Abstract
LOTUS domains are helix-turn-helix protein folds identified in essential germline proteins and are conserved in prokaryotes and eukaryotes. Despite originally predicted as an RNA binding domain, its molecular binding activity towards RNA and protein is controversial. In particular, the most conserved binding property for the LOTUS domain family remains unknown. Here, we uncovered an unexpected specific interaction of LOTUS domains with G-rich RNA sequences. Intriguingly, LOTUS domains exhibit high affinity to RNA G-quadruplex tertiary structures implicated in diverse cellular processes including piRNA biogenesis. This novel LOTUS domain-RNA interaction is conserved in bacteria, plants and animals, comprising the most ancient binding feature of the LOTUS domain family. By contrast, LOTUS domains do not preferentially interact with DNA G-quadruplexes. We further show that a subset of LOTUS domains display both RNA and protein binding activities. These findings identify the LOTUS domain as a specialized RNA binding domain across phyla and underscore the molecular mechanism underlying the function of LOTUS domain-containing proteins in RNA metabolism and regulation.
INTRODUCTION
LOTUS domains (named after Limkain, Oskar and Tudor domain-containing proteins; also known as the OST-HTH domain) are an ancient family of winged helix-turn-helix globular domains found in proteins in prokaryotes and eukaryotes (1,2). This ∼80 amino acids domain was initially predicted as a conserved fold in vertebrate germline proteins TDRD5 and TDRD7, as well as Drosophila Oskar (2). In animals, LOTUS domain-containing proteins are expressed predominantly in the germline with pivotal roles in RNA regulation, germ cell development and fertility as well as implications in human diseases (3–8).
Despite the important functions of LOTUS domain proteins in RNA biology, the binding properties of LOTUS domains remain controversial. The LOTUS domain was originally predicted as an RNA binding domain based on its similarity to an archaeal bacterial nucleic acids binding domain (1,2). However, the RNA binding activity of the LOTUS domain has not been convincingly shown (9–13). Instead, some LOTUS domains have been shown to exhibit conserved protein binding activity by interacting with an essential animal germline protein Vasa (11). However, a Vasa ortholog does not exist in bacteria, fungi and plants, making the binding targets of LOTUS domains in these organisms unclear. Currently, the most conserved binding property for the LOTUS domain family in all organisms, if any, remains unknown.
RNA G-quadruplex (G4) is a unique RNA tertiary structure prevalent in coding and noncoding RNAs from bacteria to eukaryotes (14). Formed from G-rich RNA sequences as a stacking helical structure of high thermostability, RNA G4s are biological regulators of resident RNAs in diverse cellular processes (15). The presence and regulatory roles of G4s in oncogene transcripts, telomere RNAs and other cellular mRNAs have been demonstrated in mRNA translational control, telomere homeostasis, and pre-mRNA slicing (16–19). The regulation of these processes has implications in human diseases such as cancer, aging and neurological disorders (16,19,20). In animal germ cells, abundant G4 forming sequences has been found in long non-coding RNAs (e.g. piRNA precursors) and implicated in piRNA biogenesis (21–23). For many years, G4 has been a structural curiosity due to its unconventional structural folding and high thermostability (14,15). Although globally unfolded in vivo, G4 has been proposed to be under complex regulation by G4-specific helicases and G4 RNA binding proteins (24). However, information on specialized proteins or motifs that recognize RNA G4s is limited (25–29), especially a conserved globular protein fold that specifically recognizes G4 has not been shown.
In this report, through the study of germline LOTUS domain-containing proteins TDRD5 in piRNA regulation, we uncovered an unexpected RNA binding property of the LOTUS domains to G-rich RNAs, in particular RNA G4 tertiary structure. We show that the high affinity LOTUS domain–G4 interaction is evolutionarily conserved from bacteria, plants to animals. Intriguingly, in animal germline, LOTUS domains harbor dual RNA binding and protein binding activity. These results identified the LOTUS domain as a novel G-rich and G4 RNA binding domain in prokaryote and eukaryote and provide new insights into the mechanism whereby LOTUS domain-containing proteins engage in RNA regulation.
MATERIALS AND METHODS
Bioinformatic analysis
TDRD5-CLIP and MILI-CLIP sequencing data (SRA accession number: SRP093845) (30) were used to analyze for nucleotide composition and G-quadruplex prediction. Sequenced TDRD5-CLIP and MILI-CLIP reads were processed with fastx_clipper to clip the sequencing adapter read-through. Clipped reads were filtered by length (≥15nt) and aligned to the following sets of sequences: 214 piRNA clusters, coding RNAs, noncoding RNAs, repeats and intron (30,31). Alignments were performed using Bowtie (one base mismatch allowed). The nucleotide compositions of CLIP reads from piRNA clusters, coding RNAs, noncoding RNAs, repeats and intron were calculated and normalized to the nucleotide compositions of the mouse genome. The nucleotide compositions of CLIP reads from Top 100 piRNA clusters were calculated. The nucleotide compositions of Top 100 piRNA clusters were used as a control.
G-quadruplex forming sequence prediction was performed as described (21,32). Briefly, G-quadruplex prediction was performed using a pattern: G2–4–N1–7–G2–4–N1–7–G2–4–N1–7–G2–4. G: guanosine; N: any nucleosides. The G4 numbers of TDRD5 CLIP reads from piRNA clusters, coding RNAs, noncoding RNAs, repeats and intron were calculated. G4 numbers of random reads from piRNA clusters, coding RNAs, noncoding RNAs, repeats and intron were also calculated as a control.
Protein purification
cDNA fragments of LOTUS domains were obtained by PCR amplification or gene synthesis (GenScript) and subcloned into pET28a (His-tag) vector (Supplementary Table S2). Drosophila Vasa (200–661aa) cDNA was obtained by PCR amplification and were subcloned into pGEX-4t-1 (GST-tag) vector. Plasmids were transformed into Escherichia coli (BL21 or Rosetta) and recombinant proteins were induced with 0.2 mM IPTG (Roche) at 18°C overnight. His-tagged proteins were affinity purified with Ni-NTA Agarose Resin (Thermo Scientific). GST-tagged proteins were affinity purified using Glutathione Agarose Resin (Thermo Scientific). Proteins were further purified by gel filtration chromatography with ÄKTApurifier UPC 10 (GE Healthcare). The running buffer for gel filtration chromatography is 10 mM Tris–HCl (pH 7.4) and 100 mM KCl.
Oligonucleotide annealing
To form G4 structure, 5′-biotinylated G4 RNA or DNA oligonucleotides and their mutants were synthesized by Integrated DNA Technologies (IDT) and were annealed in 10 mM Tris–HCl (pH 7.4), 100 mM KCl by heating at 95°C for 5 min and slow cooling to 16°C. To inhibit G4 formation, 5′-biotinylated G4 RNA oligonucleotides were annealed in 10 mM Tris–HCl (pH 7.4), 100 mM LiCl by heating at 95°C for 5 min and slow cooling to 16°C. To form double stranded RNA, 5′-biotinylated RNA oligonucleotides were annealed in 10 mM Tris–HCl (pH 8.0), 20 mM NaCl by heating at 95°C for 5 min and slow cooling to 16°C.
