Abstract
Glycine max is one of the most important grain and oil crops, and improvement of seed yield is one of the major objectives in soybean breeding. The AP2/ERF superfamily members are involved in regulating flower and seed development in many species, and therefore play key roles in seed yield. However, it is still unknown that how many AP2/ERF members were presented in the G. max genome and whether these AP2/ERF family members function in flower and seed development in G. max. Here, we identified 380 AP2/ERF superfamily genes in the G. max genome. Phylogenetic analysis showed that 323 members were grouped into the ERF family, and 49 into the AP2 family. Among the AP2 family, 14 members of the euAP2 lineage showed high identity with their orthologs, and eight member of the ANT lineage were expressed highly in the seeds. Furthermore, seven of them (GmAP2-1 to GmAP2-7) were successfully cloned and over-expressed in Arabidopsis thaliana. The transgenic Arabidopsis plants over-expressing these GmAP2 genes flowered earlier relative to the wild type control. The seed length and width, and seed area of these over-expression lines were increased compared with the wild type, and seed weight of over-expression lines of GmAP2-1, GmAP2-4, GmAP2-5, and GmAP2-6 were greater than those of the wild type. Furthermore, the seed number per silique of the over-expression lines for GmAP2 genes were not affected except GmAP2-5. Collectively, GmAP2-1, GmAP2-4, and GmAP2-6 played important roles in regulating seed weight by affecting seed length, width and area, and further controlling seed yield.
Keywords: Glycine max, APETALA2/ethylene-responsive element binding factor, flowering, seed weight, seed size
Introduction
Soybean (Glycine max) was domesticated in China more than 5,000 years ago, which is one of the most important grain and oil crops. Improvement of seed yield is one of the major objectives in soybean breeding. Soybean is typically a short-day sensitive plant (Fehr and Caviness, 1977; Fehr, 1987), and both the time of flowering and date of seed maturity underlie seed yield (Yuan et al., 2002). Previous studies have demonstrated that the APETALA2 (AP2) gene in Arabidopsis thaliana regulates floral development (Jofuku et al., 1994b; Chen et al., 2004; Wollmann et al., 2010; Yant et al., 2010; Dinh et al., 2012; Krogan et al., 2012; Liu et al., 2014; Huang et al., 2017), ovule and seed development, and subsequently mediates seed size and seed weight (Jofuku et al., 1994b; Jofuku et al., 2005; Ohto et al., 2005; Ohto et al., 2009). Besides AP2 in Arabidopsis, its orthologs in other plant species were also found to be able to regulate seed-related phenotypes. For an example, over-expression of LaAP2L1 from Larix led to over 200% greater seed yield in Arabidopsis, contributed by enhanced cell proliferation and prolonged growth duration (Li et al., 2013). The AP2-like genes in Petunia regulates flower and seed development (Maes et al., 2001). Over-expression of the AfAP2-2 gene from Aechmea fasciata reduces seed size and delays flowering in Arabidopsis (Lei et al., 2019). All of the above-mentioned AP2 genes belong to the APETALA2/ethylene-responsive element binding factor (AP2/ERF) superfamily, which suggested that AP2/ERF superfamily members have potential impact on seed yield in soybean as in other plant species.
The AP2/ERF superfamily is a plant-specific transcription factor family containing a large number of members with at least one AP2 domain. This superfamily can be mainly divided into ERF, RAV, and AP2 families based on the numbers of the conserved AP2 domain and other DNA binding domains (Riechmann and Meyerowitz, 1998; Mizoi et al., 2012). The ERF family members consist of two subfamilies (ERF and DREB) with a single AP2 domain. The RAV family members have a single AP2 domain and an additional B3 domain. The AP2 family members contain a double and tandemly repeated AP2 domain (Sakuma et al., 2002; Nakano et al., 2006; Mizoi et al., 2012; Shu et al., 2015).
The ERF family members mainly respond to abiotic or biotic stresses in soybean and other plant species. For example, GmSGR, a seed-specific ERF family member in G. max, reduced ABA sensitivity and enhanced salt sensitivity in the seeds of the transgenic Arabidopsis plants by regulating the expression levels of the AtEm6 and AtRD29B genes (Wang et al., 2008). Over-expression of the GmERF3 gene increased tolerances to salt, drought, and diseases in transgenic tobacco plants (Zhang et al., 2009). Both GmERF5 and GmERF113 enhance resistance to pathogen Phytophthora sojae in G. max (Dong et al., 2015; Zhao et al., 2017). GsERF6 from G. soja significantly enhanced tolerance to bicarbonate in the transgenic Arabidopsis plants (Yu et al., 2016a). GsERF71 positively regulates alkaline stress tolerance in Arabidopsis (Yu et al., 2017). GsSnRK1 interplays with GsERF7 to regulate stress resistance in G. soja (Feng et al., 2020). These studies on ERF family members showed that ERF proteins mainly involved in abiotic and biotic stresses in soybean. However, a recent study showed that one ERF family member Glyma19g192400 is involved in plant growth regulation that mainly affects plant height, while another ERF member Glyma19g163900 is related to seed weight, which is involved in maintaining seed size, embryo size, seed weight and seed yield (Assefa et al., 2019).
The RAV family members regulate diverse processes, including cold tolerance, dehydration and circadian rhythm (Matias-Hernandez et al., 2014). In Arabidopsis, TEMPRANILLO1 (TEM1) and TEM2, two members of the RAV family, were identified as repressors in floral induction (Osnato et al., 2012). In G. max, GmRAV controls photosynthesis and senescence (Zhao et al., 2008), and GmRAV1 regulates the regeneration of roots and adventitious buds in both transgenic Arabidopsis and soybean plants (Zhang et al., 2019).
The AP2 family proteins are divided into two lineages: the ANT lineage and the euAP2 lineage (Kim et al., 2006). The ANT lineage is supported by a 10-aa insertion in the AP2-R1 domain and a 1-aa insertion in the AP2-R2 domain, relative to all other members of the AP2 family (Kim et al., 2006). The euAP2 lineage contains six members (AP2, TOE1, TOE2, TOE3, SMZ, and SNZ) that are targeted by a major developmental microRNA172 (miR172), which functions as a floral promoter (Park et al., 2002; Aukerman and Sakai, 2003; Schmid et al., 2003; Chen et al., 2004). The ap2 mutant and the quadruple mutant (toe1/toe2/smz/snz) flowered earlier than the wild type, but did not flower as early as the 35S:miR172 lines. The flowering time of the hexuple mutant (ap2/toe1/toe2/toe3/smz/snz) showed no difference from the 35S:miR172 plants (Yant et al., 2010). These results indicate that all the six members of the euAP2 lineage regulate the flowering time in Arabidopsis, and they were all under the control of miR172.
Among the AP2 family members, the AP2 gene is expressed in all four floral organs including sepal, petal, stamen and carpel, and in the developing ovules, which plays an important role in the control of flower, fruit and seed development (Jofuku et al., 1994b; Ripoll et al., 2011). In addition, the AP2 gene also functions in stem cell maintenance in shoot apical meristem (Würschum et al., 2006) and the control of floral stem cells (Aukerman and Sakai, 2003; Chen, 2004). The AP2 gene also mediates seed size by affecting embryo, endosperm, and seed coat development (Jofuku et al., 2005; Ohto et al., 2005; Ohto et al., 2009).
Most advances on the AP2 genes were carried out in Arabidopsis, while only few studies characterized the function of the AP2 orthologs in other plant species. For example, the double mutant lip1/lip2 in Antirrhinum majus displays homeotic conversion from sepals to leaves, leaving petals intact, which indicated that these two genes have a partially conserved role in floral organ identity with AtAP2 (Keck et al., 2003). The ortholog of AtAP2 in Petunia, PhAP2A, is expressed in a similar pattern as AtAP2 during flower development. However, the phap2a mutant does not display the same phenotypes as the Arabidopsis ap2 mutant due to the existence of other redundant AP2 members in Petunia (Maes et al., 2001). The AtAP2 ortholog in Solanum lycopersicum, SlAP2a, is a negative regulator of tomato fruit ripening (Chung et al., 2010). In G. max, only one AP2 family member Glyma01g022500 was reported to be associated with shoot related development and it affected internode number, but the molecular mechanism of this gene associated with internode number is still unclear (Assefa et al., 2019). It is still unknown that how many AP2 members were presented in the G. max genome, and whether these AP2 family members function in flower or seed development.
In this study, the protein sequences of all putative AP2/ERF superfamily members were analyzed in comparison with those of the Arabidopsis orthologs. Among them, seven AP2 genes (GmAP2-1 to GmAP2-7) were further characterized by ectopic over-expression in Arabidopsis. The seed phenotypes including the seed length and width, seed area and size were affected at different levels in these transgenic lines. Our study established an atlas of the AP2/ERF superfamily members in soybean, which will facility in-depth investigation on functions of AP2 gene members and the utilization in molecular breeding of soybean with improved seed yield.
Materials and Methods
Plant Material and Growth Condition
Soybean material used in this study is the cultivar Williams 82. The ap2-6 mutant of AP2 gene (At4g36920) and transgenic Arabidopsis plants are in the Col-0 background. Arabidopsis seeds were chilled at 4°C in the dark for 3 days, and then grown under long-day conditions (16-h light/8-h dark) at 22°C in growth chamber and seeds were harvested when the plant was completely mature. Soybean seeds were grown under long-day conditions (16-h light/8-h dark) at 25°C in growth chamber.
