Abstract
Inhibition of phosphodiesterase 4 (PDE4) is a promising therapeutic approach for the treatment of inflammatory pulmonary disorders, i.e. asthma and chronic obstructive pulmonary disease. However, the treatment with non-selective PDE4 inhibitors is associated with side effects such as nausea and vomiting. Among the subtypes of PDE4 inhibited by these inhibitors, PDE4B is expressed in immune, inflammatory and airway smooth muscle cells, whereas, PDE4D is expressed in the area postrema and nucleus of the solitary tract. Thus, PDE4D inhibition is responsible for the emetic response. In this regard, a selective PDE4B inhibitor is expected to be a potential drug candidate for the treatment of inflammatory pulmonary disorders. Therefore, a shared feature pharmacophore model was developed and used as a query for the virtual screening of Maybridge and SPECS databases. A number of filters were applied to ensure only compounds with drug-like properties were selected. Accordingly, nine compounds have been identified as final hits, where HTS04529 showed the highest affinity and selectivity for PDE4B over PDE4D in molecular docking. The docked complexes of HTS04529 with PDE4B and PDE4D were subjected to molecular dynamics simulations for 100ns to assess their binding stability. The results showed that HTS04529 was bound tightly to PDE4B and formed a more stable complex with it than with PDE4D.
Keywords: Phosphodiesterase 4, Shared feature pharmacophore model, Virtual screening, Molecular docking, Molecular dynamics simulations, Computational chemistry, Pharmaceutical chemistry, Bioinformatics, Pharmaceutical science, Pharmacology
Phosphodiesterase 4; Shared feature pharmacophore model; Virtual screening; Molecular docking; Molecular dynamics simulations; Computational chemistry; Pharmaceutical chemistry; Bioinformatics; Pharmaceutical science; Pharmacology
1. Introduction
Cyclic nucleotide phosphodiesterases (PDEs) are a diverse family of enzymes involved in the hydrolysis of 3′,5′-cyclic adenosine monophosphate (cAMP) and 3′,5′-cyclic guanosine monophosphate (cGMP) to their inactive 5′-AMP and 5′-GMP forms, respectively (Azam and Tripuraneni, 2014). In mammals, 21 genes encoding 11 families of PDEs, namely PDE1-PDE11 (Kanes et al., 2007; O'Donnell and Zhang, 2004). The PDE4 family is the largest of the 11 PDE families that selectively hydrolyse cAMP and includes four subtypes: PDE4A, PDE4B, PDE4C and PDE4D (Baumer et al., 2007; Conti et al., 2003; Souness et al., 2000). All four PDE4 subtypes comprise a related structural organization with a highly conserved catalytic domain in the C-terminal region and upstream conserved regions (UCR1: ~55 amino acids and UCR2: ~78 amino acids) in the N-terminal which further classify the PDE4 subtypes into four classes: long, short, super-short and dead-short (Burgin et al., 2010; Conti et al., 2003; Klussmann, 2016). The absence of UCR1 renders the short forms of PDE4 monomeric (Bolger et al., 2015; Houslay, 2010; Klussmann, 2016). The UCR1 is a target of protein kinase A phosphorylation that increases the hydrolysis of cAMP by 2–4 fold over the basal levels. Whereas, the UCR2 opening and closing regulate the access of cAMP to the active site. The small inhibitors which bind to the active site of PDE4 and interact with specific residues in the UCR2 can close the UCR2 and inhibit the enzyme (Burgin et al., 2010). PDE4 subtypes express in immune and inflammatory cells at different levels where PDE4B being the most abundant in immune, inflammatory and airway smooth muscle cells (Yuasa et al., 2001). PDE4A is expressed at low levels in inflammatory cells while; PDE4C is absent. On the other hand, the PDE4D express in the area postrema and nucleus of the solitary tract (Lamontagne et al., 2001; Perez-Torres et al., 2000).
The production of pro-inflammatory cytokines and inflammatory cytokines is regulated via degradation of cAMP by PDE4. The inhibition of PDE4 results in the elevation of cAMP level and activation of PKA and exchange protein directly activated by cAMP (Epca 1/2). The activation of PKA leads to the phosphorylation of cAMP-responsive element-binding protein (CREB) and activation of the transcription factor 1 (ATF-1) which results in increasing the production of anti-inflammatory cytokines and decrease the inflammatory cytokines (Li et al., 2018). Roflumilast, a PDE4 inhibitor, was approved by the FDA for the treatment of asthma and chronic obstructive pulmonary disease (COPD). Roflumilast showed no effect on the acute phase of bronchoconstriction in patients with asthma whereas, it showed a significant improvement in the lung function, as well as a reduction in the exacerbation rate when used with long-acting bronchodilators in patients with COPD in large clinical trials. However, the maximum tolerated dose of roflumilast is near the bottom of the efficacy dose-response curve (Phillips, 2020).
The development of PDE4 inhibitors hasn't achieved much success yet due to the low therapeutic index of the existing inhibitors and gastrointestinal side effects, i.e. nausea, vomiting and diarrhoea, that severely restrict their therapeutic application. These side effects might be attributed, to the structural similarity of the two PDE isoforms, PDE4B (responsible for most of the anti-inflammatory response) and PDE4D (related to the gastrointestinal side effects). Thus to overcome these problems, a selective PDE4B inhibitor may offer a solution for maximising the therapeutic action and minimising the side effects (Fan Chung, 2006; Wang et al., 1999). Recently, the implication of pharmacophore modelling, molecular docking and molecular dynamics (MD) simulations in the discovery of subfamily-selective PDE4B inhibitors showed a lot of success (Li et al., 2016; Shubina et al., 2015). Accordingly, pharmacophore-based virtual screening, molecular docking and MD simulations have been applied in this study with the objective to identify a selective PDE4B inhibitor and unravel the crucial amino acids involved in the binding interactions with the inhibitor and its stabilisation in the active site of PDE4B.
2. Materials and methods
2.1. Development of pharmacophore model: ligand-based approach
Forty-three PDE4 inhibitors were selected to constitute the training set compounds based on the inhibitory activity values (IC50) of PDE4B and PDE4D from the reported literature (Supplementary Material-Table S1) (Goto et al., 2014; Hagen et al., 2014; Naganuma et al., 2009). The selected inhibitors have IC50 of PDE4B lower than that of PDE4D thus; they were used for the development of ligand-based (LB) pharmacophore model. ChemSketch 12.0 was used to sketch the two-dimensional (2D) structures of the training set compounds which, were later converted into their three-dimensional (3D) structures. The pharmacophore modelling was carried out using LigandScout 4.2 software (Wolber and Langer, 2005). Initially, multiple acceptable conformations for each compound present in the training set were generated using OMEGA conformation model generation method (Seidel et al., 2017; Wolber and Langer, 2005) followed by clustering the compounds according to their 3D pharmacophore characteristics. Finally, an independent pharmacophore model for each cluster was created. The pharmacophore model included only the chemical features present in all the training set compounds.
2.2. Development of pharmacophore model: structure-based approach
The structure-based (SB) pharmacophore model was created based on the structure of PDE4B (PDB ID: 3W5E) with its co-crystallised ligand using standard protocol for LigandScout 4.2 (Wolber and Langer, 2005). In the process of pharmacophore generation, the defined active site was first analysed, followed by determination of the protein-co-crystallised ligand interactions with the active site residues. Based on the interactions, the corresponding pharmacophoric features were generated. These features included hydrogen bond acceptor (HBA), hydrogen bond donor (HBD), hydrophobic area (HA), aromatic ring (AR) and the ionisable area within the receptor active site sphere. The pharmacophore model obtained in this manner was considered as the SB pharmacophore model.
2.3. Shared feature pharmacophore model generation and validation
The LB and SB pharmacophore models were superimposed to create the shared feature pharmacophore model. The pharmacophore for PDE4B (PDB ID: 3W5E) was set as the reference element. The generated model was validated for its ability to discriminate the active compounds from the decoys by screening a dataset consisting of 12 active and 600 decoy compounds. The active compounds with a known PDE4 inhibitory activity were obtained from the literature (Supplementary Materials, Tables 1-S5) to constitute the test set; while the decoy compounds (with no reported inhibitory activity for both PDE4B and PDE4D) were downloaded from PubChem. The test set and decoys were screened using the generated shared feature pharmacophore model. Validation parameters such as AUC100% (Area Under Curve), sensitivity (Se) and specificity (SP) were calculated. In addition, the discrimination efficiency of the generated pharmacophore was evaluated by calculating the enrichment factor (EF) and goodness of hit (GH) score.
| EF = (Ha/Ht)/(A/D) |
| GH score = (3/4 Ya + 1/4 Se) SP |
Ya: Yield of actives, Ha: Number of actives in the hit list, Ht: Number of hits, A: Number of actives in the database, D: Number of compounds in the database.
