Abstract
Background
Osteosarcoma (OS) is the most common bone tumor. Many studies have reported that circular RNAs (circRNAs) play an important role in the development of a variety of human cancers. However, the underlying mechanism of circ_0001721 in regulating osteosarcoma progression remains unknown.
Materials and Methods
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the levels of circ_0001721, miR-372-3p, and mitogen-activated protein kinase 7 (MAPK7) in osteosarcoma tissues and cells. Besides, glycolysis was investigated by glucose consumption, lactate production and hexokinase II (HK2) protein level. Cell proliferation and apoptosis were determined by 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide (MTT) and flow cytometry, separately. Cell migration and invasion were determined by transwell assay. Moreover, the protein levels of HK2 and epithelial-to-mesenchymal transition (EMT) markers were determined by Western blot analysis. The relationship between miR-372-3p and circ_0001721 or MAPK7 was predicated by starbase v3.0 and confirmed by dual-luciferase reporter assay or RNA binding protein immunoprecipitation (RIP) assay. Murine xenograft model was established to investigate the role of circ_0001721 in vivo.
Results
The levels of circ_0001721 and MAPK7 were upregulated in osteosarcoma tissues and cells, while miR-372-3p was downregulated. Knockdown of circ_0001721 inhibited glycolysis, cell proliferation, cell migration, invasion and epithelial-to-mesenchymal transition (EMT), and promoted apoptosis. Circ_0001721 was validated as a sponge of miR-372-3p and mediated glycolysis, cell proliferation, apoptosis, migration, invasion, and EMT of osteosarcoma cells through miR-372-3p. MAPK7 was a target of miR-372-3p and overexpression of MAPK7 attenuated anti-cancer role of miR-372-3p in OS cells. Further studies revealed that circ_0001721 regulates MAPK7 expression via sponging miR-372-3-p. Finally, knockdown of circ_0001721 inhibited tumor progression in vivo.
Conclusion
Circ_0001721 promoted osteosarcoma development through the miR-372-3p/MAPK7 axis.
Keywords: circ_0001721, miR-372-3p, MAPK7, osteosarcoma
Introduction
Osteosarcoma (OS) is a highly aggressive, metastatic bone tumor with a poor prognosis and is common in children and adolescents.1 There is no specific drug or treatment for this tumor, so it is urgent to conduct in-depth research on this tumor.2
Circular RNAs (CircRNAs) are single-stranded RNAs that form covalently closed continuous loops,3 and they are resistant to exonuclease-mediated degradation.4 It had been reported that circRNAs are associated with the development of many types of cancers,5 for example, colorectal cancer,6 prostate cancer,7 ovarian cancer,8 nasopharyngeal carcinoma,9 as well as OS.10 Lu et al reported that circrRNA hsa_circ_0006848 could be used as a new biomarker for early gastric cancer.11 Wang et al found that circular RNA SMARCA5 inhibited proliferation, migration, and invasion of NSCLC cells.12 Ying et al confirmed that circ-TSPAN4 promoted lung adenocarcinoma metastasis.13 Circular RNA PIP5K1A could promote colon cancer development by inhibiting miR-1273a.14 Li et al found that circ_0001721 was highly expressed in OS tissues and inhibited cell apoptosis in vivo.15 Yet, the regulatory mechanism of circ_0001721 in OS progression had not been adequately addressed.
MicroRNAs (MiRNAs) are short-length (about 22 nucleotides),16 highly conserved, non-coding RNAs that regulate the post-transcriptional level of messenger RNA (mRNA) by binding to the 3-untranslated region (3ʹUTR) of mRNA.17 Abnormal levels of many miRNAs had been identified in plasma samples from patients in cell experiments, clinical cancer specimens, saliva, serum, or oral malignancies.18 MiRNAs are involved in many aspects of cancer, including tumor growth,19 cell proliferation, apoptosis, migration, invasion, metastasis, glucose metabolism, radio-sensitivity, and chemosensitivity.20 Xu et al confirmed that miR-372-3p could inhibit the growth and metastasis of osteosarcoma cells by targeting FXYD6.21 Wang et al found that miR-372-3p promoted cell growth and metastasis in lung squamous cell carcinoma through targeting FGF9.22 Syring et al pointed out that circulating serum miR-372-3p can be used as a biomarker for patients with testicular germ cell carcinoma.23 Although the functional study of miR-372-3p in tumors has made some progress, the mechanism of miR-372-3p in the occurrence and development of osteosarcoma remains unclear.
Related studies showed that MAPK7 was involved in the development of a variety of cancers, including OS.24 Zhou et al proved that MAPK7 was involved in the proliferation and apoptosis of lung cancer.25 Dong et al reported that miR-143 regulates the proliferation and migration of osteosarcoma cells through targeting MAPK7.26 This study explored the role of MAPK7 and non-coding RNA interaction in OS development.
CircrRNAs can function as competing endogenous for RNAs (ceRNAs) though sponging miRNAs to modulate the genes expression of miRNAs targets.27 In our study, we evaluated the abundance of circ_0001721, miR-372-3p, and MAPK7 in OS tissues and cells. In addition, we also studied their effects on glycolysis, cell growth, apoptosis, migration, invasion, and explored the regulatory networks of circ_0001721, miR-372-3p, and MAPK7. Therefore, these findings might provide new insights and hopes for the treatment of OS.
Materials and Methods
Tissue Collection and Cell Culture
56 paired OS tissues and nearby non-cancerous tissues were collected from the Second Hospital of Shanxi Medical University. The clinical information of the patients, including age, gender, tumor size, and pulmonary metastasis was summarized in Table 1. The written informed consent was acquired from every patient. This study was authorized by the Ethics Committee of the Second Hospital of Shanxi Medical University and was performed in accordance with the Helsinki Declaration. Human osteoblastic cell line (hFOB1.19) and OS cell lines (U2OS and HOS) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). McCoy’s 5A medium (Sigma, St Louis, MO, USA), containing 5% CO2 and 10% fetal bovine serum (FBS; Sigma), was utilized to culture cells.
Table 1.
Circ_0001721 Expression and Clinicopathologic Characteristics of OS Patient
| Parameter | Case | Circ_0001721 Expression | P valuea | |
|---|---|---|---|---|
| Low (n=26) | High (n=30) | |||
| Age (years) | 0.436 | |||
| ≤60 | 32 | 18 | 15 | |
| >60 | 24 | 12 | 15 | |
| Gender | 0.410 | |||
| Male | 29 | 15 | 14 | |
| Female | 27 | 11 | 16 | |
| Tumor size | 0.003** | |||
| ≤2 cm | 31 | 20 | 11 | |
| >2 cm | 25 | 6 | 19 | |
| Pulmonary metastasis | 0.005** | |||
| Yes | 35 | 23 | 14 | |
| No | 21 | 5 | 16 | |
Note: **P< 0.01aChi-square test.
RNA Isolation and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
OS tissues and cells were collected, and total RNA was extracted using the TRIzol reagent (Vazyme, Nanjing, China). Then RNA was reversely transcribed to complementary DNA (cDNA) by the PrimeScript™ RT Master Mix kit (Takara, Dalian, China). The qRT-PCR was performed by SYBR Green PCR Master Mix (Vazyme) and data were analyzed using the 2−ΔΔCt method. Beta-actin (β-actin) and U6 were introduced as inner references. Primers in this research: circ_0001721 (forward 5ʹ-CACCTAAAGTTAGGCGGCAC-3ʹ, reverse 5ʹ-TGGGTCAAAAGTGCTCTGTG-3ʹ); miR-372-3p (forward, 5ʹ-TTTCACGACGCTGTAAACTCGCA-3ʹ, reverse 5ʹ-CAGTGCGTGTCGTGGAG-3ʹ);MAPK7 (forward, 5ʹ- TGGATGACCTGAGCCGAGTG-3ʹ, reverse 5ʹ-GCACTCTGACTGATTCTCTGG-3ʹ); β-actin (forward 5ʹ-GCACCACACCTTCTACAATG-3ʹ, reverse, 5ʹ-TGCTTGCTGATCCACATCTG-3ʹ); GAPDH (forward, 5ʹ-CGCTCTCTGCTCCTCCTGTTC-3ʹ, reverse, 5ʹ-ATCCGTTGACTCCGACCTTCAC-3ʹ); U6 (forward 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ, reverse 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ).
