Abstract
FANCJ, a DNA helicase and interacting partner of the tumor suppressor BRCA1, is crucial for the repair of DNA interstrand crosslinks (ICL), a highly toxic lesion that leads to chromosomal instability and perturbs normal transcription. In diploid cells, FANCJ is believed to operate in homologous recombination (HR) repair of DNA double-strand breaks (DSB); however, its precise role and molecular mechanism is poorly understood. Moreover, compensatory mechanisms of ICL resistance when FANCJ is deficient have not been explored. In this work, we conducted a siRNA screen to identify genes of the DNA damage response/DNA repair regime that when acutely depleted sensitize FANCJ CRISPR knockout cells to a low concentration of the DNA cross-linking agent mitomycin C (MMC). One of the top hits from the screen was RAP80, a protein that recruits repair machinery to broken DNA ends and regulates DNA end-processing. Concomitant loss of FANCJ and RAP80 not only accentuates DNA damage levels in human cells but also adversely affects the cell cycle checkpoint, resulting in profound chromosomal instability. Genetic complementation experiments demonstrated that both FANCJ’s catalytic activity and interaction with BRCA1 are important for ICL resistance when RAP80 is deficient. The elevated RPA and RAD51 foci in cells co-deficient of FANCJ and RAP80 exposed to MMC are attributed to single-stranded DNA created by Mre11 and CtIP nucleases. Altogether, our cell-based findings together with biochemical studies suggest a critical function of FANCJ to suppress incompletely processed and toxic joint DNA molecules during repair of ICL-induced DNA damage.
INTRODUCTION
Interstrand cross-links (ICLs) are a formidable type of DNA damage that interfere with cellular DNA replication and transcription (1). In replicating cells, persistent ICLs represent a lethal form of chromosomal damage because they result in highly recombinogenic DNA double-strand breaks (DSBs), causing a loss of genetic information. Historically, ICLs were known to arise from exposure to certain clastogenic compounds (e.g. nitrogen mustard) used in chemical warfare (2). Ironically, cancer clinics began to use ICL-inducing agents as chemotherapy drugs to treat leukemia and various solid tumors (3). Rapidly dividing cancer cells are hypersensitive to the DNA damaging effects of cisplatin, psoralen, mitomycin C (MMC) and other DNA cross-linking drugs; however, the cytotoxicity of such compounds for normal (noncancerous) cells has posed a significant drawback for their efficacy in combating cancer. A source of endogenous ICLs was discovered to be formaldehyde and aldehyde derivatives, which arise from cellular metabolic processes including lipid peroxidation and oxidative demethylation reactions (4,5). Experimental evidence now also points to alcohol consumption contributing to acetaldehyde accumulation, ultimately leading to macromolecule damage, including genomic DNA ICLs (6).
A primary pathway for the repair of DNA ICLs is orchestrated by a set of >20 proteins in which the corresponding mutations are linked to a chromosomal instability disorder known as Fanconi Anemia (FA) (7). The FA pathway of ICL repair is a complex multi-step process with several functional levels including DNA ICL detection, pathway activation, unhooking of the ICL, translesion DNA synthesis past the ICL remnant, and recombinational repair to restore genomic integrity (8). In the absence of a functional FA pathway, cells attempt to cope with ICL-induced DNA damage, typically DSBs, by eliciting homologous recombination (HR) or nonhomologous end-joining (NHEJ) which differ significantly in their fidelity of repair (9). The most poorly understood class of FA proteins are those dedicated to HR repair of the DSBs arising from downstream processing of the ICL-induced DNA damage. Among these proteins is the BRCA1-interacting FANCJ DNA helicase in which mono-allelic mutations are associated with cancers of the breast (10) and ovary (11). Although purified recombinant FANCJ helicase protein is known to unwind duplex (12,13) and G-quadruplex (14–16) DNA substrates in vitro, as well as remove the major strand exchange protein Rad51 from DNA in an ATP-dependent manner (17), its cellular role(s) in HR repair and maintenance of genomic stability is enigmatic. Cells deficient in FANCJ display a HR defect (18) and are hypersensitive to treatments that cause DNA ICLs or DSBs (for review, see (19)), as well as drugs that cause replication stress (14,20,21), but FANCJ’s precise substrates and mechanism(s) of action in vivo are not well characterized.
To better understand the cellular response to ICL-induced DNA damage, we screened DNA damage response/DNA repair gene targets that when depleted sensitize human FANCJ CRISPR knockout (KO) cells to a very low dose of an ICL-inducing agent. Among the leading hits is RAP80, a ubiquitin-binding protein that is believed to recruit BRCA1 and other proteins to DSBs and modulate DNA end-processing (22). The results from cell-based assays, together with the biochemical discovery of a novel DNA branch-migration activity catalyzed by FANCJ, suggest a model in which the loss of FANCJ and RAP80 cause severe defects by impairing maturation of HR repair intermediates incurred by ICL-induced DNA damage. Meanwhile, a compromised cell cycle checkpoint in these cells resulted in their progression into mitosis with unrepaired DNA damage. The potential clinical importance of our findings in terms of chromosomal instability imposed by dual loss of FANCJ and RAP80, and the efficacy of DNA cross-linking agents used in cancer therapy, is discussed.
MATERIALS AND METHODS
Cell culture, knockdown and DNA transfection
U2OS FANCJ KO and HeLa FANCJ KO cells were generated as previously described (20). U2OS RAP80 KO cells and MERIT40 KO cells were generated as previously described (23). HEK293 Abraxas KO cells were generated as previously described (24). DR-GFP U2OS (25) and EJ5 U2OS (26) were gifts from Dr Xiaofan Wang (Duke University, USA) and Dr Jeremy Stark (City of Hope, USA), respectively. All cells were cultured in DMEM medium with 10% fetal bovine serum (Sigma-Aldrich) and 1× penicillin/streptomycin (Gibco) in an atmosphere of 5% CO2 at 37°C.
For siRNA-mediated knockdown, siRNA was diluted with Opti-MEM medium (Gibco) and then mixed with INTERFERin (Polyplus transfection) according to the manufacturer's instructions. Cells were transfected with a final concentration of 30 nM siRNA. For stable expression of exogenous FANCJ, FANCJ KO cells were transfected with plasmids encoding GFP, GFP-FANCJ-WT and GFP-FANCJ-H396D by using Lipofectamine 2000 as per manufacturer's instruction. After 30 days of selection in G418 (500 μg/ml), cells expressing high levels of GFP were sorted by flow-cytometry.
RNAi screen
The primary screen was performed in triplicate in 96-well plates as described previously (27,28), utilizing a Dharmacon-DNA damage response siRNA library consisting of a pool of four siRNAs per gene and two non-targeting control siRNAs. Two-thousand WT or FANCJ CRISPR KO cells were seeded in 96-well plates and reverse transfected with the library of 25 nM gene-specific pool siRNAs using INTERFERin transfection reagent (Polyplus). Each 96-well plate contained 80 gene-specific pool siRNAs and two control siRNAs. Twenty-four hr after transfection, media was removed and cells were incubated in fresh media with or without 6.25 nM MMC at 37°C for 72 h. Cell viability was measured by the WST-1 cell proliferation assay, and percentage viability of WT or FANCJ KO cells transfected with the siRNA pool against the indicated gene was calculated by normalizing the absorbance values of the cells transfected with the gene-specific siRNA to that of the control siRNA. The screening was done in triplicate, and mean percent viability relative to control siRNA is plotted for each individual siRNA with standard deviations indicated by error bars. A minimum loss of 30% cell viability in the FANCJ KO cells (compared to the isogenic WT cells) was set as a threshold cutoff to select gene-targeted siRNA pools that sensitize U2OS cells as a function of FANCJ status. In addition, we used a threshold cutoff minimum of 55% reduction in viability of the FANCJ KO cells transfected with the target RNAi pool compared to the same cells transfected with control RNAi.
Western blot analysis
Cells were lysed in 1× SDS sample buffer (Millipore). Lysates were sonicated for 15 s and heated at 100°C for 5 min. Proteins were separated on 6–12% SDS-PAGE gels and transferred to PVDF membranes (Millipore). Membranes were blocked with 5% non-fat milk or BSA, and then incubated with primary antibodies overnight at 4°C. After washing with 1× PBS with 0.1% Tween-20, the HRP-conjugated secondary antibodies were then added to PVDF membrane. Membranes were developed with Pierce ECL or Femto Western Blotting Substrate (Thermo-Fisher) and imaged with Bio-Rad ChemiDoc XRS imaging system.
Colony formation assay
Cells were transfected with 30 nM of indicated siRNAs. After 24 h cells were re-transfected. Forty-eight hr after the first transfection, 500–1000 cells according to plating efficiency were seeded in six-well plates with media containing the specified MMC or cisplatin concentration. Cells were incubated for 6 days in MMC or cisplatin after which media was changed. The cells were allowed to recover for 14 days after seeding, and then fixed with 100% methanol for 20 min and stained with 0.5% crystal violet in 25% methanol at room temperature for 20 min. The dishes were gently washed with water and dried in the air. Colonies were counted and percent survival is expressed as the number of colonies in the MMC-treated or cisplatin-treated condition at the indicated dose of drug compared to the untreated condition. The results are presented as the mean ± standard error of mean (s.e.m.) from three independent experiments.
Annexin staining
Cells were transfected with 30 nM of indicated siRNAs. After 24 h cells were re-transfected. Forty-eight hr after the first transfection, cells were treated with 3 μM MMC or vehicle control for 1 h and allowed to recover in fresh media. After 72 h treatment, apoptosis was measured with Annexin V–FITC/PI staining kit (Abcam) by flow cytometry analysis. The results are presented as the mean ± s.e.m. from three independent experiments.
Immunofluorescence assay
Cells were cultured onto four-well Nunc chamber slides. To detect RPA, Rad51, CtIP and BRCA1 foci, we performed pre-extraction for 5 min on ice in CSK buffer followed by fixation with 4% PFA/PBS for 10 min, permeabilization in 0.5% Triton X-100/PBS for 15 min and blocking in 10% FBS/PBS for 1 h. Cells were incubated overnight with specific primary antibody: mouse anti-RPA (EMD Millipore); rabbit anti-Rad51 (Abcam); mouse anti-BRCA1 (Santa Cruz); rabbit anti-CtIP (Abcam) at 4°C in 1% BSA/PBS, followed by species-specific fluorescein-conjugated secondary antibodies (Alexa Fluor 594 Anti-Rabbit or Alexa Fluor 488 Anti-Mouse), and counterstained with DAPI. Slides were analyzed with Zeiss Axio Observer.Z1 Fluorescence Microscope. For each time point, at least 75 nuclei were scored at 63× magnification, and the results are presented as the mean ± s.e.m. from three independent experiments.
BrdU assay
Cells grown in the presence of 10 μM BrdU (Sigma) and 3 μM MMC for 16 h and immunofluorescence stained with mouse anti-BrdU antibody (Sigma) under non-denaturing conditions to detect BrdU incorporated into ssDNA as described previously (29). For each time point, at least 75 nuclei were scored at 63× magnification to determine average number of BrdU foci per nucleus. The results are presented as the mean ± s.e.m. from three independent experiments.
Laser localized ICLs
A diamond pen was used to score a cross in the center of the growth surface of a 35 mm glass bottom culture dish prior to plating cells. This facilitated identification of targeted cells during imaging of immuno-stained cells. Prior to an experiment the cells were incubated with either 6 μM trimethyl psoralen (TMP) for 20 min prior to laser treatment. The plates were placed in an environmental chamber, maintained at 37°C, 5% CO2 and 80% humidity, mounted on a Nikon Eclipse TE2000 confocal microscope. Cells were visualized with a Plan Fluor ×60/1.25 numerical aperture oil objective. The microscope was equipped with an SRS NL100 nitrogen laser-pumped dye laser (Photonics Instruments, St. Charles, IL, USA). To photoactivate the psoralens, defined regions of interest (ROI, 4 × 20 pixels, 0.16 μm/pixel) in individual nuclei, in the vicinity of the etched cross, were exposed to the laser firing 3-ns pulses at 365 nm with a repetition rate of 10 Hz. The power measured at the back aperture of the ×60 objective was 0.7 nW. The laser was controlled by Volocity-5 software (Improvision; PerkinElmer Life Sciences). The beam was oriented by galvanometer-driven displacers and fired randomly throughout the region until the entire region was exposed. The diffraction-limited spot size was ∼300 nm. Two firing cycles were applied to each region.
ssDNA assay
Cells were labeled with 20 μM CldU for 48 h prior to introducing laser localized ICLs. During a 2.5 h incubation period post targeting, the cells were exposed to 100 μM cytosine arabinoside (ara-C) to prevent repair synthesis. Fixing procedure for the assay was as follows: one 3 min treatment on ice with CSK1 buffer (10 mM Pipes, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 0.05% Triton X-100) and another 3 min treatment on ice with CSK2 buffer (10 mM Tris pH 7.4, 10 mM NaCl, 1% Tween. 0.5% deoxycholate and 3 mM MgCl2). Following detergent treatments, cells were fixed in 2% formaldehyde mixed with 2% sucrose for 10 min at room temperature, and washed with PBS-T (1× PBS, 0.05% Tween-20). To visualize CldU at laser stripe, cells were exposed to appropriate primary and secondary antibodies without acid treatment. To control for genomic DNA labeling, a second set of plates was treated with 2.5 M HCl for 1 h, followed by 200 mM Tris–HCl pH 8.0, and a wash with PBS, prior to blocking (see below).