Phylogenetic Analysis
Multiple sequence alignment was performed by using ClustalX. The neighbor-joining (NJ) method was applied to construct trees using MEGA X software (Kumar et al., 2018). Bootstrapping with 500 replications was performed. The amino acid sequences of AP2 are retrieved from the Arabidopsis database (https://www.arabidopsis.org/) and NCBI database (http://www.ncbi.nlm.nih.gov/). All the amino acid sequences of soybean are retrieved from the soybean database (http://soykb.org/).
Multiple Sequence Alignment
The multiple sequence alignment was performed by using the software DNAMAN. The accession number are as followed (shown in parenthesis): Arabidopsis AtAP2 (AT4G36920); Glycine max GmAP2-1 (Glyma01g39520.3), GmAP2-2 (Glyma05g18041.1), GmAP2-3 (Glyma15g04930.1), GmAP2-4 (Glyma13g40470.1), GmAP2-5 (Gm16g00950.2), GmAP2-6 (Glyma08g38190.2), and GmAP2-7 (Glyma15g34770.1).
Gene Expression Analysis
Total RNAs were isolated from roots, stems, leaves, flowers, and seeds at different development stages: 10, 20, 30, 40, 50, 60, 70, and 80 day after pollination (DAP) of reference cultivar Williams 82, and also isolated from the transgenic lines of each gene in Arabidopsis. Semi-quantitative RT-PCR was carried out on Applied Biosystems Veriti Thermal Cycler. The PCR conditions were 94°C for 3 min; 20-30 cycles of 94°C for 20 s, 60°C for 20 s, and 72°C for 20 s; followed by a final extension of 72°C for 10 min. Quantitative RT-PCR analyses were carried out on Applied Biosystems 7500 Real-Time PCR Systems by using the SYBR Green reagent (Takara) according to the manufacturer’s instructions. The PP2A gene (Protein Phosphatase 2A subunit A3, At1g13320) in Arabidopsis and the SUB3 gene (Soybean ubiquitin-3, Glyma20g27950) in soybean were used as the reference genes. The primer sequences used for RT-PCR were listed in Supplementary Table S2. Data were calculated from three biological replicates, and each biological replicate was examined in triplicate.
RNA Isolation, Gene Cloning, and Plasmid Construction
Total RNAs were prepared by using the TRNzol Reagent (TIANGEN) according to the manufacturer’s instructions. First strand cDNA was synthesized using the HiFiScript reverse transcriptase (CWBIO). Fragments of candidate GmAP2 genes were amplified using the cDNA templates from soybean cultivar Williams 82 seeds with the gene specific primers (Supplementary Table S1). PCRs were carried out using Phusin High-Fidelity DNA Polymerase (NEB). Reactions were performed at 98°C for 2 min, and 98°C for 10 s, 56-64°C for 20 s, 72°C for 60 s for 35 cycles, and then 72°C for 7 min. The PCR products were cloned into pENTR entry vector (Invitrogen) and then subcloned into pB7WG2D by the LR reaction to generate the plant over-expression vectors, which were further confirmed by sequencing.
Generation of the Transgenic Arabidopsis
The over-expression vectors constructed as above mentioned were transformed into the Agrobacterium tumefaciens strain GV3101, and used for the transformation using the floral-dip method (Clough and Bent, 1998). The transformed plants were grown in the growth chamber, and the seeds were collected and screened on MS medium supplemented with 10 mg/L PPT for 2-3 days at 4°C, and then transformed to a growth chamber at 22°C and a 16/8 h photoperiod for 5-7 days. The seedlings were selected and transferred to sterile soil, and grown at 22°C and a 16/8 h photoperiod. The seeds were harvested from individual T1 generation transgenic Arabidopsis plants, and DNA was isolated from leaf using CTAB method. Positive transgenic lines were further identified by PCR with gene-specific primers. Then we used the same PCR methods to select T2 generation until getting homozygous T3 generation transgenic Arabidopsis plants.
Phenotypes of the Transgenic Arabidopsis Plants
The wild-type and transgenic Arabidopsis plants were grown in the same tray to ensure the same growth condition (16-h light/8-h dark) at 22°C. The seed number was calculated from 36 siliques for each line. The rosette leaf number of flowering and bolting time of transgenic lines were detected with 10 plants for each line.
Measurement of Seed Weight, Length, Width, and Area
Measurement of seed weight, length, width, and area were performed as previously described (Jiang et al., 2013), with minor modifications. For seed weight, plants were grown concurrently under identical conditions and seeds were harvested when the seeds were completely mature. One thousand seeds per transgenic line were dried at 37°C for 6 days and weighed. Data are presented as means ± SD from at least three independent experiments. For seed length and width, dried seeds were photographed using a Leica M165FC microscope and then measured by ImageJ software. ImageJ software was also used to calculate the seed area using 40 seeds for each line.
Statistical Analysis
The averages and standard deviations were calculated by using the SigmaPlot 10.0 software (Systat Software, Inc., Chicago, IL, United States). The Student’s t-test was used for p-value generation between the wild type and each transgenic Arabidopsis lines.
Results
Phylogenetic Analysis of the AP2/ERF Superfamily in G. max
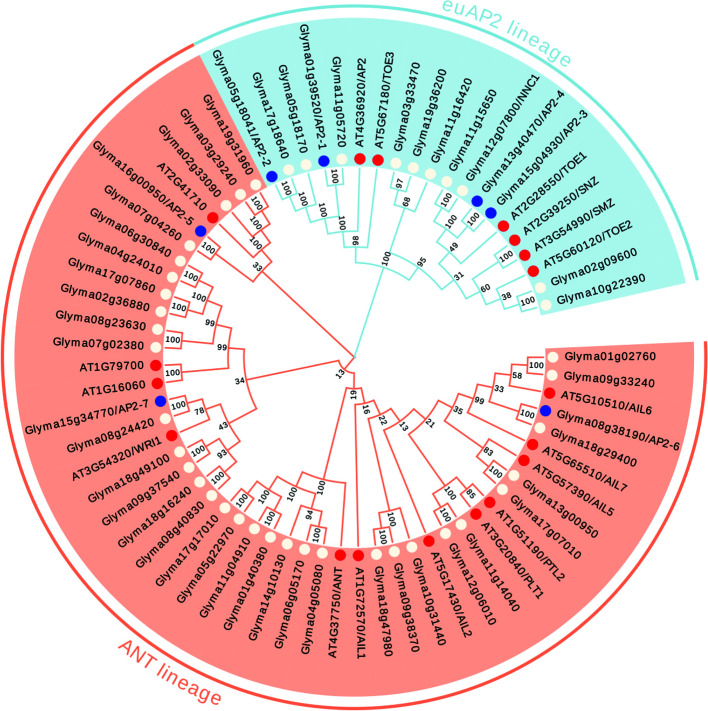
To determine the evolutionary relationships of the AP2/ERF superfamily proteins in G. max, we obtained the amino acid sequences of 380 AP2/ERF superfamily members from the soybean genome database (http://soykb.org/), and 145 AP2/ERF superfamily protein sequences were also retrieved from The Arabidopsis Information Resource (TAIR) and analyzed in parallel. The phylogenetic tree was constructed based on the alignment of the full-length amino acid sequences of all these 525 proteins (Figure 1, Supplementary Figures S1 and S2).
Figure 1.
Phylogenetic analysis of the AP2 family members in soybean and Arabidopsis. The euAP2 lineage was indicated in blue background, and the ANT lineage was indicated in orange background. The AP2 members in soybean were indicated in white dot with characterized 7 GmAP2 in blue dot, and the AP2 members in Arabidopsis were indicated in red dot. The neighbor-joining (NJ) method was applied to construct trees using MEGA X software. Bootstrapping with 1,000 replications was performed.
Among these family members, 49 proteins from G. max and 18 from Arabidopsis were assigned to the cluster of the AP2 family, due to the presence of the tandemly repeated double AP2 domain (Figure 1). Three hundred and twenty three proteins from G. max and 122 from Arabidopsis were grouped into the ERF family, and they all contain a single AP2 domain (Figure 1, Supplementary Figure S1). Eight proteins from G. max and five from Arabidopsis were clustered as the RAV family, with the presence of a single AP2 domain together with a B3 domain (Figure 1, Supplementary Figure S2).
The number of the AP2/ERF superfamily members in G. max was 2.62 folds more than that of the Arabidopsis. In particular, the numbers of the AP2 and ERF family genes in G. max were 2.72 and 2.65 folds more than those in Arabidopsis, respectively. Compared with Arabidopsis, these two family members of AP2/ERF superfamily proteins in G. max appeared to increase proportionally during evolution.
Screening of the Candidate AP2 Genes in G. max
Previous studies showed that the AP2 gene (At4g36920) in Arabidopsis regulated flower and seed development (Jofuku et al., 1994b; Jofuku et al., 2005; Ohto et al., 2005; Ohto et al., 2009). In addition, the other five genes (TOE1, TOE2, TOE3, SMZ and SNZ) in the same euAP2 lineage with AP2, also regulate flower development in Arabidopsis (Yant et al., 2010). Based on these studies and the phylogenetic relationship, we found that 14 genes from G. max (Glyma01g39520, Glyma02g09600, Glyma03g33470, Glyma05g18041, Glyma05g18170, Glyma10g22390, Glyma11g05720, Glyma11g15650, Glyma11g16420, Glyma12g07800, Glyma13g40470, Glyma15g04930, Glyma17g18640 and Glyma19g36200) were clustered together with the six genes (AP2, TOE1, TOE2, TOE3, SMZ, and SNZ) of the euAP2 lineage from Arabidopsis in the same clade (Figure 1). Therefore, these 14 genes are most likely the AP2 orthologs in G. max for the regulation of flower and seed development.
