Abstract
The development and world-wide spread of multidrug-resistant (MDR) bacteria have a high concern in the medicine, especially the extended-spectrum of beta-lactamase (ESBL) producing Escherichia coli and methicillin-resistant Staphylococcus aureus (MRSA). There are currently very limited effective antibiotics to treat infections caused by MDR bacteria. Peat-soil is a unique environment in which bacteria have to compete each other to survive, for instance, by producing antimicrobial substances. This study aimed to isolate bacteria from peat soils from South Kalimantan Indonesia, which capable of inhibiting the growth of Gram-positive and Gram-negative bacteria. Isolates from peat soil were grown and identified phenotypically. The cell-free supernatant was obtained from broth culture by centrifugation and was tested by agar well-diffusion technique against non ESBL-producing E. coli ATCC 25922, ESBL-producing E. coli ATCC 35218, methicillin susceptible Staphylococcus aureus (MSSA) ATCC 29,213 and MRSA ATCC 43300. Putative antimicrobial compounds were separated using SDS-PAGE electrophoresis and purified using electroelution method. Antimicrobial properties of the purified compounds were confirmed by measuring the minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC). In total 28 isolated colonies were recovered; three (25PS, 26PS, and 27PS) isolates produced proteins with strong antimicrobial activities against both reference strains. The substance of proteins from three isolates exerted strong antimicrobial activity against ESBL-producing E. coli ATCC 35,218 (MIC = 2,80 µg/mL (25PS), 3,76 µg/mL (26PS), and 2,41 µg/mL (27PS), and MRSA ATCC 43,300 (MIC = 4,20 µg/mL (25PS), 5,65 µg/mL (26PS), and 3,62 µg/mL (27PS), and also had the ability bactericidal properties against the reference strains. There were isolates from Indonesian peat which were potentials sources of new antimicrobials.
Keywords: Peat soil bacteria, ESBL-producing E. coli, MRSA, Antimicrobial properties
1. Introduction
The widespread use of antibiotics to treat infectious diseases causes a global increase in multidrug-resistant (MDR) bacterial populations (Bogaard et al., 2000). MDR bacteria is a major medical problem because directly related to increasing health care costs and mortality rates (Oelschlaeger, 2010). The rapid emergence of MDR bacteria is not followed by the discovery of new antimicrobials, therefore there is currently very limited effective antibiotics available to treat MDR bacteria (Ammerlaan et al., 2013). New research is urgently needed to discover and develop new antimicrobial substances which are active against MDR bacteria.
The extended-spectrum beta-lactamase (ESBL)-producing E. coli and methicillin-resistant Staphylococcus aureus are two of the most common MDR bacteria. Their incidence were high particularly in developing countries, reaching 6% in Japan (Luvsansharav et al., 2011), 30% in China (Ni et al., 2016), 30% in Thailand (Sawatwong et al., 2019), and 62.2%-85% in Indonesia (Anggraini et al., 2018, Hayati et al., 2019) for ESBL-producing E. coli, and for MRSA reaching 16.5%–23.5% in India, 7.9% in Vietnam, 3.5%-3.8% in Taiwan (Wong et al., 2018), 8.1% in Indonesia (Kuntaman et al., 2016), and greater than 70% in Korea (Chen and Huang, 2014).
One strategy for discovering new antimicrobials is by bio prospecting, which is an exploration of new compounds in unique ecological niches. Peat soil is one of the unique and extreme environments for the discovery of antimicrobial compounds from bacteria living in it (Ong, et al., 2015). Peat soil found in Kalimantan Indonesia always in wet conditions in which the decomposed organic matters has been formed and accumulated over thousands of years with acidic pH conditions (pH 2.9–4.5), limited nutrient conditions, low oxygen solubility, and temperatures around 230-32 °C, therefore the bacteria have to compete with other bacteria to survive (Yule, 2010).
Bacterial growth under extreme and unique environmental conditions is very likely to develop special adaptation mechanisms and produce unique molecules, compounds or secondary metabolites (Pettit, 2011). One example of successful bio prospection is the discovery of abyssomicins, a new antibiotic produced by Verrucosispora sp. from the sediments of the South China Sea (Wang et al., 2013). Another of bio prospection was the discovery of new bacterial strain of MSt1T isolated from tropical peat soils in Selangor Malaysia, which has antimicrobial activity against Gram-negative and positive bacteria (Aw et al., 2016). Ong et al reported about Burkholderia spp which is also isolated from peat soils with antimicrobial activity against Gram-positive bacteria, Gram-negative bacteria and yeast (Ong et al., 2016).
In addition to the discovery of bacteria as bioactive compounds producer from peat soils, bio prospection can also increase the diversity of bacteria in the databases. There are a huge diversity of microbes in tropical peat swamp forests. Metagenomic research in Thailand's tropical peat-lands showed there were 80% of bacterial communities have potency to become new species or strains (Kanokratana, et al., 2011). Currently the main concern of WHO is to find new antimicrobials, or antibiotic compounds to treat infections caused by the MDR bacteria such as ESBL-producing E. coli and MRSA infection (WHO, 2017, Pana and Zaoutis, 2018). This study aimed to isolate and investigate the ability of bacteria from peat soils from South Kalimantan Indonesia to inhibit the growth of Gram-positive and Gram-negative bacteria reference strains, either sensitive or resistant strains. In this study used two study designs, first an exploratory study that is to prove whether there are peat soil bacteria that produce antimicrobial compounds against ESBL-producing E. coli and MRSA using the well diffusion method. Second, an experimental/comparative study design that tests the antimicrobial activity produced by pure compounds from peat soil bacterial isolates against ESBL-producing E. coli and MRSA by dilution methods MIC (minimum inhibitory concentration) and MBC (minimum bactericidal concentration) compared to antibiotics standard (Meropenem and vancomycin).