Fixed cells were permeabilized with 0.5% Triton X-100, 1% bovine serum albumin, 100 mM glycine, and 0.2 mg/ml EDTA in PBS at 4°C for 10 min, digested with RNase A in PBS-EDTA (5 mM) solution for 30 min at 37°C, blocked in 10% goat serum and 5% BSA in PBS and 0.01% sodium azide for 1 h at room temperature, and then incubated with appropriate primary antibody diluted in blocking solution for 1 h at 37°C in a humid chamber [mouse anti-γ-H2AX 1:1000 (EMD Millipore, #05–636), Rat anti-BrdU (1:200) Abcam #ab6326]. γ−H2AX stained the targeted ROI, BrdU labeled the CldU labeled ssDNA. After three 10 min washes in PBS-T, cells were incubated with fluorescence-tagged secondary antibodies [AF488 goat anti-mouse, AF594 goat anti-rabbit, AF633 goat anti-rat, 1:1000, Invitrogen] for 1 h at 37°C, washed three times for 10 min in PBS-T, and mounted in Prolong + DAPI.
HR and NHEJ assays
The HR repair and NHEJ assays were performed in DR-GFP U2OS cells and EJ5 U2OS cells, respectively. Cells were transfected with 30 nM of indicated siRNAs. After 24 h cells were re-transfected. Forty-eight hours after the first transfection, cells were transfected with the plasmids expressing I-SceI endonuclease and DsRed. Four days after the first transfection, the cells were harvested and analyzed by flow cytometry for the expression of GFP and DsRed. Flow cytometry data were collected from 10 000 to 50 000 cells. The results are presented as mean ± s.e.m. from three independent experiments.
Detection of chromosomal aberrations
Cells were incubated with 100 ng/ml colcemid at 37°C for 3 h. Cells for metaphase preparation were collected according to standard procedure as previously described (30). In brief, the cellular pellet was resuspended in prewarmed 0.075 M KCl solution and incubated at 37°C for 20 min followed by addition of fixative solution (3:1 methanol/acetic acid). Cell suspension was dropped onto cold slides to make chromosome preparations. The slides were air dried overnight and stored at −20°C until analysis. Chromosomal aberrations were examined in Giemsa-stained metaphases under a Zeiss Axio Observer.Z1 microscope. At least 50 chromosomes were examined, and chromosomal abnormalities scored at 100× magnification.
DNA branch-migration assay
Oligonucleotides used for DNA substrates are listed in Table 1. To prepare substrates for 3′ → 5′ or 5′ → 3′ three-stranded branch migration, 25 pmol of DNA oligos 169 (3′ → 5′) or 193 (5′ → 3′) were labeled with 30 μCi γ32P-ATP (Perkin Elmer) and T4 Polynucleotide kinase (New England Biolabs) and unincorporated ATP removed. The radiolabeled 169 or 193 oligos were annealed to 25 pmol of 71 or 117 in TE pH 8.0 by heating at 95°C followed by slow cooling to RT. The 169/71 and 193/117 duplexes (0.5 nM) were mixed with DNA oligos 201 or 379 (1 nM), respectively in branch migration buffer containing 25 mm Tris acetate, pH 7.5, 2 mM ATP, 5 mm magnesium acetate, 2 mM DTT, 100 μg/ml BSA, 15 mM phosphocreatine, and creatine phosphokinase (30 units/ml) for 15 min at 37°C. To prepare substrate for examining DNA helicase activity the 169*/71 duplex was mixed with ssDNA oligonucleotide 248HEL as described above. Reactions were initiated by adding FANCJ (1.25 nM) for 30 min or 0–60 min for kinetics experiments. Reactions were stopped by adding 1.5% SDS and proteinase K (800 μg/ml) for 15 min at 37°C, followed by 0.1 volumes of loading dye (70% glycerol, 0.1% bromophenol blue) prior to loading the on 8% polyacrylamide 1× TBE gels. Gels were run at 135 V for 1.5 h. Gels were exposed to a phosphorscreen, visualized with a Typhoon FLA 9500 (GE Healthcare), and quantified using ImageQuant (GE Healthcare).
Table 1.
Oligonucleotides used to prepare DNA substrates for FANCJ biochemical assays
| Oligo name | Oligo length (nt) | Oligo sequence (5′→3′) |
|---|---|---|
| 169 | 93 | TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTT |
| 193 | 94 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTCACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA |
| 71 | 94 | CTTTAGCTGCATATTTACAACATGTTGACCTACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA |
| 117 | 94 | TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTGCTGAATCTGGTGCTGTAGGTCAACATGTTGTAAATATGCAGCTAAAG |
| 201 | 94 | TCCTTTTGATAAGAGGTCATTTTTGCGGATGGCTTAGAGCTTAATTATAAAATCTGGTGCTGTAGGTCAACATGTTGTAAATATGCAGCTAAAG |
| 379 | 94 | CTTTAGCTGCATATTTACAACATGTTGACCTACAGCAAAGAATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA |
| 379HEL | 94 | TCCAGTTGTACAACATTTATACGTCGATTTCACAGCACCAGATTCAGCAATTAAGCTCTAAGCCATCCGCAAAAATGACCTCTTATCAAAAGGA |
RESULTS
RNAi screen of isogenic FANCJ knock-out and wild-type cells
We conducted a siRNA screen to identify genes of the DNA damage response/DNA repair regime that when acutely depleted sensitize FANCJ KO cells to the DNA damaging effects caused by a relatively low concentration of the DNA cross-linking agent MMC. The FANCJ KO cells previously described (20) were confirmed to have complete loss of FANCJ by Western blot (Supplementary Figure S1A). A very low dose of MMC (6.25 nM) was chosen such that by itself the drug exerted only a minimal effect on viability of the FANCJ KO cells when compared to the isogenic WT cells (Supplementary Figure S1B). We used a custom siRNA library that contains 240 known DNA damage response and repair genes with a pool of four individual siRNAs per gene in separate wells of 96-well plates. The WT and FANCJ KO cells were individually transfected with the indicated pool siRNA library gene targets. Twenty-four hr later the media was changed, and cells were either left untreated or exposed to 6.25 nM MMC for 72 h. Cell viability was subsequently measured by the WST-1 cell proliferation assay, and percentage viability of the WT or FANCJ KO cells transfected with each gene-targeted siRNA pool was calculated by normalizing the absorbance values of the individual gene-targeted siRNAs to that of the control (non-specific) siRNA. The screen was completed in triplicate, and the mean percent viability is plotted for each siRNA in the absence or presence of MMC (Supplementary Figures S2 and S3).
Reduced colony survival of cells deficient in both FANCJ and RAP80 after mitomycin exposure
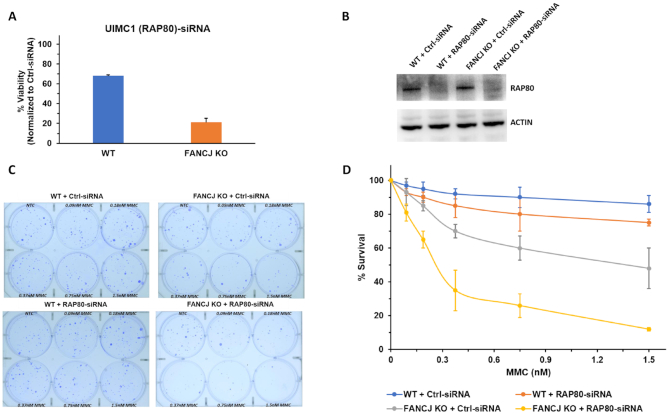
UIMC1/RAP80 (hereafter referred to as RAP80) was one of the top hits in the RNAi screen, displaying under conditions of 6.25 nM MMC exposure a 45% reduced proliferation of FANCJ KO cells compared to WT cells (Figure 1A). The reduced proliferation caused by the co-deficiency of FANCJ and RAP80 in MMC-treated cells led us to test for cell survival by a clonogenic assay. The clonogenic assay is devised to determine reproductive cell death to assess drug sensitivity (31); consequently, it provides a sensitive measurement of cell division not assessed by the RNAi screen's metabolic activity assay. Initially, the isogenic pair of FANCJ KO or WT cells were tested for sensitivity to increasing MMC concentrations to determine an optimal dose (Supplementary Figure S4), followed by clonogenic assays of the isogenic FANCJ KO and WT cells transfected with RAP80 RNAi or control siRNA (Figure 1). Cells transfected with RAP80 siRNA displayed little to no RAP80 as detected by Western blot (Figure 1B). As shown in Figure 1C and D, FANCJ KO cells depleted of RAP80 showed significantly reduced survival at an MMC concentration as low as 0.2 nM compared to FANCJ KO cells transfected with control siRNA. At 0.38 nM MMC, the RAP80-depleted FANCJ KO cells displayed 35% survival compared to 70% survival of the control siRNA transfected FANCJ KO cells. WT cells depleted of RAP80 only showed a 15% decrease in survival at 0.38 nM MMC exposure, indicating that the combined deficiency of both RAP80 and FANCJ resulted in a greater decrease in survival compared to either FANCJ or RAP80 deficiency alone. Heightened sensitivity of FANCJ/RAP80 co-deficient cells was also observed at MMC doses of 0.76 and 1.52 nM (Figure 1D).
Figure 1.
Loss of RAP80 sensitizes FANCJ KO cells to a low dose of Mitomycin C. Panel (A), RNAi screening performed as described in Materials and Methods demonstrated that UIMC1 (RAP80) was one of the ten positive hits of the 240 DNA damage response genes that in RNAi-depleted FANCJ KO cells sensitized them to 6.25 nM MMC as measured by a cell viability assay. Panel (B), Western blot showing RNAi-mediated RAP80 depletion in the isogenic FANCJ KO and WT cell lines. Panel (C), WT or FANCJ KO cells were transfected with 30 nM control siRNA or RAP80 pool siRNA. After 24 h cells were re-transfected with 30 nM control siRNA or RAP80 pool siRNA, and 500–1000 cells were seeded in six-well plates with media containing the specified MMC concentration. Cells were incubated for 6 days after which media was changed. Colonies were detected 14 days after the initial seeding using crystal violet staining. Panel (D), Quantification of colony formation from three independent experiments.
To address the specificity of the effects caused by the pool RNAi depletion of RAP80, we tested individual siRNAs against RAP80 for their sensitization of FANCJ KO cells to MMC. As shown by western blot, RAP80 siRNA#1 was very effective in depleting RAP80 (Supplementary Figure S5A). Moreover, FANCJ KO cells depleted of RAP80 by siRNA #1 behaved very similarly to those cells depleted of RAP80 by the pool siRNA in terms of their sensitization to MMC (Supplementary Figure S5B). At an MMC dose of 0.37 nM, the percentage survival of FANCJ KO cells depleted of RAP80 by RAP80 siRNA#1 was reduced by 57% compared to a combined reduction of 35% attributed to FANCJ KO (29%) and RAP80 depletion (6%). These results suggest that a combined deficiency of FANCJ and RAP80 together exerts a more severe effect on MMC survival compared to the additive effects of FANCJ deficiency and RAP80 deficiency.
Because ALT cells (such as the U2OS cell line used in the RNAi screen) are known to have aberrant HR pathways and are partially deficient due to lack of ATRX gene expression (32), we tested two non-ALT cell lines (HeLa and HEK293 (see below)) to evaluate the broader applicability of the apparent FANCJ-RAP80 genetic interaction. As shown in Supplementary Figure S5C, at an MMC concentration of 0.38 nM RAP80-depleted FANCJ KO HeLa cells displayed 48% survival compared to 75% survival for the FANCJ KO cells transfected with control siRNA or 85% survival for WT cells depleted of RAP80. A hypersensitivity of HeLa cells co-deficient in FANCJ and RAP80 to other MMC doses was also observed.
To extend the finding that cells deficient in both FANCJ and RAP80 are hypersensitive to ICL-induced DNA damage, we tested the effect of the DNA cross-linking agent cisplatin (Supplementary Figure S5D). As expected, WT cells transfected with a control siRNA displayed only modest sensitivity to a low cisplatin dose range of 7.5–60 nM as determined by colony survival assays. RAP80-depleted cells were also resistant to these concentrations of cisplatin, consistent with what was observed for the DNA cross-linking agent MMC (Figure 1 and Supplementary Figure S5B). FANCJ KO cells showed sensitivity to cisplatin in a dose-dependent manner (Supplementary Figure S5D). However, FANCJ KO cells depleted of RAP80 displayed a significantly greater sensitivity to cisplatin at all concentrations tested, consistent with the results from the MMC experiments.
We next performed annexin V-FITC and propidium iodide (PI) staining to determine the percentage of apoptotic and necrotic cells after an acute 1 h exposure to 3 μM MMC. FANCJ KO or WT cells depleted of RAP80 displayed similar percentages of death (25% and 23%, respectively) when examined 72 h after MMC exposure compared to only ∼6% apoptosis/necrosis for WT cells transfected with control siRNA (Supplementary Figure S6). In contrast, 46% of the RAP80-depleted FANCJ KO cells exposed to MMC were apoptotic or necrotic. These findings demonstrate that acute exposure of FANCJ- and RAP80-deficient cells to MMC caused a significantly greater level of death compared to cells deficient of FANCJ or RAP80.
