Nicotine is a major tobacco alkaloid in tobacco waste. Pyridine and pyrrolidine pathways are the two best-elucidated nicotine metabolic pathways; Pseudomonas geniculata N1 catabolizes nicotine via a hybrid between the pyridine and pyrrolidine pathways. The crucial enzyme, 6-hydroxypseudooxynicotine amine oxidase (HisD), links the upstream and downstream portions of the VPP pathway; however, there is little structural information about this important enzyme. In this study, we determined the crystal structure of HisD from Pseudomonas geniculata N1. Its basic insights about the structure may help us to guide the engineering of such enzymes for bioremediation and bioconversion applications.
KEYWORDS: nicotine, 6-hydroxypseudooxynicotine amine oxidase, VPP pathway
ABSTRACT
Bacteria degrade nicotine mainly using pyridine and pyrrolidine pathways. Previously, we discovered a hybrid of the pyridine and pyrrolidine pathways (the VPP pathway) in Pseudomonas geniculata N1 and characterized its key enzyme, 6-hydroxypseudooxynicotine amine oxidase (HisD). It catalyzes oxidative deamination of 6-hydroxypseudooxynicotine to 6-hydroxy-3-succinoylsemialdehyde-pyridine, which is the crucial step connecting upstream and downstream portions of the VPP pathway. We determined the crystal structure of wild-type HisD to 2.6 Å. HisD is a monomer that contains a flavin mononucleotide, an iron-sulfur cluster, and ADP. On the basis of sequence alignment and structure comparison, a difference has been found among HisD, closely related trimethylamine dehydrogenase (TMADH), and histamine dehydrogenase (HADH). The flavin mononucleotide (FMN) cofactor is not covalently bound to any residue, and the FMN isoalloxazine ring is planar in HisD compared to TMADH or HADH, which forms a 6-S-cysteinyl flavin mononucleotide cofactor and has an FMN isoalloxazine ring in a “butterfly bend” conformation. Based on the structure, docking study, and site-directed mutagenesis, the residues Glu60, Tyr170, Asp262, and Trp263 may be involved in substrate binding. The expanded understanding of the substrate binding mode from this study may guide rational engineering of such enzymes for biodegradation of potential pollutants or for bioconversion to generate desired products.
IMPORTANCE Nicotine is a major tobacco alkaloid in tobacco waste. Pyridine and pyrrolidine pathways are the two best-elucidated nicotine metabolic pathways; Pseudomonas geniculata N1 catabolizes nicotine via a hybrid between the pyridine and pyrrolidine pathways. The crucial enzyme, 6-hydroxypseudooxynicotine amine oxidase (HisD), links the upstream and downstream portions of the VPP pathway; however, there is little structural information about this important enzyme. In this study, we determined the crystal structure of HisD from Pseudomonas geniculata N1. Its basic insights about the structure may help us to guide the engineering of such enzymes for bioremediation and bioconversion applications.
INTRODUCTION
Nicotine is a major tobacco alkaloid and the primary toxin in tobacco waste. It can easily spread in the environment because it is water soluble and is emerging as a public health threat (1–4). As a result, nicotine was classified as a “toxic release inventory” chemical by the U.S. Environmental Protection Agency in 1994 (5). Microbial treatment is an important tool for removing nicotine from tobacco industry waste. Many microorganisms that use various nicotine degradation pathways have been isolated from the environment, and nicotine degradation pathways have been widely investigated by researchers. Nicotine catabolism pathways can be divided into three different types on the basis of identified intermediates: the pyridine pathway of the Gram-positive bacterium Arthrobacter sp. (6), the pyrrolidine pathway of the Gram-negative bacterium Pseudomonas putida S16 (7), and a hybrid of pyridine and pyrrolidine pathways (the VPP pathway) present in Ochrobactrum sp. strain SJY1 (8), Pseudomonas geniculata N1 (9), and Agrobacterium tumefaciens S33 (10).
The VPP pathway is congruent with the upstream part of the pyridine pathway and the downstream portion of the pyrrolidine pathway. Although the VPP pathway has been genetically characterized and most of the catalytic enzymes in this pathway have been functionally analyzed (6–11), many challenges remain in understanding the biochemistry and structural mechanism of key enzymes in the pathway. 6-Hydroxypseudooxynicotine (6-HPON) amine oxidase catalyzes the transformation of 6-HPON to 6-hydroxy-3-succinoylsemialdehyde-pyridine (HSSAP) (Fig. 1A), which links the upstream and downstream portions of the VPP pathway. Recently, 6-HPON amine oxidases have been found in Agrobacterium tumefaciens S33 (10) and Pseudomonas geniculata N1 (9); however, there is little structural information about this important enzyme.
FIG 1.
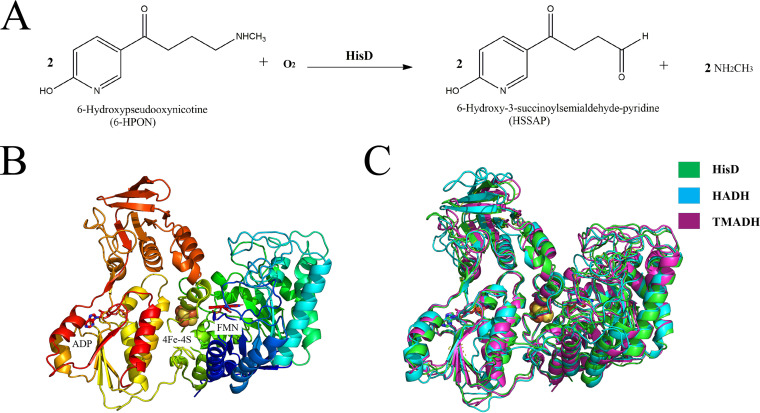
HisD structure. (A) Reaction scheme showing the HisD-catalyzed conversion of 6-hydroxypseudooxynicotine (6-HPON) to 6-hydroxy-3-succinoylsemialdehyde-pyridine (HSSAP). (B) Schematic representation of the HisD structure. The structure is colored in rainbow colors. (C) Structural comparison of HisD (green) with HADH (PDB 3K30; blue) and TMADH (PDB 1DJQ; magenta).
In this study, we determined the crystal structure of HisD from Pseudomonas geniculata N1 to a resolution of 2.6 Å. 6-HPON amine oxidase (HisD) is a homolog of trimethylamine dehydrogenase (TMADH), which catalyzes the oxidative N-demethylation of trimethylamine to form dimethylamine and formaldehyde, from Methylotrophus methylophilus and histamine dehydrogenase (HADH), which catalyzes the oxidative deamination of histamine to imidazole acetaldehyde, from Nocardioides simplex (12, 13), sharing ∼40% and ∼50% sequence identity with those proteins, respectively. However, we found significant differences that indicate HisD is a functionally distinct enzyme. We use docking study and mutation experiments to identify the key residues that might be involved in substrate binding. The structure of HisD suggests that HisD is unique compared to HADH and TMADH, and they do not share a mechanism. Structure and docking studies on HisD are needed to unveil the mechanism used by HisD to recognize its substrate and provide a basis for further understanding the molecular mechanisms of nicotine biodegradation.