2. Materials and methods
2.1. Sample collection
Peat soil samples were collected from the peat soil area of Banjarmasin district in South Kalimantan, Indonesia. Fifty grams of peat soils obtained from 50 cm depth were taken using core borer to increase the possibility of isolating various types of bacteria. Peat soil samples of medium maturity level were placed in sterile containers and transported at an environmental temperature within 18 h for analysis in the laboratory (Ong, et al., 2015).
2.2. Isolation of bacteria from peat soil
One gram of peat soil was soaked in 0.85% (w/v) saline solution and serially diluted from 10−1 to 10−6, then inoculated on trypticase soy agar (TSA) and Mac Conkey agar, and incubated at 30 °C aerobically for 2 days. The growing colonies were purified by using streak method on TSA and Mac Conkey agar, then incubated aerobically at 30 °C.
2.3. Maintenance of the isolate
A pure culture is routinely subculture and maintained on TSA media at 30 °C, while for long-term storage was done by cryopreservation with glycerol 20% and skim 10% (v/v) at −80 °C.
2.4. Morphology identification of peat soil bacteria
The isolated colonies were identified based on colony morphology, bacterial morphology on Gram staining, catalase test, and motility test.
2.5. Screening of antimicrobial activity
Each pure colony was subculture in TSA broth at 37 °C for 48 h and then centrifuged at 3500 rpm for 10 min. Cell-free supernatant was expected to contain secondary metabolites including antibacterial compounds and used for inhibitory effect screening. A hundred µL supernatant was filled into each of 6 mm well on a Mueller Hinton agar (MHA) which had previously been inoculated by surface spreading of non ESBL-producing E. coli ATCC 25922, ESBL-producing E. coli ATCC 35218, methicillin-susceptible S. aureus (MSSA) ATCC 29213, and MRSA ATCC 43,300 of 0.5 Mc Farland standard culture in TSA broth, incubated aerobically for 24 h at 37 °C. The inhibition zone diameters around the wells were measured using a digital Caliper.
2.6. Purification of antimicrobial compounds and determination of molecular weight
Peat soil isolates suspected of having antimicrobial activity were grown on TSB media for two days at 30 °C, then centrifuged at 3000 rpm for 20 min at 4 °C. The supernatant was separated from the pellet, cold absolute acetone has been added of a 1: 1 ratio and stored in the freezer for 24 h. The supernatant was centrifuged again at 7000 rpm for 20 min at 4 °C. The dried pellet was added with PBS 2 mL, mixed then stored at temperature −20 °C for running on SDS PAGE Electrophoresis as mentioned elshewere to separate the protein content of supernatant according to the molecular weight. The resulted protein bands were extracted using electroelution to obtain pure compounds.
2.7. MIC and MBC determination
Pure putative antimicrobial compounds were tested for MIC and MBC towards ESBL-producing E. coli ATCC 35,218 and MRSA ATCC 43,300 used the CLSI standards method in triplicates. Meropenem (1 µg/ml disc) and vancomycin (4 µg/ml) were used as control.
3. Results
3.1. Bacterial isolates from peat soil
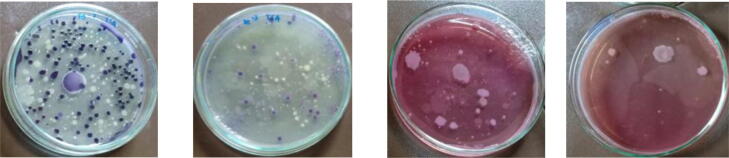
The growth of peat soil bacteria on TSA and Mac Conkey can be seen in Fig. 1. Lactose fermenter bacteria were detected on Mac Conkey agar from their pink-red color colonies.
Fig. 1.
Bacterial colonies from peat soil used media TSA and Mac Conkey Agar.
Characteristics of each colony varied with regards to shape, color, elevation, diameter, and the edge of the colony. Some colonies were white, yellowish or green on TSA agar. The elevation of the colony was shaky, flat, jagged, or concave. The diameter of the colonies ranged from 1.0 to 4.5 mm, and the edge was round or irregular.
3.2. Morphology of peat soil bacteria
In total 28 different colonies were isolated on TSA and Mac Conkey Agar media. The identification results of the colonies by using Gram staining, colony morphology, and catalase tests were shown in Table 1.
Table 1.
Phenotypic characteristics of 28 bacterial isolates from peat soil.
