Abstract
KRAS is one of the most mutated oncogenes and still considered an undruggable target. An alternative strategy would consist in targeting its gene rather than the protein, specifically the formation of G-quadruplexes (G4) in its promoter. G4 are secondary structures implicated in biological processes, which can be formed among G-rich DNA (or RNA) sequences. Here we have studied the major conformations of the commonly known KRAS 32R, or simply 32R, a 32 residue sequence within the KRAS Nuclease Hypersensitive Element (NHE) region. We have determined the structure of the two major stable conformers that 32R can adopt and which display slow equilibrium (>ms) with each other. By using different biophysical methods, we found that the nucleotides G9, G25, G28 and G32 are particularly implicated in the exchange between these two conformations. We also showed that a triad at the 3′ end further stabilizes one of the G4 conformations, while the second conformer remains more flexible and less stable.
INTRODUCTION
Cancer is one of the most predominant causes of death around the world, and according to the International Agency for Research on Cancer (IARC) represents a major issue for human health. Among the different types of cancer, some deadly versions are still without therapeutic solutions. Such examples include lung, pancreatic and colorectal cancers, where KRAS is particularly implicated (1–4). KRAS is one of the most mutated oncogenes located on chromosome 12 and is responsible for the coding of a G-protein of 23.2 kDa (5–7). It plays an important role in cell proliferation, differentiation and survival. Recent studies have also found that KRAS reprograms the metabolism and controls the redox homeostasis in pancreatic cancer (8,9). The KRAS protein promotes oncogenic events switching from an inactive form bound to GDP, to a constitutively active form bound to GTP (10,11). However, even if its implication is well documented in a broad range of cancers, KRAS is considered to be nearly undruggable and thus remains one of the least successful therapeutic targets, with limited treatments and huge side effects (12,13). A broad range of therapies against KRAS-mediated cancers have been developed in the past decades. Most of these therapies have been designed to target either the KRAS protein directly, the enzymes involved in its association to the plasma membrane, or the downstream effector pathways (9,14–17). Moreover, recent discoveries on the effect of KRAS on cancer metabolism gave new therapeutic opportunities (18). As the anti-KRAS strategies currently tested did not give satisfactory results for clinical applications (8), new therapeutic approaches focus on upstream elements, by targeting mRNA or directly the gene and its promoter regions (19).
The promoter sequence of several oncogenes, including KRAS, is highly rich in guanine nucleotides thus allowing the formation of non-canonical structures called G-quadruplexes (G4) (20,21). G4 consists of the stacking of at least two G-tetrads, or G-quartets, which are planar structures involving four guanines linked in a Hoogsteen hydrogen bonds network (22,23). These structures are fundamentally different from canonical double helical DNA; they can be highly polymorphic depending on base composition, number of strands or loop length (24–27). As they are present in several key regulatory regions of genes, G-quadruplexes structures have been tested as possible therapeutically targets (28–30), including KRAS (12,14,31–33). It is particularly attractive to hypothesize that small ligands may bind specifically to G4 structures without having any influence on double-stranded DNA. Such ligands can compete with the binding of nuclear proteins to the critical G4 in the KRAS promoter and suppress transcription (34–36). This would reduce the activity of the oncogene near the beginning of the network pathway in KRAS metabolism. In this study, we examined the major structure of the commonly known KRAS 32R, or 32R, a 32-nucleotide sequence within the KRAS Nuclease Hypersensitive Element (NHE) region (32,37). The high resolution structural information of this noncanonical DNA element may help reveal its folding mechanism and its role in KRAS regulation. In addition, the 3D structure is necessary for rational drug design and validation of new targets for KRAS-related cancers. Previous studies have already reported that the 32R sequence corresponds to the minimal interaction domain of several transcription factors and related proteins such as MAZ, PARP-1 and hnRNP A1 (38,39). The most likely function of 32R G4 is to recruit nuclear factors forming the transcription pre-initiation complex to the KRAS promoter. By means of primer extension experiments with Taq polymerase (40), it has been found that 32R forms a G4 structure involving the entire 32-mer sequence. In the presence of a G4-stabilizing ligand, 32R showed also another folding which engages only 21 of the nucleotides (sequence called 21R) (40). Both the 32R and 21R G4 structures of KRAS have been demonstrated to interact with nuclear proteins (31,39,40). Several studies have been conducted to address questions about the KRAS G4 structures, including one of our previous studies where we proposed some structural insights by resolving the structure of a G4 formed by a sequence derived from 21R (41,42), and used this DNA to screen against a chemical library for G4 ligands. Up to now, the structure proposed for the larger 32R sequence has only been obtained by dimethyl sulfate (DMS) footprinting and CD experiments (39). A striking feature coming from these studies is that 32R folds into a G4 characterized by a long loop of 11 nt, incorporating a G-run that surprisingly did not take part in the formation of the G-tetrad core. Moreover, it was discovered that 32R exhibited an extensive structural polymorphism, as the deletion of a G-run at the 5′-end resulted in a dimeric G4 structure (resolved by NMR) having the ability to inhibit oncogenic KRAS in a cancer cell line (43). In the present study, we demonstrate for the first time by high-resolution solution state NMR methods that the entire 32R sequence of the KRAS promoter folds in two G4 conformers that coexist in equilibrium. The results of our study represent an advancement in understanding the folding of a critical sequence of the KRAS promoter where the transcription pre-initiation complex is formed.
