Ergot alkaloids derived from LA or DHLA are the bases for numerous pharmaceuticals with applications in the treatment of dementia, migraines, hyperprolactinemia, and other conditions. However, extraction of ergot alkaloids from natural sources is inefficient, and their chemical synthesis is expensive. The ability to control and redirect ergot alkaloid synthesis in fungi may allow more efficient production of these important chemicals and facilitate research on novel derivatives. Our results show that Metarhizium brunneum can be engineered to efficiently produce and secrete LA and DHLA and, also, to produce a novel derivative of DHLA not previously found in nature. The engineering of dihydroergot alkaloids, including a novel species, is important because very few natural sources of these compounds are known. Our approach establishes a platform with which to use M. brunneum to study the production of other ergot alkaloids, specifically those classified as lysergic acid amides and dihydroergot alkaloids.
KEYWORDS: ergot alkaloids, Metarhizium, lysergic acid, dihydroergot alkaloids, alkaloid secretion
ABSTRACT
Ergot alkaloids are important specialized fungal metabolites that are used to make potent pharmaceuticals for neurological diseases and disorders. Lysergic acid (LA) and dihydrolysergic acid (DHLA) are desirable lead compounds for pharmaceutical semisynthesis but are typically transient intermediates in the ergot alkaloid and dihydroergot alkaloid pathways. Previous work with Neosartorya fumigata demonstrated strategies to produce these compounds as pathway end products, but their percent yield (percentage of molecules in product state as opposed to precursor state) was low. Moreover, ergot alkaloids in N. fumigata are typically retained in the fungus as opposed to being secreted. We used clustered regularly interspaced short palindromic repeat (CRISPR)–CRISPR-associated protein 9 (Cas9) and heterologous expression approaches to engineer these compounds in Metarhizium brunneum, representing an alternate expression host from a different lineage of fungi. The relative percent yields of LA (86.9%) and DHLA (72.8%) were much higher than those calculated here for previously engineered strains of N. fumigata (2.6% and 2.0%, respectively). Secretion of these alkaloids also was measured, with averages of 98.4% of LA and 87.5% of DHLA being secreted into the growth medium; both values were significantly higher than those measured for the N. fumigata derivatives (both of which were less than 5.6% secreted). We used a similar approach to engineer a novel dihydroergot alkaloid in M. brunneum and, through high-performance liquid chromatography-mass spectrometry (LC-MS) analyses, provisionally identified it as the dihydrogenated form of lysergic acid α-hydroxyethylamide (dihydro-LAH). The engineering of these strains provides a strategy for producing novel and pharmaceutically important chemicals in a fungus more suitable for their production.
IMPORTANCE Ergot alkaloids derived from LA or DHLA are the bases for numerous pharmaceuticals with applications in the treatment of dementia, migraines, hyperprolactinemia, and other conditions. However, extraction of ergot alkaloids from natural sources is inefficient, and their chemical synthesis is expensive. The ability to control and redirect ergot alkaloid synthesis in fungi may allow more efficient production of these important chemicals and facilitate research on novel derivatives. Our results show that Metarhizium brunneum can be engineered to efficiently produce and secrete LA and DHLA and, also, to produce a novel derivative of DHLA not previously found in nature. The engineering of dihydroergot alkaloids, including a novel species, is important because very few natural sources of these compounds are known. Our approach establishes a platform with which to use M. brunneum to study the production of other ergot alkaloids, specifically those classified as lysergic acid amides and dihydroergot alkaloids.
INTRODUCTION
Ergot alkaloids are a complex family of specialized metabolites produced by several species of fungi in the phylum Ascomycota. These alkaloids are historically known for causing mass poisoning events as a result of infection of grain crops by the fungus Claviceps purpurea (1, 2). Some other ergot alkaloid-producing species are symbionts of plants, where the alkaloids deter insect and mammalian herbivores (1–4). The effects of these toxins are due to their structural similarity to neurotransmitters and high binding affinity for several neurological receptors, including the adreno, dopamine, and 5-hydroxytryptamine receptors (5, 6). Many natural ergot alkaloids are derivatives of lysergic acid (LA) and possess vasoconstrictive properties. Derivatives of dihydrolysergic acid (DHLA), which are rare in nature, have been shown to possess vasorelaxant properties and are used to synthesize several ergot-alkaloid-derived pharmaceuticals (6, 7). Most ergot alkaloid-derived pharmaceuticals are semisynthetics made by hydrolyzing naturally occurring ergot alkaloids to LA (sometimes reducing LA to DHLA) and then synthesizing amide side chains (8). The availability of fungi that produce LA or DHLA as pathway end products, as opposed to transient intermediates, might facilitate pharmaceutical research and production.
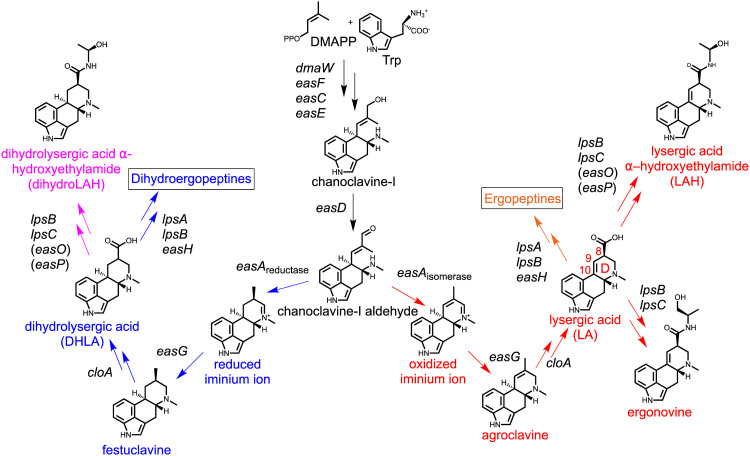
The difference between LA and DHLA is the presence of a double bond in the fourth-formed and final ring (D ring) of the ergoline nucleus (Fig. 1). Ergot alkaloid synthesis begins with a set of five genes (dmaW, easF, easC, easE, and easD), the products of which work together to prenylate tryptophan and convert it into a tricyclic intermediate, chanoclavine-I aldehyde (1, 2, 9, 10). In the next series of steps, closure of the D ring can result in either an unsaturated or saturated product, yielding agroclavine or festuclavine, respectively. Agroclavine serves as precursor to LA, whereas festuclavine is the precursor to DHLA. The path taken at this branchpoint is dependent on the allele of easA present in an organism (11–14). The isomerase allele, such as that found in the fungus Epichloë festucae (formerly called Neotyphodium lolii), encodes an enzyme that temporarily reduces the C-8–C-9 double bond of chanoclavine-I aldehyde to allow isomerization before the C-8–C-9 double bond is restored. This isomerization brings the aldehyde and amine groups into close proximity, facilitating ring closure. The reductase allele of easA found in Neosartorya fumigata (Aspergillus fumigatus) encodes an enzyme that fully reduces the double bond, allowing rotation of the aldehyde functional group such that it can react with the secondary amine. In either case, the resulting iminium ion is subsequently reduced by the product of easG (14, 15).
FIG 1.
Pathways to the synthesis of lysergic acid amides and dihydrolysergic acid amides. Both pathways begin with the same core set of genes (dmaW, easF, easE, easC, and easD) and branch at easA depending on the allele present in a species. The pathway branch to the right (with red labeling) leads to lysergic acid amides, as are found in M. brunneum (26), with a further branch to ergopeptines (labeled in orange), as found in Claviceps purpurea and several Epichloë species (1, 2, 9). Red font is used to mark the carbons present in the D ring of ergoline nucleus. The pathway branch to the left (with blue labeling) leads to dihydroergot alkaloids as found naturally in C. africana (45) or in previously engineered strains of N. fumigata (18, 19). The dihydroergot alkaloid pathway features a further branch, shown in pink, that occurs in no natural species but whose formation is a subject of this paper. Double arrows represent multiple enzymatic steps. Enzymes with hypothesized roles are marked by parentheses. Trp, l-tryptophan; DMAPP, dimethylallylpyrophosphate.
