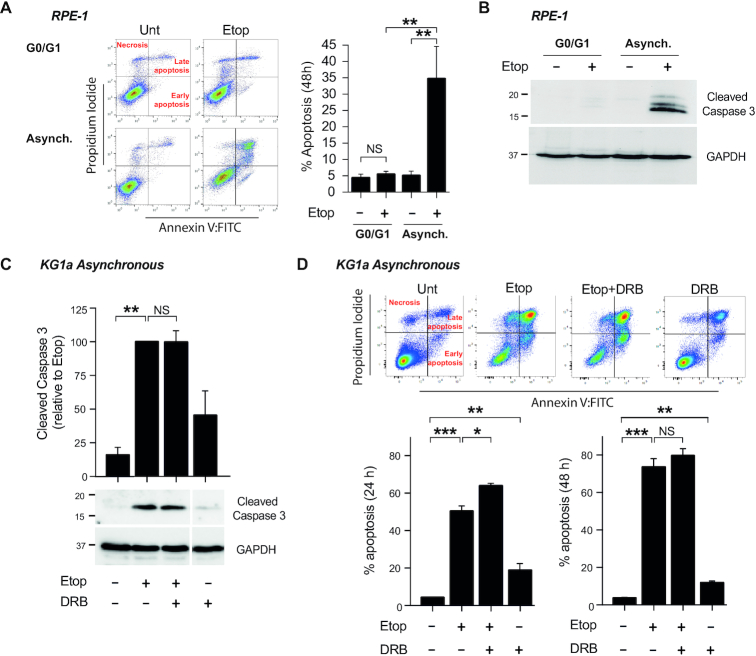
Since transcription-associated TOP2 activity is a novel source of endogenous DSBs, we next investigated whether they could induce cell death. In asynchronous RPE-1 cells, etoposide promoted 40% cell death 48 h after treatment, as measured by FACS and caspase-3 cleavage (Figure 3A and B). In contrast, etoposide did not promote any significant cell death in non-cycling RPE-1 cells (Supplementary Figure S1; Figure 3A and B). These results suggest that transcription-associated TOP2 activity causes limited or no cytotoxic DSBs in quiescent cells.
Figure 3.
DSB-induced apoptosis by etoposide is independent of transcription. (A) Apoptosis analysis by FACS of G0/G1 and asynchronous RPE-1 cells treated with 100 μM etoposide for 11 h. Following treatment cells were cultured in drug-free medium for 48 h. Left, Representative dot-plots (Annexin-V vs. Propidium Iodide). Right, Mean (± s.e.m.) of the percentage of Annexin-V positive cells. Statistical significance was determined by t-test of three independent experiments (**P< 0.01, NS, not significant). (B) Protein blots of cleaved Caspase-3 in RPE-1 cells treated as in (A). GADPH was used as a loading control. Molecular weight markers are in kDa. (C) Protein blots of cleaved Caspase-3 in cycling KG1a cells treated with 40 μM etoposide for 3 h. Where indicated, pre-incubation with DRB (100 μM) for 1 h prior to etoposide treatment. After treatment, cells were cultured in drug-free medium for 24 h. Bottom, representative images. Molecular weight markers are in kDa. Top, quantification (mean ± s.e.m.) of three independent experiments. Caspase-3 protein levels were normalized with respect to GAPDH. Statistical significance was determined by t-test (**P< 0.01, NS, not significant). (D) Apoptosis analysis of cycling KG1a cells by FACS. Cells were treated as in (C). Cells were incubated in drug-free medium for 24 and 48 h. Top, representative dot-plots (Annexin-V versus Propidium Iodide) at 24 h. Bottom, Percentage of Annexin-V positive cells (mean ± s.e.m.) of three independent experiments at 24 and 48 h. Statistical significance was determined by t-test (*P< 0.05, **P< 0.01, ***P< 0.001, NS, not significant).
In myeloid KG1a cells, caspase-3 cleavage was very strongly induced by etoposide (Figure 3C), with 50% of cells apoptotic 24 h after treatment and >70% apoptotic after 48 hours, as measured by FACS (Figure 3D). Notably, these effects were not prevented by RNAP II inhibition with DRB (Figure 3C, D), or other transcription inhibitors such as flavopiridol and cordycepin (Supplementary Figure S4C). These results suggest that, in proliferating cells, accumulation of DSBs and apoptosis resulting from TOP2 poisoning are not dependent on transcription-associated TOP2 activity but rather on other TOP2-associated processes such as replication.
Gene transcription is a source of mutations and genomic translocations in proliferating cells
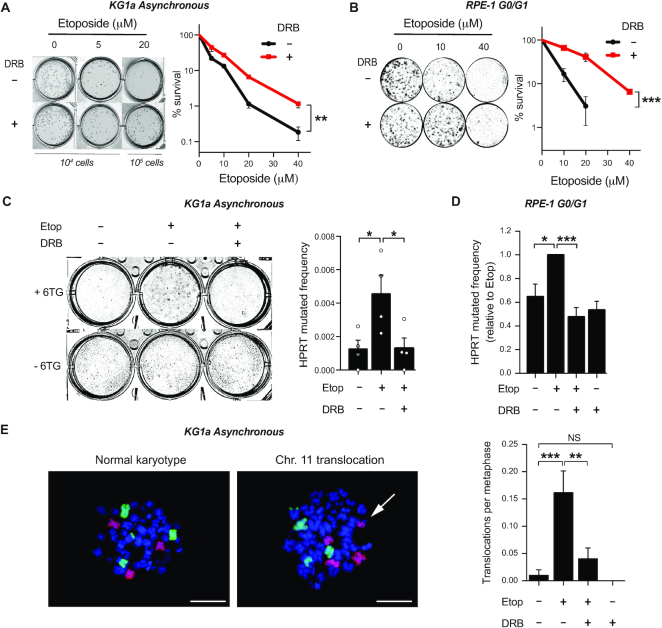
DSBs can rapidly kill cells by triggering apoptotic pathways (35). At the same time, illegitimate DSB repair can promote mutagenesis and genomic instability altering cellular homeostasis and, in the long-term, reducing cellular viability. In order to investigate whether transcription-dependent TOP2 activity influenced etoposide-induced cytotoxicity, we assessed clonogenicity in myeloblasts grown in semisolid media. Cell survival was reduced in a dose-dependent manner following treatment with increasing concentrations of etoposide, suggesting that cell death can still occur days after DSB repair is complete (Figure 4A). Strikingly, pre-treatment with DRB, that had not affected DSB induced apoptosis (Figure 3D), significantly reduced sensitivity to etoposide, suggesting that although transcription is not a major cause of rapid cell killing by TOP2 in proliferating cells, it compromises long-term cell survival (Figure 4A).
