A. baumannii is one of the most challenging pathogens for which development of new and effective antimicrobials is urgently needed. Colistin is a last-resort antibiotic, and even colistin-resistant A. baumannii strains exist. Here, we present a lysin that sensitizes A. baumannii for colistin and can revert colistin resistance to colistin susceptibility. The lysin also shows a strong, turgor pressure-dependent intrinsic antibacterial activity, providing new insights in the mode of action of lysins with intrinsic activity against Gram-negative bacteria.
KEYWORDS: Acinetobacter baumannii, antibiotic resistance, bacteriophage, colistin, endolysin
ABSTRACT
The prevalence of extensively and pandrug-resistant strains of Acinetobacter baumannii leaves little or no therapeutic options for treatment for this bacterial pathogen. Bacteriophages and their lysins represent attractive alternative antibacterial strategies in this regard. We used the extensively drug-resistant A. baumannii strain MK34 to isolate the bacteriophage PMK34 (vB_AbaP_PMK34). This phage shows fast adsorption and lacks virulence genes; nonetheless, its narrow host spectrum based on capsule recognition limits broad application. PMK34 is a Fri1virus member of the Autographiviridae and has a 41.8-kb genome (50 open reading frames), encoding an endolysin (LysMK34) with potent muralytic activity (1,499.9 ± 131 U/μM), a typical mesophilic thermal stability up to 55°C, and a broad pH activity range (4 to 10). LysMK34 has an intrinsic antibacterial activity up to 4.8 and 2.4 log units for A. baumannii and Pseudomonas aeruginosa strains, respectively, but only when a high turgor pressure is present. The addition of 0.5 mM EDTA or application of an osmotic shock after treatment can compensate for the lack of a high turgor pressure. The combination of LysMK34 and colistin results in up to 32-fold reduction of the MIC of colistin, and colistin-resistant strains are resensitized in both Mueller-Hinton broth and 50% human serum. As such, LysMK34 may be used to safeguard the applicability of colistin as a last-resort antibiotic.
IMPORTANCE A. baumannii is one of the most challenging pathogens for which development of new and effective antimicrobials is urgently needed. Colistin is a last-resort antibiotic, and even colistin-resistant A. baumannii strains exist. Here, we present a lysin that sensitizes A. baumannii for colistin and can revert colistin resistance to colistin susceptibility. The lysin also shows a strong, turgor pressure-dependent intrinsic antibacterial activity, providing new insights in the mode of action of lysins with intrinsic activity against Gram-negative bacteria.
INTRODUCTION
Multidrug-resistant (MDR) bacteria have emerged as a notorious problem that poses a real threat to global health (1). Particularly, Acinetobacter baumannii has gained increasing attention as an opportunistic pathogen, causing a wide range of hospital-acquired infections. These infections result in symptoms ranging from mild symptomatic cases to life-threatening ventilator-associated pneumonia. A. baumannii infections have a poor prognosis with high mortality. Treatment of A. baumannii infections is challenging due to their resistance to a large variety of antimicrobials (including carbapenems, aminoglycosides, tetracyclines, quinolones, and β-lactams). This rapid resistance development is a consequence of the ability of A. baumannii to acquire new genetic material, limiting the available therapeutic options (2). Colistin, also known as polymyxin E, is a last-line antibiotic used against extensively drug-resistant A. baumannii infections, in spite of its side effects. Colistin is a polycationic cyclic lipopeptide that interacts with the lipopolysaccharide (LPS) layer, resulting in membrane disruption, leakage of intracellular contents, and consequent bacterial death (3). However, colistin-resistant A. baumannii strains have been recently reported as well (4).
The appearance of pandrug-resistant bacteria such as colistin-resistant A. baumannii strains has fueled the research of antimicrobial alternatives. One promising route of investigation are bacteriophages (phages) and their lytic enzymes (lysins). Phages have been tested and used to treat bacterial infections since their discovery, and the interest in their use has been surging over the last years (5). A recent case study of successful phage therapy against a pandrug-resistant A. baumannii isolate (including colistin) getting worldwide attention is the Patterson case. In this case, the causative A. baumannii strain was used to compose a personalized cocktail of phages from available phage banks, followed by new phage isolations from the environment when phage resistance emerged. The patient’s clinical status has been improved after 2 days of intravenous administration of this personalized phage cocktail. Later on, the antibiotic minocycline was also added to his regimen. The patient was discharged home after 245 days (6).
Whereas phages have been intensively investigated to be used in therapy for a century, the antibacterial potential of phage-encoded lysins has gained increasing interest over the last 2 decades (5). Lysins include virion-associated peptidoglycan hydrolases (VAPHs) and endolysins, both degrading the peptidoglycan layer. VAPHs create a local hole in the peptidoglycan layer at the initiation of phage infection, followed by ejection of the phage genome into the cell. Endolysins act at the end of the replication cycle. They accumulate in the cytoplasm until a sudden, holin-timed permeabilization of the cytoplasmic membrane allows them to reach the peptidoglycan layer. This results in a massive degradation of the cell wall and release of the progeny phage (5). The biochemical specificity of lysins is generally broader (ranging from the genus to serovar level) compared to the one of phages, which are specific at the strain level. When specific lysins are applied exogenously to Gram-positive bacteria, they cause rapid osmotic lysis (7). However, this effect is greatly diminished in the case of Gram-negative bacteria due to the protective outer membrane that limits access to the peptidoglycan (8). Nevertheless, some lysins have intrinsic antibacterial activity against Gram-negative bacteria when added exogenously. This activity is explained by the presence of a C-terminal amphipathic helix. A. baumannii, in particular, appears to be susceptible to this group of lysins with intrinsic antibacterial activity (9–14). Many efforts have improved the antibacterial activity of lysins against A. baumannii. One example is the combination of lysin with EDTA or weak acids (citric or malic acid), which act as outer membrane destabilizers (15). In addition, artilysin Art-175 is an engineered lysin fused with an outer membrane permeabilizing peptide and is able to pass the bacterial outer membrane of A. baumannii and kill the cell after peptidoglycan degradation (16).
Phages are abundant, as are their lysins. This abundance allows the discovery of phages and lysins at local sites where drug-resistant bacteria are prevalent. Hospitals in Egypt, among many other countries, are experiencing a significant rise of MDR A. baumannii infections (17). We have isolated a set of A. baumannii strains, classified as MDR to extensively drug resistant (18), in hospitals in two nearby cities. Subsequently, we identified a highly specific phage (vB_AbaP_PMK34) in the general sewage system, which is also connected with the hospital in which we isolated the most extensively drug-resistant strain (MK34). Based on a comprehensive microbiological and genetic characterization of the phage isolated in this study, we identified its lysin. We present here an analysis of the antibacterial potential of the phage and its lysin.
RESULTS
Multidrug- and extensively drug-resistant A. baumannii strains are prevalent at Egyptian hospitals.
Ten A. baumannii strains (Table 1) were isolated from patients at the Beni-Suef and Fayoum governorate hospitals in Egypt. All of them were confirmed as A. baumannii by biochemical and microbiological assays and by the detection of the blaOXA-51-like gene encoding a class D carbapenemase, unique for A. baumannii (19). Their antibiotic resistance profile was established against a panel of 12 different antibiotics (Table 1). The results show that all strains are resistant to at least four of the antibiotics tested, with A. baumannii MK34 and MK11 being the most resistant isolates (11 of the 12 antibiotics). All strains are sensitive to colistin. Moreover, the presence of the blaOXA-23-like gene (encoding a class D carbapenemase) was present in seven out of the 11 strains. Only A. baumannii MK60, MK22, O3, and A2 lack the blaOXA-23-like gene. The presence of this gene has been reported as having the strongest correlation with high resistance levels against carbapenems such as meropenem and imipenem (20). Accordingly, strains O3 and A2, which lack the blaOXA-23-like gene, are susceptible to meropenem and imipenem. However, strains MK60 and MK22, which are also negative for the blaOXA-23-like gene, are resistant to one or both carbapenems, indicating that other carbapenemases are involved in these resistant phenotypes.
TABLE 1.
Overview of the A. baumannii strains isolated in this studya
| A. baumannii strain | Origin | Source | Antibiotic resistanceb | Class D carbapenemases |
Phage sensitivityc | |
|---|---|---|---|---|---|---|
| blaOXA-51-like | blaOXA-23-like | |||||
| MK11 | Beni-Suef University Hospital | Sputum | AMC, TZP, CTX, CAZ, FEP, MEM, IPM, GN, AK, CIP, and TE | + | + | R |
| MK22 | Beni-Suef University Hospital | Pus | AMC, TPZ, CAZ, FEP, MEM, TE, and CIP | + | – | R |
| MK34 | Beni-Suef University Hospital | Sputum | AMC, TZP, CTX, CAZ, FEP, MEM, IPM, GN, AK, CIP, and TE | + | + | S |
| MK50 | Beni-Suef University Hospital | Sputum | AMC, TPZ, CTX CAZ, FEP, MEM, IPM, TE, and CIP | + | + | R |
| MK60 | Beni-Suef University Hospital | Urine | AMC, TPZ, CTX, CAZ, FEP, MEM, IPM, TE, AK, and CIP | + | – | R |
| MK70 | Beni-Suef University Hospital | Blood | AMC, TPZ, CTX, CAZ, FEP, MEM, IPM, TE, AK, and CIP | + | + | R |
| MK73 | Beni-Suef University Hospital | Sputum | AMC, TPZ, CTX CAZ, FEP, MEM, IPM, TE, and CIP | + | + | R |
| O3 | Fayoum University Hospital | Sputum | AMC, TPZ, CAZ, and FEP | + | – | R |
| A1 | Fayoum private laboratory | Urine | AMC, TPZ, CAZ, FEP, MEM, IPM, TE, AK, and CIP | + | + | R |
| A2 | Fayoum University Hospital | Sputum | AMC, TPZ, CAZ, FEP, and CIP | + | – | R |
The hospital, source of isolation (body fluid), microbiological antimicrobial resistance profile, genetically verified presence of class D carbapenemases, and phage susceptibility are indicated for each strain.
AMC, amoxicillin-clavulanic acid; TZP, piperacillin-tazobactam; CTX, cefotaxime; CAZ, ceftazidime; FEP, cefepime; MEM, meropenem; IPM, imipenem; GN, gentamicin; AK, amikacin; CIP, ciprofloxacin; TE, tetracycline; COL, colistin.
R, resistant; S, susceptible.
Phage vB_AbaP_PMK34 possesses a strong lytic activity but narrow host range.
