Abstract
The genomes of RNA viruses contain regulatory elements of varying complexity. Many plus-strand RNA viruses employ largescale intra-genomic RNA-RNA interactions as a means to control viral processes. Here, we describe an elaborate RNA structure formed by multiple distant regions in a tombusvirus genome that activates transcription of a viral subgenomic mRNA. The initial step in assembly of this intramolecular RNA complex involves the folding of a large viral RNA domain, which generates a discontinuous binding pocket. Next, a distally-located protracted stem-loop RNA structure docks, via base-pairing, into the binding site and acts as a linchpin that stabilizes the RNA complex and activates transcription. A multi-step RNA folding pathway is proposed in which rate-limiting steps contribute to a delay in transcription of the capsid protein-encoding viral subgenomic mRNA. This study provides an exceptional example of the complexity of genome-scale viral regulation and offers new insights into the assembly schemes utilized by large intra-genomic RNA structures.
INTRODUCTION
Positive-strand RNA viruses comprise a large group of agriculturally and medically important pathogens that infect a wide range of hosts. The successful takeover of their hosts requires multiple steps that involves precise regulation and careful coordination. A critical component of this control is the modulation of different viral processes by RNA sequences and structures located within viral genomes (1–4). In some cases, the RNA-based regulation is mediated by large functional RNA folds, some of which span the entire length of a viral genome (5). Accordingly, overall RNA genome architecture and dynamics can contribute significantly to the orchestration of different phases that occur during viral infections (6,7). Notably, this large-scale form of riboregulation is employed by many significant plant and animal messenger-sensed RNA viruses, including luteoviruses (8,9), carmoviruses (10,11), umbraviruses (12,13), flaviviruses (14–17), hepacivirus (18–24) and coronaviruses (25–27).
Tombusviruses (family Tombusviridae) are important model plus-strand RNA viruses (28). Studies performed on members of this genus have resulted in pioneering discoveries (29–31) and led to significant progress in the identification of pro- and antiviral host factors (32–34). Tombusviruses have also been invaluable for investigating how global viral RNA genome structure actively controls essential viral processes (5,6). Their 4.8 kb-long coding-sensed ssRNA genomes contain a vast network of intra-genomic, base pair-mediated, long-distance RNA–RNA interactions (LDRIs) that play different critical roles during the viral reproductive cycle (7). In particular, two tombusviruses, the prototype of the genus, Tomato bushy stunt virus (TBSV), and the closely-related Carnation Italian ringspot virus (CIRV) (Figure 1A), have been instrumental in deducing the structure and function of this complex LDRI network (35).
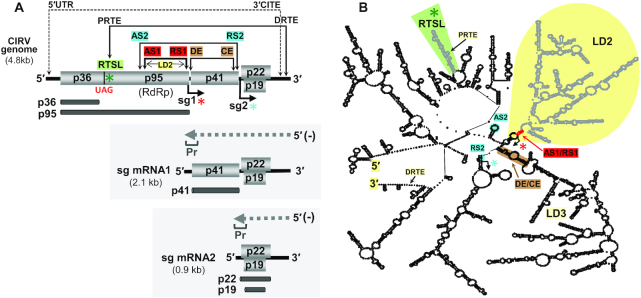
Figure 1.
Carnation Italian ringspot virus (CIRV) genome and subgenomic mRNAs. (A) Schematic representation of the CIRV genome (top) and sg mRNAs (below in shaded boxes) with encoded proteins indicated. The small black arrows below the genome indicate the initiation sites for sg mRNA1 (sg1, red asterisk) and 2 (sg2, turquoise asterisks) in the genome. Proteins translated from the genome and sg mRNAs are depicted as dark gray bars immediately below encoded proteins. The positions of the sequences that form long-distance RNA-RNA interactions (LDRIs) involved in translation (5′UTR/3′CITE), readthrough (PRTE/DRTE), sg mRNA2 transcription (DE/CE, brown and AS2/RS2, turquoise) and sg mRNA1 transcription (AS1/RS1, red) are indicated by double-headed arrows. The minus-strand intermediates in transcription are shown above each sg mRNA as dashed grey arrows, with promoter sequences (Pr) indicated at their 3′-ends. (B) Simplified model of the RNA secondary structure for the TBSV genome (48) with relevant large RNA domains (LD2 and LD3), RNA elements (RTSL, green shading, PRTE and DRTE), and LDRIs indicated. The LDRI color-coding scheme corresponds to that depicted in panel (A).
Tombusvirus RNA genomes are not 5′-capped or 3′-polyadenylated, thus they rely on an unconventional mode of translation, which has been studied extensively in CIRV (36–39). An RNA structure in CIRV′s 3′-untranslated region (3′UTR), termed the 3′-cap independent translation enhancer (3′CITE), binds to eukaryotic translation initiation factor 4F (eIF4F). The eIF4F-bound 3′CITE then simultaneously base-pairs with the 5′UTR via an LDRI, which positions eIF4F near the 5′-end of the genome, where it mediates ribosome recruitment (38) (Figure 1A). This results in translation of the auxiliary RNA replication protein, p36. Production of the p95 RNA-dependent RNA polymerase (RdRp) requires translational readthrough of the p36 stop codon. This recoding event involves an extended RNA stem-loop (SL) structure, termed the readthrough SL (RTSL), located immediately 3′ to the p36 termination codon, UAG (Figure 1A, green asterisk). The RTSL is not able to direct readthrough on its own and, to function, requires the formation of an LDRI between a bulged sequence in RTSL (the proximal readthrough element, PRTE) and a complementary sequence (the distal readthrough element, DRTE) in the 3′UTR of the genome (39) (Figure 1A). This LDRI not only promotes readthrough, it also concomitantly inhibits genomic minus-strand RNA synthesis, which would interfere with translation. Thus, the RTSL, via an LDRI, functions as a dual regulator that coordinates translational recoding and genome replication.
LDRIs are also involved in controlling the production of tombusvirus subgenomic (sg) mRNAs, which are small virus genome-derived mRNAs that are transcribed by the viral RdRp during infections (40,41). Structurally, sg mRNAs are 3′-coterminal with the viral genome, while their 5′-ends map to internal regions. Consequently, they encode 3′-proximal ORFs that are translationally silent within the context of the full-length genome. By modulating sg mRNA transcription, the virus is able to control the amount and timing of viral protein production during infections. Tombusviruses transcribe two sg mRNAs (Figure 1A). The smaller sg mRNA2 is transcribed earlier during infections, and mediates translation of both the p19 suppressor of gene silencing and the p22 cell-to-cell movement protein. The larger sg mRNA1 is transcribed later in infections, and encodes the capsid protein (CP) (42,43).
Tombusviruses (40), nodaviruses (family Nodaviridae) (44) and toroviruses (family Tobaniviridae) (45) transcribe their sg mRNAs using a premature termination mechanism (46). In this process, the viral RdRp terminates transcription prematurely while synthesizing a minus-strand from a full-length plus-strand viral RNA genome. The stalling of the RdRp occurs when it encounters an RNA element within the genome called an attenuation structure. This termination event leads to the production of a 3′-truncated minus-sense RNA species that possess a promoter sequence at its 3′-end (Figure 1A, Pr). The promoter is then recognized by the viral RdRp, which transcribes the coding-sense sg mRNAs from the truncated intermediate.
The attenuation structures that block the progression of RdRps are helical RNA structures that are located ∼2–5 nt upstream from where the copying RdRp stalls. In some viruses, the inhibitory stem is formed by LDRIs (40,44). Production of tombusvirus sg mRNA2 involves two sets of LDRIs. One occurs between activator sequence 2 (AS2) and receptor sequence 2 (RS2), spanning ∼2100 nucleotides (47), and the other involves distal element (DE) and core element (CE), traversing ∼1100 nucleotides (42) (Figure 1A, turquoise and brown). When viewed in the context of the RNA secondary structure model for the TBSV genome (48), the DE/CE interaction corresponds to the closing stem of a sizable RNA domain, termed large domain 3 (LD3), which, along with formation of the adjacent LD2, acts to unite the AS2 and RS2 sequences (Figure 1B).
Efficient sg mRNA1 transcription requires an LDRI between AS1 and RS1, which spans ∼1000 nucleotides (Figure 1A, red) and forms a helix just three nucleotides upstream of the sg mRNA1 initiation site (Supplementary Figure S1) (49). The 7 nt long AS1 sequence is the terminal loop of an RNA hairpin, designated AS1-SL, that facilitates its accessibility (Supplementary Figure S1) (49). The AS1/RS1 interaction has been verified experimentally to (i) pair and operate in the plus-strand of the genome, (ii) occur in cis and (iii) promote production of sg mRNA1 minus-strand intermediates (49). The AS1/RS1 interaction is also predicted to form the closing helix of LD2 (Figure 1B), thus accurate folding of LD2 is proposed to be important for formation of the AS1/RS1 LDRI (48,49).
In this study, we show that the attenuation structure for sg mRNA1 is far more complex than previously appreciated, with the AS1/RS1 interaction being a component of a group of critical LDRIs. Unexpectedly, the active RNA structure includes the recoding RNA element, RTSL, as well as specific subsections of LD2. Formation of a functional attenuation structure requires multiple LDRIs within LD2 that generate a discontinuous binding site for RTSL. The docking of RTSL into this binding pocket acts as a linchpin that stabilizes an active conformation of the RNA complex. Functional and structural aspects of these LDRIs are discussed and a likely path for assembly of this intra-genomic RNA attenuation structure is presented.
MATERIALS AND METHODS
Plasmid DNA construction
Nucleotide substitutions were introduced into a cloned cDNA copy of the full-length wt CIRV genome (50) through standard PCR-based site-directed mutagenesis. Each of the mutated CIRV clones was sequenced over the entire inserted PCR fragment containing the modification to confirm that only the desired change was present.