RESULTS
Overall structure of HisD.
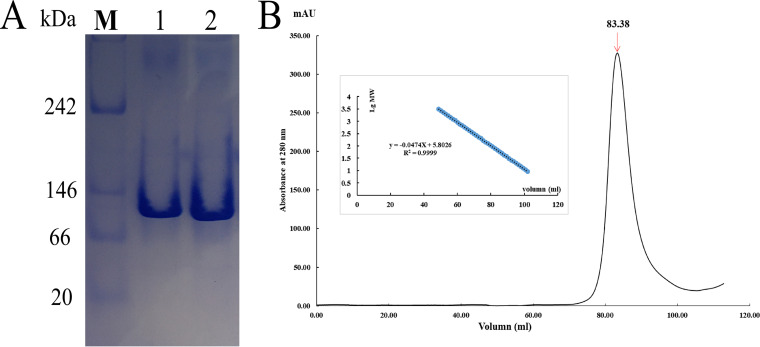
Recombinant HisD was crystallized in the space group C222 with one molecule per asymmetric unit (Fig. 1B). Analytical native PAGE and size exclusion chromatography on Superdex 200 confirmed that HisD functions as a monomer in solution (Fig. 2). Full-length HisD contains 672 residues. Residues 1 to 669 could be modeled discretely, but the last three residues were disordered. A DALI server search (14) revealed that functionally characterized proteins are histamine dehydrogenase from Nocardioides simplex (Protein Data Bank [PDB] 3K30, Z score = 57.6, root mean square deviation [RMSD] of 1.5 Å for 664 Cα atoms, and sequence identity of 50%) and trimethylamine dehydrogenase from Methylophilus methylotrophus W3A1 (PDB 1DJQ, Z score = 57.4, RMSD of 1.6 Å for 664 Cα atoms, and sequence identity of 40%). The overall structure of HisD is similar to other oxidoreductases, including 2-naphthoyl-CoA reductase (NCR; PDB 6QKG, Z score = 47.1, RMSD of 2.7 Å for 636 Cα atoms, and sequence identity of 26%) and DCR (PDB 1PS9, Z score = 45.9, RMSD of 2.5 Å for 623 Cα atoms, and sequence identity of 24%). All of the structural homologs mentioned above exhibit N-terminal domain binding to flavin mononucleotide (FMN). Both the primary sequence and the tertiary structure of HisD are similar to those of homologous amine oxidases HADH and TMADH (Fig. 1C).
FIG 2.
Analysis of the polymerization state of the HisD monomer in solution. (A) Native PAGE of purified HisD. Lane “M” represents the protein marker. Lanes 1 and 2 represent purified HisD. (B) Analysis of size exclusion chromatography on Superdex 200; the elution volume of HisD is 83.38 ml. (Inset) Log plot of the molecular masses of the standard proteins used versus their elution volume values.
The overall fold of the HisD is comprised of three domains, a large domain (residues 1 to 378), a medium domain (residues 379 to 487 and residues 617 to 669), and a small domain (residues 489 to 616). The large domain contains an N-terminal TIM barrel (15), consisting of eight parallel β-strands on the inside covered by eight α-helices on the outside that bind to FMN. The [4Fe-4S] cluster is coordinated to four cysteine residues (Cys344, Cys347, Cys350, and Cys362) and is positioned outside the barrel and close to the medium and small domains. A molecular ADP, located in the medium domain and exposed to solvent, was observed. These domains will be referred to as the N-terminal, middle, and C-terminal domains, respectively.
FMN binding site of HisD.
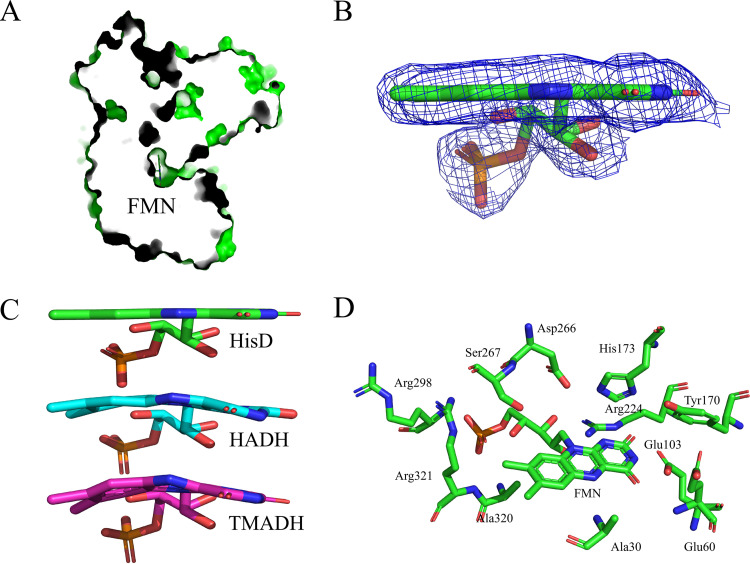
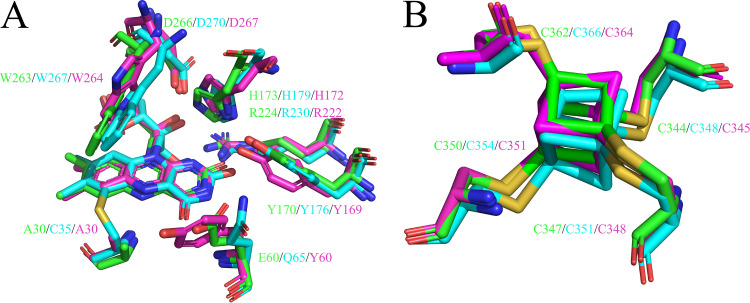
The FMN cofactor adopts an extended conformation and is deeply inserted into the N-terminal domain (Fig. 3A). The electron density for the FMN is very clear (Fig. 3B), suggesting that the isoalloxazine ring of FMN in HisD is planar rather than in the “butterfly bend” conformation that exists in TMADH and HADH (Fig. 3C). The HisD FMN-binding site is shown in Fig. 3D. The ribityl chain forms hydrogen bonds with the Arg224 guanidinium group, Asp266 carboxyl group, and Ser267 hydroxyl group. The N-1, N-3, and N-5 atoms of the isoalloxazine ring form a hydrogen bond with the Arg224 guanidinium group, Glu103 carboxyl group, and backbone nitrogen atom of Ala30, respectively. The Glu103 carboxyl group also forms hydrogen bonds with the O-2 atom of the isoalloxazine ring. The amide side chain of Tyr170 and His173 and the Glu60 backbone nitrogen atom form hydrogen bonds with the O-4 atom of the isoalloxazine ring. The FMN phosphate group is surrounded by Arg298, Ala320, and Arg321 backbone nitrogen atoms. The Arg321 guanidinium group also forms hydrogen bonds with the phosphate. The stereoviews of the FMN-binding site from HisD, HADH, and TMADH were demonstrated (Fig. 4A). Among these structures, a Tyr-His-Asp triad was located on the isoalloxazine ring of FMN. Tyr169 has been shown to stabilize the semiquinone of FMN and [4Fe-4S]+ (16). The Y170A mutant of HisD showed the loss of enzyme activity, suggesting that the corresponding residue also functions directly in HisD. The factors contributing to the butterfly bend in HADH and TMADH have not been defined. Cys35 forms a covalent bond with FMN in HADH (Fig. 4A), which is not found between corresponding residues and FMN in HisD or TMADH. The cysteinyl cross-link formation is not enough to account for the bend of FMN. We have not been able to find particular interactions that contribute to the extent of the butterfly bend in structural observations.
