Abstract
While the mechanism of CRISPR/Cas9 cleavage is understood, the basis for the large variation in mutant recovery for a given target sequence between cell lines is much less clear. We hypothesized that this variation may be due to differences in how the DNA damage response affects cell cycle progression. We used incorporation of EdU as a marker of cell cycle progression to analyze the response of several human cell lines to CRISPR/Cas9 treatment with a single guide directed to a unique locus. Cell lines with functionally wild-type TP53 exhibited higher levels of cell cycle arrest compared to lines without. Chemical inhibition of TP53 protein combined with TP53 and RB1 transcript silencing alleviated induced arrest in TP53+/+ cells. Using dCas9, we determined this arrest is driven in part by Cas9 binding to DNA. Additionally, wild-type Cas9 induced fewer 53BP1 foci in TP53+/+ cells compared to TP53−/− cells and DD-Cas9, suggesting that differences in break sensing are responsible for cell cycle arrest variation. We conclude that CRISPR/Cas9 treatment induces a cell cycle arrest dependent on functional TP53 as well as Cas9 DNA binding and cleavage. Our findings suggest that transient inhibition of TP53 may increase genome editing recovery in primary and TP53+/+ cell lines.
INTRODUCTION
Genome engineering is a powerful tool, not only for modifying cells for therapeutic uses, but also for examining endogenous expression and localization of proteins, as well as aiding in deciphering their interactions with other proteins. In terms of semi-targeted insertion into the genome, the phage integrases, particularly phiC31, have been very useful for expressing exogenous cassettes in a variety of plants and animals for both translational and basic science purposes (reviewed in 1–2). However, targeting specific, single loci in the genome relies on homologous recombination (HR), which although highly efficient in yeast and some other simple model organisms (3–5), has been much less efficient in metazoans. With the development of the zinc-finger nucleases (ZFN; 6) and the TALE-nucleases (TALENs; 7), gene editing in metazoans and cells derived from them became a more feasible method of investigation and therapeutic development. These advances in the genome engineering toolbox built on seminal work with the I-SceI endonuclease (8), and greatly increased the efficiency of HR by introduction of a targeted double-strand break (DSB) in genomic DNA, while also allowing mutagenesis through the error-prone repair pathway due to the overhang-style of break induced by these nucleases.
Whereas the ZFNs and TALENs offered an unprecedented degree of control over where to induce a DSB due their modular, programmable nature, the design and cloning of these tools is challenging. This impediment has been reduced by discovery and application of the CRISPR/Cas9 system, which has proven to be a versatile tool for genome and transcriptome engineering. Consisting of a nuclease directed to a genomic target by a guide RNA, the engineered version of Cas9 requires only cloning the 20-base targeting sequence for the variant derived from Streptococcus pyogenes, for example (9–11). The major constraint on use of Cas9 is the requirement of a protospacer adjacent motif (PAM) downstream of the target sequence. This site varies in size and sequence between the different Cas9 proteins. For SpCas9, the PAM is canonically NGG (9), and the protein makes a predominantly blunt-ended DSB three to four bases 5′ from the PAM (12,13).
Whichever nuclease is used for genome engineering, there are still challenges faced in the efficient generation of the desired clones or organisms. For example, in addition to a rather cryptic target programming system, the ZFNs are highly cytotoxic (14). This cytotoxicity is greatly reduced when using TALENs, increasing the efficiency with which the desired modifications can be obtained (14). The cytotoxicity of the CRISPR/Cas9 varies between different cell lines, but is, unfortunately, high in induced pluripotent and embryonic stem cells (15,16), among the prime candidates for targeting. Also, the frequency of off-target mutations appears to be more severe with Cas9, as a recent report found larger deletions around the DSB and more complex rearrangements than previously identified (17). However, other studies, including both wild-type and nuclease-dead Cas9, have reported extremely high specificity, a virtual lack of true off-target activity, and an absence of complex rearrangements (18–20). Further compounding these issues is the high variance in gene editing efficiency between cell types and lines for a given guide RNA (13,21).
We asked if this high variance in gene editing efficiency could be explained by cell-line-specific cell cycle delay or arrest in response to CRISPR/Cas9 treatment. We reasoned that, because Cas9 predominantly generates blunt-ended DSBs (12,13), the DNA repair machinery would precisely repair the break through canonical NHEJ, which would enable Cas9 to target and cleave the same sequence again, enabling a cycle of damage and repair. This cycle would be expected to at least delay cells from progressing through the cell cycle and possibly arrest cells (22). Consequentially, this delay would reduce the efficiency of CRISPR/Cas9-based genome editing by reducing the frequency of modified cells, and the beneficial effect of removal of Cas9 from its target sequence via destabilization of Cas9 itself would be due to reducing this cell cycle delay. Such a destabilized Cas9 has been previously generated by fusing FKBP12-L106P destabilization domain (23) fused to the N-terminus of wild-type Cas9 to generate DD-Cas9 (13,24). DD-Cas9 can be inducibly and reversibly stabilized by the addition of the small molecule Shield-1 (13,23,24).
Here, we characterize the response of several human cell lines to wild-type, destabilization domain-linked, and nuclease-dead SpCas9 targeted to the safe harbor locus H11 on chromosome 22 (25). We identify a strong cell cycle delay/arrest phenotype that appears to be dependent on the TP53 status of each cell line. We determine that inhibition and transcript silencing of TP53 reduces the effect of this delay and illustrate that Cas9 appears to block the repair machinery from recognizing the DSB. Furthermore, we show that CRISPR/Cas9 treatment arrests cells to a higher degree than TALEN treatment.
MATERIALS AND METHODS
Cell culture
hTERT RPE-1 wild-type and p53-null (from M.-F. Tsou, Memorial Sloan Kettering Cancer Center) cells, HCT-116 wild-type and p53-null cells, HEK293T cells and U2-OS cells were maintained in DMEM/F-12 media (10-092-CV, Corning, Corning, NY, USA) supplemented with 10% Cosmic calf serum (SH3008703, HyClone Laboratories, Logan, UT). Cells were passaged with 0.25% Trypsin-EDTA (25-052-CI, Corning) at a 1:10 split and maintained in 10-cm tissue culture dishes.
Western blotting
RPE1 cells plated in six-well plates one day prior were transfected with polyethylenimine (PEI, homemade) and 2 μg of plasmid encoding the H11-r1-2 sgRNA and DD-Cas9 (13) in the presence or absence of 0.5 μM Shield-1 in ethanol (632189, Clontech Laboratories, Inc., Mountain View, CA, USA). After 24 h, Shield-1 was washed out with phosphate-buffered saline (PBS, homemade, pH 7.4) and replaced with fresh media. Protein was isolated at the time of washout, 24 h later, and 48 h later by extraction with RIPA buffer (150 mM sodium chloride, 1% NP-40 alternative, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0]) and immediately flash-frozen in liquid nitrogen.
For Western blotting using RPE1 cell lysate, 10 μg of protein were denatured in the presence of 1× Laemmli buffer and 0.1 M DTT at 100°C for 5 min. Samples were then resolved on an 8% SDS-PAGE gel at 100 V in 1× Running buffer. Protein was transferred to a nitrocellulose membrane (1620112, Bio-Rad Laboratories, Hercules, CA, USA) under wet transfer conditions at 200 mA for 1 h with constant current at 4°C. Following transfer, the membrane was blocked for 1 h at room temperature with shaking in Tris-buffered saline + 0.2% Tween-20 (TBST) + 5% milk. Membranes were incubated overnight with agitation at 4°C with 1:2000 monoclonal mouse anti-alpha-tubulin (T9026, Sigma-Aldrich, St. Louis, MO, USA; clone DM1A) and 1:1000 monoclonal mouse anti-Cas9 (844302, Biolegend, San Diego, CA, USA) The following day, membranes were washed 3 times for 5 minutes each with TBST. Secondary incubation was carried out at room temperature with agitation for 1 hour with either donkey anti-mouse IgG IRdye800CW (926-32212, LI-COR Biosciences, Lincoln, NE, USA) or goat anti-mouse IgG IRdye680RD (926-68070, LI-COR Biosciences) followed by washing three times for 5 min each with TBST. Membranes were then imaged on a LI-COR Odyssey (LI-COR Biosciences).
Cell cycle progression analysis
RPE1 cells were plated one day prior to transfection on poly-l-lysine-coated glass coverslips at 2.5 × 104 cells per well in a 24-well plate. The following day cells were transfected via PEI with 500 ng of either pKER-Clover (13) or pKER-NLS-Clover or a vector encoding the H11-r1-2 or H11 r2-3 sgRNA and WT-Cas9 or DD-Cas9 (initially from 13), in the presence or absence of 0.5 μM Shield-1 in ethanol (Clontech Laboratories). Twenty four hours after transfection, Shield-1 was washed out with PBS and replaced with fresh media containing 10 μM EdU in DMSO (A10044, Thermo Fisher Scientific, Waltham, MA, USA). Approximately 45 h after EdU addition, cells were washed with cold PBS, fixed for 10 min at room temperature with 4% formaldehyde (diluted from 50-00-0, Electron Microscopy Sciences, Hatfield, PA, USA) for 10 min, and rehydrated with PBS for 10 min. Cells were then blocked and permeabilized with PBS containing 5% bovine serum albumin (A9418, Sigma-Aldrich) and 0.2% Tween-20 (PBS-BT) for 30 minutes. After blocking, the click reaction between EdU and Alexafluor-594 azide (A10270, Life Technologies) was carried out using the Click-It 594 Labeling Kit (C10086, Life Technologies) according to the manufacturer's instructions. After labeling and subsequent washing of cells three times for 5 min each with PBS + 0.2% Tween-20 (PBS-T), cells were incubated with 1:500 goat anti-GFP (600-101-215, Rockland Immunochemicals, Limerick, PA, USA) and/or 1:1000 mouse IgG1 anti-Cas9 in PBSBT overnight at 4°C. The following day cells were washed three times with PBS-T for 5 min each and incubated for 1 hour in the dark at room temperature with DAPI and either 1:1000 donkey anti-goat IgG Alexafluor-488 (A-11055, Life Technologies) or 1:1000 donkey anti-mouse IgG Alexafluor-488 (A-21202, Life Technologies) and DAPI (D9542, Sigma-Aldrich). Coverslips were then washed with PBS-T three times for 5 min each and affixed to slides via MOWIOL.