2.4. Virtual screening
The pharmacophore-based virtual screening was employed to identify potential hit compounds that could selectively inhibit PDE4B. The validated shared feature pharmacophore was used as a 3D structural query in the virtual screening of Maybridge (51,775 compounds) and SPECS (212,294 compounds) databases to retrieve novel scaffolds for PDE4B inhibition (Sakkiah and Lee, 2012; Vuorinen et al., 2014). In the first step, the two libraries were converted into 3D multi-conformational databases for virtual screening using LigandScout 4.2 advanced algorithm, idbgen, which computes conformations and annotates each conformation with pharmacophore features. Next, the pharmacophore model was added to the screening perspective in LigandScout followed by loading the two libraries as well. Finally, the shared feature pharmacophore model was employed for the screening of the two libraries at once (Erlina and Yanuar, 2018).
In the second step, the retrieved hits were filtered based on some parameters to ensure only compounds with drug-like properties were selected. The first filter that was applied was Lipinski's rule of five. According to this rule, the orally active drug should have < five hydrogen bond donors, < ten hydrogen bond acceptors, molecular weight (M.wt) < 500 Da and log p (octanol/water partition coefficient) < five (Lipinski, 2004; Lipinski et al., 1997). Then, the hits were filtered based on the molecular polar surface (PSA), whereby compounds with PSA < 140 Å2 are believed to permeate the cell membrane. Finally, the hits were further ranked based on the pharmacophore fit score (PFS); which used to measure the fit values of each hit compounds to the pharmacophoric features of PDE4B. The PFS was set according to the fit values of the test set compounds that were used to validate the pharmacophore model. Thus, only hits with PFS higher than or equal to the PFS of any of the test set compounds were retained.
2.5. Molecular docking study
2.5.1. Protein preparation
The first step in a good docking study is the selection of the most appropriate protein structure for both PDE4B and PDE4D from the numerous structures available in the PDB. The selection criteria for the protein structure was, the resolution must be higher than 2.5 Å, the completeness of the active site, the regulatory region and the control region 3 (CR3), in addition to the presence of a co-crystallised ligand in the active site. The protein structures fulfilling the above criteria were downloaded from PDB.
The selected protein structures were prepared for docking by extracting the co-crystallised ligands, removing the water molecules and optimising the initial structures (alternate the residue conformations, correct connectivity and bond order) using Accelrys Discovery Studio (DS) 2.5.5 (Accelrys, 2009). This was followed by correcting the protonation state using Propka server. Finally, the prepared protein structures were subjected to short minimisation with 250 steps of steepest descent followed by 500 steps of conjugated gradient in Sander module of AMBER14 program. A cutoff distance at 12 A° for non-bonded interactions was applied (Case et al., 2014).
2.5.2. Ligand preparation
The nine compounds obtained from the virtual screening, as well as the standard compound (rolipram), were visualised in the DS Visualizer (BIOVIA, 2017). The structures of the ligands were checked to correct the bond orders and add the missing hydrogen atoms. Finally, GaussView 6 was used to optimise the structures of the ligands.
2.5.3. Molecular docking
Three structures of PDE4B (PDB ID: 3W5E, 3KKT and 3HMV) and PDE4D (PDB ID: 3G58, 6BOJ and 6NJH), were used in molecular docking to check the affinity of the ligands for the different 3D structure of the same protein. LigandScout 4.2 was used for performing molecular docking. Initially, the protein structure was loaded; followed by locating the active site from the position of the co-crystallised ligand. After locating the active site, the ligands were loaded; and their energies were minimised using MMFF94 force field. Finally, the ligands were docked into the active site of the target protein using AutoDock Vina 1.1 implemented in LigandScout 4.2. The ligands were ranked based on their binding energy and selectivity for PDE4B over PDE4D. Poses with the most negative binding energies (favourable binding) for PDE4B were identified as best poses. Rolipram was used as a standard for evaluating the affinity and selectivity of the hits (Goto et al., 2013).
2.6. Molecular dynamics study
In order to determine the stability of the complexes and calculate the binding free energies for each complex, the docked PDE4B (PDB ID: 3HMV)-ligand complex and PDE4D (PDB ID: 3G58)-ligand complex with the lowest binding energy were selected for molecular dynamics (MD) simulations. The MD simulations were performed using Particle Mesh Ewald Molecular Dynamics (PMEMD.CUDA) from AMBER14 on NVIDIA GPUs (Quadro, 2000D) (Case et al., 2014). Each complex was prepared by using Antechamber, Parmchk and AMBER force field (ff14SB). The Antechamber and Parmchk were used to assign general AMBER force field (GAFF) parameters, calculate the restricted electrostatic potential (RESP) under Gaussian g09 and generate the prepi and frcmod files for the ligands (Xing et al., 2017). While; the ff14SB was used to describe the molecular characteristics of the complex and obtain the topology (top) and coordinate (crd) files for each complex (Pérez et al., 2007). tLeap was then used for solvation of the complex in a cubic box of TIP3P water extending at least 10 Å in each direction from the solute. Na+ ions were added to neutralize the complex, and the cutoff radius was kept to 12 Å to compute the non-bonded interactions (Harrach and Drossel, 2014; Zhu et al., 2017).
A 100 ns simulation was carried out for each complex. The simulations were performed under periodic boundary conditions. The long-range electrostatic interactions were treated using Particle Mesh Ewald (PME) method whereas the SHAKE algorithm was used to constrain bonds that involve hydrogen (Darden et al., 1993; Linse and Linse, 2014). Initially, the solvated system was subjected to a series of 500 steps of steepest descent followed by 1000 steps of conjugated gradient minimisations to relax the system. Then, each complex was heated from 0 to 310.15 K over a period of 60 ps followed by 200 ps of NPT (constant number of particles, pressure and temperature) equilibration at 310.15 K and 1 atm pressure. Eventually, an unrestrained MD simulation (no force applied on any atoms) was performed at a constant temperature of 310.15 K for 100 ns. During this phase, the structural coordinates of the system were recorded every 0.1 ps intervals to build a trajectory of the system dynamics (Kuhn and Kollman, 2000).
The evaluation of the structural properties, root mean squared deviations (RMSD) of the backbone atoms, and determination of root mean square fluctuations (RMSF) of the residues and hydrogen bond were carried out using cpptraj module of AmberTools 14. The binding free energy calculation of each complex was performed based on Molecular Mechanics-Poisson Boltzmann Surface Area (MM-PBSA) using MMPBSA.py method, and the decomposition energy of the key residues was calculated using per-residue decomposition energy module of AMBER14. The binding and decomposition energies were extracted from the trajectories of the last 20 ns of the simulations (Amir-Hassan et al., 2017; Martínez-Muñoz et al., 2017).
3. Results and discussion
3.1. Development of pharmacophore model: ligand-based approach
The pharmacophore model represents the chemical features of the molecular structure that are responsible for the interaction with the target. The generated LB pharmacophore model was based on the pharmacophoric features present in the training set compounds that were selected from the literature. The selection of these compounds was based on their structural diversity and potency. A single cluster was generated after clustering the ligands according to their 3D pharmacophore characteristics. Consequently, one pharmacophore model was developed as the LB pharmacophore model (Figure 1). The generated LB-pharmacophore model comprised of six pharmacophoric features, two HBA, one HA, and three AR.
Figure 1.
Ligand-based pharmacophore model. The pharmacophore features are colour-coded: HBA: red, HA: yellow, AR: blue.
3.2. Development of pharmacophore model: structure-based approach
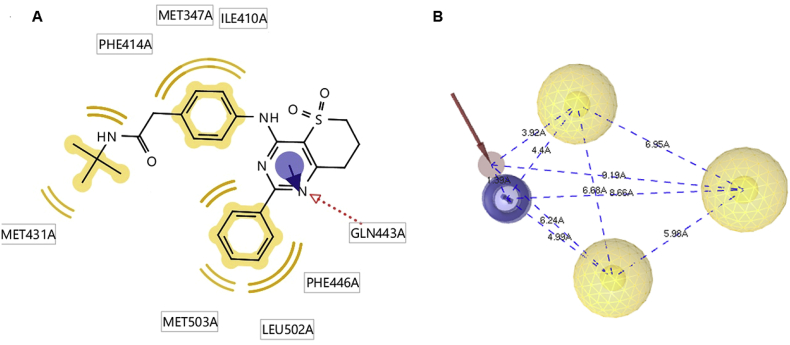
The SB pharmacophore was derived from the protein-ligand complex using the interactions observed between the ligand and the protein. In this study, the generated SB-pharmacophore model for PDE4B (PDB ID: 3W5E) consists of five features: one HBA, three HA, and one AR. In the 2D representation, the HBA feature of the co-crystallised ligand is represented by the amino group which is oriented towards GLN443A of 3W5E. The HA feature is represented by the two phenyl rings and the butyl group; while, the AR feature is represented by the pyrimidine ring. The representative pharmacophore of PDE4B-co-crystallised ligand complex is shown in Figure 2, where the red arrow indicates the HBA, the yellow colour refers to the hydrophobic interactions, and blue colour indicates the AR.
