Abstract
Sensitive assays are essential for the accurate identification of individuals infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here, we report a multiplexed assay for the fluorescence-based detection of seroconversion in infected individuals from less than 1 µl of blood, and as early as the day of the first positive nucleic acid test after symptom onset. The assay uses dye-encoded antigen-coated beads to quantify the levels of immunoglobulin G (IgG), IgM and IgA antibodies against four SARS-CoV-2 antigens. A logistic regression model trained using samples collected during the pandemic and samples collected from healthy individuals and patients with respiratory infections before the first outbreak of coronavirus disease 2019 (COVID-19) was 99% accurate in the detection of seroconversion in a blinded validation cohort of samples collected before the pandemic and from patients with COVID-19 five or more days after a positive nasopharyngeal test by PCR with reverse transcription. The high-throughput serological profiling of patients with COVID-19 allows for the interrogation of interactions between antibody isotypes and viral proteins, and should help us to understand the heterogeneity of clinical presentations.
Subject terms: Diagnostic markers, Viral infection
A multiplexed fluorescence-based assay detects seroconversion in individuals infected with SARS-CoV-2 from less than 1 µl of blood as early as the day of the first positive nasopharyngeal nucleic acid test after symptom onset.
Main
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel beta-coronavirus that has caused the coronavirus disease 2019 (COVID-19) pandemic1–3. To identify infected individuals and contain the spread of the disease, rapid and accurate population-wide screening is essential. Currently, nucleic acid amplification tests (NAATs) of nasopharyngeal swabs are the primary method used to identify SARS-CoV-2-infected individuals2. These tests can diagnose disease during a narrow window of active infection and have limited clinical sensitivity4–7. Thus, additional complementary methods are needed to identify individuals who are or have been infected with SARS-CoV-2. Robust serological assays that detect antibodies against SARS-CoV-2 can fulfil this critical need. These assays could be used to identify symptomatic infected individuals who have tested negative by NAATs. Furthermore, serological assays could be used to identify individuals who have been infected with SARS-CoV-2 and were asymptomatic or had mild symptoms, thereby providing a better understanding of how widespread the virus is within a population. This information can aid epidemiologists in determining a more accurate population prevalence of antibodies to SARS-CoV-2 and contribute to contact tracing operations, key to curbing the pandemic8. Finally, antibody testing will be necessary in order to run highly powered and accurate prevention trials9.
Many serological enzyme-linked immunosorbent assays (ELISAs) have been developed recently to detect anti-SARS-CoV-2 antibodies. However, these assays have important limitations4,8,10–19. First, most assays lack the ability to detect antibodies at early stages of infection. Second, false positive results can occur due to non-specific binding from high levels of pre-existing antibodies in blood20. Finally, these assays lack the resolution to quantify changes in the immune response over time, which may be key in understanding clinical progression.
To address these limitations, we developed an ultra-sensitive, multiplexed single molecule array (Simoa) assay for immunoglobulin isotypes against immunogenic SARS-CoV-2 proteins, providing us with detailed information about the immune response to SARS-CoV-2 at early stages and throughout the course of infection21. Simoa provides up to 1,000-fold improvement in analytical sensitivity over the standard ELISA22,23. This ultra-sensitivity enables samples to be highly diluted, resulting in significantly reduced non-specific binding. Additionally, unlike the standard ELISA, Simoa has a wide dynamic range and allows precise quantification of multiple analytes over a concentration range of four orders of magnitude. This feature is particularly important since antibody levels can vary greatly among individuals and may fluctuate significantly during different stages of infection within the same individual, and thus it is advantageous to have a single assay format that can accurately measure antibody levels over a wide concentration range.
Our multiplexed, ultra-sensitive Simoa assay detects the binding of immunoglobulin G (IgG), IgM and IgA against SARS-CoV-2 to four viral targets (spike protein, the S1 subunit, the receptor-binding domain (RBD) and nucleocapsid), enabling the quantification of 12 binding interactions. In this assay format, four types of dye-encoded paramagnetic beads are each coated with one of the four SARS-CoV-2 targets (Fig. 1 and Methods). We used this assay to measure the levels of IgG, IgM and IgA against four viral targets in plasma samples from patients who were classified as SARS-CoV-2 negative or positive by nasopharyngeal PCR with reverse transcription (RT–PCR) testing. We also measured these immunoglobulins in samples from patients with respiratory infections and from healthy individuals, collected before the start of the COVID-19 pandemic. We show that these serological assays detect anti-SARS-CoV-2 antibodies with high sensitivity and specificity at both early and late stages of infection.
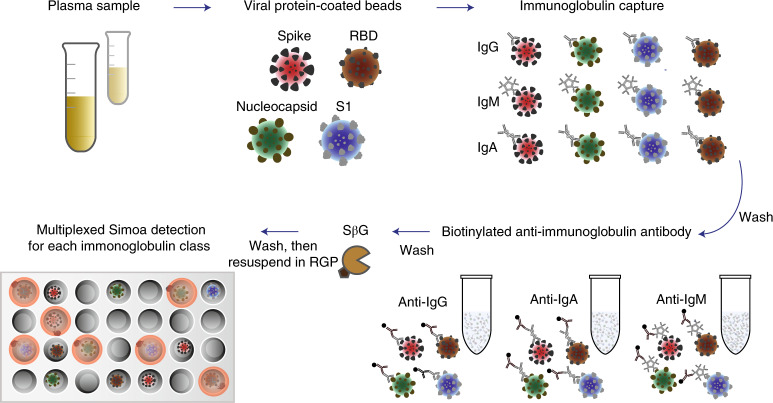
Fig. 1. Schematic of the Simoa serological assay.