As the most resistant strain to antibiotics, A. baumannii MK34 was selected in the search for a suitable phage in the central sewage system of Beni-Suef. This system also collects the wastewater from the hospital in which the strain was isolated. Raw sewage was collected and enriched with strain MK34, resulting in the isolation of a lytic phage, vB_AbaP_PMK34, herein referred to as PMK34. The isolated phage forms clear plaques with a halo that expands upon longer incubation, a feature typically associated with the presence of a phage depolymerase degrading capsular polysaccharides (21). Host range analysis of PMK34 showed that the phage has a highly specific host spectrum, infecting A. baumannii MK34 but no other strain isolated in this study (Table 1).
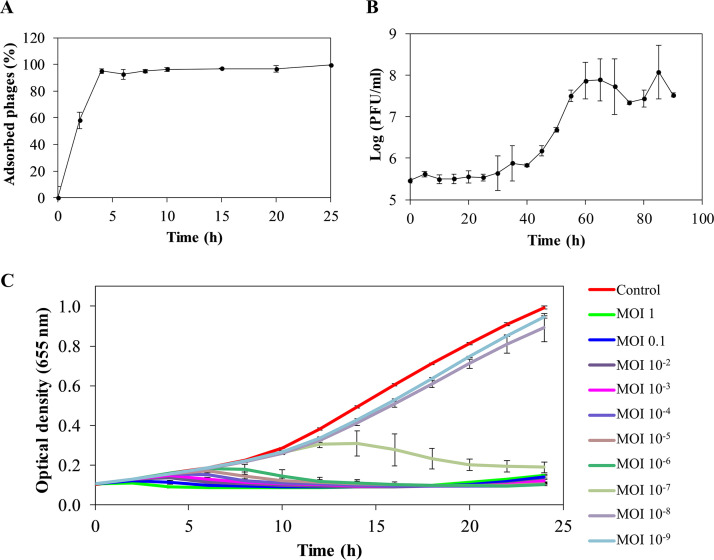
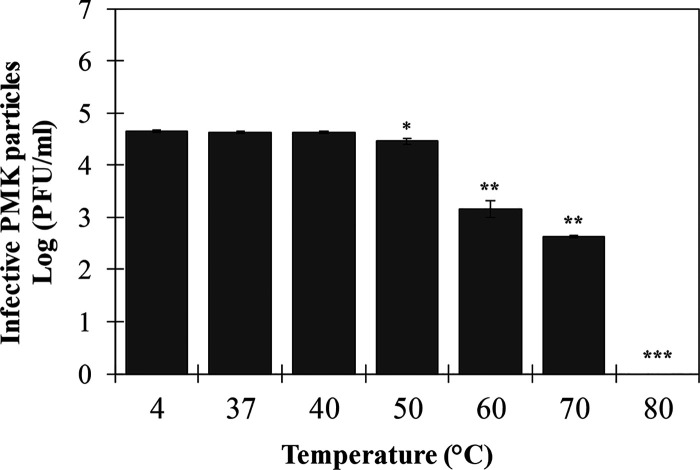
Adsorption and one-step growth curves were established to assess the infection kinetics. Approximately 40% of the phage particles are adsorbed after 2 min, and a maximum adsorption rate of ≥90% is reached after 4 min of incubation (Fig. 1A). The one-step growth curve follows the typical triphasic pattern with a latent period of 30 min and a burst size of 113 ± 32 PFU/infected cell (Fig. 1B). Furthermore, different multiplicities of infection (MOIs; 1 to 10−9) of phage PMK34 were used to kill exponential growing A. baumannii MK34 cells. Bacterial growth is completely inhibited with an MOI as low as 10−6, whereas inhibition only appeared with a delay in the case of an MOI of 10−7. MOIs of 10−8 and 10−9 showed bacterial growth comparable to the uninfected control (Fig. 1C). However, after 20 h a slight increase in the optical density at 655 nm (OD655) was observed for MOI values larger than 10−4, suggesting the emergence of mutant strains. This was confirmed by the appearance of resistant colonies within the lysis zone when spotting different phage dilutions of the higher MOI values. Phage PMK34 is relatively stable between 4 and 70°C with reductions of 1.5 and 2 log units after 1 h of exposure to 60 and 70°C (P < 0.01), respectively. A temperature of 80°C completely abolishes phage infectivity (Fig. 2).
FIG 1.
Infection kinetics of phage PMK34 against A. baumannii MK34. (A) Phage adsorption curve obtained by infecting exponentially growing bacteria (OD655 = 0.6) with PMK34 (MOI of 0.01). The results are expressed as the percentage of the maximum number of adsorbed phages. (B) One-step growth curve of PMK34 when using an MOI of 0.01 to infect a bacterial culture (106 CFU/ml). (C) Lytic effect of different MOIs of PMK34 (1 to 10−9) during a 24 h of incubation. The OD655 was monitored spectrophotometrically at 30-min intervals. All values represent the means ± the standard deviations (SD) of three biological replicates.
FIG 2.
Stability of PMK34 under various temperature conditions. The infectivity of PMK34 after exposure to a temperature ranging from 4 to 80°C for 1 h was determined. Values represent the log(PFU/ml) expressed as the means ± the SD of three independent replicates. A paired Student t test was conducted to compare significance between mean values, with a temperature of 4°C as a reference (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
In silico genome analysis of phage vB_AbaP_PMK34 and its endolysin LysMK34.
PMK34 contains a linear double-stranded DNA molecule of 41,847 bp (accession number MN433707) with a GC content of 39.34%. Neither tRNA nor antibiotic resistance genes were detected within the genome. BLASTn analysis against nonredundant database revealed the highest similarity to Acinetobacter phage IME200 (NC_028987.2) with an identity of 96.13% (coverage, 88%). Moreover, sequence identity from 93% up to 95% was also observed with phages AbKT21phiIII (MK278859.1), vB_AbaP_AS12 (KY268295.1), AbaP_B09_Aci08 (MH763831.1), vB_AbaP_AS11 (KY268296.1) and vB_ApiP_P1 (MF033350.1), all podoviruses belonging to the Autographiviridae.
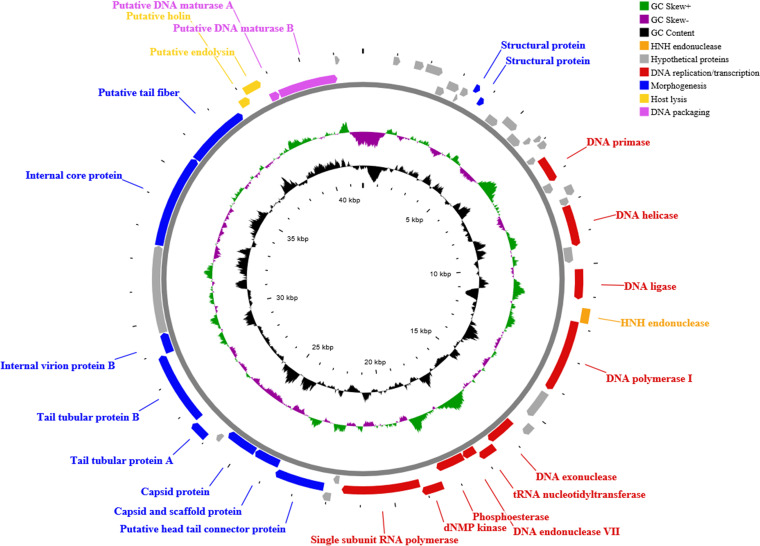
RAST annotation predicted 50 open reading frames (ORFs; see Table S1 in the supplemental material; Fig. 3) on the leading strand of the genome, showing unidirectional transcription (from left to right), and start with an ATG codon, except for orf9 and orf17, which start with TTG. BLASTx in addition to NCBI Conserved Domain Database (CDD) functional analysis assigned 25 ORFs (50% of the total) as hypothetical proteins with unknown function, whereas the remaining 25 ORFs are categorized into four clearly differentiated groups: DNA replication/transcription, morphogenesis, host lysis, and DNA packaging genes (Fig. 3; see Table S1 in the supplemental material). A fifth region, which is the least similar to homologous phages, is dominated by hypothetical genes. Three putative viral promoters with consensus sequences related to those found in the genome of the Fri1virus phages (Acinetobacter phages of family Autographiviridae) have been detected (see Table S2). Furthermore, two putative rho-independent transcription terminators were detected downstream from the RNA polymerase (orf33) and the capsid gene (orf38) (see Table S3).
FIG 3.
CG viewer representation of the PMK34 genome. The outer ring represents the 50 predicted ORFs (arrows) of the phage, with different colors representing the different modular functional groups. The middle and inner rings illustrate the GC skew and GC content, respectively. The PMK34 genome reveals a typical modular organization with DNA replication/transcription, morphogenesis, host lysis, and DNA packaging genes, apart from a region dominated by hypothetical genes. The DNA replication/transcription gene group (11 ORFs) comprises genes encoding the DNA primase/helicase (orf17 and orf21), DNA ligase (orf23), DNA polymerase I (orf25), DNA exonuclease (orf28), tRNA nucleotidyltransferase (orf29), DNA endonuclease VII (orf30), phosphoesterase (orf31), dNMP kinase (orf32), and single subunit RNA polymerase (orf33). Only a single HNH endonuclease-encoding gene (orf24) was found upstream of the DNA polymerase I (orf25) sequence. The DNA replication/transcription gene group is followed by morphogenesis gene group separated by two hypothetical proteins (orf34 and orf35). Different structural proteins encoded by this gene cluster include the head tail connector (orf36), scaffold protein (orf37), capsid (orf38), tail tubular proteins A (orf40) and B (orf41), internal virion proteins (orf42 and orf44), and a tail fiber with putative pectate lyase domain (orf45). The host cell lysis gene group comprises two overlapping genes, i.e., the holin gene (orf46) located upstream of the endolysin sequence (orf47). No spanin gene is detected. The holin protein has three predicted transmembrane helices with N-out and C-in topology, being a member of the class I holin. The final predicted proteins are the DNA packaging and maturase genes A (orf48) and B (orf49).
Of note, the gene (orf47) encoding the endolysin was found at the module of host lysis, preceded as expected by a putative class I holin gene. The predicted endolysin is a globular protein with 185 amino acids and has a predicted molecular weight of 20.9 kDa, a predicted isoelectric point of 9.48, and a predicted cationic surface charge of +7.8 at physiological pH. Moreover, its domain analysis revealed a protein with a conserved lysozyme domain (CDD accession number cl00222) (see Table S1). A multiple sequence alignment of LysMK34 with similar endolysin sequences (see Fig. S1) shows the presence of two amphipathic helices in the C terminus between amino acids 112 to 183, rich in both hydrophobic/nonpolar (alanine, phenylalanine, proline, methionine, tryptophan, leucine, and isoleucine) and basic amino acids (arginine and lysine), as shown in helical wheels (see Fig. S2). Such amphipathic helices have been related before to intrinsic antibacterial activity.
LysMK34 has a high muralytic activity and does not trigger cytotoxicity in human epithelial cell lines.