| Isolates number | Gram staining | Catalase test | Colony color | Family name |
|---|---|---|---|---|
| 1PS | Gram-negative rod | Positive | White | Enterobacteriaceae |
| 2PS | Gram-negative rod | Negative | White | Enterobacteriaceae |
| 3PS | Gram-positive rod, no spores | Positive | Milky white | Bacillaceae |
| 4PS | Gram-positive cocci | Negative | Clear white | Streptococcaceae |
| 5PS | Gram-negative rod | Positive | Yellowish | Enterobacteriaceae |
| 6PS | Gram-negative rod | Positive | Creamy white | Enterobacteriaceae |
| 7PS | Gram-positive cocci | Positive | Purple | Staphylococcacea |
| 8PS | Gram-positive rod, no spores | Positive | Whitish purple | Bacillaceae |
| 9PS | Gram-negative rod | Positive | Purple | Enterobacteriaceae |
| 10PS | Gram-positive rod, no spores | Positive | Yellow | Bacillaceae |
| 11PS | Gram-negative cocci | Negative | White | Streptococcacea |
| 12PS | Gram-positive rod, no spores | Negative | Yellow | Bacillaceae |
| 13PS | Gram-negative rod | Positive | Red | Enterobacteriaceae |
| 14PS | Gram-negative rod | Positive | White | Enterobacteriaceae |
| 15PS | Gram-positive cocci | Positive | Creamy white | Staphylococcacea |
| 16PS | Gram-negative rod | Positive | Purple white | Enterobacteriaceae |
| 17PS | Gram-negative rod | Positive | White | Enterobacteriaceae |
| 18PS | Gram-negative rod | Positive | Yellowish white | Enterobacteriaceae |
| 19PS | Gram-positive cocci | Negative | Yellow | Streptococcaceae |
| 20PS | Gram-positive rod, no spores | Negative | White | Clostridiaceae |
| 21PS | Gram-positive rod, no spores | Positive | White | Bacillaceae |
| 22PS | Gram-negative rod | Positive | Purple | Enterobacteriaceae |
| 23PS | Gram-positive rod, no spores | Positive | White | Bacillaceae |
| 24PS | Gram-positive rod, no spores | Positive | Red | Bacillaceae |
| 25PS | Gram-negative rod | Positive | White | Enterobacteriaceae |
| 26PS | Gram-negative rod | Positive | Yellowish | Enterobacteriaceae |
| 27PS | Gram-positive rod, no spores | Positive | Creamy | Bacillaceae |
| 28PS | Gram-positive cocci | Positive | Creamy white | Staphylococcaceae |
The diversity of recovered bacteria classified based on morphology and catalase test are shown in Table 2. They were for catalase-positive bacteria including Staphylococcaceae, Bacillaceae or Enterobacteriaceae, and catalase-negative bacteria including Streptococcaceae, Clostridiaceae, and Enterobacteriaceae. They were consist of gram-negative bacteria e.g. Enterobacteriaceae (46.4%) and Bacillaceae (28.6%), while gram-positive bacteria consist of Streptococcaceae (10.7%), Staphylococcaceae (10.7%) and Clostridiaceae (3.6%).
Table 2.
Diversity of 28 bacterial isolated from peat soil.
| No | Family | Number | % |
|---|---|---|---|
| 1 | Enterobacteriaceae | 13 | 46.4 |
| 2 | Bacillaceae | 8 | 28.6 |
| 3 | Streptococcaeae | 3 | 10.7 |
| 4 | Staphylococcaceae | 3 | 10.7 |
| 5 | Clostridiaceae | 1 | 3.6 |
| Total number | 28 | 100 |
3.3. Screening of antimicrobial activity
The screening tests of antimicrobial activity of all supernatant isolates showed some isolates had no inhibitory effect while others had inhibitory effects against non ESBL-producing E. coli ATCC 25922, ESBL-producing E. coli ATCC 35218, MSSA ATCC 29213, and MRSA ATCC 43300, as indicated by the presence of clear zones around the wells. At this stage of the study, no further tests have been done to confirm such antimicrobial inhibition.
Table 3 showed that there were some isolates have a clear zone inhibition with a variable diameter zone size. Diameter of inhibition of more than 10 mm was chosen to consider a potential isolate with antimicrobial activities. Four isolates, 9PS, 25PS, 26PS and 27PS, were have wider inhibition zone, which may probably the most potential have antimicrobial properties, and presented in Fig. 2, Fig. 3. Isolates number 9PS only active against gram-negative bacteria e.g. non ESBL-producing E. coli ATCC 25,922 and ESBL-producing E. coli ATCC 35218, even though with smaller inhibition zone. The isolate number 25PS, 26PS, and 27PS were able to inhibit the growth of both gram-positive bacteria and gram-negative bacteria, e.g. non ESBL-producing E. coli ATCC 25,922 and ESBL-producing E. coli ATCC 35218, and MSSA ATCC 29213, and MRSA ATCC 43300.
Table 3.
Mean of inhibition zone diameter (mm) of antimicrobial screening of peat soil isolates by well-diffusion method in triplicate.
| No isolate | non ESBL- producing E. coli ATCC 25,922 | ESBL-producing E. coli ATCC 35,218 | MSSA ATCC 29,213 | MRSA ATCC 43,300 |
|---|---|---|---|---|
| 1PS | – | – | – | – |
| 2PS | 8,3 | – | – | – |
| 3PS | – | – | – | – |
| 4PS | – | – | – | – |
| 5PS | 8,3 | – | – | – |
| 6PS | – | – | 8,6 | – |
| 7PS | 9,3 | – | – | – |
| 8PS | – | – | 7 | – |
| 9PS | 12 | 10 | 7 | – |
| 10PS | – | – | – | – |
| 11PS | – | – | – | – |
| 12PS | – | – | – | 7 |
| 13PS | – | – | – | – |
| 14PS | – | – | – | – |
| 15PS | – | – | – | – |
| 16PS | – | – | – | – |
| 17PS | – | – | 10 | 9 |
| 18PS | – | – | – | – |
| 19PS | 11,3 | – | 7 | – |
| 20PS | – | – | – | – |
| 21PS | – | – | – | – |
| 22PS | – | – | 7 | – |
| 23PS | – | – | 8 | 7 |
| 24PS | – | – | – | – |
| 25PS | 19,3 | 19 | 11,5 | 10,3 |
| 26PS | 19,6 | 19,3 | 16,4 | 17,1 |
| 27PS | 20 | 16,7 | 14,5 | 12,1 |
| 28PS | – | – | – | – |
– = no antimicrobial activity.