Oligonucleotide pull-down assay
Biotinylated oligonucleotides were bound to streptavidin agarose beads (Thermo Scientific) by shaking at room temperature for 30 min followed by incubation with purified His-tagged LOTUS domains at 4°C for 1 h. After washing three times in binding buffer, beads were boiled at 95°C in SDS sample buffer. Western blotting was performed using an anti-His antibody (Thermo Scientific) to detect bound LOTUS domain. The binding buffer for the pull-down assay contains 25 mM Tris–HCl (pH 7.4), 150 mM KCl, 0.5 mM DTT, 5 mM EDTA and 0.5% NP-40. For dot blot analysis, biotinylated oligonucleotides were spotted onto a positively charged nylon membrane (Millipore). Membrane was UV crosslinked at 120 mJ/cm2 and incubated with HRP-conjugated streptavidin and ECL substrate (Thermo Scientific) for chemiluminescent detection.
HEK293T cells were transfected with GFP-TDRD5 FL (1–1040aa), GFP-TDRD5-N (1–400aa), GFP-TDRD5-C (401–1040aa) or GFP as a negative control. After 48 hours, the transfected cells were collected and homogenized using binding buffer (25 mM Tris–HCl pH 7.4, 150 mM KCl, 0.5 mM DTT, 5 mM EDTA and 0.5% NP-40). Biotinylated oligonucleotides were bound to streptavidin agarose beads and were incubated with HEK293T protein lysates. Western blotting was performed using an anti-GFP antibody (Abcam).
Mouse adult testes were collected and homogenized using binding buffer (25 mM Tris–HCl pH 7.4, 150 mM KCl, 0.5 mM DTT, 5 mM EDTA and 0.5% NP-40). Biotinylated oligonucleotides were bound to streptavidin agarose beads and were incubated with testis lysates. Western blotting was performed using an anti-TDRD5 antibody (30). β-Actin served as a negative control.
Circular dichroism (CD) spectroscopy
200 μl of 5 μM annealed oligonucleotides were used to obtain CD spectra on Chirascan CD spectropolarimeter (Applied Photophysics) using a quartz cuvette with 1 mm path length. Scans were performed over the range of 220–320 nm using a response time of 1 s, 0.5 nm step and 1 nm bandwidth at 25°C. The absorbance of buffer was subtracted from recorded spectra. The curves were smoothed using GraphPad Prism (GraphPad software).
Enzyme-linked immunosorbent assay (ELISA)
ELISA assays were performed using standard methods. Briefly, annealed biotinylated oligonucleotides were bound to streptavidin-coated plates (Thermo Scientific) for 30 min with shaking, and incubated with purified His-tagged proteins at a series of concentrations (0–1000 nM) in 10 mM Tris–HCl (pH 7.4), 100 mM KCl or LiCl for 30 min. After washing 3 times in wash buffer (25 mM Tris–HCl, pH 7.4, 150 mM KCl or LiCl, 0.5 mM DTT, 5 mM EDTA and 0.5% NP-40), the plates were incubated with HRP-conjugated anti-His antibody (Thermo Scientific). Tetramethylbenzidine (TMB) was used as the HRP substrate (Thermo Scientific). Absorbance was measured at 450 nm using a SpectraMax Plus microplate reader (Molecular Devices).
GST pull-down assay
GST-tagged LOTUS domain proteins (5 μg) were incubated for 30 min at 25°C with 10 μg His-tagged Vasa (200–623aa) in a buffer containing 20 mM Tris, pH 7.5, 150 mM NaCl, 10% glycerol, 0.1% Tween 20 and 5 mM DTT. 30 μl of glutathion agarose beads (Thermo scientific) were added and the mixture incubated for an additional hour at 25°C. Beads were collected and washed 5 times with the incubation buffer. Proteins were eluted using SDS sample buffer and analyzed by SDS-PAGE and coomassie blue staining.
Electrophoretic mobility shift assay (EMSA)
5-FAM-labeled G4 and mutant G4 RNA oligos were synthesized by Integrated DNA Technologies (IDT) and were annealed in 10 mM Tris–HCl (pH 7.4), 100 mM KCl by heating at 95°C for 5 min and slow cooling to 16°C. To form RNA/protein complexes, annealed RNAs (250 nM) were incubated with purified His-tagged LOTUS domains at a series of concentrations at 4°C for 30 min in 20 μl binding buffer (25 mM Tris–HCl pH 7.4, 150 mM KCl, 0.5 mM DTT, 5 mM EDTA and 0.5% NP-40). After incubation, samples containing TDRD5 L1 and L3 were analyzed by native TBE-PAGE (pH 9.5) at 160 V for 40 min; samples containing TDRD5 L2 were analyzed by native TBE-PAGE (pH 10.5) at 160 V for 40 min. The fluorescent signals were captured by the ChemiDoc System (Bio-Rad).
Differential radial capillary action of ligand assay (DRaCALA)
RNA oligonucleotides were 5′-end labeled by [γ-32P] ATP followed by purification using NucAway spin columns (Invitrogen) to remove free [γ-32P] ATP. Labeled RNAs were diluted to 1 nM in 10 mM Tris–HCl (pH 7.4), 100 mM KCl, 5% glycerol and were annealed by heating at 95°C for 5 min and slowly cooling to 16°C. 1 μl protein samples were added into 9 μl annealed RNA oligonucleotides to achieve final protein concentrations of 10−10–10−5 M. After incubation for 10 min at room temperature, 4 μl mixed samples were spotted onto nitrocellulose membrane (Bio-Rad) and allowed to diffuse and air-dry for 30 min. Membranes were exposed on phosphorimager screen overnight followed by scanning on Typhoon scanner (GE Healthcare). The radioactive signals were quantified by Quantity One (Bio-Rad). The bound RNA was calculated as described (33).
Statistical analysis
Statistical analysis of experimental data was performed using a two-tailed Student's paired t-test for Figure 1. P values are shown. For ELISA, results are presented as mean ± s.e.m. of biological triplicates. Dissociation constants (Kd) were calculated using GraphPad Prism (GraphPad software).
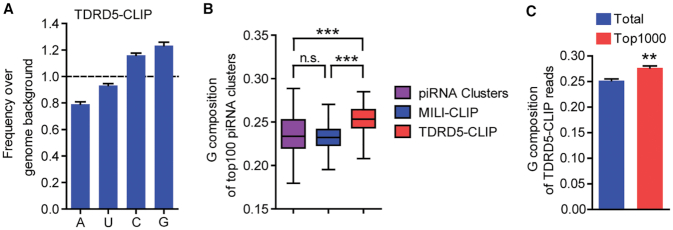
Figure 1.