Figure 4.
Transcription induces TOP2-dependent mutagenesis and translocations. (A) Clonogenic survival of KG1a cells treated with the indicated concentration of etoposide for 3 h. Where indicated cells were pre-incubated with DRB (100 μM) for 1 h prior to etoposide treatment. After that, cells were washed and cultured in methylcellulose-containing medium for 14 days. Left, representative images of cultures. Right, quantification (mean ± s.e.m.) of three independent experiments. Statistical significance was determined by two-way ANOVA (**P< 0.01). (B) Clonogenic survival of RPE-1 cells, treated while serum-starved, with the indicated concentration of etoposide for 3 h. Where indicated cells were pre-incubated with DRB (100 μM) for 1 h prior to etoposide treatment. After treatment, cells were cultured in serum containing drug-free medium for 10 days. Left, representative images. Right, quantification (mean ± s.e.m.) of three independent experiments. Statistical significance was determined by two-way ANOVA (***P< 0.001). (C) HPRT mutagenic assay in KG1a cells. Cells were treated with 40 μM etoposide for 3 h. Where indicated, cells were pre-incubated with DRB (100µM) for 1 h prior to etoposide. 7 days after treatment living cells were cultured in 6TG containing drug-free medium for 14 days. HPRT mutated frequency is calculated by comparing 6TG-resistant cells to the survival in a 6TG-free culture. Data are the mean (± s.e.m.) of four independent experiments. Statistical significance was determined by t-test (*P< 0.05, ***P< 0.001). (D) HPRT mutagenic assay in serum-starved RPE-1 cell line. Serum-starved cells were treated with 40 μM etoposide for 3 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 1 h, prior to etoposide treatment. Cells were then cultured in 6TG containing drug-free medium for 10 days. HPRT mutated frequency represented relative to etoposide-treated cultures. Other details as in (C). (E) Translocation frequencies (translocations per metaphase) in chromosome 8 and 11 were quantified in KG1a metaphase spreads prepared 12 h after 20 μM etoposide for 3 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 1 h prior etoposide. Left, FISH images of KG1a normal karyotype vs. translocation of chromosome 11. Scale bar, 10 μm. Right, quantification (mean ± s.e.m.) of two independent experiments. Translocations in chromosomes 8 and 11 are plotted together. Statistical significance was determined by t-test (**P< 0.01, ***P< 0.001, NS, not significant).
To confirm this newly revealed transcription-dependent etoposide cytotoxicity, we studied clonogenic survival of RPE-1 cells that had been treated, while quiescent, with an acute dose of etoposide. Importantly, most DSBs were repaired within a few hours of etoposide treatment (Figure 5A) (26) and no significant apoptosis was detected (Supplementary Figure S6A and B). However, clonogenic survival was clearly reduced: an effect that was significantly rescued by inhibition of RNAP II before TOP2 poisoning (Figure 4B). Overall, the combined data suggest that inhibition of transcription strongly reduces the long-term deleterious effects of repaired TOP2-dependent DSBs.
Figure 5.