During the synthesis of LA and DHLA, the P450 monooxygenase product of cloA catalyzes a 6-electron oxidation reaction, oxidizing the methyl groups of agroclavine and festuclavine to the corresponding carboxylic acids (16, 17). Robinson and Panaccione (17) showed that N. fumigata is capable of accumulating LA when the cloA allele from an Epichloë species is expressed in a mutant background that produces agroclavine, the substrate for CloA. A specialized allele of cloA from Claviceps africana or Claviceps gigantea is needed in the synthesis of DHLA. Synthesis of DHLA in N. fumigata was achieved by expressing the cloA allele from C. gigantea (18) or a synthetic version of the allele from C. africana (19) in a festuclavine-accumulating background; however, the percent yield of DHLA was poor, as approximately 2% of the precursor festuclavine was converted to DHLA in engineered strains of N. fumigata (18). The authors hypothesized that N. fumigata may have had issues with enzyme and substrate compartmentalization or with accumulating products of the reaction. Moreover, N. fumigata typically retains its ergot alkaloids in or on its conidia, with only very small quantities of ergot alkaloids found in its growth medium (20). This combination of substrate conversion and product retention issues raises doubts about the ability of N. fumigata to serve as an effective expression host for LA and DHLA derivatives. A fungus that naturally produces LA derivatives may be better suited to produce LA and its derivatives or DHLA and its derivatives as artificial end products. Moreover, a fungus that secretes these compounds, as opposed to retaining them, would facilitate alkaloid isolation and purification.
The fungus Metarhizium brunneum, a member of the Clavicipitaceae, beneficially colonizes plant roots (21, 22) and also acts as a generalist entomopathogen (23–25). This species produces several LA amides, with its pathway culminating in the production of lysergic acid α-hydroxyethylamide (LAH), along with much smaller quantities of ergonovine (also known as ergometrine) (26). The production of ergonovine occurs through the combination of the nonribosomal peptide synthetase products of lpsB and lpsC. The product of lpsB activates LA by adenylation and binds it as a thioester to prepare it for incorporation into LA amides (27, 28). Knockout mutation of lpsB in C. purpurea resulted in the accumulation of LA (27). The formation of LAH has been hypothesized to require peptide synthetases encoded by lpsB and lpsC and to also involve the products of easO and easP (1, 9, 29). Metarhizium brunneum also is noteworthy in that it secretes relatively large proportions of its ergot alkaloids compared to the levels of secretion by other ergot alkaloid-producing fungi (26). As a producer of lysergic acid amides, M. brunneum contains the isomerase allele of easA and a functional copy of lpsB.
The studies described above have elucidated the roles of EasA, CloA, and LpsB, leading to strategies for producing LA and DHLA in different fungal backgrounds through introduction or modification of the genes encoding these enzymes. We hypothesized that engineering pathways for the production of LA and DHLA in M. brunneum, which typically produces LA amides, would result in a higher conversion of substrate to the desired product than was observed in N. fumigata strains, which were engineered in a background that lacks capacity to make LA amides. We also hypothesized that since M. brunneum secretes the majority of its own LA amides (26), it would secrete the related but simpler molecules LA and DHLA when engineered as pathway end products. A final hypothesis tested in this present study was that M. brunneum could be modified to produce novel derivatives of DHLA, in particular a dihydrogenated form of LAH. Here, we show the generation of strains of M. brunneum capable of accumulating the pharmaceutically important ergot alkaloids LA and DHLA, as well as a novel dihydroergot alkaloid, and assess the percent yield and secretion of these compounds relative to these traits in previously engineered N. fumigata strains.
RESULTS
Production and secretion of lysergic acid in M. brunneum.
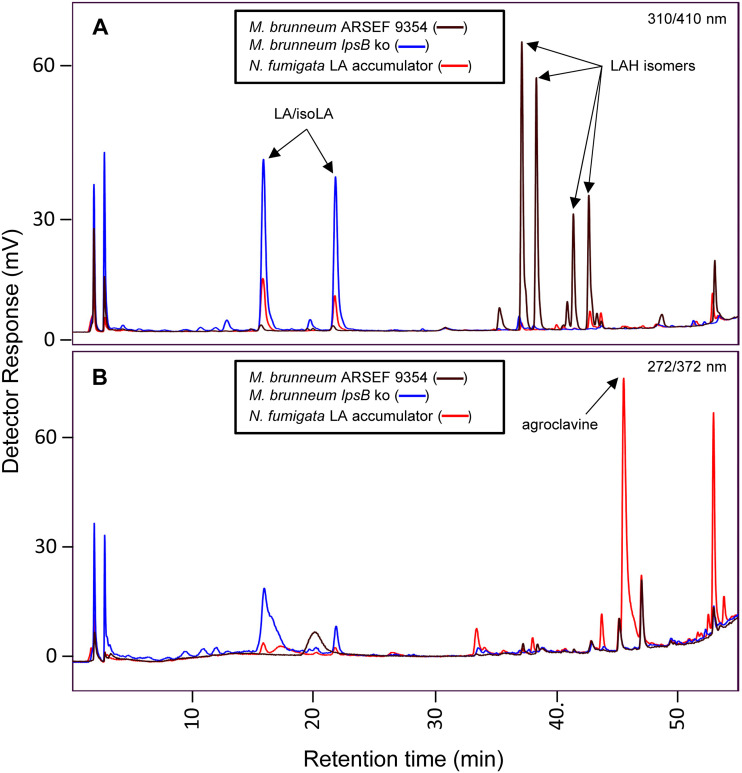
The generation of a strain of M. brunneum capable of accumulating LA was achieved through an approach in which a single guide RNA (sgRNA) specific for lpsB was complexed with Cas9 (clustered regularly interspaced short palindromic repeat [CRISPR]-associated protein 9) for transient expression in transformed protoplasts, resulting in knockout of lpsB. The sgRNA:Cas9 complex was cotransformed along with a fragment of DNA containing the bar gene, conferring resistance to phosphinothricin (Fig. S1 in the supplemental material). Sanger sequencing of the lpsB locus in the mutated strain revealed it to have recombined in such a way that after being cut by Cas9, two partial fragments of the bar construct amplicon used as the selectable marker were incorporated into the lpsB locus in the ensuing repair process (Fig. S2). Phosphinothricin resistance in the strain was provided by an additional, full copy of the bar amplicon integrated elsewhere in the genome, as confirmed through further PCR and Sanger sequencing. The lpsB knockout strain was analyzed by high-performance liquid chromatography (HPLC) with fluorescence detection and compared to wild-type M. brunneum and a strain of N. fumigata previously engineered to accumulate LA (17) as references (Fig. 2). The lpsB knockout strain lacked LAH; instead, it accumulated LA as the end product of its ergot alkaloid pathway, as evidenced by detection of peaks corresponding to LA at ∼16 min, along with its stereoisomer at ∼22 min.
FIG 2.
HPLC chromatograms of the lpsB knockout strain of M. brunneum (lpsB ko) compared to wild-type M. brunneum and LA-accumulating strain of N. fumigata. Fluorescence was detected at 410 nm after excitation at 310 nm (A) and at 372 nm after excitation at 272 nm (B). Peaks corresponding to characterized ergot alkaloids are indicated: LA/isoLA, lysergic acid/isolysergic acid; LAH, lysergic acid α-hydroxyethylamide.
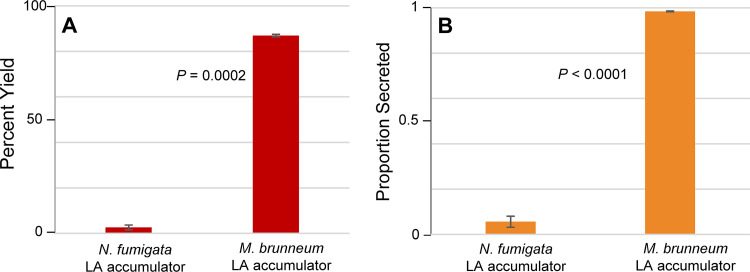
Broth cultures of the lpsB knockout mutant were grown in triplicate and compared to similarly grown cultures of the N. fumigata strain previously engineered to accumulate LA (17). Alkaloid extracts prepared from the dried fungal mats were analyzed through HPLC with fluorescence detection and compared to extracts of their respective growth medium. Percent yield of LA was approximated by comparing relative peak areas of LA and its precursor agroclavine to standard curves prepared from ergot alkaloids with similar fluorophores. The M. brunneum lpsB knockout strain had a much higher percent yield than the LA-producing strain of N. fumigata (P = 0.0002) (Fig. 3A). The difference in percent yield between the engineered M. brunneum and N. fumigata strains also is evident in the amount of the CloA substrate agroclavine accumulating in the N. fumigata strain relative to its accumulation in the M. brunneum strain, as shown in HPLC chromatograms (Fig. 2B; Table S1). Comparison of the solid- and liquid-phase extracts also revealed that the M. brunneum lpsB knockout strain secreted LA in a much higher proportion than the N. fumigata strain (P < 0.0001) (Fig. 3B).