Preparation of infectious CIRV genomic RNA
SmaI-linearized wild type (wt) and mutant full-length CIRV genome cDNAs were used as templates for in vitro transcription reactions using a T7 Flashscribe transcription kit (CellScript) to synthesize uncapped genomic CIRV RNAs, as described previously (51).
In vitro translation assay
In vitro-generated viral genomic RNAs (0.5 pmol) were assessed for translation and readthrough using a wheat germ extract (wge) in vitro translation system (Promega) and proteins were monitored by incorporation of [35S]-methionine, as described previously (36,39). Translation products were separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis, detected using a Typhoon FLA 9500 variable mode imager (GE Healthcare), and quantified using QuantityOne software (Bio-Rad). Each wge translation experiment was performed three times independently and averages and standard errors of the mean (SEM) were calculated. Readthrough levels were calculated as a ratio of the amount of p95 readthrough product relative to that of its corresponding p36 pre-readthrough product, with the ratio for the wt genome set as 100% (36,39).
Protoplast transfection and viral RNA analysis
Production of genomic and subgenomic CIRV RNAs were assessed after protoplast infections, as described previously (51). Protoplasts were prepared from the cotyledons of 6-day old cucumber plants. For each viral RNA genome tested, ∼500 000 protoplasts were transfected with 5 μg of CIRV transcript using polyethylene glycol and CaCl2 (51). Transfected protoplasts were incubated under constant fluorescent light at 22°C for 22 h. Total nucleic acids were extracted and separated by agarose gel electrophoresis and plant 25S ribosomal RNA bands were monitored as controls to ensure even loading. Total nucleic acids were then transferred to a nylon membrane and plus-strand viral RNA accumulation levels were assessed using a [γ -32P]-labeled oligonucleotide probe complementary to the 3′-end of the CIRV genome and subgenomic mRNAs (coordinates 4739–4760). Northern blots were imaged using a Typhoon FLA 9500 and RNA bands were quantified using the QuantityOne software. Relative sg mRNA1 levels were calculated as the ratio of sg mRNA1 levels to their cognate genome levels, with the wt ratio set to 100%. Each set of protoplast transfections was carried out three times independently and averages and SEM values were calculated.
Minus-strand viral RNA accumulation was analyzed as described previously (52). Briefly, total nucleic acids isolated from protoplast infections were denatured with dimethyl sulfoxide and glyoxal and separated by agarose gel electrophoresis in 10 mM sodium phosphate buffer (pH 7.0). Northern blotting, imaging, and data analysis was performed as described earlier, except that [α-32P]-UTP-labeled riboprobe, corresponding to the 3′-end of CIRV cDNA (coordinates 4381–4760), was used for detection.
Electrophoretic mobility shift assay (EMSA)
DNA fragments of RTSL and LD2 and their derivatives were generated by standard PCR that incorporated a T7 promoter upstream of the 5′-ends of the RNA-encoding region. Individual, or mixtures of, in vitro-transcribed RNA fragments (10 pmol each in 3.6 μl of water) were heated at 94°C for 3 min, then combined with 0.4 μl RNA binding buffer (final concentration: 5 mM HEPES pH7.8, 6 mM MgCl2, 100 mM KCl. 3.8% glycerol) (39,53,54). The tubes were placed at 37°C for 30 min and snap-cooled on ice for 2 min. An equal volume of sterile 20% glycerol was added to each sample and the entire contents were separated by nondenaturing 4% (or 8%) polyacrylamide gel electrophoresis in a running buffer containing 45 mM Tris, pH 8.3, 43 mM boric acid and 1 mM MgCl2 (54). Gels were then stained with 1 mg/mL ethidium bromide (39), imaged using Typhoon FLA 9500 scanner, and RNA bands were quantified using the QuantityOne software. Relative binding efficiencies were determined by quantifying the amount of shifted LD2 or LD2-core by comparing their levels in LD2-only or LD2-core-only lanes with their corresponding unbound levels in mixtures with RTSL. Thus, relative binding efficiency is presented as a percentage of shifted LD2 or LD2-core. Each EMSA experiment was conducted three times independently, with averages and SEMs provided.
In-line probing of RNA secondary structure
In vitro-generated RNA transcripts of wt LD2-core and wt RTSL were purified using two cycles of the crush-soak RNA purification method (55). Purified transcripts were then dephosphorylated using calf-intestinal phosphatase (NEB) and 5′-end labeled using [γ-32P]-ATP and T4 polynucleotide kinase (NEB). End-labeled transcripts were recovered by G-50 column chromatography and used for in-line reactions that were carried out at 25°C for 40 hours in 1x in-line reaction buffer (50 mM Tris–HCl pH 8.3, 100 mM KCl, 20 mM MgCl2) (56,57). Reactions contained labeled fragments individually (∼1 pmol) or as a mixture with their unlabeled partner fragment (30 pmol) (56,57). Labeled fragments were also used to generate untreated controls, as well as size ladders generated by alkaline hydrolysis or RNase T1 digestion. All samples were separated in 10% denaturing polyacrylamide gels (58) and imaged and quantified as described in the previous sections. In-line probing was performed twice, with consistent results. Reactivities were used to generate an in-line-guided secondary structure model for LD2-core, RTSL or a complex of both as described in Supplementary Figures S10–S12. RNA secondary structures presented were generated using RNA2Drawer software (59).
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE)
SHAPE analysis of the LD2 region of the CIRV genome was performed using 1-methyl-7-nitroisatoic anhydride (1M7), as described previously (39). Four primers were used to map the secondary structure of the LD2 (primer coordinates in the CIRV genome: 1871–1895, 2169–2191, 2444–2464 and 2800–2824). Following fluorescent capillary sequencing, the raw data was analyzed using the ShapeFinder software (60) to generate relative reactivities at single nucleotide resolution. SHAPE reactions were performed twice for each of the four primers and average reactivities were used. The reactivity data was normalized against the average of the ten highest reactivity values, as described previously (39). The RNAStructure web server was used to combine SHAPE reactivity data with thermodynamic prediction to generate a secondary structure model of LD2 as described in Supplementary Figure S2 (61). RNA secondary structures presented were generated using RNA2Drawer software (59).
RESULTS
The RTSL terminal loop (RTSL-TL) regulates sg mRNA1 accumulation via an LDRI
Translational readthrough for the CIRV genome requires a long-distance RNA–RNA interaction (LDRI) between RTSL and the 3′UTR, involving the PRTE and DRTE partner sequences, respectively (Figure 1A, B) (39). To investigate the possible involvement of other regions of the RTSL in the readthrough process, silent nucleotide substitutions were introduced into its terminal loop (mutants TC-1 and TC-2) and closing base pair (TC-3) (Figure 2A). In vitro translation of CIRV genomes containing these modifications showed that readthrough production of p95 was similar to wt, or moderately affected (∼103% to ∼87%) (Figure 2B). However, northern blot analysis of protoplasts transfected with the same mutant viral genomes revealed an unanticipated role for the terminal loop of RTSL (RTSL-TL) in facilitating the accumulation of sg mRNA1. In these infections, sg mRNA1 levels were quantified relative to the corresponding levels of their cognate genomes, with that for wt set at 100%. Both terminal loop substitutions resulted in a ∼fivefold decrease in relative sg mRNA1 accumulation, whereas alteration in the loop's closing base pair yielded wt levels (Figure 2C). Notably, the negative effects of the RTSL-TL mutants were specific for sg mRNA1, as typical levels of sg mRNA2 were maintained. Also, because the modifications introduced were not present in sg mRNA1, altered RNA stability was ruled out as a cause for the observed decreases. Instead, the results indicated a role for RTSL-TL in regulating the transcriptional efficiency of sg mRNA1.
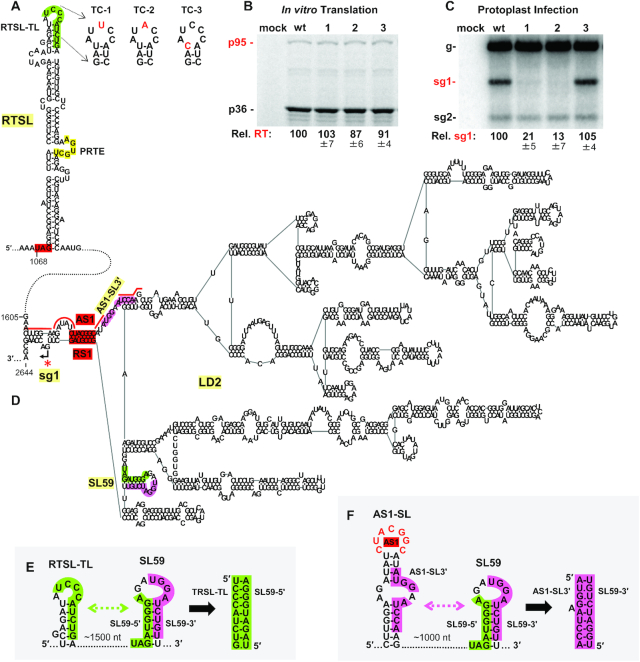
Figure 2.