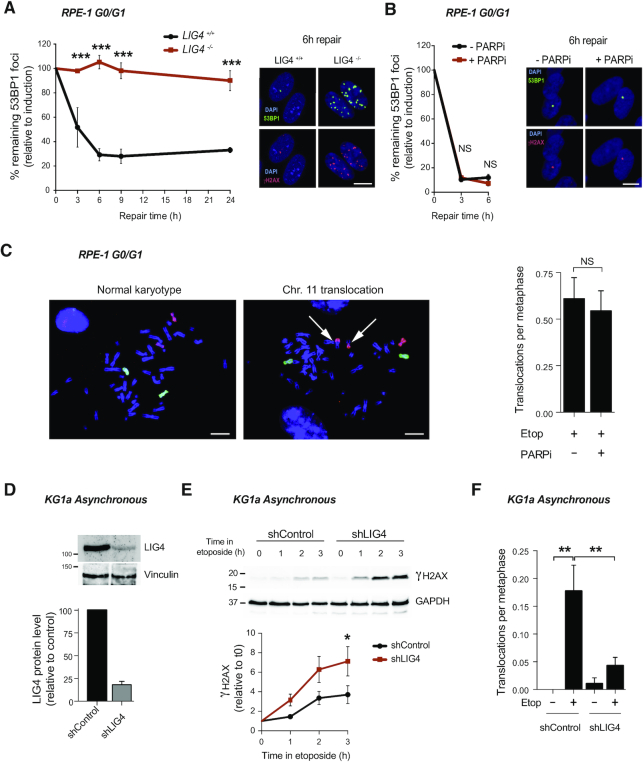
cNHEJ pathway repairs TOP2-induced DSBs. (A) 53BP1 foci in serum-starved wild-type (LIG4+/+) and LIG4−/− RPE-1 cells after 30 min treatment with 10 μM etoposide, and after the indicated repair periods in drug-free medium. Values are shown as percentage of remaining foci after induction (0 h of repair). Representative images of 53BP1 foci (green), γH2AX foci (red) and DAPI counterstain (blue) for the 6 h repair time point are shown. Scale bar, 10 μm. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by two-way ANOVA (***P< 0.001, NS, not significant). (B) 53BP1 foci in serum-starved RPE-1 cells before and 30 min after treatment with 10 μM etoposide, and after the indicated repair periods in etoposide-free medium. Where indicated, cells were pre-incubated with the PARP inhibitor KU58948 (1 μM) for 1h prior to etoposide treatment and during repair. Other details as in (A). (C) Translocation frequencies (translocations per metaphase) in chromosome 8 and 11 were quantified in serum-starved (G1/G0) RPE-1 cells in metaphase spreads prepared 48 h after etoposide treatment (1 h, 50 μM). Where indicated, cells were pre-treated with PARP inhibitor (1 μM) for 1 h prior to, during, and 4 h after etoposide treatment. Data are the mean (± s.e.m.) of two independent experiments. (D) Analysis of LIG4 protein level after the generation of a KG1a knockdown cell line by stably expression of shRNA against LIG4 (shRNA LIG4). Top, representative protein blots. Molecular weight markers are in kDa. Bottom, quantification of two independent experiments (mean ± s.e.m.). LIG4 signal normalized with respect to Vinculin. LIG4 protein relative to the control cells (shRNA control). (E) Accumulation of γH2AX in shRNA LIG4 and control cells after 20 μM etoposide for indicated time. Top, proteins blots. Bottom, quantification (mean ± s.e.m.) of three independent experiments. γH2AX protein level was normalized with respect to GAPDH. Molecular weight markers are in kDa. Data are the mean (± s.e.m) of three independent experiments. Statistical significance was determined by t-test at indicated time (*P< 0.05) (F) Translocation frequencies (translocations per metaphase) in chromosome 8 were quantified in KG1a shRNA LIG4 and control cells metaphase spreads prepared 12 h after 20 μM etoposide (3 h) treatment by chromosome 8 FISH. Data (mean ± s.e.m.) of four independent experiments are shown. Statistical significance was determined by t-test (**P< 0.01).
We hypothesized that reduction in cell survival may be more strongly associated with mutation and genome instability than with residual DSBs. To study mutagenesis induced by TOP2 poisoning, surviving cells from etoposide-treated KG1a clonogenics were split into medium containing 6-thioguanine (6TG). Survival in 6TG represents correlation with mutation frequency in transcribed genes, since inactivating mutations in the hypoxanthine phosphoribosyltransferase 1 (HPRT1) gene confer resistance to 6TG (36) (Supplementary Figure S7A). Etoposide induced a dose-dependent increase of 6TG-resistant cells, confirming that TOP2 poisoning resulted in mutations (Supplementary Figure S7B). Strikingly, the ∼4-fold increase in the number of 6TG resistant cells generated by etoposide was ablated when cells had been pre-treated with DRB (Figure 4C), suggesting that mutations induced by abortive cycles of TOP2 are completely transcription-dependent. This result was consistently observed in non-cycling RPE-1 cells, confirming that mutagenesis was not dependent on any TOP2 activity other than transcription (Figure 4D, Supplementary Figure S7C).
One of the most studied mutagenic effects of TOP2 poisoning is the formation of chromosomal translocations. We have previously shown that gene transcription in quiescent cells is a source of TOP2-induced DSBs and genomic rearrangements (21). In order to examine the transcription dependence of chromosomal translocations in cycling cells, we employed whole chromosome FISH to directly observe these events. In proliferating myeloid cells, TOP2 poisoning resulted in accumulation of translocations that were prevented by pre-incubation with DRB, implicating gene transcription in the formation of TOP2-induced DSBs that result in genomic translocations (Figure 4E).
Collectively, these results demonstrate that transcription is the major contributor to genomic instability induced by TOP2 abortive activity in cycling cells, producing mutations and chromosomal exchanges that can have deleterious effects on cell survival.
Canonical NHEJ mediates TOP2-dependent genome instability
To address the role of cNHEJ in the repair of TOP2-induced breaks, we employed non-cycling RPE-1 cells lacking Ligase 4 (LIG4), an essential core factor of cNHEJ (16,27) (Supplementary Figure S8A). These cells were hypersensitive to etoposide (Supplementary Figure 8B). In accordance with cNHEJ being the major repair pathway involved in the repair of TOP2 breaks in G0/G1, no repair was observed in LIG4−/− cells for up to 24 h after etoposide treatment (Figure 5A).
We attempted to use whole chromosome FISH to directly examine the impact of cNHEJ on the frequency of TOP2-induced chromosome translocations in LIG4−/− cells. However, we repeatedly failed to obtain metaphases from LIG4−/− cells, that had been treated with etoposide while synchronized in G0/G1, probably due to the persistence of DSBs in these cells (Figure 5A). In contrast, inhibition of PARP1, a factor that facilitates altNHEJ pathway (37–39), did not affect repair of DSBs (Figure 5B) and we were able to isolate metaphases with no difference in the formation of chromosomal translocations (Figure 5C). It is also important to note that PARP inhibition did not alter transcription (Supplementary Figure S9). These results suggest that cNHEJ is the major pathway involved in the repair of TOP2 breaks in quiescent cells.