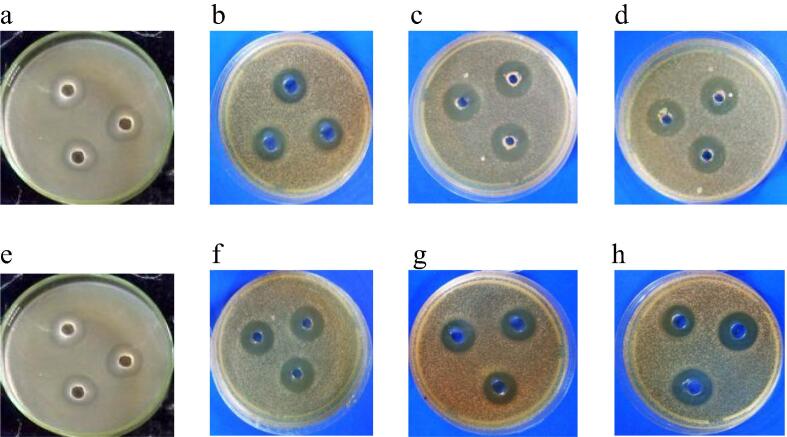
Fig. 2.
Inhibition effects of cell-free supernatants against gram-negative bacteria. a: 9PS, b: 25PS, c: 26PS, and d: 27PS against non ESBL-producing E. coli ATCC 25922, while e: 9PS, f: 25PS, g: 26PS, and h: 27PS against ESBL-producing E. coli ATCC 35218.
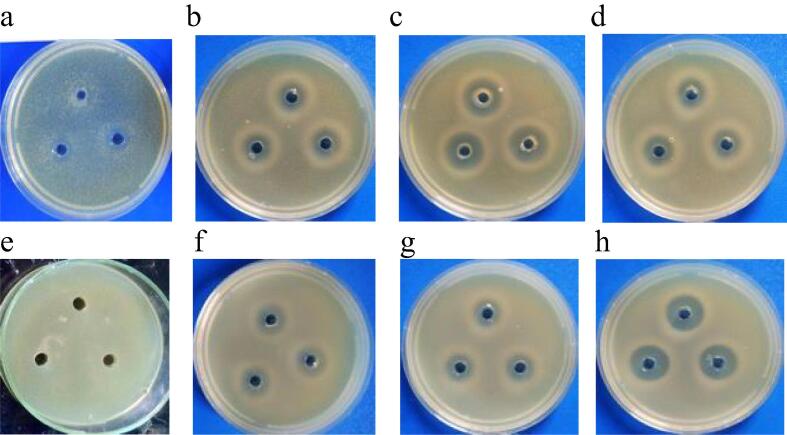
Fig. 3.
Inhibition effects of cell-free supernatants against gram-positive bacteria. a: 9PS, b: 25PS, c: 26PS, and d: 27PS against MSSA ATCC 29213, while e: 9PS, f: 25PS, g: 26PS, and h: 27PS against MRSA ATCC 43300.
Fig. 2 showed the least diameter of inhibition was found on isolates number 9PS against non ESBL-producing E. coli ATCC 25,922 (diameter of inhibition zone 12 mm) and against ESBL-producing E. coli ATCC 35,218 (inhibition zone diameter 10 mm). Cell-free supernatant of isolate number 25PS, 26PS and 27PS showed inhibitory effect against non ESBL-producing E. coli ATCC 25,922 (diameter of inhibition zone 19.3 mm, 19.6 mm and 20 mm respectively) and against ESBL-producing E coli ATCC 35,218 (inhibition zone diameter 19 mm, 19.3 mm and 16.7 mm respectively).
Fig. 3 revealed that the supernatant of 9PS showed weak inhibitory effect against S. aureus ATCC 29,213 (diameter of inhibition zone 7 mm) and no effects against MRSA ATCC 43,300 (no diameter of inhibition zone). In contrast, cell-free supernatant of 25PS, 26PS and 27PS isolates showed stronger probable inhibitory effect against MSSA ATCC 29,213 (diameter of inhibition zone 11.5 mm, 16.4 mm and 14.5 mm respectively) and against MRSA ATCC 43,300 (diameter of inhibition zone 10.3 mm, 17.1 mm and 10.3 mm respectively).
A total of 28 bacteria isolates of peat soil has been isolated and identified up to the family level. The most potential isolates to produce antimicrobial substances were 25PS (Enterobacteriaceae), 26PS (Enterobacteriaceae) and 27PS (Bacillaceae) against the four reference strain (see Table 1), while 9PS only has potential antibacterial properties against non ESBL-producing E. coli ATCC 25,922 and ESBL-producing E. coli ATCC 35218. All of these isolates need to be further identified and characterized by biochemical or pharmaceutical technique, and molecular biology technique.
3.4. Purification of antimicrobial compounds and identification of molecular weights with SDS PAGE electrophoresis
The antimicrobial compound isolate 25PS has a molecular weight of 43 KDa the same as isolate 27PS (shown with arrow), while the antimicrobial compound isolate 26PS has a molecular weight of 45 KDa (shown with arrow). Comparative compounds are from 10PS isolates that do not have the ability of antimicrobial compounds (Fig. 4).
Fig. 4.