The gene encoding the endolysin (orf47) was cloned into the pVTD3 expression vector, followed by transformation of E. coli BL21(DE3)-Codon Plus-RIL. After expression and purification of LysMK34 with Ni2+ affinity chromatography, the yield was 38 mg/liter expression volume with a purity of at least 95% as estimated by conventional SDS-PAGE (see Fig. S3). LysMK34 has a muralytic activity of 1,499.9 ± 131 U/μM when tested against standard permeabilized P. aeruginosa substrate.
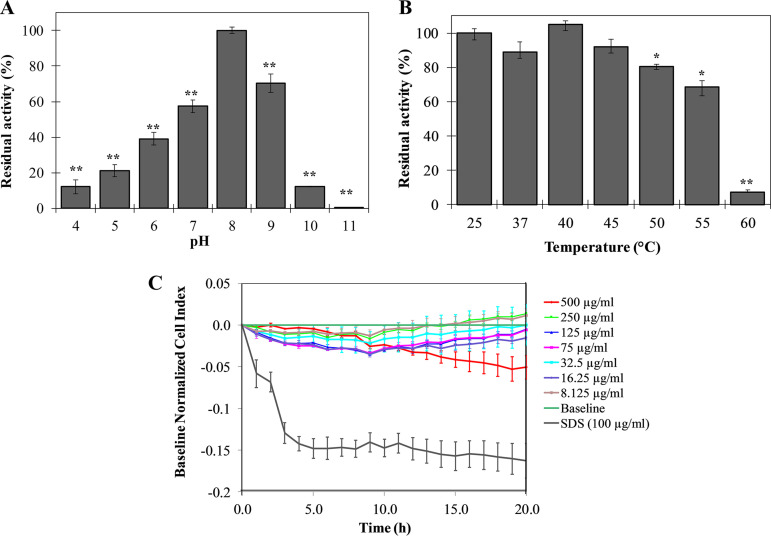
Moreover, the range of pH activity of LysKM34 is relatively broad, from at least 4 to 10, with pH 8 as optimal condition (Fig. 4A). The muralytic activity gradually decreases at lower pH values, with a minimal activity of 12% ± 4% remaining at pH 4. In the alkaline range, LysMK34 retains 70% ± 5% of its activity at pH 9, followed by a decrease to 12.0% ± 0.3% at pH 10, whereas no residual activity is present at pH 11. LysMK34 has typical mesophilic characteristics, retaining full activity up to 45°C (Fig. 4B). This activity decreases by 20 and 25%, when the temperature increases to 50 and 55°C, respectively. Only 7.28% ± 1.11% of enzymatic activity is retained when the protein is exposed to 60°C. Higher temperatures completely inactivate LysMK34.
FIG 4.
Stability (A and B) and cytotoxicity (C) of LysMK34. Stability of LysMK34 was tested by diluting 0.3 μg/ml protein in Britton-Robinson universal buffer (pH 4 to 11) (A) and treating 0.3 μg/ml protein with different temperatures (25 to 60°C) for 10 min (B). The results are expressed as the percentage of residual activity relative to activity at either optimum pH (8) or activity at 25°C. Each point is the mean ± the SD of three independent replicates. Asterisks represent statistical differences compared to the control (Student t test [*, P < 0.05; **, P < 0.01; ***, P < 0.001]). (C) Variation in the normalized cell index (CI) of HaCaT monolayers treated with different concentration of LysKM34 (8.12 to 500 μg/ml). Normalization of data was performed at 10 min after protein addition with respect to the CI observed in the control sample (value 0 in the graph). Values represent means ± the SD of three replicates.
In addition, LysMK34 does not show cytotoxicity against a preformed monolayer of human epithelial cells (HaCaT), since the treatment with increasing concentrations of the protein (up to 500 μg/ml) did not result in variation of the cell index after 20 h of exposure. This invariant cell index indicates that the monolayer of HaCaT is not morphologically altered or disrupted in the presence of LysMK34 (Fig. 4C).
LysMK34 shows a strong bactericidal effect in combination with a high intracellular pressure.
The bactericidal activity against the multidrug-resistant A. baumannii strain MK34 was evaluated in buffers with either low (20 mM HEPES-NaOH [pH 7.4]) or high tonicity (20 mM HEPES, 150 mM NaCl [pH 7.4]). The low- and high-tonicity buffers induce high and low turgor pressures inside the bacterial cells, respectively. In the low-tonicity buffer, an amount of 25 μg/ml LysMK34 causes a 3.35 ± 0.19 log unit reduction of bacterial cells within 2 h (see Fig. S4). Higher LysMK34 concentrations (50 to 1,000 μg/ml) further increase the bactericidal activity up to 4.5 log units (corresponding to the detection limit) (see Fig. S4). In contrast, a maximum bacterial killing effect of merely 0.6 ± 0.1 log unit was obtained in a buffer with high tonicity, even when using a 20-fold concentration (500 μg/ml; see Fig. S4). This lack of bactericidal activity was observed in spite of the high muralytic activity of LysMK34 (1,499.9 U/μM) in the high-tonicity buffer (20 mM HEPES, 150 mM NaCl [pH 7.4]). This suggests an interplay between the extent of required peptidoglycan degradation and the intracellular pressure that must be withheld by the peptidoglycan structure to keep the cell intact. Cells with a reduced turgor pressure may require a more extensive disruption of the peptidoglycan sacculus to achieve full lysis compared to cells exposed to peptidoglycan degradation at high intracellular pressure.
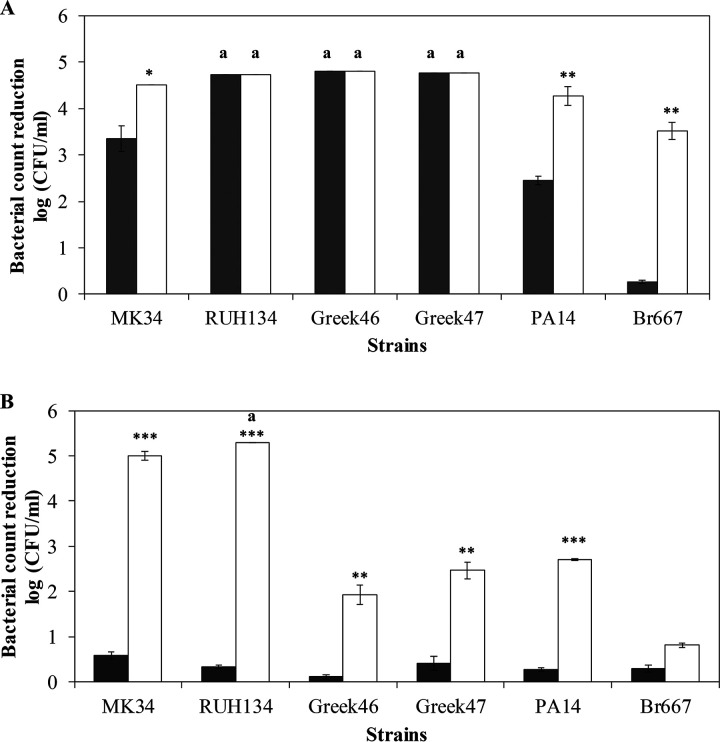
The bactericidal activity of LysMK34 was tested in the same low- and high-tonicity buffers against a broader panel of A. baumannii and P. aeruginosa strains (Fig. 5). Concentrations of 25 μg/ml (low-tonicity buffer) and 500 μg/ml (high-tonicity buffer) LysMK34 were used, respectively. Notably, the multidrug-resistant and epidemiological A. baumannii RUH134 strain and both pandrug-resistant (including colistin-resistant) A. baumannii strains Greek46 and Greek47 show a susceptibility similar to that of A. baumannii MK34, but with improved killing in low-tonicity buffer (high intracellular pressure) (Fig. 5A). The P. aeruginosa PA14 laboratory strain (with generally low antibiotic resistance) is also more susceptible at a high intracellular pressure. A notable exception is the extensively drug-resistant P. aeruginosa Br667 strain, which remains resistant to LysMK34 under both buffer conditions (Fig. 5).
FIG 5.
Antibacterial activity of LysMK34 against different A. baumannii (MK34, RUH134, Greek46, and Greek47) and P. aeruginosa (PA14 and Br667) strains. The bacterial cells were resuspended in a buffer with either low tonicity, corresponding to a high intracellular turgor pressure (20 mM HEPES-NaOH [pH 7.4] alone or supplemented with 0.5 mM EDTA; dark gray and white bars, respectively) (A), or high tonicity and thus a low intracellular turgor pressure (20 mM HEPES-NaOH, 150 mM NaCl [pH 7.4] alone or supplemented with 0.5 mM EDTA; dark gray and white bars, respectively) (B). Antibacterial activity is expressed as the reduction in the bacterial number in log(CFU/ml) after exposure to LysMK34 (25 μg/ml in buffer with low tonicity and 500 μg/ml in buffer with high tonicity) for 2 h. Each value represents the mean ± the SD of three independent replicates. Asterisks represent statistical differences compared to cells not treated with 0.5 mM EDTA (Student t test [*, P < 0.05; **, P < 0.01; ***, P < 0.001]). An “a” label indicates a reduction in the bacterial number that is under the detection limit (10 CFU/ml).
EDTA destabilizes the outer membrane by chelating the stabilizing divalent ions, thus potentially sensitizing bacteria for lysins. When 0.5 mM EDTA was added, the bactericidal activity improved under all circumstances where maximal killing was not yet achieved. The lowest killing in the presence of 0.5 mM EDTA was observed for the P. aeruginosa strains in buffer with high tonicity (low intracellular pressure) (Fig. 5B). In conclusion, the reduced killing in a high-tonicity buffer (low intracellular pressure) can be partially or completely reverted by destabilizing the outer membrane with 0.5 mM EDTA.
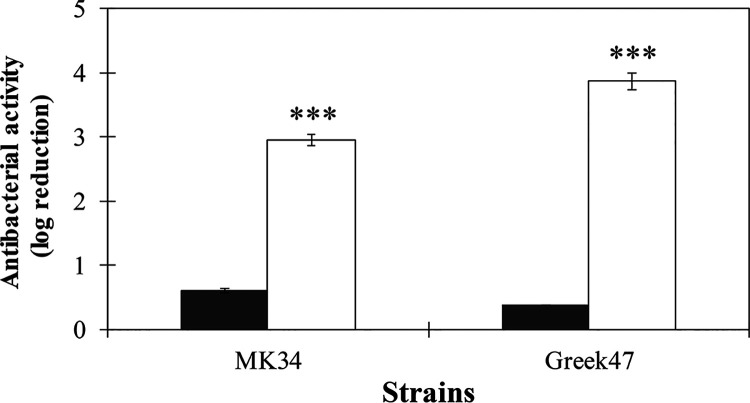
To further corroborate our hypothesis on the influence of the intracellular pressure on the bactericidal efficiency, cells suspended in a buffer with high tonicity (thus having a low intracellular pressure) were exposed to 500 μg/ml LysMK34 as before, but then washed and diluted in buffer of low tonicity to induce an osmotic shock. A. baumannii MK34 and Greek47 were selected as representatives for colistin-sensitive and -resistant strains, respectively. The cell counts obtained after this buffer change demonstrated that the bactericidal effect increases after the osmotic shock from less than 1 to 3.00 ± 0.03 and 3.86 log units for A. baumannii strain MK34 and Greek47, respectively (Fig. 6). These observations indicate that LysMK34 causes sublethal damage to the cells by peptidoglycan degradation and only come to full lysis by increasing the intracellular turgor pressure. Thus, also under conditions of low intracellular turgor pressure, LysMK34 can penetrate the outer membrane and then reach the peptidoglycan layer.