RTSL regulates sg mRNA1 transcription. (A) RNA secondary structure of RTSL (39) with the p36 stop codon highlighted in red, the PRTE depicted in yellow, and a sequence of interest (RTSL-TL) in the terminal loop colored in green. Nucleotide substitutions introduced into RTSL in genomic mutants are shown in red. All substitutions were made at wobble positions and did not alter the p95 amino acid sequence. (B) In vitro translation analysis in wheat germ extract of wt and mutant CIRV RNA genomes shown in panel (A). Samples, labeled at the top, were separated in a 12% SDS polyacrylamide gel and the positions of CIRV proteins are indicated on the left. Average p95 readthrough levels (Rel. RT) for each mutant relative to that of the wt (set as 100%) are shown below the gel, along with standard errors from three independent experiments. (C) Northern blot analysis of total nucleic acids isolated from plant protoplasts transfected with wt and mutant CIRV RNA genomes shown in panel (A). Respective viral infections, labeled at the top, were probed for plus-strand viral RNAs. The positions of the viral genome (g), sg mRNA1 (sg1) and sg mRNA2 (sg2), are shown on the left. Average sg mRNA1 accumulation levels, relative to that of the wt (set as 100%), are shown below the blot with standard errors determined from three independent experiments. (D) RNA secondary structure of the LD2 domain in CIRV deduced from structure probing analysis (Supplementary Figure S2A). The ∼400 nucleotides between RTSL and LD2 are shown as a connecting dashed line. Relevant features of LD2 include the AS1/RS1 LDRI in red, and SL59, composed of sequences that are complementary to RTSL-TL (green) and AS1-SL3′ (pink). The sg mRNA1 transcription initiation site is denoted by an arrow and a red asterisk. Sequences corresponding to the two halves of the stem in AS1-SL are indicated by red lines. (E) Sequences and structures involved in forming the RTSL-TL/SL59–5′ (green) interaction. (F) Sequences and structures involved in forming the AS1-SL3′/SL59–3′ (pink) interaction.
Modulation of sg mRNA1 transcription by RTSL-TL could occur by it interacting with a protein factor or a complementary RNA sequence in the CIRV genome. As tombusviruses are known for controlling important viral processes via intra-genomic RNA-RNA interactions, the latter possibility was deemed more probable (36,39,41,47,49,62). To regulate sg mRNA1 transcription, RTSL-TL would likely have to interact with a sequence located near the initiation site for sg mRNA1 transcription. In tombusviruses, this initiation site is positioned just downstream from the transcription-promoting AS1/RS1 interaction (Supplementary Figure S1), which forms the closing stem of the RNA domain LD2, as shown for TBSV (Figure 1B) (48,49). Corresponding structure probing analysis (63) of the CIRV genome predicted a comparable AS1/RS1-containing LD2 (Figure 2D and Supplementary Figure S2A) that structurally mimicked that in TBSV (Supplementary Figure S2B). Also, the CIRV AS1/RS1 LDRI was shown, as demonstrated previously for TBSV (49), to be necessary for sg mRNA1 transcription (Supplementary Figure S3).
CIRV’s LD2 was examined for a potential base-pairing partner for RTSL-TL, and a candidate 9 nt long segment was identified 70 nt upstream from the transcription initiation site for sg mRNA1. This sequence was present within a predicted RNA hairpin structure, SL59, located within the 3′-end of the p95 ORF, some ∼1500 nt away from RTSL-TL (Figure 2D, green). Its partner sequence, RTSL-TL, was present in the terminal loop of RTSL and extended into the adjoining 3′-stem region (Figure 2A, green); thus, for the RTSL/SL59 interaction to occur, the helical region of RTSL-TL would need to unpair. Similarly, to associate with RTSL-TL, the complementary partner sequence in SL59, comprising the 5′-half of this hairpin (herein termed SL59–5′, green), would have to unpair from its 3′-half (SL59–3′, pink) (Figure 2E). A potential base pairing partner for the displaced SL59–3′ was also identified that mapped to the 3′-half of AS1-SL, termed AS1-SL3′ (pink) (Figure 2F). Consequently, the binding of RTSL-TL to SL59–5′ (green interaction) could be accompanied by an intra-LD2 interaction (pink) (Supplementary Figure S4A), both of which (in addition to the AS1-RS1 interaction) were supported by comparative sequence analysis showing maintenance of the base pairing, despite sequence variations (Supplementary Figure S4B).
The binding of RTSL-TL to SL59–5′ was investigated functionally by introducing compensatory nucleotide substitutions into the candidate partner sequences and assessing the effects on sg mRNA1 accumulation following transfection of mutant viral RNA genomes into protoplasts (Figure 3A–C). Whenever possible, translationally neutral or conservative substitutions were used. Disruption of base pairing potential in mutants TC-6 and TC-7 diminished sg mRNA1 plus- and minus-strand levels below ∼10% of wt, while regenerating pairing capacity with alternate nucleotides in mutant TC-8 restored levels up to ∼50–62% of wt (Figure 3B, C). This correlation between base pairing stability and sg mRNA1 accumulation is consistent with a role for the interaction in mediating transcription of sg mRNA1. Notably, the low levels of accumulation of the intermediate minus-strand sg mRNA1 templates in TC-6 and TC-7 indicated that disrupting the RTSL-TL/SL59–5′ interaction hindered proper formation of the RdRp attenuation structure for sg mRNA1 (Figure 3C). Similar results were observed when comparable mutational analysis was performed to assess the proposed AS1-SL3′/SL59–3′ interaction (Figure 3D-F). Thus, in addition to AS1/RS1, two other LDRIs, RTSL-TL/SL59–5′ and AS1-SL3′/SL59–3′, are critical for generating an effective RdRp attenuation structure. Since formation of the intra-LD2 interaction (pink) would free SL59–5′ for binding to RTSL-TL (green) (Supplementary Figure S4A), the probable order of these interactions would be the former followed by the latter.
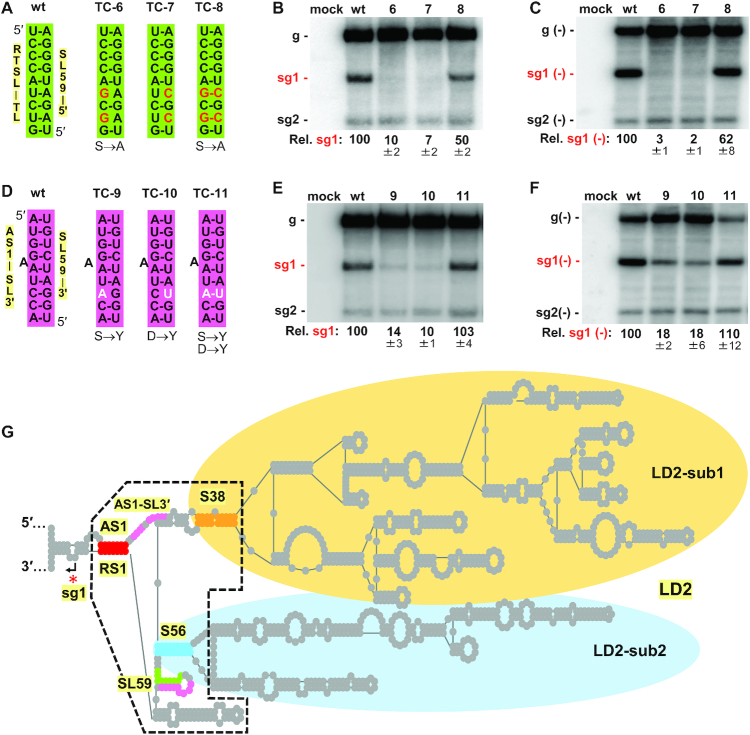
Figure 3.
Functional analysis of candidate LDRIs in regulating sg mRNA1 transcription. (A, D) Compensatory nucleotide substitutions introduced in RTSL-TL/SL59–5′ (green) and AS1-SL3′/SL59–3′ (pink) LDRIs in mutant genomes are shown as red and white nucleotides, respectively. Any corresponding changes in the p95 amino acid sequence are shown under the mutant sequences. (B, E) Northern blot analysis of plus-strand CIRV RNAs isolated from protoplasts transfected with wt and mutant CIRV RNA genomes depicted in panels (A) and (D), respectively. Identities of the viral genomes tested are indicated above the blots and positions of positive-sense genome and sg mRNAs are shown on the left. Average sg mRNA1 accumulation levels relative to that of the wt are provided below the blots with standard errors obtained from three independent experiments. (C, F) Northern blot analysis of minus-strand CIRV RNAs isolated from protoplast infections. Identities of the viral genomes tested are indicated above the blots and positions of the minus-sense genome and sg mRNAs are shown on the left. Average minus-strand sg mRNA1 accumulation levels relative to that of the wt are provided below the blots with standard errors obtained from three independent experiments. (G) Simplified RNA secondary structure depiction of CIRV’s LD2. Regulatory RNA elements important for sg mRNA1 transcription are highlighted: AS1/RS1 (red), AS1-SL3′ (pink), S38 (orange), S56 (blue), SL59–5′ (green) and SL59–3′ (pink). A core regulatory region at the base of LD2 is defined by a black dashed line. The arrow and the red asterisk show the sg mRNA1 transcription initiation site.
Additional LD2 substructures are necessary for sg mRNA1 transcription
The organisation of LD2 includes two subdomains, LD2-sub1 and LD2-sub2, which have closing stems (S38, orange, and S56, blue, respectively) that are proximal to the sequences involved in the RTSL-TL/SL59–5′ and AS1-SL3′/SL59–3′ interactions (Figure 3G). These closing stems, which are maintained in the genus (Supplementary Figure S5), could therefore influence formation of the latter-mentioned interactions. To address this question, compensatory mutational analysis was performed on S38 and S56, which yielded results supporting the importance of their helical stability (Supplementary Figure S6). Accordingly, the closing stems of both LD2 subdomains also contribute to the assembly of an effective RdRp attenuation structure. This allowed for approximate delineation of a core region of functional importance at the base of LD2 (Figure 3G, black dashed line).
