Abstract
The differentiation and regeneration of skeletal muscle from myoblasts to myotubes involves myogenic transcription factors, such as myocardin-related transcription factor A (MRTF-A) and serum response factor (SRF). In addition, post-transcriptional regulation by miRNAs is required during myogenesis. Here, we provide evidence for novel mechanisms regulating MRTF-A during myogenic differentiation. Endogenous MRTF-A protein abundance and activity decreased during C2C12 differentiation, which was attributable to miRNA-directed inhibition. Conversely, overexpression of MRTF-A impaired differentiation and myosin expression. Applying miRNA trapping by RNA affinity purification (miTRAP), we identified miRNAs which directly regulate MRTF-A via its 3′UTR, including miR-1a-3p, miR-206-3p, miR-24-3p and miR-486-5p. These miRNAs were upregulated during differentiation and specifically recruited to the 3′UTR of MRTF-A. Concomitantly, Ago2 recruitment to the MRTF-A 3′UTR was considerably increased, whereas Dicer1 depletion or 3′UTR deletion elevated MRTF-A and inhibited differentiation. MRTF-A protein expression was inhibited by ectopic miRNA expression in murine C2C12 and primary human myoblasts. 3′UTR reporter activity diminished upon differentiation or miRNA expression, whereas deletion of the predicted binding sites reversed these effects. Furthermore, TGF-β abolished MRTF-A reduction and decreased miR-486-5p expression. Our findings implicate miR-24-3p and miR-486-5p in the repression of MRTF-A and suggest a complex network of transcriptional and post-transcriptional mechanisms regulating myogenesis.
INTRODUCTION
Skeletal muscle tissue is one of the largest organ systems of all mammals and is essential for viability and motility. To guarantee constant operational capability even after severe damage, muscle can regenerate in relatively short time from satellite cells (reviewed by (1)). Upon damage or injury, satellite cells are activated from quiescence and start to divide asymmetrically to form a myoblast population (2). The differentiation of myoblasts is characterized by sequential expression of the muscle-restricted transcription factors MyoD and Myogenin, responsible for transcription of genes associated with e.g. cell-cycle arrest and muscle contractility (3,4). At late stages, myocytes accumulate myosin heavy chain polypeptides and fuse to form multinucleated myotubes. In addition, myogenesis requires post-transcriptional regulation by myogenic microRNAs (myo-miRNAs). Several myo-miRNAs are upregulated by e.g. MyoD and myogenin during myogenic differentiation including miR-1, -133 and -206, each of which inhibits various target RNAs (5,6).
In addition to muscle-restricted regulatory factors, several other factors are required for myogenic differentiation and myoblast fusion. Rho family GTPases and its downstream transcription factor, serum response factor (SRF), play important and dynamic roles during differentiation (7–11). Upon Rho-induced changes in actin dynamics, SRF-dependent transcription is controlled by its coactivator myocardin-related transcription factor A (MRTF-A) (12,13). The MRTF-SRF transcription factor module activates a plethora of targets including many muscle-specific genes (14,15). MRTF-A is implicated in skeletal muscle differentiation and homeostasis and is required for myogenesis in vivo and in vitro (16,17). Regeneration of skeletal muscle injuries correlates with increased MRTF-A protein in satellite cells (18). Yet, during myoblast fusion MRTF-A abundance declines, and overexpression of MRTF-A as well as active RhoA results in differentiation defects in vitro (11,19). The mechanism of MRTF-A regulation and its implication for myogenic differentiation is not well understood, however.
The immortalized murine cell line C2C12 is a widespread model to recapitulate skeletal muscle differentiation from myoblasts to myotubes. To identify novel mechanisms of MRTF-A regulation, we used C2C12 cells to functionally analyze the modulation of MRTF-A expression. Our study revealed post-transcriptional regulation as a crucial regulatory process for MRTF-A in myogenic differentiation. Novel miRNAs were identified using an unbiased miRNome-wide affinity purification and sequencing approach. Components of the RNAi pathway were recruited to the 3′UTR and required for MRTF-A regulation and C2C12 differentiation. We were able to validate the miRNAs miR-1a-3p, miR-206-3p, miR-24-3p and miR-486-5p for binding to cognate 3′UTR elements and inhibiting the MRTF-A protein expression in later stages of myogenic differentiation. Our results suggest a complex post-transcriptional regulation of MRTF-A abundance during myogenesis which critically affects differentiation.
MATERIALS AND METHODS
Plasmids and reagents
Primers for cloning, mutagenesis and qRT-PCR are listed in Supplementary Table S2. The SRF reporter plasmid p3D.A-Luc was described previously (12,20,21). Murine MRTF-A 3′UTR region was obtained by PCR from cDNA and cloned into pMIR-REPORT vector (Thermo Fisher, Waltham, MA, USA). Potential miRNA binding sites were deleted using SOMA mutagenesis (22) For miTRAP analysis, the murine MRTF-A 3′UTR was subcloned into pCDNA3.1-MS2 as described previously (23). For overexpression of the candidate miRNAs mirVana mimics (Thermo Fisher) were used. MRTF-A and Dicer1 knockdown was performed with ON-TARGETplus pool siRNA (Dharmacon, Lafayette, CO, USA) and RNAiMax (Thermo Fisher) following the manufacturers’ instructions. Hemagglutinin-tagged (HA) murine full length MRTF-A and a constitutively active variant (lacking the G-actin binding region derived from exons 3–5) were cloned from cDNA via PCR into pLVX-Puro (Clontech, Mountain View, USA). For transient transfection Lipofectamine 3000 was used following the manufacturer's instructions. TGF-β treatment (10 ng/ml, 6 h pretreatment) was performed using recombinant TGF-β1 (Promokine, Heidelberg, Germany).
Cell culture
Cell lines C2C12 (DSMZ – German collection of Microorganisms and Cell Cultures) and COS-7 were cultured subconfluently in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, 1 mM sodium pyruvate and antibiotic–antimycotic (Thermo Fisher) at 37°C and 5% CO2. Differentiation in C2C12 was induced by medium change to DMEM containing 2% horse serum (HS), 2 mM l-glutamine, 1 mM sodium pyruvate as well as antibiotic–antimycotic. To generate C2C12 cells carrying endogenous deletion of the MRTF-A 3′UTR, the CRISPR-Cas9 approach was used. Two guide RNAs (sg1 5′-ACAGGAGCGTACACTGGGTA-3′ and sg2 5′-GTGCTGACTCAATGCAGAGA-3′) were designed by using the tool CHOP-CHOP (24) (https://chopchop.cbu.uib.no/) to delete the majority of the 3′UTR from the MRTF-A gene locus without affecting the coding region or the polyadenylation signal. SgRNA sequences were cloned into lentiCRISPR-V2 vector (Addgene; Plasmid # 52961, a gift from Feng Zhang). C2C12 cells were lentivirally infected and selected with puromycin for one week. CRISPR and control pools of C2C12 cells were characterized for genetic deletion by PCR and sequence analysis.
Primary human skeletal muscle myoblasts (HSMM, Lonza, Basel, Switzerland; Cat. no. CC-2580) were cultured in the SkBM-2 medium (Lonza, CC-3246). Human immortalised LHCN-M2 myoblasts (Evercyte, Vienna, Austria; Cat. no. CkHT-040-231-2) were cultured in MyoUp medium (Evercyte, MHT-040). Cells were lentivirally infected with empty vector, full-length MRTF-A or a constitutively active MRTF-A variant. Following seeding after 48 h, differentiation was induced by changing the medium (DMEM/M199 4:1, HEPES 0.02 M, Zinc Sulfate 0.03 μg/ml, Vitamin B12 1.4 μg/ml, insulin 10 μg/ml and apo-transferrin 100 μg/ml) and replacing it every second day.
Reverse transcription-quantitative PCR (qRT-PCR)
RNA was extracted using the miRCURY™ RNA Isolation Kit – Cell & Plant (QIAGEN, Hilden, Germany) and miScript II RT Kit (QIAGEN) following the manufacturer's protocols. For cDNA synthesis 1 μg of RNA was used. Real-time PCR amplification and analysis was performed using a LightCycler 480 with DyNAmo ColorFlash SYBR Green (Roche, Mannheim, Germany) and the primers listed in Supplementary Table S2. For normalization, HPRT1, YWHAZ and RPS29 mRNAs were used as controls. miRNA quantification was performed by using the miRNA miScript Primer assay and miScript SYBR Green PCR Kit (QIAGEN). U6, SNORD61 and SNORD95 (QIAGEN) were used as controls. Calculations of relative changes in gene expression were done using the ΔΔ cycle threshold (Ct) method (25). For statistical analysis the unpaired two sample Student's t-test was applied, using the software GraphPad prism V 7.03 (GraphPad Software, San Diego, USA).
