Abstract
The human cytomegalovirus (HCMV), whose genome is 235 ± 1.9 kbp long, is a common herpesvirus. However, the functions of many of its genes are still unknown. HCMV is closely associated with various human diseases and infects 60–90% of the global population. It can infect various human cells, including fibroblasts, epithelial cells, endothelial cells, smooth muscle cells, and monocytes. Although HCMV infection is generally asymptomatic and causes subtle clinical symptoms, it can generate a robust immune response and establish a latent infection in immunocompromised individuals, including those with AIDS, transplant recipients, and developing fetuses. Currently available antivirals approved for the treatment of HCMV-associated diseases are limited by dose-limiting toxicity and the emergence of resistance; however, vaccines and immunoglobulins are unavailable. In this review, we have summarized the recent literature on 43 newly identified HCMV genes. We have described their novel functions on the viral replication cycle, latency, and host immune evasion. Further, we have discussed HCMV-associated diseases and current therapeutic targets. Our review may provide a foundational basis for studies aiming to prevent and develop targeted therapies for HCMV-associated diseases.
Keywords: human cytomegalovirus, functional profiling, therapeutic targets, viral genome, viral replication, anti-viral agents
Introduction
The human cytomegalovirus (HCMV), whose linear double-stranded DNA genome is 235 ± 1.9 kbp in length, has the largest genome among human herpesviruses. It has an E-type genome structure consisting of two large reverse domains called long (L) and short (S), each of which is composed of three regions: central unique, unique long, and unique short (Van Damme and Van Loock, 2014). Each region is flanked by terminal (TRL and TRS) and internal (IRL and IRS) inverted repeats (Murphy and Shenk, 2008). The HCMV genome contains various gene families, including RL11, UL14, UL18, UL25, UL82, UL120, US6, US7, US12, and US22. A study has recently demonstrated that the HCMV genome has more than 751 translated open reading frames (ORFs) (Stern-Ginossar et al., 2012; Patro, 2019). Of these, 282 viral transcripts are translationally active, 206 of which possess unique coding potential (Balazs et al., 2017) and one-third of the viral poly-A RNA transcripts are protein-coding (Gatherer et al., 2011). In addition, HCMV encodes four major long non-coding RNAs (lncRNAs) (RNA1.2, RNA2.7, RNA4.9, and RNA5.0) (Tai-Schmiedel et al., 2020) and at least 16 pre-miRNAs and 26 mature miRNAs (Pavelin et al., 2013; Ding et al., 2017; Zhang et al., 2020). Although the functions of many genes are still unknown, the roles and functions of most HCMV genes in infective stages have been identified.
HCMV infects 60–90% of the population worldwide (Stevenson et al., 2014) and can infect various human cells, including fibroblasts, epithelial cells, endothelial cells, smooth muscle cells, and monocytes (Gerna et al., 2019). In fibroblasts, HCMV produces abundant progeny (viral particles) through extensive replication and proliferation (Kalejta, 2008b). HCMV-infected monocytes carry viral particles to multiple organs via the bloodstream, thus contributing to productive and persistent infection (Smith et al., 2004, 2007; Nogalski et al., 2011; Chan et al., 2012; Stevenson et al., 2014). After induction, the HCMV-infected monocytes can differentiate into macrophages and further support the replication and expression of viral genes (Stevenson et al., 2014). Further, the inactivation of some US12 family members (e.g., US18 and US20) plays a role in cell tropism by mediating viral replication in specific cells (Cavaletto et al., 2015). The deletion of the US18 abrogates replication in human gingival tissues (Gurczynski et al., 2014), whereas the US20 affects the replication of HCMV clinical strains in endothelial cells (Cavaletto et al., 2015).
Primary HCMV infection is generally asymptomatic and causes subtle clinical symptoms in healthy individuals (Gandhi and Khanna, 2004; Reeves and Sinclair, 2008). However, in immunocompromised individuals, it can generate a robust immune response (Magro et al., 2007) and establish a latent infection via immune escape mechanisms. It can be activated from the latent state, which shows a dynamic change of “infection-latency-activation” in immunocompromimsed individuals (Reyda et al., 2014) and permits the life-long persistence of the virus. In productive infection, HCMV exhibits a temporal cascade of gene expression, and these genes can be classified into three, namely immediate early (IE), early (E), and late (L) genes (Reeves, 2011; Isomura and Stinski, 2013; Supplementary Table S1). The major IE genes (MIE), UL122 and UL123, play critical roles in subsequent viral gene expression and the efficiency of viral replication (Isomura and Stinski, 2013).
Human cytomegalovirus is closely associated with various diseases, such as tumors, circulatory system diseases, digestive system diseases, and ophthalmic diseases (idiopathic) (Samanta et al., 2003; Lindholt and Shi, 2006; Magro et al., 2007; DiMaggio et al., 2009; Chen H.P. et al., 2015; Wang H.W. et al., 2016; Wang Z. et al., 2016; Dennis et al., 2018; Guo et al., 2018; Moussawi et al., 2018; Song et al., 2018; Alston and Dix, 2019; Liu et al., 2020). The roles of HCMV genes in the occurrence and development of diseases have been previously described (Schleiss, 2011). For example, HCMV encodes a G protein-coupled receptor US28, which activates pro-migratory signaling and mediates vascular smooth muscle cell migration (Melnychuk et al., 2004; Vomaske et al., 2010). On the one hand, US28 enhances the inflammatory properties of cardiovascular diseases (Söderberg-Nauclér, 2006). On the other hand, US28 promotes the invasiveness of diseased cells in atherosclerotic plaques (Krishna et al., 2018). Antivirals licensed for the treatment of HCMV infection and diseases are limited by dose-limiting toxicity and the emergence of resistance (Sinkó, 2019). Furthermore, relevant vaccines and immunoglobulins have not been developed (Xu and Yuan, 2018). Accordingly, detailed analyses of the relationships between HCMV genes and their associated diseases are needed for the development of preventive measures and therapeutic intervention.
The associations of HCMV genes with diseases have not yet been reviewed in detail, and the current knowledge on the functions of HCMV genes on the basis of a previous report (Van Damme and Van Loock, 2014) has to be updated. Thus, in this review, we summarized the recent literature on HCMV gene functions and described the relationship between HCMV genes and their associated diseases in detail. Particularly, we focused on the novel functions of 43 HCMV genes, namely UL7, UL8, UL10, UL21A, UL23, UL25, UL26, UL27, UL31, UL34, UL35, UL37.1, UL38, UL45, UL47, UL48, UL50, UL53, UL56, UL71, UL76, UL77, UL112, UL116, UL130, UL131A, UL133, UL136, UL138, UL141, UL146, UL148, UL148D, US12, US18, US20, US21, US25, US28, US29, US31,US33, and RNA4.9. However, those of 39 genes (RL1, RL5A, RL8A, RL9A, RL9, UL2, UL3, UL6, UL12, UL13, UL14, UL15, UL15A, UL19, UL21, UL22, UL42, UL59, UL62, UL90, UL120, UL121, UL127, UL129, UL140, UL147A, UL148A, UL148B, UL148C, UL149, US1, US13, US14, IS15, US26, US34A, RNA1.2, RNA5.0, and IRL14) remain unknown, and further studies are needed to elucidate their biological function. Overall, our comprehensive review provides a foundational basis for future research aimed at the prevention and targeted treatment of HCMV-associated diseases.
Gene Function Annotation Data
Human cytomegalovirus genes are involved in diverse processes, including latency, immunomodulation, assembly, maturation, egress, viral protein, gene expression and regulation, replication, cell tropism and cell type-specific replication, modulation of the host cell cycle and protein synthesis, viral growth, cellular tracking, nucleotide repair and modification, entry, viral spread, manipulation of host cell signaling pathways, apoptosis, angiogenesis, and tumor formation (Van Damme and Van Loock, 2014; Supplementary Table S2).
According to their role in viral growth, HCMV genes may be classified as essential or dispensable, and their deletion can cause moderate or severe growth defects or enhanced growth (Schwartz and Stern-Ginossar, 2019). In terms of the virus life cycle, HCMV gene functions can be divided into entry, replication, assembly, maturation, and egress. Six genes (UL35, UL73, UL74, UL75, UL100, and UL115) are involved in cell entry. Following entry, a temporal cascade begins, leading to the expression of immediate early (IE), early (E), and late (L) genes. There are 36 genes are necessary for DNA replication, ten of which (UL26, UL31, UL34, UL37.1, UL133, UL136, UL141, US29, US33, and RNA4.9) are recently identified. UL36, UL37, UL37.1, UL37.3, UL38, UL61, UL123, UL138, UL148D, US1, US2, US3, TRS1, and IRS are IE genes, whereas there are 59 E genes including UL87, UL104, UL112, and UL114. There are 55 early-late (E/L) genes (such as UL3, UL40, UL46, UL72) and at least 75 L genes, such as UL2, UL7, and UL14, which are expressed following the onset of viral DNA replication. When a new genome is produced, DNA is packaged in a newly formed capsid. Immature viruses complete the maturation process, and finally, the virus is released. Thirty-five genes are involved in assembly, maturation, and egress, including 8 newly identified genes (UL35, UL47, UL48, UL50, UL53, UL56, UL71, and UL77). After viral egress, HCMV can now cause various diseases.
We further classified the genes based on their roles in immunomodulation, apoptosis, and angiogenesis/tumor formation. We found 61 genes related to the latent infection state, only two of which (miR-UL148D and miR-US29) are newly discovered (Meshesha et al., 2016; Pan et al., 2016). HCMV genes related to immunomodulation and apoptosis are also determinants of virus survival time. A total of 57 genes are related to immune regulation, whereas 14 are related to apoptosis. Of these, 15 genes (UL8, UL10, UL23, UL26, UL31, UL45, UL48, UL112, UL130, UL131, UL148, US12, US18, US20, and US31) related to immunomodulation and six genes (UL37.1, UL38, UL76, UL138, UL148D, and US21) related to apoptosis are newly identified. UL7, UL112, US28, and RNA4.9 are involved in angiogenesis/tumor formation. Lastly, the number of recently discovered genes related to gene expression/regulation, cellular trafficking, nucleotide repair/modification, and virion stability is 29, 8, 5, and 1, respectively.