Slides were imaged on an Axioskop 200M microscope. Three 423 μm × 324 μm windows were analyzed per coverslip in ImageJ by counting total number of nuclei, number of GFP+/Cas9+ cells, and number of GFP+/Cas9+ EdU+ nuclei and the average was calculated for each coverslip.
For flow cytometry-based analysis, cells were plated at 5 × 104 to 7.5 × 104 cells per well in triplicate in 24-well plates and transfected 24 h later as described above. All plasmids included an EF-1α–driven Clover expression cassette with an N-terminal nuclear localization sequence, one of the guides described above and WT-Cas9, DD-Cas9, dCas9, or APEX2-X-dCas9. One additional plasmid encoded the NLS-Clover cassette and WT-SpCas9 alone. Where appropriate, cells were treated with 20 μM pifithrin-α in DMSO (P4359, Sigma-Aldrich; PFTα) from 24 h pre-transfection through the end of the experiment, with replacement every 24 h. Shield-1, where appropriate, and EdU were added as described above. Roughly 45 h post-EdU-addition, cells were harvested via trypsinization into 1.75 ml microcentrifuge tubes, centrifuged at 300 × g for 5 min at 4°C, washed with 800 μl PBS, centrifuged at 300 × g for 5 min at 4°C, and fixed for 10 min with 200 μl 4% formaldehyde. All subsequent centrifugations were carried out at 300 × g for 5 min at 4°C. Cells were then washed twice with 800 μl PBS before blocking and permeabilization as described above. Click chemistry and immunofluorescence were carried out as a described above staining for Clover, Cas9 and EdU with the following changes: Click chemistry was carried out before primary antibody incubation, anti-Cas9 was used at 1:500, all stainings were carried out for 30 min at room temperature in the dark, two washes were carried out between click chemistry, primary staining, and secondary staining with addition of 500 μl of PBS-T followed by centrifugation at 300 × g for 5 min at 4°C. Secondary staining used the following antibodies and stains: 1:1000 donkey anti-goat IgG Alexafluor-488 (A-11055, Life Technologies), 1:1000 donkey anti-mouse IgG Alexafluor-680 (A10038, Life Technologies), and DAPI. Following the final wash after secondary staining, cell pellets were resuspended in 200 μL of PBS and transferred to polypropylene FACS tubes (35002, BD Falcon) for analysis.
For experiments involving short hairpin RNAs, the list of transfected plasmids also included plasmids expressing nuclear-localized Clover, WT-Cas9, and either a single short-hairpin expression cassette directed against TP53 or a scramble shRNA control, or two short-hairpin expression cassettes directed against TP53 and RB1. The TP53 hairpin cassette was cloned unchanged via PCR from pCXLE-hOCT3/4-shp53-F, a gift from Shinya Yamanaka (41, Addgene plasmid # 27077). The RB1 short hairpin cassette was cloned via PCR unchanged from pMKO.1 puro RB shRNA, a gift from William Hahn (42, Addgene plasmid # 10670). The scramble shRNA cassette was cloned unchanged via PCR from a scramble shRNA plasmid, a gift from David Sabatini (43, Addgene plasmid #1864). After cloning into the Cas9 plasmids, the sequence integrity of the short-hairpin cassettes were checked by Sanger sequencing. The sequence of the TP53 hairpin is 5′- gactccagtggtaatctacttcaagagagtagattaccactggagtc-3′. The sequence of the RB hairpin is 5′-ccggcagagatcgtgtattgagattctcgagaatctcaatacacgatctctgtttttgaatt-3′. The sequence of the scramble hairpin is 5′-cctaaggttaagtcgccctcgctcgagcgagggcgacttaaccttagg-3′.
Live imaging of transfected RPE-1 cells
150,000 wild-type RPE-1 cells were plated on an ibidi 35mm μ-Dish (81156, ibidi GmbH, Germany). Twenty four hours later, cells were transfected with 2 μg of the plasmid encoding the H11r2-3 sgRNA, NLS-Clover, and WT-Cas9 or the plasmid encoding NLS-Clover with PEI. At 24 h post-transfection, cells were imaged for brightfield and Clover expression on a Keyence BZ-X710 fluorescence microscope (Keyence, Osaka, Japan) with an S Plan Fluor 20×/0.45 objective (Nikon, Tokyo, Japan) in a humidified chamber with 5% CO2. Cells were imaged every 15 min over 45–50 h. Additionally, some transfections were imaged for 20 h beginning 4 days post-transfection. Individual imaging windows were analyzed in ImageJ for number of mitoses of transfected and untransfected cells.
shRNA and pifithrin-α validation
The short-hairpin expression cassettes for shScramble, shTP53 and shRB described above were amplified via PCR and cloned in the pJet plasmid backbone vector via the CloneJet PCR cloning kit (K1232, ThermoFisher). Each expression cassette was verified by Sanger sequencing. To validate the efficiency of these shRNAs in our RPE-1 cells, we plated 50 000 cells per well in a 24-well plate on poly-d-lysine-coated coverslips one day before transfection with 500 ng of plasmid via Lipofectamine 3000 transfection reagent (L300000, ThermoFisher) according to the manufacturer's instructions. Forty eight hours post-transfection, cells were fixed via methanol at −20°C for 10 min, rehydrated for 10 min, and blocked and stained as described above. TP53 and RB1 were stained for on separate coverslips. Primary antibodies used were 1:500 mouse IgG2a anti-TP53 (DO-1, 645701, BioLegend) and 1:100 mouse IgG2a anti-RB1 (H-2, sc-74570, Santa Cruz Biotechnology, Dallas, TX). The secondary antibody used was 1:1000 donkey anti-mouse IgG AlexaFluor-568 (A10037, ThermoFisher). Nuclei were stained with DAPI. Coverslips were then imaged on a Keyence BZ-X710 fluorescence microscope. Levels of TP53 and RB1 were quantified in ImageJ. Background fluorescence correction was calculated via averaging of (integrated density/area) for three regions per image. Corrected fluorescence was calculated as integrated density – (area × background fluorescence correction factor).
RPE-1 cells were plated on poly-d-lysine-coated glass coverslips at 50 000 cells per coverslips in individual wells of a 24-well in the presence of 20 μM pifithrin-α or DMSO. Twenty four hours later, media was replaced with media containing 50 μM Etoposide (E1383, Sigma-Aldrich) and either 20 μM pifithrin-α or DMSO. Cells were incubated for 48 h with media replacement at 24 h. Following the incubation, cells were fixed with methanol at −20°C for 10 min, rehydrated for 10 min, and blocked and stained as described above. The primary antibodies used were 1:500 mouse IgG2b anti-53BP1 (612522, BD Biosciences) and 1:200 rabbit anti-p21 (14-6715-81, eBioscience). The secondary antibodies used were 1:1000 donkey anti-rabbit IgG AlexaFluor-488 (A-21206, ThermoFisher) and 1:1000 donkey anti-mouse IgG AlexaFluor-568. Nuclei were stained with DAPI. Coverslips were then imaged on a Keyence BZ-X710 fluorescence microscope and cells were scored based on gross presence or absence of 53BP1 foci.
Flow cytometry
Flow cytometric analysis for this project was done on instruments in the Stanford Shared FACS Facility. Flow cytometric analysis was carried out on an LSRII-class analyzer (BD Biosciences, San Jose, CA, USA). DAPI was detected via excitation with a 405-nm violet laser and 450 ± 50 nm BP filter. AlexFluor-488/Clover was detected via excitation with a 488-nm blue laser, a 505 nm LP splitter, and 525 ± 50 nm BP filter. AlexaFluor-594 was detected via excitement with 532-nm green laser, a 600 nm LP splitter, and a 610 ± 20 nm BP filter. AlexaFluor-680 was detected via excitement with a 640-nm red laser, a 685 nm LP splitter, and a 710 ± 50 nm BP filter.
For the EdU-based cell cycle assay, gates for immunofluorescence were set via a combination of completely unlabled and untransfected cells, pKER-Clover-treated-alone cells, and fluorescence-minus-one immunostained cells for each AlexaFluor (i.e. three additional controls with each fully stained with the exception of one different AlexaFluor-labeled secondary). The order of gating is as follows: initially scatter → singlets → Clover+ → [Cas9 EdU] subpopulations. Data for 10 000 cells were collected for each sample via the initial scatter gate. Additionally, we found that collecting on the Clover+ gate ultimately yielded no difference in [Cas9 EdU] subpopulation percentages compared to collecting on Scatter. All flow cytometric data were analyzed using FlowJo software (Tree Star, Ashland, OR, USA).
Double-strand break visualization
3.5 × 104, 5 × 104 or 7.5 × 104 HEK293T, WT-, or TP53−/− RPE-1 cells were plated on poly-D-lysine-coated coverslips in a 24-well plate. Twenty four hours later, cells were transfected with plasmid vectors encoding nuclear-localized Clover and either WT-Cas9 and H11-r2-3 guide or DD-Cas9 and H11-r1-2 guide in the presence of 0.5 μM Shield-1 (which was washed out 24 h post-transfection with PBS). At 3 days post-transfection, cells were washed with PBS, fixed with methanol for 10 min at −20°C, rehydrated in cold PBS at room temperature for 10 min, and blocked with PBS-BT for a minimum of 30 min. Cells were then incubated with the following primary antibodies for 1 h at room temperature: 1:500 mouse IgG2b anti-53BP1 and 1:200 rabbit anti-p21. Cells were then washed three times for 5 min each with PBS-T before secondary antibody staining in PBS-BT with the following antibodies and DAPI: 1:1000 AlexaFluor-488 donkey anti-goat IgG, 1:1000 AlexaFluor-568 donkey anti-mouse IgG, 1:1000 AlexaFluor-680 donkey anti-rabbit IgG. Cells were then washed three times for 5 min each with PBS-T, sealed to a slide with MOWIOL, and cured overnight. Slides were imaged on a Keyence BZ-X710 fluorescence microscope. Cells were then quantified by counting transfected cells categorized by the presence or absence of 53BP1 foci and the presence of nuclear p21 in ImageJ.
