Abstract
Increased blood sugar levels in prolonged diabetes lead to secondary complications such as retinopathy, neuropathy, and nephropathy, which gradually end in death. Synthesis of nano-phytomedicines from active phytoconstituents for novel emerging applications in the field of pharmaceuticals is of huge interest among researchers. In the present investigation, encapsulated ellagic acid (NEA) was synthesized at four different concentrations (0.2%, 0.3%, 0.4%, 0.5%) using ZnO nanoparticles as encapsulating agent. The surface morphology (fiber-like structures) of the nanoparticles were determined by scanning electron microscopy (SEM) and particle size (161–297 nm) and zeta potential (− 54.9–38.4 mV) were determined by dynamic light scattering technique. Further, the α-glucosidase and aldose reductase enzymes were significantly inhibited by the 0.4% of NEA compared to the other concentrations which strengthened our studies in overcoming the secondary complications of diabetes. The interaction analysis between ellagic acid and insulin receptor found Hit 1 among 10 executed ∆G score and energy of − 5.76, − 4.63 kcal/mol and formed polar bond with Arg 113 with − 1.175 Å distance. The residues Arg115, Lys116, Phe118, Ile115, Arg1131, Arg1155, Ile1157, Lys1165 and Phe1186 were found in ligand–protein interactions. ADME/T analysis of hit 1 was within the acceptable range without any toxic functional groups, providing a framework for developing novel therapeutics.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02411-1) contains supplementary material, which is available to authorized users.
Keywords: α-Glucosidase, Aldose reductase, Antioxidant, Docking, Ellagic acid, Nanopharmaceuticals, Polyphenols
Introduction
Improving the current method of diabetes control to overcome complications such as ROS reduction using sophisticated technology and herbals is the need of the hour. Polyphenols make the largest family of phytochemicals with numerous biological properties such as antimicrobial, antihyperglycemic and anticancerous activity. Besides these, having the ability to donate hydrogen atoms or electrons, polyphenols possess very strong free radical scavenging action as evidenced by many researchers (Perron and Brumaghim 2009; Kim et al. 2014). Novel drug formulation is targeted on improving life expectancy and health status of the population using herbal compounds (Ahmad et al. 2013). Ellagic acid is a natural polyphenol extensively available in fruits such as pomegranates, raspberries, strawberries and blueberries. Due to its poor solubility, the therapeutic effects could not be achieved, which in turn may be due to poor absorption and retention in the body. Hence, a carrier is needed to effectively deliver the drug and achieve therapeutic output. One method to produce such a carrier is nanotechnology, by which particles in the range of 1–1000 nm in dimension (Etheridge 2013) are synthesized by physical, chemical and physicochemical methods to form stable drug suspensions through particle size reduction approaches (Etheridge 2013; Merisko-Liversidge and Liversidge 2011). This technology improves the solubility and biological responses besides minimizing toxicity and degradation, which is supposed to be due to large surface to volume ratio. Noble metal oxide nanoparticles differ from their bulk counterparts due to their unique electronic, optical, mechanical, magnetic and chemical properties (Feng et al. 2012). However, safety analyses need to be ensured by characterization techniques, particularly of nanomedicine.
Nanoparticles display unique and tunable biophysical characteristics which are determined by their shape and size compared to their respective bulk material. The human body has an antioxidant defense system towards free radicals, but overproduction of free radicals causes an imbalance condition between the free radical and antioxidant defense in the body that leads to several diseases including inflammation, aging and diabetes (Lobo et al. 2010). Zinc oxide forms the center of research as a special material accounting for its biocompatibility in biomedical and environmental applications.
Dietary carbohydrates are broken down into simple monosaccharides by a group of digestive enzymes called α-glucosidases (AGs) and elevated postprandial hyperglycemia is due to action of-glucosidase. Postprandial hyperglycemia in diabetes can be managed by inhibiting α-glucosidase. The digestion and absorption of carbohydrates are delayed in the digestive tract by the α-glucosidase inhibitors (AGIs). In the insulin-insensitive tissues of diabetics such as lens and retina, elevated levels of sorbitol are produced due to increased availability of glucose through polyol (Williamson et al. 1992). Aldose reductase (AR), the first enzyme in the polyol pathway, reduces excess d-glucose to d-sorbitol with conversion of NADPH to NADP + . Accumulation of sugar alcohols occur first in the lens epithelium, leading to cataract (Murata et al. 2001) which can be prevented by inhibiting the aldose reductase by means of aldose reductase inhibitors (ARIs).
Elevated expression of human insulin receptor tyrosine kinase is found in diabetes. Based on the facts available from literature and molecular evidences, human insulin receptor tyrosine kinase was considered as one of the prime targets in diabetes, hence there is a need for designing a novel therapeutic compound to combat the situation. In spite of several existing drugs, search for a novel compound with improved efficacy and minimal side effects has been continuing. Many in vitro and in vivo data have been accumulated which support the role of ellagic acid in controlling diabetes. The present study was implemented to design effective antagonists, targeting human insulin receptor tyrosine kinase which sheds light on the importance of finding potent inhibitors (Mavromoustakos et al. 2011). Molecular docking dynamics of a small molecule with the receptor using computational method predicted the interactions between two molecules.
Our study deals with the encapsulation of ellagic acid at various concentrations using ZnO nanoparticles and evaluates the best out of them having radical scavenging, AG and AR inhibitory activities. The study also analyzes the stability of protein–ligand docked complex followed by molecular docking experiments to propose potential inhibitors (Mondal et al. 2015; Wright et al. 2014).
Materials and methods
Synthesis of nanoencapsulated ellagic acid
Zinc nitrate hexahydrate [Zn (NO3)2.6H2O, 96%] was obtained from Fisher Scientific and ellagic acid (95%, HPLC powder from tree bark) procured from Sigma-Aldrich. All the suspensions were prepared with deionized water.
Encapsulated ellagic acid (NEA) using ZnO nanoparticles was synthesized by Prasad et al. (2012). The percentage of entrapment efficiency was calculated as per the methodology of Papadimitriou and Bikiaris (2009). It is calculated as follows:
UV–visible spectroscopy
The bioreductant nanoparticles (EA-ZnNPs) in four different concentrations were monitored by UV–visible (UV–Vis) spectrum at various time intervals. The UV–Vis spectra of this solution were recorded (UV-2450, SHIMADZU Spectrophotometer) from 200 to 800 nm.
Particle size and zeta potential analysis of ZnO nanoparticles
The aqueous suspension of the synthesized nanoparticles was filtered through a 0.22–l m syringe driven filter unit, and the size and distribution of the nanoparticles was measured using the dynamic light scattering technique (Nanopartica, HORIBA, and SZ-100).
SEM analysis
The morphology and crystalline phase of the nanocrystals were determined by scanning electron microscopy (SEM). The sample was deposited on a carbon film for SEM analysis using Hitachi SEM (a TM 3000).