Reporter assays
4.5 × 104 COS-7 cells per 24-well were reversely co-transfected using Lipofectamine 3000 (Thermo Fisher) with 10 ng pMIR_REPORT_MRTF-A_3′UTR, 10 ng pMIR-REPORT_Beta-Gal, 480 ng pEF and 10nM mirVana mimics 48 h prior to harvest. Luciferase activity was normalized to Beta-Gal values. For C2C12, 4 × 104 cells per 12-well were seeded 24 h prior to transfection with 100 ng pMIR_Report_MRTF-A_3′UTR, 100 ng pMIR-REPORT_Beta-Gal and 800 ng pEF and incubated for 48 h followed by harvest or induction of differentiation. MRTF-A promoter activity of C2C12 cells (8.75 × 104, 12-well) over the course of myogenic differentiation was determined by using the Dual-Glo luciferase kit (Promega, Madison, WI, USA) following transfection with 500 ng p3D.A-Luc, 50 ng ptkRL and 450 ng pEF.
Western blot analysis
Western blot analysis was performed following standard protocols. Primary antibodies against MRTF-A (dilution: 1:1000, Cell Signaling), Tubulin (1:5000, Sigma), HA (1:1000, Sigma), Myogenin (1:500, Santa Cruz), Myosin Heavy chain 1/2/4/6 (1:1000, Santa Cruz), Dicer1 (1:500, Santa Cruz), Vinculin (1:1000, Sigma), AGO2 (1:1000, Abcam) and maltose binding protein (1:1000, Abcam) were incubated overnight at 4°C. Fluorophore conjugated secondary antibodies IRDye 700 or IRDye 800 (1:15000, LI-COR Biosciences) were incubated for 1 h at room temperature. Imaging and quantification was done using the Odyssey Image Scanner System with the software Image Studio V 3.1.4 (LI-COR Biosciences, Cambridge, UK), as described (26).
miTRAP (miRNA trapping by RNA in vitro affinity purification)
The miTRAP was essentially performed as described previously (23,27,28). In short, amylose beads (New England Biolabs) were incubated with purified maltose binding protein-MS2 binding protein complex. The MRTF-A 3′UTR fused to a MS2 loop was transcribed in vitro and immobilized on the MBP-MS2BP coated amylose beads prior to incubation with cell lysates of undifferentiated or differentiated C2C12 cells. Subsequently the RNA–protein complexes were eluted and purified. The final eluate was sequenced via deep sequencing using an Illumina HighScan sequencer (Illumina, San Diego, USA) with version 3 chemistry and flow cell following the manufacturer's instructions. Sequencing data processing was performed by Knut Krohn at the University of Leipzig. The 50 bp reads were demultiplexed using the software CASAVA v1.8.2 (Illumina) and trimmed with ‘cutadapt’. Reads with a length of 15–27 bases were kept and aligned to Mus musculus (UCSC mm10) mature sequences of miRBase v20 (29) using the software bowtie (30). Differential miRNA expression was analyzed using the edgeR software package (31). The weighted trimmed mean of M-values (TMM) method (32) was used for normalization of the datasets and served as basis for the calculation of ‘pseudo’ counts per million (CPM). Only miRNAs with an average abundance of more than 10 CPM were considered.
RESULTS
MRTF-A protein levels are downregulated during myogenic differentiation
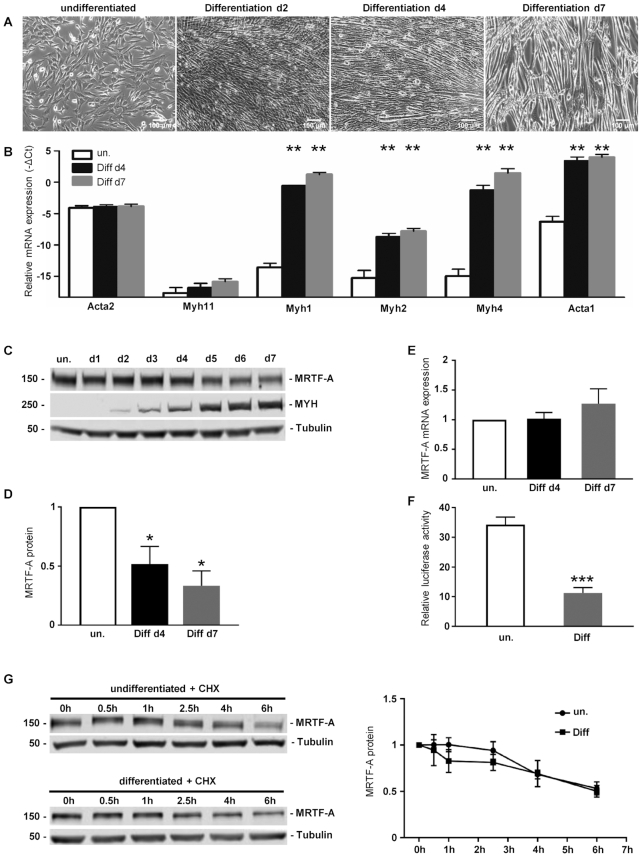
The murine skeletal muscle cell line C2C12 is able to recapitulate terminal differentiation from myoblasts to myotubes. To elucidate this process, C2C12 cells were seeded subconfluently and differentiation was induced by changing to medium that contained 2% horse serum. At day 2, the cells were increasingly spindle-shaped and aligned to each other (Figure 1A, d2). The first multinucleated myotubes were observable at day 4 (Figure 1A, d4) with matured myotubes at day 7 (Figure 1A, d7). Subsequently the differentiation process was also verified by RT-qPCR. Skeletal muscle associated genes Myh1, Myh2, Myh4 and Acta1 were hardly detectable in undifferentiated C2C12 cells (Figure 1B). After induction of differentiation the genes were upregulated, with highest levels at day 7 of myogenesis (Figure 1B). In contrast, smooth muscle cell differentiation related genes Acta2 and Myh11 showed little change of expression during myogenesis (Figure 1B). Furthermore, western blot confirmed the upregulation of myosin heavy chain polypeptides MYH1, MYH2, MYH4 and MYH6 over the course of differentiation (Figure 1C).
Figure 1.
MRTF-A protein level is downregulated over the course of myogenic differentiation. (A) Phase-contrast images (10×) of C2C12 cells upon differentiation for 2, 4 and 7 days (d2, d4, d7), in comparison to undifferentiated control cells (un.). (B) Increased expression of skeletal muscle associated genes (Myh1, Myh2, Myh4 and Acta1) in differentiating C2C12 cell. The indicated mRNAs were quantified by qRT-PCR and normalized to a set of housekeeping mRNAs. Shown is the –ΔCt value as a log2-measure for the relative mRNA expression. (C) Immunoblots of lysates from differentiating C2C12 cells, using antibodies against MRTF-A, myosin heavy chain polypeptides 1, 2, 4 and 6 (MYH), and alpha-tubulin as a control. (D) Quantification of MRTF-A immunoblots at myogenic differentiation for 4 and 7 days, normalized to undifferentiated control cells. (E) MRTF-A mRNA expression over the course of differentiation. (F) MRTF-SRF transcriptional activity in undifferentiated and differentiated cells, measured by relative luciferase activity of a MRTF-dependent promoter reporter construct. (G) Analysis of MRTF-A protein stability in undifferentiated and differentiated C2C12 by using a cycloheximide (CHX) chase assay. Left panels: representative immunoblots of MRTF-A and tubulin protein after incubation of cells with CHX for the times indicated. Right panel: Quantification of the relative MRTF-A amount of three independent experiments. Error bars, SEM (n = 3); *P< 0.05, **P< 0.01, ***P< 0.001 (Student’s t-test). Size bar, 100 μm.
Myogenic differentiation is modulated by the MRTF-SRF transcription factor module. To determine the influence of myogenic differentiation on MRTF-A expression we analyzed the expression pattern by RT-qPCR and Western blot. Interestingly MRTF-A protein levels decreased over the course of differentiation (Figure 1C and D), while in contrast mRNA expression did not change significantly (Figure 1E). Subsequently we used a MRTF-dependent luciferase reporter to determine the MRTF/SRF transcriptional activity. This promoter assay showed a myogenesis dependent decrease in activity in differentiated C2C12 compared to the undifferentiated cells (Figure 1F).
To elucidate the possible reason for the reduced MRTF-A protein level and function, we performed a cycloheximide chase assay to analyze alterations in MRTF-A protein stability during myogenesis. Undifferentiated and differentiated C2C12 cells were treated with cycloheximide to estimate the MRTF-A protein half life time. Decreased MRTF-A levels over the course of differentiation were not caused by obviously abated protein stability (Figure 1G). Together the results suggest that MRTF-A could be regulated post-transcriptionally, as mRNA expression and protein stability did not change considerably over the course of differentiation.
Deregulated MRTF-A expression inversely correlates with myogenic differentiation
To ascertain the impact of MRTF-A deregulation on myogenic differentiation, C2C12 cells transiently overexpressing HA-tagged MRTF-A were induced for differentiation. The early marker of myoblast differentiation, the transcription factor myogenin, was only slightly reduced compared to the vector control (Figure 2A). In contrast the late marker of skeletal muscle differentiation, the myosin heavy chain polypeptides, showed a significant decrease at day 4 of differentiation, indicative for reduced differentiation upon elevated MRTF-A activity (Figure 2A). Phase-contrast microscopy confirmed that myotube formation of C2C12 cells was impaired by overexpression of MRTF-A (Figure 2B).
Figure 2.
