Abstract
Background and aims
Mouse models of colitis have been used to study the pathogenesis of inflammatory bowel disease (IBD) and for pre-clinical development of therapeutic agents. Various epigenetic pathways have been shown to play important regulatory roles in IBD. Reversible N6-methyladenosine (m6A) methylation represents a new layer of post-transcriptional gene regulation that affects a variety of biological processes. We aim to study how deletion of a critical component of m6A writer complex, METTL14, in T cells affects the development of colitis.
Methods
Conditional Mettl14 was lineage specifically deleted with CD4-regulated Cre in T cells. Colitis phenotype was determined by H&E staining, colon weight-to-length ratio and cytokine expression. We additionally utilized T cell transfer model of colitis and adoptive transfer of regulatory T cells. Mice were treated with antibiotics to determine if the colitis could be attenuated.
Results
METTL14 deficiency in T cells induced spontaneous colitis in mice. This was characterized by increased inflammatory cell infiltration, increased colonic weight-to-length ratio and increased Th1 and Th17 cytokines. The colitis development was due to dysfunctional regulatory T (Treg) cells, as adoptive transfer of WT Treg cells attenuated the colitis phenotype. The METTL14-deficient Treg cells have decreased RORγt expression compared with WT controls. METTL14 deficiency caused impaired induction of naïve T cells into induced Treg cells. Antibiotic treatment notably attenuated the colitis development.
Conclusion
Here we report a new mouse model of spontaneous colitis based on perturbation of RNA methylation in T cells. The colitis is T cell-mediated and dependent on the microbiome. This model represents a new tool for elucidating pathogenic pathways, studying the contribution of intestinal microbiome and preclinical testing of therapeutic agents for inflammatory bowel disease.
Keywords: Inflammatory Bowel Disease, RNA m6A Methylation, Mouse Colitis Model, Regulatory T Cells
Abbreviations used in this paper: FACS, fluorescence-activated cell sorter; IBD, inflammatory bowel disease; IFNγ, interferon gamma; IL, interleukin; iTreg, induced regulatory T; m6A, N6-methyladenosine; mRNA, messenger RNA; OTU, operational taxonomic unit; Treg, regulatory T cell; TNFα, tumor necrosis factor alpha
Synopsis.
We have found that deletion of RNA methylation writer METTL14 in T cells induced spontaneous colitis in mice. This is characterized by a Th1/Th17 phenotype. The colitis development was due to dysfunctional regulatory T cells and is dependent on the microbiome.
The incidence and prevalence of inflammatory bowel disease (IBD) are increasing worldwide in association with increasing urbanization.1 Mouse models of colitis have been used to study the pathogenesis of IBD and for preclinical development of new therapeutic modalities. Multiple experimental models of IBD have been developed. While no single model recapitulates the full spectrum of human IBD, the spectrum of models provides insights into various aspects of the disease.2
Reversible messenger RNA (mRNA) N6-methyladenosine (m6A) methylation serves as an additional layer of post-transcriptional regulation.3,4 This abundant mRNA modification is installed by a methyltransferase complex with the catalytic core containing a heterodimer of METTL3 (methyltransferase like 3) and METTL14 (methyltransferase like 14), which also binds an accessory protein WTAP.5, 6, 7 Global deletion of Mettl14 leads to embryonic lethality early in gestation.8 Deletion of Mettl3 or Mettl14 have been shown to impact a range of developmental processes.4,9, 10, 11, 12, 13, 14, 15
T cells have been shown to be integral in the pathogenesis of IBD. Several currently approved biologic therapies, including anti-integrin therapy and anti-IL12/23 therapy, act by preventing recruitment or activation of T cells.16 It has been reported that a deletion of RNA methylation writer enzyme Mettl3 in T cells leads to dysfunction in both naïve T cells and Treg cells, with naïve T cells losing their ability to induce inflammation and Treg cells losing their immune suppressive capacity.14,17 Using a conditional genetic deletion approach, we selectively deleted Mettl14, another component of the methyltransferase complex key to the methylation, in T cells. This leads to a global loss of mRNA m6A in T cells. Here, we show that deletion of Mettl14 in T cells induces development of spontaneous colitis in mice.
Results
CD4-Cre+/TgMettl14FL/FL Conditional Knockout Mice Develop Spontaneous Colitis
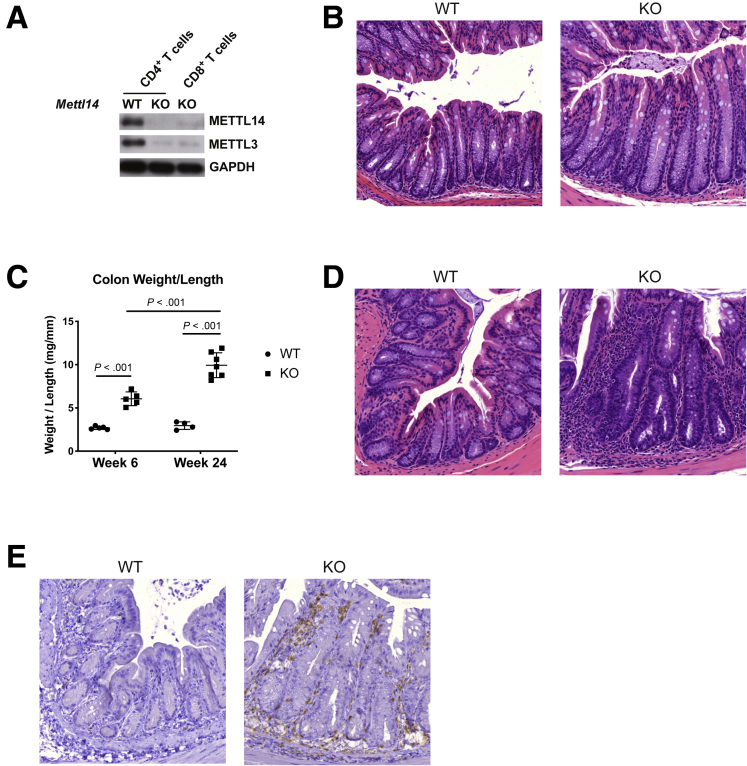
We generated the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice by crossing the CD4-Cre+/Tg mice with the Mettl14FL/FL mice. In these mice, the Mettl14 allele is selectively deleted in T cells. Analysis of the T cells by Western blot showed absence of METTL14 protein and significantly decreased expression of the associated METTL3 protein (Figure 1A). The mice null for T cell METTL14 are normal at 4 weeks of age with normal colonic histology (Figure 1B). By 6 weeks of age, the CD4-Cre+/Tg Mettl14FL/FL mice developed colitis characterized by increased colonic weight-to-length ratio (Figure 1C). The colitis became progressively more severe with a much higher colonic weight-to-length ratio by 24 weeks compared with 6 weeks (Figure 1C). Histology showed significant mucosal damage and inflammation characterized by disruption of crypt architecture, crypt branching, crypt abscess formation, marked increase in mucosal thickness, and lymphocyte infiltration in the epithelial and submucosal layers in the CD4-Cre+/Tg Mettl14FL/FL mice (Figure 1D). Most of the lymphocytes were CD4+ as seen on immunochemistry staining (Figure 1E).
Figure 1.
Colonic weight-to-length ratios and histology of CD4-Cre+/TgMettl14FL/FL conditional knockout mice. (A) CD4+ and CD8+ T cells were isolated by FACS sorting and analyzed by Western blot for METTL14 and associated METTL3 expression. GAPDH is used as an internal control. (B) Hematoxylin and eosin stain of colon at 4 weeks of age. (C) Colonic weight-to-length ratio of mice at 6 and 24 weeks of age. Data are represented as mean ± SD. (D) Hematoxylin and eosin stain of colon at 24 weeks of age. (E) Anti-CD4 immunohistochemistry staining of colon cross-section at 24 weeks of age. n = 4–8 mice per group. Data representative of 2 independent experiments with similar results. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout mouse.