Genes Involved in the Viral Life Cycle
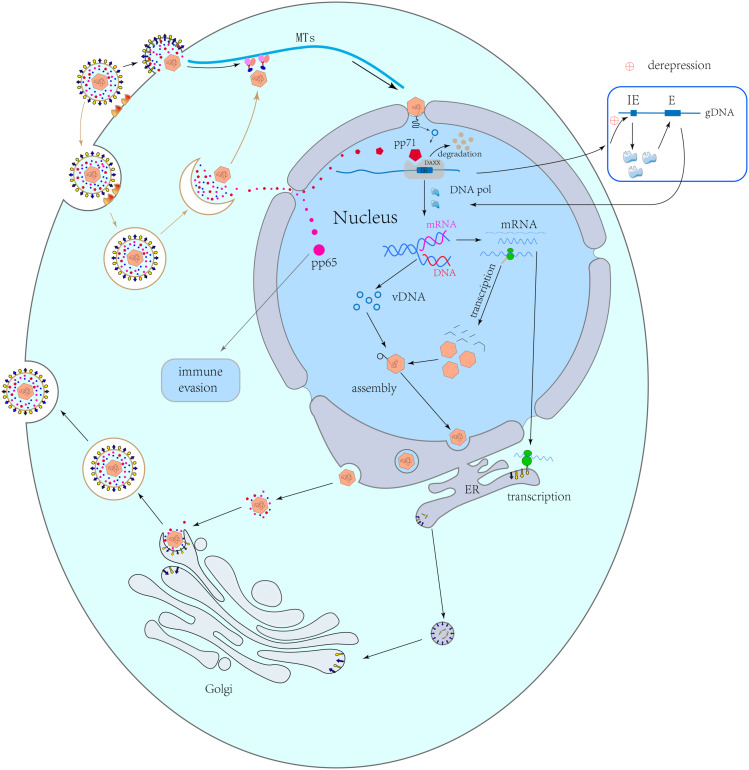
Most HCMV genes play important roles in its viral life cycle, which involves four stages, namely entry, replication, assembly and maturation, and egress (Figure 1).
FIGURE 1.
Viral life cycle. Capsid-related proteins transport capsids along the microtubules to the nuclear pores. Capsids remain in the cytoplasm and release DNA into the nucleus. pp71 can be independently transported into the nucleus, where it combines with Daxx to cause its degradation, resulting in the derepression of viral immediate early (IE) genes and the activation of early (E) genes after transcription and translation. The viral early proteins induce viral genome replication. Following late gene transcription and translation, the viral structural proteins are synthesized and are assembled with the replicated genome to form a new naked virus, which passes through the nuclear membrane via an envelopment-de-envelopment pathway. The naked virus migrates to an assembly site on a Golgi-derived vesicle, where it obtains the final envelope with viral glycoproteins. Finally, virions are released by budding.
Entry
HCMV infects host cells through two mechanisms, namely membrane fusion and endocytosis, that involve different envelope glycoprotein complexes (Nguyen and Kamil, 2018). The complexes include a trimer complex composed of gH/gL/gO (UL75/UL115/UL74) and a pentamer complex composed of gH/gL/UL128/UL130/UL131 (Huber and Compton, 1998; Wang and Shenk, 2005). Trimers interact with the platelet-derived growth factor receptor α (PDGFRα) and subsequently mediate the pH-independent membrane fusion in fibroblasts (Wu et al., 2017). In contrast, pentamers interact with neuropilin-2 (Nrp2) via low pH-dependent endocytosis in epithelial and endothelial cells (Martinez-Martin et al., 2018; Nguyen and Kamil, 2018). In addition, several reports also found that CD147, CD46, and OR14I1 serve as epithelial entry factors (Nguyen and Kamil, 2018; Vanarsdall et al., 2018; Stein et al., 2019; Xiaofei et al., 2019). The transforming growth factor β receptor type 3 (TGFβRIII) and neuregulin-2 (NRG2) are additional hits for trimers to promote viral entry in fibroblasts (Nguyen and Kamil, 2018; Vanarsdall et al., 2018; Gerna et al., 2019). Viral cell tropism is correlated with the levels of gH/gL/gO and gH/gL/UL128/UL130/UL131 (Zhou et al., 2013). Zhou et al. (2013) found that the levels of gH/gL/gO are higher than those of gH/gL/UL128/UL130/UL131 in TR, TB40/e, AD169, and PH virions, whereas those of gH/gL/UL128/UL130/UL131 are higher in Merlin virions. In epithelial-derived viruses, the ratio of gH/gL/UL128/UL130/UL131 to gH/gL/gO is increased by approximately two-fold (Wang et al., 2007). The surface gM/gN (UL100/UL73) dimer and the gB (UL55) trimer are also involved in viral entry (Gardner and Tortorella, 2016; Foglierini et al., 2019). gM/gN plays an important role in adhesion by interacting with heparin sulfate proteoglycans on the cell surface, whereas the gB trimer acts as a proximal mediator during membrane fusion (Eisenberg et al., 2012; Nguyen and Kamil, 2018).
Replication and Gene Expression
After passing through the membrane, HCMV transports its capsid along the microtubules to the nuclear pore by hijacking the intracellular machinery (Kalejta, 2008a). In this process, some tegument proteins remain in the cytoplasm or bind tightly to the capsid and mediate the nuclear-pore transport. Other proteins, such as pp65 (pUL83) and pp71 (pUL82), are transported into the nucleus independently (Kalejta, 2008b). The former (pp65) contributes to viral replication, whereas the latter (pp71) mainly affects gene expression (Biolatti et al., 2018a; Kalejta and Albright, 2020). Additionally, the interaction between the pUL47/pUL48 dimer and the capsid can promote the former’s transfer to the nuclear pore complex (Kalejta, 2008a). The capsid is finally dissociated on the microtubule, docks to the nuclear pore, and releases DNA into the nucleus (Kalejta, 2008b). After pp71 entry into the nucleus, it combines with death domain-associated protein (Daxx), which binds to histone deacetylases (HDACs) to repress transcription, thereby causing the degradation of Daxx and inducing the activation of the IE genes (Saffert and Kalejta, 2006). Then, as replication begins, the proteins in IE (pIE) can act as activators to promote the expression of early and late genes via their interaction with cytokines (Adamson and Nevels, 2020). Early proteins are mainly transcription factors and polymerases involved in viral DNA replication, transcription, and protein synthesis, whereas late proteins are mainly structural proteins synthesized after viral DNA replication (Gruffat et al., 2016). A wide range of viral genes are involved in viral replication. For instance, HCMV UL34 encodes a sequence-specific DNA binding protein (pUL34), which interacts with pUL84, pIE2, and pUL44 and contributes to the establishment of a nuclear environment necessary for viral gene expression and DNA replication (Rana and Biegalke, 2014; Slayton et al., 2018). pUL44 is an obligate nuclear-resident, non-structural viral protein for HCMV DNA replication (Neo et al., 2019). Recent studies have shown that UL133, UL136, UL141, US29, and US33 are also involved in viral DNA replication (Caviness et al., 2014, 2016; Shen et al., 2014; Guo et al., 2015; Zou et al., 2018).
Assembly, Maturation, and Egress
There are four core components of HCMV capsids, namely major capsid protein (MCP/pUL86), minor capsid binding protein (TRI1/pUL46), minor capsid protein (TRI2/pUL85), and smallest capsid protein (SCP/pUL48A), assembled with the help of pUL80 (Gibson, 2008; Tandon and Mocarski, 2012). During the maturation of virion particles, other capsid-related proteins bind to the capsid. pUL77 and pUL93 stabilize DNA packaging (Köppen-Rung et al., 2016). After capsid maturation, an enzyme complex called a terminase, consisting of pUL51, pUL52, pUL56, pUL77, pUL89, and pUL93, binds to concatemeric viral DNA and cleaves it into unit-length genomes for DNA packaging (Ligat et al., 2018). Neuber et al. (2017) suggested that the mutual interplay between terminase proteins (pUL51, pUL56, and pUL89) can protect the viral or cellular pathways which are vital for the HCMV life cycle from the detrimental effects of incomplete terminase assembly. After assembly, the naked virus leaves the nucleus with the help of the nuclear egress complex (NEC) in an envelopment-deenvelopment-reenvelopment manner (Mettenleiter et al., 2006; Kalejta, 2008b). The core NEC is composed of pUL50 and pUL53, whereas the HCMV-specific multi-component NEC is mainly composed of pUL97, p32/gC1qR, emerin, protein kinase C (PKC), and additional proteins (Milbradt et al., 2018). Milbradt et al. proposed one potential mechanism by which pUL53 and the NEC-related kinase pUL97 bind to the nucleocapsid at the inner nuclear membrane. By complexing with pUL53 and the membrane-anchored pUL50, pUL97 may cause the virus to enter the perinuclear space via phosphorylating the remaining nuclear lamin A/C to disrupt the nuclear lamina (Sharma et al., 2015; Milbradt et al., 2018). Additionally, pUL93 interacts with both components of the NEC (pUL50 and pUL53), suggesting that pUL93 is related to the NEC (DeRussy et al., 2016).
Following nuclear egress, additional tegumentation and final envelopment occur once the capsid is transported to the virion assembly compartment (vAC) in the cytoplasm. This is followed by its excretion from the cell (Das et al., 2007; Das and Pellett, 2011; Alwine, 2012; Hook et al., 2014). The vAC is formed by the reorganization of Golgi bodies via HCMV miRNAs, thus providing additional structural proteins to the nucleocapsid for virion maturation (Alwine, 2012; Hook et al., 2014). pUL48, pUL94, and pUL103 play roles in virion tegumentation by contributing to cytoplasmic vAC biogenesis (Das et al., 2014). Furthermore, pUL48 directs pUL47 to the vAC to promote secondary envelopment and tegumentation (Cappadona et al., 2015). The TB-47stop virus, whose pUL47 expression is blocked, showed severe growth defects in host cells and accumulated non-enveloped capsids in the cytoplasm (Cappadona et al., 2015). Finally, the HCMV life cycle is completed for approximately 72 h (Lee and Grey, 2020).