Figure 2.
A) PDE4B-co-crystallised ligand interactions. B) The corresponding pharmacophore model. The pharmacophore features are colour-coded: HBA: red, HA: yellow, AR: blue.
3.3. Shared feature pharmacophore model generation and validation
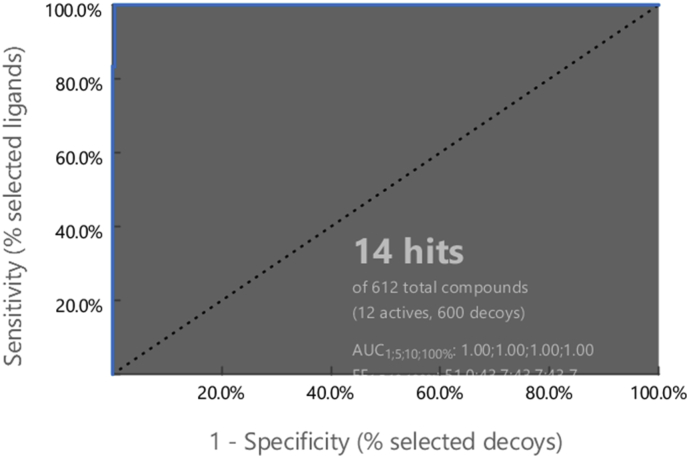
Following the generation of LB and SB pharmacophores, the shared feature pharmacophore was developed. This model was later used in virtual screening for identification of new PDE4B inhibitors. The new model consists of three features, one HBA, one HA, and one AR, as shown in Figure 3. The shared feature pharmacophore model was refined by decreasing the tolerance for some of the selected features; then it was validated using the test set and decoys. The validation results (Table 1) and the ROC (Receiver Operating Characteristic) curve (Figure 4), clearly suggest that shared feature pharmacophore model recognised all the active compounds in the data set, while predicting two inactives to be active compounds (false positive). The AUC100% value = 1, GH score (0.87) and EF (43.70), indicate its ability to recognise the true positives in the dataset. Thus, the shared feature pharmacophore model is the optimal model to be used for virtual screening and identifying selective PDE4B inhibitors.
Figure 3.
Shared feature pharmacophore model. The pharmacophore features are colour-coded: HBA: red, HA: yellow, AR: blue.
Table 1.
Validation parameters of the shared feature pharmacophore model.
| Hits | 14 |
| AUC100% | 1 |
| FN | 0 |
| FP | 2 |
| %YA | 85.71% |
| SE | 1 |
| SP | 0.99 |
| GH score | 0.87 |
| EF | 43.70 |
False negative (FN), False positive (FP).
Figure 4.
ROC curve of the validated pharmacophore model.
3.4. Virtual screening
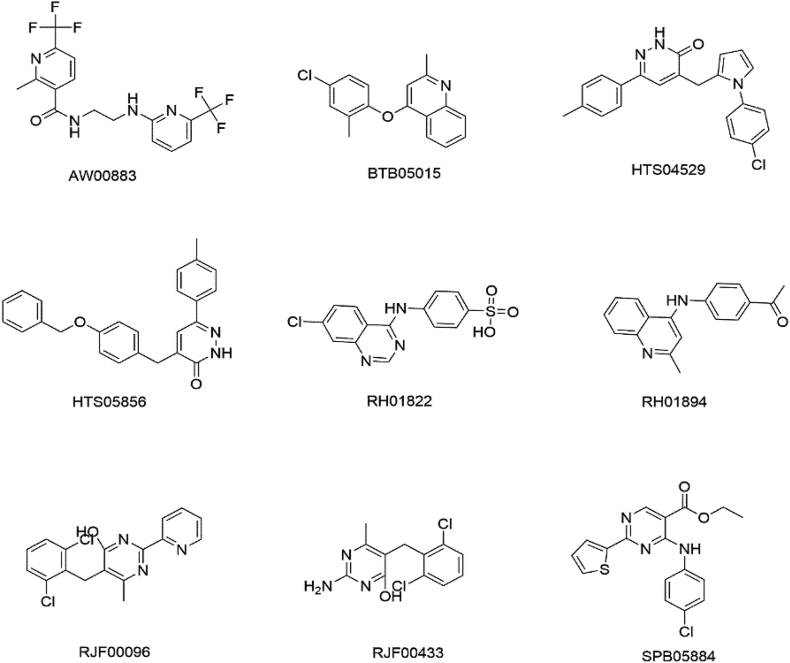
Virtual screening was performed to retrieve hits from the two databases for further evaluation. Based on the pharmacophore similarity, a total of 12063 hits were obtained from Maybridge and SPECS databases after the first round of screening. The hits were subjected to filtering by applying Lipinski's rule of five for their likelihood to be orally bioavailable. Accordingly, 7841 drug-like hits were obtained. These hits were further narrowed down by applying the PFS > 38.9 filter, this resulted in a total of nine compounds. These nine compounds were selected for molecular docking studies (Figure 5) (Table 2).
Figure 5.
The 2D structures of the retrieved compounds from the virtual screening.
Table 2.
Computed standard properties of the hits based on Lipinski's Rule of Five, polar surface area and pharmacophore fit score.
| Compound ID | M.wt | Log P | No. of HBD | No. of HBA | PSA | PFS |
|---|---|---|---|---|---|---|
| AW00883 | 392.30 | 2.79 | 2 | 5 | 66.91 | 38.93 |
| BTB05015 | 283.76 | 4.82 | 0 | 2 | 22.12 | 38.91 |
| HTS04529 | 375.86 | 3.49 | 1 | 4 | 50.68 | 38.93 |
| HTS05856 | 382.46 | 4.66 | 1 | 4 | 54.98 | 38.91 |
| RH01822 | 335.77 | 1.30 | 2 | 6 | 92.18 | 38.92 |
| RH01894 | 276.34 | 2.61 | 1 | 3 | 41.99 | 38.94 |
| RJF00096 | 346.22 | 3.84 | 1 | 4 | 58.9 | 38.97 |
| RJF00433 | 284.14 | 3.36 | 2 | 4 | 72.03 | 38.91 |
| SPB05884 | 359.83 | 3.88 | 1 | 5 | 64.11 | 38.94 |
3.5. Analysis the results of molecular docking study
Molecular docking was performed to study the binding modes of the hit compounds within the active site of both receptors. The standard and the nine compounds that passed Lipinski's rule of five, PSA and PFS filters were docked into the active site of both receptors (Goto et al., 2013). The docking results were analysed in term of binding energy and binding mode. The binding energy of the best binding mode for each ligand with both receptors and the amino acids involved in the protein-ligand interactions are summarised in Table 3 and Supplementary Material-Table S2 and Table S3.
Table 3.
The binding energy of the standard and the nine hits to PDE4B and PDE4D.
| Compounds | Binding energy with PDE4B (kcal/mol) |
Binding energy with PDE4D (kcal/mol) |
||||
|---|---|---|---|---|---|---|
| 3O0J | 4KP6 | 3W5E | 3G58 | 6IM6 | 2FM0 | |
| Roilipram | -16.20 | -16.70 | -19.90 | -19.10 | -16.70 | -17.90 |
| HTS04529 | -20.00 | -20.30 | -23.40 | -19.70 | -20.30 | -19.60 |
| HTS05856 | -21.50 | -20.30 | -23.40 | -20.20 | -16.90 | -21.50 |
| RH01894 | -17.70 | -21.30 | -20.20 | -18.20 | 17.50 | -20.20 |
| RJF00096 | -15.30 | -16.00 | -20.70 | -17.20 | -16.70 | -17.10 |
| RJF00433 | -17.50 | -19.50 | -16.50 | -16.50 | -17.10 | -18.00 |
| RH01822 | -16.70 | -19.00 | -19.40 | -17.60 | -18.60 | -19.00 |
| SPB05884 | -15.90 | -17.60 | -18.70 | -17.50 | -17.10 | -17.10 |
| BTB05015 | -17.40 | -18.10 | -19.40 | -17.60 | -16.40 | -18.20 |
| AW00883 | -17.60 | -18.40 | -19.50 | -17.20 | -18.10 | -17.60 |
As can be seen from Table 3, all the inhibitors exhibited comparable binding energies for both receptors. Two inhibitors displayed the lowest binding energy with PDE4B when compared to other compounds and more selectivity for PDE4B over PDE4D. These inhibitors are HTS04529 and HTS05856, with binding energies < -20.00 kcal/mol. Therefore, the underlying binding interactions of these two compounds with both receptors were further analysed to explore the structural features that contribute to the PDE4B selectivity.