Radical scavenging activity
The free radical scavenging activity of the four concentrations of the NEA was determined by using DPPH according to Kavitha et al. (2012). The ability to scavenge the stable free radical DPPH is measured by a decrease in the absorbance at 517 nm.
Qualitative analysis (thin layer chromatography): All the concentrations of NEA were applied on a TLC plate as a spot (1–2 mg/ml) using mobile phase (ethyl acetate: formic acid: water at 17:2:3) (Mohammad et al. 2010). The plates were left for 20–30 min, to develop the chromatogram, allowed to dry and exposed to iodine vapors in a closed chamber. The color change (yellow spot) on the TLC plate demonstrates the presence of antioxidant activity.
Quantitative analysis: Each sample (concentration) was dissolved in methanol, 10 mg/10 mL, and used as a stock solution. From the stock solutions, 1 mL of each compound solution with different concentrations (0.25–1.00 g) was added to 3 mL of methanolic DPPH (0.004%) solution. After 30 min, the absorbance of the test compounds were taken at 517 nm using UV–VIS spectrophotometer, which was compared with the corresponding absorbance of standard BHT (butylated hydroxytoluene) concentrations (0.25–1.00 g). DPPH solution was used as control without the test compounds, and methanol as blank. The percentage of scavenging activity of DPPH free radical was measured by using the following formula,
where A0 is the absorbance of the control and At the absorbance of the sample. IC50 values were calculated for compounds, which exhibited significant activity. IC50 is defined as concentration sufficient to obtain 50% of maximum scavenging activity.
In vitro α-glucosidase inhibition assay and aldose reductase inhibition assay
The AGI assay and ARI assay were carried out in vitro following the methods of Dahlqvist and Borgstrom (1961) and Hayman and Kinoshita (1965), respectively.
Preparation of AG: Rats were killed; small intestines were removed and cleaned in ice cold 80 mM phosphate buffer (pH 7.0). The intestine was then cut open, mucosa was scraped off and homogenized with four parts (v/w) of ice cold phosphate buffer (pH 7.0). Nuclei and cell debris were removed by centrifugation at 3000 rpm for 10 min and the supernatant was used as the source of enzyme. We used acarbose as positive control for the AG assay.
Preparation of AR: The eyeballs were removed from 9-week-old male rats. Lenses were dissected by the posterior approach and homogenized in 10 volumes of 100 mM potassium phosphate buffer, pH 6.2. The homogenate was centrifuged at 15,000×g for 30 min at 4 °C and the resulting supernatant was used as the source of AR. We used quercetin as the positive control.
Enzyme inhibition assays: The inhibitory activities of NEA on AG and AR are expressed as percentage of inhibition which was calculated as
where Ac is the absorbance of the control (without NEA) and As is the absorbance of the sample (with NEA). The concentration of the inhibitor required to inhibit 50% of enzyme activity under the assay conditions is defined as IC50.
In silico vetting
STRING analysis and interactive residue prediction
The knowledge of protein–protein interactions is crucial to understand the physiology of expression on proteins (Stark et al. 2006). This was addressed by implementing STRING networking tool, which currently covers 5,214,234 genes/proteins from 1133 organisms derived from four sources, such as genomic contexts, high-throughput experiments, conserved coexpression and literature studies (Szklarczyk et al. 2014; Kushwaha and Shakya 2010). The analysis was performed with confidence score more than 0.400, interactions not more than 50 to achieve global networking of target/receptor on gene ontology, pathways and a hierarchical clustering of association network with other biologically important proteins in the function.
After spotting the vital role of protein, the 3D structure of human insulin receptor tyrosine kinase was retrieved from the protein data bank (PDB). The LigPrep, PDBsum and swissBIF tools were applied to identify the active site cavities of the protein human insulin receptor tyrosine kinase. The results were cross-validated with the CASTp server (https://sts.bioengr.uic.edu/castp/calculation.php) to compute the correctness of active cavities on human insulin receptor tyrosine kinase.
Protein preparation
The structure of phosphorylated insulin receptor tyrosine kinase (1IR3) was retrieved from the PDB and the protein was minimized. Essential hydrogen atoms, Kollman united atom type charges and solvation parameters were added and the 3D structure was then processed. The identified active site cavities of the protein human insulin receptor tyrosine kinase [Ali Khosravi et al. 2019; Pradeep Pant and Jayaram 2019; Liang et al. 1998] were also assigned in the grid cubic box.
Ligand selection
The ligand 2D and 3D structures were retrieved from PubChem and prepared interactively using AutoDockTools (ADT) with Python script and the file was saved in PDBQT format file to be used as input (Bikadi and Hazai 2009). The structure of ellagic acid was drawn using Marvin Sketch to obtain SMILES and SDF format to be utilized in the study. The ellagic acid residues for human insulin receptor tyrosine kinase were selected from PDB sum and LigPrep analysis which were well correlated with the literature mining data. Lastly, the files were prepared for docking and the optimization of the ligand was carried out in Auto dock.
Docking simulations
The analysis was performed using Lamarckian genetic algorithm (LGA) and Solis & Wets local search method (Duhovny et al. 2002). Initial position, orientation, and torsions of the ligand molecules were set randomly and all rotatable torsions were released during docking. Each docking experiment was derived from ten different runs that were set to terminate after a maximum of 250,000 energy evaluations and the population size was set to 150. Docking simulations were performed using the Lamarckian genetic algorithm (LGA) and the Solis & Wets local search method and AMBER was used. A temperature constant of 298.15 K and atmospheric pressure of 1 were used during simulation studies (Kanipakam et al. 2020). During the search, a translational step of 0.2 Å, and quaternion and torsion steps of 5 were applied. The docking analysis was further cross checked with Docking server, PatchDock (Shukla et al. 2017) and 1 click-dock server tools to dock the ellagic acid with proteins to refine the results. The dimension of the grid box was set at 25 × 25 × 25 with the default spacing of 0.375 Å between grid points including all the important interactive sites of amino acids (Hema et al. 2015). A shape comparison filter is combined with a Monte Carlo conformational search for generating ligand poses consistent with the binding site. Candidate poses are minimized in the context of the binding site using a grid-based method for evaluating receptor–ligand interaction energies. Therefore, the binding orientations of the proposed molecules were predicted through docking simulations by analyzing the trajectory data obtained.
Free energy calculations
MM-GBSA is a computationally efficient method for estimating the binding free energy (ΔGbind) of protein–ligand complexes (Kollman et al. 2000). Autodock scoring function based on the AMBER force field was used in the present study to estimate the free energy of binding of a ligand to its target. In this method, ΔG bind is expressed through the well-known equation:
where (Gcomplex), (Greceptor) and (Gligand) are the average values of Gibbs free energy for the complex, receptor and ligand, respectively (Li et al. 2011). Separate simulation of the complex and its two components can be used, but due to difficulties in obtaining converged free energies, it is more common to extract configurations of the free enzyme and ligand from simulation of the complex (Tripathi et al. 2013; Du et al. 2011).