To further identify additional members in the AP2 family that are potentially involved in seed development in G. max, we also retrieved the expression profiles of all the ANT lineage genes from the online database (https://soybase.org/soyseq/), and found that eight genes (Glyma01g02760, Glyma07g04260, Glyma08g24420, Glyma08g38190, Glyma13g00950, Glyma15g34770, Glyma16g00950, and Glyma18g29400) were expressed significantly higher in seeds than in other tissues (Table 1). The typical expression profile indicated that these eight genes of the ANT lineage may be also involved in regulating seed development in G. max.
Table 1.
The relative transcript level of AP2 family genes in different tissues of soybean.
| Gene locus | Gene name | Lineage | YL | R | N | F | P | PS10 | PS14 | S10 | S14 | S21 | S25 | S28 | S35 | S42 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Glyma01g39520 | GmAP2-1 | euAP2 lineage | 169 | 200 | 19 | 116 | 135 | 130 | 85 | 42 | 35 | 13 | 45 | 39 | 45 | 22 |
| Glyma05g18041 | GmAP2-2 | no | no | no | no | no | no | no | no | no | no | no | no | no | no | |
| Glyma15g04930 | GmAP2-3 | 24 | 289 | 37 | 23 | 0 | 1 | 1 | 2 | 2 | 1 | 0 | 1 | 1 | 1 | |
| Glyma13g40470 | GmAP2-4 | 55 | 171 | 79 | 12 | 3 | 0 | 2 | 5 | 6 | 5 | 12 | 14 | 13 | 3 | |
| Glyma12g07800 | GmNNC1 | 184 | 50 | 5 | 43 | 29 | 38 | 30 | 4 | 5 | 12 | 18 | 6 | 12 | 5 | |
| Glyma02g09600 | 47 | 64 | 6 | 127 | 15 | 13 | 26 | 3 | 0 | 0 | 2 | 1 | 4 | 0 | ||
| Glyma03g33470 | 25 | 104 | 24 | 55 | 80 | 53 | 56 | 6 | 25 | 15 | 10 | 8 | 10 | 4 | ||
| Glyma05g18170 | 117 | 81 | 26 | 137 | 128 | 89 | 43 | 42 | 51 | 27 | 37 | 23 | 24 | 11 | ||
| Glyma10g22390 | 13 | 8 | 0 | 8 | 2 | 3 | 6 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma11g05720 | 169 | 130 | 7 | 183 | 246 | 204 | 117 | 90 | 97 | 39 | 72 | 59 | 55 | 28 | ||
| Glyma11g15650 | 243 | 114 | 16 | 18 | 10 | 6 | 5 | 0 | 4 | 2 | 3 | 3 | 3 | 2 | ||
| Glyma11g16420 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma17g18640 | 110 | 106 | 96 | 235 | 166 | 137 | 82 | 132 | 90 | 27 | 37 | 17 | 54 | 19 | ||
| Glyma19g36200 | 9 | 197 | 83 | 87 | 15 | 17 | 20 | 12 | 32 | 34 | 38 | 20 | 36 | 33 | ||
| Glyma16g00950 | GmAP2-5 | ANT lineage | 2 | 0 | 0 | 14 | 4 | 3 | 0 | 18 | 38 | 29 | 76 | 51 | 156 | 88 |
| Glyma08g38190 | GmAP2-6 | 24 | 1 | 0 | 4 | 10 | 5 | 1 | 34 | 72 | 16 | 52 | 43 | 60 | 31 | |
| Glyma15g34770 | GmAP2-7 | 11 | 30 | 21 | 1 | 1 | 1 | 0 | 14 | 42 | 26 | 108 | 60 | 125 | 27 | |
| Glyma01g02760 | 21 | 43 | 6 | 2 | 2 | 4 | 3 | 30 | 41 | 26 | 70 | 42 | 72 | 24 | ||
| Glyma01g40380 | 7 | 16 | 0 | 9 | 12 | 19 | 4 | 3 | 0 | 0 | 1 | 1 | 5 | 0 | ||
| Glyma02g33090 | 37 | 14 | 10 | 20 | 37 | 24 | 7 | 11 | 26 | 21 | 22 | 10 | 12 | 3 | ||
| Glyma02g36880 | 0 | 6 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma03g29240 | 32 | 4 | 9 | 16 | 20 | 25 | 10 | 17 | 29 | 13 | 21 | 18 | 38 | 35 | ||
| Glyma04g05080 | 239 | 55 | 9 | 83 | 59 | 48 | 12 | 8 | 13 | 8 | 9 | 3 | 10 | 0 | ||
| Glyma04g24010 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma05g22970 | 179 | 39 | 0 | 42 | 58 | 44 | 12 | 43 | 69 | 33 | 55 | 19 | 29 | 17 | ||
| Glyma06g05170 | 108 | 31 | 5 | 47 | 52 | 50 | 3 | 22 | 21 | 8 | 11 | 3 | 2 | 0 | ||
| Glyma06g30840 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma07g02380 | 20 | 11 | 2 | 26 | 13 | 31 | 20 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma07g04260 | 7 | 0 | 0 | 54 | 41 | 24 | 4 | 9 | 53 | 48 | 167 | 116 | 229 | 115 | ||
| Glyma08g23630 | 18 | 18 | 52 | 10 | 13 | 17 | 9 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma08g24420 | 3 | 24 | 6 | 0 | 1 | 2 | 0 | 5 | 34 | 44 | 112 | 63 | 133 | 37 | ||
| Glyma08g40830 | 4 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 1 | 8 | 2 | 12 | 3 | ||
| Glyma09g33240 | 35 | 74 | 113 | 4 | 11 | 6 | 2 | 17 | 79 | 60 | 106 | 66 | 95 | 48 | ||
| Glyma09g37540 | 0 | 0 | 7 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma09g38370 | 0 | 31 | 0 | 2 | 0 | 1 | 2 | 0 | 9 | 9 | 6 | 6 | 15 | 7 | ||
| Glyma10g31440 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | ||
| Glyma11g04910 | 3 | 15 | 0 | 9 | 3 | 3 | 2 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | ||
| Glyma11g14040 | 0 | 19 | 0 | 1 | 0 | 0 | 0 | 3 | 8 | 5 | 7 | 1 | 5 | 5 | ||
| Glyma12g06010 | 0 | 29 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 1 | 3 | 1 | 3 | 1 | ||
| Glyma13g00950 | 74 | 51 | 7 | 50 | 36 | 31 | 14 | 22 | 46 | 36 | 180 | 115 | 199 | 125 | ||
| Glyma14g10130 | 33 | 21 | 35 | 10 | 9 | 8 | 0 | 9 | 19 | 8 | 11 | 1 | 11 | 0 | ||
| Glyma17g07010 | 50 | 88 | 8 | 54 | 71 | 60 | 25 | 27 | 44 | 31 | 81 | 34 | 61 | 43 | ||
| Glyma17g07860 | 0 | 7 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Glyma17g17010 | 167 | 28 | 8 | 41 | 39 | 17 | 4 | 138 | 135 | 46 | 82 | 49 | 80 | 44 | ||
| Glyma18g16240 | 6 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 7 | 1 | ||
| Glyma18g29400 | 6 | 1 | 4 | 3 | 9 | 4 | 5 | 28 | 42 | 40 | 89 | 102 | 147 | 83 | ||
| Glyma18g47980 | 0 | 20 | 0 | 3 | 1 | 2 | 0 | 3 | 6 | 6 | 20 | 7 | 19 | 12 | ||
| Glyma18g49100 | 0 | 0 | 158 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | ||
| Glyma19g31960 | 47 | 17 | 5 | 35 | 19 | 25 | 7 | 3 | 7 | 1 | 20 | 14 | 19 | 11 |
Data were retrieved from https://soybase.org/soyseq/.
YL, young leave; R, roots; N, nodule; F, flower; P, one cm pod; PS10, pod shell at 10 days after pollination; PS14, pod shell at 14 days after pollination; S10, seeds 10 days after pollination; S14, seeds 14 days after pollination; S21, seeds 21 days after pollination; S25, seeds 25 days after pollination; S28, seeds 28 days after pollination; S35, seeds 35 days after pollination; S42, seeds 42 days after pollination.
We therefore initially amplified all the above mentioned 22 genes by RT-PCR, but only eight of them were successfully cloned and used for further studies. These eight genes (Glyma12g07800, Glyma01g39520, Glyma05g18041, Glyma15g04930, Glyma13g40470, Glyma16g00950, Glyma08g38190, and Glyma15g34770) were further designated in order as GmNNC1, GmAP2-1, GmAP2-2, GmAP2-3, GmAP2-4, GmAP2-5, GmAP2-6, and GmAP2-7, respectively. Among them, GmNNC1 were recently found to negatively regulate nodule number by another group (Wang et al., 2014), which will not be further pursued in this study. Therefore, these seven AP2 genes (GmAP2-1 to GmAP2-7) from G. max were further characterized in this study.