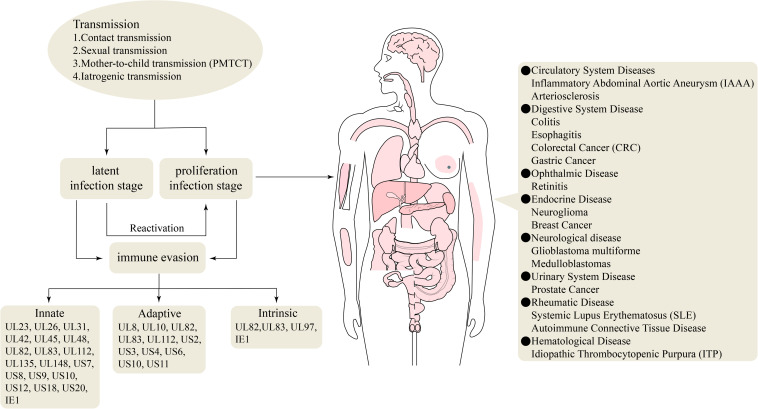
Latency
Although most virus can be effectively eliminated upon the activation of host immune system, HCMV can still establish life-long latency once infected. There are 61 genes related to latency, namely RL2, RL4, RL6, UL4, UL5, UL17, UL28, UL30, UL32, UL37, UL38, UL39, UL40, UL44, UL50, UL52, UL61, UL64, UL65, UL67, UL68, UL70, UL73, UL75, UL76, UL79, UL80, UL81ast/LU NA, UL8/pp71, UL84, UL87, UL95, UL98, UL99/pp28, UL105, UL108, UL110, UL111A, UL114, UL115, UL122, UL123, UL124, UL125, UL126A, UL128, UL132, UL133, UL135, UL138, UL144, UL145, UL148D, UL150, US17, US28, US29, US32, US34, RNA2.7, and RNA4.9.
The transcript of the UL115.5A region, expressed during latent infection and homologous to interleukin 10 (IL-10), can help the virus escape immune recognition (Jenkins et al., 2004). The UL111A and UL111.5A genes encode IL-10 that also play a role as immunomodulatory cytokine (Jenkins et al., 2004). IE1 expression can prevent the apoptosis induced by US28, which may function as a proapoptotic factor (Jenkins et al., 2004). Furthermore, US28 causes potential infections by weakening a variety of cellular signal transduction factors, including mitogen-activated protein kinase and nuclear factor kappa-B (NF-κB) (Krishna et al., 2017). UL138 has long and short isoforms, both of which suppress major IE gene transcription and promote latency (Lee et al., 2016). Hcmv-miR-UL36 down-regulates HCMV UL138, indicating that hcmv-miR-UL36 may be a new latent period-related determinant of viral miRNAs contributing to HCMV replication (Huang et al., 2013; Zhang et al., 2020). The protein isoforms encoded by UL136 regulates the latent and replication states of infection, whereas the complex interaction between UL136 subtypes balances the replication and latency period of HCMV (Caviness et al., 2016). miR-UL148D and US29 are recently discovered loci associated with latency (Lau et al., 2016; Pan et al., 2016). By targeting the host cell’s immediate early response gene 5 (IER5) and inhibiting the activin A-triggered secretion of IL-6, miR-UL148D facilitates viral infection during latent infection (Lau et al., 2016; Pan et al., 2016). In addition, the star “passenger” form of miR-US29 is preferentially expressed during latency, as evidenced by a shift in the abundance of the two arms of miR-US29 between the productive and latency stages (Meshesha et al., 2016). A recent study has also suggested that UL76 has a dominant negative effect on replication, which may be related to latency (Wang et al., 2004).
Immune Evasion
HCMV also encodes abundant genes and proteins closely related to immunoregulation (Gatherer et al., 2011). During its long-term coevolution with the host, HCMV has developed a variety of immune escape strategies. The immune system therefore cannot completely clear HCMV (Söderberg-Nauclér, 2008).
Innate Response
The innate immune system is the host’s first line of defense against invading pathogens (Akira et al., 2006). When the human body is infected by microbes, pattern recognition receptors (PRRs) detect pathogen-associated molecular patterns (PAMPs) and induce the production of type I interferons (IFNs) and pro-inflammatory cytokines (Medzhitov, 2007). Among the PRRs, the cyclic guanosine monophosphate-adenosine monophosphate (cGAMP) synthase (cGAS) has been shown to act as a DNA sensor that transmits DNA virus infection signals to various cell lines (Medzhitov, 2007; Li et al., 2013). cGAS is capable of recognizing double-stranded DNA and catalyzing the synthesis of cGAMP, which functions as a second messenger in innate inmune (Wu et al., 2013). Subsequently, cGAMP activates the mediator of IRF3 activation (MITA), which triggers innate antiviral response (Zhong et al., 2008). There are several mechanisms by which HCMV antagonizes this signaling pathway. For example, the tegument protein UL82 inhibits the cellular transport and activation of MITA to evade antiviral immunity (Fu et al., 2017). UL31 also mediates immune invasion through the inhibition of DNA sensing by cGAS (Huang et al., 2018). Further, UL83 interacts with cGAS and interferon gamma-inducible protein 16 (IFI16) in the nucleus, thereby inhibiting the induction of IFN production (Biolatti et al., 2018b). The IE1 and IE2 proteins, which are synthesized after HCMV infection, can inhibit the IFN signal transduction and thereby inhibit the innate immune response (Paulus and Nevels, 2009). In addition, it has been suggested that US10 attenuates IFN-β (Park et al., 2019). The inhibition of cGAS activation and MITA trafficking by UL42 is contributes to the evasion of immune antiviral immunity by HCMV (Fu et al., 2019).
The innate immune system is primarily composed of natural killer (NK) cells, dendritic cells, and macrophages (Takeuchi and Akira, 2010). NK cells are capable of recognizing virus-infected cells and play an important role in the host defense against HCMV (Dell’Oste et al., 2020). US9 can mediate IFN-β production and block NK-cell activation (Seidel et al., 2015). pUL135 remodels the actin cytoskeleton and weakens the effects of F-actin filaments during synapse formation (Stanton et al., 2014). It can thus weaken the function of cytotoxic immune effector cells and subsequently evade immune recognition in infected cells (Stanton et al., 2014). Hcmv-miR-UL112 attenuates NK-cell activity by inhibiting type I interferon secretion (Huang et al., 2015). UL148 increases degranulation in both cytotoxic T lymphocytes and NK cells against HCMV-infected cells, including NK-driven antibody-dependent cellular cytotoxicity (Wang et al., 2018). US12 family genes control immune ligands and suppresses NK-cell activation (Fielding et al., 2017).
Adaptive Response
Adaptive immunity is vital for controlling primary HCMV infection. After infection, the host immune response can be enhanced by increasing the number of highly differentiated cells, such as CD4+T and CD8+T cells (Manandhar et al., 2019). The US2-US11 region encodes at least five glycoproteins (US2, US3, US6, US10, and US11), which are specifically used to interfere with the expression of the antigen peptides of CD8+T cells and inhibit MHC class I antigen presentation (Kim et al., 2011). In addition, US2 and US11 products are expressed during the early phase of infection and down-regulate class I molecules during viral infections (Hansen and Bouvier, 2009; Gabor et al., 2020). Meanwhile, HCMV miR-US4-1 inhibits CD8+T cell response by targeting the endoplasmic reticulum aminopeptidase 1, which plays an important role in the production of antigenic peptides and blocks the production of viral epitopes (Kim et al., 2011). HCMV miR-UL112-5p also targets the endoplasmic reticulum aminopeptidase 1 to inhibit the processing and presentation of the HCMV pp65495–503 peptide to specific cytotoxic T lymphocytes (Kim et al., 2011; Huang et al., 2015; Romania et al., 2017). Additional gene products involved in immunomodulation have also been identified. After the infection of THP-1 cells, pUL8 decreases the levels of proinflammatory factors to inhibit inflammation (Perez-Carmona et al., 2018). The interaction of pUL10 with leukocytes mediates immunosuppression by reducing T-cell proliferation and cytokine production (Bruno et al., 2016).
Intrinsic Response
The mammalian immune system consists of the innate and adaptive immune responses. However, a number of studies have suggested the existence of a special form of innate immunity known as intrinsic response, which forms the third branch of the immune system (Bieniasz, 2004). The intrinsic immune system is composed of a variety of cellular proteins termed restriction factors (RFs) (Bieniasz, 2003). These cellular proteins, including the IFN-γ inducible protein 16, a 100-kDa speckled protein (Sp100), promyelocytic leukemia protein (PML), and the human death domain-associated protein 6 (hDaxx), can interfere with the various stages of the virus replication cycle (Glass and Everett, 2013). Sp100, hDaxx, and PML also form a subnuclear substructure known as nuclear domain 10 (ND10), which can restrict the expression of immediate-early gene and influence the virus replication cycle (Glass and Everett, 2013). However, HCMV is capable of overcoming these RFs by encoding viral proteins, such as pp71, IE1, UL97, and pp65. For instance, UL97 and pp65 (pUL83) antagonize IFI16, resulting in the mislocalization of IFI16 into the cytoplasm (Dell’Oste et al., 2014; Biolatti et al., 2016). IE1 and pp71 can block or reverse the inhibitory effects of ND10 during viral replication through the modification or destruction of nuclear domain 10 proteins (Hofmann et al., 2002; Cantrell and Bresnahan, 2005).
HCMV-Related Infection and Diseases
Humans are the only host of HCMV, and most human organs and tissues can be infected. Upon infection, a long-term HCMV infection can be established, which correlates with chronic inflammation that influences the development of cardiovascular diseases and some types of cancers in otherwise immunocompetent individuals (Söderberg-Nauclér, 2006, 2008; Nogalski et al., 2014). Primary and active infection causes serious diseases in immunotolerant individuals. Active infection can lead to various diseases, such as arteriosclerosis, colitis, esophagitis, colorectal cancer (CRC), gastric cancer (GC), idiopathic thrombocytopenic purpura (ITP), prostate cancer, systemic lupus erythematosus (SLE), systemic sclerosis (SSc), autoimmune connective tissue disease, neuroglioma, breast cancer, and retinitis (Samanta et al., 2003; Magro et al., 2007; DiMaggio et al., 2009; Chen H.P. et al., 2015; Wang H.W. et al., 2016; Wang Z. et al., 2016; Arcangeletti et al., 2018; Dennis et al., 2018; Guo et al., 2018; Moussawi et al., 2018; Song et al., 2018; Alston and Dix, 2019; Liu et al., 2020; Figure 2). In immunocompromised individuals, including those with acquired immunodeficiency syndrome (AIDS), transplant recipients, and developing fetuses, the incidence of HCMV infection and mortality are elevated (Crough and Khanna, 2009; Nogalski et al., 2014).