Cell Culture and Transfection
The small interfering RNA against-circ_0001721 (si-circ_0001721), miR-372-3p (anti-miR-372-3p) and its negative control (si-NC), the mimic or inhibitor of miR-372-3-p (miR-372-3p or anti-miR-372-3p) and their negative control (miR-NC or anti-miR-NC), the pcDNA3.0-MAPK7 overexpression plasmid (MAPK7) were bought from Ribobio (Guangzhou, China). Lentivirus harboring short hairpin RNA targeting circ_0001721 (sh-circ_0001721) and negative control (sh-NC) were constructed by GeneCopoeia (Rockville, MD, USA). According to the recommendations, HOS and U2OS cells were transfected using the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA).
Glucose Consumption and Lactate Production
The HOS and U2OS cells (1 × 105/well), which were transfected or non-transfected, were seeded into 6-well plates overnight and then incubated for 48 h. Glucose Assay Kit and Lactate Assay Kit (Sigma) were used for the detection of glucose consumption and lactate production, respectively, following the manufacturer’s protocol. The relative levels of glucose consumption and lactate production repeated three times.
Western Blot Assay
At 48 h post-transfection, OS tissues and cells were collected and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, Shanghai, China) containing phenylmethanesulfonyl fluoride (PMSF; Beyotime) to obtain total protein. The protein concentrations of the total cellular lysates were quantified using a bicinchoninic acid (BCA) protein assay kit (Beyotime). An equal amount (40 µg) of protein was resolved by 810% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After that, the proteins were transferred to polyvinylidene fluoride (PVDF) membranes (0.2 μm, Beyotime). Next, all membranes were blocked using the 5% non-fat milk and then probed with the following primary antibodies: antibodies against hexokinase II (HK2) (1:5000, ab227198, 102 kDa), E-cadherin (1:500, ab15148, Abcam), Vimentin (1:2500, ab92547, Abcam), β-actin (1:2500, ab8227, Abcam), MAPK7 (1:200, ab92547, Abcam) and GAPDH (1:2000, ab37168, Abcam). Subsequently, all membranes were maintained in HRP-conjugated anti-rabbit/mouse IgG (Sangon Biotech). Finally, all protein bands were observed using the enhanced chemiluminescence (ECL) system (GEHeathcare, Waukesha, WI, USA). The protein abundances were normalized with GAPDH and β-actin, and ImageJ software was applied to evaluate the density.
3-(4,5-Dimethyl-2-Thiazolyl)-2,5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
MTT assay was performed to detect cellular growth ability. HOS and U2OS cells (5 × 103 cells/well) were plated into 96-well plates and incubated for 48 h. Then 20 μL MTT solution (5 mg/mL) was added into each well. After incubating for 4 h at room temperature, 150 μL DMSO was added into each well. The absorbance at 490 nm was determined using a microplate reader.
Flow Cytometry
HOS and U2OS cells (3.5×105/well) were seeded into six-well plates for 48 h. The ratio of apoptotic cells was detected using the V-fluorescein isothiocyanate propidium iodide (Annexin V-FITC/PI) kit (BD Pharmingen, San Diego, CA, USA) and analyzed using a FACSCalibur flow cytometer with Cell Quest software (BD Biosciences, Franklin Lakes, NJ, USA)
Cell Migration Assay and Cell Invasion Assay
Cell (HOS and U2OS) migration and invasion assays were carried out using a 24-well insert (8 μm pores, Corning Incorporated, Corning, NY, USA). For migration, HOS and U2OS cells re-suspended in serum-free DMEM medium (100 μL) were placed into the top chamber (non-coated membrane). For invasion, HOS and U2OS cells re-suspended in serum-free DMEM medium (100 μL) were placed in the top chamber with the Matrigel-coated membrane. DMEM containing 10% FBS (600 μL) would be placed into the bottom chamber. The cells (remaining on top chambers) were gently wiped using a cotton swab following incubation for 24 h. After that, migrated or invaded cells from the bottom surfaces were fixed using 4% paraformaldehyde and subsequently stained using 0.1% crystal violet in darkness for 20 min. Then, the cells were imaged and counted by a microscope (Leica, Solms, Germany).
Dual-Luciferase Reporter Assay
Potential binding sites between circ_0001721 and miR-372-3p, or sites between miR-372-3p and MAPK7 were provided by starBase v3.0 or targetscan, separately. The circ_0001721 or MAPK7-3ʹUTR fragment containing putative and mutated circ_0001721 or MAPK7 binding site was cloned into the pmirGlO luciferase reporter vector (Promega, Madison, WI, USA), namely circ_0001721-WT and MAPK7-WT or circ_0001721-MUT and MAPK7-MUT. The circ_0001721-WT and circ_0001721-MUT with miR-NC reporter plasmids, and MAPK7-3ʹUTR-WT and MAPK7-3ʹUTR-MUT with miR-NC reporter plasmids were co-transfected into HOS and U2OS cells. At 48 h post-transfection, a dual-luciferase reporter assay system (Promega) was employed to assess the luciferase activity, followed by normalizing to Renilla luciferase activity.
RNA Immunoprecipitation (RIP) Assay
Magna RIP Kit (Millipore) was used for RIP assay. Briefly, HOS and U2OS cells were lysed by the RIP buffer supplemented with magnetic beads and then conjugated using the anti-argonaute 2 (anti-Ago2) or IgG antibodies (negative control) overnight at 4°C. After that, the protein was digested through proteinase K buffer, followed by RNA purification. Finally, qRT-PCR was carried out to measure the abundance of circ_0001721 and miR-372-3p.
Murine Xenograft Model in vivo
To measure the effects of circ_0001721 in vivo, we injected HOS cells or control transduced cells with sh-circ_0001721 into 5-week-old female nude mice (NU/NU Crl: NU-Fox1nu, Charles River Laboratories; Sulzfeld, Germany). Briefly, 1 × 106 cells were resuspended in phosphate-buffered saline (PBS; 1:1 mixed with matrigel, Corning Incorporated) and subcutaneously injected into the mammary fat pad of the mice. Cells treated with sh-circ_0001721 were infected cells were injected subcutaneously into the flank of the mice, and control mice were injected with non-transformed cells at the same site. A total of five mice were used for the in vivo experiments. Tumor growth was monitored every 6 days by caliper measurements, and animals were sacrificed before tumors reached a diameter of 10 mm in the 30th day. Tumors were harvested for histological analyses, and tumor volumes were calculated by the equation V (mm3) = (width)2 × length/2. The animal experiment was approved by the Animal Care and Use Committee of the Second Hospital of Shanxi Medical University and carried out following the guidelines of the national animal protection and ethics institute.
Statistical Analysis
Experimental data were calculated by GraphPad Prism (GraphPad, La Jolla, CA, USA) and presented as mean ± standard deviation (SD). Two independent groups were compared by using Student’s t-test. For more than two groups, the one-way analysis of variance (ANOVA) was utilized to evaluate the difference. Pearson’s correlation coefficient was utilized to analyze the correlation between circ_0001721, miR-372-3p, and MAPK7 in OS tissues. Every experiment was repeated at least three times independently. P < 0.05 represented statistical significance.
