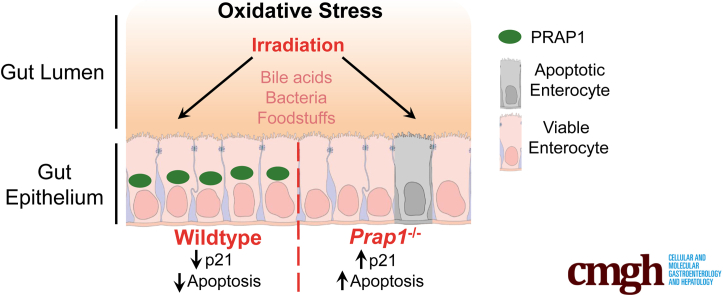
Abstract
Background & Aims
The intestinal epithelium must be resilient to physiochemical stress to uphold the physiological barrier separating the systemic compartment from the microbial and antigenic components of the gut lumen. Identifying proteins that mediate protection and enhancing their expression is therefore a clear approach to promote intestinal health. We previously reported that oral ingestion of the probiotic Lactobacillus rhamnosus GG not only induced the expression of several recognized cytoprotective factors in the murine colon, but also many genes with no previously described function, including the gene encoding proline-rich acidic protein 1 (PRAP1). PRAP1 is a highly expressed protein in the epithelium of the gastrointestinal tract and we sought to define its function in this tissue.
Methods
Purified preparations of recombinant PRAP1 were analyzed biochemically and PRAP1 antisera were used to visualize localization in tissues. Prap1-/- mice were characterized at baseline and challenged with total body irradiation, then enteroids were generated to recapitulate the irradiation challenge ex vivo.
Results
PRAP1 is a 17-kilodalton intrinsically disordered protein with no recognizable sequence homology. PRAP1 expression levels were high in the epithelia of the small intestine. Although Prap1-/- mice presented only mild phenotypes at baseline, they were highly susceptible to intestinal injury upon challenge. After irradiation, the Prap1-/- mice showed accelerated death with a significant increase in apoptosis and p21 expression in the small intestinal epithelium.
Conclusions
PRAP1 is an intrinsically disordered protein highly expressed by the gastrointestinal epithelium and functions at exposed surfaces to protect the barrier from oxidative insult.
Keywords: Oxidative Stress, Intrinsically Disordered Proteins, p21, Small Intestine
Abbreviations used in this paper: BSA, bovine serum albumin; CD, circular dichroism; FITC, fluorescein isothiocyanate; IDP, intrinsically disordered protein; IF, immunofluorescence; IL, interleukin; MTT, 3-(4,5-dimethylthiazol-2-yl)-diphenyltetrazolium bromide; PBS, phosphate-buffered saline; pCMV, cytomegalovirus expression vector; PCR, polymerase chain reaction; PRAP1, proline-rich acidic protein 1; 6xHis, hexahistadine tag; TBI, total body irradiation; TBST, Tris-buffered saline with 0.1% Tween
Graphical abstract
Summary.
Although highly expressed in the gastrointestinal tract, the function of proline-rich acidic protein 1 (PRAP1) is unknown. By using Prap1 null mice, we show that PRAP1 protects the gastrointestinal epithelium from irradiation-induced apoptosis and signifcantly limits p21 expression.
Induction of cellular-protective pathways and the associated effector molecules is particularly important in tissues that frequently are exposed to environmental xenobiotics and oxidative stress, such as the gastrointestinal epithelium.1, 2, 3 Such enterocytes are in intimate contact with the microbiota and their products as well as digestive components and ingested foodstuffs, requiring the epithelial cells perform their absorptive function while tolerating and responding to these exogenous stressors. When cells encounter exogenous insult that results in heightened oxidative stress and DNA damage, several pathways downstream of p53 activation determine cell fate and whether the cells will repair the DNA damage and continue in the cell cycle. In the case of overwhelming damage, they enact programmed cell death to safely eliminate injured cells.4,5 Improper response to exogenous insult can lead to compromised barrier integrity, allowing luminal components to traverse the epithelial layer to subepithelial compartments where they induce heightened localized or systemic inflammation that can result in a variety of pathologic states.6, 7, 8 Therefore, identifying proteins that promote a proper epithelial response to exogenous insult and enhancing their expression is a subject of intense investigative focus.
It increasingly is appreciated that the normal resident gut microbiota play a role in eliciting cytoprotection.9 Corroborating this notion are studies showing that the intestines of germ-free mice that lack a microbiota are more susceptible to exogenous insult.2,10 Furthermore, ingestion of putatively beneficial bacteria, also known as probiotics, including Lactobacillus rhamnosus GG, are known to elicit cytoprotection in the gut.11 To identify potential novel cytoprotective genes induced by Lactobacillus rhamnosus GG in the murine intestine, transcriptomic analysis was performed on the colon 4 hours after oral gavage. Among the highest induced transcripts was a transcript coding for proline-rich acidic protein 1 (PRAP1).2 PRAP1 is a 17-kilodalton secreted protein with no recognizable sequence homology.12 Although Prap1 originally was discovered to be highly expressed in the pregnant mouse uterus and later in the murine small intestine,13,14 its function in the host remains largely unexplored. Studies using cultured transformed cell lines proposed that PRAP1 functions downstream of p53 signaling after DNA damage, with the disruption of PRAP1 function in this system rendering neoplastic cells more susceptible to chemotherapeutic agents.15 Other studies described injection of pregnant mouse uteri with PRAP1 antisera and detected dysregulation in the expression of multiple proteins involved in apoptosis and inflammation, ultimately affecting embryo implantation.16 Although PRAP1 has been implicated in cell survival, apoptosis, and response to injury, the in vivo function of PRAP1 in the gastrointestinal tract remains unknown.
We first aimed to characterize the structure and expression pattern of PRAP1 in vivo. We generated PRAP1 recombinant protein and PRAP1 antisera and found that PRAP1 is an intrinsically disordered protein highly expressed in the epithelia of the gastrointestinal tract in both mice and human beings. To determine the function of PRAP1 we challenged germ line Prap1-/- mice with irradiation to show that PRAP1 protected the enterocytes from irradiation-induced apoptosis and that PRAP1 expression prolonged the survival of the mice after irradiation. Finally, manipulation of PRAP1 expression in both enteroids and an epithelial cell line showed that PRAP1 significantly decreased epithelial expression of p21 and improved cell viability after irradiation. Together, these data show that PRAP1 functions in vivo to protect the gastrointestinal epithelium from oxidative insult.
Results
PRAP1 Is an Intrinsically Disordered Protein Conserved in Placental Mammals
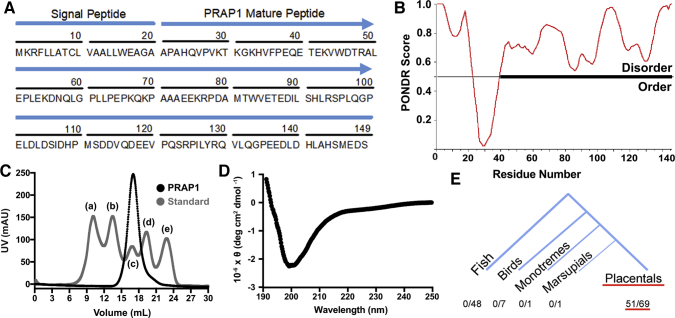
PRAP1 is a 17-kilodalton secreted protein composed of 149 amino acids. The first 20 amino acids on the N terminus serve as a signal peptide while the remaining amino acids form the secreted portion of the protein (Figure 1A). The amino acid sequence of the secreted protein does not have any detectable sequence homology with other proteins using the National Institutes of Health Basic Local Alignment Search Tool analysis,12 and analysis of PRAP1 using Predictors of Natural Disordered Regions17 predicts that PRAP1 has a high disordered score throughout the secreted portion of the protein and thus is predicted to be predominantly disordered (Figure 1B). Analysis of recombinant PRAP1 on size exclusion chromatography showed that PRAP1 eluted at a considerably larger functional size than its predicted molecular weight (16,570 daltons) (Figure 1C). This suggests that PRAP1 is either multimeric or has an extended conformation that is a fundamental characteristic of intrinsically disordered proteins. We next used circular dichroism (CD) to identify secondary structures within PRAP1, which showed a strong negative band near 200 nm, yet no significant signals around 208, 215, or 222 nm, indicating the absence of α helix or β sheet in its secondary structure (Figure 1D).18 Together, these data show that PRAP1 is intrinsically disordered, lacking any defined 3-dimensional structure. To identify species that express PRAP1 orthologs, the human PRAP1 amino acid sequence was queried in the Comparative Genomics feature of Ensembl (Cambridgeshire, UK).19 Of the 126 species considered, PRAP1 orthologs were present in 51 species, all of which were placental mammals (Figure 1E). These data confirm that PRAP1 is indeed an intrinsically disordered protein, has no homology to other more functionally defined proteins, and evolved relatively late in evolution, at the emergence of placental mammals.