High G-contents in TDRD5-bound RNA in vivo. (A) TDRD5-CLIP reads contain higher G-contents than mouse genome. Nucleotide compositions of TDRD5 CLIP reads were normalized to that of the mouse genome. (B) G composition in TDRD5-CLIP reads is higher than that of piRNA clusters and MILI-CLIP reads. G compositions in top-100 piRNA clusters, MILI-CLIP or TDRD5-CLIP reads were shown. n.s., Not significant; ***P < 10−20. (C) G composition of top-1000 and total TDRD5-CLIP reads. **P < 0.01.
RESULTS
High G-composition in testicular TDRD5-bound RNA
We previously reported that TDRD5 is an RNA-binding protein directly associating with piRNA precursors using high-throughput sequencing of RNA isolated by cross-linking immunoprecipitation (HITS-CLIP or CLIP-seq) (30). TDRD5 CLIP reads were primarily mapped to piRNA clusters (30). To further explore the RNA binding property of TDRD5, we analyzed TDRD5 CLIP reads and compared the nucleotide composition to that of the mouse genome. Interestingly, we found an enrichment of guanine (G) nucleotides in TDRD5 CLIP reads over the average of the mouse genome (Figure 1A). This correlates with previous reports that intergenic piRNA clusters are enriched in G nucleotides compared with other intergenic regions of the mouse genome (Supplementary Figure S1A) (21). Given that TDRD5 selectively binds with piRNA precursors, the higher G contents in TDRD5 CLIP reads could simply be explained by the selective binding of TDRD5 to piRNA precursors over other RNAs. We analyzed the G compositions of TDRD5 CLIP reads from different genomic regions. TDRD5 CLIP reads from different genomic regions all showed higher G contents compared with the genomic background (Supplementary Figure S1B). In addition, when mapping the TDRD5 CLIP reads to the top-100 most piRNA-producing piRNA clusters (accounting for >80% of all piRNAs produced in adult testis) (30,31), surprisingly, we found statistically significant G enrichment in TDRD5 CLIP reads compared with MILI CLIP reads (representing mature piRNAs) and the piRNA cluster background (Figure 1B and Supplementary Figure S1C). Next, we analyzed the most abundant TDRD5-bound sequences and found a G-enrichment for the top-1000 reads compared with the entire TDRD5 CLIP reads (Figure 1C). Together, these data suggest that TDRD5 preferentially interacts with G-rich sequences within piRNA precursor RNAs, a phenomenon also observed for piRNA biogenesis factor MOV10L1 (21).
LOTUS domains selectively bind G-rich RNA
We next investigated how TDRD5 engages in RNA binding. TDRD5 contains one Tudor domain and three LOTUS domains. The Tudor domain displays conserved binding to PIWI proteins in animal germ cells (34). The LOTUS domain is a putative RNA binding domain whose RNA binding property is still controversial (9–11). To explore the RNA binding potential of LOTUS domains, we expressed three individual recombinant LOTUS domains of mouse TDRD5 (named L1, L2 and L3) in E. coli and tested purified LOTUS domains binding to Poly(A), Poly(U), Poly(G) and Poly(C) RNA oligonucleotides using a biotin-oligo pull-down assay (Figure 2A and B, Supplementary Table S1 and S2). While L1, L2 and L3 of TDRD5 showed no binding to poly(A), poly(U) and poly(C) RNA oligos, strikingly, L1 and L2 exhibited selective binding to poly(G) RNA oligo (Figure 2C). We next performed an ELISA assay to estimate the affinity of LOTUS domain-RNA interaction. In the ELISA assay, biotin-labeled RNA oligos were immobilized onto the streptavidin-coated plates and were incubated with purified soluble TDRD5 LOTUS domains. Consistent with the RNA pull-down assay, ELISA showed that TDRD5 L1 and L2 displayed specific binding to poly(G) RNA oligos with high affinity at ∼10 nM while no detectable binding to poly(A), poly(U) or poly(C) oligos (Figure 2D). TDRD5 L3 lacked interactions to all oligos tested (Figure 2D). To test whether TDRD5 has specific interaction with poly(G) RNA in a physiological setting, we next performed biotin-oligo pull-down assay using adult mouse testis lysates. Both endogenous TDRD5 isoform 1 (containing L1, L2 and L3) and isoform 2 (only containing L1 and L3) was specifically pulled down by poly(G) RNA, but not by poly(A), poly(U), or poly(C) RNA oligos (Figure 2E). These results suggest that LOTUS domains of TDRD5 preferentially bind to G-rich RNAs in vitro and in vivo.
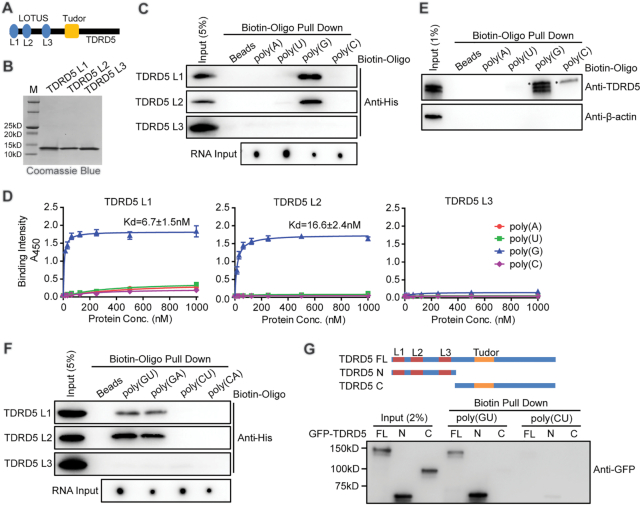
Figure 2.
TDRD5 LOTUS domains bind to G-rich RNA. (A) Domain architecture of mammalian TDRD5. Three LOTUS domains are abbreviated as L1, L2 and L3, respectively. (B) Purification of mouse TDRD5 LOTUS domains. His-tagged LOTUS domains were purified by affinity chromatography followed by gel filtration chromatography. (C) LOTUS domains specifically bind to poly(G) RNA. Biotin-labeled oligonucleotides were bound to streptavidin beads. After incubating with purified His-tagged LOTUS domains, Western blotting using an anti-His antibody was performed to detect bound LOTUS domains. Biotin-labeled oligonucleotide inputs were measured by dot blot analysis. (D) LOTUS domains specifically bind to poly(G) RNA by ELISA assays. n = 3, error bars represent s.e.m. Dissociation constants (Kd) are indicated. (E) poly(G) RNA binds endogenous TDRD5 from mouse testes. Biotin-labeled poly(G) RNA oligonucleotides were bound to streptavidin beads. After incubating with adult mouse testis lysates, Western blotting was performed using TDRD5 and β-actin antibodies. TDRD5 isoform1 and isoform2 were shown. Asterisk shows a non-specific band. (F) LOTUS domains bind to G-rich RNA. Biotin-labeled oligonucleotides were bound to streptavidin beads. After incubating with purified His-tagged LOTUS domains, Western blotting using an anti-His antibody was performed to detect bound LOTUS domains. Biotin-labeled oligonucleotide inputs were measured by dot blot analysis. (G) LOTUS domains but not the Tudor domain of TDRD5 bind to G-rich RNA. HEK293T cells were transfected with plasmids coding for GFP-tagged full-length (FL) or truncated (N or C) TDRD5. Biotin-labeled oligonucleotides were bound to streptavidin beads. After incubating with HEK293T cell lysates, Western blotting was performed using an anti-GFP antibody.