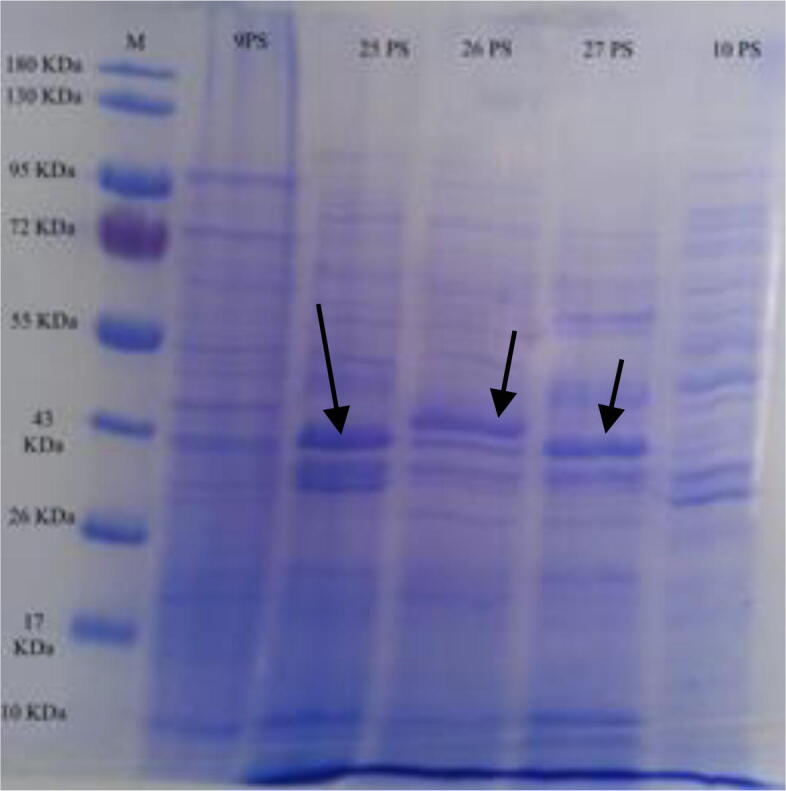
Molecular weight of compounds from peat soil bacterial isolates that produce antimicrobial proteins.
3.5. Minimum inhibition concentration and minimum bactericidal concentration
MIC test of antimicrobial compound from peat soil bacterial isolates against ESBL- producing E. coli ATCC 35,218 showed that the minimum inhibitory concentration of the three compounds from peat soil bacterial isolates obtained a concentration of 2.80 µg/ml (25 PS), 3.76 µg/ml (26PS) and 2.41 μg/ml (27 PS). MBC test on ESBL-producing E. coli ATCC 35,218 showed that the minimum kill concentration was 4.20 μg/mL (25PS), 5.65 μg/mL (26PS), and 3.62 μg/mL (27PS). MIC and MBC obtained by the three compounds of peat soil bacterial isolates were significantly different from meropenem (p = 0.012) and also proved in the Mann Whitney test the difference occurred in each compound against meropenem (p = 0.025) (Table 4).
Table 4.
Results of MIC (Minimum Inhibitor Concentration) and MBC (Minimum Bactreicidal Concentration) antimicrobial compounds from peat soil bacteria against ESBL-producing E. coli ATCC 35,218 compared with meropenem antibiotics.
| Compounds | MIC concentration (µg/mL) | P** | Concentration of MBC (µg/mL) | P** |
|---|---|---|---|---|
| purity | purity | |||
| 26PS (45 kDa) | 3,76 | 0,025 | 5,65 | 0,025 |
| 25PS (43 kDa) | 2,8 | 0,025 | 4,20 | 0,025 |
| 27PS (43 kDa) | 2,41 | 0,025 | 3,62 | 0,025 |
| Meropenem | 1 | 1 | ||
| P* | 0,012 | 0,012 |
Kruskal Wallis Tests.
Mann Whitney Tests.
MIC test of antimicrobial protein compound from peat soil bacterial isolates against MRSA ATCC 43,300 bacteria showed that the minimum inhibitory concentration was 4.20 µg/mL (25PS), 5.65 µg/mL (26PS), and 3.62 µg/mL (27PS)). The MBC against MRSA ATCC 43,300 is 5.60 μg/mL (25PS), 7.53 μg/mL (26PS), and 4.83 μg/mL (27PS). From the MBC to MIC ratio that is less than 3 times it can be concluded that the antimicrobial compounds of the three peat soil bacterial isolates are bactericidal. MIC and MBC values of the three antimicrobial compounds of peat soil bacterial isolates were significantly higher than vancomycin (p = 0.012), with the difference occurring in each compound against vancomycin (p = 0.025) (Table 5).
Table 5.
Results of MIC (Minimum Inhibitor Concentration) and MBC (Minimum Bactericidal Concentration) antimicrobial compounds from peat soil bacteria against MRSA ATCC 43,300 compared with vancomycin antibiotics.
| Compounds | MIC concentrations (µg/mL) | P** | Senyawa | Concentration of MBC (µg/mL) | P** |
|---|---|---|---|---|---|
| purity | purity | ||||
| 26PS (45 kDa) | 5,65 | 0,025 | 26PS (45 kDa) | 7,53 | 0,025 |
| 25PS (43 kDa) | 4,20 | 0,025 | 25PS (43 kDa) | 5,60 | 0,025 |
| 27PS (43 kDa) | 3,62 | 0,025 | 27PS (43 kDa) | 4,83 | 0,025 |
| Vancomycin | 4 | Vancomycin | 4 | ||
| P* | 0,012 | P* | 0,012 |
Kruskal Wallis tests.