Figure 1.
PRAP1 is an intrinsically disordered protein conserved in placental mammals. (A) Amino acid sequence of human PRAP1 with signal peptide and secreted portion of the protein labeled. (B) Analysis of the PRAP1 amino acid sequence using the Predictor of Natural Disordered Regions (PONDR) software. The predicted ordered and disordered regions are plotted for each residue. (C) Size exclusion chromatogram of purified recombinant PRAP1 protein compared with a molecular weight standard: (a) thyroglobulin (670,000 daltons), (b) γ-globulin (158,000 daltons), (c) ovalbumin (44,000 daltons), (d) myoglobin (17,000 daltons), and (e) vitamin B12 (1350 daltons). (D) Circular dichroism spectra of recombinant PRAP1. (E) Analysis of the human PRAP1 amino acid sequence using the comparative genomics feature of Ensembl software. The number of PRAP1 orthologs identified in each taxonomic clade are indicated.
PRAP1 Is Highly Expressed in the Epithelium of the Gastrointestinal Tract in Mice and Human Beings
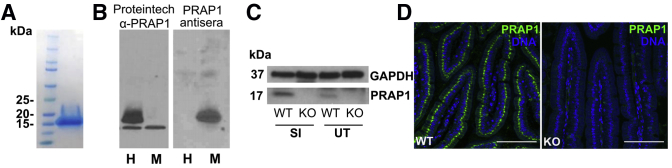
To fully define the spatial and temporal expression of PRAP1, we generated antisera specific for mouse PRAP1. We first produced and purified recombinant mouse PRAP1 protein with a hexahistadine tag on the N terminus (6xHis-PRAP1) (Figure 2A). By using this recombinant 6xHis-PRAP1 protein, we generated antisera in rabbits. To validate specificity, we used this PRAP1 antisera to blot for PRAP1 in lysate from cells that overexpressed either human or mouse PRAP1. Although the commercially available PRAP1 polyclonal antibody (Proteintech, Rosemont, IL) produced a strong band at 20 kilodaltons for the cell lysate overexpressing human PRAP1, only our PRAP1 antisera was able to detect the overexpression of mouse PRAP1 (Figure 2B). We further validated our PRAP1 antisera by blotting whole tissue lysate collected from the small intestine and uterus of wild-type and Prap1-/- mice. The antisera detected a band at 17 kilodaltons in the wild-type tissue but not the Prap1-/- tissue (Figure 2C). Furthermore, immunofluorescence (IF) staining using our PRAP1 antisera in the small intestine of wild-type and Prap1-/- mice showed high specificity, with no detectable signal in the Prap1-/- tissue (Figure 2D). In summary, using recombinant mouse PRAP1 protein, we generated PRAP1 antisera suitable for the detection of PRAP1 in murine tissue.
Figure 2.
Generation and validation of PRAP1 recombinant protein, PRAP1 antisera, and Prap1-/-mice. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis with Coomassie staining of recombinant 6xHis-PRAP1 expressed in E coli and purified by a Ni-NTA affinity chromatography column followed by a size exclusion column. (B) A human colonic epithelial cell line (SK-CO15) was transfected to overexpress human (H) and mouse (M) PRAP1. Cell lysates were blotted with a commercially available antibody specific for human PRAP1 (Proteintech, first blot) or with PRAP1 rabbit antisera generated using 6xHis-PRAP1 (second blot). (C) Western blot of small intestine and uterine whole tissue from wild-type and Prap1-/- mice, blotted with PRAP1 antisera introduced in panel B. (D) Immunofluorescence staining of wild-type and Prap1-/- duodenum using PRAP1 antisera. Whole-body knockout mice were procured from MMRRC-UC Davis and backcrossed to obtain a fully congenic C57BL/6 background. Scale bar: 100 μm. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KO, knockout, SI, small intestine; WT, wild-type; UT, uterine tissue.
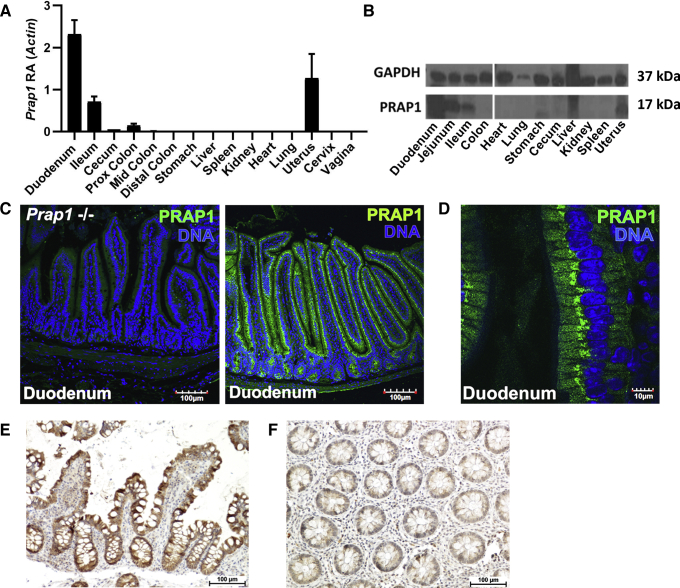
Immunoblot analysis and measurement of transcript levels in the mouse showed that Prap1 is highly abundant in the small intestine, with the relative abundance being 2-fold when compared with β-actin (Figure 3A and B). Prap1 expression was highest in the proximal small intestine with expression diminishing along the caudal axis, becoming nearly undetectable in the distal large intestine. By using PRAP1 antisera, we detected abundant PRAP1 protein expressed exclusively in the gut epithelium (Figure 3C), with PRAP1 protein localization strongest in the perinuclear compartment of the cell (Figure 3D). To determine whether the PRAP1 expression pattern is similar in both mice and human beings, immunohistochemistry was performed on diagnostic biopsy specimens of the human ileum (Figure 3E). Immunohistochemical staining showed high PRAP1 expression in the enterocytes of the human small intestine, consistent with the expression pattern we detected in mice. A human colonic biopsy specimen served as a negative control and showed dramatically lower levels of PRAP1 staining in this tissue (Figure 3F). Together, these data show that PRAP1 is highly expressed in the epithelium of the small intestine in both mice and human beings.
Figure 3.
PRAP1 is highly expressed by the epithelium of the gastrointestinal tract in mice and human beings. (A) Quantification of Prap1 transcript measured via quantitative PCR in the indicated tissues from 8-week-old wild-type C57BL/6 mice (n = 3 mice). (B) Western blot analysis for the detection of PRAP1 protein abundance in the indicated tissues dissected from 8-week-old wild-type C57BL/6 mice. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. Images are representative of 3 mice per tissue collected. (C) Immunofluorescence for the detection of PRAP1 (green) in the duodenum of 8-week-old wild-type C57BL/6 mice or Prap1-/- mice. Images are representative of the analysis of 5 mice per tissue collected. (D) Immunofluorescence staining of PRAP1 (green) in the duodenum of 8-week-old wild-type mice at 60× magnification. (E and F) Immunohistochemistry staining for the detection of PRAP1 (brown) in the human ileum (E) and colon (F). Image is representative of 3 subjects. Prox, proximal.