We next examined whether TDRD5 LOTUS domains interact with other G-rich or non-G-containing RNAs. Biotin-labeled poly(GU), poly(GA), poly(CU) and poly(CA) RNA oligos were used to perform the RNA pull-down assay and ELISA assay (Supplementary Table S1). Biotin-oligo pull-down assay showed that L1 and L2 of TDRD5 selectively bound with G-rich RNAs poly(GU) and poly(GA), but not non-G-containing RNAs poly(CU) and poly(CA) (Figure 2F). L3 did not show binding to all oligos tested. ELISA assay confirmed this result by showing L1 and L2 of TDRD5 selectively binding to poly(GU) and poly(GA) and the affinity was at 100–200 nM range (Supplementary Figure S2). Interestingly, the binding affinity of LOTUS domains with poly(GU) and poly(GA) was lower than that with poly(G), suggesting that the G-composition is positively correlated with the affinity of LOTUS with RNAs (Figure 2D and Supplementary Figure S2).
To test whether the RNA binding activity of TDRD5 depends on LOTUS domains and/or other segments of the protein, full-length and truncated segments of TDRD5 were expressed in HEK293T cells and lysates were used to perform pull-down assay using biotinylated poly(GU) and poly(CU) RNA oligos. Full-length TDRD5 and TDRD5 N-terminal region containing three LOTUS domains selectively interacted with poly(GU) RNA, while the TDRD5 C-terminal region containing the Tudor domain did not interact with RNA oligos tested (Figure 2G). TDRD5 N-terminal region also showed weak interaction with poly(CU) RNA, suggesting that multiple LOTUS domains may enhance overall RNA binding activity to substrate RNAs (Figure 2G). Taken together, we found that LOTUS domains are a novel RNA binding domain that selectively binds to G-rich RNAs with high affinity.
LOTUS domains do not bind double-stranded RNA (dsRNA)
Since LOTUS domains of TDRD5 specifically bound with G-rich single-stranded RNA (ssRNA) oligos tested, we next tested whether LOTUS domains bind double-stranded RNA (dsRNA) with the same G-rich sequences. We mixed and annealed two complementary ssRNA oligos to form dsRNA. Among them, poly(GU)–poly(CA) or poly(GA)–poly(CU) mixtures would form dsRNA, while non-complementary poly(GU)–poly(CU) or poly(GA)–poly(CA) mixtures would not form dsRNA, and thus remaining as ssRNA mixture. We performed ELISA using TDRD5 L1 and dsRNA or ssRNA oligo mixtures. TDRD5 L1 showed no binding to the mixture of poly(GU)–poly(CU) and poly(GA)–poly(CA) mixtures that represent dsRNAs (Supplementary Figure S3). In contrast, TDRD5 L1 showed binding activity with poly(GU)–poly(CU) and poly(GA)–poly(CA) mixtures that could not form dsRNAs (Supplementary Figure S3). These results indicate that LOTUS domains preferentially interact with G-rich ssRNA, but not dsRNA.
LOTUS domains bind G-quadruplex RNA
RNA G-quadruplexes (G4s) are unique G-rich RNA tertiary structures found in coding and noncoding RNAs from bacteria to eukaryotes (14) (Figure 3A). Increasing evidence shows that G4 forming sequences are enriched in intergenic piRNA clusters that produce piRNA precursor RNAs (21). Since TDRD5 directly binds piRNA precursors (30) and LOTUS domains selectively recognize G-rich RNA, we hypothesize that TDRD5 LOTUS domains bind RNA G4s. To test LOTUS domain-G4 binding, we performed oligo pull-down assay using three biotin-labeled RNA G4s and their respective mutant forms (Figure 3A). Of these, telomeric repeat-containing RNA (TERRA) G4 is a well-characterized RNA G4 structure; cluster G4–1 (present at an average of 27 times in TDRD5 CLIP libraries) and cluster G4–2 were two G4 forming sequences derived from mouse piRNA cluster sequences. These RNA oligos were annealed to fold into G4 tertiary structures, which were validated by circular dichroism (CD) spectroscopy. All three RNA G4s displayed the characteristic spectrum of parallel G4 with a positive peak at ∼260 nm and a negative peak at ∼240 nm (Supplementary Figure S4) (35). As expected, all mutant G4s in which GGG were replaced with CCG to prevent G4 structure formation did not show such peak patterns, indicating failure of these mutant sequences to fold into G4 tertiary structures (Supplementary Figure S4). Using oligo pull-down assay, we showed that L1 and L2 of TDRD5 selectively bound to all three G4s, but not their respective G4 mutants. Similar to previous results, TDRD5 L3 did not show any interactions with any RNA tested (Figure 3B). We further performed EMSA assays using 5-carboxyfluorescein (5-FAM) labeled G4 and G4 mutant RNAs. Results showed that TDRD5 L1 and L2 but not L3 displayed specific gel shift with RNA G4s, consistent with the results from RNA pull-down assays (Supplementary Figure S5A–C). We also performed ELISA and showed that high affinity interactions of LOTUS domains with G4s were in the 20–50 nM range (Figure 3C). To further confirm the binding affinity of the LOTUS–G4 interaction, we performed DRaCALA RNA binding assay (Supplementary Figure S5D). TDRD5 L1 exhibited a nanomolar binding affinity to G4, consistent with the ELISA results. Together, these data demonstrate that the TDRD5 LOTUS domains bind to RNA G4s with high affinity.
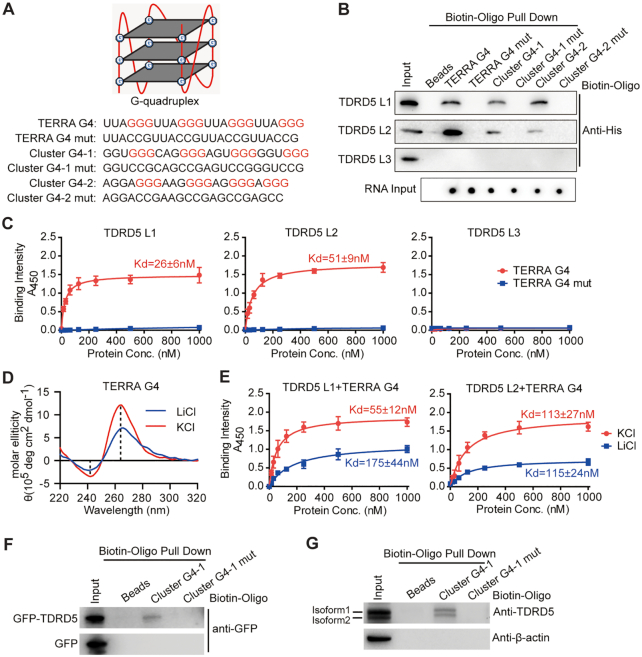
Figure 3.