FIGURE 2.
Genes involved in the immune evasion and HCMV-associated diseases.
Neoplastic Diseases
Although the mechanism of HCMV in many cancers is unclear, some studies have shown that HCMV plays a role in tumor regulation and influences cancer development (Michaelis et al., 2011). Taher et al. (2013, 2014) reported that HCMV has a high prevalence in the brain metastases of patients with primary breast and colorectal cancers, which suggests its involvement in tumor metastasis. Jin et al. (2014) found that UL135 and UL136 transcripts are only detected in gastric cancer patients, especially in the tissues of those in stage III. Roman abraheta et al. suggested that HCMV may up-regulate the adhesion of prostate cancer cells to the endothelial cells and extracellular matrix proteins, accompanied by the overexpression of integrin (Blaheta et al., 2006).
In addition, HCMV encodes pUL123 and pUL122 that prevent glioblastoma cell death by promoting angiogenesis, which is vital for tumor growth and survival in glioblastoma multiforme (Herbein, 2018). US28 induces angiogenesis and activates Gαq-linked signaling pathways, thereby increasing the expression of vascular endothelial growth factors in HCMV-infected glioblastoma cells (Maussang et al., 2009; Heukers et al., 2018). US28 induces the angiogenic phenotype in patients with colorectal cancer (Maussang et al., 2006). Meanwhile, US33 activates the proliferation and angiogenesis and inflammatory signaling pathways in glioblastoma cells (Heukers et al., 2018; van Senten et al., 2019). UL111A affects the survival time of gastric carcinoma patients by altering the number of infiltrating T cells (Liu et al., 2020).
Non-neoplastic Diseases
Frequent and active CMV infection plays a vital role in the induction and acceleration of chronic inflammatory reactions in the aortas of patients with inflammatory abdominal aortic aneurysm (Lindholt and Shi, 2006; Liu et al., 2020). By aggravating the apoptosis of endothelial cells induced by oxidized low-density lipoprotein, miR-US25-1 may promote atherosclerosis (Fan et al., 2014). Some studies reported that HCMV infection is associated with the pathogenesis of SLE. The titers of anti-HCMV IgG and IgM antibodies have been demonstrated to be significantly higher in SLE patients than in healthy individuals. Furthermore, the positivity rate of HCMV UL55 was significantly higher in SLE patients than in healthy controls, suggesting that UL55 may play an important role in the development of SLE (Chen J. et al., 2015). Another study suggested the association between HCMV infection and SLE via the human antibody response targeting UL44 (Neo et al., 2019). UL44 is redistributed to the cell surface during virus-induced apoptosis as part of a complex with these autoantigens like nucleolin and dsDNA (Neo et al., 2019). Meanwhile, US31 induces inflammation in mono-macrophages by promoting NF-κB2 activation, which may also contribute to the progression of SLE (Guo et al., 2018). HCMV encodes UL94 related to SSc, which may function in cellular gene expression and induce apoptosis in human endothelial cells (Lunardi et al., 2000).
Potential Therapeutic Targets
Available anti-virals for HCMV-related diseases include ganciclovir, foscarnet, and cidofovir (Sinkó, 2019). Ganciclovir acts as a competitive inhibitor of deoxyguanosine (Upadhyayula and Michaels, 2013). Its greater accumulation in CMV-infected cells suggests that ganciclovir has better antiviral activity than acyclovir (Marshall and Koch, 2009). Meanwhile, foscarnet reversibly inhibits viral DNA polymerases (De Clercq and Li, 2016). In CMV retinitis patients administered with induction therapy with foscarnet, ocular symptoms were initially relieved but recurred soon after stopping the treatment (Heiden et al., 2019). Cidofovir competitively inhibits DNA elongation (Britt and Prichard, 2018) and is helpful for treating colitis (Baniak and Kanthan, 2016). Cottone et al. (2001) showed that antiviral treatment can relieve the clinical symptoms of patients (among five patients, three with ganciclovir and two with foscarnet) with ulcerative colitis. The eradication of CMV with antiviral drugs can improve the symptoms of severe and refractory ITP (DiMaggio et al., 2009). However, the apparent toxic effects of these drugs should also be considered. Neutropenia is the most common toxicity associated with ganciclovir (Venton et al., 2014), whereas foscarnet may cause kidney toxicity (Goldfarb and Coe, 1998). Renal toxicity is associated with cidofovir (Moyle, 2000). These drugs can cause significant adverse events, as well as long-term toxicity in children.
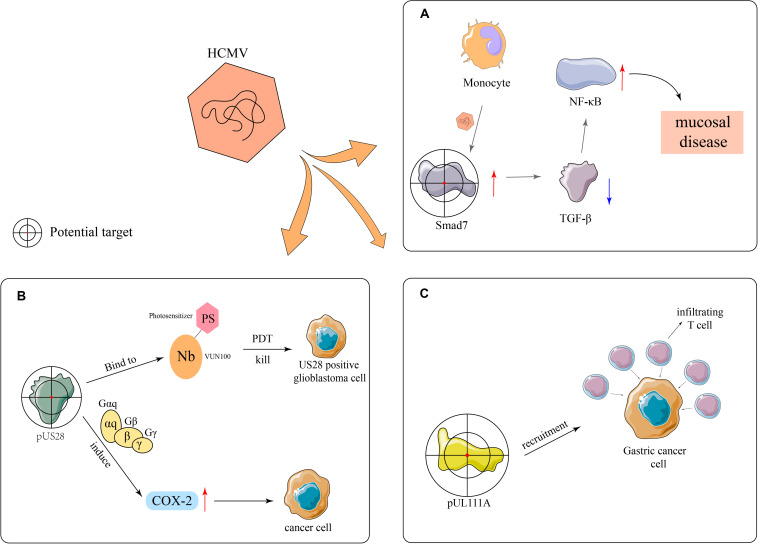
HCMV has the ability to upregulate the expression of Smad7, an intracellular antagonist of the transforming growth factor beta (TGF-β) signaling, in monocytes and attenuate the inhibitory effect of stromal TGF-β on NF-κB, thus promoting mucosal inflammation (Dennis et al., 2018; Figure 3A). In addition, Dennis et al. (2018) found that the expression of Smad7 antisense oligonucleotide in monocytes restores susceptibility to stromal TGF-β-induced inflammation anergy, which indicates that Smad7 may be used as a potential therapeutic target for CMV mucosal disease. VUN100, a kind of nanobody, consists of antibody fragments extracted from heavy-chain antibodies from the family Camelidae and can bind to the N-terminus and extracellular loop 3 (ECL3) of US28. It can selectively kill US28-expressing tumor cells during photodynamic therapy (De Groof et al., 2019). Additionally, US28 can induce the expression of inflammatory proteins, such as cyclooxygenase-2 (COX-2), by activating Gαq and Gβγ, and subsequently the synthesis of PGE2, which promotes the onset of inflammation and initiates or aggravates tumor formation (Maussang et al., 2009). Specific COX-2 inhibitors can prevent HCMV replication and reduce the expression of US28 in tumor cells, suggesting that HCMV plays an important role in medulloblastoma and US28 is a potential therapeutic target (Maussang et al., 2009; Baryawno et al., 2011; Figure 3B). The overexpression of the UL111A protein may result in the recruitment of multiple infiltrating T cells, thereby inhibiting the growth and invasion of gastric cancer cells. Therefore, UL111A may be another potential therapeutic target (Liu et al., 2020; Figure 3C).
FIGURE 3.
Potential therapeutic targets. (A) HCMV upregulates the expression of the TGF-β antagonist Smad7 in monocytes, thereby reducing the activation effects of TGF-β toward NF-κB and ultimately resulting in mucosal diseases. (B) VUN100 can bind the viral pUS28 during photodynamic therapy and can selectively kill US28-expressing glioblastoma cells. In addition, pUS28 promotes the expression of inflammatory factors, such as COX-2, to catalyze inflammatory processes through the activation of Gαq and Gβγ and participates in tumor onset and development. (C) The overexpression of UL111A leads to the recruitment of infiltrating T cells in gastric cancer.
Promising therapeutic targets aimed at the treatment of HCMV-related diseases have recently emerged, such as the viral terminase complex and the viral pUL97 kinase (Piret and Boivin, 2019). Currently, breakthrough anti-HCMV infection drugs based on these two targets include letermovir and maribavir (Britt and Prichard, 2018). Letermovir inhibits the formation of mature HCMV virus particles by targeting the pUL56 subunit of the viral terminase complex (Britt and Prichard, 2018) and has showed efficacy in a Phase III clinical trial. It has been approved for use in hematopoietic stem cell transplant recipients who are seropositive for HCMV as it can prevent the reactivation and re-infection of CMV (Piret and Boivin, 2019). Meanwhile, maribavir can influence the key processes involved in CMV replication, specifically the formation of viral envelopes and the discharge of viral particles, through the targeted inhibition of the protein kinase UL97 in CMV (Britt and Prichard, 2018). The drug showed good treatment efficacy in a Phase II trial and may potentially satisfy the treatment needs of patients with antiviral drug resistance. Maribavir is currently being tested in a Phase III trial, during which its efficacy will be further evaluated (Winston et al., 2008; Piret and Boivin, 2019). Studies on the treatment of HCMV infection have progressed relatively slowly and some of these aspects remain to be explored. The advent of letermovir and maribavir will promote the development of other antiviral drugs for the prevention and treatment of HCMV infection, thus improving the treatment and prognosis of HCMV-infected patients (Britt and Prichard, 2018).