Plasma is incubated with four types of dye-encoded beads that are each coupled to one of four viral targets (spike, S1, RBD and nucleocapsid). IgG, IgA and IgM antibodies specific to the SARS-CoV-2 targets bind to the viral antigen-coated beads. After washing, beads are introduced to biotinylated anti-human immunoglobulin antibodies to label either IgG, IgM or IgA in the different reactions. After additional washes, the enzyme SβG is introduced. The beads are washed, resuspended in fluorogenic RGP and loaded into a 216,000-microwell array for multicolour imaging.
Results
Detection of anti-SARS-CoV-2 antibodies in a training cohort
After testing a small discovery cohort in which our assay showed a clear separation between patients with COVID-19 and the pre-pandemic cohort (Supplementary Fig. 5), we further validated our method using a training cohort of 472 samples that included pre-pandemic samples collected from patients with other viral respiratory infections before 1 October 2019. This cohort is a critical control for validating any SARS-CoV-2 serological assay, as cross-reactivity with other anti-viral antibodies is a likely source of false positive results in SARS-CoV-2 serological tests. This training cohort consists of four groups: (1) pre-pandemic plasma samples from healthy adults with no recorded respiratory infection in their medical history at the time of collection (100 individuals); (2) pre-pandemic samples collected from adults with a documented respiratory infection (including bacterial and viral pneumonia) within the preceding 2 months (100 individuals); (3) samples from adults who tested positive for SARS-CoV-2 by nasopharyngeal RT–PCR (172 samples from 91 individuals taken at multiple time points); and (4) samples from symptomatic adults who tested negative for SARS-CoV-2 by nasopharyngeal RT–PCR (100 samples from 95 individuals taken at multiple time points).
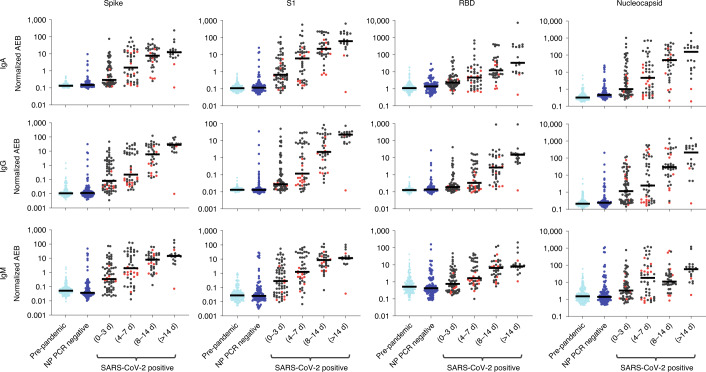
Figure 2 shows IgG, IgM and IgA normalized mean average enzymes per bead (AEB) levels in SARS-CoV-2 nasopharyngeal RT–PCR-positive samples, which were divided into four subgroups based on the number of days after symptom onset. In addition, Fig. 2 shows the immunoglobulin levels in the pre-pandemic group and the SARS-CoV-2 nasopharyngeal RT–PCR-negative group. All measurements were made in duplicate and the coefficient of variation values are plotted in the Supplementary Fig. 6. Normalization was performed using a standard set of calibrators in each run. These calibrators were produced by serially diluting a large volume of blood from seroconverted patients. All four RT–PCR-confirmed SARS-CoV-2-positive subgroups (0–3 d, 4–7 d, 8–14 d and >14 d after positive nasopharyngeal RT–PCR) showed higher median IgG, IgM and IgA levels in SARS-CoV-2-positive samples compared with the pre-pandemic group (see Supplementary Table 6 for all Mann–Whitney U-test results). As in the discovery cohort, all 12 antigen–antibody isotype combinations showed an increase in median immunoglobulin levels over the course of the disease for those who tested positive for SARS-CoV-2 by nasopharyngeal RT–PCR but were not considered immunocompromised. Interestingly, some symptomatic individuals who tested negative for SARS-CoV-2 by nasopharyngeal RT–PCR displayed anti-SARS-CoV-2 antibody signals well above those in the pre-pandemic group; these may represent individuals infected with SARS-CoV-2 who falsely tested negative by RT–PCR.
Fig. 2. Profiling seroconversion in COVID-19.
Simoa serological assay results for IgG, IgM and IgA against the four viral targets (spike, S1 subunit, RBD and nucleocapsid) for pre-pandemic samples (light blue; n = 200), nasopharyngeal (NP) PCR-negative samples (dark blue; n = 100) and SARS-CoV-2-positive samples (black (immunocompetent; n = 141) or red (immunosuppressed; n = 31)). The SARS-CoV-2-positive samples were divided into four groups according to time since since the first positive nasopharyngeal RT–PCR test. Black lines indicate the median normalized AEB value of each population.
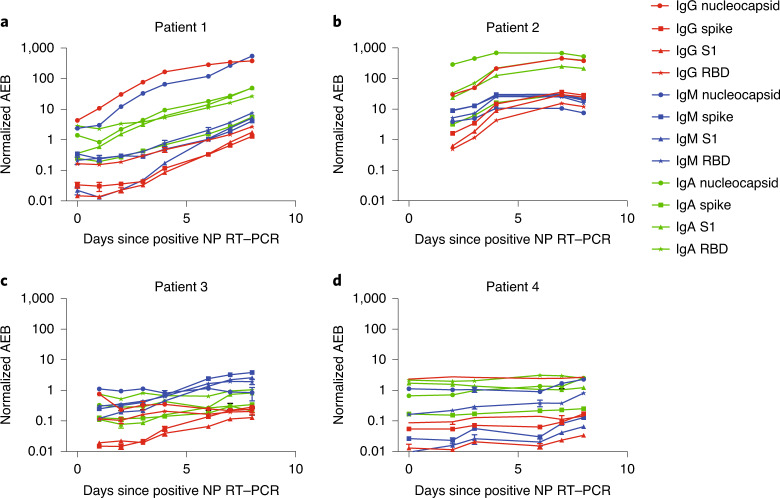
Included in the SARS-CoV-2 validation cohort were four patients who were sampled longitudinally over the course of 8 d (Fig. 3). Within this group, the two patients who survived longer (patient 1 (eventual recovery) and patient 2 (20-d survival after COVID-19 diagnosis)) had on average 10–100 times higher anti-SARS-CoV-2 antibody levels 1 week following infection compared with the other two patients (8- and 11-d survival after COVID-19 diagnosis). Furthermore, the only patient who ultimately recovered (patient 1) mounted a robust and diverse humoral immune response (characterized by a 10- to 100-fold increase in all immunoglobulin isotypes against all four viral antigens) within the first week of infection, while the immune response in patients who died from COVID-19 appeared substantially weaker in terms of absolute anti-SARS-CoV-2 antibody levels (patients 3 and 4), as did the rate and diversity of antibody production (patients 2, 3 and 4). These observations suggest that the magnitude and antibody specificity of the humoral immune response against SARS-CoV-2 may influence disease outcome and highlights the ability of the Simoa assay to profile the dynamics of this response at high resolution throughout the course of infection.