Results
Circ_0001721 Was Upregulated in Osteosarcoma Tissues and Cells
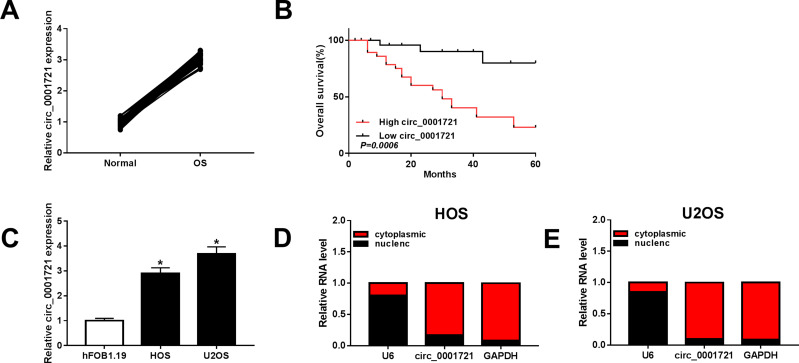
The RT-PCR results of 56 pairs of tissue samples showed that the expression of circ_0001721 was upregulated in OS tissues than normal tissues (Figure 1A). Then, we analyzed the potential clinical significance of circ_0001721 expression in OS patients, and all patients were divided into two groups (High and Low) by using the median value of circ_0001721 expression level. As presented in Table 1, the high expression of circ_0001721 was significantly associated with Tumor size (p=0.003) and Pulmonary metastasis (p=0.005), implying that circ_0001721 may influence the clinical prognosis of OS patients. Then, overall survival was evaluated with the use of a stratified Log rank test and summarized with the use of Kaplan-Meier methods. The results suggested that the OS patients with higher circ_0001721 expression had poorer overall survival than those with low circ_0001721 expression (Figure 1B). The expression of circ_0001721 was higher in HOS and U2OS cells than that in hFOB 1.19 (Figure 1C). The results showed that circ_0001721 was more distributed in the cytoplasm than in the nucleus in HOS and U2OS cells (Figure 1D and E).
Figure 1.
Circ_0001721 was upregulated in OS tissues and cells. (A) The expression of circ_0001721 in OS tissues (n=56) and paratumor tissues (n=56) was measured by qRT-PCR analysis. (B) Statistics of overall survival of large circ_0001721 group and low circ_0001721 group. (C) The level of circ_0001721 in OS cells (HOS and U2OS) and normal osteoblast cell line (hFOB1.19) were determined by qRT-PCR analysis. (D and E) QRT-PCR assay was conducted to evaluate the abundance of circ_0001721 in the cytoplasm and the nucleus in OS cells. *P<0.05.
Knockdown of Circ_0001721 Inhibited Glycolysis, Cell Proliferation, Cell Migration, and Invasion and Epithelial-to-Mesenchymal Transition (EMT), and Promoted Apoptosis
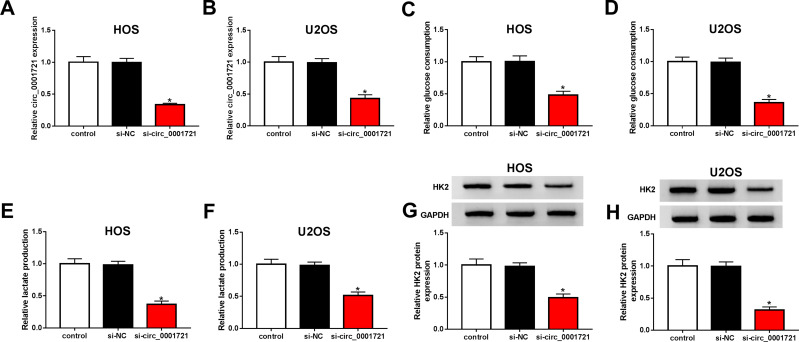
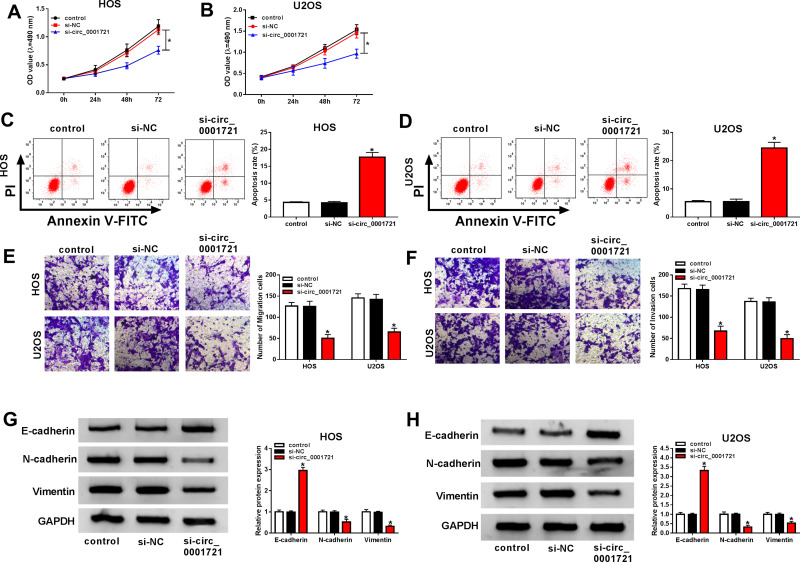
The transfection efficiency was detected by qRT-PCR. The expression of circ_0001721 in cells (HOS and U2OS) treated with si-circ_0001721 is lower than that treated with si-NC and control (Figure 2A and B). Compared with the transfected HOS and U2OS cells with si-NC and control, qRT-PCR and Western blot showed that the glucose consumption, lactic acid production, and HK2 expression were all decreased in the transfected HOS and U2OS cells with si-circ_0001721 (Figure 2CH). In other words, knockdown of circ_0001721 inhibited glycolysis in cells. MTT assay (Figure 3A and B) revealed that knockdown of circ_0001721 reduced cell proliferation. Flow cytometry (Figure 3C and D) showed that si-circ_0001721 increased the apoptotic rate of HOS and U2OS cells. Transwell tests indicated that the number of cell migration (Figure 3E) or invasion (Figure 3F) was reduced in HOS and U2OS cells with circ_0001721 knockdown. Moreover, the Western blot proved that si-circ_0001721 inhibited EMT, specifically, promoting the expression of E-cadherin protein and inhibiting the expression of N-cadherin and Vimentin proteins in OS cells (HOS and U2OS) (Figure 3G and H).
Figure 2.
Knockout circ_0001721 inhibited glycolysis in OS cells. HOS and U2OS cells were transfected with control, si-NC or si-circ_0001721. (A and B) QRT-PCR was used to detect the transfection efficiency. (C and D) QRT-PCR was used to detect glucose consumption in HOS and U2OS cells. (E and F) QRT-PCR was used to detect lactic acid production in HOS and U2OS cells. (G and H) QRT-PCR was used to detect the expression of HK2 in HOS and U2OS cells. *P<0.05.
Figure 3.
Knockdown of circ_0001721 inhibited OS cell proliferation, migration and, invasion, and promoted cell apoptosis and EMT. HOS and U2OS cells were transfected with control, si-NC or si-circ_0001721. (A and B) MTT was used to detect cell proliferation of cell lines (HOS and U2OS). (C and D) Flow cytometry was used to detect cell apoptosis. (E and F) Transwell assay was employed to explore HOS and U2OS cells migration and invasion abilities. (G and H) Western blot assay was applied to test the protein levels of E-cadherin, N-cadherin, and Vimentin in HOS and U2OS cells. *P<0.05.