Prap1-/- Mice Have an Altered Gut Microbiota and Increased Inflammation
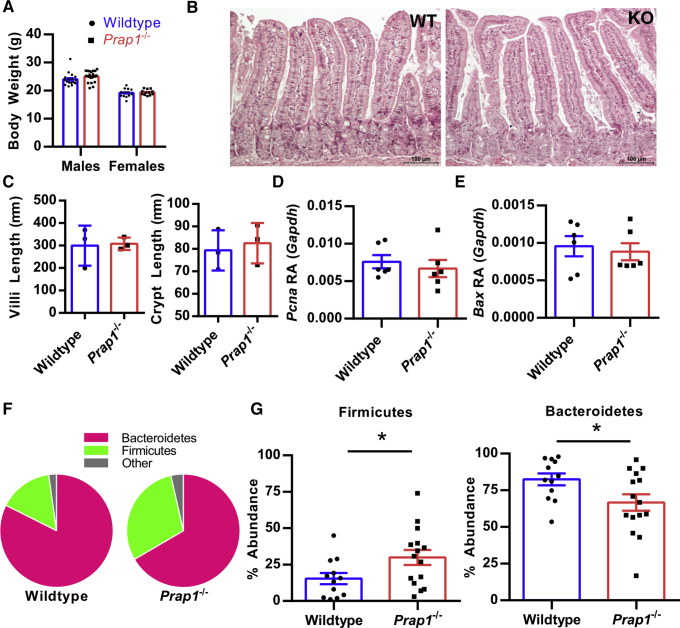
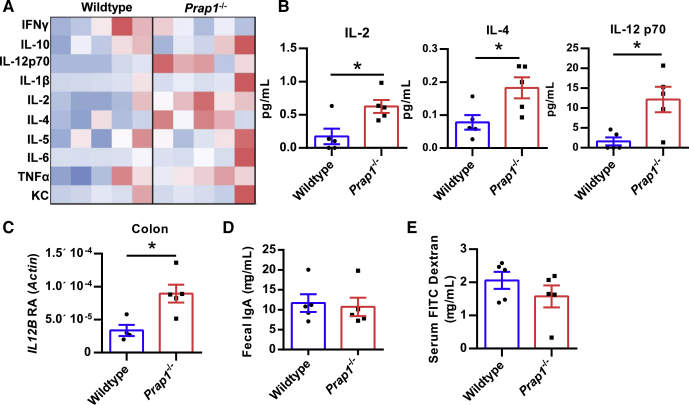
Prap1 germline null mice were maintained in a specific pathogen-free barrier vivarium and monitored for any symptoms of spontaneous disease. Prap1-/- mice were aged successfully to 20 weeks and no spontaneous disease was apparent. At 10 weeks of age, the Prap1-/- mice showed no difference in body weight, gut barrier architecture, or in the expression levels of proliferative or pro-apoptotic proteins in the small intestine (Figure 4A–E). To investigate whether the Prap1-/- mice have an altered gut microbiota, we sequenced the 16S ribosomal RNA gene from microbial DNA isolated from the small intestinal lumen of Prap1-/- and wild-type littermate controls. We detected a change in the ratio of Bacteroidetes:Firmicutes in which wild-type mice had an average ratio of 82:15 and Prap1-/- mice had a statistically significant shift in this ratio, with an average ratio of 66:29 (Figure 4F and G). Because Prap1-/- mice had a significant shift in the abundances of dominant bacterial phyla, we next sought to determine whether the Prap1-/- mice had any significant inflammatory differences. To detect systemic inflammation at baseline, we measured the levels of proinflammatory cytokines in the sera using a multiplex enzyme-linked immunosorbent assay that simultaneously detects the level of 10 proinflammatory cytokines. Sera collected from Prap1-/- mice had increased levels of proinflammatory cytokines (Figure 5A), with significantly higher interleukin (IL)2, IL4, and IL12p70 (Figure 5B). Along with increased systemic cytokines, the Prap1-/- mice also had an increased IL12β transcript in colonic tissue (Figure 5C). To determine the extent of local inflammation in the gut we measured the amount of secreted IgA and found there was no difference in the amount of secreted fecal IgA between wild-type and Prap1-/- littermates (Figure 5D). To determine whether the altered microbiota and increased cytokine expression could be attributed to a defective gut barrier, we orally gavaged adult wild-type and Prap1-/- littermates with fluorescein isothiocyanate (FITC) dextran and collected the sera 4 hours later. There was no difference in serum FITC dextran, indicating no significant alteration in gut permeability (Figure 5E). In summary, these data show that although Prap1-/- mice have an altered microbiota and increased cytokine expression, they do not show any serious mucosal defects when unchallenged.
Figure 4.
Prap1-/-mice have an altered microbiota in the small intestine. (A) The body weight of wild-type and Prap1-/- littermates at 10 weeks old. (B) H&E staining of wild-type and Prap1-/- small intestine. Images are representative of 3 mice per group. (C) Quantification of villi and crypt length in the small intestine of wild-type and Prap1-/- mice. Significance was determined using an unpaired t test (n = 3 mice). ∗P < .05. (D and E) Quantitative PCR analysis of Pcna (D) and Bax (E) expression in whole tissue from the small intestine of wild-type and Prap1-/- mice. Expression levels are relative to Gapdh. Significance was determined using an unpaired t test. ∗P < .05 (n = 6 mice). (F) Pie chart comparison of the average Bacteriodetes:Firmicutes phyla ratio in the small intestine of wild-type and Prap1-/- littermates measured via 16S ribosomal RNA sequencing (n ≥ 12 mice). (G) Relative abundance of Firmicutes and Bacteroidetes in the small intestine of wild-type and Prap1-/- littermates. Significance was determined using an unpaired t test (n ≥ 12 mice). ∗P < .05. All data are graphed as the means ± SEM. Bax RA, Bax relative abundance; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KO, knockout; Pcna RA, Pcna relative abundance; WT, wild-type.
Figure 5.
Prap1-/-mice have increased inflammation but no significant intestinal barrier defect. (A) A multiplex enzyme-linked immunosorbent assay (ELISA) was used for the detection of 10 proinflammatory cytokines in sera of 10-week-old wild-type and Prap1-/- littermates. Data are shown as a heat map, with red indicating a higher than average concentration. Each column represents 1 mouse. (B) Graphic representation of significantly different cytokine levels shown in panel A, including IL2, IL4, and IL12p70. Statistical significance was determined using an unpaired t test (n = 5 mice). ∗P < .05. (C) Quantification of IL12β transcript levels measured via quantitative PCR in the colon of 10-week-old wild-type and Prap1-/- mice relative to the abundance of β-actin. Statistical significance was determined using an unpaired t test (n = 5 mice). ∗P < .05. (D) Quantification of IgA levels in fecal pellets collected from 10-week-old wild-type and Prap1-/- mice, measured via ELISA. Statistical significance was determined using an unpaired t test (n = 5 mice). (E) Quantification of FITC dextran in the sera of unchallenged 10-week-old wild-type and Prap1-/- mice 4 hours after oral gavage with 4 kilodaltons FITC dextran. Statistical significance was determined using an unpaired t test (n = 5 mice). IFN, interferon; KC, keratinocyte chemoattractant; RA, relative abundance; TNF, tumor necrosis factor.
Prap1-/- Mice Are More Susceptible to Radiologic Challenge
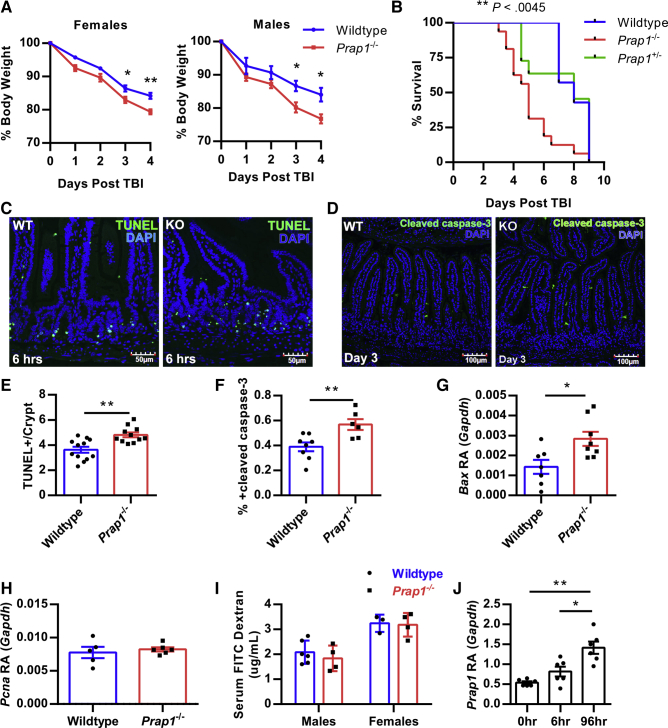
Total body irradiation (TBI) introduces a significant amount of cellular oxidative stress that results in DNA damage and rapid apoptosis of dividing stem cells in the gastrointestinal epithelium. To investigate whether Prap1-/- mice were more susceptible to this exogenous insult, we challenged mice with a lethal dose of TBI. Prap1-/- females and males lost significantly more body weight when compared with wild-type littermate controls starting at 3 days after irradiation (Figure 6A). Furthermore, Prap1-/- mice had significantly reduced viability after irradiation, with a median survival of 5 days, whereas the wild-type controls had a median survival of 8 days (Figure 6B). To compare early cellular injury in the small intestine of wild-type and Prap1-/- mice, apoptotic cells were labeled with terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL) staining 6 hours after irradiation (Figure 6C). At the tissue level, Prap1-/- mice had significantly more apoptotic cells per crypt compared with wild-type mice (Figure 6E). To compare injury at a later time point, wild-type and Prap1-/- mice were killed 72 hours after irradiation and the amount of apoptosis in the small intestine was measured via cleaved caspase-3 immunofluorescence. Again, the Prap1-/- mice had significantly higher levels of apoptosis in the epithelium of the small intestine compared with wild-type controls (Figure 6D and F). By 96 hours after irradiation when Prap1-/- mice were approaching 75% initial body weight, their small intestine had a significantly higher amount of Bax expression, indicating a significant increase in apoptosis compared with wild-type controls (Figure 6G). Although the Prap1-/- mice had increased apoptosis, they did not have any significant changes in Pcna expression, indicating no significant change in proliferation compared with wild-type controls (Figure 6H). To determine whether the increased apoptosis in the Prap1-/- mice coincided with increased gut barrier permeability, we orally gavaged wild-type and Prap1-/- mice with FITC dextran 72 hours after TBI. Quantification of serum FITC dextran levels showed no difference between wild-type and Prap1-/- mice, suggesting no significant change in gut barrier permeability at this particular time point (Figure 6I). Lastly, wild-type mice significantly increased Prap1 expression in the small intestine 96 hours after irradiation compared with unchallenged wild-type mice or mice 6 hours after irradiation (Figure 6J). Taken together, these data show that PRAP1 expression protects the epithelium from irradiation-induced apoptosis 6 hours after challenge and this protection persists for days later, coinciding with increased expression of PRAP1 by the gut epithelium.