The architecture of the binding site of PDE4 enzyme achieves all the required criteria for a druggable binding site. It consists of three subdomains, a) M pocket represents the area in the enzyme that contains the metal ions (Zn+2 and Mg+2) that are important for catalysis the hydrolysis of cAMP. b) S pocket, signifies the solvent filled side pocket which has polar residues and, c) Q pocket which is known as an inhibitor pocket and further divided into hydrophobic clamp (P-clamp) and conserved purine-selective glutamine. The Q pocket flanked by narrow hydrophobic pockets known as Q1 (deep small hydrophobic pocket with tightly bound water) and Q2 (protruding hydrophobic pocket) (Card et al., 2004; Huai et al., 2003). This region includes the hydrophobic residues that are less conserved than M pocket. A number of studies have shown that the PDE4 inhibitors share two features, a planar ring structure held within the hydrophobic residues of the P-clamp (PHE446 in PDE4B vs PHE538 in PDE4D at the roof of the P-clamp and ILE410 and PHE414 in PDE4B vs ILE502 and PHE506 in PDE4D on the floor of the binding site) (Tripuraneni and Azam, 2016) and a hydrogen bonding interaction with the invariant glutamine residue (GLN443 in PDE4B vs GLN535 in PDE4D) (Card et al., 2004; Xing et al., 2017). The cAMP is recognized by enzyme upon the formation of a hydrogen bond with the Q pocket. As a consequence, the phosphate moiety forms a complex with the metal in the M pocket, promoting the hydrolysis of cAMP (Houslay et al., 2005; Spina, 2008). Therefore, the PDE4 inhibitor should be able to occupy the Q and M pockets to block the entry and hydrolysis of cAMP.
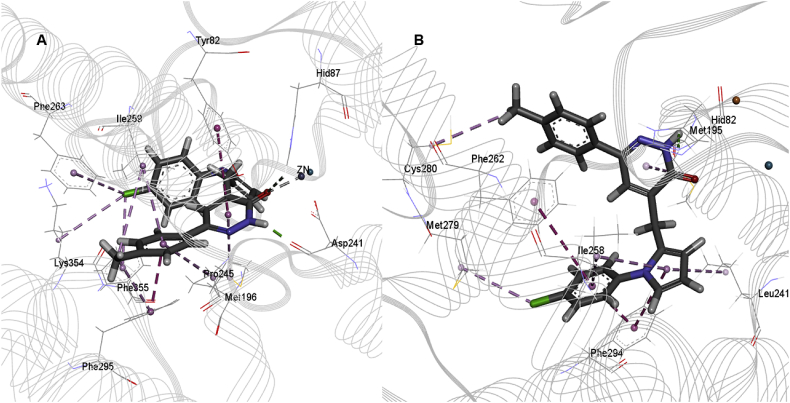
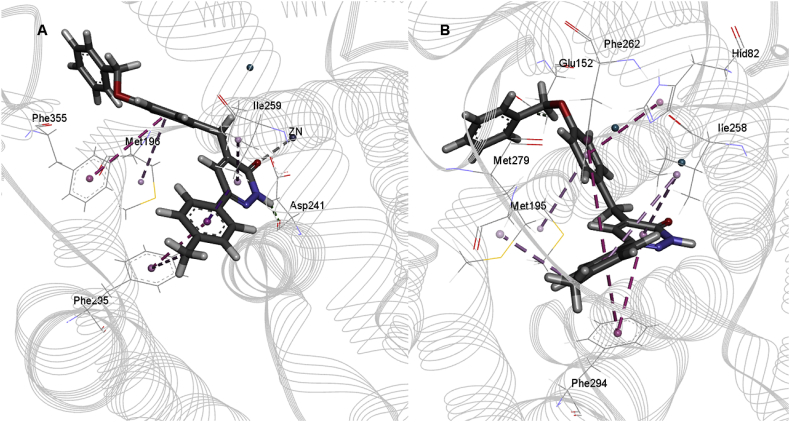
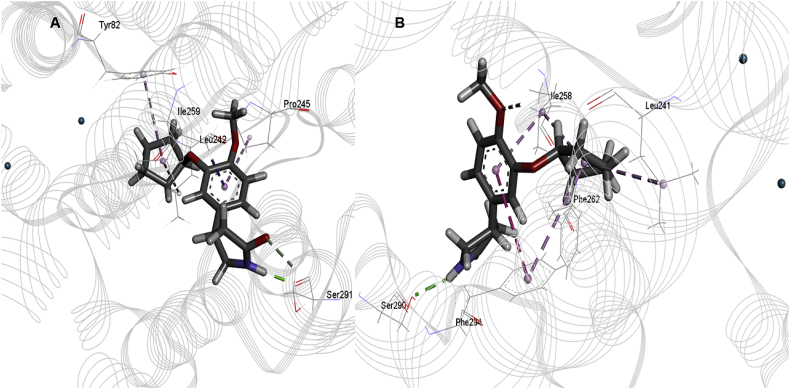
The docking study has revealed that the interactions between PDE4B and the two inhibitors, HTS04529 and HTS05856, were dominated by the Pi-Pi stacking interactions between pyridazinone and the substituted phenyl rings. The interactions were seen in the region surrounding TYR82-ILE299 (PDB ID: 3HMV, the residue's numbering starts from 1) due to the presence of the active site in that region. The two inhibitors interacted with the catalytic domain of PDE4B. Their central pyridazinone ring stacked between PHE295 and ILE259 in an ideal position to hydrogen bond with the ASP241. These interactions represent the interactions involving Q switch, and P-clamp observed among all PDE4 inhibitors (Card et al., 2004). Moreover, the carbonyl group exhibited a coordinate bond with the Zn+2 cation, which stabilised the inhibitors further in the catalytic domain (Figures 6A and 7A). The hydrogen-bonding and the hydrophobic interactions, as well as the proximity of the inhibitors to Zn+2, showed that HTS04529 and HTS5856 have an orientation within the active site that would inhibit the entry of cAMP into the catalytic domain. This finding points towards the higher activity of these two inhibitors in comparison to the other inhibitors in the series. For the highly active inhibitors, HTS04529 and HTS05856, residue TYR82, MET196, ASP241, ILE259, PHE263 and PHE295 became more stable due to the hydrophobic and hydrogen-bonding interactions. While region HID199-ARG229 became more flexible due to the absence of interactions. These results are consistent with previous studies and postulate that the region ASN253-ILE299 is favourable for binding and region LEU249-ILE299 is involved in hydrophobic interactions (Li et al., 2016; Tripuraneni and Azam, 2016; Xu et al., 2000). In contrast to HTS04529 and HTS05856, only hydrophobic and hydrogen-bonding interactions were observed between rolipram and PDE4B. These interactions are with the residues SER291, PRO245, ILE259, TYR82 and LEU242 (Figure 8A). The ligand did not involve in any aromatic interaction or coordinate bonds with the metal ion; which explains the lower affinity of rolipram for PDE4B than HTS04529 and HTS05856.
Figure 6.
The best binding pose and the interactions of HTS04529 within the active site of A) PDE4B and B) PDE4D, H-bond is presented by green, hydrophobic interactions by pink, Pi-Pi stacking interactions by magenta and metal coordinate by grey dotted lines.
Figure 7.
The best binding pose and interactions of HTS05856 within the active site of A) PDE4B and B) PDE4D, H-bond is presented by green, hydrophobic interactions by pink, Pi-Pi stacking interactions by magenta and metal coordinate by grey dotted lines.
Figure 8.
The best binding pose and interactions of rolipram within the active site of A) PDE4B and B) PDE4D, H-bond is presented by green, hydrophobic interactions by pink and Pi-Pi stacking interactions by magenta dotted lines.