Circ_0001721 Targeted miR-372-3p, and miR-372-3p Silencing Could Restore the Inhibition of Circ_0001721 Knockdown on Glycolysis, Cell Proliferation, Invasion, and Migration and EMT and Promotion of Cell Apoptosis
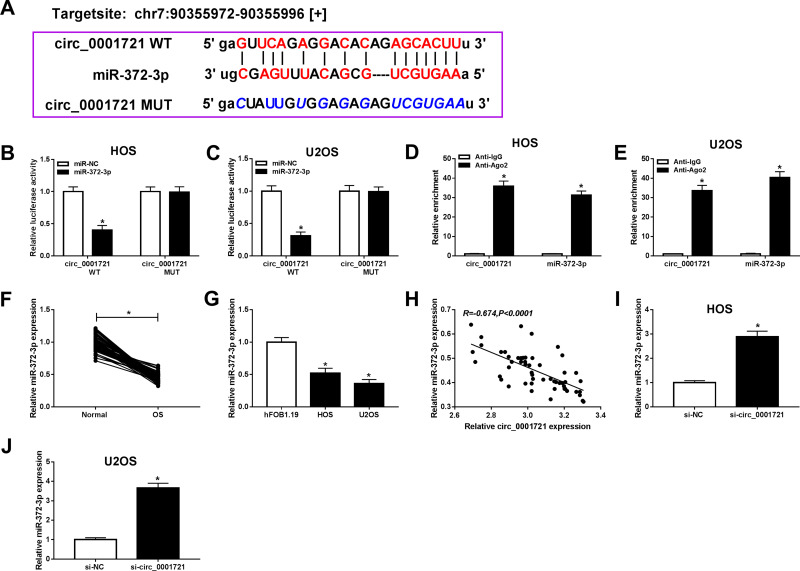
To indicate the connection of circ_0001721 and miR-372-3pin OS cells (HOS and U2OS), starBase v3.0 predicted the presumed associated sequence between circ_0001721 and miR-372-3p, the online analysis exhibited that miR-181a-5p had a presumed associative sequence with circ_0001721 (Figure 4A). Furthermore, dual-luciferase reporter assay was performed and confirmed that the luciferase activity of circ_0001721-WT was decreased by miR-372-3p, while the activity of circ_0001721-MUT did not change in OS cells (HOS and U2OS)(Figure 4B and C). In other words, circ_0001721 could target and bind to miR-372-3p. In addition, the RIP assay showed that enrichment of circ_0001721 and miR-372-3p was markedly enhanced in the Ago2 group compared with that in the IgG group (Figure 4D and E). The expression of miR-372-3p in OS tissues and normal tissues was detected by qRT-PCR. And the results showed that the expression of miR-372-3p in OS tissues was lower than that in normal tissues (Figure 4F), this expression pattern also existed in OS cell lines (HOS and U2OS) compared with the standard cell lines (hFOB1.19) (Figure 4G), but the expression of miR-372-3p was increased in HOS and U2OS cells treated with circ_0001721 knockdown (Figure 4I and J). QRT-PCR proved that circ_0001721 was negatively correlated with the expression of miR-372-3p (Figure 4H). This evidence proved that circ_0001721 might play an essential role in OS cells by interacting with miR-372-3p.
Figure 4.
Circ_0001982 directly interacted with miR-372-3p in OS cells. Starbase v3.0 predicted the putative site between circ_0001721 and miR-372-3p. (B and C) The dual-luciferase reporter assay was measured in HOS, and U2OS cells co-transfected with circ_0001721-WT or circ_0001721-MUT and miR-372-3p or miR-NC. (D and E) The enrichment of circ_0001721 or miR-372-3p was measured by RIP assay in HOS and U2OS cells incubated with Ago2 or IgG. (F and G) The expression of miR-372-3p was measured by qRT-PCR in OS tissues and HOS and U2OS cells. (H) The expression of miR-372-3p was negatively correlated with MAPK7 (R=−0.674). (I and J) The level of miR-1287-5p was determined using the qRT-PCR analysis in HOS and U2OS transfected with si-NC or si-circ_0001982. *P<0.05.
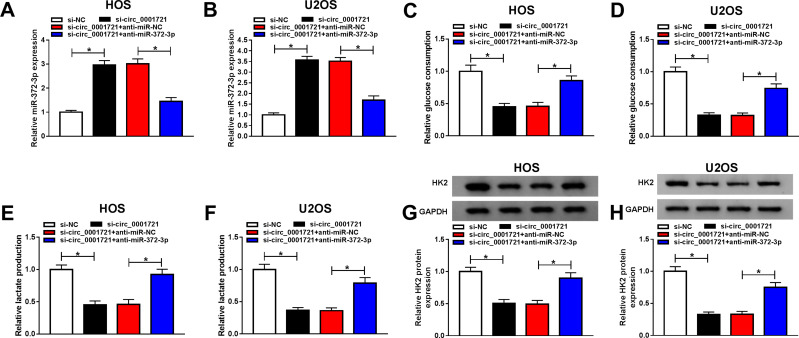
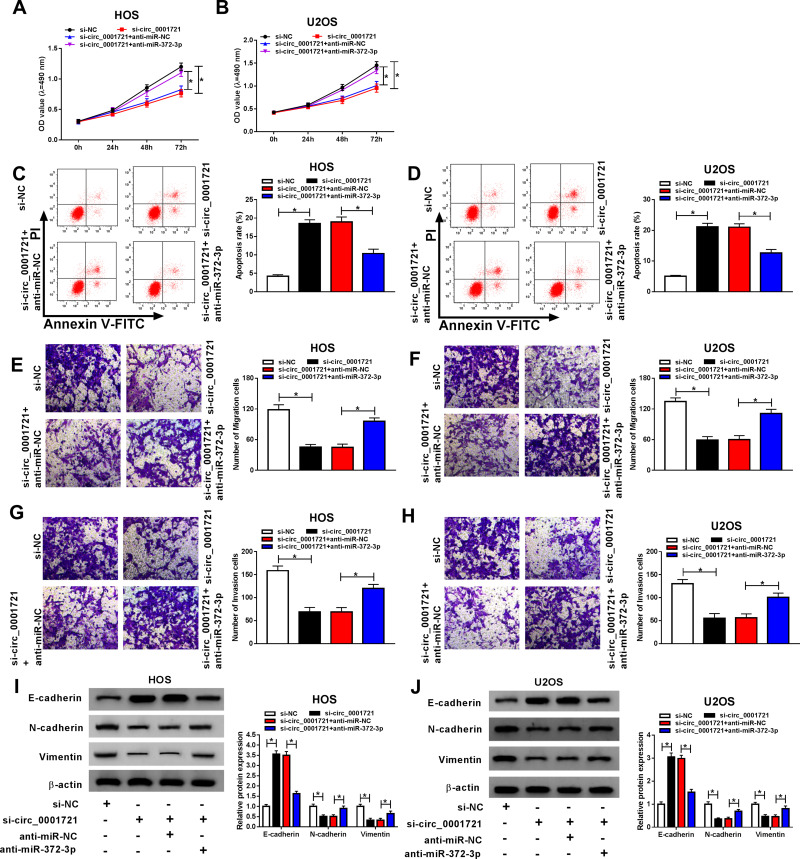
The Silencing of miR-372-3p Reversed Circ-0001721 Knockdown Inhibition of Cell Cancer Progression OS Cells (HOS and U2OS Cells)
Although the interaction between circ_0001721 and miR-372-3p has been confirmed, the biological behaviors of circ_0001721 and miR-372-3p regulating OS remain to be determined. QRT-PCR was used to detect the transfection efficiency and showed that the expression of miR-372-3p was abnormally gone up in HOS and U2OS cells transfected with si-circ_0001721 compared with that in those cells transfected with si-NC, but the downregulation of miR-372-3p reversed the promoting effect of circ_0001721 knockdown on the expression of miR-372-3p in HOS and U2OS cells (Figure 5A and B). Furthermore, knockdown of miR-372-3p weakened the inhibition of glucose consumption (Figure 5C and D), lactic acid formation (Figure 5E and F), and HK2 protein expression (Figure 5G and H) induced by circ_0001721 interference in HOS and U2OS cells. In other words, knockdown of circ_0001721 inhibited glycolysis and knockdown of miR-372-3p reverses this inhibitory effect. MTT assay indicated that the downregulation of miR-372-3p reversed the inhibitory effect of circ_0001721 knockdown on the proliferation of HOS and U2OS cells (Figure 6A and B). Flow cytometry confirmed that the silencing of miR-372-3p weakened the pro-apoptosis, anti-migration, and anti-invasion effects caused by circ_0001721 knockdown in HOS and U2OS cells (Figure 6C and D). Transwell assay demonstrated that the silencing of miR-372-3p reversed the inhibitory effect of circ-0001721 knockdown on migration (Figure 6E and F) and invasion (Figure 6G and H). What’s more, Western blot results showed that the silencing of miR-138-5p reversed the inhibition of circ_0001721 knockdown on EMT, specifically inhibiting the expression of E-cadherin protein and inhibiting the expression of N-cadherin and Vimentin proteins in HOS and U2OS cells (Figure 6I and J). Collectively, our data revealed that knockdown of circ_0001721 inhibited the progression of OS cells by up-regulating miR-372-3p. Downregulation of miR-372-3p reversed the inhibitory effects of circ_0001721 knockdown on the progression of OS cells.