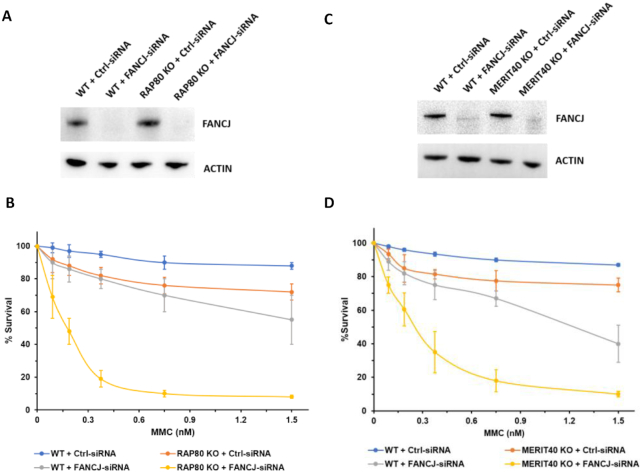
To confirm the findings from the colony survival experiments with FANCJ KO cells (Figure 1), we performed reciprocal experiments with RAP80 KO cells (Figure 2B). RAP80 KO cells depleted of FANCJ displayed greater MMC sensitivity than the combined MMC sensitivity of FANCJ-depleted WT cells and RAP80 KO cells treated with control siRNA. Taken together with the results shown in Figure 1, we conclude that the combined loss of both FANCJ and RAP80 renders cells more sensitive to MMC than the additive effects of FANCJ deficiency and RAP80 deficiency.
Figure 2.
RAP80 KO cells or Merit40 KO cells depleted of FANCJ are hypersensitive to MMC. Panels (A) and (C), western blot showing RNAi-mediated FANCJ depletion in the isogenic RAP80 KO or Merit40 KO and WT cell lines. Panels (B) and (D), WT and RAP80 KO or Merit40 KO cells were transfected with 30 nM control siRNA or FANCJ siRNA. After 24 h cells were re-transfected with 30 nM control siRNA or FANCJ siRNA, and 500–1000 cells were seeded in 6-well plates with media containing the specified MMC concentration. Cells were incubated for 6 days after which media was changed. Colonies were detected 14 days after the initial seeding using crystal violet staining. Quantification of colony formation is from three independent experiments.
The apparent synergism between FANCJ and RAP80 due to loss of one by CRISPR KO and the other by RNAi depletion led us to attempt to CRISPR double KO of both FANCJ and RAP80. After several attempts, we were unsuccessful in creating FANCJ/RAP80 double KO’s with U2OS cells, indicating that the combined complete loss of both FANCJ and RAP80 is lethal. While we were able to isolate a limited number of small surviving colonies that we believe to be knocked out for both FANCJ and RAP80 in HeLa, the cells were extremely slow growing (again consistent with a strong synthetic sickness), making it technically difficult to perform any reliable experiments.
Merit40 deficiency exacerbates MMC sensitivity of FANCJ-depleted cells
Merit40 resides in a RAP80 protein complex that facilitates the targeting of BRCA1 to DNA lesions (33–35). To determine if Merit40 deficiency behaves in a manner similar to RAP80 in terms of its genetic interaction with FANCJ, we performed MMC survival assays with isogenic Merit40 KO or WT cells transfected with control siRNA or FANCJ siRNA. We observed that the reduction in colony formation for Merit40 KO cells was less than that observed for WT cells depleted of FANCJ (Figure 2D). Merit40 KO cells depleted of FANCJ showed even further reduced survival throughout the MMC titration range (0.075–1.5 nM) compared to the additive loss in cell survival upon MMC exposure for Merit40 KO cells transfected with control siRNA and WT cells transfected with FANCJ siRNA (Figure 2D). For example, Merit40 KO cells displayed a 20% reduction in survival at an MMC dose of 0.75 nM, and FANCJ-depleted WT cells showed a 32% reduction in survival, whereas the combined loss of the two resulted in an 81% reduced colony survival. These results suggest that like RAP80, Merit40 deficiency combined with FANCJ deficiency exacerbates MMC sensitivity as measured by colony survival assays.
FANCJ catalytic activity is required to suppress heightened MMC sensitivity in a RAP80-deficient background
We next wanted to determine if the synergistic effect of RAP80 and FANCJ co-deficiency on MMC sensitivity was dependent on FANCJ catalytic activity. To address this, we asked whether expression of a clinically relevant FANCJ ATPase domain mutant designated FANCJ-H396D, which we previously determined to be severely compromised in its ATP hydrolysis and completely dead as a helicase (36), in a FANCJ KO background depleted of RAP80 could rescue the MMC sensitivity. FANCJ KO cells were stably transfected with a GFP expressing empty vector, GFP-WT-FANCJ, or GFP-FANCJ-H396D. Western blot analysis confirmed that the expression level of FANCJ-H396D mutant was nearly the same as that of WT-FANCJ normalized to the loading control actin (Supplementary Figure S7A). FANCJ KO cells stably expressing GFP-WT-FANCJ, GFP-FANCJ-H396D or empty vector transfected with RAP80 siRNA displayed little detectable RAP80 as assessed by Western blot (Supplementary Figure S7B)
At an MMC concentration of 0.18 nM, the RAP80-depleted FANCJ KO cell line expressing FANCJ-H396D showed a 57% reduction in survival, comparable to what was observed in RAP80-depleted FANCJ KO cells transfected with a GFP construct (60% reduction) (Supplementary Figure S7C). This reduction was significantly greater than the 28% reduction in FANCJ KO cells transfected with control siRNA. WT cells depleted of RAP80 only showed 6% reduction in survival. Therefore, the additive effect of RAP80 depletion and FANCJ KO was ∼1.7-fold less than that of the combined deficiency of FANCJ and RAP80, confirming their synergistic interaction. Moreover, expression of the catalytically defective FANCJ-H396D in the RAP80-depleted FANCJ KO cells did not alter this effect, suggesting that FANCJ’s intact ATPase/helicase activity is required for MMC resistance even when RAP80 is depleted.
Concomitant deficiency of FANCJ and RAP80 causes ICL-induced ssDNA accumulation and elevated RPA and RAD51 foci in MMC-treated cells
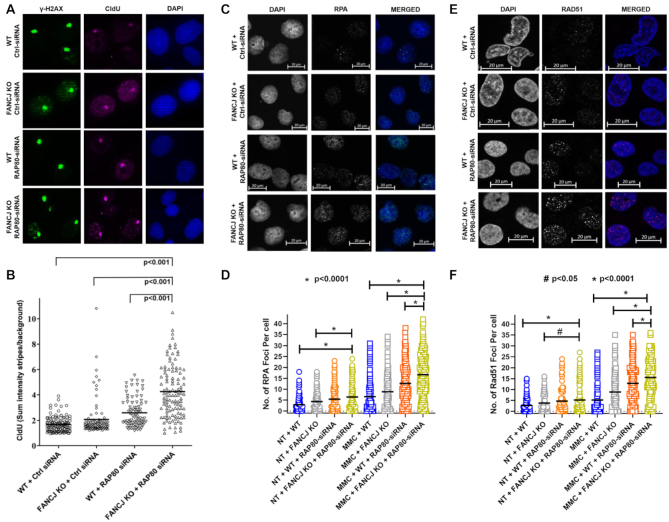
Several reports suggest that RAP80 recruits and sequesters BRCA1 to the HR repressive BRCA1-A complex (37–39). RAP80 represses HR in response to DSBs by blocking DNA end-resection (40,41). BRCA1, which resides with FANCJ in a distinct complex from that of RAP80 (39), is also required to promote HR via its interaction with FANCJ (18). We hypothesized that depletion of RAP80 would result in accumulation of single-stranded (ss) DNA due to excessive end-resection and loss of FANCJ would further inhibit processing of the excessively resected DNA. Therefore, we asked if there was an increased amount of single-stranded DNA persisting at sites of laser-activated Pso-ICL 150 min post laser targeting. As shown in Figure 3A and B, the FANCJ KO cells depleted of RAP80 displayed ∼2.3-fold greater staining for CldU-marked ssDNA detected in γ-H2AX stripes compared to the isogenic WT cells transfected with control siRNA. These results suggest the accumulation of single-stranded DNA over time in cells co-deficient of FANCJ and RAP80 that are subjected to ICL-induced damage.
Figure 3.
Dual loss of FANCJ and RAP80 results in ssDNA accumulation at ICL-induced DNA damage, and elevated RPA and RAD51 foci in MMC-treated cells. Panels (A) and (B), Pso-ICL CldU ssDNA accumulation. WT or FANCJ KO cells were transfected with 30 nM control siRNA or RAP80 siRNA#1. After 24 h cells were re-transfected with 30 nM control siRNA or RAP80 siRNA#1. Cells were kept in 20 μM CldU for 48 hr. Forty-eight hr after the first transfection, cells were incubated with psoralen followed by laser treatment (1%) to induce Pso-ICLs. Cells were fixed and stained with anti-γ-H2AX and anti-CldU antibody. Panel (A), representative images of γ−H2AX and CldU stripes. Panel (B), The mean intensity of CldU localized to ICL stripes were quantified in single cells at 150 min after laser treatment. Panels (C–F), Number of RPA and RAD51 foci per cell are elevated in the non-treated and MMC-treated cells doubly deficient in RAP80 and FANCJ. WT or FANCJ KO cells were transfected with 30 nM control siRNA or RAP80 siRNA#1. After 24 hr cells were re-transfected with 30 nM control siRNA or RAP80 siRNA#1. Forty-eight hours after the first transfection, cells were either left untreated or were treated with 3 μM MMC for 1 h and allowed to recover in fresh media. Sixteen hours later, RPA (Panels C, D) and Rad51 foci (Panels E, F) were determined after removing the soluble proteins and fixing chromatin bound proteins. Shown are representative images of the MMC-treated cells and quantitative analyses of both the untreated and MMC-treated cells. Over 150 cells were examined, and the results are mean ± standard error of mean (s.e.m.) from three biological repeats, with P-values determined by Student's t-tests.
In the experiments with MMC, heightened nuclear Replication Protein A (RPA) and RAD51 foci may be attributed to the over-abundant DNA end-resection and lack of subsequent maturation of the RPA-coated and RAD51-coated ssDNA molecules. Indeed, our experimental data suggest this to be the case. FANCJ KO cells acutely exposed to 3 μM MMC for 1 h followed by a 16 h incubation showed a 1.3-fold increase in the average number of RPA foci per cell compared to WT cells, whereas depletion of RAP80 in WT cells elevated the number RPA foci per cell by 2.1-fold (Figure 3C and D). However, loss of both FANCJ and RAP80 resulted in a 2.9-fold increase in number of RPA foci per cell. For RAD51 staining, deficiency of FANCJ or RAP80 in the MMC-treated cells resulted in 1.8-fold and 2.5-fold increases in number of cells with Rad51 foci per cell, whereas deficiency of both FANCJ and RAP80 resulted in a 3.2-fold increase in RAD51 staining (Figure 3E-F). In the absence of MMC treatment, depletion of FANCJ and/or RAP80 also resulted in significantly higher number of RPA and Rad51 foci per cell (Figure 3D, F).
We performed a time-course experiment to examine the effect of FANCJ and/or RAP80 depletion on the appearance and disappearance of RPA and Rad51 foci to determine their temporal dynamics of foci formation during ICL repair. FANCJ KO cells displayed similar abundance of RPA and Rad51 foci in response to MMC treatment at both the 6 and 12 h time-points compared to control siRNA transfected cells (Supplementary Figure S8A and B). However, there was a 2-fold greater number of cells with >10 RPA foci in FANCJ KO cells compared to isogenic WT cells at the 24 h time-point post MMC exposure (Supplementary Figure S8A). A significantly 4-fold greater number of FANCJ-KO cells with 10 or more RPA foci persisted at 36 h compared to the WT cells which showed a decline compared to the 24-h timepoint. RAP80-depleted FANCJ KO or WT cells showed comparably elevated percentage with greater than 10 RPA foci at the 6 or 12 h time-point compared to cells not depleted of RAP80. Notably, FANCJ KO cells depleted of RAP80 displayed nearly 70% with >10 RPA foci at 24 h post MMC exposure and the elevated level persisted at 36 h, significantly more than the WT cells depleted of RAP80. Based on these results, we conclude that throughout the time course of recovery from 6–36 h, RPA foci persisted to the greatest extent in cells deficient of both FANCJ and RAP80 (Supplementary Figure S8A).
We next examined the percentage of cells in the FANCJ/RAP80 series that displayed >10 Rad51 foci (Supplementary Figure S8B). Here, we observed a similar pattern to that of cells with abundant RPA foci. Rad51 foci were enriched in RAP80-depleted FANCJ KO or WT cells throughout the time-course; however, notably the percentage of RAP80-depleted FANCJ KO cells with >10 Rad51 foci persisted at the 24 and 36 h time-points post MMC exposure, reaching a very high level of ∼70% (Supplementary Figure S8B). We conclude from these data that like RPA foci, Rad51 foci persisted in the most abundant manner in those cells deficient in both FANCJ and RAP80. Taken together, these results suggest that ICL repair intermediates bound by RPA and Rad51 persist in cells co-deficient in FANCJ and RAP80.