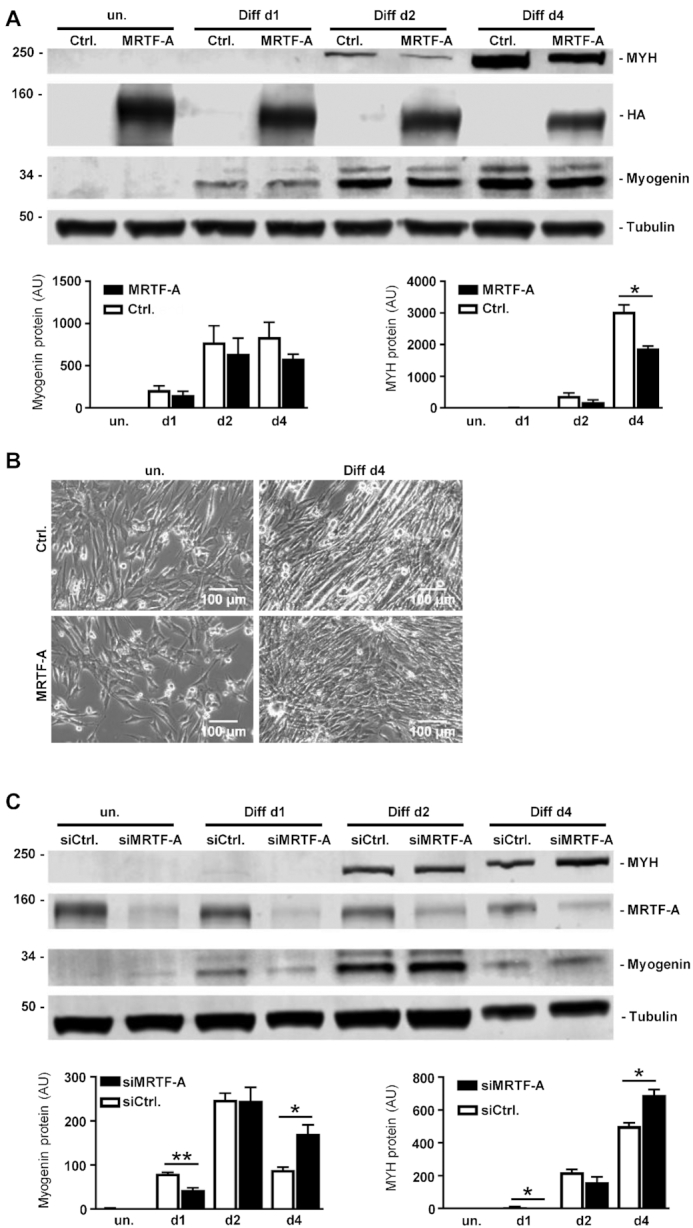
Experimental changes of MRTF-A expression inversely correlate with myogenic differentiation. (A) C2C12 cells were transfected with HA-tagged MRTF-A or control vector (Ctrl.) 48 h prior to induction of myogenic differentiation. (B) Phase contrast images of cells overexpressing HA-tagged MRTF-A. (C) C2C12 differentiation upon partial depletion of MRTF-A using siRNA. Shown are immunoblots for myosin heavy chain polypeptides, HA-tag, myogenin and tubulin of lysates collected at the times indicated. Quantification of relative fluorimetric intensities of three independent experiments are expressed below by bar charts. Error bars, SD (n = 3); *P< 0.05, **P< 0.01 (Student’s t-test). Size bar, 100 μm.
To test whether human myoblasts respond comparably and to achieve MRTF-A overexpression more persistently, human immortalized LHCN myoblasts were lentivirally infected with MRTF-A-HA or a constitutively active version thereof. Genetic fusion with GFP via a self-cleaving peptide (P2A) enabled us to verify a high transduction efficiency and persistence of MRTF-A overexpression (data not shown). Similar to C2C12 murine myoblasts, differentiation of human LHCN cells was impaired by elevated MRTF-A activity, as evident by reduced myogenic marker expression and lack of myotube formation during 6 days of differentiation (Supplementary Figure S1).
Conversely, we partially depleted MRTF-A in undifferentiated C2C12 myoblasts using siRNA, followed by myogenic differentiation. The myogenin level was decreased in the early phase of differentiation, whilst at later differentiation myogenin protein was increased upon MRTF-A knockdown compared to control (Figure 2C). The same pattern was observable for the myosin heavy chain isoforms during myogenesis. While in the early phase of differentiation the myosin heavy chains were not detectable in the knockdown, higher protein levels were ascertainable in the knockdown at day 4 compared to the control (Figure 2C). These results indicate delayed differentiation kinetics upon MRTF-A knockdown, affecting both early and late markers of myoblast-to-myotube differentiation. Together, a tight temporal regulation of MRTF-A appears to be crucial for proper myogenic differentiation.
Identification of miRNAs regulating MRTF-A
Since the MRTF-A mRNA expression and protein stability was not changed over the course of differentiation, we investigated other explanations for the observed changes in MRTF-A protein amounts. To elucidate the role of post-transcriptional regulation by miRNAs on MRTF-A, miRNA trapping by RNA affinity purification (miTRAP) was used. The entire in vitro transcribed MRTF-A 3′UTR containing a MS2-loop sequence functioned as a bait for MRTF-A regulating miRNA candidates in lysates of differentiated versus non-differentiated C2C12 cells (Figure 3A). To control for the specificity of the pulldown and analyze a potential recruitment of the RISC complex to the 3′UTR of MRTF-A, lysates and eluates were immunoblotted with antibodies against AGO2, vinculin and the maltose binding protein (MBP). Strikingly, AGO2 binding to the MRTF-A 3′UTR was strongly increased upon differentiation, indicating elevated RISC-dependent regulation of MRTF-A expression by miRNAs upon differentiation (Figure 3B). Moreover, AGO2 recruitment was specific for the 3′UTR and hardly detectable in the MS2-loop control, whilst the total AGO2 level did not alter considerably in undifferentiated and differentiated C2C12 cells. For validation, maltose binding protein as part of the amylose bead complex was solely and equally detectable in the eluates, while vinculin as a non-RNA binding protein was exclusively detectable in the lysates (Figure 3B).
Figure 3.
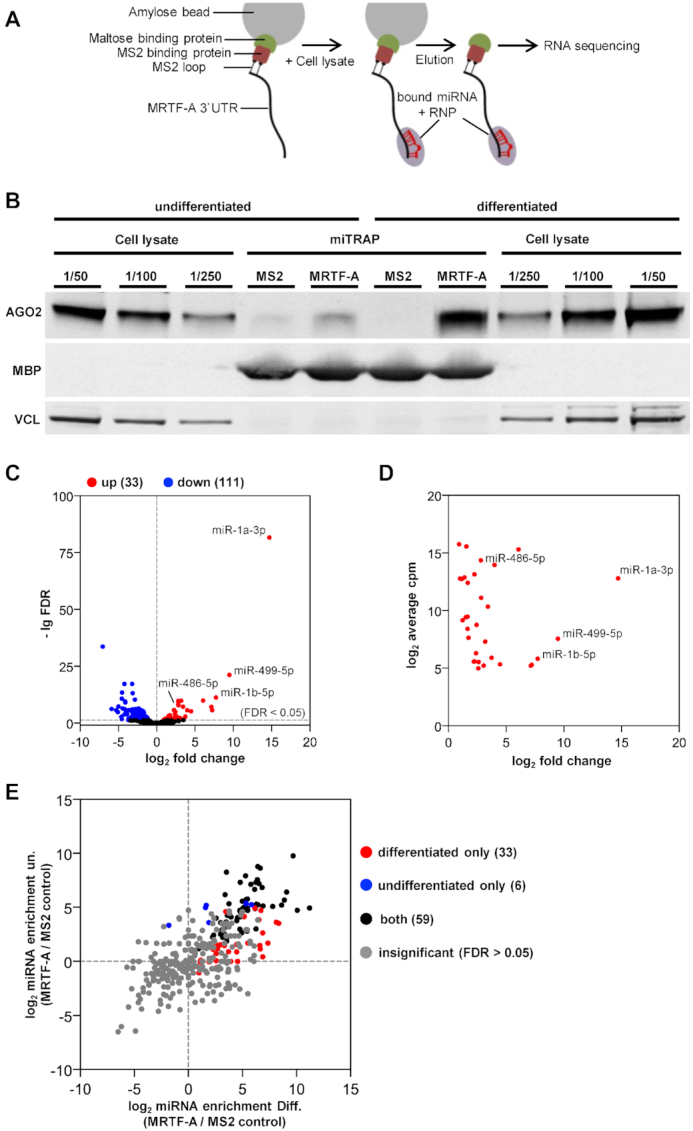
Identification of miRNAs and Ago2 protein recruited to the 3′UTR of MRTF-A using miTRAP. (A) Schematic depiction of the miTRAP approach. The 3′UTR of MRTF-A was immobilized to amylose beads and incubated with cell lysates from undifferentiated and differentiated C2C12 cells followed by elution of the bound miRNAs and RNPs. Eluates of three independent biological replicates were sequenced via miRNA sequencing and bioinformatically analyzed to identify differentially enriched miRNAs. (B) Total cell lysates (decreasing amounts of 1/50, 1/100, 1/250) and eluates (miTRAP) of the immobilized 3′UTR (MRTF-A) and the MS2 loop control (MS2) were immunoblotted for AGO2. Maltose binding protein (MBP) and the non-RBP vinculin (VCL) were blotted to control for equal binding and loading. (C) Volcano plot of sequenced miRNAs up- or downregulated in total lysates of differentiated C2C12 cells. A threshold false discovery rate (FDR) of 5% is indicated. (D) Cellular abundance of sequenced miRNAs induced upon differentiation. The averaged counts per million (CPM) are plotted against the fold change. (E) Enrichment analysis of sequenced miRNAs following miTRAP. Ratios of individual miRNA bound to MRTF-A 3′UTR versus MS2 control in undifferentiated (un.) and differentiated (Diff.) were plotted. Red and blue dots indicate miRNAs specifically recruited to MRTF-A 3′UTR in differentiated and undifferentiated conditions, respectively. Black dots represent miRNA significantly enriched in both, while gray dots were not significantly enriched at a threshold FDR of 5% (n = 3).