FIG 6.
Effect of changing intracellular turgor pressure on the antibacterial activity of LysMK34 against A. baumannii MK34 (a colistin-sensitive strain) and A. baumannii Greek47 (a colistin-resistant strain). Exponentially grown bacterial cells were suspended in high tonicity, corresponding to a low intracellular turgor pressure (20 mM HEPES-NaOH, 150 mM NaCl [pH 7.4]) and then treated with 500 μg/ml LysMK34 for 2 h at room temperature. The treated cells were washed twice using high-tonicity buffer and then either diluted prior to plating in high-tonicity (20 mM HEPES-NaOH, 150 mM NaCl [pH 7.4]; black bar) or low-tonicity (20 mM HEPES-NaOH [pH 7.4]; white bars) buffer, the latter condition causing an osmotic shock. The antibacterial activity was calculated as the ratio of reduction in log units [log10(N0/Ni)], where N0 is the number of LysMK34 untreated cells, and Ni is the number of cells after treatment. Each value represents the mean ± the SD of three independent replicates. Asterisks represent statistical differences compared to cells diluted in in high tonicity (20 mM HEPES-NaOH, 150 mM NaCl [pH 7.4]). *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student t test).
LysMK34 acts synergistically with colistin and reverts colistin resistance in colistin-resistant A. baumannii strains.
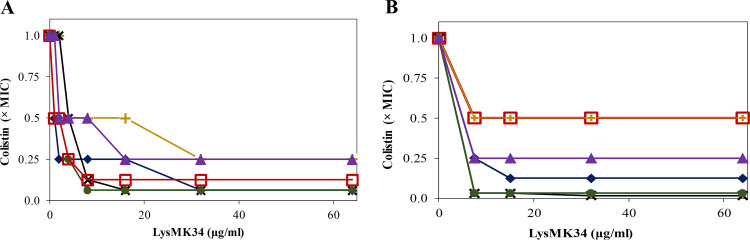
The antibacterial activity of LysMK34 was further evaluated by determining MIC values against the panel of A. baumannii and P. aeruginosa strains. Notably, no growth inhibition was observed in Mueller-Hinton (MH) broth (MIC > 1,000 μg/ml) for any strain. Subsequently, we evaluated whether colistin could potentiate the susceptibility of LysMK34, or vice versa, whether LysMK34 could decrease the required dose of colistin to inhibit the different strains. The MIC values for colistin are 1 μg/ml for P. aeruginosa PA14 and 2 μg/ml for A. baumannii MK34, RUH134, and P. aeruginosa Br667, respectively. As expected, the colistin-resistant A. baumannii strains Greek46 and Greek47 show elevated MIC values of 8 and 32 μg/ml (EUCAST breakpoint for resistance is 4 μg/ml), respectively. When combining LysMK34 and colistin a synergistic action was observed (Fig. 7A). An amount of 32 μg/ml LysMK34 reduced the MIC values of colistin in a strain-dependent manner to 1/4, 1/8, or 1/16 of the original colistin MIC values. The synergistic effect was most outspoken for the colistin-resistant A. baumannii strains Greek46 and Greek47, for which the MIC values of colistin were reduced 16-fold (to 0.5 and 2 μg/ml, respectively) starting from concentrations of 16 and 8 μg/ml LysMK34, respectively (Fig. 7A). Thus, LysMK34 resensitizes colistin-resistant strains to colistin by making them responsive to colistin concentrations below the breakpoint for resistance (4 μg/ml).
FIG 7.
Isobologram representation of MIC values of LysMK34 in combination with colistin in MH broth (A) or in 50% human serum (B) against A. baumannii MK34 (blue filled diamond; colistin MIC values of 2 μg/ml in both MH broth and serum), RUH134 (red open square; colistin MIC values of 2 and 4 μg/ml in MH broth and serum, respectively), Greek46 (black cross; colistin MIC values of 8 and 32 μg/ml in MH broth and serum, respectively), and Greek47 (green filled circle; colistin MIC value of 32 and 64 μg/ml in MH broth and serum, respectively) and P. aeruginosa Br667 (yellow cross; colistin MIC values of 2 and 4 μg/ml in MH broth and serum, respectively) and PA14 (purple filled triangle; colistin MIC values of 1 and 2 μg/ml in MH broth and serum, respectively). In panel B, the curves of RUH134 and Br667 completely overlap. The plots represent inhibitory combinations of LysMK34 (expressed in μg/ml) and colistin (expressed in reduced MIC fold, with 1× MIC corresponding to the above-mentioned values). Values represent the means of three independent replicates.
Human serum also contains compounds that may interfere with outer membrane permeability. Therefore, the combinations of LysMK34 and colistin were also evaluated in 50% human serum instead of MH broth (Fig. 7B). Similarly, no growth inhibition by LysMK34 was observed for any strain up to 1,000 μg/ml. The MIC values for colistin in human serum are 2 μg/ml for both A. baumannii MK34 and P. aeruginosa PA14. Interestingly, A. baumannii RUH134 and P. aeruginosa Br667 show doubled colistin MIC values compared to those observed in MH broth (4 μg/ml). Also, the colistin-resistant A. baumannii strains Greek46 and Greek47 showed a 2-fold increase in MIC for colistin in human serum (32 and 64 μg/ml, respectively). In the presence of 8 μg/ml LysMK34, an up to 32-fold reduction of the MIC of colistin (to 1 and 2 μg/ml) was observed for the colistin-resistant A. baumannii strains Greek46 and Greek47, respectively. For the other strains, MIC reductions of colistin of 2- to 8-fold were observed at this LysMK34 concentration (Fig. 7B).
For all of the conditions described here, all minimum bactericidal concentrations (MBCs) were found to be equal to the MICs, validating the bactericidal mode of action of both LysMK34 and colistin.
Role of the fatty acid tail in synergetic effects between colistin and LysMK34.
The bactericidal action of colistin is based on two different actions. First, the positively charged cyclic heptapeptide moiety of colistin destabilizes the outer membrane by displacing divalent cations (Ca2+, Mg2+) that bridge adjacent negatively charged lipopolysaccharide molecules. Second, colistin depolarizes the cytoplasmic membrane via its fatty acyl tail, leading to bacterial death (3). A possible hypothesis for the observed synergy in colistin-susceptible strains is that colistin enables access of LysMK34 to the peptidoglycan layer by disrupting (locally) the outer membrane. Polymyxin B nonapeptide (PMBN) is a derivative of polymyxin B, which is highly similar to colistin (polymyxin E). PMBN retains the cationic cyclic peptide that permeabilizes the outer membrane but lacks the fatty acid tail that is responsible for the bactericidal effect of polymyxins B and E (colistin). An experiment similar to that using colistin was repeated with PMBN. PMBN shows no inhibitory activity against any of the A. baumannii strains, either alone or in combination with increasing concentrations of LysMK34 (data not shown). These different observations for PMBN and colistin indicate an essential role for the fatty acid tail of colistin in the observed synergistic effects for A. baumannii. In addition, these observations suggest that our hypothesis is invalid and that the synergy between colistin and LysMK34 for the colistin-susceptible strains may rely not on colistin-induced permeabilization of the outer membrane (alone) but rather on the co-acting activities of both colistin and LysMK34.
The molecular mechanisms of synergy between LysMK34 and colistin are different for colistin-sensitive and colistin-resistant strains.
To further investigate the synergistic effects of LysMK34 and colistin, cells were exposed sequentially to either LysMK34 or colistin, instead of simultaneously. Specifically, colistin-sensitive A. baumannii MK34 and colistin-resistant A. baumannii Greek47 cells brought to low intracellular turgor pressure were pretreated with LysMK34 (100 μg/ml) for 2 h, followed by washing using MH broth and exposure to colistin (0 to 32 μg/ml) in a standard MIC assay. Different responses were observed for each strain (see Fig. S5). In the case of the colistin-sensitive strain A. baumannii MK34, the MIC of colistin was reduced 16-fold (from 2 to 0.125 μg/ml) compared to the control pretreated with buffer alone. However, the MIC of colistin in the case of the colistin-resistant A. baumannii Greek47 did not show any reduction compared to the control. Pretreatment with LysMK34 thus sensitized the colistin-sensitive strain for colistin in a persistent way but not the colistin-resistant strain. Conversely, when cells were pretreated with 0.25 × MIC of colistin, no measurable MIC was noted for LysMK34 up to 1 mg/ml for both strains (data not shown).
DISCUSSION
Based on an initial assessment to evaluate antibiotic resistance among A. baumannii isolates in two Egyptian cities, we isolated the new lytic bacteriophage named PMK34 from raw sewage, using the most drug-resistant strain MK34 (resistant to 11 of 12 antibiotics) as host. PMK34 presents a genome organization consistent with phage members of the Fri1virus genus of the recently established Autographiviridae family (22). Within the genome, the highest dissimilarities were found within the early cluster (between 0 and 4 kb) of hypothetical genes. These unique proteins are thought to be involved in redirecting host synthetic machinery to the phage’s needs at the beginning of infection cycle (23). In addition, the tail fiber gene (orf45) shows low conservation, particularly in its C terminus, which is involved in receptor binding, whereas the N terminus anchors the tail fiber to the phage particle. The infection parameters of PMK34 with a relatively large burst size (113 ± 32 phages/infected cell) and short latent period (30 min) are in accordance with A. baumannii phages PD-6A3, vB_AbaP_AS12, and vB_AbaP_Acibel004 (23–25). Many A. baumannii infecting phages have a relatively broad host range which reaches up to 68% of the tested strains (26). In contrast, PMK34 has a restricted host range infecting only one of the strains tested, i.e., strain MK34 used for phage enrichment and propagation. A similarly narrow spectrum was also observed for phages vB_AbaS_Loki and AB1 (27, 28). We suspect that the single tail fiber (orf45) with a predicted pectate lyase domain is highly specific for the capsular type of MK34. More than 100 distinct capsule types have been identified for A. baumannii (29). Such capsule depolymerizing activity is also responsible for the expanding halos surrounding plaques (21), which we have observed for PMK34. The observation that only 1 of 13 tested strains is susceptible might limit the applicability of this phage as a therapeutic phage. However, as part of a phage cocktail it may prove valuable because of its beneficial infection kinetics and the absence of known virulence or antimicrobial resistance genes. In addition, PMK34 may prove useful for capsular typing of A. baumannii.