Conclusion
We have provided a summary of recent literature on HCMV genes involved in the viral replication cycle, immune evasion, and HCMV-related diseases. Further studies are needed to characterize the functions of multiple newly identified HCMV ORFs, such as UL121 and UL120. To identify potential new targets for antiviral drug discovery and development, studies elucidating the molecular mechanisms by which HCMV genes contribute to disease development are warranted.
Author Contributions
LY, GG, and XX planned the work. LY, YQ, and WY drafted the manuscript. YQ, WY, GG, HW, and XX revised the manuscript. LY, YQ, WY, GG, HW, and XX participated in the literature search and discussion. WY arranged figures. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- AIDS
Acquired immunodeficiency syndrome
- cGAMP
cyclic guanosine monophosphate-adenosine monophosphate
- cGAS
cyclic guanosine adenosine synthase
- COX-2
cyclooxygenase-2
- CRC
colorectal cancer
- Daxx
death domain associated protein
- E
early
- E/L
early-late
- ECL3
extracellular loop 3
- GC
gastric cancer
- HCMV
human cytomegalovirus
- HDACs
histone deacetylases
- hDaxx
human death domain-associated protein 6
- IE
immediate early
- IFI16
interferon gamma-inducible protein 16
- IFNs
type I interferons
- IL-10
interleukin 10
- ITP
idiopathic thrombocytopenic purpura
- L
late
- MIE
major immediate early
- MITA
mediator of IRF3 activation
- ND10
nuclear domain 10
- NEC
nuclear egress complex
- NF- κ B
nuclear factor kappa-B
- NK
natural killer cells
- NRG2
neuregulin-2
- Nrp2
neuropilin-2
- PAMPs
pathogen-associated molecular patterns
- PDGFR α
platelet-derived growth factor receptor α
- PKC
protein kinase C
- PML
promyelocytic leukemia protein
- PRRs
pattern recognition receptors
- RFs
restriction factors
- SLE
systemic lupus erythematosus
- Sp100
100-kDa speckled protein
- SSc
systemic sclerosis
- TGF β RIII
transforming growth factor β receptor type 3
- TGF- β
transforming growth factor beta
- vAC
virion assembly compartment.
Footnotes
Funding. This study was funded by the Key R&D Program of Zhejiang Province (grant number: 2020C03029); the National Natural Science Foundation of China (grant numbers: 81472308 and 81672707); and the Projects of Wenzhou Municipal Science and Technology Bureau (grant numbers: Y20150054 and Y20150057).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02104/full#supplementary-material
References
- Adamson C. S., Nevels M. M. (2020). Bright and early: inhibiting human cytomegalovirus by targeting major immediate-early gene expression or protein function. Viruses 12:110. 10.3390/v12010110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akira S., Uematsu S., Takeuchi O. (2006). Pathogen recognition and innate immunity. Cell 124 783–801. [DOI] [PubMed] [Google Scholar]
- Alston C. I., Dix R. D. (2019). SOCS and herpesviruses, with emphasis on cytomegalovirus retinitis. Front. Immunol. 10:732. 10.3389/fimmu.2019.00732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine J. C. (2012). The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog. 8:e1002878. 10.1371/journal.ppat.1002878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arcangeletti M.-C., Maccari C., Vescovini R., Volpi R., Giuggioli D., Sighinolfi G., et al. (2018). A paradigmatic interplay between human cytomegalovirus and host immune system: possible involvement of viral antigen-driven CD8+ T cell responses in systemic sclerosis. Viruses 10:508. 10.3390/v10090508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazs Z., Tombacz D., Szucs A., Csabai Z., Megyeri K., Petrov A. N., et al. (2017). Long-read sequencing of human cytomegalovirus transcriptome reveals RNA isoforms carrying distinct coding potentials. Sci. Rep. 7:15989. 10.1038/s41598-017-16262-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baniak N., Kanthan R. (2016). Cytomegalovirus colitis: an uncommon mimicker of common colitides. Archiv. Pathol. Lab. Med. 140 854–858. 10.5858/arpa.2015-0176-RS [DOI] [PubMed] [Google Scholar]
- Baryawno N., Rahbar A., Wolmer-Solberg N., Taher C., Odeberg J., Darabi A., et al. (2011). Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J. Clin. Invest. 121 4043–4055. 10.1172/JCI57147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz P. D. (2003). Restriction factors: a defense against retroviral infection. Trends Microbiol. 11 286–291. [DOI] [PubMed] [Google Scholar]
- Bieniasz P. D. (2004). Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5 1109–1115. [DOI] [PubMed] [Google Scholar]
- Biolatti M., Dell’Oste V., De Andrea M., Landolfo S. (2018a). The human cytomegalovirus tegument protein pp65 (pUL83): a key player in innate immune evasion. New Microbiol. 41 87–94. [PubMed] [Google Scholar]
- Biolatti M., Dell’Oste V., Pautasso S., Gugliesi F., von Einem J., Krapp C., et al. (2018b). Human cytomegalovirus tegument protein pp65 (pUL83) dampens Type I interferon production by inactivating the DNA sensor cGAS without affecting STING. J. Virol. 92:e01774-17. 10.1128/JVI.01774-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biolatti M., Dell’Oste V., Pautasso S., von Einem J., Marschall M., Plachter B., et al. (2016). Regulatory interaction between the cellular restriction factor IFI16 and viral pp65 (pUL83) modulates viral gene expression and IFI16 protein stability. J. Virol. 90 8238–8250. 10.1128/JVI.00923-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaheta R. A., Weich E., Marian D., Bereiter-Hahn J., Jones J., Jonas D., et al. (2006). Human cytomegalovirus infection alters PC3 prostate carcinoma cell adhesion to endothelial cells and extracellular matrix. Neoplasia 8 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt W. J., Prichard M. N. (2018). New therapies for human cytomegalovirus infections. Antiv. Res. 159 153–174. 10.1016/j.antiviral.2018.09.003 [DOI] [PubMed] [Google Scholar]
- Bruno L., Cortese M., Monda G., Gentile M., Calo S., Schiavetti F., et al. (2016). Human cytomegalovirus pUL10 interacts with leukocytes and impairs TCR-mediated T-cell activation. Immunol. Cell Biol. 94 849–860. 10.1038/icb.2016.49 [DOI] [PubMed] [Google Scholar]
- Cantrell S. R., Bresnahan W. A. (2005). Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J. Virol. 79 7792–7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadona I., Villinger C., Schutzius G., Mertens T., von Einem J. (2015). Human cytomegalovirus pUL47 modulates tegumentation and capsid accumulation at the viral assembly complex. J. Virol. 89 7314–7328. 10.1128/JVI.00603-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavaletto N., Luganini A., Gribaudo G. (2015). Inactivation of the human cytomegalovirus US20 gene hampers productive viral replication in endothelial cells. J. Virol. 89 11092–11106. 10.1128/jvi.01141-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness K., Bughio F., Crawford L. B., Streblow D. N., Nelson J. A., Caposio P., et al. (2016). Complex interplay of the UL136 isoforms balances cytomegalovirus replication and latency. mBio 7:e01986. 10.1128/mBio.01986-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviness K., Cicchini L., Rak M., Umashankar M., Goodrum F. (2014). Complex expression of the UL136 gene of human cytomegalovirus results in multiple protein isoforms with unique roles in replication. J. Virol. 88 14412–14425. 10.1128/JVI.02711-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan G., Nogalski M. T., Stevenson E. V., Yurochko A. D. (2012). Human cytomegalovirus induction of a unique signalsome during viral entry into monocytes mediates distinct functional changes: a strategy for viral dissemination. J. Leukoc. Biol. 92 743–752. 10.1189/jlb.0112040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H. P., Jiang J. K., Chan C. H., Teo W. H., Yang C. Y., Chen Y. C., et al. (2015). Genetic polymorphisms of the human cytomegalovirus UL144 gene in colorectal cancer and its association with clinical outcome. J. Gen. Virol. 96 3613–3623. 10.1099/jgv.0.000308 [DOI] [PubMed] [Google Scholar]
- Chen J., Zhang H., Chen P., Lin Q., Zhu X., Zhang L., et al. (2015). Correlation between systemic lupus erythematosus and cytomegalovirus infection detected by different methods. Clin. Rheumatol. 34 691–698. 10.1007/s10067-015-2868-3 [DOI] [PubMed] [Google Scholar]
- Cottone M., Pietrosi G., Martorana G., Casà A., Pecoraro G., Oliva L., et al. (2001). Prevalence of cytomegalovirus infection in severe refractory ulcerative and Crohn’s colitis. Am. J. Gastroenterol. 96 773–775. [DOI] [PubMed] [Google Scholar]
- Crough T., Khanna R. (2009). Immunobiology of human cytomegalovirus: from bench to bedside. Clin. Microbiol. Rev. 22 e34–e38. 10.1128/CMR.00034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Ortiz D. A., Gurczynski S. J., Khan F., Pellett P. E. (2014). Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J. Virol. 88 9086–9099. 10.1128/JVI.01141-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Pellett P. E. (2011). Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J. Virol. 85 5864–5879. 10.1128/JVI.00155-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Vasanji A., Pellett P. E. (2007). Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81 11861–11869. 10.1128/JVI.01077-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E., Li G. (2016). Approved antiviral drugs over the Past 50 Years. Clin. Microbiol. Rev. 29 695–747. 10.1128/CMR.00102-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groof T. W. M., Mashayekhi V., Fan T. S., Bergkamp N. D., Sastre Toraño J., van Senten J. R., et al. (2019). Nanobody-targeted photodynamic therapy selectively kills viral GPCR-expressing glioblastoma cells. Mol. Pharm. 16 3145–3156. 10.1021/acs.molpharmaceut.9b00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Oste V., Biolatti M., Galitska G., Griffante G., Gugliesi F., Pasquero S., et al. (2020). Tuning the orchestra: HCMV vs. innate immunity. Front. Microbiol. 11:661. 10.3389/fmicb.2020.00661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell’Oste V., Gatti D., Gugliesi F., De Andrea M., Bawadekar M., Lo Cigno I., et al. (2014). Innate nuclear sensor IFI16 translocates into the cytoplasm during the early stage of in vitro human cytomegalovirus infection and is entrapped in the egressing virions during the late stage. J. Virol. 88 6970–6982. 10.1128/JVI.00384-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E. A., Smythies L. E., Grabski R., Li M., Ballestas M. E., Shimamura M., et al. (2018). Cytomegalovirus promotes intestinal macrophage-mediated mucosal inflammation through induction of Smad7. Mucosal. Immunol. 11 1694–1704. 10.1038/s41385-018-0041-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRussy B. M., Boland M. T., Tandon R. (2016). Human cytomegalovirus pUL93 links nucleocapsid maturation and nuclear egress. J. Virol. 90 7109–7117. 10.1128/jvi.00728-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMaggio D., Anderson A., Bussel J. B. (2009). Cytomegalovirus can make immune thrombocytopenic purpura refractory. Br. J. Haematol. 146 104–112. 10.1111/j.1365-2141.2009.07714.x [DOI] [PubMed] [Google Scholar]
