Bifidobacterium is a natural inhabitant of the human gastrointestinal (GI) tract. Morphological observations revealed that extracellular appendages of bifidobacteria in complex microbial communities are important for understanding its adaptations to the GI tract environment. We identified dynamic extracellular vesicle (EV) production by Bifidobacterium longum in bacterium-free fecal fermentation broth that was strongly suggestive of differing bifidobacterial extracellular appendages in the GI tract. In addition, export of the adhesive moonlighting proteins mediated by EVs may promote bifidobacterial colonization. This study provides new insight into the roles of EVs in bifidobacterial colonization processes as these bacteria adapt to the GI environment.
KEYWORDS: Bifidobacterium, adhesion, colonization, extracellular vesicle, fecal fermentation, symbiosis
ABSTRACT
Extracellular proteins are important factors in host-microbe interactions; however, the specific factors that enable bifidobacterial adhesion and survival in the gastrointestinal (GI) tract are not fully characterized. Here, we discovered that Bifidobacterium longum NCC2705 cultured in bacterium-free supernatants of human fecal fermentation broth released a myriad of particles into the extracellular environment. The aim of this study was to characterize the physiological properties of these extracellular particles. The particles, approximately 50 to 80 nm in diameter, had high protein and double-stranded DNA contents, suggesting that they were extracellular vesicles (EVs). A proteomic analysis showed that the EVs primarily consisted of cytoplasmic proteins with crucial functions in essential cellular processes. We identified several mucin-binding proteins by performing a biomolecular interaction analysis of phosphoketolase, GroEL, elongation factor Tu (EF-Tu), phosphoglycerate kinase, transaldolase (Tal), and heat shock protein 20 (Hsp20). The recombinant GroEL and Tal proteins showed high binding affinities to mucin. Furthermore, the immobilization of these proteins on microbeads affected the permanence of the microbeads in the murine GI tract. These results suggest that bifidobacterial exposure conditions that mimic the intestine stimulate B. longum EV production. The resulting EVs exported several cytoplasmic proteins that may have promoted B. longum adhesion. This study improved our understanding of the Bifidobacterium colonization strategy in the intestinal microbiome.
IMPORTANCE Bifidobacterium is a natural inhabitant of the human gastrointestinal (GI) tract. Morphological observations revealed that extracellular appendages of bifidobacteria in complex microbial communities are important for understanding its adaptations to the GI tract environment. We identified dynamic extracellular vesicle (EV) production by Bifidobacterium longum in bacterium-free fecal fermentation broth that was strongly suggestive of differing bifidobacterial extracellular appendages in the GI tract. In addition, export of the adhesive moonlighting proteins mediated by EVs may promote bifidobacterial colonization. This study provides new insight into the roles of EVs in bifidobacterial colonization processes as these bacteria adapt to the GI environment.
INTRODUCTION
The gastrointestinal (GI) microbiota is composed of highly diverse microorganisms (1). Associations between the microbes in the GI environment contribute to development of the complex, stable, and specific microbiota (2–4), suggesting the involvement of several molecular mechanisms to establish and maintain symbiotic microbial relationships. Bacterial surface adhesion molecules (adhesion factors) are the predominant bacterial structures involved in host-microbe interactions (5–7).
Bifidobacteria are Gram-positive rod-shaped anaerobes that are natural inhabitants of the human GI tract (8) and have various reported health benefits (9, 10). In our previous study, we showed that several Bifidobacterium strains are capable of adhering to porcine colonic mucins (11). Various types of bifidobacterial extracellular appendages promote adherence to the host mucosal surface, such as fimbriae (12–15), exo-alpha-sialidase (16), transaldolase (Tal) (17), enolase (18), and DnaK (19).
Turroni et al. (13) showed that the expression of genes encoding extracellular and membrane-spanning proteins in Bifidobacterium bifidum PRL2010 is modified in response to epithelial cell contact. Meanwhile, a study employing a unique approach showed that probiotic Propionibacterium freudenreichii in a dialysis membrane bag adapted to piglet intracolonic conditions exhibits dramatic transcriptomic differences from an in vitro control (20). These results strongly suggest that bacterial gene expression changes or adapts to the surrounding conditions, including those encountered in the GI tract. Although adhesion factors might be key functional determinants of stable colonization processes of Bifidobacterium in the GI tract, currently, studies examining the extracellular appendages in complex symbiotic microbial communities via electron microscopy are lacking.
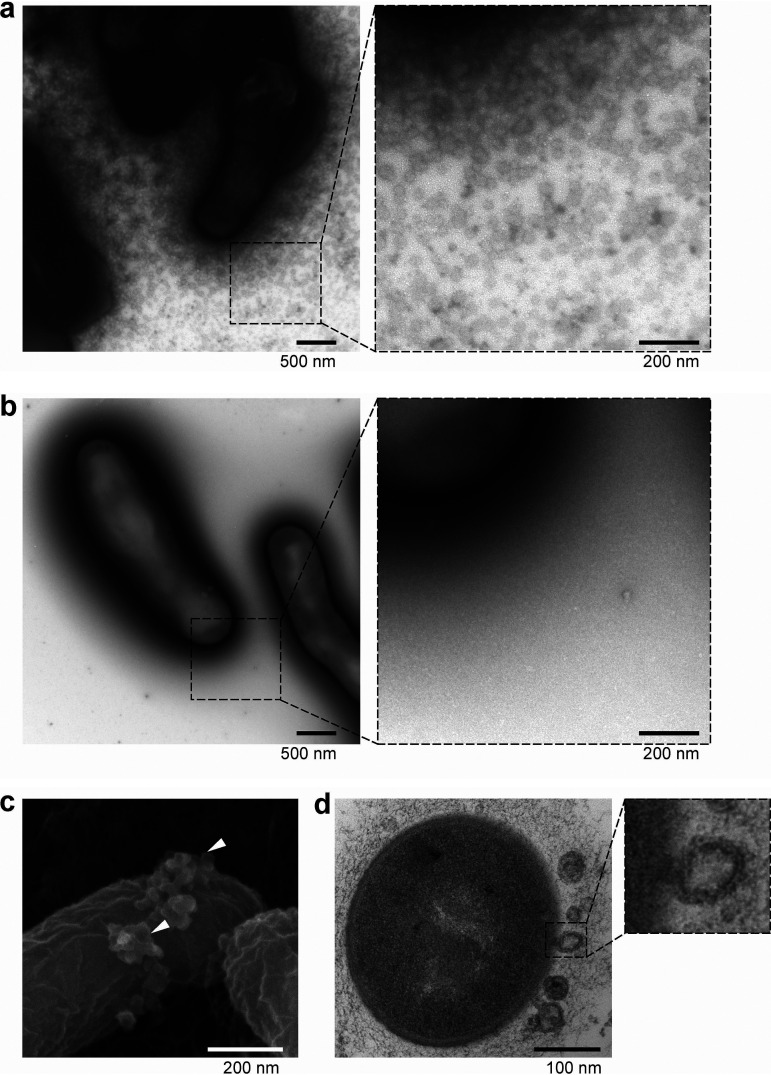
To this end, here we employed the single-batch anaerobic culture system (referred to as the Kobe University human intestinal microbiota model [KUHIMM]), which is composed of a jar fermentor that simulates human colonic microbiota from fecal samples (21, 22). Subsequently, the Bifidobacterium longum subsp. longum NCC2705 was identified within the bacterium-free supernatants of KUHIMM fecal fermentation broth (referred to as KUHIMM broth). This species produces abundant extracellular particles, as determined by electron microscopy (Fig. 1a). In addition, the particles contained several mucin-binding cytoplasmic proteins. The objective of this study was to characterize the physiological properties of these extracellular particles and their role in Bifidobacterium host adhesion.
