Abstract
N 6-methylation of 2′-O-methyladenosine (Am) in RNA occurs in eukaryotic cells to generate N6,2′-O-dimethyladenosine (m6Am). Identification of the methyltransferase responsible for m6Am catalysis has accelerated studies on the function of m6Am in RNA processing. While m6Am is generally found in the first transcribed nucleotide of mRNAs, the modification is also found internally within U2 snRNA. However, the writer required for catalyzing internal m6Am formation had remained elusive. By sequencing transcriptome-wide RNA methylation at single-base-resolution, we identified human METTL4 as the writer that directly methylates Am at U2 snRNA position 30 into m6Am. We found that METTL4 localizes to the nucleus and its conserved methyltransferase catalytic site is required for U2 snRNA methylation. By sequencing human cells with overexpressed Mettl4, we determined METTL4’s in vivo target RNA motif specificity. In the absence of Mettl4 in human cells, U2 snRNA lacks m6Am thereby affecting a subset of splicing events that exhibit specific features such as 3′ splice-site weakness and an increase in exon inclusion. These findings suggest that METTL4 methylation of U2 snRNA regulates splicing of specific pre-mRNA transcripts.
INTRODUCTION
N 6-methylation of adenosine generates RNA modifications that regulate the expression and metabolism of the host RNA without changing the underlying sequence (1). One resultant RNA modification is N6-methyladenosine (m6A) that is found internally within various RNA species including mRNAs and rRNAs (2–5). Each of the several identified m6A methyltransferases (writers) that catalyze N6-methylation of adenosine have different target sequence specificities. For example, METTL3 catalyzes N6-methylation of DRACH to give DRm6ACH (D = A/G/U, R = A/G, H = A/C/U) (2,3,6,7). METTL3 also functions in a complex with co-factors including METTL14, WTAP, KIAA1429 and RBM15/RBM15B (8–12). This complex mostly targets mRNAs in the region proximal to the stop codon. Another m6A writer is METTL16 that methylates ‘UACAGAGAA’ to give ‘UACm6AGAGAA’. Besides U6 snRNA, METTL16 also targets the Mat2a 3′UTR (13). Finally, METTL5 and ZCCHC4 have been identified as the writers responsible for m6A catalysis in 18S rRNA and 28S rRNA respectively (14–16).
Adenosine N6-methylation of 2′-O-methyladenosine (Am) results in the RNA modification N6,2′-O-dimethyladenosine (m6Am) that is found in the first transcribed nucleotide adjacent to the RNA methylguanosine cap (Supplementary Figure S1A) (17,18). Given its predicted N6-methyladenine methyltransferase domain and its interaction with the phosphorylated C-terminal tail of RNA polymerase II during RNA transcription, PCIF1 was a strong candidate as a N6-methyladenine writer (19,20). Subsequently, PCIF1 was established as the writer responsible for the N6-methylation of Am at the transcriptional start site (TSS) to generate m6Am (21–25). This facilitated the characterization of TSS-associated m6Am as a regulator of mRNA resistance to DCP2-mediated decapping, and potentially mRNA translation and cell growth (17,21,23,24).
m6Am sites have also been mapped to internal RNA sites, specifically at U2 snRNA position 30 (18,26). The functional importance of m6Am in U2 snRNA was not well-characterized, likely because the writer responsible for methylating U2 snRNA Am30 was previously unknown. In our work here, we utilize a primarily sequencing approach to establish methyltransferase-like 4 (Mettl4) as the U2 snRNA m6Am writer. By overexpressing Mettl4, we uncovered both the in vivo target sequence preference of METTL4 and its ability to catalyze m6Am formation internally within mRNAs. Finally, by knocking out Mettl4 from cells, we demonstrate that the resulting loss of U2 snRNA m6Am perturbs splicing of target mRNAs with distinctive features such as weak 3′ splice sites.
MATERIALS AND METHODS
m6ACE-seq
m6ACE-seq libraries were constructed as previously described (25): Poly(A) RNA was purified using Poly(A)Purist Mag kit (Thermofisher AM1922) according to manufacturer's instructions, then fragmented to ∼120nt by incubating in RNA fragmentation buffer (Ambion AM8740) for 7.5 min at 70°C. Fragmented RNA was treated with 10 U T4 PNK (NEB M0201) for 30 min at 37°C before adding 1 mM ATP and incubating for an additional 30 min at 37°C, then purified using Oligo Clean & Concentrator (Zymo D4060). 3′ ligation was performed as described (27,28), where 200 pmol 5′-adenylated,3-dideoxyC DNA adapters (Supplementary Table S1) were ligated with 400 U truncated T4 RNA ligase 2 (NEB M0242) in 1X ATP-free T4 RNA ligase buffer [50 mM Tris pH 7.5, 60 μg ml−1 BSA, 10 mM MgCl2, 10 mM DTT, 12.5% PEG8000] for 2 h at 25°C. Ligated RNA was purified with Ampure XP beads (Beckman Coulter A63881). 200 pg of 3′-ligated methylated RNA spike-in (Supplementary Table S1) was added to 1 μg of ligated Poly(A) RNA and the mixture was denatured for 5 min at 65°C before incubating for 2 min on ice. This denatured RNA mixture was incubated overnight at 4°C with 8 μg anti-m6A antibody (Synaptic Systems 202003) in 1× IP buffer [150 mM LiCl, 10 mM Tris pH 7.4, 0.1% IGEPAL CA-630 (Sigma I8896)] supplemented with 1 U μl−1 RNasin Plus (Promega N2611). In parallel, 1.2 mg Dynabeads-Protein-A was blocked overnight at 4°C in 1× IP buffer supplemented with 0.5 mg ml−1 BSA (Sigma A7906). The antibody–RNA mixture was split into 50 μl aliquots on ice and crosslinked with 0.15 J cm−2 254 nm UV radiation six times. The antibody–RNA mixture was recombined and 1% of it was set aside as input-RNA and the remainder (designated as m6ACE-RNA) was mixed with decanted BSA-blocked Dynabeads-Protein-A for 1.5 h at 4°C. Beads bound with crosslinked RNA were then washed with 250 μl of the following cold buffers in this order: Wash buffer 1 [1 M NaCl, 50 mM HEPES–KOH pH 7.4, 1% Triton X-100, 0.1% sodium deoxycholate, 2 mM EDTA], Wash buffer 2 [0.5 M NaCl, 50 mM HEPES–KOH pH 7.4, 1% IGEPAL, 0.1% sodium deoxycholate, 2 mM EDTA], Wash buffer 3 [1% sodium deoxycholate, 25 mM LiCl, 10 mM Tris pH 8, 1% Triton X-100, 2 mM EDTA], TE [10 mM Tris pH 8, 1 mM EDTA] and finally 10 mM Tris pH 8. m6ACE RNA was then denatured in 10 μl 10 mM Tris pH 8 for 5min at 65°C and for 2 min on ice. m6ACE RNA was digested with 1 U XRN-1 (NEB M0338) in XRN-1 buffer [100 mM LiCl, 45 mM Tris pH 8, 10 mM MgCl2, 1 mM DTT] and 1 U μl−1 RNasin Plus shaking at 1krpm for 1 h at 37°C. The m6ACE RNA-bead mixture was then washed with Wash buffer 1, Wash buffer 2, Wash buffer 3, TE and 10 mM Tris pH 8. Both input and m6ACE RNAs were eluted in elution buffer [1% SDS, 200 mM NaCl, 25 mM Tris pH 8, 2 mM EDTA, 1 mg ml−1 Proteinase K (Thermo Scientific EO0491)], shaking at 1 krpm for 1.5 h at 50°C. RNAs were ethanol-precipitated and ligated to 5 pmol 5′ adapters (Supplementary Table S1) with 10 U T4 RNA ligase (Ambion AM2140) supplemented with 12.5% PEG8000 and 2 U μl−1 RNasin Plus for 16 h at 16°C before being purified with Oligo Clean & Concentrator. 5 pmol of reverse transcription primer (Supplementary Table S1) was annealed (72°C 2 min, ice 2 min) and reverse transcription was performed with 200 U SuperscriptIII (Invitrogen 18080) for 1 h at 50°C, with the reaction stopped by incubating for 15 min at 70°C. The cDNA was PCR amplified with Phusion High-fidelity PCR mastermix (NEB M0530) and Truseq PCR primers. Finally, primer–dimer and adapter–dimers were removed with Ampure XP beads before undergoing PE75 sequencing on the Illumina Nextseq platform.