Mann Whitney tests.
4. Discussion
ESBL-producing bacteria have become a worldwide problem with serious consequences on limited treatment options of infectious diseases and increases the cost of treatment due to the use of more expensive treatment, longer hospitalization, increased morbidity, and mortality (Ong et al., 2015, WHO, 2017). Twenty-eight colonies have been isolated from peat soil from South Kalimantan, Indonesia consist of Enterobacteriaceae (46.4%), Bacillaceae (28.6%), Streptococcaceae (10.7%), Staphylococcaceae (10.7%) and Clostridiacecae (3.6%).
The screening test of inhibitory effects of all isolates against non ESBL- producing E. coli ATCC 25922, ESBL-producing E. coli ATCC 35218, MSSA ATCC 29213, and MRSA ATCC 43,300 showed there were no inhibitory effects by 14 isolates (50%); very small inhibitory effects (diameter of inhibition of less than 10 mm) by 10 isolates (36%) and inhibitory effects with diameter of inhibition zone of ≥10 mm by 4 isolates, consist of 9PS (Enterobacteriaceae), 25PS (Enterobacteriaceae), 26PS (Enterobacteriaceae) and 27PS (Bacillaceae). Isolate 9PS only active against Gram-negative bacteria, while 25PS, 26PS, 27PS showed antimicrobial potency against both Gram negative and Gram positive bacteria.
In this study, cell-free supernatant of 25PS (Enterobacteriaceae), 26PS (Enterobacteriaceae) and 27PS (Bacillaceae) isolates of peat soil contained substances resulted from their metabolism that had ability as an antimicrobials. The supernatant was not only able to inhibit the growth of sensitive type bacteria such as non ESBL-producing E. coli ATCC 25,922 and MSSA ATCC 29213, but also inhibit multidrug-resistant bacteria such as ESBL-producing E. coli ATCC 35,218 and MRSA ATCC 43300.
MIC and MBC test showed that antimicrobial protein compounds from peat soil bacterial isolates showed bactericidal against ESBL -producing E.coli and MRSA at concentrations of less than 8 µg/ml. This means that antimicrobial compounds found in the study have a broad spectrum (Gram negative and Gram positive) and potential for use in the treatment of infections by Gram positive and Gram negative multiresistant bacteria, specifically compounds produced by the isolate 27PS, which have MIC and MIC MBC is smaller than the compound produced by the isolate 25PS and 26PS. The results of statistical analysis with the Kruskal Wallis test showed that the MIC and MBC values of the three antimicrobial compounds were significantly different from the antibiotics meropenem and vancomycin with p = 0.012 and the difference occurred in each antimicrobial compound against the antibiotics meropenem and vancomycin (p = 0.025) with the test Mann Whitney. This means that its effectiveness is still below meropenem and vancomycin but it still has the potential to be used in the treatment of infections by multiresistant Gram-positive and Gram-negative germs.
This study found three potential isolates producing antimicrobial compounds, namely 25PS isolate, 26PS isolate, and 27PS isolate. Of the three isolates, there was one isolate that was the most potential because it had MIC and MBC values less than 4 µg/mL, namely 27PS isolate. Antimicrobial compounds produced respectively have sizes of 43 kDa, 45 kDa and 43 kDa.
A study from peat soils in Malaysia isolated Bulkholderia paludis which has antimicrobial activity against strains of S. aureus and three strains of E. faecalis (MIC = 3.33 µg/mL and 6.29 µg/mL) (Ong et al., 2016) and Paenibacillus tyrfis against E. coli (MIC = 1.5 µg / mL), MRSA (MIC = 25 µg/mL) and Candida albicans IMR (MIC = 12.5 µg / mL) (Aw et al., 2016); however, the study did not examine the antimicrobial effect of multiresistant strains such as ESBL-producing which is far more common in Southeast Asia than MRSA (Suwantarat and Carroll, 2016, Chen et al., 2019). Thus the results of this study are more applicable in Indonesia and Southeast Asia, where there are far more cases of infection by ESBL-producing strains.
Bacteria could produce antimicrobials in the natural environment to compete with other bacterial competitors. Tropical peat soil is a unique wetland ecosystem, formed by the accumulation layers of decomposed organic matters. Therefore, it is worthwhile to explore bacteria from tropical peat swamps for the discovery of novel microorganisms that have antibacterial inhibitory effects (Ong, et al., 2015).
Previous studies suggested that there is a huge diversity of microbes in tropical peat swamp forests. However, most research on tropical peat swamp forests mainly focuses on bacteria related to nutrient recycling such as methanotrophic and acidophilic bacteria, but no study focus and the antimicrobial producing bacteria (Voglmayr and Yule, 2006, Jauhiainen et al., 2008, Yule, 2010). A study regarding with peat soil bacteria in Malaysia detected some bacterial species which were able to produce antimicrobials against Gram-negative and Gram-positive bacteria, as well as yeasts with various diameter zones ranging from 1.7 mm to 13.3 mm (Ong, et al., 2015).
Bacteria live in various unique environmental conditions, such as pH, temperature, and several source of nutrition’s in which bacteria therefore could produce bioactive compounds as products of their metabolism or as secondary metabolites, one of which has antibiotic properties to maintain bacterial populations and protect themselves from competing microorganisms (Martin, Casqueiro and Liras, 2005).