Fig. 3. Profiling the seroconversion time course in COVID-19.
a–d, Normalized AEB over the 10 d since a positive nasopharyngeal RT–PCR for patients 1 (a), 2 (b), 3 (c) and 4 (d). Patient 1 was a 67-year-old man who recovered 10 d after diagnosis with COVID-19. Patient 2 was a 50-year-old man with multiple comorbidities who died from acute respiratory distress syndrome 20 d after diagnosis with COVID-19. He received remdesivir from days 1–5. Patient 3 was a 50-year-old man with pancytopenia and B-cell acute lymphoblastic leukaemia. He died from acute respiratory distress syndrome 8 d after diagnosis with COVID-19. Patient 4 was an 89-year-old man who died from hypoxaemic respiratory failure 8 d after diagnosis with COVID-19. He received hydroxychloroquine from days 1–5. The circle, square, triangle and star represent the mean of two replicate measurements, while the error bars represent the standard deviation.
To determine the ability of the serological Simoa assays to detect early seroconversion, we separated patients with nasopharyngeal RT–PCR-confirmed COVID-19 into two subgroups based on whether samples were collected: (1) within the first week following a positive nasopharyngeal RT–PCR test (classified as early stage); or (2) after the first week (classified as late stage). Our findings in the discovery cohort prompted us to exclude immunocompromised individuals in order to build a more robust seroconversion classification model (31 time points, representing ten patients). We assessed the performance of the Simoa assays using multivariate analyses comparing early-stage cases, late-stage cases or all positive cases against the pre-pandemic group. A noteworthy feature of the Simoa serological assay is that a single sample provides information on the antibody response to four viral targets, each across three immunoglobulin isotypes. This feature allowed us to train and validate a logistic regression model with all 12 parameters for each of the three case groups. To assess the possibility of classifying seroconversion using a smaller set of parameters, two additional models that included only a subset of the parameters, chosen carefully using cross-validation, were developed (Supplementary Tables 7 and 8 and Methods). The final subset of markers selected were IgA S1, IgA nucleocapsid, IgG nucleocapsid and IgG spike for the early-stage model and IgA S1 for the late-stage model.
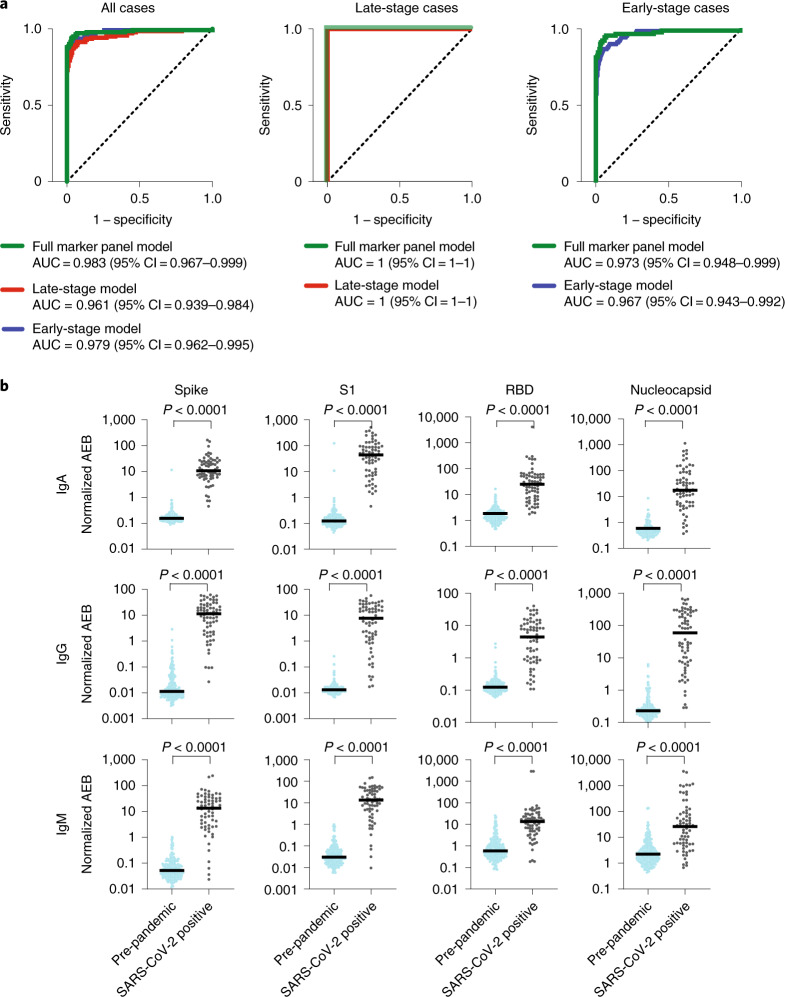
Figure 4a shows the receiver operating characteristic (ROC) curves for each of the tested models for all cases, only early-stage cases and only late-stage cases. For the early-stage group, the area under the curve (AUC) was 0.9732 for the model using the full panel of markers and 0.9675 for the early-stage model. The AUC was 1.00 for the late-stage group, for both the late-stage model and the full panel of markers. Thus, the 12-parameter model had a sensitivity of 81%, with 100% specificity within the first week of a positive nasopharyngeal RT–PCR test and 100% sensitivity and specificity after the first week of a positive nasopharyngeal RT–PCR test. The lower sensitivity during the first week undoubtedly stems from a delayed immune response in some individuals. Furthermore, we assessed whether the group of symptomatic patients who were SARS-CoV-2 negative by nasopharyngeal RT–PCR (all of whom were initially tested given clinical suspicion for COVID-19) produced antibodies against SARS-CoV-2. Interestingly, 14 out of 100 patients within this group were above the seroconversion threshold. It is possible that these patients with negative nasopharyngeal RT–PCR results were actually infected with SARS-CoV-2 and in the process of seroconversion.