RTSL-TL and SL59-5′ interact physically by intra-genomic association
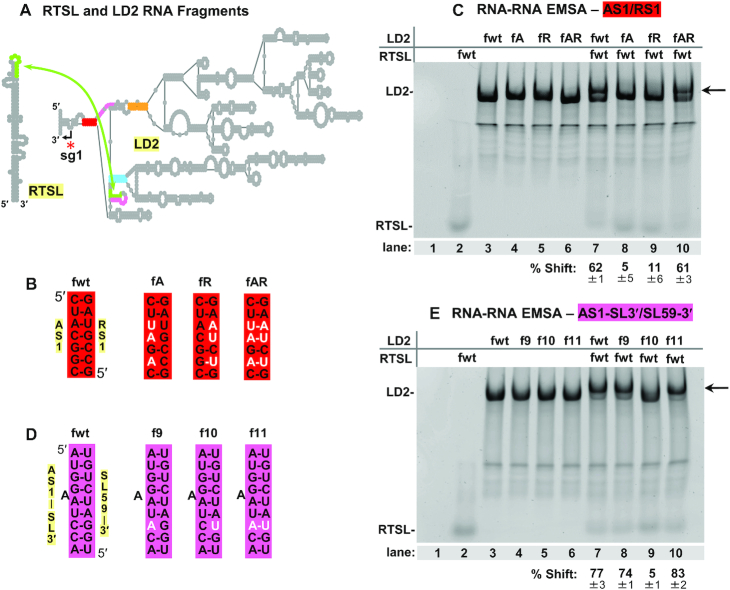
Having obtained in vivo genetic evidence for the RTSL-TL/SL59–5′ interaction, we next sought physical support for this pairing event. To achieve this, fragments of RTSL (106 nt) and LD2 (1040 nt) (Figure 4A) that contained the same compensatory mutations in RTSL-TL and SL59–5′ that were tested earlier (Figure 3A) were used in RNA-RNA electrophoretic mobility shift assays (EMSAs) (Figure 4B). Incubation of wt fragments of RTSL and LD2 led to the formation of an RNA–RNA complex, observed as an upward shift of the LD2 fragment (Figure 4B, compare lane 4 with 6). Combinations of fragments in which the RTSL-TL/SL59–5′ interaction was destabilized diminished shifting, while restoration of pairing regenerated the shift (Figure 4B, lanes 7 & 8 and lane 9, respectively). Thus, formation of an RTSL/LD2 complex is dependent on the RTSL-TL/SL59–5′ (green) interaction.
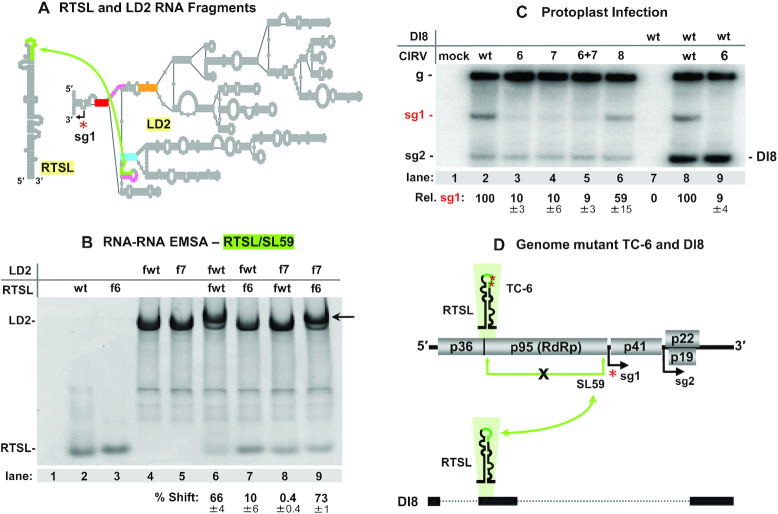
Figure 4.
RTSL-TL and SL59–5′ sequences interact physically and via an intragenomic interaction. (A) Simplified depiction of CIRV RTSL (106 nt) and LD2 (1040 nt) RNA fragments tested in RNA–RNA EMSAs. (B) Ethidium bromide-stained native 4% polyacrylamide gel of EMSA with the RNA fragments containing mutations shown in Figure 3A. The contents of each lane are indicated above the gel, with the fragment type shown to the far left. Lane 1 represents a mock lane containing only RNA binding buffer and glycerol. The black arrow on the right side of the gel points to the position where the RTSL/LD2 complexes migrate. The percentages with standard errors of shifted LD2 RNAs are provided below and were obtained from three independent EMSA experiments. (C) Northern blot analysis of protoplast co-transfection and single transfections. Identities of the viral genomic and DI RNAs tested are indicated above the gel. Modifications in the RNAs are shown in Figure 3A. Positions of positive-sense genome, sg mRNAs, and DI8 are indicated on the left and right of the blot. Average sg mRNA1 accumulation levels relative to that of the wt are provided below the blot with standard errors obtained from three independent experiments. (D) Schematic depiction of the co-transfection involving mutant TC-6 CIRV genome (top) and wt DI8 (bottom, horizontal black bars correspond to regions of the viral genome present in DI8). TC-6 modifications (two burgundy asterisks) in RTSL prevents formation of an intragenomic RTSL-TL/SL59–5′ LDRI (green double-headed arrow with a black X). The DI8 RNA contains wt RTSL in its sequence that, when co-inoculated with TC-6, can potentially base pair in trans with the wt SL59–5′ sequence in TC-6 (the curved green arrow connects DI8’s wt RTSL and TC-6’s wt SL59).
The demonstration of an in trans interaction in vitro raised the possibility that the same could be true during viral infections. To address this prospect, virus genome mutants TC-6 and TC-7 (Figure 3A), each of which was unable to form the RTSL-TL/SL59–5′ interaction in cis, but could potentially form it between each other in trans, were co-transfected into protoplasts. Levels of sg mRNA1 in the co-transfection (TC-6+TC-7) were similar to those for the individual transfections, i.e. ∼10%, and well below the ∼59% observed for the compensatory mutant TC-8 (Figure 4C, lanes 3–6). As a further test, a small noncoding CIRV genome-derived RNA replicon, DI8, containing a wt RTSL was utilized (Figure 4D, lower section). DI8 replicates only in the presence of the CIRV genome, which provides RdRp for its reproduction (39). Thus, DI8 amplification is limited to cells occupied by both the replicon and the CIRV genome. Co-transfection of DI8 and TC-6 (containing a mutated RTSL-TL and a wt SL59–5′ compatible with the wt RTSL in DI8) resulted in high levels of accumulation of both viral RNAs (Figure 4C, lane 9). However, despite robust co-accumulation, no increase in sg mRNA1 levels was observed (Figure 4C, compare lane 9 with lane 3). Collectively, these results indicate that during CIRV infections, the RTSL-TL/SL59–5′ interaction occurs as an intra-genomic event.
RTSL-TL/SL59-5′ complex formation requires both AS1/RS1 and AS1-SL3′/SL59-3′ interactions
Three key interactions are involved in efficient formation of the RdRp attenuation structure, RTSL-TL/SL59–5′ (green), AS1-SL3′/SL59–3′ (pink) and AS1/RS1 (red). To investigate the order in which these binding events occur, additional RNA–RNA EMSAs were performed (Figure 5). When the AS1/RS1 interaction corresponding to the closing stem of LD2 (Figure 5A, red) was assessed via compensatory mutations (Figure 5B), the results indicated its requirement for RTSL-TL binding to SL59–5′ (Figure 5C, lanes 7 to 10). Interestingly, disruption of AS1/RS1 in the LD2 fragment led to a slight decrease in its mobility, suggesting a more open conformation, consistent with AS1/RS1′s role in stabilizing the basal region of this large RNA domain (Figure 5C, compare lanes 4 and 5 with lanes 3 and 6). A dependence on the AS1-SL3′/SL59–3′ (pink) interaction was also observed, as RTSL/LD2 complex formation was inhibited by the CU mismatch in mutant f10 (Figure 5D, E lane 9). In contrast, the AG mismatch in f9 allowed for complex formation (Figure 5D, E lane 8). In the CIRV genome, this modification led to strong inhibition of sg mRNA1 levels in protoplast transfections (Figure 3D–F). The differing results observed for the AG mismatch in the EMSA is likely the consequence of this common non-canonical base pair being less destabilizing under the higher salt conditions of the assay. Notwithstanding, inhibition of complex formation with the CU mismatch and its recovery with the AU pair indicates that the AS1-SL3′/SL59–3′ (pink) interaction is indeed required for RTSL-TL binding to SL59–5′ (Figure 5D, 5E). These results, when considered along with the requisite for partner sequence accessibility and proximity, support the following sequential order for the formation for the three critical interactions. The AS1/RS1 (red) interaction would occur first and position AS1-SL3′ proximal to SL59–3′. Next, formation of the AS1-SL3′/SL59–3′ (pink) interaction would concurrently liberate SL59–5′ (green). Lastly, pairing of SL59–5′ with RTSL-TL (green) would complete assembly of the attenuation structure.
Figure 5.
Formation of the RTSL/LD2 complex is dependent on both the AS1/RS1 and AS1-SL3′/SL59–3′ interactions. (A) Simplified depiction of CIRV RTSL (106 nt) and LD2 (1040 nt) RNA fragments tested in RNA-RNA EMSAs. (B, D) Compensatory nucleotide substitutions (in white) that were introduced, respectively, into the AS1/RS1 (red) and AS1-SL3′/SL59–3′ (pink) complementary partner sequences. (C, E) Ethidium bromide-stained native 4% polyacrylamide gels of EMSAs testing the RNA fragments containing mutations shown in panels (B) and (D), respectively. The contents of each lane are indicated above the gels, with the fragment type shown to the far left. Lane 1 represents a mock lane containing just the RNA binding buffer and glycerol. The black arrows on the right side of the gels point to the position where the RTSL/LD2 complexes migrate. The percentages with standard errors of shifted LD2 RNAs are provided below and were obtained from three independent EMSA experiments.