Next we performed miRNA sequencing of the total cell lysates and the miTRAP eluates of three independent biological replicates. On average, we obtained around 502 000 and 392 000 sequences in total cell libraries of undifferentiated and differentiated C2C12 cells, respectively. Upon normalization and statistical analysis using a false discovery rate (FDR) of 0.05, 111 miRNAs were down- and 33 were upregulated during differentiation (Figure 3C; Supplementary Table S1A, B). Significantly upregulated miRNAs included many well-known myo-miRs which were highly abundant, e.g. miR-1a-3p, miR-1b-5p, miR-499-5p and miR-486-5p (Figure 3D). In the miTRAP eluates, a total of 98 miRNAs were significantly enriched (FDR < 0.05; CPM > 10) by binding to the MRTF-A 3′UTR compared to the MS2-loop control (Figure 3E, Supplementary Table S1C). Of these, 33 (red dots) were specifically recruited to MRTF-A only when lysates of differentiated cells were used. In addition, 59 miRNAs (black dots) were selectively enriched from both differentiated and undifferentiated cell lysates. Only six miRNAs associated with the MRTF-A 3′UTR exclusively in undifferentiated cell lysates. This suggests that the majority of MRTF-A specific miRNAs is found either in both or in differentiated conditions.
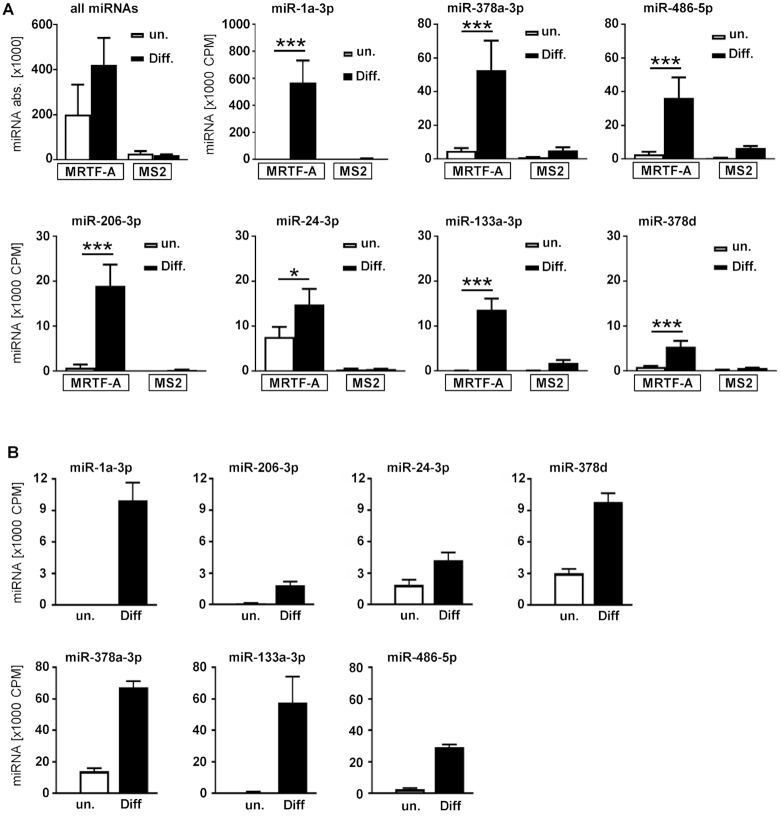
Notably, 16 miRNAs associating under both conditions or only in differentiated cell lysates were significantly upregulated upon differentiation, whereas 32 were significantly decreased (Supplementary Table S1A–C). However, the number of sequenced miRNAs bound to the 3′UTR of MRTF-A more than doubled in eluates of differentiated C2C12 cells, consistent with an increased post-transcriptional regulation (Figure 4A, top left). To identify individual miRNAs which likely regulate MRTF-A upon differentiation and are amenable to experimental characterization, three criteria were applied: Firstly, miRNAs should show at least a twofold enrichment in total lysates of differentiated cells. Secondly, candidate miRNAs should be enriched at least 1.5-fold in eluates from differentiated versus undifferentiated cells following binding to the MRTF-A 3′UTR. Finally, to avoid false positive results caused by nonspecific miRNA binding, the potential candidate's enrichment to the MRTF-A 3′UTR compared to the MS2-loop control was set to fourfold.
Figure 4.
Enrichment of miRNAs by binding to the MRTF-A 3′UTR and by myogenic differentiation. (A) Number of sequenced miRNA fragments in miTRAP eluates of undifferentiated or differentiated cells in comparison to the MS2 control. All miRNAs (upper left) and the selected miRNAs are presented independently. (B) Abundance of miRNA sequences in total lysates of undifferentiated and differentiated C2C12. Shown is the expression change of the candidate miRNA during differentiation of C2C12. CPM; counts per million. Error bars, SD (n = 3); *P< 0.05, **P< 0.01, ***P< 0.001 (Student’s t-test).
Of the 92 miRNAs which were significantly enriched to the MRTF-A 3′UTR upon differentiation (Figure 3E), seven conformed to these additional criteria, including miR-1a-3p, miR-206-3p, miR-24-3p, miR-133a-3p, miR-378a-3p, miR-378d and miR-486-5p. Characterization of these selected miRNAs showed that they were highly present in the miTRAP eluates, but much reduced when undifferentiated cells were investigated (Figure 4A). Except for miR-24-3p, which is not yet known as a myo-miR, this may partially reflect their induced expression upon myogenic differentiation (Figure 4B). However, miR-24-3p, miR-206-3p, miR-378a-3p, miR-378d and miR-486-5p were also expressed at low levels in undifferentiated cells, explaining their significant miTRAP enrichment over the MS2 control in both conditions, as shown in Figure 3E (black dots). Nevertheless, quantification by both miRNA sequencing and qRT-PCR showed that the selected miRNAs increased manyfold in differentiated C2C12 lysates, consistent with an inducible role of miRNA-mediated MRTF-A regulation during myogenesis (Figure 4B; Supplementary Figure S2).
MRTF-A reduction and myogenic differentiation requires post-transcriptional regulation via the 3′UTR
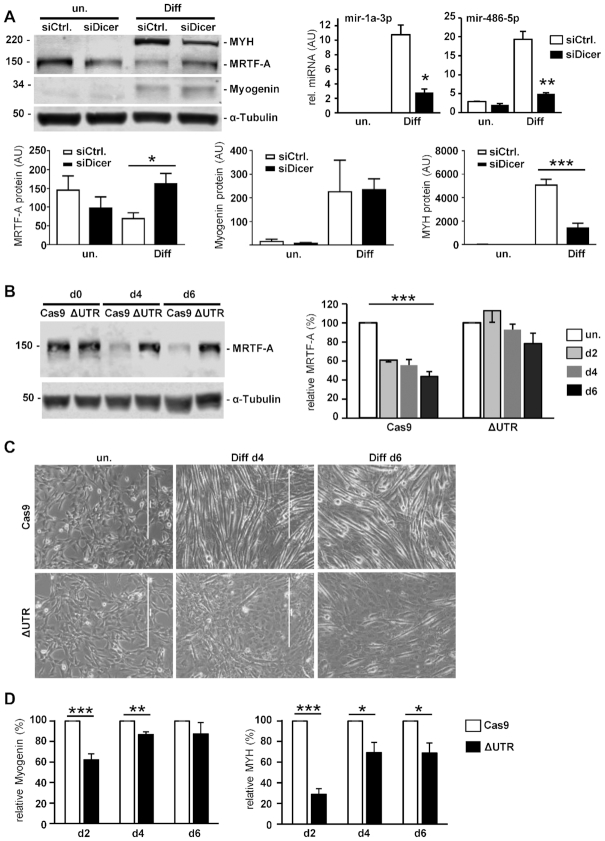
To further analyze a possible post-transcriptional regulation of MRTF-A and its role in myogenic differentiation, we performed a siRNA-mediated knockdown of Dicer1 as a crucial member of the RNAi pathway. Dicer1 knockdown correlated with decreased levels of miR-1a-3p and miR-486-5p. The depletion of Dicer1 lead to an increased MRTF-A protein level and impaired myogenic differentiation, as demonstrated by reduced myosin heavy chain expression (Figure 5A). In contrast, myogenin expression as an earlier marker of myoblast differentiation was not visibly reduced. These findings corroborate the enhanced Ago2 recruitment to the MRTF-A 3′UTR (Figure 3B) and implicate post-transcriptional regulation via miRNAs in skeletal muscle differentiation.