Fig. 4. Classification of COVID-19 using Simoa serological assays.
a, Left: ROC curves for all positive cases (n = 141) in the pre-pandemic cohort (n = 199) using either the full marker panel model (green), the early-stage model (blue) or the late-stage model (red). Middle: ROC curves for late-stage cases (more than 1 week after positive nasopharyngeal PCR test; n = 50) in the pre-pandemic cohort (n = 199) using either the full marker panel model (green) or the late-stage model (red). Right: ROC curves for early-stage cases (first week after positive nasopharyngeal PCR test; n = 91) in the pre-pandemic cohort (n = 199) using either the full marker panel model (green) or the early-stage model (blue). AUC values and 95% confidence intervals (CIs) are shown for each graph. b, Simoa serological assay results for IgG, IgM and IgA against the four viral targets (spike, S1 subunit, RBD and nucleocapsid) for pre-pandemic samples (light blue; n = 232) and nasopharyngeal RT–PCR-positive samples (grey; n = 68). Statistical significance was determined using Mann–Whitney U-tests. All U-tests were two tailed and did not correct for multiple comparisons. Black lines indicate the median normalized AEB value of each population.
Accuracy of Simoa serological assay in a blinded validation cohort
To further evaluate the classification accuracy of the three models, we tested a final independent set of blinded samples. This validation set consisted of 300 samples from: (1) 68 samples from 28 individuals who tested positive for SARS-CoV-2 using nasopharyngeal RT–PCR within the past 5–34 d; and (2) 232 pre-pandemic controls all collected before 1 October 2019, including 132 patients with a documented respiratory infection (including bacterial and viral pneumonia) within the preceding 31 d, as well as 100 healthy individuals with no recorded history of viral respiratory infection. Figure 4b shows IgG, IgM and IgA normalized mean AEB levels in SARS-CoV-2 nasopharyngeal RT–PCR-positive samples and the pre-pandemic controls. We classified these samples using the three models we trained on the training set (all cases versus all controls for each model). The signal threshold for a positive test result was determined based on the cut-off that yielded 100% specificity in the training set. Table 1 shows the sensitivity and specificity, as well as the positive and negative predictive values, for a population prevalence of 22.6%. The early model consisting of four parameters performed best, with 99% sensitivity and specificity, followed by the late model, which used only a single parameter. The full marker panel gave lower accuracy, with 100% sensitivity and 94% specificity.
Table 1.
Sensitivity, specificity, positive predictive value and negative predictive value for the validation set using the models created on the training set
| Sensitivity (%) | Specificity (%) | Positive predictive value (%) | Negative predictive value (%) | |
|---|---|---|---|---|
| Early-stage model | 99 | 99 | 97 | 100 |
| Late-stage model | 99 | 99 | 96 | 100 |
| Full marker panel model | 100 | 94 | 83 | 100 |
Discussion
Quantitative and sensitive SARS-CoV-2 serological assays are necessary to address unmet needs in the clinical setting and epidemiological studies, as well as therapeutic and vaccine development. Using the Simoa technology, we developed a highly sensitive serological assay for the detection of IgG, IgM and IgA in patients with COVID-19 against four SARS-CoV-2 targets in a high-throughput format. The ultra-sensitivity enables plasma to be diluted 4,000×, greatly reducing the degree of non-specific circulating immunoglobulin binding. Furthermore, at this dilution factor, our dynamic range spans four orders of magnitude, allowing for accurate quantification of the full range of disease from early to late stage. This dilution factor requires a minute sample size of less than 1 μl plasma. Although our Simoa serological assay is a powerful analytical tool with which to understand the host immune response of the COVID-19 pandemic, it requires a specialized instrument that is not widely available.
While the clinical sensitivity of current SARS-CoV-2 nasopharyngeal RT–PCR tests is still under scrutiny, a recent meta-analysis concluded that up to 29% of initially negative tests are false negatives6. Therefore, it was crucial to establish a true negative sample group in addition to the negative nasopharyngeal RNA test group when evaluating our serological assay for the COVID-19 pandemic. Our analysis includes samples collected from US adults presenting before 1 October 2019, and thus before the introduction of SARS-CoV-2 into the United States and the first documented case in the world. This cohort included 100 patients presenting with a recent respiratory infection. Although our pre-pandemic control samples were not assessed for other coronavirus infections at the time of their illness, we assessed them for antibodies to the four common human coronaviruses and observed that several samples had particularly high levels for one or more of these species (Supplementary Fig. 7). Our results suggest no significant cross-reactivity of antibodies to our selected target antigens before the SARS-CoV-2 outbreak, indicating minimal cross-reactivity among coronaviridae, or other respiratory viruses, in our assay.
In the training cohort, we were able to detect seroconversion as early as the day of the first positive nasopharyngeal PCR test after symptom onset. In a blinded validation set, which began at 5 d after a positive RT–PCR, both our one- and four-parameter models were 99% specific and sensitive while the 12-parameter model was 100% sensitive but only 94% specific. Additionally, the resolution of our method enables us to probe small variations in the immune response between patients and within a single patient.