Figure 5.
Circ_0001721 affected glycolysis of osteosarcoma cells through miR-372-3p. (A and B) The expression of miR-372-3p and the transfection efficiency was evaluated using qRT-PCR in HOS and U2OS cells. (C and D) QRT-PCR was used to detect glucose consumption in HOS and U2OS. (E and F) QRT-PCR was used to detect lactic acid production in HOS and U2OS cells. (G and H) QRT-PCR was used to detect the expression of HK2 in HOS and U2OS cells. *P<0.05.
Figure 6.
Silencing miR-372-3p reversed the effect of circ_0001982 knockdown on OS cells. The HOS and U2OS cells were transfected with si-NC, or si-circ_0001721, si-circ_0001721 + anti-miR-NC, or si-circ_0001982 + anti-miR-372-3p. (A and B) Cell proliferation was assessed by MTT assay in HOS and U2OS cells. (C and D) Flow cytometry was used to detect cell apoptosis. (E and F) Transwell assay was employed to explore HOS and U2OS cells migration ability. (G and H) Transwell assay was applied to assess HOS and U2OS cells invasion ability. (I and J) Western blot assay was applied to test the protein levels of E-cadherin, N-cadherin and Vimentin in HOS and U2OS cells. *P<0.05.
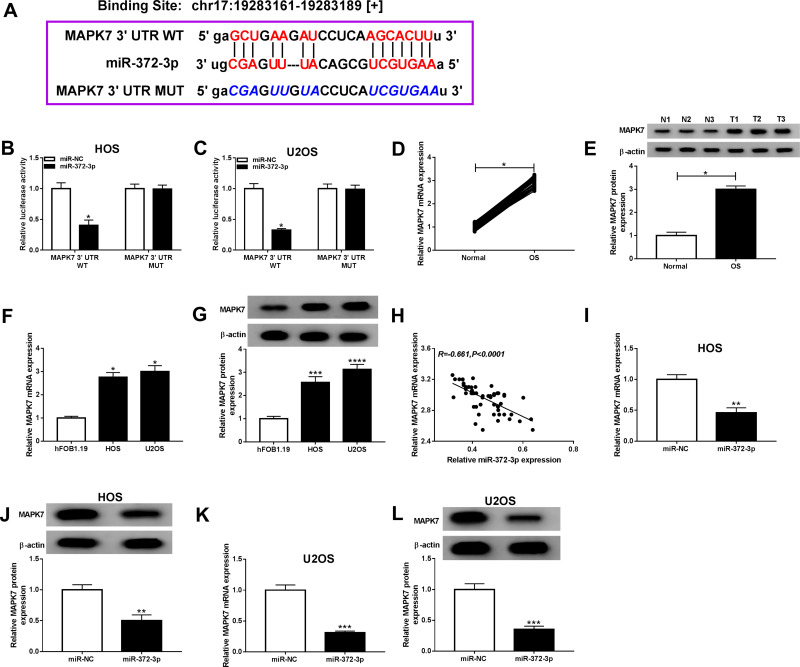
MiR-372-3p Targeted MAPK7 and Was Negatively Correlated with MAPK7
To elucidate the potential mechanism of miR-372-3p-mediated progression in OS, we used starbase v3.0 to search the direct target of MAPK7. As presented in Figure 7A, MAPK7 might be a target of miR-372-3p. Furthermore, dual-luciferase reporter assay was performed and confirmed that the luciferase activity of MAPK7-WT was decreased by miR-372-3p, while the activity of MAPK7-MUT did not change in OS cells (HOS and U2OS) (Figure 7B and C). QRT-PCR and Western blot was used to detect the mRNA and protein expression of MAPK7 in and OS tissue samples and cells, miR-372-3p was upregulated in OS tissue samples (Figure 7D and E) and in OS cell lines (HOS and U2OS) compared with the normal cell lines (hFOB1.19) (Figure 7F and G). The above results showed that miR-372-3p was negatively correlated with MAPK7 (Figure 7H). But the mRNA and protein expression of MAPK7 was decreased in HOS and U2OS cells with miR-372-3p overexpression (Figure 7IL).
Figure 7.
MiR-372-3p directly interacted with MAPK7 in BC cells. The HOS and U2OS cells were co-transfected with MAPK7 3ʹUTR-WT+miR-NC, MUC19 3ʹUTR-WT+miR-372-3p, MAPK7 3ʹUTR-MUT+miR-NC, 3ʹUTR-MUT+miR-372-3p. (A) Starbase v3.0 predicted the putative site between miR-372-3p and MAPK7. (B and C) The luciferase activity was measured in HOS and U2OS cells. (D-E) The expression of MAPK7 was measured by qRT-PCR and Western blot in OS tissues. (F and G) The expression of MAPK7 was measured by qRT-PCR and Western blot in hFOB1.19, HOS and U2OS cells. (H) The expression of miR-372-3p was negatively correlated with MAPK7 (R=−0.661). (I and J) The level of MAPK7 was determined using the qRT-PCR and Western blot analysis in HOS cells transfected with pcDNA3.0-miR-372-3p. (K and L) MAPK7 level was determined using the qRT-PCR and Western blot analysis in U2OS cells transfected with pcDNA3.0-miR-372-3p.*P<0.05, **P<0.01, ***P<0.001, **** P<0.0001.