Ara-C-based proliferation assay
RPE-1 WT and p53-null cells were plated at 1.25 × 105 cells per well in each well of a 12-well tissue culture plate. The following day, cells were transfected in triplicate with PEI and plasmid vectors encoding nuclear-localized Clover alone or (i) WT-Cas9 and H11r2-3 guide, (ii) one of a pair of TALENs targeting the human H11 locus or (iii) both TALENs. Twenty four hours after transfection, cytosine arabinoside (ara-C; C-1768, Sigma-Aldrich) was added to two wells of each condition at a concentration of 100 μM. The third replicate of each condition was treated with a volume of water equivalent to the volume of ara-C added to the media. Ara-C treatment was maintained for 5 days with replacement every 2 days. After 5 days of treatment, the cells were imaged on an IncuCyte Zoom (Essen BioScience, Ann Arbor, Michigan), with nine images taken per well. The IncuCyte's analysis software was then used to calculate the number of transfected cells per image. Afterwards, the ratio of surviving Clover+ cells in each ara-C replicate relative to the water control for that condition was calculated and compared to a theoretical ratio of 1 using a one-sample t-test.
Statistics
All statistical analysis was carried out using Graphpad Prism 7 or 8 for Windows (Graphpad Software, Inc.). For all Cas9 EdU subpopulation analyses, ordinary two-way ANOVAs followed by post-hoc Dunnett's multiple comparisons tests were used with each subpopulation category constituting a separate family and each treatment being compared to the WT-RPE-1 treatment. To analyze the percentage of observed mitoses for Clover alone versus WT-Cas9, a one-way ANOVA followed by Sidak's multiple comparisons test was employed. To compare EdU+ percentages between RPE-1 cells treated with WT-Cas9 in the presence or absence of a guide RNA, we employed a one-way ANOVA followed by Sidak's multiple comparisons test. We used the same statistical approach to analyse the percentage of transfected cells between Clover-alone, WT-Cas9 with a guide RNA, and WT-Cas9 without a guide RNA. To analyze the percentage of 53BP1 foci between different Cas9 treatments and cell types, we employed an ordinary two-way ANOVA followed by a post-hoc Dunnett's multiple comparisons test comparing WT-Cas9-treated RPE-1 cells against the other treatments. To analyze the effect of ara-C on Cas9-treated cells, we compared the ratio of transfected cells in ara-C-treatment over vehicle to the theoretical value of 1 using a one-sample t-test. For examining the percentage of transfected cells as well as EdU positivity via immunofluorescence microscopy, we employed ordinary one-way ANOVAs followed by post-hoc Tukey's multiple comparisons tests making all possible comparisons. The knockdown efficacy of shRNAs was analyzed via two-tailed, unpaired t-tests. To determine the effect of pifithrin-α on RPE-1 cells, we utilized a two-way ANOVA followed by a post-hoc Sidak's multiple comparisons test. For all analyses, we used α = 0.05.
RESULTS
Wild-type SpCas9 leads to dilution of transfected cells in the population by limiting S-Phase progression in RPE-1 cells
Having previously characterized the behaviour of destabilized variants of SpCas9 in HEK293T cells (13), we sought to characterize the kinetics of destabilized SpCas9 in a more primary-like immortalized cell line, RPE-1, with the goal of optimizing genome engineering in these cells. In this experiment, we transfected RPE-1 cells with the DD-Cas9 and WT-Cas9 vectors in the presence or absence of the stabilizing small molecule ligand Shield-1 for 24 h before washout. We examined three time points: immediately after washout (24 h post-transfection), 4 h post-washout, and 24 h post-washout. Western blotting revealed little change in DD-Cas9 levels by 24 h post-washout, regardless of Shield-1 treatment, as would be expected due to continued production of the protein and its large size (Supplementary Figure S1A). However, we observed that WT-Cas9 level decreased more rapidly than the destabilization variants by 24 h post-washout, which is consistent with dilution of transfected cells within the population (Supplementary Figure S1A).
We remained concerned about the apparent dilution of cells transfected with wild-type Cas9 as compared to the DD-Cas9-transfected cells. We reasoned that this dilution could be caused by delayed cell cycle progression or cell cycle arrest generated from repeated cycles of cleavage and repair by Cas9. To this end, we transfected RPE-1 cells with a vector encoding the H11-r1-2 sgRNA and WT-Cas9, or DD-Cas9, in the presence or absence of Shield-1 for 24 h with and without washout and visualized newly synthesized DNA in transfected cells with EdU for approximately two doublings (45-48 hours) following washout. pKER-Clover (13), a vector encoding Clover fluorescent protein under the control of the EF1α promoter, was used as a transfection control. Fluorescence microscopy revealed that the percentage of transfected cells was not significantly different between Cas9 treatments (Supplementary Figure S1B), but there were significant differences compared to Clover transfected cells (Supplementary Figure S1C; summarized in Supplementary Table S1), and that there were significant differences between treatments in the percent of cycling cells within the Cas9+ population as visualized by EdU incorporation (Supplementary Figure S1D-E; summarized in Supplementary Table S2), overall indicating a severe delay of cell cycle progression associated with WT-Cas9 and suggesting that the observed decrease in WT-Cas9 protein levels by Western blot is due to dilution of the signal by actively dividing non-transfected cells.
CRISPR/Cas9-associated cell cycle delay is highly variable between different cell types
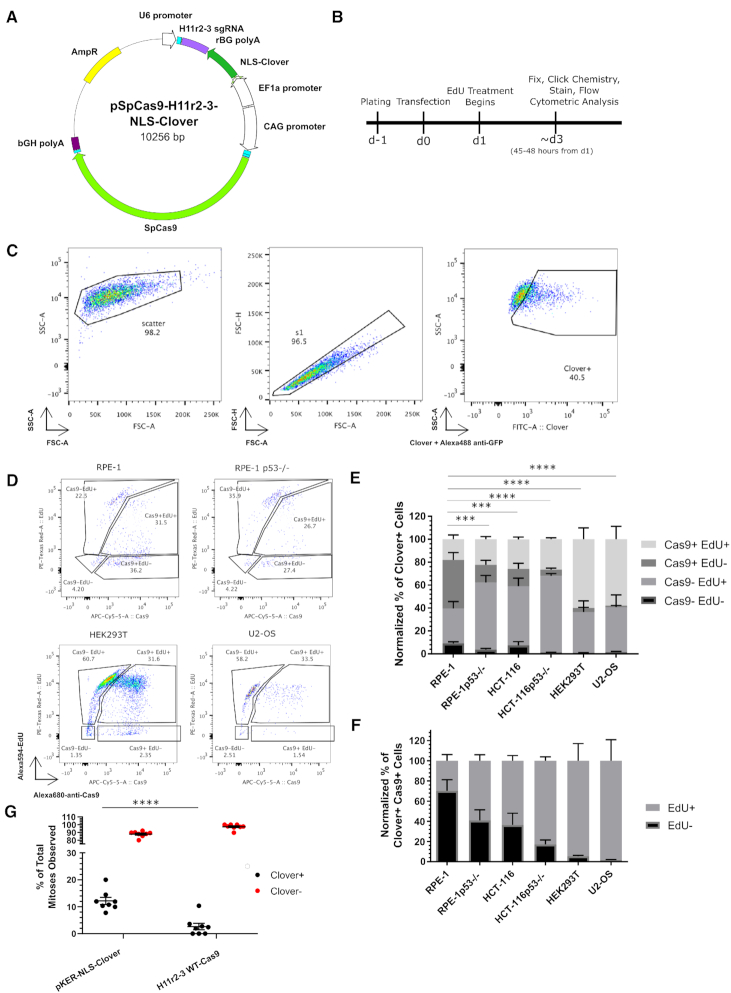
Given the low level of EdU+ cells observed in the CRISPR/Cas9-treated cells and the relatively low-throughput of our immunofluorescence approach, we repeated our experiment in wild-type RPE-1 cells using flow cytometry. Additionally, we included HEK293T, U2-OS, TP53−/− RPE-1 and wild-type and TP53−/- HCT-116 cells in our panel. To control for transfection efficiency, we included a nuclear-localized Clover (NLS-Clover) reporter cassette on the CRISPR/Cas9 vector under the control of the EF1α promoter (Figure 1A). We followed the same experimental timeline as we used for the immunofluorescence analysis (Figure 1B). After gating on Clover+ cells (Figure 1C), we further analyzed cells based on Cas9 and EdU positivity (Figure 1D). We first categorized the Clover+ population into [Cas9+ EdU+], [Cas9+ EdU-], [Cas9- EdU+] and [Cas9- EdU-] subpopulations, and noted that transfected HEK293T and U2-OS cells displayed larger percentages of [Cas9+ EdU+] cells (59.92 ± 9.95 and 57.99 ± 11.22% respectively) than both types of RPE-1 and HCT-116 cells ([18.03 ± 3.71% for RPE-1 cells], [22.34 ± 2.32% for TP53−/− RPE-1], [25.88 ± 1.97% for wild-type HCT-116], and [26.54 ± 1.29% for TP53−/− HCT-116]; Figure 1E). Next, we compared each subpopulation between wild-type RPE-1 and all other cell types. For the [Cas9+ EdU-] subpopulation, we found that RPE-1 cells were significantly different from the other cell types ([42.35 ± 6.51% RPE-1] versus [15.36 ± 3.99% TP53−/− RPE-1], [15.12 ± 4.93% HCT-116], [5.45 ± 1.40% TP53−/− HCT-116], [3.54 ± 0.52% HEK293T], and [1.13 ± 0.22% U2-OS]; P < 0.001 for all comparisons). The comparisons for the other subpopulations appear in Supplementary Table S3. For clarity, we then considered the Cas9+ subpopulations alone, reasoning that transfected cells expressing Cas9 at detectable levels would be most likely to be cells possessing DSBs (Figure 1F). After normalization of the Cas9+ population, we observed that WT RPE-1 cells displayed the lowest percentage of EdU+ cells (29.78 ± 6.12% of Cas9+ cells), whereas HEK293T and U2-OS cells displayed the highest amounts of EdU+ cells (94.51 ± 17.01 and 98.18 ± 20.88%, respectively). We also observed that the TP53−/− RPE-1 and HCT-116 cell lines displayed higher percentages of EdU+ cells than their wild-type counterparts (Figure 1F). These results indicated that cell cycle progression is inhibited to varying degrees in response to CRISPR/Cas9 treatment in different cell types, and that TP53 mediates this inhibition of cell cycle progression.