TDRD5 LOTUS domains bind to folded RNA G-quadruplex structure. (A) Sequences of different RNA G4s. Schematic representation of a RNA G-quadruplex (G4) formed by a G-rich sequence was shown at the top. (B) In vitro RNA pull-down analysis of TDRD5 LOTUS domains. Biotin-labeled RNA G4 oligonucleotides and their respective mutants (mut) were used in the RNA pull-down assay as described in (A). (C) TDRD5 LOTUS domains bind RNA G4 with high affinity. ELISA assay was performed with three individual TDRD5 LOTUS domains (L1, L2 and L3) and indicated RNA oligos. The concentration (conc.) of LOTUS domains used is shown on the x-axis of each graph. TDRD5 L1 and L2 but not L3 bind RNA G4. n = 3, error bars represent s.e.m. Dissociation constants (Kd) are indicated. (D) Circular dichroism spectroscopy of TERRA G4 RNA oligos in KCl or LiCl buffers. (E) ELISA assay was performed with TDRD5 LOTUS domains and TERRA G4 RNA oligos in KCl or LiCl buffers. n = 3, error bars represent s.e.m. Dissociation constants (Kd) are indicated. (F) RNA G4 binds ectopically expressed full-length TDRD5. HEK293T cells were transfected with GFP-TDRD5 plasmid or GFP empty plasmid as a negative control. Biotin-labeled RNA G4 oligonucleotides were annealed and bound to streptavidin beads. After incubating with cell lysates, Western blotting was performed with an anti-GFP antibody. (G) RNA G4 binds endogenous TDRD5 from mouse testes. Biotin-labeled G4 oligonucleotides were annealed and bound to streptavidin beads. After incubating with adult mouse testis lysates, Western blotting was performed with TDRD5 and β-actin antibodies. TDRD5 isoform1 and isoform2 are shown.
We next used another approach to validate that LOTUS domain-G4 interaction depends on G4 tertiary structure rather than primary sequences. K+ is required for the stabilization and maintenance of G4 structure, while Li+ does not stabilize G4 structure (28). After annealing TERRA G4 oligo in buffer containing KCl or LiCl, CD spectroscopy confirmed that TERRA G4 oligo displayed the characteristic spectrum of parallel G4 in KCl buffer and this characteristic spectrum was diminished in the presence of LiCl (Figure 3D). We then performed ELISA assay to detect the LOTUS–G4 binding in the presence of KCl or LiCl. Both L1 and L2 of TDRD5 bound TERRA G4 with a high affinity in KCl buffer, while their interactions were significantly reduced in LiCl buffer (Figure 3E). As a control, the interaction of LOTUS domain with poly(GU) RNA, which could not fold into G4 structure, was not affected in the presence of LiCl (Supplementary Figure S6A). These results indicate that LOTUS domains recognize G4 tertiary structure.
We next examined the effect of various types of TERRA G4 mutations on LOTUS domain binding (Supplementary Figure S6B). We disrupted G4 formation by changing G-contents or the positions of Gs in mutant G4 oligos. CD spectroscopy showed that all four TERRA G4 mutants could not fold into G4 structure (Supplementary Figure S6C). ELISA results showed that TERRA G4 mutants displayed either no interaction or reduced interaction with TDRD5 L1 (Supplementary Figure S6D). It is worth noting that TERRA G4 mut4 had reduced binding to TDRD5 L1 despite having the same G content as wild-type TERRA G4, suggesting that G4 tertiary structure promotes LOTUS domain in recognition of G-rich RNAs.
Since individual TDRD5 LOTUS domains interact with RNA G4s, we wondered whether endogenous full-length TDRD5 protein is capable of interacting with RNA G4s. We analyzed G4 forming RNA sequences in TDRD5 CLIP reads and found that TDRD5 CLIP reads had a mild enrichment for G4 forming sequences compared with random reads from the mouse genome (Supplementary Figure S7). To directly test whether full-length TDRD5 interacts with RNA G4s, we expressed GFP-tagged TDRD5 in HEK293T cells and used cell lysates to perform the G4 oligo pull-down assay. Full-length TDRD5 interacted with G4 derived from piRNA clusters, but not with G4 mutant (Figure 3F). The same oligo pull-down assay using adult mouse testis lysates showed the same result, suggesting that TDRD5 could bind piRNA precursors through recognition of G4 sequences in vivo (Figure 3G). Taken together, we conclude that TDRD5 LOTUS domains interact with RNA G4s and the folded G4 tertiary structure enhances LOTUS domain binding.
LOTUS domain preferentially recognizes RNA G4 but not DNA G4
Both RNA and DNA sequences can fold into G4 structures. While RNA only forms parallel G4 structure, DNA can form parallel G4 or anti-parallel G4 structure (35,36). We next examined whether LOTUS domains interact with G4 structure formed by DNA sequences. We used DNA oligos of TERRA G4, cluster G4–1 and cluster G4–2 and their respective mutants described above by annealing to promote DNA G4 structure formation. CD spectroscopy revealed that cluster G4–1 DNA and cluster G4–2 DNA oligonucleotides folded into parallel G4, while TERRA G4 DNA formed anti-parallel G4 (Supplementary Figure S8A) (37). G4 mutant DNA oligos did not form G4 structure (Supplementary Figure S8A). ELISA binding assay revealed that TDRD5 L1 bound with TERRA RNA G4, but not TERRA DNA G4, suggesting LOTUS domains preferentially recognize RNA G4, but not DNA G4 (Supplementary Figure S8B). Interestingly, LOTUS domains showed very weak but detectable binding to cluster G4–1 and cluster G4–2 DNA G4 that folded into parallel G4 structure (Supplementary Figure S8B). This result suggests that tertiary parallel G4 structures play a role in enhancing the binding of the LOTUS domain to its substrate.