Secondary metabolites are compounds that are not essential for microbial growth or reproduction but provide survival functions in a variety of natural conditions (Martin, Casqueiro and Liras, 2005). These environmental conditions such as acidity, salinity, oxygen availability, nutrients source and environmental temperature needed to carry out metabolism. Many of the secondary metabolites are regulated by complex synthesis mechanisms within the bacteria itself which include polyketide synthase (PKS) and non-ribosomal polyketide synthase (NRPS) (Donadio, Monciardini and Sosio, 2007).
Other sources for producing antibiotics from secondary metabolites were Streptomyces (Bérdy, 2005). The use of bacteria as sources of antimicrobial, instead of a fungus such as Streptomyces, may offer more benefits as the incubation time for bacteria is much shorter, therefore, the antimicrobial compounds may be harvested much earlier. A research reported that bacteria associated with sponges Pseudomonas fluorescens H40 and H41 and Pseudomonas aeruginosa H51 showed antimicrobial activity against Gram-negative and Gram-positive bacteria, including vancomycin resistant Enterococcus faecium (VRE) and multidrug resistant bacteria such as Klebsiella pneumonia (Santos et al., 2010).
In Southeast Asia, there is rising of bacterial infections due MDR bacteria and become a huge problem to manage such infections, mostly due to ESBL-producing E. coli and methicillin-resistant Staphylococcus aureus (Kuntaman et al., 2016, Suwantarat and Carroll, 2016, Chen et al., 2019).
From peat soil in South Kalimantan, we were able to isolate 28 aerobic bacteria which 3 of them have an inhibitory effect against both sensitive and resistant gram-negative and gram-positive bacteria. The strength of this research is that the production of antimicrobials has extensive antimicrobial activity against Gram-negative and Gram-positive bacteria, so that it is expected to be applied on an industrial scale, in the pharmaceutical and medical fields. Limitations in this study have not been identified the types of antimicrobial compounds produced from peat soil bacteria, and identification of the mechanism of action of antimicrobial compounds has not been done. Proteomics study would reveal the weight molecule of the antimicrobial compounds, while molecular biology analysis to identify the gene responsible for the production of the antimicrobials.
In this study also reported that the active compound responsible is the protein group. So far, the secondary metabolites responsible for antibiotics have not been many primary metabolites/ proteins have antimicrobial activity. However, it still needs to be examined again whether the proteins that have been successfully purified so that they do not contain other compounds are necessary to identify and ensure the structure of the resulting protein. In this research, the certainty of the compounds produced is protein based on the method used.
The results of this study are expected to be an alternative in the discovery of antimicrobial protein compounds from bacteria in the era of the increasing number of resistant pathogenic bacteria.
5. Conclusion
This research shows that there are at least three species or strains of bacteria potentially as produce antimicrobial protein compounds that have bactericidal ability on a broad range of susceptible and MDR bacteria. 27PS isolates have the most potential to be developed into new antimicrobials because they have the lowest MIC and MBC.
Declarations
Author contribution statement
Dede Mahdiyah: performed the experiments; wrote the paper.
Winarto Reki: designed the experiment, analyzed, interpreted the data, and final proofreading.
Helmia Farida: supervise the experiment, analyzed and interpreted the data, and proof reading.
Ign Riwanto: analyzed and interpreted the data.
Hendro Wahyono: analyzed and interpreted the data.
Mustofa: designed the experiments, analyzed and interpreted data.
Tri Laksana Nugroho: analyzed and interpreted the data.
6. Funding statement
The work was supported by a grant of the Indonesian Endowment Fund for Education (LPDP) within the Ministry of Finance, Indonesia, number PRJ-6362/LPDP.3/2016.
7. Additional information
No additional information is available for this paper.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors would like to thanks to my promotor and co-promotor, who have helped my study, and Ministry of Finance, Indonesia for funding number PRJ-6362/LPDP.3/2016.
Footnotes
Peer review under responsibility of King Saud University.
References
- Ammerlaan H.S.M. ‘Secular trends in nosocomial bloodstream infections: antibiotic-resistant bacteria increase the total burden of infection. Clin. Infect. Dis. 2013;56(6):798–805. doi: 10.1093/cid/cis1006. [DOI] [PubMed] [Google Scholar]
- Anggraini D. Prevalence and susceptibility profile of ESBL-Producing Enterobacteriaceae in Arifin Achmad General Hospital Pekanbaru. J. Kedok. Brawijaya. 2018;30(1):47–52. [Google Scholar]
- Aw, Y., K., et al. 2016. Newly isolated Paenibacillus tyrfis sp. nov., from Malaysian tropical peat swamp soil with broad spectrum antimicrobial activity. Front. Microbiol. 7 (MAR), 1–9. doi: 10.3389/fmicb.2016.00219. [DOI] [PMC free article] [PubMed]
- Bérdy J. Bioactive microbial metabolites. J. Antibiotics. 2005;58(1):1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Bogaard, A., E., Van Den, and Stobberingh, E., E., 2000. Epidemiology of resistance to antibiotics Links between animals and humans. Inter. J. Antimicrob. Agents. 14, 327–335. [DOI] [PubMed]