FIG 1.
Electron microscope images of B. longum NCC2705. B. longum was cultured with fecal fermentation “KUHIMM” broth (a, c, and d) or basal GAM broth (b). (a and b) Whole bacterial cells were negatively stained with uranyl acetate and examined under a TEM. (c) The bacterial cells were coated with a thin osmium layer and examined under an SEM. SEM images showed extracellular vesicle (EV)-like particles (indicated by arrowheads) at the bacterial cell surface. (d) Ultrathin sections of B. longum examined under a TEM revealed an EV-like particle released from the cell membrane.
RESULTS
Characterization of extracellular particles produced by B. longum NCC2705.
To explore Bifidobacterium in the human colonic microbiota, B. longum NCC2705 was cultured on basal Gifu anaerobic medium (GAM) agar containing KUHIMM broth (fecal sample code, M60). Transmission electron microscope (TEM) images of B. longum NCC2705 revealed a myriad of particles in the KUHIMM broth-cultured cells (Fig. 1a) but not in those cultured in the basal GAM (Fig. 1b). No particles were detected in control bacterium-free GAM and KUHIMM broths (data not shown). Scanning electron microscope (SEM) images showed the particles budding from the KUHIMM broth-cultured B. longum NCC2705 cell surface (Fig. 1c). Furthermore, cell membrane protrusions were observed in the ultrathin B. longum NCC2705 cell sections (Fig. 1d). The viable cell counts of B. longum NCC2705 did not differ between cultures in the KUHIMM and GAM broths for bacterial suspensions of the same optical density (see Fig. S1 in the supplemental material). Therefore, we concluded that these particles were actively produced by B. longum NCC2705, as the level of B. longum NCC2705 particle production also differed between cultures containing cell-free culture supernatants from individual fecal samples (see Fig. S2).
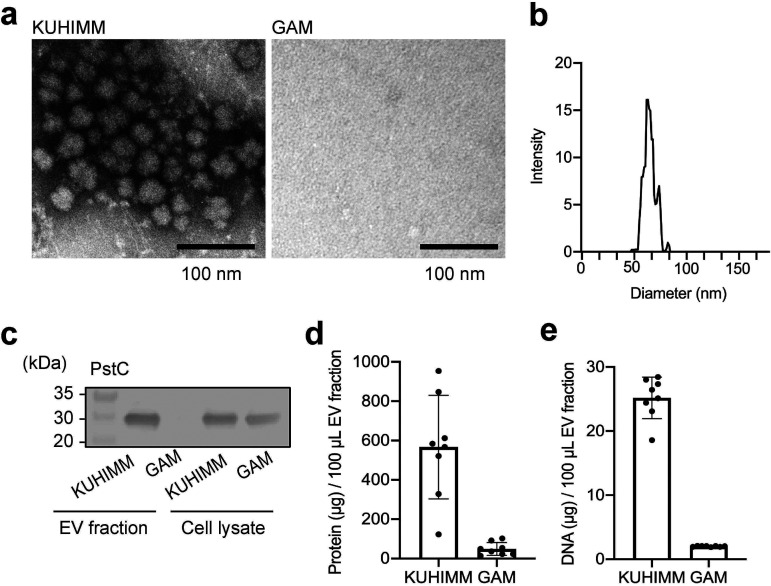
Next, we characterized the physiological properties of these particles. Since bacteria release extracellular particles, referred to as extracellular vesicles (EVs) (23), ranging from 20 to 400 nm in diameter (24), we isolated the high-molecular-weight fraction, designated the crude EV fraction, from the B. longum NCC2705 suspension using ultrafiltration filters (pore size approximately 35 nm). TEM images of the EV fraction showed the presence of numerous particles in the KUHIMM broth-cultured B. longum NCC2705 (Fig. 2a). The particles were approximately 50 to 80 nm in diameter (Fig. 2b). EVs reportedly are composed of the expected membrane proteins along with phospholipids, nucleic acids, and cytoplasmic proteins (24–26). The membrane protein, phosphate ABC transporter permease (PstC, BL0314), was also detected in the KUHIMM broth-cultured B. longum NCC2705 EV fraction, yet was absent from the GAM-cultured EV fraction, indicating that the EV isolation procedure was successful (Fig. 2c). To examine whether these particles carried bacterial DNA and proteins, purified fractions were analyzed using the bicinchoninic acid (BCA) protein assay and a fluorescent DNA-binding dye. Protein and double-stranded DNA (dsDNA) quantities were higher in KUHIMM broth-cultured B. longum NCC2705 EV fractions than in the GAM-cultured EV fractions (Fig. 2d and e). Therefore, we concluded that many of the particles observed were, in fact, EVs.
FIG 2.
Physicochemical characterization of particles produced by B. longum NCC2705. The crude extracellular vesicle (EV) fraction was isolated from the B. longum suspension. (a) EV fraction was negatively stained with uranyl acetate, and images were obtained using a TEM. (b) EV size distribution was assessed using a NanoSight nanoparticle characterization system. (c) Crude EV fraction and whole-cell lysate were analyzed using Western blotting with an anti-PstC antibody to detect membrane protein. Protein (d) and DNA (e) concentrations were determined using a bicinchoninic acid protein assay and QuantiFluor dsDNA system, respectively. Each data point represents one EV fraction (n = 8). The error bars indicate the means ± standard deviations (SDs).
Protein composition of B. longum NCC2705 EVs.