m6ACE-seq analysis
m6ACE-seq analysis was performed as previously described (25): Fastq sequences were first filtered for a quality score of 20, then trimmed of 5′ and 3′ adapter sequences and poly(A) tails using Cutadapt (29). The 8-mer UMI located at the first 8 nucleotides of read 1 was registered and trimmed. Any complementary UMI sequence in read 2 was also trimmed. Reads were mapped to the methylated spike-in (Supplementary Table S1) using Bowtie2, or to the hg38 assembly transcriptome (Gencode v28 comprehensive gene annotations) using STAR (30,31). Aligned pairs that had the same mapping coordinates and UMIs were filtered out as PCR duplicates. Read-start coordinates in hg38-mapped reads that began with an adenosine nucleotide, and had a minimum mean read count of 1 across the triplicate samples were collated. m6A or m6Am sites were identified as read starts that were at least 2-fold enriched in m6ACE libraries than in the corresponding input libraries. This enrichment was calculated using DESeq2 (32) performed on A-only sites across triplicate pairs of m6ACE and corresponding input libraries (FDR < 0.1, padj < 0.05). Identified sites that were 1–4 nucleotides upstream of another identified significant Rm6AC site or sites found within clustered read-starts were filtered out.
To calculate the relative methylation level (RML) of each site in each sample: The read-start counts at positions –4 to 0 of each site in the m6ACE library were summed and divided by the read-start counts at positions –51 to 0 of the same site in the input library to give ‘X’. Similarly, the read-start counts at positions -4 to 0 of the spike-in m6A site in the m6ACE library were summed and divided by the read-start counts at positions –21 to 0 of the same spike-in m6A site in the input library to give ‘Y’. X was normalized to Y to give RML. RML values of each site was averaged across triplicates for each sample condition. A site was denoted as differentially methylated between METTL4WT-rescue and METTL4CD-rescue if the average RML differs between sample conditions with a log2fold-change cutoff of 2.0, as well as a one-tailed Student's T-test P-value cutoff of <0.041 (false discovery rate, FDR < 0.1). Consensus motif analysis was performed using Meme-chip (33). Metagene analysis was performed using MetaPlotR (34). To identify overlap between de novo methylation sites and Am sites previously identified by Nm-seq, we used the HEK NM-seq dataset from NCBI’s Gene Expression Omnibus (GEO) under accession number GSE90164.
RNAseq
A total of 10μg of RNA was treated with RQ1 RNase-Free DNase (Promega M6101) as per manufacturer's protocol. Poly(A)+ cDNA libraries were subsequently generated from the DNase (Promega M610A) treated RNA using the TruSeq Stranded mRNA Library Prep Kit (Illumina RS-122-2101, RS-122-2102 and RS-122–2103). Libraries were sequenced on an Illumina HiSeq 2500 platform.
RNAseq splicing and gene expression analysis
The quality of the RNAseq datasets was inspected using FastQC. Read alignment to the genome was then performed using STAR (2.7.0a) in two-pass mode using the hg38 genome assembly and GENCODE v30 gene annotations (31).
Alternative splicing differences were analysed by an in-house pipeline using replicate MATS (rMATS 4.0.2) and filtered based on cut-offs of FDR ≤ 0.05, an absolute change in percent spliced in (|ΔPSI|) ≥ 0.1 (35,36). In addition, events with a mean junction count <20 for either condition, as well as those with <10 inclusion or spliced junction counts for both conditions, were filtered out to enhance the reliability of our splicing dataset. The validation of several differential alternative splicing events was performed using an RT-PCR assay (36,37). The validation criterion for the RT-PCR assays was a minimum of 5% ΔPSI in the direction of change corresponding to the rMATS findings.
Differentially expressed genes between the WT and Mettl4-KO datasets were analysed using a combination of RSEM (38) and limma (39), with cut-offs of FDR ≤ 0.05 and an absolute log2(fold-change) ≥ 1.
Differential alternative splicing event feature analysis
Splice-site strengths were calculated using MaxEntScan (37). Comparisons were made between the exon features of the differentially spliced cassette exons and a transcriptome-wide background based on all known human internal exons (classified into non-cassette and cassette exons based on the USCS hg38 Alt Events table). Cut-offs of 500nt and 12 500nt were applied on the exon and intron length datasets respectively to reduce data skewing caused by extreme outliers.
The frequency of the ‘HMAGKD’ motif was calculated for the differentially spliced cassette exons using exonic sequences, as well as intronic regions of up to 250nt upstream and downstream of the exons (with the splice sites excluded). Transcriptome-wide ‘HMAGKD’ occurrence frequencies were also generated using hg38 GENCODEv30 exon annotations. The GC content of exonic and splice site proximal intronic regions was also examined in a similar manner, but using a rolling window of 10nt with 1nt intervals, restricted to 48nt upstream and 50nt downstream to accommodate for the shortest introns present in the cassette exon dataset.
The statistical significance of feature differences was calculated using Wilcoxon's rank-sum test (or the Kruskal–Wallis test for comparisons across all groups) and P-values were adjusted using the Benjamini–Hochberg correction when necessary.
Splice-site and RNA binding protein motif sequence analysis
The Jensen–Shannon Divergences of splice site sequence subsets and associated gamma distribution tests for statistical significance were calculated using the R package DiffLogo (40). All sequence logos were generated using ggseqlogo (41). The differentially spliced cassette exons were scanned for RNA binding protein motifs using rMAPS2 (42). The enrichment of putative 6–8nt long, strand-specific RNA sequence features within 125nt stretches up- and downstream of the exon (excluding the splice sites) were also examined using MEME, with background nucleotide frequencies generated from the corresponding regions of the human transcriptome (33). The sequences generated by MEME were then fed into TOMTOM to search for RNA binding proteins with similar binding motifs. The biasness in the distribution of the detected motifs between the short (<250nt) and long introns (≥250nt) of the input dataset was examined using the Fisher exact test with Benjamini-Hochberg corrections.
RT-PCR assay
1 μg of total RNA was primed with oligo-dT(20) and reverse transcribed with SuperscriptIII as per manufacturer's protocol. 1 μl of 5 U/ul RNAse H (ThermoFisher Scientific EN0201) was then added to each reaction and incubated at 37°C for 20 min. The cDNA was used for PCR amplification for 35 cycles using DreamTaq DNA polymerase (Thermo Scientific K1081) and respective primers (Supplementary Table S1) before being assessed via agarose gel electrophoresis.