LOTUS domain–G4 RNA interactions are evolutionarily conserved
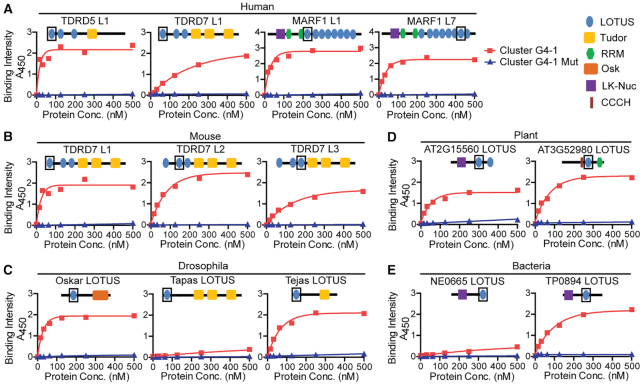
The LOTUS domain fold is highly conserved in bacteria, plants and animals with a topology of three α-helices and two β-sheets that adopt a winged helix-turn-helix conformation (Supplementary Figure S9). We next tested whether the RNA binding activity of LOTUS domains to G4 is evolutionarily conserved. In mammals, only three genes encode LOTUS domain-containing proteins: TDRD5, TDRD7 and MARF1. Both TDRD5 and TDRD7 have three tandem LOTUS domains while MARF1 contains eight tandem LOTUS domains. Given that mouse TDRD5 L1 and L2 have G4 RNA binding activity, we further purified more mammalian LOTUS domains including mouse TDRD7 L1, L2 and L3, human TDRD5 L1, human TDRD7 L1, human MARF1 L1 and L7 to test their RNA binding activities (Supplementary Figure S10 and Supplementary Table S2). ELISA assay revealed that all of these LOTUS domains bound with both cluster G4–1 and TERRA G4 RNAs, but not their respective G4 mutants (Figure 4A and B, Supplementary Figure S11A and S11B), indicating conserved G4 binding activity among different mammalian LOTUS domain proteins. We next tested G4 RNA binding activities of three Drosophila LOTUS domains from three different Drosophila proteins Oskar, Tejas and Tapas, each containing a single LOTUS domain (Supplementary Figure S10). ELISA assay showed that LOTUS domains from Oskar and Tejas specifically bound with RNA G4, but not G4 mutants (Figure 4C, Supplementary Figure S11C). However, Tapas LOTUS domain showed minimal G4 RNA binding activity (Figure 4C and Supplementary Figure S11C). We further purified two representative LOTUS domains from plants (AT2G15560-LOTUS and AT3G52980-LOTUS) and two from bacteria (TP0894-LOTUS and NE0665-LOTUS) (Supplementary Figure S10) (2). Both plant and bacterial LOTUS domains specifically bound with G4s, but not G4 mutants. One exception was the bacterial NE0665-LOTUS domain, which showed no significant G4 RNA binding activity (Figure 4D and E, Supplementary Figure S11D and S11E). Taken together, these results demonstrate that LOTUS domain–G4 RNA interaction is an ancient and widespread protein-RNA interaction in both prokaryotes and eukaryotes that may have broad biological implications.
Figure 4.
LOTUS domain–RNA G4 interaction is evolutionarily conserved. (A–E) Binding of representative LOTUS domains from diverse species to RNA G4 (Cluster G4–1 and its mutant) was determined by ELISA assay. A representative graph of 2–3 technical repeats is shown for each LOTUS domain. (A) Human LOTUS domains. (B) Mouse LOTUS domains. (C) Drosophila LOTUS domains. (D) Plant LOTUS domains. (E) Bacterial LOTUS domains. His-tagged LOTUS domains were used to bind immobilized biotin-G4 or its G4 mutant in the ELISA assay. The concentration (conc.) of LOTUS domains used is shown on the x-axis of each graph. The position of each LOTUS domain used is shown as a box in cartoons of protein architecture.
Ancient RNA binding and newly evolved protein binding of the LOTUS domain family
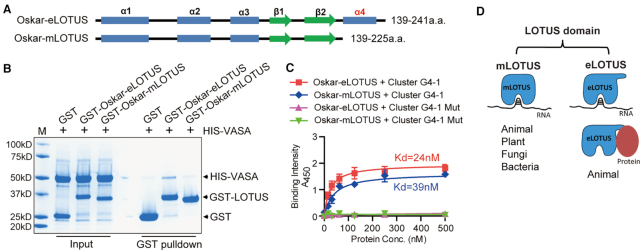
The LOTUS domain family is divided into two subclasses: the extended LOTUS (eLOTUS) and minimal LOTUS (mLOTUS) based on the presence or absence of a C-terminal helix (Figure 5A and Supplementary Figure S12) (11). eLOTUS, but not mLOTUS, exhibits conserved protein binding activity toward germ cell RNA helicase Vasa/DDX4 involving this C-terminal extension (11). In particular, the eLOTUS of Drosophila Oskar shows binding activity to Vasa, although no RNA binding activity has been detected (9–11). In contrast, we show from above results that the eLOTUS of Drosophila Oskar displayed high affinity binding to RNA G4 (Figure 4C). However, it is unclear whether eLOTUS and mLOTUS of Drosophila Oakar both display RNA binding activity as well as protein binding activity. To test this, we expressed and purified both eLOTUS and mLOTUS from Drosophila Oskar and performed protein binding assay as well as RNA binding assay (Figure 5A). Consistent with published results, Oskar eLOTUS interacted with Vasa while mLOTUS that lacks the C-terminal extended helix did not interact with Vasa by GST pull-down assay (Figure 5B) (11). We next tested the RNA binding activity for Oskar eLOTUS and mLOTUS. ELISA assay showed that both Oskar eLOTUS and mLOTUS interacted with G4 RNA with high affinity, but not with mutant G4s, suggesting the C-terminal extended helix in eLOTUS is not required for RNA binding activity of LOTUS domains (Figure 5C). Given that mLOTUS lacking the C-terminal extension exist in bacteria, fungi, plants and animals while eLOTUS are only present in animals (Supplementary Figure S12), these data suggest that RNA binding, rather than protein binding, is the most ancient binding activity of the LOTUS domain family and that eLOTUS harbor both RNA and protein binding capacity for cellular function (Figure 5D).
Figure 5.
Drosophila Oskar eLOTUS exhibits both RNA and protein binding activity. (A) Schematic illustration of secondary structures of Oskar extended LOTUS (eLOTUS) and minimal LOTUS (mLOTUS). (B) Oskar eLOTUS but not mLOTUS interacts with Vasa. GST pull-down assay was performed using GST, GST-Oskar eLOTUS or GST-Oskar mLOTUS against His-Vasa (200–661aa). (C) Both Oskar eLOTUS and mLOTUS interact with RNA G4. ELISA assay was performed with Oskar eLOTUS or mLOTUS and indicated RNA oligos. n = 2. Dissociation constants (Kd) are indicated. (D) A proposed model illustrating different binding modes of mLOTUS and eLOTUS subfamilies.
DISCUSSION
We reveal here unexpected RNA binding property of the LOTUS domain family conserved in bacteria, plants to animals. These data suggest that LOTUS domain proteins are a novel class of RNA binding proteins capable of directly engaging RNA regulation through LOTUS domains. We show that the LOTUS core domains of mLOTUS and eLOTUS from prokaryotes to eukaryotes have previously unrecognized binding preference to RNA G4 structure. Remarkably, we demonstrate that animal eLOTUS with the C-terminal extension have both RNA binding and protein binding properties. Together these findings unify the current controversial views on LOTUS domain binding property by revealing the most conserved binding as RNA binding.