Colitis in CD4-Cre+/TgMettl14FL/FL Conditional Knockout Mice Showed a Th1/Th17 Predominant Phenotype
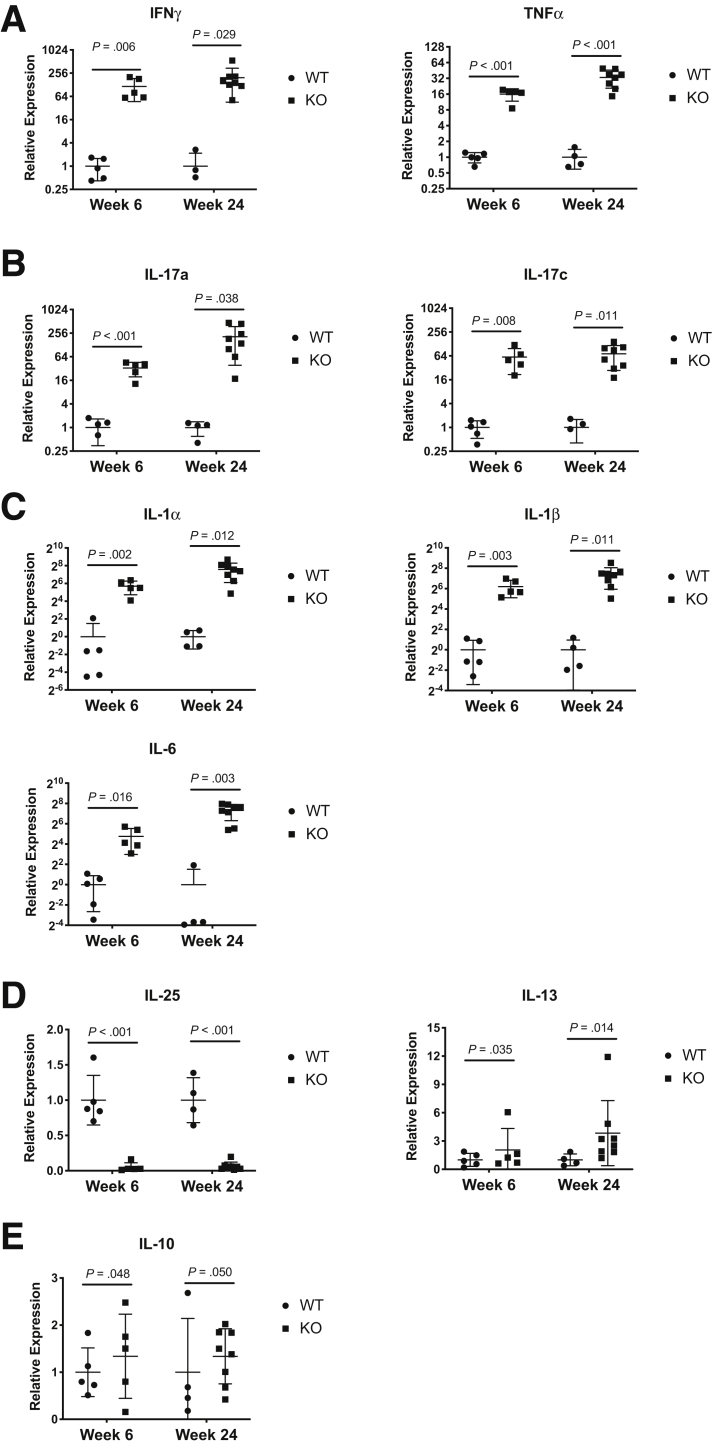
To determine whether the colitis was mediated by a Th1 or Th2 or Th17 response, we measured the cytokine profile from colonic epithelial scrapping of CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice compared with littermate control mice. CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice had a marked increase in Th1 cytokines characterized by increased interferon gamma (IFNγ) and tumor necrosis factor alpha (TNFα) (Figure 2A). The CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice also exhibited a marked increase in Th17 cytokines characterized by increased interleukin (IL)-17a and IL-17c (Figure 2B). Other cytokines associated with inflammation were also increased including increased IL-1α, IL-1β, and IL-6 (Figure 2C). In contrast, the level of Th2-potentiating cytokine IL-25 was significantly reduced, and the level of Th2 cytokine IL-13 was similar between the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice and littermate control mice (Figure 2D). The level of inhibitory cytokine IL-10 was similar between the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice and littermate control mice (Figure 2E).
Figure 2.
Cytokine expression profile of CD4-Cre+/TgMettl14FL/FL Conditional knockout mice. Relative expression of (A) Th1 cytokines IFNγ and TNFα; (B) Th17 cytokines IL-17a and IL-17c; (C) cytokines associated with inflammation including IL-1α, IL-1β, and IL-6; (D) Th2-potentiating cytokine IL-25 and Th2 cytokine IL-13; and (E) inhibitory cytokine IL-10. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. Log2 scale was used on the y-axis for panels A–C; linear scale was used on the y-axis for panels D and E. Data are represented as mean ± SD. n = 4–8 mice per group. Data representative of 2 independent experiments with similar results.
Mettl14-Deficient Treg Cells Are Unable to Suppress Naïve T Cell–Induced Colitis
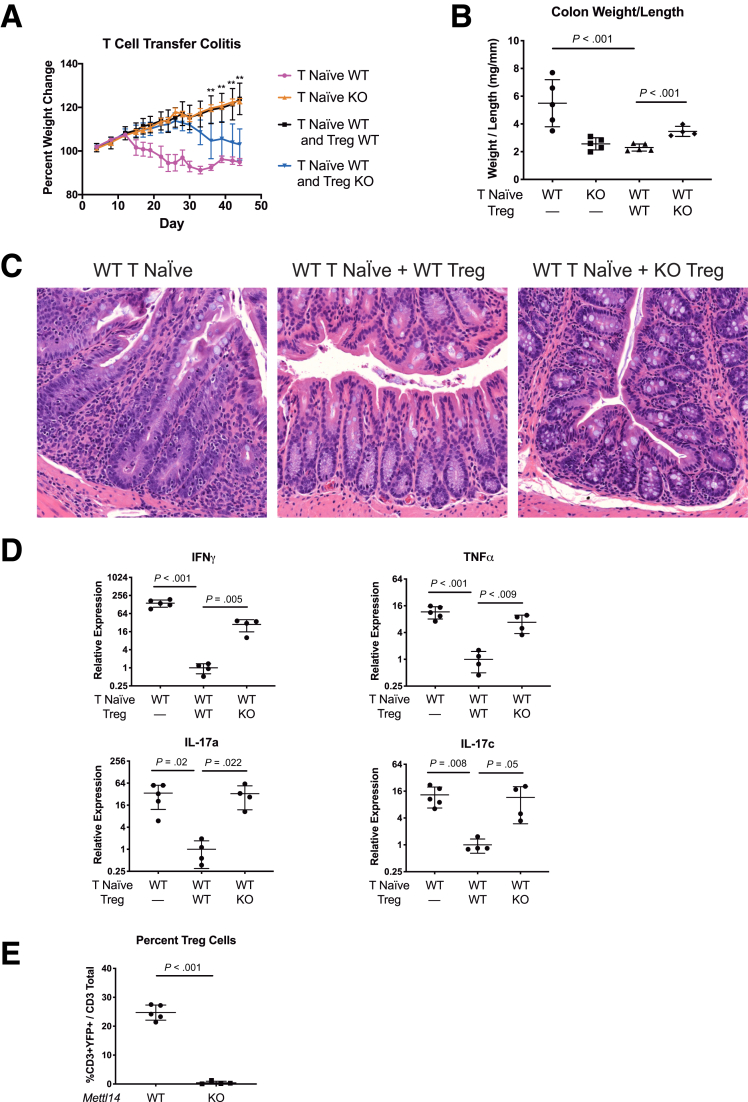
The colitis seen in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice could be due to either hyperactive proinflammatory T cells or defective Treg cells that are unable to maintain homeostasis and prevent spontaneous inflammation. To test these hypotheses, we utilized a T cell transfer colitis model where naïve T cells were transferred either alone or together with Treg cells into lymphopenic Rag1–/– mice. It has been reported that when wild-type (WT) naïve T cells are transferred into the lymphopenic Rag1–/– mice, the Rag1–/– mice develop colitis after 4–8 weeks. Co-transferring Treg cells has been shown to suppress the naïve T cell–induced colitis in this model.18 We found that transferring WT naïve T cells into Rag1–/– mice induced colitis in the Rag1–/– mice characterized by progressive weight loss over time (Figure 3A). However, transferring Mettl14–/– naïve T cells was unable to induce weight loss in the recipient Rag1–/– mice (Figure 3A). When WT naïve T cells were transferred with WT Treg cells into Rag1–/– mice, the WT Treg cells were able to completely suppress the naïve T cell–induced colitis with no weight loss seen after 7 weeks (Figure 3A). When we transferred Mettl14-deficient Treg cells with the WT naïve T cells into the Rag1–/– mice, the Mettl14-deficient Treg cells were unable to suppress naïve T cell–induced colonic inflammation. The mice developed progressive weight loss (Figure 3A), with increased colonic weight-to-length ratio compared with mice that received wildtype Treg cells (Figure 3B). These data support that the colitis phenotype seen in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice was predominantly due to dysfunctional Treg cells.