Figure 1.
CRISPR/Cas9 treatment decreases cell cycle progression. (A) Schematic of transfected plasmid, identifying the H11 r2-3 gRNA cassette, the NLS-Clover cassette, and the WT-Cas9 cassette. (B) Experimental design schematic. (C) Flow cytometry gating strategy for identifying the transfected (Clover+) subpopulation. (D) Representative flow cytometric data of transfected cells for Cas9 and EdU positivity for the indicated cell lines. (E) Bar graph of the normalized percentages of four Cas9 EdU subpopulations in transfected cells for the six indicated cell lines. n = at least two independent experiments consisting of three technical replicates for each transfection. Data are shown as the mean ± SEM. Data were analyzed via an ordinary two-way ANOVA followed by a post-hoc Dunnett's multiple comparisons test comparing each cell type to RPE-1 cells. Significance is shown for the [Cas9+ EdU-] subpopulation comparisons. ***P < 0.001, ****P < 0.0001. (F) Bar graph of the Cas9+ normalized percentages of EdU+ and EdU- from panel E. Data is shown as the mean ± SEM. (G) Quantification of observed mitoses during live imaging of transfected and untransfected RPE-1 cells over a roughly two-day period. Each dot represents an individual movie and are from a total of 2 biological replicates. Transfected cells are indicated in black and untransfected cells are indicated in red. Each black dot has a corresponding red dot. Mean ± SEM are indicated by bars. ****P < 0.001. Data were analyzed by a two-way ANOVA followed by Sidak's multiple comparisons test.
Additionally, we chose to directly examine if WT-Cas9-tranfected cells would divide, using live-cell fluorescence microscopy. We transfected WT RPE-1 cells with either pKER-NLS-Clover or the WT-Cas9 plasmid encoding NLS-Clover and the H11r2-3 sgRNA and began imaging 24 h post-transfection for approximately two days. We then quantified the number of mitoses for both transfected (Clover+) and untransfected (Clover-) cells in both treatments (Figure 1G, Movies S1–S2). We observed that mitoses in transfected Cas9-treated cells were a smaller percentage of total mitoses than that of the transfected pKER-NLS-Clover-treated cells (2.63 ± 1.21% versus 12.13 ± 1.31%; P < 0.0001). This result indicates that CRISPR/Cas9 treatment leads to increased cell cycle arrest across a population of cells.
Mediators of CRISPR/Cas9-associated cell cycle arrest
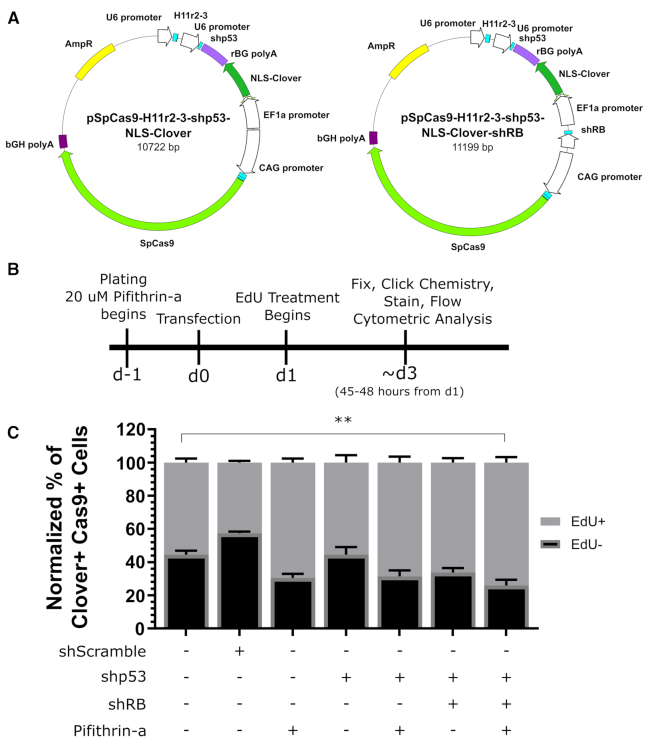
Our comparison of cell cycle progression in different cell lines suggested that TP53 status may mediate cell cycle arrest in response to CRISPR/Cas9 treatment, similar to the conclusion reached by other groups (16,26). We then investigated the effect of abrogating TP53 functionality in wild-type RPE-1 cells. We took a two-pronged approach of transcript knockdown via the addition of a short-hairpin expression cassette under the control of the U6 promoter against TP53 to our WT-Cas9 plasmid targeting the H11 locus (Figure 2A) and functional protein inhibition via the small molecule pifithrin-α (27). We considered additional factors affecting cell cycle progression in CRISPR/Cas9-treated cells based on the cell lines used. Because U2-OS cells are reported to lack functional RB1 (28), which is activated in response to DNA damage (29), we constructed a WT-Cas9-expression plasmid containing a short-hairpin expression cassette targeting RB1 in addition to the TP53-targeting hairpin. We also constructed an H11-targeting, WT-Cas9-expression plasmid containing a scrambled shRNA expression cassette as a control. Additionally we verified the functionality of the short hairpins and pifithrin-α in RPE-1 cells. For the short hairpins, we transfected RPE-1 cells with the shScramble, shTP53, or shRB1 expression cassettes cloned into the pJET plasmid and used immunofluorescence to quantify TP53 or RB1 intensity. For both TP53 and RB1, we observed significant knockdown relative to Scramble control (Supplementary Figure S2A, B; P < 0.0001 for both). To test the efficacy of pifithrin-α in RPE-1 cells, we treated the cells with 20 μM pifithrin-α or DMSO for 24 h before replacing the media with fresh pifithrin-α or DMSO and 50 μM Etoposide for 48 h, at which point we visualized 53BP1 foci via immunofluorescence (Supplementary Figure S2C). Upon quantifying nuclei by presence or absence of 53BP1 foci, we observed that pifithrin-α treatment significantly increased the percentage of nuclei lacking 53BP1 foci (30.71 ± 7.26% for treated versus 8.09 ± 1.45% for DMSO; P = 0.0361), indicative of pifithrin-α inhibiting TP53 function (Supplementary Figure S2D).
Figure 2.
Inhibition of TP53 can alleviate CRISPR/Cas9-associated cell cycle arrest. (A) Schematic of Cas9 reporter plasmids containing a short-hairpin cassette against TP53 or two short hairpin cassettes against TP53 and RB1. (B) Schematic of experimental design. (C) Bar graph of Cas9+ normalized percentages of EdU+ and EdU- cells for indicated treatments. n = at least two independent experiments each consisting of three technical replicates for each transfection. Data is shown as the mean ± SEM. **P < 0.009. Data were analyzed via an ordinary two-way ANOVA followed by a post-hoc Dunnett's multiple comparisons test, where each treatment was compared to wild-type RPE-1 cells treated with wild-type Cas9.
We then theorized that that the combination of transcript knockdown and functional protein inhibition would alleviate CRISPR/Cas9-inudced cell cycle arrest. To test this, we used our flow cytometry-based assay, with the only changes being the pretreatment of cells with pifithrin-α 24 hours before transfection and the presence of pifithrin-α throughout the experiment (Figure 2B). We found that the only the combination of pifithrin-α and both short hairpin cassettes significantly increased the EdU+ subpopulation of the transfected Cas9+ cells (an increase of 18.54 ± 5.87%; P = 0.009; Figure 2C). Additionally, we noted that treatment of wild-type RPE-1 cells with pifithrin-α and the TP53 short hairpin cassette resulted in similar levels of CRISPR/Cas9-associated cell cycle arrest as untreated p53-null RPE-1 cells (Supplementary Figure S2E; 58.52 ± 5.43% EdU+ for treated WT RPE-1 versus 63.67 ± 4.51% EdU+ for untreated TP53−/− RPE-1, P = 0.9767). However, combined treatment or treatment with only the TP53 short hairpin significantly increased CRISPR/Cas9-associated cell cycle arrest in TP53−/− RPE-1 cells (Supplementary Figure S2E; [36.33 ± 4.51% EdU- for no treatment] versus [56.26 ± 6.67% for TP53 shRNA] and [55.93 ± 4.91% EdU- for combined treatment], respectively; P < 0.05 for both), suggesting the presence of a low-affinity, off-target transcript only revealed in the absence of TP53 message. These results demonstrate that CRISPR/Cas9-associated cell cycle arrest can be partially alleviated by inhibiting TP53.