To surpass the limitation of unrepaired DSBs and study chromosomal translocations in asynchronous cultures, we depleted LIG4 in KG1a cells (Figure 5D). Notably, LIG4 depletion resulted in etoposide induced DSBs accumulation, confirming the implication of cNHEJ in the repair of TOP2 breaks in myeloid cells (Figure 5E). Interestingly, LIG4 deficiency ablated the induction of translocations (Figure 5F). These results suggest that genomic translocations induced by TOP2 during transcription are cNHEJ-dependent.
cNHEJ mediates MLL repair
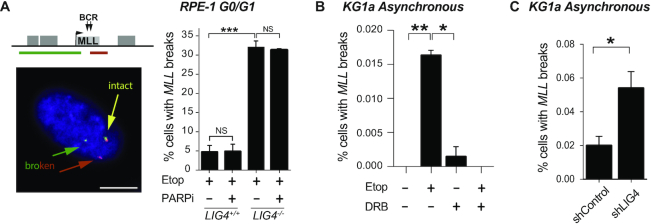
We have previously shown that MLL breakage due to TOP2 abortive activity is transcription-dependent (21). To examine the role of cNHEJ in the repair of TOP2-induced MLL breakage, we employed FISH probes flanking this locus so that we could detect etoposide-induced breaks via their impact on the spatial proximity of these probes (21) (Figure 6A). Etoposide-induced MLL breakage was increased 6-fold in G0/G1 LIG4−/− RPE-1 cells compared with wild-type cells (Figure 6A). PARP inhibition did not alter MLL breakage, even in the absence of LIG4 (Figure 6A), demonstrating that altNHEJ is not required for repair of MLL breaks. MLL breakage was also LIG4 and transcription-dependent in cycling KG1a cells (Figure 6B and C), demonstrating that cNHEJ mediates the repair of TOP2-induced breaks during transcription of MLL in myeloid cells.
Figure 6.
cNHEJ pathway repairs transcription-dependent DSBs at MLL. (A) Detection of broken MLL loci by FISH in serum-starved wild-type and LIG4−/− RPE-1 cells following mock-treatment (DMSO) or treatment with 100 μM etoposide for 6 h. Where indicated, cells were pre-incubated with PARP inhibitor KU58948 (1 μM) for 1 h prior to etoposide treatment. Left, representative images of intact (yellow) and broken (red and green) MLL loci. Scale bar 10 μm. The position of the FISH probes (red and green lines in top cartoon) flanking the MLL breakpoint cluster are shown. Right, data are the mean (± s.e.m.) of at least two independent experiments. Statistical significance was determined by t-test (***P< 0.001, NS, not significant). (B) Detection of broken MLL loci by FISH in KG1a cells following treatment with 100 μM etoposide for 3 h. Where indicated, cells were pre-incubated with DRB (100 μM) for 1 h prior to etoposide treatment. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by t-test (*P< 0.05, **P< 0.01). (C) Detection of broken MLL loci by FISH in KG1a shRNA LIG4 and control cells following treatment with 100 μM etoposide for 3 h. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by t-test (*P< 0.05).
Homologous recombination prevents TOP2-dependent genome instability
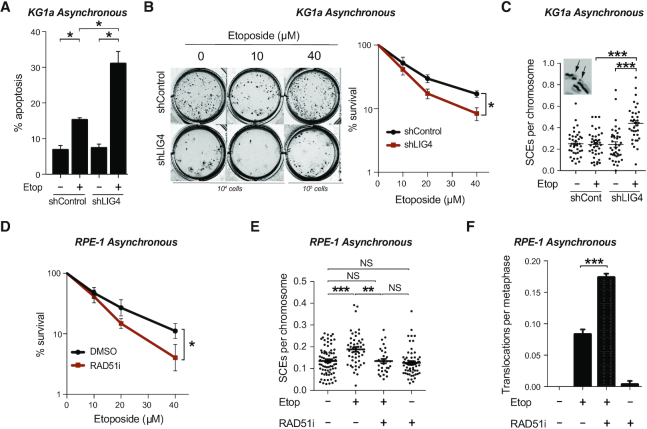
We next tested the effect of LIG4 depletion on the sensitivity of cycling cells to etoposide. In LIG4-deficient asynchronous KG1a cells, etoposide treatment resulted in a very drastic (up to 50%) increase in cell death, measured by FACS, compared with wild type cells (Figure 7A). In the long-term, clonogenic cell survival assays also showed hypersensitivity of LIG4-deficient cells to increasing concentrations of etoposide (Figure 7B and Supplementary Figure S10). We hypothesized that the repair of TOP2 breaks is increasingly dependent on HR in the absence of cNHEJ. To address this question, we analysed sister chromatid exchanges (SCEs), a well-established hallmark of HR. With low doses of etoposide, SCEs were increased considerably in LIG4-depleted cells (Figure 7C), indicative of increased HR, in agreement with a greater participation of a translocation-suppressing repair pathway in the absence of cNHEJ.
Figure 7.