Sequence Analyses of the GmAP2 Genes
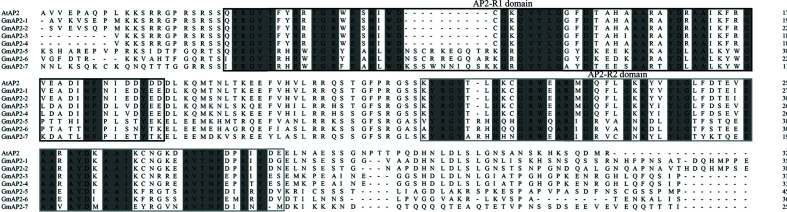
To further analyze the seven candidate GmAP2 genes, their amino acid sequences were aligned with AP2 gene of Arabidopsis. These GmAP2 proteins contained two AP2 domains (Figure 2, Supplementary Figure S3), which supported the fact that they belonged to the AP2 family. The deduced GmAP2-1, GmAP2-2, GmAP2-3, GmAP2-4 proteins showed 60.5%, 77.01%, 79.33%, and 57.19% identity with AtAP2 at amino acid level, respectively, and they were clustered with AtAP2 in the phylogenetic tree (Figure 1, Supplementary Table S2). The deduced GmAP2-5, GmAP2-6, and GmAP2-7 proteins showed relatively low identity with AtAP2 (54.44%, 53.85%, and 52.33%, respectively, Supplementary Table S1). Meanwhile, the identity among GmAP2-1, GmAP2-2 and GmAP2-3 is more than 73% at amino acid level, more than 90% between GmAP2-3 and GmAP2-4, and more than 70% among GmAP2-5, GmAP2-6, and GmAP2-7 (Supplementary Table S1). It was obvious that four proteins (GmAP2-1, GmAP2-2, GmAP2-3, and GmAP2-4) shared higher sequence identity with AtAP2 than the other three GmAP2 proteins (GmAP2-5, GmAP2-6, and GmAP2-7).
Figure 2.
Sequence alignment of the candidate AP2 proteins in soybean with those from Arabidopsis. The alignment was performed by using software DNAMAN. The two AP2 domains were boxed, with black color for AP2-R1 domain and grey color for AP2-R2 domain. All the sequences for soybean used in the figure are retrieved from the Soybean Database (http://soykb.org/) and AtAP2 are retrieved from the Arabidopsis Database (https://www.arabidopsis.org/). The accession number are as followed (shown in parenthesis): Arabidopsis thaliana: AtAP2 (At4g36920); Glycine max: GmAP2-1 (Glyma01g39520.3), GmAP2-2 (Glyma05g18041.1), GmAP2-3(Glyma15g04930.1), GmAP2-4 (Glyma13g40470.1), GmAP2-5 (Gm16g00950.2), GmAP2-6 (Glyma08g38190.2), and GmAP2-7 (Glyma15g34770.1).
Their AP2 domain regions (two AP2 domains and the linker sequence between them) were also aligned, and the corresponding sequence of GmAP2-1, GmAP2-2, GmAP2-3, and GmAP2-4 showed 96.77%, 96.77%, 86.45%, and 87.74% identity with AtAP2, respectively. The sequence identity in the AP2 domain regions was much higher than in the other regions of these four proteins. However, the sequence identity of the AP2 domain regions in GmAP2-5, GmAP2-6, and GmAP2-7, respectively, were lower as aligned with their full-length amino acid sequence.
Expression Profiles of the Candidate GmAP2 Genes
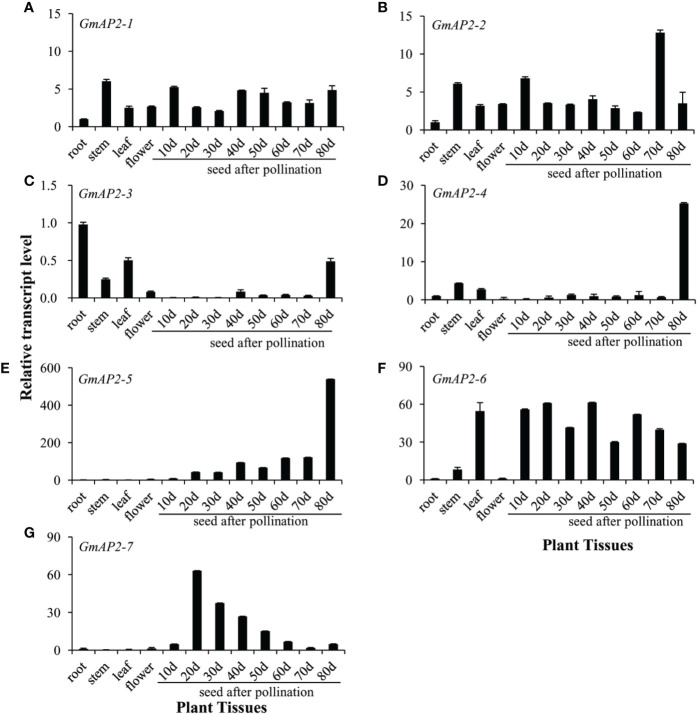
In order to further analyze the expression pattern of these candidate GmAP2 genes, RNAs were extracted from roots, stems, leaves, flowers, and seeds at different developing stages (10, 20, 30, 40, 50, 60, 70, and 80 days after pollination-DAP). The relative expression levels of these eight GmAP2 genes in different tissues were detected by real time quantitative PCR. The transcript levels of GmAP2-1 and GmAP2-2 were relatively stable in all tested tissues (Figures 3A, B). GmAP2-3 was expressed higher in roots, leaves and in 80-DAP seeds than in other tissues (Figure 3C). The transcript levels of GmAP2-4 were much higher in 80-DAP seeds than in other tissues (Figure 3D). The transcript levels of GmAP2-5 were relatively low in roots, stems, leaves and flowers, but its expression gradually increased with seed maturity. In particular, the transcript level of GmAP2-5 reached the highest level in 80-DAP seeds (Figure 3E). The transcript levels of GmAP2-6 were significantly higher in leaves and seeds than in roots, stems or flowers (Figure 3F). GmAP2-7 was mainly expressed in seeds, and its transcripts reached the highest level in 20-DAP seeds, and then gradually decreased afterwards (Figure 3G).
Figure 3.
Expression profiles of the candidate AP2 genes in different tissues of G. max. Total RNAs were isolated from roots, stems, leafs and flowers, 10 days seeds after pollination (10 days), 20, 30, 40, 50, 60, 70, and 80 days. The relative transcript level in roots for each gene was set as a value of 1.0. The SUB3 gene (Soybean ubiquitin-3, Glyma20g27950) in G. max was used as the reference gene. (A–G) The relative transcript level of GmAP2-1 (A), GmAP2-2 (B), GmAP2-3 (C), GmAP2-4 (D), GmAP2-5 (E), GmAP2-6 (F), and GmAP2-7 (G), respectively.
Collectively, these results suggested that GmAP2-1 and GmAP2-2 might function in different tissues of G. max; GmAP2-3 and GmAP2-4 might play a major role during late seed developmental stage; GmAP2-5 might be primarily involved in seed development, especially late stage. By contrast, GmAP2-7 might function in early seed developmental stage and GmAP2-6 might play a key role in both leaves and seed development.
Over-Expression of Seven GmAP2 Genes in Arabidopsis
To functionally characterize these GmAP2 genes, we individually transformed them into the wild type Arabidopsis (Columbia-0). More than 70 over-expression lines for every gene were obtained, and the transgenic plants with a 3:1 (resistant:sensitive) segregation ratio based on the Bar resistance were preselected. The T3 homozygous transgenic lines were verified by RT-PCR with gene-specific primers (Supplementary Figure S4), and at least six homozygous over-expression lines were further analyzed for each transgene.
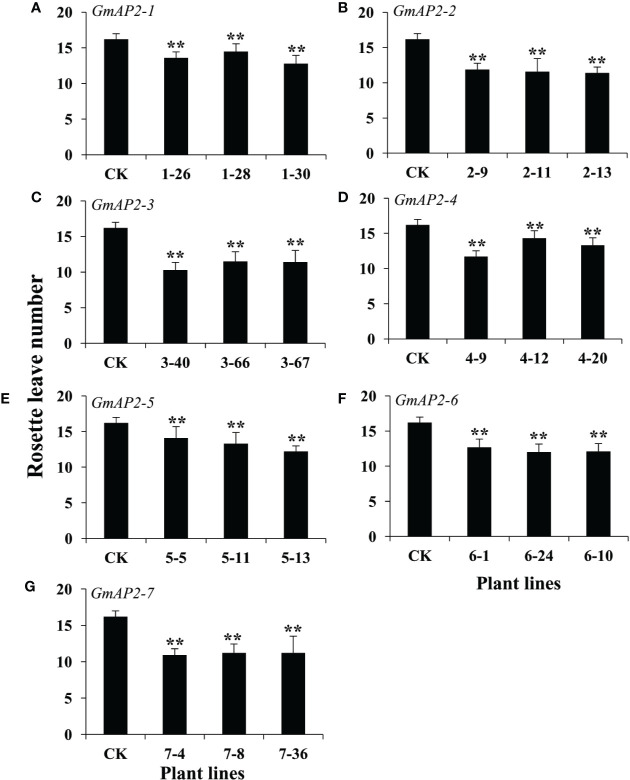
Due to their potential roles in the regulation of flowering time, we evaluate the flower time phenotype of these transgenic lines by counting the number of rosette leaves at the beginning of bolting. The number of rosette leaves in the transgenic lines over-expressing GmAP2-1, GmAP2-2, GmAP2-3, GmAP2-4, GmAP2-5, GmAP2-6, and GmAP2-7, were 12.8-14.5, 11.2-11.9, 10.3-11.5, 11.7-14.3, 12.2-14.1, 12.1-;12.7, and 10.9-11.2, respectively, which were all less than that of the wild type control (16.2) (Figure 4). Meanwhile, we also observed that these transgenic lines flowered early than the wild type control (Supplementary Figure S5). These results indicated that over-expression of these seven GmAP2 genes led to the early flowering, and these GmAP2 genes were involved in the regulation of flowering time.