We further explored the application potential of the lysin of PMK34. orf47 encodes a globular endolysin belonging to the GH19 subfamily of the lysozyme superfamily, which is commonly shared by endolysins of other phages infecting A. baumannii (10, 13, 14, 23). However, protein sequence alignment of these lysins (see Fig. S1) showed specific changes, including charged amino acids, which may account for differences in properties. The muralytic activity of LysMK34 (1,499.9 ± 131 U/μM) is 1.1- to 16-fold higher than those of previously described globular endolysins carrying a GH19 domain (ABgp46, LysAci7 [10, 30]), a GH24 domain (Lys68 [31]), a transglycosylase domain (PsP3gp10, P2gp09, and BcepC6Bgp22 [32]), or an amidase domain (K11gp3.5, KP32gp15, and CR8gp3.5 [30, 32]) analyzed with the same method. Yet, modular endolysins such as Gp110, OBPgp279, PVP-SE1gp146, EL188, and KZ144 offer a further increased muralytic activity (1.4- to 22.8-fold), which can be attributed to the presence of a cell binding domain bringing the enzymatically active domain close to its substrate (31–34). LysMK34 is active in a wide pH range (4 to 10) and relatively stable up to 55°C, compatible with most applications. Since LysMK34 has no cytotoxic effect on human keratinocytes, topical skin applications such as injured skin or burn wound infections can be considered as well.
LysMK34 has an intrinsic antibacterial activity against A. baumannii strains and P. aeruginosa PA14 but not against the MDR Br667 strain. Recently, a growing number of lysins with intrinsic antibacterial activity has been reported, particularly against A. baumannii (9–14, 35). The most plausible explanation for this beneficial property is the presence of amphipathic helices in the C terminus, as is the case for LysMK34 (see Fig. S2). Such helices are known to interfere with the stabilizing ionic and hydrophobic forces of the LPS layer, facilitating uptake of the lysin (13, 36).
Reducing the turgor pressure has a large negative effect on the intrinsic antibacterial activity, showing the importance of a concerted action of peptidoglycan degradation and osmotic pressure to induce osmotic lysis. Particularly, under conditions of low turgor pressure, cells may be only sublethally injured and recover easily under favorable conditions, such as plating on rich nutrients (Luria-Bertani [LB] agar). Peptidoglycan degradation and osmotic pressure must not act simultaneously, since we have shown that applying an osmotic shock shortly after lysin exposure also results in a significantly increased antibacterial activity. An alternative explanation for similar observations may be the lack of activity under a high salt concentration (11). However, this does not apply to LysMK34 specifically, since its unexpectedly high muralytic activity was observed under conditions with 150 mM NaCl. The effect of these experimental conditions, such as the choice of the suspension buffer and the diluent used, is often neglected in studies on lysins with intrinsic antibacterial activity, for which the diluent is not often specified (9, 10, 12–14, 35). In these cases, results are difficult to compare, or misinterpretation of the observations may occur. For example, some studies evaluating the effect of salt concentration on the antibacterial activity of the lysin do not specify the diluent (9, 12, 35), while using a diluent different from the suspension buffer may have a strong effect on the outcome. It is also recommended to use the same diluent as the suspension buffer of the cells to avoid osmotic changes. For example, Antonova et al. used a high-tonicity buffer (phosphate-buffered saline) as a diluent but a low-tonicity buffer (i.e., 20 mM Tris-HCl [pH 7.5]) as a resuspension buffer (11). The observed antibacterial activity is most likely due to the high turgor pressure during the lysin exposure.
Inclusion of a chelating agent (0.5 mM EDTA) at least partially releases the dependency for turgor pressure to induce osmotic lysis, indicating that more LysMK34 molecules can penetrate the outer membrane under this condition, resulting in more extensive peptidoglycan degradation. In the case of the MDR P. aeruginosa strain Br667, 0.5 mM EDTA even sensitizes the cells for the antibacterial activity of LysMK34. These observations underscore that the stabilizing ionic forces between divalent cations and the LPS phosphate groups remain a relevant hurdle for LysMK34 and putatively for other lysins with intrinsic antibacterial activity. We should note that all experiments were performed under laboratory conditions and that the physiological state of the outer membrane may not reflect in vivo conditions.
Instead of considering lysins as an alternative for antibiotics, it is attractive to evaluate lysins in combination with existing antibiotics. Lysins have been reported to resensitize resistant strains to specific antibiotics (37) or to display synergistic effects (38, 39). As a novel class of antibiotics, combined therapies of well-known small-molecule antibiotics and lysins may also find easier entrance to the clinic based on ethical clinical trial designs (antibiotic and lysin versus antibiotic alone) and a more rapid acceptance by clinicians. Lysin/antibiotic combinations have been extensively studied both in vitro and in vivo to combat Gram-positive bacteria. However, the exploration of this concept is still in its infancy for Gram-negative bacteria. Previously, in an analysis combining A. baumannii phage lysin LysABP-01 with different antibiotics, only colistin emerged as a synergistic compound against colistin-sensitive A. baumannii strains (38). Based on this finding, we expanded this approach to investigate whether a combination of LysMK34/colistin could revert colistin resistance in A. baumannii strains. This would be particularly welcomed since colistin is a last-resort antibiotic for the treatment of A. baumannii and P. aeruginosa infections. Despite the substantial killing in buffer, LysMK34 has no MIC up to 1 mg/ml in MH broth. Sykilinda et al. (40) explained the same observation for AcLys, which also has intrinsic antibacterial activity against A. baumannii, by a rapid inactivation of AcLys in MH broth. However, based on our earlier observations, LysMK34 may (partially) degrade peptidoglycan, resulting in the formation of spheroplasts that do not undergo osmotic lysis due to a sufficiently isotonic environment, followed by a regrowth due to favorable growth and nutrient conditions in culture media. Spheroplast-mediated antibiotic tolerance has been reported before for cell wall-acting antibiotics (41). For colistin-sensitive A. baumannii strains, we observed that sub-MIC (1/4 to 1/16) amounts of colistin sensitize the cells in MH broth for LysMK34 or vice versa. Comparable synergy of colistin with LysABP-01 was previously attributed to the outer membrane permeabilizing activity of colistin (38). However, the lack of synergy with combinations of LysMK34 and PMBN indicates that mere outer membrane permeabilization is not sufficient to obtain synergy. Indeed, also the fatty acid-mediated bactericidal effect of colistin contributes to the overall antibacterial effect of LysMK34/colistin combinations.
We found that the combination of LysMK34/colistin against colistin-resistant A. baumannii strains results in a reversion to the colistin-sensitive phenotype. Colistin resistance in A. baumannii is caused by the addition of phosphoethanolamine to the lipid A moiety. This dampens the negative charge of the LPS molecule and consequently the affinity for colistin or mutations in the lipid A biosynthesis. The final result of this is the complete loss of LPS (3). The latter mechanism results in reduced virulence and increased sensitivity for lysozyme, which was not observed for LysMK34 (Fig. 6). Therefore, a more plausible explanation for the reversion of colistin-resistance of strains Greek46 and Greek47 is that the self-penetrating properties of the bulky LysMK34 assist the small-molecule colistin, which lost its binding site, in penetrating through the outer membrane. In sum, combining colistin with LysMK34 may represent a viable approach to safeguard the applicability of this last-resort antibiotic, while also lowering the required doses and possible side effects.
MATERIALS AND METHODS
Bacterial strains, antimicrobial resistance determination, and culture conditions.
Ten A. baumannii strains were isolated from hospitalized patients of the Beni-Suef and Fayoum governorate hospitals in Egypt (Table 1) as described before (42). The isolates were confirmed as Acinetobacter species using conventional biochemical tests, identification in CHROMagar Acinetobacter (CHROMagar, Paris, France). Species-level identification was achieved using Microscan (Siemens Healthineers) (42). The sensitivity of each strain to antibiotics (amoxicillin-clavulanic acid, piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, meropenem, imipenem, gentamicin, amikacin, ciprofloxacin, tetracycline, and colistin) was analyzed using the Kirby-Bauer disk diffusion method (43). Furthermore, the presence of plasmid-mediated class D carbapenemase blaOXA-51-like and blaOXA-23-like genes was verified by PCR as previously described (42).
In addition, the previously characterized colistin-resistant A. baumannii strains Greek46 and Greek47, A. baumannii RUH134 (44), and Pseudomonas aeruginosa PAO1, PA14, and Br667 (45) were used in this work. Escherichia coli BL21(DE3)-Codon Plus-RIL (Agilent Technologies, Inc.) was used as an expression host for protein production.
All strains were grown in LB broth (1% tryptone, 0.5% yeast extract, and 1% NaCl) at 37°C with shaking (250 rpm) or on LB agar plates (LB broth supplemented with 2% agar). For proper selection of E. coli clones, LB broth supplemented with 50 μg/ml kanamycin and 25 μg/ml chloramphenicol was used.
Phage isolation and propagation.
Raw sewage water samples (200 ml) were collected from a general water treatment system in Beni-Suef city and used as a source for phage isolation. Subsequently, phage isolation was carried out by enrichment (46), followed by a spot assay in a double layer to determine the presence of lytic phages, as previously described (47). For this, A. baumannii MK34 was used as a host strain. The selection and purification of the obtained transparent lysis plaques were performed by collecting the lysis plaques in 900 μl of SM buffer (10 mM Tris-HCl, 10 mM MgSO4, and 100 mM NaCl [pH 7.5]), followed by a double-layer assay (48) repeated at least five times. In addition, phage stock preparation and concentration were performed using standard procedures (49), followed by digestion of bacterial DNA and RNA using 1 μg/ml DNase I (Sigma-Aldrich, UK) and 1 μg/ml of RNase A (Sigma-Aldrich) for 30 min at 37°C. Finally, phage concentrated stock was filtrated using 0.22-μm polyvinylidene difluoride membrane filters (VWR, Leuven, Belgium) and stored at 4°C.
Phage host range determination, antimicrobial activity, and thermal stability.
To determine the host range of the phage, a panel of 10 A. baumannii strains (Table 1), in addition to A. baumannii RUH134, Greek46, and Greek47, were used to prepare bacterial lawns on which 10 μl of the phage suspension was dropped using the double-layer agar technique (47). Plates were then incubated for 16 h at 37°C and examined for the presence of a lysis zone.