MATERIALS AND METHODS
DNA samples preparation
Uniformly {13C, 15N}-labeled KRAS 32R WT DNA was purchased from Silantes GmbH. G9T and G25T samples were produced in our lab using M-MuLV enzyme from New England Biolabs and dNTPs from Eurisotop. Non-labelled samples were purchased from IDT (Integrated DNA Technologies; USA). They were synthesized on the 250 nmol scale and purified by reverse-phase HPLC. Samples labelled site-specifically at a level of 5% 15N, 13C were synthesized in our laboratory (INSERM U1212, Bordeaux) on an automated Expedite 8909 DNA synthesizer at 1 μmol scale on a 1000-Å primer support (Link Technologies SynBase CPG). All the standard phosphoramidites (dABz, dT, dGiBu, dCAc), reagents, and solvents used during the synthesis were purchased from Glen Research. The dGiBu-phosphoramidite (U-13C10, 98%; U-15N5, 98%; CP 95%) was purchased from Cambridge Isotope Laboratories. After the synthesis, the oligonucleotides were cleaved from the support and the nucleobases were deprotected with ammonium hydroxide at 55°C for 16 h and then lyophilized. Oligonucleotide solutions were prepared in 1× buffer (10 mM K2HPO4/KH2PO4; 50 mM KCl; pH 6,6). In order to form G-quadruplexes, samples are heated at 95°C for 5 min then quickly cooled in ice. This process was repeated four times and the final sample kept at 4°C. All NMR samples presented in the manuscript were re-synthesized and each spectra repeated more than once.
Circular dichroism
All circular dichroism (CD) experiments used a Jasco J-1500 CD spectropolarimeter using Spectra Manager software. Quartz cuvettes were used and contained 500 μl of 5 μM oligonucleotide samples in 1× buffer. The CD spectra were measured in the region between 220 and 320 nm with a scan speed of 50 nm/min and a response time of 1 s. Three scans were collected and averaged. All experiments were done at 37°C. CD melting studies were performed on 5 μM DNA sample. CD melting was performed in the single wavelength mode. The data were collected at several wavelengths. Temperature was changed from 10 to 90°C at 10°C intervals, with a rate of 0.4°C/min between intervals. These parameters lead to an overall acquisition time of 1 h per 10°C temperature change.
Native gel electrophoresis
Native gel experiments were performed at 4°C using 8% acrylamide/bisacrylamide (19:1 ratio) gels containing 10 mM KCl and 1× TBE buffer with a running buffer also containing 10 mM KCl and 1× TBE buffer. A low molecular weight DNA ladder from New England Biolabs (range from 25 to 766 bp) was used for each gel. Each sample was prepared at a final concentration at 25 μM in 20 μl of 1× buffer with 6 μl sucrose. Before loading samples, the gel was pre-run for 1 h in order to eliminate impurities. The samples were then loaded and migrated for 3 h at a constant voltage 110 V. Stains-All dye was used to reveal the oligonucleotide bands.
NMR spectroscopy
NMR spectra were recorded on Bruker Advance 700, 850 and 950 MHz spectrometers equipped with cryogenically cooled probes. Experiments were performed at 37°C. For solution NMR, standard 3 mm NMR tubes were used. The samples were prepared in 1× buffer with 10% D2O added for the lock. The concentration of DNA samples was between 0.35 and 2 mM. 15N and 13C HSQC experiments were performed using fully-labelled samples or with 5% low-enrichment 15N, 13C site-specific labeling for assignment of imino protons. To unambiguously assign the intranucleotide exchangeable imino to the nonexchangeable purine H8 and pyrimidine H6 protons through-bond connectivities, we used a HCCNH-TOCSY spectrum in uniformly {13C, 15N}-labeled DNA (37,38). Melting experiments were performed using 500 μM of oligonucleotides. The NMR tube was put into the spectrometer at a start temperature of 4°C (277K). After 5 min, enough time for temperature stabilization, a 1H spectrum was recorded. The same process was repeated at increasing temperatures until a maximum temperature of 60°C (333 K) was reached.
Structure calculation
32R G9T and G25T G-quadruplex structures were calculated using ARIA2/CNS based on NMR-derived distance and dihedral constraints (39,40). Distance constraints were manually extracted from peak volumes integrated in Sparky and CCPNMR software and classified as weak (4.0–6.5 Å), medium (2.5–4.5 Å), and strong (1.8–3 Å). Dihedral angles were constrained based on intraresidue H1′–H8 NOE peak intensities, and the canonical B-DNA backbone conformation was used for the stem–loop. After calculation, two different structures which had lower violations were chosen for AMBER refinement. More detailed protocols are provided in Supplementary Information.
RESULTS AND DISCUSSION
KRAS 32R WT forms two major distinct conformers and different topologies
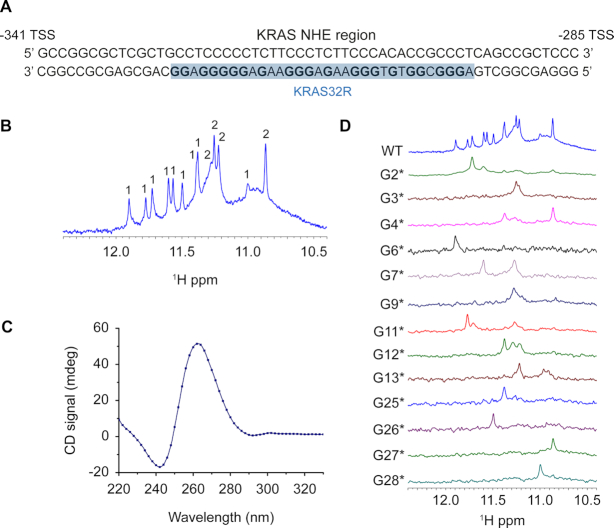
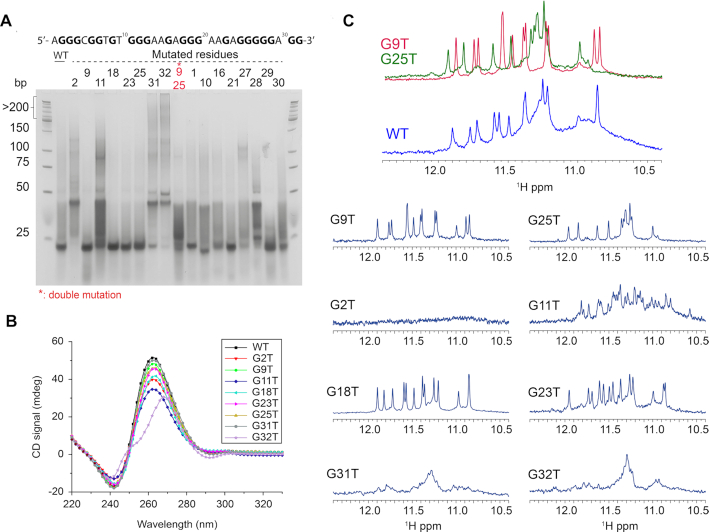
In this work, we have used solution NMR spectroscopy to determine the high-resolution structure of the 32R sequence containing six tracts of guanines from the KRAS NHE region (Figure 1A). The 1D 1H NMR spectrum of 32R is depicted in Figure 1B. The spectrum shows seven well-resolved single peaks and four double-imino peaks. The double imino peaks were later resolved by inspection of 2D {1H–1H} NOESY spectra (such as in Figure 3). In addition, other non-resolved broad regions are present such as near 11 ppm. Taken together, the number of imino peaks exceed the number expected for a single conformation, and the peak broadening also suggests structural polymorphism. We next used CD spectroscopy to inspect the polarity of the chain. The CD spectrum shows a clear maximum at 260 nm and a minimum at 240 nm (41,42) (Figure 1C), indicative of a parallel G4 structure. Next, we used 1D 15N-labeling filtered NMR experiments with a series of 32R samples, each bearing a different site-specific labelled guanine residue. The site-specific 15N- isotope enrichment at ∼5% (43), was sufficient for individual identification of specific guanines. This set of several interesting results were obtained from the individual spectra (Figure 1D). Some nucleobases produced clear