To validate that the increased RPA foci formation in cells exposed to MMC is attributed to elevated ssDNA, we examined for BrdU foci in cells prelabeled with the nucleotide analogue with an antibody that specifically recognizes BrdU in the single-stranded state, following a published protocol (29) (Supplementary Figure S9). We found that a deficiency of FANCJ or RAP80 in the MMC-treated cells resulted in 1.4-fold and 2.1-fold increases in the number of BrdU foci per cell, respectively. Deficiency of both FANCJ and RAP80 resulted in a 2.7-fold increase in the number of BrdU foci per cell, as measured by these parameters. These results are consistent with the increase in RPA and RAD51 foci which we observed in these cells after treatment with MMC.
We next performed a reciprocal set of experiments and assessed the number of RPA and RAD51 foci per cell in MMC-treated RAP80 KO cells that were depleted of FANCJ (Supplementary Figure S10). As expected, loss of RAP80 alone resulted in a significant (1.8-fold) increase in the average number of RPA foci per cell (Supplementary Figure S10A). FANCJ depletion in the WT cells also showed a significant 1.4-fold increase in the average number of RPA foci per cell. FANCJ-depleted RAP80 KO cells showed 2.6-fold increase in RPA foci per cell compared to the WT cells. For RAD51 staining, deficiency of FANCJ or RAP80 in the MMC-treated cells resulted in 1.7-fold and 2-fold increases in the average number of Rad51 foci per cell, respectively, whereas depletion of both FANCJ and RAP80 resulted in a 3-fold increase in the number of RAD51 foci per cell (Supplementary Figure S10B). Thus, in either the RAP80 KO background or the FANCJ KO background (Figure 3), loss of the additional player (FANCJ or RAP80, respectively) exacerbated the level of MMC-induced RPA and RAD51 staining, suggesting the elevated accumulation of ssDNA and RAD51 protein filaments in MMC-treated cells when both FANCJ and RAP80 are deficient.
Strand resection provoked by MMC-induced DNA damage is dependent on CtIP and Mre11 nucleases
The accumulation of RPA and Rad51 foci in MMC-treated cells co-deficient in FANCJ and RAP80 suggested to us that the overly active DNA end-resection at sites of ICL-induced DNA damage resulted in ssDNA accumulation. To provide mechanistic insight, we sought to address if known DNA end-resection nucleases are involved. First, we examined foci formation of CtIP, a known nucleolytic processing enzyme that operates at DSBs in mammalian cells (42–44). CtIP foci formation were visualized after a 1 h exposure to 3 μM MMC followed by recovery for 16 hr (Supplementary Figure S11A). Under these conditions, 35% of WT cells and 42% of FANCJ KO cells displayed greater than 10 CtIP foci per cell. WT cells depleted of RAP80 displayed a significant 34% increase in CtIP immuno-staining, whereas FANCJ KO cells depleted of RAP80 showed a 41% increase compared to WT cells.
We next sought to address if CtIP was responsible for ssDNA production in the MMC-treated cells, given our observations that CtIP foci were significantly enriched in the MMC-treated cells. CtIP depletion in FANCJ KO cells, RAP80-depleted WT cells and FANCJ/RAP80 double-deficient cells showed 22%, 28% and 30% of cells with >10 BrdU, respectively (Supplementary Figure S11B). The significant reduction in BrdU foci for all these cell lines indicates that CtIP is important for strand resection of MMC-induced DNA damage in these genetic backgrounds.
We next asked if these same cells pretreated with 50 μM Mirin, a compound that inhibits the DNA end-processing nuclease Mre11 (45), and then exposed to MMC under the same conditions as described above, would display altered BrdU foci abundance (Supplementary Figure S11B). Strikingly, we observed only 7% of the Mirin-treated cells depleted of both FANCJ and RAP80 to have >10 BrdU foci. Mirin also suppressed BrdU foci in WT cells, as well as those cells depleted of FANCJ or RAP80. These results suggest that the ssDNA formed in the FANCJ/RAP80 double-deficient cells is dependent on both the CtIP and Mre11 nucleases.
Co-deficiency of FANCJ and RAP80 impairs double-strand break repair
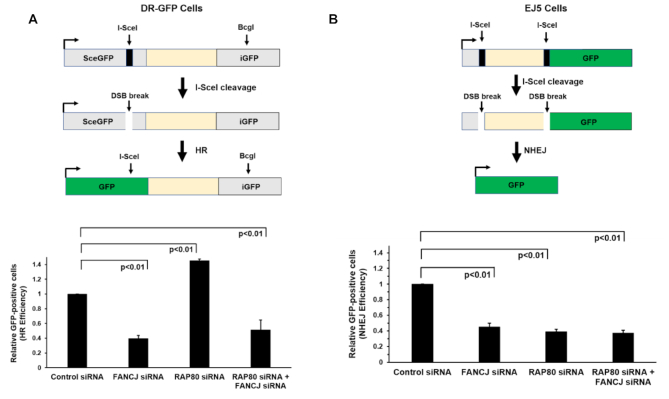
HR-mediated DSB repair is an important downstream event in ICL repair; therefore, we sought to address the potential interactive roles of FANCJ and RAP80 in DSB repair. To assess the effect of deficiency in FANCJ, RAP80 or both in DSB repair, we performed I-SceI-induced DSB repair assays with cells depleted of FANCJ and/or RAP80 using a chromosomal integrated GFP reporter construct to evaluate efficiency of HR repair or NHEJ (46). HR repair of the I-SceI DSB was reduced 2.5-fold in FANCJ-depleted cells and increased 1.4-fold in RAP80 cells (Figure 4A), consistent with previous reports for FANCJ (18) and RAP80 (40,41). Interestingly, co-depletion of FANCJ and RAP80 suppressed the elevated HR repair observed in singly depleted RAP80 cells, bringing it to a level approximating that observed in the FANCJ-depleted cells. Thus, the elevated HR repair of I-SceI-induced DSBs observed in RAP80-depleted cells was suppressed by a co-deficiency of FANCJ.
Figure 4.
Cells depleted of FANCJ and RAP80 display reduced double-strand break repair by homologous recombination or nonhomologous end-joining. The I-SceI DSB repair assay was used to measure HR repair or NHEJ of the indicated RNAi-depleted cell lines as described in Materials and Methods. Panel (A), HR efficiency in siRNA transfected DR-GFP cells. Panel (B), NHEJ efficiency in siRNA transfected EJ5 cells. The results are mean ± standard error of mean (s.e.m.) from three biological repeats, with P-values determined by Student's t-tests.
Next, we measured NHEJ using the I-SceI DSB repair assay (Figure 4B). Depletion of FANCJ or RAP80 alone reduced NHEJ by nearly 2.5-fold, consistent with findings from previous studies of FANCJ (47,48) and RAP80 (40). Co-depletion of both FANCJ and RAP80 resulted in a similarly reduced NHEJ, suggesting that they may behave in an epistatic manner. Collectively, these results demonstrate that co-depletion of FANCJ and RAP80 resulted in a significant reduction in both HR and NHEJ, the two major DSB repair pathways in the cell.
MMC-induced BRCA1 foci formation is compromised by the Co-deficiency of FANCJ and RAP80
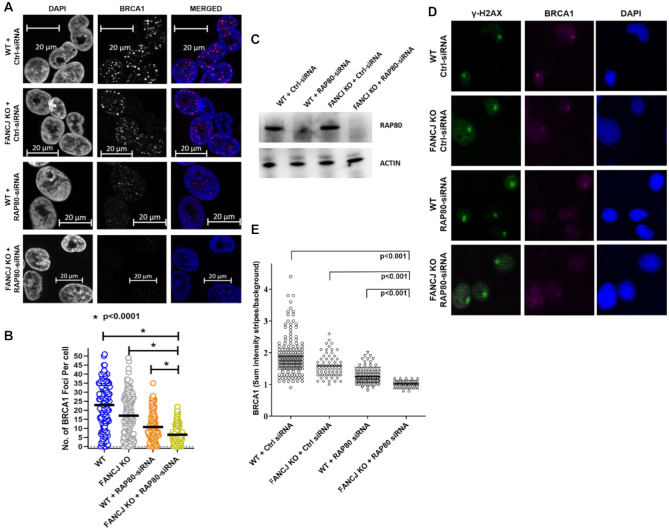
Given that BRCA1 plays a critical role in the response to ICL-induced DNA damage and that both FANCJ and RAP80 interact with BRCA1 in distinct sub-complexes (49), we examined BRCA1 foci formation in FANCJ- and/or RAP80-deficient cells after a 1 h exposure to 3 μM MMC and recovery for 16 h (Figure 5A-B). Upon exposure to MMC the average number of BRCA1 foci per cell was 23 in the WT cells. FANCJ depletion reduced the average number of BRCA1 foci per cell to 17. RAP80 depletion in the WT cells exerted a greater effect by reducing the average number of BRCA1 foci per cell to 11. Combined deficiency of FANCJ and RAP80 led to the greatest reduction in the average number of BRCA1 foci per cell to 6, indicating an additive effect on ICL damage induced BRCA1 localization caused by the loss of the two proteins. These results suggest that FANCJ and RAP80 act upstream and at least partly independently to elicit BRCA1 foci formation during the ICL-induced DNA damage response.
Figure 5.
FANCJ and RAP80 act upstream and independently to elicit BRCA1 foci during the cellular response to mitomycin C and recruitment of BRCA1 to laser-induced psoralen cross-links. Panel (A), WT or FANCJ KO cells were transfected with 30 nM control siRNA or RAP80 siRNA#1. After 24 h, cells were re-transfected with 30 nM control siRNA or RAP80 siRNA#1. Forty-eight hr after the first transfection, cells were treated with 3 μM MMC for 1 h and allowed to recover in fresh media. Sixteen hours later, BRCA1 foci were examined after removing the soluble proteins and fixing chromatin bound proteins. Representative images for BRCA1 foci in the indicated cell lines are shown. Panel (B), Quantitative analysis of MMC-induced BRCA1 foci per cell. Over 100 cells were examined, and the results are mean ± standard error of mean (s.e.m.) from three biological repeats, with P-values determined by Student's t-tests. Panel (C), Western blot showing RNAi-mediated RAP80 depletion in the isogenic FANCJ KO or WT cell lines. Cells were incubated with psoralen followed by laser treatment (1%) to induce Pso-ICLs. Cells were fixed and stained with anti-γ−H2AX and anti-BRCA1 antibody Panel (D), representative images of γ−H2AX and BRCA1 stripes. Panel (E), Mean intensity of BRCA1 localized to ICL stripes were quantified in single cells at 20 min after laser treatment. The results are mean ± standard error of mean (s.e.m.) from three biological repeats, with P-values determined by Student's t-tests.
BRCA1 recruitment to laser-induced psoralen-interstrand cross-links is significantly reduced by a Co-deficiency of FANCJ and RAP80
The functional involvement of FANCJ and RAP80 in the DNA damage response and regulation of recombinational repair led us to interrogate their importance in modulating BRCA1 recruitment to laser-induced Pso-ICLs. Using the isogenic FANCJ KO and WT cells transfected with RAP80-RNAi or control siRNA, endogenous BRCA1 recruitment to the Pso-ICL was detected in γ-H2AX marked stripes and quantified as sum intensity strip over background. As shown in Figure 5D and E, the FANCJ KO or RAP80-depleted WT cells both showed a significant diminishment of BRCA1 recruitment intensity, suggesting they have roles upstream of BRCA1 in the ICL-induced DNA damage response. Combined deficiency of FANCJ and RAP80 resulted in the greatest reduction of BRCA1 recruitment to the Pso-ICL, indicating that their effects on BRCA1 localization to the ICL-induced DNA damage are at least in part separate from one another.
DNA damage induced checkpoint severely compromised in cells deficient in both RAP80 and FANCJ
Persistent DNA damage leads to checkpoint activation to delay cell cycle progression and promote repair of the damaged DNA (50). Activated cell cycle checkpoints prevent cells from entering mitosis with the damaged DNA and avoid mitotic catastrophe. BRCA1 recruitment is crucial for the activation of the S- and G2/M-checkpoint (51,52). BRCA1 forms at least 3 distinct complexes (BRCA1-A, BRCA1-B and BRCA1-C). BRCA1-A complex is comprised of RAP80, Abraxas, BRCC36, BRCC45, BARD1, BRCA1 and MERIT40. BRCA1-B complex is composed of BRCA1, TopBP1 and FANCJ. BRCA1-C contains BRCA1, CtIP, and MRN complex (39,49). Based upon these findings and our own observations that FANCJ and RAP80 play distinct roles in ICL-induced BRCA1 foci formation and BRCA1 recruitment to ICL-induced DNA damage, we hypothesized that RAP80 (as a member of BRCA1-A) and FANCJ (as a member of BRCA1-B) act in two parallel pathways to activate cell cycle checkpoints elicited by MMC exposure as marked by increase in phosphorylation of Chk1 at S317 and S345 (53,54).