Toxicity properties
High oral availability is often an important consideration for the development of bioactive molecules as therapeutic agents. The selected optimized molecules were studied for their drug-like properties. An in silico study was carried out for the prediction of pharmacokinetic properties using PreADME web-based approach (Lee et al. 2002). Properties such as blood–brain penetration, skin permeability, MDCK cell permeability and rule of five were used to judge the compound’s overall potentiality as a drug molecule. The overall pharmacological properties of the proposed molecules were analyzed for the normal ranges without any violations and biologically active without any toxic functional groups (Hema et al. 2015).
Results
The entrapment efficiencies of 0.4% and 0.5% were found to be significant (23% and 25%, respectively) compared to the other concentrations (supplementary data).
UV–visible spectral analyses
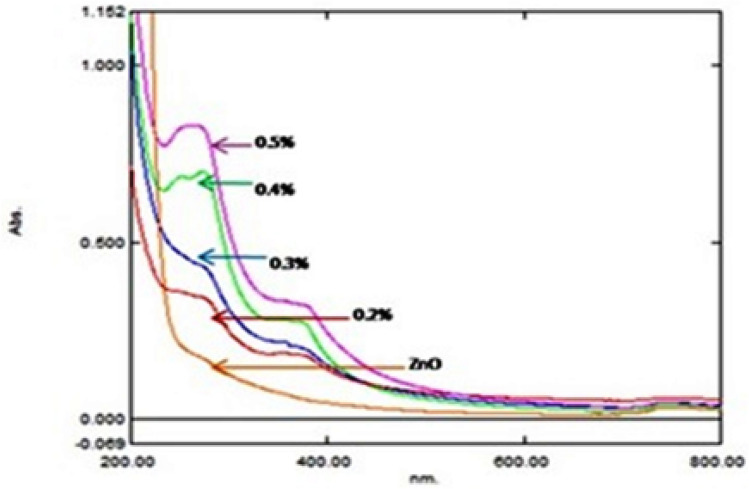
The optical properties of nano-sized particles were examined by means of UV–visible absorption spectroscopy. The spectrum showed the absorbance peak at 360 nm corresponding to the characteristic LSPR band of ZnO nanoparticles (Fig. 1).
Fig. 1.
The spectra of 0.2%, 0.3%, 0.4% and 0.5% represent the absorbance peaks at 360 nm, corresponding to the characteristic LSPR band of ZnO nanoparticles, confirming the encapsulation with the reference ZnO characteristic spectrum
Particles of sizes between a few nanometers to 1000 nm are generally considered as nanoparticles in the pharmaceutical field (Junghanns and Muller 2008). By our studies, we can conclude that the 0.4% and 0.5% concentrations are more reliable compared to the other concentrations with respect to the size, stability and the radical scavenging activity, further confirming the drug as a promising antioxidant in the biological system. The biomatrix present in ellagic acid may be responsible for change in the absorbance UV–VIS micrograph (Gunalan et al. 2012; Revina et al. 2007) The peak also indicates the broad nano-sized particle distribution of NEA–ZnO nanoparticles, while the absorption peak at 230 nm represents that of ellagic acid corresponding to the cyclic structures. The results are in agreement with those of Brahmam et al. (2014).
Particle size analysis
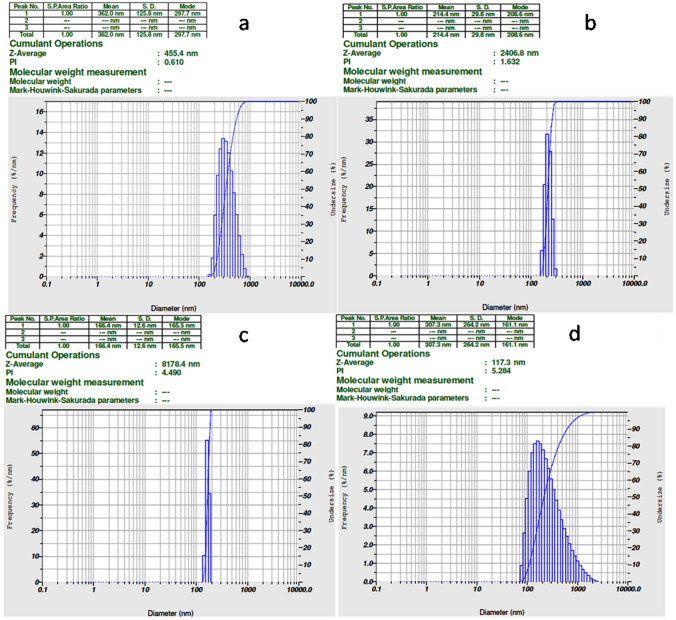
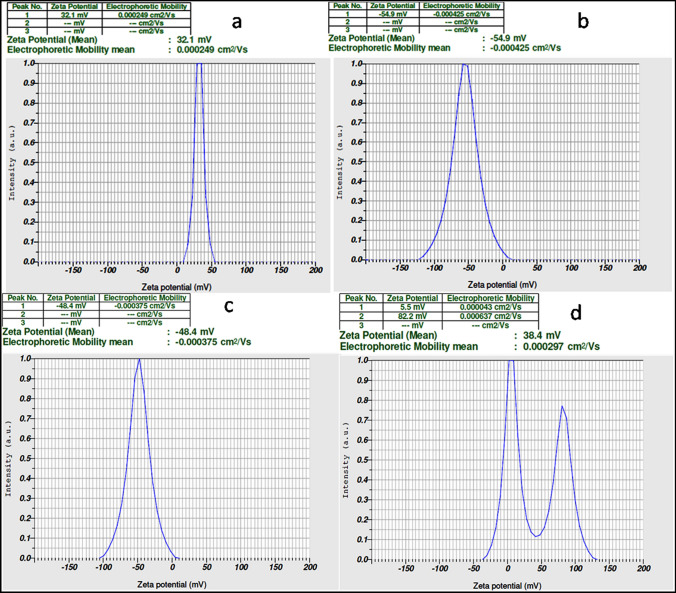
The particle size distribution spectra for the zinc oxide nanoparticles were recorded as diameter (nm) versus frequency (%/nm) spectra with diameter (nm) on the x axis and frequency (%/nm) on the y axis. The zeta potential spectra for the zinc oxide nanoparticles were recorded as zeta potential versus intensity spectra with zeta potential (mV) on the x axis and intensity (a.u) on the y axis. Particle sizes of 297.4 nm, 208.6 nm, 168.5 nm and 161.1 nm (Fig. 2a–d) and zeta potentials of 32.1 mV, − 54.9 mV, − 48.4 mV and 38.4 mV for 0.2%, 0.3%, 0.4% and 0.5%, respectively, were recorded (Fig. 3a–d).
Fig. 2.
Particle sizes of NEA (a − 0.2%, b − 0.3%, c − 0.4%, d − 0.5%) corresponding to 297.7 nm, 208.6 nm, 165.5 nm, 161.1 nm, respectively, satisfy the fact that particles of sizes between 100 and 1000 nm are generally considered as nanoparticles in the pharmaceutical field
Fig. 3.