To test the antimicrobial activity of the phage, an overnight culture of A. baumannii MK34 strain was diluted 100-fold in LB broth and incubated at 37°C while shaking (at 250 rpm) to the exponential growth phase (OD655 = 0.6; 109 CFU/ml). Bacteria were then diluted in LB broth to 108 CFU/ml, and different phage concentrations were added to achieve an MOI from 1 to 10−9. The OD655 was monitored for 24 h at 30-min intervals using a microtiter plate reader (Infinite 200 PRO NanoQuant; Tecan, Switzerland). Bacterial culture mixed with sterile SM buffer was used as a negative control. In parallel, phage were spotted on a lawn A. baumannii MK34 with final MOIs from 1 to 10−9 to detect the presence of resistant mutants.
The thermal stability of the phage was tested against a range of temperatures (from 4 to 80°C) (50). Briefly, thermal stability was tested by incubating phage suspensions (105 PFU/ml) at the tested temperatures in SM buffer pH 7 for 1 h. Then, the phage titer was determined using the double-layer agar technique and the lawn of A. baumannii MK34.
Adsorption rate and one-step growth curve.
The adsorption assay and one-step growth curve were carried out as described previously (51). For the adsorption assay, A. baumannii MK34 was grown to the exponential phase (OD655 = 0.6), diluted to 106 CFU/ml, and then mixed with the phage (MOI = 0.01). The mixture was then incubated at 37°C, and 100-μl samples were taken at 2-min intervals for the first 10 min, followed by 5-min intervals for 25 min. At each time point, 100 μl of the mixture were 10-fold diluted in cold LB broth and centrifuged at 12,000 × g for 5 min, and then the number of nonadsorbed phages was determined in the supernatant using the double-layer agar technique. The experiment was repeated in three independent biological replicates.
The one-step growth curve was plotted to determine the latent period and phage burst size. Briefly, 10 ml of exponentially growing (OD655 = 0.6) A. baumannii MK34 was centrifuged, diluted to 107 CFU/ml, and then mixed with phage at an MOI of 0.01. The mixture was incubated at 37°C for 5 min to facilitate adsorption. Bacterial cells with adsorbed phages were collected by centrifugation (16,000 × g for 10 min) and resuspended in 10 ml of LB broth for incubation at 37°C. Samples were taken at 5-min intervals for 90 min to count the number of phage particles.
Genomic DNA isolation, sequencing, annotation, and bioinformatic analysis.
Phage genomic DNA (gDNA) was extracted using the phenol-chloroform/isoamyl alcohol protocol as described earlier (52). Afterward, DNA was precipitated by the addition of 3 volumes of 95% ethanol and 1/10 volume of 3 M sodium acetate (pH 5) and then incubated in ice for 5 min. The sample was then centrifuged at 16,000 × g for 15 min, and the pellet was washed twice with 70% ethanol and finally resuspended in 50 μl of deionized water. The DNA concentration was measured using a DS-11 spectrophotometer (DeNovix, Inc.).
The gDNA was sequenced on a MinION device using a FLO-MIN106 flow cell (R9.4 Chemistry; Oxford Nanopore Technology [ONT]). The sequencing library was created using a 1D ligation approach kit (ONT), with a step of end repair/dA tailing (New England Biolabs, UK). The sequencing proceeded for 4 h using the control software MinKNOW v2.1 (ONT). Base-calling of the raw sequencing data was achieved using Albacore v2.3.4. The reads were verified for quality using NanoFilt v1.6 to remove reads shorter than 1,000 bases and with quality scores of Q < 8. The genome assembly was conducted using 9,872 reads (totaling 59M bases) with the long-read assembler Canu v1.7 and subsequently polished using the software Nanopolish v0.9 to improve the assembly accuracy.
The phage genome was annotated using the RAST online server (53), manually curated using BLASTx (54) and the NCBI Conserved Domain Database (CDD) (55), and visualized with the CG view server (56). The presence of putative tRNA sequences was detected with tRNAscan-SEv.2.0 (57). Putative viral promoters and rho-independent terminators were detected using MEME (58) and ARNold (59) online tools, respectively, followed by visual inspection. Genomic comparative analysis with other homologous phages was conducted using BRIG software (60). The predicted lysin sequence was analyzed with InterPro (61). Multiple sequence/structural alignment (MSA) of the predicted lysin was done with PROMALS3D (62). The isoelectric points (pI) and molecular weights of the putative lysins were predicted by the ProtParam tool of the Expasy server (https://web.expasy.org/protparam/) (63), whereas the presence of transmembrane helices in the detected proteins was analyzed with TMHMM v.2.0 program (64).
Cloning, expression, and purification of lysin LysMK34.
The gene encoding the putative endolysin (orf47), referred to as LysMK34, was amplified using specific primers (forward [5′-GTCGGTCTCACCATGATTCTGACTAAAGACGGGTTTAG-3′] and reverse [5′-GTCGGTCTCATACTTTAAACTCCGTAGAGCGCG-3′]; IDT, Leuven, Belgium) flanked by the BsaI restriction sites (underlined). Subsequently, the amplicon was digested with 1 U of BsaI (Thermo Fisher Scientific, MA) and cloned into the expression vector pVTD3 (65), which introduces the coding sequence of a C-terminal 6×His tag. E. coli BL21(DE3)-Codon Plus-RIL cells (Agilent Technologies, Inc.) were then transformed using thermal heat shock with pVTD3-lysMK34. Finally, LysKM34 was overexpressed by induction of exponentially grown (OD655 = 0.6) with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG; Thermo Fisher Scientific), followed by overnight incubation at 30°C. Induced E. coli cells were disrupted by freezing-thawing cycles, followed by sonication, as described previously (65). Afterward, C-terminal His-tagged proteins were purified using the His GraviTrap kit (GE Healthcare) and dialyzed by using 3.5K MWCO Slide-A-Lyzer MINI Dialysis Devices (Thermo Fisher Scientific) against 20 mM HEPES-NaOH–150 mM NaCl (pH 7.4) buffer (65). Finally, the purity of the protein was analyzed by 12% SDS-PAGE. Protein concentration was estimated using a DS-11 spectrophotometer (DeNovix, Inc.) using an extinction coefficient of 33,350 M−1 cm−1 and a molecular mass of 22.1 kDa.
Muralytic activity assay and stability of LysMK34 to pH and temperature.
Pseudomonas aeruginosa PAO1 cells were prepared as the substrate for muralytic assays as previously described (66) and adjusted to an OD655 of 1.5 using 20 mM HEPES-NaOH–150 mM NaCl (pH 7.4). The muralytic activity of LysMK34 (0 to 150 nM) was determined by a conventional turbidity reduction assay (67) by monitoring the OD655 at room temperature in intervals of 30 s for 30 min in a microtiter reader plate (Bio-Rad, CA). The muralytic activity (U/μM) was calculated in triplicate as reported previously. Substrate mixed with buffer was used as a negative control.
Similarly, the effect of pH and temperature were tested using the turbidity reduction assay, as described previously (32), with some modifications. To test the effect of pH, substrate cells were suspended in Britton-Robinson universal buffer (0.04 M H3PO4, 0.04 M H3BO3, 0.04 M CH3COOH, and 0.15 M NaCl) adjusted to different pH values (4 to 11) and then mixed with LysMK34 to a final concentration of 0.3 μg/ml. The muralytic activity was again calculated as described above. Thermal stability was assessed by incubating 0.3 μg/ml LysMK34 (diluted in universal buffer [pH 8]) in a water bath at different temperatures (25 to 60°C) for 10 min, followed by cooling to room temperature, followed in turn by a turbidity reduction assay. All results are expressed as the percentage of residual activity related to the control (25°C, universal buffer [pH 8]).
Antibacterial activity assay of LysMK34 against A. baumannii and P. aeruginosa.
Antibacterial activity of the purified lysin was assessed in both low (20 mM HEPES-NaOH [pH 7.4])- and high (20 mM HEPES-NaOH, 150 mM NaCl [pH 7.4])-tonicity buffers (68). To test the activity of LysKM34, A. baumannii MK34 was initially used to optimize the protein concentration. For the assay, A. baumannii MK34 was grown to an OD655 of 0.6, washed twice with either low- or high-tonicity buffer and then suspended in 1:100 of the initial volume with the corresponding buffer. Subsequently, 100-μl portions of the cells were mixed with 50-μl portions of different concentrations of the purified protein (25 to 1,000 μg/ml) and 50-μl portions of either low- or high-tonicity buffer or 50-μl portions of either low- or high-tonicity buffer supplemented with 0.5 mM EDTA. Next, the mixture was incubated at 25°C for 2 h. Finally, cell counts were calculated by making serial dilutions in the same buffer, followed by plating on LB agar and incubation at 37°C for 18 h. The log reduction was calculated relative to a corresponding control cell suspension under the same conditions (low- or high-tonicity buffer, with or without 0.5 mM EDTA). The addition of 0.5 mM EDTA alone had a small effect of ∼0.02-log reduction. In addition, the experiment was carried out with different strains of A. baumannii (RUH134, Greek46, and Greek47) and P. aeruginosa (Br667 and PA14) using an optimized concentration for each buffer system (25 μg/ml in low-tonicity buffer and 500 μg/ml for high-tonicity buffer). The assays were performed in three independent replicates.
The effect of the tonicity of the buffer on the antibacterial activity of the lysin was tested by exposing exponentially growing A. baumannii MK34 and Greek47 strains to 500 μg/ml of LysMK34 in 20 mM HEPES-NaOH–150 mM NaCl (pH 7.4) for 2 h at room temperature. Next, the remaining bacteria were washed twice and diluted using either 20 mM HEPES-NaOH or 20 mM HEPES-NaOH–150 mM NaCl (pH 7.4). Bacteria were then plated on LB agar and incubated at 37°C for 18 h.
MIC checkerboard analysis.
A conventional broth microdilution assay was performed in MH broth to determine MIC values of LysMK34 (0 to 1,000 μg/ml), colistin (0 to 64 μg/ml), and a combination of both (up to 64 μg/ml of LysMK34 and up to 2× MIC of colistin) against A. baumannii MK34, RUH134, Greek46, and Greek47 and P. aeruginosa Br667 and PA14 (69). The MICs were determined by visual inspection as the lowest concentration without bacterial growth after overnight incubation at 37°C. The MBC was determined by spotting 10-μl portions of the cultures of the wells of the MIC assay on LB agar plates, followed by incubation for 24 h at 37°C. The MBC corresponds to the lowest concentration that showed no growth on plates. All assays were performed in independent triplicates. In addition, a checkboard MIC assay was also carried out using up to 64 μg/ml polymyxin B nonapeptide (PMBN; Sigma-Aldrich Company, UK) in combination with LysMK34 (0 to 64 μg/ml).