FIG 3.
FMN binding sites. (A) Surface presentation of the FMN binding cavity. (B) FMN in HisD: electron density map around FMN; the 2FO-FC electron density map is contoured at 1σ. (C) Stereoview of FMN from HisD (carbon shown in green) and the orientation of the isoalloxazine ring system compared to that in HADH (carbon shown in magenta) and TMADH (carbon shown in yellow). (D) Stereoview of the FMN binding sites.
FIG 4.
Stereoviews of the active site of HisD (green), HADH (cyan), and TMADH (magenta). The superimposition was done on the whole molecule. (A) Stereoview of the FMN of HisD, HADH, and TMADH. (B) Stereoview of the [4Fe-4S] of HisD, HADH, and TMADH.
[4Fe-4S] cluster and ADP binding site of HisD.
The HisD [4Fe-4S] cluster appeared in the crystal structure as a cysteine-coordinated [4Fe–4S] cluster, and the stereoview of the [4Fe–4S] binding site from HisD, HADH, and TMADH showed that the residues involved in binding the [4Fe-4S] cluster are conserved (Fig. 4B). The [4Fe-4S] cluster is coordinated to four Cys residues (Cys344, Cys347, Cys350, and Cys362) in HisD. The distance between the [4Fe-4S] iron and the FMN 8α-methyl carbon is 5.6 Å, and the distance to the closest oxygen atom of ADP phosphate group is 15 Å. It has been proposed that electrons from the substrate-reduced flavin pass from the 8α-methyl to the closest Cys ligand upon deoxidation in TMADH (17). HisD may use an electron transfer mechanism.
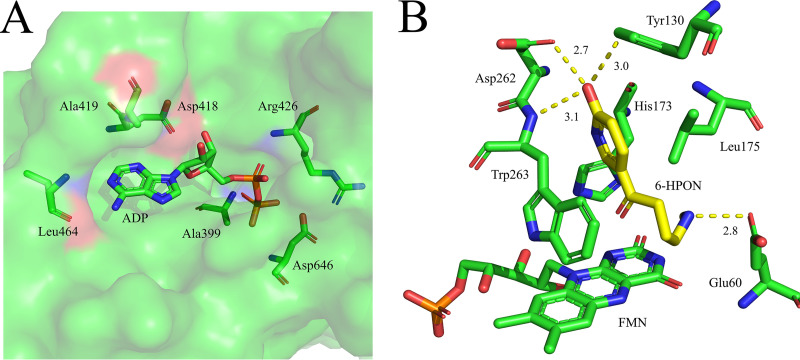
The ADP is exposed to solvent, the adenosine ring N-1 and N-6 atoms form a hydrogen bond with nitrogen and carbonyl oxygen atoms in the Leu464 backbone (Fig. 5A). ADP’s ribose forms a hydrogen bond with the Asp418 carboxyl group. Ala399, Arg426, and Asp646 backbone nitrogen atoms form hydrogen bonds with the ADP phosphate groups. The apparent 6-HPON Km and kcat values for HisD were 168.4 ± 7.2 μM and 0.23 ± 0.11 s−1, respectively.
FIG 5.
Stereoviews of the ADP and 6-hydroxypseudooxynictine binding sites in HisD. (A) Closeup view of the ADP-binding site on HisD. (B) Stereoview of 6-hydroxypseudooxynicotine modeled into the HisD active site.
Substrate binding by HisD.
To understand the molecular basis for the recognition of substrate 6-HPON, we performed molecular docking calculations of 6-HPON binding to HisD. 6-HPON is positioned over the FMN isoalloxazine ring (Fig. 5B). The positively charged 6HPON amino group forms salt bridges or hydrogen bonding interactions with Glu60 carboxylate groups (2.8 Å). The Asn262 carboxylate group hydrogen bonds with the hydroxyl of 6HPON (2.7 Å), as does the Tyr130 amide side chain and N atom on the peptide bond forms of Asn262 and Trp263 (3.0 and 3.1 Å). In addition to the hydrogen bonding residues, hydrophobic side chains of Leu175 and Trp263 line the binding pocket and have van der Waals contact with 6-HPON. Leu175 interacts with 6-HPON through the side chain pyridine ring via a CH-π interaction, and the Trp263 aromatic ring is involved in a π-stacking interaction with the 6-HPON pyridine ring. This positioning places the protonated 6-HPON amino group close to the isoalloxazine ring, as required for catalysis. To corroborate our docking study, we created point mutants of critical residues on the active site of HisD and measured the kinetic constants of mutants of HisD. In comparison with values for HisD, the catalytic activity of the Y130A mutant was reduced by half and the Km value was increased by 1.6-fold. In addition, point mutations of E60A, D262A, and W263A of HisD resulted in the complete loss of enzymatic activity (Table 1).
TABLE 1.
Kinetic analysis of HisD wild-type and mutantsa
| Enzyme | Mean ± SEM |
|
|---|---|---|
| kcat (s−1) | Km (μM) | |
| Wild type | 0.23 ± 0.11 | 168.4 ± 7.2 |
| E60A | ND | ND |
| Y130A | 0.12 ± 0.02 | 273.6 ± 19.1 |
| Y170A | ND | ND |
| D262A | ND | ND |
| W263A | ND | ND |
ND, not detected (the kinetic constants were beneath the measurable limits of detection and thus were not detectable).