Figure 6.
Prap1-/-mice are more susceptible to radiologic challenge and have increased apoptosis in the intestinal epithelium. (A) Percentage body weight loss of wild-type C57BL/6 and littermate Prap1-/- mice after 10 Gy TBI. Statistical analysis represents a comparison of wild-type vs Prap1-/- on each respective day using 2-way analysis of variance, Bonferroni multiple comparisons test (n = 4 mice). ∗P < .05, ∗∗P < .01. (B) Survival of wild-type, Prap1+/- or Prap1-/- mice after 10 Gy TBI. Statistical significance was determined using the log-rank test (n ≥ 11 mice). ∗∗P < .01. (C) Representative images of terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL)-positive cells (green) within the small intestine of 8-week-old wild-type and Prap1-/- littermates 6 hours after receiving 10 Gy TBI. (D) Representative images of cleaved caspase-3–positive cells (green) within the small intestine of 8-week-old wild-type and Prap1-/- littermates 72 hours after receiving 10 Gy TBI. (E) Quantification of TUNEL-positive cells in panel C. Significance was determined using an unpaired t test (n ≥ 11 mice). ∗∗P < .01. (F) Quantification of cleaved caspase-3–positive cells in panel F. Significance was determined using an unpaired t test (n ≥ 6 mice). ∗∗P < .01. (G) Quantification of Bax transcript via quantitative PCR on whole tissue from the small intestine of wild-type and Prap1-/- littermates 96 hours after 10 Gy TBI. Significance was determined using an unpaired t test (n = 6 mice). ∗P < .05. (H) Quantification of Pcna transcript via quantitative PCR on whole tissue from the small intestine of wild-type and Prap1-/- littermates 96 hours after 10 Gy TBI. Significance was determined using an unpaired t test (n = 6 mice). (I) Quantification of serum FITC dextran in wild-type and Prap1-/- littermates after oral gavage with 4 kilodaltons FITC dextran 72 hours after 10 Gy TBI. Significance was determined using an unpaired t test (n ≥ 3 mice). All data are graphed as the means ± SEM. (J) Quantification of Prap1 transcript via quantitative PCR on whole tissue from the small intestine of wild-type mice at different time points after 10 Gy TBI. Significance was determined using 1-way analysis of variance, Tukey multiple comparisons test (n = 6 mice). ∗∗P < .005. Bax RA, Bax relative abundance; DAPI, 4′,6-diamidino-2-phenylindole; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; KO, knockout; Pcna RA, Pcna relative abundance; WT, wild-type.
PRAP1 Protects Enteroids From Irradiation-Induced Apoptosis by Limiting p21 Expression
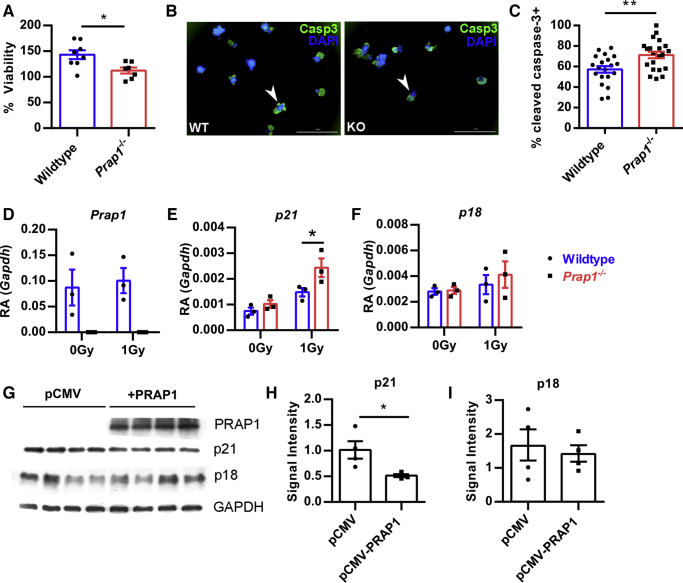
To determine whether PRAP1 protection from irradiation could be observed in an isolated epithelial model, we harvested crypts from the small intestine of wild-type and Prap1-/- mice to culture enteroids ex vivo. On day 5 of culture, the enteroids were irradiated with 2 Gy and viability of the enterocytes was measured via the metabolic reduction of 3-(4,5-dimethylthiazol-2-yl)-diphenyltetrazolium bromide (MTT) to formazan. At 48 hours after irradiation, the Prap1-/- enteroids had a significant decrease in the percentage of viability compared with wild-type controls (Figure 7A). To determine whether this decrease in viability could be attributed to an increase in apoptosis, we stained the enteroids for cleaved caspase-3 at 24 hours after irradiation (Figure 7B). Enumeration of cleaved caspase-3–positive enteroids showed Prap1-/- enteroids had a significantly higher percentage undergoing apoptosis after irradiation (Figure 7C). In summary, these data show that PRAP1 is capable of protecting the epithelium from irradiation-induced apoptosis in an isolated epithelial system.
Figure 7.
PRAP1 protects enteroids from irradiation-induced apoptosis by limiting p21 expression. (A) The percentage of viability of wild-type or Prap1-/- enteroids 48 hours after 2 Gy. Cell viability was measured using the addition of MTT and the percentage of viability was calculated using the cell viability measured before irradiation. Significance was determined using an unpaired t test (n ≥ 7 wells). ∗P < .05. (B) Representative immunofluorescence images for the detection of cleaved caspase-3 in wild-type and Prap1-/- enteroids 24 hours after 2 Gy. Examples of cleaved caspase-3–positive enteroids are indicated by a white arrowhead. Scale bar: 1000 μm. (C) Quantification of cleaved caspase-3–positive enteroids in panel B. Each data point represents the percentage of cleaved caspase-3–positive enteroids in a well. Data were pooled from 4 independent experiments. Significance was determined via unpaired t test (n = 4 mice per group). ∗∗P < .01. (D–F) Quantification of Prap1 (D), p21 (E), and p18 (F) transcript via quantitative PCR in wild-type and Prap1-/- enteroids 24 hours after 0 Gy and 1 Gy. Significance was determined via an unpaired t test. ∗P < .05. Each data point represents enteroids harvested from a unique mouse (n = 3 mice per group). (G) Protein levels were determined via Western blot from epithelial cells transfected with an empty cytomegalovirus expression vector (pCMV) pCMV or a cytomegalovirus expression vector encoding PRAP1 (pCMV-PRAP1) 48 hours after 8 Gy. (H and I) Quantification of p21 (H) and p18 (I) protein levels in panel G determined via signal intensity relative to GAPDH. Significance was determined using an unpaired t test (n = 4). ∗P > .05. All data are graphed as means ± SEM. Casp3, caspase-3; DAPI, 4′,6-diamidino-2-phenylindole; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KO, knockout; pCMV, empty cytomegalovirus expression vector; WT, wild-type
Previous literature has reported that PRAP1 acts downstream of p53, protects neoplastic cells from chemotherapeutic agents, and induces cell-cycle arrest.15 To further identify the mechanism by which PRAP1 protects cells from DNA-damaging agents, we collected RNA from wild-type and Prap1-/- enteroids 24 hours after irradiation and compared the expression of PRAP1 and other proteins known to be important for cell-cycle arrest. As expected, the wild-type enteroids had strong PRAP1 expression both before and after irradiation (Figure 7D). Interestingly, although both wild-type and Prap1-/- enteroids showed an increase in p21WAF1/Cip1 expression after irradiation, the Prap1-/- enteroids had significantly higher p21 expression compared with wild-type controls (Figure 7E). Expression of p18Ink4c, another protein implicated in cell-cycle arrest, was not different between wild-type and Prap1-/- enteroids (Figure 7F). To determine whether this difference in p21 expression could be observed in a different epithelial model we transiently overexpressed human PRAP1 in a human epithelial cell line. After 24 hours of PRAP1 overexpression, the cells were irradiated with 8 Gy and cell lysate was collected 48 hours later (Figure 7G). Consistent with our findings in the enteroids, Western blot analysis showed the cells overexpressing PRAP1 had significantly less p21 protein after irradiation (Figure 7H) and no difference in p18 (Figure 7I). Taken together, these data show that PRAP1 protects the epithelium from irradiation-induced apoptosis and significantly limits the amount of p21 expression after challenge.