Figure 5.
Dicer and the 3′UTR are required for regulation of MRTF-A and myogenic differentiation. (A) Transient knockdown of Dicer1 using siRNA. After 48 h of incubation, myogenic differentiation was induced in C2C12 when indicated. miR-1a-3p and miR-486-5p were quantified by qRT-PCR for validation (top right). The proteins MYH, MRTF-A, myogenin and tubulin were compared to a control transfection (siCtrl.) by immunoblotting (top left). Quantification of relative protein amounts is depicted below. Error bars, SD (n = 3) (B–D) The 3′UTR at the endogenous locus of MRTF-A was deleted using CRISPR-Cas9. Selected pools of C2C12 cells were analysed for protein expression and differentiation. (B) Immunoblot and quantification of MRTF-A protein level in cells with deleted 3′UTR (ΔUTR) compared to control cells lacking the sgRNA (Cas9) (C) Phase contrast images (10x). (D) Quantified myogenin and MYH protein expression, shown as percentage from the Cas9 control cells at d2, d4 and d6 of differentiation Error bars, SEM (n = 4); *P< 0.05, **P< 0.01, ***P< 0.001 (Student’s t-test). Size bar, 400 μm.
To investigate whether MRTF-A is directly regulated post-transcriptionally, the 3′UTR was deleted at the endogenous MRTF-A locus by CRISPR–Cas9. Pools of infected and selected C2C12 cells were validated by PCR and sequencing, showing that the majority of the 3′UTR between the STOP and the polyadenylation signal was successfully deleted (not shown). These ΔUTR C2C12 cells showed an elevated MRTF-A protein level over the course of the differentiation, compared to the Cas9 control cells with significantly decreasing MRTF-A expression (Figure 5B). In addition, myotube formation was impaired in cells with deleted MRTF-A 3′UTR (Figure 5C). Markers of myogenic differentiation were reduced in the 3′UTR deleted cells, most pronounced for MYH and at earlier time points of differentiation (Figure 5D). This suggests that posttranscriptional regulation of the MRTF-A 3′UTR is critical during myogenic differentiation.
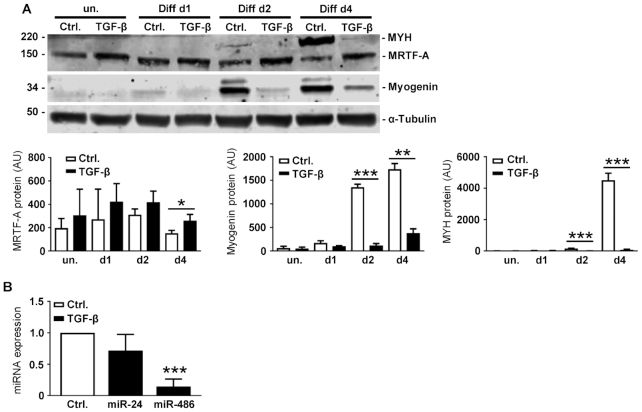
C2C12 differentiation can also be inhibited by TGF-β signaling. TGF-β acts as a repressor for premature skeletal muscle regeneration after damage and stimulates pro-inflammatory responses (33,34). We therefore incubated C2C12 cells with recombinant TGF-β1, followed by induction of differentiation. TGF-β treatment increased MRTF-A protein expression and prevented its drop upon differentiation (Figure 6A). Concomitantly, the early and late differentiation markers myogenin and MYH were effectively inhibited by TGF-β pretreatment. In addition, we analyzed whether TGF-β also affects expression of the miRNA identified by miTRAP. In undifferentiated C2C12, only miR-24-3p and miR-486-5p were endogenously expressed at detectable levels. In comparison to the untreated control, TGF-β1 decreased the expression of miR-24-3p only slightly, but a significant 80% reduction of miR-486-5p was observable in undifferentiated C2C12 (Figure 6B). This raises the possibility that TGF-β affects MRTF-A expression and myogenic differentiation partially by miR inhibition (see Discussion).
Figure 6.
Involvement of TGF-β pathways in regulating MRTF-A and myogenic differentiation. (A) C2C12 cells were treated with TGF-β1 or DMSO (Ctrl.) 6 h prior to induction of myogenic differentiation. Shown are representative immunoblots of lysates after the differentiation time indicated. Quantification is depicted below. (B) Undifferentiated C2C12 cells were treated with TGF-β1. After 48 h of incubation the cells were harvested and expression of miR-24-3p and miR-486-5p was determined. Expressed as normalized changes in comparison to untreated control cells. Error bars, SD (n = 3); *P< 0.05, **P< 0.01, ***P< 0.001 (Student’s t-test).
miR-1a-3p, miR-24-3p, miR-206-3p and miR-486-5p reduce MRTF-A expression
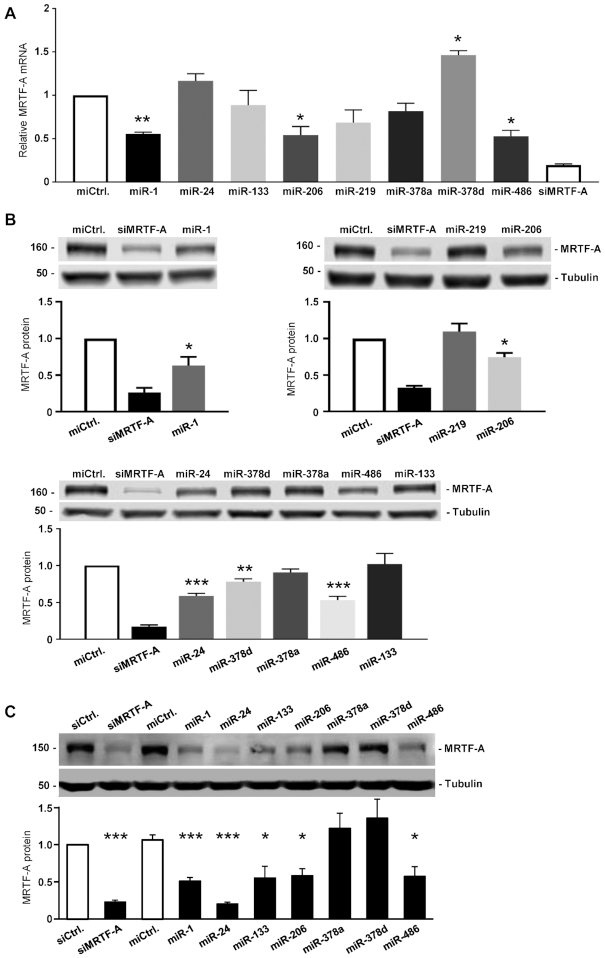
To elucidate the roles of the miRNAs identified via miTRAP, murine and human myoblasts were transfected with miRNA mimics and analyzed by RT-qPCR and Western blot. The miTRAP identified candidate miRNAs miR-1a-3p, miR-206-3p and miR-486-5p decreased endogenous MRTF-A mRNA levels upon overexpression in undifferentiated C2C12 cells (Figure 7A). For comparison, a synthetic siRNA targeting the MRTF-A ORF was used as a positive control. miR-1a-3p, miR-206-3p and miR-486-5p also efficiently reduced the MRTF-A protein amounts analyzed by western blotting (Figure 7B). In contrast to the RT-qPCR, the western blotting revealed that miR-24-3p and miR-378d also had a decreasing effect on the MRTF-A protein level (Figure 7B). However, the mouse miRNAs miR-378a-3p and miR-378d are identical in the seed region and share the same predicted binding sites, arguing against a biological relevance of the slight MRTF-A reduction observed in murine C2C12 cells. For analyzing the human system, primary human skeletal myoblasts were transfected with the corresponding human miRNAs. Expression of MRTF-A protein was reduced by miR-1a-3p, miR-24-3p, miR-133a-3p, miR-206-3p and miR-486-5p in primary human myoblasts, whilst two negative controls and miR-378a/d did not show any effect (Figure 7C). Taken together, the results indicate that miR-1/miR-206, miR-24-3p and miR-486-5p are robust and conserved regulators of MRTF-A in both murine and primary human myoblasts, whereas miR-378a/d, miR-133 and miR-219 are not.
Figure 7.