As shown in Supplementary Figure S12, Chk1-S317 and Chk1-S345 became phosphorylated in WT cells exposed to 3 μM MMC. Depletion of RAP80 in the WT cells resulted in a partial decrease in phosphorylation of Chk1-S317 or Chk1-S345, as normalized to total Chk1. These results are consistent with a previous report that Chk1 activation is defective in RAP80-depleted cells exposed to ionizing radiation (37). FANCJ KO cells displayed a more modest reduction in MMC-induced Chk1-S317 or Chk1-S345 phosphorylation, consistent with previous work suggesting a moderate decrease in camptothecin-induced Chk1 activation (55). However, combined deficiency of both FANCJ and RAP80 practically eliminated Chk1-S317 and Chk1-S345 phosphorylation, indicating a severely compromised ICL-induced DNA damage checkpoint.
In response to IR-induced DNA damage, ATM phosphorylates Chk2 at residue T68 (56). A previous study showed that camptothecin-treated FANCJ mutant cells display similar p-Chk2-T68 levels as FANCJ-WT cells (55). In another study, it was reported that depletion of Abraxas (a component of the BRCA1 complex A) also did not cause reduction of p-Chk2-T68 in IR-treated cells (24). In the current study focused on ICL-induced DNA damage, we observed that Chk2-T68 phosphorylation was induced in the WT cells exposed to 3 μM MMC. In comparison to WT cells, loss of FANCJ, depletion of RAP80, or a deficiency in both FANCJ and RAP80 showed similar levels of phosphorylated Chk2-T68 (Supplementary Figure S12). These results suggest that unlike the ATR signaling pathway, the loss of FANCJ and/or RAP80 did not significantly alter the ATM-dependent checkpoint response compared to WT cells exposed to the ICL-inducing agent MMC.
Although individual loss of FANCJ (57) or RAP80 (37) has been reported to cause a defect in G2/M checkpoint activation in response to IR-induced DNA damage, analysis of the effect of combined loss of both FANCJ and RAP80 in the G2/M checkpoint response has not been previously reported. To understand the dual importance of FANCJ and RAP80 in the MMC-induced checkpoint response, RAP80 was depleted in FANCJ KO cells and cells were stained with phospho-H3 Ser10 which is a marker of condensed chromosomes in mitotic cells (58). In the absence of exogenously induced DNA damage, loss of FANCJ or RAP80 alone resulted in a 2-fold increase in mitotic index whereas loss of both FANCJ and RAP80 together resulted in a 6-fold increase (Supplementary Figure S13). Combined deficiency of FANCJ and RAP80 in cells exposed to 3 μM MMC for 1 h followed by 18 h recovery resulted in an 18-fold increase in mitotic index compared to only a 2-fold increase for MMC-treated RAP80-depleted cells and a 7-fold increase for MMC-treated FANCJ-depleted cells (Supplementary Figure S13). These results suggest a synergistic effect of FANCJ and RAP80 loss on the G2/M checkpoint response.
Abraxas/CCDC98 mediates the interaction of BRCA1 with other components of the BRCA1-A complex, which include BRCC36, BRE, MERIT40 and RAP80 (35,59–61). The Chen laboratory had shown that depletion of Abraxas does not affect the formation of RAP80 foci, but reduces the formation of BRCA1 foci after IR-induced DNA damage (59). Thus, Abraxas functions downstream of RAP80, serving as a bridge to recruit BRCA1. To confirm that the phenotypes observed are dependent on BRCA1, we performed MMC colony survival assays with Abraxas KO cells depleted of FANCJ. At an MMC dose of 0.38 nM, FANCJ-depleted Abraxas KO cells displayed 55% survival compared to 92% survival for the Abraxas KO cells transfected with control siRNA and 81% survival of WT cells depleted of FANCJ (Supplementary Figure S14A). Therefore, the combined deficiency of both FANCJ and Abraxas resulted in a synergistic increase in MMC sensitivity. Similar results were obtained with greater MMC doses (Supplementary Figure S14A).
Phosphorylation of FANCJ at S990 is important for the BRCA1-FANCJ interaction (57,62), which mediates the DNA damage-induced G2/M checkpoint (57). RAP80 has also been shown to be important for the G2/M checkpoint as well as BRCA1 recruitment to IR-induced DSBs (37,38,63). We hypothesized that FANCJ and RAP80 act in parallel pathways to recruit BRCA1 to activate the G2/M checkpoint; therefore, we wanted to ask if RAP80-depleted cells expressing the FANCJ-S990A mutant protein would show elevated sensitivity to MMC. At an MMC dose of 0.18 nM, the RAP80-depleted cells expressing FANCJ-S990A were reduced by 20% compared to a 7% reduction of RAP80-proficent cells expressing the FANCJ-S990A mutant protein and 5% reduction for RAP80-depleted cells (Supplementary Figure S14B). It is plausible that a distinct role of FANCJ in ICL resistance is elicited when BRCA1 is not efficiently recruited to ICL-induced DNA damage, analogous to its involvement in a switch from HR repair to translesion-dependent synthesis by polymerase η (62).
Elevated chromosomal instability in cells doubly deficient for FANCJ and RAP80
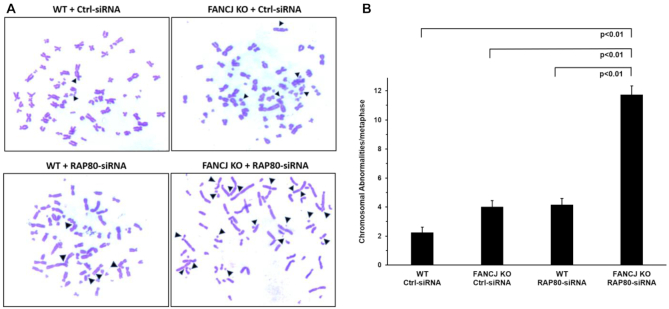
The increased level of DNA damage and defective checkpoint response in cells that were deficient in both FANCJ and RAP80 prompted us to assess if chromosomal instability would be elevated due to cells progressing into mitosis with DNA aberrations. Microscopic analysis of metaphase spreads demonstrated a 5.5-fold greater number of chromosomal abnormalities per metaphase in FANCJ KO cells depleted of RAP80 compared to WT cells transfected with control siRNA (Figure 6). FANCJ KO cells transfected with control siRNA or WT cells depleted of RAP80 by RNAi each displayed a 1.8-fold increase in chromosome aberrations per metaphase, indicating that the co-deficiency of both FANCJ and RAP80 has a significantly greater effect on chromosomal instability than the additive loss of either protein alone.
Figure 6.
Elevated spontaneous chromosomal instability in cells deficient of FANCJ and RAP80. WT or FANCJ KO cells were treated with 30 nM control or RAP80 siRNA#1. After 24 h cells were re-transfected with 30 nM control or RAP80 siRNA#1. Forty-eight hr after the first transfection, cells were treated with 200 ng/ml of Colcemid for 3 hr and swollen in 75 mM KCl for 25 min at 37°C. Cells were fixed on ice with a 3:1 methanol/acetic acid solution. Metaphases were dropped onto slides preheated at 42°C, allowed to dry, and stained with Giemsa. The numbers of chromosomal abnormalities per metaphase were counted. Panel (A), Representative images of metaphase spreads. Panel (B), Quantification of chromosome abnormalities are mean ± standard error of mean (s.e.m.) from three independent experiments, with P-values determined by Student's t-tests.
To determine whether structural chromosomal aberrations accumulate due to a combined deficiency of FANCJ and RAP80, metaphase spreads were analyzed cytogenetically for chromosomal alterations. Table 2 lists the types and number of chromosomal abnormalities from metaphase spreads. In addition to fragmentations, there were elevated breaks, dicentrics, and fusions in cells doubly deficient for FANCJ and RAP80, along with a 2.5-fold increase in the number of radial chromosomes compared to additive deficiency. Overall, these results demonstrate an increased number of spontaneous chromosomal aberrations in the FANCJ- and RAP80-depleted cells compared to their combined single deficiencies.
Table 2.
Lists the types and number of chromosomal abnormalities from metaphase spreads
| Instability (N = 50) | WT + Ctrl-siRNA | FANCJ KO + Ctrl-siRNA | WT + RAP80-siRNA | FANCJ KO + RAP80-siRNA |
|---|---|---|---|---|
| No. of fragmented chromosomes | 18 | 35 | 32 | 145 |
| No. of breaks | 20 | 42 | 35 | 101 |
| No. of dicentric | 18 | 32 | 31 | 68 |
| No. of fusions | 46 | 130 | 136 | 250 |
| No. of multiradial chromosomes | 0 | 8 | 1 | 22 |
| Total no. of anomalies per spread | 2 | 4.9 | 4.7 | 11.7 |
FANCJ promotes three-stranded branch-migration in the 5′ to 3′ direction
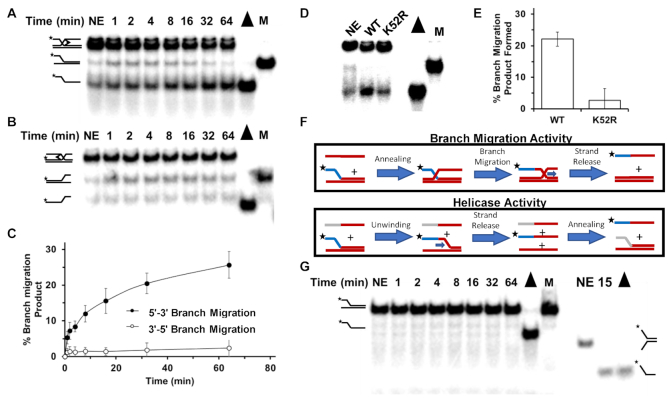
The results from our experiments with cells deficient in both FANCJ and RAP80 suggest that ICL-induced DNA damage is improperly processed, resulting in overly resected DNA ends that are not efficiently repaired by HR. This may reflect the accumulation of strand invasion intermediates that represent incompletely matured recombinant DNA molecules due to lack of FANCJ helicase, an enzyme that is implicated in HR repair but its role(s) is poorly understood. Previously, it was reported that FANCJ can unwind forked duplex and three-stranded non-mobile D-loop substrates (13) and displace RAD51 bound to single-stranded DNA (17) in an ATP-dependent manner; however, the ability of FANCJ to branch-migrate three-stranded DNA structures that represent early intermediates of HR repair had not been assessed. Using purified recombinant FANCJ protein and mobile three-way junction DNA substrates, we assayed FANCJ’s ability to branch-migrate the DNA structures in a 5′ to 3′ or 3′ to 5′ direction. These substrates were previously used to characterize the 3′ to 5′ branch-migration directionality by the human RECQ1 DNA helicase (64) and the 5′ to 3′ directionality of the human Twinkle mitochondrial DNA helicase (65). The results from these assays, shown in Figure 7A–C, demonstrate that FANCJ branch-migrates in the 5′ to 3′ direction with minimal detection of the expected product from 3′ to 5′ branch-migration.
Figure 7.
FANCJ promotes three-stranded DNA branch-migration in the 5′ to 3′ direction. DNA products of branch-migration reaction mixtures containing a radiolabeled three-stranded DNA substrate FANCJ, and ATP as indicated in Materials and Methods were incubated for the indicated times and resolved by electrophoresis on non-denaturing 8% polyacrylamide gels and visualized by autoradiography. DNA substrates were designed to detect 5′ to 3′ branch-migration activity (Panel A) or 3′ to 5′ branch-migration activity (Panel B). Representative results from at least three independent experiments are shown. Quantitation of DNA products for Panels (A) and (B) is shown in Panel (C). Standard deviations are indicated by error bars. Panel (D), FANCJ-WT or FANCJ-K52R (ATPase-deficient mutant) was incubated with 5′ to 3′ branch-migratable substrate in presence of ATP for 30 min and products resolved by gel electrophoresis. Panel (E), Quantitation of at least three experiments from Panel (D). Panel (F), Depiction of experiments to distinguish whether helicase or branch-migration is responsible for the product observed in Panel (A). The ssDNA added to the branch-migratable substrate used in Panel (A) anneals to the upstream template strand (both entirely red). However, for the helicase assay the 5′ sequence (grey) of the ssDNA oligonucleotide was changed such that it can only anneal after the 2-stranded DNA substrate was unwound first. Panel (G), FANCJ-WT was incubated with the helicase substrate in the presence of ATP and a third partially complementary ssDNA oligonucleotide as depicted in the helicase activity schematic shown in Panel (F). Note duplex length of substrate on left is 63 bp; as a control for FANCJ catalytic activity, a 19 bp forked duplex substrate was tested for unwinding by FANCJ on the right.
To determine if the observed branch-migration activity by FANCJ was dependent on its intrinsic catalytic functions, we compared the activity of FANCJ-WT with a site-directed FANCJ mutant protein, the engineered Walker A box FANCJ-K52R ATPase-dead variant (13). The FANCJ-K52R mutant protein was significantly compromised in its ability to perform branch-migration, suggesting that ATP hydrolysis and strand separation activity typically ascribed to ATP-dependent DNA unwinding are required for efficient branch-migration catalyzed by FANCJ (Figure 7D and E).