Defining the structural requirements for the RTSL-TL/SL59-5′ interaction
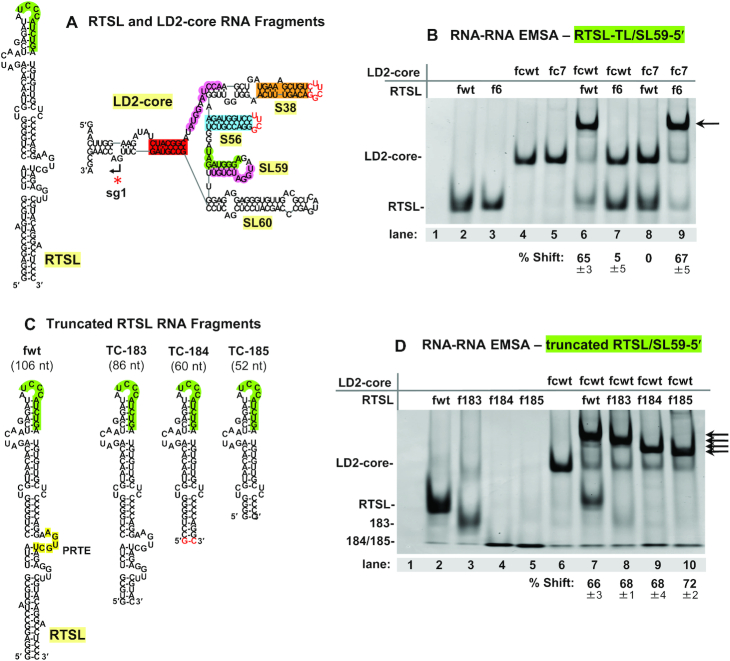
Infections with CIRV genome mutants revealed that efficient activation of sg mRNA1 transcription required the basal region of LD2, bounded by AS1/RS1, S38 and S56 (Figure 3G, black dotted line). To determine if more distal sequences or structures in LD2-sub1 or LD2-sub2 were required for RTSL-TL binding, a 188 nt-long RNA fragment containing only the core region of LD2 was constructed (Figure 6A, LD2-core). In LD2-core, the subdomain sequences beyond S38 and S56 were replaced with ultra-stable UNCG-type tetraloops. When LD2-core and RTSL fragments containing compensatory mutations in the RTSL-TL/SL59–5′ interaction (Figure 3A) were tested by EMSA, the results were equivalent to those observed with the complete LD2 fragment (compare Figure 6B, lanes 6–9, with Figure 4B, lanes 6–9). LD2-core thus accurately recapitulated the binding activity of the full-length LD2, implying that all determinants for efficient RTSL binding are present in this smaller fragment. RTSL binding to LD2-core also exhibited equivalent binding activities compared to full-length LD2 in terms of dependence on the AS1/RS1 (Supplementary Figure S7B, lanes 7–10, with Figure 5C, lanes 7–10) and AS1-SL3′/SL59–3′ interactions (Supplementary Figure S7C, lanes 7–10, with Figure 5E, lanes 7–10). Thus, LD2-core behaves comparably to full length LD2. Additionally, the potential involvement of SL60 in RTSL binding was assessed by deleting it from LD2-core and the results indicated no role for this substructure in complex formation (Supplementary Figure S8).
Figure 6.
Structural requirements for RTSL/LD2 complex formation. (A, C) Secondary structures of CIRV RTSL (106 nt) and LD2-core (188 nt) RNA fragments tested in RNA-RNA EMSAs. The red nucleotides in the LD2-core secondary structure represent the added UNCG-type tetraloops that replaced LD2 sub1 and sub2 beyond S38 and S56, respectively. The G–C pair shown in red in RTSL mutant TC-184 was added to allow for its transcription from PCR templates using T7 RNA polymerase. (B, D) Ethidium bromide-stained 8% native polyacrylamide gels of EMSAs testing the RNA fragments containing modifications shown in Figure 3A and panel C, respectively. The contents of each lane are indicated above the gels, with the fragment type shown to the far left. Lane 1 represents a mock lane containing only RNA binding buffer and glycerol. The black arrows on the right side of the gels point to the positions where the RTSL/LD2 complexes migrate. The percentages with standard errors of shifted LD2-core RNAs are provided below the gels and were obtained from three independent EMSA experiments.
The portion of RTSL required for binding to LD2-core was also sought by generating fragments with increasingly larger truncations of its lower region (Figure 6C) The EMSA results revealed that the bottom half of RTSL, including the PRTE, was dispensable for binding (Figure 6D). Therefore, the portion of RTSL essential for translational readthrough (i.e. the PRTE) is not required for formation of the RTSL-TL/SL59–5′ interaction.
In-line probing analysis of LD2-core and RTSL reveals a second key binding site
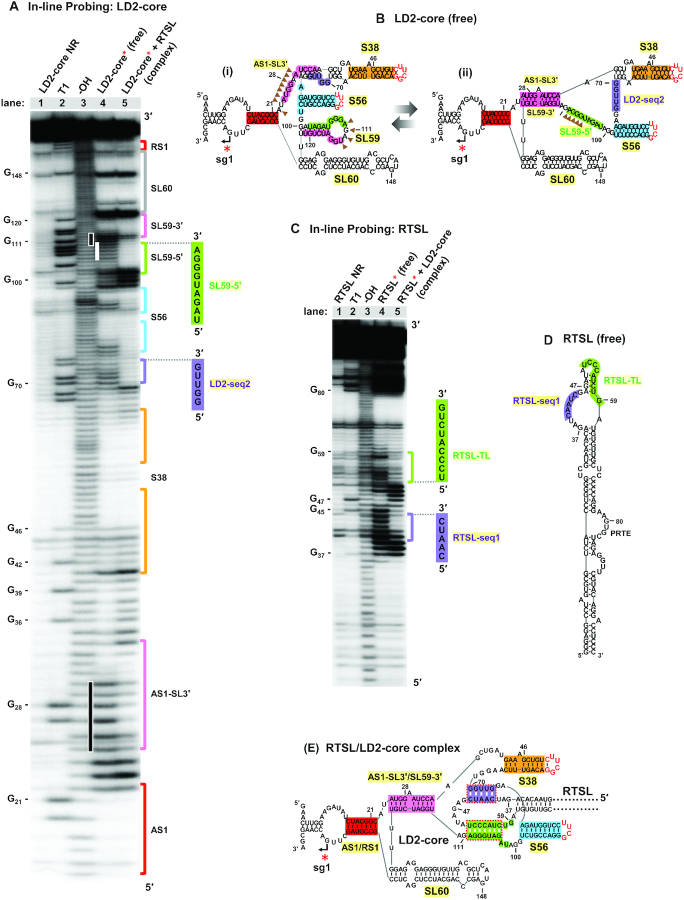
With both genetic and physical evidence supporting the formation and function of the RTSL/LD2 interaction, we next sought to gain additional insights into the nature of this RNA complex through solution structure probing analysis. To this end, in-line probing was used to assess the RNA structure of RTSL and LD2-core, both individually and in complex. Under the assay conditions, residues that are flexible, and thus likely single-stranded, undergo spontaneous hydrolysis (56). Information gained from the analysis is then used to build structural models consistent with the chemical reactivity data.
LD2-core was assessed first (Figure 7A). In its free state, the structural status of SL59–5′ (green), its adjacent partner sequence SL59–3′ (pink), and the alternate partner of the latter, AS1-SL3′ (pink), were of particular interest. The reactivity data (Figure 7A, lane 4) suggested that unbound LD2-core likely exists as a conformational mixture that includes SL59 (Figure 7Bi) and the mutually-exclusive AS1-SL3′/SL59–3′ (pink) interaction (Figure 7Bii). Probing results with free LD2-core that were consistent with the formation of SL59 included (i) high reactivity in the 5′-portion of AS1-SL3′ (pink, coordinates 25 to 30), indicating that a proportion of this sequence does not pair with SL59–3′ (Figure 7A, lower black bar and 7Bi, brown-shaded triangles) and (ii) high reactivity in the loop residues in SL59, which would be reactive in the context of SL59 (Figure 7A, upper black bar and 7Bi, brown-shaded triangles). Further evidence for SL59′s functional relevance and structural existence was provided, respectively, by comparative sequence analysis supporting its conservation (Supplementary Figure S9) and RNA structure modelling, guided by the in-line reactivity data, that predicted its presence in the optimally-folded LD2-core (Supplementary Figure S10A). Conversely, the moderate reactivity of residues in SL59–5′ (green, 106–110), which indicated an unpaired state in a proportion of the structural population, was consistent with an alternative non-SL59-containing structure (Figure 7A, white bar and 7Bii, brown-shaded triangles); a concept bolstered by the prerequisite for the SL59–5′-freeing AS1-SL3′/SL59–3′ (pink) interaction for complex formation (Figure 5E and Supplementary Figure S7C). Collectively, these data suggest that, when unbound, the core region of LD2 is comprised of a mixture that includes the two structural conformations presented (Figure 7B), however other configurations are also plausible (Supplementary Figure S10B).
Figure 7.
In-line structural probing analysis of CIRV LD2-core and RTSL RNAs. (A, C) Sequencing gel analysis following in-line probing of radiolabeled LD2-core (LD2-core*) and RTSL (RTSL*) RNA fragments, respectively. Lane 1 contains untreated LD2-core* or RTSL* RNA samples (NR, no reaction). Lane 2 contains the RNase T1-digested LD2-core* or RTSL* RNA samples to generate G ladders. Lane 3 contains LD2-core* or RTSL* RNA samples that were subjected to alkaline hydrolysis reaction (–OH) to generate cleavages at every nucleotide position. Lane 4 contains in-line reactions from free LD2-core* or free RTSL* RNA fragments (free). Lane 5 shows in-line reactions when LD2-core* or RTSL* was incubated with unlabeled RTSL and LD2-core, respectively, to generate a complex. Nucleotide positions of selected G residues are indicated on the left. Different regulatory sequences are color coded and labeled on the right side of the gels. Black bars on the left of lane 4 in panel (A) indicate SL59 and AS1-SL3′ sequences that show high cleavage levels in free LD2-core. The white bar on the left of lane 4 shows moderate cleavage levels for SL59–5′ in free LD2-core. (B) Two alternative RNA secondary structure conformations for free LD2-core. The structure on the left (i) was deduced as the optimal structure by in-line-guided folding of LD2-core, as described in Supplementary Figure S10A. Areas of notable reactivity are indicated by brown arrowheads (which correspond to vertical black bars in panel A). The structure on the right (ii) was generated with folding constraints that maintained nucleotides 106–110 as unpaired (brown arrowheads, which correspond to the vertical white bar in panel A). (D) RNA secondary structure of free RTSL was deduced as the optimal structure by in-line-guided folding of RTSL, as described in Supplementary Figure S11A. RTSL-TL and RTSL-seq1 are shown in green and purple, respectively. (E) RNA secondary structure of the RTSL/LD2-core complex, deduced by in-line probing results from analysis of the RNA complex (Supplementary Figure S12).