Voltammetric evaluation of the redox potentials of HisD.
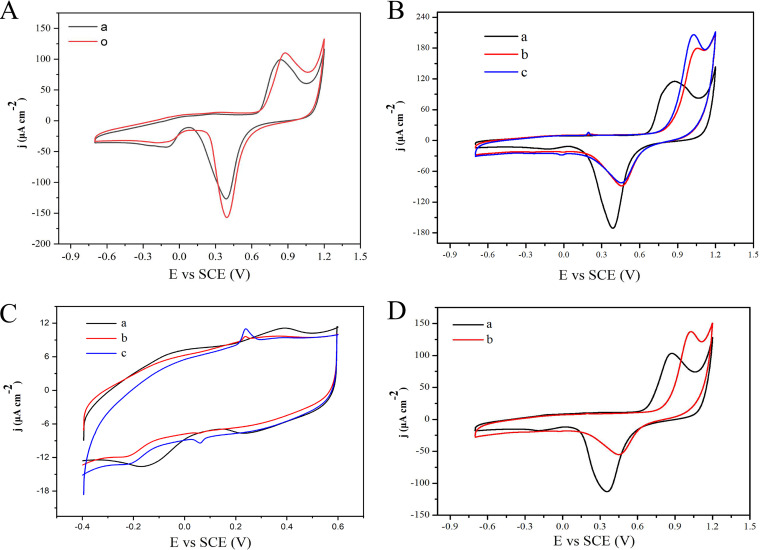
We tried to obtain an electrochemical signal for HisD using cyclic voltammetry. According to “curve o” (red) in Fig. 6A, the NPG electrode has a pair of redox peaks (oxidation peak potential, +0.9V; reduction peak potential, +0.4V). The redox peak of NPG in “curve a” (black) was lower than that before enzyme modification, which proved that the enzyme binding obscured some active sites of NPG, leading to the reduction of its redox peak. In order to detect the redox potential of HisD, 50 and 100 μM 6-HPON were added, and the results showed that the substrates caused emergence of a new redox peak with an oxidation peak potential of +0.2 V and a reduction peak potential of 0.0V (Fig. 6B and C). In order to further confirm that the appearance of this peak was caused by HisD, we repeated the above-described experiment with the NPG/GCE electrode, and the results showed that no new peak appeared (Fig. 6D). Based on these results, the redox potential of HisD was evaluated to be +0.2 V at pH 7.4.
FIG 6.
Cyclic voltammogram (CV) of HisD. (A) CV of the NPG/GCE electrode (curve o) and HisD/NPG/GCE electrode (curve a) in PBS (50 mM [pH 7.4]). (B) CV of the HisD/NPG/GCE electrode in PBS (50 mM [pH 7.4]) with 0 μM 6-HPON (curve a), 50 μM 6-HPON (curve b), and 100 μM 6-HPON (curve c) in a wide range of assays. (C) CV of the HisD/NPG/GCE electrode in PBS (50 mM [pH 7.4]) with 0 μM 6-HPON (curve a), 50 μM 6-HPON (curve b) and 100 μM 6-HPON (curve c) in a small range of assays. (D) CV of the NPG/GCE electrode in PBS (50 mM [pH 7.4]) with 0 μM 6-HPON (curve a) and 100 μM 6-HPON (curve b).
DISCUSSION
Several crystal structures for enzymes involved in the pyridine and pyrrolidine nicotine degradation pathways have been reported (18–22). However, as of now, there are no available crystal structures for enzymes participating in the VPP pathway. In this study, we determined the structure of full-length HisD, which is involved in removing methylamine from 6-hydroxypseudooxynicotine in the VPP pathway. Sequence alignment and structural comparisons with homologous enzymes revealed that the spatial arrangements of cofactors are similar in these enzymes and that the residues involved in binding cofactors are conserved (Fig. 4 and 7).
FIG 7.
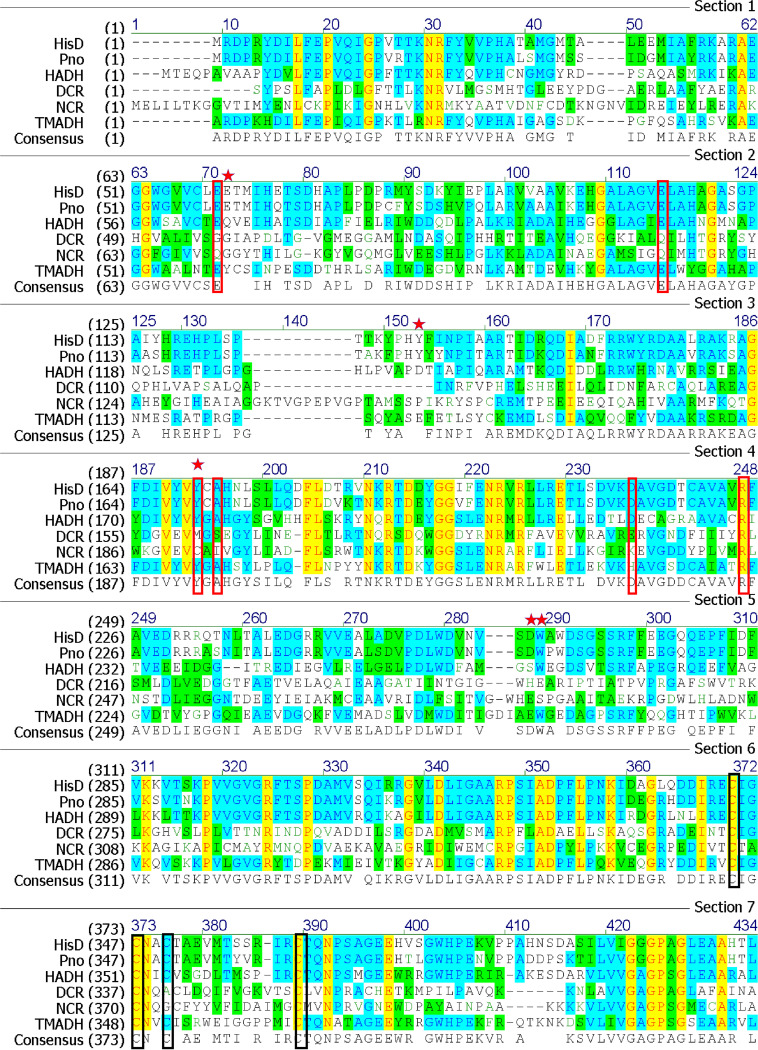
Sequence alignment of HisD, Pno (NCBI accession no. WP_024899819.1), HADH (PDB 3K30), TMADH (PDB 1DJQ), NCR (PDB 6QKG), and DCR (PDB 1PS9). The alignment was determined using Clustal W, and the result was generated with ESPript. The FMN binding sites are outlined in red, [4Fe-4S] cluster binding sites are outlined in black, and the mutation sites in HisD are marked with a pentagon.