To address the possibility that the DNA product generated from reaction mixtures containing FANCJ and the mobile three-stranded DNA substrate shown in Figure 7A is attributed to FANCJ helicase activity and not its branch-migration activity, we performed an additional experiment. The pre-existing forked duplex component of the 3-stranded substrate was incubated with FANCJ, ATP and the single-stranded oligonucleotide complementary to the nucleotide sequence in the unlabeled strand annealed to the radiolabeled strand in the original forked duplex (Figure 7F). In this scenario, the unlabeled strand product of the forked duplex substrate unwound by FANCJ could potentially anneal to the single-stranded oligonucleotide originally in the reaction mixture, resulting in the release of the radiolabeled single-stranded oligonucleotide. As shown in Figure 7G, no such product was detected. In a control reaction mixture, the radiolabeled forked duplex DNA substrate was denatured by heat in the presence of the third oligonucleotide; in this case the radiolabeled single-strand was detected. The inability of FANCJ to unwind the 63 base pair duplex of the substrate is consistent with our previous observations that while FANCJ is capable of unwinding shorter duplex DNA substrates (e.g. 19-bp (Figure 7G, (13)) or 25-bp (66), the helicase is poorly processive and inefficient at unwinding DNA duplexes of 50-bp or more unless the single-stranded DNA binding protein RPA is present in the reaction mixture (67). Altogether, these results suggest that FANCJ branch-migration activity of the three-way junction was responsible for the single-stranded product detected in Figure 7A.
DISCUSSION
Understanding the comprehensive network of proteins and their partners responding to genomic DNA damage occurring endogenously or induced by genotoxic compounds, environmental pollutants, ionizing or ultraviolet light radiation or cancer chemotherapeutic drugs deserves attention as the consequences of inefficiently repaired DNA damage can be disastrous for human health in the case of unintended exposure to caustic agents or clinically beneficial in terms of potentially shrinking tumors and eliminating cancerous cells from the body. In this study, we have focused on the genetic interactions of the FANCJ helicase implicated in ICL repair with other DNA damage response/DNA repair factors that modulate the sensitivity of human cells to the ICL-inducing agent MMC. Given that ICLs are highly toxic and are used prolifically in the clinic to target rapidly dividing and metabolizing cancer cells, the results from such studies should provide insight from both basic science and clinical application perspectives.
Using an exquisitely low dose of MMC in which even FANCJ KO cells are hardly sensitive by proliferation or colony survival, we interrogated the library of DNA damage response/DNA repair genes for MMC sensitization. This led to identification of 10 gene targets broadly distributed into four functional categories: (i) DNA damage sensing, signaling and transducer factors (RAP80) (68), ATM (69), OBFC2A(SSB2) (70), RAD23A (71)); (ii) TLS (Polk, PolH, PCNA) (72); (iii) HR intermediate resolution (GEN1) (73); (iv) DNA repair (TDP1) (74) and reactive oxygen species damage regulation (SOD1) (75). The broad spectrum of pathway functions suggests the accumulated DNA damage requires attention by cellular nucleic acid processing machinery to avoid genotoxicity. Models for replication-coupled ICL repair depict the collaborative action of proteins involved in NER, TLS with the FA pathway (8). We suspect that RAP80 (which regulates DNA end-resection) helps FANCJ-deficient cells elicit alternative forms of damage repair and activate cell cycle checkpoints. Identification of RAP80 as a gene target was reinforced by our observation that depletion of BRCC3/BRCC36 (another member of the RAP80 complex (60)) in the MMC-treated FANCJ KO cells caused their reduced proliferation by 25% (Supplementary Figure S3).
With the connection of both FANCJ and RAP80 to the tumor suppressor BRCA1 and their dual involvement in the metabolism of DSBs, we focused the current work on characterizing how the co-deficiency of FANCJ and RAP80 leads to MMC hypersensitivity. As might be expected, FANCJ’s function was dependent on its catalytic ATPase/helicase function suggesting that its ability to unwind duplex DNA and/or displace the major strand exchange protein RAD51 in an ATPase-dependent manner are important. However, a key intermediate in HR repair (in which FANCJ is believed to operate) is the three-stranded displacement (D)-loop structure arising from single-strand invasion into recipient homologous duplex mediated by RAD51 in replicating (diploid) cells. A recent review from the Heyer lab nicely describes the critical importance of the strand invasion D-loop DNA intermediate as a decision mode for HR (76). Helicases and topoisomerases control the fate of such mobile three-stranded DNA structures in multiple ways including (i) removal of toxic joint molecules; (ii) suppression of cross-overs; (iii) prevention of multi-invasion induced rearrangements; (iv) assurance of homology search accuracy. This led us to perform biochemical assays to determine if FANCJ could catalyze branch-migration of mobile three-way junction substrates. FANCJ’s ability to perform branch-migration with a defined directionality (5′ to 3′) distinguishes its activity from that of the previously described RECQ1 DNA helicase which operates in a 3′ to 5′ manner (64). It is plausible that ATPase-dependent FANCJ branch-migration activity characterized in the present study may play a role in the repair of ICL-induced DNA damage or HR repair of a DSB; however, this hypothesis requires further investigation. FANCJ’s ability to catalyze branch-migration in a 5′ to 3′ direction is distinguished from that of the DNA translocase RAD54 (77) or the BLM helicase (78), which were shown to strongly branch-migrate with a 3′ to 5′ polarity on longer DNA substrates (79). Consistent with a unique role of FANCJ in HR, its deficiency resulted in reduced HR repair of an I-SceI-induced chromosomal DSB ((18); this study), a lesion that is predominantly repaired by a homology-directed repair pathway (80).
Purified recombinant RTEL1, related to FANCJ by its sequence homology in the helicase core domain and Fe-S cluster (81,82), was reported to disrupt a preformed plasmid-based D-loop DNA substrate (83), and shown to displace a third strand (characterized by a 3′ strand invasion) of a non-mobile D-loop substrate in an RPA-dependent manner (84); however its activity on branch-migrating three-stranded DNA substrates was not assessed. Moreover, recent work from the Sekelsky lab demonstrated that RTEL1 depletion or BLM depletion increased SDSA in human cells (85), suggesting that either RTEL1 or BLM act to suppress SDSA and that another DNA branch-migrating enzyme is responsible for promoting SDSA. HR via the SDSA pathway allows for non-crossover recombination, the most conservative outcome of HR that avoids structural alterations or extensive genetic changes in daughter cells (76). Consistent with a role of FANCJ to suppress cross-overs, we previously determined that FA-J fibroblasts displayed a significantly greater level of sister chromatid exchange (SCE) than wild-type cells (21).
Despite the experimental evidence that FANCJ facilitates DSB repair by facilitating HR, the mechanistic details are lacking. In this study, we present biochemical data showing FANCJ branch-migrates mobile three-stranded DNA structures (that represent a model strand invasion intermediate of HR) in an ATP-dependent manner in vitro. Given FANCJ’s importance in the ICL response, we postulate that FANCJ enables the timely maturation of the HR intermediate in a manner dependent on its catalytic function. In the future, separation-of-function mutants that specifically inactivate the branch-migration activity of FANCJ (but leave intact helicase activity) would be useful to explore the relevance of FANCJ’s specific catalytic activities in human cells. The relationship of BRCA1 to FANCJ or RAP80 in the ICL DNA damage response is complex, given that BRCA1 can independently form sub-complexes with either protein, as well as CtIP. Our findings demonstrate that ssDNA which accumulates in MMC-treated cells co-deficient in FANCJ and MRE11 is dependent on MRE11/CtIP nuclease processing of the DNA damage.
Our HR reporter results are consistent with a model in which an increase in HR upon RAP80 depletion is likely attributed to an increase in DNA end-resection at DSBs and creation of single-stranded DNA overhangs (40,41). Loss of both FANCJ and RAP80 resulted in a significant reduction in HR to nearly the level of FANCJ-depleted cells despite our observations that RPA and Rad51 foci in these cells were elevated. We interpret these findings to mean that RAP80 deficiency results in an increased resection and single-stranded DNA generation; however, the concomitant loss of FANCJ in these cells compromises repair of the HR intermediates.
RAP80 behaves like by 53BP1 by acting as a barrier to the end-resection process (86). Indeed, while RAP80 deficiency was reported to reduce IR-induced BRCA1 foci, it ironically increases long-strand resection (41,86). Similarly, loss of 53BP1 has been shown to promote DNA end-resection in the absence of BRCA1, resulting in Rad51 foci formation in cells exposed to IR or MMC (87–90). Our studies suggest that under conditions of a RAP80 deficiency, CtIP may act either in complex with residual BRCA1 or independently of BRCA1 at sites of ICL-induced DNA damage to aid in strand resection.
The HR phenotypes (scored by the restriction digest chromosomal recombination assay) due to either a FANCJ deficiency (resulting in decreased HR) or RAP80 deficiency (resulting in elevated HR) is consistent with what was reported by others. The combined FANCJ and RAP80 deficiency suppresses the elevated HR seen with RAP80 deficiency. However, it must be stressed that a restriction enzyme induced DSB is distinct from ICL-induced DNA damage, the latter not only leading to DSBs but also interfering with transcription and replication. Moreover, the processing steps involved in ICL repair are considerably more involved than repair of DSBs via either HR or NHEJ. Therefore, while findings at a frank DSB for FANCJ or RAP80 (as well as other DNA damage response proteins) are informative, at ICL-induced lesions the range of functions could be expanded or distinct. This would apply to the resection step and its relationship to the processing of specific forms of DNA damage, as well as more broadly the DNA damage response overall when forks are prevented from proceeding normally. The focus of the current study was the cellular response to ICL-induced DNA damage imposed by MMC, as defined by our original RNAi screen which led to the identification of RAP80 as a genetic interacting factor with FANCJ. Stalled and reversed forks, as well as processed forks and DSBs, are all DNA structures that arise in vivo due to ICL-induced damage. The elevated RPA and RAD51 foci in cells that are co-deficient in FANCJ and RAP80 after MMC exposure and recovery under the conditions of the RNAi screen are likely to reflect aberrant hyper-recombination and ineffective processing of HR intermediates in addition to improperly metabolized stalled or regressed replication forks. The new experimental data showing kinetics of RPA and RAD51 foci formation in response to MMC exposure support the findings that these DNA damage proteins are altered in their subcellular localization as a function of FANCJ/RAP80 status.
Previous studies demonstrated that BRCA1 forms separate and distinct complexes with RAP80, FANCJ and CtIP (91). Loss of the RAP80–BRCA1 interaction in RAP80-deficient cells was reported to increase the BRCA1-CtIP interaction which can enhance end-resection leading to elevated RAD51 and RPA foci in IR-treated cells (41). This is consistent with our observations that there is increased CtIP foci formation in MMC-treated RAP80-depleted cells (Supplementary Figure S11A), and that CtIP depletion resulted in a significant reduction of BrdU foci (Supplementary Figure S11B). Altogether, the results suggest that CtIP (and Mre11) is responsible for the end-resection in MMC-treated cells that are deficient in RAP80.
The roles of CtIP in the DNA damage response appear to be complex. CtIP deficiency negatively affects the response of DNA damage response proteins to laser induced ICLs (92). In contrast, the same study found that γH2AX and phosphorylated ATM recruitment to laser generated DSBs are unaffected by CtIP status. Recently, Nath et al. reported that a FANCJ deficiency caused a reduction in CtIP recruitment to AsiSI-induced DSBs (93). FANCJ S900 phosphorylation and K1249 acetylation were found to be important for interaction with CtIP and DSB end-resection at AsiSI-induced DSBs. In our work, we determined that CtIP foci formation in FANCJ-deficient cells after MMC exposure and recovery was not significantly different from WT cells. CtIP’s involvement in genomic stability appears to be multi-faceted, depending in part on the type of DNA damage. Experimental evidence suggests that monoubiquitinated FANCD2 aids CtIP in channeling DSBs arising from ICL processing into the HR pathway; conversely, CtIP suppresses illegitimate recombination in FA-deficient cells (94). Understanding FANCJ’s crosstalk with CtIP in the DNA damage response will require further study.
Cells co-deficient in FANCJ and RAP80 acutely exposed to MMC and allowed to recover displayed significantly elevated RPA and Rad51 foci compared to a single deficiency in either FANCJ or RAP80. The increased RPA and RAD51 staining suggest that FANCJ and RAP80 play at least partly independent functions in HR repair of MMC-induced DNA damage. Earlier it was reported that Fancj−/− mouse embryonic fibroblasts exposed to MMC displayed elevated RAD51 positive staining during the recovery period (95), consistent with the findings in this study. FANCJ may help to mature HR intermediates and suppress the persistence of RAD51 protein filaments via its dual roles in DNA branch-migration (this study) and ability to displace Rad51 bound to DNA and modulate Rad51 strand exchange (17). The accumulation of ssDNA tracts at laser-activated Pso-ICLs in cells co-deficient of FANCJ and RAP80 suggests a model in which ICL-induced DNA damage leads to DSBs which over time become overly resected and not matured by the HR pathway. A previous study demonstrated that depletion of the DNA2 helicase/nuclease suppressed cisplatin sensitivity of FANCD2-deficient cells, suggesting that DNA2-mediated over-resection is responsible for aberrant DNA repair (96). Our study implicates FANCJ as a key player to suppress ICL-induced DNA damage by helping to process single-stranded DNA via maturation of HR intermediates, thereby ensuring chromosomal stability. Interestingly, the BLM helicase (which interacts with FANCJ (21)) is proposed to suppress excessive HR by its recruitment to hydroxyurea-induced stalled replication forks; RAP80’s interaction with ubiquitylated BLM is required for its recruitment (97).