FIG 3.
Percent yield and secretion of LA in N. fumigata mutant compared to M. brunneum mutant. (A) Mean percent yields of lysergic acid relative to those of precursor agroclavine between strains. (B) Mean proportions of lysergic acid secreted into growth medium by strains. Standard errors of the means for both data sets are shown by the error bars. P values from analysis of variance (ANOVA) are shown between bars.
Production and secretion of DHLA in M. brunneum.
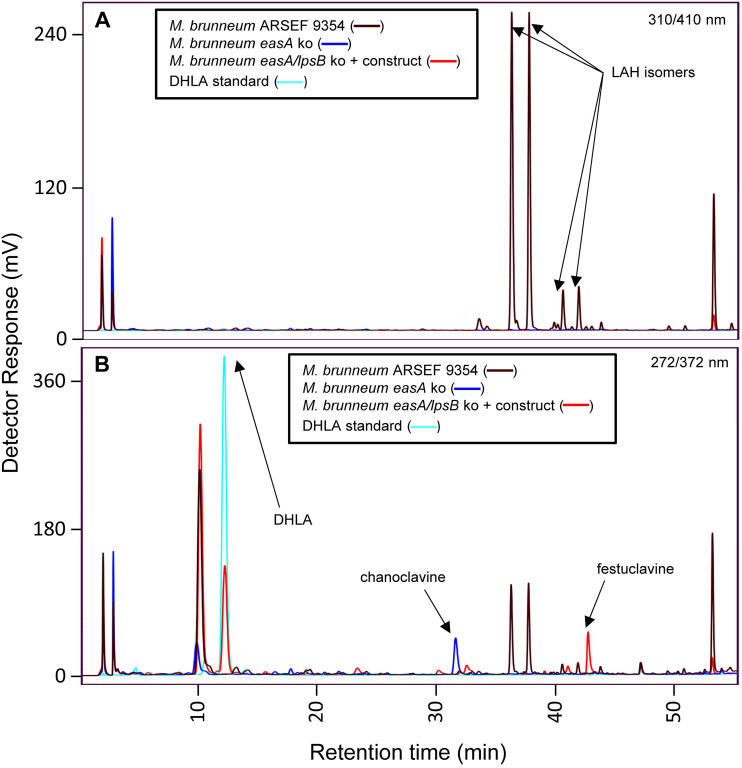
Generation of a strain of M. brunneum producing DHLA was achieved through two successive transformations (Fig. S1). In the first transformation, the easA locus of wild-type M. brunneum was targeted for knockout in an approach similar to that described for the lpsB knockout. The resulting mutant recombined at the easA locus, incorporating a full copy of the bar construct selectable marker (Fig. S3) and disrupting the easA coding sequences. HPLC analysis of this strain indicated a lack of LAH and accumulation of chanoclavine-I, as observed previously in an N. fumigata easA knockout (13) (Fig. 4).
FIG 4.
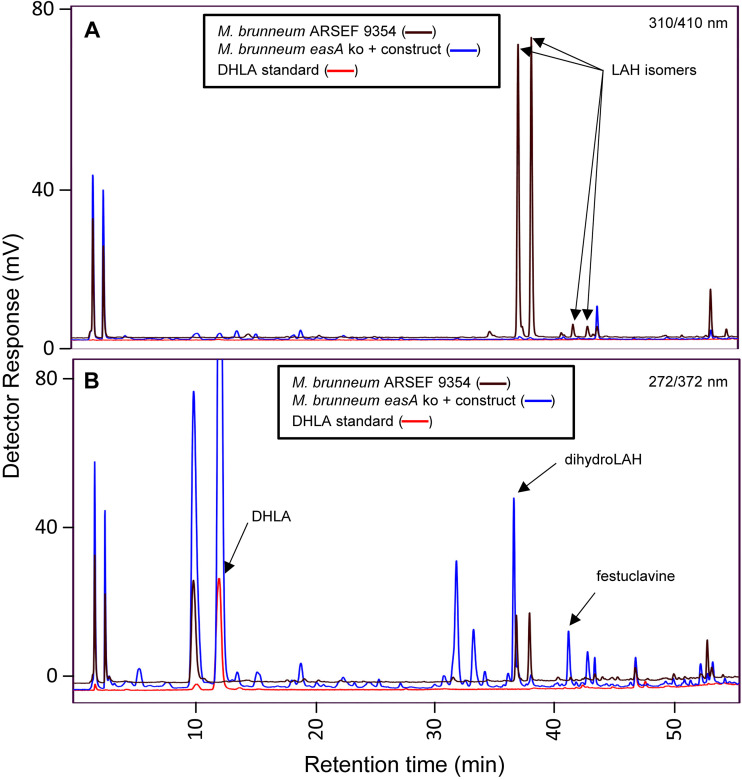
HPLC chromatograms of M. brunneum mutants compared to wild-type M. brunneum and DHLA standard. The “construct” noted in the key contains N. fumigata easA and C. africana cloA. Fluorescence was detected at 410 nm after excitation at 310 nm (A) and at 372 nm after excitation at 272 nm (B). LAH, lysergic acid α-hydroxyethylamide.
In the second transformation, with the easA knockout strain as the recipient, the lpsB locus was knocked out by the CRISPR-based strategy described above, and at the same time, the strain was augmented by the introduction of a construct containing two genes previously shown to be required for synthesis of DHLA. Fusion PCR was used to generate a dihydroergot alkaloid expression construct, which contained the easA allele from N. fumigata (to complement the easA knockout in the recipient but with an allele that produces the dihydroergot alkaloid substrate festuclavine) (13) and a previously generated synthetic cloA allele based on the protein sequence found in C. africana (to oxidize festuclavine to DHLA) (19). The two genes were divergently transcribed under the control of a bidirectional dmaW/easG promoter from the M. brunneum eas cluster (Fig. S4). This “dihydro” construct was introduced into the easA knockout strain of M. brunneum in pBCHygro (30), as a selectable marker, along with an sgRNA:Cas9 complex targeting lpsB for knockout. Knockout of lpsB was assessed by a strategy similar to that used for the previously discussed knockouts (Fig. S3). The mutated locus incorporated a fragment of the pBChygro vector in the lpsB coding sequences. Resistance to hygromycin and expression of the dihydro construct were conferred by a full construct integrated elsewhere. Extracts of this strain were analyzed by HPLC and compared with extracts of wild-type M. brunneum, the easA knockout strain of M. brunneum, and a chemical standard for DHLA as references (Fig. 4). The mutant strain lacked LAH and did not accumulate detectable levels of chanoclavine-I. Instead, the mutant accumulated DHLA, as indicated by the presence of a peak that aligned with the standard at ∼12 min.
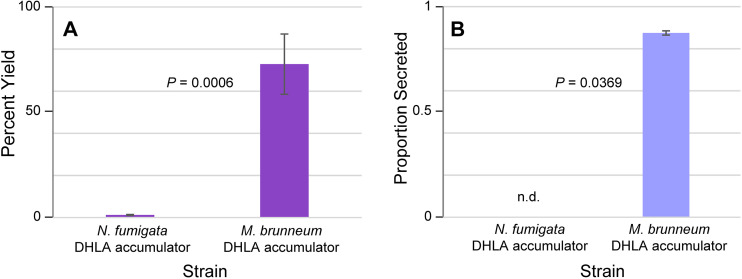
Broth-based cultures of the mutant were grown in triplicate and compared to similarly grown cultures of a previously described strain of N. fumigata that accumulates DHLA (18). Dried fungal mats were extracted and compared to extracts of their respective growth medium by HPLC with fluorescence detection, as described above for the LA accumulators. The DHLA-accumulating M. brunneum strain converted festuclavine substrate to DHLA to give a much higher percent yield than did the previously engineered strain of N. fumigata (P = 0.0006) (Fig. 5A; Table S1). The M. brunneum DHLA-accumulating strain also secreted DHLA in a very high proportion, whereas no DHLA was detectable in the spent culture fluids of the N. fumigata strain (Fig. 5B).
FIG 5.