For PDE4D (PDB ID: 3G58, the residue's numbering starts from 1), the binding modes of HTS04529 and HTS05856 with the receptor showed that the two inhibitors occupied the P-clamp and interacted with the key residues ILE258, PHE262 and PHE294. The phenyl ring of the two inhibitors stacked between PHE262 and PHE294. Furthermore, the phenyl ring of HTS05856 showed Pi-Pi interactions with HID82 (Figures 6B and 7B) while HTS04529 was involved in hydrogen bonding interaction with the receptor. This hydrogen bond formed between the nitrogen atom of pyridazinone and HID82. There was no interaction with either Zn+2 or Mg+2. The position of rolipram within the active site of PDE4D showed that the ligand formed two hydrophobic interactions with LEU241 and ILE258, one hydrogen bond with SER290 and Pi-Pi interactions with PHE294 (Figure 8B).
The similarity between the binding sites of PDE4B and PDE4D makes the elucidation of selectivity and designing of selective inhibitor a challenging task. Although many reported studies have tried to identify the main differences between the binding sites of these two isoforms using experimental tests, the reason behind the selective inhibition of specific PDE4 isoform is still not fully clear (Feng et al., 2018; Wang et al., 2007). Thus, computational methods have been used to explore the structures of PDE4 isoforms, particularly PDE4B and PDE4D, and find the residues that are crucial for selectivity. The residues that determine PDE4B/4D selectivity are located in the CR3 region outside the catalytic pocket. These residues can be targeted for the design of PDE4B subtype-selective inhibitors. The CR3 is a flexible region that can adopt multiple orientations. Each small compound presents different chemical features that stabilise different poses of CR3. Fox et al. tried to understand the role of CR3 in regulating the activity of PDE4. They compared the binding mode of several compounds to the CR3 of either PDE4B or PDE4D, whereby they found that each compound can interact with different residues along the CR3 helix resulting in multiple “closed” conformations (Fox et al., 2014). Several non-conserved residues can be found within the CR3 of PDE4B and PDE4D, that could contribute to the subtype-selectivity (Table 4). However, only three of these residues protrude into the binding pocket. These residues are LEU502 in PDE4B vs GLU502 in PDE4D, PHE505 and PHE506 in both PDE4B and PDE4D (Kranz et al., 2009) (see Table 4).
Table 4.
The sequence of CR3 in PDE4B and PDE4D. The non-conserved residues are highlighted in bold.
| PDE4B | ∗498/347CQGLMEKFQFE509/358 |
| PDE4D | 593/349GQTEKFQFELT603/359 |
The residue number in PDB/The residue number in our study.
In the crystal structures of PDE4B (PDB ID: 4MYQ and 3HMV) complexed with A-33 and THBT, both ligands displayed common binding schema with the catalytic site like most inhibitors, i.e. occupying the P-clamp and H-bonding with the hydrophobic residues (Fox et al., 2014; Kranz et al., 2009). Additionally, the thienyl group on A-33 interacted with the LEU in the CR3 of PDE4B (Fox et al., 2014). Comparison of the binding mode of HTS04529 with A-33 and THBT showed that HTS04529 formed similar interactions with the catalytic domain of PDE4B. The stacking of pyridazinone ring of HTS04529 between PHE295 and ILE259 and the interaction with the Zn+2 allow the ligand to occupy the Q and M pocket; which would result in blocking the entry of cAMP. Furthermore, the hydrophobic interactions between the chloro-substituted phenyl group of the inhibitor and the residues LYS354 and PHE355 of the CR3 of PDE4B stabilise a unique conformation of CR3. These results show that it is possible to design highly selective PDE4B inhibitors by exploiting the non-conserved residues outside the catalytic domain. Accordingly, HTS04529 was selected for MD simulations to illustrate the changes in the profile of interactions and evaluate the stability of the ligand in the active site of PDE4B.
3.6. Analysis of the results of molecular dynamics simulations
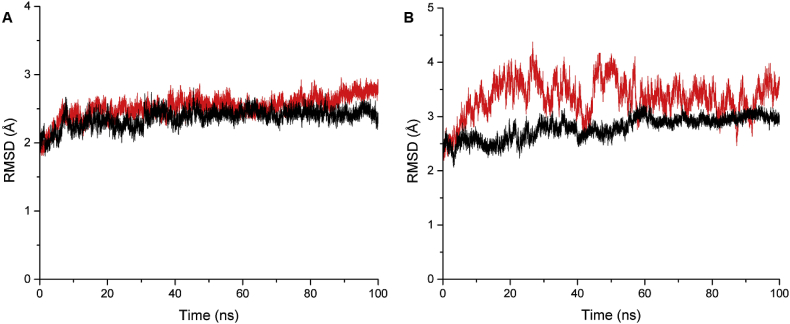
MD simulations were performed for 100 ns using the docked conformation of PDE4B- rolipram, PDE4B-HTS04529, PDE4D-rolipram and PDE4D-HTS04529 complexes to investigate the atomic details of molecular interactions. The RMSD was calculated for the backbone atoms of PDE4B and PDE4D complexes relative to the docked structures to evaluate the dynamic stabilisation in the time scale of the simulation period. From the RMSD plot shown in Figure 9A, it can be seen that the simulation reached convergence at 15 ns and the PDE4B complexes attained a stable RMSD value of 2–2.75 Å. This means no significant conformational changes took place in the protein structure upon binding of the ligands. While for PDE4D complexes, the PDE4D-HTS04529 presented greater instability than the PDE4D-rolipram complex (Figure 9B). The PDE4D-rolipram complex reached equilibration after 50ns and did not show any variation higher than 3 Å. In contrast, the PDE4D-HTS04529 complex has fluctuated during the entire simulation time between 2.5 and 4.25 Å; which means the binding of the ligand affects the stability of the receptor.
Figure 9.
RMSD of the backbone atoms of PDE4B and PDE4D complexes. A) PDE4B-HTS04529 (red line) and PDE4B-Rolipram (black line). B) PDE4D-HTS04529 (red line) and PDE4D-Rolipram (black line).
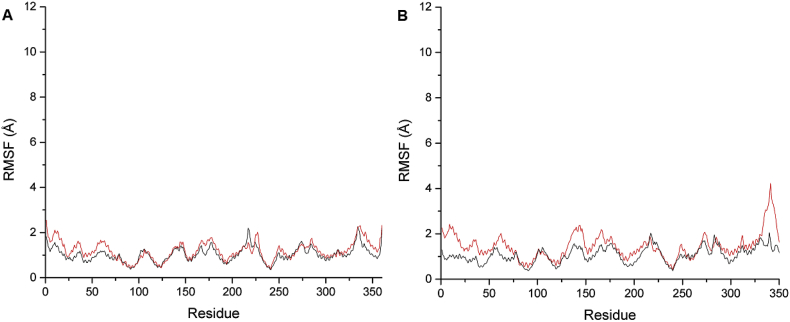
The RMSF analysis was performed to assess the mobility of the residues upon binding of the ligands. It is observed from the plots in Figure 10, the residues in PDE4B complexes have lower mobility in comparison to the residues in PDE4D complexes. The residues in the catalytic domain of PDE4B (region TYR82-ILE299) showed RMSF value in the range of 0.34–1.69 Å indicating the low fluctuations of these residues and supporting the docking findings that binding of the ligand stabilises the protein structure. As expected, the backbone residues with high fluctuations (above 2Å) were those in the flexible region SER217-THR227. Furthermore, the residue positioned between PRO334-ASP342 showed fluctuation above 2Å as well, but the highest fluctuation was observed in residue SER1, which showed RMSF value of 2.5Å. Whereas for PDE4D complexes, the residues of PDE4D-HTS04529 complex displayed the highest mobility. The RMSF values of the region GLN334-ASP342 were above 2.5 Å with the highest RMSF value of 4.2Å showed by the residue ASP341. In contrast, the binding of rolipram to the PDE4D reduced the flexibility of the protein's residues where only two residues displayed RMSF values higher then 2Å (SER217, RMSF 2.02Å and PRO340, RMSF 2.05Å). Therefore, the binding of rolipram made the complex more stable.
Figure 10.
RMSF of the amino acid residues of PDE4B and PDE4D complexes. A) PDE4B-HTS04529 (red line) and PDE4B-Rolipram (black line). B) PDE4D-HTS04529 (red line) and PDE4D-Rolipram (black line).