Discussion
We report that PRAP1 is an intrinsically disordered protein that is highly expressed in the small intestine of mice and human beings. At homoeostasis, Prap1 null mice show mild physiological differences, with increased cytokine levels and an altered gut microbiota. However, Prap1 null mice are significantly more susceptible to oxidative insult by ionizing radiation, showing accelerated mortality and enterocyte apoptosis. In addition, Prap1-/- enteroids are more susceptible to irradiation-induced apoptosis and have increased p21 expression. These data show that PRAP1 functions as an important component of the epithelial response to oxidative insult.
The study of intrinsically disordered protein (IDP) structure and function is inherently complex because the extended structure of IDPs is flexible and dependent on their environment and subcellular location.20 Several IDPs have been shown to regulate cell signaling pathways because their flexible conformation allows them to bind regulatory proteins with disparate functions. The ability of IDPs to weakly bind multiple proteins allows them to facilitate the assembly of cellular signaling complexes that control pathway signaling.20 For example, p21 is a 21-kilodalton intrinsically disordered protein that has the ability to bind and regulate several cyclin-dependent kinase (CDK)-cyclin complexes and ultimately regulates cell-cycle progression.21 The ability to bind several proteins affords multiple functions and the regulation of a variety of complexes. In addition, IDPs also have been implicated in the formation of hydrogels by regulating liquid phase transitions, as well as the formation of non–membrane-bound intracellular granules.22 Given the intracellular perinuclear localization of PRAP1 in the intestinal epithelial cells, it is possible that PRAP1 functions intracellularly to assemble proteins that function in cell signaling, ultimately regulating the cellular response to oxidative stress and cell-cycle progression. In addition, there is a signal peptide on the N terminus of PRAP1, and our laboratory and others have found that PRAP1 is secreted at relatively high levels into the supernatant of cultured cells.23 It may be possible that similar to many other IDPs, PRAP1 has multiple functions that are dictated by cellular location and the presence of interacting proteins.
The discovery of novel effector proteins that protect from exogenous insults is a challenging endeavor because the function of these proteins is not apparent in vivo at homeostatic conditions. For example, experimental rodents generally are maintained in conditions that minimize exposure to xenobiotic agents or pathogenic microbes.24 As a result, mice that are null for proteins that confer protection often do not show any spontaneous phenotypes. Rather, it is only in response to exogenous insults that these null mice show phenotypes, typically manifested as increased susceptibility, or failure to recover after insult.25,26 Although Prap1-/- mice showed normal epithelial proliferation, apoptosis, and gut permeability at baseline, there were signs of suboptimal intestinal function because inflammatory markers were increased and the upper gastrointestinal microbiota was altered. After challenge with irradiation, underlying suboptimal intestinal functions were exaggerated with a significant increase in epithelial apoptosis, without a corresponding increase in proliferation. Although irradiation is not a naturally occurring challenge for the intestinal epithelium, the mucosa commonly encounters exogenous stressors that increase oxidative stress, and this is especially true for the small intestinal epithelium where PRAP1 expression is highest. The small intestine must tolerate a myriad of stressors that are introduced by digestive processes, nutrient absorption, and microbial contact.27 Importantly, these stressors have the potential to significantly increase with a change in diet or gastrointestinal infection.28, 29, 30 Discovering proteins critical to the epithelial response to oxidative insult has the potential to identify therapeutic targets to prevent tissue damage that ultimately results in a leaky gut phenotype. This occurs after the breakdown of the epithelial barrier of the gastrointestinal tract, increasing translocation of luminal antigens, microbes, and their products, which leads to increased local and systemic inflammation.31 In this study, identifying a protective function for PRAP1 in the gut may allow for the development of effective therapeutics that augment the function of PRAP1, and thereby enhance protection against exogenous stressors.
To elucidate the protective role of PRAP1 in the intestine, we used TBI as a model of exogenous challenge to the intestinal epithelium. TBI generates a large amount of oxidative stress, inducing rapid apoptosis of vulnerable proliferating cells in the bone marrow and intestinal crypts.32 Loss of the cellular immune system and a compromised intestinal barrier is a combination that leads to rapid weight loss, lack of fluid retention, and increases the risk of developing systemic bacterial infection.32,33 Although we did not detect an increase in gut permeability 72 hours after irradiation via FITC dextran in the Prap1-/- mice, we did observe a significant prolonged survival in the wild-type mice after a lethal dose of irradiation compared with the Prap1-/- littermates. It is probable that a significant defect in the gut barrier as a result of excessive apoptosis immediately precedes necessary end points for humane euthanasia of the mouse, therefore making differences in gut permeability difficult to measure accurately. Although apoptotic cells in the epithelium can be replaced quickly by neighboring epithelial cells,5 we did not see a corresponding increase in Pcna expression in the Prap1-/- tissue. In addition, we found a significant increase in the expression of p21, a protein critical to regulating cell-cycle progression. In summary, the Prap1-/- epithelium shows increased apoptosis for days after irradiation in combination with dysregulation of a critical cell-cycle regulating protein, ultimately affecting survival of the Prap1-/- mice.
The limitations of the current study are that the complete molecular mechanism whereby PRAP1 confers protection from irradiation remains enigmatic. Because PRAP1 is an IDP, it is likely that PRAP1 has multiple functions and has many interacting proteins based on the localization and disease context. In an in vitro model, Huang et al15 have shown PRAP1 to be downstream of p53 activation and that PRAP1 was required to arrest cells in the cell cycle after treatment with fluorouracil, ultimately resulting in less DNA damage and less caspase-dependent apoptosis.15 Consistently, we observed decreased viability and increased apoptosis in the Prap1-/- enteroids after irradiation. Recapitulating the in vivo phenotype with ex vivo Prap1-/- enteroids shows that the mechanism of PRAP1 protection is intrinsic to the epithelium and does not require other systemic host processes to function. Interestingly, both the enteroids and an epithelial cell line had decreased p21 expression in the presence of PRAP1 after irradiation. Although p21 has been shown to bind and affect several different signaling complexes and pathways (attributed to the fact that p21 is also an IDP34), the existing literature consistently has shown that p21 can have dual effects regarding the induction of apoptosis. In the context of UVB irradiation, overexpression of p21 led to increased apoptosis in keratinocytes.35 In addition, multiple studies have shown that p21 overexpression increased apoptosis of cancer cells and rendered them more susceptible to chemotherapeutic drugs.36,37 Here, our data show that PRAP1 significantly limits p21 expression, thereby preventing irradiation-induced apoptosis. Future studies involving the identification of PRAP1 interacting proteins in intestinal epithelial cells will be extremely valuable in defining the mechanism by which PRAP1 limits p21 expression and protects from oxidative insult.
Methods
Production of Recombinant PRAP1
The coding sequence for the secreted form of mouse PRAP1 protein (AA 21–149) was cloned into a pET28a vector with a hexahistadine tag (6xHis) tag on the N terminus of the protein generating pET28a-6xHis-PRAP1. This plasmid was transformed into Escherichia coli and expression of PRAP1 was induced using 1 mol/L isopropyl β-d-1-thiogalactopyranoside at 30°C for 4 hours. E coli was lysed using 6 mol/L guanidine and solubilized with 10 mol/L urea. 6xHis-PRAP1 protein was purified using nickel-nitrilotriacetic acid (Ni-NTA) agarose and the buffer was exchanged using phosphate-buffered saline (PBS). For further purification and analysis, PRAP1 protein was loaded onto a size-exclusion chromatography column S200 10/300 at 0.5 mL/min and equilibrated in PBS. The S200 10/300 column was calibrated with a molecular weight standard containing thyroglobulin (670,000 daltons), γ-globulin (158,000 daltons), ovalbumin (44,000 daltons), myoglobin (17,000 daltons), and vitamin B12 (1350 daltons) (Bio-Rad, Hercules, CA) (Figure 1C).
Circular Dichroism
Circular dichroism (CD) spectroscopy was monitored by a J-810 CD spectropolarimeter (Jasco, Easton, MD) to examine PRAP1 secondary structure. PRAP1 protein was dialyzed overnight in phosphate buffer (50 mmol/L NaPO4, 15 mmol/L NaCl, pH 6.8) at a concentration of 0.44 mg/mL for all measurements. The CD signal and molar ellipticity was measured from 190 to 260 nm with a 1-mm quartz cuvette at 25°C. Data shown are the average of 3 spectral scans and after buffer subtraction.