Figure 3.
Mettl14-deficient Treg cells are unable to suppress naïve T cell–induced colitis in Rag1–/– adoptive transfer T cell model of colitis. WT naïve T cells, Mettl14–/– naïve T cells, WT naïve T cells + WT Treg cells, or WT naïve T cells + Mettl14–/– Treg cells were transferred to Rag1–/– mice. (A) Weight change. ∗∗P < .01 comparing mice that received WT naïve T cells + WT Treg cells vs WT naïve T cells + Mettl14–/– Treg cells. (B) Colon weight-to-length ratio. (C) Hematoxylin and eosin stain of colon. (D) Relative expression of Th1 cytokine IFNγ/TNFα and Th17 cytokine IL-17a/IL-17c. Log2 scale was used on the y-axis. (E) Flow cytometry analysis of percent of Treg cell in mesenteric lymph nodes. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. Data are represented as mean ± SD; n = 4–5 mice per group. Data representative of 2 independent experiments with similar results.
We subsequently focused our analysis on mice that were co-transferred with Treg cells. Histology showed that mice that were co-transferred with WT Treg cells developed no inflammation while marked inflammation was seen in the mice that were co-transferred with Mettl14-deficient Treg cells (Figure 3C). Cytokine analysis showed that while WT Treg cells were able to suppress naïve T cell–induced Th1 cytokines IFNγ/TNFα and Th17 cytokines IL-17a/IL-17c, the Mettl14-deficient Treg cells were unable to suppress naïve T cell–induced Th1 and Th17 cytokine production (Figure 3D). Flow cytometry analysis showed that mice received Mettl14-deficient Treg cells have a marked decrease of Treg cells in the mesenteric lymph nodes compared with mice that received WT Treg cells (Figure 3E).
Adoptive Transfer of Treg Cells Attenuates the Colitis in CD4-Cre+/TgMettl14FL/FL Conditional Knockout Mice
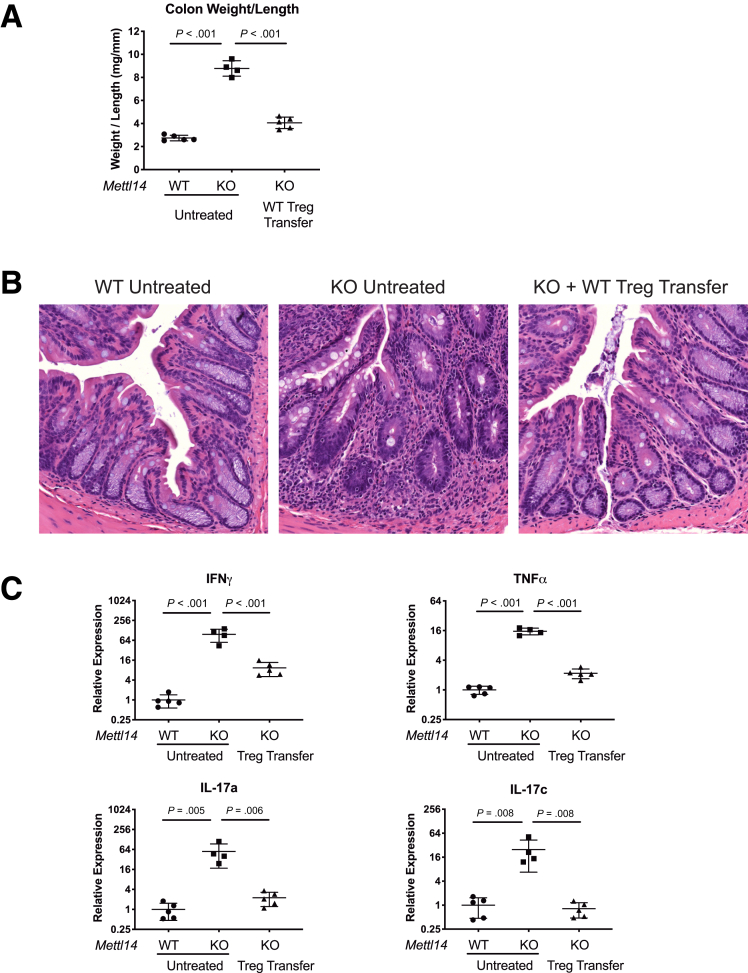
To further test whether the colitis in CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice was due to dysfunctional Treg cells, we adoptively transferred WT Treg cells into the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice to determine whether the colitis phenotype could be attenuated. We found that transferring WT Treg cells into the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice attenuated the colitis phenotype as assessed by decreased colonic weight-to-length ratio (Figure 4A) and decreased inflammatory cell infiltration (Figure 4B) compared with untreated CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice. Transferring WT Treg cells into the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice also attenuated the levels of Th1 and Th17 cytokines compared with untreated CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice (Figure 4C).
Figure 4.
Adoptive transfer of WT Treg cells attenuates the colitis in CD4-Cre+/TgMettl14FL/FL conditional knockout mice. (A) Colonic weight-to-length ratio, (B) Hematoxylin and eosin stain of colon, (C) relative expression of Th1 cytokine IFNγ/TNFα and Th17 cytokine IL-17a/IL-17c in WT mice, CD4-Cre+/TgMettl14FL/FL conditional knockout mice and CD4-Cre+/TgMettl14FL/FL conditional knockout mice adoptively transferred with WT Treg cells. Log2 scale was used on the y-axis. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. Data are represented as mean ± SD; n = 4–5 mice per group. Data representative of 2 independent experiments with similar results.
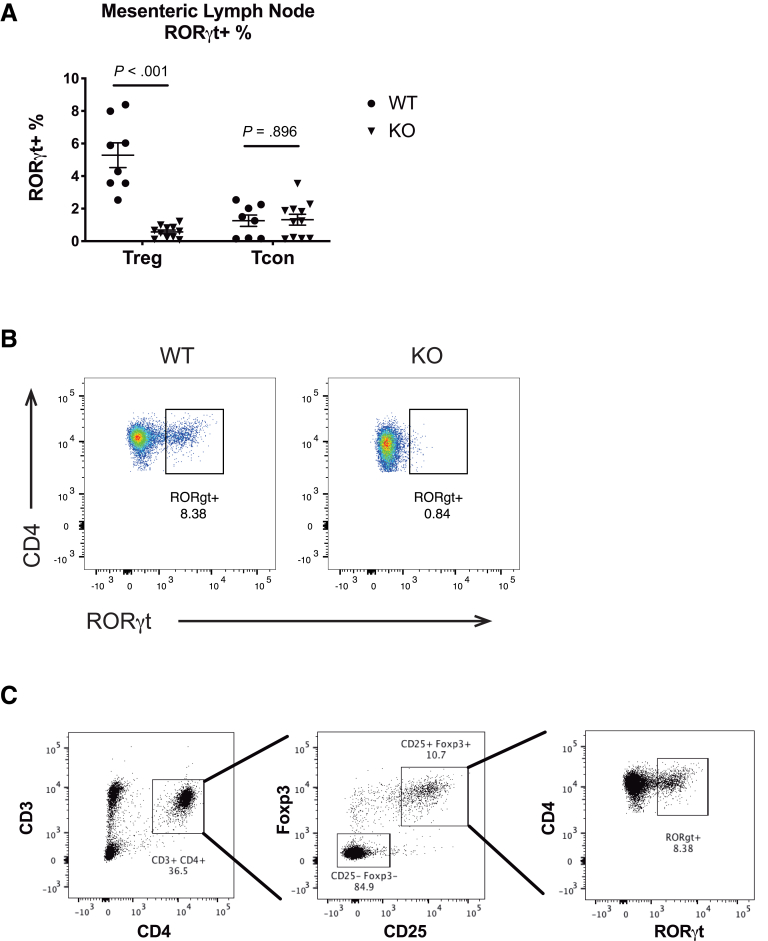
Mettl14-Deficient Treg Cells Have Decreased RORγt Expression
The Foxp3+ RORγt+ Treg cells have been shown to be a distinct stable subset of regulatory T cells that exhibit enhanced suppressive capacity in vivo.19 Because the CD4-Cre+/Tg Mettl14FL/FL mice have deficient Treg cells, we asked whether the expression of RORγt was affected. Flow cytometry analysis of mesenteric lymph nodes showed that the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice had significantly reduced Foxp3+ RORγt+ Treg cells compared with littermate control animals (Figure 5A and B). In contrast, the RORγt+ fraction is unchanged in conventional T cells (Figure 5A). The flow cytometry gating strategy is shown in Figure 5C.