Zeta potential of NEA (a − 0.2%, b − 0.3%, c − 0.4%, d − 0.5%) demonstrate the fact that the hydrosols with a large negative (− 54.9 mV corresponding to 0.3% NEA & − 48.4 mV corresponding) or positive zeta potential tend to repel each other, avoiding particle agglomeration and are considered to be more stable compared to the rest
The obtained particle size and zeta potential signify the presence of repulsive electrostatic forces among the synthesized zinc nanoparticles leading to monodispersity of the particles. The hydrosols with a large negative or positive zeta potential tend to repel each other and hence there is no agglomeration of the particles. For the zinc nanoparticles synthesized from EA, least particle size and high zeta potential values were recorded. However, if the particles have low zeta potential values, then there will be no force to prevent the particles coming together and flocculating (Roy and Bhattacharya 2013).
SEM
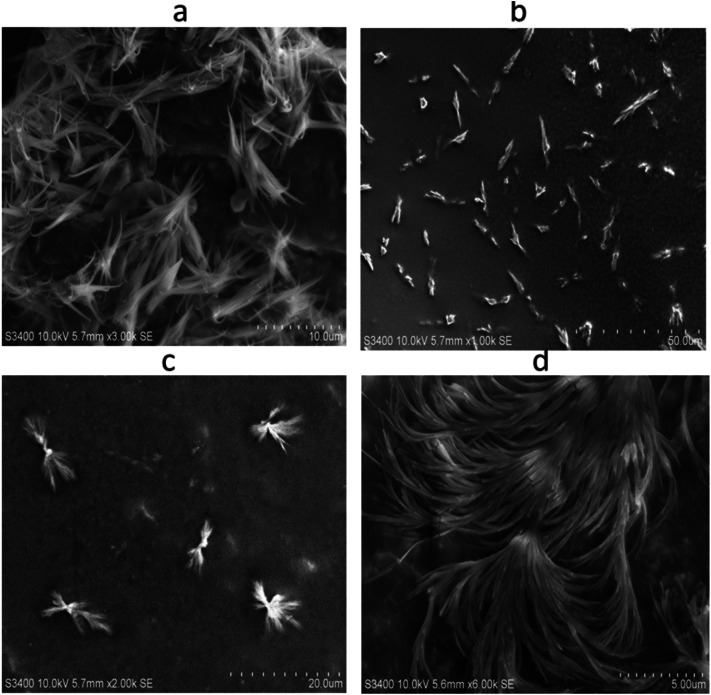
The SEM images of synthesized NEA are shown in Fig. 4 and the analysis showed that the synthesized nanoparticles have a uniform, needle-like morphology (Fig. 4a–d), with a narrow particle size distribution and smooth surfaces. Only very few numbers of spherical, micron-size particles, with a broad particle size distribution and a smooth surface can be seen in Fig. 4c sample corresponding to 0.4%.
Fig. 4.
SEM analyses of NEA (a − 0.2%, b − 0.3%, c − 0.4%, d − 0.5%): The nanoparticles show uniform, needle-like morphology (a–d), with a narrow particle size distribution and smooth surfaces. Dense particle size distribution and smooth surface with feathery appearance in the (d) corresponding to the 0.5% concentration could be responsible for the efficient release of drug so as the activity
The aggregation of particles in SEM images might be due to the large surface area and high surface energy of ZnO nanoparticles (Raoufi 2013). The microscopy results are in good agreement with that of Rasha N. Moussawi and Digambara Patra 2016 (Rasha and Patra 2016), where the curcumin-doped ZnO particles were fiber-like, almost similar to that of the 0.5% NEA (Fig. 4d).
Radical-scavenging activity
Qualitative analysis: The chromatograms represent distinct characteristic spots of the drug on exposure to iodine vapors, which indicates that all the four concentrations of NEA possess antioxidant activity (Fig. 3 of supplementary data). The Rf values calculated through thin layer chromatography were in the range of 0.7–0.77.
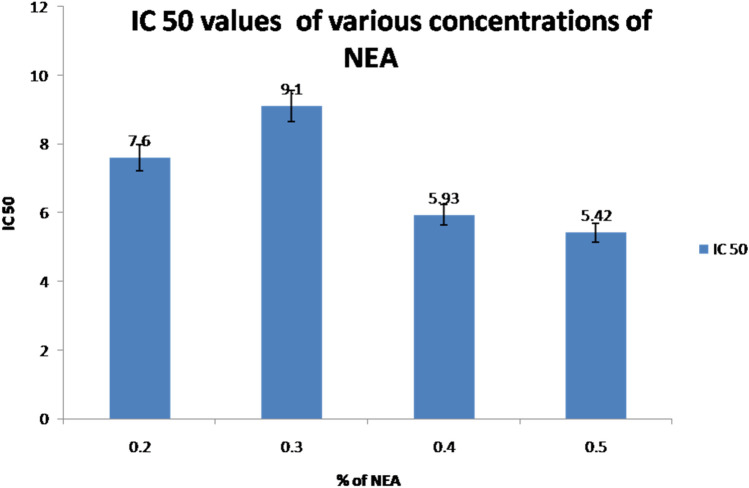
Quantitative assay: The DPPH antioxidant assay is based on the ability of decolorization of DPPH (a stable free radical containing an odd electron responsible for absorbance at 517 nm) in the presence of antioxidants. The results of the study revealed that 0.4% of NEA showed better antioxidant activity when compared to the other concentrations and the IC50 values, as shown in (Fig. 5).
Fig. 5.
Antioxidant activity vs. % NEA. Lowest IC 50 value of 0.5% NEA represents the concentration with higher antioxidant activity, followed by 0.4% NEA. Values are represented as mean ± S.D. n = 6 at each concentration
The number and position of hydrogen-donating hydroxyl groups on the aromatic ring of the phenol molecules is responsible for the radical-scavenging activity of polyphenols. The hydroxyl functional group scavenges free radicals and prevents the oxidation of biological molecules by converting more ROS by donating a hydrogen atom into inactive species (Zafar et al. 2016). The principle for the reduction of DPPH free radicals is that the antioxidant reacts with stable free radical DPPH and converts it into 1,1-diphenyl-2-picrylhydrazine. When DPPH accepts an electron donated by an antioxidant compound, the DPPH is decolorized and can be quantitatively measured by changes in absorbance.