- Ding M., Wang X., Wang C., Liu X., Zen K., Wang W., et al. (2017). Distinct expression profile of HCMV encoded miRNAs in plasma from oral lichen planus patients. J. Transl. Med. 15:133. 10.1186/s12967-017-1222-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg R. J., Atanasiu D., Cairns T. M., Gallagher J. R., Krummenacher C., Cohen G. H. (2012). Herpes virus fusion and entry: a story with many characters. Viruses 4 800–832. 10.3390/v4050800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J., Zhang W., Liu Q. (2014). Human cytomegalovirus-encoded miR-US25-1 aggravates the oxidised low density lipoprotein-induced apoptosis of endothelial cells. Biomed. Res. Int. 2014:531979. 10.1155/2014/531979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielding C. A., Weekes M. P., Nobre L. V., Ruckova E., Wilkie G. S., Paulo J. A., et al. (2017). Control of immune ligands by members of a cytomegalovirus gene expansion suppresses natural killer cell activation. eLife 6:22206. 10.7554/eLife.22206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foglierini M., Marcandalli J., Perez L. (2019). HCMV envelope glycoprotein diversity demystified. Front. Microbiol. 10:1005. 10.3389/fmicb.2019.01005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-Z., Guo Y., Zou H.-M., Su S., Wang S.-Y., Yang Q., et al. (2019). Human cytomegalovirus protein UL42 antagonizes cGAS/MITA-mediated innate antiviral response. PLoS Pathog. 15:e1007691. 10.1371/journal.ppat.1007691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y.-Z., Su S., Gao Y.-Q., Wang P.-P., Huang Z.-F., Hu M.-M., et al. (2017). Human cytomegalovirus tegument protein UL82 inhibits STING-mediated signaling to evade antiviral immunity. Cell Host Microb. 21 231–243. 10.1016/j.chom.2017.01.001 [DOI] [PubMed] [Google Scholar]
- Gabor F., Jahn G., Sedmak D. D., Sinzger C. (2020). Downregulation of MHC class I molecules by HCMV occurs during all phases of viral replication but is not always complete. Front. Cell Infect. Microbiol. 10:283. 10.3389/fcimb.2020.00283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M. K., Khanna R. (2004). Human cytomegalovirus: clinical aspects, immune regulation, and emerging treatments. Lancet Infect. Dis. 4 725–738. 10.1016/S1473-3099(04)01202-2 [DOI] [PubMed] [Google Scholar]
- Gardner T. J., Tortorella D. (2016). Virion glycoprotein-mediated immune evasion by human cytomegalovirus: a sticky virus makes a slick getaway. Microbiol. Mol. Biol. Rev. 80 663–677. 10.1128/MMBR.00018-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatherer D., Seirafian S., Cunningham C., Holton M., Dargan D. J., Baluchova K., et al. (2011). High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. U.S.A. 108 19755–19760. 10.1073/pnas.1115861108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerna G., Kabanova A., Lilleri D. (2019). Human cytomegalovirus cell tropism and host cell receptors. Vaccines 7:70. 10.3390/vaccines7030070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. (2008). Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 325 187–204. 10.1007/978-3-540-77349-8_11 [DOI] [PubMed] [Google Scholar]
- Glass M., Everett R. D. (2013). Components of promyelocytic leukemia nuclear bodies (ND10) act cooperatively to repress herpesvirus infection. J. Virol. 87 2174–2185. 10.1128/JVI.02950-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb D. S., Coe F. L. (1998). Foscarnet crystal deposition and renal failure. Am. J. Kidney Dis. 32 519–520. [DOI] [PubMed] [Google Scholar]
- Gruffat H., Marchione R., Manet E. (2016). Herpesvirus late gene expression: a viral-specific pre-initiation complex is key. Front. Microbiol. 7:869. 10.3389/fmicb.2016.00869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G., Ye S., Xie S., Ye L., Lin C., Yang M., et al. (2018). The cytomegalovirus protein US31 induces inflammation through mono-macrophages in systemic lupus erythematosus by promoting NF-κB2 activation. Cell Death Dis. 9:104. 10.1038/s41419-017-0122-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Qi Y., Huang Y., Liu Z., Ma Y., Shao Y., et al. (2015). Human cytomegalovirus miR-US33-5p inhibits viral DNA synthesis and viral replication by down-regulating expression of the host Syntaxin3. FEBS Lett. 589 440–446. 10.1016/j.febslet.2014.12.030 [DOI] [PubMed] [Google Scholar]
- Gurczynski S. J., Das S., Pellett P. E. (2014). Deletion of the human cytomegalovirus US17 gene increases the ratio of genomes per infectious unit and alters regulation of immune and endoplasmic reticulum stress response genes at early and late times after infection. J. Virol. 88 2168–2182. 10.1128/JVI.02704-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen T. H., Bouvier M. (2009). MHC class I antigen presentation: learning from viral evasion strategies. Nat. Rev. Immunol. 9 503–513. 10.1038/nri2575 [DOI] [PubMed] [Google Scholar]
- Heiden D., Tun N., Smithuis F. N., Keenan J. D., Oldenburg C. E., Holland G. N., et al. (2019). Active cytomegalovirus retinitis after the start of antiretroviral therapy. Br. J. Ophthalmol. 103 157–160. 10.1136/bjophthalmol-2018-312406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbein G. (2018). The human cytomegalovirus, from oncomodulation to oncogenesis. Viruses 10:408. 10.3390/v10080408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heukers R., Fan T. S., de Wit R. H., van Senten J. R., De Groof T. W. M., Bebelman M. P., et al. (2018). The constitutive activity of the virally encoded chemokine receptor US28 accelerates glioblastoma growth. Oncogene 37 4110–4121. 10.1038/s41388-018-0255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Sindre H., Stamminger T. (2002). Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J. Virol. 76 5769–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook L. M., Grey F., Grabski R., Tirabassi R., Doyle T., Hancock M., et al. (2014). Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microb. 15 363–373. 10.1016/j.chom.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Chen D., He J., Cai J., Shen K., Liu X., et al. (2015). Hcmv-miR-UL112 attenuates NK cell activity by inhibition type I interferon secretion. Immunol. Lett. 163 151–156. 10.1016/j.imlet.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Huang Y., Qi Y., Ma Y., He R., Ji Y., Sun Z., et al. (2013). Down-regulation of human cytomegalovirus UL138, a novel latency-associated determinant, by hcmv-miR-UL36. J. Biosci. 38 479–485. 10.1007/s12038-013-9353-4 [DOI] [PubMed] [Google Scholar]
- Huang Z.-F., Zou H.-M., Liao B.-W., Zhang H.-Y., Yang Y., Fu Y.-Z., et al. (2018). Human cytomegalovirus protein UL31 inhibits DNA sensing of cGAS to mediate immune evasion. Cell Host Microb. 24:7. 10.1016/j.chom.2018.05.007 [DOI] [PubMed] [Google Scholar]
- Huber M. T., Compton T. (1998). The human cytomegalovirus UL74 gene encodes the third component of the glycoprotein H-glycoprotein L-containing envelope complex. J. Virol. 72 8191–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isomura H., Stinski M. F. (2013). Coordination of late gene transcription of human cytomegalovirus with viral DNA synthesis: recombinant viruses as potential therapeutic vaccine candidates. Expert Opin. Therap. Targets 17 157–166. 10.1517/14728222.2013.740460 [DOI] [PubMed] [Google Scholar]
- Jenkins C., Abendroth A., Slobedman B. (2004). A novel viral transcript with homology to human interleukin-10 is expressed during latent human cytomegalovirus infection. J. Virol. 78 1440–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Hu C., Wang P., Chen J., Wu T., Chen W., et al. (2014). Latent infection of human cytomegalovirus is associated with the development of gastric cancer. Oncol. Lett. 8 898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta R. F. (2008a). Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr. Top. Microbiol. Immunol. 325 101–115. 10.1007/978-3-540-77349-8_6 [DOI] [PubMed] [Google Scholar]
- Kalejta R. F. (2008b). Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72 249–265. 10.1128/MMBR.00040-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta R. F., Albright E. R. (2020). Expanding the known functional repertoire of the human cytomegalovirus pp71 protein. Front. Cell. Infect. Microbiol. 10:95. 10.3389/fcimb.2020.00095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee S., Shin J., Kim Y., Evnouchidou I., Kim D., et al. (2011). Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat. Immunol. 12 984–991. 10.1038/ni.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köppen-Rung P., Dittmer A., Bogner E. (2016). Intracellular distribution of capsid-associated pUL77 of human cytomegalovirus and interactions with packaging proteins and pUL93. J. Virol. 90 5876–5885. 10.1128/JVI.00351-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna B. A., Miller W. E., O’Connor C. M. (2018). US28: HCMV’s swiss army knife. Viruses 10:445. 10.3390/v10080445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna B. A., Poole E. L., Jackson S. E., Smit M. J., Wills M. R., Sinclair J. H. (2017). Latency-associated expression of human cytomegalovirus US28 attenuates cell signaling pathways to maintain latent infection. mBio 8:e01754-17. 10.1128/mBio.01754-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau B., Poole E., Krishna B., Sellart I., Wills M. R., Murphy E., et al. (2016). The expression of human cytomegalovirus MicroRNA MiR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci. Rep. 6:31205. 10.1038/srep31205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-H., Grey F. (2020). Systems virology and human cytomegalovirus: using high throughput approaches to identify novel host-virus interactions during lytic infection. Front. Cell. Infect. Microbiol. 10:280. 10.3389/fcimb.2020.00280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. H., Caviness K., Albright E. R., Lee J.-H., Gelbmann C. B., Rak M., et al. (2016). Long and short isoforms of the human cytomegalovirus UL138 protein silence IE transcription and promote latency. J. Virol. 90 9483–9494. 10.1128/JVI.01547-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-D., Wu J., Gao D., Wang H., Sun L., Chen Z. J. (2013). Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341 1390–1394. 10.1126/science.1244040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligat G., Cazal R., Hantz S., Alain S. (2018). The human cytomegalovirus terminase complex as an antiviral target: a close-up view. FEMS Microbiol. Rev. 42 137–145. 10.1093/femsre/fuy004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindholt J. S., Shi G. P. (2006). Chronic inflammation, immune response, and infection in abdominal aortic aneurysms. Eur. J. Vasc. Endovasc. Surg. 31 453–463. [DOI] [PubMed] [Google Scholar]