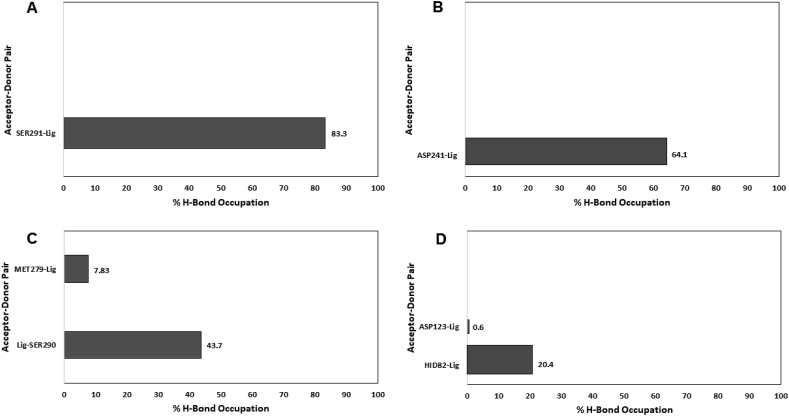
The number of intermolecular hydrogen bonds, as well as the hydrogen bonds occupancies, were calculated to determine the strength of binding of rolipram and HTS04529 to the target receptors. Both ligands formed one hydrogen bond with PDE4B and two hydrogen bonds with PDE4D (Figure 11). These results were consistent with the hydrogen bond interactions obtained by molecular docking.
Figure 11.
Hydrogen bonds occupancies between both PDEs (PDE4B and PDE4D) and the ligands. A) PDE4B-rolipram. B) PDE4B-HTS04529. C) PDE4D-rolipram. D) PDE4D-HTS04529.
For PDE4B complexes, the hydrogen bond formed between SER291 and rolipram and ASP241 and HTS04529 with occupancies of 83.3% and 64.1%, respectively. The amino acids acted as hydrogen bond acceptors, whereas the ligands acted as hydrogen bond donors. The hydrogen bonds were stable; they formed at the beginning of the simulations and remained stable during the entire simulation time.
In contrast, the hydrogen bonds between PDE4D and the two ligands were of intermediate and weak strengths, respectively. The first hydrogen bond was formed between SER290 and rolipram and HID82 and HTS04529 with occupancies of 43.7% and 20.4%, respectively. The HID82 acted as hydrogen bond acceptor while SER290 acted as hydrogen bond donor. These hydrogen bonds were of intermediate strength where they formed at the beginning of the simulations and were disrupted and formed again during the simulation process. Weak hydrogen bonds were formed between MET279 and rolipram and ASP123 and HTS04529 with occupancies of 7.83% and 0.6%, respectively. The two residues acted as hydrogen bond acceptors where they accepted the hydrogen atoms from the ligands. The two hydrogen bonds were not seen in the original complex obtained by docking. However, due to the weakness and instability of these hydrogen bonds, they formed for a short time and broke readily.
The analysis of MD trajectories showed a similar binding interaction between HTS04529 and PDE4B as predicted by the docking study. In addition to the hydrogen bonds, the hydrophobic interactions with the stable region of the catalytic domain TYR82-ILE299 and the residues LYS354 and PHE355 of the CR3 was preserved in more 80% of the trajectories (Supplementary Material-Figure S1). While the region HID199-ARG229 was more flexible as no interaction was observed in that region. The metal coordination bond with Zn+2 and the Pi-Pi stacking interaction with TYR82 was preserved in 100% of the MD trajectory. Whereas, the Pi-Pi stacking interaction with PHE295 was preserved in 75% of the MD trajectories. These results support the stability of the complex as no dramatic changes observed in the mode of the binding interactions.
On the other hand, the analysis of MD trajectories of HTS04529-PDE4D complex indicated the presence of several Pi-Pi stacking and hydrophobic interactions between the inhibitor and the receptor with high to low frequencies. Among these interactions, the hydrophobic interactions with CYS280, LEU241 and the three main residues in the P-clamp (ILE258, PHE262 and PHE294) were preserved in 50–100% of the MD trajectories. The interactions with MET195 and ASP123 were preserved in only 20% of the trajectories. While; the Pi-Pi stacking interactions were observed with different residues during the simulations with modest to low frequencies (Supplementary Material-Figure S2). These results indicate the instability of the ligand within the active site of the receptor as the binding interactions were continuously changing.
The binding free energy calculations were performed using MMPBSA.py method with the default settings. The values of the total binding free energies for the last 20ns of simulations and the energy contribution by each component are presented in Table 5. The results have shown that HTS04529 has a higher affinity for PDE4B (lower binding energy) than PDE4D, which is consistent with the molecular docking results. While, rolipram displayed much higher affinity for PDE4D over PDE4B. In the case of HTS04529 binding to the two targets, the electrostatic interactions had a higher contribution to the total binding energy than the van der Waals interactions. Whereas, the van der Waals interactions predominate in the case of rolipram binding to PDE4B and PDE4D. The non-polar solvation energy was similar in the four complexes and favourable for binding but, the polar solvation energy was the same in PDE4B-HTS04529 and PDE4D-HTS04529 complexes and different in PDE4B-rolipram and PDE4D-rolipram complexes. The polar solvation energy contributed more in the total binding energy of PDE4B-rolipram complex than PDE4D-rolipram complex. However, the polar solvation energy is unfavourable for binding; thus, it reduced the affinity of rolipram for PDE4B.
Table 5.
Total binding free energy of PDEB and PDE4D complexes and the energy contribution by each component during the last 20ns of simulations.
| Binding energy components | PDE4B-rolipram | PDE4D-rolipram | PDE4B-HTS04529 | PDE4D-HTS04529 |
|---|---|---|---|---|
| VDW | -40.0232 ± 0.0654 | -49.7979 ± 0.0757 | -48.0598 ± 0.1033 | -36.7434 ± 0.0905 |
| EEL | -11.1343 ± 0.0769 | -5.9711 ± 0.1533 | -51.1525 ± 0.1240 | -49.2828 ± 0.1440 |
| Non-polar solvation energy | -3.6203 ± 0.0034 | -3.9909 ± 0.0029 | -4.6528 ± 0.0027 | -3.5533 ± 0.0034 |
| Polar solvation energy | 29.8817 ± 0.0539 | 24.2153 ± 0.0029 | 76.6634 ± 0.0892 | 76.0847 ± 0.0965 |
| ΔG Total | -24.8962 ± 0.0647 | -35.5447 ± 0.0816 | -27.2017 ± 0.0876 | -13.4948 ± 0.1083 |
VDW: van der Waals interactions, EEL: electrostatic interactions, ΔG Total: total binding free energy.
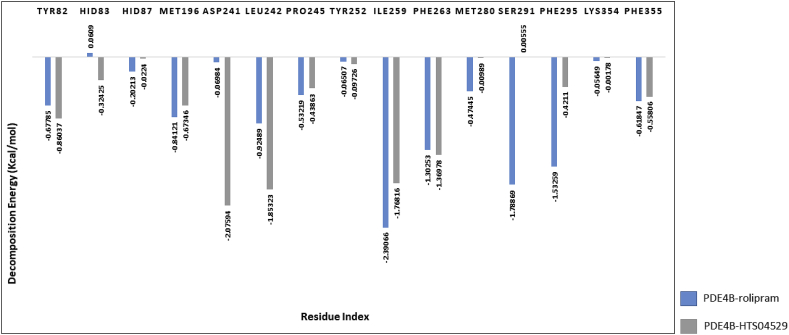
In order to investigate the role of key residues in the binding interface and assess the energetic contribution of each residue involved in the hydrogen bond interactions and other non-bonded interactions with rolipram and HTS04529, the per-residue decomposition energy was calculated. In PDE4B-HTS04529 complex, ASP241 was the highest contributor to the binding energy (decomposition energy -2.07594 kcal/mol) along with LEU242, ILE259 and PHE263 with favourable energy < -1 kcal/mol (Figure 12). While in PDE4B-rolipram complex, ILE259 was the highest contributor to the binding energy with decomposition energy -2.39066 kcal/mol, in addition to the SER291 and PHE295 with decomposition energy < -1 kcal/mol.
Figure 12.
Decomposition energy of the key residues of PDE4B complexes.
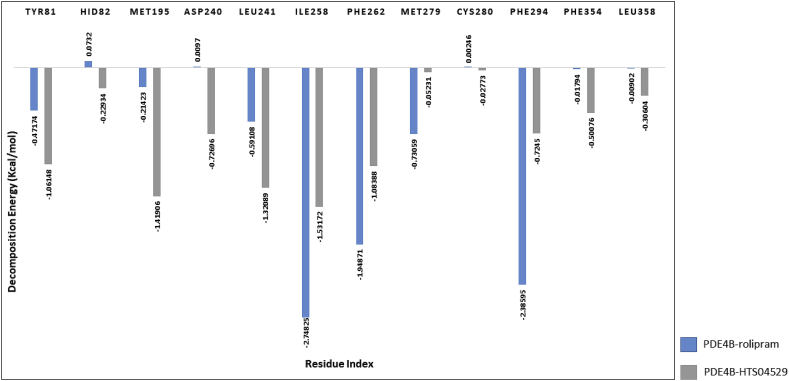
With regard to PDE4D complexes, two residues contributed significantly in the total binding energy of the two complexes. These residues are ILE258 and PHE262, with decomposition energy < -1 kcal/mol (Figure 13). The residues TYR81, MET195 and LEU241, were favourable for binding in PDE4D-HTS04529 complex where the decomposition energies were -1.06148, -1.41906 and -1.32089, respectively. While PHE294 was favourable for binding in PDE4D-rolipram complex in which the decomposition energy was -2.38595. In contrast, the positive energy contribution by HID83 in PDEB-rolipram complex and HID82 in PDE4D-rolipram complex were unfavourable for binding; thus, they could play an opposite role in the two complexes and reduce the ligand stability.