CRISPR/Cas9-associated cell cycle arrest is partially dependent on cleavage
Having investigated the possibility of alleviating CRISPR/Cas9-associated cell cycle arrest, we next sought to identify the molecular mechanism of the arrest. To this end, we reasoned that there are two non-exclusive mechanisms of action: (i) cleavage by Cas9 and (ii) targeting and binding DNA by Cas9. We investigated these possibilities using the EdU-based flow cytometry assay with transfected cells.
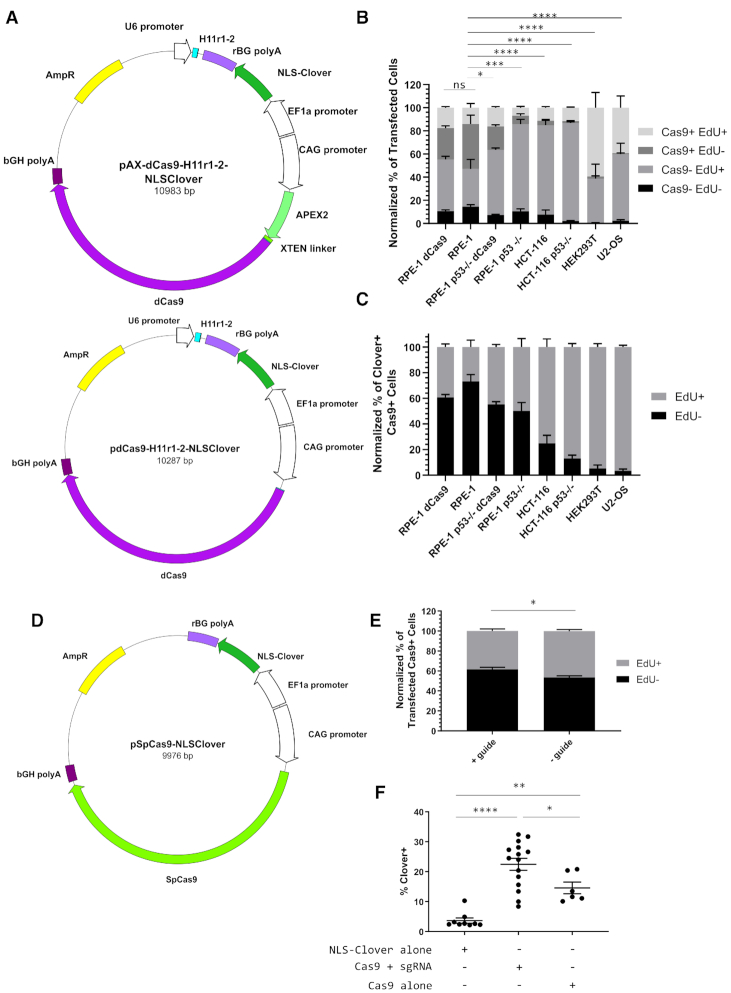
To address if cleavage by Cas9 contributed to CRISPR/Cas9-associated cell cycle arrest, we investigated the cell cycle response to targeted nuclease-dead Cas9 (dCas9) using our EdU-based flow cytometry assay. We repurposed an APEX2-tagged variant of dCas9 (AX-dCas9) that we had originally constructed for labelling and proteome mapping of genomic loci, much like the C-BERST approach (30). We then transfected HEK293T, U2-OS, wild-type and TP53−/− HCT-116, and wild-type and TP53−/− RPE-1 cells with a plasmid encoding NLS-Clover fluorescent protein, AX-dCas9, and a guide RNA cassette targeting the H11 locus (Figure 3A). Additionally, we constructed a dCas9 version of the same plasmid to control for non-specific effects resulting from APEX2. HEK293T (59.41 ± 13.18%) and U2-OS (39.28 ± 10.17%) displayed high levels of the [Cas9+ EdU+] subpopulation (Figure 3B; summarized in Supplementary Table S4). However, we observed an increase in the percent of [Cas9- EdU+] TP53−/− RPE-1 cells (75.33 ± 4.25%) compared to wild-type RPE-1 cells (32.75 ± 8.48%; P < 0.0001), indicative of Cas9 binding to DNA contributing to cell cycle arrest in a TP53-dependent manner and the existence of a threshold for Cas9 detection in our assay (Figure 3B). This observation also holds true when considering the [Cas9+ EdU-] subpopulation where, for example, both dCas9- and AX-dCas9-treated TP53−/− RPE-1 cells (20.21 ± 1.48% and 7.33 ± 1.73%, respectively) had significantly less non-cycling Cas9+ cells than RPE-1 cells (38.76 ± 7.63%, P < 0.05 and < 0.0005, respectively). We also observed no significant difference in cell cycle progression between dCas9- and AX-dCas9-treated RPE-1 cells (P = 0.3067 for [Cas9- EdU+] and 0.994 for [Cas9+ EdU+]). Examining the Cas9+ fraction of transfected cells emphasized that TP53-deficient cell lines display higher relative levels of EdU+ cells compared to their wild-type counterparts (Figure 3C). Taken together, these results reinforce the role of TP53 in mediating CRISPR/Cas9-associated cell cycle arrest and show that this arrest is only partially dependent on Cas9-mediated cleavage as dCas9-treated cells still arrest in a TP53-dependent manner.
Figure 3.
CRISPR/Cas9-associated cell cycle arrest is mediated by Cas9 binding. (A) Schematics of APEX2-dCas9 and dCas9 plasmids encoding the H11 r1-2 gRNA and an NLS-Clover cassette. (B) Bar graph of the Clover+-normalized percentages of four Cas9 EdU subpopulations in AX-dCas9- or dCas9-transfected cells for the six indicated cell lines. n = at least two independent experiments consisting of three technical replicates for each transfection. Data are shown as the mean ± SEM. Data were analyzed via an ordinary two-way ANOVA followed by a post-hoc Dunnett's multiple comparisons test comparing each cell type to AX-dCas9-treated RPE-1 cells. Significance is shown for the [Cas9+ EdU-] subpopulation comparisons. ns = not significant, **P = 0.0342, ***P = 0.0007, ****P < 0.0001. (C) Bar graph of the Cas9+ normalized percentages of EdU+ and EdU- from panel B. Data is shown as the mean ± SEM. (D) Schematic of plasmid encoding WT-Cas9 and NLS-Clover without a gRNA cassette. (E) Bar graph of Cas9+ normalized percentages of EdU+ and EdU- transfected wild-type RPE-1 cells with and without the H11r2-3 gRNA. n = at least two independent experiments consisting of at least two technical replicates each. Data are shown as the mean ± SEM. Data were analyzed via a one-way ANOVA followed by a post-hoc Sidak's multiple comparisons test. *P = 0.0117. (F) Dot plot of percentage of Clover+ cells for indicated transfections. n = three independent experiments consisting of at least two technical replicates each. Bars indicate mean ± SEM. *P = 0.0381, **P = 0.0065, ****P < 0.0001. Data were analyzed via a one-way ANOVA followed by a post-hoc Sidak's multiple comparisons test.
CRISPR/Cas9-associated cell cycle arrest is partially dependent on the Cas9 protein itself
To address whether CRISPR/Cas9-associated cell cycle arrest is dependent on binding of Cas9 to genomic DNA, we generated a WT-Cas9-expressing plasmid lacking a guide RNA expression cassette under the control of the CAG promoter and an NLS-Clover expression cassette under the control of the EF-1α promoter (Figure 3D). We compared wild-type RPE-1 cells transfected with this plasmid (- guide) or the H11-targeting WT-Cas9 plasmid (+ guide) and found a slight difference in the Cas9+ population for EdU incorporation (46.47 ± 1.47% versus 38.43 ± 2.15%; P = 0.0117; Figure 3E). Upon examining the percentage of cells that were fluorescent-protein-positive three days post-transfection in our flow cytometry-based assay, we observed that the fluorescence protein+ percentage of Cas9-alone treated cells was more similar to Clover-alone transfected cells than to Cas9+sgRNA-treated cells (Figure 3F; a difference of 10.92 ± 3.22% [P = 0.065] versus a difference of 18.78 ± 2.58% [P < 0.0001]). Additionally, Cas9+guide-treated cells had a significantly higher percentage of fluorescent protein+ cells than Cas9-alone treated cells (22.45 ± 2.00% versus 14.58 ± 1.96; P = 0.0381). Taken together, these results indicate CRISPR/Cas9-associated cell cycle arrest is also induced by the Cas9 protein alone, albeit to a lesser degree than Cas9 complexed with a guide.
Bound CRISPR/Cas9 complex blocks DSB recognition
Having identified that Cas9 bound to DNA contributes to CRISPR/Cas9-associated cell cycle arrest, we hypothesized that Cas9 may prevent DNA damage recognition proteins, such as 53BP1, from detecting resulting DSB. Such a hypothesis is attractive because of Cas9’s exceptionally long DNA occupancy time (32) and recent evidence that high levels of transcription through the targeted area may lead to higher levels of editing, presumably through dislodging of Cas9 from the target DNA (33). Additionally, we reasoned that the presence of 53BP1 foci would also indicate a degree of cell cycle progression (34).