Probing results for LD2-core when in complex with RTSL revealed a notable reduction in reactivity of SL59–5′ (green), consistent with it base-pairing with RTSL-TL (Figure 7A, compare lanes 4 and 5). Correlative results were observed when RTSL was probed individually (Figure 7C, lane 4, 7D, and Supplementary Figure S11) or in complex, the latter of which showed a corresponding reduction in reactivity of RTSL-TL (green) in the bound state (Figure 7C, compare lanes 4 and 5). The probing results also revealed a potential second inter-fragment interaction involving two 5 nt-long complementary sequences (i.e. corresponding reduced reactivities in the bound states) (Figure 7A, C, purple) located between S38 and S56 in LD2-core and in a bulged region of RTSL (Figure 7B and D, purple, respectively). Thus, in addition to the RTSL-TL/SL59–5′ (green) interaction, a second interaction between RTSL and LD2 (purple) could also be functionally relevant, as structurally modeled (Figure 7E and Supplementary Figure S12).
The potential second interaction was initially assessed in protoplast infections with CIRV genomes containing compensatory mutations in the partner sequences. The results indicated that base pairing of these sequences was required for both sg mRNA1 plus- and minus-strand synthesis (Supplementary Figure S13A–C). EMSA analysis of the same mutations in the context of the LD2-core and RTSL fragments indicated that complex formation was dependent upon complementarity of the sequences in RTSL and LD2, termed RTSL-seq1 and LD2-seq2, respectively (Supplementary Figure S13D). Additionally, RTSL-seq1/LD2-seq2 pairing was found to be well conserved among the members of the genus Tombusvirus (Supplementary Figure S13E). These findings support a critical role for the RTSL-seq1/LD2-seq2 (purple) interaction in mediating formation of an effective attenuation structure for sg mRNA1 transcription (Figure 7E).
DISCUSSION
Global architecture of viral RNA genomes can contribute significantly to the regulation of critical viral functions. Accordingly, there is considerable interest in understanding how these genome-level RNA structures assemble and function. Our investigation of a tombusvirus led to the discovery of a novel intra-genomic RNA complex that activates sg mRNA1 transcription. Notably, this RNA-based attenuation structure is comparatively complex and provides new perspectives into this higher-order level of viral riboregulation.
The initial goal of this study was to investigate the possible role of the apical region of RTSL in the readthrough process, thus its observed involvement in sg mRNA1 transcription was unexpected. This additional function hinted at possible regulatory cross-talk between readthrough and transcription activities. Indeed, the presence of transcriptional regulatory sequences in the RdRp coding region of the viral genome would require the suppression of readthrough to allow for unimpeded transcription. Though an appealing possibility, in vitro translation analysis showed either no effect or minor decreases in readthough when the RTSL-TL/SL59–5′ interaction was disrupted (Figure 2B), whereas a notable increase (i.e. derepression) would have been expected if it was involved in coordinating the two processes. Nonetheless, the new transcriptional function uncovered adds to its previously known roles in promoting readthrough and inhibiting minus-strand RNA synthesis and classifies RTSL as a unique multifunctional RNA element controlling three distinct viral processes.
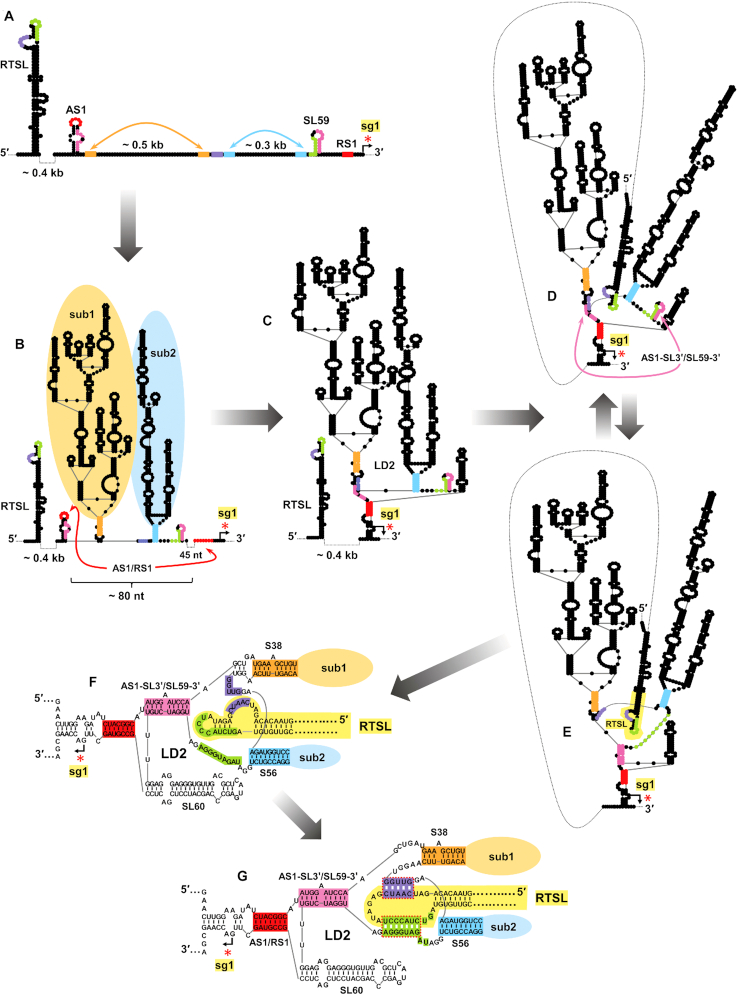
The complexity of the RdRp attenuation signal formed between RTSL and LD2 provided a unique opportunity to explore the assembly of this functional RNA complex. The comparatively smaller and localized components involved in the interaction, RTSL, AS1-SL and SL59, are anticipated to fold independently and relatively rapidly after their emergence during progeny viral RNA genome synthesis (Figure 8A). In contrast, formation of larger and more complex structures, such as subdomain-1 and -2 of LD2, would likely require additional time (Figure 8B). A role for these subdomains in assembly of the functional complex is supported by the observed importance of their closing stems for mediating efficient transcription (Supplementary Figure S6). Notably, the establishment of these subdomains unites AS1 and RS1 (red) to within ∼80 nt, thereby markedly reducing their ∼1000 nt distance of separation in the linear genome (compare Figure 8A with B). This colocalization would in turn facilitate base pairing of AS1 and RS1 (Figure 8B) and complete formation of LD2 (Figure 8C).
Figure 8.
Proposed RNA genome folding pathway leading to activation of sg mRNA1 transcription in CIRV. Note, this is a highly simplified folding pathway based on conjectured temporally-distinct transitions dictated by differences in stability, complexity and the relative spatial positions of the subcomponents. The relative timescales for transitions are not known and the structures shown represent approximations of probable intermediates within ensemble populations. (A) Schematic depiction of partially folded sections of the CIRV genome including RTSL, AS1-SL and SL59. AS1 and RS1 are separated by ∼1000 nucleotides in the linear sequence. Orange and blue double-headed arrows point to the complementary sequences involved in formation of S38 of sub1 and S56 of sub2, respectively. Small secondary structures within sub1 and sub2 are not shown, but are anticipated to form on a similar timescale as RTSL, AS1 and SL59. (B) RNA secondary structure after folding of LD2 sub1 (orange) and sub2 (blue), which are closed by stems S38 (orange) and S56 (blue), respectively. In this conformation AS1 and RS1 are brought within ∼80 nt from one another, which facilitates their base pairing (red double headed arrow). (C) Formation of the AS1/RS1 interaction completes folding of LD2 and its basal core region. (D) SL59, when present in the basal core region, prevents the docking of RTSL into the binding pocket by sequestering its partner sequences (refer also to Figure 7B(i)). Base pairing between AS1-SL3′ and SL59–3′ (pink double-headed arrow) leads to a conformational change in the basal core region. (E) Formation of the AS1-SL3′/SL59–3′ interaction (pink) frees the key binding pocket sequences (purple and green) and results in a functional binding site for RTSL. (F) Detailed pre-docking depiction of RTSL and the functional binding pocket in LD2. (G) Detailed post-docking depiction of RTSL and LD2 resulting in formation of an active attenuation structure.
The AS1/RS1 (red) interaction also mediates formation of the core region of LD2 that ultimately forms the binding pocket for RTSL. Probing data suggests that this core region likely exists as a conformational ensemble that includes incompatible and compatible forms, with respect to RTSL binding (Figure 8D and E, respectively). The presence of SL59 precludes formation of the essential AS1-SL3′/SL59–3′ (pink) interaction (Figure 8D), while formation of the latter is needed to free SL59–5′ (green) and LD2-seq2 (purple) to allow their binding with partner sequences in RTSL (Figure 8E). SL59 thus represents an integral but transient component in the folding process. Functionally, the formation of SL59 could prevent its critical halves from interacting with non-cognate complementary sequences that would interfere with correct folding of the binding pocket. In this capacity, SL59 would provide a safe, temporary, storage form for its component sequences until their requirement for binding pocket formation, initiated by the AS1/RS1 interaction.