Percent yield and secretion of DHLA in N. fumigata mutant compared to those of M. brunneum mutant. (A) Mean percent yields of DHLA relative to precursor festuclavine between strains. (B) Average proportions of DHLA secreted into growth medium by strains. Standard errors of the means for both sets are shown by the error bars. The P value from ANOVA is shown between bars in panel A, whereas the P value for the data shown in panel B was derived from a Wilcoxon rank-sum test because DHLA was not detected in N. fumigata culture filtrates. n.d., not detected.
Production and secretion of a novel lysergic acid amide in M. brunneum.
The ability of M. brunneum to produce and secrete additional, novel dihydroergot alkaloids was demonstrated by substituting alleles at the easA locus (to produce the dihydroergot alkaloid intermediate festuclavine) and augmenting with a festuclavine-oxidizing allele of cloA. The easA and cloA alleles were introduced via the previously described easA/cloA dihydro construct while targeting the easA locus of wild-type M. brunneum for knockout. The strategy was thus similar to that used as described above to generate the DHLA mutant, but in this case, the recipient fungus retained a functional copy of lpsB such that the DHLA produced might be incorporated into amides of DHLA. The easA knockout strain incorporated a full copy of the expression construct as cloned into pBChygro (Fig. S5). HPLC analysis of this strain revealed a lack of peaks corresponding to the stereoisomers of LAH (Fig. 6A). Instead the engineered strain contained a novel peak eluting at ∼37 min in the 272-/372-nm fluorescence chromatogram, corresponding to a retention time slightly shorter than that of LAH, which was observed in the wild type and which fluoresces maximally with excitation at 310 nm and emission at 410 nm (Fig. 6). The change in fluorescence properties is consistent with a shift from an unsaturated D ring typical of LA derivatives (fluorescing more strongly at 310 nm/410 nm) to the saturated D ring of dihydroergot alkaloids (fluorescing more intensely at 272 nm/372 nm) (31).
FIG 6.
HPLC chromatograms of M. brunneum mutant with novel alkaloid compared to those of parental strains from which it was derived: wild-type M. brunneum and easA knockout (easA ko) of M. brunneum. Fluorescence was detected at 410 nm after excitation at 310 nm (A) and at 372 nm after excitation at 272 nm (B). The wild-type (black line) peaks eluting just later than dihydro-LAH correspond to the most abundant LAH isomers (marked in panel A) fluorescing under suboptimal excitation and emission wavelengths. A DHLA standard was also included. LAH, lysergic acid α-hydroxyethylamide; DHLA, dihydrolysergic acid; dihydro-LAH, provisional dihydrolysergic acid α-hydroxyethylamide.
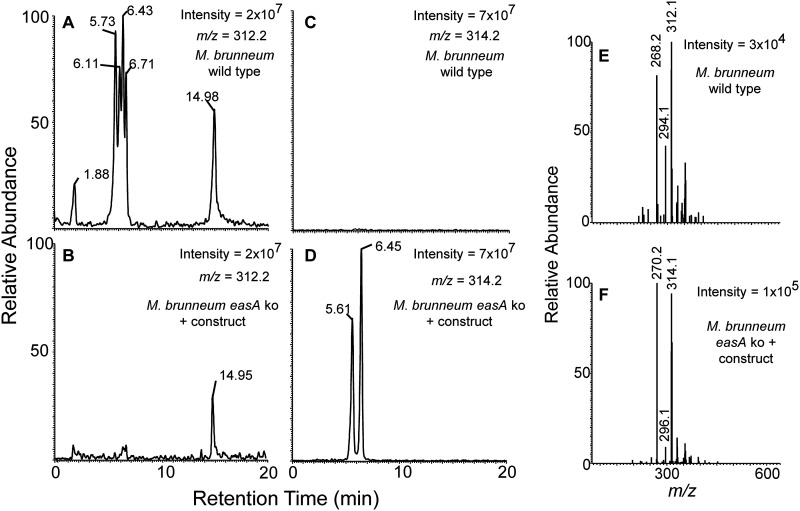
Extracts of this strain were then compared further with those of the wild type through liquid chromatography-mass spectrometry (LC-MS) (Fig. 7). Parent ions of m/z 312.2 were found in the wild-type strain, with four peaks eluting at ∼5.7 min to ∼6.7 min. The observed m/z values of these four ions are consistent with those expected for the protonated ions of the four stereoisomers of LAH. These stereoisomers result from alternate stereochemistry at the site where the alanine derivative attaches to LA and from the keto-enol tautomerization that occurs at C-8 of LA, where the amide side branch attaches from the ergoline ring system (Fig. 1) (26, 31, 32). None of these LAH stereoisomer peaks were detected in the mutant strain; instead, the mutant accumulated parent ions of m/z 314.2, which is consistent with the [M + H]+ of the dihydrogenated form of LAH (dihydro-LAH). The presence of two peaks (∼5.6 min and at ∼6.5 min) indicates the presence of stereoisomers, which would be expected from the chiral carbon at the attachment site of the alanine-derived residue of dihydro-LAH. Dihydrogenated ergot alkaloids lack the stereoisomers from the keto-enol tautomerization that occurs at C-8 (18, 19, 31, 33).
FIG 7.
LC-MS analyses of M. brunneum mutant with novel alkaloid compared to wild-type M. brunneum. (A to D) Extracted ion chromatograms of wild-type M. brunneum analyzed at indicated m/z values (A, C) compared to the easA knockout strain expressing the dihydro expression construct (B, D). (E, F) Fragmentation spectra obtained from parent ions shown in panels A and D. Instrument resolution accurate to 0.1 Da.
Fragmentation analyses revealed a close relationship between the m/z 312.2 ions of wild-type M. brunneum and the m/z 314.2 ions of the engineered M. brunneum mutant (Fig. 7). Major ions of m/z 268 and m/z 294 were obtained from the m/z 312 ion. The m/z 268 ion corresponds to ergine, the simple amide of LA and spontaneous hydrolysis product of LAH, and the m/z 294 ion is consistent with [LAH − H2O]+. The fragmentation pattern of the m/z 314 ion aligned with that of LAH but with each fragment 2 Da greater, consistent with the presence of a saturated D ring in the ergoline nucleus of the molecule. Collectively, the data strongly suggest that the novel metabolites accumulating in the engineered strain are stereoisomers of dihydro-LAH, a compound not previously found in nature. Cultures of this mutant were grown similarly to those described for the previous strains, and the mean percent yield of the putative dihydro-LAH (relative to DHLA as precursor) was calculated to be 16.9% (±4.3%) (±standard error [SE]). A mean of 95.9% (±0.61%) of the provisional dihydro-LAH was secreted from the fungus into the growth medium.
DISCUSSION
Our data show that M. brunneum can be engineered to accumulate LA and DHLA with higher percent yields than were calculated for strains of N. fumigata previously engineered to produce these same ergot alkaloids (17–19). Moreover, we have found that the natural ability of M. brunneum to secrete its own alkaloids (26) allows these engineered strains to secrete the majority of each of these products. We also demonstrated that M. brunneum can be engineered to produce a novel derivative of DHLA, which evidence indicates is dihydro-LAH, and that the fungus secretes almost the entire proportion of this product. The increased percent yields of products relative to those observed in N. fumigata, abundant secretion of LA, DHLA, and their derivatives, and amenability to engineering of novel compounds make M. brunneum an effective platform for ergot alkaloid research and, potentially, production.
Previous experiments have shown that N. fumigata can be engineered to accumulate LA through the expression of the isomerase allele of easA along with cloA in an appropriate mutant background (17). While conversion of agroclavine to LA was not measured in that previous study, the same strain was used in our experiments, and we observed yield of LA from agroclavine with a relatively low percentage. Our strain of M. brunneum, which normally produces derivatives of LA, had no difficulty accumulating LA as the terminal product of its pathway. The M. brunneum strain had a much higher percent yield than N. fumigata.
Additional previously published experiments have shown that N. fumigata can be engineered to produce DHLA when alleles of cloA from species that naturally produce this compound are introduced into a festuclavine-accumulating mutant (18, 19). Bragg et al. (18) tested the allele from the maize ergot pathogen C. gigantea in an N. fumigata mutant and determined that conversion of festuclavine to DHLA was very low. A similar result was obtained when Arnold and Panaccione (19) used a cloA allele from C. africana that was codon optimized for N. fumigata. The authors suspected that the low conversion could have been due to issues related to enzyme and substrate compartmentalization or accumulation of the reaction products. Metarhizium brunneum appeared to have no significant problem in converting substrate to end product, although the reasons for the difference in results between expression hosts have yet to be elucidated. Secretion was not tested in the previous studies, because N. fumigata typically retains its alkaloids (20); however, secretion from the relevant N. fumigata strain was tested here for comparison purposes and found to be low for LA and nondetectable for DHLA. In contrast, the engineered M. brunneum strain secreted the vast majority of its LA and DHLA.