Figure 13.
Decomposition energy of the key residues of PDE4D complexes.
4. Conclusion
A hierarchical virtual screening approach based on the pharmacophore modelling and molecular docking has been carried out to identify new selective PDE4B inhibitor and to provide insights into the structural basis of the mechanism of the identified inhibitor. As a consequence, one compound, HTS04529, exhibited low binding energy and selectivity for PDE4B over PDE4D. The binding mode of HTS04529 within the active site of PDE4B has achieved all the requirements to inhibit the entry of cAMP; thus, block its hydrolysis. Furthermore, the MD simulations have been performed to confirm the binding affinity and the stability of the HTS04529 within the active site of PDE4B. The low RMSD and RMSF values showed that the PDE4B-HTS04529 complex attained the equilibration at an early stage of the simulations, and the binding of the ligand did not alter the enzyme structure significantly. The predicted interactions by the molecular docking (hydrogen bond, Pi-Pi stacking and the hydrophobic interactions as well as the metal coordinate with Zn+2) were preserved in more than 90% of the MD trajectories. In addition, the inhibitor exhibited the most promising binding free energy with PDE4B. These results were consistent with the docking findings. Accordingly, HTS04529 can be considered as an interesting lead compound for the design of highly selective PDE4B inhibitors.
Declarations
Author contribution statement
A. Gaurav:Conceived and designed the experiments; Analyzed and interpreted the data.
M. Al Nema: performed the experiments; wrote the paper.
V. Sanghiran Lee: Analyzed and interpreted the data; contributed reagents, materials, analysis tools or data.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Authors would like to acknowledge the computational facilities from Data Intensive Computer Center (DICC), https://www.dicc.um.edu.my/, University of Malaya.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- Accelrys D.S. Accelrys, Inc.; San Diego, CA, USA: 2009. Version 2.5. 5. [Google Scholar]
- Amir-Hassan A., Lee V.S., Baharuddin A., Othman S., Xu Y., Huang M., Yusof R., Rahman N.A., Othman R. Conformational and energy evaluations of novel peptides binding to dengue virus envelope protein. J. Mol. Graph. Model. 2017;74:273–287. doi: 10.1016/j.jmgm.2017.03.010. [DOI] [PubMed] [Google Scholar]
- Azam M.A., Tripuraneni N.S. Selective phosphodiesterase 4B inhibitors: a review. Sci. Pharm. 2014;82:453–481. doi: 10.3797/scipharm.1404-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumer W., Hoppmann J., Rundfeldt C., Kietzmann M. Highly selective phosphodiesterase 4 inhibitors for the treatment of allergic skin diseases and psoriasis. Inflamm. Allergy – Drug Targets. 2007;6:17–26. doi: 10.2174/187152807780077318. [DOI] [PubMed] [Google Scholar]
- BIOVIA D.S. Dassault Systèmes; San Diego, CA, USA: 2017. Discovery Studio Modeling Environment. [Google Scholar]
- Bolger G.B., Dunlop A.J., Meng D., Day J.P., Klussmann E., Baillie G.S., Adams D.R., Houslay M.D. Dimerization of cAMP phosphodiesterase-4 (PDE4) in living cells requires interfaces located in both the UCR1 and catalytic unit domains. Cell. Signal. 2015;27:756–769. doi: 10.1016/j.cellsig.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin A.B., Magnusson O.T., Singh J., Witte P., Staker B.L., Bjornsson J.M., Thorsteinsdottir M., Hrafnsdottir S., Hagen T., Kiselyov A.S., Stewart L.J., Gurney M.E. Design of phosphodiesterase 4D (PDE4D) allosteric modulators for enhancing cognition with improved safety. Nat. Biotechnol. 2010;28:63–70. doi: 10.1038/nbt.1598. [DOI] [PubMed] [Google Scholar]
- Card G.L., England B.P., Suzuki Y., Fong D., Powell B., Lee B., Luu C., Tabrizizad M., Gillette S., Ibrahim P.N., Artis D.R., Bollag G., Milburn M.V., Kim S.H., Schlessinger J., Zhang K.Y. Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure. 2004;12:2233–2247. doi: 10.1016/j.str.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Case D.A., Babin V., Berryman J.T., Betz R.M., Cai Q., Cerutti D.S., Cheatham T.E., III, Darden T.A., Duke R.E., Gohlke H., Goetz A.W., Gusarov S., Homeyer N., Janowski P., Kaus J., Kolossváry I., Kovalenko A., Lee T.S., LeGrand S., Luchko T., Luo R., Madej B., Merz K.M., Paesani F., Roe D.R., Roitberg A., Sagui C., Salomon-Ferrer R., Seabra G., Simmerling C.L., Smith W., Swails J., Walker R.C., Wang J., Wolf R.M., Wu X., Kollman P.A. AMBER 14. University of California; San Francisco: 2014. [Google Scholar]
- Conti M., Richter W., Mehats C., Livera G., Park J.Y., Jin C. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J. Biol. Chem. 2003;278:5493–5496. doi: 10.1074/jbc.R200029200. [DOI] [PubMed] [Google Scholar]
- Darden T., York D., Pedersen L. Particle mesh Ewald: an N⋅ log (N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- Erlina L., Yanuar A. Pharmacophore-based virtual screening from Indonesian herbal database to finding new inhibitor of HDAC4 and HDAC7. J. Young Pharm. 2018;10:7. [Google Scholar]
- Fan Chung K. Phosphodiesterase inhibitors in airways disease. Eur. J. Pharmacol. 2006;533:110–117. doi: 10.1016/j.ejphar.2005.12.059. [DOI] [PubMed] [Google Scholar]
- Feng X., Wang H., Ye M., Xu X.-T., Xu Y., Yang W., Zhang H.-T., Song G., Ke H. Identification of a PDE4-specific pocket for the design of selective inhibitors. Biochemistry. 2018;57:4518–4525. doi: 10.1021/acs.biochem.8b00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox D., 3rd, Burgin A.B., Gurney M.E. Structural basis for the design of selective phosphodiesterase 4B inhibitors. Cell. Signal. 2014;26:657–663. doi: 10.1016/j.cellsig.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T., Shiina A., Murata T., Tomii M., Yamazaki T., Yoshida K.-i., Yoshino T., Suzuki O., Sogawa Y., Mizukami K. Identification of the 5, 5-dioxo-7, 8-dihydro-6H-thiopyrano [3, 2-d] pyrimidine derivatives as highly selective PDE4B inhibitors. Bioorg. Med. Chem. Lett. 2014;24:893–899. doi: 10.1016/j.bmcl.2013.12.076. [DOI] [PubMed] [Google Scholar]
- Goto T., Shiina A., Yoshino T., Mizukami K., Hirahara K., Suzuki O., Sogawa Y., Takahashi T., Mikkaichi T., Nakao N., Takahashi M., Hasegawa M., Sasaki S. Identification of the fused bicyclic 4-amino-2-phenylpyrimidine derivatives as novel and potent PDE4 inhibitors. Bioorg. Med. Chem. Lett. 2013;23:3325–3328. doi: 10.1016/j.bmcl.2013.03.104. [DOI] [PubMed] [Google Scholar]
- Hagen T.J., Mo X., Burgin A.B., Fox D., 3rd, Zhang Z., Gurney M.E. Discovery of triazines as selective PDE4B versus PDE4D inhibitors. Bioorg. Med. Chem. Lett. 2014;24:4031–4034. doi: 10.1016/j.bmcl.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrach M.F., Drossel B. Structure and dynamics of TIP3P, TIP4P, and TIP5P water near smooth and atomistic walls of different hydroaffinity. J. Chem. Phys. 2014;140:174501. doi: 10.1063/1.4872239. [DOI] [PubMed] [Google Scholar]
- Houslay M.D. Underpinning compartmentalised cAMP signalling through targeted cAMP breakdown. Trends Biochem. Sci. 2010;35:91–100. doi: 10.1016/j.tibs.2009.09.007. [DOI] [PubMed] [Google Scholar]
- Houslay M.D., Schafer P., Zhang K.Y. Keynote review: phosphodiesterase-4 as a therapeutic target. Drug Discov. Today. 2005;10:1503–1519. doi: 10.1016/S1359-6446(05)03622-6. [DOI] [PubMed] [Google Scholar]