Homologous recombination prevents chromosomal instability induced by TOP2. (A) Apoptosis analysis by FACS of KG1a shRNA LIG4 and control cells. Asynchronous cells were treated with 40 μM etoposide for 3h and incubated in drug-free medium for 48 h after treatment. Mean (± s.e.m.) of the percentage of Annexin-V positive cells of three independent experiments. Statistical significance was determined by t-test (*P< 0.05). (B) Clonogenic survival of KG1a shRNA LIG4 and control cells treated with the indicated concentration of etoposide for 3 h. After treatment, cells were washed and cultured in methylcellulose-containing medium for 14 days. Left, representative images of cultures. Right, quantification (mean ± s.e.m.) of three independent experiments. Statistical significance was determined by two-way ANOVA (*P< 0.05). (C) Sister chromatid exchanges of KG1a shRNA LIG4 and control cells treated with 10 μM etoposide for 30 min. After that, cells were washed and cultured in drug-free medium for 8 h, previous to metaphase spread preparation. See insets for representative image of SCEs. Mean (± s.e.m.) of SCE events per chromosome per metaphase from two independent experiments. Statistical significance was determined by t-test (***P< 0.001). (D) Clonogenic survival of RPE-1 cells with the indicated concentration of etoposide for 3 h. Where indicated cells were pre-incubated with RAD51 inhibitor (10μM) for 30 minutes prior to, during and 20 h after etoposide treatment. After treatment, cells were cultured in serum containing drug-free medium for 8 days. Data are the mean (± s.e.m.) of three independent experiments. Statistical significance was determined by two-way ANOVA (*P< 0.05). (E) Sister chromatid exchange of RPE-1 treated with 5 μM etoposide for 30 min. Where indicated cells were pre-incubated with RAD51 inhibitor (10 μM) for 30 min prior to, during and after etoposide treatment. Cells were cultured in drug-free medium for 6 h previous to metaphase spread preparation. Mean (± s.e.m.) of SCE events per chromosome per metaphase from two independent experiments. Statistical significance was determined by T-test (**P < 0.01, ***P< 0.001, NS, not significant). (F) Translocation frequencies (translocations per metaphase) in chromosome 8 and 11 were quantified in asynchronous RPE-1 cells in metaphase spreads prepared 24 h after etoposide treatment (1 h, 25 μM). Where indicated, cells were pre-treated with RAD51 inhibitor (10 μM) for 30 min prior to, during, and 6 h after etoposide treatment. Data are the mean (± s.e.m.) from three independent experiments. Statistical significance was determined by t-test (***P< 0.001).
Similar to previous observations in KG1a cells (Figure 5F), etoposide-induced chromosomal translocations were significantly reduced in LIG4-depleted asynchronous RPE-1 cells (Supplementary Figure S11A and C), whereas SCEs increased (Supplementary Figure S11B). On the other hand, disruption of HR by chemical inhibition of RAD51 with RI-1 (40) sensitized cells to etoposide (Figure 7D). Strikingly, RAD51 inhibition reduced etoposide-induced SCEs (Figure 7E) and significantly increased etoposide-induced chromosomal translocations (Figure 7F). Together these results demonstrate that cNHEJ mediates transcription-induced TOP2-dependent translocations while HR prevents them.
DISCUSSION
In this work, we have studied the TOP2 abortive activity linked to cellular transcription. Isolation of TOP2cc revealed that RNAP II inhibition leads to reduced levels of trapped TOP2ß in the presence of etoposide, suggesting that TOP2ß activity is closely associated with transcription in both cycling and non-cycling cells. These results support recent genome-wide studies that determined prominent TOP2 activity in gene bodies (6,41). Notably, reduction in TOP2 activity resulted in a proportional decrease in the accumulation of etoposide-induced DSBs, even in the absence of replication, demonstrating that transcription-dependent TOP2 activity can be a source of abortive DNA breaks. Although we cannot exclude residual transcription due to incomplete inhibition of RNAP II, the fact that DRB did not completely ablate DSB formation in quiescent cells suggests that there may be replication-independent processes other than transcription that can promote abortive TOP2cc. For example, these may be related to spatial organisation of the genome, since recent studies have shown that loop anchors, bound by CTCF and cohesin, can mediate TOP2ß abortive breaks (9,42,43). To what extent this activity accounts for transcription-induced DSBs remains controversial (14).
Inhibition of RNAP I did not modify TOP2ß activity nor DSB formation in quiescent cells. This result was unexpected, considering that nucleolar transcription accounts for the majority of RNAP activity in eukaryotic cells, and that both TOP2α and ß localize to nucleolus, contributing to RNAP I initiation and elongation (44,45). Interestingly, TOP2ß also co-localizes with CTCF and RAD21 in the spacer promoter region of rDNA (46). Further studies are warranted to investigate whether TOP2 functions differently in the nucleolus and at the sites of protein-coding genes.
Chemotherapeutic TOP2 poisons kill tumour cells by inducing high numbers of DSBs. The efficacy of these agents depends on proliferation rates (17,47). An emerging role of TOP2 in transcription regulation would perhaps predict a higher contribution of transcription to cell death than previously considered (48). However, here we have shown that transcription-induced TOP2 DSBs contribute very poorly to rapid etoposide-induced cell death. Rather, we observed long-term, transcription dependent, genotoxic effects of TOP2 poisoning. Notably, the long-term decrease in cell survival correlated with dose-dependent inactivating mutations in HPRT1, a surrogate marker for genome-wide mutagenesis. This was prevented by RNAP II inhibition, placing transcription as the main source of TOP2-induced genome instability. Our results suggest a model in which the deleterious effects of etoposide can persist after DSB repair (Supplementary Figure S12).
Distinction between long and short-term cell death induced by TOP2 poisons and elucidation contribution of transcription to genome instability (Supplementary Figure S12) has important implications for chemotherapy. We have also observed the formation of genomic translocations as a mutagenic outcome of DSB. Recurrent genomic translocations are of great interest, because these are key events in the development of some solid tumors and leukemias (49). Here we have shown, by direct scoring of chromosomal rearrangements, that TOP2-dependent chromosomal translocations are transcription-dependent, in accordance with our previous report in non-cycling RPE-1 cells (21) and a recent study by Gothe et al. in other cellular models (43).