Figure 4.
Rosette leave numbers of the transgenic lines over-expressing seven individual GmAP2 genes in Arabidopsis. The number of rosette leaves was calculated at the beginning of bolting to evaluate the flowering time of the transgenic lines. Ten plants for each line were selected to count the number of the rosette leaves. CK indicates the wild type Arabidopsis. (A–G) The over-expression lines for GmAP2-1 (A), GmAP2-2 (B), GmAP2-3 (C), GmAP2-4 (D), GmAP2-5 (E), GmAP2-6 (F), GmAP2-7 (G). Data are presented as mean ± SD, Student’s t test (n = 10, **P < 0.01).
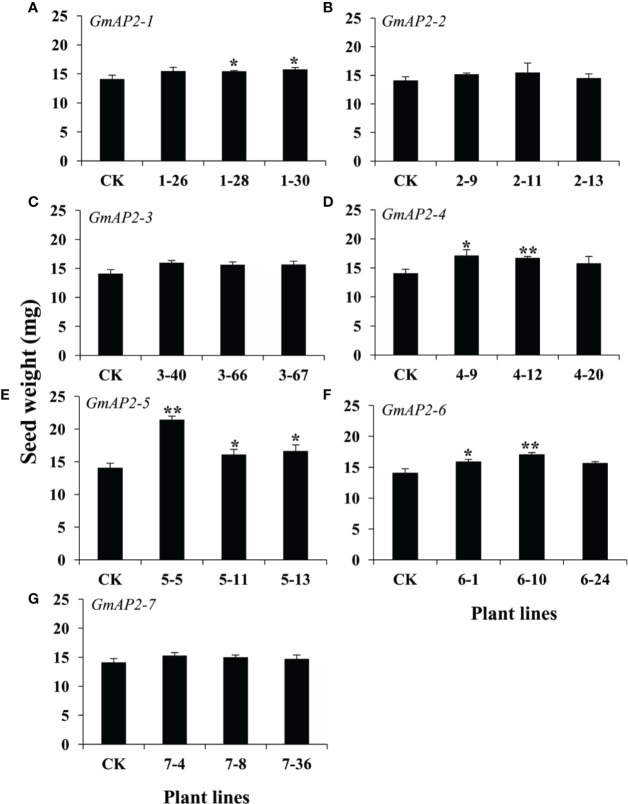
Besides the regulation of flowering time, AP2 and AP2 orthologs are also involved in seed development (Jofuku et al., 1994b; Maes et al., 2001; Jofuku et al., 2005; Ohto et al., 2005; Ohto et al., 2009; Li et al., 2013; Lei et al., 2019). Therefore, we also detected the seed phenotype of these transgenic lines, and found that the thousand seed weight of the GmAP2-1, GmAP2-4, GmAP2-5, and GmAP2-6 over-expression (OE) lines were 10%, 18%-21%, 14%-52%, and 11%-21% greater, respectively, than the wild type (Figures 5A, D–F). But the seed weight of the GmAP2-2, GmAP2-3, and GmAP2-7 OE lines showed no significant difference from the wild type (Figures 5B, C, G).
Figure 5.
Seed weight of the transgenic lines over-expressing individual GmAP2 genes in Arabidopsis. Seeds were harvested from 10 plants for each line, and seed weight per 1,000 dried mature seeds was measured. CK indicates the wild type Arabidopsis. (A–G) The over-expression lines for GmAP2-1 (A), GmAP2-2 (B), GmAP2-3 (C), GmAP2-4 (D), GmAP2-5 (E), GmAP2-6 (F), and GmAP2-7 (G). Data are presented as mean ± SD, Student’s t test (n = 3, *P < 0.05, **P < 0.01).
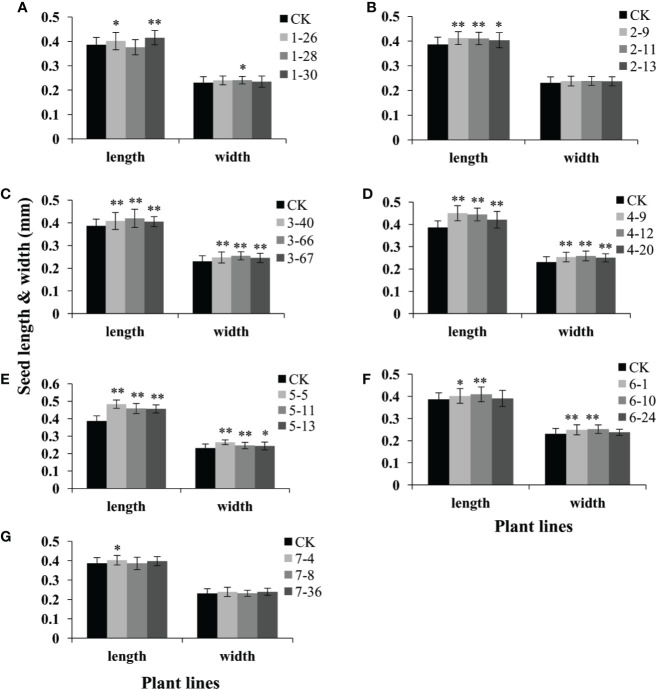
We further found that seed length of the transgenic lines over-expressing these seven genes (GmAP2-1 to GmAP2-7) were 3.9%-7.5%, 4.5%-6.6%, 4.9%-8.6%, 8.9%-16.5%, 18.0%-25.1%, 4.0%-6.0%, and 4.1% longer than the wild type (Figure 6). In particular, seed length of the GmAP2-5 OE lines was 18.0%-25.1% longer than the wild type (Figure 6E). The seed width of the OE lines of five genes (GmAP2-1, GmAP2-3, GmAP2-4, GmAP2-5, and GmAP2-6) were 4.4%, 6.3%-10.3%, 8.4%-12.1%, 5.3%-14.6%, and 7.9%-9.3% longer than the wild type, whereas the OE lines for GmAP2-2 and GmAP2-7 genes showed no difference with the wild type in term of seed width (Figure 6).
Figure 6.
Seed length and width of the transgenic lines over-expressing individual GmAP2 genes in Arabidopsis. Seeds were harvested from 10 plants for each line, and seed length and width of 40 seeds for each line were measured. CK indicates the wild type Arabidopsis. (A–G) The over-expression lines for GmAP2-1 (A), GmAP2-2 (B), GmAP2-3 (C), GmAP2-4 (D), GmAP2-5 (E), GmAP2-6 (F), and GmAP2-7 (G). Data are presented as mean ± SD, Student’s t test (n = 40, *P < 0.05, **P < 0.01).
In addition, the seed area of the OE lines of these seven genes were increased by 10.8-12.9%, 3.4%-6.5%, 7.6%-12.7%, 9.3%-13.2%, 6.0%-23.5%, 6.0%-19.0%, and 3.9%-9.5% compared with the wild type (Supplementary Figure S6). These results together implied that GmAP2-1, GmAP2-4, GmAP2-5, and GmAP2-6 functioned in regulating seed weight by affecting seed length, width and area.
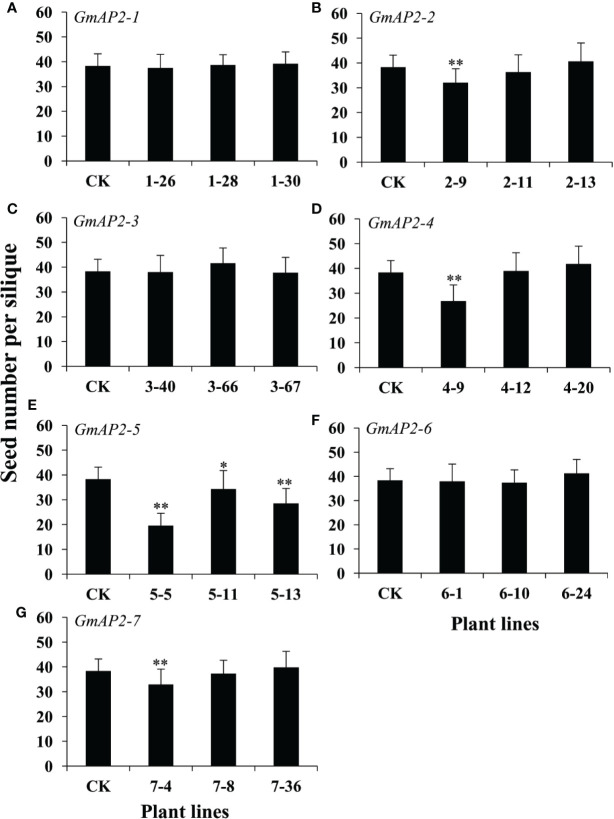
It is well known that seed number in each silique is important for seed yield. Therefore, we further calculated seed number per silique of the transgenic lines, and found that seed number per silique in the transgenic lines did not change for the six genes, but that for GmAP2-5 was obviously reduced compared with the wild type (Figure 7). These results showed that these AP2 genes did not affect seed number per silique except GmAP2-5, indicating that these AP2 genes did not affect plant fertility except GmAP2-5. Taken together, three AP2 genes, GmAP2-1, GmAP2-4, and GmAP2-6 control seed yield by affecting seed weight and size in Arabidopsis.