In HisD, the FMN isoalloxazine ring is planar, rather than in the “butterfly bend” conformation that is seen in TMADH and HADH (Fig. 3C). The FMN isoalloxazine ring is tethered to the polypeptide through a covalent bond between Cys35 and the FMN C-6 atom, whereas the FMN is not covalently attached in either TMADH or HisD. Further, Ala30 replaces the thiol residue of HADH (Fig. 4A). Binding sites for ADP were confirmed in HisD as Ala399, Asp418, Ala419, Arg426, Leu464, and Asp646 (Fig. 5A).
In TMADH, the Tyr-His-Asp triad, comprising Tyr169, His172, and Asp267, has been proposed to stabilize the semiquinone of FMN and [4Fe-4S]+ (16). Charge repulsion from the negative charge on the Tyr hydroxyl group pushes the unpaired electron density on the FMN toward [4Fe-4S] and mediates a spin-spin interaction. The His residue remains neutral to achieve the maximum rate of 6-S-Cys-FMN reduction (23). Corresponding to Tyr176-His179-Asp270 residues in HADH, Tyr170-His173-Asp266 observed in HisD are located close to the pyrimidine moiety of the FMN isoalloxazine ring. Residues involved in binding the [4Fe-4S] cluster are highly conserved in HisD, HADH, TMADH, NCR, and DCR (Fig. 7). The spatial arrangement of the [4Fe-4S] cluster in HisD is very similar to that in TMADH and HADH (Fig. 1C).
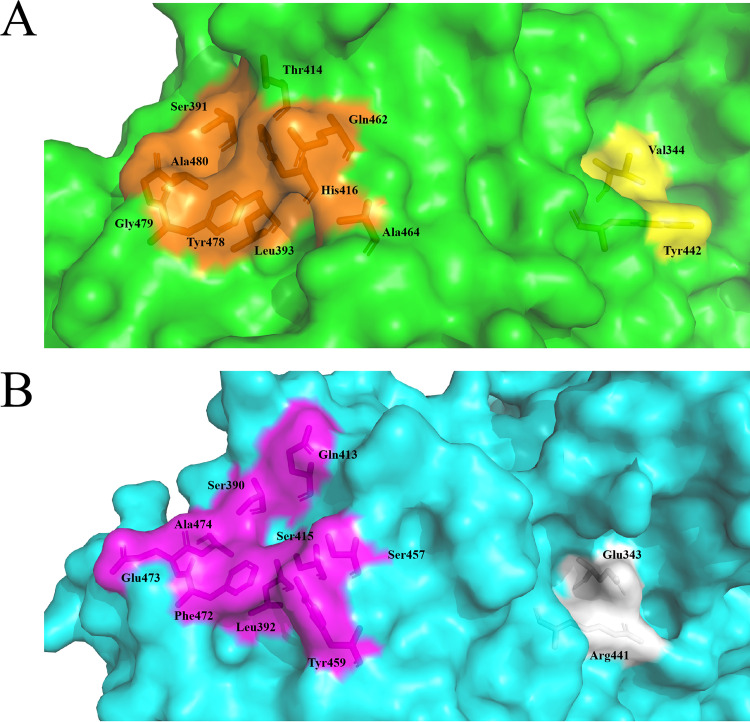
Crystal structures of the TMADH-ETF complex revealed that the proposed recognition peptide sequence of ETF (24) interacts with a shallow surface groove on TMADH, comprised of nine amino acids (Ser391, Leu393, Thr414, His416, Gln462, Ala464, Tyr478, Gly479, and Ala480) (Fig. 8A). Leu194 in the ETF subunit is proposed to act as an anchor that positions ETF in the TMADH groove via van der Waals interactions with Leu393 and Tyr478 (25). Although the presence of an ETF-like protein has not yet been determined in Pseudomonas geniculata N1, inspection of the HisD crystal structure suggests there is a similar shallow groove on the surface (Fig. 8B). From the sequence alignment, the corresponding residues in HisD are Ser390, Leu392, Gln413, Ser415, Ser457, Tyr459, Phe472, Glu473, and Ala474. The residues Ser390, Leu392, and Ala480 are conserved, and Phe472 is homologous to Tyr478 in HisD. The hydrophobic patch in the groove, comprising Ser391, Leu393, Ala464, Tyr478, and Ala480, that interacts with Leu194 in the ETF recognition loop also exists in HisD (Ser390, Leu392, Tyr459, Phe472, and Ala474). However, four other residues in HisD are charged (Gln413, Ser415, Ser457, and Glu473), in contrast with the corresponding residues in TMADH (Thr414, His416, Gln462, and Gly479). HADH also seems to have a similar shallow groove on the surface, with the corresponding residues being Arg393, Leu395, Asp416, Val418, Tyr462, Arg463, Phe475, Gly476, and Phe477 (13).
FIG 8.
(A) Surface grooves of TMADH involved in recognition loop of the subunit of ETF through van der Waals interaction electron transfer to ETF (orange) and the electron-transfer reaction (yellow). (B) Corresponding HisD surface grooves are shown in magenta and white, respectively.
In TMADH, Val344 is located at the bottom of the surface groove where Tyr442 resides and engages in van der Waals interactions with Cys345 and Tyr442. Val344 has been shown to participate in electron transfer to ferricenium (25). In HisD, the residue corresponding to the Val344 in TMADH is Glu343, and a Glu is also present in HADH. Comparing these three structures, we found that the surface grooves shown in TMADH involved in ETF binding and electron transfer are not strictly conserved in these three enzymes, since there are similarities and differences in groove residues in the three enzymes. For TMADH, there are two proposed pathways for electron transfer from the [4Fe-4S] to the surface Tyr442-Cys345-Glu439-Tyr442 that transfers electrons to ETF or Cys345-Val344 that transfer electrons to ferricenium (17). HADH could also use two analogous electron-transfer pathways. Interestingly, the analogous pair of choices found in HisD are Cys344-Ala438-Arg441 and Cys344-Glu343. Although we have not identified an ETF-like protein in Pseudomonas geniculata N1, it is possible that HisD utilizes the Cys344-Ala438-Arg441 pathway with an electron acceptor protein partner.
The redox potential of HisD has been determined to be +0.2 V, whereas the reported values of HADH and TMADH are +34 and +44 mV, respectively (26, 27). Theoretical studies on lumiflavin and C-6-methylsulfanyllumiflavin suggested that the butterfly bend raises the two electron reduction potentials, making the flavin more reactive to substrate oxidation (28). Compared to those of HADH and TMAHD, the redox potential of HisD is much more positive, which might account for FMN being planar in HisD more than in a “butterfly bend” conformation.