Figure 5.
Mettl14-deficient Treg cells have decreased RORγt expression. (A) RORγt percentage in the mesenteric lymph nodes of Treg and conventional T cells. (B) Representative flow cytometry image showing decreased RORγt expression in Treg cells. The cells were gated from a parent population of CD25+Foxp3+ cells. (C) Gating strategy of flow cytometry analysis of RORγt+ cells. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. Data are represented as mean ± SD; n = 8–11 mice per group. Combined data from 3 independent experiments shown.
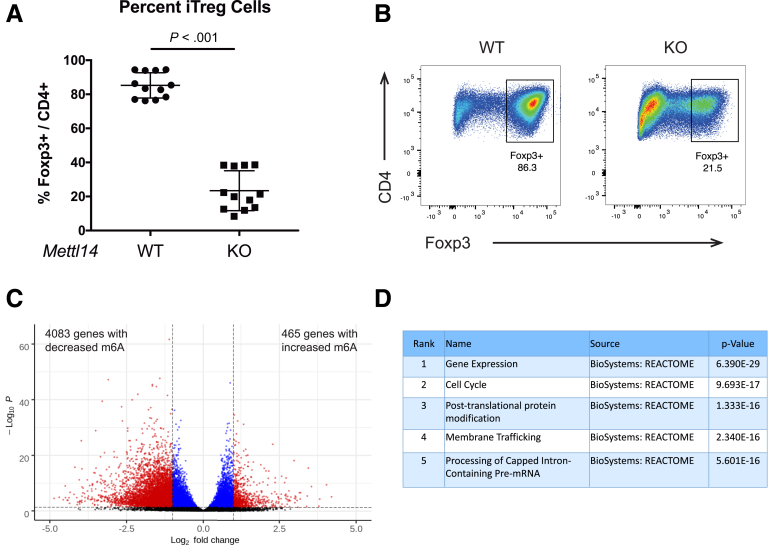
Mettl14 Deficiency Causes Impaired Induction of Naïve T Cells Into Induced Treg Cells
Induced Treg (iTreg) cells has been reported to act synergistically with natural Treg cells to control experimental colitis.20 We asked whether Mettl14 deficiency affects induction of iTreg from naïve T cells. While WT naïve T cells could be induced in vitro into iTreg cells with 85% efficiency as evidenced by Foxp3 expression, Mettl14-deficient naïve T cells have a significantly reduced induction efficacy, with only 24% of cells expressing Foxp3 (Figure 6A and B). The m6A expression profile of WT iTreg cells were compared with that of WT naïve T cells to determine genes with differential m6A levels. We found that there are 465 genes with increased m6A levels and 4083 genes with decreased m6A levels in iTreg cells compared with naïve T cells (Figure 6C). Pathway analysis showed that the most significantly affected pathways include gene expression, cell cycle, and post-translational protein modification (Figure 6D).
Figure 6.
Mettl14 deficiency causes impaired induction of naïve T cells into iTreg cells. (A) Induction efficiency of iTreg cells from naïve T cells as evidenced by Foxp3 expression. (B) Representative flow cytometry image showing percent of Foxp3+ iTreg cells after induction from naïve T cells. The cells were gated from a parent population of CD4+ cells. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. Data are represented as mean ± SD; n = 12 per group. Combined data from 3 independent experiments shown. (C) m6A profiling of iTreg cells compared with naïve T cells. Black color indicates nonsignificant; blue color indicates P < .05 and absolute Log2 fold change <1; red color indicates P < .05 and absolute Log2 fold change >1. (D) Significant pathways represented by genes with differential m6A levels. The top 5 most significant pathways were shown.
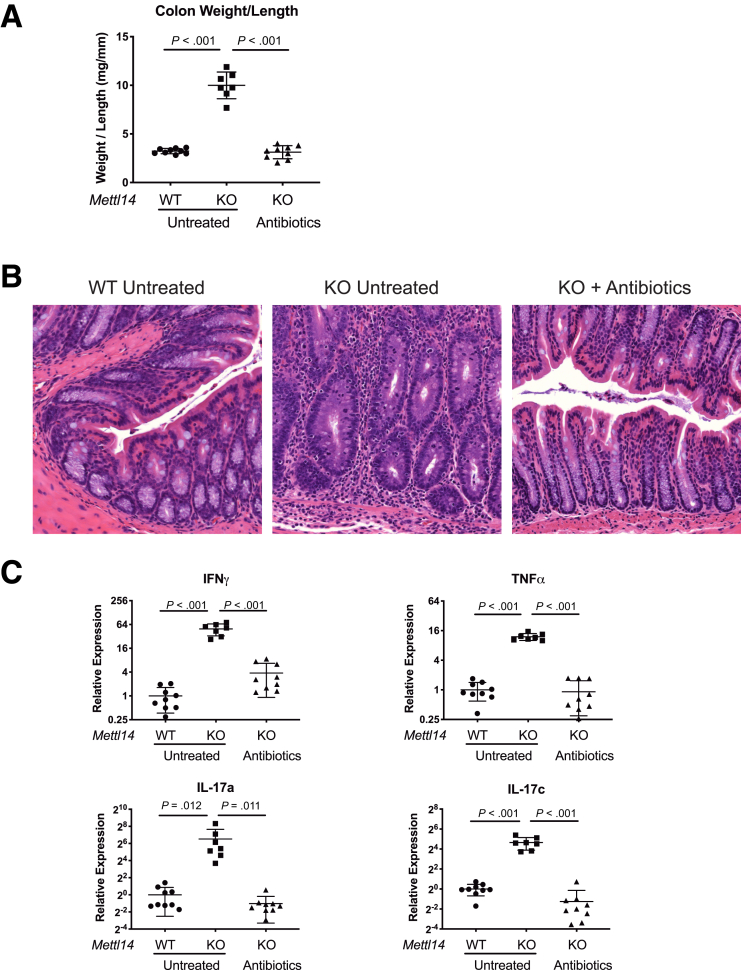
Antibiotics Treatment Attenuates the Colitis Phenotype in CD4-Cre+/TgMettl14FL/FL Conditional Knockout Mice
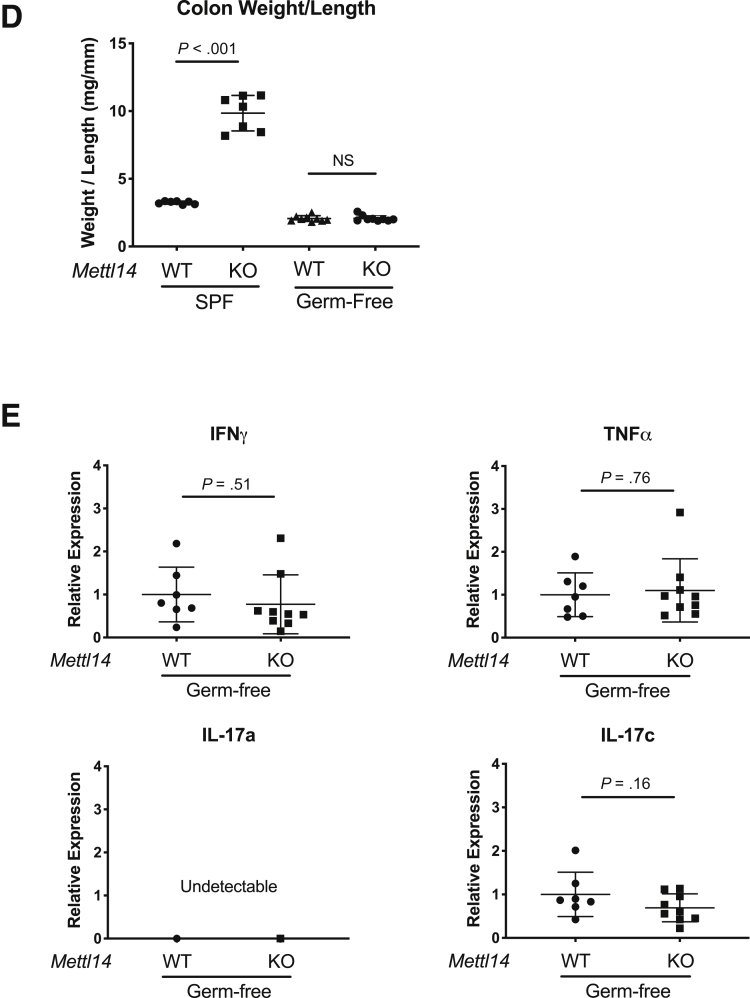
The colon is constantly exposed to microbial antigens. One of the functions of Treg cells is to prevent inflammation induced by bacterial antigens and maintain homeostasis. Because we found that the colitis phenotype is likely due to dysfunctional Treg cells in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice, we determined whether the colitis phenotype could be attenuated by antibiotics treatment. We used a combination of ciprofloxacin and metronidazole which are the 2 antibiotics currently used for treatment of human IBD. We found that treating the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice with ciprofloxacin + metronidazole prevented the development of colitis phenotype as assessed by normal colonic weight-to-length ratio (Figure 7A) and eliminated inflammatory cell infiltration (Figure 7B). The antibiotic-treated CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice also had normal levels of Th1 and Th17 cytokines (Figure 7C). In contrast, the untreated CD4-Cre+/Tg Mettl14 FL/FL conditional knockout mice developed colitis characterized by significantly increased colonic weight-to-length ratio (Figure 7A), inflammatory cell infiltration (Figure 7B), and elevated Th1 and Th17 cytokines (Figure 7C). We have further rederived the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice and WT littermate control mice in a germ-free environment. Under the germ-free environment, the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice did not develop colitis as evidenced by normal colonic weight-to-length ratio (Figure 7D) and normal cytokine expression profile (Figure 7E).