Lps2 (also called LpsB), the product of lpsB, is required for the formation of ergopeptines (27, 28, 34) and ergonovine (28). A similar requirement for this enzyme in the biosynthesis of LAH was reasonable to hypothesize, but a direct role for LpsB in LAH biosynthesis had not been demonstrated until the present study. We showed that the presence of this gene is necessary not only for LAH, which was not detectable in the lpsB knockout, but also for production of its dihydrogenated form, which accumulated when the dihydro expression construct was introduced into the easA knockout but not when introduced into the lpsB knockout. Our data indicate that LpsB from M. brunneum accepts DHLA as a substrate, but the relatively low rate of conversion of DHLA to provisional dihydro-LAH (16.9%) may indicate a lower affinity for DHLA as a substrate. This enzyme substrate combination was tested previously in C. purpurea by Riederer et al. (34), who showed that LpsB (Lps2) of C. purpurea accepted DHLA with the same Km with which it accepted LA. In our dihydro-LAH example, however, the low relative percent yield may reflect factors other than the affinity of LpsB for DHLA as a substrate, because several activities downstream from LpsB are required for the conversion of DHLA into dihydro-LAH.
The reasons why M. brunneum can produce LA and DHLA more efficiently than did N. fumigata are an area yet to be explored. The presence of transporter proteins allowing easy movement of substrates and reaction products would be one potential explanation for the higher percent yield and secretion; however, no candidate alkaloid transporter genes have yet been identified in M. brunneum, and no genes in the eas cluster resemble transporter-encoding genes. Research on the biosynthesis of other fungal toxins has shown the importance of cellular compartmentalization and transport of pathway intermediates or end products between compartments (35–37). Compartmentalization and/or interaction of enzymes relative to one another may also be more conducive for product formation in M. brunneum, which naturally produces similar ergot alkaloids, as opposed to N. fumigata, which produces ergot alkaloids from a different branch of the pathway (9, 10).
Although the M. brunneum strains more efficiently produced and secreted higher proportions of LA and DHLA than similarly engineered N. fumigata strains, the absolute quantities of ergot alkaloids the fungus produced in culture (Table S1) were low in comparison to the quantities observed when the fungus colonizes Galleria mellonella larvae (26). Culture conditions or genetic modifications that promote increased and more reliable yields of ergot alkaloids in culture would help make this fungus an excellent platform for ergot alkaloid modification and production. The great differences in the accumulation of ergot alkaloids in insects compared to their accumulation in cultures (26) indicates strong regulation of the pathway. The eas clusters of Metarhizium species lack genes that would appear to encode transcription factors; thus, genes controlling the expression of eas genes may be encoded elsewhere in the genome. The increased accumulation of ergot alkaloids in insects also indicates a role for ergot alkaloids in fungus-insect interaction. The mutants generated in this present study may be helpful in assessing potential effects of different ergot alkaloids in fungus colonization of insects.
The use of hygromycin as a selectable marker for transformed M. brunneum strains was facilitated through the utilization of a very high concentration of the chemical. Previous literature (38, 39) has mentioned the insensitivity of Metarhizium spp. to hygromycin as an impediment to using this compound as a selectable marker for transformation. Nonetheless, we tested concentrations from 200 μg/ml to 800 μg/ml and found that a concentration of 600 μg/ml allowed the formation of transformed colonies that were distinguishable from background growth compared to the growth observed on no-DNA control plates.
Overall, our work shows that M. brunneum has the potential to serve as a platform in which to produce pharmaceutically relevant ergot alkaloids and novel derivatives of these compounds. The approaches used here also may facilitate functional analysis of additional genes involved in LA amide synthesis, which is ongoing in our laboratory. Additional future work will focus on identification of factors responsible for the higher percent yield and secretion observed in this fungus.
MATERIALS AND METHODS
Growth and maintenance of fungi.
Cultures of wild-type M. brunneum (ARSEF 9354; U.S. Department of Agriculture ARS Collection of Entomopathogenic Fungal Cultures, Ithaca, NY) and its transformants were maintained on sucrose-yeast extract (SYE) agar medium (per liter, 20 g sucrose, 10 g yeast extract, 1 g magnesium sulfate-heptahydrate, and 15 g agar). Cultures were grown at 30°C for at least 7 days before testing for ergot alkaloids. Cultures of the previously engineered N. fumigata strains (17, 18) were maintained at 37°C on malt extract agar medium (per liter, 6.0 g malt extract, 1.8 g maltose, 6.0 g dextrose, 1.2 g yeast extract, and 15 g agar).
Design of CRISPR sgRNA and Cas9 complexing.
Single guide RNA (sgRNA) sequences targeting the easA (GenBank accession number XM_014685465.1) and lpsB (GenBank accession number XM_014685466.1) loci were chosen based on sequence recommendations for Cas9 cutting efficiency (40). A 20-nucleotide target sequence (underlined in the primers, listed below) was chosen for both genes, and primers containing these were used for sgRNA synthesis, with TTCTAATACGACTCACTATAGGACAGGAAATAGACTCGGCGTTTTAGAGCTAGA used as the primer for lpsB sgRNA synthesis and TTCTAATACGACTCACTATAGGACAAGAAGCCAATCTTGCGTTTTAGAGCTAGA used for easA. Both primers were diluted to 1 μM before being included in sgRNA synthesis reactions involving the EnGen sgRNA synthesis kit (New England Biolabs [NEB], Ipswich, MA). sgRNA products were cleaned using the Monarch RNA cleanup kit (NEB). Quality of sgRNA was confirmed by SDS-PAGE. Prior to fungal transformation, sgRNAs were diluted to 20 μM and complexed with EnGen spy Cas9 NLS (NEB) by combining 2 μl of sgRNA, 2 μl of Cas9 enzyme, 0.5 μl of 10× buffer 3.1 (NEB), and 0.5 μl of nuclease-free water. The reaction mixture was incubated at room temperature for 30 min to allow complexing before being moved to ice until needed.
Preparation of transformation constructs.
A synthetic version of the phosphinothricin N-acetyltransferase gene coding sequence (GenBank accession number MT350122), which confers resistance to bialaphos, was codon optimized for Metarhizium anisopliae, placed under the control of the glycerol-3-phosphate dehydrogenase (GPDH) promoter from M. brunneum ARSEF 3297 (nucleotides 1,625,569 to 1,626,574 in the record under GenBank accession number NW_014574712.1), and purchased from GenScript (Piscataway, NJ) in a pUC57 construct. The promoter and coding portion of this construct, referred to as the bar construct or bar amplicon, were amplified through PCR. All PCRs were performed with Phusion green hot start II high-fidelity PCR master mix (Thermo Scientific, Waltham, MA) and followed similar protocols, with an initial denaturation at 98°C (15 s), annealing at a temperature specific to each primer as indicated in Table 1 (15 s), extension at 72°C for the time interval specified in Table 1, and a final extension at 72°C for 60 s. PCRs were conducted in a 20-μl reaction mixture containing 10 μl of PCR master mix (deoxynucleoside triphosphates [dNTPs], buffer, polymerase, and gel loading dye), primers at a final concentration of 0.5 μM each, and approximately 20 to 100 ng of template DNA. The optimized GPDH promoter-bar fusion construct was amplified with primer combination 1. A DNA Clean & Concentrator kit (Zymo Research, Irvine, CA) was used to purify the amplicon.
TABLE 1.