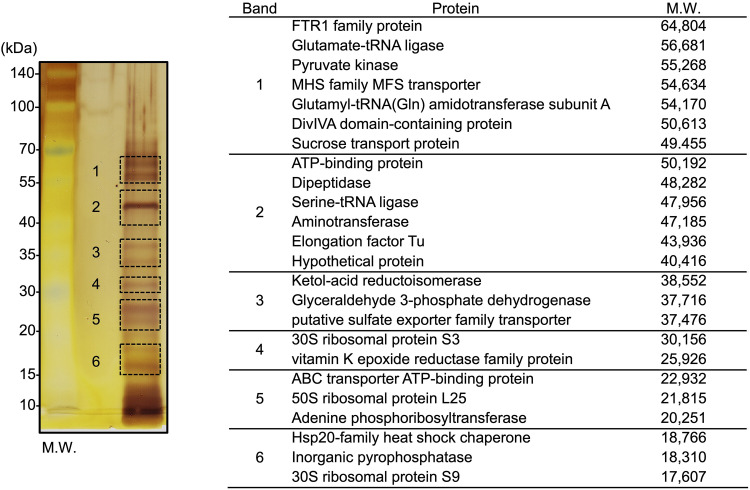
The protein composition of KUHIMM broth-cultured B. longum NCC2705 EVs was determined using mass spectrometry. The proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) from the purified EV-containing fractions in cultures were visualized using silver staining (Fig. 3). The six major bands were then excised for in-gel trypsin digestion, and proteins were identified using matrix-assisted laser desorption ionization–time of flight mass spectroscopy (MALDI-TOF MS). B. longum EVs were found to contain various proteins, of which, cytoplasmic proteins were the most abundant (Fig. 3; see Table S1). In addition, the band pattern and most identified proteins differed between the EV fractions and B. longum whole-cell lysates (see Fig. S3a) from the bacterial cell-free KUHIMM broth (Fig. S3b). However, elongation factor Tu (EF-Tu; WP_007051202.1) and pyruvate kinase (WP_007055467.1) were detected in both fractions (EV fraction and whole-cell lysate). These results suggest that most proteins identified in the EV fraction were secreted by B. longum NCC2705 EVs.
FIG 3.
Proteomic analysis of the EV fraction from B. longum NCC2705. SDS-PAGE separated proteins from the crude EV fraction, which were visualized using silver staining (left). Six bands fragments were excised for protein identification by MALDI-TOF MS. Identified proteins and calculated molecular weights are indicated on the right. Additional information is presented in Table S1 in the supplemental material.
Identification of mucin-binding proteins in the EV fraction.
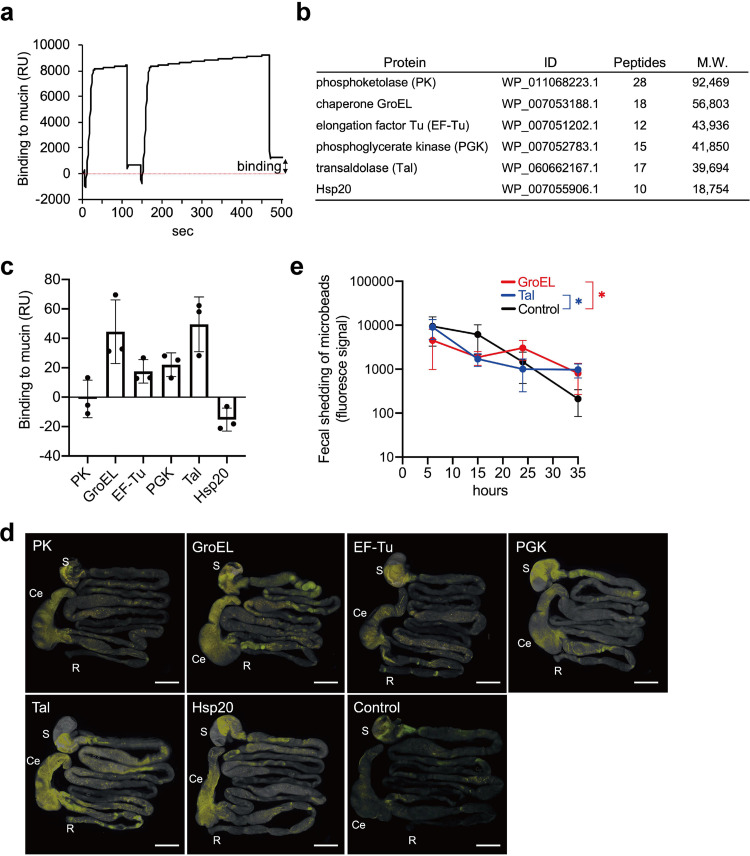
We next searched for mucin-binding proteins in B. longum EVs. To this end, biomolecular interaction analysis-mass spectrometry (BIA-MS) technology was used to identify proteins bound to mucin (27). Purified porcine gastric mucin (PGM) (11) was immobilized on CM5 sensor chips. EV fractions were twice injected into the Biacore apparatus to obtain the association phase. Bound proteins (approximately 1,200 resonance units [RU] per flow cell) (representative signal is shown in Fig. 4a) were collected from a total of four flow cell lines and identified using nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS) after tryptic digestion. Proteins from the EVs fraction bound to the PGM are outlined in Fig. 4b and include chaperone GroEL, EF-Tu, phosphoglycerate kinase (PGK), Tal, and heat shock protein 20 (Hsp20), all of which have previously described roles in commensal or pathogenic bacterial colonization (17, 28–36). Additionally, we selected the most abundant phosphoketolase (PK) as a candidate binding protein. These proteins have known functions in essential cellular processes—for example, Tal and PK are enzymes in the pentose phosphate pathway, GroEL and Hsp20 are molecular chaperones, EF-Tu contributes to protein synthesis, and PGK is involved in glycolysis and energy generation. All six proteins lack typical Sec and Tat signal sequences (see Fig. S4).
FIG 4.
Identification and characterization of mucin-binding proteins in EVs from B. longum NCC2705. (a) Representative signal of Biacore. Isolated protein samples from B. longum NCC2705 EV fractions were injected into the porcine gastric mucin (PGM)-immobilized sensor chip on the Biacore system to obtain the association phase. (b) The PGM-bound proteins are listed in this table, as identified by nano-LC-MS/MS. (c) Binding of recombinant proteins to PGM. Resonance unit (RU) values were measured at the end of the association period. The error bars indicate the SDs; data are based on three biological replicates. (d) Persistence of recombinant-protein-immobilized microbeads in the mouse whole gastrointestinal tract (n = 3 mice per group). Bars, 1 cm. (e) Fecal shedding of GroEL- and transaldolase (Tal)-immobilized microbeads. Means were compared using one-way ANOVA (control [noncoated beads] versus GroEL- or Tal-immobilized beads); the error bars indicate the SDs. *, P < 0.05; Ce, cecum; R, rectum; S, stomach.