In vitro α-glucosidase inhibition assay and aldose reductase inhibition assay
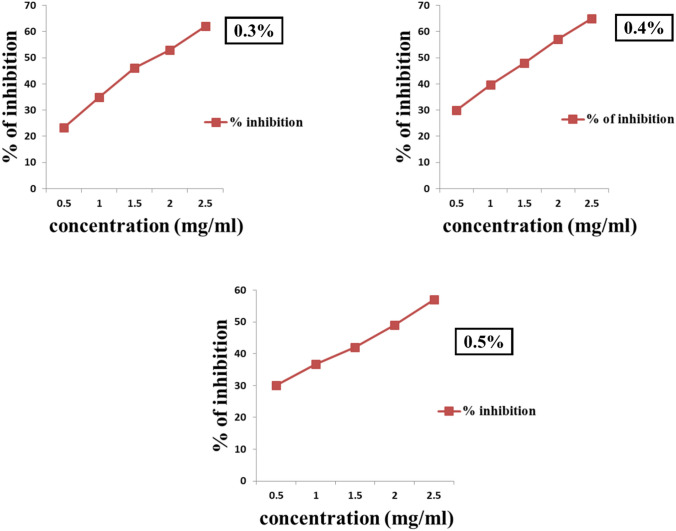
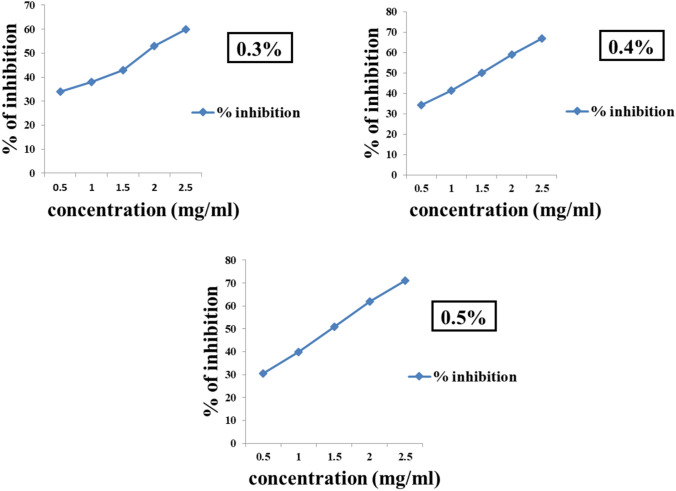
The 0.2% NEA showed no significant inhibition of AR and AG and the other three concentrations are represented in Figs. 6 and 7, respectively. The inhibitory activities of quercetin and acarbose, used as reference standards for AR and AG are represented in the supplementary data (Fig. 4).
Fig. 6.
Aldose reductase assay: the 0.4% NEA demonstrating higher inhibition on the enzyme activity, i.e., close to 65%, followed by 0.5% NEA demonstrating inhibition close to 60% at a concentration of 2.5 mg/ml
Fig. 7.
α-Glucosidase assay: 0.5% NEA inhibited the enzyme activity significantly close to 75%, followed by 0.4% NEA demonstrating inhibition close to 70% at a concentration of 2.5 mg/ml
Our investigations on enzyme inhibition coincide with that of Marella et al. (2016) where the Mcy protein, a potent antidiabetic active principle , inhibited the AR and AG enzymes in in vitro studies. As the concentration of NEA increased, the percentage of inhibition was concomitantly improved.
In silico studies
Network analysis and binding site determination
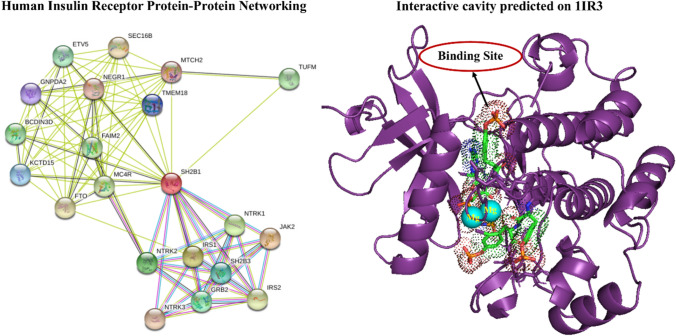
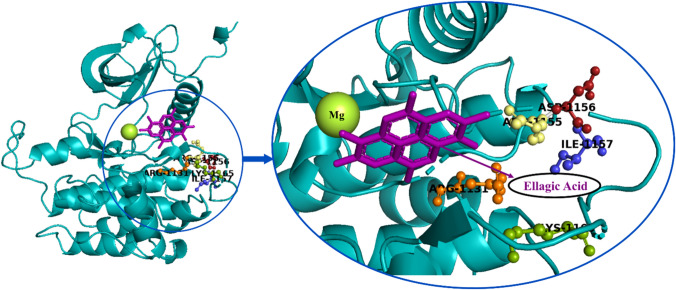
STRING resulted with interacting genes/proteins to construct protein–protein interaction networks. The protein human insulin receptor tyrosine kinase has confidence scores of > 0.900 with STRING v9.1 analysis, and hence considered as functionally important in protein network highly interacting with other proteins (Fig. 8). The STRING analysis revealed a great association between the insulin receptor binding to ellagic acid, which is involved in a major pathway projecting the crucial strength of the receptor at the protein level.
Fig. 8.
Protein–protein networking showing interaction and gene relationship associated with human insulin receptor, the series of subsequent phenotypes were kept with nominal nodes using STRING analysis (left) and the interactive binding cavity of native human insulin receptor–1IR3 depicting the presence of ellagic acid with magnesium ions (right) visualized using PyMOL tool
Active site analysis provides a significant insight into the docking simulation study. Active site analysis tools could also be used to calculate the number, boundary of mouth openings of every pocket, molecular reachable surface and area (Knox et al. 2011). Active site analysis of the insulin receptor was performed using, Swiss PDB Viewer and PDB ligand Explorer and PDBsum. The ellagic acid residues, namely Arg 1155, Phe 1156, Arg 1131, Lys 1165 and Ile 1157 were selected from PDB sum as active site cavity in the current study after the cross validation with literature mining with in silico tools (Fig. 8; Table 1).
Table 1.