Quantitative Reverse-Transcription Polymerase Chain Reaction Analysis
Murine tissue was homogenized in TRIzol reagent (Invitrogen, Carlsbad, CA) and RNA was isolated using the phenol-chloroform extraction method. Complementary DNA was made using the iScript Reverse Transcription Supermix (Bio-Rad). Real-time polymerase chain reaction (PCR) analysis then was performed using the iQ SYBR Green Supermix on a MyiQ real-time PCR system (Bio-Rad). β-actin and Gapdh were used as housekeeping genes where indicated and all quantification is shown as relative abundance to β-actin or Gapdh using the ΔΔCT analysis method. The primer sequences used were as follows: Prap1: 5’-ATCTACAGCTTCGCCATTCG-3’, 5’-GTTTGCCTTTGGTCTTGACAG-3’; Gapdh: 5’-TCTCCCTCACAATTTCCATCC-3’, 5’-GGGTGCAGCGAACTTTATTG-3’; Actin: 5’-ACCTTCTACAATGAGCTGCG-3’, 5’-CTGGATGGCTACGTACATGG-3’; Pcna: 5’-GGCTCTCAAAGACCTCATCAA-3’, 5’-GAGTAAGCTGTACCAAGGAGAC-3’; Bax: 5’-CAAGAAGCTGAGCGAGTGTC-3’, 5’-GTCCACGTCAGCAATCATCC-3’; IL12: 5’-GATGTGTCCTCAGAAGCTAACC-3’, 5’-CCAGTCCACCTCTACAACATAAA-3’; p21: 5’-GTTCCTTGCCACTTCTTACCT-3’, 5’-TCATCCTAGCTGGCCTTAGA-3’; and p18: 5’-TAGCCTGATGGAGGCAAATG-3’, 5’-CGGACAGCCAACCAACTAA-3’.
Production of PRAP1 Antisera
Full-length mouse PRAP1 recombinant protein was injected into rabbits after a 70-day protocol, whereupon antibodies were generated and serum was collected following the standard polyclonal package protocol performed by Pocono Rabbit Farms (Canadensis, PA). Rabbit PRAP1 antisera was aliquoted and stored at -80°C. PRAP1 specificity was confirmed via immunofluorescence and immunoblot analysis of tissue samples from wild-type and Prap1-/- mice.
Prap1 Whole-Body Knockout Mice
Prap1 whole-body knockout mice (strain: B6;129S5-Prap1tm1Lex/Mmucd, 032532-UCD), originally generated by Genentech (South San Francisco, CA), have all 5 exons of the Prap1 gene targeted by homologous recombination. The mice were resuscitated by the Mutant Mouse Resource and Research Center at UC Davis (MMRC-UC Davis) (Davis, CA). Prap1 heterozygous mice then were shipped from MMRRC-UC Davis and housed within a specific pathogen-free facility at Emory University (Atlanta, GA). Prap1 heterozygous mice then were backcrossed with wild-type C57BL/6 mice (Jackson Laboratories, Bar Harbor, ME) until the mice were determined to be fully congenic using the Speed Congenics 128 SNP Panel (Charles River, Wilmington MA). The Prap1-/- mouse colony was maintained with heterozygous breeding pairs to provide wild-type littermate controls for all experiments. All animal procedures were approved by the Institutional Animal Care and Use Committee of Emory University.
Immunoblot Analysis
Immunoblot analysis using the PRAP1 antisera was performed on harvested murine tissues as previously described.38 PRAP1 protein was detected using a 1:1000 dilution of PRAP1 antisera generated by the laboratory (see earlier) in 5% milk made in Tris-buffered saline plus 0.1% Tween (TBST) (Thermo Fisher Scientific, Waltham, MA).
Immunofluorescence
Murine tissues were harvested and immediately fixed in methacarn solution to preserve any luminal material. Tissues were embedded in paraffin and sectioned at 5 μm. Sections were rehydrated in xylene and ethanol baths before undergoing a blocking step with 5% bovine serum albumin (BSA) in PBS. The primary antibody was diluted in 5% BSA and added to the section overnight at 4°C. The secondary antibody conjugated to an Alexa Fluor was diluted 1:1000 in 5% BSA and added to the sections for 1 hour at room temperature. 4′,6-diamidino-2-phenylindole diluted 1:1000 in 5% BSA was added as a counterstain for 5 minutes before the sections were mounted with Prolong Antifade Mountant (Invitrogen) and stored long term at -80°C. Slides were washed 3 times in PBS after each staining step. Images were captured on an FV1000 confocal microscope (Olympus, Tokyo, Japan).
Immunohistochemistry Staining on Human Sections
Diagnostic curettings of de-identified human tissue were obtained in collaboration with Dr Krisztina Hanley and Dr Brian Robinson (Emory University, Atlanta, GA). Immunohistochemistry was performed by the Cancer Tissue and Pathology Core at Emory University. Human PRAP1 was visualized using a commercially available polyclonal antibody (Proteintech) diluted 1:400.
Histologic Assessment of Intestinal Architecture
Murine proximal small intestine was fixed with formalin and embedded in paraffin before sectioning and staining with H&E. Approximately 20 independent villi and crypt lengths were obtained from bright-field microscope images using ImageJ (National Institutes of Health, Bethesda, MD).
Microbiota Analysis
Luminal content from the distal ileum of mice was collected and microbial DNA was isolated using the QIAmp Stool DNA mini kit (Qiagen, Hilden, Germany). Preparation of 16S samples was performed as reported previously.39 PCR amplification of the 16S ribosomal RNA V4 region was performed using the 515F/806R primer pair (GTGCCAGCMGCCGCGGTAA and GGACTACHVGGGTWTCTAAT) with a unique 12-base Golay barcode on each reverse primer. DNA concentration was measured using a Qubit fluorometer (Thermofisher, Waltham, MA) and run on a bioanalyzer to confirm a single 16S band. The DNA was pooled at equimolar ratios and then sequenced using an Illumina Miseq sequencer (San Diego, CA) at the Emory Integrated Genomics Core (Emory University). Read counts for each sample were generated by uploading the raw sequence files to Illumina’s 16S Metagenomics Application on the BaseSpace Sequence Hub platform.
Measurement of Cytokines and Fecal IgA
Cytokine levels in whole serum from 10-week-old mice were measured using the V-PLEX Proinflammatory Panel 1 Mouse Kit (Meso Scale Discovery, Rockville, MD) following the manufacturer’s protocol with the help of the Emory Multiplexed Immunoassay Core (Emory University). For Fecal IgA, fecal pellets were collected from adult mice and vortexed in PBS with 5 mmol/L EDTA to a final concentration of 0.1 mg feces/mL. The samples then were centrifuged at 6000 g for 10 minutes and the supernatant was removed. An IgA enzyme-linked immunosorbent assay kit (Invitrogen) was used to quantify the amount of mouse IgA present in the supernatant following the manufacturer’s protocol.
Measurement of Intestinal Permeability
Adult mice (8 weeks old) were fasted for 12 hours and orally gavaged with 10 mg FITC-conjugated dextran (4 kilodaltons) dissolved in PBS. After 4 hours the mice were killed and serum was diluted 1:4 with PBS. The quantity of FITC-dextran was determined by using a spectrophotometer capable of 485-nm excitation and 528-nm emission. The final concentration was calculated by comparison with a FITC-dextran standard curve. For irradiation experiments, mice were fasted for 4 hours on day 3 before oral gavage with FITC-dextran.
TBI Challenge
Prap1-/- and wild-type littermate controls at 8 weeks of age received 10 G radiation (225 kV and 17.7 mA) using a RS2000 X-ray irradiator (Rad Source Technologies, Buford, GA). Weight loss was monitored daily and the mice were killed once they reached 75% of their initial body weight. Apoptotic cells were visualized in tissue sections collected 6 hours after radiation challenge using the In Situ Cell Death detection kit (Roche, Basel, Switzerland) and quantified by counting the number of positive cells per crypt in 10 random fields of view per mouse (n ≥ 9 mice) at 400× total magnification on an FV1000 confocal fluorescence microscope (Olympus). Cleaved caspase-3–positive cells were visualized 72 hours after radiation challenge using the polyclonal cleaved caspase-3 antibody from Cell Signaling (Danvers, MA). By using a total magnification of 200×, the number of positive cells in 120 random villi was enumerated for each mouse (n ≥ 6 mice), and, assuming an average of 86 enterocytes per villi, the proportion of positive cells was calculated.
Generation of Enteroids
Enteroids from wild-type and Prap1-/- littermates were generated using the IntestiCult Organoid Growth Medium (mouse) from StemCell Technologies (Vancouver, Canada) following the manufacturer’s provided protocol. Briefly, the entire length of the small intestine was dissected, washed, and digested for the collection of small intestinal crypts. The crypts were enumerated and combined with a 1:1 ratio of Intesticult Organoid Growth Medium and Matrigel (Corning, Tewksbury, MA). Media was refreshed every 2 days and enteroids were passaged every 7 days. For irradiation experiments, enteroids were irradiated on days 4–5 of culture.
Assessment of Enteroid Viability
Enteroid viability was assessed by quantification of MTT reduction to formazan following the protocol described by Grabinger et al.40 Briefly, after treatment with either irradiation or a staurosporine-positive control, sterile MTT was added at a final concentration of 500 ug/mL for 1 hour at 37°C. Cell media then was replaced with 2% sodium dodecyl sulfate to digest the Matrigel and then dimethyl sulfoxide was added to solubilize the formazan. Each well then was read at 563 nm using a spectrophotometer and the positive control optical density was subtracted from each reading. The percentage of viability was calculated by dividing the optical density of the treatment wells by the optical density of untreated wells, multiplied by 100.