Figure 7.
Antibiotic treatment attenuates the colitis in CD4-Cre+/TgMettl14FL/FL conditional knockout mice. The CD4-Cre+/TgMettl14FL/FL conditional knockout mice were treated with ciprofloxacin + metronidazole for 3 weeks and compared with untreated mice. (A) Colonic weight-to-length ratio, (B) hematoxylin and eosin stain of colon, and (C) relative expression of Th1 cytokine IFNγ/TNFα and Th17 cytokine IL-17a/IL-17c. Log2 scale was used on the y-axis. (D) Colonic weight-to-length ratio of mice in specific pathogen–free (SPF) facility compared with germ-free facility. (E) Relative expression of Th1 cytokine IFNγ/TNFα and Th17 cytokine IL-17a/IL-17c of mice in germ-free facility. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. Data are represented as mean ± SD; n = 7–9 mice per group. Combined data from 2 independent experiments shown.
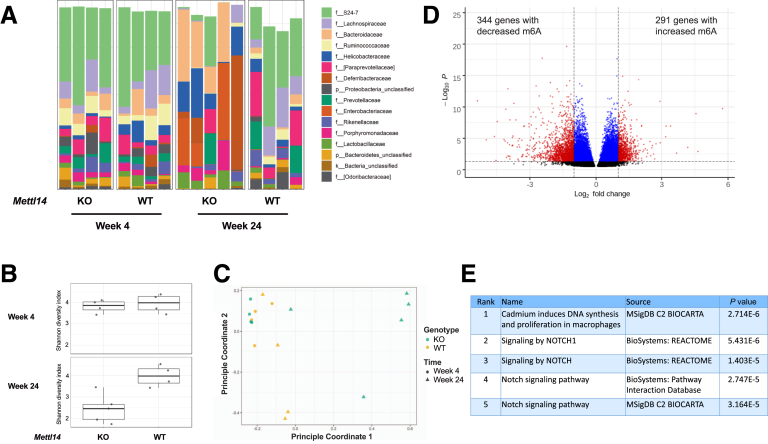
Microbiome Analysis of the CD4-Cre+/TgMettl14FL/FL Conditional Knockout Mice
We analyzed the microbiome of the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice that had developed severe colitis and compared them to the microbiome of WT littermate control mice that do not have colitis. The CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice and littermate control mice were housed together in the same cage to minimize cage differences. We found that at week 4, there was no difference in taxonomic composition between WT and CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice (Figure 8A). At week 24, the taxonomic composition of the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice showed a reduction in the families S24-7 and Lachnospiraceae, and an increase in Bacteroidaceae, Helicobacteraceae, Deferribacteraceae, and Enterobacteriaceae compared with WT control animals (Figure 8A). The alpha diversity was substantially reduced at week 24 in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice compared with WT control mice (Figure 8B). Principle coordinate analysis showed that there was a significant difference in beta diversity at week 24 in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice compared with WT control mice (Figure 8C). Neither alpha diversity nor beta diversity was different at week 4 between the CD4-Cre+/Tg Mettl14FL/FL conditional knockout and WT control mice (Figure 8B and C). We additionally determined whether the microbiome affected m6A expression in CD4+ T cells. The m6A profile of CD4+ T cells isolated from germ-free mice were compared with that of CD4+ T cells isolated from mice from a specific pathogen–free facility. We found that there are 291 genes with increased m6A levels and 344 genes with decreased m6A levels in germ-free CD4+ T cells compared with T cells from specific pathogen free facility (Figure 8D). Pathway analysis showed NOTCH signaling as the most significantly affected pathway (Figure 8E).
Figure 8.
Microbiome analysis of the CD4-Cre+/TgMettl14FL/FL conditional knockout mice. (A) Taxonomic composition, (B) alpha diversity, and (C) principal coordinate analysis plot of beta diversity of CD4-Cre+/TgMettl14FL/FL conditional knockout mice compared with control animals at week 4 and week 24. WT indicates wild-type littermate control. KO indicates CD4-Cre+/TgMettl14FL/FL conditional knockout. n = 4–5 mice per group. (D) m6A profiling of CD4+ T cells from germ-free mice compared with CD4+ T cells from specific-pathogen-free mice. Black color indicates nonsignificant; blue color indicates P < .05 and absolute Log2 fold change < 1; red color indicates P < .05 and absolute Log2 fold change > 1. (E) Significant pathways represented by genes with differential m6A levels. The top 5 most significant pathways were shown.
Discussion
Here, we show that deletion of RNA m6A methyltransferase component METTL14 in T cells leads to spontaneous colitis. The development of colitis is predominantly due to dysfunctional Treg cells. The Mettl14-deficient Treg cells were unable to suppress naïve T cell–induced inflammation in a T cell adoptive transfer model of colitis. By adoptively transferring WT Treg cells into mice with Mettl14-deficient T cells, we were able to suppress the colitis development. We showed that Mettl14 deficiency caused an impaired induction of naïve T cells into iTreg cells. We also found that the colitis is dependent on the microbiome as we were able to suppress colitis development in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice with antibiotic treatment. While there is a significant difference in microbiome between WT and CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice at week 24, there is no difference in microbiome at week 4 before the development of colitis. The microbiome changes at week 24 may reflect alterations due to colonic inflammation.
One of the commonly used colitis models is the IL-10–/– mice. The IL-10–/– mice develop spontaneous colitis with a Th1 response.21 The development of colitis in the IL10–/– mice is dependent on the genetic background and intestinal microbiota. It has been previously reported that in our facility the IL-10–/– mice develops spontaneous colitis only 25%–30% of the time by week 24.22 Our colitis model with the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice has 100% penetrance with all mice developing early signs of colitis by week 6, and the colitis phenotype becomes progressively more severe over time. While the IL-10 model is ideal for testing pathobiont due to its incomplete penetrance, our colitis model is ideal for testing protective microbiota that could potentially attenuate colitis development. While we showed that antibiotics treatment induced remission in our colitis model, the role of antibiotics to induce or maintain remission in IBD is limited. This is a limitation of our knockout mouse model to reflect IBD pathogenesis.