- Huai Q., Colicelli J., Ke H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- Kanes S.J., Tokarczyk J., Siegel S.J., Bilker W., Abel T., Kelly M.P. Rolipram: a specific phosphodiesterase 4 inhibitor with potential antipsychotic activity. Neuroscience. 2007;144:239–246. doi: 10.1016/j.neuroscience.2006.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klussmann E. Protein-protein interactions of PDE4 family members - functions, interactions and therapeutic value. Cell. Signal. 2016;28:713–718. doi: 10.1016/j.cellsig.2015.10.005. [DOI] [PubMed] [Google Scholar]
- Kranz M., Wall M., Evans B., Miah A., Ballantine S., Delves C., Dombroski B., Gross J., Schneck J., Villa J.P., Neu M., Somers D.O. Identification of PDE4B over 4D subtype-selective inhibitors revealing an unprecedented binding mode. Bioorg. Med. Chem. 2009;17:5336–5341. doi: 10.1016/j.bmc.2009.03.061. [DOI] [PubMed] [Google Scholar]
- Kuhn B., Kollman P.A. Binding of a diverse set of ligands to avidin and streptavidin: an accurate quantitative prediction of their relative affinities by a combination of molecular mechanics and continuum solvent models. J. Med. Chem. 2000;43:3786–3791. doi: 10.1021/jm000241h. [DOI] [PubMed] [Google Scholar]
- Lamontagne S., Meadows E., Luk P., Normandin D., Muise E., Boulet L., Pon D.J., Robichaud A., Robertson G.S., Metters K.M., Nantel F. Localization of phosphodiesterase-4 isoforms in the medulla and nodose ganglion of the squirrel monkey. Brain Res. 2001;920:84–96. doi: 10.1016/s0006-8993(01)03023-2. [DOI] [PubMed] [Google Scholar]
- Li H., Zuo J., Tang W. Phosphodiesterase-4 inhibitors for the treatment of inflammatory diseases. Front. Pharmacol. 2018;9:1048. doi: 10.3389/fphar.2018.01048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhou N., Liu W., Li J., Feng Y., Wang X., Wu C., Bao J. Discover natural compounds as potential phosphodiesterase-4B inhibitors via computational approaches. J. Biomol. Struct. Dyn. 2016;34:1101–1112. doi: 10.1080/07391102.2015.1070749. [DOI] [PubMed] [Google Scholar]
- Linse B., Linse P. Tuning the smooth particle mesh Ewald sum: application on ionic solutions and dipolar fluids. J. Chem. Phys. 2014;141:184114. doi: 10.1063/1.4901119. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A. Lead-and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004;1:337–341. doi: 10.1016/j.ddtec.2004.11.007. [DOI] [PubMed] [Google Scholar]
- Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- Martínez-Muñoz A., Bello M., Romero-Castro A., Rodríguez-Fonseca R.A., Rodrigues J., Sánchez-Espinosa V.A., Correa-Basurto J. Binding free energy calculations using MMPB/GBSA approaches for PAMAM-G4-drug complexes at neutral, basic and acid pH conditions. J. Mol. Graph. Model. 2017;76:330–341. doi: 10.1016/j.jmgm.2017.07.017. [DOI] [PubMed] [Google Scholar]
- Naganuma K., Omura A., Maekawara N., Saitoh M., Ohkawa N., Kubota T., Nagumo H., Kodama T., Takemura M., Ohtsuka Y. Discovery of selective PDE4B inhibitors. Bioorg. Med. Chem. Lett. 2009;19:3174–3176. doi: 10.1016/j.bmcl.2009.04.121. [DOI] [PubMed] [Google Scholar]
- O'Donnell J.M., Zhang H.T. Antidepressant effects of inhibitors of cAMP phosphodiesterase (PDE4) Trends Pharmacol. Sci. 2004;25:158–163. doi: 10.1016/j.tips.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Perez-Torres S., Miro X., Palacios J.M., Cortes R., Puigdomenech P., Mengod G. Phosphodiesterase type 4 isozymes expression in human brain examined by in situ hybridization histochemistry and[3H]rolipram binding autoradiography. Comparison with monkey and rat brain. J. Chem. Neuroanat. 2000;20:349–374. doi: 10.1016/s0891-0618(00)00097-1. [DOI] [PubMed] [Google Scholar]
- Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T.E., III, Laughton C.A., Orozco M. Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J.E. Inhaled phosphodiesterase 4 (PDE4) inhibitors for inflammatory respiratory diseases. Front. Pharmacol. 2020;11:259. doi: 10.3389/fphar.2020.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakkiah S., Lee K.W. Pharmacophore-based virtual screening and density functional theory approach to identifying novel butyrylcholinesterase inhibitors. Acta Pharmacol. Sin. 2012;33:964. doi: 10.1038/aps.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel T., Bryant S.D., Ibis G., Poli G., Langer T. 3D pharmacophore modeling techniques in computer-aided molecular design using LigandScout. Tutorials Chemoinform. 2017;281:279–309. [Google Scholar]
- Shubina V., Niinivehmas S., T Pentikainen O. Reliability of virtual screening methods in prediction of PDE4Binhibitor activity. Curr. Drug Discov. Technol. 2015;12:117–126. doi: 10.2174/1570163812666150702123018. [DOI] [PubMed] [Google Scholar]
- Souness J.E., Aldous D., Sargent C. Immunosuppressive and anti-inflammatory effects of cyclic AMP phosphodiesterase (PDE) type 4 inhibitors. Immunopharmacology. 2000;47:127–162. doi: 10.1016/s0162-3109(00)00185-5. [DOI] [PubMed] [Google Scholar]
- Spina D. PDE4 inhibitors: current status. Br. J. Pharmacol. 2008;155:308–315. doi: 10.1038/bjp.2008.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripuraneni N.S., Azam M.A. Pharmacophore modeling, 3D-QSAR and docking study of 2-phenylpyrimidine analogues as selective PDE4B inhibitors. J. Theor. Biol. 2016;394:117–126. doi: 10.1016/j.jtbi.2016.01.007. [DOI] [PubMed] [Google Scholar]
- Vuorinen A., Engeli R., Meyer A., Bachmann F., Griesser U.J., Schuster D., Odermatt A. Ligand-based pharmacophore modeling and virtual screening for the discovery of novel 17β-hydroxysteroid dehydrogenase 2 inhibitors. J. Med. Chem. 2014;57:5995–6007. doi: 10.1021/jm5004914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Peng M.-S., Chen Y., Geng J., Robinson H., Houslay M.D., Cai J., Ke H. Structures of the four subfamilies of phosphodiesterase-4 provide insight into the selectivity of their inhibitors. Biochem. J. 2007;408:193–201. doi: 10.1042/BJ20070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Wu P., Ohleth K.M., Egan R.W., Billah M.M. Phosphodiesterase 4B2 is the predominant phosphodiesterase species and undergoes differential regulation of gene expression in human monocytes and neutrophils. Mol. Pharmacol. 1999;56:170–174. doi: 10.1124/mol.56.1.170. [DOI] [PubMed] [Google Scholar]
- Wolber G., Langer T. LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 2005;45:160–169. doi: 10.1021/ci049885e. [DOI] [PubMed] [Google Scholar]
- Xing M., Akowuah G.A., Gautam V., Gaurav A. Structure-based design of selective phosphodiesterase 4B inhibitors based on ginger phenolic compounds. J. Biomol. Struct. Dyn. 2017;35:2910–2924. doi: 10.1080/07391102.2016.1234417. [DOI] [PubMed] [Google Scholar]
- Xu R.X., Hassell A.M., Vanderwall D., Lambert M.H., Holmes W.D., Luther M.A., Rocque W.J., Milburn M.V., Zhao Y., Ke H. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- Yuasa K., Kanoh Y., Okumura K., Omori K. Genomic organization of the human phosphodiesterase PDE11A gene. Evolutionary relatedness with other PDEs containing GAF domains. Eur. J. Biochem. 2001;268:168–178. doi: 10.1046/j.1432-1327.2001.01866.x. [DOI] [PubMed] [Google Scholar]
- Zhu J., Lv Y., Han X., Xu D., Han W. Understanding the differences of the ligand binding/unbinding pathways between phosphorylated and non-phosphorylated ARH1 using molecular dynamics simulations. Sci. Rep. 2017;7:12439. doi: 10.1038/s41598-017-12031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.