individual peaks, such as G25–G28, while the majority of the guanine bases including G3, G7 or G13 clearly depicted two or more peaks. All samples presented in the figure were re-synthesized and each respective spectra repeated twice. Taken together, the NMR results support either the formation of an asymmetric dimer, the existence of a large G-tetrad core composed by more than three tetrads, or a mixture of conformers. Native polyacrylamide electrophoresis for the native sequence 32R and single G→T substitutions is shown in Figure 2A. The native gels allowed us to exclude dimer formation, except for 9–25* that represents a double substituent. The corresponding CD spectrum for each mutant is displayed in Figure 2B. We also observed that the rest of the guanines did not have observable 15N-filtered imino peaks (Supplementary Figure S1). Cogoi et al. (44,45) previously used DMS-footprinting to demonstrate a parallel G-quadruplex structure composed by three G-tetrads and three loops and excluded a dimer possibility. Although some mutants (G2T, G11T, G31T, G32T) showed a gel-band corresponding to a hypothetical dimer, these sequences presented a 1D 1H NMR spectra significantly different from what was observed for the 32R native sequence in Figure 2C. These mutated DNA sequences have a very complicated 1D NMR spectrum with broad peaks that support the presence of oligomerization. Altogether, the mutation results imply important structural rearrangements upon G→T changes emphasizing the importance of those respective guanines. The exception is G2T which does not display any imino peak in contrast to that observed for the other G→T mutants. The results also indicate that high order and/or unstable species not observed in the NMR time scale are present, since the CD signal and the native gel indicate the presence of parallel G4 structures with exception for the CD spectrum of G32T. Other base mutations (X→T) were tested (Supplementary Figure S2) in order to understand the role of each residue in the structure of 32R. The respective 1H NMR spectrum of the imino regions showed different results depending on the mutated residue and we could distinguish four different categories of mutations (Supplementary Figure S2). (a) Mutations of residues already known to be important for the folding of the G-quadruplex core, confirmed by 15N-filtered experiments. These substitutions lead expectedly to a dramatic effect on the G-quadruplex imino region. For example, the G2 or G11 mutations seem to disrupt completely the imino folding pattern that otherwise should be responsible for the existence of a single conformation in the sample, but when analyzed by CD an all-parallel conformer seems to exist. The CD spectra therefore reveal a broad mixture of parallel conformers, which reduce significantly the signal in the NMR spectra giving the impression that the G4 is almost inexistent. The respective native gel sample displays a band compatible with a dimer or some other high order species. (b) Mutations of some of the guanine bases not implicated in the tetrad core, such as G18 and G23, resulted in much cleaner spectra with sharper peaks. We presume that these mutations lead to the elimination of minor conformation sub-states. (c) The last three residues (A30 to G32) mutations unexpectedly had a great impact in the imino peak-pattern, although not as severe as G2 or G11 mutations. A G-quadruplex imino region was clearly visible but without sharp peaks, indicating that a large number of different conformations still co-existed. (d) The last type of G→T mutations were those corresponding to residues identified to be part of the tetrad core, such as G9 and G25, and showed well-resolved spectra. Among those, we can distinguish two patterns of imino peaks. One group composed of G9, G18–G23 and G29, and a second group containing guanines G25, G26, G27 and G28. Hereafter, and for simplification, we refer to those two groups of mutants as G9T and G25T respectively. The spectrum of G9T showed thirteen well-resolved imino peaks. For G25T, it was more difficult to correctly quantify the number of peaks due to the superposition around 11.3 ppm. Both mutants had a parallel signature in CD spectra (Figure 2B) and the native gel showed a unique band with the same migration as the wild type. The imino peaks profile for G25T is more complex and differs from G9T. We can easily identify an overlapped region around 11.2 ppm similar to the one we can observe for 32R WT (Figure 2C). The formation of these two principal groups of imino-peak patterns indicate that two major distinct conformers may coexist among other minor populations, together forming the species observed in the 32R WT sample. We estimated from isolated cross peaks belonging to H2′-H2′ (Supplementary Figure S17) in isolated bases from each conformer, a population distribution of 65–69% for G9T and 35–31% of G25T of folded species. To test the possibility that KRAS 32R WT adopts alternative stable conformers in addition to the G9T and G25T structures, we performed double mutations and inspected these using NMR and CD spectroscopy (Supplementary Figure S3). We observed that the double mutant G9TG25T could not form a proper G4, although multiple peaks with much smaller intensities are visible in the imino region. As observed before, replacing G2 for a thymine severely affects both G4 conformers. We hypothesize that G2 may be important for the folding initiation and stabilization as observed before with WT sequence. Overall, G9T is unique because no other guanine can take its place and give rise to a single well-folded conformer. For example, G9TG32T and G9TG31T double mutants present multiple imino peaks characteristic of multiple conformations. On the other hand, the G25TG26T double mutant still produces a clear imino pattern with twelve sharp peaks similar to what we found with G25T (Supplementary Figure S6). The results indicate that there is a sliding in the last track to move from G25–G27 to G27–G29, which produces a less-stable conformer with an almost identical imino pattern.