Formation of the RTSL binding pocket requires both global folding of the large RNA domain LD2 and detailed conformational arrangements within its basal core region. Key features of the resulting docking site includes two discontinuous sequences (LD2-seq2, purple and SL59–5′, green) that map to either side of S56, the closing stem of subdomain-2 (Figure 8F). The docking of RTSL, via bipartite binding of RTSL-seq1 and RTSL-TL with these sites, acts as a linchpin in the formation and stabilization of the higher-order RNA complex capable of blocking progression of the viral RdRp (Figure 8G). This final docking step could confer its effect by bolstering the AS1/RS1 (red) interaction by either direct or allosteric means. In the latter case, RTSL pairings could stabilize the adjacent AS1-SL3′/SL59–3′ (pink) interaction, which, in turn, could structurally support the juxtaposed AS1/RS1 (red) helix (Figure 8G). Alternatively, direct, presumably non-canonical, interactions between RTSL and AS1/RS1 could function to stabilize the latter. A third possibility is that an additional part(s) of the RNA complex, in addition to the AS1/RS1 helix, contacts the RdRp and contributes to the stalling activity. Future, higher-resolution structural analysis will be required to investigate further the precise mode of RNA-based inhibition of the RdRp.
The formation and stability of the RNA attenuation structure is highly cooperative, as verified by the strong inhibitory effects of nucleotide mismatches in any of its component interactions (Figure 3 and Supplementary Figures S3, S6, and S13). Although our analyses indicate that these interactions can occur spontaneously in vitro (Figure 4 and Figure 5), viral or host proteins could also assist in folding of the attenuation RNA complex during infections (e.g. RNA chaperones (64)). Assembly of the RNA complex follows a multistep folding pathway involving the spatial unification of numerous distant regions of the genome (Figure 8). In this folding scheme, two steps in particular are likely to be rate limiting, and thus determinants of the timing of active complex formation leading to sg mRNA1 transcription. The first is the generation of LD2, including formation of the binding pocket, which would depend on overall domain folding and subsequent refinement of the docking site. A second restrictive step would be the docking event, the rate of which would be determined by the facility with which RTSL stochastically and productively encounters the binding pocket. Together, these steps could function to delay the production of CP from sg mRNA1 to later in the infection, when packaging is required. Indeed, time course experiments of viral RNA accumulation with tombusviruses show that, compared with that for sg mRNA2, sg mRNA1 transcription is delayed (42,43).
Intra-genomic LDRIs are also used to control sg mRNA transcription in other genera of the family Tombusviridae, including aureusviruses (65) and pelarspoviruses (66), while dianthoviruses utilize an inter-genomic interaction (67). However, in these cases the attenuation RNA structures are less complex than that described here; though, based on our unexpected results, further analyses may be warranted. This mode of sg mRNA regulation via LDRIs also extends beyond plant viruses to include plus-strand RNA viruses that infect insects and mammals. The sole sg mRNA produced by the insect-infecting Flock house virus (family Nodaviridae) is produced via a premature termination mechanism that utilizes an RNA-based attenuation structure composed of a three-helix junction formed by distant sequences (44). In contrast, coronaviruses use an alternate discontinuous transcription mechanism for sg mRNA production, where 3′ and then 5′ segments of the viral genome are copied discontinuously during minus-strand synthesis (68). In Transmissible gastroenteritis coronavirus, the discontinuous step for the production of the mRNA encoding the nucleocapsid protein is facilitated by an LDRI in the genome that unites the regions where the viral RdRp dissociates and reinitiates (69). Other plus-strand RNA viruses that infect humans and animals also depend on LDRIs for regulating viral processes, most notably flaviviruses (e.g. dengue virus (14,16,17) and zika virus (15)), hepaciviruses (e.g. hepatitis C virus (18,70)) and aphthoviruses (e.g. foot-and-mouth disease virus (71,72)). Moreover, other categories of RNA virus such are retroviruses (e.g. HIV (73)) and negative-strand RNA viruses (e.g. influenza virus (74)) also rely on LDRIs.
Understanding the molecular mechanisms of large-scale RNA circuits and their structural and functional integration is key to determining how RNA viruses regulate their infectious cycles. Deciphering such LDRI networks, however, has remained a challenge because many reside in coding regions and have multiple functions, as illustrated herein. In this study, we uncovered a new function for the folding of a large viral RNA domain in creating a distinctive binding pocket, and showed that subsequent docking of a distal RNA structure into this binding site acts as a linchpin that stabilizes an RNA complex required for viral transcription. We also proposed a plausible multistep pathway for the formation of the active intra-genomic RNA complex, an area of LDRI research that remains largely unexplored. These novel findings reinforce the importance and often overlooked underlying role of global RNA structure in viruses. Indeed, in many instances viral RNA genomes should be viewed as large complex RNA switches, and tombusviruses, with no fewer than eight functional LDRIs, serve as valuable prototypes for understanding this intriguing category of RNA-mediated regulation.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our laboratory for reviewing the manuscript and Baodong Wu for assistance during the early stages of this work.
Contributor Information
Tamari Chkuaseli, Department of Biology, York University, Toronto, Ontario M3J 1P3, Canada.
K Andrew White, Department of Biology, York University, Toronto, Ontario M3J 1P3, Canada.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council (NSERC) of Canada Discovery Grant (to K.A.W); NSERC Graduate Fellowship (to T.C.). Funding for open access charge: York University Open Access Author Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1. Liu Z.Y., Qin C.F.. Structure and function of cis-acting RNA elements of flavivirus. Rev. Med. Virol. 2020; 30:e2092. [DOI] [PubMed] [Google Scholar]
- 2. Madhugiri R., Fricke M., Marz M., Ziebuhr J.. Coronavirus cis-acting RNA elements. Adv. Virus Res. 2016; 96:127–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Y., Wimmer E., Paul A.V.. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim. Biophys. Acta. 2009; 1789:495–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Newburn L.R., White K.A.. Cis-acting RNA elements in positive-strand RNA plant virus genomes. Virology. 2015; 479–480:434–443. [DOI] [PubMed] [Google Scholar]
- 5. Miller W.A., White K.A.. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Annu. Rev. Phytopathol. 2006; 44:447–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nicholson B.L., White K.A.. Functional long-range RNA-RNA interactions in positive-strand RNA viruses. Nature Rev. Microbiol. 2014; 12:493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chkuaseli T., White K.A.. Intragenomic long-distance RNA-RNA interactions in plus-strand RNA plant viruses. Front Microbiol. 2018; 9:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Guo L., Allen E.M., Miller W.A.. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001; 7:1103–1109. [DOI] [PubMed] [Google Scholar]
- 9. Barry J.K., Miller W.A.. A -1 ribosomal frameshift element that requires base pairing across four kilobases suggests a mechanism of regulating ribosome and replicase traffic on a viral RNA. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:11133–11138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuhlmann M.M., Chattopadhyay M., Stupina V.A., Gao F., Simon A.E.. An RNA element that facilitates programmed ribosomal readthrough in turnip crinkle virus adopts multiple conformations. J. Virol. 2016; 90:8575–8591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chattopadhyay M., Shi K., Yuan X., Simon A.E.. Long-distance kissing loop interactions between a 3′ proximal Y-shaped structure and apical loops of 5′ hairpins enhance translation of Saguaro cactus virus. Virology. 2011; 417:113–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gao F., Simon A.E.. Multiple cis-acting elements modulate programmed -1 ribosomal frameshifting in Pea enation mosaic virus. Nucleic Acids Res. 2016; 44:878–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Du Z., Alekhina O.M., Vassilenko K.S., Simon A.E.. Concerted action of two 3′ cap-independent translation enhancers increases the competitive strength of translated viral genomes. Nucleic Acids Res. 2017; 45:9558–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sanford T.J., Mears H.V., Fajardo T., Locker N., Sweeney T.R.. Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res. 2019; 47:9789–9802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li P., Wei Y., Mei M., Tang L., Sun L., Huang W., Zhou J., Zou C., Zhang S., Qin C.F. et al.. Integrative analysis of Zika virus genome RNA structure reveals critical determinants of viral infectivity. Cell Host Microbe. 2018; 24:875–886. [DOI] [PubMed] [Google Scholar]