RESULTS
U2 snRNA m6Am is absent in Mettl4-KO cells
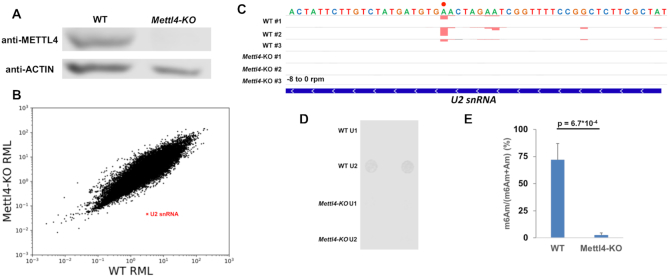
Mettl4 is one of a diverse family of proteins that are homologous to the MT-A70 subunit of the m6A writer Mettl3 (43). In order to assess Mettl4 as a potential writer, we performed m6A-crosslinking-exonuclease-sequencing (m6ACE-seq), which is capable of quantitatively mapping precise N6-methyladenine methylomes in the form of m6A or m6Am (Supplementary Figure S1A) (25). Comparison of the RNA methylomes between wildtype (WT) and Mettl4-KO HEK293T cells revealed a single site that exhibited almost 70-fold reduced relative methylation level (RML) in the absence of METTL4 (Figure 1A, B). The single-base-resolution of m6ACE-seq allowed us to map this site to position 30 of U2 snRNA (Figure 1C). To verify that this METTL4-dependent adenosine methylation is located within U2 snRNA, we isolated various snRNAs for anti-m6A dot blotting (Supplementary Figures S1B, C). As expected, we detected an RNA methylation signal specific only to WT U2 snRNA but not in Mettl4-KO U2 snRNA or any U1 snRNA (Figure 1D).
Figure 1.
METTL4 mediates m6Am modification in U2 snRNA. (A) Western blotting of human METTL4 in WT versus Mettl4-KO lysate with ACTIN as a loading control. (B) Scatterplot of average relative methylation level (RML) of sites identified via m6ACE-seq of WT versus Mettl4-KO RNA. U2 snRNA is denoted with a red ‘X’. (C) m6ACE (red) and Input (black) read-start counts (in reads per million mapped or RPM) mapped to U2 snRNA. U2 snRNA m6Am position identified in (B) is denoted by a red dot. Sequence corresponds to the same strand as the m6Am site. Blue horizontal bar represents transcript. (D) Anti-m6A dot blotting of various snRNAs purified from WT versus Mettl4-KO RNA. Duplicate dots are shown. (E) Nucleoside HPLC–MS/MS of m6Am as a percentage of total m6Am and Am in U2 snRNA purified from WT versus Mettl4-KO RNA. Displayed are average and standard deviation error of biological triplicates. One-tailed Student's t-test P-value is shown.
Since the anti-m6A antibody is not able to distinguish between the structurally similar m6A and m6Am modifications (Supplementary Figure S1A), isolated U2 snRNA was digested and subjected to nucleoside high performance liquid chromatography coupled with tandem mass spectrometry (HPLC–MS/MS) to verify the identity of the RNA modification within U2 snRNA. While there were no appreciable m6A levels in U2 snRNA, we instead found that the majority of Am are in the N6−methylated m6Am form (Figure 1E, Supplementary Figures S1D, E). More importantly, m6Am was not detected in Mettl4-KO U2 snRNA (Figure 1E, Supplementary Figure S1D). We note that the HPLC–MS/MS m6Am signal was derived internally from U2 snRNA as we did not use any decapping enzyme in digesting U2 snRNA, thereby preventing the quantification of any cap-adjacent RNA nucleotide. Furthermore, previous and current RNA methylomes have demonstrated that U2 snRNA lacks a TSS-associated adenosine N6-methylation (Figure 1C) (25). Altogether, this demonstrates that METTL4 is necessary for m6Am formation within U2 snRNA.
N 6-methylation of U2 snRNA-internal Am requires the METTL4 DPPW catalytic motif
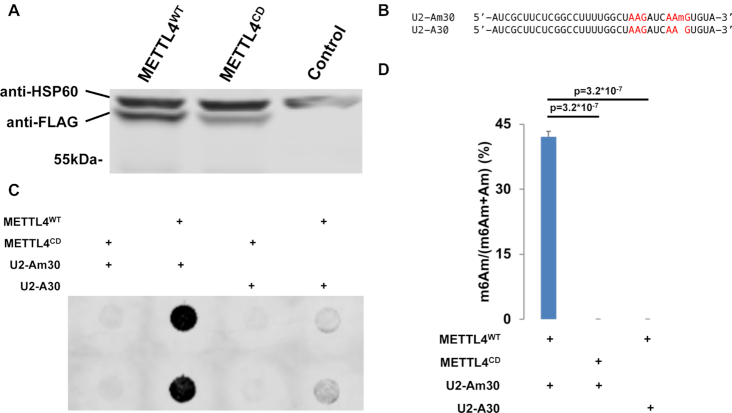
METTL4 can either directly catalyze the N6-methylation of Am or act as an indirect co-factor to the actual catalytic writer. To investigate METTL4’s role in RNA methylation, we expressed and purified recombinant C-terminal-3x-FLAG-tagged human METTL4 (METTL4WT) (Figure 2A, Supplementary Figure S2A). Incubation of METTL4WT with a RNA oligonucleotide matching the 35nt 5′ end of U2 snRNA (U2-Am30, Figure 2B, Supplementary Table S1) generated a N6-methyladenosine signal detectable via anti-m6A dot blotting (Figure 2C). Using HPLC-MS/MS, we verified this methylated signal as m6Am and not m6A (Figure 2D). Additionally, METTL4 contains a ‘DPPW’ motif, which are catalytic residues required for methyltransferase activity (43). Therefore, we mutated the METTL4 ‘DPPW’ to ‘APPA’ to generate a catalytically-dead METTL4 (METTL4CD, Figure 2A, Supplementary Figure S2A). Using the same in vitro methylation assay, we found that loss of the ‘DPPW’ catalytic site in METTL4CD abrogates its ability to methylate RNA Am into m6Am (Figure 2C, D). This demonstrates that human METTL4 directly catalyzes the N6-methylation of Am to m6Am in vitro.
Figure 2.
METTL4 directly catalyzes m6Am formation in vitro. (A) Western blotting of cell lysate from Mettl4-KO HEK293T transfected with METTL4WT or METTL4CD or untransfected (control). Expected size of purified protein is 57kDa. HSP60 (∼61 kDa) is displayed as a comparison. (B) Sequence of U2-Am30 and U2-A30 RNA substrates used for in vitro methylation. ‘AAG’ sites are highlighted in red. (C) Anti-m6A dot blotting of RNA substrates in vitro methylated by METTL4WT or METTL4CD. Duplicate dots are shown. (D) Nucleoside HPLC–MS/MS of m6Am as a percentage of total m6Am and Am in RNA substrates in vitro methylated by METTL4WT or METTL4CD. Displayed are average and standard deviation error of triplicates. One-tailed Student's t-test P-value is shown.
Given the structural similarity between Am and A, we next tested if METTL4 is also able to methylate A into m6A (Supplementary Figure S1A). We subjected a similar U2 snRNA-based RNA substrate with the single Am replaced with A (U2-A30, Supplementary Table S1), to in vitro methylation by METTL4WT (Figure 2B). This led to quantifiable m6A signals, albeit at a lower level than that of METTL4WTin vitro methylation of U2-Am30 (Supplementary Figure S2B). Expectedly, this methylation activity was absent for METTL4CD. Both U2-A30 and U2-Am30 contain adenosine in 2 adenosines in ‘AAG’ motifs but only adenosine in position 30 within the ‘CAAGUG’ context is N6-methylated by METTL4 (Figure 2B–D, Supplementary Figure S2B). This suggests A in the middle of ‘CAAGUG’ to be METTL4’s target substrate sequence, with a preference for Am over A for adenine-N6-methylation.