Similarly, a combination of LysMK34 (0 to 500 μg/ml) and colistin (0 to 64 μg/ml) was evaluated in 50% human serum (Hypo-Opticlear Human Sera; Sigma-Aldrich Company, Ltd., UK). Briefly, exponentially grown A. baumannii and P. aeruginosa strains were diluted in 100% human serum (106 CFU/ml) and then exposed to LysMK34 (0 to 500 μg/ml diluted in MH broth) and colistin (0 to 64 μg/ml diluted in MH broth) for a final concentration of 50% human serum. MIC values were calculated as described above.
Finally, a variation of the MIC assay was performed using pretreated cells as a substrate for the MIC assay. Thus, exponentially growing A. baumannii strains MK34 and Greek47 were washed twice using 20 mM HEPES-NaOH–150 mM NaCl (pH 7.4). The cells were subsequently treated with either 100 μg/ml LysMK34 or 0.25× MIC colistin, followed by incubation for 2 h at room temperature. These pretreated cells were washed twice and diluted in MH broth (106 CFU/ml) for a conventional MIC assay with colistin (0 to 32 μg/ml) in the case of cells pretreated with LysMK34 or with LysMK34 (0 to 1,000 μg/ml) for cells pretreated with colistin. Untreated cells were used as a control.
Cytotoxicity assay in human keratinocytes.
To test the potential cytotoxicity of LysMK34, a cytotoxicity model of a epithelial cell line (HaCaT) was used as previously described (70). Briefly, the real-time cell analyzer (RTCA-DP) xCelligence (ACEA Bioscience, Inc., San Diego, CA) was used to monitor in real time the growth of the HaCaT cell line, measuring the impedance signal, expressed as cell index (CI) every 15 min. When the cells reached the confluent growth after 20 h, different concentrations of the protein (8.13 to 500 μg/ml) were added, and the CI was measured for an additional 20 h. Values are finally expressed as the baseline normalized CI, calculated as previously described (70). SDS at a final concentration of 100 μg/ml was used as a positive cytotoxic control.
Statistical analysis.
SPSS Statistics for Windows v22.0 (IBM Corp.) was used for all calculations. A Student t test was used to compare the differences between the treated and untreated bacterial cultures at a level of significance of P < 0.05 (unless otherwise stated).
Accession number(s).
The PMK34 full genome sequence was deposited in the NCBI GenBank database under accession number MN433707.
Supplementary Material
ACKNOWLEDGMENTS
K.A. was funded by the Missions Sector, Ministry of Higher Education, Egypt. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication C.L. is supported by a PhD fellowship from FWO Vlaanderen (1S64720N). This study was supported by the Research Foundation Flanders (FWO) Research project G066919N.
We thank S. Malhotra-Kumar (Laboratory of Medical Microbiology, Vaccines, and Infectious Disease, University of Antwerp, Antwerp, Belgium) and J. P. Pirnay (Laboratory for Molecular and Cellular Technology, Queen Astrid Hospital, Neder-over-Heembeek, Belgium) for kindly providing the colistin-resistant A. baumannii strains (Greek46 and Greek47) and for A. baumannii RUH134 and P. aeruginosa PA14 and Br667, respectively.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Matsuzaki S, Rashel M, Uchiyama J, Sakurai S, Ujihara T, Kuroda M, Ikeuchi M, Tani T, Fujieda M, Wakiguchi H, Imai S. 2005. Bacteriophage therapy: a revitalized therapy against bacterial infectious diseases. J Infect Chemother 11:211–219. doi: 10.1007/s10156-005-0408-9. [DOI] [PubMed] [Google Scholar]
- 2.Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. 2007. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 51:3471–3484. doi: 10.1128/AAC.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trimble MJ, Mlynárčik P, Kolář M, Hancock RE. 2016. Polymyxin: alternative mechanisms of action and resistance. Cold Spring Harb Perspect Med 6:a025288. doi: 10.1101/cshperspect.a025288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valencia R, Arroyo LA, Conde M, Aldana JM, Torres MJ, Fernández-Cuenca F, Garnacho-Montero J, Cisneros JM, Ortiz C, Pachón J, Aznar J. 2009. Nosocomial outbreak of infection with pan–drug-resistant Acinetobacter baumannii in a tertiary care university hospital. Infect Control Hosp Epidemiol 30:257–263. doi: 10.1086/595977. [DOI] [PubMed] [Google Scholar]
- 5.Abdelkader K, Gerstmans H, Saafan A, Dishisha T, Briers Y. 2019. The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses 11:96. doi: 10.3390/v11020096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schooley RT, Biswas B, Gill JJ, Hernandez-Morales A, Lancaster J, Lessor L, Barr JJ, Reed SL, Rohwer F, Benler S, Segall AM, Taplitz R, Smith DM, Kerr K, Kumaraswamy M, Nizet V, Lin L, McCauley MD, Strathdee SA, Benson CA, Pope RK, Leroux BM, Picel AC, Mateczun AJ, Cilwa KE, Regeimbal JM, Estrella LA, Wolfe DM, Henry MS, Quinones J, Salka S, Bishop-Lilly KA, Young R, Hamilton T. 2017. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant Acinetobacter baumannii infection. Antimicrob Agents Chemother 61:e00954-17. doi: 10.1128/AAC.00954-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fischetti VA. 2008. Bacteriophage lysins as effective antibacterials. Curr Opin Microbiol 11:393–400. doi: 10.1016/j.mib.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Briers Y, Walmagh M, Van Puyenbroeck V, Cornelissen A, Cenens W, Aertsen A, Oliveira H, Azeredo J, Verween G, Pirnay J, Miller S, Volckaert G, Lavigne R. 2014. Engineered endolysin-based “Artilysins” to combat multidrug-resistant Gram-negative pathogens. mBio 5:e01379-14. doi: 10.1128/mBio.01379-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, Schuch R, Fischetti VA. 2015. Novel phage lysin capable of killing the multidrug-resistant Gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother 59:1983–1991. doi: 10.1128/AAC.04641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliveira H, Vilas Boas D, Mesnage S, Kluskens LD, Lavigne R, Sillankorva S, Secundo F, Azeredo J. 2016. Structural and enzymatic characterization of ABgp46, a novel phage endolysin with broad anti-Gram-negative bacterial activity. Front Microbiol 7:208. doi: 10.3389/fmicb.2016.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonova NP, Vasina DV, Lendel AM, Usachev EV, Makarov VV, Gintsburg AL, Tkachuk AP, Gushchin VA. 2019. Broad bactericidal activity of the Myoviridae bacteriophage lysins LysAm24, LysECD7, and LysSi3 against Gram-negative ESKAPE pathogens. Viruses 11:284. doi: 10.3390/v11030284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Larpin Y, Oechslin F, Moreillon P, Resch G, Entenza JM, Mancini S. 2018. In vitro characterization of PlyE146, a novel phage lysin that targets Gram-negative bacteria. PLoS One 13:e0192507. doi: 10.1371/journal.pone.0192507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai M-J, Lin N-T, Hu A, Soo P-C, Chen L-K, Chen L-H, Chang K-C. 2011. Antibacterial activity of Acinetobacter baumannii phage ϕAB2 endolysin (LysAB2) against both Gram-positive and Gram-negative bacteria. Appl Microbiol Biotechnol 90:529–539. doi: 10.1007/s00253-011-3104-y. [DOI] [PubMed] [Google Scholar]
- 14.Huang G, Shen X, Gong Y, Dong Z, Zhao X, Shen W, Wang J, Hu F, Peng Y. 2014. Antibacterial properties of Acinetobacter baumannii phage Abp1 endolysin (PlyAB1). BMC Infect Dis 14:681. doi: 10.1186/s12879-014-0681-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerstmans H, Criel B, Briers Y. 2018. Synthetic biology of modular endolysins. Biotechnol Adv 36:624–640. doi: 10.1016/j.biotechadv.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Briers Y, Lavigne R. 2015. Breaking barriers: expansion of the use of endolysins as novel antibacterials against Gram-negative bacteria. Future Microbiol 10:377–390. doi: 10.2217/fmb.15.8. [DOI] [PubMed] [Google Scholar]
- 17.El-Sayed MA, Amin MA, Tawakol WM, Loucif L, Bakour S, Rolain JM. 2015. High prevalence of blaNDM-1 carbapenemase-encoding gene and 16S rRNA armA methyltransferase gene among Acinetobacter baumannii clinical isolates in Egypt. Antimicrob Agents Chemother 59:3602–3605. doi: 10.1128/AAC.04412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 19.Turton JF, Woodford N, Glover J, Yarde S, Kaufmann ME, Pitt TL. 2006. Identification of Acinetobacter baumannii by detection of the blaOXA-51-like carbapenemase gene intrinsic to this species. J Clin Microbiol 44:2974–2976. doi: 10.1128/JCM.01021-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han L, Lei J, Xu J, Han S. 2017. blaOXA-23-like and blaTEM rather than blaOXA-51-like contributed to a high level of carbapenem resistance in Acinetobacter baumannii strains from a teaching hospital in Xi’an, China. Medicine 96:e8965. doi: 10.1097/MD.0000000000008965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Mi Z, Mi L, Huang Y, Li P, Liu H, Yuan X, Niu W, Jiang N, Bai C, Gao Z. 2019. Identification and characterization of capsule depolymerase Dpo48 from Acinetobacter baumannii phage IME200. PeerJ 7:e6173. doi: 10.7717/peerj.6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner D, Ackermann HW, Kropinski AM, Lavigne R, Sutton JM, Reynolds DM. 2017. Comparative analysis of 37 Acinetobacter bacteriophages. Viruses 10:5. doi: 10.3390/v10010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Popova A, Lavysh D, Klimuk E, Edelstein M, Bogun A, Shneider M, Goncharov A, Leonov S, Severinov K. 2017. Novel Fri1-like viruses infecting Acinetobacter baumannii-vB_AbaP_AS11 and vB_AbaP_AS12-characterization, comparative genomic analysis, and host-recognition strategy. Viruses 9:188. doi: 10.3390/v9070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu M, Hu K, Xie Y, Liu Y, Mu D, Guo H, Zhang Z, Zhang Y, Chang D, Shi Y. 2018. A novel phage PD-6A3, and its endolysin Ply6A3, with extended lytic activity against Acinetobacter baumannii. Front Microbiol 9:3302. doi: 10.3389/fmicb.2018.03302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merabishvili M, Vandenheuvel D, Kropinski AM, Mast J, De Vos D, Verbeken G, Noben JP, Lavigne R, Vaneechoutte M, Pirnay JP. 2014. Characterization of newly isolated lytic bacteriophages active against Acinetobacter baumannii. PLoS One 9:e104853. doi: 