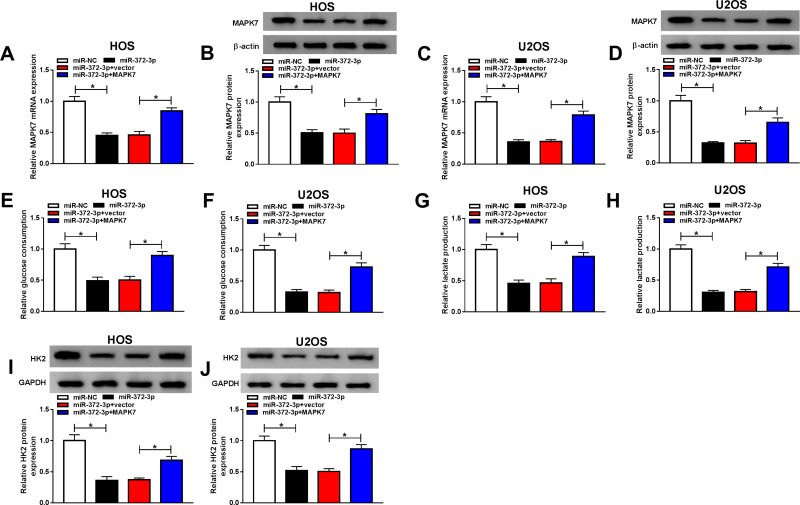
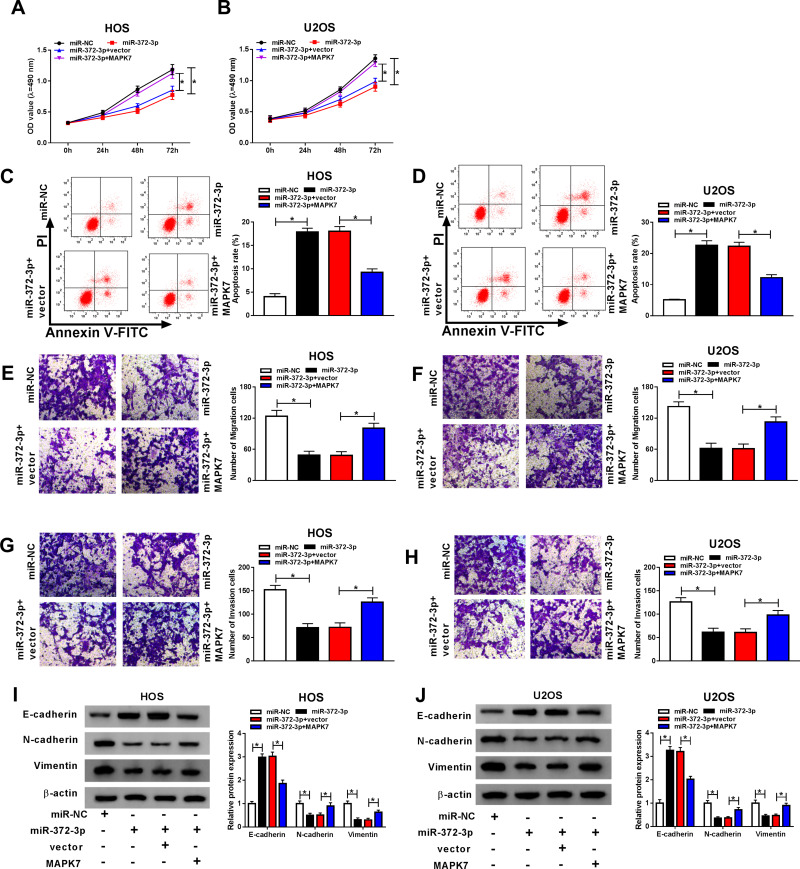
Overexpression of MAPK7 Attenuated Anti-Cancer Role of miR-372-3p in OS Cells
To make clear the influence of interaction between miR-372-3p, and MAPK7 on OS cell progression, miR-372-3p mimic and MAPK7 overexpression plasmid were co-transfected into HOS and U2OS cells. QRT-PCR and Western blot analysis results revealed that the level of MAPK7 was suppressed by miR-372-3p up-regulation in HOS and U2OS cells, while was promoted by the introduction of MAPK7 (Figure 8AD). QRT-PCR was used to detect the glucose consumption, lactic acid formation, and Western blot used to detect HK2 protein expression in HOS and U2OS cells treated with miR-NC, miR-372-3P, miR-372-3P+vector, or miR-372-3P+pcDNA3.0-MAPK7. The data showed that overexpression of MAPK7 attenuated inhibition role of silencing of miR-372-3p in glucose consumption, lactic acid formation, and HK2 protein expression (Figure 8EJ). These results suggested that MAPK7 overexpression attenuated the inhibitory effect of miR-372-3p silencing on glycolysis. MTT assay indicated that the overexpression of MAPK7 reversed the inhibitory effect of miR-372-3p on the proliferation of HOS and U2OS cells (Figure 9A and B). Flow cytometry and transwell assay showed that overexpression of MAPK7 weakened the pro-apoptosis, anti-migration, and anti-invasion effects caused by miR-372-3p interference in HOS and U2OS cells (Figure 9CH). Moreover, Western blot results showed that the overexpression of MAPK7 reversed the inhibition of miR-372-3p mimic on EMT, specifically inhibiting the expression of E-cadherin protein and promoting the expression of N-cadherin and Vimentin proteins in OS cells (HOS and U2OS) (Figure 9I and J). Collectively, our data showed that the overexpression of MAPK7 reversed the inhibitory effect of miR-372-3p.
Figure 8.
Overexpression MAPK7 reversed the inhibitory effects of MAPK7 on the progression of OS cells glycolysis. The HOS and U2OS cells were transfected with miR-NC, or miR-372-3P, miR-372-3P + vector, or miR-372-3P + pcDNA3.0-MAPK7. (A and B) RT-PCR was used to detect the transfection efficiency in HOS cells. (C and D) RT-PCR was performed to examine the transfection efficiency in U2OS cells. (E and F) RT-PCR was used to detect glucose consumption in HOS and U2OS cells. (G and H) RT-PCR was used to detect lactic acid production in HOS and U2OS cells. (I and J) Western blot was used to detect the expression of HK2 in HOS and U2OS cells. *P<0.05.
Figure 9.
Overexpression of MAPK7 reversed the effects of miR-372-3P on OS cells. The HOS and U2OS cells were transfected with miR-NC, or miR-372-3P, miR-372-3P+vector, or miR-372-3P+pcDNA3.0-MAPK7. (A and B) Cell proliferation was assessed by MTT assay in HOS and U2OS cells. (C and D) Flow cytometry was used to detect cell apoptosis. (E and F) Transwell assay was employed to explore HOS and U2OS cells migration ability. (G and H) Transwell assay was carried out to detect HOS and U2OS cells invasion ability. (I and J) Western blot assay was applied to test the protein levels of E-cadherin, N-cadherin, and Vimentin in HOS and U2OS cells. *P<0.05.
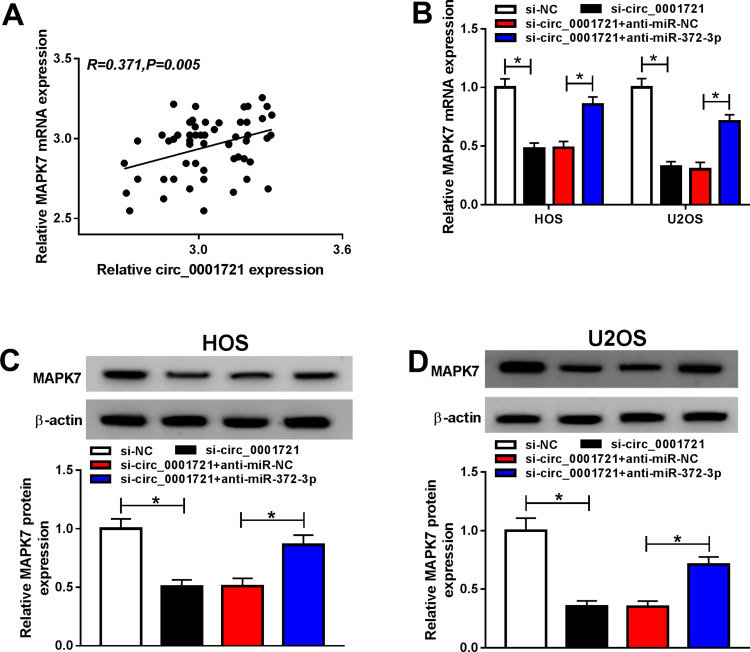
Circ_0001721 Promotes the Development of OS Tumors by Regulating MAPK7 Through miR-372-3p
The mRNA expression and protein expression of MAPK7 were detected by RT-PCR and Western blot and the data showed that the expression of miR-372-3p was gone down in HOS and U2OS cells transfected with si-circ_0001721 compared with that in those cells transfected with si-NC, but the downregulation of miR-372-3p reversed the inhibition effect of circ_0001721 knockdown on the expression of MAPK7 in HOS and U2OS cells (Figure 10B-D). Circ_0001721 was positively correlated with MAPK7 (Figure 10A), and circ_0001721 regulated MAPK7 through miR-372-3p.