Whole-Mount Immunofluorescence Staining of Enteroids
The immunofluorescence staining protocol was adapted from O’Rourke et al41 and modified for staining and imaging in a 96-well plate. Enteroids were grown and treated in 96-well plates and on the day of staining media was replaced with 80 μL 4% paraformaldehyde and fixed for 20 minutes at room temperature. Enteroids then were washed with IF buffer and permeabilized for 20 minutes at room temperature with 80 μL 0.5% Triton X-100 (Thermo Fisher Scientific) in PBS. By this point, the Matrigel was dissolved and the enteroids remained attached to the bottom of the well. After washing with IF buffer, the enteroids were blocked for 30 minutes at room temperature with 5% BSA in PBS. The primary antibody specific for cleaved caspase-3 (Cell Signaling) was diluted 1:100 in blocking solution and incubated overnight at 4°C. After washing with IF buffer, secondary antibody specific for rabbit IgG was diluted 1:1000 in blocking buffer for 1 hour at room temperature, protected from light. Secondary antibody was removed and 4′,6-diamidino-2-phenylindole diluted 1:1000 in PBS was added for 10 minutes protected from light. The enteroids then were washed with IF buffer and then with PBS. PBS was left in the well during imaging on a Lionheart FX Automated Microscope (BioTek, Winooski, VT).
Preparation of Enteroids for RNA Isolation
Enteroids were grown in 24-well plates and collected using 1 mL ice-cold PBS. Enteroids from at least 4 wells were pooled into a 15-mL conical tube and spun at 500 × g for 5 minutes, at 4°C. The supernatant was carefully collected while also collecting as much Matrigel as possible above the enteroid pellet. One milliliter of fresh cold PBS was slowly added to the tube to lift any remaining Matrigel off the enteroid pellet. All remaining PBS/Matrigel was carefully collected, leaving the enteroid pellet undisturbed. The enteroid pellet was collected with 300 μL TRIzol and transferred to a 1.7-mL Eppendorf tube. The enteroids were sonicated twice for 5 seconds, resting on ice in between. The RNA was isolated and quantitative PCR was performed as described earlier.
Overexpression of Human PRAP1
A human colonic epithelial cell line (SK-CO15) was grown to confluence in a 24-well plate before the addition of 2 μL Lipofectamine 2000 (Invitrogen) with 1 μg plasmid DNA. Cells were transfected with either an empty cytomegalovirus expression plasmid with a c-terminal myc tag (pCMV-myc), pCMV-PRAP1-myc to transiently overexpress human PRAP1, or pCMV-green fluorescent protein (GFP) to monitor the success of the transfection. After an overnight transfection, media was refreshed and 6 hours later subjected to 8 Gy of radiation (225 kV and 17.7 mA) using a RS2000 X-ray irradiator (Rad Source Technologies). After 48 hours the cells were washed with cold PBS and collected with RIPA buffer (Thermo Fisher Scientific) and Western blot was performed as previously described.38 PRAP1 was detected by immunoblot with the commercially available anti-PRAP1 antibody (Proteintech) diluted 1:1000 in 5% milk and TBST. Glyceraldehyde-3-phosphate dehydrogenase was detected using a rabbit monoclonal antibody (Cell Signaling) diluted 1:5000 in 5% milk and TBST while all other proteins were detected using the Cell Cycle Regulation Antibody Sampler Kit from Cell Signaling following the manufacturer’s protocol. Quantification of bands was performed by measuring the signal intensity with ImageJ software, and calculated as the signal intensity relative to glyceraldehyde-3-phosphate dehydrogenase.
Statistical Analysis
Cumulative data were graphed as means ± SEM, with significance determined via the Student unpaired t test or analysis of variance if comparing more than 2 groups. Multiple comparisons used either the Tukey multiple comparisons test or the Dunnett multiple comparisons test when appropriate. Survival curves were compared using the log-rank (Mantel–Cox) test. All statistical tests were performed using GraphPad Prism 8.0.1 (San Diego, CA). A P value of .05 or less was considered significant.
Access to Data
All authors had access to the study data and reviewed and approved the final manuscript.
CRediT Authorship Contributions
Alexandra A. Wolfarth (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Validation: Lead; Visualization: Lead; Writing vestigation: Lead; Methodology: Lead; Validation: Lead; Vis; Xu Liu, PhD (Data curation: Supporting; Formal analysis: Supporting; Methodology: Supporting; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting); Trevor M. Darby, PhD (Data curation: Supporting; Formal analysis: Supporting; Validation: Supporting); Darra J. Boyer (Data curation: Supporting); Jocelyn B. Spizman (Data curation: Supporting); Joshua A. Owens (Data curation: Supporting; Visualization: Supporting); Bindu Chandrasekharan, PhD (Data curation: Supporting; Writing orting)orting;Writi Supporting); Crystal R. Naudin, PhD (Data curation: Supporting; Methodology: Supporting); Krisztina Z. Hanley, MD (Resources: Supporting); Brian S. Robinson, MD, Ph.D. (Resources: Supporting); Eric A. Ortlund, PhD (Project administration: Supporting; Supervision: Supporting; Writing roject administration: Supporti; Rheinallt M. Jones, PhD (Conceptualization: Supporting; Methodology: Supporting; Supervision: Supporting; Writing : Supporting; Methodology: Supporting;ng;porting;ng –t.ll Supporting); Andrew Neish, MD (Conceptualization: Lead; Funding acquisition: Lead; Project administration: Lead; Resources: Lead; Supervision: Lead; Writing – original draft: Lead; Writing : Lead; Resources: Lead;.
Footnotes
Author contributions Andrew S. Neish and Alexandra A. Wolfarth conceived the study; Andrew S. Neish, Eric A. Ortlund, and Alexandra A. Wolfarth designed the experiments; Alexandra A. Wolfarth, Xu Liu, Trevor M. Darby, Darra J. Boyer, Jocelyn B. Spizman, Joshua A. Owens, Bindu Chandrasekharan, and Crystal R. Naudin conducted the experiments; Alexandra A. Wolfarth, Xu Liu, and Darra J. Boyer acquired and analyzed the data; Krisztina Z. Hanley and Brian S. Robinson provided reagents; and Andrew S. Neish, Rheinallt M. Jones, and Alexandra A. Wolfarth drafted the manuscript.
Conflicts of interest The authors disclose no conflicts.
Funding Supported by American Heart Association postdoctoral fellowship 17POST33660110 (X.L.); by the Crohn's and Colitis Foundation Award (T.M.D.); by American Heart Association fellowship 19POST34370006 (C.R.N.); and supported in part by National Institutes of Health grants R01DK095750 (E.A.O.), R01DK098391 and R01CA179424 (R.M.J.), and R01DK071604 and R01AI064462 (A.S.N.).