Identified proteins from B. longum NCC2705 were expressed as recombinant His6-tagged proteins in Escherichia coli. The expression and purification of recombinant proteins were verified using SDS-PAGE, followed by staining with Coomassie brilliant blue (CBB) (see Fig. S5). A Biacore experiment evaluated the binding of recombinant proteins to PGM. The recombinant proteins exhibited unique variation in the ability to bind to PGM, ranging from −21.2 to 69.5 RU (Fig. 4c). GroEL and Tal showed high binding affinities to PGM. In contrast, PK and Hsp20 exhibited more durable binding to the blank cell (nonimmobilized) than to the mucin-immobilized flow cell, with negative RUs (Fig. 4c). Therefore, among the six proteins, GroEL, EF-Tu, PGK, and Tal were identified as the candidate mucin-binding proteins, while PK and Hsp20 were postulated to likely interact with the CM5 dextran surface rather than PGM.
The six individual recombinant proteins were then chemically coupled with fluorescent microbeads to mimic a bacterial cell surface-localized protein to confirm whether the identified proteins affect the persistence of microorganisms in vivo (37). At 24 h after administration, mouse GI tracts were harvested and treated with optical clearing agents (see Fig. S6). The localization of microbeads depended on the immobilized target protein. There was high retention of recombinant-protein-immobilized microbeads in the murine cecum (Fig. 4d). In particular, GroEL- and Tal-immobilized microbeads exhibited diffusion throughout the stomach up to the rectum. Hsp20-immobilized microbeads also localized in the cecum (Fig. 4d). We monitored the fecal shedding of GroEL- and Tal-immobilized microbeads for 35 h (Fig. 4e) and quantified the fluorescence signals of both protein-immobilized microbeads through to the end of the study; however, the fluorescence intensity of control beads significantly diminished. These results are indicative of a potential persistence of the B. longum EV-derived mucin-adhesive proteins, particularly Tal and GroEL, in the GI tract.
DISCUSSION
Bifidobacterial extracellular appendages, including fimbriae, surface exopolysaccharides, and several other adhesion factors, are involved in the initial physical contact with the host mucosal surface (12, 13, 16–19, 38, 39). These extracellular appendages determine the individual nature of strains and provide insight into the GI environment survival strategies. Accordingly, observations of the bifidobacterial cell surface structure via electron microscopy, following exposure to the symbiotic microbial community, can serve to significantly improve the current understanding regarding bifidobacterial colonization and survival within the GI tract.
Surprisingly, B. longum NCC2705 cultured with bacterium-free supernatants of human fecal fermentation broth (KUHIMM broth) exported many vesicles to the extracellular environment. Since we did not detect any such particles in the GAM or KUHIMM broths, we concluded that the myriad of EVs were produced by B. longum NCC2705. Bacterial cell lysis and bubbling cell death are well-characterized mechanisms underlying EV formation (24, 40). A previous study showed that Gram-positive bacterium Bacillus subtilis EV production occurs through cell membrane bubbling dependent upon peptidoglycan damage (41). A similar membrane protrusion was observed in KUHIMM broth-cultured B. longum NCC2705 (Fig. 1d), suggesting the occurrence of similar biological processes. Interestingly, the EV production pattern of B. longum NCC2705 differed among the individual fecal samples (Fig. 1a; see also Fig. S2a and b in the supplemental material). Several studies have indicated that environmental conditions influence the formation of EVs, including medium composition, temperature, iron concentration, oxygen availability, exposure to genotoxic stress, and phage infection (24, 42). Since the compositions of the microbiota and metabolites produced differ in each KUHIMM broth (21, 22), we hypothesize that intestinal microbiota-derived metabolites directly, or indirectly, stimulate the active production of B. longum NCC2705 EVs. Importantly, the bifidobacterial cell surface phenotype (such as EV production) may differ substantially in vivo and in vitro.
Previous studies have reported the therapeutic applications of bifidobacterial EVs. B. bifidum LMG13195-derived EVs induce regulatory T (Treg) cell polarization. These EVs may, therefore, be useful adjuvants for immunotherapy (43). Although B. longum KACC 91563-derived EVs, containing a family of five extracellular solute-binding proteins, were found to reduce the occurrence of diarrhea in a food allergy murine model (44), the functional role of GI tract EVs in Bifidobacterium colonies remains poorly understood.
Using the BIA-MS approach, mucin-binding cytoplasmic proteins (PK, GroEL, EF-Tu, PGK, Tal, and Hsp20) were isolated from B. longum NCC2705 EV fractions. Immobilization of these proteins with microbeads affected the prevalence of microbeads in the mouse GI tract, suggesting that B. longum EVs export various cytoplasmic proteins to the extracellular environment as a transporter. Some of these proteins (particularly, Tal and GroEL) exert additional functions, such as adhesion to the host mucosal surface, which might promote the colonization of B. longum in the GI tract as an “adhesive moonlighting protein” (45). Moreover, in bifidobacteria, certain cell surface-localized adhesive moonlighting proteins have been described. Specifically, B. bifidum A8 Tal promotes adhesion and aggregation to mucin (17), while Bifidobacterium animalis subsp. lactis BI07 DnaK and enolase bind to surface-exposed human plasminogen (18, 19). The secretory mechanism for these cytoplasmic proteins is mostly unknown. Therefore, elucidating the export processes of the adhesive moonlighting proteins mediated by EVs can provide important insights for further understanding bifidobacterial colonization of the GI tract.
However, as we did not determine the mechanism underlying protein export from EVs, further analyses are needed to evaluate the localization of these proteins on the cell surface, which can be accomplished via Western blotting with protein-specific antibodies. Additionally, as observed in GI tract tissue clearing, the localization of microbeads was dependent on the immobilized protein type, suggesting that adhesion properties such as ligand-specific and nonspecific binding differ among proteins. The Hsp20-immobilized microbeads affected the permanence of microbeads in the murine GI tract despite not binding to mucin, suggesting that Hsp20 binds to another ligand molecule. Therefore, the mucosal-surface-binding properties of individual proteins should be analyzed further.
In conclusion, we detected dynamic EV production by B. longum NCC2705 in bacterium-free fecal fermentation broth that was strongly suggestive of differing bifidobacterial extracellular appendages in the GI tract. In addition, the export of the adhesive moonlighting proteins mediated by EVs is vital for understanding the bifidobacterial GI tract colonization strategy. These results provide new insight into the role of EVs in bifidobacterial GI tract colonization strategies based on communication between complex symbiotic intestinal microbes via microbiota-derived metabolites.