Figure 1.
KRAS32R characterization with (A) its localization within KRAS NHE sequence in the promoter region; (B) its 1D proton NMR imino region at 37°C in 700 MHz spectrometer with several countable peaks; (C) its CD spectrum at 37°C with a positive peak around 260 nm and a negative peak around 240 nm characteristic of parallel G4 and (D) determination of the guanines implicated in KRAS 32R tetrads using 15N-filtered NMR spectra of samples containing ≈5% of 15N-enriched isotope. All experiments were performed in buffer 1× (50 mM KCl; 10 mM KPi; pH 6.66).
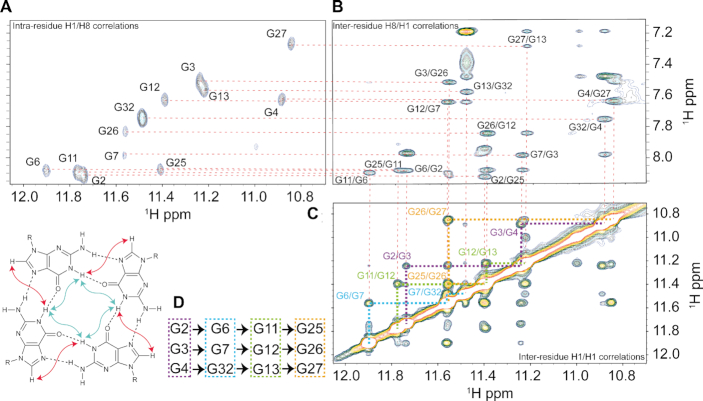
Figure 3.
(A) NMR HCCNH-TOCSY experiment of KRAS 32R G9T showing intra-residue H1/H8 correlations in order to identify H8 of guanines implicated in G9T G-quadruplex and {1H–1H} NOESY inter-residue H8/H1 correlations in (B) and in (C) {1H–1H} NOESY inter-residue H1/H1 correlations were used to determine tetrads pattern in (D). All experiments were performed at 37°C in buffer 1× (50 mM KCl; 10 mM KPi; pH 6.66).
Figure 2.
(A) Native gel experiment of KRAS 32R and several simple mutants and one double mutant showing that some mutants harbor the same conformation as wild type whereas several mutants such as G2T or G32T severely impact G4 formation; (B) the CD spectra of KRAS 32R mutants at 37°C show that most of the mutations do not affect G4 conformation, showing all parallel conformation with characteristic peaks around 260 nm (positive) and 240 nm (negative), except for G32T showing signal similar to hybrid-type G4; (C) KRAS 32R mutants imino region at 37°C showing different effects of mutations in agreement with results obtained in native gel demonstrating the existence of two major conformations and the role of the 3′end residues. Imino region comparison between KRAS 32R wild type (blue), G9T and G25T showing that profiles from G9T and G25T can be found in wild type spectra as an evidence of the model. All experiments were performed in buffer 1× (50 mM KCl; 10 mM KPi; pH 6.66).
Both KRAS 32R G9T and G25T adopt parallel conformations with propeller loop topologies. G9T contains a syn fold-back 3′ end motif that increases its stability
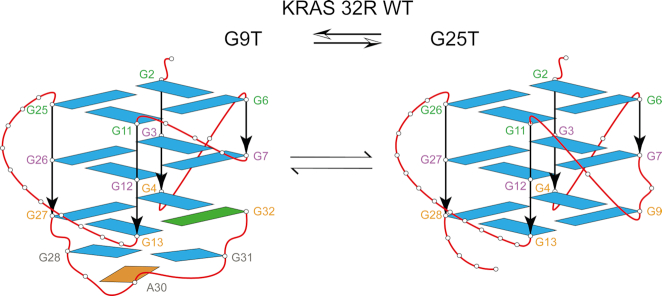
In order to get insight in the topologies adopted by G9T and G25T, we performed additional CD and NMR experiments. CD and 1D 1H NMR spectra were used to monitor melting experiments and obtain Tm values (Supplementary Figure S4A). The CD melting results show that 32R WT, G9T and G25T melt with temperatures of 59, 57.5 and 56°C, respectively, for the forward (heating) experiment, and 50, 52 and 48°C, respectively, for the backward (cooling) experiments. The melting followed by analysis of the ratio of the imino versus the aromatic peaks in the 1D 1H NMR experiments (Supplementary Figure S4B), gives a gross estimation about each conformer folding stability. These results confirm that G25T is the least stable conformer. The CD melting results and the 1D 1H spectra support our hypothesis that the G9T conformer is more stable than the G25T conformer. We proceeded to identify each guanine implicated in the G9T and G25T conformers using site-specific low-enrichment 15N-labeling (Supplementary Figure S5). The results revealed that each imino proton in the G9T mutant was implicated in a single conformation. In the case of the G25T mutant, some of the imino peaks were involved in multiple conformations, such as G4, G7 and G11. Interestingly, the 1D 1H NMR spectra of G9T presented thirteen peaks instead of the twelve expected for a G-quadruplex with three tetrads of guanines. Together with through-bond HCCNH-TOCSY and the through-space {1H–1H} NOESY spectra (250 ms as mixing time), we were able to identify all imino protons and stablish the correlations with the respective aromatic protons (H8) for G9T and G25T mutants, (Figure 3 and Supplementary Figure S6 respectively). From there, all of the H6/8-H1′ NOE sequential connectivities could be mapped for all 32 bases (Supplementary Figures S7 and S8 respectively). Additional experiments with mutants by 1D 1H NMR, CD spectroscopy and PAGE (Supplementary Figures S9, S10 for G9T and S11, S12 for G25T) were combined with 2D classical NMR experiments, such as {31P–1H} HSQC, {13C–1H} HSQC, {1H–1H} TOCSY and COSY, to interpret and support the assignments. We identified that the three tetrad-layered guanine core was composed of G2–G6–G11–G25, G3–G7–G12–G26, G4–G32–G13–G27 for G9T and by G2–G6–G11–G26, G3–G7–G12–G27, G4–G9–G13–G28 for G25T. According to the H1′–H8 correlations observed in the {1H–1H} NOESY spectrum, all guanine bases, except G32, were in the anti glycosidic position (43). In G9T, G32 folds back within the third strand and completes the tetrad formed at the 3′ tail. In addition to the tetrad core with a folded-back guanine, G9T showed NOEs connectivities between G28, A30 and G31 compatible with the formation of a triad (46). Some of the inspected NOEs included correlations between A30 (H8) and G28 (NH2), between A30 (H8) and G31 (H8), and between G31 (H8) and G28 (NH2), among other diverse sugar correlations. Both mutants have three tetrads of guanines forming the central core, with parallel strands connected differently in terms of loops. G9T has a short one-base propeller loop (C5), one three-base propeller loop (T8–T10) and a long propeller loop composed by eleven nucleotides (A14–G24). Finally, G28 through G31 form a