10.1371/journal.pone.0104853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jansen M, Wahida A, Latz S, Krüttgen A, Häfner H, Buhl EM, Ritter K, Horz HP. 2018. Enhanced antibacterial effect of the novel T4-like bacteriophage KARL-1 in combination with antibiotics against multidrug resistant Acinetobacter baumannii. Sci Rep 8:1. doi: 10.1038/s41598-018-32344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner D, Wand ME, Briers Y, Lavigne R, Sutton JM, Reynolds DM. 2017. Characterization and genome sequence of the lytic Acinetobacter baumannii bacteriophage vB_AbaS_Loki. PLoS One 12:e0172303. doi: 10.1371/journal.pone.0172303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H, Liang L, Lin S, Jia S. 2010. Isolation and characterization of a virulent bacteriophage AB1 of Acinetobacter baumannii. BMC Microbiol 10:131. doi: 10.1186/1471-2180-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh JK, Adams FG, Brown MH. 2018. Diversity and function of capsular polysaccharide in Acinetobacter baumannii. Front Microbiol 9:3301. doi: 10.3389/fmicb.2018.03301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Rubio L, Gerstmans H, Thorpe S, Mesnage S, Lavigne R, Briers Y. 2016. DUF3380 domain from a Salmonella phage endolysin shows potent N-acetylmuramidase activity. Appl Environ Microbiol 82:4975–4981. doi: 10.1128/AEM.00446-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oliveira H, Thiagarajan V, Walmagh M, Sillankorva S, Lavigne R, Neves-Petersen MT, Kluskens LD, Azeredo J. 2014. A thermostable Salmonella phage endolysin, Lys68, with broad bactericidal properties against Gram-negative pathogens in presence of weak acids. PLoS One 9:e108376. doi: 10.1371/journal.pone.0108376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walmagh M, Boczkowska B, Grymonprez B, Briers Y, Drulis-Kawa Z, Lavigne R. 2013. Characterization of five novel endolysins from Gram-negative infecting bacteriophages. Appl Microbiol Biotechnol 97:4369–4375. doi: 10.1007/s00253-012-4294-7. [DOI] [PubMed] [Google Scholar]
- 33.Walmagh M, Briers Y, Dos Santos SB, Azeredo J, Lavigne R. 2012. Characterization of modular bacteriophage endolysins from Myoviridae phages OBP, 201ϕ2-1 and PVP-SE1. PLoS One 7:e36991. doi: 10.1371/journal.pone.0036991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Briers Y, Volckaert G, Cornelissen A, Lagaert S, Michiels CW, Hertveldt K, Lavigne R. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages ϕKZ and EL. Mol Microbiol 65:1334–1344. doi: 10.1111/j.1365-2958.2007.05870.x. [DOI] [PubMed] [Google Scholar]
- 35.Guo M, Feng C, Ren J, Zhuang X, Zhang Y, Zhu Y, Dong K, He P, Guo X, Qin J. 2017. A novel antimicrobial endolysin, LysPA26, against Pseudomonas aeruginosa. Front Microbiol 8:293. doi: 10.3389/fmicb.2017.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Düring K, Porsch P, Mahn A, Brinkmann O, Gieffers W. 1999. The non‐enzymatic microbicidal activity of lysozymes. FEBS Lett 449:93–100. doi: 10.1016/s0014-5793(99)00405-6. [DOI] [PubMed] [Google Scholar]
- 37.Nair S, Poonacha N, Desai S, Hiremath D, Tuppad D, Mohan T, Chikkamadaiah R, Durgaiah M, Kumar S, Channabasappa S, Vipra A, Sharma U. 2018. Restoration of sensitivity of a diverse set of drug-resistant Staphylococcus clinical strains by bactericidal protein P128. J Med Microbiol 67:296–307. doi: 10.1099/jmm.0.000697. [DOI] [PubMed] [Google Scholar]
- 38.Thummeepak R, Kitti T, Kunthalert D, Sitthisak S. 2016. Enhanced antibacterial activity of Acinetobacter baumannii bacteriophage ØABP-01 endolysin (LysABP-01) in combination with colistin. Front Microbiol 7:1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wittekind M, Schuch R. 2016. Cell wall hydrolases and antibiotics: exploiting synergy to create efficacious new antimicrobial treatments. Curr Opin Microbiol 33:18–24. doi: 10.1016/j.mib.2016.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Sykilinda N, Nikolaeva A, Shneider M, Mishkin D, Patutin A, Popov V, Boyko K, Klyachko N, Miroshnikov K. 2018. Structure of an Acinetobacter broad-range prophage endolysin reveals a C-Terminal α-helix with the proposed role in activity against live bacterial cells. Viruses 10:309. doi: 10.3390/v10060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cross T, Ransegnola B, Shin JH, Weaver A, Fauntleroy K, VanNieuwenhze MS, Westblade LF, Dörr T. 2019. Spheroplast-mediated carbapenem tolerance in Gram-negative pathogens. Antimicrob Agents Chemother 63:e00756-19. doi: 10.1128/AAC.00756-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sofy KA, Saafan AE, AbdelGhani SM, Amin MA. 2015. Phenotypic and genotypic characterization of different classes of β-lactamases amongst Acinetobacter spp. isolated from Egyptian hospitals. N Egypt J Microbiol 42:36. [Google Scholar]
- 43.Hudzicki J. 2009. Kirby-Bauer disk diffusion susceptibility test protocol. American Society for Microbiology, Washington, DC. [Google Scholar]
- 44.De Vos D, Pirnay JP, Bilocq F, Jennes S, Verbeken G, Rose T, Keersebilck E, Bosmans P, Pieters T, Hing M, Heuninckx W, De Pauw F, Soentjens P, Merabishvili M, Deschaght P, Vaneechoutte M, Bogaerts P, Glupczynski Y, Pot B, van der Reijden TJ, Dijkshoorn L. 2016. Molecular epidemiology and clinical impact of Acinetobacter calcoaceticus-baumannii complex in a Belgian burn wound center. PLoS One 11:e0156237. doi: 10.1371/journal.pone.0156237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pirnay JP, De Vos D, Cochez C, Bilocq F, Pirson J, Struelens M, Duinslaeger L, Cornelis P, Zizi M, Vanderkelen A. 2003. Molecular epidemiology of Pseudomonas aeruginosa colonization in a burn unit: persistence of a multidrug-resistant clone and a silver sulfadiazine-resistant clone. J Clin Microbiol 41:1192–1202. doi: 10.1128/jcm.41.3.1192-1202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattila S, Ruotsalainen P, Jalasvuori M. 2015. On-demand isolation of bacteriophages against drug-resistant bacteria for personalized phage therapy. Front Microbiol 6:1271. doi: 10.3389/fmicb.2015.01271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams MH. 1959. Bacteriophages. Interscience Publishers, Inc, New York, NY. [Google Scholar]
- 48.Kropinski AM, Mazzocco A, Waddell TE, Lingohr E, Johnson RP. 2009. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol Biol 501:69–76. doi: 10.1007/978-1-60327-164-6_7. [DOI] [PubMed] [Google Scholar]
- 49.Maniatis T, Fritsch EF, Sambrook F, Engel J. 1982. Molecular cloning: a laboratory manual. Acta Biotechnol 49:411. [Google Scholar]
- 50.Capra M, Quiberoni A, Reinheimer J. 2004. Thermal and chemical resistance of Lactobacillus casei and Lactobacillus paracasei bacteriophages. Lett Appl Microbiol 38:499–504. doi: 10.1111/j.1472-765X.2004.01525.x. [DOI] [PubMed] [Google Scholar]
- 51.Clokie MRJ, Kropinski AM. 2009. Bacteriophages in materials and methods and protocols, vol 2: molecular and applied aspects. Humana Press, Totowa, NJ. [Google Scholar]
- 52.Peng F, Mi Z, Huang Y, Yuan X, Niu W, Wang Y, Hua Y, Fan H, Bai C, Tong Y. 2014. Characterization, sequencing and comparative genomic analysis of vB_AbaM-IME-AB2, a novel lytic bacteriophage that infects multidrug-resistant Acinetobacter baumannii clinical isolates. BMC Microbiol 14:181. doi: 10.1186/1471-2180-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 55.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Tasneem A, Thanki N, Yamashita RA, Zhang D, Zhang N, Bryant SH. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res 37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. 2011. ARNold: a web tool for the prediction of Rho-independent transcription terminators. RNA Biol 8:11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 60.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mitchell AL, Attwood TK, Babbitt PC, Blum M, Bork P, Bridge A, Brown SD, Chang H-Y, El-Gebali S, Fraser MI, Gough J, Haft DR, Huang H, Letunic I, Lopez R, Luciani A, Madeira F, Marchler-Bauer A, Mi H, Natale DA, Necci M, Nuka G, Orengo C, Pandurangan AP, Paysan-Lafosse T, Pesseat S, Potter SC, Qureshi MA, Rawlings ND, Redaschi N, Richardson LJ, Rivoire C, Salazar GA, Sangrador-Vegas A, Sigrist CJA, Sillitoe I, Sutton GG, Thanki N, Thomas PD, Tosatto SCE, Yong S-Y, Finn RD. 2019. InterPro in 2019: improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res 47:D351–D360. doi: 10.1093/nar/gky1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pei J, Kim B-H, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300. doi: 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 65.Gerstmans H, Grimon D, Gutierrez Fernandez D, Lood C, Rodríguez A, van Noort V, Lammertyn J, Lavigne R, Briers Y. 2020. A VersaTile driven platform for rapid hit-to-lead development of engineered lysins. Sci Adv 6:eaaz1136. doi: 10.1126/sciadv.aaz1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lavigne R, Briers Y, Hertveldt K, Robben J, Volckaert G. 2004. Identification and characterization of a highly thermostable bacteriophage lysozyme. Cell Mol Life Sci 61:2753–2759. doi: 10.1007/s00018-004-4301-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Briers Y, Lavigne R, Volckaert G, Hertveldt K. 2007. A standardized approach for accurate quantification of murein hydrolase activity in high-throughput assays. J Biochem Biophys Methods 70:531–533. doi: 10.1016/j.jbbm.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Briers Y, Walmagh M, Grymonprez B, Biebl M, Pirnay JP, Defraine V, Michiels J, Cenens W, Aertsen A, Miller S, Lavigne R. 2014. Art-175 is a highly efficient antibacterial against multidrug-resistant strains and persisters of Pseudomonas aeruginosa. Antimicrob Agents Chemother 58:3774–3784. doi: 10.1128/AAC.02668-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weinstein MP. 2018. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. National Committee for Clinical Laboratory Standards, Wayne, PA. [Google Scholar]
- 70.Valdés L, Gueimonde M, Ruas-Madiedo P. 2015. Monitoring in real time the cytotoxic effect of Clostridium difficile upon the intestinal epithelial cell line HT29. J Microbiol Methods 119:66–73. doi: 10.1016/j.mimet.2015.09.022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.