MATERIALS AND METHODS
Bacterial culture.
Human intestinal bacteria were cultured with the KUHIMM system, as described previously (22), using a pH-controlled multichannel jar fermentor (Bio Jr.8; ABLE, Tokyo, Japan) into which was added a human fecal sample. Briefly, each vessel, containing 100 ml of GAM (05422; Nissui Pharmaceutical Co., Tokyo, Japan), was held at 37°C with constant stirring at 300 rpm under anaerobic conditions (N2/CO2 [80:20]) during fermentation. Each vessel was inoculated with 100 μl of fecal suspension to initiate the KUHIMM and cultured for 30 h. These fecal samples were obtained from healthy human volunteers who were individuals of Japanese descent, with nonsmoking status and in good health and physical condition. Fecal samples were designated M27 (male, age 27), F40 (female, age 40), and M60 (male, age 60). Each fecal sample was stored in an anaerobic swab culture. This study was performed under the guidelines of Kobe University Hospital and was approved by the institutional ethics review board of Kobe University (approval no. 1902). All methods were performed in accordance with the Declaration of Helsinki.
After fermentation using KUHIMM, the debris was removed by centrifugation (6,010 × g, 10 min, 4°C), and the supernatant was obtained by suction filtration (0.45-μm pore size; Advantech, Taipei, Taiwan). The collected samples were then centrifuged (20,000 × g, 30 min, 10°C), and the supernatants were heat inactivated at 60°C for 30 min. The samples were maintained at −80°C as “cell-free KUHIMM broth” until subsequent analyses. B. longum NCC2705 was anaerobically precultured in GAM at 37°C for 15 h using the AnaeroPack system (Mitsubishi Gas Chemical Co., Tokyo, Japan). Fresh GAM broth (10 ml) containing 3% (wt/vol) agar was mixed with an equal volume of cell-free KUHIMM broth (as a control, GAM broth) and was poured into petri dishes. Next, 20 μl of B. longum NCC2705 culture broth (after 15 h) was spread on the agar plate and cultured at 37°C for 30 h under anaerobic conditions. The bacterial culture and relevant experiments were performed in triplicate (for M60 fecal sample) or duplicate (for M27 and F40 fecal samples), each with a different fecal donor.
Electron microscopy.
B. longum NCC2705 colonies were fixed with 4% paraformaldehyde (PFA) for 10 min on an agar plate and applied to a Formvar carbon film on copper grids (300 mesh; Nisshin EM, Tokyo, Japan). The grids were negatively stained with 2% (wt/vol) uranyl acetate. Images were obtained using an H-7600 TEM (Hitachi Co. Ltd., Tokyo, Japan). For ultrathin sections, PFA-fixed bacterial cells were gently washed in phosphate-buffered saline (PBS) and fixed successively in PBS containing osmium tetroxide (1% [wt/vol]). The cells were embedded in a 1.5% (wt/vol) agarose gel and dehydrated by ethanol. Next, the sample was covered with epoxy resin, sliced with a microtome (Leica Ultracut UCT ultramicrotome) to a thickness of 60 nm, and examined using an H-7600 TEM.
For SEM imaging, B. longum colonies were fixed with 4% PFA solution in PBS on an agar plate. Samples were washed in PBS and dehydrated by successive immersions in increasing concentrations of ethanol. The samples were critical point dried and sputter coated with a thin osmium layer using a model HPC-1SW osmium coater (Nisshin EM Corp., Tokyo, Japan). Images were obtained using an S-4700 field emission (FE) SEM (Hitachi Co. Ltd., Tokyo, Japan).
Crude EV preparation from B. longum NCC2705 and MALDI-TOF MS.
B. longum NCC2705 colonies were scraped and suspended in 5 ml of PBS adjusted to an optical density at 600 nm (OD600) of 5.0. The bacterial suspension was centrifuged (10,000 × g, 5 min, 4°C), and the supernatants were passed through a 0.45-μm cellulose membrane filter (Advantech). The sample was ultracentrifuged at 150,000 × g for 60 min at 15°C (Himac CS100FNX; Koki Holdings, Tokyo, Japan), and the supernatants were carefully decanted. The pellet was resuspended in 1 ml of PBS and concentrated using a 100,000-molecular-weight-cutoff and Nanosep 100K ultrafiltration filters (Pall; final volume, approximately 100 μl); this was designated the “crude EV fraction.” The crude EV fraction was fixed with 4% PFA and applied to grids. Images were obtained using an H-7600 TEM, as described above. Protein and nucleic acid (dsDNA) concentrations were determined using a bicinchoninic acid (BCA) protein assay kit (TaKaRa BIO, Kusatsu, Japan) and QuantiFluor dsDNA system (Promega, Madison, WI, USA). The EV size distribution was obtained using a NanoSight LM10 nanoparticle characterization system (NanoSight, Salisbury, UK). The results are presented as averages of values from two independent experiments.
For SDS-PAGE, the crude EV fraction was mixed with 2× Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) (with 5% [vol/vol] β-mercaptoethanol [β-ME]) and boiled for 5 min. Next, 20 μl of the sample was separated on an acrylamide gel (MULTIGEL II mid 4/20; Cosmo Bio Co., Ltd., Tokyo, Japan). The gel was subsequently stained using a silver staining kit (Integrale, Tokushima, Japan). The significant bands (as indicated in Fig. 3) were identified using MALDI-TOF MS (Shimadzu, Kyoto, Japan). Searches identified the peptides against the Mascot database (Matrix Science, London, UK).
Preparation of B. longum NCC2705 whole-cell lysates and Western blotting.
The B. longum suspension (0.5 ml; described above) was centrifuged (4,000 × g, 5 min), and pelleted cells were suspended in 0.3 ml PBS containing 25 U mutanolysin (Sigma-Aldrich, M9901), followed by incubation for 1 h at 37°C. The suspension was subjected to bead beating for 180 s at 4,800 rpm using 0.3 g of 0.1-mm zirconia-silica beads (Bio Spec Products, Bartlesville, OK, USA). Cell debris was removed by centrifugation at 6,000 × g and 4°C for 60 s. After centrifugation, the supernatant, designated the whole-cell lysate fraction (10 μg protein/well), was separated using a 4% to 20% Mini-PROTEAN TGX precast gel (Bio-Rad). The gel was subsequently stained using Bullet CBB Stain One (Nacalai Tesque, Kyoto, Japan). The significant bands (as indicated in Fig. S3a in the supplemental material) were identified by MALDI-TOF MS as described above.