diagonal loop that encases a triad formed by G28–A30–G31. Together with the diagonal loop, the triad caps and stabilizes (51,52) the 3′ tail of the G9T conformer. In the case of G25T conformer, it contains two short one-nucleotide propeller loops (C5) and (T10), a bulge (T8), and a long propeller loop composed by twelve nucleotides (A14–T25). A simple folding topology of both models is depicted in Figure 4. A possible conformation exchange between both structures is achieved in G9T by G25 base sliding-out from the tetrad core and simultaneously G26 takes its place; G27 slides to the central tetrad and finally G28 sliding-in forming part of the tetrad core in G25T. This sliding effect in the same direction is also present in the case of a double mutant G25T–G26T where the imino peak of G29, which is not observed in G25T, is well visible in G25T–G26T (Supplementary Figure S13). One of the consequences was that G32 could not fold-back and be part of the guanine tetrad core of the G25T conformer. A second important consequence is the loss of the triad (G28, A30 and G31) at the 3′-end in the G25T mutant. Overall, G25T has less stable structural elements, in agreement with CD and NMR melting experiments. In addition, we observed from different samples preparations containing 32R that before and after thermal melts we always got the same peak pattern and intensities in {1H–1H} NOESY spectra evidencing the same relative populations between the both conformers.
Figure 4.
KRAS 32R wild type contains two conformers in equilibrium, presenting different structural features depicted here by two families of mutants. Both mutants are formed by a three-layered tetrad core of guanines. G9T (left) has a fold-back guanine in syn glyosidic conformation (G32, green) and it is capped in the 3′-end with a triad (G28, A30 and G31). Conformer G25T (right) has all guanines in anti-position and contains three regular propeller loops.
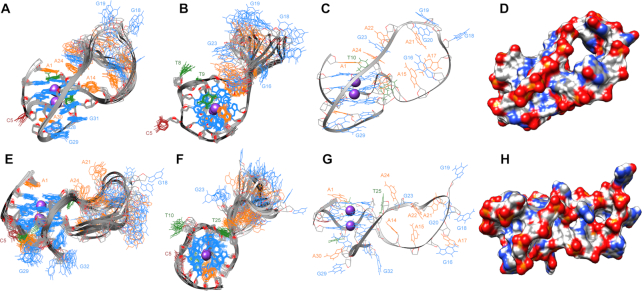
NMR solution structure of KRAS 32R G9T and G25T
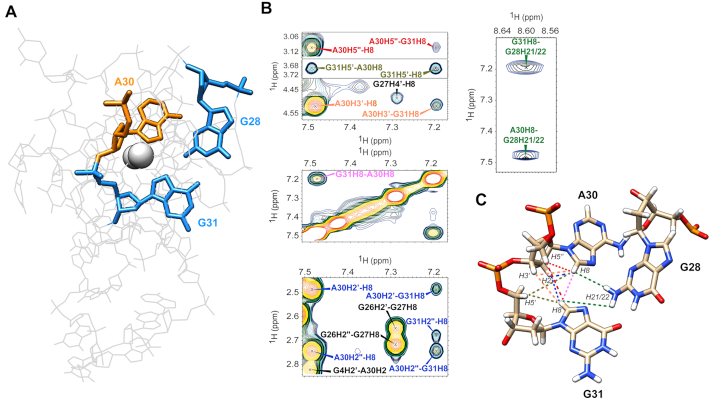
Unless otherwise specified, the NMR solution structures of both G9T and G25T mutants were calculated with spectra collected at 37°C. The necessary restraints were extracted from {1H–1H} NOESY experiments with a mixing time of 250 ms. A total of 873 and 564 NOE-derived restraints were obtained for G9T and G25T, respectively (Supplementary Tables S1 and S2). Figure 5 depicts the 10 lowest-energy structures superimposed obtained after MD simulations for structure refinement purposes in presence of water and K+ counter-ions (cf. Materials and Methods). For G9T, all principal structural motifs (loops and tetrad-core) converged during the MD simulation, with a final heavy-atom RMSD of 1.5 Å. On the other hand, due to an increased peak broadening and lower number of NOEs, G25T has greater variation in the ensemble, with a heavy-atom RMSD of 1.9 Å. This variation was most pronounced in the long propeller loop and in the 3′-end. Compared to G25T, the long propeller loop in G9T presents more π-π interactions between bases such as A24, G23 and A22 and less dispersion of the nucleotides (Figure 5). In addition, the bases from A1, T10 and A24 nucleotides in G9T also participate in π-π interactions, further stabilizing the top of the first tetrad (G2–G6–G11–G25). At the 3′-end, G32 folds back and participates to the third tetrad (G4–G32–G13–G27). In the case of G25T, only A1 and T8 seem to provide the same sort of stabilization over the tetrad core. The triad observed in G9T runs diagonally and connects two opposite tracts composed by G6–G7–G32 and G25–G26–G27 respectively. The triad structure is stabilized by π–π interactions with the closest tetrad of guanines and by a network of hydrogen bonds including one formed between G28 with A30 and two between G28 and G31. In the case of G28–A3, one of the hydrogens (H21) of the amine group acts as donor and N6 from A30 is the acceptor. Two other hydrogen bonds can also be observed between G28 and G31, involved the same type of atoms, and between G28 and G31 using H1 and O6, respectively. In addition, we observed that G28 and G31 have their respective H1 protons outwards in exchange with the solvent, and that may explain why we could not observe the corresponding imino peaks in 15N-filtered 1D NMR experiments at 37°C (Supplementary Figure S1), nor when spectra were acquired at 4°C. The formation of the triad was also supported from observed inter-NOEs corresponding to H8 protons from both A30 and G31, which come closer and share several NOEs with respective sugar protons nearby. The triad structure has the inter-proton NOEs color mapped depicted in Figure 6. NMR spectra collected at 47°C retained the respective H8-H8 inter-base NOEs between A30 and G31 (data not shown). In addition, when we inspected the {1H–1H} NOESY spectrum of 32R WT we could identify all of the same cross peaks correlations, with the exception of G31H8–G28H21/22, indicating that the triad is well preserved in the WT sample, including G28 with A30 and between G28 and G31 (Supplementary Figure S14). The triad we describe in the 3′-terminal of G9T resembles the one found at the 3′-end in the structure of the Pu24I G-quadruplex (PDB: 2A5P) published by Phan et al. in 2005. A fold back guanine (G24) was shown to support the structural elements for the formation of a GAG triad composed by G20–A22–G23 (54).