Expression of miRNA candidates affects the MRTF-A level in murine and human myoblasts. (A) MRTF-A mRNA expression in undifferentiated C2C12 cells transfected with the indicated miRNA for 48 h. A control miRNA (miCtrl.) and a MRTF-A-specific siRNA (siMRTF-A) were used as a negative and positive control, respectively. (B) MRTF-A protein level upon transient miRNAs transfection in C2C12 cells. (C) MRTF-A protein level upon miRNAs transfection for 72 h in primary human HSMM skeletal muscle myoblasts. Normalized protein quantification is shown below each blot. Error bars, SEM (n = 3); *P< 0.05, **P< 0.01, ***P< 0.001 (Student’s t-test).
miR-1a-3p, miR-24-3p and miR-486-5p specifically regulate the 3′UTR of MRTF-A
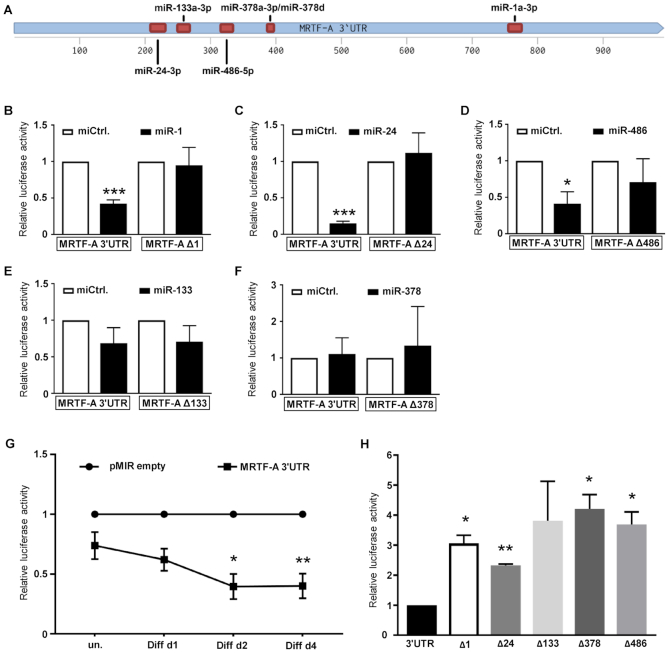
To evaluate the specificity of miR-dependent regulation of MRTF-A expression, the 3′UTR of the murine MRTF-A gene was cloned into the pMIR-REPORT luciferase reporter system (Figure 8A). COS-7 cells were co-transfected with the reporter constructs and the miRNA. miR-1a-3p, miR-24-3p and miR-486-5p significantly decreased the 3′UTR-dependent luciferase activity (Figure 8B–D), whilst miR-133-3p and miR-378d failed to affect reporter activity (Figure 8E and F). These results were consistent with the analysis in C2C12 cells. Since miR-206-3p is largely identical, functionally redundant to miR-1a-3p and previously reported, the effect of miR-206-3p was not tested further. Analysis of the MRTF-A 3′UTR revealed binding sites for miR-1a-3p/miR-206-3p, miR-24-3p and miR-486-5p including their seed sequences (Figure 8A). Deletion of the predicted miRNA binding sites largely abolished the repression of the 3′UTR reporter by their respective miRNAs, demonstrating a molecular connection to the MRTF-A regulation observed (Figure 8B–D).
Figure 8.
miR-1a-3p, miR-24-3p and miR-486-5p regulate MRTF-A via recognition sites on its 3′UTR. (A) Schematic of the 3′UTR of murine MRTF-A with the STOP-codon at position 1 and the polyadenylation signal at 985. Deleted miRNA binding sites are indicated by red boxes. (B–F) COS-7 cells were transiently transfected with pMIR-REPORT luciferase reporters, harboring the 3′UTR of murine MRTF-A or mutants thereof with a deletion of the predicted binding sites. The indicated miRNAs were co-transfected and their effects compared to a control miRNA (miCtrl.). After 48 h luciferase activity was determined and normalized to co-transfected galactosidase activity. (G) Decrease of 3′UTR-controlled luciferase activity upon differentiation of C2C12. Cells were transiently transfected with the indicated reporters 48 h prior to myogenic differentiation. Shown is the reporter activity over the course of differentiation, normalized to the vector control (pMIR empty). (H) Relative 3′UTR luciferase reporter activity of the deletion constructs in C2C12 cells following myogenic differentiation for four days. Shown in comparison to MRTF-A wild type 3′UTR reporter which is set as 1. Error bars, SD (n = 3); *P< 0.05, **P< 0.01, ***P< 0.001 (Student’s t-test).
To characterize the kinetics of post-transcriptional regulation of MRTF-A during myogenesis, C2C12 cells were transfected with the 3′UTR luciferase reporter system. Compared to the empty vector control, the luciferase activity decreased over the course of C2C12 differentiation to ∼40% (Figure 8G), consistent with the observed MRTF-A protein reduction and a 3′UTR mediated repression during myogenesis. In contrast, deleting the individual miRNA binding sites elevated the 3′UTR reporter activity in differentiated C2C12 in comparison to the wild type reporter (Figure 8H). Reporter activities in undifferentiated C2C12 were unaffected by specific deletions, presumably due to largely absent miRNAs (Supplementary Figure S3). Together, the results infer that the miRNAs miR-1a-3p/206-3p, miR-24-3p and miR-486-5p post-transcriptionally repress MRTF-A protein expression via specific interactions with its 3′UTR. In addition, miR-133-3p and miR-378a/d bound the MRTF-A 3′UTR and affected its expression in some experimental setups.
DISCUSSION
In the present study we show a critical role of miRNA-mediated regulation of MRTF-A during myogenic differentiation. Based on an unbiased miRNome-wide screening approach, we characterized miR-1a-3p/206-3p, miR-24-3p and miR-486-5p as regulators of C2C12 myoblast differentiation via 3′UTR-mediated MRTF-A repression. Our results are consistent with a model in which concerted action of both transcriptional and post-transcriptional mechanisms orchestrate the differentiation into skeletal muscle myotubes (Figure 9). Myogenesis-induced upregulation of miR-1a-3p/206-3p, miR-24-3p and miR-486-5p represses translation of MRTF-A, which in turn releases its inhibitory effect on late stages of differentiation. Thereby, execution of the myogenic program is stabilized by a positive feedback loop.
Figure 9.
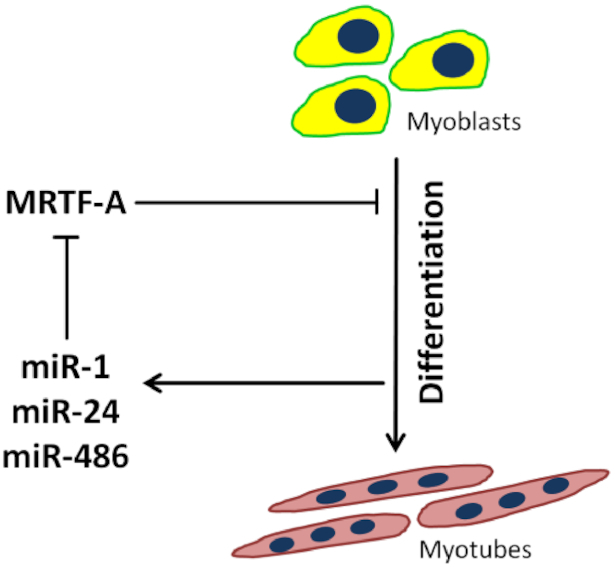
Model depicting the regulatory circuitry of myogenic C2C12 differentiation via post-transcriptional MRTF-A regulation. The inhibitory effect of the transcription factor MRTF-A is attenuated by the myogenesis-induced miRNAs miR-1a-3p, miR-24-3p and miR-486-5p, which bind the MRTF-A 3′UTR.
MRTF-A is crucial for skeletal myogenesis, and its deregulation in the form of overexpression or knockout is associated with differentiation defects (16,17,19) (Figure 2). Myogenesis is characterized by decreasing MRTF-A levels and activity over the course of differentiation (Figure 1). Interestingly, the MRTF-A activator RhoA shows similar expression and activity patterns, and RhoA activation results in differentiation defects after constitutive overexpression (19). Taken together, this suggests a positive role for the RhoA-MRTF-SRF signaling axis in early myogenic events, whereas MRTF-A activity needs to be dampened at later stages of differentiation.
While MRTF-A protein levels decreased over the course of differentiation, mRNA abundance and the rate of protein turnover did not obviously change (Figure 1). This pointed towards a major role of post-transcriptional regulation of MRTF-A by translational repression during myogenic differentiation. In general, miRNA-mediated post-transcriptional regulation is known to play a pivotal role in muscle differentiation. Various myo-miRs have been identified previously, including miR-1a/b, miR-133, miR-206, miR-208b, miR-486 and miR-499, each with multiple targets (5). Our study confirmed these and extended the potential myo-miRNome to 33 miRNAs which were significantly upregulated upon the differentiation of C2C12 cells (Figure 3C). However, of the previously reported MRTF-A regulating miRNAs, miR-1a-3p, miR-206-3p and miR-219a-5p (35–37), only miR-1a-3p and miR-206-3p were significantly enriched with the MRTF-A 3′UTR and selectively enhanced upon differentiation. Our data clearly indicate the pivotal role of post-transcriptional control during myogenic differentiation. In our C2C12 cell model, Dicer1 knockdown resulted in downregulation of myogenin and myosin heavy chain, while MRTF-A protein levels were elevated. This supports previous studies indicating an essential role of Dicer1 in the post-transcriptional regulation of skeletal myogenesis in vivo (38). Moreover, RISC/miRNA-association with the MRTF-A 3′UTR was markedly increased upon differentiation, and deletion of the endogenous 3′UTR reversed MRTF-A reduction and impaired differentiation (Figure 5). Together, this strongly indicates a miRNA-mediated regulation via the 3′UTR, although we cannot exclude an involvement of RNA binding proteins.