Primers and PCR protocol information
| Primer pair | Sequence (5′ to 3′)a | Product (length in bp) | Annealing temp (°C), extension time (s) |
|---|---|---|---|
| 1 | GGCAGCTTGGAGTATGTCTG | Optimized bar with GPDH promoter (1,981) | 62, 60 |
| CCTTGCTTGAGAAGGTTTTGG | |||
| 2 | CTCAGCGGCCGCGCACCATGTCAAGAAGTAGC | N. fumigata easA plus overlap with M. brunneum easG/dmaW promoter (1,543) | 62, 45 |
| CCATCTCGGAAAAGAAAAATGCGAGAAGAACCGTCCT | |||
| 3 | GAGGACGGTTCTTCTCGCATTTTTCTTTTCCGAGATGG | M. brunneum easG/dmaW promoter plus overlaps for N. fumigata easA and synthetic C. africana cloA (1,345) | 60, 45 |
| GTACAGCCAAAGCTGAGACATCGTAAACCAGAGTATTATG | |||
| 4 | CATAATACTCTGGTTTACGATGTCTCAGCTTTGGCTGTAC | Synthetic C. africana cloA plus overlap with M. brunneum easG/dmaW promoter (1,903) | 64, 60 |
| CGTAGTCGACACAGCAAAGTCTGATAAGTG | |||
| 5 | CTCAGCGGCCGCGCACCATGTCAAGAAGTAGC | N. fumigata easA–M. brunneum easG/dmaW promoter–synthetic C. africana cloA fusion product (4,713) | 70, 150 |
| CGTAGTCGACACAGCAAAGTCTGATAAGTG | |||
| 6 | CCGCGCACCATGTCAAGAAG | N. fumigata easA–promoter–C. africana cloA in pBChygro (4,705) | 66, 150 |
| CGAGGTCGACACAGCAAAGTC | |||
| 7 | GAGCAACAAGGAGCGTCAATG | M. brunneum lpsB internal sequence flanking CRISPR cut site (425)b | 65, 30 |
| GCTTATCGCTTGGCCAAGAG | |||
| 8 | GTCGGTATGAAGCTGTCCACC | M. brunneum easA internal sequence flanking CRISPR cut site (423)b | 65, 30 |
| GTATCCCTCGTCCGAGAACG | |||
| 9 | GTCGGTATGAAGCTGTCCACC | N. fumigata easA C. africana cloA–pBChygro construct in easA cut site (same orientation, 5′ side) (variable length) | 65, 150 |
| CGAGGTCGACACAGCAAAGTC | |||
| 10 | CCGCGCACCATGTCAAGAAG | N. fumigata easA–C. africana cloA–pBChygro construct in easA cut site (same orientation, 3′ side) (variable length) | 64, 150 |
| GTATCCCTCGTCCGAGAACG | |||
| 11 | GTCGGTATGAAGCTGTCCACC | N. fumigata easA–C. africana cloA–pBChygro construct in easA cut site (opposite orientation, 5′ side) (no product) | 65, 150 |
| CCGCGCACCATGTCAAGAAG | |||
| 12 | CGAGGTCGACACAGCAAAGTC | N. fumigata easA–C. africana cloA–pBChygro construct in easA cut site (opposite orientation, 3′ side) (no product) | 64, 150 |
| GTATCCCTCGTCCGAGAACG |
Underlines indicate unique restriction sites used for cloning fusion PCR product: GTCGAC, SalI; GCGGCCGC, NotI.
Expected product size based on wild-type locus and subject to insertions/deletions following CRISPR.
A construct for expressing the N. fumigata easA allele (13) together with the synthetic, intron-free C. africana cloA allele (19) under the control of the bidirectional easG/dmaW promoter from M. brunneum (nucleotides 206,186 to 207,489 in the record under GenBank accession number NW_014574698.1) was generated using a three-way fusion PCR. The individual fragments were amplified using primer sets designed to include overlaps to allow fusion, with both gene portions including roughly 150 bp of their native 3′ untranslated region (3′ UTR). The primers used in the final fusion PCR included recognition sites for NotI and SalI to allow digestion and ligation into pBChygro (Fungal Genetics Stock Center, Manhattan, KS) (30). Genomic DNA was extracted from N. fumigata strain Af293 and wild-type M. brunneum samples according to the GeneClean spin protocol (MP Biomedicals, Solon, OH). These templates were used with primer combinations 2 and 3 (Table 1), respectively, to generate the easA and promoter fragments. Primer combination 4 was used together with a stock of the synthetic cloA allele plasmid (19). The fragments (consisting of 1,543, 1,345, and 1,903 bp, respectively) were gel purified using a ZymoClean gel DNA recovery kit (Zymo). Equimolar ratios of each fragment were then combined in a subsequent PCR using primer combination 5. A DNA Clean & Concentrator kit (Zymo) was used to clean the fusion product before both it and pBChygro were subjected to double digestion. These fragments were then ligated following a cleanup step, and the final product, N. fumigata easA–C. africana cloA–pBChygro (referred to as the “dihydro construct”) was used to transform competent Escherichia coli cells (NovaBlue; Sigma-Aldrich, St. Louis, MO). Transformed cells were plated on LB medium (per liter, 10 g tryptone, 5 g yeast extract, 5 g NaCl, and 15 g agar) supplemented with chloramphenicol (25 μg/ml). Plasmid products were harvested and purified using a Zyppy plasmid miniprep kit (Zymo), and correct assembly was verified through double digestion with NotI/SalI.
Protoplast preparation and fungal transformation.
Protoplasts of M. brunneum were prepared as described previously for N. fumigata (41). Protoplasts were then transformed with complexed sgRNA:Cas9 and selectable markers according to established methods (13, 41). Our CRISPR-Cas9 approach differed from that employed by Chen et al. (42) for the entomopathogenic fungus Beauveria bassiana in that we opted for transient expression of introduced, sgRNA-complexed Cas9 as opposed to stably transforming the gene encoding Cas9 into the fungus. Moreover, since our transformation reactions contained 50 million protoplasts, each containing variable numbers of nuclei, we cotransformed selectable marker genes along with sgRNA:Cas9 complexes to help identify colonies arising from protoplasts that took up introduced elements. In order to generate an easA knockout strain of M. brunneum, 5 μl of complexed sgRNA:Cas9 targeting easA was added together with 1 μg of the optimized bar amplicon (in a maximum volume of 5 μl). These transformants were plated in bar transformation medium (per liter, 310 g sucrose, 2.5 g ammonium nitrate, 1.7 g amino acid-free yeast nitrogen base, 1 g magnesium sulfate-heptahydrate, 0.5 g monobasic potassium phosphate, 0.5 g dibasic potassium phosphate, 0.5 g potassium chloride, 0.05 g chloramphenicol, and 7 g agarose) supplemented with phosphinothricin (Gold Biotechnology, St. Louis, MO) at 200 μg/ml and incubated at 30°C. Upon surfacing, colonies were transferred to bar maintenance medium (per liter, 20 g sucrose, 2.5 g ammonium nitrate, 1.7 g amino acid-free yeast nitrogen base, 1 g magnesium sulfate-heptahydrate, 0.5 g monobasic potassium phosphate, 0.5 g dibasic potassium phosphate, 0.5 g potassium chloride, 0.05 g chloramphenicol, and 15 g agar) supplemented with phosphinothricin at 200 μg/ml and incubated at 30°C to allow further growth under selection. In order to generate an LA-accumulating strain of M. brunneum, the above-described protocol was repeated with the substitution of the sgRNA targeting lpsB.
To produce M. brunneum strains capable of producing DHLA and dihydro-LAH, the wild-type and easA knockout strains were subjected to a slightly modified transformation. For a DHLA producer, the easA knockout strain was transformed with 5 μl of complexed sgRNA:Cas9 targeting lpsB and 1 μg of the dihydro expression construct (in a maximum volume of 5 μl). Transformants were plated in TM102 medium (per liter, 310 g sucrose, 10 g malt extract, 10 g peptone, 2 g yeast extract, 1 g magnesium sulfate-heptahydrate, 0.5 g monobasic potassium phosphate, 0.5 g dibasic potassium phosphate, 0.5 g potassium chloride, 0.05 g chloramphenicol, and 15 g agar) supplemented with hygromycin (InvivoGen, San Diego, CA) at 600 μg/ml and incubated at 30°C. To generate a dihydro-LAH-producing strain, this protocol was repeated but with the wild-type M. brunneum strain and sgRNA targeting easA.
PCR and Sanger sequencing were used to assess recombination at the target loci. Genomic DNA was extracted from transformant lines (as described above) and primer combinations 7 and 8 were used in order to assess easA and lpsB loci, respectively. Primer set 5 was used to determine the presence or absence of the dihydro expression construct. Upon failure to generate a product from those targeted at easA with the dihydro expression construct, we hypothesized that the entire ∼11.4-kb construct had integrated, making amplification across the modified locus impractical. To amplify portions of the easA or lpsB loci and determine the orientation and extent of the dihydro expression construct integration, primer combinations 9 and 10 were tried together with primer combinations 11 and 12. PCR products were generated from primer combinations 9 and 10, due to the polarity of the inserted fragment. These fragments were cleaned, concentrated, and sequenced by Sanger technology at Eurofins Genomics (Louisville, KY).