Experimental data from several labs demonstrated that RAP80 helps recruit BRCA1 to sites of DSBs where BRCA1 modulates cell cycle checkpoint and DNA damage response (37–39). Our results showing that deficiency in RAP80 (and FANCJ) resulted in reduced MMC-induced BRCA1 foci suggest that both FANCJ and RAP80, in their distinct complexes with BRCA1, aid in its recruitment to ICL DNA damage and DSBs. The dysregulation of BRCA1 localization to sites of DNA damage due to co-deficiency of FANCJ and RAP80 is likely to contribute to the aberrant cell cycle checkpoint function. This hypothesis led us to directly test if BRCA1’s recruitment to ICL-induced DNA damage is compromised by loss of RAP80 and/or FANCJ by examining BRCA1 nuclear localization to the site of a laser-induced Pso-ICL. The significant reduction in BRCA1 staining intensity at the site of irradiated damage confirmed that both FANCJ and RAP80 are important for BRCA1 recruitment to the ICL damage. Altogether, our results suggest that the defective localization of BRCA1 to sites of ICL damage contributes to the reduced DNA repair and chromosomal instability characteristic of RAP80- and FANCJ-deficiency. Consistent with a coordinate of BRCA1 and FANCJ to help cells deal with DNA damage, the complex formation of FANCJ with BRCA1 is required for HR repair (98) and repression of mutagenic end-joining of DSBs (47). It is likely that FANCJ-defective cells elicit the activity of other helicases to deal with either frank DSBs or those incurred by ICLs. Likewise, the apparently dual roles of FANCJ in DSB repair by HR and NHEJ suggest that compensatory pathways are evoked to help cells cope with endogenous or exogenously induced DSBs; however, this significant line of investigation is beyond the scope of the present study. FANCJ’ s newly discovered biochemical activity to branch-migrate mobile DNA junctions in an ATP-dependent manner is likely to be important for modulating HR repair of ICL-induced DNA damage, a pathway observed to be dependent on a threshold of FANCJ catalytic activity (36).
Beyond their direct involvement in DNA repair, both FANCJ and RAP80 play roles in checkpoint activation. However, it has not been determined if the two proteins act in the same pathway or not. Our results clearly demonstrate the DNA damage induced checkpoint elicited by MMC exposure is severely compromised in cells deficient in both FANCJ and RAP80 compared to the single loss of either. Thus, the co-deficiency of FANCJ and RAP80 represents a double-edged sword as both repair of ICL-induced DNA damage and the checkpoint response are crippled, allowing cells with toxic DNA damage to proceed into mitosis. The resulting chromosomal instability and mitotic catastrophe leading to cell loss suggests that both DNA repair modulation and checkpoint activation by FANCJ and RAP80 are crucial for survival when cells are challenged with even a relatively low level of a DNA cross-linking agent.
The synergism of RAP80 (and other DNA damage response factors (e.g. Merit40)) with FANCJ raises the possibility their genetic interaction may be informative from a clinical perspective. Yan et al. found that RAP80 depletion by RNAi increased radio-sensitivity of the breast cancer cell line MCF-7 (99). Screening of BRCA1/BRCA2 mutation negative breast cancer cases revealed numerous genetic alterations in RAP80, including a single amino acid deletion mutation in a ubiquitin interaction motif that impaired DSB localization and recruitment of other DNA repair proteins, and was associated with chromosomal instability (100). Similarly, a screen of FANCJ mutation-associated cancers may be informative to assess RAP80 mutational status, given our observations that chromosomal instability is significantly elevated in cells deficient in both FANCJ and RAP80.
A recent study reported that RAP80 expression in invasive ductal breast cancer tissues is significantly lower than in paired normal breast tissues (101). It is plausible that the efficacy of DNA damaging chemotherapy drugs or radiation and the outcome of breast or ovarian cancer patients with deficiency in FANCJ or related tumor suppressors is dependent on the expression level of RAP80 and other factors that modulate DNA repair or the DNA damage response. Supporting this idea, depletion of RAP80 by RNA interference induced apoptosis, down-regulated invasiveness/migration, and enhanced cisplatin sensitivity of breast cancer cells (101).
Devising improved chemotherapeutic drug or radiation strategies will require a better understanding of the overlapping versus unique pathways that tumor suppressor proteins like FANCJ and RAP80 operate with other factors to stabilize the genome. Achieving a therapeutic threshold may be enhanced by a comprehensive assessment of gene interactions relevant to the DNA damage response.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Jeremy Stark for DR-GFP U2OS cells and Dr Xiaofan Wang for EJ5 U2OS cells. We thank Drs Roger Greenberg and Qinqin Jiang for the U2OS RAP80 KO cells and MERIT40 KO cells. We also thank Dr Bin Wang for the Abraxas KO HEK293 cell.
Contributor Information
Sanket Awate, Laboratory of Molecular Gerontology, National Institute on Aging, NIH, Baltimore, MD, USA.
Joshua A Sommers, Laboratory of Molecular Gerontology, National Institute on Aging, NIH, Baltimore, MD, USA.
Arindam Datta, Laboratory of Molecular Gerontology, National Institute on Aging, NIH, Baltimore, MD, USA.
Sumeet Nayak, Department of Cancer Biology, University of Massachusetts Medical School – UMASS Memorial Cancer Center, Worcester, MA, USA.
Marina A Bellani, Laboratory of Molecular Gerontology, National Institute on Aging, NIH, Baltimore, MD, USA.
Olivia Yang, Department of Biophysics and Biophysical Chemistry, Johns Hopkins University, Baltimore, MD, USA.
Christopher A Dunn, Flow Cytometry Unit, National Institute on Aging, NIH, Baltimore, MD, USA.
Claudia M Nicolae, Department of Biochemistry and Molecular Biology, Penn State College of Medicine, Hershey, PA, USA.
George-Lucian Moldovan, Department of Biochemistry and Molecular Biology, Penn State College of Medicine, Hershey, PA, USA.
Michael M Seidman, Laboratory of Molecular Gerontology, National Institute on Aging, NIH, Baltimore, MD, USA.
Sharon B Cantor, Department of Cancer Biology, University of Massachusetts Medical School – UMASS Memorial Cancer Center, Worcester, MA, USA.
Robert M Brosh, Jr., Laboratory of Molecular Gerontology, National Institute on Aging, NIH, Baltimore, MD, USA
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program, National Institute on Aging, National Institutes of Health to M.S., R.B., R01 ES026184 to G.-L.M., R01 CA176166-01A1 to S.C., R01 CA225018-01A1 to S.C., and Basser Center for BRCA Research to S.C.. Funding for open access charge: Intramural Research Program, National Institute on Aging, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hashimoto S., Anai H., Hanada K.. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes Environ. 2016; 38:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kohn K.W. Beyond DNA cross-linking: history and prospects of DNA-targeted cancer treatment–fifteenth Bruce F. Cain Memorial Award Lecture. Cancer Res. 1996; 56:5533–5546. [PubMed] [Google Scholar]
- 3. Deans A.J., West S.C.. DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer. 2011; 11:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pontel L.B., Rosado I.V., Burgos-Barragan G., Garaycoechea J.I., Yu R., Arends M.J., Chandrasekaran G., Broecker V., Wei W., Liu L. et al.. Endogenous formaldehyde is a hematopoietic stem cell genotoxin and metabolic carcinogen. Mol. Cell. 2015; 60:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voulgaridou G.P., Anestopoulos I., Franco R., Panayiotidis M.I., Pappa A.. DNA damage induced by endogenous aldehydes: current state of knowledge. Mutat. Res. 2011; 711:13–27. [DOI] [PubMed] [Google Scholar]
- 6. Garaycoechea J.I., Crossan G.P., Langevin F., Mulderrig L., Louzada S., Yang F., Guilbaud G., Park N., Roerink S., Nik-Zainal S. et al.. Alcohol and endogenous aldehydes damage chromosomes and mutate stem cells. Nature. 2018; 553:171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheung R.S., Taniguchi T.. Recent insights into the molecular basis of Fanconi anemia: genes, modifiers, and drivers. Int. J. Hematol. 2017; 106:335–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datta A., Brosh R.M. Jr.. Holding all the cards-How Fanconi Anemia proteins deal with replication stress and preserve genomic stability. Genes. 2019; 10:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kottemann M.C., Smogorzewska A.. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013; 493:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cantor S.B., Guillemette S.. Hereditary breast cancer and the BRCA1-associated FANCJ/BACH1/BRIP1. Future Oncol. 2011; 7:253–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rafnar T., Gudbjartsson D.F., Sulem P., Jonasdottir A., Sigurdsson A., Jonasdottir A., Besenbacher S., Lundin P., Stacey S.N., Gudmundsson J. et al.. Mutations in BRIP1 confer high risk of ovarian cancer. Nat. Genet. 2011; 43:1104–1107. [DOI] [PubMed] [Google Scholar]
- 12. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D.M.. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. PNAS. 2004; 101:2357–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gupta R., Sharma S., Sommers J.A., Jin Z., Cantor S.B., Brosh R.M. Jr.. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 2005; 280:25450–25460. [DOI] [PubMed] [Google Scholar]
- 14. Bharti S.K., Sommers J.A., George F., Kuper J., Hamon F., Shin-ya K., Teulade-Fichou M.P., Kisker C., Brosh R.M. Jr.. Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 2013; 288:28217–28229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. London T.B., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., Hiom K.. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008; 283:36132–36139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu Y., Shin-ya K., Brosh R.M. Jr.. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008; 28:4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sommers J.A., Rawtani N., Gupta R., Bugreev D.V., Mazin A.V., Cantor S.B., Brosh R.M. Jr.. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J. Biol. Chem. 2009; 284:7505–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P.R., Cantor S.B.. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005; 8:255–265. [DOI] [PubMed] [Google Scholar]
- 19. Brosh R.M. Jr., Cantor S.B.. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet. 2014; 5:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peng M., Cong K., Panzarino N.J., Nayak S., Calvo J., Deng B., Zhu L.J., Morocz M., Hegedus L., Haracska L. et al.. Opposing roles of FANCJ and HLTF protect forks and restrain replication during stress. Cell Rep. 2018; 24:3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Suhasini A.N., Rawtani N.A., Wu Y., Sommers J.A., Sharma S., Mosedale G., North P.S., Cantor S.B., Hickson I.D., Brosh R.M. Jr.. Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 2011; 30:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Harding S.M., Greenberg R.A.. Choreographing the double strand break response: ubiquitin and SUMO control of nuclear architecture. Front. Genet. 2016; 7:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jiang Q., Paramasivam M., Aressy B., Wu J., Bellani M., Tong W., Seidman M.M., Greenberg R.A.. MERIT40 cooperates with BRCA2 to resolve DNA interstrand cross-links. Genes Dev. 2015; 29:1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu Q., Paul A., Su D., Mehmood S., Foo T.K., Ochi T., Bunting E.L., Xia B., Robinson C.V., Wang B. et al.. Structure of BRCA1-BRCT/Abraxas complex reveals phosphorylation-dependent BRCT dimerization at DNA damage sites. Mol. Cell. 2016; 61:434–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang Q., Goldstein M., Alexander P., Wakeman T.P., Sun T., Feng J., Lou Z., Kastan M.B., Wang X.F.. Rad17 recruits the MRE11-RAD50-NBS1 complex to regulate the cellular response to DNA double-strand breaks. EMBO J. 2014; 33:862–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bennardo N., Cheng A., Huang N., Stark J.M.. Alternative-NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet. 2008; 4:e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tyner J., Druker B.J.. RNAi screen for therapeutic target in leukemia. Cell Cycle. 2009; 8:2144. [PubMed] [Google Scholar]
- 28. Tyner J.W., Walters D.K., Willis S.G., Luttropp M., Oost J., Loriaux M., Erickson H., Corbin A.S., O’Hare T., Heinrich M.C. et al.. RNAi screening of the tyrosine kinome identifies therapeutic targets in acute myeloid leukemia. Blood. 2008; 111:2238–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mukherjee B., Tomimatsu N., Burma S.. Immunofluorescence-based methods to monitor DNA end resection. Methods Mol. Biol. 2015; 1292:67–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Awate S., Dhar S., Sommers J.A., Brosh R.M. Jr.. Cellular assays to study the functional importance of human DNA repair helicases. Methods Mol. Biol. 2019; 1999:185–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Franken N.A., Rodermond H.M., Stap J., Haveman J., van Bree C.. Clonogenic assay of cells in vitro. Nat. Protoc. 2006; 1:2315–2319. [DOI] [PubMed] [Google Scholar]
- 32. Juhász S., Elbakry A., Mathes A., Löbrich M.. ATRX promotes DNA repair synthesis and sister chromatid exchange during homologous recombination. Mol. Cell. 2018; 71:11–24. [DOI] [PubMed] [Google Scholar]
- 33. Feng L., Huang J., Chen J.. MERIT40 facilitates BRCA1 localization and DNA damage repair. Genes Dev. 2009; 23:719–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao G., Patterson-Fortin J., Messick T.E., Feng D., Shanbhag N., Wang Y., Greenberg R.A.. MERIT40 controls BRCA1-Rap80 complex integrity and recruitment to DNA double-strand breaks. Genes Dev. 2009; 23:740–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang B., Hurov K., Hofmann K., Elledge S.J.. NBA1, a new player in the Brca1 A complex, is required for DNA damage resistance and checkpoint control. Genes Dev. 2009; 23:729–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bharti S.K., Sommers J.A., Awate S., Bellani M.A., Khan I., Bradley L., King G.A., Seol Y., Vidhyasagar V., Wu Y. et al.. A minimal threshold of FANCJ helicase activity is required for its response to replication stress or double-strand break repair. Nucleic Acids Res. 2018; 46:6238–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim H., Chen J., Yu X.. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007; 316:1202–1205. [DOI] [PubMed] [Google Scholar]