The ten top binding cavities predicted on insulin receptor tyrosine kinase (1IR3)
| S. no | Predicted site | Amino acid residue name | Bind scores (kcal/mol) |
|---|---|---|---|
| 1 | Cavity I | IRAFESVGMKNLDHYPQW | − 18.570 |
| 2 | Cavity II | WEKGVIDLFTMYRCSAHP | − 31.540 |
| 3 | Cavity III | ETDFIKRAVYNLSPHM | − 34.845 |
| 4 | Cavity IV | ELKIHGRVMTASPDFQY | − 31.166 |
| 5 | Cavity V | KAMVEGDRLQYNHTSFP | − 37.418 |
| 6 | Cavity VI | PDESWKVFGAILTMYRCH | − 31.540 |
| 7 | Cavity VII | RDYKQLWMPFGAIETSCN | − 28.570 |
| 8 | Cavity VIII | LISDRGNFTAVKEMHYPW | − 44.479 |
| 9 | Cavity IX | YQRGNDASKLPEIT | − 29.974 |
| 10 | Cavity X | IRKLGEVTANHMDSFQYP | − 40.545 |
Analysis of molecular docking simulations
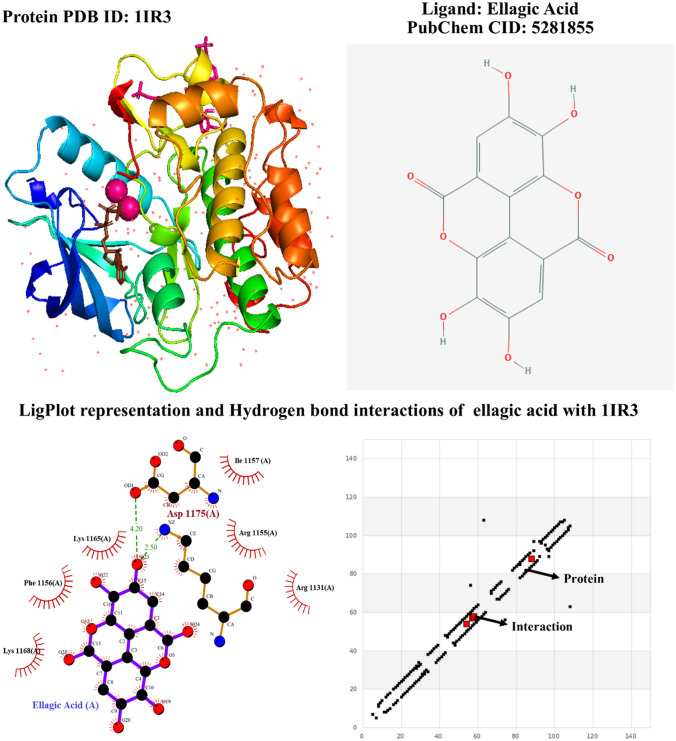
Docking was performed between insulin receptor tyrosine kinase (1IR3) and ellagic acid (Fig. 9) using AutoDock, binding energy values were evaluated and interactions were analyzed. The AutoDock results revealed ten best hits against insulin receptor tyrosine kinase, where they have resulted in good binding energies of − 5.76 kcal/mol, − 5.32 kcal/mol, − 5.31 kcal/mol, − 4.71 kcal/mol, − 4.53 kcal/mol, − 4.38 kcal/mol, − 4.27 kcal/mol, − 3.75 kcal/mol, − 3.75 kcal/mol, − 3.53 kcal/mol, and − 3.30 kcal/mol, respectively. The active site consists of residues: Arg 1155, Phe 1156, Arg 1131, Lys 1165, and Ile 1157 of receptor, found to interact with the residues of ellagic acid predicting the complex to be more stable (Table 2).
Fig. 9.
Depiction of crystal structure of human insulin receptor and ellagic acid retrieved from PDB and visualized with PyMOL and PubChem, respectively (top). The detailed representation of amino acid residues formed between the docked complex of human insulin receptor and ellagic acid using LigPrep along with hydrogen bond interactions plot (bottom)
Table 2.
ADME/T properties of hit 1
| ID | Value |
|---|---|
| Blood brain penetration (c.brain/c.blood) | 0.321339 |
| Buffer_solubility_mg_L | 8382.42 |
| Caco2 | 20.4888 |
| CYP_2C19_inhibition | Inhibitor |
| CYP_2C9_inhibition | Inhibitor |
| CYP_2D6_inhibition | Non |
| CYP_2D6_substrate | Non |
| CYP_3A4_inhibition | Inhibitor |
| CYP_3A4_substrate | Non |
| Human intestinal absorption (HIA %) | 61.395127 |
| MDCK cell permeability | 17.2974 (nm/s) |
| Pgp_inhibition | Non |
| Plasma_Protein_Binding (%) | 88.402085 |
| Pure_water_solubility_mg_L | 922.464 |
| Skin_Permeability Logkp.cm/hour | − 4.86783 |
| SKlogD_value | 1.86206 |
| SKlogP_value | 1.86206 |
| SKlogS_buffer | − 1.55692 |
| SKlogS_pure | − 2.51534 |
Among the ten, hit 1 showed the highest docking score and binding energies with superior number of H-bonds and van der Waals interactions with ellagic acid. Arg 113 showed polar interaction with ellagic acid with a distance of − 1.17 Å. Arg 115, Lys 116, Phe 118, Ile 115 formed four hydrogen bonds with a distance of − 3.0456, − 1.2038, − 0.5475 and − 0.4051, respectively. The amino acid residues Arg 1131, Arg 1155, Ile 1157, Lys 1165, Lys 1165 and Phe 1186 were also involved in the interactive site of receptor–ligand complex (Fig. 9).
Molecular docking using PatchDock and Docking Server based on AutoDock parameters was performed for the docked complex structures of protein–ellagic acid to know the binding energy values and interactions. The PatchDock results revealed the ten best hits that were refined using fire dock module and found hit 1 with best binding affinity and pose with ACE score of − 75.5 kcal/mol with the RMSD parameter set to 1.00 Å (Fig. 10). The residues found in the interactive site, polar region and hydrophobic showed very good affinity towards the receptor (Table 3). The molecular docking results showed that the binding was highly stable and thus underlined with good docking score, with binding energies thereby exerting a prominent effect on the 3D structure of the protein. The molecular docking analysis results proved that the binding was highly stable. Therefore, the study showed novel insight into the natural dynamics by revealing the presence of hydrogen bonds on different timescales docking complex in solution. Hence, hit 1 has good pharmacological properties with high stability, can act as potential competitive inhibitor receptor to block its activity and would be useful for developing new therapeutics.
Fig. 10.
Representation of molecular side of ellagic acid that bound to human insulin receptor (left), depiction of amino acid residues responsible for the activation of protein levels found in the active pocket of ellagic acid and 1IR3 complex (right)
Table 3.
The ten top docked poses of 1IR3 and ellagic acid by PatchDock
| Rank | Solution number | Global energy | Attractive van der Waal forces | Repulsive van der Waal forces | Atomic contact energy (kcal/mol) |
|---|---|---|---|---|---|
| 1 | 6 | − 22.24 | − 14.09 | 2.38 | − 3.42 |
| 2 | 4 | − 22.42 | − 15.01 | 2.48 | − 1.96 |
| 3 | 2 | − 22.10 | − 15.17 | 5.22 | − 2.48 |
| 4 | 1 | − 20.50 | − 14.32 | 3.06 | − 1.74 |
| 5 | 3 | − 19.89 | − 14.13 | 4.05 | − 2.27 |
| 6 | 8 | − 19.84 | − 15.72 | 5.14 | − 1.79 |
| 7 | 5 | − 17.61 | − 14.42 | 11.06 | − 3.34 |
| 8 | 7 | − 11.88 | − 7.61 | 1.62 | − 3.37 |
| 9 | 10 | − 11.12 | − 8.81 | 2.98 | − 1.08 |
| 10 | 9 | − 8.86 | − 8.25 | 2.26 | 0.14 |
Absorption digestion metabolism excretion toxicity (ADMET) properties
The ADMET properties were analyzed using PreADME tool which is a PC-based program that detected the drug likeliness and drug-like properties of ellagic acid and the consecutive hits selected in the present study. The compounds which are labeled as drug-like resemble the existing drug molecules and exhibit the property cutoff values listed in Table 2. The compounds exceeding the cutoff values tend to have solubility and permeability problems, which would lead to poor oral bioavailabity (Lipinski and Hopkins 2004).