In vivo target sequence preference of METTL4
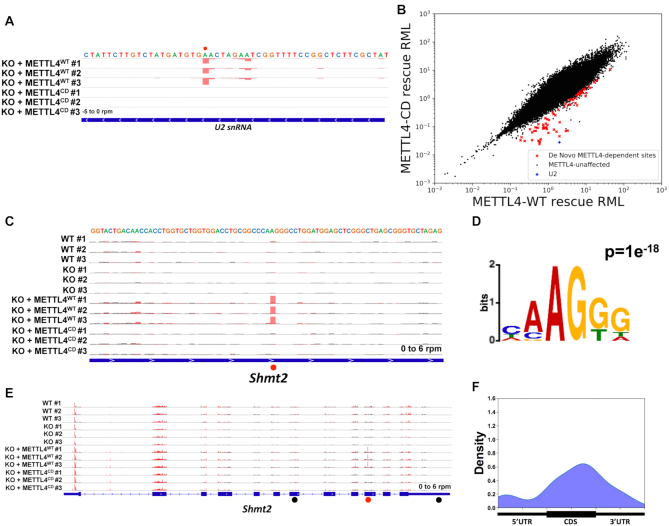
Since METTL4 catalyzes Am N6-methylation in vitro, we next investigated if METTL4 can also do so in vivo by rescuing METTL4 expression in Mettl4-KO cells. Similar to all other known m6A and m6Am writers, exogenous METTL4 mainly localizes to the nucleus though in rare cases, exogenous METTL4 was also found in the cytoplasm (Supplementary Figure S3A). More importantly, overexpressing METTL4WT rescued the loss of m6Am in U2 snRNA of Mettl4-KO cells (Figure 3A). On the other hand, overexpressing METTL4CD did not result in the same rescue effect, indicating that METTL4WT is directly catalyzing N6-methylation of U2 snRNA Am in vivo (Figure 3A).
Figure 3.
In vivo target sequence preference of METTL4. (A, C, E) m6ACE (red) and Input (black) read-start counts mapped to various genes. Red dots denote identified N6-methyladenine positions. Black dots denote ‘HMAGKD’ sites that are not identified as methylated by m6ACE-seq. Sequence corresponds to the same strand as the methylation site. Blue horizontal bar represents transcript. (C) is a magnified version of (E). (B) Scatterplot of average RML of sites identified via m6ACE-seq of RNA isolated from Mettl4-KO cells overexpressing METTL4WT (METTL4WT-rescue) or METTL4CD (METTL4CD-rescue). De novo METTL4-dependent are sites with RML reductions of at least log2fold of 2 (FDR < 0.1) in METTL4CD-rescue compared to METTL4WT-rescue cells. (D, F) MEME analysis (D) and metagene distribution profile (F) of METTL4-dependent de novo methylation sites.
While we had previously described the preferred RNA target for METTL4 catalysis, we envisioned that treating an extensive variety of RNA sequences with METTL4 will provide a clearer picture of its target sequence preference. Fortunately, the overexpression of METTL4WT in human cells affords such an opportunity: since METTL4WT is capable of methylating U2 snRNA in vivo, exposing METTL4WT to the entire human transcriptome allows us to simulate an in vivo methylation assay and determine METTL4’s target sequence motif by identifying additional sites of specific RNA methylation. As such, we compared the global methylomes of the METTL4WT- and METTL4CD-rescue cells to determine if METTL4 also mediates adenosine-N6-methylation of other RNAs besides U2 snRNA. This revealed that overexpressing METTL4WT, but not METTL4CD, led to the appearance of multiple N6-methyl-adenine sites in several mRNAs (Figure 3B, Supplementary Table S2). Since these sites are absent in WT cells, we denote them as de novo RNA methylation sites (Figure 3C, Supplementary Figure S3B). Amongst these de novo methylation sites, we identified the dominant motif sequence to be ‘HMAGKD’ (Figure 3D, H = A/C/U, M = A/C, K = G/U, D = A/G/U, A is methylation site). Since the U2 snRNA ‘CAm6AmGUG’ motif is a subset of the ‘HMAGKD’, this further validates ‘HMAGKD’ to be METTL4’s target sequence motif.
We note that the same mRNA can possess multiple ‘HMAGKD’ sequences but only have one being de novo methylated (Figure 3E, Supplementary Figure S3C). This argues that the presence of a ‘HMAGKD’ sequence is not the sole criterion for de novo methylation by METTL4WT, and that there exists other cis-acting elements and/or trans-acting factors that guide this process. Given that multiple ‘HMAGKD’ sites derived from the same mRNA are all expressed at the same level yet are not all de novo methylated, this eliminates mRNA expression level as a deciding criterion. We next considered that METTL4 preferred to methylate Am over A, suggesting that all de novo methylated targets initially contained ‘HMAmGKD’. As such, we analyzed Am methylomes mapped using Nm-seq but found no overlap between METTL4WT targeted sites and previously mapped Am sites within the human transcriptome (44). However, we do not completely rule out Am presence as a criterion since Am sites mapped by Nm-seq have yet to be fully validated and there could be multiple false negatives (45). Finally, we assessed if de novo N6-methylated adenosine exhibited specific positional patterns along the mRNA length. This revealed a slight preference of METTL4WT-dependent methylation to be within the coding sequence (CDS) (Figure 3F). This suggests that cis-acting elements or trans-acting factors associated with the CDS help to guide de novo methylation by METTL4WT.
METTL4 regulates pre-messenger RNA splicing
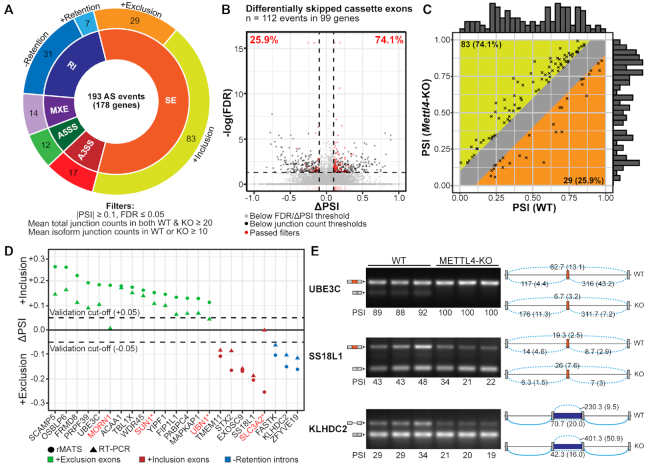
We next investigated if METTL4 methylation of U2 snRNA affected other cellular processes and initially found that loss of Mettl4 had no appreciable effect on overall U2 snRNA levels or cell growth rate (Supplementary Figures S4A, B). Given the role of U2 in pre-mRNA splicing, we next performed RNA sequencing on WT and Mettl4-KO HEK293T cells to search for splicing changes. By applying a stringent criterion to identify the most reliable splicing changes (see Materials and Methods), we detected a total of 193 splicing events in 178 genes that change upon KO of Mettl4. We found a majority of cassette exons over other alternative splicing types, and more alternative 3′ splice sites than 5′ splice sites (Figure 4A). Retained introns were more spliced out upon Mettl4-KO: among the total of 112 affected cassette exons, there are 29 (∼26%) events with enhanced exclusion and 83 (74%) with increased inclusion in Mettl4-KO cells (Figure 4B).