Figure 7.
Seed number in each silique of the transgenic lines over-expressing individual GmAP2 genes in Arabidopsis. Thirty-six yellow siliques were collected from 10 plants each line, and seed number per silique was counted. CK indicates the wild type Arabidopsis. (A–G) The over-expression lines for GmAP2-1 (A), GmAP2-2 (B), GmAP2-3 (C), GmAP2-4 (D), GmAP2-5 (E), GmAP2-6 (F), and GmAP2-7 (G). Data are presented as mean ± SD, Student’s t test (n = 36, *P < 0.05, **P < 0.01).
Complementation Assays of GmAP2 Genes in the Arabidopsis ap2-6 Mutant
Previous studies showed that the Arabidopsis ap2-6 mutant flowered earlier than the wild type (Yant et al., 2010), and seed weight and seed size of the ap2 mutant were increased compared with the wild type (Jofuku et al., 2005; Ohto et al., 2005), with reduced fertility in the strong mutant alleles of AP2 (ap2-6) (Ohto et al., 2005). To determine whether these seven GmAP2 genes can restore the defective phenotypes (flowering time, seed weight and seed number per silique) of the ap2-6 mutant, they were individually over-expressed in the ap2-6 mutant. Our data showed that the flower structure of all the transgenic lines in the ap2-6 mutant background was similar to the wild type (Supplementary Figure S7), indicating that these seven GmAP2 was able to rescue the defective phenotype of ap2-6 mutant.
The number of rosette leaves in the over-expression lines of the seven GmAP2 genes in the ap2-6 mutant showed no significant difference from the wild type (Figure 8A). We further found that seed weight of the individual over-expression lines of these GmAP2 genes in the ap2-6 mutant were same as the wild type (Figure 8B), but they were far lower than those of the ap2-6 mutant (Jofuku et al., 2005; Ohto et al., 2005). Their seed number per silique was also the same as the wild type (Figure 8C), but was higher than 23 ± 8.1 seeds of the ap2-6 mutant (Ohto et al., 2005). Taken together, these results indicated that these seven GmAP2 genes could restore the defective phenotypes of early flowering, large seed and less seed number of the Arabidopsis ap2-6 mutant.
Figure 8.
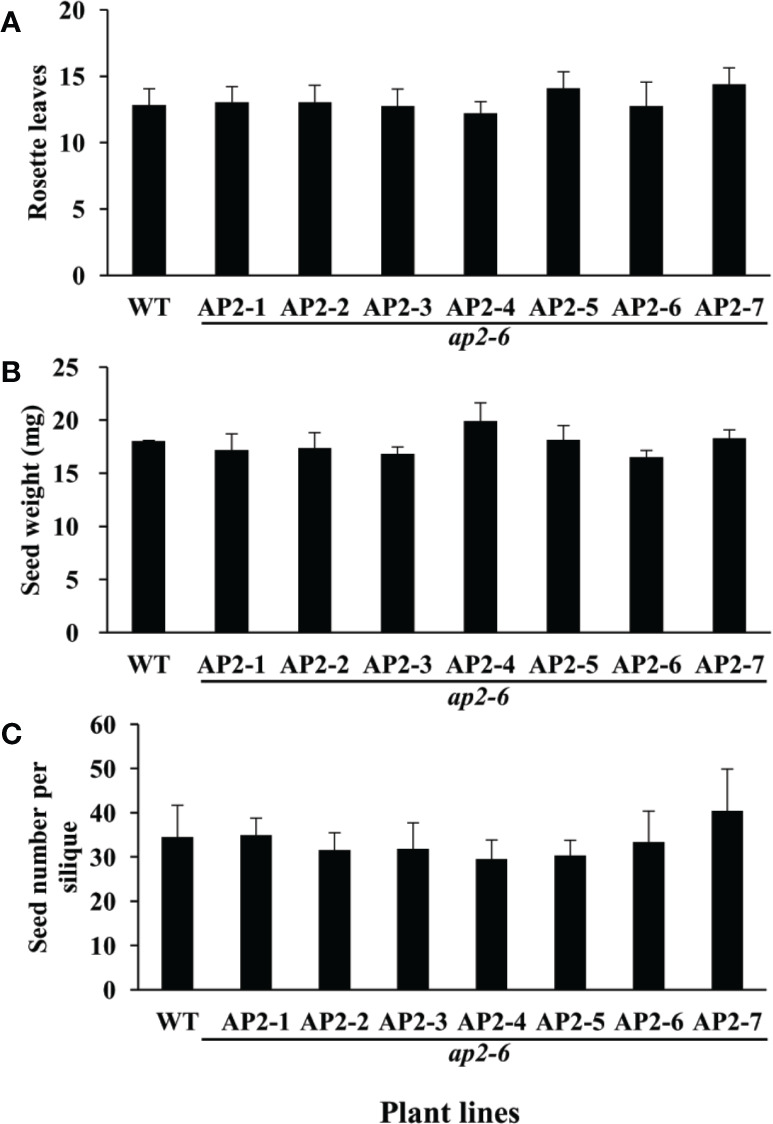
Rosette leave number, seed weight, and seed number in each silique of the transgenic lines over-expressing individual GmAP2 genes in the ap2-6 mutant. (A) The number of rosette leaves was calculated at the beginning of bolting to evaluate the flowering time of each transgenic line. Twenty plants for each line were selected to count the number of the rosette leaves. Data are presented as mean ± SD, Student’s t test (n = 20, *P < 0.05, **P < 0.01). (B) Seeds were harvested from 10 plants for each line, and seed weight per 1,000 dried mature seeds was measured. Data are presented as mean ± SD, Student’s t test (n = 3, *P < 0.05, **P < 0.01). (C) Twenty yellow siliques were collected from 10 plants each line, and seed number per silique was counted. WT indicates the wild type Arabidopsis. Data are presented as mean ± SD, Student’s t test (n = 20).
Discussion
Soybean is the world’s largest single source of vegetable protein and oil, accounting for more than 50% of the world edible oil (Soystats 2013, http://www.soystats.com). Same as for the other major crops, grain yield is always the most important trait in soybean breeding and cultivation, and improvement of seed yield is one of the major objectives in soybean breeding. Previous studies showed that both the time of flowering and date of seed maturity underlie seed yield (Yuan et al., 2002), and the ovule and seed development control final seed weight (Jofuku et al., 1994b; Jofuku et al., 2005; Ohto et al., 2005; Ohto et al., 2009). The APETALA2 (AP2) gene in Arabidopsis, as a member of AP2/ERF superfamily, regulates floral development (Jofuku et al., 1994a; Yant et al., 2010), and ovule and seed development, and subsequently mediates seed size and seed weight (Jofuku et al., 1994b; Jofuku et al., 2005; Ohto et al., 2005; Ohto et al., 2009).
In this study, we identified all 380 AP2/ERF superfamily members in G. max, which is more than two folds of the numbers in Arabidopsis. Meanwhile, the numbers of the ERF and the AP2 family in G. max are more than doubled than in Arabidopsis, respectively (Figure 1, Supplementary Figures S1 and S2). These results indicated that the AP2/ERF superfamily members appeared to increase proportionally, compared with Arabidopsis. We also found that the euAP2 lineage in G. max (Figure 1), and the number of miR172 in G. max that can target the euAP2 lineage (Wang et al., 2014), are more than doubled than the number of Arabidopsis. Compared with Arabidopsis, the numbers of miR172 family and their target genes (the genes of the euAP2 lineage) in G. max also appeared to increase proportionally. The same phenomenon was found for other large transcription factor families. Previous studies showed that the members of WRKY and R2R3-MYB family in G. max, are about two folds of the number in Arabidopsis (Chen et al., 2006; Du et al., 2012; Phukan et al., 2016; Yu et al., 2016b). The gene numbers of the same family were increased, which suggested that some genes developed new function during evolution.
Recent studies showed that miR172c in soybean targets the AP2 transcription factor NNC1 to activate the expression of ENOD40 gene (Wang et al., 2014), and miR172 in Arabidopsis targets all the six members of the euAP2 lineage (AP2, TOE1, TOE2, TOE3, SMZ and SNZ) to regulate flowering time (Yant et al., 2010). Another study showed that typical AP2 genes firstly appear in gymnosperms and diverged in angiosperms, following expansion of group members and functional differentiation (Wang et al., 2016). The gene numbers of the same family were increased, which also suggested that more genes functioned redundantly. miRNAs often showed conserved function in different species, and miR172 in soybean and their target genes had more members than those of Arabidopsis, indicating that more euAP2 genes functioned redundantly in soybean. Our study showed that at least four AP2 family members in soybean were involved in seed weight and seed size, in contrast, only single AP2 members in Arabidopsis regulates seed weight and seed size (Jofuku et al., 2005; Ohto et al., 2005). Compare with Arabidopsis, more AP2 family members in soybean played redundant role in regulating seed weight and seed size. Based on these evidences, it is believed that AP2/ERF superfamily members in G. max possibly are redundant with more diverse functions compared with Arabidopsis.