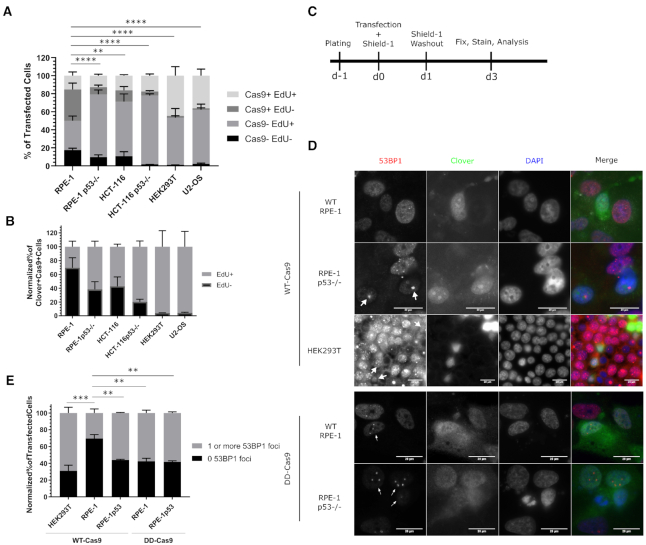
To these ends, we made use of DD-Cas9 (13) as a way to remove Cas9 from DNA and expose the DSB. We carried out our flow-cytometry-based assay using a similar experimental design as our previous experiments, and also used this experimental scheme to assay cells for the presence of DSBs via immunofluorescence-based microscopy at the end point of 3 days post-transfection (Figure 4A–C). To identify transfected cells, we modified the DD-Cas9 plasmid by inserting a self-contained nuclear-localized Clover expression cassette. The guide RNA in this plasmid was directed against the H11 locus.
Figure 4.
Wild-type Cas9 blocks recognition of Cas9-induced double-strand breaks. (A) Bar graph of the normalized percentages of four Cas9 EdU subpopulations in cells transfected with a plasmid encoding DD-Cas9, NLS-Clover, and the H11r1-2 gRNA for the six indicated cell lines. n = at least two independent experiments consisting of three technical replicates for each transfection. Data are shown as the mean ± SEM. Data were analyzed via an ordinary two-way ANOVA followed by a post-hoc Dunnett's multiple comparisons test comparing each cell type to RPE-1 cells. Significance is shown for the [Cas9+ EdU-] subpopulation comparisons. **P = 0.0048, ****P < 0.0001. (B) Bar graph of the Cas9+ normalized percentages of EdU+ and EdU- from panel A. Data is shown as the mean ± SEM. (C) Experimental design schematic. (D) Representative immunofluorescence of transfected wild-type and TP53−/− RPE-1 and HEK293T cells for 53BP1 foci. White arrowheads indicate Clover+ cells possessing one or more 53BP1 foci. Scale bar indicates 20 μm. (E) Bar graph illustrating percentages of given cell lines transfected with the indicated vector that possessed either zero 53BP1 foci or one or more 53BP1 foci. n = at least two independent experiments consisting of at least 60 transfected cells per experiment. Data are shown as the mean ± SEM. ***P < 0.001, ****P = 0.0001. Data were analyzed via an ordinary two-way ANOVA followed by a post-hoc Dunnett's multiple comparisons test, where each treatment was compared to wild-type RPE-1 cells treated with wild-type Cas9.
We then investigated the cell cycle response to targeted DD-Cas9 in wild-type and TP53−/− RPE-1, wild-type and TP53−/− HCT-116, HEK293T and U2-OS cells. In this experiment, Shield-1 was present for 24 hours following transfection before washout and addition of EdU. We observed that the [Cas9+ EdU-] subpopulation was significantly lower for every cell type when compared to wild-type RPE-1 cells (Figure 4A; [34.62 ± 6.97% for WT RPE-1] versus [7.86 ± 2.26% for TP53−/− RPE-1], [12.62 ± 4.22% for HCT-116], [4.49 ± 0.86% for TP53−/− HCT-116], [1.92 ± 0.17% for HEK293T] and [1.71 ± 0.46% for U2-OS]; P < 0.005 for all; summarized in Supplementary Table S5), which was emphasized when considering the Cas9+ subpopulations alone (Figure 4B). However, the [Cas9- EdU+] subpopulation was significantly larger in all cell types compared to wild-type RPE-1 cells (Figure 4A; [32.67 ± 5.15% for WT-RPE-1] versus [69.63 ± 4.74% for TP53−/− RPE-1; P < 0.0001], [60.26 ± 8.67% for HCT-116; P = 0.0002], [76.32 ± 2.03% for TP53−/− HCT-116; P < 0.0001], [52.48 ± 10.08% for HEK293T; P = 0.0136] and [59.77 ± 6.35% for U2-OS; P = 0.0007]). Taken together, these results further support the role of TP53 in mediating CRISPR/Cas9-associated cell cycle arrest and are consistent with the recent report that DSBs generated by DD-Cas9 lead to cell cycle delay even after repair (35). These results also raise the question of to what degree are RPE-1 cells capable of recognizing and repairing Cas9-induced DSBs.
To examine whether cells are capable of recognizing and repairing Cas9-induced DSBs, we transfected HEK293T, wild-type RPE-1, and TP53−/− RPE-1 cells with vectors encoding a nuclear-localized Clover expression cassette, a guide RNA expression cassette targeting the H11 locus, and either WT-Cas9 or DD-Cas9 in the presence of Shield-1 for 24 hours before washout before examining the cells via immunofluorescent microscopy at roughly 3 days post-transfection (Figure 4C). We then identified transfected cells by Clover/GFP immunofluorescence and categorized cells by the presence or absence of 53BP1 foci within the nucleus (Figure 4D). Additionally, we examined cells for significant levels of nuclear p21, which would indicate canonical TP53-mediated cell cycle arrest, and found very few nuclear p21+ transfected cells across all conditions (Supplementary Figure S3). We observed that wild-type RPE-1 cells transfected with wild-type Cas9 displayed the highest level of cells lacking 53BP1 foci (69.49 ± 4.93%), whereas TP53−/− RPE-1 cells transfected with wild-type Cas9 displayed higher levels of cells possessing one or more 53BP1 foci (55.94 ± 0.77%; Figure 4E). We additionally observed that the choice of DD-Cas9 had no difference in TP53−/− RPE-1 cells (57.54 ± 3.57% possessing one or more 53BP1 foci with DD-Cas9), but resulted in a significantly higher percentage of cells possessing one or more 53BP1 foci in wild-type RPE-1 cells compared to wild-type Cas9 treatment (57.54 ± 3.57% for DD-Cas9 versus 30.51 ± 4.93% for WT-Cas9; P = 0.0033). Consistent with the results from the cell cycle progression assay, HEK293T cells transfected with wild-type Cas9 displayed the highest percentage of cells with 53BP1 foci (68.92 ± 6.81%). These results demonstrate that wild-type Cas9 prevents recognition of nuclease-induced DSBs, and that this effect can be ameliorated either by elimination of TP53, or by destabilization of Cas9.
Additionally, we compared the [Cas9 Edu] subpopulations across the varied Cas9 treatments against WT-Cas9 for wild-type and TP53−/− RPE-1 cells (Supplementary Figure S4A, B). For wild-type RPE-1 cells, we found no significant difference between treatments for any subpopulation (Supplementary Figure S4A, Supplementary Table S6). For TP53−/− RPE-1 cells, DD-Cas9 treatment was significantly different than WT-Cas9 for all subpopulations except the [Cas9- EdU-] subpopulation, with 69.63 ± 4.74% versus 48.25 ± 4.94% for [Cas9- EdU+] (P < 0.0001), 7.86 ± 2.26% versus 20.29 ± 2.84% for [Cas9+ EdU-] (P = 0.0076), and 12.76 ± 1.65% versus 27.51 ± 2.18% for [Cas9+ EdU+] (P < 0.0012) (Supplementary Figure S4B, Supplementary Table S7). Additionally, AX-dCas9 treatment was significantly different from WT-Cas9 for the [Cas9- EdU+] and [Cas9+ EdU+] subpopulations, dCas9 treatment was only significantly different for the [Cas9+ EdU+] subpopulation (16.18 ± 0.94% versus 27.51 ± 2.18%; Supplementary Table S7), indicative of both Cas9 binding inducing a lesser degree of cell cycle arrest and that APEX2-dCas9 has less of an effect, although we have not explored the nature of this effect.
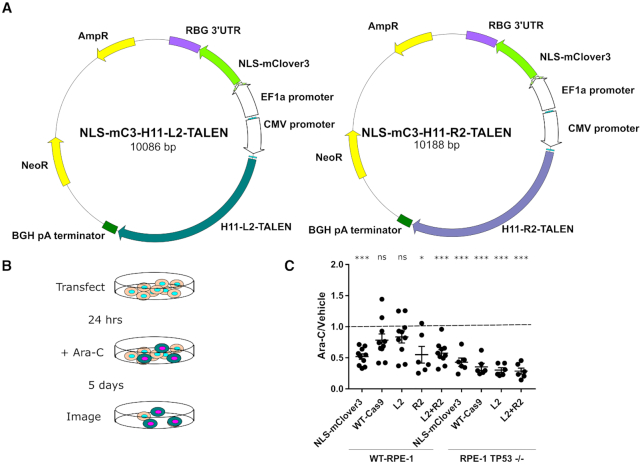
CRISPR/Cas9 treatment arrests cells to a greater extent than TALEN treatment
Having identified a cell cycle progression defect in CRISPR/Cas9-treated cells through lack of EdU incorporation, we chose to further investigate the nature of this arrest and whether the defect is unique to the CRISPR/Cas9 system. We modified vectors encoding a pair of TALENs targeted against the human H11 locus (25), but at a different site in the locus, with a self-contained nuclear-localized mClover3 expression cassette (Figure 5A). This pair of TALENs was chosen because they have been previously well-characterized (25). For the experimental design, we transfected wild-type and TP53−/− RPE-1 cells with (i) a vector encoding a nuclear-localized mClover3 (31), (ii) a vector encoding WT-Cas9 targeted against the H11 locus and a nuclear-localized Clover expression cassette, (iii and iv) either of the previously described TALEN plasmids or (v) both H11 TALEN plasmids 24 h after plating. After 24 h, we began treating the cells with 100 μM cytosine arabinoside (ara-C), a cytosine analog that kills cells actively synthesizing DNA, for 5 days, at which point we quantified surviving transfected cells using an Incucyte Zoom live cell analyser (Figure 5B). We then used the Incucyte's software to automate counting of Clover+ cells in each image to compare control-treated to ara-C-treated wells. In our analysis, we calculated the ratio of Clover+ cells in ara-C treatment to control, reasoning that a ratio not significantly different from 1 would indicate a degree of cell cycle arrest in the transfected cells, whereas ratios significantly different than 1 would indicate cell cycle progression in the case where the ratio is less than 1. We observed that TP53−/− RPE-1 cells, regardless of vector, displayed significant cell cycle progression, whereas the wild-type RPE-1 cells displayed significant cell cycle progression only in the presence of mClover3 alone, both TALENs together, and the R2 TALEN alone, but not for targeted wild-type Cas9 or the L2 TALEN alone (Figure 5C). These results indicate that TALEN treatment leads to less cell cycle arrest than CRISPR/Cas9 treatment, and underscores the requirement of TP53 in mediating this arrest. Additionally, these results combined with the extended duration of ara-C treatment provide further evidence that CRISPR/Cas9-treated cells do indeed undergo a cell cycle arrest.