- 38. Sobhian B., Shao G., Lilli D.R., Culhane A.C., Moreau L.A., Xia B., Livingston D.M., Greenberg R.A.. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science (New York, N.Y.). 2007; 316:1198–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P., Elledge S.J.. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science (New York, N.Y.). 2007; 316:1194–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coleman K.A., Greenberg R.A.. The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J. Biol. Chem. 2011; 286:13669–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu Y., Scully R., Sobhian B., Xie A., Shestakova E., Livingston D.M.. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 2011; 25:685–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010; 17:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yun M.H., Hiom K.. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009; 459:460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Marini F., Rawal C.C., Liberi G., Pellicioli A.. Regulation of DNA double strand breaks processing: focus on barriers. Front. Mol. Biosci. 2019; 6:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dupré A., Boyer-Chatenet L., Sattler R.M., Modi A.P., Lee J.H., Nicolette M.L., Kopelovich L., Jasin M., Baer R., Paull T.T. et al.. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat. Chem. Biol. 2008; 4:119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weinstock D.M., Nakanishi K., Helgadottir H.R., Jasin M.. Assaying double-strand break repair pathway choice in mammalian cells using a targeted endonuclease or the RAG recombinase. Methods Enzymol. 2006; 409:524–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dohrn L., Salles D., Siehler S.Y., Kaufmann J., Wiesmuller L.. BRCA1-mediated repression of mutagenic end-joining of DNA double-strand breaks requires complex formation with BACH1. Biochem. J. 2012; 441:919–926. [DOI] [PubMed] [Google Scholar]
- 48. Suhasini A.N., Sommers J.A., Muniandy P.A., Coulombe Y., Cantor S.B., Masson J.Y., Seidman M.M., Brosh R.M. Jr.. Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol. Cell. Biol. 2013; 33:2212–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang B. BRCA1 tumor suppressor network: focusing on its tail. Cell Biosci. 2012; 2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Langerak P., Russell P.. Regulatory networks integrating cell cycle control with DNA damage checkpoints and double-strand break repair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011; 366:3562–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Xu B., Kim S., Kastan M.B.. Involvement of Brca1 in S-phase and G(2)-phase checkpoints after ionizing irradiation. Mol. Cell. Biol. 2001; 21:3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yarden R.I., Pardo-Reoyo S., Sgagias M., Cowan K.H., Brody L.C.. BRCA1 regulates the G2/M checkpoint by activating Chk1 kinase upon DNA damage. Nat. Genet. 2002; 30:285–289. [DOI] [PubMed] [Google Scholar]
- 53. Smits V.A., Gillespie D.A.. DNA damage control: regulation and functions of checkpoint kinase 1. FEBS J. 2015; 282:3681–3692. [DOI] [PubMed] [Google Scholar]
- 54. Tapia-Alveal C., O’Connell M.J.. Methods for studying checkpoint kinases - Chk1. Methods Mol. Biol. 2011; 782:171–179. [DOI] [PubMed] [Google Scholar]
- 55. Xie J., Peng M., Guillemette S., Quan S., Maniatis S., Wu Y., Venkatesh A., Shaffer S.A., Brosh R.M. Jr., Cantor S.B.. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet. 2012; 8:e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Matsuoka S., Rotman G., Ogawa A., Shiloh Y., Tamai K., Elledge S.J.. Ataxia telangiectasia-mutated phosphorylates Chk2 in vivo and in vitro. PNAS. 2000; 97:10389–10394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yu X., Chini C.C., He M., Mer G., Chen J.. The BRCT domain is a phospho-protein binding domain. Science (New York, N.Y.). 2003; 302:639–642. [DOI] [PubMed] [Google Scholar]
- 58. Wei Y., Mizzen C.A., Cook R.G., Gorovsky M.A., Allis C.D.. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. PNAS. 1998; 95:7480–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kim H., Huang J., Chen J.. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat. Struct. Mol. Biol. 2007; 14:710–715. [DOI] [PubMed] [Google Scholar]
- 60. Wang B., Elledge S.J.. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. PNAS. 2007; 104:20759–20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hu X., Kim J.A., Castillo A., Huang M., Liu J., Wang B.. NBA1/MERIT40 and BRE interaction is required for the integrity of two distinct deubiquitinating enzyme BRCC36-containing complexes. J. Biol. Chem. 2011; 286:11734–11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Xie J., Litman R., Wang S., Peng M., Guillemette S., Rooney T., Cantor S.B.. Targeting the FANCJ-BRCA1 interaction promotes a switch from recombination to poleta-dependent bypass. Oncogene. 2010; 29:2499–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cho H.J., Oh Y.J., Han S.H., Chung H.J., Kim C.H., Lee N.S., Kim W.J., Choi J.M., Kim H.. Cdk1 protein-mediated phosphorylation of receptor-associated protein 80 (RAP80) serine 677 modulates DNA damage-induced G2/M checkpoint and cell survival. J. Biol. Chem. 2013; 288:3768–3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Bugreev D.V., Brosh R.M. Jr., Mazin A.V.. RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 2008; 283:20231–20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Khan I., Crouch J.D., Bharti S.K., Sommers J.A., Carney S.M., Yakubovskaya E., Garcia-Diaz M., Trakselis M.A., Brosh R.M. Jr.. Biochemical characterization of the human mitochondrial replicative Twinkle helicase: substrate specificity, DNA branch migration, and ability to overcome blockades to DNA unwinding. J. Biol. Chem. 2016; 291:14324–14339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Suhasini A.N., Sommers J.A., Mason A.C., Voloshin O.N., Camerini-Otero R.D., Wold M.S., Brosh R.M. Jr.. FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J. Biol. Chem. 2009; 284:18458–18470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gupta R., Sharma S., Sommers J.A., Kenny M.K., Cantor S.B., Brosh R.M. Jr.. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007; 110:2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lombardi P.M., Matunis M.J., Wolberger C.. RAP80, ubiquitin and SUMO in the DNA damage response. J. Mol. Med. 2017; 95:799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Clouaire T., Marnef A., Legube G.. Taming tricky DSBs: ATM on duty. DNA Repair (Amst.). 2017; 56:84–91. [DOI] [PubMed] [Google Scholar]
- 70. Shi W., Vu T., Boucher D., Biernacka A., Nde J., Pandita R.K., Straube J., Boyle G.M., Al-Ejeh F., Nag P. et al.. Ssb1 and Ssb2 cooperate to regulate mouse hematopoietic stem and progenitor cells by resolving replicative stress. Blood. 2017; 129:2479–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tan X., Liang R.Y., Chuang S.M.. hHR23A is required to control the basal turnover of Chk1. Cell. Signal. 2015; 27:2304–2313. [DOI] [PubMed] [Google Scholar]
- 72. Vaisman A., Woodgate R.. Translesion DNA polymerases in eukaryotes: what makes them tick. Crit. Rev. Biochem. Mol. Biol. 2017; 52:274–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. West S.C., Blanco M.G., Chan Y.W., Matos J., Sarbajna S., Wyatt H.D.. Resolution of recombination intermediates: mechanisms and regulation. Cold Spring Harb. Symp. Quant. Biol. 2015; 80:103–109. [DOI] [PubMed] [Google Scholar]
- 74. Pommier Y., Huang S.Y., Gao R., Das B.B., Murai J., Marchand C.. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2). DNA Repair (Amst.). 2014; 19:114–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang Y., Branicky R., Noe A., Hekimi S.. Superoxide dismutases: dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018; 217:1915–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Piazza A., Heyer W.D.. Moving forward one step back at a time: reversibility during homologous recombination. Curr. Genet. 2019; 65:1333–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bugreev D.V., Hanaoka F., Mazin A.V.. Rad54 dissociates homologous recombination intermediates by branch migration. Nat. Struct. Mol. Biol. 2007; 14:746–753. [DOI] [PubMed] [Google Scholar]
- 78. Bachrati C.Z., Borts R.H., Hickson I.D.. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006; 34:2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Mazina O.M., Rossi M.J., Deakyne J.S., Huang F., Mazin A.V.. Polarity and bypass of DNA heterology during branch migration of Holliday junctions by human RAD54, BLM, and RECQ1 proteins. J. Biol. Chem. 2012; 287:11820–11832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Moynahan M.E., Pierce A.J., Jasin M.. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol. Cell. 2001; 7:263–272. [DOI] [PubMed] [Google Scholar]
- 81. Estep K.N., Brosh R.M. Jr.. RecQ and Fe-S helicases have unique roles in DNA metabolism dictated by their unwinding directionality, substrate specificity, and protein interactions. Biochem. Soc. Trans. 2018; 46:77–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rudolf J., Makrantoni V., Ingledew W.J., Stark M.J., White M.F.. The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell. 2006; 23:801–808. [DOI] [PubMed] [Google Scholar]
- 83. Barber L.J., Youds J.L., Ward J.D., McIlwraith M.J., O’Neil N.J., Petalcorin M.I., Martin J.S., Collis S.J., Cantor S.B., Auclair M. et al.. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell. 2008; 135:261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Youds J.L., Mets D.G., McIlwraith M.J., Martin J.S., Ward J.D., NJ O.N., Rose A.M., West S.C., Meyer B.J., Boulton S.J.. RTEL-1 enforces meiotic crossover interference and homeostasis. Science (New York, N.Y.). 2010; 327:1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Zapotoczny G., Sekelsky J.. Human cell assays for synthesis-dependent strand annealing and crossing over during double-strand break repair. G3 (Bethesda, Md.). 2017; 7:1191–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Daley J.M., Sung P.. 53BP1, BRCA1, and the choice between recombination and end joining at DNA double-strand breaks. Mol. Cell. Biol. 2014; 34:1380–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bouwman P., Aly A., Escandell J.M., Pieterse M., Bartkova J., van der Gulden H., Hiddingh S., Thanasoula M., Kulkarni A., Yang Q. et al.. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat. Struct. Mol. Biol. 2010; 17:688–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Bunting S.F., Callen E., Kozak M.L., Kim J.M., Wong N., Lopez-Contreras A.J., Ludwig T., Baer R., Faryabi R.B., Malhowski A. et al.. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Mol. Cell. 2012; 46:125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lord C.J., Ashworth A.. Mechanisms of resistance to therapies targeting BRCA-mutant cancers. Nat. Med. 2013; 19:1381–1388. [DOI] [PubMed] [Google Scholar]
- 90. Nakada S., Yonamine R.M., Matsuo K.. RNF8 regulates assembly of RAD51 at DNA double-strand breaks in the absence of BRCA1 and 53BP1. Cancer Res. 2012; 72:4974–4983. [DOI] [PubMed] [Google Scholar]
- 91. Huen M.S., Sy S.M., Chen J.. BRCA1 and its toolbox for the maintenance of genome integrity. Nat. Rev. Mol. Cell Biol. 2010; 11:138–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Duquette M.L., Zhu Q., Taylor E.R., Tsay A.J., Shi L.Z., Berns M.W., McGowan C.H.. CtIP is required to initiate replication-dependent interstrand crosslink repair. PLos Genet. 2012; 8:e1003050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nath S., Nagaraju G.. FANCJ helicase promotes DNA end resection by facilitating CtIP recruitment to DNA double-strand breaks. PLoS Genet. 2020; 16:e1008701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Murina O., von Aesch C., Karakus U., Ferretti L.P., Bolck H.A., Hänggi K., Sartori A.A.. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Rep. 2014; 7:1030–1038. [DOI] [PubMed] [Google Scholar]
- 95. Matsuzaki K., Borel V., Adelman C.A., Schindler D., Boulton S.J.. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes Dev. 2015; 29:2532–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Karanja K.K., Lee E.H., Hendrickson E.A., Campbell J.L.. Preventing over-resection by DNA2 helicase/nuclease suppresses repair defects in Fanconi anemia cells. Cell Cycle. 2014; 13:1540–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Tikoo S., Madhavan V., Hussain M., Miller E.S., Arora P., Zlatanou A., Modi P., Townsend K., Stewart G.S., Sengupta S.. Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination. EMBO J. 2013; 32:1778–1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Cantor S.B., Bell D.W., Ganesan S., Kass E.M., Drapkin R., Grossman S., Wahrer D.C., Sgroi D.C., Lane W.S., Haber D.A. et al.. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001; 105:149–160. [DOI] [PubMed] [Google Scholar]
- 99. Yan J., Kim Y.S., Yang X.P., Li L.P., Liao G., Xia F., Jetten A.M.. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007; 67:6647–6656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nikkila J., Coleman K.A., Morrissey D., Pylkas K., Erkko H., Messick T.E., Karppinen S.M., Amelina A., Winqvist R., Greenberg R.A.. Familial breast cancer screening reveals an alteration in the RAP80 UIM domain that impairs DNA damage response function. Oncogene. 2009; 28:1843–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jin G., Mao X., Qiao Z., Chen B., Jin F.. RAP80 expression in breast cancer and its relationship with apoptosis in breast cancer cells. OncoTargets Ther. 2019; 12:625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.