Figure 5.
Side view of the superposition of the ten lowest-energy structures at 37°C based on all heavy-atoms for 32R G9T and G25T in (A) and (E), respectively, and the corresponding up view in (B) and (F) (with guanines in blue, adenines in orange, thymines in green and the only cytosine in brown). (C) and (G) show only one structure of G9T and G25T, respectively, and the geometric arrangement of the loop in each conformer. (D) and (H) show same structure as (C) and (G) with surfaces.
Figure 6.
Representation of the triad within the KRAS 32R G9T calculated structure in (A) with guanines 28 and 31 in blue and adenine 30 in orange. Ten cross peaks proving the existence of the triad at 37°C found in KRAS 32R G9T {1H–1H} NOESY spectrum are show in (B) involving aromatic and sugar protons from the three bases implicated in the triad and the nearby residues. The corresponding restraints are shown in the calculated triad in (C) with the corresponding colors.
CONCLUSIONS
G-quadruplexes have emerged in the last decade as possible new therapeutic targets for developing new anticancer strategies (47–49). Validating the G4 structures as possible targets requires a deeper understanding of their organization, structure and mode of action. In this study, we have determined the high-resolution structures of two G-quadruplexes existing in a 32-mer G-rich sequence in the NHE region of the KRAS promoter sequence, determined at physiological temperature and with K+ ions in solution. We found that two G4s coexist in equilibrium giving rise to two different G4 structures. Both structures are parallel-stranded and G9T is slightly more stable than G25T. Both conformers contain long loops that have little influence in the global stability of each G4 conformer. One of the two G4s is characterized by a folded-back guanine and a 3′-end triad that increases the thermal stability. The structure of the two G4s of 32R, determined by the present NMR study, is nicely in keeping with the DMS footprinting pattern previously obtained with KRAS 32R sequence. This present study also advances our understanding of the folding of 32R, a critical promoter sequence recognized by nuclear factors that activate transcription (44). The probable function of 32R G4 is to recruit proteins to the promoter and to assemble the transcription pre-initiation complex. A ChIP seq analysis of transcriptionally active chromatin has shown that the KRAS promoter is indeed folded (50). Our study demonstrates that the folding of 32R is rather complex, as the resulting G4 structure exists in equilibrium between two conformers, each characterized by a defined structure. The fingerprint of both conformers can be well visualized from cross peaks found in the {1H–1H} NOESY spectra of G9T and G25T when overlaid with 32R WT spectra (Supplementary Figure S15), and was also used to help in the identification of the majority of iminos cross peaks in 32R WT spectra attributed to two different conformers (Supplementary Figure S16). The next challenging steps will be to establish whether (i) both G4 conformers of 32R are biologically relevant with respect to the transcription initiation process; (ii) shifting the equilibrium towards one of the two G4 forms modulates the binding of the proteins to the promoter. This last point is supported by the fact that the regulatory 32R sequence can fold into different G4 conformations: the two conformers described in this study as well as a more compact G4 structure (G4 21R) formed by a 21-nt portion of sequence 32R (51). These G4s may look similar according to certain biochemical methods, but high resolution NMR structures unveiled important structural differences, which are worth exploring in the development of small molecules for therapeutic purposes. As a final remark, we would like to point out that a truncated sequence of 32R was found to dimerize into a head-to-head G4 conformation defined by NMR (52). In this work we did not found evidence for the formation of a dimer complex. We also think that such structure is unlikely to form in vivo within the KRAS promoter, but it shows bioactivity when used as a decoy to inhibit KRAS in cancer cells (53).
DATA AVAILABILITY
The NMR chemical shifts of 32R G9T and G25T have been deposited in the Biological Magnetic Resonance Bank (accession code 34431 and 34441) and their coordinates have been deposited in the Protein Data Bank (accession codes 6SUU for G9T and 6T2G for G25T).
Supplementary Material
ACKNOWLEDGEMENTS
The work on the 800 MHz NMR spectrometer (IECB) was possible thanks to support from TGIR-RMN-THC Fr3050 CNRS, CNRS UMS3033, Inserm US001, Univ. Bordeaux. We appreciate the numerous discussion with Radovan Fiala and the access to the National NMR Centre at CEITEC MU, Brno, Czech Republic. Finally, we appreciate the support from La ligue contre le Cancer, comité de Gironde. We would also like to express our gratitude to Brune Vialet for the synthesis of some oligonucleotides.
Author contributions: The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Contributor Information
Julien Marquevielle, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
Coralie Robert, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
Olivier Lagrabette, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
Mona Wahid, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
Anne Bourdoncle, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
Luigi E Xodo, Department of Medicine, Laboratory of Biochemistry, 33100 Udine, Italy.