Figure 10.
Circ_0001721 regulates MAPK7 through miR-372-3p. The HOS and U2OS cells were transfected with si-NC, or si-circ_0001721, si-circ_0001721+ anti-miR-NC, or si-circ_0001721+anti-miR-372-3P. (A) Circ_0001721 expression is positively correlated with MAPK7 (R=0.371) (B-D) The levels of mRNA and protein expression of MAPK7 were evaluated using qRT-PCR and Western blot in HOS and U2OS cells. *P < 0.05.
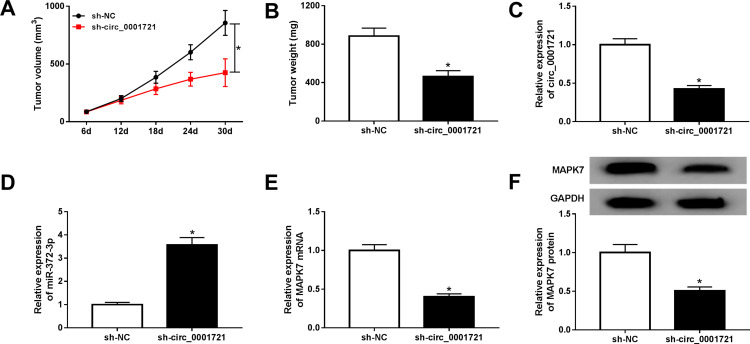
To further investigate the anti-cancer role of circ_0001721 silence, HOS cells, stably transfected with sh-circ_0001721 or sh-NC cells, were used to establish xenograft model in vivo. After cell injection for 30 days, tumor volume and weight were significantly reduced in a sh-circ_0001721 group compared with those in the sh-NC group (Figure 11A and B). Meanwhile, circ_0001721 expression was notably decreased in the sh-circ_0001721 group compared with those in the sh-NC (Figure 11C). Moreover, the expression of miR-372-3p was increased in the sh-circ_0001721 group when compared with that in the sh-NC group (Figure 11D). However, the levels of the protein and mRNA of MAPK7 were decreased in the sh-circ_0001721 group in comparison to those in the sh-NC group (Figure 11E and F). In conclusion, circ_0001721 could promote tumor development in vivo.
Figure 11.
Circ_0001721 knockdown inhibited tumor development in vivo. (A) Volume analysis of xenograft tumors. (B) Weight analysis of xenograft tumors. (C-E) The mRNA levels of circ_0001721, miR-372-3P, MAPK7 mRNA, and MAPK protein in xenograft tumors treated with HOS cells stably expressing sh-circ_0001721 or sh-NC were quantified by qRT-PCR. (E) QRT-PCR was carried out to determine the protein expression level of MAPK7 in xenograft tumors. (F) Western blot was carried out to determine the protein expression level of MAPK7 in xenograft tumors.*P <0.05.
Discussion
As a robust metastatic tumor in children and adolescents, osteosarcoma is highly invasive.28 The poor clinical outcome of OS patients is a massive problem in clinical treatment. Therefore, it is necessary to find new molecular targets and study their potential mechanism of action.
Many studies showed that circrRNAs were involved in regulating the progression of many cancers.29 CircRNAs served as competitive endogenous RNA recognition and characterization of miRNA-mRNA.30 Pei et al found that circ_0000218 played a carcinogenic role in the progression of colorectal cancer.31 Lu et al reported that circRNAs HIPK3 induced proliferation and inhibited apoptosis in non-small cell lung cancer cells.32 Lu et al confirmed that circ_0021977 inhibited the proliferation, migration, and invasion of colorectal cancer cells.6
To explore the function of circ_0001721, miR-372-3p and MAPK7, we checked its expression level and found that circ_0001721 was conspicuously upregulated,15 miR-372-3p was low expressed,21 and MAPK7 was highly expressed in OS tissues and cells,24 which was in line with a previous report. Our experimental results showed that the down-regulation of circ_0001721 effectively inhibited tumor occurrence. Specifically, the down-regulation of circ_0001721 inhibited glycolysis, cell proliferation, migration, invasion and EMT, and promoted apoptosis of OS cells. Previous studies on miR-372-3p have been numerous. For example, Wang et al reported that miR-372-3p promoted the growth and metastasis of squamous cell carcinoma.22 Xu et al confirmed that miR-372-3p inhibited the growth and metastasis of osteosarcoma cells by targeting FXYD6.21 Starbase predicted the targeting relationship between circ_0001721 and miR-372-3p and verified the relationship by dual-luciferase reporter assay and RIP. The results showed that miR-372-3p was negatively correlated with circ_0001721 expression in cell lines. The knockdown of circ_0001721 promoted the expression of miR-372-3p. The knockdown of circ_0001721 inhibited glycolysis, cell proliferation, migration, invasion, EMT, and promoted apoptosis through miR-372-3p.
To deeply explore the mechanism of miR-372-3p in OS, its target genes were predicted. And MAPK7 was confirmed to be a target of miR-372-3p. We then checked the protein level of MAPK7 mRNA and miR-372-3p in OS cells and found that resulted in decreased MAPK7 expression, which is miR-372-3p negatively regulated the expression of MAPK7. Overexpression of MAPK7 attenuated the anti-cancer effect of miR-372-3p in OS cells, specifically by reversing the miR-372-3p-mediated inhibition of glycolysis, cell proliferation, migration, invasion and EMT, and promotion of apoptosis. Circ_0001721 negatively regulated MAPK7 through miR-372-3p, which was confirmed by qRT-PCR and Western blot. Experiments in vivo with circ_0001721 knockdown had proved that circ_0001721 knockdown significantly inhibited tumor growth and reduced the expression of MAPK7 mRNA, MAPK7 protein, and miR-372-3p. Thus, circ_0001721 has a significant role in promoting the development of osteosarcoma.
Conclusion
Our research disclosed that circ_0001721 was strikingly upregulated in OS tissues and cells. Also, circ_0001721 could regulate the glycolysis, cell proliferation, migration, invasion and EMT, and apoptosis of OS cells by the miR-372-3p/MAPK7 axis. This novel mechanism might provide new effective therapeutic methods for OS.
Funding Statement
There is no funding to report.