References
- 1.Wallace J.L. Prostaglandins, NSAIDs, and gastric mucosal protection: why doesn't the stomach digest itself? Physiol Rev. 2008;88:1547–1565. doi: 10.1152/physrev.00004.2008. [DOI] [PubMed] [Google Scholar]
- 2.Jones R.M., Desai C., Darby T.M., Luo L., Wolfarth A.A., Scharer C.D., Ardita C.S., Reedy A.R., Keebaugh E.S., Neish A.S. Lactobacilli modulate epithelial cytoprotection through the Nrf2 pathway. Cell Rep. 2015;12:1217–1225. doi: 10.1016/j.celrep.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aw T.Y. Intestinal glutathione: determinant of mucosal peroxide transport, metabolism, and oxidative susceptibility. Toxicol Appl Pharmacol. 2005;204:320–328. doi: 10.1016/j.taap.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Beyfuss K., Hood D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018;23:100–117. doi: 10.1080/13510002.2017.1416773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edelblum K.L., Yan F., Yamaoka T., Polk D.B. Regulation of apoptosis during homeostasis and disease in the intestinal epithelium. Inflamm Bowel Dis. 2006;12:413–424. doi: 10.1097/01.MIB.0000217334.30689.3e. [DOI] [PubMed] [Google Scholar]
- 6.Soderholm J.D., Perdue M.H. Stress and gastrointestinal tract. II. Stress and intestinal barrier function. Am J Physiol Gastrointest Liver Physiol. 2001;280:G7–G13. doi: 10.1152/ajpgi.2001.280.1.G7. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff S.C., Barbara G., Buurman W., Ockhuizen T., Schulzke J.D., Serino M., Tilg H., Watson A., Wells J.M. Intestinal permeability--a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 9.Abella V., Scotece M., Conde J., Pino J., Gonzalez-Gay M.A., Gomez-Reino J.J., Mera A., Lago F., Gomez R., Gualillo O. Leptin in the interplay of inflammation, metabolism and immune system disorders. Nat Rev Rheumatol. 2017;13:100–109. doi: 10.1038/nrrheum.2016.209. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Chirlaque C., Aranda C.J., Ocon B., Capitan-Canadas F., Ortega-Gonzalez M., Carrero J.J., Suarez M.D., Zarzuelo A., Sanchez de Medina F., Martinez-Augustin O. Germ-free and antibiotic-treated mice are highly susceptible to epithelial injury in DSS colitis. J Crohns Colitis. 2016;10:1324–1335. doi: 10.1093/ecco-jcc/jjw096. [DOI] [PubMed] [Google Scholar]
- 11.Ciorba M.A., Riehl T.E., Rao M.S., Moon C., Ee X., Nava G.M., Walker M.R., Marinshaw J.M., Stappenbeck T.S., Stenson W.F. Lactobacillus probiotic protects intestinal epithelium from radiation injury in a TLR-2/cyclo-oxygenase-2-dependent manner. Gut. 2012;61:829–838. doi: 10.1136/gutjnl-2011-300367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J., Rajkumar N., Hooi S.C. Characterization and expression of the mouse pregnant specific uterus protein gene and its rat homologue in the intestine and uterus. Biochim Biophys Acta. 2000;1492:526–530. doi: 10.1016/s0167-4781(00)00135-4. [DOI] [PubMed] [Google Scholar]
- 14.Kasik J., Rice E. A novel complementary deoxyribonucleic acid is abundantly and specifically expressed in the uterus during pregnancy. Am J Obstet Gynecol. 1997;176:452–456. doi: 10.1016/s0002-9378(97)70514-3. [DOI] [PubMed] [Google Scholar]
- 15.Huang B.H., Zhuo J.L., Leung C.H., Lu G.D., Liu J.J., Yap C.T., Hooi S.C. PRAP1 is a novel executor of p53-dependent mechanisms in cell survival after DNA damage. Cell Death Dis. 2012;3:e442. doi: 10.1038/cddis.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiong G.F., Zhang Y.S., Han B.C., Chen W., Yang Y., Peng J.P. Estradiol-regulated proline-rich acid protein 1 is repressed by class I histone deacetylase and functions in peri-implantation mouse uterus. Mol Cell Endocrinol. 2011;331:23–33. doi: 10.1016/j.mce.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Xue B., Dunbrack R.L., Williams R.W., Dunker A.K., Uversky V.N. PONDR-FIT: a meta-predictor of intrinsically disordered amino acids. Biochim Biophys Acta. 2010;1804:996–1010. doi: 10.1016/j.bbapap.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenfield N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrero J., Muffato M., Beal K., Fitzgerald S., Gordon L., Pignatelli M., Vilella A.J., Searle S.M., Amode R., Brent S., Spooner W., Kulesha E., Yates A., Flicek P. Ensembl comparative genomics resources. Database (Oxford) 2016;2016:1–17. doi: 10.1093/database/baw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright P.E., Dyson H.J. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon M.K., Mitrea D.M., Ou L., Kriwacki R.W. Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans. 2012;40:981–988. doi: 10.1042/BST20120092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weber S.C., Brangwynne C.P. Getting RNA and protein in phase. Cell. 2012;149:1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J., Wong H., Ramanan S., Cheong D., Leong A., Hooi S.C. The proline-rich acidic protein is epigenetically regulated and inhibits growth of cancer cell lines. Cancer Res. 2003;63:6658–6665. [PubMed] [Google Scholar]
- 24.Rosshart S.P., Vassallo B.G., Angeletti D., Hutchinson D.S., Morgan A.P., Takeda K., Hickman H.D., McCulloch J.A., Badger J.H., Ajami N.J., Trinchieri G., Pardo-Manuel de Villena F., Yewdell J.W., Rehermann B. Wild mouse gut microbiota promotes host fitness and improves disease resistance. Cell. 2017;171:1015–1028 e13. doi: 10.1016/j.cell.2017.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Fouts D.E., Starkel P., Hartmann P., Chen P., Llorente C., DePew J., Moncera K., Ho S.B., Brenner D.A., Hooper L.V., Schnabl B. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe. 2016;19:227–239. doi: 10.1016/j.chom.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mashimo H., Wu D.C., Podolsky D.K., Fishman M.C. Impaired defense of intestinal mucosa in mice lacking intestinal trefoil factor. Science. 1996;274:262–265. doi: 10.1126/science.274.5285.262. [DOI] [PubMed] [Google Scholar]
- 27.Madesh M., Benard O., Balasubramanian K.A. Apoptotic process in the monkey small intestinal epithelium: 2. Possible role of oxidative stress. Free Radic Biol Med. 1999;26:431–438. doi: 10.1016/s0891-5849(98)00218-4. [DOI] [PubMed] [Google Scholar]
- 28.Sodhi C.P., Fulton W.B., Good M., Vurma M., Das T., Lai C.S., Jia H., Yamaguchi Y., Lu P., Prindle T., Ozolek J.A., Hackam D.J. Fat composition in infant formula contributes to the severity of necrotising enterocolitis. Br J Nutr. 2018;120:665–680. doi: 10.1017/S0007114518001836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X., Wei X., Sun Y., Du J., Li X., Xun Z., Li Y.C. High-fat diet promotes experimental colitis by inducing oxidative stress in the colon. Am J Physiol Gastrointest Liver Physiol. 2019;317:G453–G462. doi: 10.1152/ajpgi.00103.2019. [DOI] [PubMed] [Google Scholar]
- 30.Ma'ayeh S.Y., Knorr L., Skold K., Garnham A., Ansell B.R.E., Jex A.R., Svard S.G. Responses of the differentiated intestinal epithelial cell line Caco-2 to infection with the Giardia intestinalis GS isolate. Front Cell Infect Microbiol. 2018;8:244. doi: 10.3389/fcimb.2018.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68:1516–1526. doi: 10.1136/gutjnl-2019-318427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams J.P., Brown S.L., Georges G.E., Hauer-Jensen M., Hill R.P., Huser A.K., Kirsch D.G., Macvittie T.J., Mason K.A., Medhora M.M., Moulder J.E., Okunieff P., Otterson M.F., Robbins M.E., Smathers J.B., McBride W.H. Animal models for medical countermeasures to radiation exposure. Radiat Res. 2010;173:557–578. doi: 10.1667/RR1880.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vriesendorp H.M., Vigneulle R.M., Kitto G., Pelky T., Taylor P., Smith J. Survival after total body irradiation: effects of irradiation of exteriorized small intestine. Radiother Oncol. 1992;23:160–169. doi: 10.1016/0167-8140(92)90326-p. [DOI] [PubMed] [Google Scholar]
- 34.Follis A.V., Galea C.A., Kriwacki R.W. Intrinsic protein flexibility in regulation of cell proliferation: advantages for signaling and opportunities for novel therapeutics. Adv Exp Med Biol. 2012;725:27–49. doi: 10.1007/978-1-4614-0659-4_3. [DOI] [PubMed] [Google Scholar]
- 35.Chen A., Huang X., Xue Z., Cao D., Huang K., Chen J., Pan Y., Gao Y. The role of p21 in apoptosis, proliferation, cell cycle arrest, and antioxidant activity in UVB-irradiated human HaCaT keratinocytes. Med Sci Monit Basic Res. 2015;21:86–95. doi: 10.12659/MSMBR.893608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sheikh M.S., Rochefort H., Garcia M. Overexpression of p21WAF1/CIP1 induces growth arrest, giant cell formation and apoptosis in human breast carcinoma cell lines. Oncogene. 1995;11:1899–1905. [PubMed] [Google Scholar]
- 37.Choi Y.H., Yoo Y.H. Taxol-induced growth arrest and apoptosis is associated with the upregulation of the Cdk inhibitor, p21WAF1/CIP1, in human breast cancer cells. Oncol Rep. 2012;28:2163–2169. doi: 10.3892/or.2012.2060. [DOI] [PubMed] [Google Scholar]
- 38.Matthews J.D., Owens J.A., Naudin C.R., Saeedi B.J., Alam A., Reedy A.R., Hinrichs B.H., Sumagin R., Neish A.S., Jones R.M. Neutrophil-derived reactive oxygen orchestrates epithelial cell signaling events during intestinal repair. Am J Pathol. 2019;189:2221–2232. doi: 10.1016/j.ajpath.2019.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alam A., Leoni G., Quiros M., Wu H., Desai C., Nishio H., Jones R.M., Nusrat A., Neish A.S. The microenvironment of injured murine gut elicits a local pro-restitutive microbiota. Nat Microbiol. 2016;1:15021. doi: 10.1038/nmicrobiol.2015.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grabinger T., Delgado E., Brunner T. Analysis of cell death induction in intestinal organoids in vitro. Methods Mol Biol. 2016;1419:83–93. doi: 10.1007/978-1-4939-3581-9_7. [DOI] [PubMed] [Google Scholar]
- 41.O'Rourke K.P., Dow L.E., Lowe S.W. Immunofluorescent staining of mouse intestinal stem cells. Bio Protoc. 2016;6 doi: 10.21769/bioprotoc.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]