These seven AP2 orthologs from soybean have a partially conserved role in floral organ identity with AtAP2 as in other plant species. Previous studies showed that the AP2 gene is expressed in all four floral organs including sepal, petal, stamen and carpel, and affects floral organ identity specification (Drews et al., 1991; Jofuku et al., 1994a). It was observed that the homeotic transformation of sepals into carpels in ap2-5, ap2-6, and ap2-7; of petals to stamens in ap2-5; of petals to carpels in ap2-6 (Kunst et al., 1989). Our study showed that these 7 GmAP2 genes were expressed in flowers (Figure 3) and were able to rescue the defective phenotype of ap2-6 mutant, but their over-expression did not produce excess floral organs (Supplementary Figure S7), indicating that they retained partially conserved function in floral organ identity. Other studies showed that many AP2 orthologs in other species were also expressed in floral organs; and the mutation of AP2 orthologs led to the defective floral organ, and their over-expression produce excess floral organs; these AP2 orthologs generally rescue the defective phenotype of floral organ identity in the Arabidopsis ap2 mutant (Keck et al., 2003; Nilsson et al., 2007; Liu et al., 2012; Luo et al., 2012; Yan et al., 2012; Zhang et al., 2018). Therefore, the AP2 genes of different plant species, including these 7 GmAP2 genes, may share conserved roles in floral organ identity.
The Arabidopsis AP2 and AP2 orthologs in other species were reported to repress the flowering time, and their mutants led to earlier flowering, and their over-expression lines flower later compared with the wild type. For example, the ap2 mutants and other members of the euAP2 lineage in Arabidopsis flowered earlier than the wild type (Yant et al., 2010). The flowering time was delayed by 8-9 days in plants over-expressed LaAP2L1 from Larix compared with the vector control plants (Li et al., 2013). Over-expression of the AfAP2-2 gene from Aechmea fasciata delays flowering in Arabidopsis (Lei et al., 2019). The transgenic PaAP2L2 plants flowered later than control plants (Nilsson et al., 2007). Our data indicated that these seven genes were able to rescue the earlier flowering phenotype in ap2-6 mutant. However, in contrast to the late flowering phenotype in the over-expression plants of the AP2 orthologs (LaAP2L1, AfAP2-2, and PaAP2L2), the over-expression of these seven GmAP2 genes led to early flowering. Although these seven GmAP2 genes were able to rescue the early flowering time phenotype of ap2-6 mutants, which is different from the other AP2 genes, indicating that they had no similar role in regulating flowering time with other AP2 genes. The molecular mechanism of functional differentiation between GmAP2 and other AP2 orthologs (LaAP2L1, AfAP2-2 and PaAP2L2) needs further investigation.
It is well known that AP2 in Arabidopsis and AP2 orthologs in other species play key roles in seed development. It was indicated that AP2 and AP2 orthologs in Petunia hybrid, Aechmea fasciata and Brassica napus negatively regulated seed weight and seed size. For example, AP2 negatively regulates seed weight and seed size in Arabidopsis (Jofuku et al., 2005; Ohto et al., 2005). In addition, PhAp2A is capable of restoring the seed defective phenotype of the Arabidopsis ap2-1 mutant (Maes et al., 2001); and the seed size of Arabidopsis over-expressing AfAP2-2 gene from Aechmea fasciata was reduce (Lei et al., 2019). In the RNAi lines for the BnAP2 gene, seeds showed defects in shape, structure and development and larger seed size (Yan et al., 2012). However, another study reported that over-expression of LaAP2L1 from Larix led to larger organ size and over 200% greater seed yield in Arabidopsis, due to enhanced cell proliferation and prolonged growth duration (Li et al., 2013), indicating that the AP2 ortholog from Larix positively regulated seed weight and seed size. Our data suggested that GmAP2-1, GmAP2-4, GmAP2-5, and GmAP2-6 positively regulated seed weight and seed size, which is the same as LaAP2L1.
In addition, the seed weight of individual over-expression lines of these GmAP2 genes in ap2-6 mutant was not obviously different from that of the wild type (Figure 8B), and was significantly decreased compared with that of the ap2-6 mutant, suggesting that these GmAP2 genes have a partial conserved negative role in regulating seed weight and seed size. In comparison, the function of GmAP2 in regulating seed yield and size in Arabidopsis was positive by over-expression and negative in the complimentary assay, which is different from AP2 orthologs of other plant species.
Our studies implied that GmAP2-1, GmAP2-4, GmAP2-5, and GmAP2-6 function in regulating seed weight by affecting seed length, width and area, but GmAP2-1, GmAP2-4 and GmAP2-6 did not affect seed number per silique (Figure 7). Taken together, GmAP2-1, GmAP2-4, and GmAP2-6 genes control seed yield by affecting seed weight and seed size, and they could be utilized as potential target genes for seed yield breeding in soybean.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
WJ and YP conceived the project, designed the experiments. WJ, XZ, and XS performed the experiments and analyzed the data. WJ and XZ prepared the manuscript. JY and WJ performed phylogenetic analysis. YP revised the manuscript and supervised the project.
Funding
This work was supported by the National Key Research and Development Plan (2016YFD0101005). This work was also supported by the National Natural Science Foundation of China (31870281).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.566647/full#supplementary-material
References
- Assefa T., Otyama P., II, Brown A. V., Kalberer S. R., Kulkarni R. S., Cannon S. B. (2019). Genome-wide associations and epistatic interactions for internode number, plant height, seed weight and seed yield in soybean. BMC Genomics 20 (1), 527. 10.1186/s12864-019-5907-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman M. J., Sakai H. (2003). Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15 (11), 2730–2741. 10.1105/tpc.016238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Yang X. Y., He K., Liu M. H., Li J. G., Gao Z. F., et al. (2006). The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60 (1), 107–124. 10.1007/s11103-005-2910-y [DOI] [PubMed] [Google Scholar]
- Chen X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 (5666), 2022–2025. 10.1126/science.1088060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung M. Y., Vrebalov J., Alba R., Lee J., McQuinn R., Chung J. D., et al. (2010). A tomato (Solanum lycopersicum) APETALA2/ERF gene, SlAP2a, is a negative regulator of fruit ripening. Plant J. 64 (6), 936–947. 10.1111/j.1365-313X.2010.04384.x [DOI] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 (6), 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Dinh T. T., Girke T., Liu X., Yant L., Schmid M., Chen X. (2012). The floral homeotic protein APETALA2 recognizes and acts through an AT-rich sequence element. Development 139 (11), 1978–1986. 10.1242/dev.077073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong L., Cheng Y., Wu J., Cheng Q., Li W., Fan S., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66 (9), 2635–2647. 10.1093/jxb/erv078 [DOI] [PubMed] [Google Scholar]
- Drews G. N., Bowman J. L., Meyerowitz E. M. (1991). Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell 65 (6), 991–1002. 10.1016/0092-8674(91)90551-9 [DOI] [PubMed] [Google Scholar]
- Du H., Yang S. S., Liang Z., Feng B. R., Liu L., Huang Y. B., et al. (2012). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12, 106. 10.1186/1471-2229-12-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr W. R., Caviness C. E. (1977). Stages of soybean development. Iowa Agric. Exp. Station Special Rep. 80. [Google Scholar]
- Fehr W. R. (1987). “Principles of cultivar development,” in Theory and technique, vol. 1 (New York: Macmillan Press; ). [Google Scholar]
- Feng X., Feng P., Yu H., Yu X., Sun Q., Liu S., et al. (2020). GsSnRK1 interplays with transcription factor GsERF7 from wild soybean to regulate soybean stress resistance. Plant Cell Environ. 43 (5), 1192–1211. 10.1111/pce.13726 [DOI] [PubMed] [Google Scholar]
- Huang Z., Shi T., Zheng B., Yumul R. E., Liu X., You C., et al. (2017). APETALA2 antagonizes the transcriptional activity of AGAMOUS in regulating floral stem cells in Arabidopsis thaliana. New Phytol. 215 (3), 1197–1209. 10.1111/nph.14151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. B., Huang H. Y., Hu Y. W., Zhu S. W., Wang Z. Y., Lin W. H. (2013). Brassinosteroid regulates seed size and shape in Arabidopsis. Plant Physiol. 162 (4), 1965–1977. 10.1104/pp.113.217703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K. D., Boer B. G. W. D., Okamuro M. J. K. (1994. a). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 (9), 1211–1225. 10.1105/tpc.6.9.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K. D., den Boer B. G., Van Montagu M., Okamuro J. K. (1994. b). Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell 6 (9), 1211–1225. 10.1105/tpc.6.9.1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jofuku K. D., Omidyar P. K., Gee Z., Okamuro J. K. (2005). Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc. Natl. Acad. Sci. U. S. A. 102 (8), 3117–3122. 10.1073/pnas.0409893102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck E., McSteen P., Carpenter R., Coen E. (2003). Separation of genetic functions controlling organ identity in flowers. EMBO J. 22 (5), 1058–1066. 10.1093/emboj/cdg097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Soltis P. S., Wall K., Soltis D. E. (2006). Phylogeny and domain evolution in the APETALA2-like gene family. Mol. Biol. Evol. 23 (1), 107–120. 10.1093/molbev/msj014 [DOI] [PubMed] [Google Scholar]