Despite numerous attempts of cocrystallization with 6-HPON, none of the analyzed crystals showed any electron density for bound substrate. Therefore, we modeled the substrate in the HisD active site using the AutoDock method. According to the docking study and mutation experiment, residues Glu60, Tyr170, Asp262, and Try263 are key amino acids. Amino acid sequence alignment showed that residues Glu60, Tyr170, Asp262, and Try263 are conserved in HADH, TMADH, and Pno; these residues may stabilize the substrate (Fig. 7). Further analysis revealed that only Pno and HisD are completely conserved, as they are isoenzymes. However, HADH and TMADH still have some slight differences due to the difference in their substrates.
In summary, we determined the structure of HisD, an enzyme in the VPP nicotine degradation pathway. The overall structure, domain organization, and the spatial arrangement of FMN and [4Fe-4S] are very similar to those of TMADH and HADH. From the docking study, we predict that 6-HPON preferentially binds to the active site in the following manner. The anionic residue Glu60 stabilizes the protonated amino group of 6-HPON by forming a salt bridge, whereas His173, Leu175, and Tyr263 stabilize the pyridine ring via van der Waals and hydrogen bond interactions. In addition to predicting the 6-HPON binding model, this structure will be of further aid in protein engineering of HisD for potential bioremediation and industrial applications.
MATERIALS AND METHODS
Chemical reagents.
l-Nicotine (purity >99%) was bought from Fluka Chemie GmbH (Switzerland). 6-HPON was prepared as previously described (10). Crystallization screens were obtained from Hampton Research. Source 15Q and Superdex 10/300 GL were obtained from GE Healthcare. All other reagents and solvents used in this study were of analytical grade and are commercially available.
Expression and purification of HisD.
The mutants of HisD used in this study were constructed by site-directed PCR mutagenesis, and the constructs were verified via gene sequencing. These plasmids were transformed into P. putida KT2440 by electroporation using the following conditions: 1 to 2 μg plasmid DNA was added to 100 μl of electrocompetent cells of strain KT2440, and the mixture was electroporated at 12 kV/cm, 200 Ω, and 25 μF using a Bio-Rad Gene-Pulser X-cell (Bio-Rad Laboratories, Hercules, CA). The primers are listed in Table 2. HisD and its mutants were expressed in P. putida KT2440 using a pVLT33 vector and purified as described previously (9). Cells were grown to exponential phase at 30°C, induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and then grown at 25°C for 20 h. HisD was purified using Ni2+-NTA affinity chromatography (Qiagen) followed by separation of the eluted fractions by ion-exchange chromatography with a Source 15Q 4.6/100 PE (GE Healthcare) column and Superdex 200 gel filtration chromatography (GE Healthcare). The gel filtration buffer contained 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, and 2 mM dithiothreitol. Subsequently, fractions with putative recombinant HisD proteins (∼74 kDa, assessed via SDS-PAGE) were combined, concentrated to 14 mg/ml, and then flash frozen in liquid nitrogen for storage at –80°C. SDS-PAGE and native PAGE were carried out as previously described (29).
TABLE 2.
Strains, plasmids, and primers used in this study
| Strain, plasmid, or primer | Description or primer sequencea | Source or reference |
|---|---|---|
| Strain | ||
| P. putida KT2440 | Metabolically versatile saprophytic soil bacterium | 9 |
| Plasmids | ||
| pVLT33 | Kanr, expression vector in Pseudomonas | 9 |
| pVLT33-hisD | Kanr, pVLT33 containing hisD | This study |
| Primers | ||
| HisD-E60A-f | GTGCCTCGAGGCAACGATGATACACGAGACGTCG | This study |
| HisD-E60A-r | GTGTATCATCGTTGCCTCGAGGCACACGACC | This study |
| HisD-Y130A-f | AGTATCCCCACGCATTTATCAACCCGATCGCCG | This study |
| HisD-Y130A-r | GGTTGATAAATGCGTGGGGATACTTCGTCGTCG | This study |
| HisD-Y170A-f | CGTGTACGTCGCATGCGCGCACAACCTGTCGCTGC | This study |
| HisD-Y170A-r | TTGTGCGCGCATGCGACGTACACGATGTCAAACC | This study |
| HisD-D262A-f | GAACGTCAGCGCATGGGCTTGGGACAGTGGCAGCAGC | This study |
| HisD-D262A-r | TCCCAAGCCCATGCGCTGACGTTCACGTCCCAAAGG | This study |
| HisD-W263A-f | CGTCAGCGACGCAGCTTGGGACAGTGGCAGCAGCC | This study |
| HisD-W263A-r | CTGTCCCAAGCTGCGTCGCTGACGTTCACGTCCC | This study |
Underlining indicates mutation sites.
Enzyme activity assays.
HisD activity was determined spectrophotometrically as previously described (30), with some modifications. The initial rate of reaction at 30°C was measured by monitoring 2,6-dichlorophenolindophenol (DCIP) reduction at 600 nm in the presence of phenazine methosulfate (PMS). The reaction mixture contained 50 mM Tris-HCl buffer (pH 8.0), 500 μM PMS, 50 μM DCIP, and a 0.5 mM mixture of 6-hydroxy-N-methylmyosmine and 6-hydroxypseudooxynictine. One unit of enzyme activity was defined as the amount of enzyme catalyzing the oxidation of 1 μmol of DCIP per min.
Crystallization, data collection, processing, and refinement.
HisD crystals were grown from a 1:1 (vol/vol) mixture of a HisD protein solution (14 mg/ml) and reservoir solution (0.5 M ammonium sulfate, 0.1 M sodium citrate dihydrate [pH 5.6], and 1.0 M lithium sulfate monohydrate) using the hanging-drop vapor diffusion method at 20°C. Crystallization conditions included 25% (vol/vol) glycerol that was subsequently used as a cryoprotectant, and thus crystals were rapidly looped and transferred into cryoprotectant prior to flash freezing in liquid nitrogen. All 0.9789-Å wavelength data were collected at 100K using a Dectris PILATUS3 6M detector at beamline BL19U1 of the Shanghai Synchrotron Radiation Facility (SSRF). Data were processed with HKL3000 (31). The crystal structure was resolved by the molecular replacement method with the program Phaser (32) using the Nocardioides simplex histamine dehydrogenase (PDB 3K30) structure as the search model. Structure refinement was achieved with the Coot (33) and Refmac5 (34) programs. All molecular figures were generated with PyMOL. The HisD structure has been deposited in the PDB under access code 6L6J. Data collection and refinement statistics for HisD are summarized in Table 3.