HPLC and LC-MS analyses.
In order to obtain large quantities of ergot alkaloids for chemical analyses, Galleria mellonella larvae were inoculated as described previously (26, 43) and incubated at room temperature for 7 days. Twenty microliters of each methanol-extracted sample was analyzed for ergot alkaloids by HPLC with fluorescence detection by methods described in detail previously (31). The column was a 150- by 4.6-mm inner-diameter, 5-μm particle size Prodigy C18 column (Phenomenex, Torrance, CA), and the mobile phase was a 55-min, binary, multilinear gradient of 5% acetonitrile to 75% acetonitrile in 50 mM aqueous ammonium acetate. Ergot alkaloids were detected with two serially connected fluorescence detectors, one set with excitation and emission wavelengths of 310 and 410 nm, respectively, to detect LA and its derivatives and the other at 272 and 372 nm to detect DHLA and its derivatives.
Extracts of wild-type M. brunneum and engineered N. fumigata strains (17, 18) served as chemical references for LA, DHLA, and LAH. A previously prepared DHLA standard (19) was also used as a reference at a concentration of 10 μg/ml. For percent yield and secretion analyses, alkaloids were approximately quantified by comparing their peak areas to standard curves prepared from ergot alkaloids with identical fluorophores: dihydroergocristine (Sigma) for alkaloids fluorescing at 272 and 372 nm and ergotamine (Sigma) for alkaloids fluorescing at 310 and 410 nm. LC-MS analysis was conducted according to previously established methods (44).
Percent yield and secretion.
In order to determine percent yield and secretion of individual alkaloids, 15-ml cultures of SYE broth were inoculated with 106 spores of each of the mutant strains of M. brunneum. Neosartorya fumigata strains that accumulate either LA (17) or DHLA (18) were grown similarly, with the substitution of malt extract broth (promoting ergot alkaloid production in N. fumigata) as the medium. All cultures were grown in triplicate at room temperature and out of direct sunlight so as to prevent photochemical modification. After 10 days, fungal mats were separated from the remaining liquid via vacuum filtration through preweighed, 0.2-μm nylon filters and were allowed to dry. Dried portions were weighed, while the volumes of liquid portions of cultures were recorded. Alkaloids were extracted with methanol based on previously described methods (26). Percent yield was calculated as observed product relative to a theoretical yield wherein all observable precursor (e.g., agroclavine for LA) would have been converted to product (Table S1). Data from these analyses had variances that were normally distributed and so were analyzed by analysis of variance (ANOVA). In the case of DHLA, since no DHLA was detected in the broth of N. fumigata cultures, a Wilcoxon’s rank sum test was used to compare treatments. All statistical analyses were performed with the JMP software package (SAS, Cary, NC).
Data availability.
The sequence of the construct containing the codon-optimized phosphinothricin N-acetyltransferase gene coding sequence, which confers resistance to bialaphos, under the control of the GPDH promoter from M. brunneum ARSEF 3297 has been deposited in GenBank under accession number MT350122.
Supplementary Material
ACKNOWLEDGMENTS
This research was funded by NIH grant number 2R15-GM114774-2, with additional support from USDA Hatch project number NC1183.
Experiments with lysergic acid-producing strains were conducted in accordance with licenses from the West Virginia Board of Pharmacy (TI0555042) and the U.S. Drug Enforcement Agency (RP0463353).
Footnotes
Supplemental material is available online only.
This paper is published with the approval of the West Virginia Agriculture and Forestry Experiment Station as article 3379.
REFERENCES
- 1.Florea S, Panaccione DG, Schardl CL. 2017. Ergot alkaloids of the family Clavicipitaceae. Phytopathology 107:504–518. doi: 10.1094/PHYTO-12-16-0435-RVW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haarmann T, Rolke Y, Giesbert S, Tudzynski P. 2009. Ergot: from witchcraft to biotechnology. Mol Plant Pathol 10:563–577. doi: 10.1111/j.1364-3703.2009.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potter DA, Stokes JT, Redmond CT, Schardl CL, Panaccione DG. 2008. Contribution of ergot alkaloids to suppression of a grass-feeding caterpillar assessed with gene knockout endophytes in perennial ryegrass. Entomol Exp Appl 126:138–147. doi: 10.1111/j.1570-7458.2007.00650.x. [DOI] [Google Scholar]
- 4.Panaccione DG, Cipoletti JR, Sedlock AB, Blemings KP, Schardl CL, Machado C, Seidel GE. 2006. Effects of ergot alkaloids on food preference and satiety in rabbits, as assessed with gene-knockout endophytes in perennial ryegrass (Lolium perenne). J Agric Food Chem 54:4582–4587. doi: 10.1021/jf060626u. [DOI] [PubMed] [Google Scholar]
- 5.Pertz H, Eich E. 1999. Ergot alkaloids and their derivatives as ligands for serotoninergic, dopaminergic, and adrenergic receptors, p 411–440. In Kren V, Cvak L (ed), Ergot: the genus Claviceps. Harwood Academic Publishers, Amsterdam, The Netherlands. [Google Scholar]
- 6.Østergaard JR, Mikkelsen E, Voldby B. 1981. Effects of 5-hydroxytryptamine and ergotamine on human superficial temporal artery. Cephalalgia 1:223–228. doi: 10.1046/j.1468-2982.1981.0104223.x. [DOI] [PubMed] [Google Scholar]
- 7.Iliff LD, Du Boulay GH, Marshall J, Russell RW, Symon L. 1977. Effect of nicergoline on cerebral blood flow. J Neurol Neurosurg Psychiatry 40:746–747. doi: 10.1136/jnnp.40.8.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Jia Y. 2017. Ergot alkaloids: synthetic approaches to lysergic acid and clavine alkaloids. Nat Prod Rep 34:411–432. doi: 10.1039/c6np00110f. [DOI] [PubMed] [Google Scholar]
- 9.Robinson SL, Panaccione DG. 2015. Diversification of ergot alkaloids in natural and modified fungi. Toxins (Basel) 7:201–218. doi: 10.3390/toxins7010201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallwey C, Li S-M. 2011. Ergot alkaloids: structure diversity, biosynthetic gene clusters and functional proof of biosynthetic genes. Nat Prod Rep 28:496–510. doi: 10.1039/c0np00060d. [DOI] [PubMed] [Google Scholar]
- 11.Cheng JZ, Coyle CM, Panaccione DG, O’Connor SE. 2010. A role for old yellow enzyme in ergot alkaloid biosynthesis. J Am Chem Soc 132:1776–1777. doi: 10.1021/ja910193p. [DOI] [PubMed] [Google Scholar]
- 12.Cheng JZ, Coyle CM, Panaccione DG, O’Connor SE. 2010. Controlling a structural branch point in ergot alkaloid biosynthesis. J Am Chem Soc 132:12835–12837. doi: 10.1021/ja105785p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coyle CM, Cheng JZ, O’Connor SE, Panaccione DG. 2010. An old yellow enzyme gene controls the branch point between Aspergillus fumigatus and Claviceps purpurea ergot alkaloid pathways. Appl Environ Microbiol 76:3898–3903. doi: 10.1128/AEM.02914-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallwey C, Matuschek M, Xie X-L, Li S-M. 2010. Ergot alkaloid biosynthesis in Aspergillus fumigatus: conversion of chanoclavine-I aldehyde to festuclavine by the festuclavine synthase FgaFS in the presence of the old yellow enzyme FgaOx3. Org Biomol Chem 8:3500–3508. doi: 10.1039/c003823g. [DOI] [PubMed] [Google Scholar]
- 15.Matuschek M, Wallwey C, Xie X-L, Li S-M. 2011. New insights into ergot alkaloid biosynthesis in Claviceps purpurea: an agroclavine synthase EasG catalyses, via a non-enzymatic adduct with reduced glutathione, the conversion of chanoclavine-I aldehyde to agroclavine. Org Biomol Chem 9:4328–4335. doi: 10.1039/c0ob01215g. [DOI] [PubMed] [Google Scholar]