While antibody testing is clearly key in curbing the COVID-19 pandemic, several limitations of antibody-based testing should be considered24,25. First, serological testing cannot replace NAATs for the diagnosis of COVID-19 due to the later onset of seroconversion. Second, until a concrete link between levels of anti-SARS-CoV-2 antibodies and immunity from reinfection can be established, it is not possible to make clinical decisions or to develop public health policy based on this type of testing. Finally, even once a link between antibody levels and immunity can be established, it will need to be determined how long immunity lasts for proper clinical decision-making.
Despite these limitations, the Simoa serological assay provides a powerful analytical tool that will advance the understanding of host immunity to SARS-CoV-2 by enabling analysis of the antibody response throughout the course of infection with high resolution. We demonstrated this capability by profiling multiple immunoglobulin isotypes across a panel of SARS-CoV-2 antigens using blood samples collected at serial time points from individual patients with COVID-19. This type of comprehensive analysis of the immune response to SARS-CoV-2 can provide critical insights necessary for the development of therapeutics and vaccines. In future studies, we aim to use this method to address unanswered questions such as which specific antigen–antibody interactions are important and how they relate to long-term immunity, how long these antibodies remain in the bloodstream and how community immunity can affect the spread of SARS-CoV-2.
Methods
Plasma samples for the discovery cohort
Clinical samples were obtained from patients presenting to the Massachusetts General Hospital with viral respiratory symptoms. Plasma collection was performed with a variety of tube types. SARS-CoV-2 infection was confirmed by RT–PCR on nasopharyngeal specimens (n = 19 RT–PCR-negative samples and n = 42 RT–PCR-positive samples from 21 individuals taken at two time points). Additional plasma samples collected from healthy adults pre-pandemic (January to December 2019) before international travel were also included in the analysis (n = 20). All samples were collected under approval of the Institutional Review Board for Human Subjects Research at Massachusetts General Hospital. Human plasma samples were heat inactivated (56 °C for 60 min). Samples were diluted in Homebrew Sample Diluent (Quanterix) to their final dilution factor, which varied for each experiment, as described in the Results. All sample measurements had two technical replicates, which were averaged.
Plasma samples for the training cohort
Clinical samples were obtained from patients presenting to Brigham and Women’s Hospital with viral respiratory symptoms (n = 172 samples from 91 individuals taken at different time points who tested positive and n = 100 samples from 95 individuals taken at different time points who tested negative). Of the SARS-CoV-2-positive cohort, ten patients were immunosuppressed, encompassing 31 time points. SARS-CoV-2 infection was confirmed by RT–PCR on nasopharyngeal specimens. Additional samples were obtained from the Partners Biobank, which includes pre-pandemic samples from before 1 October 2019 from individuals who presented with a recent respiratory infection (n = 100) or patients who had no record of viral or bacterial infection in their medical record (n = 100). All samples were collected under approval of the Partners Institutional Review Board for Human Subjects Research. In this training cohort, all plasma was collected in purple-top tubes and samples were not heat inactivated. Samples were diluted in Homebrew Sample Diluent to their final dilution factor.
Plasma samples for the validation cohort
In total, 68 samples were from 28 adults who tested positive for SARS-CoV-2 using nasopharyngeal RT–PCR within the past 5–34 d. The remaining 232 samples were from pre-pandemic controls and were collected before 1 October 2019, including: (1) 20 patients with bacterial pneumonia in the past 31 d; (2) 20 patients with unspecified pneumonia in the past 31 d; (3) 50 patients with upper respiratory infection within the past 31 d; (4) 42 patients with a confirmed viral respiratory illness within the past 31 d; and (5) 100 healthy individuals with no recorded respiratory viruses in their medical history. In this validation cohort, all plasma was collected in purple-top tubes and samples were not heat inactivated. Samples were diluted in Homebrew Sample Diluent to their final dilution factor. All samples were collected under approval of the Partners Institutional Review Board for Human Subjects Research.
RBD expression and purification
SARS-2 RBD (GenBank; MN975262.1) was cloned into pVRC vector for mammalian expression (FreeStyle 293F or Expi293F suspension cells). The construct contains a human rhinovirus 3C-cleavable carboxy (C)-terminal streptavidin-binding peptide-His8× tag. Supernatants were harvested 5 d post-transfection and passaged directly over Cobalt-TALON resin (Takara) followed by size exclusion chromatography on Superdex 200 Increase (GE Healthcare) in 1× phosphate-buffered saline. Typical yields from FreeStyle 293F cells are approximately 9 mg l−1 culture. Affinity tags were removed using human rhinovirus 3 C protease (Thermo Fisher Scientific) and the protein was repurified using Cobalt-TALON resin to remove the protease, tag and non-cleaved protein. RBD sequence and purification validation are provided in Supplementary Fig. 1.
Preparation of a stabilized ectodomain of spike protein
To express a stabilized ectodomain of spike protein, a synthetic gene encoding residues 1–1,208 of SARS-CoV-2 spike with the furin cleavage site (residues 682–685) replaced by a GGSG sequence, proline substitutions at residues 986 and 987 and a foldon trimerization motif followed by a C-terminal His6× tag was created and cloned into the mammalian expression vector pCMV-IRES-puro (Codex BioSolutions). The expression construct was transiently transfected in HEK 293T cells using polyethylenimine. Protein was purified from cell supernatants using Ni-NTA resin (Qiagen) and the eluted fractions containing S protein were pooled, concentrated and further purified by gel filtration chromatography on a Superose 6 column (GE Healthcare) following a protocol described previously26. Negative stain electron microscopy analysis was performed as described27. Additional information is provided in Supplementary Fig. 2.