Jean-Louis Mergny, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
Gilmar F Salgado, European Institute of Chemistry and Biology (IECB), ARNA laboratory, INSERM U1212 - CNRS UMR 5320, University of Bordeaux, France.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
iNEXT [653706] funded by the Horizon 2020 programme of the European Commission; L.E.X. was supported by AIRC [IG-2017, Project code 19898]. Funding for open access charge: INSERM laboratory [U1212].
Conflict of interest statement. None declared.
REFERENCES
- 1. Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018; 68:394–424. [DOI] [PubMed] [Google Scholar]
- 2. Siegel R.L., Miller K.D., Jemal A.. Cancer statistics, 2018. CA Cancer J. Clin. 2018; 68:7–30. [DOI] [PubMed] [Google Scholar]
- 3. Riely G.J., Kris M.G., Rosenbaum D., Marks J., Li A., Chitale D.A., Nafa K., Riedel E.R., Hsu M., Pao W. et al.. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin. Cancer Res. 2008; 14:5731–5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Karnoub A.E., Weinberg R.A.. Ras oncogenes: split personalities. Nat. Rev. Mol. Cell Biol. 2008; 9:517–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Riely G.J., Ladanyi M.. KRAS mutations: an old oncogene becomes a new predictive biomarker. J. Mol. Diagn. 2008; 10:493–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tan C., Du X.. KRAS mutation testing in metastatic colorectal cancer. World J. Gastroenterol. 2012; 18:5171–5180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Krasinskas A.M., Chiosea S.I., Pal T., Dacic S.. KRAS mutational analysis and immunohistochemical studies can help distinguish pancreatic metastases from primary lung adenocarcinomas. Mod. Pathol. 2014; 27:262–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cox A.D., Fesik S.W., Kimmelman A.C., Luo J., Der C.J.. Drugging the undruggable RAS: Mission possible. Nat. Rev. Drug Discov. 2014; 13:828–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L. et al.. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012; 149:656–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang M.H., Nickerson S., Kim E.T., Liot C., Laurent G., Spang R., Philips M.R., Shan Y., Shaw D.E., Bar-Sagi D. et al.. Regulation of RAS oncogenicity by acetylation. PNAS. 2012; 109:10843–10848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Maurer T., Garrenton L.S., Oh A., Pitts K., Anderson D.J., Skelton N.J., Fauber B.P., Pan B., Malek S., Stokoe D. et al.. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. PNAS. 2012; 109:5299–5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bournet B., Buscail C., Muscari F., Cordelier P., Buscail L.. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur. J. Cancer. 2016; 54:75–83. [DOI] [PubMed] [Google Scholar]
- 13. Ryan M.B., Corcoran R.B.. Therapeutic strategies to target RAS-mutant cancers. Nat. Rev. Clin. Oncol. 2018; 15:709–720. [DOI] [PubMed] [Google Scholar]
- 14. Xie C., Li Y., Li L.L., Fan X.X., Wang Y.W., Wei C.L., Liu L., Leung E.L., Yao X.J.. Identification of a new potent inhibitor targeting KRAS in non-small cell lung cancer cells. Front. Pharmacol. 2017; 8:823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Q., Burke J.P., Phan J., Burns M.C., Olejniczak E.T., Waterson A.G., Lee T., Rossanese O.W., Fesik S.W.. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew. Chem. 2012; 51:6140–6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leung E.L.H., Luo L.X., Liu Z.Q., Wong V.K.W., Lu L.L., Xie Y., Zhang N., Qu Y.Q., Fan X.X., Li Y. et al.. Inhibition of KRAS-dependent lung cancer cell growth by deltarasin: blockage of autophagy increases its cytotoxicity. Cell Death. Dis. 2018; 9:216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cox A.D., Der C.J., Philips M.R.. Targeting RAS membrane association: back to the future for anti-RAS drug discovery. Clin. Cancer Res. 2015; 21:1819–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pupo E., Avanzato D., Middonti E., Bussolino F., Lanzetti L.. KRAS-driven metabolic rewiring reveals novel actionable targets in cancer. Front. Oncol. 2019; 9:848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim W., Lee S., Kim H.S., Song M., Cha Y.H., Kim Y.H., Shin J., Lee E.S., Joo Y., Song J.J. et al.. Targeting mutant KRAS with CRISPR-Cas9 controls tumor growth. Genome Res. 2018; 28:374–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bedrat A., Lacroix L., Mergny J.L.. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016; 44:1746–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong H.M., Stegle O., Rodgers S., Huppert J.L.. A toolbox for predicting g-quadruplex formation and stability. J. Nucleic Acids. 2010; 2010:564946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sannohe Y., Sugiyama H.. Overview of formation of G-quadruplex structures. Curr. Protoc. Nucleic Acid Chem. 2010; doi:10.1002/0471142700.nc1702s40. [DOI] [PubMed] [Google Scholar]
- 23. Stegle O., Payet L., Mergny J.L., MacKay D.J., Leon J.H.. Predicting and understanding the stability of G-quadruplexes. Bioinformatics. 2009; 25:i374–i382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tippana R., Xiao W., Myong S.. G-quadruplex conformation and dynamics are determined by loop length and sequence. Nucleic Acids Res. 2014; 42:8106–8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cang X., Sponer J., Cheatham T.E. 3rd.. Explaining the varied glycosidic conformational, G-tract length and sequence preferences for anti-parallel G-quadruplexes. Nucleic Acids Res. 2011; 39:4499–4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Le H.T., Dean W.L., Buscaglia R., Chaires J.B., Trent J.O.. An investigation of G-quadruplex structural polymorphism in the human telomere using a combined approach of hydrodynamic bead modeling and molecular dynamics simulation. J. Phys. Chem. B. 2014; 118:5390–5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dai J., Carver M., Yang D.. Polymorphism of human telomeric quadruplex structures. Biochimie. 2008; 90:1172–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bochman M.L., Paeschke K., Zakian V.A.. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lipps H.J., Rhodes D.. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009; 19:414–422. [DOI] [PubMed] [Google Scholar]