Multiple other colitis models have been used to study IBD. Besides the IL-10–/– mice, The most commonly used models include DSS (dextran sulfate sodium)-induced colitis, TNBS (2,4,6-trinitrobenzenesulfonic acid)-induced colitis, oxalazone-induced colitis, and T cell adoptive transfer colitis.2 Each of these models induce a different type of response and have specific advantages and disadvantages. DSS induces a colitis by disrupting the colonic epithelial barrier and is a useful model for studying epithelial injury, restitution, and innate immune mechanisms. TNBS is a hapten-carrier model that renders colonic proteins immunogenic with a Th1 predominant response, although the type of cytokine response is strongly strain-dependent. Oxalazone-induced colitis is also a hapten-carrier model, but it induces a Th2 predominant response.2 The T cell adoptive transfer colitis model is useful for studying T cell subpopulations transferred into immunodeficient mice.18 Our colitis model has several advantages including spontaneous rapid onset, 100% penetrance, progressively severe phenotype, and defined Th1/Th17-predominant cytokine profile. We have bred over 200 CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice, and all of them showed weight loss and colitis by 4 months of age and scarified due to the development of colitis. We believe the our spontaneous colitis model will be a valuable tool to investigate the impact of impaired T cell function on gut homeostasis with specific focus on m6A methylation in regulation regulatory T cell function. In our model, the inflammation is in the colon and not the small bowel, and it may be a suitable model to study ulcerative colitis rather than Crohn’s disease, as ulcerative colitis has inflammation limited to the colon and Crohn’s disease frequently has both small and large bowel inflammation. While the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice do not seem to develop any other pathology, this does not rule out a possible immune deficiency–like phenotype. It would be of future interest to investigate whether this model would be suitable to study immune deficiency syndromes that primarily affect the gut.
The mRNA m6A methylation has been shown to be an important post-transcriptional gene regulation mechanism.3,23,24 This modification is added to the mRNAs by a methyltransferase complex with the catalytic core containing METTL3 and METTL14. It has previously been reported that lineage specific deletion of Mettl3 in mouse T cells leads to chronic inflammation in the intestine after 3 months of age.17 This is similar to the phenotype we observed in our mice with lineage specific deletion of Mettl14 in T cells, except that our mice developed colitis as early as week 6, and the inflammation became progressively more severe over time. Additionally, it was reported that mice with Foxp3-mediated deletion of Mettl3 in regulatory T cells develop a severe systemic autoimmune response after weaning and start to die in 8–9 weeks.17 The regulatory T cells deficient in Mettl3 lost their ability to suppress naïve T cell proliferation. In our colitis model, the deletion of Mettl14 also leads to Treg dysfunction with loss of suppressive capacity. While there also appears to be a dysfunction in the naïve T cells similar to that reported in the Mettl3-deficient naïve T cells,14 the Treg dysfunction is predominant because the mice developed a proinflammatory colitis phenotype that could be reversed by adoptively transferring WT Treg cells.
The causative role between intestinal microbiota and development of IBD is not yet established. However, multiple evidences support the gut microbiome’s involvement in IBD pathogenesis. Many studies have shown that patients with IBD have reduced bacterial diversity, but no causative microorganism has been consistently implicated to cause IBD. In patients with IBD that have their fecal stream diverted through a loop ileostomy, their colitis often improves.25 Patients with Crohn’s disease treated with antibiotics may experience a modest improvement in their disease activity.26 In patients with ulcerative colitis, an intensive multidonor fecal microbiota transplant over an 8-week period has been shown to improve their ulcerative colitis.27 The fecal microbiota transplantation was associated with a significantly increased fecal microbiota diversity.27 Prepared donor fecal microbiota transplantation has been shown to result in a higher likelihood of remission at 8 weeks compared with autologous fecal microbiota transplantation in patients with ulcerative colitis.28 In our colitis model, there is a decrease in both alpha and beta diversity associated with the development of colitis. The fecal microbiota are necessary for the development of colitis because the mice did not develop colitis when housed in a germ-free facility. We have additionally found that there are over 600 genes with differential m6A level in CD4+ T cells isolated from germ-free mice compared with CD4+ T cells isolated from specific pathogen–free mice. This shows that the gut microbiota can regulate the m6A epitranscriptome in CD4+ T cells. Similar results have been shown at the organ level in which the microbiome has been reported to affect m6A level in brain, intestine, and liver.29 The most significantly affected pathway is the NOTCH signaling pathway, which has been shown to regulate T cell homeostasis and differentiation.30 It is not clear whether the dysbiosis leads to the inflammation or is a secondary phenomenon. This would be an area of further investigation.
Thousands of mRNAs could be modified with m6A. It was previously reported that in the Mettl3-deficient regulatory T cells, the Treg dysfunction is potentially mediated by increased SOCS level that suppresses the IL-2-Stat5 signaling pathway.17 There are likely additional pathways affected by the m6A modification that are important in maintaining Treg function. One of the subsets of Treg cells are the Foxp3+ RORγt+ Treg cells that exhibit enhanced suppressive capacity in vivo.19 We have found that the Mettl14-deficient Treg cells have decreased RORγt expression that likely contributes to their decreased suppressive capacity in vivo. The iTreg cells have been shown to act synergistically with natural Treg cells to control experimental colitis.20 We showed that Mettl14 deficiency causes impaired induction of iTreg cells from naïve T cells in vitro. This could be a mechanism that contributes to the Treg dysfunction seen in the CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice. We found over 4000 genes with differential m6A expression in the iTreg cells compared with naïve T cells. Significantly affected pathways include gene expression, cell cycle and post-translational protein modification. Other pathways affected by m6A modifications in Treg cells would be interesting areas of future investigation.
In summary, we have identified a new spontaneous colitis model mediated by T cell deficiency of METTL14, a component of an m6A writer. The colitis development is dependent on dysfunctional Treg cells and the intestinal microbiota. This model likely represents a new tool for elucidating pathogenic mechanisms, studying the contribution of intestinal microbiome and preclinical testing of therapeutic agents for IBD.
Materials and Methods
Mice
The generation of Mettl14FL/FL mice have been described previously.13 The Mettl14FL/FL mice were crossed with CD4-Cre+/Tg mice (The Jackson Laboratory, Bar Harbor, ME) to generate CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice and littermate control mice. The Mettl14FL/FL mice were crossed with Foxp3YFP–Cre mice (The Jackson Laboratory) to generate Foxp3YFP–Cre Mettl14FL/FL conditional knockout mice and littermate control mice. Littermate control animals were used for all experiments. Animals were housed under either specific pathogen–free conditions or in our gnotobiotic facility. The conditional knockout mice and littermate control mice were housed together to minimize cage differences in microbiome. The Institutional Animal Care and Use Committee of the University of Chicago approved the use of animals in these experiments.
T Cell Transfer Colitis Model
The T cell transfer colitis model has been described previously.18 Briefly, for transfer of CD4+ naïve T cells, splenic CD4+ cells from CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice and littermate control mice were isolated with CD4+ T cell isolation kit II (Miltenyi Biotec, Bergisch Gladbach, Germany) according to manufacturer’s protocol. The isolated CD4+ T cells were then stained with CD4-FITC, CD25-PE, CD62L-BV421 and CD44-APC (BioLegend, San Diego, CA). Naïve T cells were fluorescence-activated cell sorter (FACS) sorted as CD4+, CD25–, CD62L+, and CD44– cells. Each recipient Rag1–/– mice was injected with 5 × 105 naïve T cells. For co-transfer of CD4+ naïve T cells and Treg cells, naïve T cells were isolated as described previously from C57BL6-CD45.1 mice in which the CD45.1 marker can be used as a donor marker for naïve T cells. For isolation of Treg cells, splenic CD4+ cells from Foxp3YFP–Cre Mettl14FL/FL conditional knockout mice and littermate control mice were isolated with CD4+ T cell isolation kit II (Miltenyi Biotec). The T cells were then stained with CD4-APC-Fire750 (BioLegend). Treg cells were sorted as YFP+ CD4+ cells. Recipient Rag1–/– mice were injected with naïve T cells and Treg cells at a 2:1 ratio. The mice were then weighed every 3–4 days. Mice were scarified 7–8 weeks after T cell transfer.
FACS Analysis
Mouse spleen or mesenteric lymph node was collected, and single-cell preparation was made by grinding the spleen or lymph node between 2 glass slides. Red blood cell lysis was performed with ACK (ammonium-chloride-potassium) lysing buffer according to manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). Cells are filtered with a 70-μm filter and stained for FACS analysis according to previously published protocols.31 The CD3-BUV395, CD45.2-PE-CF594, and RORγt-BV421 antibodies are from BD Biosciences (Franklin Lakes, NJ). The CD4-FITC, CD25-PE, CD62L-BV421, CD44-APC, CD45.1-BUV785 antibodies are from BioLegend.
Histology
Mouse tissues were collected and fixed in 10% neutral buffered formalin (Thermo Fisher Scientific). Tissues were embedded in paraffin and stained with hematoxylin and eosin.