Our results were further strengthened as the proposed ten hits also showed very good affinity toward the receptor with minimal range of estimated inhibition constant, Ki electrostatic energy frequency interaction surface. The subsequent ΔG binding calculations of the proposed lead molecules and the analyses of binding determinants and hydrogen bond were calculated to better understand the main energy components contributing to the formation of different enzyme–ligand complexes which include van der Waals, electrostatic, polar solvation, non-polar solvation and entropy contributions of the receptor–ligand docking complex.
In addition to the rule of five, in silico filters (Jorgensen et al. 1996) can also find the toxicity parameters that are essential to discard the compounds without any drug-like characteristics and undesirable toxic effects before screening. The ten hits showed minimal range of values without toxic groups, obeying pharmacological parameters.
The availability of existing inhibitors in combination with bioinformatics tools and databases is of great assistance in reducing the problem of searching for potential inhibitors/activators in a large pool of gene/protein polls. The proposed hit 1 molecule represented a panel of broad functional relevance in drug discovery research in developing powerful drug molecules for the treatment regimen in diabetes.
Nanotechnology promises to re-engineer the man-made world, molecule by molecule, sparking a wave of novel revolutionary commercial products from machines to medicine. Still, their biological effects need to be evaluated and all relevant exposure routes have to be assessed. Hence nano EA can be explored as a new source of alternative medicine in the treatment of diabetes and its secondary complications in particular. Considering the molecular docking dynamics studies, hit 1 was highly stable with good binding affinity and, hence, we conclude that hit 1 can be suggested as a novel antagonist against human selected receptor, which could decrease and inhibit the enhanced levels of human insulin receptor tyrosine kinase.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The project was supported by the Department of Science and Technology (DST)—Science and Engineering Research Board (SERB) under National Post Doctoral Fellowship (PDF/2015/000534).
Compliance with ethical standards
Conflict of interest
The authors declare no competing financial interest.
Contributor Information
Saritha Marella, Email: sarimarella@gmail.com.
T. N. V. K. V. Prasad, Email: tnvkvprasad@gmail.com
References
- Ahmad N, Umar S, Ashafaq M, Akhtar M, Iqbal Z, Samim M, Ahmad FJ. A comparative study of PNIPAM nanoparticles of curcumin, demethoxycurcumin, and bisdemethoxycurcumin and their effects on oxidative stress markers in experimental stroke. Protoplasma. 2013;250(6):1327–1338. doi: 10.1007/s00709-013-0516-9. [DOI] [PubMed] [Google Scholar]
- Bikadi Z, Hazai E. Application of PM6 semi-empirical method to modeling protein enhances docking accuracy of AutoDock. J Cheminform. 2009;11:1–15. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmam NH, Ajay KSR, Wasi K, Alim H, Naqvi BRJ. Biosynthesis of stable antioxidant ZnO nanoparticles by Pseudomonas aeruginosa rhamnolipids. PLoS ONE. 2014;9(9):e106937. doi: 10.1371/journal.pone.0106937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlqvist A, Borgstrom B. Digestion and absorption of disaccharides in man. Biochem J. 1961;81:411–418. doi: 10.1042/bj0810411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Sun H, Xi L, Li J, Yang Y, Liu H, Yao X. Molecular modeling study of checkpoint kinase 1 inhibitors by multiple docking strategies and prime/MM-GBSA calculation. J Comput Chem. 2011;32:2800–2809. doi: 10.1002/jcc.21859. [DOI] [PubMed] [Google Scholar]
- Duhonvy D, Nussinov R, Wolfson HJ (2002) Efficient unbound docking of rigid molecules, in Gusfield wt al., Ed proceeding of the 2nd workshop on algorithm in Bioinformatics (WABI) Rome, Italy, Lecture Notes in Computer Science, 2452, pp.185–200, Springer Verlag.
- Etheridge ML, Campbell SA, Erdman AG, Christy LH, Susan MW. The big picture on nanomedicine: the state of investigational and approved nanomedicine products. Nanomedicine. 2013;9:1–14. doi: 10.1016/j.nano.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Zhang C, Gao G, Cui D. Facile synthesis of hollow Cu2O octahedral and spherical nanocrystals and their morphology-dependent photocatalytic properties. Nanoscale Res Lett. 2012;7:1–10. doi: 10.1186/1556-276X-7-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunalan S, Rajeshwari S, Venkatesh R. Green synthesized ZnO nanoparticles against bacterial and fungal pathogens. Prog Nat Sci. 2012;22:693–700. [Google Scholar]
- Hayman S, Kinoshita JH. Isolation and properties of lens aldose reductase. J Biol Chem. 1965;240:877–882. [PubMed] [Google Scholar]
- Hema K, Vani Priyadarshini I, Sandeep S, Pradeep N, Chiranjeevi P, Umamaheswari A. Subunit vaccine design against pathogens causing atherosclerosis. J Biomol Struct Dyn. 2015;33(supplement 1):135–136. [Google Scholar]
- Hema K, Sandeep S, Pradeep N, Umamaheswari A. Identification of effectual inhibitors against human insulin like growth factor binding protein-2. J Inform Data Min. 2016;2(1):1–10. [Google Scholar]
- Hema K, Sandeep S, Pradeep N, Umamaheswari A. In silico identification of leads targeting Interleukin-6 against pathogenesis of atherosclerosis. J Biomol Struct Dyn. 2019;1:32–33. [Google Scholar]
- Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- Junghanns JU, Müller RH. Nanocrystal technology, drug delivery and clinical applications. Int J Nanomedicine. 2008;3(3):295–309. doi: 10.2147/ijn.s595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanipakam H, Sharma K, Thinlas T, Mohammad G, Pasha MAQ. Structural and functional alterations of nitric oxide synthase 3 due to missense variants associate with high-altitude pulmonary edema through dynamic study. J Biomol Struct Dyn. 2020;17:1–16. doi: 10.1080/07391102.2019.1711190. [DOI] [PubMed] [Google Scholar]
- Kavitha P, Saritha M, Laxma Reddy K. Synthesis, structural characterization, fluorescence, antimicrobial, antioxidant and DNA cleavage studies of Cu(II) complexes of formyl chromone Schiff bases. Spectrochim Acta A Mol Biomol Spectrosc. 2012;102:159–168. doi: 10.1016/j.saa.2012.10.037. [DOI] [PubMed] [Google Scholar]
- Khosravi A, Jayaram B, Goliaei B, Masoudi-Nejad A. Active repurposing of drug candidates for melanoma based on GWAS, PheWAS and a wide range of omics data. Mol Med. 2019;25(1):30. doi: 10.1186/s10020-019-0098-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Quon MJ, Kim JA. New insights into the mechanisms of polyphenols beyond antioxidant properties; lessons from the green tea polyphenol, epigallocatechin 3-gallate. Redox Biol. 2014;2:187–195. doi: 10.1016/j.redox.2013.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox C, Law V, Jewison T, Liu P, Ly S, Frolkis A. DrugBank 3.0: a comprehensive resource for ‘omics’ research on drugs. Nucleic Acids Res. 2011;39:D1035–1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, et al. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- Kushwaha SK, Shakya M. Protein interaction network analysis—approach for potential drug target identification in Mycobacterium tuberculosis. J Theor Biol. 2010;262:84–294. doi: 10.1016/j.jtbi.2009.09.029. [DOI] [PubMed] [Google Scholar]
- Lee S, Lee IH, Kim HJ, Chang GS, Chung JE, No KT (2002) The Pre ADMET Approach: Web based program for rapid prediction of physico-chemical, drug absorption and drug-like properties. Euro QSAR 2002-designing drugs and crop, Protectants: processes problems and solutions, 1: 418–412.