Data Sharing Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for Publication
Not applicable.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Tabone MD, Terrier P, Pacquement H, et al. Outcome of radiation-related osteosarcoma after treatment of childhood and adolescent cancer: a study of 23 cases. J Clin Oncol. 1999;17(9):2789–2795. doi: 10.1200/JCO.1999.17.9.2789 [DOI] [PubMed] [Google Scholar]
- 2.Qu Q, He Z, Jiang Y, et al. C(1)(8)H(1)(7)NO(6) inhibits invasion and migration of human MNNG osteosarcoma cells via the PI3K/AKT signaling pathway. Med Sci Monit. 2019;25(75):27–37. doi: 10.12659/MSM.918431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 4.Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang BQ, Li ML, Quan HY, et al. Functional roles of circular RNAs during epithelial-to-mesenchymal transition. Mol Cancer. 2019;18(1):138. doi: 10.1186/s12943-019-1071-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C, Jiang W, Hui B, et al. The circ_0021977/miR-10b-5p/P21 and P53 regulatory axis suppresses proliferation, migration, and invasion in colorectal cancer. J Cell Physiol. 2020;235(3):2273–2285. doi: 10.1002/jcp.29135 [DOI] [PubMed] [Google Scholar]
- 7.Shen Z, Zhou L, Zhang C, Xu J. Reduction of circular RNA Foxo3 promotes prostate cancer progression and chemoresistance to docetaxel. Cancer Lett. 2020;468:88–101. doi: 10.1016/j.canlet.2019.10.006 [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Wu A, Yang B, Zhu X, Teng Y, Ai Z. Profiling and bioinformatics analyses reveal differential circular RNA expression in ovarian cancer. Gene. 2020;724:144150. doi: 10.1016/j.gene.2019.144150 [DOI] [PubMed] [Google Scholar]
- 9.Zhong Q, Huang J, Wei J, Wu R. Circular RNA CDR1as sponges miR-7-5p to enhance E2F3 stability and promote the growth of nasopharyngeal carcinoma. Cancer Cell Int. 2019;19:252. doi: 10.1186/s12935-019-0959-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi: 10.1186/s12943-019-1007-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J, Zhang PY, Xie JW, et al. Circular RNA hsa_circ_0006848 related to ribosomal protein L6 acts as a novel biomarker for early gastric cancer. Dis Markers. 2019;2019:3863458. doi: 10.1155/2019/3863458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Li H, Lu H, Qin Y. Circular RNA SMARCA5 inhibits the proliferation, migration, and invasion of non-small cell lung cancer by miR-19b-3p/HOXA9 axis. Onco Targets Ther. 2019;12:7055–7065. doi: 10.2147/OTT.S216320 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Ying X, Zhu J, Zhang Y. Circular RNA circ-TSPAN4 promotes lung adenocarcinoma metastasis by upregulating ZEB1 via sponging miR-665. Mol Genet Genomic Med. 2019;7(12):e991. doi: 10.1002/mgg3.991 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol. 2019;25(35):5300–5309. doi: 10.3748/wjg.v25.i35.5300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Guo L, Yin G, Yu G, Zhao Y, Pan Y. Upregulation of circular RNA circ_0001721 predicts unfavorable prognosis in osteosarcoma and facilitates cell progression via sponging miR-569 and miR-599. Biomed Pharmacother. 2019;109:226–232. doi: 10.1016/j.biopha.2018.10.072 [DOI] [PubMed] [Google Scholar]
- 16.Xie Q, Chen X, Lu F, et al. Aberrant expression of microRNA 155 may accelerate cell proliferation by targeting sex-determining region Y box 6 in hepatocellular carcinoma. Cancer. 2012;118(9):2431–2442. doi: 10.1002/cncr.26566 [DOI] [PubMed] [Google Scholar]
- 17.Loh HY, Norman BP, Lai KS, Rahman N, Alitheen NBM, Osman MA. The regulatory role of micrornas in breast cancer. Int J Mol Sci. 2019;20(19):4980. doi: 10.3390/ijms20194940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang C, Li Y. Prospective applications of microRNAs in oral cancer. Oncol Lett. 2019;18(4):3974–3984. doi: 10.3892/ol.2019.10751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, Dong J, He Y, et al. miR-218 inhibited tumor angiogenesis by targeting ROBO1 in gastric cancer. Gene. 2017;615:42–49. doi: 10.1016/j.gene.2017.03.022 [DOI] [PubMed] [Google Scholar]
- 20.Sadri Nahand J, Moghoofei M, Salmaninejad A, et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int J Cancer. 2020;146(2):305–320. doi: 10.1002/ijc.32688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu SY, Xu PF, Gao TT. MiR-372-3p inhibits the growth and metastasis of osteosarcoma cells by targeting FXYD6. Eur Rev Med Pharmacol Sci. 2018;22(1):62–69. doi: 10.26355/eurrev_201801_14101 [DOI] [PubMed] [Google Scholar]
- 22.Wang Q, Liu S, Zhao X, Wang Y, Tian D, Jiang W. MiR-372-3p promotes cell growth and metastasis by targeting FGF9 in lung squamous cell carcinoma. Cancer Med. 2017;6(6):1323–1330. doi: 10.1002/cam4.1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Syring I, Bartels J, Holdenrieder S, Kristiansen G, Muller SC, Ellinger J. Circulating serum miRNA (miR-367-3p, miR-371a-3p, miR-372-3p and miR-373-3p) as biomarkers in patients with testicular germ cell cancer. J Urol. 2015;193(1):331–337. doi: 10.1016/j.juro.2014.07.010 [DOI] [PubMed] [Google Scholar]
- 24.Hou Y, Feng H, Jiao J, et al. Mechanism of miR-143-3p inhibiting proliferation, migration and invasion of osteosarcoma cells by targeting MAPK7. Artif Cells Nanomed Biotechnol. 2019;47(1):2065–2071. doi: 10.1080/21691401.2019.1620252 [DOI] [PubMed] [Google Scholar]
- 25.Zhou N, Yan HL. MiR-24 promotes the proliferation and apoptosis of lung carcinoma via targeting MAPK7. Eur Rev Med Pharmacol Sci. 2018;22(20):6845–6852. doi: 10.26355/eurrev_201810_16153 [DOI] [PubMed] [Google Scholar]
- 26.Dong X, Lv B, Li Y, Cheng Q, Su C, Yin G. MiR-143 regulates the proliferation and migration of osteosarcoma cells through targeting MAPK7. Arch Biochem Biophys. 2017;630:47–53. doi: 10.1016/j.abb.2017.07.011 [DOI] [PubMed] [Google Scholar]
- 27.Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging. 2019;11(19):8182–8203. doi: 10.18632/aging.102312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sapio L, Salzillo A, Illiano M, et al. Chlorogenic acid activates ERK1/2 and inhibits proliferation of osteosarcoma cells. J Cell Physiol. 2020;235(4):3741–3752. doi: 10.1002/jcp.29269 [DOI] [PubMed] [Google Scholar]
- 29.Bai H, Lei K, Huang F, Jiang Z, Zhou X. Exo-circRNAs: a new paradigm for anticancer therapy. Mol Cancer. 2019;18(1):56. doi: 10.1186/s12943-019-0986-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan W, Peng S, Wang J, et al. Identification and characterization of circRNAs as competing endogenous RNAs for miRNA-mRNA in colorectal cancer. PeerJ. 2019;7:e7602. doi: 10.7717/peerj.7602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pei FL, Cao MZ, Li YF. Circ_0000218 plays a carcinogenic role in colorectal cancer progression by regulating miR-139-3p/RAB1A axis. J Biochem. 2019;167(1):55–65. doi: 10.1093/jb/mvz078 [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Han X, Ren J, Ren K, Li Z, Sun Z. Circular RNA HIPK3 induces cell proliferation and inhibits apoptosis in non-small cell lung cancer through sponging miR-149. Cancer Biol Ther. 2020;21(2):113–121. doi: 10.1080/15384047.2019.1669995 [DOI] [PMC free article] [PubMed] [Google Scholar]