For identifying the MRTF-A specific miRNome in myoblasts, our previously established miTRAP method to discover candidate miRNAs which interact with a transcript of interest was applied (23,39). Confirming the suitability and experimental power of this approach, we also identified previously reported MRTF-A specific myo-miRs, including miR-1a-3p (35). To validate known and novel candidates, three approaches were used: the effect of miRNA overexpression on MRTF-A (Figure 7), 3′UTR reporter assays with and without co-expression of miRNAs, and analysis of the deletions of predicted binding sites in differentiating C2C12 cells (Figure 8). Together, these studies revealed miR-24-3p and miR-486-5p as novel post-transcriptional MRTF-A regulators.
The muscle enriched miR-24-3p promotes myogenesis and is negatively regulated by TGF-β1 (40). In addition miR-24 inhibits the skeletal muscle fibrosis-inducing TGF-β/Smad signaling pathway by repressing Smad2 (41). Our findings suggest that miR-24-3p downregulates MRTF-A during differentiation, thereby promoting myogenesis. Interestingly, in keratinocytes miR-24 was identified as an inducer of differentiation and a negative regulator of the actin dynamics (42). Embryonal stem cells (ESC) react to miR-24 overexpression with loss of self-renewal and terminal differentiation (43). Since miR-24 is not restricted to the myogenic lineage, it remains to be investigated whether the differentiation-supporting role of miR-24 in these cell types also involves MRTF-A regulation.
We identified miR-486-5p as a novel MRTF-A regulator by its binding to the 3′UTR, by functionally reducing MRTF-A protein, and by its inhibition of the 3′UTR via a novel recognition site. MiR-486 is not exclusively expressed in muscle and also plays important roles in hematopoietic stem cell proliferation and erythroid differentiation (44,45). Nevertheless, miR-486 is classified as a myo-miR because of a crucial role in myogenesis. It was shown to be a posttranscriptional regulator of PTEN, FOXO1 and Pax7 (45–47). While Pax7 is a repressor of MyoD, it was shown that MyoD induces the expression of miR-486 (47). Overexpression of miR-486 resulted in a milder phenotype in muscular dystrophy-associated mouse models (48). Interestingly, miR-486 is transcriptionally upregulated by MRTF-A/SRF (46). Together with our finding that MRTF-A is regulated by miR-486-5p, this suggests that MRTF-A/SRF and miR-486 form a regulatory feedback loop. Here MRTF-A/SRF and MyoD induce the expression of miR-486 in undifferentiated muscle cells. In turn miR-486-5p represses MRTF-A post-transcriptionally, thus leading to a balanced MRTF-A expression needed for proper myogenic differentiation. During myogenesis miR-1a-3p, miR-24-3p and miR-486-5p are further upregulated resulting in decreasing MRTF-A protein levels and function. In line with this hypothesis, the promoters of miR-1, miR-24 and miR-486 contain E-box sequences, which may facilitate collective induction by myogenic helix-loop-helix transcription factors such as MyoD and myogenin.
We showed that TGF-β treatment of undifferentiated C2C12 decreases miR-486-5p expression (Figure 6B). Thus we speculate that MRTF-A is subject to anti-differentiation signals from TGF-β via reduced levels of miR-486-5p. Previously an anti-fibrotic effect of miR-486 mediated Smad2 regulation was described (49). Together, there might be a regulatable antagonism between TGF-β/Smad signaling and MRTF-A reduction which is mediated by miR-486-5p and, possibly, miR-24-3p. Upon increased TGF-β/Smad2 signaling, MRTF-A becomes de-repressed by miR-486-5p/miR-24-3p downregulation, resulting in impaired myogenesis. However, the consequences of TGF-β signaling on the miRNA-mediated MRTF-A regulation described here await further investigation.
We showed that MRTF-A overexpression blocks myogenesis also in human cells, and that miR-1a-3p/206-3p, miR-24-3p and miR-486-5p suppress endogenous MRTF-A expression in primary human skeletal muscle myoblasts, consistent with the murine C2C12 model. Whilst the sequences of the miRNAs identified here are evolutionarily highly conserved, the predicted target sites of the 3′UTR for miR-24-3p and miR-486-5p are not. Nevertheless, the strong effect of miR-24 on MRTF-A expression in human myoblasts cells indicates the existence of another target site. Indeed, algorithmic predictions suggest two more binding sites on the human 3′UTR of MRTF-A. Thus we would like to propose functional conservation for the posttranscriptional regulation of MRTF-A during myoblast differentiation, which includes miR-24-3p and miR-486-5p.
DATA AVAILABILITY
Complete datasets and cloning details are available from the corresponding author on reasonable request. MiRNA-Seq data of differentiated C2C12 cells and the miTRAP approach has been deposited in NCBI’s Gene Expression Omnibus database under series accession number GSE136956.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Anja Weber for technical assistance, and Knut Krohn (University of Leipzig) for NGS. Open Access Publishing is supported within a funding programme by the DFG.
Author contributions: I.H. designed, performed and analyzed experiments and wrote the manuscript. A.K.S. designed, performed and analyzed the experiments concerning CRISPR deletion and human cells and wrote the manuscript. F.P. and J.B. performed experiments and analyzed the data. L.L. performed experiments. D.M. analyzed the RNAseq data. S.H. analyzed experiments and wrote the manuscript. G.P. conceived the study, designed and analyzed the experiments and wrote the manuscript. All authors read and approved the final manuscript.
Notes
Present address: Ingo Holstein, Institute of Human Genetics, University of Ulm and University of Ulm Medical Center, Ulm, Germany.
Contributor Information
Ingo Holstein, Institute for Physiological Chemistry, Medical Faculty, Martin Luther University Halle-Wittenberg, Hollystrasse 1, 06114 Halle (Saale), Germany.
Anurag Kumar Singh, Institute for Physiological Chemistry, Medical Faculty, Martin Luther University Halle-Wittenberg, Hollystrasse 1, 06114 Halle (Saale), Germany.
Falk Pohl, Institute for Physiological Chemistry, Medical Faculty, Martin Luther University Halle-Wittenberg, Hollystrasse 1, 06114 Halle (Saale), Germany.
Danny Misiak, Institute of Molecular Medicine, Medical Faculty, Martin Luther University Halle-Wittenberg, Charles Tanford Protein Center, Kurt-Mothes-Straße 3a, 06120 Halle (Saale), Germany.
Juliane Braun, Institute of Molecular Medicine, Medical Faculty, Martin Luther University Halle-Wittenberg, Charles Tanford Protein Center, Kurt-Mothes-Straße 3a, 06120 Halle (Saale), Germany.
Laura Leitner, Institute for Physiological Chemistry, Medical Faculty, Martin Luther University Halle-Wittenberg, Hollystrasse 1, 06114 Halle (Saale), Germany.
Stefan Hüttelmaier, Institute of Molecular Medicine, Medical Faculty, Martin Luther University Halle-Wittenberg, Charles Tanford Protein Center, Kurt-Mothes-Straße 3a, 06120 Halle (Saale), Germany.
Guido Posern, Institute for Physiological Chemistry, Medical Faculty, Martin Luther University Halle-Wittenberg, Hollystrasse 1, 06114 Halle (Saale), Germany.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Training Group 1591 (Post-transcriptional control in gene expression), funded by the Deutsche Forschungsgemeinschaft (DFG). Funding for open access charge: DFG.
Conflict of interest statement. None declared.