- Liu X., Lin K., Huang X., Xie W., Xiang D., Ding N., et al. (2020). Overexpression of the human cytomegalovirus UL111A is correlated with favorable survival of patients with gastric cancer and changes T-cell infiltration and suppresses carcinogenesis. J. Cancer Res. Clin. Oncol. 146 555–568. 10.1007/s00432-019-03092-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardi C., Bason C., Navone R., Millo E., Damonte G., Corrocher R., et al. (2000). Systemic sclerosis immunoglobulin G autoantibodies bind the human cytomegalovirus late protein UL94 and induce apoptosis in human endothelial cells. Nat. Med. 6 1183–1186. [DOI] [PubMed] [Google Scholar]
- Magro C. M., Crowson A. N., Ferri C. (2007). Cytomegalovirus-associated cutaneous vasculopathy and scleroderma sans inclusion body change. Hum. Pathol. 38 42–49. 10.1016/j.humpath.2006.06.002 [DOI] [PubMed] [Google Scholar]
- Manandhar T., Hò G.-G. T., Pump W. C., Blasczyk R., Bade-Doeding C. (2019). Battle between host immune cellular responses and HCMV immune evasion. Intern. J. Mol. Sci. 20:626. 10.3390/ijms20153626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall B. C., Koch W. C. (2009). Antivirals for cytomegalovirus infection in neonates and infants: focus on pharmacokinetics, formulations, dosing, and adverse events. Paediatr. Drugs 11 309–321. 10.2165/11316080-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Martinez-Martin N., Marcandalli J., Huang C. S., Arthur C. P., Perotti M., Foglierini M., et al. (2018). An unbiased screen for human cytomegalovirus identifies Neuropilin-2 as a central viral receptor. Cell 174:28. 10.1016/j.cell.2018.06.028 [DOI] [PubMed] [Google Scholar]
- Maussang D., Langemeijer E., Fitzsimons C. P., Stigter-van Walsum M., Dijkman R., Borg M. K., et al. (2009). The human cytomegalovirus-encoded chemokine receptor US28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res. 69 2861–2869. 10.1158/0008-5472.CAN-08-2487 [DOI] [PubMed] [Google Scholar]
- Maussang D., Verzijl D., van Walsum M., Leurs R., Holl J., Pleskoff O., et al. (2006). Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 103 13068–13073. 10.1073/pnas.0604433103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R. (2007). Recognition of microorganisms and activation of the immune response. Nature 449 819–826. [DOI] [PubMed] [Google Scholar]
- Melnychuk R. M., Streblow D. N., Smith P. P., Hirsch A. J., Pancheva D., Nelson J. A. (2004). Human cytomegalovirus-encoded G protein-coupled receptor US28 mediates smooth muscle cell migration through Galpha12. J. Virol. 78 8382–8391. 10.1128/JVI.78.15.8382-8391.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshesha M. K., Bentwich Z., Solomon S. A., Avni Y. S. (2016). In vivo expression of human cytomegalovirus (HCMV) microRNAs during latency. Gene 575 101–107. 10.1016/j.gene.2015.08.040 [DOI] [PubMed] [Google Scholar]
- Mettenleiter T. C., Klupp B. G., Granzow H. (2006). Herpesvirus assembly: a tale of two membranes. Curr. Opin. Microbiol. 9 423–429. [DOI] [PubMed] [Google Scholar]
- Michaelis M., Baumgarten P., Mittelbronn M., Driever P. H., Doerr H. W., Cinatl J. (2011). Oncomodulation by human cytomegalovirus: novel clinical findings open new roads. Med. Microbiol. Immunol. 200 1–5. 10.1007/s00430-010-0177-7 [DOI] [PubMed] [Google Scholar]
- Milbradt J., Sonntag E., Wagner S., Strojan H., Wangen C., Lenac Rovis T., et al. (2018). Human cytomegalovirus nuclear capsids associate with the core nuclear egress complex and the viral protein kinase pUL97. Viruses 10:35. 10.3390/v10010035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussawi F. A., Kumar A., Pasquereau S., Tripathy M. K., Karam W., Diab-Assaf M., et al. (2018). The transcriptome of human mammary epithelial cells infected with the HCMV-DB strain displays oncogenic traits. Sci. Rep. 8:12574. 10.1038/s41598-018-30109-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyle G. (2000). Toxicity of antiretroviral nucleoside and nucleotide analogues: is mitochondrial toxicity the only mechanism? Drug Saf. 23 467–481. [DOI] [PubMed] [Google Scholar]
- Murphy E., Shenk T. (2008). Human cytomegalovirus genome. Curr. Top. Microbiol. Immunol. 325 1–19. 10.1007/978-3-540-77349-8_1 [DOI] [PubMed] [Google Scholar]
- Neo J. Y. J., Wee S. Y. K., Bonne I., Tay S. H., Raida M., Jovanovic V., et al. (2019). Characterisation of a human antibody that potentially links cytomegalovirus infection with systemic lupus erythematosus. Sci. Rep. 9:9998. 10.1038/s41598-019-46329-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuber S., Wagner K., Goldner T., Lischka P., Steinbrueck L., Messerle M., et al. (2017). Mutual interplay between the human cytomegalovirus terminase subunits pUL51, pUL56, and pUL89 promotes terminase complex formation. J. Virol. 91:e02384-16. 10.1128/JVI.02384-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen C. C., Kamil J. P. (2018). Pathogen at the gates: human cytomegalovirus entry and cell tropism. Viruses 10:704. 10.3390/v10120704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalski M. T., Chan G., Stevenson E. V., Gray S., Yurochko A. D. (2011). Human cytomegalovirus-regulated paxillin in monocytes links cellular pathogenic motility to the process of viral entry. J. Virol. 85 1360–1369. 10.1128/jvi.02090-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogalski M. T., Collins-McMillen D., Yurochko A. D. (2014). Overview of human cytomegalovirus pathogenesis. Methods Mol. Biol. 1119 15–28. 10.1007/978-1-62703-788-4_2 [DOI] [PubMed] [Google Scholar]
- Pan C., Zhu D., Wang Y., Li L., Li D., Liu F., et al. (2016). Human cytomegalovirus miR-UL148D facilitates latent viral infection by targeting host cell immediate early response Gene 5. PLoS Pathog. 12:e1006007. 10.1371/journal.ppat.1006007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park A., Ra E. A., Lee T. A., Choi H. J., Lee E., Kang S., et al. (2019). HCMV-encoded US7 and US8 act as antagonists of innate immunity by distinctively targeting TLR-signaling pathways. Nat. Commun. 10:4670. 10.1038/s41467-019-12641-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro A. R. K. (2019). Subversion of immune response by human cytomegalovirus. Front. Immunol. 10:1155. 10.3389/fimmu.2019.01155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulus C., Nevels M. (2009). The human cytomegalovirus major immediate-early proteins as antagonists of intrinsic and innate antiviral host responses. Viruses 1 760–779. 10.3390/v1030760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavelin J., Reynolds N., Chiweshe S., Wu G., Tiribassi R., Grey F. (2013). Systematic microRNA analysis identifies ATP6V0C as an essential host factor for human cytomegalovirus replication. PLoS Pathog. 9:e1003820. 10.1371/journal.ppat.1003820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Carmona N., Martinez-Vicente P., Farre D., Gabaev I., Messerle M., Engel P., et al. (2018). A prominent role of the human cytomegalovirus UL8 glycoprotein in restraining proinflammatory cytokine production by myeloid cells at late times during Infection. J. Virol. 92:e02229-17. 10.1128/JVI.02229-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret J., Boivin G. (2019). Clinical development of letermovir and maribavir: overview of human cytomegalovirus drug resistance. Antivir. Res. 163:11. 10.1016/j.antiviral.2019.01.011 [DOI] [PubMed] [Google Scholar]
- Rana R., Biegalke B. J. (2014). Human cytomegalovirus UL34 Early and late proteins are essential for viral replication. Virus. Basel 6 476–488. 10.3390/v6020476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M., Sinclair J. (2008). Aspects of human cytomegalovirus latency and reactivation. Curr. Top. Microbiol. Immunol. 325 297–313. [DOI] [PubMed] [Google Scholar]
- Reeves M. B. (2011). Chromatin-mediated regulation of cytomegalovirus gene expression. Virus Res. 157 134–143. 10.1016/j.virusres.2010.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyda S., Büscher N., Tenzer S., Plachter B. (2014). Proteomic analyses of human cytomegalovirus strain AD169 derivatives reveal highly conserved patterns of viral and cellular proteins in infected fibroblasts. Viruses 6 172–188. 10.3390/v6010172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romania P., Cifaldi L., Pignoloni B., Starc N., D’Alicandro V., Melaiu O., et al. (2017). Identification of a genetic variation in ERAP1 aminopeptidase that prevents human cytomegalovirus miR-UL112-5p-mediated immunoevasion. Cell Rep. 20 846–853. 10.1016/j.celrep.2017.06.084 [DOI] [PubMed] [Google Scholar]
- Saffert R. T., Kalejta R. F. (2006). Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J. Virol. 80 3863–3871. 10.1128/JVI.80.8.3863-3871.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M., Harkins L., Klemm K., Britt W. J., Cobbs C. S. (2003). High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J. Urol. 170 998–1002. 10.1097/01.ju.0000080263.46164.97 [DOI] [PubMed] [Google Scholar]