We showed that cNHEJ is the major pathway involved in the repair of TOP2-dependent breaks in G0/G1, in agreement with the epistasis of Ku70 and TDP2 in etoposide sensitivity in avian cells (15). The role of canonical and alternative NHEJ pathways in the formation of genomic translocations in human cells has been recently revised (23), with nuclease and ionizing radiation-induced translocations being mediated by cNHEJ in human cells (22–24), and by altNHEJ in mouse cells (50,51). Strikingly, LIG4-deficiency suppressed transcription-induced genomic translocations in both cycling and asynchronous cells, demonstrating that genomic translocations induced by TOP2 during transcription result from cNHEJ-dependent repair. These results are contradictory to recently-published data that showed increased MLL translocations in LIG4−/− cells (43). Instead of using flanking probes to assess translocation, our study used whole chromosome FISH, as a direct measurement of chromosomal translocations, avoiding possible interference from phenomena such as DSB clustering (52). Other potentially significant differences in cell lines and experimental details may also explain the apparent contradiction between these two studies. Nevertheless, although we observe a major dependence on cNHEJ, we cannot totally exclude the contribution of altNHEJ, especially in the absence of a functional cNHEJ (45).
LIG4-deficient cells were hypersensitive to TOP2 poisons. Increases in SCEs indicates that the DNA breaks were repaired after DNA replication in these cells, in agreement with cNHEJ being an essential pathway for DSB repair, even in the presence of a sister chromatid. Still, minimal mutagenic effects of cNHEJ are very likely suppressed by HR, which is increased in the absence of LIG4. In accordance, HR suppression promoted TOP2-induced chromosomal translocations.
Therapy-associated chromosomal translocations are the cause of secondary leukemias. Translocations involving the breakpoint cluster region (BCR) of MLL are amongst the most common chromosome rearrangements observed in leukemia (20). Breakpoints in MLL are associated with transcription-dependent TOP2 activity (9,19,46) and depend on TDP2 activity for repair (21). We show here that transcription-dependent TOP2 breaks in MLL accumulate in LIG4-deficient cells revealing the role of cNHEJ in the repair of this oncogenic hot spot.
In summary, abortive TOP2 activity is a major endogenous threat to genome stability and cell survival (9,21,26), and transcription is associated with high levels of this activity (41–43). Our data demonstrate the marginal contribution of gene transcription to cell death induced by TOP2 poisons, but reveals a high risk of genome instability, promoted by the repair of transcription-dependent DSBs by cNHEJ. Our results suggest that combination therapy with TOP2 poisons plus transcription inhibitors may potentially result in reduced mutagenic side effects without significant loss in antineoplastic efficacy.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Sebastián Chavez, Jesús de la Cruz and Iván V. Rosado laboratories for reagents and discussion and Keith W. Caldecott and William Gittens for critical reading of the manuscript. Editorial assistance was provided by Stuart L. Rulten.
Authors contribution: J.J.O.-P. performed all experiments on KG1a cells. D.R.-C. performed all experiments on RPE-1 cells. J.O.-P. and D.R.C. made figures and revised the manuscript. F.G.-H. conceived the study, designed the experiments, interpreted the results and wrote the manuscript.
Contributor Information
Joaquín Olmedo-Pelayo, Instituto de Biomedicina de Sevilla (IBiS), Hospital Virgen del Rocío-CSIC-Universidad de Sevilla, 41013 Seville, Spain; Departamento de Genética, Universidad de Sevilla, 41080 Seville, Spain.
Diana Rubio-Contreras, Instituto de Biomedicina de Sevilla (IBiS), Hospital Virgen del Rocío-CSIC-Universidad de Sevilla, 41013 Seville, Spain; Departamento de Genética, Universidad de Sevilla, 41080 Seville, Spain.
Fernando Gómez-Herreros, Instituto de Biomedicina de Sevilla (IBiS), Hospital Virgen del Rocío-CSIC-Universidad de Sevilla, 41013 Seville, Spain; Departamento de Genética, Universidad de Sevilla, 41080 Seville, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Spanish Ministry of Science and Innovation [BFU2016-76446-P, RYC-2014-16665]; youth employment plan of the Andalusian government and the University of Seville (to J.O.-P.); predoctoral fellowship from the University of Seville [VI PPIT-US to D.R.-C.]. Funding for open access charge: Spanish Ministry of Science and Innovation [BFU2016-76446-P].
Conflict of interest statement. None declared.