- 16.Haarmann T, Ortel I, Tudzynski P, Keller U. 2006. Identification of the cytochrome P450 monooxygenase that bridges the clavine and ergoline alkaloid pathways. Chembiochem 7:645–652. doi: 10.1002/cbic.200500487. [DOI] [PubMed] [Google Scholar]
- 17.Robinson SL, Panaccione DG. 2014. Heterologous expression of lysergic acid and novel ergot alkaloids in Aspergillus fumigatus. Appl Environ Microbiol 80:6465–6472. doi: 10.1128/AEM.02137-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bragg PE, Maust MD, Panaccione DG. 2017. Ergot alkaloid biosynthesis in the maize (Zea mays) ergot fungus Claviceps gigantea. J Agric Food Chem 65:10703–10710. doi: 10.1021/acs.jafc.7b04272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arnold SL, Panaccione DG. 2017. Biosynthesis of the pharmaceutically important fungal ergot alkaloid dihydrolysergic acid requires a specialized allele of cloA. Appl Environ Microbiol 83:e00805-17. doi: 10.1128/AEM.00805-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mulinti P, Allen NA, Coyle CM, Gravelat FN, Sheppard DC, Panaccione DG. 2014. Accumulation of ergot alkaloids during conidiophore development in Aspergillus fumigatus. Curr Microbiol 68:1–5. doi: 10.1007/s00284-013-0434-2. [DOI] [PubMed] [Google Scholar]
- 21.Hu S, Bidochka MJ. 30 October 2019. Root colonization by endophytic insect-pathogenic fungi. J Appl Microbiol doi: 10.1111/jam.14503. [DOI] [PubMed] [Google Scholar]
- 22.Liao X, O’Brien TR, Fang W, St Leger RJ. 2014. The plant beneficial effects of Metarhizium species correlate with their association with roots. Appl Microbiol Biotechnol 98:7089–7096. doi: 10.1007/s00253-014-5788-2. [DOI] [PubMed] [Google Scholar]
- 23.Russell CW, Ugine TA, Hajek AE. 2010. Interactions between imidacloprid and Metarhizium brunneum on adult Asian longhorned beetles (Anoplophora glabripennis (Motschulsky)) (Coleoptera: Cerambycidae). J Invertebr Pathol 105:305–311. doi: 10.1016/j.jip.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 24.Dogan YO, Hazir S, Yildiz A, Butt TM, Cakmak I. 2017. Evaluation of entomopathogenic fungi for the control of Tetranychus urticae (Acari: Tetranychidae) and the effect of Metarhizium brunneum on the predatory mites (Acari: Phytoseiidae). Biological Control 111:6–12. doi: 10.1016/j.biocontrol.2017.05.001. [DOI] [Google Scholar]
- 25.Castrillo LA, Griggs MH, Ranger CM, Reding ME, Vandenberg JD. 2011. Virulence of commercial strains of Beauveria bassiana and Metarhizium brunneum (Ascomycota: Hypocreales) against adult Xylosandrus germanus (Coleoptera: Curculionidae) and impact on brood. Biolog Control 58:121–126. doi: 10.1016/j.biocontrol.2011.04.010. [DOI] [Google Scholar]
- 26.Leadmon CE, Sampson JK, Maust MD, Macias AM, Rehner SA, Kasson MT, Panaccione DG. 2020. Several Metarhizium species produce ergot alkaloids in a condition-specific manner. Appl Environ Microbiol 86:e00373-20. doi: 10.1128/AEM.00373-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Correia T, Grammel N, Ortel I, Keller U, Tudzynski P. 2003. Molecular cloning and analysis of the ergopeptine assembly system in the ergot fungus Claviceps purpurea. Chem Biol 10:1281–1292. doi: 10.1016/j.chembiol.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 28.Ortel I, Keller U. 2009. Combinatorial assembly of simple and complex d-lysergic acid alkaloid peptide classes in the ergot fungus Claviceps purpurea. J Biol Chem 284:6650–6660. doi: 10.1074/jbc.M807168200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schardl CL, Young CA, Pan J, Florea S, Takach JE, Panaccione DG, Farman ML, Webb JS, Jaromczyk J, Charlton ND, Nagabhyru P, Chen L, Shi C, Leuchtmann A. 2013. Currencies of mutualisms: sources of alkaloid genes in vertically transmitted Epichloae. Toxins (Basel) 5:1064–1088. doi: 10.3390/toxins5061064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silar P. 1995. Two new easy to use vectors for transformations. Fungal Genet Rep 42:23. doi: 10.4148/1941-4765.1353. [DOI] [Google Scholar]
- 31.Panaccione DG, Ryan KL, Schardl CL, Florea S. 2012. Analysis and modification of ergot alkaloid profiles in fungi. Methods Enzymol 515:267–290. doi: 10.1016/B978-0-12-394290-6.00012-4. [DOI] [PubMed] [Google Scholar]
- 32.Flieger M, Sedmera P, Vokoun J, R˘ic̄icovā A, R˘ehác˘ek Z. 1982. Separation of four isomers of lysergic acid α-hydroxyethylamide by liquid chromatography and their spectroscopic identification. J Chromatogr 236:441–452. doi: 10.1016/S0021-9673(00)84895-5.6277972 [DOI] [Google Scholar]
- 33.Mantle PG, Waight ES. 1968. Dihydroergosine: a new naturally occurring alkaloid from the sclerotia of Sphacelia sorghi (McRae). Nature 218:581–582. doi: 10.1038/218581a0. [DOI] [Google Scholar]
- 34.Riederer B, Han M, Keller U. 1996. d-Lysergyl peptide synthetase from the ergot fungus Claviceps purpurea. J Biol Chem 271:27524–27530. doi: 10.1074/jbc.271.44.27524. [DOI] [PubMed] [Google Scholar]
- 35.Roze LV, Chanda A, Linz JE. 2011. Compartmentalization and molecular traffic in secondary metabolism: a new understanding of established cellular processes. Fungal Genet Biol 48:35–48. doi: 10.1016/j.fgb.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Menke J, Dong Y, Kistler HC. 2012. Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol Plant Microbe Interact 25:1408–1418. doi: 10.1094/MPMI-04-12-0081-R. [DOI] [PubMed] [Google Scholar]
- 37.Kistler HC, Broz K. 2015. Cellular compartmentalization of secondary metabolism. Front Microbiol 6:68. doi: 10.3389/fmicb.2015.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bernier L, Cooper RM, Charnley AK, Clarkson JM. 1989. Transformation of the entomopathogenic fungus Metarhizium anisopliae to benomyl resistance. FEMS Microbiol Lett 60:261–265. doi: 10.1111/j.1574-6968.1989.tb03483.x. [DOI] [Google Scholar]
- 39.Goettel MS, Leger RJ, Bhairi S, Jung MK, Oakley BR, Roberts DW, Staples RC. 1990. Pathogenicity and growth of Metarhizium anisopliae stably transformed to benomyl resistance. Curr Genet 17:129–132. doi: 10.1007/BF00312857. [DOI] [Google Scholar]
- 40.Liu X, Homma A, Sayadi J, Yang S, Ohashi J, Takumi T. 2016. Sequence features associated with the cleavage efficiency of CRISPR/Cas9 system. Sci Rep 6:19675. doi: 10.1038/srep19675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bilovol Y, Panaccione DG. 2016. Functional analysis of the gene controlling hydroxylation of festuclavine in the ergot alkaloid pathway of Neosartorya fumigata. Curr Genet 62:853–860. doi: 10.1007/s00294-016-0591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen J, Lai Y, Wang L, Zhai S, Zou G, Zhou Z, Cui C, Wang S. 2017. CRISPR/Cas9-mediated efficient genome editing via blastospore-based transformation in entomopathogenic fungus Beauveria bassiana. Sci Rep 8:45763. doi: 10.1038/srep45763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Panaccione DG, Arnold SL. 2017. Ergot alkaloids contribute to virulence in an insect model of invasive aspergillosis. Sci Rep 7:8930. doi: 10.1038/s41598-017-09107-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan KL, Akhmedov NG, Panaccione DG. 2015. Identification and structural elucidation of ergotryptamine, a new ergot alkaloid produced by genetically modified Aspergillus nidulans and natural isolates of Epichloë species. J Agric Food Chem 63:61–67. doi: 10.1021/jf505718x. [DOI] [PubMed] [Google Scholar]
- 45.Barrow KD, Mantle PG, Quigley FR. 1974. Biosynthesis of dihydroergot alkaloids. Tetrahedron Lett 15:1557–1560. doi: 10.1016/S0040-4039(01)93135-1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequence of the construct containing the codon-optimized phosphinothricin N-acetyltransferase gene coding sequence, which confers resistance to bialaphos, under the control of the GPDH promoter from M. brunneum ARSEF 3297 has been deposited in GenBank under accession number MT350122.