TABLE 3.
Data collection and refinement statistics for HisD
| Parameter | Value for HisD |
|---|---|
| Data collection statistics | |
| Space group | C222 |
| Wavelength (Å) | 0.97918 |
| Cell dimensions | |
| a, b, c (Å) | 114.95, 212.99, 79.35 |
| α, β, γ (°) | 90.00, 90.00, 90.00 |
| Molecules per asymmetric unit | 1 |
| Resolution range (Å) (outer shell) | 50.00–2.60 (2.69–2.60) |
| Completeness (%) (outer shell) | 99.7 (97.9) |
| Redundancy (outer shell) | 9.3 (8.2) |
| Total no. of observations | 276,129 |
| No. of unique reflections | 29,678 |
| Rmerge (%) (outer shell) | 16.4 (60.1) |
| I/σI (outer shell) | 12.75 (2.67) |
| Refinement statistics | |
| Resolution range (Å) | 39.94–2.60 |
| Rwork/Rfree (%) | 20.10/23.98 |
| No. of reflections | 28,917 (2,029) |
| RMSD | |
| Bond length (Å) | 0.005 |
| Bond angle (°) | 1.410 |
| Ramachandran plot (%) | |
| Most favored | 94.0 |
| Allowed | 5.4 |
| Disallowed | 0.6 |
| No. of atoms | |
| Protein | 5,360 |
| ADP | 27 |
| FMN | 31 |
| [4Fe-4S] | 8 |
| Water | 123 |
| Avg B-factor (Å2) | |
| Protein | 34.91 |
| ADP | 34.98 |
| FMN | 28.73 |
| [4Fe-4S] | 28.49 |
| Water | 33.49 |
Preparation of HisD/NPG/GCE electrode and cyclic voltammetry.
A glassy carbon electrode (GCE) was polished on a piece of chamois leather with 0.3-μm gamma alumina powder (CH Instruments). The electrodes were then cleaned ultrasonically in absolute ethyl alcohol and ultrapure water for 60 s each. Nanoporous gold (NPG) was made by dealloying silver from 12-carat white gold leaves (5 mm × 5 mm, Au50Ag50 wt%; Sepp Leaf Products) in concentrated HNO3 at 30°C for 30 min and then washed with ultrapure water three times. The GCE was also immersed in water and then slowly covered with NPG leaf to form an NPG/GCE electrode. The electrochemical activity region was detected by cyclic voltammetry (CV) in 0.5 M H2SO4 aqueous solution at a scanning rate of 50 mV s−1. The HisD/NPG/GCE electrode was prepared by immersing an NPG/GCE electrode in HisD enzyme solution (100 μg/ml) for 24 h and was stored at 4°C for future use. The HisD/NPG/GCE electrode was tested as a control in phosphate-buffered saline (PBS; 50 mM [pH 7.4]), and then substrate was added. All electrochemical measurements were carried out in a conventional three-electrode system using a CHI 760E electrochemical workstation (Shanghai Chenhua Apparatus Corporation, China).
Docking study.
Molecular docking was carried out using the software AutoDock (35), and the substrate was docked into the active site of HisD. The coordinates of the central grid (47.442, 50.643, and 22.056) were determined based on the location of the FMN, and the grid box size was set as 40 × 40 × 40 Å3. The programs AutoGrid 4 and AutoDock 4 were employed with default parameters.
Data availability.
The structure of HisD has been deposited in the PDB database under access code 6L6J.
ACKNOWLEDGMENTS
This study was supported by grants from the National Key Research and Development Project (2018YFA0901200) and the Science and Technology Commission of Shanghai Municipality (17JC1403300), by the “Shuguang Program” (17SG09) supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission, and by grants from the Chinese National Science Foundation for Excellent Young Scholars (31422004).
We thank Xia Wang (State Key Laboratory of Microbial Technology, Shandong University, People’s Republic of China) for analyzing the cyclic voltammetry data. We also thank Wen-ming Qin and other staff members at the beamline BL19U1 at the Shanghai Synchrotron Radiation Facility (Shanghai, China) for experimental support.
REFERENCES
- 1.Henningfield JE, Miyasato K, Jasinski DR. 1985. Abuse liability and pharmacodynamic characteristics of intravenous and inhaled nicotine. J Pharmacol Exp Ther 234:1–12. [PubMed] [Google Scholar]
- 2.Benowitz NL, Hukkanen J, Peyton J III. 2009. Nicotine chemistry, metabolism, kinetics and biomarkers. Handb Exp Pharmacol 192:29–60. doi: 10.1007/978-3-540-69248-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Civilini M, Domenis C, Sebastianutto N, Bertoldi MD. 1997. Nicotine decontamination of tobacco agro-industrial waste and its degradation by micro-organisms. Waste Manag Res 15:349–358. doi: 10.1177/0734242X9701500403. [DOI] [Google Scholar]
- 4.Henningfield JE, Benowitz NL, Slade J, Houston TP, Davis RM, Deitchman SD. 1998. Reducing the addictiveness of cigarettes. Tob Control 7:281–293. doi: 10.1136/tc.7.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Novotny TE, Zhao F. 1999. Consumption and production waste: another externality of tobacco use. Tob Control 8:75–80. doi: 10.1136/tc.8.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandsch R. 2006. Microbiology and biochemistry of nicotine degradation. Appl Microbiol Biotechnol 69:493–498. doi: 10.1007/s00253-005-0226-0. [DOI] [PubMed] [Google Scholar]
- 7.Tang HZ, Wang LJ, Wang WW, Yu H, Zhang KZ, Yao YX, Xu P. 2013. Systematic unraveling of the unsolved pathway of nicotine degradation in pseudomonas. PLoS Genet 9:e1003923. doi: 10.1371/journal.pgen.1003923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Tang HZ, Zhu XY, Li YY, Xu P. 2015. Molecular mechanism of nicotine degradation by a newly isolated strain, Ochrobactrum sp. strain SJY1. Appl Environ Microbiol 81:272–281. doi: 10.1128/AEM.02265-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang WW, Zhu XY, Liu X, Wu W, Xu P, Tang HZ. 2019. Cloning and characterization the nicotine degradation enzymes 6-hydroxypseudooxynicotine amine oxidase and 6-hydroxy-3-succinoylpyridine hydroxylase in Pseudomonas geniculata N1. Int J Biodeter Biodegrad 142:83–90. doi: 10.1016/j.ibiod.2019.05.003. [DOI] [Google Scholar]