For Western blotting, protein samples from the crude EV fraction and whole-cell lysates were transferred to a polyvinylidene difluoride membrane that was blocked with 5% (wt/vol) skim milk in Tris-buffered saline (TBS)-0.05% (vol/vol) Tween 20 (TBS-T) for 15 h at 4°C. After the membrane was washed with TBS-T, it was incubated with the rabbit anti-PstC antibody diluted 1:800 in TBS-T at room temperature for 1 h. We then washed and incubated the membrane with alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma-Aldrich) diluted 1:1,000 in TBS-T for 1 h. After washing, we developed signals with a 5-bromo-4-chloro-3-indolylphosphate (BCIP)-nitroblue tetrazolium (NBT) liquid substrate system (Nacalai Tesque). Antisera against the PstC (BL0314) synthetic peptide (CDQAANSQTVSSFTGGK) were raised in rabbits by using routine immunization procedures.
BIA-MS.
To identify adhesive proteins in the crude EV fraction, we performed BIA-MS (27) using a surface plasmon resonance (SPR) biosensor (Biacore 1000; GE Healthcare, Chicago, IL, USA) and mass spectrometry, as previously described (46). We immobilized porcine mucin type II (PGM; Sigma, St. Louis, MO, USA) on a CM5 dextran sensor chip (approximately 2,500 to 4,200 resonance units [RU] in all flow cells [FC1 to FC4]; GE Healthcare). The experiment was performed at a flow rate of 10 μl/min in HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.005% [vol/vol] surfactant P-20; GE Healthcare) at 25°C. For analyte preparation, the crude EV fraction (100 μl) was suspended 1:10 in 0.1 mM Triton X-100 and incubated for 30 min at 25°C. We concentrated the mixtures and changed the buffer to HBS-EP using a 10,000-molecular-weight (MW)-cutoff Amicon Ultra filter (Merck, Kenilworth, NJ, USA). The sample was injected into FC1 to FC4 at a flow rate of 10 μl/min until saturation. We undocked the sensor chip from the Biacore system, and 5 μl of 2 M NaCl was dropped on the SPR biosensor’s surface and incubated for 30 min. The collected samples containing proteins were identified by nano-LC-MS/MS (Shimazu). Searches against the Mascot database identified the obtained peptides. SignalP-5.0 (http://www.cbs.dtu.dk/services/SignalP/) was used to assess the presence or absence of secretory signals.
Expression and purification of recombinant proteins in E. coli.
Six identified proteins (Fig. 4b) were expressed in E. coli and purified using N-terminal His tags. Genes (bl0959, groEL, tuf, pgk, tal, and bl0576) were amplified by PCR with Ex Taq DNA polymerase (TaKaRa BIO) using B. longum NCC2705 genomic DNA as a template (primer pairs are listed in Table S2). The amplified fragments were inserted into pET28b (Novagen, Madison, WI, USA) at NdeI and HindIII restriction sites. Sequencing confirmed all resulting plasmids, and their DNA was introduced into E. coli BL21(DE3). Transformed cells were grown in Luria-Bertani medium at 28°C or 37°C with shaking. When the OD600 reached 0.4, we added isopropyl-β-d-thiogalactopyranoside (0.3 mM) to induce protein expression. Recombinant proteins were purified as previously described (37) using a HisTrap HP column (GE Healthcare) in line with the ÄKTA start system (GE Healthcare) according to the standard operating procedures. Purified proteins were dialyzed in 20 mM HEPES buffer containing 150 mM NaCl (pH 7.0).
The Biacore 1000 assessed the interaction of recombinant proteins with PGM. Immobilized PGM (approximately 3,700 RUs) was on a CM5 (flow cell 2) dextran sensor chip, and flow cell 1 was used as a blank. Recombinant proteins (1,000 nM) were injected on the CM5 sensor chip. Experiments were performed at a flow rate of 10 μl/min in HBS-EP buffer at 25°C. We measured RU at the end of the association and corrected signals for nonspecific binding by subtracting the value of the blank.
Immobilization of recombinant proteins on microbeads.
Next, recombinant proteins were coupled with Fluoresbrite YG carboxylate microbeads (1.5 μm; Polysciences, Inc., Warrington, PA, USA) using the PolyLink protein coupling kit (Polysciences, Inc.) as described previously (37), and microbeads (12.5 mg) were suspended in the PolyLink coupling buffer. The EDAC solution was added and mixed gently to activate the beads for protein coupling. After adding 300 μg of each purified recombinant protein, we incubated the mixture with gentle shaking for 60 min. The beads were washed twice, resuspended in 400 μl of 50 mM phosphate buffer containing 150 mM NaCl and 0.05% (wt/vol) bovine serum albumin (pH 7.5), and stored at 4°C until use. Uncoated activated beads without protein coupling were used as a control.
Animals.
Eight-week-old C57BL/6JJcl male mice were obtained from CLEA Japan, Inc. (Tokyo, Japan). All mice were housed under specific-pathogen-free (SPF) conditions at 22 ± 2°C under a 12-h light/dark cycle with ad libitum access to food and water. The animal care and experiments were approved by the Institutional Animal Care and Use Committee of Kitasato University (approval no. 18-040), and all animal experiments were performed in accordance with the approved guidelines.
Mouse experiment.
Before treatment, mice were fasted for 8 h. SPF mice (C57BL/6JJcl, 8 weeks) were administered 100 μl of gently stirred recombinant protein beads or control beads (approximately 107 particles) via gavage. After treatment, the mice were allowed ad libitum access to food and water. Intestinal tissue collection was performed 24 h after bead administration. Tissue clearing was performed as described previously using the water-based optical clearing agent SeeDB (47), with several modifications to optimize the method for GI tissues (37). Whole-gut images were acquired with a fluorescence stereomicroscope (M205, charge-coupled-device [CCD] camera, DFC7000T; Leica, Wetzlar, Germany) using a 395- to 455-nm mercury short arc lamp. A series of images were aligned to construct a single image using Photoshop CC 2020 (Adobe Systems, San Jose, CA, USA).
For the quantification of microbead fecal shedding, 100 μl of recombinant GroEL- or Tal-immobilized beads or control beads (approximately 108 particles) was administered per mouse by gavage. Next, 10 mg of feces was suspended in 200 μl of 1 M NaCl containing 0.05% Tween 20. The samples were centrifuged at 300 × g for 1 min to remove digestive contents. Then, 50 μl of the supernatant was mixed 1:3 with PBS. The suspension was filtered through nylon mesh (70 μm), and the filtrate (100 μl) was transferred into wells of a black 96-well plate. The fluorescence (excitation, 441 nm; emission, 486 nm) was measured using a TECAN Infinite 200 PRO microplate reader (Tecan Inc., Männedorf, Switzerland). Microbeads from nontreated mouse feces were used as blank samples.