Bead coupling and verification
Recombinant SARS-CoV-2 antigens (nucleocapsid, RBD, S1 and spike) were coupled to four types of dye-encoded 2.7-µm carboxylated paramagnetic beads (Quanterix) using 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride) (EDC) chemistry (Thermo Fisher Scientific; 77149). Recombinant SARS-CoV-2 nucleocapsid protein with a C-terminal His tag was purchased from RayBiotech (230-30164). It was derived from transfected human HEK293 cells. SARS-CoV-2 S1 protein with a C-terminal His tag was purchased from Sino Biological (V0591-V08H). A DNA sequence encoding the S1 subunit (Val 16–Arg 685) was expressed in HEK293 cells. The spike and RBD antigens were produced in the laboratories of B.C. and A.G.S., respectively, as described above. A total of 2.8 × 108 beads were washed three times with 200 µl Bead Wash Buffer (Quanterix) and three times with 200 µl Bead Conjugation Buffer (Quanterix), then resuspended in 300 µl Bead Conjugation Buffer. Immediately before use, 10 mg EDC was reconstituted in 1 ml Bead Conjugation Buffer. To activate the beads for conjugation, EDC was added to the bead suspension and the beads were agitated on a HulaMixer (Thermo Fisher Scientific) for 30 min at 4 °C. After activation, the beads were washed once with 200 μl Bead Conjugation Buffer, then resuspended in 300 μl Bead Conjugation Buffer containing the antigen. Beads were agitated on the HulaMixer for 2 h at 4 °C. The antigen-conjugated beads were washed twice with 200 μl Bead Wash Buffer, then blocked with bovine serum albumin for 30 min at room temperature in 200 μl Bead Blocking Buffer (Quanterix). The blocked beads were washed with 200 μl Bead Wash Buffer and 200 μl Bead Diluent (Quanterix) and resuspended in 200 μl Bead Diluent. The amounts of recombinant protein and EDC used in each reaction were as follows: nucleocapsid (20 μg antigen and 2 μl EDC); RBD (20 μg antigen and 6 μl EDC); S1 (20 μg antigen and 6 μl EDC); and spike (16.67 μg antigen and 6 μl EDC). Nucleocapsid, spike, S1 and RBD were conjugated to 488, 647, 700 and 750 nm dye-encoded beads, respectively. The beads were counted using a Beckman Coulter Particle Counter and stored at 4 °C.
Antigen coupling to the beads was confirmed by an anti-His tag assay for spike, S1 and nucleocapsid beads and by an anti-RBD assay for RBD beads (Supplementary Fig. 3). For spike, S1 and nucleocapsid, confirmation of antigen attachment to the beads was demonstrated by Simoa with His tag experiments using a biotinylated anti-His tag antibody (Thermo Fisher Scientific; MA121315BTI) on the HD-X Analyzer (Quanterix). The anti-His tag antibody was plated at concentrations of 0.1–10,000 pg ml−1 using tenfold dilutions. RBD was provided without a His tag; therefore, RBD beads were used as a control against the anti-His tag antibody assay. RBD conjugation to beads was confirmed by Simoa with an anti-RBD antibody (clone CR3022) and a biotinylated anti-human IgG antibody (Bethyl Laboratories; A80-148B). Nucleocapsid beads were used as a control for these experiments due to the ability of anti-RBD antibody binding to RBD, S1 and spike protein but not nucleocapsid.
Biotinylation
Detection antibodies for IgA, IgG and IgM were purchased from Thermo Fisher Scientific, Bethyl Laboratories, Abcam, BioLegend and R&D Systems (see the section ‘Immunoglobulin Simoa assay format’) and were biotinylated for use in Simoa assays as described previously by Cohen et al.28. Briefly, the antibodies were passed through an Amicon filter three times in Biotinylation Reaction Buffer (Quanterix). Antibody concentrations were determined using a NanoDrop One spectrophotometer. Antibodies were conjugated to biotin using EZ-Link NHS-PEG4-Biotin (Thermo Fisher Scientific) by resuspending NHS-PEG4-Biotin in dionized H2O. For all immunoglobulins, NHS-PEG4-Biotin was added in 40× molar excess and incubated for 30 min. All biotinylated antibodies were then purified using three washes in an Amicon filter.
Immunoglobulin Simoa assay format
Simoa experiments were performed in an automated three-step assay format onboard the HD-X Analyzer (Quanterix), as described by Rivnak et al.29. Human plasma samples were diluted in Homebrew Detector/Sample Diluent (Quanterix). Anti-human immunoglobulin antibodies were diluted in Homebrew Detector/Sample Diluent to final concentrations of 7.73 ng ml−1 IgG (Bethyl Labratories; A80-148B), 216 ng ml−1 IgM (Thermo Fisher Scientific; MII0401) and 150 ng ml−1 IgA (Abcam; ab214003). Streptavidin-β-galactosidase (SβG) concentrate (Quanterix) was diluted to 30 pM in SβG Diluent (Quanterix). System Wash Buffer 1, System Wash Buffer 2, resorufin β-d-galactopyranoside (RGP) and Simoa Sealing Oil were purchased from Quanterix and loaded onto the HD-X Analyzer per the manufacturer’s instructions. In the first step of the assay, 25 μl of the four SARS-CoV-2 antigen-coupled multiplex beads were incubated with 100 μl of diluted human plasma for 15 min. The total number of beads used per reaction was 475,000 (125,000 each of nucleocapsid, RBD and S1 beads and 100,000 spike beads). After incubation, six wash steps were performed with System Wash Buffer 1. In the second step, the beads were resuspended in 100 μl of the respective biotinylated anti-human immunoglobulin antibody and incubated for 5.25 min, then washed six times with System Wash Buffer 1. In the third step, the beads were resuspended in 100 μl of SβG, incubated for 5.25 min and washed six times. The beads were resuspended in 25 μl of RGP before being loaded into the microwell array for analysis. Following bead loading, the microwell array was sealed with oil and imaged in five optical channels. AEB values were calculated by the software in the HD-X Analyzer. The assay was validated as described in the Supplementary Information.