REFERENCES
- 1. Buckingham M., Rigby P.W.. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev. Cell. 2014; 28:225–238. [DOI] [PubMed] [Google Scholar]
- 2. Kuang S., Kuroda K., Le Grand F., Rudnicki M.A.. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 2007; 129:999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Davie J.K., Cho J.-H., Meadows E., Flynn J.M., Knapp J.R., Klein W.H.. Target gene selectivity of the myogenic basic helix–loop–helix transcription factor myogenin in embryonic muscle. Dev. Biol. 2007; 311:650–664. [DOI] [PubMed] [Google Scholar]
- 4. Hollenberg S.M., Cheng P.F., Weintraub H.. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. U.S.A. 1993; 90:8028–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mok G.F., Lozano-Velasco E., Munsterberg A.. microRNAs in skeletal muscle development. Semin. Cell Dev. Biol. 2017; 72:67–76. [DOI] [PubMed] [Google Scholar]
- 6. Rao P.K., Kumar R.M., Farkhondeh M., Baskerville S., Lodish H.F.. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. U.S.A. 2006; 103:8721–8726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takano H., Komuro I., Oka T., Shiojima I., Hiroi Y., Mizuno T., Yazaki Y.. The Rho family G proteins play a critical role in muscle differentiation. Mol. Cell. Biol. 1998; 18:1580–1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gauthier-Rouviere C., Vandromme M., Tuil D., Lautredou N., Morris M., Soulez M., Kahn A., Fernandez A., Lamb N.. Expression and activity of serum response factor is required for expression of the muscle-determining factor MyoD in both dividing and differentiating mouse C2C12 myoblasts. Mol. Biol. Cell. 1996; 7:719–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei L., Zhou W., Croissant J.D., Johansen F.E., Prywes R., Balasubramanyam A., Schwartz R.J.. RhoA signaling via serum response factor plays an obligatory role in myogenic differentiation. J. Biol. Chem. 1998; 273:30287–30294. [DOI] [PubMed] [Google Scholar]
- 10. Castellani L., Salvati E., Alema S., Falcone G.. Fine regulation of RhoA and Rock is required for skeletal muscle differentiation. J. Biol. Chem. 2006; 281:15249–15257. [DOI] [PubMed] [Google Scholar]
- 11. Charrasse S., Comunale F., Grumbach Y., Poulat F., Blangy A., Gauthier-Rouviere C.. RhoA GTPase regulates M-cadherin activity and myoblast fusion. Mol. Biol. Cell. 2006; 17:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Miralles F., Posern G., Zaromytidou A.I., Treisman R.. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003; 113:329–342. [DOI] [PubMed] [Google Scholar]
- 13. Olson E.N., Nordheim A.. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 2010; 11:353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Descot A., Hoffmann R., Shaposhnikov D., Reschke M., Ullrich A., Posern G.. Negative regulation of the EGFR-MAPK cascade by actin-MAL-mediated Mig6/Errfi-1 induction. Mol. Cell. 2009; 35:291–304. [DOI] [PubMed] [Google Scholar]
- 15. Esnault C., Stewart A., Gualdrini F., East P., Horswell S., Matthews N., Treisman R.. Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 2014; 28:943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cenik B.K., Liu N., Chen B., Bezprozvannaya S., Olson E.N., Bassel-Duby R.. Myocardin-related transcription factors are required for skeletal muscle development. Development. 2016; 143:2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Selvaraj A., Prywes R.. Megakaryoblastic leukemia-1/2, a transcriptional co-activator of serum response factor, is required for skeletal myogenic differentiation. J. Biol. Chem. 2003; 278:41977–41987. [DOI] [PubMed] [Google Scholar]
- 18. Mokalled M.H., Johnson A.N., Creemers E.E., Olson E.N.. MASTR directs MyoD-dependent satellite cell differentiation during skeletal muscle regeneration. Genes Dev. 2012; 26:190–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwasaki K., Hayashi K., Fujioka T., Sobue K.. Rho/Rho-associated kinase signal regulates myogenic differentiation via myocardin-related transcription factor-A/Smad-dependent transcription of the Id3 gene. J. Biol. Chem. 2008; 283:21230–21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Posern G., Sotiropoulos A., Treisman R.. Mutant actins demonstrate a role for unpolymerized actin in control of transcription by serum response factor. Mol. Biol. Cell. 2002; 13:4167–4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sotiropoulos A., Gineitis D., Copeland J., Treisman R.. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999; 98:159–169. [DOI] [PubMed] [Google Scholar]
- 22. Pfirrmann T., Lokapally A., Andreasson C., Ljungdahl P., Hollemann T.. SOMA: a single oligonucleotide mutagenesis and cloning approach. PLoS One. 2013; 8:e64870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Braun J., Misiak D., Busch B., Krohn K., Huttelmaier S.. Rapid identification of regulatory microRNAs by miTRAP (miRNA trapping by RNA in vitro affinity purification). Nucleic. Acids. Res. 2014; 42:e66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Labun K., Montague T.G., Krause M., Torres Cleuren Y.N., Tjeldnes H., Valen E.. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019; 47:W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001; 29:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Werner S., Lutzkendorf J., Muller T., Muller L.P., Posern G.. MRTF-A controls myofibroblastic differentiation of human multipotent stromal cells and their tumour-supporting function in xenograft models. Sci. Rep. 2019; 9:11725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kohn M., Lederer M., Wachter K., Huttelmaier S.. Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA. 2010; 16:1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Muller S., Glass M., Singh A.K., Haase J., Bley N., Fuchs T., Lederer M., Dahl A., Huang H., Chen J. et al.. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019; 47:375–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kozomara A., Griffiths-Jones S.. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2013; 42:D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Langmead B., Trapnell C., Pop M., Salzberg S.L.. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009; 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McCarthy D.J., Chen Y., Smyth G.K.. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012; 40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinson M.D., Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010; 11:R25–R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ashcroft G.S., Yang X., Glick A.B., Weinstein M., Letterio J.J., Mizel D.E., Anzano M., Greenwell-Wild T., Wahl S.M., Deng C. et al.. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat. Cell Biol. 1999; 1:260–266. [DOI] [PubMed] [Google Scholar]
- 34. Liu D., Black B.L., Derynck R.. TGF-beta inhibits muscle differentiation through functional repression of myogenic transcription factors by Smad3. Genes Dev. 2001; 15:2950–2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Minami T., Kuwahara K., Nakagawa Y., Takaoka M., Kinoshita H., Nakao K., Kuwabara Y., Yamada Y., Yamada C., Shibata J. et al.. Reciprocal expression of MRTF‐A and myocardin is crucial for pathological vascular remodelling in mice. EMBO J. 2012; 31:4428–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhuang C., Yuan Y., Song T., Wang H., Huang L., Luo X., He H., Huo L., Zhou H., Wang N. et al.. miR-219a-5p inhibits breast cancer cell migration and epithelial-mesenchymal transition by targeting myocardin-related transcription factor A. Acta Biochim. Biophys. Sin. (Shanghai). 2017; 49:1112–1121. [DOI] [PubMed] [Google Scholar]
- 37. Zhang W.L., Lv W., Sun S.Z., Wu X.Z., Zhang J.H.. miR-206 inhibits metastasis-relevant traits by degrading MRTF-A in anaplastic thyroid cancer. Int. J. Oncol. 2015; 47:133–142. [DOI] [PubMed] [Google Scholar]
- 38. O’Rourke J.R., Georges S.A., Seay H.R., Tapscott S.J., McManus M.T., Goldhamer D.J., Swanson M.S., Harfe B.D.. Essential role for Dicer during skeletal muscle development. Dev. Biol. 2007; 311:359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Busch B., Bley N., Muller S., Glass M., Misiak D., Lederer M., Vetter M., Strauss H.G., Thomssen C., Huttelmaier S.. The oncogenic triangle of HMGA2, LIN28B and IGF2BP1 antagonizes tumor-suppressive actions of the let-7 family. Nucleic Acids Res. 2016; 44:3845–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sun Q., Zhang Y., Yang G., Chen X., Zhang Y., Cao G., Wang J., Sun Y., Zhang P., Fan M. et al.. Transforming growth factor-beta-regulated miR-24 promotes skeletal muscle differentiation. Nucleic Acids Res. 2008; 36:2690–2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sun Y., Wang H., Li Y., Liu S., Chen J., Ying H.. miR-24 and miR-122 negatively regulate the transforming growth Factor-β/Smad signaling pathway in skeletal muscle fibrosis. Mol. Ther. Nucleic Acids. 2018; 11:528–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Amelio I., Lena A.M., Viticchie G., Shalom-Feuerstein R., Terrinoni A., Dinsdale D., Russo G., Fortunato C., Bonanno E., Spagnoli L.G. et al.. miR-24 triggers epidermal differentiation by controlling actin adhesion and cell migration. J. Cell Biol. 2012; 199:347–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ma Y., Yao N., Liu G., Dong L., Liu Y., Zhang M., Wang F., Wang B., Wei X., Dong H. et al.. Functional screen reveals essential roles of miR-27a/24 in differentiation of embryonic stem cells. EMBO J. 2015; 34:361–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang L.S., Li L., Li L., Chu S., Shiang K.D., Li M., Sun H.Y., Xu J., Xiao F.J., Sun G. et al.. MicroRNA-486 regulates normal erythropoiesis and enhances growth and modulates drug response in CML progenitors. Blood. 2015; 125:1302–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang L.-S., LI L., Li L., Shiang K.-D., Li M., Bhatia R.. MicroRNA-486-5p targets Foxo1 and regulates human hematopoietic stem cell proliferation and erythroid differentiation. Blood. 2010; 116:3871–3871. [Google Scholar]
- 46. Small E.M., O’Rourke J.R., Moresi V., Sutherland L.B., McAnally J., Gerard R.D., Richardson J.A., Olson E.N.. Regulation of PI3-kinase/Akt signaling by muscle-enriched microRNA-486. Proc. Natl. Acad. Sci. U.S.A. 2010; 107:4218–4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dey B.K., Gagan J., Dutta A.. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011; 31:203–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alexander M.S., Casar J.C., Motohashi N., Vieira N.M., Eisenberg I., Marshall J.L., Gasperini M.J., Lek A., Myers J.A., Estrella E.A. et al.. MicroRNA-486–dependent modulation of DOCK3/PTEN/AKT signaling pathways improves muscular dystrophy–associated symptoms. J. Clin. Invest. 2014; 124:2651–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ji X., Wu B., Fan J., Han R., Luo C., Wang T., Yang J., Han L., Zhu B., Wei D. et al.. The anti-fibrotic effects and mechanisms of MicroRNA-486-5p in pulmonary fibrosis. Sci. Rep. 2015; 5:14131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Complete datasets and cloning details are available from the corresponding author on reasonable request. MiRNA-Seq data of differentiated C2C12 cells and the miTRAP approach has been deposited in NCBI’s Gene Expression Omnibus database under series accession number GSE136956.