Figure 4.
Summary of splicing changes upon Mettl4-KO. (A) Breakdown of the 193 differential alternative splicing events between wild-type and Mettl4-KO HEK293T, as reported by rMATS (Supplementary Table S3), and with the indicated cutoffs. SE, skipped or cassette exon; A5SS, alternative 5′ splice sites; A3SS, alternative 3′ splice sites; MXE, mutually exclusive exons; RI, retention of introns. (B) Volcano plot of differentially spliced cassette exons, with events that passed the filters denoted as red dots. Dashed horizontal and vertical lines respectively depict the cutoffs in significance (FDR) and magnitude of splicing change (ΔPSI). (C) Plot of initial PSI (WT) against final PSI (Mettl4-KO). The marginal histograms represent the relative distribution of initial and final PSI values. (D) Plot of the ΔPSI values reported by rMATS and the corresponding RT-PCR assays for 20 cassette exons and three introns. Red gene names indicate splicing events that were not validated. Asterisks indicate events that were discretionally unvalidated based on high between-sample variations or inability to resolve one of the isoforms. (E) RT-PCR validations of two differential alternative splicing events for cassette exons in the UBE3C and SS18L1 genes, as well as an intron retention event in KLHDC2, in three biological replicates per WT or KO. The identity of splicing products by exon inclusion or exclusion is schematically shown next to the gels, along with mean junction reads (standard deviation within parentheses) across WT or KO samples. The PSI values obtained via RT-PCR are expressed in PSI values multiplied by 100.
Most of the splicing events affected in Mettl4-KO cells had a high splicing level or PSI (percentage spliced in), which was further increased in knockout cells (Figure 4C). As shown before, the magnitude of splicing change (ΔPSI) was the largest for events with intermediate starting PSI (Supplementary Figure S4C) (46). Genes with METTL4-dependent splicing events also did not overlap with METTL4-dependent differentially expressed genes (Supplementary Figure S4D). We attempted validation by RT-PCR of twenty representative cassette exons and three intron retention events, with an overall validation rate of 83% (19/23 cases) (Figure 4D, E, Supplementary Figure S4E).
Features of splicing events affected by Mettl4-KO
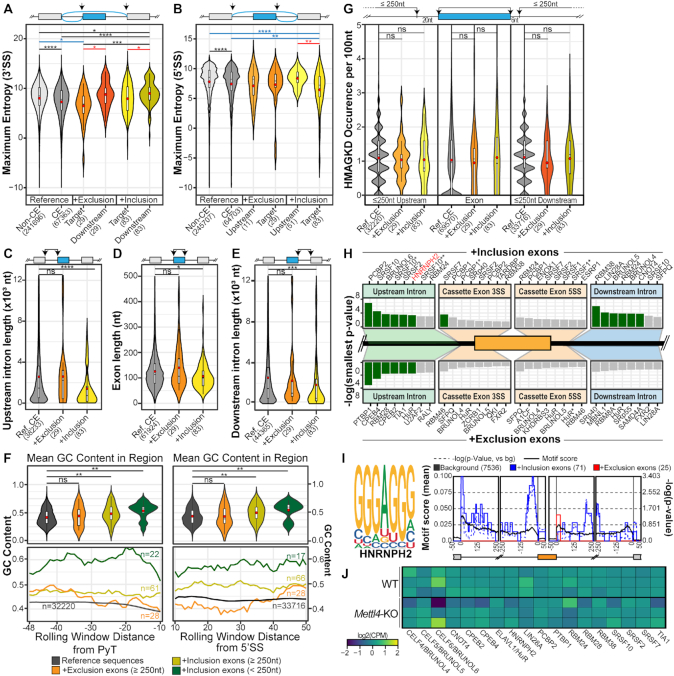
Analysis of splice-site strength using the MAXENT algorithm revealed that the downregulated cassette exons in Mettl4-KO have weak 3′ splice sites compared to all constitutive exons as references, and compared to exons downstream of the cassette exon (Figure 5A) (37). Comparison of the 3′ splice-site motifs by Jensen-Shannon divergence revealed that the polypyrimidine tract motif for upregulated exons exhibits two positions (–6 and –18) with a significantly higher incidence of cytosines than purines, while polypyrimidine tracts for downregulated exons showed significantly higher purines at four positions (–6, –7, –11 and –20) (Supplementary Figure S4F). Upregulated cassette exons exhibited weak 5′ splice-site scores compared to references and upstream exons, although the results are somewhat less clear-cut (Figure 5B), and motif divergence was detected at position +5 with an excess of non-consensus C/U for exons with increased exclusion in Mettl4-KO (Supplementary Figure S4G). Upregulated cassette exons in Mettl4-KO were flanked by far shorter introns compared to exons in other categories, plus these exons were themselves slightly shorter than reference exons (Figure 5C–E). The introns flanking the exons with increased inclusion in Mettl4-KO also exhibited a high GC content, compared to corresponding regions across the human transcriptome and other METTL4-regulated exons (Figure 5F). The elevated GC content is particularly pronounced in the shorter flanking introns, as a known feature of mammalian and avian genomes (47).
Figure 5.
Features of affected METTL4-dependent splicing events. (A, B) Violin plot of the 3′ and 5′ splice site maximum entropy score of all human non-cassette exons (‘non-CE’), all human cassette exons (‘CE’), exons undergoing increased inclusion or exclusion (‘Target’), as well as their downstream exons for 3′ splice-site strength analysis and upstream exons for 5′ splice-site strength analysis. Mean values are indicated as red circles and group sizes are indicated within parentheses along axis labels. Statistical significance was calculated between all groups using the Kruskal-Wallis test with the Benjamini–Hochberg correction. (C–E) Violin plot of exonic and flanking intronic lengths of all human cassette exons (‘Ref. CE’), and exons undergoing increased inclusion or exclusion. Mean values are indicated as red circles and group sizes are indicated within parentheses. Statistical significance was calculated against the background cassette exons subset using the Wilcoxon Ranked Sum test with the Benjamini-Hochberg correction. (F) Rolling window line plots (bottom) of the GC content in the 48nt region upstream of the polypyrimidine tract (PyT) and 50nt region downstream of the 5′ splice site 6nt conserved sequence. The four datasets include all cassette exons in the human transcriptome, the downregulated exons with flanking intron lengths ≥250nt and the upregulated exons with flanking intron lengths of either ≥250nt or <250nt. Violin plots represent the mean overall GC content of these regions per dataset. Mean values are indicated as red circles and group sizes are indicated within parentheses. Statistical significance was calculated against the background cassette-exon subset using the Wilcoxon Ranked Sum test with the Benjamini-Hochberg correction. (G) Violin plots of the ‘HMAGKD’ motif occurrence frequency in exonic and up to 250nt of upstream and downstream intronic sequences. Datasets include all human cassette exons (‘Ref. CE’), and the exons undergoing increased exclusion or inclusion. Mean values are indicated as red circles and group sizes are indicated within parentheses. Statistical significance was calculated against the background cassette exons subset using the Wilcoxon Ranked Sum test with the Benjamini-Hochberg correction. (H) List of enriched RNA binding proteins in the plotted regions as detected by rMAPS. Green bars indicate statistically significant hits after Bonferroni correction. Exons undergoing increased inclusion (top-half, n = 71 after filtering by rMAPS) and those undergoing increased exclusion (bottom-half, n = 25 after internal filtering by rMAPS). Asterisks indicate distinct binding motifs of RBPs that are known to recognise multiple sequences. (I) Binding motif of HNRNP-H2 and the rMAPS derived regions of enrichment for this motif within the differentially spliced cassette exons. (J) Gene expression heatmap of RBPs identified by rMAPS, plotted as log2(counts per million) values. ns, *, **, *** and **** respectively denote P > 0.05, P ≤ 0.05, P < 0.01, P < 0.001 and P < 0.0001.