We uncovered a unique binding preference for the LOTUS domain family to G-rich primary sequences and G4 tertiary structure. The binding of LOTUS domains to other non-G-rich RNAs tested thus far was negative. This special binding feature could explain why previous reports studying Drosophila Oskar eLOTUS domain failed to detect LOTUS domain RNA binding ability but rather identified a binding to animal Vasa protein (9–11). We show the same Drosophila Oskar eLOTUS domain has a high affinity to RNA while exhibiting binding to Vasa protein. The ability of LOTUS domain to associate with Vasa is eLOTUS C-terminal extension dependent because mLOTUS shows only RNA binding but not protein binding activity. Based on the novel bimodal binding capacity for eLOTUS domains, we propose that the capability for LOTUS domains to engage both RNA and protein provides a new mode of action for animal eLOTUS domain proteins such as Oskar, TDRD5, and TDRD7 (Supplementary Figure S12). This mechanism could efficiently couple RNA binding to protein complexes involving RNA regulation to promote germ plasm/nuage formation for germ cell development.
We discovered that RNA binding is the most ancient binding for the entire LOTUS domain family by comprehensive testing both mLOTUS and eLOTUS across diverse species. This is consistent with the original prediction that first defined the LOTUS domains (1,2). Unexpectedly, this RNA bind activity shows selective specificity to G-rich and G4 containing RNA. This ancient RNA binding feature can be traced back to bacterial mLOTUS of MARF1-like proteins that contain mLOTUS and NYN nuclease domains (2). We also show that plant MARF1-like protein and animal MARF1 retain this conserved RNA binding feature. In contrast to bacterial and plant MARF1-like proteins, mammalian MARF1 contains multiple tandem mLOTUS domains that are suggested to bind ssRNA and dsRNA substrates (12). We show individual mLOTUS of MARF1 has high affinity binding to G4 RNA and propose that tandem mLOTUS configuration may enhance the overall RNA binding activity to substrate RNA. Consistent with this, three tandem LOTUS domains of TDRD5 showed weak but detectable interaction with poly(CU) RNA oligos (Figure 2G). This also explains why TDRD5-bound RNA sequence reads have significant but mild G and G4 enrichment from TDRD5 CLIP-seq experiments (Figure 1 and Supplementary Figure S7). Some LOTUS domain proteins also harbor other type of RNA binding motifs. It is conceivable that tandem LOTUS domains in combination with distinct RNA binding motifs could together expand the RNA binding variety for LOTUS domain-containing proteins to exert their unique functions in different biological contexts.
Our results suggest a strong link between LOTUS domain proteins and RNA G4 regulation. This is particularly relevant in the context of mammalian piRNA biogenesis in which we detected specific LOTUS domain-G4 RNA interaction from piRNA biogenesis factor TDRD5 (30). piRNA precursors harbor numerous G4 forming sequences and our results indicate that LOTUS domains of TDRD5 bind to G4s derived from piRNA precursors. Emerging evidence has shown that the relative positioning and frequency of G4 sequences along piRNA precursors create ‘hot spots’ for piRNA processing into mature piRNAs (21,23). This raises the possibility that G4 may exist as a structural mediator for the processing of piRNA precursors. We propose that the piRNA precursor processing is driven by the binding of piRNA precursors to the LOTUS domains of TDRD5 via the G-rich/G4 structural motifs. This binding stabilizes precursor association with the piRNA processing machinery, promoting its processing into piRNAs. The prevalence of G4 sequences in TDRD5-regulated piRNA precursors and the ability of TDRD5 and its LOTUS domains to directly bind G4s in vitro and ex vivo suggest that TDRD5 functions as a new class of RNA binding protein in piRNA precursor processing through direct RNA structural motif recognition. Interestingly, another piRNA biogenic RNA helicase MOV10L1 directly binds G-rich and G4 forming sequences. MOV10L1 unwinds RNA structures including G4 to promote the endonucleolytic cleavage of piRNA precursors (23). We therefore speculate that TDRD5 acts as a G4 binding protein to coordinate with MOV10L1 in directing piRNA precursor unwinding and processing. This underscores an important involvement of recognizing and dissolving RNA elements during piRNA biogenesis.
Together, our study reveals that LOTUS domains are a novel family of protein folds with conserved high affinity binding to RNA G4. It is currently unclear how LOTUS domains preferentially recognize G-rich sequences and G4 structures. Our data indicate that both primary sequence and tertiary structure of RNA are involved in LOTUS-G4 interaction. Future studies on the structural basis and biological significance of this specific LOTUS domain-RNA interaction will shed light on the molecular mechanism whereby LOTUS domain proteins play important roles in posttranscriptional RNA regulation in diverse prokaryotic and eukaryotic species.
Supplementary Material
ACKNOWLEDGEMENTS
We thank X. Cheng for critical reading of the manuscript, G. Smith, J. Ireland for sharing equipment, and H. Kim, Y. Wu, T. Zhang, D. Sui for technical assistance.
Contributor Information
Deqiang Ding, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA; Clinical and Translational Research Center of Shanghai First Maternity and Infant Hospital, Frontier Science Center for Stem Cell Research, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Chao Wei, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA.
Kunzhe Dong, USDA Agricultural Research Service, Avian Disease and Oncology Laboratory, East Lansing, MI 48823, USA.
Jiali Liu, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA; State Key Laboratory for Agrobiotechnology, College of Biological Sciences, China Agricultural University, Beijing 100193, China.
Alexander Stanton, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA.
Chao Xu, Division of Molecular and Cellular Biophysics, Hefei National Laboratory for Physical Sciences at the microscale, School of Life Sciences, University of Science and Technology of China, Hefei, Anhui 230027, China.
Jinrong Min, Structural Genomics Consortium, University of Toronto, Toronto, Ontario M5G 1L7, Canada.
Jian Hu, Department of Biochemistry and Molecular Biology, Michigan State University, East Lansing, MI 48824, USA.