Figure 5.
CRISPR/Cas9 treatment leads to a greater degree of cell cycle arrest than TALEN treatment. (A) Schematic of H11 L2 and R2 TALEN plasmids encoding NLS-mClover3. (B) Schematic of experimental design of ara-C-based cell cycle arrest assay. (C) Dot plot of ratios of Clover+ cells after 5 days of ara-C treatment versus vehicle for indicated cell lines and transfected vectors. n = three independent experiments each consisting of two ara-C-treated technical replicates and one vehicle-treated control. Error bars indicate SEM. *P < 0.05, ***P < 0.0005. Data were analyzed with a one-sample t-test comparing the actual mean against a theoretical mean of 1.
DISCUSSION
In this work, we find that CRISPR/Cas9 treatment leads to extended cell cycle arrest in some human cell lines. This arrest requires, in part, functional TP53 and RB1, and can be alleviated to a degree by manipulation of TP53 activity. Additionally, we found that this arrest is mediated by DNA cleavage, DNA binding and the Cas9 protein itself, in descending order of importance. Furthermore, we provide evidence that the propensity of Cas9 to remain bound to DNA after cleavage reduces the ability of cells to recognize and repair the double-strand break, and that this too is dependent on functional TP53.
Our work demonstrating an extended cell cycle arrest in response to CRISPR/Cas9 treatment has several implications for the planning, conduct, and interpretation of gene editing and genome engineering experiments. First, and consistent with other recent studies (16,26,35–37), our work suggests that caution is required in the interpretation of CRISPR/Cas9-based screens. Currently, the outcomes of such screens are usually assessed by the retention/loss of guides targeting specific genes from larger pools of guides, and are interpreted as identifying the sufficiency or necessity of these specific genes for the phenotype in question (38). Our work raises the possibility of such screens unintentionally selecting for pre-existing cells with non-functional TP53 or for cells possessing pre-existing mutations in the target locus, rendering the target locus uneditable. Such a possibility would bias results of the screen, leading to false positives or false negatives, depending on the nature of the screen. Selection of pre-existing mutants might be acceptable in fulfilling a screen, unless one is interested specifically in alleles derived from Cas9-mediated cutting. Second, our work identifies TP53 inhibition as a potential means to increase desired clone recovery in directed genome engineering CRISPR/Cas9 experiments. This inhibition can accomplished in several ways, including mutation of TP53, treatment with a small-molecule inhibitor, and/or transcript silencing by RNAi. Given that even temporary inhibition of TP53 can result in an increase genome instability, an alternative is to use an inducible destabilized Cas9, which somewhat mitigates the detrimental effect of Cas9 expression (13,24,35). Finally, our work raises the question of what, exactly, is Cas9 treatment selecting for in the variety of experiments in which it used. Our results suggest that investigators should take into account that CRISPR/Cas9 treatment is likely selecting for cells that are untargetable due to pre-existing endogenous variation at that locus, and thus are somewhat resistant to the growth inhibition effect of Cas9 treatment.
Our results are largely consistent with two recently-published works that also identified TP53 as being important in both a CRISPR/Cas9-induced DNA damage response and an induced cytoxicity response (16,26). In the work of Haapaniemi et al. (26), TP53 was identified as mediating a DNA-damage response in RPE-1 cells; their screen also identified GATA6, CDKN1A/P21 and RB1. They reasoned that the effect of these proteins was related to cell cycle arrest rather than cell death, based on the lack of cleaved caspase 3 in treated cells. Their work focused on the role of this TP53-mediated response in homology-directed repair (HDR) and demonstrated that the addition of ectopic MDM2, a negative regulator of TP53, does increase the percentage of HDR-resolved cells in the assay. The authors interpreted this result as a specific effect on HDR, however an alternative explanation is that MDM2 downregulation of TP53 simply allows the proliferation of the initial targeted cells, consistent with our observation of a Cas9-mediated arrest. The work of Ihry and colleagues (16) identified a TP53-dependent cytotoxicity phenotype in response to CRISPR/Cas9 treatment via differential expression analysis from RNA-seq data in human embryonic and induced pluripotent stem cells. An inducible version of DD-Cas9 was used in these experiments, and the assay identified p21 as the most differentially expressed gene. This is a surprising finding given the lack of increase of nuclear p21 in CRISPR/Cas9-treated RPE-1 cells in our experiments. While the identification of TP53 as the mediator in this cytotoxicity phenotype is interesting, this general phenotype has been previously observed (15).
A third recent study has proposed that CRISPR/Cas9 treatment only delays cell cycle progression (35). Interestingly, this delay was observed using a doxycycline-inducible, integrated version of DD-Cas9. As use of an inducibly-expressed, reversibly-stabilized Cas9 would be expected to limit the genomic occupancy time of Cas9, their results are consistent with our findings regarding DD-Cas9 and Shield-1 in addition to demonstrating a possible limit on the number of simultaneous DSBs that wild-type cells can recover from. This study examined a much earlier time point than our work (1 day as opposed to approximately 3 days post-transfection/induction) and relies on fluorescent reporters of cell cycle stage instead of incorporation of a nucleotide analog over an extended period of time, somewhat restricting the resolution of their analysis of cell cycle effect. This limitation is especially relevant in light of the observation that DNA damage in G1 can lead to arrest in the subsequent G2 phase (39). A conclusion of this paper (35) was that a small number of Cas9-induced DSBs do not lead to permanent cell cycle arrest, however, this must be viewed in light of the use of DD-Cas9 in their experiments, and that removal of DD-Cas9 from the locus would limit the inhibitory effect of Cas9, as we also observed. Thus, their results and ours support the preferential use of an inducible DD-Cas9 over wild-type for genome engineering applications.
The presence of [Cas9+ EdU+] wild-type RPE-1 cells in the experiments shown in Figure 1 suggests that these cells were able to progress through the cell cycle despite the Cas9 treatment. We noticed in analyzing our flow cytometry data that this population appears to be two subpopulations: (i) cells that have progressed through one cell cycle since onset of EdU incubation, (ii) cells that have progressed through two cell cycles. Considering our experimental design, the first subpopulation most likely progressed through no more than two cell cycles before arresting, the first occurring sometime after transfection but before EdU incubation, and the second after EdU incubation. The majority of the second subpopulation most likely progressed through two cell cycles, beginning after the start of EdU incubation, before arresting after the second complete cell cycle. The remaining minority most likely reflects edited cells. The possibility that a substantial fraction of [Cas9+ EdU+] cells arrested after progressing through at least one cell cycle is supported by the recent finding that inducing DNA damage in G2 leads to cells arresting in the following G1 phase, whereas damage induced in G1 phase leads to arrest in the following G2 (39). Interestingly, that study also describes a diminished ability of U2-OS cells to permanently arrest in response to damage, which is consistent with our findings. This report also noted that damage in G1 results in a gradual switch from temporary to permanent arrest as the damage level increases, raising the possibility that persistent DNA damage, such as that induced by CRISPR/Cas9, might gradually lead to permanent arrest, which also could explain the [Cas9+ EdU+] fraction of cells.
More recently, another study has demonstrated that zinc finger nuclease (ZFN)- and Cas9-mediated DSBs made in human CD34+ hematopoietic stem and progenitor cells also induce a TP53-mediated DNA damage response (44). Interestingly, ZFN-induced DSBs are reported to resolve faster than those generated by Cas9 ribonucleoprotein complexes and appear to resolve the effects of the DNA damage response quicker, consistent with our observations of TALENs. These findings strongly support a role of occupancy time in the resolution of designer-nuclease-induced DSBs. Additionally, the authors report an attenuation of the TP53-mediated damage response with the dominant negative truncated TP53 peptide GSE56. This transient inhibition is reported to be especially important when using AAV as a source of repair templates for HDR, as the use of AAV appears to greatly inhibit the function, transplantation, and engraftment of these CD34+ cells. Coupled with the finding that Cas9 protein alone can lead to unintended selection for TP53-inactivating mutations (45), these findings, as well as our data, strongly advocate for the inclusion of transient TP53 inhibition in CRISPR/Cas9-based genome engineering applications.
In considering that Cas9 may occlude the DSBs it itself generates, one should also reflect on what occurs when using DD-Cas9. Once the stabilizing small molecule Shield-1 is withdrawn, DD-Cas9 becomes destabilized and susceptible to degradation. Thus, the DSB should then become available to repair. This availability appears to be reflected in the fact that the proportion of Cas9- cells increased in the population of DD-Cas9 transfected TP53−/− RPE-1 cells (Figure 4A), but not in transfected wild-type RPE-1 cells. However, availability of the DSB to repair is evident by the presence of nuclear 53BP1 foci in transfected wild-type RPE-1 cells (Figure 4E).