- Chen C., Huang Y. New epidemiology of Staphylococcus aureus infection in Asia. Europ. Soc. of Clin. Infect. Dis. 2014;20(7):605–623. doi: 10.1111/1469-0691.12705. [DOI] [PubMed] [Google Scholar]
- Chen S. The higher prevalence of extended spectrum beta-lactamases among Escherichia coli ST131 in Southeast Asia is driven by expansion of a single, locally prevalent subclone. Nat. Res. 2019;9(March):1–14. doi: 10.1038/s41598-019-49467-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donadio S., Monciardini P., Sosio M. Polyketide synthases and nonribosomal peptide synthetases: the emerging view from bacterial genomics. Nat. Prod. Rep. 2007;24:1073–1109. doi: 10.1039/b514050c. [DOI] [PubMed] [Google Scholar]
- Hayati Z., Rizal S., Putri R. Isolation of extended-spectrum b-lactamase (esbl) producing Escherichia coli and Klebsiella pneumoniae from DR Zainoel Abidin. General Hospital, Aceh. Inter.l J. Trop. Vet. Biomed. Res. 2019;4:16–22. [Google Scholar]
- Jauhiainen J. Carbon dioxide and methane fluxes in drained tropical peat before and after hydrological restoration. Ecol. 2008;89(12):3503–3514. doi: 10.1890/07-2038.1. [DOI] [PubMed] [Google Scholar]
- Kanokratana P. Insights into the phylogeny and metabolic potential of a primary tropical peat swamp forest microbial community by metagenomic analysis. Microbial. Ecol. 2011;61(3):518–528. doi: 10.1007/s00248-010-9766-7. [DOI] [PubMed] [Google Scholar]
- Kuntaman K. Prevalence of methicillin resistant Staphylococcus aureus from nose and throat of patients on admission to medical wards of Dr Soetomo Hospital, Surabaya. Indonesia. South. Asian. J. Trop. Med. Pub. Health. 2016;47(1):66–70. [PubMed] [Google Scholar]
- Luvsansharav U. Prevalence of fecal carriage of extended-spectrum b-lactamase-producing Enterobacteriaceae among healthy adult people in Japan. J. Infect. Chemoter. 2011;17:722–725. doi: 10.1007/s10156-011-0225-2. [DOI] [PubMed] [Google Scholar]
- Martin J., Casqueiro J., Liras P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr. Op. Microbiol. 2005;8:282–293. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Ni Q. Prevalence and quinolone resistance of fecal carriage of extended-spectrum beta-lactamase-producing Escherichia coli in six communities and two physical examination centers population in Shanghai, China. Diagnos. Microbiol. Infect. Dis. 2016;86(4):428–433. doi: 10.1016/j.diagmicrobio.2016.07.010. [DOI] [PubMed] [Google Scholar]
- Oelschlaeger T. Mechanisms of probiotic actions: a review. Inter. J. Med. Microbiol. 2010;300(1):57–62. doi: 10.1016/j.ijmm.2009.08.005. Epub 2009 Sep 23. [DOI] [PubMed] [Google Scholar]
- Ong, K., et al., 2016. Burkholderia paludis sp. nov., an antibiotic-siderophore producing novel Burkholderia cepacia complex species, isolated from malaysian tropical peat swamp soil. Front. in Microbiol. 7 (DEC), 1–14. doi: 10.3389/fmicb.2016.02046. [DOI] [PMC free article] [PubMed]
- Ong, K., S., Catherine, M., and Lee, M., S., 2015. Antimicrobial producing bacteria isolated from tropical peat swamp soil. Malay. J. of Microbiol. 11 (2), 170–175. doi: 10.21161/mjm.12914.
- Pana, Z., and Zaoutis, T., 2018. Treatment of extended-spectrum β-lactamase-producing Enterobacteriaceae (ESBLs) infections : what have we learned until now ? F1000Research. 7 (0), 1–9. doi: 10.12688/f1000research.14822.1. [DOI] [PMC free article] [PubMed]
- Pettit R. Culturability and secondary metabolite diversity of extreme microbes: expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Marine. Biotechnol. 2011;13(1):1–11. doi: 10.1007/s10126-010-9294-y. [DOI] [PubMed] [Google Scholar]
- Santos O. Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res. Microbiol. 2010;161(7):604–612. doi: 10.1016/j.resmic.2010.05.013. [DOI] [PubMed] [Google Scholar]
- Sawatwong P. High burden of extended-spectrum β-lactamase–producing Escherichia coli and Klebsiella pneumoniae bacteremia in older adults : a seven-year study in two Rural Thai Provinces. Am. J. Trop. Med. Hyg. 2019;100(4):943–951. doi: 10.4269/ajtmh.18-0394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwantarat N., Carroll K. Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob. Resist. Infect. Cont. 2016;5(15):1–8. doi: 10.1186/s13756-016-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H., Yule C. Polyancora globosa gen. sp. nov., an aeroaquatic fungus from Malaysian peat swamp forests. Mycological. Res. 2006;110:1242–1252. doi: 10.1016/j.mycres.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Wang Q. Abyssomicins from the South China sea deep-sea sediment Verrucosispora sp: natural thioether michael addition adducts as antitubercular prodrugs. Angew. Chemie Inter. Ed. 2013;52:1231–1234. doi: 10.1002/anie.201208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO, 2017. Global priority list of antibiotic-resistant bacteria to guide research, discovery, and development of new antibiotics.1-7.
- Wong J. Prevalence and risk factors of community- associated methicillin-resistant Staphylococcus aureus carriage in Asia-Pacific region from 2000 to 2016: a systematic review and meta-analysis. Clin. Epidemiol. 2018;10:1489–1501. doi: 10.2147/CLEP.S160595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yule C. Loss of biodiversity and ecosystem functioning in Indo-Malayan peat swamp forests. Biodivers. Conserv. 2010;19:393–409. doi: 10.1007/s10531-008-9510-5. [DOI] [Google Scholar]