Chen Chen, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA; Department of Obstetrics, Gynecology and Reproductive Biology, Michigan State University, Grand Rapids, MI 49503, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
MSU AgBioResearch funds and NIH [R01HD084494, R01GM132490 to C.C., in part]; Fundamental Research Funds for the Central Universities of China [22120200057 to D.D.]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1. Callebaut I., Mornon J.P.. LOTUS, a new domain associated with small RNA pathways in the germline. Bioinformatics. 2010; 26:1140–1144. [DOI] [PubMed] [Google Scholar]
- 2. Anantharaman V., Zhang D., Aravind L.. OST-HTH: a novel predicted RNA-binding domain. Biol. Direct. 2010; 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lehmann R., Nusslein-Volhard C.. Abdominal segmentation, pole cell formation, and embryonic polarity require the localized activity of oskar, a maternal gene in Drosophila. Cell. 1986; 47:141–152. [DOI] [PubMed] [Google Scholar]
- 4. Lachke S.A., Alkuraya F.S., Kneeland S.C., Ohn T., Aboukhalil A., Howell G.R., Saadi I., Cavallesco R., Yue Y., Tsai A.C. et al.. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011; 331:1571–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanaka T., Hosokawa M., Vagin V.V., Reuter M., Hayashi E., Mochizuki A.L., Kitamura K., Yamanaka H., Kondoh G., Okawa K. et al.. Tudor domain containing 7 (Tdrd7) is essential for dynamic ribonucleoprotein (RNP) remodeling of chromatoid bodies during spermatogenesis. PNAS. 2011; 108:10579–10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yabuta Y., Ohta H., Abe T., Kurimoto K., Chuma S., Saitou M.. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J. Cell Biol. 2011; 192:781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Su Y.Q., Sugiura K., Sun F., Pendola J.K., Cox G.A., Handel M.A., Schimenti J.C., Eppig J.J.. MARF1 regulates essential oogenic processes in mice. Science. 2012; 335:1496–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Breitwieser W., Markussen F.H., Horstmann H., Ephrussi A.. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev. 1996; 10:2179–2188. [DOI] [PubMed] [Google Scholar]
- 9. Jeske M., Bordi M., Glatt S., Muller S., Rybin V., Muller C.W., Ephrussi A.. The crystal structure of the Drosophila germline inducer Oskar identifies two domains with distinct Vasa helicase- and RNA-binding activities. Cell Rep. 2015; 12:587–598. [DOI] [PubMed] [Google Scholar]
- 10. Yang N., Yu Z., Hu M., Wang M., Lehmann R., Xu R.M.. Structure of Drosophila Oskar reveals a novel RNA binding protein. PNAS. 2015; 112:11541–11546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jeske M., Muller C.W., Ephrussi A.. The LOTUS domain is a conserved DEAD-box RNA helicase regulator essential for the recruitment of Vasa to the germ plasm and nuage. Genes Dev. 2017; 31:939–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yao Q., Cao G., Li M., Wu B., Zhang X., Zhang T., Guo J., Yin H., Shi L., Chen J. et al.. Ribonuclease activity of MARF1 controls oocyte RNA homeostasis and genome integrity in mice. PNAS. 2018; 115:11250–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhu L., Kandasamy S.K., Liao S.E., Fukunaga R.. LOTUS domain protein MARF1 binds CCR4-NOT deadenylase complex to post-transcriptionally regulate gene expression in oocytes. Nat. Commun. 2018; 9:4031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Millevoi S., Moine H., Vagner S.. G-quadruplexes in RNA biology. Wiley Interdiscipl. Rev. RNA. 2012; 3:495–507. [DOI] [PubMed] [Google Scholar]
- 15. Agarwala P., Pandey S., Maiti S.. The tale of RNA G-quadruplex. Org. Biomol. Chem. 2015; 13:5570–5585. [DOI] [PubMed] [Google Scholar]
- 16. Kumari S., Bugaut A., Huppert J.L., Balasubramanian S.. An RNA G-quadruplex in the 5' UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007; 3:218–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sexton A.N., Collins K.. The 5' guanosine tracts of human telomerase RNA are recognized by the G-quadruplex binding domain of the RNA helicase DHX36 and function to increase RNA accumulation. Mol. Cell. Biol. 2011; 31:736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Didiot M.C., Tian Z., Schaeffer C., Subramanian M., Mandel J.L., Moine H.. The G-quartet containing FMRP binding site in FMR1 mRNA is a potent exonic splicing enhancer. Nucleic Acids Res. 2008; 36:4902–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Phan A.T., Kuryavyi V., Darnell J.C., Serganov A., Majumdar A., Ilin S., Raslin T., Polonskaia A., Chen C., Clain D. et al.. Structure-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nat. Struct. Mol. Biol. 2011; 18:796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. et al.. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014; 507:195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vourekas A., Zheng K., Fu Q., Maragkakis M., Alexiou P., Ma J., Pillai R.S., Mourelatos Z., Wang P.J.. The RNA helicase MOV10L1 binds piRNA precursors to initiate piRNA processing. Genes Dev. 2015; 29:617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ibrahim F., Maragkakis M., Alexiou P., Mourelatos Z.. Ribothrypsis, a novel process of canonical mRNA decay, mediates ribosome-phased mRNA endonucleolysis. Nat. Struct. Mol. Biol. 2018; 25:302–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang X., Yu L., Ye S., Xie J., Huang X., Zheng K., Sun B.. MOV10L1 binds RNA G-quadruplex in a structure-specific manner and resolves it more efficiently than MOV10. iScience. 2019; 17:36–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Guo J.U., Bartel D.P.. RNA G-quadruplexes are globally unfolded in eukaryotic cells and depleted in bacteria. Science. 2016; 353:aaf5371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vasilyev N., Polonskaia A., Darnell J.C., Darnell R.B., Patel D.J., Serganov A.. Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP. PNAS. 2015; 112:E5391–E5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Heddi B., Cheong V.V., Martadinata H., Phan A.T.. Insights into G-quadruplex specific recognition by the DEAH-box helicase RHAU: solution structure of a peptide-quadruplex complex. PNAS. 2015; 112:9608–9613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Takahama K., Takada A., Tada S., Shimizu M., Sayama K., Kurokawa R., Oyoshi T.. Regulation of telomere length by G-quadruplex telomere DNA- and TERRA-binding protein TLS/FUS. Chem. Biol. 2013; 20:341–350. [DOI] [PubMed] [Google Scholar]
- 28. Zheng S., Vuong B.Q., Vaidyanathan B., Lin J.Y., Huang F.T., Chaudhuri J.. Non-coding RNA generated following Lariat debranching mediates targeting of AID to DNA. Cell. 2015; 161:762–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thandapani P., Song J., Gandin V., Cai Y., Rouleau S.G., Garant J.M., Boisvert F.M., Yu Z., Perreault J.P., Topisirovic I. et al.. Aven recognition of RNA G-quadruplexes regulates translation of the mixed lineage leukemia protooncogenes. eLife. 2015; 4:e06234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ding D., Liu J., Midic U., Wu Y., Dong K., Melnick A., Latham K.E., Chen C.. TDRD5 binds piRNA precursors and selectively enhances pachytene piRNA processing in mice. Nat. Commun. 2018; 9:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li X.Z.G., Roy C.K., Dong X.J., Bolcun-Filas E., Wang J., Han B.W., Xu J., Moore M.J., Schimenti J.C., Weng Z.P. et al.. An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol. Cell. 2013; 50:67–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lombardi E.P., Londono-Vallejo A.. A guide to computational methods for G-quadruplex prediction. Nucleic Acids Res. 2020; 48:1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Roelofs K.G., Wang J., Sintim H.O., Lee V.T.. Differential radial capillary action of ligand assay for high-throughput detection of protein-metabolite interactions. PNAS. 2011; 108:15528–15533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen C., Nott T.J., Jin J., Pawson T.. Deciphering arginine methylation: tudor tells the tale. Nat. Rev. Mol. Cell Biol. 2011; 12:629–642. [DOI] [PubMed] [Google Scholar]
- 35. Fay M.M., Lyons S.M., Ivanov P.. RNA G-quadruplexes in biology: principles and molecular mechanisms. J. Mol. Biol. 2017; 429:2127–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brazda V., Haronikova L., Liao J.C., Fojta M.. DNA and RNA quadruplex-binding proteins. Int. J. Mol. Sci. 2014; 15:17493–17517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Biffi G., Tannahill D., McCafferty J., Balasubramanian S.. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013; 5:182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.