Finally, we searched for enriched motifs in the intronic fragments flanking the affected splice sites and exons. We did not find a specific enrichment of the ‘HMAGKD’ METTL4 consensus motif in the regulated exons and up to an extended 250nt segment of flanking introns excluding splice sites (Figure 5G), suggesting that METTL4 does not regulate splicing by pre-mRNA modification. In fact, probing the genome for this motif revealed that it becomes more frequent towards the 3′ end of the transcript, yet there are no differences between cassette and non-cassette internal exons (Supplementary Figure S4H). We identified a few RNA Binding Protein (RBP) motifs, which are mostly GC rich and enriched either in the upstream or downstream intronic regions, but not in the cassette exon body (Figure 5H). Among these, the GGGAGGG consensus motif corresponding to the binding site of hnRNP–H2 protein (33), was found by rMAPS to peak around the U2 snRNA binding site in the upstream intron (Figure 5I). The enrichment of this motif was also corroborated by similar results from a MEME de novo motif analysis of 125nt regions around the upregulated cassette exons (Supplementary Figures S4I, J), and is disproportionately found in the previously described subset of short upstream flanking introns within this dataset (Supplementary Figure S4K). There were a few more enriched motifs including C-rich upstream intronic sequences that could be bound by proteins like PCBP2 (Supplementary Figures S4I, L). The RBPs that could bind to the motifs identified by rMAPS did not show gene expression changes in Mettl4-KO (Figure 5J). Overall, the METTL4 modification appears to repress splicing as its loss of function increases cassette exon inclusion or splicing of retained introns, and these splicing events exhibit particular features like splice-site weakness and short introns. Mettl4-KO affected splicing events may possibly be linked to U2 snRNA modification, yet other mechanisms mediated by RBPs may also apply.
DISCUSSION
Previous studies have investigated the function of the human DNA modification N6-methyl-deoxyadenosine (6mA) (48–50). Based on a past phylogenetic prediction of N6-methylation writers (43), METTL4 was reported to be the writer that mediates 6mA methylation (51,52). Our work here demonstrates that METTL4 functions as a RNA N6-methylation writer. Specifically, nucleotide 30 of U2 snRNA is an adenosine known to be ribose-2′O-methylated to give Am (26), and was also recently shown to be N6-methylated to m6Am (18). By knocking out human Mettl4 and sequencing the resulting loss of RNA methylation, we confirmed that METTL4 is required for N6-methylation at adenosine 30 of U2 snRNA. We also used nucleoside HPLC-MS/MS to validate the identity of the resultant modification as m6Am. By overexpressing recombinant METTL4, we demonstrated the necessity of METTL4’s catalytic site in its methylation activity and that METTL4 is able to directly methylate U2 snRNA both in vitro and in vivo.
N 6-methylation writers such as METTL3 and PCIF1 have multiple in vivo targets and thus by comparing the difference in precise RNA methylomes before and after depleting these writers in cells, we can determine the in vivo target RNA preference for each writer (25). METTL4 however, only has 1 clear in vivo target, making such an analysis non-trivial. We circumvented this limitation by overexpressing either METTL4WT or METTL4CD in HEK293T cells to simulate an in vivo methylation assay. Comparing differences between the RNA methylomes in the 2 overexpressed cells revealed ectopic METTL4WT-dependent de novo methylation sites, allowing us to determine METTL4’s in vivo target preference to be ‘HMAGKD’. We note that presence of the ‘HMAGKD’ sequence alone does not guarantee in vivo de novo methylation by METTL4. This indicates that other cis-acting elements, such as RNA secondary structure or Am presence, or trans-acting factors, such as METTL4 co-factors help to guide this process. During the preparation of this manuscript, another study also reported METTL4 as a U2 snRNA m6Am writer (53) but due to a lack of m6Am-sequencing, could not conduct the extensive in vivo target RNA preference analysis of our study. Therefore, our findings will facilitate further studies on how METTL4 specifically targets U2 snRNA.
As opposed to U2 pseudouridines, which in yeast were shown to affect either U2 snRNP biogenesis, particle stability, splicing function and even growth phenotypes (54), the role of U2 m6Am function remains largely uncharacterized. Our transcriptomic analysis revealed a limited yet reliable set of splicing events affected in Mettl4-KO cells. Another study (53) found more splicing events affected by Mettl4-KO but the limited information of these splicing events did not allow us to directly compare our results. These differences could be accounted for by different cutoffs for RNA-seq analysis. Splicing events regulated by METTL4 had a typical distribution of subtypes (cassette exons, etc) as compared to most cellular processes or alterations of splicing factors (55). Our meta-analysis of METTL4-dependent differential alternative splicing events suggests that the splicing alterations are governed by 3′-splice site features: (i) A slight overabundance of changes in alternative 3′ splice sites over 5′ splice sites; (ii) an overall weakness of 3′ splice-site strength for cassette exons downregulated in Mettl4-KO cells; (iii) enrichment of motifs upstream the 3′ splice sites, such as that for hnRNP-H2 in upregulated cassette exons. We did not perform Branch Point Sequence (BPS) analyses because of the high degeneracy of this motif in humans, and because only 25% of our splicing targets have mapped BPS (56,57). These data hint that the METTL4-dependent splicing events mainly act via the 3′ splice site, which is directly linked to U2 snRNA and likely to its modifications.
In human major spliceosome introns (>99% of all introns), 3′ splice sites are recognized by the pre-formed and stable U2 Auxiliary Factor (U2AF) heterodimer, in which the large U2AF65 subunit binds to the polypyrimidine tract while the small U2AF35 subunit binds a sequence motif around the intron-terminal AG dinucleotide (58–63). Similar to other splicing signals, 3′ splice sites are highly heterogeneous in humans. Hence, the relative contribution of the two subunits to 3′ splice-site recognition is highly variable. This had previously led to the model of AG-dependent versus AG-independent introns, as the latter introns have long polypyrimidine tracts that make U2AF35 binding dispensable for 3′ splice-site recognition (64). U2AF binding to 3′ splice sites together with Splicing Factor 1 (SF1) to the BPS then recruits the U2 snRNP in an ATP-dependent manner to displace SF1, such that U2 snRNA base pairs to the BPS and bulges out an A to perform the first transesterification step of splicing (65,66). As the U2–30 position is not in the BPS recognition sequence yet 5′-adjacent to it, we hypothesize that the m6Am modification may affect the recruitment of U2 snRNP by U2AF in different manners depending on the pre-mRNA substrate, thereby affecting the overall splicing patterns to a small degree. Surprisingly, we found that Mettl4-KO tends to increase inclusion of cassette exons and splicing of retained introns. As a possible explanation, the U2 snRNA modification may mainly act in a repressive manner, perhaps by reducing binding of U2 snRNA to certain BPS. The short length of introns flanking the exons with increased inclusion upon Mettl4-KO, together with their high GC content, suggest that the U2 modification may particularly affect recognition of BPS by intron definition (67). Further studies should aim to identify the mechanistic details of splicing events affected by METTL4 depletion, likely by in vitro splicing reactions with reconstituted U2 snRNP particles without and with U2 snRNA position 30 m6Am modification, so as to derive the respective U2 interactomes, degree of U2 association with pre-mRNAs, and efficiency of spliceosome formation. Furthermore, the enrichment of GGGAGGG motif in the upstream flanking introns of several METTL4-dependent splicing events suggests that some such events might be regulated by the cognate RBP that recognizes this motif, which is hnRNP-H2 (68), perhaps in competition with U2 snRNA. The RBPs with enriched motifs in METTL4-dependent splicing events did not undergo any major expression or splicing change at RNA level upon Mettl4-KO, so if these factors account for a fraction of splicing events, they may be regulated by translation efficiency or post-translational modifications. Further studies should address the connection between METTL4 and hnRNP-H2 or other RBPs. Finally, the splicing events affected by Mettl4-KO did not reveal any significant enrichment in functional groups of genes (not shown). Nevertheless, it is possible that METTL4 could play tissue or developmental-specific roles, such as recently reported for adipocyte differentiation (69) and perhaps in other cells to be characterized in the future. In summary, our work here on the discovery of METTL4 as a U2 m6Am writer provides a framework to explain how METTL4 regulates pre-mRNA splicing, thereby proposing a new pathway for epitranscriptomic modifications to regulate RNA processing.