REFERENCES
- 1. Baranello L., Kouzine F., Levens D.. DNA topoisomerases. Transcription. 2014; 4:232–237. [DOI] [PubMed] [Google Scholar]
- 2. Pommier Y., Sun Y., Huang S.-Y.N., Nitiss J.L.. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat. Rev. Mol. Cell Biol. 2016; 17:703–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Madabhushi R. The roles of DNA topoisomerase IIβ in transcription. IJMS. 2018; 19:1917–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ju B.G. A topoisomerase II -mediated dsDNA break required for regulated transcription. Science. 2006; 312:1798–1802. [DOI] [PubMed] [Google Scholar]
- 5. Madabhushi R., Gao F., Pfenning A.R., Pan L., Yamakawa S., Seo J., Rueda R., Phan T.X., Yamakawa H., Pao P.-C. et al.. Activity-induced DNA breaks govern the expression of neuronal early-response genes. Cell. 2015; 161:1592–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dellino G.I., Palluzzi F., Chiariello A.M., Piccioni R., Bianco S., Furia L., Conti G., Bouwman B.A.M., Melloni G., Guido D. et al.. Release of paused RNA polymerase II at specific loci favors DNA double-strand-break formation and promotes cancer translocations. Nat. Genet. 2019; 1011–1023. [DOI] [PubMed] [Google Scholar]
- 7. Nitiss J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009; 9:327–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ashour M.E., Atteya R., El-Khamisy S.F.. Topoisomerase-mediated chromosomal break repair: an emerging player in many games. Nat. Rev. Cancer. 2015; 15:137–151. [DOI] [PubMed] [Google Scholar]
- 9. Canela A., Maman Y., Jung S., Wong N., Callen E., Day A., Kieffer-Kwon K.-R., Pekowska A., Zhang H., Rao S.S.P. et al.. Genome organization drives chromosome fragility. Cell. 2017; 170:507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Capranico G., Tinelli S., Austin C.A., Fisher M.L., Zunino F.. Different patterns of gene expression of topoisomerase II isoforms in differentiated tissues during murine development. Biochim. Biophys. Acta. 1992; 1132:43–48. [DOI] [PubMed] [Google Scholar]
- 11. Hoa N.N., Shimizu T., Zhou Z.W., Wang Z.-Q., Deshpande R.A., Paull T.T., Akter S., Tsuda M., Furuta R., Tsusui K. et al.. Mre11 is essential for the removal of lethal topoisomerase 2 covalent cleavage complexes. Mol. Cell. 2016; 64:580–592. [DOI] [PubMed] [Google Scholar]
- 12. Cortés-Ledesma F., El-Khamisy S.F., Zuma M.C., Osborn K., Caldecott K.W.. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009; 461:674–678. [DOI] [PubMed] [Google Scholar]
- 13. Zeng Z., Cortes-Ledesma F., Khamisy El S.F., Caldecott K.W.. TDP2/TTRAP Is the Major 5′-Tyrosyl DNA phosphodiesterase activity in vertebrate cells and is critical for cellular resistance to topoisomerase II-induced DNA damage. J. Biol. Chem. 2010; 286:403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gómez-Herreros F. DNA double strand breaks and chromosomal translocations induced by DNA topoisomerase II. Front. Mol. Biosci. 2019; 6:3347–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gómez-Herreros F., Romero-Granados R., Zeng Z., Álvarez-Quilón A., Quintero C., Ju L., Umans L., Vermeire L., Huylebroeck D., Caldecott K.W. et al.. TDP2–dependent non-homologous end-joining protects against topoisomerase II–induced DNA breaks and genome instability in cells and in vivo. PLoS Genet. 2013; 9:e1003226-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R.. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017; 18:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013; 8:82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Markovits J., Pommier Y., Kerrigan D., Covey J.M., Tilchen E.J., Kohn K.W.. Topoisomerase II-mediated DNA breaks and cytotoxicity in relation to cell proliferation and the cell cycle in NIH 3T3 fibroblasts and L1210 leukemia cells. Cancer Res. 1987; 47:2050–2055. [PubMed] [Google Scholar]
- 19. Cowell I.G., Sondka Z., Smith K., Lee K.C., Manville C.M., Sidorczuk-Lesthuruge M., Rance H.A., Padget K., Jackson G.H., Adachi N. et al.. Model for MLL translocations in therapy-related leukemia involving topoisomerase IIβ-mediated DNA strand breaks and gene proximity. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8989–8994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wright R.L., Vaughan A.T.M.. A systematic description of MLL fusion gene formation. Crit. Rev. Oncol./Hematol. 2014; 91:283–291. [DOI] [PubMed] [Google Scholar]
- 21. Gómez-Herreros F., Zagnoli-Vieira G., Ntai I., Martínez-Macías M.I., Anderson R.M., Herrero-Ruíz A., Caldecott K.W.. TDP2 suppresses chromosomal translocations induced by DNA topoisomerase II during gene transcription. Nat. Commun. 2017; 8:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. So C.C., Martin A.. DSB structure impacts DNA recombination leading to class switching and chromosomal translocations in human B cells. PLoS Genet. 2019; 15:e1008101-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghezraoui H., Piganeau M., Renouf B., Renaud J.-B., Sallmyr A., Ruis B., Oh S., Tomkinson A.E., Hendrickson E.A., Giovannangeli C. et al.. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. Mol. Cell. 2014; 55:829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biehs R., Steinlage M., Barton O., Juhász S., Künzel J., Spies J., Shibata A., Jeggo P.A., Löbrich M.. DNA double-strand break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination. Mol. Cell. 2017; 65:671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haffner M.C., Aryee M.J., Toubaji A., Esopi D.M., Albadine R., Gurel B., Isaacs W.B., Bova G.S., Liu W., Xu J. et al.. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat. Genet. 2010; 42:668–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gómez-Herreros F., Schuurs-Hoeijmakers J.H.M., McCormack M., Greally M.T., Rulten S., Romero-Granados R., Counihan T.J., Chaila E., Conroy J., Ennis S. et al.. TDP2 protects transcription from abortive topoisomerase activity and is required for normal neural function. Nat. Genet. 2014; 46:516–521. [DOI] [PubMed] [Google Scholar]