- 30. Gomez-Marquez J. DNA G-quadruplex: structure, function and human disease. FEBS J. 2010; 277:3451. [DOI] [PubMed] [Google Scholar]
- 31. Amato J., Madanayake T.W., Iaccarino N., Novellino E., Randazzo A., Hurley L.H., Pagano B.. HMGB1 binds to the KRAS promoter G-quadruplex: a new player in oncogene transcriptional regulation. Chem. Commun. 2018; 54:9442–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cogoi S., Rapozzi V., Cauci S., Xodo L.E.. Critical role of hnRNP A1 in activating KRAS transcription in pancreatic cancer cells: a molecular mechanism involving G4 DNA. Biochim. Biophys. Acta Gen. Subj. 2017; 1861:1389–1398. [DOI] [PubMed] [Google Scholar]
- 33. Carvalho J., Pereira E., Marquevielle J., Campello M.P.C., Mergny J.L., Paulo A., Salgado G.F., Queiroz J.A., Cruz C.. Fluorescent light-up acridine orange derivatives bind and stabilize KRAS-22RT G-quadruplex. Biochimie. 2018; 144:144–152. [DOI] [PubMed] [Google Scholar]
- 34. Neidle S. The structures of quadruplex nucleic acids and their drug complexes. Curr. Opin. Struct. Biol. 2009; 19:239–250. [DOI] [PubMed] [Google Scholar]
- 35. Cogoi S., Paramasivam M., Filichev V., Geci I., Pedersen E.B., Xodo L.E.. Identification of a new G-quadruplex motif in the KRAS promoter and design of pyrene-modified G4-decoys with antiproliferative activity in pancreatic cancer cells. J. Med. Chem. 2009; 52:564–568. [DOI] [PubMed] [Google Scholar]
- 36. Pattanayak R., Basak P., Sen S., Bhattacharyya M.. Interaction of KRAS G-quadruplex with natural polyphenols: a spectroscopic analysis with molecular modeling. Int. J. Biol. Macromol. 2016; 89:228–237. [DOI] [PubMed] [Google Scholar]
- 37. Sklenar V., Dieckmann T., Butcher S.E., Feigon J.. Through-bond correlation of imino and aromatic resonances in 13C-, 15N-labeled RNA via heteronuclear TOCSY. J. Biomol. NMR. 1996; 7:83–87. [DOI] [PubMed] [Google Scholar]
- 38. Sklenar V., Masse J.E., Feigon J.. HCCCH experiment for through-bond correlation of thymine resonances in 13C-labeled DNA oligonucleotides. J. Magn. Reson. 1999; 137:345–349. [DOI] [PubMed] [Google Scholar]
- 39. Brunger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S. et al.. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta crystallog D, Biol Crystallogr. 1998; 54:905–921. [DOI] [PubMed] [Google Scholar]
- 40. Brunger A.T. Version 1.2 of the crystallography and NMR system. Nat. Protoc. 2007; 2:2728–2733. [DOI] [PubMed] [Google Scholar]
- 41. Randazzo A., Spada G.P., da Silva M.W.. Circular dichroism of quadruplex structures. Top. Curr. Chem. 2013; 330:67–86. [DOI] [PubMed] [Google Scholar]
- 42. Vorlickova M., Kejnovska I., Bednarova K., Renciuk D., Kypr J.. Circular dichroism spectroscopy of DNA: from duplexes to quadruplexes. Chirality. 2012; 24:691–698. [DOI] [PubMed] [Google Scholar]
- 43. Webba da Silva M. NMR methods for studying quadruplex nucleic acids. Methods. 2007; 43:264–277. [DOI] [PubMed] [Google Scholar]
- 44. Cogoi S., Paramasivam M., Spolaore B., Xodo L.E.. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008; 36:3765–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cogoi S., Xodo L.E.. G-quadruplex formation within the promoter of the KRAS proto-oncogene and its effect on transcription. Nucleic Acids Res. 2006; 34:2536–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Phan A.T., Kuryavyi V., Gaw H.Y., Patel D.J.. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005; 1:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sengupta P., Chattopadhyay S., Chatterjee S.. G-Quadruplex surveillance in BCL-2 gene: a promising therapeutic intervention in cancer treatment. Drug Discov. Today. 2017; 22:1165–1186. [DOI] [PubMed] [Google Scholar]
- 48. Francisco A.P., Paulo A.. Oncogene expression modulation in cancer cell lines by DNA G-Quadruplex-Interactive small molecules. Curr. Med. Chem. 2017; 24:4873–4904. [DOI] [PubMed] [Google Scholar]
- 49. Asamitsu S., Obata S., Yu Z., Bando T., Sugiyama H.. Recent progress of targeted G-Quadruplex-Preferred ligands toward cancer therapy. Molecules. 2019; 24:doi:10.3390/molecules24030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hansel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M. et al.. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016; 48:1267–1272. [DOI] [PubMed] [Google Scholar]
- 51. Paramasivam M., Cogoi S., Xodo L.E.. Primer extension reactions as a tool to uncover folding motifs within complex G-rich sequences: analysis of the human KRAS NHE. Chem. Commun. (Camb.). 2011; 47:4965–4967. [DOI] [PubMed] [Google Scholar]
- 52. Podbevsek P., Plavec J.. KRAS promoter oligonucleotide with decoy activity dimerizes into a unique topology consisting of two G-quadruplex units. Nucleic Acids Res. 2016; 44:917–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cogoi S., Zorzet S., Rapozzi V., Geci I., Pedersen E.B., Xodo L.E.. MAZ-binding G4-decoy with locked nucleic acid and twisted intercalating nucleic acid modifications suppresses KRAS in pancreatic cancer cells and delays tumor growth in mice. Nucleic Acids Res. 2013; 41:4049–4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Phan A.T., Kuryavyi V., Gaw H.Y., Patel D.J.. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005; 1:67–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NMR chemical shifts of 32R G9T and G25T have been deposited in the Biological Magnetic Resonance Bank (accession code 34431 and 34441) and their coordinates have been deposited in the Protein Data Bank (accession codes 6SUU for G9T and 6T2G for G25T).