Data analysis
Duplicate measurements per sample were obtained for each of the 12 immunoglobulin and viral target combinations. The average of duplicate measurements was calculated. For cohort 2, in each run, four calibrators were included for each assay. These calibrators were produced by serial dilution of samples from SARS-CoV-2-positive individuals who were not included in either cohort 1 or 2. These calibrators were used to normalize samples run between batches. For modelling, the mean of marker duplicate values was calculated and then log-transformed. The logistic regression outcome reference group comprised pre-pandemic control samples only. In different models, the outcome index group comprised all SARS-CoV-2-positive cases, early-stage cases or late-stage cases.
Before modelling, fivefold cross-validation was performed to identify subsets of predictive markers. Two cross-validations were run: first, early-stage cases and pre-pandemic controls; and second, late-stage cases and all controls. For each cross-validation, every included participant was randomly assigned to one of five groups. For each fivefold cross-validation, one group was held out as a test set and the other four groups combined were the training set. Using PROC ADAPTIVEREG in SAS, each training set started from a model of all 12 markers and worked backwards to an intercept-only model. Of all the models run on a given training set, the one yielding the smallest cross-validation error was identified as the fold-specific model. The generalized cross-validation criterion (GCV) was the measure of the fold-specific model’s predictive accuracy. The contribution of each variable to the fold-specific model was measured by its importance, defined as the square root of the GCV value of the fold-specific model from which all basis functions involving the variable had been removed, minus the square root of the GCV value of the selected model, then scaled to set the largest importance value to 100. To validate the data, in each test set, predicted probabilities of the outcome were calculated using the markers with an importance of at least 60 in the respective training set. Predicted probabilities from the various test sets were then merged together to estimate the area under the ROC curve across the test sets.
The final markers selected from each cross-validation were those with an importance of at least 60 in at least three of the five training sets. Thus, there were three panels of markers: the full model of all 12 markers measured; a subset identified by the cross-validation of controls and early-stage cases; and a subset identified by the cross-validation of controls and late-stage cases.
Model discrimination was assessed by the area under the ROC curve. Calibration was evaluated using LOESS-smoothed calibration plots of observed probability (0 or 1) versus estimated probability of the outcome. To assess the classification accuracy of particular models, those with a probability of being a case of greater than 50% were predicted to be cases, while those with a probability below 50% were predicted to be controls (Supplementary Table 9). The predicted case status for a given model was then cross-tabulated with the observed case status.
Analyses were run using SAS 9.4 (SAS Institute) and R 4.0.0. ROC curves were calculated using MATLAB R2019a. The statistical tests were performed in GraphPad Prism 7. All figures were plotted in GraphPad Prism 7 and Adobe Illustrator 2015.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Supplementary Figs. 1–7, Tables 1–9 and descriptions for Supplementary Codes 1 and 2.
Cross-validation and model training.
Classification of the blinded validation set according to the models.
Acknowledgements
We thank L. Xie and E. Wood Karlson for helpful discussions regarding the experimental design. Funding for this work came from a generous donation from B. Hostetter and A. Hostetter and the Chleck Family Foundation. This work was also largely funded through a grant from the Massachusetts Consortium on Pathogen Readiness. In addition, support came from the Global TravEpiNet (GTEN) system sponsored by the US Centers for Disease Control and Prevention (grant no. U01CK000490; to E.T.R. and R.C.C.), as well as grants from NIGMS (no. T32GM007753) and NIAID (nos. T32AI007245 and R01AI146779).
Author contributions
M.N., T.G., A.F.O., A.M.M., L.C. and D.R.W. conceived of the approach. M.N., T.G., A.F.O. and A.M.M. performed the experiments. M.N., T.G., E.L.B., L.C. and R.L. analysed the data. Y.C., J.Z., J.E.F., B.M.H., T.M.C., B.C. and A.G.S. produced and purified the antigens. R.C.C. and E.T.R. collected the samples for the discovery cohort. C.-P.M. assisted with chart review for the training cohort. M.N., T.G., A.F.O., L.C., A.M.M. and D.R.W. co-wrote the paper. All authors were involved with designing the experiments and reviewing and discussing the data. All authors commented on the manuscript.
Data availability
The main data supporting the results of this study are available within the paper and its Supplementary Information. All data generated in this study, including source data and the data used to produce the figures, are available from Mendeley at 10.17632/2f73pzdkr4.1 (ref. 30).
Code availability
The custom codes used in this study are available as Supplementary Codes 1 and 2.
Competing interests
D.R.W. has a financial interest in Quanterix Corporation, a company that develops an ultra-sensitive digital immunoassay platform. He is an inventor of the Simoa technology and a founder of the company and also serves on its Board of Directors. D.R.W.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Partners HealthCare in accordance with their policies on competing interests. The assays in this publication have been licensed by Brigham and Women’s Hospital to Quanterix Corporation. M.N., T.G., A.F.O., A.M.M. and D.R.W. have filed intellectual property on the technology described in this manuscript. L.C., E.L.B., R.L., C.-P.M., Y.C., J.Z., J.E.F., B.M.H., T.M.C., B.C., A.G.S., G.A., R.C.C. and E.T.R. declare no competing interests.
Footnotes
Peer review information Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Maia Norman, Tal Gilboa.
Supplementary information
is available for this paper at 10.1038/s41551-020-00611-x.
References
- 1.Zhou P, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vashist, S. K. In vitro diagnostic assays for COVID-19: recent advances and emerging trends. Diagnostics (Basel)10.3390/diagnostics10040202 (2020). [DOI] [PMC free article] [PubMed]
- 3.Cascella, M., Rajnik, M., Cuomo, A., Dulebohn, S. C. & Di Napoli, R. Features, Evaluation, and Treatment of Coronavirus (COVID-19) (StatPearls, 2020). [PubMed]
- 4.Liu, R. et al. The comparative superiority of IgM-IgG antibody test to real-time reverse transcriptase PCR detection for SARS-CoV-2 infection diagnosis. Preprint at https://www.medrxiv.org/content/10.1101/2020.03.28.20045765v1 (2020).