Statistical analysis.
Data were analyzed using GraphPad Prism (GraphPad Software, San Diego, CA, USA). Significant differences were determined using Student’s t test or one-way analysis of variance (ANOVA). P values of <0.05 were considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hiromi Ikadai (Kitasato University) for help with microscopic techniques. We also thank Hanae Fukasawa, Tatsunari Yokoi, and Yuka Kushida (Kitasato University) for technical assistance.
This work was supported by grants-in-aid for young scientists (B; no. 17K15249 and 20K15438 to K.N. and 19K15965 to M.S.) from the Japan Society for the Promotion of Science. This work was also partially supported by the Morinaga Milk Industry Co., Ltd.
We declare no conflict of interest. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. 2007. Development of the human infant intestinal microbiota. PLoS Biol 5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. 2013. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501:426–429. doi: 10.1038/nature12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sung J, Kim S, Cabatbat JJT, Jang S, Jin YS, Jung GY, Chia N, Kim PJ. 2017. Global metabolic interaction network of the human gut microbiota for context-specific community-scale analysis. Nat Commun 8:15393. doi: 10.1038/ncomms15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pizarro-Cerdá J, Cossart P. 2006. Bacterial adhesion and entry into host cells. Cell 124:715–727. doi: 10.1016/j.cell.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Kline KA, Fälker S, Dahlberg S, Normark S, Henriques-Normark B. 2009. Bacterial adhesins in host-microbe interactions. Cell Host Microbe 5:580–592. doi: 10.1016/j.chom.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 7.Nishiyama K, Sugiyama M, Mukai T. 2016. Adhesion properties of lactic acid bacteria on intestinal mucin. Microorganisms 4:34. doi: 10.3390/microorganisms4030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odamaki T, Kato K, Sugahara H, Hashikura N, Takahashi S, Xiao JZ, Abe F, Osawa R. 2016. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol 16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fukuda S, Toh H, Hase K, Oshima K, Nakanishi Y, Yoshimura K, Tobe T, Clarke JM, Topping DL, Suzuki T, Taylor TD, Itoh K, Kikuchi J, Morita H, Hattori M, Ohno H. 2011. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 469:543–547. doi: 10.1038/nature09646. [DOI] [PubMed] [Google Scholar]
- 10.Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Man Lei Y, Jabri B, Alegre M-L, Chang EB, Gajewski TF. 2015. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350:1084–1089. doi: 10.1126/science.aac4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishiyama K, Kawanabe A, Miyauchi H, Abe F, Tsubokawa D, Ishihara K, Yamamoto Y, Mukai T. 2014. Evaluation of bifidobacterial adhesion to acidic sugar chains of porcine colonic mucins. Biosci Biotechnol Biochem 78:1444–1451. doi: 10.1080/09168451.2014.918491. [DOI] [PubMed] [Google Scholar]
- 12.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JAM, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc Natl Acad Sci U S A 108:11217–11222. doi: 10.1073/pnas.1105380108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turroni F, Serafini F, Foroni E, Duranti S, O'Connell Motherway M, Taverniti V, Mangifesta M, Milani C, Viappiani A, Roversi T, Sánchez B, Santoni A, Gioiosa L, Ferrarini A, Delledonne M, Margolles A, Piazza L, Palanza P, Bolchi A, Guglielmetti S, van Sinderen D, Ventura M. 2013. Role of sortase-dependent pili of Bifidobacterium bifidum PRL2010 in modulating bacterium-host interactions. Proc Natl Acad Sci U S A 110:11151–11156. doi: 10.1073/pnas.1303897110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki K, Nishiyama K, Miyajima H, Osawa R, Yamamoto Y, Mukai T. 2016. Adhesion properties of a putative polymorphic fimbrial subunit protein from Bifidobacterium longum subsp. longum. Biosci Microbiota Food Health 35:19–27. doi: 10.12938/bmfh.2015-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milani C, Mangifesta M, Mancabelli L, Lugli GA, Mancino W, Viappiani A, Faccini A, van Sinderen D, Ventura M, Turroni F. 2017. The sortase-dependent fimbriome of the genus bifidobacterium: extracellular structures with potential to modulate microbe-host fialogue. Appl Environ Microbiol 83:e01295-17. doi: 10.1128/AEM.01295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishiyama K, Yamamoto Y, Sugiyama M, Takaki T, Urashima T, Fukiya S, Yokota A, Okada N, Mukai T. 2017. Bifidobacterium bifidum extracellular sialidase enhances adhesion to the mucosal surface and supports carbohydrate assimilation. mBio 8:e00928-17. doi: 10.1128/mBio.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González-Rodríguez I, Sánchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A. 2012. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol 78:3992–3998. doi: 10.1128/AEM.08024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Candela M, Biagi E, Centanni M, Turroni S, Vici M, Musiani F, Vitali B, Bergmann S, Hammerschmidt S, Brigidi P. 2009. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155:3294–3303. doi: 10.1099/mic.0.028795-0. [DOI] [PubMed] [Google Scholar]
- 19.Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. 2010. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156:1609–1618. doi: 10.1099/mic.0.038307-0. [DOI] [PubMed] [Google Scholar]
- 20.Saraoui T, Parayre S, Guernec G, Loux V, Montfort J, Cam AL, Boudry G, Jan G, Falentin H. 2013. A unique in vivo experimental approach reveals metabolic adaptation of the probiotic Propionibacterium freudenreichii to the colon environment. BMC Genomics 14:911. doi: 10.1186/1471-2164-14-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takagi R, Sasaki K, Sasaki D, Fukuda I, Tanaka K, Yoshida K, Kondo A, Osawa R. 2016. A single-batch fermentation system to simulate human colonic microbiota for high-throughput evaluation of prebiotics. PLoS One 11:e0160533. doi: 10.1371/journal.pone.0160533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sasaki D, Sasaki K, Ikuta N, Yasuda T, Fukuda I, Kondo A, Osawa R. 2018. Low amounts of dietary fibre increase in vitro production of short-chain fatty acids without changing human colonic microbiota structure. Sci Rep 8:435. doi: 10.1038/s41598-017-18877-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Witwer KW, Théry C. 2019. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J Extracell Vesicles 8:1648167. doi: 10.1080/20013078.2019.1648167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Toyofuku M, Nomura N, Eberl L. 2019. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol 17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 25.Berleman J, Auer M. 2013. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ Microbiol 15:347–354. doi: 10.1111/1462-2920.12048. [DOI] [PubMed] [Google Scholar]