DATA AVAILABILITY
WT HEK293T m6ACE-seq data was downloaded from NCBI’s Gene Expression Omnibus (GEO) under accession number GSE124509 (25). Data generated in this work was deposited in NCBI’s GEO under accession number GSE144745.
CODE AVAILABILITY
The custom Python scripts used in this study are available on request from the corresponding author.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the Lezhava laboratory at GIS as well as D. Moses and J. Dacanay at NTU for assistance with sequencing.
Author contributions: W.S.S.G. conceived the study. Y.T.G., C.W.Q.K. and D.Y.S. performed the experiments. D.Y.S. and W.S.S.G. performed bioinformatics analysis. X.R. and W.S.S.G. supervised the experiments and wrote the manuscript with input from all authors.
Contributor Information
Yeek Teck Goh, Genome Institute of Singapore, 60 Biopolis Street, Singapore 138672, Singapore.
Casslynn W Q Koh, Genome Institute of Singapore, 60 Biopolis Street, Singapore 138672, Singapore.
Donald Yuhui Sim, School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore.
Xavier Roca, School of Biological Sciences, Nanyang Technological University, Singapore 637551, Singapore.
W S Sho Goh, Genome Institute of Singapore, 60 Biopolis Street, Singapore 138672, Singapore.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biomedical Research Council (A*STAR) Young Investigator Grant [1610151037 to W.S.S.G.]; Genome Institute of Singapore Independent Fellowship (to W.S.S.G.); Singapore's Ministry of Education Academic Research Fund Tier 2 [MOE2016-T2-2-104(S) to X.R]; Tier 1 [RG33/15 to X.R.]. Funding for open access charge: GIS Independent fellowship.
Conflict of interest statement. W.S.S.G. has filed a technology disclosure to the institutional technology transfer office and the office has filed a provisional patent application in Singapore on the use of photo-crosslinking RNA-modification-specific antibodies and exoribonucleases to sequence RNA modifications at high resolution. The other authors declare no competing interests.
REFERENCES
- 1. Zaccara S., Ries R.J., Jaffrey S.R.. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019; 20:608–624. [DOI] [PubMed] [Google Scholar]
- 2. Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R.. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012; 149:1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al.. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. [DOI] [PubMed] [Google Scholar]
- 4. Maden B.E.H. Identification of the locations of the methyl groups in 18 S ribosomal RNA from Xenopus laevis and man. J. Mol. Biol. 1986; 189:681–699. [DOI] [PubMed] [Google Scholar]
- 5. Maden B.E.H. Locations of methyl groups in 28 S rRNA of Xenopus laevis and man: Clustering in the conserved core of molecule. J. Mol. Biol. 1988; 201:289–314. [DOI] [PubMed] [Google Scholar]
- 6. Bokar J.A., Rath-Shambaugh M.E., Ludwiczak R., Narayan P., Rottman F.. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J. Biol. Chem. 1994; 269:17697–17704. [PubMed] [Google Scholar]
- 7. Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M.. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997; 3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X. et al.. A METTL3–METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2013; 10:93–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S.. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014; 24:177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C. et al.. Structural basis of N6-adenosine methylation by the METTL3–METTL14 complex. Nature. 2016; 534:575–578. [DOI] [PubMed] [Google Scholar]
- 11. Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. et al.. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014; 8:284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R.. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016; 537:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pendleton K.E., Chen B., Liu K., Hunter O.V., Xie Y., Tu B.P., Conrad N.K.. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017; 169:824–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma H., Wang X., Cai J., Dai Q., Natchiar S.K., Lv R., Chen K., Lu Z., Chen H., Shi Y.G. et al.. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat. Chem. Biol. 2019; 15:88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinto R., Vågbø C.B., Jakobsson M.E., Kim Y., Baltissen M.P., O’Donohue M.-F., Guzmán U.H., Małecki J.M., Wu J., Kirpekar F. et al.. The human methyltransferase ZCCHC4 catalyses N6-methyladenosine modification of 28S ribosomal RNA. Nucleic Acids Res. 2019; 21:174–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Tran N., Ernst F.G.M., Hawley B.R., Zorbas C., Ulryck N., Hackert P., Bohnsack K.E., Bohnsack M.T., Jaffrey S.R., Graille M. et al.. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019; 330:1203–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.-J., Chen Q. et al.. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017; 541:371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mauer J., Sindelar M., Despic V., Guez T., Hawley B.R., Vasseur J.-J., Rentmeister A., Gross S.S., Pellizzoni L., Debart F. et al.. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019; 15:340–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iyer L.M., Abhiman S., Aravind L.. Natural history of eukaryotic DNA methylation systems. Prog. Mol. Biol. Transl. Sci. 2011; 101:25–104. [DOI] [PubMed] [Google Scholar]
- 20. Iyer L.M., Zhang D., Aravind L.. Adenine methylation in eukaryotes: apprehending the complex evolutionary history and functional potential of an epigenetic modification. Bioessays. 2016; 38:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O. et al.. Cap-specific terminal N6-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science. 2018; 44:eaav0080-13. [DOI] [PubMed] [Google Scholar]
- 22. Sun H., Zhang M., Li K., Bai D., Yi C.. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019; 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Boulias K., Toczydłowska-Socha D., Hawley B.R., Liberman N., Takashima K., Zaccara S., Guez T., Vasseur J.-J., Debart F., Aravind L. et al.. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell. 2019; 75:631–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sendinc E., Valle-Garcia D., Dhall A., Chen H., Henriques T., Navarrete-Perea J., Sheng W., Gygi S.P., Adelman K., Shi Y.. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell. 2019; 75:620–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koh C.W.Q., Goh Y.T., Goh W.S.S.. Atlas of quantitative single-base-resolution N6-methyl-adenine methylomes. Nat. Commun. 2019; 10:5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Karijolich J., Yu Y.-T.. Spliceosomal snRNA modifications and their function. RNA Biol. 2010; 7:192–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goh W.S.S., Seah J.W.E., Harrison E.J., Chen C., Hammell C.M., Hannon G.J.. A genome-wide RNAi screen identifies factors required for distinct stages of C. elegans piRNA biogenesis. Genes Dev. 2014; 28:797–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Goh W.S.S., Falciatori I., Tam O.H., Burgess R., Meikar O., Kotaja N., Hammell M., Hannon G.J.. piRNA-directed cleavage of meiotic transcripts regulates spermatogenesis. Genes Dev. 2015; 29:1032–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17:3–12. [Google Scholar]