- Krogan N. T., Hogan K., Long J. A. (2012). APETALA2 negatively regulates multiple floral organ identity genes in Arabidopsis by recruiting the co-repressor TOPLESS and the histone deacetylase HDA19. Development 139 (22), 4180–4190. 10.1242/dev.085407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018). MEGA x: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35 (6), 1547–1549. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L., Klenz J. E., Martinez-Zapater J., Haughn G. W. (1989). AP2 gene determines the identity of perianth organs in flowers of Arabidopsis thaliana. Plant Cell 1 (12), 1195–1208. 10.1105/tpc.1.12.1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Li Z. Y., Wang J. B., Fu Y. L., Xu L. (2019). Ectopic expression of the Aechmea fasciata APETALA2 gene AfAP2-2 reduces seed size and delays flowering in Arabidopsis. Plant Physiol. Biochem. 139, 642–650. 10.1016/j.plaphy.2019.03.034 [DOI] [PubMed] [Google Scholar]
- Li A., Zhou Y., Jin C., Song W., Chen C., Wang C. (2013). LaAP2L1, a heterosis-associated AP2/EREBP transcription factor of Larix, increases organ size and final biomass by affecting cell proliferation in Arabidopsis. Plant Cell Physiol. 54 (11), 1822–1836. 10.1093/pcp/pct124 [DOI] [PubMed] [Google Scholar]
- Liu Z., Gu C., Chen F., Jiang J., Yang Y., Li P., et al. (2012). Identification and expression of an APETALA2-like gene from Nelumbo nucifera. Appl. Biochem. Biotechnol. 168 (2), 383–391. 10.1007/s12010-012-9782-9 [DOI] [PubMed] [Google Scholar]
- Liu X., Dinh T. T., Li D., Shi B., Li Y., Cao X., et al. (2014). AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 80 (4), 629–641. 10.1111/tpj.12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H., Chen S., Jiang J., Teng N., Chen Y., Chen F. (2012). The AP2-like gene NsAP2 from water lily is involved in floral organogenesis and plant height. J. Plant Physiol. 169 (10), 992–998. 10.1016/j.jplph.2012.02.018 [DOI] [PubMed] [Google Scholar]
- Maes T., Van de Steene N., Zethof J., Karimi M., D’Hauw M., Mares G., et al. (2001). Petunia Ap2-like genes and their role in flower and seed development. Plant Cell 13 (2), 229–244. 10.1105/tpc.13.2.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matias-Hernandez L., Aguilar-Jaramillo A. E., Marin-Gonzalez E., Suarez-Lopez P., Pelaz S. (2014). RAV genes: regulation of floral induction and beyond. Ann. Bot. 114 (7), 1459–1470. 10.1093/aob/mcu069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoi J., Shinozaki K., Yamaguchi-Shinozaki K. (2012). AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819 (2), 86–96. 10.1016/j.bbagrm.2011.08.004 [DOI] [PubMed] [Google Scholar]
- Nakano T., Suzuki K., Fujimura T., Shinshi H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140 (2), 411–432. 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson L., Carlsbecker A., Sundas-Larsson A., Vahala T. (2007). APETALA2 like genes from Picea abies show functional similarities to their Arabidopsis homologues. Planta 225 (3), 589–602. 10.1007/s00425-006-0374-1 [DOI] [PubMed] [Google Scholar]
- Ohto M. A., Fischer R. L., Goldberg R. B., Nakamura K., Harada J. J. (2005). Control of seed mass by APETALA2. Proc. Natl. Acad. Sci. U. S. A. 102 (8), 3123–3128. 10.1073/pnas.0409858102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohto M. A., Floyd S. K., Fischer R. L., Goldberg R. B., Harada J. J. (2009). Effects of APETALA2 on embryo, endosperm, and seed coat development determine seed size in Arabidopsis. Sex Plant Reprod. 22 (4), 277–289. 10.1007/s00497-009-0116-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osnato M., Castillejo C., Matias-Hernandez L., Pelaz S. (2012). TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 3, 808. 10.1038/ncomms1810 [DOI] [PubMed] [Google Scholar]
- Park W., Li J., Song R., Messing J., Chen X. (2002). CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 12 (17), 1484–1495. 10.1016/S0960-9822(02)01017-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phukan U. J., Jeena G. S., Shukla R. K. (2016). WRKY transcription factors: molecular regulation and stress responses in plants. Front. Plant Sci. 7, 760. 10.3389/fpls.2016.00760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann J. L., Meyerowitz E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379 (6), 633–646. 10.1515/bchm.1998.379.6.633 [DOI] [PubMed] [Google Scholar]
- Ripoll J. J., Roeder A. H., Ditta G. S., Yanofsky M. F. (2011). A novel role for the floral homeotic gene APETALA2 during Arabidopsis fruit development. Development 138 (23), 5167–5176. 10.1242/dev.073031 [DOI] [PubMed] [Google Scholar]
- Sakuma Y., Liu Q., Dubouzet J. G., Abe H., Shinozaki K., Yamaguchi-Shinozaki K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290 (3), 998–1009. 10.1006/bbrc.2001.6299 [DOI] [PubMed] [Google Scholar]
- Schmid M., Uhlenhaut N. H., Godard F., Demar M., Bressan R., Weigel D., et al. (2003). Dissection of floral induction pathways using global expression analysis. Development 130 (24), 6001–6012. 10.1242/dev.00842 [DOI] [PubMed] [Google Scholar]
- Shu Y., Liu Y., Zhang J., Song L., Guo C. (2015). Genome-Wide analysis of the AP2/ERF superfamily genes and their responses to abiotic stress in Medicago truncatula. Front. Plant Sci. 6, 1247. 10.3389/fpls.2015.01247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang H., Zhang J., Chen S. (2008). A seed-specific AP2-domain transcription factor from soybean plays a certain role in regulation of seed germination. Sci. China C Life Sci. 51 (4), 336–345. 10.1007/s11427-008-0044-6 [DOI] [PubMed] [Google Scholar]
- Wang Y., Wang L., Zou Y., Chen L., Cai Z., Zhang S., et al. (2014). Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 26 (12), 4782–4801. 10.1105/tpc.114.131607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Cheng T., Lu M., Liu G., Li M., Shi J., et al. (2016). Expansion and functional divergence of AP2 group genes in spermatophytes determined by molecular evolution and Arabidopsis mutant analysis. Front. Plant Sci. 7, 1383. 10.3389/fpls.2016.01383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollmann H., Mica E., Todesco M., Long J. A., Weigel D. (2010). On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 137 (21), 3633–3642. 10.1242/dev.036673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würschum T., Groß-Hardt R., Laux T. (2006). APETALA2 regulates the stem cell niche in the Arabidopsis shoot meristem. Plant Cell 18 (2), 295–307. 10.1105/tpc.105.038398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Zhang L., Chen B., Xiong Z., Chen C., Wang L., et al. (2012). Functional identification and characterization of the Brassica napus transcription factor gene BnAP2, the ortholog of Arabidopsis thaliana APETALA2. PloS One 7 (3), e33890. 10.1371/journal.pone.0033890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant L., Mathieu J., Dinh T. T., Ott F., Lanz C., Wollmann H., et al. (2010). Orchestration of the floral transition and floral development in Arabidopsis by the bifunctional transcription factor APETALA2. Plant Cell 22 (7), 2156–2170. 10.1105/tpc.110.075606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Liu A., Duan X., Wang S., Sun X., Duanmu H., et al. (2016. a). GsERF6, an ethylene-responsive factor from Glycine soja, mediates the regulation of plant bicarbonate tolerance in Arabidopsis. Planta 244 (3), 681–698. 10.1007/s00425-016-2532-4 [DOI] [PubMed] [Google Scholar]
- Yu Y., Wang N., Hu R., Xiang F. (2016. b). Genome-wide identification of soybean WRKY transcription factors in response to salt stress. Springerplus 5 (1), 920. 10.1186/s40064-016-2647-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Duan X., Ding X., Chen C., Zhu D., Yin K., et al. (2017). A novel AP2/ERF family transcription factor from Glycine soja, GsERF71, is a DNA binding protein that positively regulates alkaline stress tolerance in Arabidopsis. Plant Mol. Biol. 94 (4-5), 509–530. 10.1007/s11103-017-0623-7 [DOI] [PubMed] [Google Scholar]
- Yuan J., Njiti V. N., Meksem K., Iqbal M. J., Triwitayakorn K., Kassem M. A., et al. (2002). Quantitative trait loci in two soybean recombinant inbred line populations segregating for yield and disease resistance. Crop Sci. 42 (1), 271–277. 10.2135/cropsci2002.2710 [DOI] [PubMed] [Google Scholar]
- Zhang G., Chen M., Li L., Xu Z., Chen X., Guo J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60 (13), 3781–3796. 10.1093/jxb/erp214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Huang S., Wang X., Liu J., Guo X., Mu J., et al. (2018). Defective APETALA2 genes lead to sepal modification in Brassica crops. Front. Plant Sci. 9, 367. 10.3389/fpls.2018.00367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Zhao L., Yang X., Li M., Sun J., Wang K., et al. (2019). GmRAV1 regulates regeneration of roots and adventitious buds by the cytokinin signaling pathway in Arabidopsis and soybean. Physiol. Plant 165 (4), 814–829. 10.1111/ppl.12788 [DOI] [PubMed] [Google Scholar]
- Zhao L., Luo Q., Yang C., Han Y., Li W. (2008). A RAV-like transcription factor controls photosynthesis and senescence in soybean. Planta 227 (6), 1389–1399. 10.1007/s00425-008-0711-7 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Chang X., Qi D., Dong L., Wang G., Fan S., et al. (2017). A novel soybean ERF transcription factor, GmERF113, increases resistance to phytophthora sojae infection in Soybean. Front. Plant Sci. 8, 299. 10.3389/fpls.2017.00299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.