- 26.Guerrero-Mandujano A, Hernández-Cortez C, Ibarra JA, Castro-Escarpulli G. 2017. The outer membrane vesicles: secretion system type zero. Traffic 18:425–432. doi: 10.1111/tra.12488. [DOI] [PubMed] [Google Scholar]
- 27.Natsume T, Nakayama H, Isobe T. 2001. BIA-MS-MS: biomolecular interaction analysis for functional proteomics. Trends Biotechnol 19:28–33. doi: 10.1016/S0167-7799(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 28.Dallo SF, Kannan TR, Blaylock MW, Baseman JB. 2002. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol Microbiol 46:1041–1051. doi: 10.1046/j.1365-2958.2002.03207.x. [DOI] [PubMed] [Google Scholar]
- 29.Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthésy-Theulaz IE. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect Immun 72:2160–2169. doi: 10.1128/iai.72.4.2160-2169.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthésy-Theulaz IE. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect Immun 74:425–434. doi: 10.1128/IAI.74.1.425-434.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Floto RA, MacAry PA, Boname JM, Mien TS, Kampmann B, Hair JR, Huey OS, Houben ENG, Pieters J, Day C, Oehlmann W, Singh M, Smith KGC, Lehner PJ. 2006. Dendritic cell stimulation by mycobacterial Hsp70 is mediated through CCR5. Science 314:454–458. doi: 10.1126/science.1133515. [DOI] [PubMed] [Google Scholar]
- 32.Kunert A, Losse J, Gruszin C, Hühn M, Kaendler K, Mikkat S, Volke D, Hoffmann R, Jokiranta TS, Seeberger H, Moellmann U, Hellwage J, Zipfel PF. 2007. Immune evasion of the human pathogen Pseudomonas aeruginosa: elongation factor Tuf is a factor H and plasminogen binding protein. J Immunol 179:2979–2988. doi: 10.4049/jimmunol.179.5.2979. [DOI] [PubMed] [Google Scholar]
- 33.Kinnby B, Booth NA, Svensäter G. 2008. Plasminogen binding by oral streptococci from dental plaque and inflammatory lesions. Microbiology 154:924–931. doi: 10.1099/mic.0.2007/013235-0. [DOI] [PubMed] [Google Scholar]
- 34.Katakura Y, Sano R, Hashimoto T, Ninomiya K, Shioya S. 2010. Lactic acid bacteria display on the cell surface cytosolic proteins that recognize yeast mannan. Appl Microbiol Biotechnol 86:319–326. doi: 10.1007/s00253-009-2295-y. [DOI] [PubMed] [Google Scholar]
- 35.Boone TJ, Tyrrell GJ. 2012. Identification of the actin and plasminogen binding regions of group B streptococcal phosphoglycerate kinase. J Biol Chem 287:29035–29044. doi: 10.1074/jbc.M112.361261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiyama K, Ochiai A, Tsubokawa D, Ishihara K, Yamamoto Y, Mukai T. 2013. Identification and characterization of sulfated carbohydrate-binding protein from Lactobacillus reuteri. PLoS One 8:e83703. doi: 10.1371/journal.pone.0083703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishiyama K, Sugiyama M, Yamada H, Makino K, Ishihara S, Takaki T, Mukai T, Okada N. 2019. A new approach for analyzing an adhesive bacterial protein in the mouse gastrointestinal tract using optical tissue clearing. Sci Rep 9:4371. doi: 10.1038/s41598-019-41151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fanning S, Hall LJ, Cronin M, Zomer A, MacSharry J, Goulding D, Motherway MO, Shanahan F, Nally K, Dougan G, van Sinderen D. 2012. Bifidobacterial surface-exopolysaccharide facilitates commensal-host interaction through immune modulation and pathogen protection. Proc Natl Acad Sci U S A 109:2108–2113. doi: 10.1073/pnas.1115621109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Connell Motherway M, Houston A, O'Callaghan G, Reunanen J, O'Brien F, O'Driscoll T, Casey PG, de Vos WM, van Sinderen D, Shanahan F. 2019. A bifidobacterial pilus-associated protein promotes colonic epithelial proliferation. Mol Microbiol 111:287–301. doi: 10.1111/mmi.14155. [DOI] [PubMed] [Google Scholar]
- 40.Turnbull L, Toyofuku M, Hynen AL, Kurosawa M, Pessi G, Petty NK, Osvath SR, Cárcamo-Oyarce G, Gloag ES, Shimoni R, Omasits U, Ito S, Yap X, Monahan LG, Cavaliere R, Ahrens CH, Charles IG, Nomura N, Eberl L, Whitchurch CB. 2016. Explosive cell lysis as a mechanism for the biogenesis of bacterial membrane vesicles and biofilms. Nat Commun 7:11220. doi: 10.1038/ncomms11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Toyofuku M, Cárcamo-Oyarce G, Yamamoto T, Eisenstein F, Hsiao CC, Kurosawa M, Gademann K, Pilhofer M, Nomura N, Eberl L. 2017. Prophage-triggered membrane vesicle formation through peptidoglycan damage in Bacillus subtilis. Nat Commun 8:481. doi: 10.1038/s41467-017-00492-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orench-Rivera N, Kuehn MJ. 2016. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 18:1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.López P, González-Rodríguez I, Sánchez B, Gueimonde M, Margolles A, Suárez A. 2012. Treg-inducing membrane vesicles from Bifidobacterium bifidum LMG13195 as potential adjuvants in immunotherapy. Vaccine 30:825–829. doi: 10.1016/j.vaccine.2011.11.115. [DOI] [PubMed] [Google Scholar]
- 44.Kim JH, Jeun EJ, Hong CP, Kim SH, Jang MS, Lee EJ, Moon SJ, Yun CH, Im SH, Jeong SG, Park BY, Kim KT, Seoh JY, Kim YK, Oh SJ, Ham JS, Yang BG, Jang MH. 2016. Extracellular vesicle-derived protein from Bifidobacterium longum alleviates food allergy through mast cell suppression. J Allergy Clin Immunol 137:507.e8–516.e8. doi: 10.1016/j.jaci.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 45.Kainulainen V, Korhonen T, Kainulainen V, Korhonen TK. 2014. Dancing to another tune–adhesive moonlighting proteins in bacteria. Biology (Basel) 3:178–204. doi: 10.3390/biology3010178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hayano T, Yamauchi Y, Asano K, Tsujimura T, Hashimoto S, Isobe T, Takahashi N. 2008. Automated SPR-LC-MS/MS system for protein interaction analysis. J Proteome Res 7:4183–4190. doi: 10.1021/pr700834n. [DOI] [PubMed] [Google Scholar]
- 47.Ke MT, Fujimoto S, Imai T. 2013. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 16:1154–1161. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.