- 27. Balmus G., Pilger D., Coates J., Demir M., Sczaniecka-Clift M., Barros A.C., Woods M., Fu B., Yang F., Chen E. et al.. ATM orchestrates the DNA-damage response to counter toxic non-homologous end-joining at broken replication forks. Nat. Commun. 2018; 10:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Thomson G., Watson A., Caldecott K., Denneny O., Depledge P., Hamilton N., Hopkins G., Jordan A., Morrow C., Raoof A. et al.. Generation of assays and antibodies to facilitate the study of human 5′-tyrosyl DNA phosphodiesterase. Anal. Biochem. 2013; 436:145–150. [DOI] [PubMed] [Google Scholar]
- 29. Schneider C.A., Rasband W.S., Eliceiri K.W.. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012; 9:671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nitiss J.L., Soans E., Rogojina A., Seth A., Mishina M.. Topoisomerase assays. Curr. Protoc. Pharmacol. 2012; doi:10.1002/0471141755.ph0303s57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Woessner R.D., Mattern M.R., Mirabelli C.K., Johnson R.K., Drake F.H.. Proliferation- and cell cycle-dependent differences in expression of the 170 kilodalton and 180 kilodalton forms of topoisomerase II in NIH-3T3 cells. Cell Growth Differ. 1991; 2:209–214. [PubMed] [Google Scholar]
- 32. Peltonen K., Colis L., Liu H., Trivedi R., Moubarek M.S., Moore H.M., Bai B., Rudek M.A., Bieberich C.J., Laiho M.. A targeting modality for destruction of RNA polymerase I that possesses anticancer activity. Cancer Cell. 2014; 25:77–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rogakou E.P., Boon C., Redon C., Bonner W.M.. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999; 146:905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang A., Lyu Y.L., Lin C.-P., Zhou N., Azarova A.M., Wood L.M., Liu L.F.. A protease pathway for the repair of topoisomerase II-DNA covalent complexes. J. Biol. Chem. 2006; 281:35997–36003. [DOI] [PubMed] [Google Scholar]
- 35. Roos W.P., Thomas A.D., Kaina B.. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer. 2016; 16:20–33. [DOI] [PubMed] [Google Scholar]
- 36. Bhattacharyya N.P., Skandalis A., Ganesh A., Groden J., Meuth M.. Mutator phenotypes in human colorectal carcinoma cell lines. Proc. Natl. Acad. Sci. U.S.A. 1994; 91:6319–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang M., Wu W., Wu W., Rosidi B., Zhang L., Wang H., Iliakis G.. PARP-1 and Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006; 34:6170–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Audebert M., Salles B., Weinfeld M., Calsou P.. Involvement of polynucleotide kinase in a Poly(ADP-ribose) polymerase-1-dependent DNA double-strand breaks rejoining pathway. J. Mol. Biol. 2006; 356:257–265. [DOI] [PubMed] [Google Scholar]
- 39. Robert I., Dantzer F., Reina-San-Martin B.. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 2009; 206:1047–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Budke B., Logan H.L., Kalin J.H., Zelivianskaia A.S., Cameron McGuire W., Miller L.L., Stark J.M., Kozikowski A.P., Bishop D.K., Connell P.P.. RI-1: a chemical inhibitor of RAD51 that disrupts homologous recombination in human cells. Nucleic Acids Res. 2012; 40:7347–7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gittens W., Johnson D.J., Allison R.M., Cooper T.J., Thomas H., Neale M.J.. A nucleotide resolution map of Top2-linked DNA breaks in the yeast and human genome. Nat. Commun. 2019; 10:4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Canela A., Maman Y., Huang S.-Y.N., Wutz G., Tang W., Zagnoli-Vieira G., Callen E., Wong N., Day A., Peters J.-M. et al.. Topoisomerase II-induced chromosome breakage and translocation is determined by chromosome architecture and transcriptional activity. Mol. Cell. 2019; 75:252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gothe H.J., Bouwman B.A.M., Gusmao E.G., Piccinno R., Petrosino G., Sayols S., Drechsel O., Minneker V., Josipovic N., Mizi A. et al.. Spatial chromosome folding and active transcription drive DNA fragility and formation of oncogenic MLL translocations. Mol. Cell. 2019; 75:267–283. [DOI] [PubMed] [Google Scholar]
- 44. Onoda A., Hosoya O., Sano K., Kiyama K., Kimura H., Kawano S., Furuta R., Miyaji M., Tsutsui K., Tsutsui K.M.. Nuclear dynamics of topoisomerase IIβ reflects its catalytic activity that is regulated by binding of RNA to the C-terminal domain. Nucleic Acids Res. 2014; 42:9005–9020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ray S., Panova T., Miller G., Volkov A., Porter A.C.G., Russell J., Panov K.I., Zomerdijk J.C.B.M.. Topoisomerase IIα promotes activation of RNA polymerase I transcription by facilitating pre-initiation complex formation. Nat. Commun. 2013; 4:1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Uusküla-Reimand L., Hou H., Samavarchi-Tehrani P., Rudan M.V., Liang M., Medina-Rivera A., Mohammed H., Schmidt D., Schwalie P., Young E.J. et al.. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016; 17:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nitiss J.L. Targeting DNA topoisomerase II in cancer chemotherapy. Nat. Rev. Cancer. 2009; 9:338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fan J.-R., Peng A.-L., Chen H.-C., Lo S.-C., Huang T.-H., Li T.-K.. Cellular processing pathways contribute to the activation of etoposide-induced DNA damage responses. DNA Repair (Amst.). 2008; 7:452–463. [DOI] [PubMed] [Google Scholar]
- 49. Mertens F., Johansson B., Fioretos T., Mitelman F.. The emerging complexity of gene fusions in cancer. Nat. Rev. Cancer. 2015; 15:371–381. [DOI] [PubMed] [Google Scholar]
- 50. Simsek D., Brunet E., Wong S.Y.-W., Katyal S., Gao Y., McKinnon P.J., Lou J., Zhang L., Li J., Rebar E.J. et al.. DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation. PLoS Genet. 2011; 7:e1002080-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhang Y., Jasin M.. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat. Struct. Mol. Biol. 2011; 18:80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aymard F., Aguirrebengoa M., Guillou E., Javierre B.M., Bugler B., Arnould C., Rocher V., Iacovoni J.S., Biernacka A., Skrzypczak M. et al.. Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 2017; 24:353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.