Quantitative Reverse-Transcription Polymerase Chain Reaction
Total RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific). Samples were analyzed by TaqMan quantitative reverse-transcription polymerase chain reaction. TaqMan gene expression master mix and all primer/probe sets were obtained from Thermo Fisher Scientific. Relative expression was calculated using the comparative CT method.32
Antibiotics Treatment
A combination of ciprofloxacin (200 mg/L) and metronidazole (600 mg/L) was added to drinking water of CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice. The antibiotics doses have previously been shown to attenuate experimental colitis in mice.33
Adoptive Transfer of Treg Cells
Treg cells were isolated by FACS sorting from Foxp3YFP mice based on YFP expression. The CD4-Cre+/Tg Mettl14FL/FL conditional knockout mice were adoptively transferred 1 × 106 Treg cells on day 0, day 7, and day 14. Control mice were injected with phosphate-buffered saline on these days. Mice were sacrificed at day 25.
Culture and FACS Analysis of iTreg Cells
Culture of iTreg cells were performed according to previously described protocol.34 Briefly, CD4+ naïve T cells were cultured in Advanced RPMI 1640 + 10% fetal bovine serum + 1% penicillin/streptomycin + 2-mM L-glutamine + 20-ng/mL transforming growth factor beta + 40-ng/mL IL-2 + 1-nM all-trans retinoic acid. Cells were collected after 96 hours of culture. Foxp3 expression was analyzed by intracellular FACS staining with eBioscience mouse regulatory T cell staining kit 1 (Thermo Fisher Scientific) according to manufacturer’s protocol.
m6A Sequencing and Analysis
m6A sequencing was performed according to previously published protocol.35 RNA was extracted by TRIZOL reagent and Direct-zol RNA miniprep (Zymo Research, Irvine, CA). PolyA mRNA was enriched with a Dynabeads mRNA DIRECT Purification Kit (Thermo Fisher Scientific). RNA fragmentation was performed with Bioruptor Pico sonicator (Diagenode, Denville, NJ). m6A-IP was performed using EpiMark N6-Methyladenosine Enrichment Kit (New England Biolabs, Ipswich, MA). Library preparation was done with SMARTer Stranded Total RNA-Seq Kit v2 (Takara, Kyoto, Japan). Sequencing was performed by the University of Chicago Genomics Facility on an Illumina HiSeq4000 (Illumina, San Diego, CA). For data analysis, reads were mapped to the mouse genome and transcriptome mm10 using HISAT (version 2.1.0).36 m6A peaks in the mapped reads were called by RADAR.37 Parameters of P value <.05 and absolute (log2 fold change) >1 was applied to call significant peaks. Pathway analysis was performed using the ToppGene suite (toppgene.cchmc.org).38 The sequencing data was deposited into the Gene Expression Omnibus database (Accession number: GSE152234).
Microbiome Analysis
Fecal pellets were placed into a MP Bio PowerMag Soil DNA Isolation Bead Plate (MP Bio, Santa Ana, CA). DNA was extracted following MP Bio’s instructions on a KingFisher robot. Bacterial 16S rRNA genes were polymerase chain reaction–amplified with dual-barcoded primers targeting the V4 region.39 Amplicons were sequenced with an Illumina MiSeq using the 300-bp paired-end kit. Sequences were denoised, taxonomically classified using Greengenes (v. 13.8) as the reference database, and clustered into operational taxonomic units (OTUs) with 97% similarity using the Mothur software package (v. 1.39.5).40 Alpha diversity was estimated with the Shannon index on raw OTU abundance tables. Differences in the significance of diversity were tested with an analysis of variance. To estimate beta diversity across samples, we excluded OTUs occurring in fewer than 10% of the samples with a count of <3 and computed Bray-Curtis indices. We visualized beta diversity, emphasizing differences across samples, using principal coordinate analysis ordination. All analyses were conducted in the R environment (v. 3.5.1) (R Foundation for Statistical Computing, Vienna, Austria). The DNA extraction and microbiome analysis were performed by Microbiome Insights, Inc (Vancouver, Canada).
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, San Diego, CA). Student’s t test and 1-way analysis of variance were used where appropriate. P values <.05 were considered statistically significant. All authors had access to the study data and had reviewed and approved the final manuscript.
Footnotes
CRediT Authorship Contributions Thomas X. Lu (Conceptualization: Equal; Data curation: Lead; Formal analysis: Lead; Investigation: Equal; Methodology: Equal; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Equal) Zhong Zheng (Conceptualization: Equal; Data curation: Equal; Formal analysis: Supporting; Investigation: Equal; Writing – original draft: Supporting) Linda Zhang (Data curation: Supporting; Formal analysis: Equal; Visualization: Supporting; Writing – review & editing: Supporting) Hui-Lung Sun (Data curation: Supporting; Formal analysis: Supporting) Marc Bissonnette (Conceptualization: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting) Haochu Huang (Conceptualization: Supporting; Funding acquisition: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting) Chuan He, Ph.D. (Conceptualization: Equal; Funding acquisition: Lead; Project administration: Lead; Supervision: Lead; Writing – original draft: Supporting; Writing – review & editing: Equal)
Conflicts of Interest This author discloses the following: Chuan He is a scientific founder and a member of the scientific advisory board of Accent Therapeutics, Inc. The remaining authors disclose no conflicts.
Funding This work was supported by the research training grant in digestive health, diseases, and nutrition (National Institutes of Health [NIH] T32DK007404 [to Thomas X. Lu]) and TL1 training grant in translational science and medical informatics (NIHTL1TR002388 [to Thomas X. Lu]). Additionally, this work was supported by NIHR21AI128469 (to Chuan He), RM1HG008935 (to Chuan He), the University of Chicago Digestive Diseases Research Center (NIHP30DK42086), and the Duchossois Family Institute. Chuan He is a Howard Hughes Medical Institute Investigator.
References
- 1.Ng S.C., Shi H.Y., Hamidi N., Underwood F.E., Tang W., Benchimol E.I., Panaccione R., Ghosh S., Wu J.C.Y., Chan F.K.L., Sung J.J.Y., Kaplan G.G. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet. 2018;390:2769–2778. doi: 10.1016/S0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Kiesler P., Fuss I.J., Strober W. Experimental models of inflammatory bowel diseases. Cell Mol Gastroenterol Hepatol. 2015;1:154–170. doi: 10.1016/j.jcmgh.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frye M., Harada B.T., Behm M., He C. RNA modifications modulate gene expression during development. Science. 2018;361:1346–1349. doi: 10.1126/science.aau1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y., Li Y., Toth J.I., Petroski M.D., Zhang Z., Zhao J.C. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., Dai Q., Chen W., He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meng T.G., Lu X., Guo L., Hou G.M., Ma X.S., Li Q.N., Huang L., Fan L.H., Zhao Z.H., Ou X.H., OuYang Y.C., Schatten H., Li L., Wang Z.B., Sun Q.Y. Mettl14 is required for mouse postimplantation development by facilitating epiblast maturation. FASEB J. 2019;33:1179–1187. doi: 10.1096/fj.201800719R. [DOI] [PubMed] [Google Scholar]