One major caveat of our work is that we chose to target a single non-transcribed locus. However, we would expect from recent work (33) that CRISPR/Cas9-associated arrest also occurs when targeting transcribed loci, with the degree of arrest related to the level of transcription through the locus. Similar TP53-dependent effects were observed in iPSCs when targeting transcribed loci (16), providing support for this being a general phenomenon in mammalian somatic cells. It is possible that embryos may be an exception due to their reliance for DNA damage repair on DNA Pol θ, which is relatively error-prone and might more frequently result in non-targetable products after an initial break (40). Going forward, determining how Cas9 is removed from DNA in the context of gene editing and how cells perceive Cas9-mediated double-strand breaks will aid in increasing the efficiency of genome engineering for experimental and therapeutic purposes.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the members of the Stearns lab for helpful and insightful commentary. Additionally, we thank the staff of the Stanford Shared FACS Facility, particularly Cathy Crumpton and Ometa Herman for sharing their expertise.
Contributor Information
Jonathan M Geisinger, Department of Biology, Stanford University, Stanford, CA 94305, USA.
Tim Stearns, Department of Biology, Stanford University, Stanford, CA 94305, USA; Department of Genetics, Stanford University Medical School, Stanford, CA 94305, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [F32GM122214 to J.M.G., R35GM130286 to T.S.]. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. The authors declare that this work is free from any conflicts of interest.
REFERENCES
- 1. Calos M. Genome editing techniques and their therapeutic applications. Clin. Pharmacol. Ther. 2017; 101:42–51. [DOI] [PubMed] [Google Scholar]
- 2. Geisinger J.M., Calos M.P.. Renault S., Duchateau P.. Site-specific recombination using PhiC31 Integrase. Site-Directed Insertion of Transgenes. 2013; 23:Dordrecht: Springer Netherlands; 211–239. [Google Scholar]
- 3. Orr-Weaver T.L., Szostak J.W., Rothstein R.J.. Yeast transformation: a model system for the study of recombination. Proc. Natl. Acad. Sci. U.S.A. 1981; 78:6354–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B. et al.. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002; 418:387–391. [DOI] [PubMed] [Google Scholar]
- 5. Kilian O., Benemann C.S.E., Niyogi K.K., Vick B.. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:21265–21269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kim Y.G., Cha J., Chandrasegaran S.. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 1996; 93:1156–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Christian M., Cermak T., Doyle E.L., Schmidt C., Zhang F., Hummel A., Bogdanove A.J., Voytas D.F.. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010; 186:757–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rouet P., Smih F., Jasin M.. Introduction of double-strand breaks into the genome of mouse cells by expression of a rare-cutting endonuclease. Mol. Cell. Biol. 1994; 14:8096–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E.. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A. et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J.. RNA-programmed genome editing in human cells. eLife. 2013; 2:e00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H.. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014; 289:21312–21324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Geisinger J.M., Turan S., Hernandez S., Spector L.P., Calos M.P.. In vivo blunt-end cloning through CRISPR/Cas9-facilitated non-homologous end-joining. Nucleic Acids Res. 2016; 44:e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pruett-Miller S.M., Reading D.W., Porter S.N., Porteus M.H.. Attenuation of zinc finger nuclease toxicity by small-molecule regulation of protein levels. PLos Genet. 2009; 5:e1000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Byrne S.M., Ortiz L., Mali P., Aach J., Church G.M.. Multi-kilobase homozygous targeted gene replacement in human induced pluripotent stem cells. Nucleic Acids Res. 2015; 43:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ihry R.J., Worringer K.A., Salick M.R., Frias E., Ho D., Theriault K., Kommineni S., Chen J., Sondey M., Ye C. et al.. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med. 2018; 24:939–946. [DOI] [PubMed] [Google Scholar]
- 17. Kosicki M., Tomberg K., Bradley A.. Repair of double-strand breaks induced by CRISPR–Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018; 36:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang L., Grishin D., Wang G., Aach J., Zhang C.-Z., Chari R., Homsy J., Cai X., Zhao Y., Fan J.-B. et al.. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat. Commun. 2014; 5:5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thakore P.I., D’Ippolito A.M., Song L., Safi A., Shivakumar N.K., Kabadi A.M., Reddy T.E., Crawford G.E., Gersbach C.A.. Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods. 2015; 12:1143–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Akcakaya P., Bobbin M.L., Guo J.A., Malagon-Lopez J., Clement K., Garcia S.P., Fellows M.D., Porritt M.J., Firth M.A., Carreras A. et al.. In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 2018; 561:416–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lin S., Staahl B.T., Alla R.K., Doudna J.A.. Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014; 3:e04766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Demeter J., Lee S.E., Haber J.E., Stearns T.. The DNA damage checkpoint signal in budding yeast is nuclear limited. Mol. Cell. 2000; 6:487–492. [DOI] [PubMed] [Google Scholar]
- 23. Banaszynski L.A., Chen L., Maynard-Smith L.A., Ooi A.G.L., Wandless T.J.. A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell. 2006; 126:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Senturk S., Shirole N.H., Nowak D.G., Corbo V., Pal D., Vaughan A., Tuveson D.A., Trotman L.C., Kinney J.B., Sordella R.. Rapid and tunable method to temporally control gene editing based on conditional Cas9 stabilization. Nat. Commun. 2017; 8:14370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhu F., Gamboa M., Farruggio A.P., Hippenmeyer S., Tasic B., Schüle B., Chen-Tsai Y., Calos M.P.. DICE, an efficient system for iterative genomic editing in human pluripotent stem cells. Nucleic Acids Res. 2014; 42:e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haapaniemi E., Botla S., Persson J., Schmierer B., Taipale J.. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018; 24:927–930. [DOI] [PubMed] [Google Scholar]
- 27. Komarov P.G., Komarova E.A., Kondratov R.V., Christov-Tselkov K., Coon J.S., Chernov M.V., Gudkov A.V.. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999; 285:1733–1737. [DOI] [PubMed] [Google Scholar]
- 28. Broceno C., Wilkie S., Mittnacht S.. RB activation defect in tumor cell lines. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:14200–14205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knudsen K.E., Booth D., Naderi S., Sever-Chroneos Z., Fribourg A.F., Hunton I.C., Feramisco J.R., Wang J.Y.J., Knudsen E.S.. RB-dependent S-phase response to DNA damage. Mol. Cell. Biol. 2000; 20:7751–7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao X.D., Tu L.-C., Mir A., Rodriguez T., Ding Y., Leszyk J., Dekker J., Shaffer S.A., Zhu L.J., Wolfe S.A. et al.. C-BERST: defining subnuclear proteomic landscapes at genomic elements with dCas9–APEX2. Nat. Methods. 2018; 15:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bajar B.T., Wang E.S., Lam A.J., Kim B.B., Jacobs C.L., Howe E.S., Davidson M.W., Lin M.Z., Chu J.. Improving brightness and photostability of green and red fluorescent proteins for live cell imaging and FRET reporting. Sci. Rep. 2016; 6:20889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A.. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014; 507:62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clarke R., Heler R., MacDougall M.S., Yeo N.C., Chavez A., Regan M., Hanakahi L., Church G.M., Marraffini L.A., Merrill B.J.. Enhanced bacterial immunity and mammalian genome editing via RNA-polymerase-mediated dislodging of Cas9 from double-strand DNA breaks. Mol. Cell. 2018; 71:42–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lukas C., Savic V., Bekker-Jensen S., Doil C., Neumann B., Sølvhøj Pedersen R., Grøfte M., Chan K.L., Hickson I.D., Bartek J. et al.. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011; 13:243–253. [DOI] [PubMed] [Google Scholar]
- 35. van den Berg J., Manjón A.G., Kielbassa K., Feringa F.M., Freire R., Medema R.H.. A limited number of double-strand DNA breaks is sufficient to delay cell cycle progression. Nucleic Acids Res. 2018; 46:10132–10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aguirre A.J., Meyers R.M., Weir B.A., Vazquez F., Zhang C.-Z., Ben-David U., Cook A., Ha G., Harrington W.F., Doshi M.B. et al.. Genomic copy number dictates a gene-independent cell response to CRISPR/Cas9 targeting. Cancer Discov. 2016; 6:914–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yuen G., Khan F.J., Gao S., Stommel J.M., Batchelor E., Wu X., Luo J.. CRISPR/Cas9-mediated gene knockout is insensitive to target copy number but is dependent on guide RNA potency and Cas9/sgRNA threshold expression level. Nucleic Acids Res. 2017; 45:12039–12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hill A.J., McFaline-Figueroa J.L., Starita L.M., Gasperini M.J., Matreyek K.A., Packer J., Jackson D., Shendure J., Trapnell C.. On the design of CRISPR-based single-cell molecular screens. Nat. Methods. 2018; 15:271–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chao H.X., Poovey C.E., Privette A.A., Grant G.D., Chao H.Y., Cook J.G., Purvis J.E.. Orchestration of DNA damage checkpoint dynamics across the human cell cycle. Cell Syst. 2017; 5:445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Thyme S.B., Schier A.F.. Polq-mediated end joining is essential for surviving DNA double-strand breaks during early Zebrafish development. Cell Rep. 2016; 15:707–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. et al.. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011; 8:409–412. [DOI] [PubMed] [Google Scholar]
- 42. Boehm J.S., Hession M.T., Bulmer S.E., Hahn W.C.. Transformation of human and murine fibroblasts without viral oncoproteins. Mol. Cell. Biol. 2005; 25:6464–6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sarbassov D.D., Guertin D.A., Ali S.M., Sabatini D.M.. Phosphorylation and regulation of Akt/PKB by the Rictor-mTOR complex. Science. 2005; 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- 44. Schiroli G., Conti A., Ferrari S., della Volpe L., Jacob A., Albano L., Beretta S., Calabria A., Vavassori V., Gasparini P. et al.. Precise gene editing preserves hematopoietic stem cell function following transient p53-mediated DNA damage response. Cell Stem Cell. 2019; 24:551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Enache O.M., Rendo V., Abdusamad M., Lam D., Davison D., Pal S., Currimjee N., Hess J., Pantel S., Nag A. et al.. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 2020; 52:662–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.