- 30. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:31–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Machanick P., Bailey T.L.. MEME-ChIP: motif analysis of large DNA datasets. Bioinformatics. 2011; 27:1696–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Olarerin-George A.O., Jaffrey S.R.. MetaPlotR: a Perl/R pipeline for plotting metagenes of nucleotide modifications and other transcriptomic sites. Bioinformatics. 2017; 33:1563–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lorenzini P.A., Chew R.S.E., Tan C.W., Yong J.Y., Zhang F., Zheng J., Roca X.. Human PRPF40B regulates hundreds of alternative splicing targets and represses a hypoxia expression signature. RNA. 2019; 25:905–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shen S., Park J.W., Lu Z.-X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. U.S.A. 2014; 111:E5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yeo G., Burge C.B.. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004; 11:377–394. [DOI] [PubMed] [Google Scholar]
- 38. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nettling M., Treutler H., Grau J., Keilwagen J., Posch S., Grosse I.. DiffLogo: a comparative visualization of sequence motifs. BMC Bioinformatics. 2015; 16:387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wagih O. ggseqlogo: a versatile R package for drawing sequence logos. Bioinformatics. 2017; 33:3645–3647. [DOI] [PubMed] [Google Scholar]
- 42. Hwang J.Y., Jung S., Kook T.L., Rouchka E.C., Bok J., Park J.W.. rMAPS2: an update of the RNA map analysis and plotting server for alternative splicing regulation. Nucleic Acids Res. 2020; 48:W300–W306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bujnicki J.M., Feder M., Radlinska M., Blumenthal R.M.. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA:m6A methyltransferase. J. Mol. Evol. 2002; 55:431–444. [DOI] [PubMed] [Google Scholar]
- 44. Dai Q., Moshitch-Moshkovitz S., Han D., Kol N., Amariglio N., Rechavi G., Dominissini D., He C.. Nm-seq maps 2′-O-methylation sites in human mRNA with base precision. Nat. Methods. 2017; 14:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Grozhik A.V., Jaffrey S.R.. Distinguishing RNA modifications from noise in epitranscriptome maps. Nat. Chem. Biol. 2018; 14:215–225. [DOI] [PubMed] [Google Scholar]
- 46. Baeza-Centurion P., Miñana B., Schmiedel J.M., Valcárcel J., Lehner B.. Combinatorial genetics reveals a scaling law for the effects of mutations on splicing. Cell. 2019; 176:549–563. [DOI] [PubMed] [Google Scholar]
- 47. Amit M., Donyo M., Hollander D., Goren A., Kim E., Gelfman S., Lev-Maor G., Burstein D., Schwartz S., Postolsky B. et al.. Differential GC content between exons and introns establishes distinct strategies of splice-site recognition. Cell Rep. 2012; 1:543–556. [DOI] [PubMed] [Google Scholar]
- 48. Wu T.P., Wang T., Seetin M.G., Lai Y., Zhu S., Lin K., Liu Y., Byrum S.D., Mackintosh S.G., Zhong M. et al.. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016; 532:329–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U. et al.. N6-methyladenine DNA modification in glioblastoma. Cell. 2018; 175:1228–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koh C.W.Q., Goh Y.T., Toh J.D.W., Neo S.P., Ng S.B., Gunaratne J., Gao Y.-G., Quake S.R., Burkholder W.F., Goh W.S.S.. Single-nucleotide-resolution sequencing of human N6-methyldeoxyadenosine reveals strand-asymmetric clusters associated with SSBP1 on the mitochondrial genome. Nucleic Acids Res. 2018; 14:204–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Greer E.L., Blanco M.A., Gu L., Sendinc E., Liu J., Aristizábal-Corrales D., Hsu C.-H., Aravind L., He C., Shi Y.. DNA methylation on N(6)-adenine in C. elegans. Cell. 2015; 161:868–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kweon S.-M., Chen Y., Moon E., Kvederaviciutė K., Klimasauskas S., Feldman D.E.. An adversarial DNA N6-methyladenine-sensor network preserves polycomb silencing. Mol. Cell. 2019; 74:1138–1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chen H., Gu L., Orellana E.A., Wang Y., Guo J., Liu Q., Wang L., Shen Z., Wu H., Gregory R.I. et al.. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 2020; 30:544–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao Y., Dunker W., Yu Y.-T., Karijolich J.. The role of noncoding RNA pseudouridylation in nuclear gene expression events. Front. Bioeng. Biotechnol. 2018; 6:966–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nilsen T.W., Graveley B.R.. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010; 463:457–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mercer T.R., Clark M.B., Andersen S.B., Brunck M.E., Haerty W., Crawford J., Taft R.J., Nielsen L.K., Dinger M.E., Mattick J.S.. Genome-wide discovery of human splicing branchpoints. Genome Res. 2015; 25:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taggart A.J., Lin C.-L., Shrestha B., Heintzelman C., Kim S., Fairbrother W.G.. Large-scale analysis of branchpoint usage across species and cell lines. Genome Res. 2017; 27:639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sheth N., Roca X., Hastings M.L., Roeder T., Krainer A.R., Sachidanandam R.. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006; 34:3955–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zamore P.D., Green M.R.. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J. 1991; 10:207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Merendino L., Guth S., Bilbao D., Martínez C., Valcárcel J.. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature. 1999; 402:838–841. [DOI] [PubMed] [Google Scholar]
- 61. Wu S., Romfo C.M., Nilsen T.W., Green M.R.. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature. 1999; 402:832–835. [DOI] [PubMed] [Google Scholar]
- 62. Zamore P.D., Patton J.G., Green M.R.. Cloning and domain structure of the mammalian splicing factor U2AF. Nature. 1992; 355:609–614. [DOI] [PubMed] [Google Scholar]
- 63. Zorio D.A.R., Blumenthal T.. Both subunits of U2AF recognize the 3′ splice site in Caenorhabditis elegans. Nature. 1999; 402:835–838. [DOI] [PubMed] [Google Scholar]
- 64. Reed R. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 1989; 3:2113–2123. [DOI] [PubMed] [Google Scholar]
- 65. Berglund J.A., Chua K., Abovich N., Reed R., Rosbash M.. The splicing factor BBP interacts specifically with the Pre-mRNA branchpoint sequence UACUAAC. Cell. 1997; 89:781–787. [DOI] [PubMed] [Google Scholar]
- 66. Parker R., Siliciano P.G., Guthrie C.. Recognition of the TACTAAC box during mRNA splicing in yeast involves base pairing to the U2-like snRNA. Cell. 1987; 49:229–239. [DOI] [PubMed] [Google Scholar]
- 67. De Conti L., Baralle M., Buratti E.. Exon and intron definition in pre-mRNA splicing. WIREs RNA. 2013; 4:49–60. [DOI] [PubMed] [Google Scholar]
- 68. Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A. et al.. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013; 499:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Z., Hou Y., Wang Y., Gao T., Ma Z., Yang Y., Zhang P., Yi F., Zhan J., Zhang H. et al.. Regulation of adipocyte differentiation by METTL4, a 6 mA methylase. Sci. Rep. 2020; 10:8285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
WT HEK293T m6ACE-seq data was downloaded from NCBI’s Gene Expression Omnibus (GEO) under accession number GSE124509 (25). Data generated in this work was deposited in NCBI’s GEO under accession number GSE144745.