- 10.Li HL, Xie KB, Yu WJ, Hu LJ, Huang HY, Xie HJ, Wang SN. 2016. Nicotine dehydrogenase complexed with 6-hydroxypseudooxynicotine oxidase involved in the hybrid nicotine-degrading pathway in Agrobacterium tumefaciens S33. Appl Environ Microbiol 82:1745–1755. doi: 10.1128/AEM.03909-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Tang HZ, Li YY, Xu P. 2015. Molybdenum-containing nicotine hydroxylase genes in a nicotine degradation pathway that is a variant of the pyridine and pyrrolidine pathways. Appl Environ Microbiol 81:8330–8338. doi: 10.1128/AEM.02253-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barber MJ, Neame PJ, Lim LW, White S, Matthews FS. 1992. Correlation of x-ray deduced and experimental amino acid sequences of trimethylamine dehydrogenase. J Biol Chem 267:6611–6619. [PubMed] [Google Scholar]
- 13.Timothy R, Gerald HL, Xia Y, Hidehiko H, DeAnna MT, Minae M, Emily ES, Limburg JL. 2010. Crystal structure of histamine dehydrogenase from Nocardioides simplex. J Biol Chem 285:25782–25791. doi: 10.1074/jbc.M109.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liisa H, Sander C. 1995. Dali: a network tool for protein structure comparison. Trends Biochem Sci 20:478–480. doi: 10.1016/S0968-0004(00)89105-7. [DOI] [PubMed] [Google Scholar]
- 15.Wierenga RK. 2001. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett 492:193–198. doi: 10.1016/s0014-5793(01)02236-0. [DOI] [PubMed] [Google Scholar]
- 16.Basran J, Jang MH, Sutcliffe MJ, Hille R, Scrutton NS. 1999. The role of Tyr-169 of trimethylamine dehydrogenase in substrate oxidation and magnetic interaction between FMN cofactor and the 4Fe/4S center. J Biol Chem 274:13155–13161. doi: 10.1074/jbc.274.19.13155. [DOI] [PubMed] [Google Scholar]
- 17.Lim LW, Shamala N, Mathews FS, Steenkamp DJ, Hamlin R, Xuong NH. 1986. Three-dimensional structure of the iron-sulfur flavoprotein trimethylamine dehydrogenase at 2.4-Å resolution. J Biol Chem 261:15140–15146. [PubMed] [Google Scholar]
- 18.Chen DD, Tang HZ, Lv Y, Zhang ZY, Shen KL, Lin K, Zhao YL, Wu G, Xu P. 2013. Structural and computational studies of the maleate isomerase from Pseudomonas putida S16 reveal a breathing motion wrapping the substrate inside. Mol Microbiol 87:1237–1244. doi: 10.1111/mmi.12163. [DOI] [PubMed] [Google Scholar]
- 19.Wu G, Chen DD, Tang HZ, Ren YL, Chen QH, Lv Y, Zhang ZY, Zhao YL, Yao YX, Xu P. 2014. Structural insights into the specific recognition of N-heterocycle biodenitrogenation-derived substrates by microbial amide hydrolases. Mol Microbiol 91:1009–1021. doi: 10.1111/mmi.12511. [DOI] [PubMed] [Google Scholar]
- 20.Wagener N, Pierik AJ, Ibdah A, Hille R, Dobbek H. 2009. The Mo-Se active site of nicotinate dehydrogenase. Proc Natl Acad Sci U S A 106:11055–11060. doi: 10.1073/pnas.0902210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treiber N, Schulz GE. 2008. Structure of 2,6-dihydroxypyridine 3-hydroxylase from a nicotine-degrading pathway. J Mol Biol 379:94–104. doi: 10.1016/j.jmb.2008.03.032. [DOI] [PubMed] [Google Scholar]
- 22.Schleberger C, Sachelaru P, Brandsch R, Schulz GE. 2007. Structure and action of a C-C bond cleaving α/β-hydrolase involved in nicotine degradation. J Mol Biol 367:409–418. doi: 10.1016/j.jmb.2006.12.068. [DOI] [PubMed] [Google Scholar]
- 23.Basran J, Sutcliffe MJ, Scrutton NS. 2001. Optimizing the Michaelis complex of trimethylamine dehydrogenase: identification of interactions that perturb the ionization of substrate and facilitate catalysis with trimethylamine base. J Biol Chem 276:42887–42892. doi: 10.1074/jbc.M108296200. [DOI] [PubMed] [Google Scholar]
- 24.Leys D, Basran J, Talfournier F, Sutcliffe MJ, Scrutton NS. 2003. Extensive conformational sampling in a ternary electron transfer complex. Nat Struct Biol 10:219–225. doi: 10.1038/nsb894. [DOI] [PubMed] [Google Scholar]
- 25.Basran J, Chohan KK, Sutcliffe MJ, Scrutton NS. 2000. Differential coupling through Val-344 and Tyr-442 of trimethylamine dehydrogenase in electron transfer reactions with ferricenium ions and electron transferring flavoprotein. Biochemistry 39:9188–9200. doi: 10.1021/bi0006868. [DOI] [PubMed] [Google Scholar]
- 26.Tsutsumi M, Fujieda N, Tsujimura S, Shirai O, Kano K. 2008. Thermodynamic redox properties governing the half-reduction characteristics of histamine dehydrogenase from Nocardioides simplex. Biosci Biotechnol Biochem 72:786–796. doi: 10.1271/bbb.70665. [DOI] [PubMed] [Google Scholar]
- 27.Pace CP, Stankovich MT. 1991. Oxidation-reduction properties of trimethylamine dehydrogenase: effect of inhibitor binding. Arch Biochem Biophys 287:97–104. doi: 10.1016/0003-9861(91)90393-w. [DOI] [PubMed] [Google Scholar]
- 28.Dixon DA, Lindner DL, Branchaud B, Lipscomb WN. 1979. Conformations and electronic structures of oxidized and reduced isoalloxazine. Biochemistry 18:5770–5775. doi: 10.1021/bi00593a004. [DOI] [PubMed] [Google Scholar]
- 29.Tang HZ, Yao YX, Zhang DK, Meng XZ, Wang LJ, Yu H, Ma LY, Xu P. 2011. A novel NADH-dependent and FAD-containing hydroxylase is crucial for nicotine degradation by Pseudomonas putida. J Biol Chem 286:39179–39187. doi: 10.1074/jbc.M111.283929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ara SJ, Mohammed SS, Shigeo T, Eiichi S, Takamitsu Y. 2000. Purification and characterization of histamine dehydrogenase from Nocardioides simplex IFO 12069. FEMS Microbiol Lett 189:183–187. [DOI] [PubMed] [Google Scholar]
- 31.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. 2006. HKL-3000: the integration of data reduction and structure solution: from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr 62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. 2004. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr 60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 35.Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The structure of HisD has been deposited in the PDB database under access code 6L6J.