- 9.Wu Y., Xie L., Wang M., Xiong Q., Guo Y., Liang Y., Li J., Sheng R., Deng P., Wang Y., Zheng R., Jiang Y., Ye L., Chen Q., Zhou X., Lin S., Yuan Q. Mettl3-mediated m(6)A RNA methylation regulates the fate of bone marrow mesenchymal stem cells and osteoporosis. Nat Commun. 2018;9:4772. doi: 10.1038/s41467-018-06898-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C.X., Cui G.S., Liu X., Xu K., Wang M., Zhang X.X., Jiang L.Y., Li A., Yang Y., Lai W.Y., Sun B.F., Jiang G.B., Wang H.L., Tong W.M., Li W., Wang X.J., Yang Y.G., Zhou Q. METTL3-mediated m6A modification is required for cerebellar development. PLoS Biol. 2018;16 doi: 10.1371/journal.pbio.2004880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., Li Y., Yue M., Wang J., Kumar S., Wechsler-Reya R.J., Zhang Z., Ogawa Y., Kellis M., Duester G., Zhao J.C. N(6)-methyladenosine RNA modification regulates embryonic neural stem cell self-renewal through histone modifications. Nat Neurosci. 2018;21:195–206. doi: 10.1038/s41593-017-0057-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K., Carter A.C., Flynn R.A., Zhou C., Lim K.S., Dedon P., Wernig M., Mullen A.C., Xing Y., Giallourakis C.C., Chang H.Y. m(6)A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L., Kim S., Wang X., Dore L.C., Jin P., Regot S., Zhuang X., Canzar S., He C., Ming G.L., Song H. Temporal control of mammalian cortical neurogenesis by m(6)A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H.B., Tong J., Zhu S., Batista P.J., Duffy E.E., Zhao J., Bailis W., Cao G., Kroehling L., Chen Y., Wang G., Broughton J.P., Chen Y.G., Kluger Y., Simon M.D., Chang H.Y., Yin Z., Flavell R.A. m(6)A mRNA methylation controls T cell homeostasis by targeting the IL-7/STAT5/SOCS pathways. Nature. 2017;548:338–342. doi: 10.1038/nature23450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S., Ben-Haim M.S., Eyal E., Yunger S., Pinto Y., Jaitin D.A., Viukov S., Rais Y., Krupalnik V., Chomsky E., Zerbib M., Maza I., Rechavi Y., Massarwa R., Hanna S., Amit I., Levanon E.Y., Amariglio N., Stern-Ginossar N., Novershtern N., Rechavi G., Hanna J.H. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 16.A SPECIAL MEETING REVIEW EDITION: CCFA/Advances in Inflammatory Bowel Diseases 2015: Highlights in Ulcerative Colitis and Crohn’s Disease: A Review of Selected Presentations From the CCFA/Advances in Inflammatory Bowel Diseases 2015 Clinical and Research Conference ∗ December 10-12, 2015 ∗ Orlando, FloridaSpecial Reporting on:∗ New and Future Adhesion Molecule-Based Therapies in IBD∗ Efficacy and Safety of Vedolizumab for Inflammatory Bowel Disease in Clinical Practice∗ A Multicenter, Double-Blind, Placebo-Controlled Phase 3 Study of Ustekinumab, a Human IL-12/23p40 Monoclonal Antibody, in Moderate-Severe Crohn’s Disease Refractory to Anti-TNFalpha: UNITI-1∗ Intravenous Iron Sucrose for Treatment of Iron Deficiency Anemia in Pediatric Inflammatory Bowel Disease∗ Does Vedolizumab Affect Postoperative Outcomes in Patients Undergoing Abdominal Operations for Inflammatory Bowel Disease?PLUS Meeting Abstract Summaries With Expert Commentary by: Gary R. Lichtenstein, MD. Professor of Medicine. Director, Center for Inflammatory Bowel Disease. University of Pennsylvania Health System. Hospital of the University of Pennsylvania. Philadelphia, Pennsylvania. Gastroenterol Hepatol (N Y) 2016;12(2 Suppl 1):1–20. [PMC free article] [PubMed] [Google Scholar]
- 17.Tong J., Cao G., Zhang T., Sefik E., Amezcua Vesely M.C., Broughton J.P., Zhu S., Li H., Li B., Chen L., Chang H.Y., Su B., Flavell R.A., Li H.B. m(6)A mRNA methylation sustains Treg suppressive functions. Cell Res. 2018;28:253–256. doi: 10.1038/cr.2018.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ostanin D.V., Bao J., Koboziev I., Gray L., Robinson-Jackson S.A., Kosloski-Davidson M., Price V.H., Grisham M.B. T cell transfer model of chronic colitis: concepts, considerations, and tricks of the trade. Am J Physiol Gastrointest Liver Physiol. 2009;296:G135–G146. doi: 10.1152/ajpgi.90462.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang B.H., Hagemann S., Mamareli P., Lauer U., Hoffmann U., Beckstette M., Fohse L., Prinz I., Pezoldt J., Suerbaum S., Sparwasser T., Hamann A., Floess S., Huehn J., Lochner M. Foxp3(+) T cells expressing RORgammat represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 2016;9:444–457. doi: 10.1038/mi.2015.74. [DOI] [PubMed] [Google Scholar]
- 20.Haribhai D., Lin W., Edwards B., Ziegelbauer J., Salzman N.H., Carlson M.R., Li S.H., Simpson P.M., Chatila T.A., Williams C.B. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keubler L.M., Buettner M., Hager C., Bleich A. A multihit model: colitis lessons from the interleukin-10-deficient mouse. Inflamm Bowel Dis. 2015;21:1967–1975. doi: 10.1097/MIB.0000000000000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devkota S., Wang Y., Musch M.W., Leone V., Fehlner-Peach H., Nadimpalli A., Antonopoulos D.A., Jabri B., Chang E.B. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10–/– mice. Nature. 2012;487:104–108. doi: 10.1038/nature11225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y., Hsu P.J., Chen Y.S., Yang Y.G. Dynamic transcriptomic m(6)A decoration: writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74:640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Russell T.A., Dawes A.J., Graham D.S., Angarita S.A.K., Ha C., Sack J. Rescue diverting loop ileostomy: an alternative to emergent colectomy in the setting of severe acute refractory IBD-colitis. Dis Colon Rectum. 2018;61:214–220. doi: 10.1097/DCR.0000000000000985. [DOI] [PubMed] [Google Scholar]
- 26.Nitzan O., Elias M., Peretz A., Saliba W. Role of antibiotics for treatment of inflammatory bowel disease. World J Gastroenterol. 2016;22:1078–1087. doi: 10.3748/wjg.v22.i3.1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paramsothy S., Kamm M.A., Kaakoush N.O., Walsh A.J., van den Bogaerde J., Samuel D., Leong R.W.L., Connor S., Ng W., Paramsothy R., Xuan W., Lin E., Mitchell H.M., Borody T.J. Multidonor intensive faecal microbiota transplantation for active ulcerative colitis: a randomised placebo-controlled trial. Lancet. 2017;389:1218–1228. doi: 10.1016/S0140-6736(17)30182-4. [DOI] [PubMed] [Google Scholar]
- 28.Costello S.P., Hughes P.A., Waters O., Bryant R.V., Vincent A.D., Blatchford P., Katsikeros R., Makanyanga J., Campaniello M.A., Mavrangelos C., Rosewarne C.P., Bickley C., Peters C., Schoeman M.N., Conlon M.A., Roberts-Thomson I.C., Andrews J.M. Effect of fecal microbiota transplantation on 8-week remission in patients with ulcerative colitis: a randomized clinical trial. JAMA. 2019;321:156–164. doi: 10.1001/jama.2018.20046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jabs S., Biton A., Becavin C., Nahori M.A., Ghozlane A., Pagliuso A., Spano G., Guerineau V., Touboul D., Giai Gianetto Q., Chaze T., Matondo M., Dillies M.A., Cossart P. Impact of the gut microbiota on the m(6)A epitranscriptome of mouse cecum and liver. Nat Commun. 2020;11:1344. doi: 10.1038/s41467-020-15126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandstadter J.D., Maillard I. Notch signalling in T cell homeostasis and differentiation. Open Biol. 2019;9:190187. doi: 10.1098/rsob.190187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holmes K., Lantz L.M., Fowlkes B.J., Schmid I., Giorgi J.V. Preparation of cells and reagents for flow cytometry. Curr Protoc Immunol. 2001 doi: 10.1002/0471142735.im0503s44. Chapter 5:Unit 5 3. [DOI] [PubMed] [Google Scholar]
- 32.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Madsen K.L., Doyle J.S., Tavernini M.M., Jewell L.D., Rennie R.P., Fedorak R.N. Antibiotic therapy attenuates colitis in interleukin 10 gene-deficient mice. Gastroenterology. 2000;118:1094–1105. doi: 10.1016/s0016-5085(00)70362-3. [DOI] [PubMed] [Google Scholar]
- 34.Karlsson F., Robinson-Jackson S.A., Gray L., Zhang S., Grisham M.B. Ex vivo generation of regulatory T cells: characterization and therapeutic evaluation in a model of chronic colitis. Methods Mol Biol. 2011;677:47–61. doi: 10.1007/978-1-60761-869-0_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu P.J., He C. Identifying the m(6)A methylome by affinity purification and sequencing. Methods Mol Biol. 2018;1649:49–57. doi: 10.1007/978-1-4939-7213-5_3. [DOI] [PubMed] [Google Scholar]
- 36.Kim D., Langmead B., Salzberg S.L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Z., Zhan Q., Eckert M., Zhu A., Chryplewicz A., De Jesus D.F., Ren D., Kulkarni R.N., Lengyel E., He C., Chen M. RADAR: differential analysis of MeRIP-seq data with a random effect model. Genome Biol. 2019;20:294. doi: 10.1186/s13059-019-1915-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kozich J.J., Westcott S.L., Baxter N.T., Highlander S.K., Schloss P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79:5112–5120. doi: 10.1128/AEM.01043-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., Sahl J.W., Stres B., Thallinger G.G., Van Horn D.J., Weber C.F. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]