- Schleiss M. R. (2011). Congenital cytomegalovirus infection: molecular mechanisms mediating viral pathogenesis. Infect. Disord. Drug Targets 11 449–465. 10.2174/187152611797636721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz M., Stern-Ginossar N. (2019). The transcriptome of latent human cytomegalovirus. J. Virol. 93:e0047-19. 10.1128/JVI.00047-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel E., Le V. T. K., Bar-On Y., Tsukerman P., Enk J., Yamin R., et al. (2015). Dynamic Co-evolution of host and pathogen: HCMV downregulates the prevalent allele MICA∗008 to escape elimination by NK cells. Cell Rep. 10 968–982. 10.1016/j.celrep.2015.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Bender B. J., Kamil J. P., Lye M. F., Pesola J. M., Reim N. I., et al. (2015). Human cytomegalovirus UL97 phosphorylates the viral nuclear egress complex. J. Virol. 89 523–534. 10.1128/JVI.02426-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z. Z., Pan X., Miao L. F., Ye H. Q., Chavanas S., Davrinche C., et al. (2014). Comprehensive analysis of human cytomegalovirus microRNA expression during lytic and quiescent infection. PLoS One 9:e88531. 10.1371/journal.pone.0088531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkó J. (2019). Current treatment modalities of immunocompromised patients with cytomegalovirus infection. II. Therapeutic options and management strategies. Orv. Hetil. 160 363–369. 10.1556/650.2019.31320 [DOI] [PubMed] [Google Scholar]
- Slayton M., Hossain T., Biegalke B. J. (2018). pUL34 binding near the human cytomegalovirus origin of lytic replication enhances DNA replication and viral growth. Virology 518 414–422. 10.1016/j.virol.2018.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. S., Bentz G. L., Alexander J. S., Yurochko A. D. (2004). Human cytomegalovirus induces monocyte differentiation and migration as a strategy for dissemination and persistence. J. Virol. 78 4444–4453. 10.1128/jvi.78.9.4444-4453.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. S., Bivins-Smith E. R., Tilley A. M., Bentz G. L., Chan G., Minard J., et al. (2007). Roles of phosphatidylinositol 3-kinase and NF-kappaB in human cytomegalovirus-mediated monocyte diapedesis and adhesion: strategy for viral persistence. J. Virol. 81 7683–7694. 10.1128/jvi.02839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderberg-Nauclér C. (2006). Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J. Intern. Med. 259 219–246. [DOI] [PubMed] [Google Scholar]
- Söderberg-Nauclér C. (2008). HCMV microinfections in inflammatory diseases and cancer. J. Clin. Virol. 41 218–223. 10.1016/j.jcv.2007.11.009 [DOI] [PubMed] [Google Scholar]
- Song J., Ma Q., Hu M., Qian D., Wang B., He N. (2018). The inhibition of miR-144-3p on cell proliferation and metastasis by targeting TOP2A in HCMV-positive glioblastoma cells. Molecules 23:3259. 10.3390/molecules23123259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R. J., Prod’homme V., Purbhoo M. A., Moore M., Aicheler R. J., Heinzmann M., et al. (2014). HCMV pUL135 remodels the actin cytoskeleton to impair immune recognition of infected cells. Cell Host Microb. 16 201–214. 10.1016/j.chom.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein K. R., Gardner T. J., Hernandez R. E., Kraus T. A., Duty J. A., Ubarretxena-Belandia I., et al. (2019). CD46 facilitates entry and dissemination of human cytomegalovirus. Nat. Commun. 10:2699. 10.1038/s41467-019-10587-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N., Weisburd B., Michalski A., Le V. T., Hein M. Y., Huang S. X., et al. (2012). Decoding human cytomegalovirus. Science 338 1088–1093. 10.1126/science.1227919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson E. V., Collins-McMillen D., Kim J. H., Cieply S. J., Bentz G. L., Yurochko A. D. (2014). HCMV reprogramming of infected monocyte survival and differentiation: a Goldilocks phenomenon. Viruses 6 782–807. 10.3390/v6020782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher C., de Boniface J., Mohammad A.-A., Religa P., Hartman J., Yaiw K.-C., et al. (2013). High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One 8:e56795. 10.1371/journal.pone.0056795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher C., Frisk G., Fuentes S., Religa P., Costa H., Assinger A., et al. (2014). High prevalence of human cytomegalovirus in brain metastases of patients with primary breast and colorectal cancers. Transl. Oncol. 7 732–740. 10.1016/j.tranon.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai-Schmiedel J., Karniely S., Lau B., Ezra A., Eliyahu E., Nachshon A., et al. (2020). Human cytomegalovirus long noncoding RNA4.9 regulates viral DNA replication. PLoS Pathog. 16:e1008390. 10.1371/journal.ppat.1008390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O., Akira S. (2010). Pattern recognition receptors and inflammation. Cell 140 805–820. 10.1016/j.cell.2010.01.022 [DOI] [PubMed] [Google Scholar]
- Tandon R., Mocarski E. S. (2012). Viral and host control of cytomegalovirus maturation. Trends Microbiol. 20 392–401. 10.1016/j.tim.2012.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyayula S., Michaels M. G. (2013). Ganciclovir, foscarnet, and cidofovir: antiviral drugs not just for cytomegalovirus. J. Pediatr. Infect. Dis. Soc. 2 286–290. 10.1093/jpids/pit048 [DOI] [PubMed] [Google Scholar]
- Van Damme E., Van Loock M. (2014). Functional annotation of human cytomegalovirus gene products: an update. Front. Microbiol. 5:218. 10.3389/fmicb.2014.00218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Senten J. R., Bebelman M. P., Fan T. S., Heukers R., Bergkamp N. D., van Gasselt P., et al. (2019). The human cytomegalovirus-encoded G protein-coupled receptor UL33 exhibits oncomodulatory properties. J. Biol. Chem. 294 16297–16308. 10.1074/jbc.RA119.007796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanarsdall A. L., Pritchard S. R., Wisner T. W., Liu J., Jardetzky T. S., Johnson D. C. (2018). CD147 promotes entry of pentamer-expressing human cytomegalovirus into epithelial and endothelial cells. mBio 9:e00781-18. 10.1128/mBio.00781-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venton G., Crocchiolo R., Fürst S., Granata A., Oudin C., Faucher C., et al. (2014). Risk factors of Ganciclovir-related neutropenia after allogeneic stem cell transplantation: a retrospective monocentre study on 547 patients. Clin. Microbiol. Infect. 20 160–166. 10.1111/1469-0691.12222 [DOI] [PubMed] [Google Scholar]
- Vomaske J., Varnum S., Melnychuk R., Smith P., Pasa-Tolic L., Shutthanandan J. I., et al. (2010). HCMV pUS28 initiates pro-migratory signaling via activation of Pyk2 kinase. Herpesviridae 1:2. 10.1186/2042-4280-1-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Shenk T. (2005). Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U.S.A. 102 18153–18158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Yu Q. C., Schroer J., Murphy E., Shenk T. (2007). Human cytomegalovirus uses two distinct pathways to enter retinal pigmented epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 104 20037–20042. 10.1073/pnas.0709704104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E. C. Y., Pjechova M., Nightingale K., Vlahava V. M., Patel M., Ruckova E., et al. (2018). Suppression of costimulation by human cytomegalovirus promotes evasion of cellular immune defenses. Proc. Natl. Acad. Sci. U.S.A. 115 4998–5003. 10.1073/pnas.1720950115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. W., Kuo C. J., Lin W. R., Hsu C. M., Ho Y. P., Lin C. J., et al. (2016). The clinical characteristics and manifestations of cytomegalovirus esophagitis. Dis. Esophag. 29 392–399. 10.1111/dote.12340 [DOI] [PubMed] [Google Scholar]
- Wang Z., Cai J., Zhang M., Wang X., Chi H., Feng H., et al. (2016). Positive expression of human cytomegalovirus phosphoprotein 65 in atherosclerosis. Biomed. Res. Int. 2016:4067685. 10.1155/2016/4067685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.-K., Duh C.-Y., Wu C.-W. (2004). Human cytomegalovirus UL76 encodes a novel virion-associated protein that is able to inhibit viral replication. J. Virol. 78 9750–9762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston D. J., Young J.-A. H., Pullarkat V., Papanicolaou G. A., Vij R., Vance E., et al. (2008). Maribavir prophylaxis for prevention of cytomegalovirus infection in allogeneic stem cell transplant recipients: a multicenter, randomized, double-blind, placebo-controlled, dose-ranging study. Blood 111 5403–5410. 10.1182/blood-2007-11-121558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Sun L., Chen X., Du F., Shi H., Chen C., et al. (2013). Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science 339 826–830. 10.1126/science.1229963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Prager A., Boos S., Resch M., Brizic I., Mach M., et al. (2017). Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog. 13:e1006281. 10.1371/journal.ppat.1006281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaofei E., Meraner P., Lu P., Perreira J. M., Aker A. M., McDougall W. M., et al. (2019). OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. Proc. Natl. Acad. Sci. U.S.A. 116 7043–7052. 10.1073/pnas.1814850116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W. F., Yuan T. M. (2018). A review on the prevention and treatment of congenital cytomegalovirus infection in mothers and infants. Zhongguo Dang Dai Er Ke Za Zhi 20 870–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Yu J., Liu Z. (2020). MicroRNAs expressed by human cytomegalovirus. Virol. J. 17:34. 10.1186/s12985-020-1296-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B., Yang Y., Li S., Wang Y.-Y., Li Y., Diao F., et al. (2008). The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29 538–550. 10.1016/j.immuni.2008.09.003 [DOI] [PubMed] [Google Scholar]
- Zhou M., Yu Q., Wechsler A., Ryckman B. J. (2013). Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope. J. Virol. 87 9680–9690. 10.1128/JVI.01167-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou F., Lu Z. T., Wang S., Wu S., Wu Y. Y., Sun Z. R. (2018). Human cytomegalovirus UL141 protein interacts with CELF5 and affects viral DNA replication. Mol. Med. Rep. 17 4657–4664. 10.3892/mmr.2018.8419 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.