- Li J, Abel R, Zhy K, Cao Y, ZhaoFriensner SRA. The VSGB 2.0 model: the next generation energy model for high resolution protein structure modeling. Proteins. 2011;79:2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S. Analytical shape computation of macromolecules: II. Identification and computation of inaccessible cavities in proteins. Proteins. 1998;33:18–29. [PubMed] [Google Scholar]
- Lipinski CA, Hopkins A. Navigating chemical space for biology and medicine. Nature. 2004;432:855–861. doi: 10.1038/nature03193. [DOI] [PubMed] [Google Scholar]
- Lobo V, Patil A, Phatak A, Chandra N. Free radicals, antioxidants and functional foods impact on human health. Pharmacogn Rev. 2010;4:118–216. doi: 10.4103/0973-7847.70902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marella S, Dilip Rajasekhar M, Kumar EGTV, Krishna Tilak T, Kameswara Rao B, Apparao Ch. Mcy protein, a potential antidiabetic agent: evaluation of carbohydrate metabolic enzymes and antioxidant status. Int J Biol Macromol. 2016;86:481–488. doi: 10.1016/j.ijbiomac.2016.01.062. [DOI] [PubMed] [Google Scholar]
- Mavromoustakos T, Durdagi S, Koukoulitsa C, Simcic M, Papadopoulos MG, Hodoscek M, Grdadolnik SG. Strategies in the rational drug design. Curr Med Chem. 2011;18:2517–2530. doi: 10.2174/092986711795933731. [DOI] [PubMed] [Google Scholar]
- Merisko-Liversidge and Liversidge Nanosizing for oral and parenteral drug delivery: a perspective on formulating poorly-water soluble compounds using wet media milling technology. Adv Drug Deliv Rev. 2011;63:427–440. doi: 10.1016/j.addr.2010.12.007. [DOI] [PubMed] [Google Scholar]
- Mohammad A, Bhawani SA, Sharma S. Analysis of herbal products by thin layer chromatography: a review. Int J Pharma Bio Sci. 2010;1(2):10. [Google Scholar]
- Mondal SI, Ferdous S, Jewel NA, Akter A, Mahmud Z, Islam MM, Afrin T, Karim N. Identification of potential drug targets by subtractive genome analysis of Escherichia coli O157:H7: an in silico approach. Adv Appl Bioinform Chem. 2015;8:49–63. doi: 10.2147/AABC.S88522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M, Ohta N, Sakurai S, Alam S, Tsai J, Kador PF, Sato S. The role of aldose reductase in sugar cataract formation: aldose reductase plays a key role in lens epithelial cell death (apoptosis) Chem Biol Interact. 2001;130–132(1–3):617–625. doi: 10.1016/s0009-2797(00)00289-1. [DOI] [PubMed] [Google Scholar]
- Papadimitriou S, Bikiaris D. Novel self-assembled core–shell nanoparticles based on crystalline amorphous moieties of aliphatic copolyesters for efficient controlled drug release. J Control Release. 2009;138(2):177–184. doi: 10.1016/j.jconrel.2009.05.013. [DOI] [PubMed] [Google Scholar]
- Perron NR, Brumaghim JL. A review of the antioxidant mechanisms of polyphenol compounds related to iron binding. Cell Biochem Biophys. 2009;53:75–100. doi: 10.1007/s12013-009-9043-x. [DOI] [PubMed] [Google Scholar]
- Pradeep P, Jayaram B. C5’ omitted DNA enhances bendability and protein binding. Biochem Biophys Res Commun. 2019 doi: 10.1016/j.bbrc.2019.05.051. [DOI] [PubMed] [Google Scholar]
- Prasad TNVKV, Sudhakar P, Sreenivasulu Y, Latha P, Munaswamy V, Raja Reddy K, Sreeprasad TS, Sajanlal PR, Pradeep TJ. Effect of titanium dioxide and zinc oxide nanoparticles on germination and vigour in tomato (Solanum lycopersicum L.) seeds. Plant Nutr. 2012;35(6):851–869. [Google Scholar]
- Raoufi D. Synthesis and photoluminescence characterization of ZnO nanoparticles. J Lumin. 2013;134:213–219. [Google Scholar]
- Rasha M, Patra D. Modification of nanostructured ZnO surfaces with curcumin: fluorescence-based sensing for arsenic and improving arsenic removal by ZnO. RSC Adv. 2016;6:17256–17268. [Google Scholar]
- Revina AA, Oksentyuk EV, Fenin AA. Synthesis and properties of zinc nanoparticles: the role and prospects of radiation chemistry in the development of modern nanotechnology. Prot Met. 2007;43:613–618. [Google Scholar]
- Roy PS, Bhattacharya SK. Size-controlled synthesis and characterization of polyvinyl alcohol-coated platinum nanoparticles: role of particle size and capping polymer on the electrocatalytic activity. Catal Sci Technol. 2013;3:1314–1323. [Google Scholar]
- Shukla VK, Singh JS, Vispute N, Ahmad B, Kumar A, Hosur RV. Unfolding of CPR3 gets initiated at the active site and proceeds via two intermediates. Biophys. 2017;112:605–619. doi: 10.1016/j.bpj.2016.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz BJ, Reguly T, Boucher I, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:535–539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi SK, Muttineni R, Singh SK. Extra precision docking, free energy calculation and molecular dynamics simulation studies of CDK2 inhibitors. J Theor Biol. 2013;334:87–100. doi: 10.1016/j.jtbi.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Williamson J, Kilo C, Tilton RG. Mechanism of glucose and diabetes-induced vascular dysfunction. In: Ruderman N, Williamson J, Brownlee M, editors. Hyperglycemia, Diabetes and Vascular Disease. New York, NY: American Physiological Society; 1992. pp. 107–132. [Google Scholar]
- Wright LP, Rohwer JM, Ghirardo A, Hammerbacher A, Ortiz-Alcaide M. Deoxyxylulose 5-phosphate synthase controls flux through the methylerythritol 4-phosphate pathway in arabidopsis. Plant Physiol. 2014;165:1488–1504. doi: 10.1104/pp.114.245191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar S, Ahmed R, Khan R. Biotransformation; a green and efficient way of antioxidant synthesis. Free Radic Res. 2016;27:1–10. doi: 10.1080/10715762.2016.1209745. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.