- 5.Bullis, S. S. M., Crothers, J. W., Wayne, S. & Hale, A. J. A cautionary tale of false-negative nasopharyngeal COVID-19 testing. IDCases10.1016/j.idcr.2020.e00791 (2020). [DOI] [PMC free article] [PubMed]
- 6.Arevalo-Rodriguez, I. et al. False-negative results of initial RT–PCR assays for COVID-19: a systematic review. Preprint at https://www.medrxiv.org/content/10.1101/2020.04.16.20066787v2 (2020). [DOI] [PMC free article] [PubMed]
- 7.Berenger, B. M., Fonseca, K., Schneider, A. R., Hu, J. & Zelyas, N. Sensitivity of nasopharyngeal, nasal and throat swab for the detection of SARS-CoV-2. Preprint at https://www.medrxiv.org/content/10.1101/2020.05.05.20084889v1 (2020).
- 8.Long Q, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26:845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 9.Lipsitch, M., Kahn, R. & Mina, M. J. Antibody testing will enhance the power and accuracy of COVID-19-prevention trials. Nat. Med.10.1038/s41591-020-0887-3 (2020). [DOI] [PubMed]
- 10.Pan, Y. et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J. Infect.10.1016/j.jinf.2020.03.051 (2020). [DOI] [PMC free article] [PubMed]
- 11.Okba, N. M. A. et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis.10.3201/eid2607.200841 (2020). [DOI] [PMC free article] [PubMed]
- 12.Liu L, et al. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients. Microbes Infect. 2020;22:206–211. doi: 10.1016/j.micinf.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao, J. et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis.10.1093/cid/ciaa344 (2020). [DOI] [PMC free article] [PubMed]
- 14.Amanat F, et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat. Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, Z. et al. Elevated serum IgM levels indicate poor outcome in patients with coronavirus disease 2019 pneumonia: a retrospective case-control study. Preprint at https://www.medrxiv.org/content/10.1101/2020.03.22.20041285v1 (2020).
- 16.Lin, D. et al. Evaluations of the serological test in the diagnosis of 2019 novel coronavirus (SARS-CoV-2) infections during the COVID-19 outbreak. Eur. J. Clin. Microbiol. Infect. Dis.10.1007/s10096-020-03978-6 (2020). [DOI] [PMC free article] [PubMed]
- 17.Lou, B. et al. Serology characteristics of SARS-CoV-2 infection after exposure and post-symptom onset. Eur. Resp. J.10.1183/13993003.00763-2020 (2020). [DOI] [PMC free article] [PubMed]
- 18.Guo, L. et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19). Clin. Infect. Dis.10.1093/cid/ciaa310 (2020). [DOI] [PMC free article] [PubMed]
- 19.Saijo M, et al. Recombinant nucleocapsid protein-based IgG enzyme-linked immunosorbent assay for the serological diagnosis of SARS. J. Virol. Methods. 2005;125:181–186. doi: 10.1016/j.jviromet.2005.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wild, D. The Immunoassay Handbook: Theory and Applications of Ligand Binding, ELISA, and Related Techniques 4th edn (Elsevier, 2013).
- 21.Rissin DM, et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 2010;28:595–599. doi: 10.1038/nbt.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen L, Walt DR. Single-molecule arrays for protein and nucleic acid analysis. Annu. Rev. Anal. Chem. 2017;10:345–363. doi: 10.1146/annurev-anchem-061516-045340. [DOI] [PubMed] [Google Scholar]
- 23.Wu, C., Maley, A. M. & Walt, D. R. Single-molecule measurements in microwells for clinical applications. Crit. Rev. Clin. Lab. Sci.10.1080/10408363.2019.1700903 (2019). [DOI] [PubMed]
- 24.Weinstein, M. C., Freedberg, K. A., Hyle, E. P. & Paltiel, A. D. Waiting for certainty on COVID-19 antibody tests—at what cost? N. Engl. J. Med.10.1056/NEJMp2017739 (2020). [DOI] [PubMed]
- 25.Jacofsky D, et al. Understanding antibody testing for COVID-19. J. Arthroplasty. 2020;35:S74–S81. doi: 10.1016/j.arth.2020.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kovacs JM, et al. HIV-1 envelope trimer elicits more potent neutralizing antibody responses than monomeric gp120. Proc. Natl Acad. Sci. USA. 2012;109:12111–12116. doi: 10.1073/pnas.1204533109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaik MM, et al. Structural basis of coreceptor recognition by HIV-1 envelope spike. Nature. 2019;565:318–323. doi: 10.1038/s41586-018-0804-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohen L, Walt DR. Evaluation of antibody biotinylation approaches for enhanced sensitivity of single molecule array (Simoa) immunoassays. Bioconjug. Chem. 2018;29:3452–3458. doi: 10.1021/acs.bioconjchem.8b00601. [DOI] [PubMed] [Google Scholar]
- 29.Rivnak AJ, et al. A fully-automated, six-plex single molecule immunoassay for measuring cytokines in blood. J. Immunol. Methods. 2015;424:20–27. doi: 10.1016/j.jim.2015.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norman, M. et al. Dataset for Ultra-Sensitive High-Resolution Profiling of Anti-SARS-CoV-2 Antibodies for Detecting Early Seroconversion in COVID-19 Patients (Mendeley, 2020); 10.17632/2f73pzdkr4.1
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1–7, Tables 1–9 and descriptions for Supplementary Codes 1 and 2.
Cross-validation and model training.
Classification of the blinded validation set according to the models.
Data Availability Statement
The main data supporting the results of this study are available within the paper and its Supplementary Information. All data generated in this study, including source data and the data used to produce the figures, are available from Mendeley at 10.17632/2f73pzdkr4.1 (ref. 30).
The custom codes used in this study are available as Supplementary Codes 1 and 2.