- 16. de Borba L., Villordo S.M., Iglesias N.G., Filomatori C.V., Gebhard L.G., Gamarnik A.V.. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol. 2015; 89:3430–3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V.. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006; 20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Romero-López C., Berzal-Herranz A.. The role of the RNA-RNA interactome in the hepatitis C virus life cycle. Int. J. Mol. Sci. 2020; 21:1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rance E., Tanner J.E., Alfieri C.. Genomic-scale interaction involving complementary sequences in the hepatitis C virus 5′UTR domain IIa and the RNA-dependent RNA polymerase coding region promotes efficient virus replication. Viruses. 2018; 11:E17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Romero-López C., Barroso-Deljesus A., García-Sacristán A., Briones C., Berzal-Herranz A.. End-to-end crosstalk within the hepatitis C virus genome mediates the conformational switch of the 3′X-tail region. Nucleic Acids Res. 2014; 42:567–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Romero-López C., Berzal-Herranz A.. The functional RNA domain 5BSL3.2 within the NS5B coding sequence influences hepatitis C virus IRES-mediated translation. Cell. Mol. Life Sci. 2012; 69:103–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Romero-López C., Berzal-Herranz A.. A long-range RNA–RNA interaction between the 5′ and 3′ ends of the HCV genome. RNA. 2009; 15:1740–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tuplin A., Struthers M., Simmonds P., Evans D.J.. A twist in the tail: SHAPE mapping of long-range interactions and structural rearrangements of RNA elements involved in HCV replication. Nucleic Acids Res. 2012; 40:6908–6921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shetty S., Stefanovic S., Mihailescu M.R.. Hepatitis C virus RNA: molecular switches mediated by long-range RNA–RNA interactions. Nucleic Acids Res. 2013; 41:2526–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moreno J.L., Zúñiga S., Enjuanes L., Sola I.. Identification of a coronavirus transcription enhancer. J. Virol. 2008; 82:3882–3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mateos-Gómez P.A., Zuñiga S., Palacio L., Enjuanes L., Sola I.. Gene N proximal and distal RNA motifs regulate coronavirus nucleocapsid mRNA transcription. J. Virol. 2011; 85:8968–8980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mateos-Gomez P.A., Morales L., Zuñiga S., Enjuanes L., Sola I.. Long distance RNA–RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013; 87:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. White K.A., Nagy P.D.. Advances in the molecular biology of tombusviruses: gene expression, genome replication and recombination. Prog. Nucleic Acids Res. Mol. Biol. 2004; 78:187–226. [DOI] [PubMed] [Google Scholar]
- 29. Harrison S.C., Olson A.J., Schutt C.E., Winkler F.K., Bricogne G.. Tomato bushy stunt virus at 2.9 A resolution. Nature. 1978; 276:368–373. [DOI] [PubMed] [Google Scholar]
- 30. Hillman B.I., Carrington J.C., Morris T.J.. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell. 1987; 51:427–433. [DOI] [PubMed] [Google Scholar]
- 31. Ye K., Malinina L., Patel D.J.. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003; 426:874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jaag H.M., Pogany J., Nagy P.D.. A host Ca2+/Mn2+ ion pump is a factor in the emergence of viral RNA recombinants. Cell Host Microbe. 2010; 7:74–81. [DOI] [PubMed] [Google Scholar]
- 33. Chuang C., Prasanth K.R., Nagy P.D.. The glycolytic pyruvate kinase is recruited directly into the viral replicase complex to generate ATP for RNA synthesis. Cell Host Microbe. 2017; 22:639–652. [DOI] [PubMed] [Google Scholar]
- 34. Nagy P.D. Tombusvirus-host interactions: co-opted evolutionarily conserved host factors take center court. Annu. Rev. Virol. 2016; 3:491–515. [DOI] [PubMed] [Google Scholar]
- 35. Nicholson B.L., White K.A.. Exploring the architecture of viral RNA genomes. Curr. Opin. Virol. 2015; 12:66–74. [DOI] [PubMed] [Google Scholar]
- 36. Nicholson B.L., White K.A.. Context-influenced cap-independent translation of Tombusvirus mRNAs in vitro. Virology. 2008; 380:203–212. [DOI] [PubMed] [Google Scholar]
- 37. Nicholson B.L., Zaslaver O., Mayberry L.K., Browning K.S., White K.A.. Tombusvirus Y-shaped translational enhancer forms a complex with eIF4F and can be functionally replaced by heterologous translational enhancers. J. Virol. 2013; 87:1872–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nicholson B.L., Wu B., Chevtchenko I., White K.A.. Tombusvirus recruitment of translational machinery via the 3′UTR. RNA. 2010; 16:1402–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cimino P.A., Nicholson B.L., Wu B., Xu W., White K.A.. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog. 2011; 12:e1002423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jiwan S.D., White K.A.. Subgenomic mRNA transcription in Tombusviridae. RNA Biol. 2011; 8:287–294. [DOI] [PubMed] [Google Scholar]
- 41. Wu B., White K.A.. Uncoupling RNA virus replication from transcription via the polymerase: functional and evolutionary insights. EMBO J. 2007; 26:5120–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang G., Slowinski V., White K.A.. Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA. 1999; 5:550–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qiu W., Scholthof H.B.. Effects of inactivation of the coat protein and movement genes of Tomato bushy stunt virus on early accumulation of genomic and subgenomic RNAs. J. Gen. Virol. 2001; 82:3107–3114. [DOI] [PubMed] [Google Scholar]
- 44. Lindenbach B.D., Sgro J.Y., Ahlquist P.. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 2002; 76:3905–3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Vliet A.L., Smits S.L., Rottier P.J., de Groot R.J.. Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J. 2002; 21:6571–6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. White K.A. The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology. 2002; 304:147–154. [DOI] [PubMed] [Google Scholar]
- 47. Lin H.X., White K.A.. A complex network of RNA-RNA interactions controls subgenomic mRNA transcription in a tombusvirus. EMBO J. 2004; 23:3365–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu B., Grigull J., Ore M.O., Morin S., White K.A.. Global organization of a positive strand RNA virus genome. PLoS Pathog. 2013; 9:e1003363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi I.R., White K.A.. An RNA activator of subgenomic mRNA1 transcription in Tomato bushy stunt virus. J. Biol. Chem. 2002; 277:3760–3766. [DOI] [PubMed] [Google Scholar]
- 50. Rubino L., Burgyan J., Russo M.. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch. Virol. 1995; 140:2027–2203. [DOI] [PubMed] [Google Scholar]
- 51. White K.A., Morris T.J.. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 1994; 68:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Choi I.R., Ostrovsky M., Zhang G., White K.A.. Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J. Biol. Chem. 2001; 276:41761–41768. [DOI] [PubMed] [Google Scholar]
- 53. Fabian M.R., White K.A.. Analysis of a 3′-translation enhancer in a tombusvirus: a dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006; 12:1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lafuente E., Ramos R., Martínez-Salas E.. Long-range RNA-RNA interactions between distal regions of the hepatitis C virus internal ribosome entry site element. J. Gen. Virol. 2002; 83:1113–1121. [DOI] [PubMed] [Google Scholar]
- 55. Petrov A., Wu T., Viani Puglisi E., Puglisi J.D.. RNA purification by preparative polyacrylamide gel electrophoresis in Laboratory methods in enzymology: RNA. Methods Enzymol. 2013; 530:315–330. [DOI] [PubMed] [Google Scholar]
- 56. Nahvi A., Green R.. Structural analysis of RNA backbone using in-line probing in laboratory methods in enzymology: RNA. Methods Enzymol. 2013; 530:381–397. [DOI] [PubMed] [Google Scholar]
- 57. Regulski E.E., Breaker R.R.. In-line probing analysis of riboswitches in Post transcriptional gene regulation. Methods Mol. Biol. 2008; 419:53–67. [DOI] [PubMed] [Google Scholar]
- 58. Slatko B.E., Albright L.M.. Denaturing gel electrophoresis for sequencing. Curr. Protoc. Mol. Biol. 1992; 16:7.6.1–7.6.13. [DOI] [PubMed] [Google Scholar]
- 59. Johnson P.Z., Kasprzak W.P., Shapiro B.A., Simon A.E.. RNA2Drawer: geometrically strict drawing of nucleic acid structures with graphical structure editing and highlighting of complementary subsequences. RNA Biol. 2019; 16:1667–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vasa S.M., Guex N., Wilkinson K.A., Weeks K.M., Giddings M.C.. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008; 14:1979–1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H.. RNAstructure: web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013; 41:W471–W474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wu B., Pogany J., Na H., Nicholson B.L., Nagy P.D., White K.A.. A discontinuous RNA platform mediates RNA virus replication: building an integrated model for RNA-based regulation of viral processes. PLoS Pathog. 2009; 5:e1000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Boerneke M.A., Ehrhardt J.E., Weeks K.M.. Physical and functional analysis of viral RNA genomes by SHAPE. Annu. Rev. Virol. 2019; 6:93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stork J., Kovalev N., Sasvari Z., Nagy P.D.. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology. 2011; 409:338–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu W., White K.A.. Subgenomic mRNA transcription in an aureusvirus: down-regulation of transcription and evolution of regulatory RNA elements. Virology. 2008; 371:430–438. [DOI] [PubMed] [Google Scholar]
- 66. Blanco-Pérez M., Hernández C.. Evidence supporting a premature termination mechanism for subgenomic RNA transcription in Pelargonium line pattern virus: identification of a critical long-range RNA-RNA interaction and functional variants through mutagenesis. J. Gen. Virol. 2016; 97:1469–1480. [DOI] [PubMed] [Google Scholar]
- 67. Sit T.L., Vaewhongs A.A., Lommel S.A.. RNA-mediated trans-activation of transcription from a viral RNA. Science. 1998; 281:829–832. [DOI] [PubMed] [Google Scholar]
- 68. Sola I., Almazán F., Zúñiga S., Enjuanes S.. Continuous and discontinuous RNA synthesis in coronaviruses. Annu. Rev. Virol. 2015; 2:265–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mateos-Gomez P.A., Morales L., Zuñiga S., Enjuanes L., Sola I.. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J. Virol. 2013; 87:177–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mauger D.M., Golden M., Yamane D., Williford S., Lemon S.M., Martin D.P., Weeks K.M.. Functionally conserved architecture of hepatitis C virus RNA genomes. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Serrano P., Pulido M.R., Sáiz M., Martínez-Salas E.. The 3′ end of the foot-and-mouth disease virus genome establishes two distinct long-range RNA-RNA interactions with the 5′ end region. J. Gen. Virol. 2006; 87:3013–3022. [DOI] [PubMed] [Google Scholar]
- 72. Diaz-Toledano R., Lozano G., Martinez-Salas E.. In-cell SHAPE uncovers dynamic interactions between the untranslated regions of the foot-and-mouth disease virus RNA. Nucleic Acids Res. 2017; 45:1416–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ooms M., Abbink T.E., Pham C., Berkhout B.. Circularization of the HIV-1 RNA genome. Nucleic Acids Res. 2007; 35:5253–5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tomescu A.I., Robb N.C., Hengrung N., Fodor E., Kapanidis A.N.. Single-molecule FRET reveals a corkscrew RNA structure for the polymerase- bound influenza virus promoter. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E3335–E3342. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.